ABSTRACT
The main components of the essential cellular process of eukaryotic ribosome biogenesis are highly conserved from yeast to humans. Among these, the U3 Associated Proteins (UTPs) are a small subunit processome subcomplex that coordinate the first two steps of ribosome biogenesis in transcription and pre-18S processing. While we have identified the human counterparts of most of the yeast Utps, the homologs of yeast Utp9 and Bud21 (Utp16) have remained elusive. In this study, we find that NOL7 is the likely ortholog of Bud21. Previously described as a tumour suppressor through regulation of antiangiogenic transcripts, we now show that NOL7 is required for early pre-rRNA accumulation and pre-18S rRNA processing in human cells. These roles lead to decreased protein synthesis and induction of the nucleolar stress response upon NOL7 depletion. Beyond Bud21’s nonessential role in yeast, we establish human NOL7 as an essential UTP that is necessary to maintain both early pre-rRNA levels and processing.
KEYWORDS: Ribosome biogenesis, pre-ribosomal RNA processing, RNA polymerase I, nucleolus, rRNA, oncogene, tumour suppressor
Introduction
Ribosome biogenesis is an essential and conserved process in all living organisms. In eukaryotes ribosome biogenesis begins in the nucleolus with the transcription of the ribosomal DNA (rDNA) by RNA polymerase I (RNAP1) to synthesize the polycistronic pre-rRNA precursor. This pre-rRNA then undergoes a series of modification, processing, and maturation steps while associating with ribosomal proteins to yield the mature small 40S subunit (18S rRNA) and large 60S subunit (28S, 5.8S, and RNAPIII-transcribed 5S rRNAs) [17,18]. Much work has been completed in baker’s yeast, Saccharomyces cerevisiae (S. cerevisiae), to understand the fundamentals of eukaryotic ribosome biogenesis [19]. More recent work has highlighted the increased complexities of this process in human cells with new factors, functions, and connections to other cellular processes being uncovered.
The small subunit (SSU) processome is the earliest stable intermediate pre-ribosome complex that forms co-transcriptionally and is marked by the presence of the U3 small nucleolar ribonucleoprotein (U3 snoRNP). Within the SSU processome, there are multiple subcomplexes that are distinguished by their order of assembly [20–22]. The first subcomplex to form around the pre-rRNA, the transcription U3 associated proteins (t-Utps) or UtpA subcomplex, is required for both transcription and 5’-external transcribed spacer sequence (5’ETS) processing of the primary transcript pre-rRNA to yield the 18S rRNA. The UtpA subcomplex was first discovered and studied in yeast as a group of essential proteins [22–24]. Other Utps, including the UtpB and UtpC subcomplexes, associate with the small subunit processome later and thus are only required for pre-18S processing [25]. Recent work has focused on highlighting the role of the SSU processome’s components through cryo-EM structures of both yeast [4] and humans [5].
To date, most of the human orthologs of the Utps have been identified except for the t-Utp, Utp9, and a non-essential early associating Utp, Bud21 [called Utp16 in [23] (Figure 1A). As a t-Utp, Utp9 coordinates both pre-rRNA transcription and pre-rRNA processing [24]. In part, a direct association with Utp8 is required for its role in pre-18S processing and for an additional role in protein synthesis through direct regulation of mature tRNA nuclear re-export [26,27]. Bud21 was first discovered as a non-essential protein required for bud site selection in S. cerevisiae. Bud21 is among the set of proteins to associate with the 5’ETS but has not been shown to be a part of any of the SSU subcomplexes in yeast [20]. Bud21 is also known to be a Ty1 retrotransposon host factor [28]. Deletion of Bud21 renders increased hypoxia tolerance [29] and improved xylose carbon source utilization [30], while Bud21 expression is upregulated upon acetic acid challenge [31]. Overall, not much is known about the molecular functions of Utp9 and Bud21 in ribosome biogenesis, even in the well-studied model organism S. cerevisiae.
Figure 1.
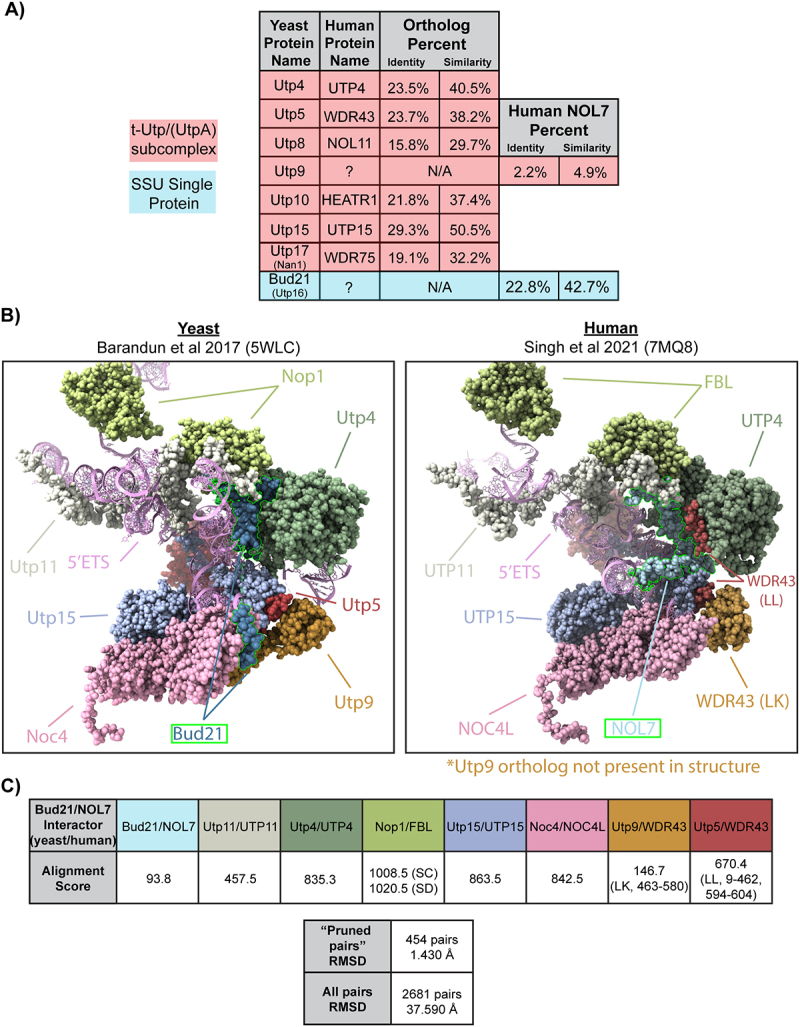
NOL7 is the likely Bud21 (Utp16) ortholog.
A. Human NOL7 contains the highest sequence similarity to yeast Bud21 (Utp16). Table of yeast and human t-UTP/UTPA subcomplex proteins and their orthologs including the yeast t-Utp interacting protein Bud21. Pairwise sequence alignments were performed using EMBOSS Needle2. Percent similarity and identity of NOL7 with two yeast Utp members that do not currently have a known human ortholog and established orthologous protein pairs are indicated.
B. Side-by-side structure comparison of Bud21 (Utp16) and NOL7 with their interaction partners in the small subunit processome. (Left) S. cerevisiae4 (pdb: 5WLC) and (Right) humans5 (pdb: 7MQ8). UCSD ChimeraX software was used to visualize Bud21 and NOL7 interacting proteins (space filling) and 5’-external transcribed spacer (5’ETS). Orthologous proteins are shown in same colouring and labelled. Bud21 and NOL7 are outlined in green. Utp9 is only present in the yeast structure, but WDR43 (chain LK) takes a similar position in the human structure.
C. Alignment score for Bud21 (Utp16) and NOL7 and their associated interacting proteins in the small subunit processome (SSU) to show similarities between analogous proteins from yeast to humans. Alignment scores for the indicated proteins were calculated based on BLOSUM-62 matrix scoring and secondary structure assignments. Scores for proteins with multiple chains are indicated by their abbreviations. Root-mean square deviation (RMSD) is reported for both ‘prune pairs’ and the entire yeast vs. human structures of proteins that interact with Bud21 and NOL7 in angstroms (Å).
Here, we describe human NOL7 as a likely yeast Bud21 ortholog and establish its function in human cells. NOL7 is required to maintain a normal number of nucleoli in MCF10A cells, an established predictive indicator of proteins that are involved in ribosome biogenesis [10,32]. More precisely, NOL7 is required for early pre-rRNA accumulation and pre-18S rRNA processing, but not for rDNA promoter activity in human cells. Its depletion leads to decreased mature 18S levels and reduced global protein synthesis, and subsequent induction of the nucleolar stress response. Our results present a new role for NOL7 in human ribosome biogenesis that extends beyond Bud21’s defined role in yeast to emphasize the increasing complexities of early ribosome biogenesis in human cells.
Methods
Publicly available expression datasets
Genotype-Tissue Expression (GTEx) unmatched normal and The Cancer Genome Atlas (TCGA) matched normal and tumour expression datasets were obtained through the Xena platform (https://xena.ucsc.edu/) [1]. RNA-seq by Expectation-Maximization (RSEM) LOG2 fold expression levels for NOL7 were subtracted from the mean of the overall normal and tumour tissues combined for graphical visualization.
Sequence alignment and structure analyses
Human NOL7 and Saccharomyces cerevisiae Utp9 and Bud21 protein sequences were used for pairwise alignment using EMBOSS Needle [2] to report percent identity and similarity (Figure 1). Vertebrate protein sequences for NOL7 were used for a multiple sequence alignment along with S. cerevisiae Bud21 using Clustal Omega [3].
S. cerevisiae (PDB ID: 5WLC) [4] and human (PDB ID: 7MQ8) [5] small subunit processome cryo-EM structures were used to analyse Bud21 and NOL7 protein-protein and 5’ETS protein-RNA interactions, respectively. Interface residues buried fraction (>0) was considered an interaction using Biojava-structure-6.0.4 for a given position on the 5’ETS (Figure 3A) and Bud21 or NOL7 (Figure S3D). UCSF ChimeraX software was used to superimpose the modelled residues of Bud21 (5WLC) and NOL7 (7MQ8) (Figure S3) and their interacting partners using the matchmaker tool. Needleman-Wunch sequence alignment was used, and Cα root mean square deviation (RMSD) values were reported. Alignment scores for specific chains were calculated based on BLOSUM-62 matrix scoring and secondary structure assignments with default parameters.
Figure 3.
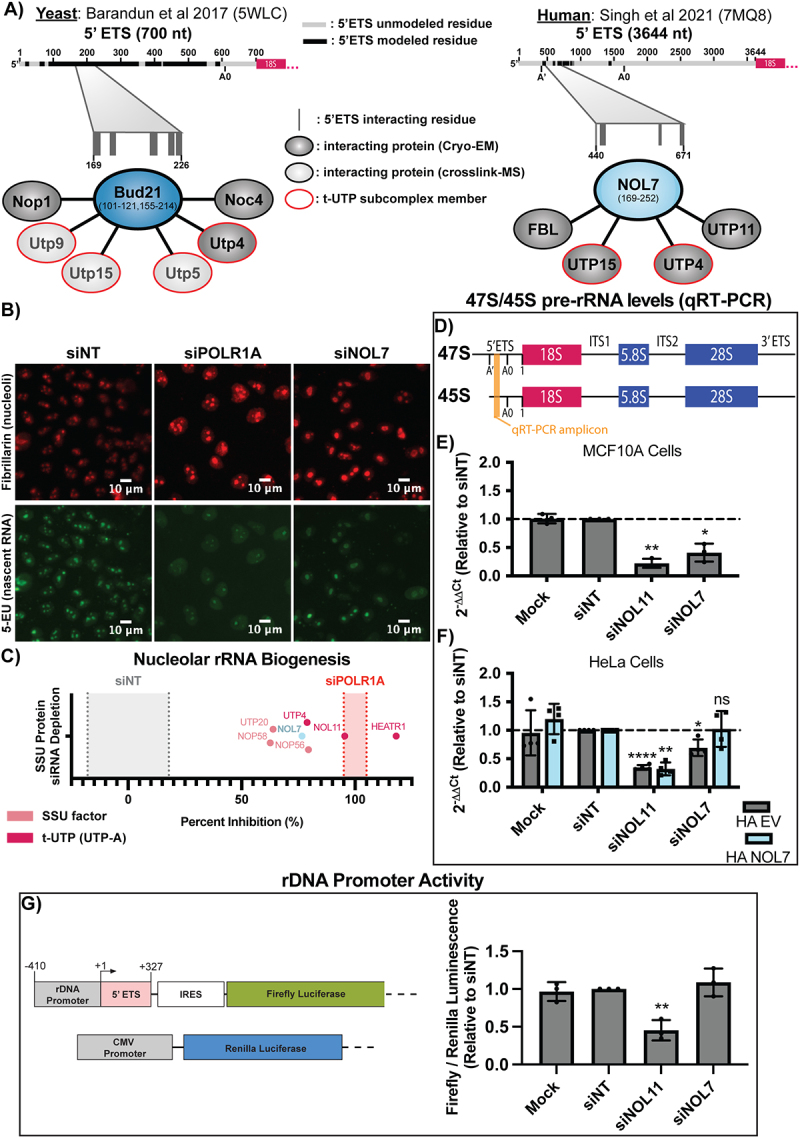
NOL7 is required for early pre-rRNA accumulation in human tissue culture cells.
A. Structure interaction summary of Bud21 (Utp16) and NOL7 within the small subunit processome of (Left) S. cerevisiae4 (pdb: 5WLC) and (Right) humans5 (pdb: 7MQ8). Data were mined from the indicated structure datasets. 5’external transcribed spacer (5’ETS) residues that were modelled (black) or not modelled (grey) are reported. 5’ETS residues that interact with Bud21 (left) or NOL7 (right) are shown by grey lines within the indicated 5’ETS region. Modelled residues on Bud21 and NOL7 are reported. Bud21 and NOL7 interacting proteins are presented that were identified within the cryo-EM structures (dark grey/black) or crosslink-mass spectrometry (MS) (light grey/black, yeast only). Red outline on proteins indicates they are an established member of the t-Utp subcomplex of the small subunit (SSU) processome.
B. NOL7 siRNA depletion decreases nucleolar rRNA biogenesis. MCF10A cells were depleted with siRNAs for 72 h and pulse labelled with 5-EU for 1 h. Fixed cells were stained for nucleoli (fibrillarin, FBL, red) and click chemistry was performed to conjugate AF488 azide to labelled nascent RNA (5-EU, green). siPOLR1A is a positive control and siNT is a negative control.
C. Quantification of the percent inhibition of nucleolar rRNA biogenesis (median nascent RNA (5-EU) signal within the nucleolus) after siNOL7 treatment in MCF10A cells from (B). The % nucleolar inhibition upon depletion of other small subunit processome (SSU) factors [(t-UTPs: HEATR1, NOL11, and UTP4) (other SSU factors: NOP56, UTP20, and NOP58)] are also indicated.11 siPOLR1A positive control (red shading) treatment set at 100%, siNT negative control (grey shading) set at 0%. 3 biological replicates, mean ± SD shown for controls indicated by shaded area, mean only shown for other factors.
D-F. qRT-PCR to measure 47S/45S pre-rRNA primary transcript levels after NOL7 depletion.
D. Schematic of 47S primary and 45S pre-rRNA transcripts with qRT-PCR amplicon locations indicated (orange). The 45S precursor arises after cleavage at the A’ site and is more readily detectable than the 47S rRNA by qRT-PCR.
E. NOL7 siRNA depletion reduces 47S/45S pre-rRNA levels in MCF10A cells. qRT-PCR measuring the primary 47S/45S pre-rRNA transcript levels. 2−ΔΔCt were measured relative to 7SL internal control and siNT negative control sample. siNOL11 is a positive control. 3 technical replicates of 3 biological replicates, plotted mean ± SD. Data were analysed by one-way ANOVA with Dunnett’s multiple comparisons test, * p ≤ 0.05, ** p ≤ 0.01.
F. The reduction of 47S/45S pre-rRNA levels after NOL7 siRNA depletion can be rescued by introduction of an si-resistant version of NOL7. qRT-PCR measuring the primary 47S/45S pre-rRNA transcripts in HeLa cells either expressing empty vector (HA EV) or siNOL7 resistant HA-tagged NOL7 (HA NOL7). 2−ΔΔCt were measured relative to 7SL internal control and siNT negative control sample. siNOL11 is a positive control. 3 technical replicates of 4 biological replicates, plotted mean ± SD. Data were analysed by one-way ANOVA with Dunnett’s multiple comparisons test, * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, ns = not significant.
G. NOL7 siRNA depletion does not change rDNA promoter activity. (Top) Schematic of reporter plasmids used: Firefly (pHrD-IRES-Luc, rDNA promoter reporter) and Renilla (CMV, transfection control) luciferase plasmids14. (Bottom) Quantification of rDNA promoter activity. Firefly luminescence measured relative to Renilla luminescence and siNT negative control. siNOL11 is a positive control. 3 technical replicates of 3 biological replicates, plotted mean ± SD. Data were analysed by one-way ANOVA with Dunnett’s multiple comparisons test, ** p ≤ 0.01.
Enrichment analyses
A list of NOL7 interacting proteins was obtained from TheBioGrid [6], hu.Map 2.0 [7], and a cryo-EM structure of the small subunit processome human pre-A0 cleavage [5] (Table S1). A list of Bud21 interacting proteins was obtained from TheBioGrid [6] and a cryo-EM structure of the S. cerevisiae small subunit processome [4] (Table S1). These lists were used for PANTHER 17.0 gene ontology overrepresentation analysis (Fisher’s Exact test, reported top main categories where fold enrichment > 2, FDR < 0.05, p < 0.05).
Plasmids
Full-length cDNA for human NOL7 was subcloned into Gateway entry vector pDONR221 (Life Technologies) per manufacturer’s instructions. Site-directed mutagenesis to generate synonymous mutations to render NOL7 resistant to siNOL7 #3 (Table S2) was performed using the Change-ITTM Multiple Mutation Directed Mutagenesis Kit (Affymetrix, 78480) per manufacturer’s instructions. NOL7 siRNA-resistant cDNA was subcloned into the pLX301 vector (Addgene, 25895) containing an N-terminal HA epitope tag with an LR reaction (Life Technologies) per manufacturer’s instructions.
Cell culture, media, and lentiviral transduction
MCF10A cells (ATCC, CRL-10317) were subcultured in Dulbecco’s modified Eagles’ medium/Nutrient mixture F-12 (Gibco, 1130–032) containing horse serum (Gibco, 16050), 10 μg/mL insulin (Sigma, I1882), 0.5 μg/mL hydrocortisone (Sigma H0135), 100 ng/mL cholera toxin (Sigma, C8052), and 20 ng/mL epidermal growth factor (Peprotech, AF-100-15). HeLa cells (ATCC, CCL-2) cells were grown in DMEM (Gibco, 41965–062) with 10% foetal bovine serum (FBS, Gibco, 10438026). Cell lines were maintained at 37°C, in a humidified atmosphere with 5% CO2.
For the high-throughput nucleolar number and 5-EU assays, 3,000 cells/well were reverse transfected in 384-well plates on day 0. For RNA or protein isolation, 100,000 cells/well were seeded into 6-well plates on day 0. For the dual-luciferase assay, 100,000 cells/well were seeded into 12-well plates on day 0.
Lentiviral production and transduction were performed as in [8]. Lentiviruses were packaged in the HEK293FT cell line by co-transfecting pVSV-G, psPAX2, and modified pLX301 vectors [9] containing either a siRNA-resistant version of HA-NOL7 CDS or empty vector in a 1:9:10 ratio (1 μg:9 μg:10 μg for a 10-cm plate) with Lipofectamine 2000 (Life Technologies, 11668019) per manufacturer’s instructions. HeLa cells were transduced with NOL7 or empty vector containing lentivirus under puromycin antibiotic selection (2 μg/mL).
RNAi
All siRNAs were purchased from Horizon Discovery Biosciences (Table S2). For the 5-EU nucleolar rRNA biogenesis assay of NOL7 ON-TARGETplus pools were used except for the NOL11 positive control where we used the siGENOME SMARTpool siRNAs. For screen validation by deconvolution, the individual ON-TARGET set of four siRNAs that comprised the pool was used. The ON-TARGETplus pools were used in the remaining functional analysis except for northern blotting experiments where siGENOME SMARTpool siRNAs were utilized. Pools of siRNAs were used in all experiments except where indicated that individual siRNAs were tested. siRNA transfection was performed using Lipofectamine RNAiMAX Transfection Reagent (Invitrogen, 13778150) per manufacturer’s instructions with a final siRNA concentration of 20 nM for 384-well plate high-throughput assays and 33 nM for all other assays. For the high-throughput screens, cells were reverse transfected in 384-well plates on day 0 where siRNA controls were added to 16 wells each and siNOL7 was added to 1 well for each replicate. For other assays, cells were transfected 24 h after plating.
Nucleolar number assay
We followed the [10] protocol for counting nucleolar number in MCF10A cells in high-throughput. For siRNA pool treatments, including siONT targeting NOL7, a hit was called based on a stringent cut-off if it produced a mean one-nucleolus percent effect greater than or equal to + 3 standard deviations (SD) above the mean percent effect for the entire genome-wide screen (122% effect) (Data S1). For siONT deconvolution, an individual siRNA targeting NOL7 was considered validated if it produced a mean one-nucleolus percent effect greater than or equal to + 3 SD above the siNT mean, using the siNT SD within each replicate (Data S2).
5-EU incorporation assay
We monitored nucleolar rRNA biogenesis using a high-throughput protocol we developed in the laboratory [11]. Briefly, following 72 h siRNA depletion, MCF10A cells were treated with 1 mM 5-ethynyl uridine (5-EU) for 1 h to label nascent RNA. Then cells were fixed, stained for nucleoli [72B9 anti-fibrillarin antibody [12], Alexa Fluor 647 goat anti-mouse IGG secondary] and nuclei (Hoëchst), and we performed click chemistry to attach an AF488 azide for 5-EU visualization. Images were acquired with IN Cell 2200 imaging system (GE Healthcare) and a custom CellProfiler pipeline was used for analysis (Data S3).
Puromycin incorporation assay
Global protein synthesis was assessed as in [13]. Following 72 h siRNA depletion, 1 μM puromycin (or 0.5 μM puromycin for Mock 0.5 control) was added to the media for 1 h to label nascent polypeptides. Then we proceeded with western blotting as below.
Western blotting
Total protein was harvested from cells by scraping followed by PBS rinse. Cells were lysed using AZ lysis buffer (50 mM Tris pH 7.5, 250 mM NaCl, 1% Igepal, 0.1% SDS, 5 mM EDTA pH 8.0) with protease inhibitors (cOmpleteTM Protease Inhibitor Cocktail, Roche, 11697498001) for 15 minutes at 4°C by vortexing. Lysed cells were spun at 21,000 RCF for 15 minutes at 4°C, supernatant was harvested and protein concentration was determined by the Bradford assay (Bio-Rad). Either 50 μg or 25 μg (puromycin blots only) of total protein was separated by SDS-PAGE and transferred to a PVDF membrane (Bio-Rad, 1620177) for blotting. The following primary antibodies were used: α-p53 (Santa Cruz, sc-126), α-puromycin (Kerafast, EQ0001), α-HA (clone 12CA5), and α-β-actin (Sigma Aldrich, A1978), α-POLR1A (Santa Cruz, sc-48,385), α-FBL (Abcam, ab226178), α-UBTF (Santa Cruz, sc-13,125). For detection of primary antibodies, secondary α-mouse-HRP conjugated antibody (GE Healthcare, Life Sciences NA931) was used. Images were acquired using Bio-Rad Chemidoc (12003153) and analysed using ImageJ software.
qRT-PCR analysis
Total cellular RNA was extracted using TRIzol (Life Technologies, 5596018) per manufacturer’s instructions. Prior to cDNA synthesis, all A260/230 values were above 1.7 by NanoDrop (ThermoFisher, ND2000CLAPTOP). cDNA was made from 1 μg total RNA using iScript gDNA clear cDNA synthesis kit (Bio-Rad, 1725035) with random primers. qPCR was performed with iTaq Universal SYBR Green Supermix (Bio-Rad, 1725121). The following amplification parameters were used: initial denaturation 95 ºC for 30 s, 40 cycles of 95 ºC for 15 s and 60 ºC for 30 s. Subsequent melt curve analysis was performed to ensure a single product, 95 ºC for 15 s, then gradual (0.3 ºC/15 s) increase from 60 ºC to 94.8 ºC. Gene-specific primers were used (Table S3). Amplification of 7SL RNA was used as an internal control and relative RNA levels were determined using comparative CT method (ΔΔCT). Three technical replicates were performed for each biological replicate.
Dual-luciferase reporter assay
After 48 h of siRNA depletion, MCF10A cells were co-transfected with 1000 ng of pHrD-IRES-Luc plasmid [14] and 0.1 ng of a CMV-Renilla constitutive internal control plasmid using Lipofectamine 3000 (Thermo Fisher Scientific, L3000015) per manufacturer’s instructions. Twenty-four h post plasmid transfection (72 h of siRNA depletion), cells were harvested and luminescence was measured by a Dual-luciferase Reporter Assay System (Promega, E1910) per manufacturer’s instructions using a GloMax Discover Microplate Reader (Promega, GM3000). The ratio of pHrD-IRES-Luc/Renilla activity was calculated to control for plasmid transfection efficiency.
Northern blotting
Total cellular RNA was extracted using TRIzol (Life Technologies, 5596018) per manufacturer’s instructions. For each sample, 3 μg of total RNA was resolved on a denaturing 1% agarose/1.25% formaldehyde gel using Tri/Tri buffer (Mansour and Pestov [15]2013) and transferred to a Hybond-XL membrane (GE Healthcare, RPN 303S). UV-crosslinked membranes were stained with methylene blue (0.025% w/v) and imaged. Blots were hybridized to 32P radiolabeled DNA oligonucleotide probes (5’ETS 5’- CCTCTCCAGCGACAGGTCGCCAGAGGACAGCGTGTCAGC-3’) or (P3 (ITS1) 5’-AAGGGGTCTTTAAACCTCCGCGCCGGAACGCGCTAGGTAC-3’) and detected by phosphorimager (Amersham™ Typhoon™, 29187194). Images were analysed using ImageJ, ratio-analysis of multiple precursors (RAMP) was performed [16].
BioAnalyzer
For each sample, 100 ng/μL total RNA in nuclease-free water was submitted for Agilent BioAnalyzer analysis, performed by the Yale Center for Genome Analysis.
Statistical analyses
Statistical analyses were performed using GraphPad Prism 9.3.1 (GraphPad Software). Tests are described in the associated figure legends.
Results
NOL7 is the likely yeast Bud21 ortholog
We hypothesized that NOL7 might be the human ortholog to either Utp9 or Bud21 (Utp16) due to its nucleolar localization [33] and because of its interactions with components of the SSU processome. We obtained a list of NOL7-associated proteins from high-throughput interactome datasets [6] and cryo-EM structural analysis [5] and performed Gene Ontology overrepresentation tests for both biological processes and cellular components using PANTHER (Log2 >1, p < 0.05) [34]. NOL7-associated factors’ most enriched categories included positive regulation of rRNA processing (GO:2000234) and transcription by RNA polymerase I (GO:0045943) processes, and the t-UTP complex (GO:0034455) and small-subunit processome (GO:0032040) components (Figure S1A, Table S1). NOL7 protein-protein associations are enriched for factors present in the early SSU processome complex, indicative of a t-UTP subcomplex member.
To address whether Utp9 or Bud21 was the more likely NOL7 ortholog, we performed protein sequence alignments using EMBOSS Needle [2]. Interestingly, human NOL7 best aligned in both percent identity and similarity with S. cerevisiae Bud21 (22.8% identity, 42.7% similarity) compared to Utp9 (2.2% identity, 4.9% similarity) (Figure 1A). The level of conservation between NOL7 and Bud21 is similar to that of other UTPs (Figure 1A), such as human UTP4 with yeast Utp4 [35] and NOL11 with the single-celled eukaryote species Capsaspora owczarzaki Utp8 [36]. We completed a multiple sequence alignment using CLUSTAL Omega [3] to visualize the conservation of vertebrate NOL7 with Bud21 across various species. We observed conservation throughout evolution of the Bud21 sequence (Figure S2). Unexpectedly, we were unable to identify NOL7 orthologs in either Drosophila melanogaster [37] (DIOPT Version 8.5 2021, https://www.flyrnai.org/cgi-bin/DRSC_orthologs.pl) or in Caenorhabditis elegans [38] (OrthoList2, http://ortholist.shaye-lab.org/). Furthermore, the overrepresented Gene Ontology categories of interacting proteins were strikingly similar between Bud21 and NOL7 (Figure S1A and S1B, Table S1).
Using the available yeast (5WLC) [4] and human (7MQ8) [5] SSU processome cryo-EM structures, we explored the level of structural conservation of the Bud21 and NOL7 residues modelled within them. Our analysis was limited because only about one-third of the Bud21 (38%) and NOL7 (32%) residues were modelled in these structures (Figure 1B,C). However, the C-terminal portions of both structures are modelled and possess a greater overlap with each other after superimposition using UCSF Chimera X matchmaker tool (RMSD = 4.86–7.60 Å) (Figure S3A and S3B). The portions of Bud21 and NOL7 superimposed with the lowest RMSD were residues in contact with or close proximity to the 5’ETS (Figure S3C and S3D). Bud21 and NOL7’s similar contact positions with protein interacting partners are revealed when looking at these structures side-by-side. Further supporting the idea that NOL7 is the likely Bud21 ortholog, only the yeast structure contains Utp9, while there is no Utp9 present in the human structure. From our structure alignment, it is possible that the presence of two copies of WDR43 have compensated to some extent for the lack of Utp9, as has been speculated previously [5]. Due to Bud21 and NOL7’s lower level of conservation based on the alignment score of the modelled residues compared to other SSU components, we emphasize why it is important to study NOL7’s role in human cells, which has the potential to be different compared Bud21 in yeast. Based on conservation of both sequence, structure, and interaction partners, we conclude that NOL7 is most likely the human ortholog of yeast Bud21 and is a component of the SSU processome.
NOL7 regulates nucleolar function in human cells
NOL7’s functional role in human ribosome biogenesis has not been yet firmly established. As a first pass, we took advantage of our laboratory’s previously established siRNA screening methodology to identify novel regulators of nucleolar function [10,32]. Briefly, MCF10A cells normally harbour 2–3 nucleoli per cell nucleus. However, upon depletion of ribosome biogenesis factors, this number decreases to 1 or increases to 5 or more nucleoli on average. Since siRNAs against NOL7 were not present in the original genome-wide siRNA library that we had screened and published [10], we specifically depleted NOL7 using siRNAs (si-ONTARGET pool, Horizon Discovery) in high throughput, applying our workflow pipeline. We observed a significant increase in MCF10A cells with 1 nucleolus (40.7% one-nucleolus cells, percent effect = 193.8%) after NOL7 depletion compared to the negative control non-targeting siRNA (siNT) (19.2% one-nucleolus cells, percent effect = 0%). Notably, NOL7 depletion produced a greater proportion of one-nucleolus harbouring cells than depletion of the positive control, the t-UTP, NOL11 (29.2% one nucleolus cells, percent effect = 100%) (Figure 2A, Data S1).
Figure 2.
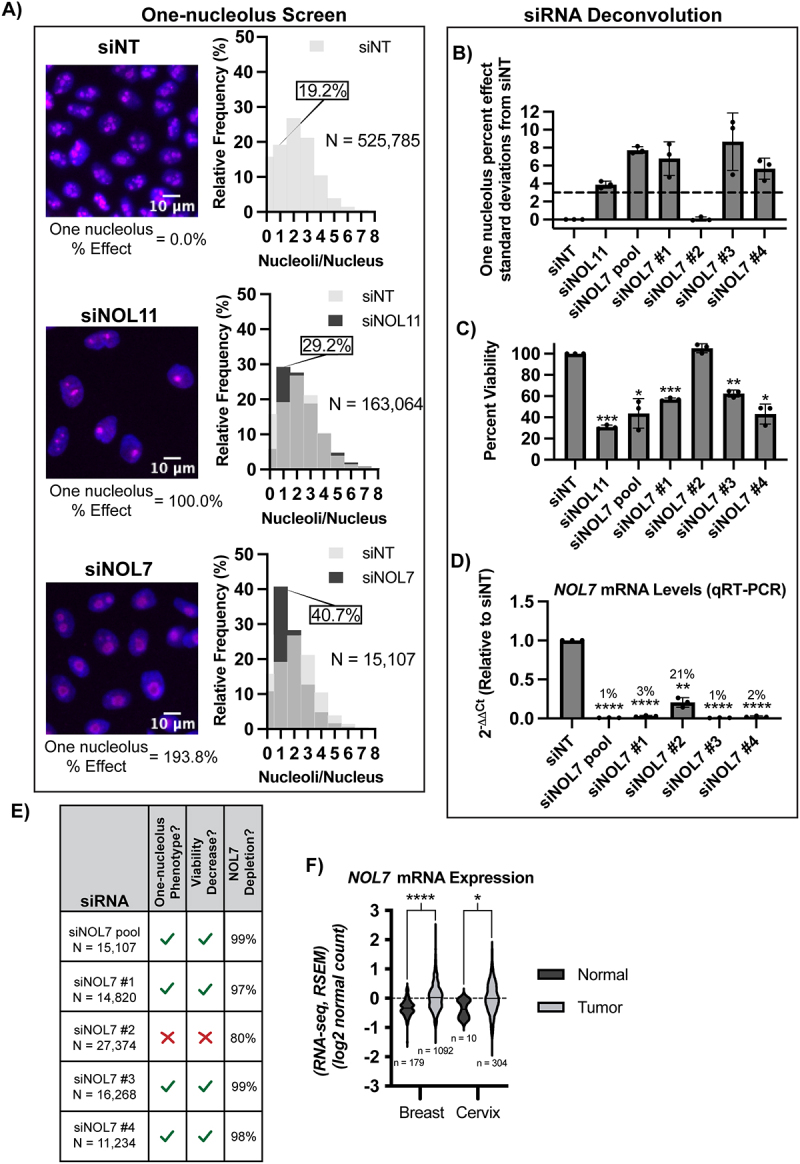
NOL7 regulates nucleolar function in human cells.
A. NOL7 depletion by an siRNA pool reduces nucleolar number in MCF10A cells. (Left) Representative merged images of nucleoli stained with fibrillarin (magenta) and nuclei stained with Hoechst (blue). siNT was used as a negative control (average of 2-3 nucleoli per nucleus), siNOL11 was used as a positive control (average of 1 nucleolus per nucleus). (Right) Histograms of relative frequency of nucleoli per nucleus from quantification of images. Light grey indicates siNT negative control, dark grey indicates either siNOL11 positive control or siNOL7 treated cells. Overlap with siNT indicated by an intermediate grey colour. Percent of one-nucleolus harbouring cells indicated. N = number of cells analysed.
B-D. Deconvolution of the 4 individual NOL7 siRNAs that constitute the NOL7 siRNA pool in MCF10A cells.
B. Testing of the 4 individual NOL7 siRNAs for the one nucleolus percent effect. siNOL7 pool and 3 out of 4 individual siRNAs targeting NOL7 produce an increase in the one-nucleolus percent effect in MCF10A cells. 3 biological replicates, plotted mean ± SD. One-nucleolus percent effect is set relative to standard deviations from the negative control, siNT. siNOL11 is a positive control. A 3 standard deviation cut-off (dashed line) was used to consider an siRNA pool treatment to be a hit and for an individual siRNA treatment to pass the deconvolution criteria of producing the one-nucleolus phenotype.
C. Testing of the 4 individual NOL7 siRNAs for cell viability. siNOL7 pool and 3 out of 4 individual siRNAs targeting NOL7 reduce MCF10A cell viability. 3 biological replicates, plotted mean ± SD. Cell viability where siNT, negative control, is set to 100%. siNOL11 is a positive control. Data were analysed by one-way ANOVA with Dunnett’s multiple comparisons test, ** p ≤ 0.01, *** p ≤ 0.001, ** p ≤ 0.01, * p ≤ 0.05.
D. Testing of the 4 individual NOL7 siRNAs for NOL7 mRNA levels. siNOL7 pool and 3 out of 4 individual siRNAs targeting NOL7 reduce NOL7 mRNA transcript levels greater than 95% in MCF10A cells. qRT-PCR measuring the primary NOL7 mRNA transcript levels. 2−ΔΔCt were measured relative to 7SL internal control and siNT negative control sample. 3 technical replicates of 3 biological replicates, plotted mean ± SD. Data were analysed by one-way ANOVA with Dunnett’s multiple comparisons test, ** p ≤ 0.01, **** p ≤ 0.0001. Relative percent of NOL7 levels compared to siNT (100%) is indicated.
E. Summary table of siNOL7 pool deconvolution in (C), (D), and (E). Reduction in NOL7 mRNA levels greater than 95% leads to reduction in nucleolar number and cell viability. N = number of cells analysed sum of 3 independent replicates. One-nucleolus percent effect passes 3 standard deviations from negative control siNT cut-off from panel (B). Viability decreased considered if significance was reached from panel (C). The percentage of NOL7 mRNA level depletion is calculated from panel (D).
F. Violin plots showing increased expression at the mRNA level for NOL7 in breast and cervical cancer. Data are from Genotype-Tissue Expression (GTEx) unmatched normal and The Cancer Genome Atlas (TCGA) matched normal and tumour RNA-seq by Expectation-Maximization (RSEM) LOG2 fold expression levels for NOL7 subtracted from the mean. NOL7 expression in normal and tumour breast tissue (left), normal and tumour cervical tissue (right). NOL7 expression in all normal and tumour tissues. Dashed line (set at 0) indicates mean of entire dataset for both normal and tumour expression, black lines indicate mean of individual normal or tumour expression dataset. Data were analysed by Student’s t-test, **** p ≤ 0.0001, * p ≤ 0.05.
To validate NOL7 as a likely hit, we deconvoluted the si-ONTARGET pool targeting NOL7 to test the ability of each individual siRNA to produce one-nucleolus containing cells and to mitigate possible off-target effects. We observed a reduction in nucleolar number using 3 of the 4 individual siRNAs that targeted NOL7, using > 3 standard deviations from siNT negative control as a stringent cut-off (Figure 2B, Data S2). As expected with inhibition of ribosome biogenesis, this one-nucleolus percent effect produced by individual siNOL7 treatments was inversely correlated with cell viability as measured by number of Höechst-stained nuclei remaining after treatment (Figure 2C). Moreover, the same 3 siRNAs that produced the one nucleolus phenotype also led to the greatest reduction (>95%) in NOL7 mRNA levels as measured by qRT-PCR (Figure 2D). Because the other siRNA (siNOL7 #2) did not produce the one-nucleolus phenotype or decrease cell viability, it is possible that ~ 20% of normal NOL7 mRNA levels is enough to maintain normal cellular function (Figure 2E). These deconvolution experiments indicate NOL7 depletion of > 95% is responsible for the hypothesized ribosome biogenesis defects based on the reduction in nucleolar number and in cell viability.
To further demonstrate that the results that we have obtained are due to depletion of NOL7, and not due to an off-target effect of siRNAs, we performed rescue experiments. We made a stable HeLa cell line expressing an siRNA-resistant and N-terminally HA-tagged version of NOL7 (resistant to siNOL7 #3 of the siON-TARGET siRNA pool) and observed that NOL7 mRNA and protein levels depleted by an siRNA can be rescued by overexpressing the si-resistant NOL7. Rescue was detected at both the mRNA level by qRT-PCR and at the protein level by western blotting using an HA antibody (Figure S4). HeLa cells were used for these experiments below to highlight the expected universal role of NOL7 in making ribosomes. Additionally, MCF10A is a non-cancerous and a ‘near-normal’ cell line [39], thus using HeLa cells allows for a more direct evaluation if NOL7 is necessary to drive ribosome biogenesis within the context of cancer.
Because NOL7 has been described as a tumour suppressor in previous studies [33,40–42], we analysed NOL7’s mRNA expression levels in breast and cervical cancer (based on cell lines utilized in this study) in Genotype-Tissue Expression (GTEx) unmatched normal and The Cancer Genome Atlas (TCGA) matched normal and tumour samples [1]. At odds with these previous studies that NOL7 is a tumour suppressor, but consistent with its hypothesized role in making ribosomes, NOL7 mRNA expression is significantly increased in both breast and cervical cancer tissue compared to normal (Figure 2F). This highlights the relevance of both cell lines used in this study, MCF10A (breast epithelial) and HeLa (cervical cancer). Moreover, we expect NOL7’s role in making ribosomes to be conserved to not only these cell lines but in all human cell types. Concordant with these results, a recent study found that NOL7 expression increases in and helps drive melanoma proliferation and metastasis [43]. These initial results point towards NOL7 having an important functional role in making ribosomes, and therefore in cell proliferation.
NOL7 is required for early pre-rRNA accumulation
Recent cryo-EM structures have offered insight into the location and potential function of Bud21 in yeast [4] and NOL7 in humans [5] within the SSU processome (pre-A0 5’ETS cleavage). We summarized the consistent interactions of Bud21 and NOL7 from these structures, showing that both proteins make contacts with the 5’ portion of the 5’ETS, the t-UTPs, UTP4 and UTP15, and the U3 snoRNP methyltransferase Nop1/fibrillarin (FBL; Figures 3A and B). We hypothesized that these direct interactions and the lack of Utp9 in the human SSU processome structure would give NOL7 the ability to function in pre-rRNA transcription. Although Bud21 has not been found to be required for rDNA transcription in yeast, it is possible that it was overlooked, since Bud21 is non-essential in yeast [23].
To gain insight into NOL7’s potential role in pre-rRNA transcription, we employed an imaging assay developed in our lab to measure nascent nucleolar rRNA biogenesis by 5-ethynyl uridine (5-EU) incorporation and biocompatible click chemistry in MCF10A cells [11]. By specifically observing the 5-EU signal residing within the nucleolus, we can measure the amount of nucleolar (pre-)rRNA produced over a 1-h time period as a readout of pre-rRNA transcription and stability. Cells depleted of NOL7 using siRNAs had an inhibition of nucleolar rRNA biogenesis of 76.6%, which is 3/4 of the positive control, depletion of the largest subunit of RNAP1, POLR1A (100.0% inhibition; Figure 3B and C, Data S3). While nucleolar rRNA biogenesis is inhibited when NOL7 is depleted it is not inhibited to the same extent as the known t-UTPs, like NOL11(Utp8 in yeast; 95.5%), HEATR1 (Utp10 in yeast; 118.1%), or to the same extent as depletion of the POLR1A (100%) (Figure 3C, Data S3) [11]. However, NOL7 depletion does have a consistent or greater nucleolar rRNA biogenesis percent inhibition compared to a disputed t-UTP [44], UTP4 (78.9%), and other SSU processome components NOP56 (82.1%), UTP20 (63.9%), and NOP58 (62.6%) (Figure 3C, Data S3). However, because this assay is a readout of both pre-rRNA transcription and stability, with this extent of inhibition of rRNA biogenesis we cannot definitively conclude that NOL7 is required for pre-rRNA transcription.
To more precisely define NOL7’s role in pre-rRNA transcription and stability, we measured steady state levels of the primary RNAP1 transcript by qRT-PCR in MCF10A cells depleted of NOL7 (Figure 3D). Although this assay reports the steady-state levels of the primary rRNA transcripts, which are already processed, it is often used as a read out for pre-rRNA transcription [45–48]. RNAP1 transcribes the polycistronic 47S pre-rRNA precursor. Cleavage at site A’ in the 5’ETS yields the 45S pre-rRNA precursor [49] that is more readily detectable by qRT-PCR (Figure 3D) [50]. NOL7 siRNA depletion resulted in a significant decrease in 47S/45S pre-rRNA levels compared to siNT, similar to the level of depletion of the positive control, NOL11 (Figure 3E). Again, we observed that the decrease in 47S/45S pre-rRNA after NOL7 depletion can be rescued by the expression of an siRNA-resistant version of NOL7, but not an empty vector control in HeLa cells (Figure 3F). By itself, and based on others’ prior conclusions, the 47S/45S qRT-PCR results point towards NOL7 playing a role in the transcription of the pre-rRNA.
Because NOL7 depletion reduces the steady state levels of early pre-rRNA precursors (Figure 3D-F), we tested if this was a result of reduced rDNA promoter activity in siNOL7 treated MCF10A cells. We used a dual-luciferase rDNA promoter (−410 to + 327) activity assay [14] to assess pre-rRNA transcription after NOL7 depletion in MCF10A cells. In contrast to the results in Figures 3E and F, NOL7 siRNA depletion did not reduce rDNA promoter activity compared to siNT, while the positive control siNOL11 did (Figure 3G). We also measured protein levels of RPA194 and UBTF by western blot to ask if NOL7 depletion reduced their abundance and thus pre-rRNA transcription. However, NOL7 depletion did not change either RPA194 or UBTF protein levels (Figure S5). NOL7 is thus not required to maintain rDNA promoter (−410 to + 327) activity in our reporter system. Taken together, we have evidence that NOL7 is required to maintain early pre-rRNA levels, but it is unclear if this is through direct regulation of pre-rRNA transcription, stability, or both.
NOL7 is required for U3 snoRNP mediated 5’ETS pre-rRNA processing to produce the small subunit rRNA
SSU processome factors are required for the processing of the pre-18S rRNA [24,44,49]. We tested NOL7’s role in pre-rRNA processing, specifically of the 5’ETS, which is mediated by the U3 snoRNP (Figure 4A) [51,52]. Due to NOL7’s interaction with the C/D box snoRNP component, FBL, we hypothesized that depletion of NOL7 could impact the stability of the U3 snoRNA and thus its function in 5’ETS processing. We measured U3 and U8 snoRNA levels by qRT-PCR in MCF10A cells following NOL7 siRNA depletion. U8 is a metazoan-specific C/D box snoRNA involved in large subunit processing [51,53]. We observed a modest, yet significant, decrease in U3 snoRNA levels upon NOL7 depletion, but not upon depletion of NOL11. The small changes in U8 snoRNA levels upon NOL7 depletion were not significant (Figure 4B). These decreases in U3 snoRNA levels were not associated with any changes in FBL protein levels as measured by western blotting (Figure S5). These results suggest a role for NOL7 in maintaining the stability of the U3 snoRNA.
Figure 4.
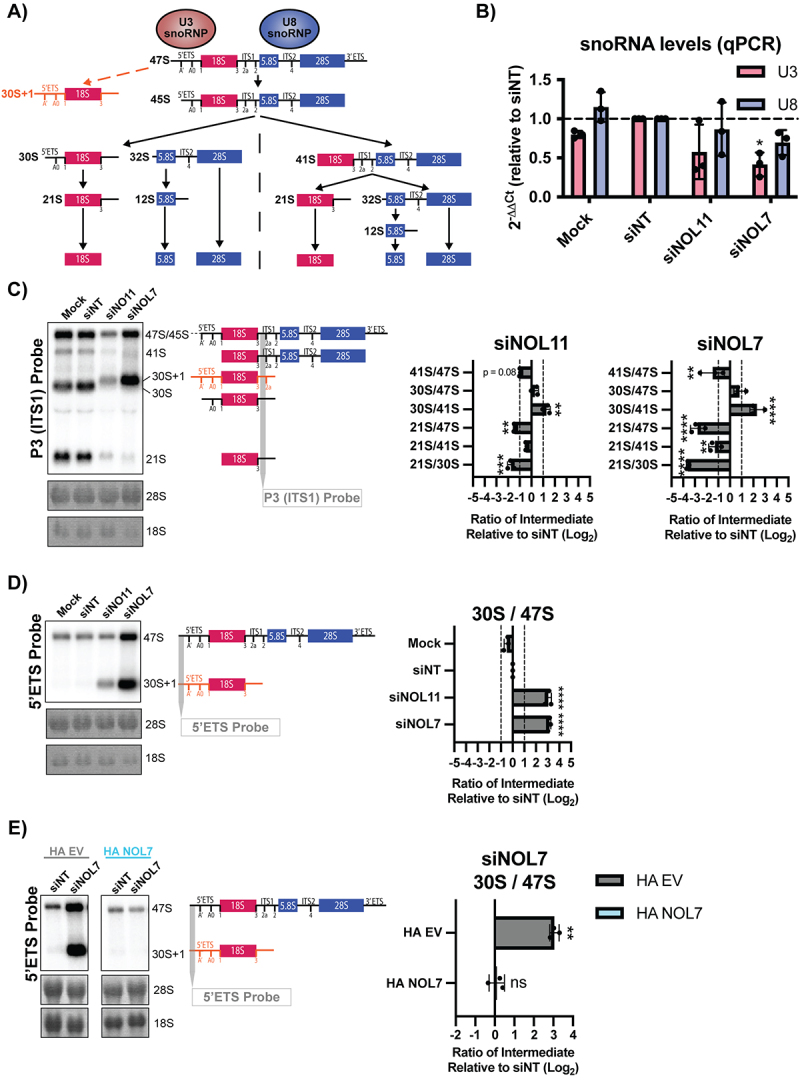
NOL7 is required for U3 snoRNA-mediated small subunit pre-rRNA processing in human tissue culture cells.
A. pre-rRNA processing in human cells. The 47S pre-rRNA precursor undergoes a series of modification and processing steps to yield the mature 18S, 5.8S, and 28S rRNAs. Upon inhibition of pre-rRNA cleavage in the 5’ETS, mediated by the U3 snoRNP, accumulation of an 5’ETS extended 30S + 1 precursor occurs (orange). The steps that produce the 18S rRNA (small subunit) are indicated in red, and the steps that produce the 5.8S and 28S rRNAs (large subunit) are indicated in blue. The U8 snoRNA is essential for processing of the rRNAs that make the large ribosomal subunit.
B. NOL7 siRNA depletion in MCF10A cells results in reduced U3 snoRNA steady-state levels. qRT-PCR measuring U3 and U8 snoRNA transcript levels in MCF10A cells. 2−ΔΔCt were measured relative to 7SL internal control and siNT negative control sample. 3 technical replicates of 3 biological replicates, plotted mean ± SD, dotted line at siNT value y = 1. Data were analysed by one-way ANOVA with Dunnett’s multiple comparisons test, * p ≤ 0.05.
C. NOL7 is required for pre-18S rRNA processing in MCF10A cells. (Left) Representative northern blot using a P3 ITS1 probe (indicated in grey) measuring steady state levels of pre-rRNA precursors leading to the 18S rRNA. The 47S/45S precursors are not separated on these blots but are both detected with this probe. Methylene blue staining of 28S and 18S was used to show even loading. siNT is a negative control and siNOL11 is a positive control. (Right) Quantification of northern blots using ratio analysis of multiple precursors (RAMP) 15 relative to siNT negative control. 3 biological replicates, plotted mean ± SD. Data were analysed by two-way ANOVA, ** p ≤ 0.01, *** p ≤ 0.001, **** ≤ 0.0001. The 47S precursor ratios on this graph are a combination of the quantification of the indicated 47S/45S band (left) since these precursors are not separated on these blots.
D. NOL7 is required for 5’ETS cleavage in MCF10A cells. (Left) Representative northern blot using a 5’ETS probe (upstream of the A’ site, indicated in grey) measuring steady state levels of 47S and 30S + 1 pre-rRNA precursors. Methylene blue staining of 28S and 18S was used to show even loading. siNT is a negative control and siNOL11 is a positive control. (Right) Quantification of northern blots using ratio analysis of multiple precursors (RAMP) 15 relative to siNT negative control. 3 biological replicates, plotted Log2 (mean ± SD). Data were analysed by one-way ANOVA with Dunnett’s multiple comparisons test, **** p ≤ 0.0001.
E. Accumulation of the 30S + 1 pre-rRNA after siNOL7 depletion can be rescued by introduction of an si-resistant version of NOL7 in Hela cells. (Left) Representative northern blot using a 5’ETS probe (upstream of the A’ site, indicated in grey) measuring steady state levels of 47S and 30S + 1 pre-rRNA precursors upon NOL7 depletion and si-resistant rescue with either empty vector (HA EV) or HA-tagged NOL7 (HA NOL7) in HeLa cells. Methylene blue staining of 28S and 18S show even loading. siNT is a negative control. (Right) Quantification of northern blots using ratio analysis of multiple precursors (RAMP) 15 relative to siNT negative control. 3 biological replicates, plotted Log2 (mean ± SD). Data were analysed by Student’s t-test, ** p ≤ 0.05, ns = not significant.
NOL7 was previously identified in a screen for human proteins required for pre-rRNA processing, where its depletion led to a pre-18S processing defect [54]. To further validate those findings, we performed northern blots to assess pre-18S rRNA processing in HeLa cells and quantified these results using ratio analysis of multiple precursors (RAMP) [16] to detect defects within the processing pathway. First, we probed ITS1 upstream of cleavage site 2 (probe P3) to measure changes in pre-rRNA intermediates leading to the mature 18S rRNA (Figure 4A). Upon siRNA depletion of NOL7 and the positive control NOL11, we observed a build-up of 30S pre-rRNA intermediates and a subsequent decrease in 21S pre-rRNA precursors, indicating an inhibition in 5’ETS processing (Figure 4C). Moreover, NOL7 depletion results in a significant decrease in 41S levels compared to 47S/45S primary pre-rRNA precursors, likely attributed to the requirement for 5’ETS cleavage to occur to produce the 41S pre-rRNA as well.
Interestingly, in siNOL7 and siNOL11 treated lanes in the northern blots (Figure 4C) we noticed an upwards shift in the mobility of the 30S pre-rRNA. It is known that upon depletion of factors required for processing of the 5’ETS, there is a build-up of an aberrant 30S + 1 intermediate, with an extended 5’ end due to the lack of A’ cleavage compared to the 30S intermediate (Figure 4A) [44,49,55,56]. To more specifically detect this defect, we used a 5’ETS probe upstream of the A’ cleavage site to measure the levels of the 47S and 30S + 1 pre-rRNAs. As expected, we observed a significant increase in the 30S + 1 intermediate compared to the upstream 47S primary transcript when both NOL7 and the positive control NOL11 were depleted (Figure 4D). This 5’ETS processing defect could be rescued in HeLa cells expressing an siRNA-resistant version of NOL7, but not by an empty vector control (Figure 4E). We conclude from these results that NOL7 is required for 5’ETS pre-rRNA processing in human tissue culture cells.
Since NOL7 is required for pre-18S processing, we tested whether NOL7 depletion would lead to a downstream reduction in the levels of mature 18S rRNA by Agilent BioAnalyzer analysis. There was a significant increase in the 28S/18S rRNA ratio upon NOL7 depletion in MCF10A cells (Figure 5A), pointing towards an 18S rRNA maturation defect. When quantifying 18S and 28S overall levels specifically, it was revealed that this increased ratio was indeed due to only decreases in 18S levels and not due to increases in 28S levels (Figure 5B and C, S6). These results indicate that NOL7’s role is specific to the maturation of the 18S rRNA. The increase in the 28S/18S ratio that was a result of decreases in 18S levels could also be rescued with the introduction of an si-resistant version of NOL7 but not an empty vector control in HeLa cells (Figure 5D-F). NOL7 plays a role in the 5’ETS pre-rRNA cleavage that leads to the 18S rRNA, through a combination of its direct presence within the SSU processome to mediate this cleavage and a likely more modest role in additionally maintaining normal U3 snoRNA steady-state levels.
Figure 5.
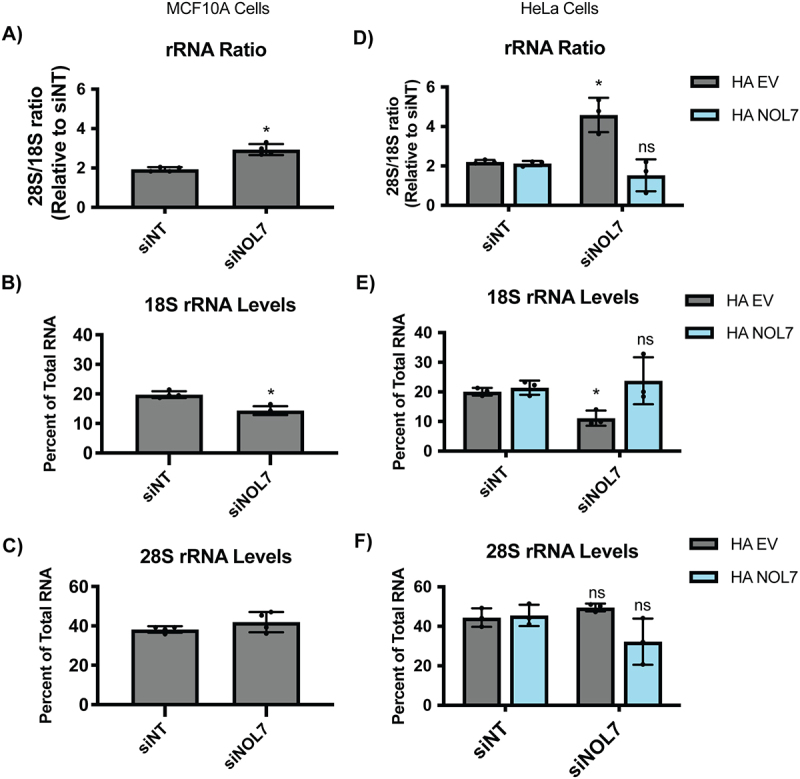
NOL7 is required to maintain mature 18S rRNA levels in human tissue culture cells.
A. NOL7 is required for production of the normal 28S/18S mature rRNA ratio in MCF10A cells. Agilent BioAnalyzer analysis of ratio of mature 28S to 18S rRNAs from MCF10A cells depleted of NOL7. 3 biological replicates, plotted mean ± SD. Data were analysed by Student’s t-test relative to siNT negative control, * p ≤ 0.05.
B-C. NOL7 is small subunit (18S) specific in its role in rRNA maturation in MCF10A cells.
B. NOL7 is required for the production of the 18S rRNA. Agilent BioAnalyzer analysis of percent of overall RNA levels for 18S rRNA in MCF10A cells depleted of NOL7. 3 biological replicates, plotted mean ± SD. Data were analysed by Student’s t-test relative to siNT negative control, * p ≤ 0.05.
C. NOL7 is not required for the production of the 28S rRNA. Panel as above (B) except that the large subunit (28S) rRNA was measured.
D. siNOL7 increase in 28S/18S mature rRNA ratio can be rescued by introduction of an si-resistant version of NOL7 in Hela cells. Agilent BioAnalyzer analysis of ratio of mature 28S to 18S rRNAs upon NOL7 depletion and si-resistant rescue with either empty vector (HA EV) or HA-tagged NOL7 (HA NOL7) in HeLa cells. 3 biological replicates, plotted mean ± SD. Data were analysed by Student’s t-test relative to siNT negative control, * p ≤ 0.05, ns = not significant.
E-F. siNOL7 small subunit (18S) specific role in rRNA maturation can be rescued by introduction of an si-resistant version of NOL7 in HeLa cells.
E. siNOL7 decreases in 18S rRNA levels can be rescued by introduction of an si-resistant version of NOL7. Agilent BioAnalyzer analysis of percent of overall RNA levels upon NOL7 depletion and si-resistant rescue with either empty vector (HA EV) or HA-tagged NOL7 (HA NOL7) in HeLa cells. 3 biological replicates, plotted mean ± SD. Data were analysed by Student’s t-test relative to siNT negative control, * p ≤ 0.05, ns = not significant.
F. siNOL7 and si-resistant version of NOL7 rescue do not change 28S levels. Panel as above in (E) except that large subunit (28S) rRNA was measured.
NOL7 is required for normal levels of protein synthesis and its depletion leads to induction of the nucleolar stress response
We hypothesized that the role for NOL7 in early pre-rRNA accumulation and pre-18S processing that we have observed would lead to downstream effects on ribosome function. Therefore, we performed a puromycin incorporation assay to measure changes in global protein synthesis in MCF10A cells depleted of NOL7 [10,13]. After siRNA knockdown, cells were treated with 1 μM puromycin, which is incorporated into the nascent polypeptide chain over a time period of 1 h. As expected, depletion of NOL7 led to a significant decrease in global protein synthesis to a similar extent as that of the positive control siNOL11 (Figure 6A and B).
Figure 6.
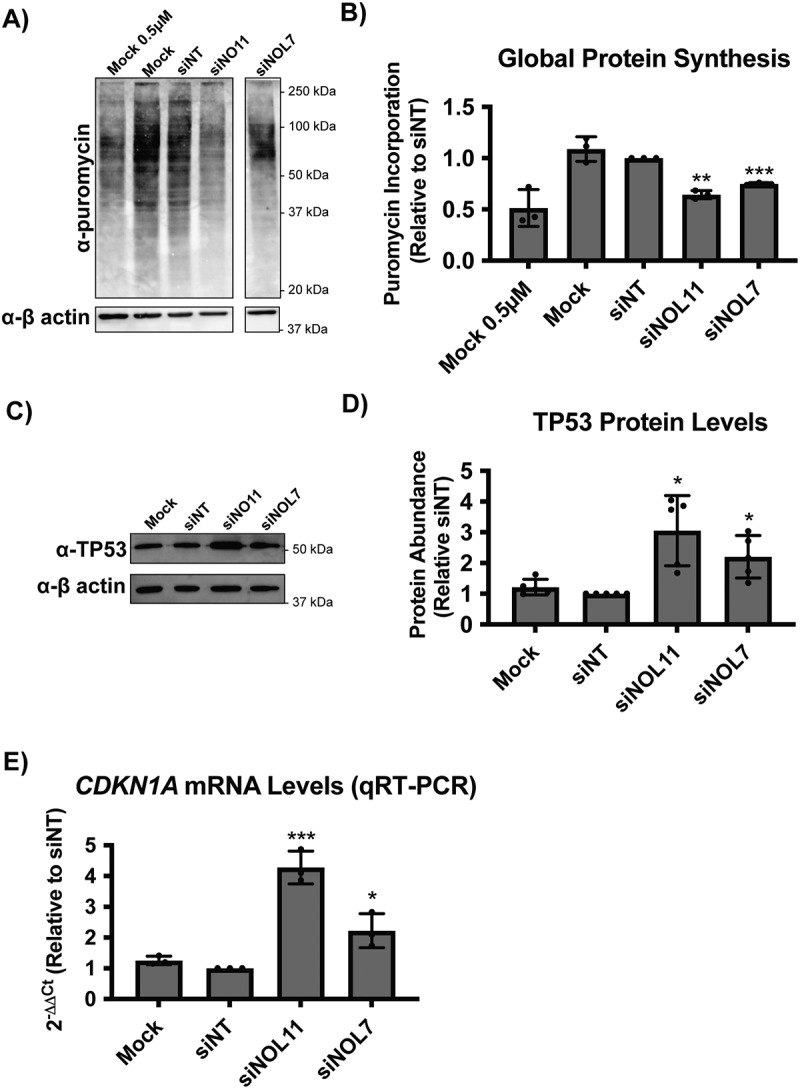
NOL7 siRNA depletion reduces global protein synthesis and causes the nucleolar stress response in MCF10A cells.
A. NOL7 siRNA depletion reduces global protein synthesis. MCF10A cells were depleted with siRNAs for 72 h, followed by a 1 h treatment of 1 µM puromycin. Total protein was analysed by western blot. Representative western blot shown where Mock and siNT are negative controls, siNOL11 is a positive control. Mock 0.5 µM was included to confirm robustness of the signal quantification. β-actin was used as a loading control.
B. Quantification of the decrease in global protein synthesis after NOL7 siRNA depletion in MCF10A cells. Quantification of western blots from (A) normalized to β-actin signal and relative to siNT negative control. 3 biological replicates, plotted mean ± SD. Data were analysed by one-way ANOVA with Dunnett’s multiple comparisons test, ** p ≤ 0.01.
C-E. siNOL7 depletion induces the nucleolar stress response in MCF10A cells.
C. NOL7 siRNA depletion leads to increased TP53 protein levels in MCF10A cells. Representative western blot for TP53 shown where Mock and siNT are negative controls, siNOL11 is a positive control. β-actin is a loading control.
D. Quantification of the increase in TP53 levels after NOL7 siRNA depletion in MCF10A cells. Quantification of western blots from (C) normalized to β-actin signal and relative to siNT negative control. 5 biological replicates, plotted mean ± SD. Data were analysed by one-way ANOVA with Dunnett’s multiple comparisons test, * p ≤ 0.05.
E. NOL7 siRNA depletion significantly increases CDKN1A (p21) mRNA levels in MCF10A cells. qRT-PCR measuring CDKN1A mRNA levels. 2− ΔΔCt were measured relative to 7SL internal control and siNT negative control sample. siNOL11 is a positive control. 3 technical replicates of 3 biological replicates, plotted mean ± SD. Data were analysed by one-way ANOVA with Dunnett’s multiple comparisons test, *** p ≤ 0.001, * p ≤ 0.05.
Because we have defined a role of NOL7 in ribosome biogenesis and in maintaining cell viability, we reasoned that depletion of NOL7 would lead to the nucleolar stress response. As a result of impaired ribosome biogenesis, cells can undergo the nucleolar stress response [57,58]. This leads to changes in nucleolar morphology and the presence of free ribosome components not incorporated into mature ribosomes [namely RPL5 (uL18), RPL11 (uL5), and the 5S rRNA], which results in induction of TP53 levels, downstream induction of CDKN1A (p21) expression, and ultimately cell cycle arrest and apoptosis [58,59]. To check for the induction of the TP53 mediated nucleolar stress response, we measured TP53 levels in MCF10A cells by western blot after NOL7 siRNA depletion. As expected, we observed a significant increase of TP53 levels in NOL7 depleted cells (Figure 6C and D). Additionally, since TP53 is a transcription factor for CDKN1A, which plays an important role in inducing cell cycle arrest [60], we measured CDKN1A mRNA levels by qRT-PCR. After depletion of NOL7 or our positive control NOL11, CDKN1A mRNA levels were also strikingly increased (Figure 6E). These results are consistent with the observed increase in CDKN1A and another cyclin-dependent kinase inhibitor, CDKN1B (p27), in melanoma cell lines after NOL7 knockdown [43].
Discussion
We have shown here that the nucleolar protein, NOL7, plays a critical role in early ribosome biogenesis, specifically in maintaining early pre-rRNA levels and in pre-18S processing, in human cells. siRNA depletion of NOL7 siRNA leads to a reduction in 47S/45S pre-rRNA levels, inhibition of 5’ETS processing, and reduced mature 18S levels. Each of these endpoints were rescued by the introduction of an si-resistant version of NOL7. These defects lead to a downstream reduction in protein synthesis and to the induction of the nucleolar stress response. These results and its presence within the SSU processome indicate NOL7’s role to be consistent with that of the yeast Bud21 ortholog in human cells.
Some of the roles for NOL7 in human ribosome biogenesis that we have found were expected based on its orthologous yeast protein, Bud21, and some were not. Previously, we had discovered Bud21 as a member of the SSU processome in yeast, but Bud21 was not previously tested for a role in pre-rRNA transcription in this organism [23,24]. Bud21 is not essential in yeast and its depletion leads to cold-sensitivity [23], a known hallmark of non-essential proteins involved in making ribosomes [61]. In contrast, NOL7 is essential in most human cancer cell lines as found in the DepMap project (https://depmap.org/) by both shRNA and CRISPR genetic perturbation screens across hundreds of cell lines [62,63]. NOL7’s newfound role in ribosome biogenesis is consistent with it being regarded as an essential protein across cancer cell lines. The relevance of NOL7’s function in cancer is further emphasized by cancer cells’ increased reliance on ribosome biogenesis, leading to new therapeutics being developed to target ribosome biogenesis, specifically RNAP1 (i.e. BMH-21) [64,65]. It is possible, given the differences in 5’ETS expansion sequences, interacting proteins, and the lack of Utp9 that human NOL7 has gained a more impactful function in pre-rRNA transcription and early pre-rRNA stability compared to yeast Bud21 that underlies its essential nature in humans.
While most of our NOL7 findings are consistent with what we know about the function of other t-UTPs, there are some distinct differences that suggest NOL7’s role is not completely aligned with them. NOL7 is necessary for early pre-rRNA accumulation and 5’ETS processing that leads to the production of the SSU. More specifically, we discovered that NOL7 is required to maintain 47S/45S pre-rRNA levels and for nucleolar rRNA biogenesis to a modest level. Each of these assays measures not only pre-rRNA transcription but also pre-rRNA stability. In contrast, in a reporter gene assay for rDNA promoter activity, NOL7 depletion had no effect. Perhaps sequences downstream of the rDNA promoter reporter gene sequences (−410 to + 327) are required for NOL7’s function [14]. Consistent with this, only a fraction of the entire 5’ETS (post A’ cleavage) is modelled in the human SSU processome structure [5], none of which is included in our rDNA reporter gene. Combining the results together, we can conclude that NOL7 is required for accumulation of the pre-rRNA, but we cannot conclude NOL7 is a t-UTP required for pre-rRNA transcription due to our conflicting results.
Our discovery of NOL7’s essential role in making ribosomes is at odds with several previous studies on NOL7’s role as a tumour suppressor. These studies focused on NOL7’s role in post-transcriptional regulation of mRNAs, more precisely mRNA stabilization through direct interactions with mRNAs and their processing machinery within the nucleus [33,40–42]. These interactions lead to a tumour suppressor effect due to preferential stabilization of antiangiogenic transcripts, of which TSP-1 has been validated [42]. It is worth noting that within our PANTHER overrepresentation analysis of Bud21 interacting proteins, but not for the shorter list of known NOL7 interactors, we discovered an enrichment of GO biological processes consistent with this other NOL7 function including: intracellular mRNA localization (GO:0008298), positive regulation of mRNA catabolic process (GO:0061014), regulation of mRNA stability (GO:0043488), and RNA transport (GO:0050658) (Figure S1B). Similarly, there is precedent for other ribosome biogenesis factors having functions in cellular processes outside the nucleolus, including nucleolin and FANCI, in which their roles are regulated by their interacting partners and subcellular localization [8,66]. It is unsurprising that the ubiquitous process of making ribosomes has cross-talk mechanisms to coordinate itself with other cellular processes. Our data and previously published work [43] strongly suggest NOL7’s role to be important for tumour growth and progression, but previous results indicate a role opposing this as well [33,40–42]. It remains to be further explored to what extent and in which contexts NOL7 functions outside of ribosome biogenesis, particularly through modulation of mRNA subsets.
We discovered that depletion of NOL7 reduces cell viability and increases TP53 and CDKN1A levels, likely as a result of the nucleolar stress response. These results agree with previously published work [43] in which NOL7 depletion increases CDKN1A levels and leads to cell cycle S-phase arrest in melanoma cell lines. While the nucleolar stress response occurs as a result of impaired ribosome biogenesis [58] (reviewed in [59]), it is also plausible that NOL7 could act in a more direct manner to modulate the cell cycle. Regulation of cell cycle (GO:0051726) was one of the top overrepresented biological processes categories among NOL7 interacting proteins (Figure S1A). While our data and analysis emphasize NOL7’s role in ribosome biogenesis, it remains to be studied if NOL7’s interaction with these proteins involved in the cell cycle has a more direct impact on cell cycle regulation and overall cell viability.
NOL7 is another example of the increased complexities of ribosome biogenesis in humans as compared to yeast. We have demonstrated this through NOL7’s role in early pre-rRNA accumulation and processing. It is also exemplified by NOL7’s other known function outside the nucleolus in regulation of nuclear mRNA stability [33,40–42], adding even more layers of complexity. These results and future studies will certainly help uncover the finer details of human ribosome biogenesis, specifically pre-rRNA transcription and SSU processome function, that will lead to better understanding of therapeutic design options in diseases such as cancer and ribosomopathies.
Supplementary Material
Acknowledgments
We thank the Yale Center for Genome Analysis (YCGA) for performing Agilent BioAnalyzyer analysis. We thank members of the Baserga laboratory for their helpful discussions and insight during the preparation and writing of this manuscript as well. We acknowledge the use of CellProfiler for image analysis (http://www.cellprofiler.org/).
Funding Statement
This work was supported by the following grants from the National Institutes of Health (NIH): R35GM131687 (to S.J.B.), 1F31DE030332 (to M.A.M.), F31AG058405 (to L.M.O.), F31DE026946 (to K.I.F.), T32GM007223 (to C.J.B., K.I.F., L.M.O., M.A.M., and S.J.B.), and T32GM007205, T32HD007149, and F30DK109582 (to S.B.S.).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15476286.2023.2217392
References
- [1].Goldman MJ, Craft B, Hastie M, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Needleman SB, Wunsch CD.. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. [DOI] [PubMed] [Google Scholar]
- [3].Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Barandun J, Chaker-Margot M, Hunziker M, et al. The complete structure of the small-subunit processome. Nat Struct Mol Biol. 2017;24:944–953. [DOI] [PubMed] [Google Scholar]
- [5].Singh S, Vanden Broeck A, Miller L, et al. Nucleolar maturation of the human small subunit processome. Science. 2021;373:eabj5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Oughtred R, Rust J, Chang C, et al. The BioGRID database: a comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021;30:187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Drew K, Wallingford JB, Marcotte EM. Hu.Map 2.0: integration of over 15,000 proteomic experiments builds a global compendium of human multiprotein assemblies. Mol Syst Biol. 2021;17:e10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sondalle SB, Longerich S, Ogawa LM, et al. Fanconi anemia protein FANCI functions in ribosome biogenesis. Proc Natl Acad Sci U S A. 2019;116:2561–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yang X, Boehm JS, Yang X, et al. A public genome-scale lentiviral expression library of human ORFs. Nat Methods. 2011;8:659–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Farley-Barnes KI, McCann KL, Ogawa LM, et al. Diverse regulators of human ribosome biogenesis discovered by changes in nucleolar number. Cell Rep. 2018;22:1923–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bryant CJ, McCool MA, Abriola L, et al. A high-throughput assay for directly monitoring nucleolar rRNA biogenesis. Open Biol. 2022;12:210305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Reimer G, Pollard KM, Penning CA, et al. Monoclonal autoantibody from a (New Zealand black x New Zealand white)F1 mouse and some human scleroderma sera target an Mr 34,000 nucleolar protein of the U3 RNP particle. Arthritis Rheum. 1987;30:793–800. [DOI] [PubMed] [Google Scholar]
- [13].Schmidt EK, Clavarino G, Ceppi M, et al. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6:275–277. [DOI] [PubMed] [Google Scholar]
- [14].Ghoshal K, Majumder S, Datta J, et al. Role of human ribosomal RNA (rRNA) promoter methylation and of methyl-CpG-binding protein MBD2 in the suppression of rRNA gene expression. J Biol Chem. 2004;279:6783–6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mansour FH, Pestov DG. Separation of long RNA by agarose-formaldehyde gel electrophoresis. Anal Biochem. 2013;441:18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang M, Anikin L, Pestov DG. Two orthogonal cleavages separate subunit RNAs in mouse ribosome biogenesis. Nucleic Acids Res. 2014;42:11180–11191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Aubert M, O’Donohue MF, Lebaron S, et al. Pre-ribosomal RNA processing in human cells: from mechanisms to congenital diseases. Biomolecules. 2018;8:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bohnsack KE, Bohnsack MT. Uncovering the assembly pathway of human ribosomes and its emerging links to disease. Embo J. 2019;38:e100278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Woolford JL Jr., Baserga SJ. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195:643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chaker-Margot M, Hunziker M, Barandun J, et al. Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis. Nat Struct Mol Biol. 2015;22:920–923. [DOI] [PubMed] [Google Scholar]
- [21].Perez-Fernandez J, Roman A, De Las Rivas J, et al. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol Cell Biol. 2007;27:5414–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Krogan NJ, Peng WT, Cagney G, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol Cell. 2004;13:225–239. [DOI] [PubMed] [Google Scholar]
- [23].Dragon F, Gallagher JE, Compagnone-Post PA, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gallagher JE, Dunbar DA, Granneman S, et al. RNA polymerase I transcription and pre-Rrna processing are linked by specific SSU processome components. Genes Dev. 2004;18:2506–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Phipps KR, Charette J, Baserga SJ. The small subunit processome in ribosome biogenesis-progress and prospects. Wiley Interdiscip Rev RNA. 2011;2:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Huang YC, Tseng SF, Tsai HJ, et al. Direct interaction between Utp8p and Utp9p contributes to rRNA processing in budding yeast. Biochem Biophys Res Commun. 2010;393:297–302. [DOI] [PubMed] [Google Scholar]
- [27].Eswara MB, McGuire AT, Pierce JB, et al. Utp9p facilitates Msn5p-mediated nuclear reexport of retrograded tRnas in Saccharomyces cerevisiae. Mol Biol Cell. 2009;20:5007–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Risler JK, Kenny AE, Palumbo RJ, et al. Host co-factors of the retrovirus-like transposon Ty1. Mob DNA. 2012;3:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shah AN, Cadinu D, Henke RM, et al. Deletion of a subgroup of ribosome-related genes minimizes hypoxia-induced changes and confers hypoxia tolerance. Physiol Genomics. 2011;43:855–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Usher J, Balderas-Hernandez V, Quon P, et al. Chemical and synthetic genetic array analysis identifies genes that suppress xylose utilization and fermentation in Saccharomyces cerevisiae. G3 (Bethesda). 2011;1:247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cheng Y, Zhu H, Du Z, et al. Eukaryotic translation factor eIF5A contributes to acetic acid tolerance in Saccharomyces cerevisiae via transcriptional factor Ume6p. Biotechnol Biofuels. 2021;14:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ogawa LM, Buhagiar AF, Abriola L, et al. Increased numbers of nucleoli in a genome-wide RNAi screen reveal proteins that link the cell cycle to RNA polymerase I transcription. Mol Biol Cell. 2021;32:956–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhou G, Doci CL, Lingen MW. Identification and functional analysis of NOL7 nuclear and nucleolar localization signals. BMC Cell Biol. 2010;11:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mi H, Huang X, Muruganujan A, et al. PANTHER version 11: expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Freed EF, Baserga SJ. The C-terminus of Utp4, mutated in childhood cirrhosis, is essential for ribosome biogenesis. Nucleic Acids Res. 2010;38:4798–4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Freed EF, Prieto JL, McCann KL, et al. NOL11, implicated in the pathogenesis of North American Indian childhood cirrhosis, is required for pre-rRNA transcription and processing. PLoS Genet. 2012;8:e1002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hu Y, Flockhart I, Vinayagam A, et al. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinf. 2011;12:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kim W, Underwood RS, Greenwald I, et al. OrthoList 2: a new comparative genomic analysis of human and caenorhabditis elegans genes. Genetics. 2018;210:445–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Soule HD, Maloney TM, Wolman SR, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- [40].Hasina R, Pontier AL, Fekete MJ, et al. NOL7 is a nucleolar candidate tumor suppressor gene in cervical cancer that modulates the angiogenic phenotype. Oncogene. 2006;25:588–598. [DOI] [PubMed] [Google Scholar]
- [41].Mankame TP, Lingen MW. The RB tumor suppressor positively regulates transcription of the anti-angiogenic protein NOL7. Neoplasia. 2012;14:1213–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Doci CL, Zhou G, Lingen MW. The novel tumor suppressor NOL7 post-transcriptionally regulates thrombospondin-1 expression. Oncogene. 2013;32:4377–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Li Y, Zhong C, Wang J, et al. NOL7 facilitates melanoma progression and metastasis. Signal Transduct Target Ther. 2021;6:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Prieto JL, McStay B. Recruitment of factors linking transcription and processing of pre-Rrna to nor chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev. 2007;21:2041–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bywater MJ, Poortinga G, Sanij E, et al. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell. 2012;22:51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kang CW, Hannan KM, Blackburn AC, et al. The therapeutic potential of RNA Polymerase I transcription inhibitor, CX-5461, in uterine leiomyosarcoma. Invest New Drugs. 2022;40:529–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cui C, Tseng H. Estimation of ribosomal RNA transcription rate in situ. Biotechniques. 2004;36:134–138. [DOI] [PubMed] [Google Scholar]
- [48].Sorino C, Catena V, Bruno T, et al. Che-1/AATF binds to RNA polymerase I machinery and sustains ribosomal RNA gene transcription. Nucleic Acids Res. 2020;48:5891–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sloan KE, Bohnsack MT, Schneider C, et al. The roles of SSU processome components and surveillance factors in the initial processing of human ribosomal RNA. RNA. 2014;20:540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Woolnough JL, Atwood BL, Liu Z, et al. The regulation of rRNA gene transcription during directed differentiation of human embryonic stem cells. PLoS ONE. 2016;11:e0157276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Langhendries JL, Nicolas E, Doumont G, et al. The human box C/D snoRNAs U3 and U8 are required for pre-rRNA processing and tumorigenesis. Oncotarget. 2016;7:59519–59534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hughes JM, Ares M Jr.. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5’ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. Embo J. 1991;10:4231–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Peculis BA, Steitz JA. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993;73:1233–1245. [DOI] [PubMed] [Google Scholar]
- [54].Tafforeau L, Zorbas C, Langhendries JL, et al. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol Cell. 2013;51:539–551. [DOI] [PubMed] [Google Scholar]
- [55].Mullineux ST, Lafontaine DL. Mapping the cleavage sites on mammalian pre-rRNAs: where do we stand? Biochimie. 2012;94:1521–1532. [DOI] [PubMed] [Google Scholar]
- [56].Lazdins IB, Delannoy M, Sollner-Webb B. Analysis of nucleolar transcription and processing domains and pre-rRNA movements by in situ hybridization. Chromosoma. 1997;105:481–495. [DOI] [PubMed] [Google Scholar]
- [57].Boulon S, Westman BJ, Hutten S, et al. The nucleolus under stress. Mol Cell. 2010;40:216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. Embo J. 2003;22:6068–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Lindstrom MS, Bartek J, Maya-Mendoza A. P53 at the crossroad of DNA replication and ribosome biogenesis stress pathways. Cell Death Differ. 2022;29:972–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].el-Deiry WS, Harper JW, O’Connor PM, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- [61].Guthrie C, Nashimoto H, Nomura M. Structure and function of E. coli ribosomes. 8. Cold-sensitive mutants defective in ribosome assembly. Proc Natl Acad Sci U S A. 1969;63:384–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Tsherniak A, Vazquez F, Montgomery PG, et al. Defining a cancer dependency map. Cell. 2017;170:564–76 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].McFarland JM, Ho ZV, Kugener G, et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat Commun. 2018;9:4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Pelletier J, Thomas G, Volarevic S. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer. 2018;18:51–63. [DOI] [PubMed] [Google Scholar]
- [65].Harold CM, Buhagiar AF, Cheng Y, et al. Ribosomal RNA transcription regulation in breast cancer. Genes (Basel). 2021;12:502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Berger CM, Gaume X, Bouvet P. The roles of nucleolin subcellular localization in cancer. Biochimie. 2015;113:78–85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


