Abstract
Background:
Ovarian cancer is one of the most widespread malignant diseases of the female reproductive system worldwide. The plurality of ovarian cancer is diagnosed with metastasis in the abdominal cavity. Epithelial-mesenchymal transition (EMT) exerts a vital role in tumor cell metastasis. However, it remains unclear whether long non-coding RNA (lncRNA) are implicated in EMT and influence ovarian cancer cell invasion and metastasis. This study was designed to investigate the impacts of lncRNA AC005224.4 on ovarian cancer.
Methods:
LncRNA AC005224.4, miR-140-3p, and snail family transcriptional repressor 2 (SNAI2) expression levels in ovarian cancer and normal ovarian tissues were determined using real-time quantitative polymerase chain reaction (qRT-PCR). Cell Counting Kit-8 (CCK-8) and Transwell (migration and invasion) assays were conducted to measure SKOV3 and CAOV-3 cell proliferation and metastasis. E-cadherin, N-cadherin, Snail, and Vimentin contents were detected using Western blot. Nude mouse xenograft assay was utilized to validate AC005224.4 effects in vivo. Dual-luciferase reporter gene assay confirmed the targeted relationship between miR-140-3p and AC005224.4 or SNAI2.
Results:
AC005224.4 and SNAI2 upregulation and miR-140-3p downregulation were observed in ovarian cancer tissues and cells. Silencing of AC005224.4 observably moderated SKOV3 and CAOV-3 cell proliferation, migration, invasion, and EMT process in vitro and impaired the tumorigenesis in vivo. miR-140-3p was a target of AC005224.4 and its reduced expression level was mediated by AC005224.4. miR-140-3p mimics decreased the proliferation, migration, and invasion of ovarian cancer cells. SNAI2 was identified as a novel target of miR-140-3p and its expression level was promoted by either AC005224.4 overexpression or miR-140-3p knockdown. Overexpression of SNAI2 also facilitated ovarian cancer cell viability and metastasis.
Conclusion:
AC005224.4 was confirmed as an oncogene via sponging miR-140-3p and promoted SNAI2 expression, contributing to better understanding of ovarian cancer pathogenesis and shedding light on exploiting the novel lncRNA-directed therapy against ovarian cancer.
Keywords: Epithelial-mesenchymal transition, LncRNA AC005224.4, Ovarian cancer, SNAI2, miR-140-3p, Tumor metastasis
Introduction
Ovarian cancer, as one of the most frequent gynecological malignancies among women worldwide, possesses high morbidity and mortality, and it is coupled with great difficulty in early detection.[1–3] Approximately 80% of the patients are first diagnosed with regional or distant metastasis in the advanced stage.[4] Maximal cytoreductive surgery followed by platinum-taxane chemotherapy remains the standard first-line therapy for ovarian cancer.[5,6] Despite the advancements in surgery and chemotherapy, the prognosis of ovarian cancer is still unsatisfactory, with an overall 5-year survival rate of about 30% in the advanced stage.[7,8] High-grade serous ovarian cancer, accepted as the most prevalent type of ovarian cancer, is an aggressive disease with high relapse and abdominal metastasis rates at late stages.[9] Therefore, it is urgently needed to conduct more extensive research to better understand the molecular changes that induce tumor cell metastasis in ovarian cancer.
Growing evidence has revealed that epithelial-mesenchymal transition (EMT) plays a critical role in cancer cell metastatic dissemination events, which endow cancer cells with a more motile and invasive phenotype.[10–12] During carcinoma progression, the occurrence of EMT makes tumor cells more aggressive. Meanwhile, after EMT, tumor cells gain more metastatic and invasion potential, similar to embryonic mesenchymal cells, together with an increased ability to invade adjacent stroma to form new tumor foci.[13–15] Moreover, the emergence of new evidence has suggested that ovarian cancer cell acquiring invasive properties is accompanied by epithelial feature loss and mesenchymal feature gain.[16,17] Furthermore, a previous study has pointed out that EMT is regulated by a cluster of zinc-finger transcription factors, including ZEB1, ZEB2, snail family transcriptional repressor 1 (SNAI1), and SNAI2.[18] As has been evidenced previously, SNAI2 protein is closely involved in EMT, which is implicated in the development of diverse tumors with its antiapoptotic activity.[19] Li et al[20] have suggested that SNAI2-3′UTR promotes ovarian cancer cell invasion via upregulating myristoylated alanine-rich C-kinase substrate (MARCKS) expression. However, the effective mechanism of SNAI2 in ovarian cancer, especially its reciprocities with non-coding RNAs, remains vague.
The non-coding RNAs, including long non-coding RNAs (lncRNAs, >200 nt) and microRNAs (miRNAs, about 20–22 nt), are important constituents of the mammalian transcriptome, which has already been the hottest research subject.[21] Multiple studies have previously manifested that the abnormal expression of lncRNAs is closely related to cancer progression, which leads to the dysregulation of related gene products.[22,23] Liang et al[24] have evidenced that lncRNA PTAR accelerates the invasion-metastasis and EMT in serous ovarian cancer via targeting miR-101-3p. Lin et al[25] have demonstrated that lncRNA DANCR facilitates tumor angiogenesis and growth in ovarian cancer via binding to miR-145. According to our microarray analysis and The Cancer Genome Atlas (TCGA)-ovarian cancer database, five lncRNAs (AC005224.4, AC009093.2, UG0898H09, AC015912.3, and AL139081.1) associated with the EMT process and prognosis of ovarian cancer patients were screened. Furthermore, lncRNA AC005224.4 exhibited the most upregulated expression in ovarian tumor tissues. Therefore, we speculated that lncRNA AC005224.4 may exert pivotal effects on ovarian cancer cell EMT and metastasis. The mutual effects of miRNAs and lncRNAs resulted in the reduction of targeted lncRNAs, playing significant roles in target gene regulation. In the present study, bioinformatics analysis (starBase v3.0, TargetScan v7.2, and HMDD v3.2 databases) was conducted to predict the targeted miRNAs for lncRNA AC005224.4 or SNAI2. miR-140-3p was found to show the most marked decrease in ovarian cancer tissue samples relative to that in normal samples. Previous research has also disclosed that miR-140-3p is tightly implicated in multitudinous cancers. For instance, Jiang et al[26] have reported that miR-140-3p inhibits cell proliferation and promotes apoptosis in colorectal cancer. Huang et al[27] have verified that miR-140-3p functions as a tumor suppressor in squamous cell lung cancer. Nevertheless, little is known about the biological function and underlying molecular mechanism of miR-140-3p in ovarian cancer, which need further clarification.
In the present study, through in-vivo and in-vitro experiments, we attempted to reveal the biological function and the underlying molecular mechanism of lncRNA AC005224.4 in ovarian cancer. It was observed that AC005224.4 knockdown inhibited ovarian cancer cell proliferation, metastasis, and EMT via sponging miR-140-3p to suppress SNAI2 expression level. Our experimental data provided substantial evidence to reveal a novel lncRNA–miRNA–mRNA signaling pathway regulatory network. The lncRNA AC005224.4/miR-140-3p/SNAI2 axis was demonstrated to play crucial roles in ovarian cancer, which offered an important insight concerning the regulatory mechanism of lncRNAs in ovarian cancer progression and new therapeutic options for ovarian cancer.
Methods
Ethical approval
All procedures performed in studies involving human participants were in accordance with the 1964 Helsinki declaration. This study was approved by the Ethics Committee of Affiliated Tumor Hospital, Xinjiang Medical University (No. K-201813). Written informed consent was obtained from all the participants.
Sample collection
Ovarian cancer specimens (>5 cm away from cancer edge) were obtained from 47 ovarian cancer patients in Affiliated Tumor Hospital, Xinjiang Medical University. The inclusion criteria for ovarian cancer patients were: (1) diagnosed as serous ovarian cancer by post-operative pathology; (2) >18 years old; (3) received ovarian cancer resection; (4) did not receive chemotherapy, radiotherapy, immunotherapy, and other treatments before surgery; (5) no history of other tumors or ovary-related diseases. The exclusion criteria for ovarian cancer patients were: (1) lesions invading the ovary; (2) substandard RNA quality. On the other hand, normal ovarian tissue specimens were obtained from 21 cases, which were confirmed as benign lesions of the uterus by post-operative pathology. Normal ovarian tissues were harvested from patients with uterine fibroids and had attained the age of menopause, who have undergone surgery to remove the uterus, along with the removal of the ovaries, and who provided consent for their organs to be used in the study, at Affiliated Tumor Hospital, Xinjiang Medical University. All tissue samples were directly stockpiled in liquid nitrogen and kept at –80°C until use.
Cell lineages and cultures
Human ovarian cancer cell lines (SNU119, SKOV3, CAOV-3, and HO8910 cells) and human ovarian epithelial cells (HOSEpiC) were purchased from BeNa Culture Collection (Beijing, China). SNU119 and CAOV-3 cells were cultivated in high-glucose Dulbecco's Modified Eagle Medium (DMEM) (GIBCO BRL, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, Waltham, MA, USA). SKOV3, HO8910, and HOSEpiC cells were cultured in RPMI-1640 medium appended with 2 mmol/L L-glutamine and 10% FBS. All cells were cultivated in a humidified environment (5% CO2, 37°C).
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA of tissues and cells was extracted using TRIZOL™ (Invitrogen, Carlsbad, CA, USA). Next, 2.0 μg of total RNA was used for reverse transcription using PrimeScript® Stra Strand Synthesis Kit (TaKaRa, Tokyo, Japan). Quantitative PCR was performed using QuantiTect® SYBR® Green RT-PCR Kit (QIAGEN, Dusseldorf, Germany). Glyceraldehyde-3-phosphate dehydrogenase (for lncRNA and mRNA) and U6 (for miRNA) served as endogenous controls. The primer sequences are listed in [Supplementary Table 1]. The relative fold changes were detected using the 2–ΔΔCT method.
RNA sequencing analysis
Sample sequencing of this experimental project was conducted with the help of Guanglian Biomedical Technology Co., Ltd. (Tianjin, China). LncRNAs’ and mRNAs’ expression differences in non-neoplastic ovarian tissue specimens and ovarian cancer tissue samples (n = 5) were analyzed using Significant Analysis of Microarray software (Stanford University, Stanford, CA, USA). Total RNA of ovarian cancer tissues and normal ovarian tissue samples were extracted and RNA purity and integrity were detected. After hybridization, different intensity fluorescence values of lncRNAs and mRNAs were obtained by chip hybridization. The fluorescence intensity values were obtained through image scanning. The aberrantly expressed lncRNAs were screened using R software (version 3.6.3; R Foundation, Vienna, Austria), with fold change-values >2 and P values <0.05 as the screening standard. Differentially expressed lncRNAs and mRNAs were displayed using a volcano plot and heat-map.
Cell transfection
The interference sequence of AC005224.4 was designed by BLOCK-iT™ RNAi Designer (Invitrogen). pcDNA3.1-AC005224.4, pcDNA3.1-SNAI2, pcDNA3.1 plasmid vector, siRNA-AC005224.4-#1, siRNA-AC005224.4-#2, siRNA-AC005224.4-#3, scrambled siRNA, miR-140-3p mimics, mimics control, miR-140-3p inhibitor, and inhibitor control were commercially obtained from GenePharma Technology Co., Ltd. (Shanghai, China). SKOV3 and CAOV-3 cells (1 × 106 cells/well) were plated in six-well plates and underwent 24-h culturing in an incubator (37°C, 5% CO2) until the cells grew to 80% to 90% confluence. All the transfection was performed using Lipofectamine™ 3000 (Life Technologies Corporation, Gaithersburg, MD, USA) following the manufacturer's instructions. Cells were allocated into 10 groups: pcDNA3.1 plasmid vector group, pcDNA3.1-AC005224.4 group, si-NC group (transfected with scrambled siRNA), si-AC005224.4-#1 group, si-AC005224.4-#2 group, si-AC005224.4-#3 group, mimics-NC + vector group (transfected with blank vector of inhibitor), miR-140-3p mimics group, pcDNA3.1-SNAI2 group, and mix group (transfected with miR-140-3p mimics + pcDNA3.1-SNAI2). After transfection, all cells were harvested for subsequent experiments.
Western blot assay
Total protein of ovarian cancer tissues, and SKOV3 and CAOV-3 cells, were extracted using radioimmunoprecipitation assay (RIPA) lysate (Beyotime, Shanghai, China). After protein quantification using the bicinchoninic acid method (Waltham, MA, USA), 80 μg protein was subject to sodium dodecyl sulfate-polyacrylamide gel electrophoresis for separation, and was then transferred onto polyvinylidene difluoride membranes. After that, the membranes were blocked (1 h) in TBS-T buffer containing 5% skim milk. Next, the membranes were subject to an overnight incubation (4°C) with the following primary antibodies (all from Abcam, Cambridge, MA, USA): anti-E Cadherin antibody (Cat# ab40772, 1/10000), anti-N Cadherin antibody (Cat# ab18203, 1/1000), anti-SNAIL antibody (Cat# ab53519, 1/1000), anti-Vimentin antibody (Cat# ab8978, 1/1000), and anti-3-phosphate glyceraldehyde dehydrogenase (GAPDH) antibody (Cat# ab8245, 1/500). After three Tris-Buffered Saline Tween-20 (TBST) washes, the membranes were hybridized (room temperature, 1.5 h) with the horseradish peroxidase-linked secondary antibody rabbit anti-mouse IgG H&L (Cat# ab6728, 1/2000, Abcam). Signal detection was performed using the enhanced chemiluminescence (ECL) system (Life Technologies Corporation); the relative protein levels were calculated by normalization to GAPDH.
Cell Counting Kit-8 (CCK-8) assay
CCK-8 (Dojindo, Kumamoto, Japan) was used to detect the effect of lncRNA AC005224.4, miR-140-3p, and SNAI2 expressions on cell proliferation. Briefly, SKOV3 and CAOV-3 cells (2 × 103 cells/well) were seeded in 96-well plates and incubated for 0, 24, 48, and 72 h, respectively. At the indicated time point, 10 μL CCK-8 was added, followed by a further 4-h incubation. SpectraMax M5 microplate reader (Molecular Devices, Sunnyvale, CA, USA) was applied to measure the absorbance value at 450 nm. All procedures were repeated at least three times.
Transwell (migration and invasion) assay
For migration assay, the upper chamber of transwell was not covered with membrane. For invasion assay, Matrigel (Corning Incorporated, Corning, NY, USA) was placed in transwell inserts for 45 min at 37°C. In the two assays, SKOV3 and CAOV-3 cells (4 × 105 cells) were cultivated in the upper chamber in the serum-free medium; 20% FBS (Thermo Fisher Scientific) was added to the lower chamber. After 24-h cell culturing, the non-migrant or non-invasive cells were wiped off using a cotton swab. Next, the cells that migrated to the lower surface of the membrane were stained using 0.1% crystal violet staining solution (Sangon Biotech, Shanghai, China). The sub-membrane cells of five different microscope views were counted and the average number was calculated.
Construction of lentiviral vectors for shRNA-AC005224.4
AC005224.4 shRNA or scrambled shRNA was constructed into BLOCK-iT™ lentiviral RNAi expression system (Invitrogen), which was designed to suppress AC005224.4 production in SKOV3 cells. Briefly, SKOV3 cells (3 × 105 cells/dish) were seeded onto 35-mm dishes. Next, 1 day after seeding, cultures were added with lentiviral particles (200 μL) in a 2-mL RPMI-1640 medium containing 10% FBS, followed by 24-h incubation (5% CO2, 37 °C). The cells were successfully infected by lentiviral particles (Lv-sh-NC or Lv-sh-AC005224.4) for 48 h. After that, the cells were harvested and subjected to qRT-PCR analysis to determine the efficiency of lentiviral particles in suppressing AC005224.4 expression.
Nude mouse xenograft assay in vivo
Pathogen-free conditions were maintained through the lifetime of 12 female BALB/c nude mice (aged 4 weeks). The xenograft in-vivo assay was approved by the Affiliated Tumor Hospital, Xinjiang Medical University. First, serum-free cell suspension of the untreated SKOV3 cells (1 × 106 cells) was subcutaneously injected into the posterior flank of nude mice, followed by further feeding until the 7th day. Subsequently, SKOV3 cells (1 × 106 cells) transfected with lentiviral-sh-AC005224.4 or lentiviral-sh-NC were injected into the tumors of nude mice. Tumor volume was measured using a caliper following the length × width2/2 formula. The average volume of the tumor was measured three times every 3 days. At the termination of the experiment (the 25th day), mice were euthanized and the tumor was excised from each mouse to measure the average volume and weight.
Dual-luciferase reporter gene assay
The targeted relationship between lncRNA AC005224.4 and miR-140-3p was predicted using ENCORI database (http://starbase.sysu.edu.cn). The targeted relationship between SNAI2 and miR-140-3p was predicted using TargetScan v7.2 database (http://www.targetscan.org/vert_72/). PmirGLO, pmirGLO-AC005224.4-wt/pmirGLO-AC005224.4-mut, and pmirGLO-SNAI2-wt/pmirGLO-SNAI2-mut were commercially obtained from Youbio (Changsha, China), and these were then co-transfected with miR-140-3p mimics/inhibitor or mimics/inhibitor control into 293 T cells using Lipofectamine-mediated gene transfer. After transfection for 48 h, cells were harvested and the luciferase activity was determined according to the protocol of dual-luciferase reporting assay system (Promega, Madison, WI, USA).
RNA immunoprecipitation (RIP) assay
The relationship between lncRNA AC005224.4 and miR-140-3p was determined using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Merck Millipore, Billerica, MA, USA). Antibodies used for RIP assay included anti-AGO2 and control IgG (Merck Millipore). The co-precipitated RNAs were used for cDNA synthesis and evaluated by qRT-PCR.
Statistical analysis
The animal experiments were performed at least six times. The cell experiments were performed at least three times. All data were generated from three independent experiments and then presented as the mean ± standard deviation. Statistical calculation was carried out using GraphPad Prism 6.0 software (La Jolla, CA, USA). Differences between two groups were compared using Student's t test, and differences among three or more groups were analyzed using one-way analysis of variance. A P value of <0.05 was considered as a statistically significant difference.
Results
Expression and prognosis significance of lncRNA AC005224.4 in ovarian cancer
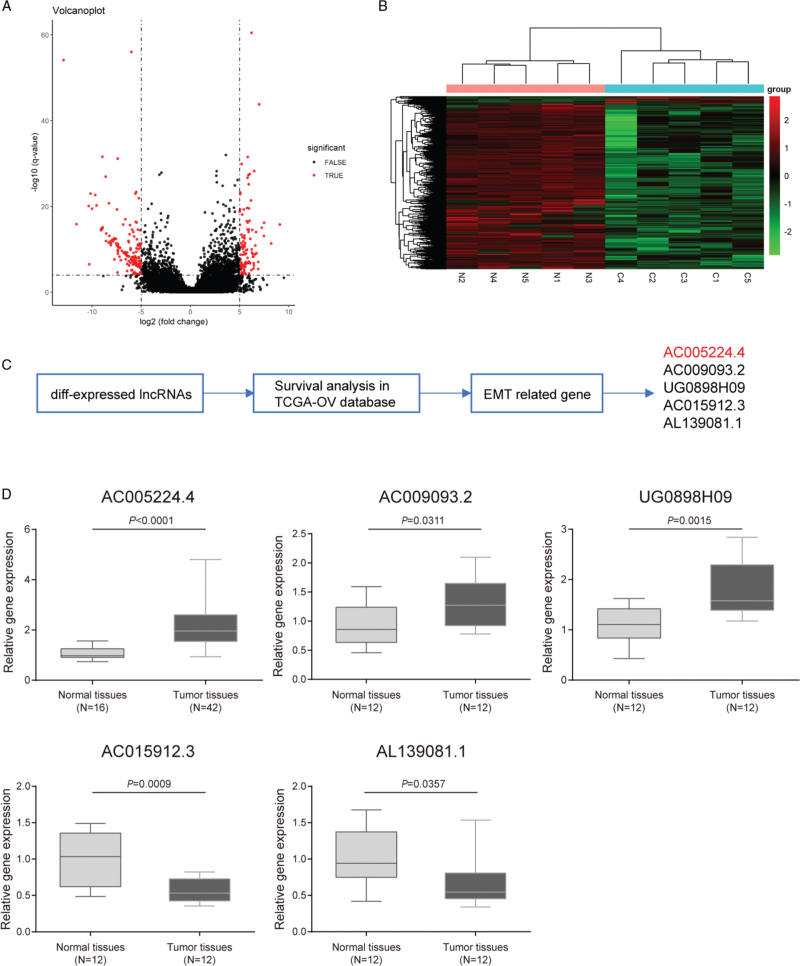
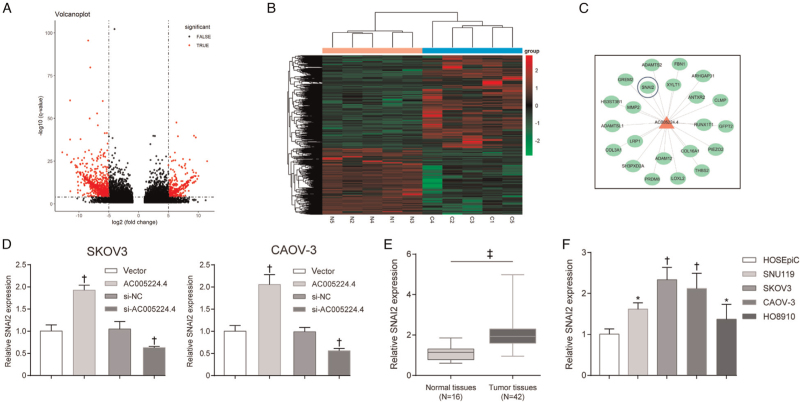
According to microarray analysis, the expression profile of lncRNAs was notably changed in ovarian cancer tissues compared with that in normal ovarian tissue specimens, and lncRNAs with fold change-values >1 and P values <0.05 were visualized using a volcano plot [Figure 1A] and heat-map [Figure 1B]. Next, Gene Ontology enrichment analysis was performed, and it was revealed that most of the differential expression genes (DEGs) in ovarian cancer tissues in our microarray and TCGA-ovarian cancer database were classified into one or more subcategories, which indicated the important molecular mechanisms within ovarian cancer development [Supplementary Figure 1]. As shown by the results, DEGs in ovarian cancer in our microarray were remarkably enriched and activated in EMT and UV response according to Biological Process [Supplementary Figure 1A]. Similarly, the DEGs in the TCGA-ovarian cancer database were drastically enriched and activated in allograft rejection, EMT, hypoxia, IL2-STAT5 signaling, and UV response allograft rejection [Supplementary Figure 1C]. Both in our microarray and TCGA-ovarian cancer database, EMT was markedly activated [Supplementary Figure 1B, D]. EMT is one of the most important features that promote tumor cell invasion and migration in various malignant tumors, including ovarian cancer. Therefore, five lncRNAs (AC005224.4, AC009093.2, UG0898H09, AC015912.3, and AL139081.1) were screened [Supplementary Figure 1C]. These five lncRNAs showed differential expression in our microarray (log2 fold change [FC] > 1, P < 0.05), and were related to the overall survival of TCGA-ovarian cancer patients (hazard ratio >1 or <1) [Supplementary Figure 2]; these five lncRNAs were differentially expressed in TCGA interstitial ovarian cancer cells compared to those in epithelial cells (P < 0.05) [Supplementary Figure 2]. Furthermore, PCR assay was performed to detect the expression of these five lncRNAs in the collected ovarian cancer and normal tissues [Figure 1D]. It was observed that among these five lncRNAs, AC005224.4 exhibited the most significantly upregulated expression in ovarian cancer tissues relative to that in normal tissues (N = 42, P < 0.001). The correlation between AC005224.4 expression and the clinicopathologic features in ovarian cancer is listed in [Table 1]. No significant association between AC005224.4 expression and age, histological subtype, or peritoneal cytology was found, while patients with high AC005224.4 expression were more likely to experience the higher pathological grade, advanced International Federation of Gynecology and Obstetrics (FIGO) stage, and larger tumor size (P < 0.05). Moreover, little is known about the role of lncRNA AC005224.4 in ovarian cancer; hence, lncRNA AC005224.4 was adopted for further analysis.
Figure 1.
LncRNA AC005224.4 was upregulated in ovarian cancer. (A, B) The expression profile of lncRNAs in five pairs of ovarian cancer tissues and normal ovarian tissues was analyzed. Differentially expressed lncRNAs were displayed in the volcano plot (A) and heat-map (B) and the threshold value was set to P values <0.05 and fold change-values >1. (C) Five alternative lncRNAs (AC005224.4, AC009093.2, UG0898H09, AC015912.3, and AL139081.1) were screened. (D) qRT-PCR assay was performed to detect the expression level of these five lncRNAs in the collected ovarian cancer tissues and normal ovarian tissues. LncRNA: Long non-coding RNA.
Table 1.
The relationship between AC005224.4 expression and clinicopathological parameters in ovarian cancer.
| AC005224.4 expression level (T/N)# | ||||
| Clinicopathological parameters | Number | Average value | SD | P |
| Age | ||||
| <50 years | 18 | 1.9546 | 0.9030 | 0.1259 |
| ≥50 years | 24 | 2.3972 | 0.9117 | |
| Histological subtype | ||||
| Serous | 32 | 2.1804 | 0.9041 | 0.8435 |
| Others | 10 | 2.2944 | 1.0192 | |
| Pathological grade | ||||
| G1–G2 | 9 | 1.6050 | 0.5246 | 0.0264∗ |
| G3 | 33 | 2.3719 | 0.9536 | |
| Peritoneal cytology | ||||
| Negative | 29 | 2.1635 | 0.8539 | 0.6490 |
| Positive | 13 | 2.3058 | 1.0854 | |
| Tumor size | ||||
| <10 cm | 19 | 1.8587 | 0.8883 | 0.0244∗ |
| ≥10 cm | 23 | 2.4957 | 0.8705 | |
| FIGO stage | ||||
| Early stage (I-II) | 12 | 1.6184 | 0.6982 | 0.0076∗ |
| Advanced (III–IV) | 30 | 2.4432 | 0.9122 | |
| CA125 | ||||
| <1000 U/mL | 32 | 2.1720 | 0.8684 | 0.6599 |
| ≥1000 U/mL | 10 | 2.3212 | 1.1108 | |
The relative level of AC005224.4 is the ratio of tumor tissue to normal tissue.
P < 0.05 was recognized as a statistically significant difference; FIGO: InternationalFederation of Gynecology and Obstetrics; CA125: Carbohydrate antigen 125.
Effects of lncRNA AC005224.4 on ovarian cancer cell proliferation, metastasis, and EMT
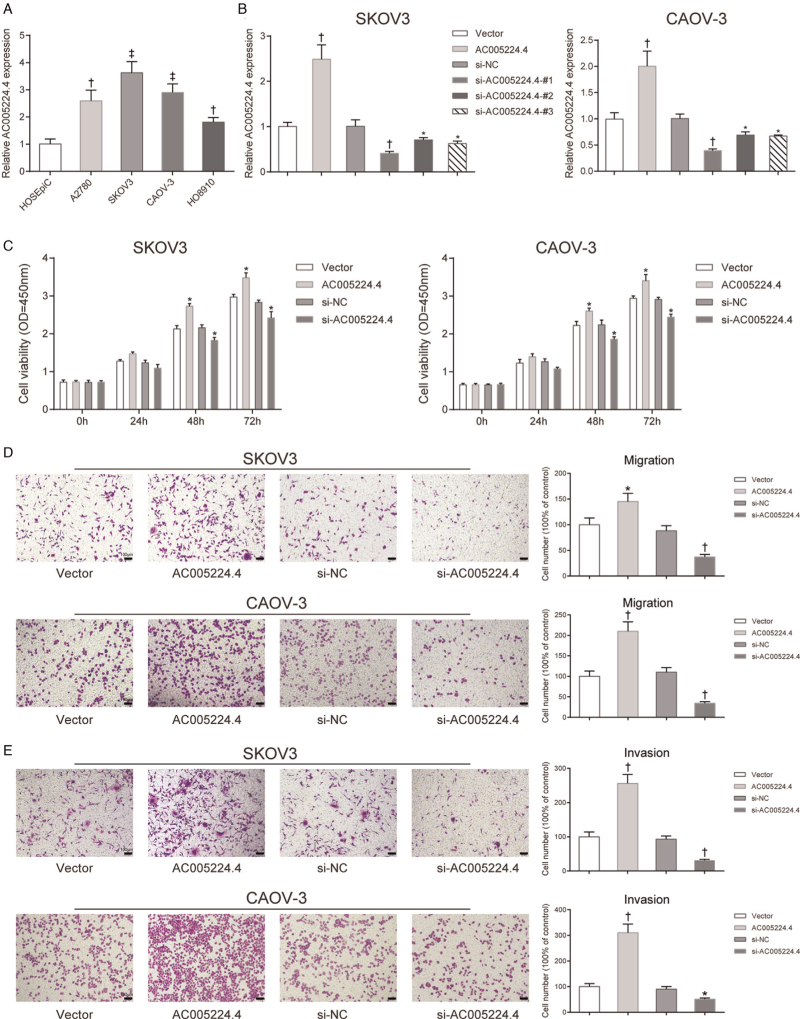
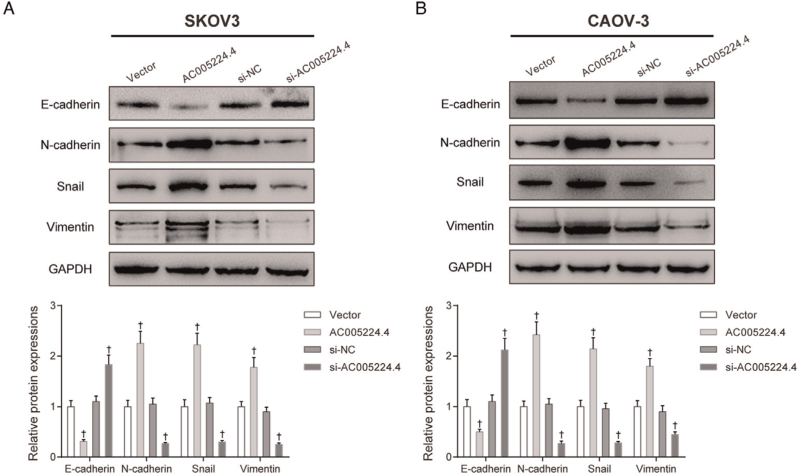
First, a different lncRNA AC005224.4 expression level in ovarian carcinoma cell lines (SNU119, SKOV3, CAOV-3, and HO8910 cells) was detected compared with that in HOSEpiC. It was found that SKOV3 and CAOV-3 cells exhibited higher AC005224.4 expression among these four cancer cells (P < 0.01; Figure 2A). Therefore, SKOV3 and CAOV-3 cells were selected as our experimental subjects for subsequent experiments. To investigate the role of AC005224.4 in ovarian cancer cell progression, pcDNA3.1-AC005224.4 and siRNA of AC005224.4 were transfected into SKOV3 and CAOV-3 cells for AC005224.4 overexpression and knockdown, respectively. The results revealed that AC005224.4 expression was elevated 150% in the AC005224.4 overexpression group, whereas the AC005224.4 expression was significantly decreased in three siRNA-AC005224.4 groups, with the si-AC005224.4-#1 group showing the lowest AC005224.4 expression (reduced by 60%) (P < 0.05; Figure 2B). Therefore, the si-AC005224.4-#1 group was selected as the experimental group in the subsequent experiments. According to CCK-8 assay results, the proliferation ability of both SKOV3 and CAOV-3 cells in the AC005224.4 overexpression group was drastically enhanced, while SKOV3 and CAOV-3 cell proliferation was decreased after silencing AC005224.4 (P < 0.05; Figure 2C). Transwell assay results indicated that AC005224.4 overexpression observably facilitated the migration and invasion of both SKOV3 and CAOV-3 cells, while AC005224.4 downregulation notably decreased SKOV3 and CAOV-3 cell migration and invasion (P < 0.01; Figure 2D, E). Furthermore, lncRNA AC005224.4 effects on EMT-related proteins (E-cadherin, N-cadherin, Snail, and Vimentin) in SKOV3 and CAOV-3 cells were detected using Western bolt assay [Figure 3]. It was observed that overexpression of AC005224.4 led to decreased E-cadherin expression, and increased expressions of N-cadherin, Snail, and Vimentin in SKOV3 and CAOV-3 cells, while AC005224.4 knockdown had the opposite effects. Taken together, these results suggested that lncRNA AC005224.4 could promote ovarian cancer cell proliferation, metastasis, and EMT in vitro.
Figure 2.
LncRNA AC005224.4 regulated ovarian cancer cell proliferation and apoptosis in vitro. (A) qRT-PCR assay was used to detect AC005224.4 expression in different ovarian carcinoma cell lines (SNU119, SKOV3, CAOV-3, and HO8910 cells) and HOSEpiC. N = 3; †P < 0.01, ‡P < 0.001, compared with HOSEpiC cells. (B) After cells were transfected with pcDNA3.1-AC005224.4, siRNA- AC005224.4 (siRNA#1, siRNA#2, and siRNA#3) and AC005224.4 expression level in SKOV3 and CAOV-3 cells were detected using qRT-PCR analysis. (C) Tissue damage was assessed using H&E staining. (D) CCK-8 assay was used to determine the proliferation of SKOV3 and CAOV-3 cells in different transfection groups. (E) Transwell assay was performed to investigate changes of SKOV3 and CAOV-3 cell migration using crystal violet staining; scale bar = 100 μm. (F) Transwell assay was performed to evaluate changes of SKOV3 and CAOV-3 cell invasion using crystal violet staining; scale bar = 100 μm. N = 3; ∗P < 0.05, †P < 0.01, ‡P < 0.001, compared with the vector group or si-NC group. CCK-8: Cell Counting Kit-8; HOSEpiC: Human ovarian epithelial cells; LncRNA: Long non-coding RNA.
Figure 3.
LncRNA AC005224.4 regulated ovarian cancer cell EMT in vitro. Western bolt assay was applied to detect the effects of lncRNA AC005224.4 on EMT-related proteins (E-cadherin, N-cadherin, Snail, and Vimentin) in (A) SKOV3 and (B) CAOV-3 cells. N = 3; ∗P < 0.01, compared with the vector group or si-NC group. EMT: Epithelial-mesenchymal transition; LncRNA: Long non-coding RNA.
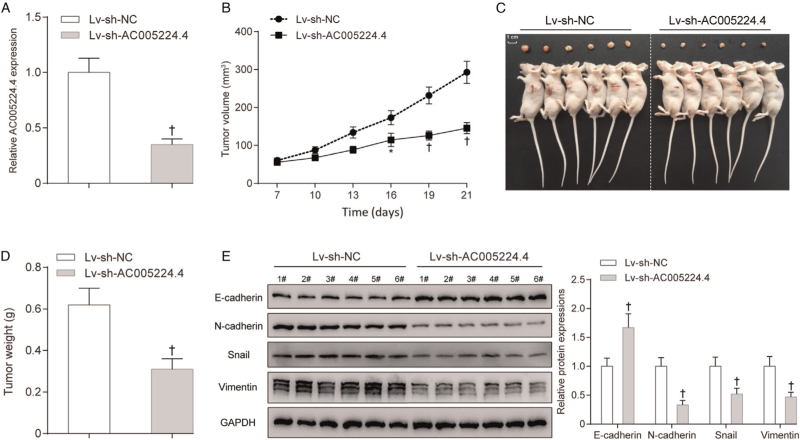
Knockdown of AC005224.4 suppressed SKOV3 cell growth in vivo
A mouse tumor xenograft model was established to investigate AC005224.4 effects on ovarian cancer in vivo. SKOV3 cells pre-transfected with lentivirus-mediated shRNA-AC005224.4 or shRNA-control were subcutaneously injected into the back of nude mice. As shown by PCR assay results, AC005224.4 expression was noticeably downregulated by about 60% in the sh-AC005224.4 group compared with that in the NC group, which indicated the successful transfection (P < 0.01; Figure 4A). After 7 days of the experiment, tumor volume was measured every 3 days, and at each point, the SKOV3 cells with AC005224.4 silencing showed smaller tumors compared to those in the sh-NC group (P < 0.05; Figure 4B). At the termination of the experiment (the 21st day), mice were euthanized and the tumor of each mouse was excised [Figure 4C]. It was found that the sh-AC005224.4 group showed notably less tumor weight than the sh-NC group (P < 0.01; Figure 4D). AC005224.4 knockdown prominently motivated E-cadherin expression while restraining the expressions of N-cadherin, Snail, and Vimentin in tumor (P < 0.01; Figure 4E). From all of the above, it can be inferred that AC005224.4 knockdown efficiently impaired the tumorigenesis ability and EMT process of ovarian cancer cells in vivo.
Figure 4.
Knockdown of AC005224.4 inhibited tumor formation in nude mice. (A) The AC005224.4 expression level in thelv-sh-AC005224.4 group was markedly decreased compared with that in the lv-sh-NC group; SKOV3 cells transfected with lentivirus-mediated shRNA targeting AC005224.4 were allocated into the Lv-sh-AC005224.4 group; SKOV3 cells transfected with shRNA lentiviral particles with non-effective scrambled shRNA sequences were allocated into the Lv-sh-NC group. (B, C) Silencing of AC005224.4 drastically suppressed ovarian cancer tumor growth by comparison to that in the lv-sh-NC group; scale bar = 1 cm. (D) Tumor weight was measured on the 21st day at the end of the experiment. (E) The effects of silencing AC005224.4 on EMT-related proteins (E-cadherin, N-cadherin, Snail, and Vimentin) of tumors were detected using Western blot. N = 6; ∗P < 0.05, †P < 0.01, compared with the Lv-sh-NC group. EMT: Epithelial-mesenchymal transition.
AC005224.4 impacted the expression of SNAI2
mRNA expression patterns were also examined in five pairs of ovarian cancer tissue specimens and normal ovarian tissues using microarray analysis. The differently expressed mRNAs were selected with the criterion of fold change-values >1 and P values <0.05, which were visualized using a volcano plot [Figure 5A] and heat-map [Figure 5B]. To explore the key genes regulated by AC005224.4 in ovarian cancer development based on our microarray and TCGA-ovarian cancer database, the co-expression network of AC005224.4 and mRNAs was established. Important transcription factors of EMT such as SNAI2 and downstream markers MMP2, ADAMST2, and COL3A1 showed markedly positive correlations with AC005224.4 [Figure 5C]. In a previous study, the transcription factor SNAI2 is characterized as a prototypical EMT transcription factor, which participates in diverse biological processes, including tumor metastasis.[19] Therefore, SNAI2 was selected for further experiments. According to PCR assay results, AC005224.4 overexpression noticeably elevated SNAI2 expression level, whereas AC005224.4 knockdown significantly reduced SNAI2 expression level in both SKOV3 and CAOV-3 cells (P < 0.01; Figure 5D). The results revealed that the SNAI2 expression level was significantly increased in 42 ovarian cancer patient tissue samples compared with that in normal ovarian tissues samples (P < 0.0001; Figure 5E). SNAI2 expression in ovarian cancer cell lines (SNU119, SKOV3, CAOV-3, and HO8910 cells) was markedly higher than that in ovarian epithelial cells HOSEpiC (P < 0.05; Figure 5F). The above results proved that SNAI2 was upregulated in both ovarian cancer tissues and cells, and positively correlated with AC005224.4.
Figure 5.
AC005224.4 impacted the expression of SNAI2. (A, B) The expression profiles of mRNAs in five pairs of ovarian cancer tissues and normal ovarian tissues were analyzed. Differentially expressed lncRNAs were displayed in the volcano plot (A) and heat-map (B) and the threshold value was set to P values <0.05 and fold change-values >1. (C) The co-expression network of AC005224.4 and mRNAs based on our microarray and TCGA-ovarian cancer database was established. (D) The effect of AC005224.4 on SNAI2 expression in SKOV3 and CAOV-3 cells was explored. N = 3; †P < 0.01, compared with the vector group or si-NC group. (E) The expression level of SNAI2 in ovarian cancer and normal tissues was detected. ‡P < 0.001, compared with normal tissues. (F) SNAI2 expression level in different ovarian carcinoma cell lines (SNU119, SKOV3, CAOV-3, and HO8910 cells) and HOSEpiC was detected. N = 3; ∗P < 0.05, †P < 0.01 compared with HOSEpiC cells. HOSEpiC: Human ovarian epithelial cells; LncRNA: Long non-coding RNA.
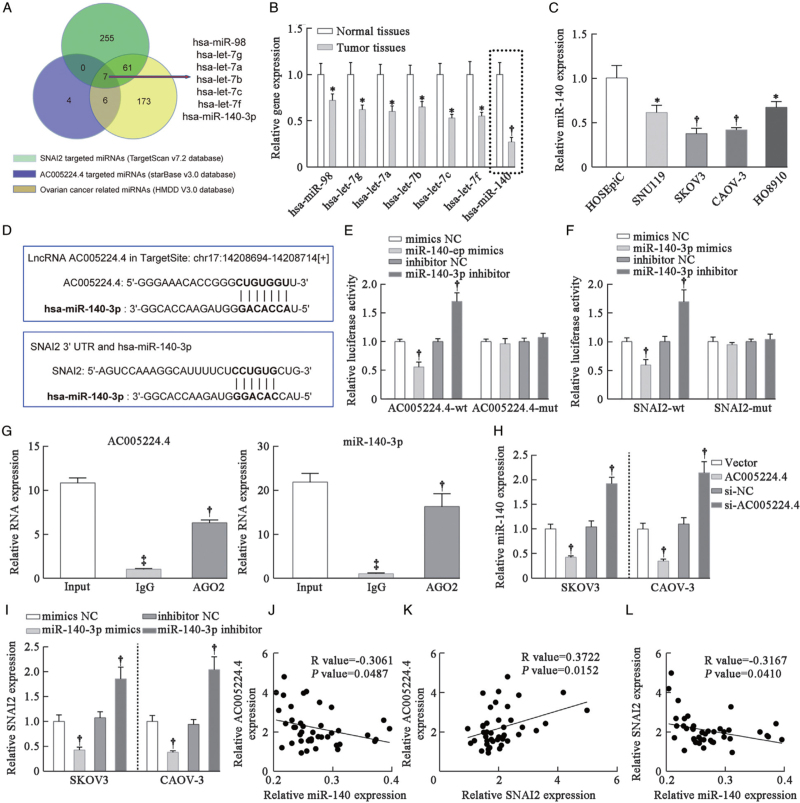
The regulatory network of AC005224.4/miR-140-3p/SNAI2 axis
Bioinformatics analysis (starBase v3.0 and TargetScan v7.2 databases) was used to predict targeted miRNAs for AC005224.4 or SNAI2. Meanwhile, the Human microRNA Disease Database (v3.2) was applied to predict ovarian cancer progression-related miRNAs. As a result, seven miRNAs (hsa-miR-98, hsa-let-7g, hsa-let-7a, hsa-let-7b, hsa-let-7c, hsa-let-7f, and hsa-miR-140-3p) that targeted both AC005224.4 and SNAI2 in ovarian cancer were identified [Figure 6A]. Among them, miR-140-3p exhibited the most marked decrease (about 70%) in ovarian cancer patient tissue samples compared with that in normal samples (P < 0.01; Figure 6B). Hence, miR-140-3p was selected for subsequent experiments. The results suggested that miR-140-3p expression was notably lower in ovarian cancer cell lines (SNU119, SKOV3, CAOV-3, and HO8910 cells) than that in ovarian epithelial cells’ HOSEpiC (P < 0.05; Figure 6C). Potential binding sites between miR-140-3p and AC005224.4/SNAI2 were predicted [Figure 6D]. Next, the targeted relationships between miR-140-3p and AC005224.4/SNAI2 were validated using a dual-luciferase reporter gene assay [Figure 6E, F]. According to the AGO2 immunoprecipitation assay results, the AGO2 antibody was able to pull down both endogenous lncRNA AC005224.4 and miR-140-3p [Figure 6G], which further validated their binding potential. As shown in Figure 6H, AC005224.4 overexpression noticeably inhibited, whereas AC005224.4 silencing significantly facilitated miR-140-3p expression level in both SKOV3 and CAOV-3 cells (P < 0.01). miR-140-3p mimics observably inhibited SNAI2 expression level, whereas miR-140-3p inhibitor markedly motivated SNAI2 expression level in SKOV3 and CAOV-3 cells (P < 0.01; Figure 6I). Moreover, linear correlation was applied to analyze the relationship among AC005224.4, miR-140-3p, and SNAI2 expression in 42 ovarian cancer patient tissue samples. A negative correlation between AC005224.4 and miR-140-3p was observed (R = −0.3061, P = 0.0487; Figure 6J). AC005224.4 expression was positively correlated with SNAI2 level (R = 0.3722, P = 0.0152; Figure 6K). miR-140-3p expression was negatively correlated with SNAI2 level (R = –0.3167, P = 0.0410; Figure 6L). The AC005224.4/miR-140-3p/SNAI2 regulatory axis may contribute to the tumorigenesis of ovarian cancer.
Figure 6.
The regulatory network of the AC005224.4/miR-140-3p/SNAI2 axis. (A) Seven miRNAs (hsa-miR-98, hsa-let-7g, hsa-let-7a, hsa-let-7b, hsa-let-7c, hsa-let-7f, and hsa-miR-140-3p) that targeted both AC005224.4 and SNAI2 in ovarian cancer were identified according to starBase v3.0, TargetScan v7.2, and HMDD v3.2 databases. (B) qRT-PCR assay was performed to detect the expression levels of these seven miRNAs in ovarian cancer tissues and normal ovarian tissues. N = 10; ∗P < 0.05, †P < 0.01, compared with normal tissues. (C) miR-140-3p expression level in different ovarian carcinoma cell lines (SNU119, SKOV3, CAOV-3, and HO8910 cells) and HOSEpiC was detected. N = 3; ∗P < 0.05, †P < 0.01 compared to HOSEpiC cells. (D) The binding sites of miR-140-3p with AC005224.4 or SNAI2 were predicted. (E, F) The targeting relationship between miR-140-3p and AC005224.4 (E) or SNAI2 (F) was verified using dual-luciferase reporter gene assay in 293 T cells. N = 3; †P < 0.01 compared with the mimics-NC group or inhibitor NC group. (G) Anti-AGO2 RIP was performed in HEK293T cells, followed by qRT-PCR to detect the expression of lncRNA AC005224.4 or miR-140-3p associated with AGO2. N = 3; ‡P < 0.001, compared with the Input group; †P < 0.01, compared with the IgG group. (H) The effect of AC005224.4 on miR-140-3p expression in SKOV3 and CAOV-3 cells was verified using qRT-PCR assay. N = 3; †P < 0.01, compared with the Vector group or si-NC group. (I) The effect of miR-140-3p on SNAI2 expression in SKOV3 and CAOV-3 cells was verified using qRT-PCR assay. N = 3; †P < 0.01 compared to the mimics-NC group or inhibitor NC group. (J) The linear analysis revealed a negative correlation between AC005224.4 and miR-140-3p expression in ovarian cancer tissues (N = 42). (K) The linear analysis revealed a positive correlation between AC005224.4 and SNAI2 expression in ovarian cancer tissues (N = 42). (L) The linear analysis revealed a negative correlation between SNAI2 and miR-140-3p expression in ovarian cancer tissues (N = 42). HMDD: Human microRNA Disease Database; HOSEpiC: Human ovarian epithelial cells; LncRNA: Long non-coding RNA; RIP: RNA immunoprecipitation.
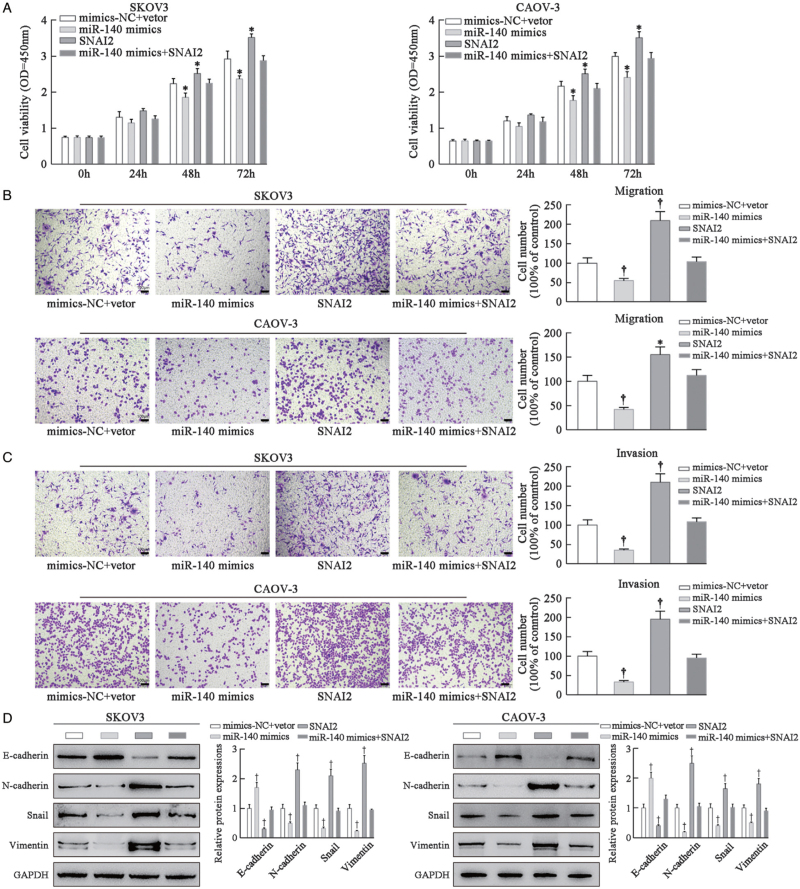
MiR-140-3p inhibits ovarian cancer progression via inhibiting SNAI2
SKOV3 and CAOV-3 cells were pre-transfected and allocated into four groups: mimics-NC + vector group (transfected with empty plasmid), miR-140-3p mimics group (transfected with miR-140-3p mimics), SNAI2 group (transfected with pcDNA3.1-SNAI2), and miR-140-3p mimics + SNAI2 group (transfected with miR-140-3p mimics + pcDNA3.1-SNAI2). As shown by CCK-8 assay results, SNAI2 overexpression drastically promoted SKOV3 and CAOV-3 cell proliferation, while miR-140-3p mimics caused the opposite effects on cell proliferation and abolished the promoting effects of SNAI2 on cell proliferation, which restored the viability of cancer cells (P < 0.01; Figure 7A). Transwell assay results showed that SNAI2 overexpression promoted, while miR-140-3p mimics inhibited tumor cell migration and invasion; miR-140-3p mimics restrained SNAI2 effects on tumor cells and reduced tumor cell metastasis (P < 0.01; Figure 7B, C). Based on Western blot assay results, miR-140-3p mimics prominently promoted E-cadherin protein expression while inhibiting the expressions of N-cadherin, Snail, and Vimentin in SKOV3 and CAOV-3 cells; SNAI2 showed the opposite trend to miR-140-3p mimics in cancer cells (P < 0.01; Figure 7D). Taken together, sufficient evidence confirmed that SNAI2 facilitated ovarian cancer cell growth, metastasis, and EMT process, whereas miR-140-3p abolished the promotive effects of SNAI2 on ovarian cancer.
Figure 7.
MiR-140-3p inhibits ovarian cancer progression via inhibiting SNAI2. SKOV3 and CAOV-3 cells were pre-transfected and allocated into four groups: mimics-NC + vector group (cells transfected with empty plasmid), miR-140-3p mimics group (cells transfected with miR-140-3p mimics), SNAI2 group (cells transfected with pcDNA3.1-SNAI2), and miR-140-3p mimics + SNAI2 group (cells transfected with miR-140-3p mimics + pcDNA3.1-SNAI2). (A) CCK-8 assay was used to determine the proliferation of SKOV3 and CAOV-3 cells in different transfection groups. (B) Transwell assay was performed to investigate the changes of SKOV3 and CAOV-3 cell migration using crystal violet staining; scale bar = 100 μm. (C) Transwell assay was performed to research the changes of SKOV3 and CAOV-3 cell invasion using crystal violet staining; scale bar = 100 μm. (D) Western blot assay was applied to detect EMT-related protein (E-cadherin, N-cadherin, Snail, and Vimentin) changes of SKOV3 and CAOV-3 cells. N = 3; ∗P < 0.05, †P < 0.01, compared with the mimics-NC + vector group. CCK-8: Cell Counting Kit-8; EMT: Epithelial-mesenchymal transition.
Discussion
Growing studies have manifested that quite multiple lncRNAs modulate protein-coding gene expression at both the post-transcriptional and transcriptional levels and are tightly involved in the crucial regulation processes of multiple human diseases, including ovarian cancer.[22,28,29] In the current study, for the first time, we identified lncRNA AC005224.4 as a key regulator of MET, which participated in the ovarian cancer invasion-metastasis cascade. LncRNA AC005224.4 expression level was observably upregulated in ovarian cancer tissues compared with that in normal ovarian tissues, which indicated that AC005224.4 may function as a cancer-promoting gene. Consistently, Tian et al[30] have reported that AC005224.4 is upregulated in abdominal aortic aneurysm. Next, the biological function of lncRNA AC005224.4 in ovarian cancer was investigated in ovarian cancer cells in vitro and xenograft mice in vivo. LncRNA AC005224.4 drastically promoted ovarian cancer cell growth, metastasis, and EMT process in vitro. Meanwhile, in-vivo experiments revealed that AC005224.4 knockdown efficiently impaired the tumorigenesis ability and EMT process of ovarian cancer. All these results suggested that lncRNA AC005224.4 was closely related to ovarian cancer progression.
Recent researches have illuminated that lncRNAs could act as an endogenous miRNA sponge and participate in post-transcriptional regulation through interaction with miRNAs.[31–33] In this study, AC005224.4 binding to miR-140-3p promoted ovarian cancer cell metastasis and EMT. Previous studies have provided evidence that miR-140-3p has different functions in various cancers. For example, Zhang et al[34] have emphasized that miR-140-3p confers suppression of the MAPK signaling pathway via targeting GRN, thereby inhibiting the EMT, invasion, and metastasis in hepatocellular carcinoma. Zhou et al[35] have argued that miR-140-3p inhibits breast cancer cell proliferation and migration via directly regulating the expression of tripartite motif 28. Dong et al[36] have demonstrated that miR-140-3p suppressed cell growth and invasion via downregulating ATP8A1 expression in non-small cell lung cancer. Similarly, this study proved that miR-140-3p inhibited ovarian cancer cell growth, metastasis, and EMT process. Importantly, miR-140-3p downregulation almost reversed the effects of AC005224.4 silencing. Furthermore, based on the sequence complementarity between AC005224.4 and miR-140-3p, it is believed that miR-140-3p is a potential direct target of AC005224.4. These results indicated that miR-140-3p is downstream of AC005224.4 in a signaling cascade that regulated ovarian cancer progression. Another point that need to be noted is that many other factors affected the interaction between RNA molecules and lncRNAs; therefore, we cannot exclude the possibility of AC005224.4 targeting other miRNAs.
To further understand the molecular regulatory mechanism of miR-140-3p in ovarian cancer progression, we considered the potential targeted genes. The upregulation of AC005224.4 led to an elevated expression of SNAI2 (a transcriptional repressor of E-cadherin) via competitively binding to miR-140-3p. Through bioinformatics, SNAI2 was predicted to be a directly targeted gene of miR-140-3p at its 3′-UTR mRNA, which has also been proposed to be associated with the progression of various cancers, including ovarian cancer. For instance, Jiang et al[37] have demonstrated that SNAI2 promotes cancer cell migration and invasion in clear cell renal cell carcinoma. Du et al[38] have elucidated that SOX13 promotes colorectal cancer metastasis via transactivating SNAI2. Fan et al[39] have proposed that the non-canonical signaling pathway of SNAI2 induces EMT in ovarian cancer cells via suppressing miR-222-3p transcription and upregulating PDCD10. In the current study, functional experiments revealed that SNAI2 overexpression noticeably facilitated ovarian cancer cell growth, metastasis, and EMT process. Therefore, we could conclude that AC005224.4 co-expressed with SNAI2 in ovarian cancer and positively regulated SNAI2 expression via competitively sponging miR-140-3p.
Several limitations still exist in this research. First, other AC005224.4-sponged miRNAs, as well as other gene targets of miR-140-3p, may contribute to the progression of ovarian cancer. Moreover, the mechanisms of the transcriptional regulation of AC005224.4 in ovarian cancer were not investigated deeply in this research.
Conclusion
Ongoing research identified that lncRNA AC005224.4 was highly expressed in ovarian cancer tissues. Next, sufficient evidence was provided to highlight that AC005224.4 promotes ovarian cancer cell growth, metastasis, and EMT via sponging miR-140-3p and regulating SNAI2. These findings facilitated us to better understand the development of ovarian cancer, providing novel therapeutic strategies for this fatal disease.
Conflicts of interest
None.
Funding
This work was supported by the Xinjiang Uygur Autonomous Region Science and Technology Supporting Xinjiang Project (2017E0263); the Tianjin Science and Technology Support Program Project (18YFZCSY00100); the Program for New Century Excellent Talents in University in China (NCET-11-1066) and Training Plan of leading subject talents in Tianjin colleges and universities; the National Natural Science Foundation of China (81972572); and CAMS Innovation Fund for Medical Sciences (2016-I2M-1-001).
Supplementary Material
Supplementary Material
Footnotes
How to cite this article: Xiong T, Wang Y, Zhang Y, Yuan J, Zhu C, Jiang W. lncRNA AC005224.4/miR-140-3p/SNAI2 regulating axis facilitates the invasion and metastasis of ovarian cancer through epithelial-mesenchymal transition. Chin Med J 2023;136:1098–1110. doi: 10.1097/CM9.0000000000002201
Supplemental digital content is available for this article.
References
- 1.Menon U, Karpinskyj C, Gentry-Maharaj A. Ovarian cancer prevention and screening. Obstet Gynecol 2018; 131:909–927.. doi: 10.1097/AOG.0000000000002580. [DOI] [PubMed] [Google Scholar]
- 2.Natanzon Y, Goode EL, Cunningham JM. Epigenetics in ovarian cancer. Semin Cancer Biol 2018; 51:160–169.. doi: 10.1016/j.semcancer.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Webb PM, Jordan SJ. Epidemiology of epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol 2017; 41:3–14.. doi: 10.1016/j.bpobgyn.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 4.La Vecchia C. Ovarian cancer: epidemiology and risk factors. Eur J Cancer Prev 2017; 26:55–62.. doi: 10.1097/CEJ.0000000000000217. [DOI] [PubMed] [Google Scholar]
- 5.Odunsi K. Immunotherapy in ovarian cancer. Ann Oncol 2017; 28: (Suppl_8): viii1–viii7.. doi: 10.1093/annonc/mdx444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orr B, Edwards RP. Diagnosis and treatment of ovarian cancer. Hematol Oncol Clin N Am 2018; 32:943–964.. doi: 10.1016/j.hoc.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 7.Barnett R. Ovarian cancer. Lancet 2016; 387:1265.doi: 10.1016/s0140-6736(16)30024-1. [DOI] [PubMed] [Google Scholar]
- 8.Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin 2018; 68:284–296.. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lisio MA, Fu L, Goyeneche A, Gao ZH, Telleria C. High-grade serous ovarian cancer: basic sciences, clinical and therapeutic standpoints. Int J Mol Sci 2019; 20:952–984.. doi: 10.3390/ijms20040952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol 2019; 20:69–84.. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 11.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell 2016; 166:21–45.. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Weinberg RA. Epithelial-to-mesenchymal transition in cancer: complexity and opportunities. Front Med 2018; 12:361–373.. doi: 10.1007/s11684-018-0656-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diepenbruck M, Christofori G. Epithelial-mesenchymal transition (EMT) and metastasis: yes, no, maybe? Curr Opin Cell Biol 2016; 43:7–13.. doi: 10.1016/j.ceb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Lu W, Kang Y. Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev Cell 2019; 49:361–374.. doi: 10.1016/j.devcel.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mittal V. Epithelial mesenchymal transition in tumor metastasis. Annu Rev Pathol 2018; 13:395–412.. doi: 10.1146/annurev-pathol-020117-043854. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y, Wang DD, Wu YP, Su D, Zhou TY, Gai RH, et al. MDM2 promotes epithelial-mesenchymal transition and metastasis of ovarian cancer SKOV3 cells. Br J Cancer 2017; 117:1192–1201.. doi: 10.1038/bjc.2017.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lupia M, Angiolini F, Bertalot G, Freddi S, Sachsenmeier KF, Chisci E, et al. CD73 regulates stemness and epithelial-mesenchymal transition in ovarian cancer-initiating cells. Stem Cell Rep 2018; 10:1412–1425.. doi: 10.1016/j.stemcr.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goossens S, Vandamme N, Van Vlierberghe P, Berx G. EMT transcription factors in cancer development re-evaluated: beyond EMT and MET. Biochim Biophys Acta Rev Cancer 2017; 1868:584–591.. doi: 10.1016/j.bbcan.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Zhou W, Gross KM, Kuperwasser C. Molecular regulation of Snai2 in development and disease. J Cell Sci 2019; 132:jcs235127-235139 doi: 10.1242/jcs.235127. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Wang J, Yue H, Lu X. SNAI2 3’untranslated region promotes the invasion of ovarian cancer cells by inducing MARCKS expression. J Cancer 2019; 10:2480–2487.. doi: 10.7150/jca.29489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest 2017; 127:761–771.. doi: 10.1172/JCI84424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res 2017; 77:3965–3981.. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genom Proteom Bioinform 2016; 14:42–54.. doi: 10.1016/j.gpb.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang H, Yu T, Han Y, Jiang H, Wang C, You T, et al. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol Cancer 2018; 17:119–131.. doi: 10.1186/s12943-018-0870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin X, Yang F, Qi X, Li Q, Wang D, Yi T, et al. LncRNA DANCR promotes tumor growth and angiogenesis in ovarian cancer through direct targeting of miR-145. Mol Carcinog 2019; 58:2286–2296.. doi: 10.1002/mc.23117. [DOI] [PubMed] [Google Scholar]
- 26.Jiang W, Li T, Wang J, Jiao R, Shi X, Huang X, et al. miR-140-3p suppresses cell growth and induces apoptosis in colorectal cancer by targeting PD-L1. Onco Targets Ther 2019; 12:10275–10285.. doi: 10.2147/OTT.S226465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang H, Wang Y, Li Q, Fei X, Ma H, Hu R. miR-140-3p functions as a tumor suppressor in squamous cell lung cancer by regulating BRD9. Cancer Lett 2019; 446:81–89.. doi: 10.1016/j.canlet.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 28.Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016; 539:452–455.. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell 2018; 172:393–407.. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian L, Hu X, He Y, Wu Z, Li D, Zhang H. Construction of lncRNA-miRNA-mRNA networks reveals functional lncRNAs in abdominal aortic aneurysm. Exp Ther Med 2018; 16:3978–3986.. doi: 10.3892/etm.2018.6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Militello G, Weirick T, John D, Döring C, Dimmeler S, Uchida S. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform 2017; 18:780–788.. doi: 10.1093/bib/bbw053. [DOI] [PubMed] [Google Scholar]
- 32.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014; 505:344–352.. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 2016; 17:272–283.. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 34.Zhang QY, Men CJ, Ding XW. Upregulation of microRNA-140-3p inhibits epithelial-mesenchymal transition, invasion, and metastasis of hepatocellular carcinoma through inactivation of the MAPK signaling pathway by targeting GRN. J Cell Biochem 2019; 120:14885–14898.. doi: 10.1002/jcb.28750. [DOI] [PubMed] [Google Scholar]
- 35.Zhou Y, Wang B, Wang Y, Chen G, Lian Q. Wang H. miR-140-3p inhibits breast cancer proliferation and migration by directly regulating the expression of tripartite motif 28. Oncol Lett 2019; 17:3835–3841.. doi: 10.3892/ol.2019.10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong W, Yao C, Teng X, Chai J, Yang X, Li B. MiR-140-3p suppressed cell growth and invasion by downregulating the expression of ATP8A1 in non-small cell lung cancer. Tumour Biol 2016; 37:2973–2985.. doi: 10.1007/s13277-015-3452-9. [DOI] [PubMed] [Google Scholar]
- 37.Jiang B, Chen W, Qin H, Diao W, Li B, Cao W, et al. TOX3 inhibits cancer cell migration and invasion via transcriptional regulation of SNAI1 and SNAI2 in clear cell renal cell carcinoma. Cancer Lett 2019; 449:76–86.. doi: 10.1016/j.canlet.2019.02.020. [DOI] [PubMed] [Google Scholar]
- 38.Du F, Li X, Feng W, Qiao C, Chen J, Jiang M, et al. SOX13 promotes colorectal cancer metastasis by transactivating SNAI2 and c-MET. Oncogene 2020; 39:3522–3540.. doi: 10.1038/s41388-020-1233-4. [DOI] [PubMed] [Google Scholar]
- 39.Fan L, Lei H, Zhang S, Peng Y, Fu C, Shu G, et al. Non-canonical signaling pathway of SNAI2 induces EMT in ovarian cancer cells by suppressing miR-222-3p transcription and upregulating PDCD10. Theranostics 2020; 10:5895–5913.. doi: 10.7150/thno.43198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.