SUMMARY
Background:
In the United States, the underlying reasons for racial/ethnic disparities in type 2 diabetes risk remain unclear. However, differences in genetic risk for insulin resistance and peripheral adipose tissue distribution may be contributing factors.
Objective:
To investigate racial/ethnic differences in associations of genetic risk for insulin resistance with leg fat and insulin sensitivity in a cohort of American children.
Methods:
Participants were healthy European American (n = 83), African American (n = 79), and Hispanic American (n = 74) children aged 7–12 years. Genetic risk scores were derived from published variants associated with insulin resistance phenotypes in European adults. Body composition was assessed using dual-energy X-ray absorptiometry. Insulin sensitivity was determined from the frequently-sampled intravenous glucose tolerance test and minimal modeling. Statistical models were adjusted for age, sex, pubertal stage, and body composition.
Results:
In the combined cohort, risk score was inversely associated with insulin sensitivity (p = 0.033) but not leg fat (p = 0.170). Within Hispanic Americans, risk score was inversely associated with insulin sensitivity (p = 0.027) and leg fat (p = 0.005), while associations were non-significant in European and African Americans (p > 0.200).
Conclusions:
The higher type 2 diabetes risk observed among Hispanic Americans may have a genetic basis related to an inability to store lipid in peripheral adipose tissue.
Keywords: Genetic risk score, peripheral adipose tissue, body fat distribution, insulin sensitivity, race/ethnicity
1 |. INTRODUCTION
The reason for racial/ethnic disparities in risk for type 2 diabetes is not clear. In the US, both African Americans (AA) and Hispanic Americans (HA) are at an elevated risk for type 2 diabetes relative to Caucasians, or European Americans (EA). One possible factor contributing to this disparity is insulin resistance, a major risk factor for type 2 diabetes. Research has demonstrated that AA have lower whole-body insulin sensitivity compared to their EA counterparts, independent of total and regional adiposity 1–5. While lower insulin sensitivity in HA (determined from a frequently-sampled intravenous glucose-tolerance test and minimal modeling) has been explained by body mass index (BMI), fat distribution, and lifestyle factors in some cases, the factors underlying lower insulin sensitivity in diverse groups are not clearly understood 2. Thus, insulin sensitivity may explain a portion of the ethnic disparity in type 2 diabetes risk, but the basis for racial/ethnic differences in insulin sensitivity is not well-defined.
Recent data from European cohort studies have suggested that insulin sensitivity has a genetic basis that relates to the inability to store lipid in peripheral adipose tissue 6. When using fasting insulin as a measure of insulin sensitivity, 53 genomic loci were identified as associated with insulin sensitivity. A polygenic risk score derived from these 53 single nucleotide polymorphisms (SNPs) was associated with relatively low adipose tissue in the leg and gynoid areas, as assessed with anthropometrics and dual-energy x-ray absorptiometry (DXA) scanning. Additionally, both low leg fat and a high genetic risk score predicted incident type 2 diabetes. These observations were taken to imply that an inability to store lipid in “safe” subcutaneous adipose depots may lead to the deposition of ectopic lipid in organs such as liver and muscle. This ectopic lipid could sequentially lead to insulin resistance by interfering with insulin signaling.
The distribution of ectopic lipid differs with race and ethnicity in pediatric and adult populations. For example, EA deposit more visceral adipose tissue than AA 7 whereas AA deposit more intermuscular adipose tissue (IMAT) 8. HA deposit more liver fat than EA, who in turn deposit more than AA 9. These differences are presumed to have a genetic basis, with some evidence existing for racial/ethnic difference in gene polymorphisms associated with accumulation of ectopic lipid 10–12. Thus, if insulin resistance stems from the deposition of ectopic lipid, then the specific ectopic depot responsible for insulin resistance may differ with race or ethnicity in children and adults.
This study was designed to test the hypothesis that a higher genetic risk score would be associated with both lower leg fat and lower insulin sensitivity in a tri-ethnic cohort of US children, and to determine if associations differed by race/ethnicity. We chose to examine children due to 1) their relative absence of metabolic perturbations that occur in adults with advancing age and obesity. Thus, associations among genetic risk, body fat distribution, and insulin sensitivity are less confounded within a younger population.
2 |. RESEARCH DESIGN AND METHODS
2.1 |. Study Participants
We analyzed cross-sectional data from a previous study that investigated genetic and environmental contributions to racial/ethnic differences in insulin-related outcomes in US children. A full description of recruitment methods, inclusion/exclusion criteria, and study design has been reported elsewhere 13. Briefly, participants were healthy girls and boys aged 7–12 y who self-reported as African American, European American, or Hispanic American. All participants were pubertal stage ≤ 3 as evaluated by a pediatrician and according to the criteria of Marshall and Tanner 14. Recruited individuals were screened for type 2 diabetes with a 2-h oral glucose tolerance test, and those with 2-h glucose ≥ 200 mg/dL were excluded from participation. Additional exclusion criteria included diagnoses of type 1 diabetes or any glucose or lipid disorders, and/or use of medications known to affect body composition or metabolism. The current analysis included 79 AA, 83 EA, and 74 HA children with complete genotypic, body composition, and insulin sensitivity data. All study procedures were approved by the University of Alabama at Birmingham Institutional Review Board. Both verbal and written consent were provided by children and their parents (respectively) prior to inclusion in the study.
2.2 |. Genotyping
DNA was acquired from all study participants. Participants were genotyped for 46 of 53 SNPs previously identified by Lotta et al 6 as associated with insulin resistance phenotypes (Table S1). DNA was genotyped using the Illumina Cardio-Metabochip by the Heflin Genotyping Core at the University of Alabama at Birmingham (UAB). The genotyped data report from Illumina was converted into an array using the gdmp package developed in R 15. Alleles for each SNP were dummy coded according to the presence (1) or absence (0) of the risk (insulin-raising) alleles. SNPs having two copies of the risk allele were coded as 2, SNPs with one copy were coded as 1, and SNPs with no copies of the risk allele were coded as 0. To create the risk score, the number of risk alleles from the 46 SNPs were summed for each participant. To account for missing genotypes, the risk allele sum was divided by the number of SNPs genotyped for each participant. Risk scores had a possible range between 0 and 2, with 0 representing an individual having none of the risk alleles for any of the 46 SNPs, and 2 representing an individual having both risk alleles for all 46 SNPs.
2.3 |. Anthropometrics
Participants were weighed wearing minimal clothing without shoes to the nearest 0.1 kg (Scale – tronix 6702W; Scale – tronix, Carol Stream, IL). Height was measured to the nearest 0.1 centimeter without shoes using a digital stadiometer (Heightronic 235; Measurement Concepts). BMI was calculated from measured heights and weights. BMI percentiles and obesity status were determined from Centers for Disease Control and Prevention (CDC) age- and sex-specific growth curves. Waist circumference was measured by trained technicians at the area between the ribs and iliac crest to the nearest millimenter with a flexible tape measure (Gulick II, Country Technology Inc, Gay Mills, WI).
2.4 |. Body Composition
Body composition was measured by DXA using a GE Lunar Prodigy densitometer (GE LUNAR Radiation Corp., Madison, WI, USA). During the DXA measurement, children wore loose-fitting lightweight clothing while lying supine with their arms at their side, per manufacturer recommendations. The DXA scans were analyzed using pediatric software (Encore 2002, version 6.10.029). Percent body fat was calculated from dividing DXA total fat mass (kg) by DXA weight (kg).
2.5 |. Insulin Sensitivity
Insulin sensitivity was assessed with a frequently-sampled intravenous glucose-tolerance test following an overnight stay in the UAB General Clinical Research Center (GCRC). For 3 days prior to arriving at the GCRC, participants were asked to consume at least 250 g of carbohydrate per day and were given a list of common foods to consume to ensure compliance with this goal during the first visit.
Following collection of two baseline blood samples, a bolus of glucose (50% dextrose, 300 mg/kg) was injected intravenously at time 0 and then at 20 min, insulin was injected intravenously (0.02 U/kg; Humulin, Lilly USA) over a 5-min period. A blood sample (2.0 ml) was collected at the following timepoints: 0, 2, 3, 4, 5, 6, 8, 10, 12, 15, 19, 20, 21, 22, 24, 26, 28, 30, 35, 40, 50, 60, 70 and 240 min as previously described 16. Serum glucose (10 μL) was analyzed by an Ektachem DT II chemistry analyzer system (Johnson & Johnson Clinical Diagnostics, Rochester, NY) with a mean intra- and interassay coefficient of variation of 0.61% and 1.45%, respectively. Insulin concentrations were determined by RIA (LINCO Research, St. Charles, MO) and measured in duplicate. Mean intra- and inter-assay coefficients of variation for insulin were 3.49% and 5.57%, respectively. Insulin sensitivity (SI) was calculated from glucose and insulin values using the minimal modeling method as described previously 16.
2.6 |. Socioeconomic Status
Socioeconomic status (SES) was determined using the Hollingshead 4-factor index of social class 17, which combines the educational attainment and occupational prestige for the number of working parents in the child’s family. Scores ranged from 8 to 66, with the higher score indicating higher theoretical social status.
2.7 |. Statistical Analyses
All analyses were conducted with RStudio Statistical Software (R Core Team, 2019, v.3.6.0). Figures were produced with the ggplot2, ggpubr, and visreg packages in R 18–20. Distributions for leg fat, percent body fat, and SI deviated from normal and were log-transformed for analysis. Differences in descriptive characteristics were assessed with one-way Analysis of Variance (ANOVA), Kruskal-Wallis (for non-parametric data), or Chi-square test as appropriate. Pairwise comparisons for descriptive statistics were evaluated with the Tukey’s HSD test or Wilcox test (for non-parametric data). Predictors of SI by race/ethnicity were assessed with multiple linear regression and adjusted for age, sex, and pubertal stage 14. Participants were stratified by quartiles of genetic risk score to compare distributions of race/ethnicity among quartiles. Due to sample size, genetic risk score was evaluated in all models as a continuous variable rather than by quartiles. Associations for genetic risk score with leg fat and SI were assessed with multiple linear regression, with genetic risk score included as the predictor. Models were adjusted for total fat mass (leg fat models), percent body fat (SI models), race/ethnicity, sex, age, and pubertal stage. SES had no significant effect on outcomes of interest; thus, we did not adjust for this variable in our analyses. To compare differences in genetic risk score associations among racial/ethnic groups, interaction terms between risk score and race/ethnicity were included in models with all participants, with significant interactions relative to the EA group. Analyses were further stratified by race/ethnicity. p < 0.05 was considered statistically significant.
2.8 |. Data and Resource Availability
The datasets generated and/or analyzed during the current study are available from the corresponding authors upon reasonable request. No applicable resources were generated or analyzed during the current study.
3 |. RESULTS
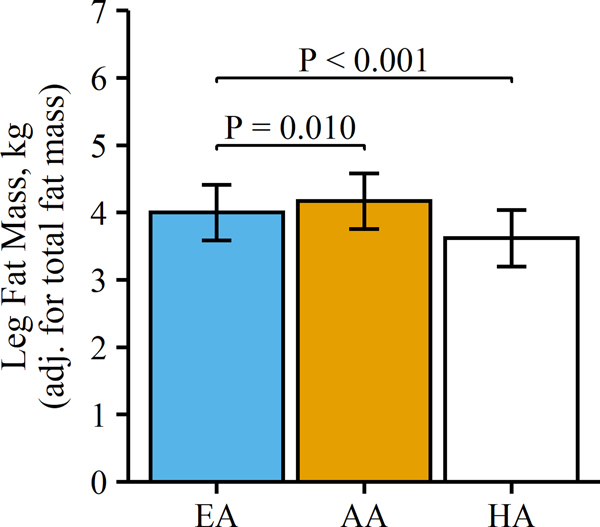
Of the 236 participants with genetic, DXA, and SI data, our study sample had a mean (SD) age of 9.52 (1.57) years, and 53.0% were male. Distribution of race/ethnicity in the sample was 35.2% EA, 33.5% AA, and 31.3% HA. Descriptive characteristics of study participants are stratified by race/ethnicity in Table 1. Age and sex distribution did not differ among EA, AA, and HA children. BMI, total body %fat, and absolute measures of fat mass (total, trunk, and leg) were higher in HA compared to EA and AA. However, HA children had the lowest leg fat mass after adjustment for overall fat mass (Table 1 and Figure 1). SI was highest in EA children and lower, but similar, between AA and HA children (Table 1). Associations of body composition variables and SES with SI, independent of age and pubertal stage, by race/ethnicity are presented in Table S2. Body mass, BMI, BMI percentile, percent body fat, total fat mass, trunk fat mass, and leg fat mass independently predicted SI in all three racial/ethnic groups. In contrast, waist-hip ratio and leg fat ratio predicted SI in HA children, but not EA or AA children.
Table 1.
Participant characteristics by race/ethnicity
| European Americans | African Americans | Hispanic Americans | p- value | |
|---|---|---|---|---|
| N | 83 | 79 | 74 | |
| Age (years) | 9.60 ± 1.62 | 9.59 ± 1.53 | 9.36 ± 1.59 | 0.566 |
| Male sex, n (%) | 45 (54.2%) | 41 (51.9%) | 39 (52.7%) | 0.956 |
| Tanner (pubertal) stage | 1.29 ± 0.55 | 1.71 ± 0.86a | 1.40 ± 0.70 | 0.001 |
| Socioeconomic status | 49.1 ± 8.86 | 37.3 ± 11.0a | 25.7 ± 11.8a,b | <0.001 |
| Body mass (kg) | 34.6 ± 8.17 | 36.4 ± 10.4 | 37.0 ± 10.1 | 0.288 |
| BMI (kg/m2) | 17.8 ± 2.67 | 18.5 ± 3.26 | 19.9 ± 3.10 a,b | <0.001 |
| BMI percentile, median (95% CI)c | 71.0 (48.0, 78.0) | 75.0 (56.0, 83.0) | 85.5 a,b (81.0, 91.0) | <0.001d |
| Obese, n (%)e | 9 (10.8%) | 9 (11.4%) | 17 (23.0%) | <0.001 |
| Waist/hip ratio | 0.855 ± 0.054 | 0.846 ± 0.093 | 0.900 ± 0.063a,b | <0.001 |
| Body fat (%) | 22.2 ± 8.62 | 20.2 ± 9.12 | 28.8 ± 8.40 a,b | <0.001 |
| Total fat mass (kg) | 8.11 ± 4.95 | 8.00 ± 6.06 | 11.2 ± 5.98 a,b | <0.001 |
| Total lean mass (kg) | 25.3 ± 4.59 | 27.0 ± 5.58 | 24.6 ± 5.06b | 0.007 |
| Trunk fat mass (kg), median (95% CI) | 2.43 (1.94, 2.87) | 2.12 (1.73, 2.64) | 4.15 a,b (3.22, 5.24) | <0.001 |
| Leg fat mass (kg) | 3.64 ± 1.93 | 3.77 ± 2.52 | 4.43 ± 2.09 a,b | 0.008 |
| Ratio of leg fat/total fat | 0.466 ± 0.045 | 0.489 ± 0.045 a | 0.409 ± 0.040a,b | <0.001 |
| Fasting plasma glucose (mg/dl) | 97.6 ± 6.40 | 95.6 ± 6.16 | 101.2 ± 6.28 a,b | <0.001 |
| Fasting plasma insulin (μU/ml) | 11.1 ± 4.34 | 12.9 ± 5.65 | 14.9 ± 8.16 a | 0.004 |
| AIRg (μU/mL·min), median (95% CI) | 487 (426, 557) | 927 a (765, 1131) | 752 a (588, 966) | <0.001d |
| SI (x10−4min−1/[μU/mL]) | 7.00 ± 4.20 | 5.01 ± 3.60 a | 5.13 ± 3.76 a | <0.001 |
| Genetic risk score | 1.04 ± 0.093 | 1.09 ± 0.085 a | 1.17 ± 0.103 a,b | <0.001 |
Data are mean ± SD unless otherwise indicated. Group differences were analyzed with one-way ANOVA and Tukey’s honest significant difference test (continuous variables) or Chi-square test (categorical variables), unless otherwise indicated.
Significantly different from European Americans (p < 0.05)
Significantly different from African Americans (p < 0.05)
As determined from CDC age- and sex-specific growth curves.
Differences analyzed with Kruskal-Wallis and Wilcox tests (non-parametric).
Cutoff for obesity is BMI ≥95th percentile according to CDC criteria.
Abbreviations: AIRg, acute insulin response to glucose; SI, insulin sensitivity index
Figure 1.
Bar plot of adjusted means (SD) for leg fat mass by race/ethnicity. European Americans (EA), n = 83; African Americans (AA), n = 79; Hispanic Americans (HA), n = 74. Multiple linear regression was used to adjust mean values for total fat mass and assess differences from EA.
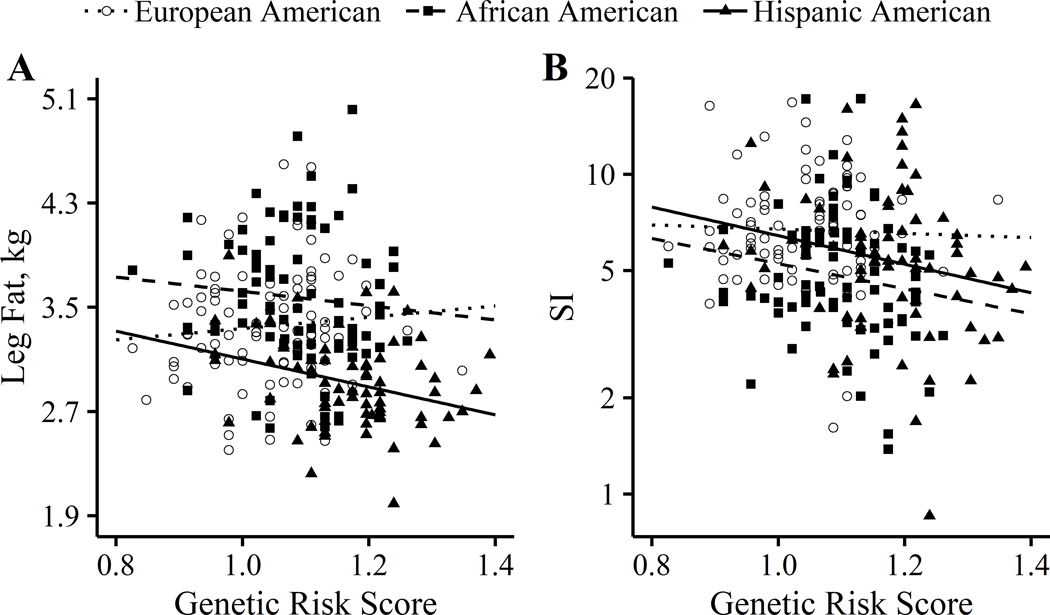
Genetic risk score ranged from 0.826 – 1.391 for the entire sample, and was highest in HA children and lowest in EA children. The distribution of race/ethnicity differed significantly across genetic risk score quartiles (p < 0.001) (Table S3). The highest proportion of participants in quartile 1 were EA children, whereas HA children made up the majority of participants in quartile 4. AA children made up the highest proportion of participants in quartiles 2 and 3. In all participants, genetic risk score significantly predicted SI, but not leg fat (Table 2). When stratified by race/ethnicity, genetic risk score predicted leg fat and SI only in HA children (Table 2 and Figure 2). The association between risk score and leg fat was stronger in HA vs. EA children (pinteraction = 0.028), but did not differ between AA and EA children (pinteraction = 0.218). On the other hand, the association between risk score and SI did not differ from EA in either HA (pinteraction = 0.275) or AA (pinteraction = 0.345) children. There was no effect of sex observed for any of the associations (pinteraction > 0.10).
Table 2.
Independent associationsa of genetic risk score (predictor) with leg fat and insulin sensitivity in all participants and by race/ethnicity
| β (SE) | p value | |
|---|---|---|
| Model 1: Leg fat | ||
| All participants (n = 236) | −0.13 (0.17) | 0.170 |
| European American (n = 84) | 0.12 (0.17) | 0.461 |
| African American (n = 79) | −0.22 (0.21) | 0.291 |
| Hispanic American (n = 74) | −0.38 (0.13)b | 0.005 |
| Model 2: Insulin sensitivity | ||
| All participants (n = 236) | −0.71 (0.33) | 0.033 |
| European American (n = 84) | −0.39 (0.50) | 0.434 |
| African American (n = 79) | −0.73 (0.64) | 0.261 |
| Hispanic American (n = 74) | −1.33 (0.59) | 0.027 |
Assessed with multiple linear regression and adjusted for total fat mass (leg fat models), percent body fat (insulin sensitivity models), sex, age, and pubertal stage.
Significantly different from European Americans (pinteraction < 0.05)
Figure 2.
Adjusted associations of genetic risk score with (A) leg fat mass and (B) SI by race/ethnicity. Multiple linear regression was used to adjust associations for total fat mass (leg fat models), percent body fat (SI models), sex, age, and pubertal stage. European Americans, n = 83; African Americans, n = 79; Hispanic Americans, n = 74. Leg fat associations: European Americans versus Hispanic Americans, pinteraction < 0.05.
4 |. DISCUSSION
This study was designed to test the hypothesis that a higher genetic risk score would be associated with both lower leg fat and lower insulin sensitivity in a cohort of US children, and to determine if associations differed by ethnicity/race. This is the first study in a US cohort that used a well-accepted measure of insulin sensitivity to test the sensitivity of a modified version of a published genetic risk score 6 to predict body fat distribution and insulin sensitivity. Further, this is the first study to determine if ethnicity/race affects the predictive ability of the genetic risk score. We found that a modified genetic risk score based on 46 of the 53 original SNPs 6 was associated with lower leg fat, specifically in HA children, and with lower insulin sensitivity, regardless of race/ethnicity. These results support the concept that a genetically determined ability to expand peripheral adipose tissue depots is protective against insulin resistance, and that a subset of HA may be missing this ability. These observations are valuable for understanding the genetic basis for insulin sensitivity, for developing screening tools and treatment paradigms to address the risk for type 2 diabetes, and for understanding, preventing, and treating racial/ethnic disparities in type 2 diabetes.
The 53-SNP genetic risk score was derived to predict fasting insulin concentrations, adjusted for BMI and high-density lipoprotein and triglyceride concentrations, as a surrogate measure for insulin resistance 6. Surrogates based on fasting values primarily reflect hepatic insulin resistance 20. In contrast, our study used the frequently-sampled intravenous glucose-tolerance test with minimal modeling, which is a well-accepted measure of whole-body insulin sensitivity that captures both hepatic and peripheral processes 21. Thus, it is likely that our results will differ from those of the original study. In addition, we used only 46 of the 53 SNPs used in the original study. However, our 46 SNPs included variants from three of the original five putative effector genes (L3MBTL3, DNAH10, and CCDC92) that were thought to explain a large portion of the predictive ability of the risk score 6. Findings from Lotta et al. indicate that these genes can affect the differentiation of adipocytes and the deposition of peripheral adipose tissue. Indeed, knockdown of these genes by siRNA showed a decrease in lipid accumulation in OP9-K cells, a preadipocyte line developed as a adipogenesis model 6, 22. However, it is likely that other SNPs in the risk score contribute to insulin resistance via other mechanisms, thus allowing the risk score to capture multiple mechanisms for insulin resistance in addition to the capacity for peripheral lipid storage.
Our results support the possibility that multiple physiologic mechanisms contribute to the risk score. In our data set, the risk score was associated with insulin sensitivity in the entire cohort combined, but with leg fat (and insulin sensitivity) only within HA children. Our sample size is relatively small for a genetics study, and thus caution must be exercised when interpreting the results. However, in the spirit of hypothesis generation, we suggest that these results may point to multiple processes for determining insulin sensitivity. Within HA children, the risk score was significantly associated with both leg fat and insulin sensitivity, such that lower relative leg fat was associated with lower insulin sensitivity. Thus, the results support the original hypothesis proposed by Lotta et al. that limited expansion of peripheral adipose tissue contributes to the etiology of genetic insulin resistance. While risk score was not associated with leg fat in the entire cohort, it was associated with insulin sensitivity. This observation may indicate that the risk score contributes to insulin sensitivity through additional mechanism(s) unrelated to the deposition of peripheral adipose tissue. These additional mechanisms may be relatively more important in individuals of non-Hispanic ethnicity, and may potentially explain differences in the etiology of type 2 diabetes among different ethnic groups.
HA exhibit a prevalence of type 2 diabetes of 12.8%, higher that than of EA, at 7.6%. However, the prevalence among AA is highest, at 13.2% 23. One of the aims of this study was to determine whether the genetic risk score shed light on the mechansims behind ethnic differences in risk for type 2 diabetes. When looking at the results stratified by ethnicity, clear differences emerged, with HA children showing the strongest associations of genetic risk score with leg fat and SI. While AA children showed similar trends in the relationships of interest to HA children (Figure 2), these associations were not statistically significant. This suggests that the genetic risk score and aspects of adipose/lipid distribution may likewise contribute to insulin sensitivity/resistance in AA, but that heterogeneity exists in the etiology, resulting in greater variance. On the other hand, the associations were qualitatively different in EA versus HA children, as reflected in the significant interactions by race/ethnicity observed for associations of risk score with both leg fat and SI. Racial/ethnic differences also were observed when considering fasting insulin, a measure primarily of hepatic insulin resistance. Although the risk score tended to predict fasting insulin within HA children (0.05 < p < 0.10; data not shown), the same was not true in the other two groups. Thus, the peripheral component of the SI index may be associated with a portion of the genetic risk score that is not associated with leg fat, and may be more common in EA and AA. Taken together, these observations suggest that the etiology of insulin resistance in EA, AA, and HA children and adults may occur through different mechanisms. Understanding the basis for racial/ethnic differences in the etiology of insulin resistance may lead to the development of a precision medicine-based approaches to preventing and/or treating type 2 diabetes.
Increasing evidence indicates that in addition to levels of overall adiposity, patterns of fat distribution and the accumulation of lipid in insulin-sensitive organs and tissues are significant determinants of insulin sensitivity 8, 24, 25. In children and adults, fat distribution and ectopic lipid deposition vary with ancestral background and may influence to some extent the heightened risk for insulin resistance among some racial/ethnic groups 1, 2, 7, 26, 27. Findings from our study and previous studies suggest that HA have a diminished ability to store lipid in gluteofemoral depots and other peripheral regions, which may negatively influence insulin sensitivity. We observed that HA children had relatively lower amounts of leg fat and higher amounts of trunk fat compared to EA and AA. Additionally, the association between leg fat/total fat ratio and insulin sensitivity was unique to HA children. Similar findings have been reported previously in Latino and HA children. A study in 150 Latino and Caucasian children found that the trunk/lower limb fat ratio was higher in Latinos, and this ratio was positively associated with both homeostatic model assessment (HOMA) and liver steatosis 28. Hetherington et al. 29 reported that in HA girls with a wide range of body fat levels, both gynoid and leg fat (calculated as a percentage of total body fat) were negatively correlated with HOMA-insulin resistance (IR). Moreover, HA girls with higher levels of total body fat had a higher percentage of android/trunk fat and lower percentages of gynoid and leg fat compared to those with lower amounts of body fat 29. From these observations, we propose that a reduced ability to store excess lipid in peripheral regions may be a driver for the increased truncal obesity and hepatic lipid accumulation observed in HA populations 25, 30, 31, consequently leading to insulin resistance.
In contrast, AA adults and children have lower truncal obesity relative to HA and EA, and as observed in our study, deposit more peripheral (glutealfemoral) subcutaneous adipose tissue 26. Thus, the elevated risk of type 2 diabetes observed in AA populations may be mediated by other fat depots or mechanisms independent of peripheral fat deposition. For instance, AA adults and children tend to accumulate more ectopic lipid in IMAT and extramycellular depots relative to EA after adjustment for total adiposity 8, 32–34. Further, a significant association between high IMAT and lower insulin sensitivity (measured by the intravenous glucose tolerance test) has been reported previously in AA women 35. It is possible that in AA, IMAT may play a larger role in insulin resistance. In addition to higher levels of IMAT, it has been hypothesized that the increased risk of insulin resistance observed in AA may also be a consequence of higher insulin secretion, decreased hepatic insulin clearance, and increased mitochondrial efficiency/oxidative stress 36, 37. It is possible that in AA children and adults, these unique metabolic/endocrine phenotypes have a larger influence on the pathogenesis of insulin resistance compared to other racial/ethnic groups.
The genetic risk score is a potentially valuable tool for type 2 diabetes prevention for several reasons. Rapid advances in technology may soon make it feasible for high throughput screening for disease risk, allowing for earlier implementation of preventive strategies. However, of perhaps greater value is the ability of the risk score to identify genetic and physiological factors that contribute to risk for disease. The association between fat distribution and insulin resistance has been recognized for years in children and adults, with a relatively central distribution being associated with risk for chronic metabolic disease 38. This line of research eventually led to the implication of visceral or intra-abdominal adipose tissue in the aetiology of type 2 diabetes. However, numerous studies continued to point to an independent “protective” effect of lower-body subcutaneous adipose tissue 39, 40. The results of these observational studies were supported by the development and use of the thiazolidinedione (TZD) class of drugs that confer increases in both direct and surrogate measures of insulin sensitivity by increasing differentiation of peripheral adipocytes 41. TZDs are PPARγ-agonists, a gene that is well established for its ability to promote adipocyte differentiation, and with polymorphisms that contribute to type 2 diabetes risk. The 53-SNP score includes polymorphisms of the PPARγ gene 6. However, the 53-SNP score also includes other genes and polymorphisms that may, with time and research, become recognized as contributing to disease risk, and could potentially be targeted by drug or lifestyle interventions.
The 53-SNP score reflects genetic insulin resistance. That is to say, it does not reflect lifestyle factors that influence insulin sensitivity/resistance, such as diet, physical activity, or weight gain. Assessment of insulin sensitivity in a research setting captures both genetic/inherent and lifestyle factors. Weight loss, regardless of whether it is induced by lifestyle, surgery, or pharmacotherapy, is considered the most effective treatment for insulin resistance 42. However, weight loss will only address acquired insulin resistance, not the genetic component. It is important from a disease prevention/treatment perspective to understand the two components of insulin resistance. Because these components are presumably additive, lifestyle and other treatment interventions will be effective in reducing total insulin resistance by reducing the acquired component, even in patients with substantial genetic insulin resistance. These concepts may be important for understanding health disparities in risk for type 2 diabetes, which are likely to contain elements of both genetic and acquired insulin resistance.
HA are at disproportionately high risk for type 2 diabetes 43. Few studies have examined insulin sensitivity in HA using accepted techniques, such as the euglycemic clamp or the intravenous glucose tolerance test. Most existing data come from the Insulin Resistance and Atherosclerosis Study (IRAS), which used the intravenous glucose tolerance test. In IRAS, insulin sensitivity was lower in HA relative to non-Hispanic whites among non-diabetic adults 2. However, this ethnic difference was statistically eliminated by adjusting for BMI, waist-hip ratio, behavioral factors, and other covariates, making it difficult to sort out whether lower insulin sensitivity among HA is innate or acquired.
In HA, impairments in insulin sensitivity may also occur at the level of the liver. In a longitudinal study conducted in HA children and young adults, increases in hepatic fat over a 2-year period were associated with higher fasting glucose, higher HbA1c values, and a 10.5% decrease in beta cell function 44. Compared to Caucasians, individuals of Mexican or Latino ancestry are prone to increased deposition of liver fat and have a higher prevalence of non-alcoholic fatty liver disease 9, 28. Greater liver fat may be due in part to a polymorphism at the PNPLA3 gene 10. However, our results here suggest that greater liver fat in HA may occur secondary to limited expansion of peripheral adipose tissue, and consequent “spill over” of lipid into the liver. It is important to note that this hypothesis invokes both a genetic component, as reflected in the 53-SNP score and the PNPLA3 polymorphism, and an acquired component (weight gain that exceeds lipid storage capacity in peripheral adipose). As such, prevention of weight gain, or implementation of a weight loss intervention, would be effective in mitigating disease risk.
Strengths of this study include extensive phenotyping of a tri-ethnic group of children using an intravenous glucose tolerance test and minimal modeling to derive a strong measure of whole-body insulin sensitivity, and using DXA to measure total and regional body composition. Use of children allows for a relatively less confounded assessment of the association between genotype and insulin sensitivity without complications introduced by long-term obesity and/or concurrent chronic disease. A limitation of our study was the relatively small sample size for a study involving genetic analyses.
In conclusion, a genetic risk score associated with fasting insulin in European adults was significantly associated with whole-body insulin sensitivity in a multiethnic group of US children. Further, the risk score was associated with leg fat in HA children, an ethnic group at high risk for developing type 2 diabetes. These results support the hypothesis that genetic insulin resistance has its roots in the inability to safely sequester fatty acids in peripheral subcutaneous adipose tissue. Further, these results suggest that the high prevalence of type 2 diabetes in HA children and adults has a genetic basis that results in a primary impairment in peripheral fat storage, and secondary increases in ectopic lipid deposition and insulin resistance. Further research is needed to determine if the genetic risk score is specifically associated with hepatic lipid in HA, and whether genetic insulin resistance has a different underlying mechanism(s) in EA and AA.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank the patients and their families for participation in the study. We would also like to thank the staff in the core laboratory and GCRC. Research funds for this study were provided by the National Institute of Diabetes and Digestive and Kidney Disease (R01 DK067426), the UAB Nutrition Obesity Research Center (P30 DK056336), the UAB Diabetes Research Center (P60 DK079626), and the National Center for Research Resources (M01RR000032 from the General Clinical Research Center). Lauren A Fowler and Sarah E Deemer were also funded by training grants from the UAB NORC Postdoctoral Training Program in Obesity Research (T32 DK062710).
Footnotes
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
REFERENCES
- 1.Tay J, Goss AM, Garvey WT, et al. Race affects the association of obesity measures with insulin sensitivity. Am J Clin Nutr. 2020;111(3):515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haffner SM, D’Agostino R Jr., Saad MF, et al. Increased insulin resistance and insulin secretion in nondiabetic African-Americans and Hispanics compared with non-Hispanic whites. The Insulin Resistance Atherosclerosis Study. Diabetes. 1996;45(6):742–748. [DOI] [PubMed] [Google Scholar]
- 3.Arslanian SA, Saad R, Lewy V, Danadian K, Janosky J. Hyperinsulinemia in African-American Children: Decreased Insulin Clearance and Increased Insulin Secretion and Its Relationship to Insulin Sensitivity. Diabetes. 2002;51(10):3014–3019. [DOI] [PubMed] [Google Scholar]
- 4.Uwaifo GI, Nguyen TT, Keil MF, et al. Differences in insulin secretion and sensitivity of Caucasian and African American prepubertal children. J Pediatr. 2002;140(6):673–680. [DOI] [PubMed] [Google Scholar]
- 5.Haffner SM, Stern MP, Dunn J, Mobley M, Blackwell J, Bergman RN. Diminished insulin sensitivity and increased insulin response in nonobese, nondiabetic Mexican Americans. Metabolism. 1990;39(8):842–847. [DOI] [PubMed] [Google Scholar]
- 6.Lotta LA, Gulati P, Day FR, et al. Integrative genomic analysis implicates limited peripheral adipose storage capacity in the pathogenesis of human insulin resistance. Nat Genet. 2017;49(1):17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katzmarzyk PT, Bray GA, Greenway FL, et al. Racial differences in abdominal depot-specific adiposity in white and African American adults. Am J Clin Nutr. 2010;91(1):7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallagher D, Kuznia P, Heshka S, et al. Adipose tissue in muscle: a novel depot similar in size to visceral adipose tissue. Am J Clin Nutr. 2005;81:903–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guerrero R, Vega GL, Grundy SM, Browning JD. Ethnic differences in hepatic steatosis: an insulin resistance paradox? Hepatology. 2009;49(3):791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis JN, Le KA, Walker RW, et al. Increased hepatic fat in overweight Hispanic youth influenced by interaction between genetic variation in PNPLA3 and high dietary carbohydrate and sugar consumption. Am J Clin Nutr. Dec 2010;92(6):1522–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yaghootkar H, Whitcher B, Bell JD, Thomas EL. Ethnic differences in adiposity and diabetes risk - insights from genetic studies. J Intern Med. 2020;288(3):271–283. [DOI] [PubMed] [Google Scholar]
- 12.Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40(12):1461–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klimentidis YC, Divers J, Casazza K, Beasley TM, Allison DB, Fernandez JR. Ancestry-informative markers on chromosomes 2, 8 and 15 are associated with insulin-related traits in a racially diverse sample of children. Hum Genomics. 2011;5(2):79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marshall WA, Tanner JM. Growth and physiological development during adolescence. Annu Rev Med. 1968;19:283–300. [DOI] [PubMed] [Google Scholar]
- 15.Abdel-Azim G. gdmp: Genomic Data Management [R package]. Version 0.2.0. 2019. Available from: https://CRAN.R-project.org/package=gdmp. Accessed 20 September 2019.
- 16.Chandler-Laney PC, Phadke RP, Granger WM, et al. Age-related changes in insulin sensitivity and β-cell function among European-American and African-American women. Obesity (Silver Spring). 2011;19(3):528–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cirino PT, Chin CE, Sevcik RA, Wolf M, Lovett M, Morris RD. Measuring socioeconomic status: reliability and preliminary validity for different approaches. Assessment. 2002;9(2):145–155. [DOI] [PubMed] [Google Scholar]
- 18.Wickham H. ggplot2: Elegant Graphics for Data Analysis. 2nd ed. New York, NY: Springer-Verlag; 2016. [Google Scholar]
- 19.Kassambara A. ggpubr: ‘ggplot2’ Based Publication Ready Plots [R package]. Version 0.2.2, 2018. Available from: https://CRAN.R-project.org/package=ggpubr. Acccessed 23 October 2019.
- 20.Breheny P, Burchett W. Visualization of Regression Models Using visreg. The R Journal. 2017;9(2):56–71._https://journal.r-project.org/archive/2017/RJ-2017-046/RJ-2017-046.pdf. Accessed 16 July 2019. [Google Scholar]
- 21.Borai A, Livingstone C, Kaddam I, Ferns G. Selection of the appropriate method for the assessment of insulin resistance. BMC Med Res Methodol. 2011;11:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergman RN. Lilly Lecture 1989. Toward physiological understanding of glucose tolerance. Minimal model approach. Diabetes. 1989;38(12):1512–1527. [DOI] [PubMed] [Google Scholar]
- 23.Lane JM, Doyle JR, Fortin JP, Kopin AS, Ordovás JM. Development of an OP9 derived cell line as a robust model to rapidly study adipocyte differentiation. PLoS One. 2014;9(11):e112123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodríguez JE, Campbell KM. Racial and Ethnic Disparities in Prevalence and Care of Patients With Type 2 Diabetes. Clin Diabetes. 2017;35(1):66–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caprio S, Perry R, Kursawe R. Adolescent Obesity and Insulin Resistance: Roles of Ectopic Fat Accumulation and Adipose Inflammation. Gastroenterology. 2017;152(7):1638–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katzmarzyk PT, Shen W, Baxter-Jones A, et al. Adiposity in children and adolescents: correlates and clinical consequences of fat stored in specific body depots. Pediatric Obesity. 2012;7(5):e42–e61. [DOI] [PubMed] [Google Scholar]
- 27.Allister-Price C, Craig CM, Spielman D, Cushman SS, McLaughlin TL. Metabolic markers, regional adiposity, and adipose cell size: relationship to insulin resistance in African-American as compared with Caucasian women. Int J Obes (Lond). 2019;43(6):1164–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cardel M, Higgins PB, Willig AL, et al. African genetic admixture is associated with body composition and fat distribution in a cross-sectional study of children. Int J Obes (Lond). 2011;35(1):60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martos-Moreno GÁ, Martínez-Villanueva J, González-Leal R, et al. Ethnicity Strongly Influences Body Fat Distribution Determining Serum Adipokine Profile and Metabolic Derangement in Childhood Obesity. Front Pediatr. 2020;8:551103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hetherington-Rauth M, Bea JW, Lee VR, et al. Relationship between fat distribution and cardiometabolic risk in Hispanic girls. Am J Hum Biol. 2018;30(5):e23149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillum RF. Distribution of waist-to-hip ratio, other indices of body fat distribution and obesity and associations with HDL cholesterol in children and young adults aged 4–19 years: The Third National Health and Nutrition Examination Survey. Int J Obes Relat Metab Disord. 1999;23(6):556–563. [DOI] [PubMed] [Google Scholar]
- 32.Karastergiou K. The Interplay Between Sex, Ethnicity, and Adipose Tissue Characteristics. Curr Obes Rep. 2015;4(2):269–278. [DOI] [PubMed] [Google Scholar]
- 33.Yim J-E, Heshka S, Albu JB, Heymsfield, Gallagher D. Femoral-gluteal subcutaneous and intermuscular adipose tissues have independent and opposing relationships with CVD risk. J Appl Physiol (1985). 2008;104(3):700–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawrence JC, Newcomer BR, Buchthal SD, et al. Relationship of intramyocellular lipid to insulin sensitivity may differ with ethnicity in healthy girls and women. Obesity (Silver Spring). Jan 2011;19(1):43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee S, Guerra N, Arslanian S. Skeletal muscle lipid content and insulin sensitivity in black versus white obese adolescents: is there a race differential? J Clin Endocrinol Metab. 2010;95(5):2426–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albu JB, Kovera AJ, Allen L, et al. Independent association of insulin resistance with larger amounts of intermuscular adipose tissue and a greater acute insulin response to glucose in African American than in white nondiabetic women. Amer J Clin Nutr. 2005;82(6):1210–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goedecke JH, Olsson T. Pathogenesis of type 2 diabetes risk in black Africans: a South African perspective. J Intern Med. 2020;288(3):284–294. [DOI] [PubMed] [Google Scholar]
- 38.Gower BA, Fowler LA. Obesity in African-Americans: The role of physiology. J Intern Med. 2020;288(3):295–304. [DOI] [PubMed] [Google Scholar]
- 39.Despres JP. Abdominal obesity as important component of insulin-resistance syndrome. Nutrition. 1993;9(5):452–459. [PubMed] [Google Scholar]
- 40.Snijder MB, Visser M, Dekker JM, et al. Low subcutaneous thigh fat is a risk factor for unfavorable glucose and lipid levels, independently of high abdominal fat. The Health ABC Study. Diabetologia. 2005;48(2):301–308. [DOI] [PubMed] [Google Scholar]
- 41.Van Pelt RE, Jankowski CM, Gozansky WS, Schwartz RS, Kohrt WM. Lower-body adiposity and metabolic protection in postmenopausal women. J Clin Endocrinol Metab. 2005;90(8):4573–4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ko KD, Kim KK, Lee KR. Does Weight Gain Associated with Thiazolidinedione Use Negatively Affect Cardiometabolic Health? J Obes Metab Syndr. 2017;26(2):102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grams J, Garvey WT. Weight Loss and the Prevention and Treatment of Type 2 Diabetes Using Lifestyle Therapy, Pharmacotherapy, and Bariatric Surgery: Mechanisms of Action. Curr Obes Rep. 2015;4(2):287–302. [DOI] [PubMed] [Google Scholar]
- 44.Cheng YJ, Kanaya AM, Araneta MRG, et al. Prevalence of Diabetes by Race and Ethnicity in the United States, 2011–2016. JAMA. 2019;322(24):2389–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gyllenhammer LE, Alderete TL, Toledo-Corral CM, Weigensberg M, Goran MI. Saturation of subcutaneous adipose tissue expansion and accumulation of ectopic fat associated with metabolic dysfunction during late and post-pubertal growth. Int J Obes (Lond). 2016;40(4):601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.