Abstract
Aims
Atrial fibrillation (AF) is one of the major causes of ischaemic stroke. In addition to clinical risk evaluated by the CHA2DS2-VASC score, the impact of genetic factors on the risk of AF-related thromboembolic stroke has been largely unknown. We found several copy number variations (CNVs) in novel genes that were associated with thromboembolic stroke risk in our AF patients by genome-wide approach. Among them, the gasdermin D (GSDMD) gene was related to inflammation. We aimed to test whether GSDMD deletion was associated with AF-related stroke.
Methods and results
A total of 400 patients with documented non-familial AF were selected, of which 100 patients were diagnosed with ischaemic stroke. The baseline characteristics of age, sex, valvular heart disease, coronary artery disease, heart failure, and CHA2DS2-VASc scores were not statistically different between cases and controls. We found that individuals who carried GSDMD homozygous deletion genotype had a higher risk for ischaemic stroke (odds ratio 2.195; 95% confidence interval, 1.24–3.90; P = 0.007), even adjusted by CHA2DS2-VASc scores. We also validated the association of GSDMD with AF stroke in a large Caucasian population (UK Biobank).
Conclusion
We found a link between the homozygous deletion of the GSDMD gene and an increased risk of stroke in patients with AF. This may implicate the use of therapy targeting GSDMD in the prevention of ischaemic stroke for AF patients.
Keywords: Atrial fibrillation • Stroke • Genetic study • GSDMD • Asia
What’s new?
This study demonstrates the association of GSDMD gene homozygous deletion with the risk of stroke in patients with atrial fibrillation (AF).
Individuals who carried GSDMD homozygous deletion genotype have a higher risk for ischaemic stroke (odds ratio 2.195).
We also validated the association of GSDMD with AF stroke in a large Caucasian population (UK Biobank).
What is already known on this topic—Atrial fibrillation (AF) is one of the major causes of ischaemic stroke. In addition to clinical risk evaluated by the CHA2DS2-VASC score, the impact of genetic factors on the risk of AF-related thromboembolic stroke has been largely unknown.
What this study adds—This is the first to demonstrate the association of GSDMD gene homozygous deletion with the risk of stroke in patients with AF.
How this study might affect research, practice, or policy—It may be possible to use therapy targeting GSDMD in the prevention of ischaemic stroke for AF patients.
Introduction
Atrial fibrillation (AF) is the most common sustained arrhythmia and is also a major public health problem in the world. AF is reported associated with substantial mortality, morbidity, and healthcare costs. The management and treatment of AF and its complications are responsible for a substantial proportion of the cost of the health care system per year all over the world.1 The most disastrous complication of AF is thromboembolism, especially thromboembolic stroke. Therefore, the management strategy for AF also focuses on the prevention of AF-related thromboembolic events, especially embolic stroke.
The contemporary 4S-AF programme, as demonstrated in the EURObservational Research Programme (EORP)-AF General Long-Term Registry, has exhibited enhanced prevention of ischaemic stroke in patients with a high risk of AF.2,3 Many clinical factors that predict the risk of stroke in patients with AF (e.g. CHADS2 or CHA2DS2-VASC score) have been proposed; however, these clinical risk factors cannot entirely identify all the AF patients who are at risk of thromboembolic events, with pooled c-statistics were only 0.658 in CHA2DS2-VASc score.4
Moreover, some AF patients with very low or even zero CHA2DS2-VASC scores still experience an embolic stroke, compared with some patients with very high CHA2DS2-VASC scores who never experience an embolic stroke. Therefore, there may be an underlying genetic factor(s) that determine an AF patient’s susceptibility to a cardioembolic event.5
Numerous studies have been conducted on the genetics of AF, aimed at improving the detection and treatment of AF.6–8 However, the impact of genetic factors or polymorphisms on the risk of thromboembolic stroke in patients with AF has been rarely addressed. Previously we have shown that genetic polymorphisms of the angiotensinogen gene and C reactive protein (CRP) gene could predict the risk of stroke for patients with AF.9,10 However, these loci only account for a small portion of the genetic risk of AF-related stroke, and it is possible that many additional genetic factors or variants have to be discovered. Regarding the genetic risk factors of thromboembolic stroke in AF patients, in addition to CRP and angiotensinogen genes, we do not know which gene(s) may be involved a priori, and in this scenario, a genome-wide approach is a powerful strategy to identify novel susceptibility genes or loci.
In the past decade, genome-wide association studies have been used to identify novel single-nucleotide polymorphisms (SNPs) or genes for human diseases, such as AF.11
Accordingly, to identify novel genes or variants that may predict thromboembolic stroke for our AF patients, we used a chip-based genome-wide approach and identified deletions in chromosomal regions 1p36.32-1p36.33, 5p15.33, 8q24.3, and 19p13.3 and amplifications in 14q11.2 that were significantly associated with AF-related stroke in the Taiwanese population. In these regions, 148 genes were involved, and using a pathway analysis, we found deletions in GNB1, PRKCZ, and GNG7 genes related to the alpha-adrenergic receptor signalling pathway playing a major role in determining the risk of AF-related stroke.12 In this preliminary genome-wide study, we also found a novel gene gasdermin D (GSDMD), the deletion of which gene was also associated with AF-related stroke, but this was not confirmed in a validation population.
It has been reported autoimmunity serves as one of the novel risk factors or mechanisms for ischaemic stroke and transient ischaemic attacks.13 Previous studies also showed inflammation might activate caspase-mediated cleavage of gasdermin D (GSDMD) and trigger pyroptosis, a lytic form of cell death that is crucial for immune defence mechanism and human diseases14 and inflammasome activation with GSDMD-mediated pyroptosis lead to cell death and blood clotting.15
Courtesy of the previous theory that inflammation-related mechanism in AF-related thromboembolic stroke, we sought to validate the association of GSDMD gene deletion with AF-related stroke in a larger population.
Methods
Study population
The study participants are from the National Taiwan University Hospital (Taipei and YunLin), National Taiwan University AF Registry (NTUAFR). This was a cross-sectional case-control study and all the study subjects were recruited at a single time point.
The selection criteria for patients involved in the study have been previously described in detail.16 All the participants were regularly followed up in the Cardiovascular Clinic at National Taiwan University Hospital (NTUH). The diagnosis of AF was determined by taking the patient's history, serial electrocardiograms (ECGs), and/or ambulatory ECG monitoring.1 Primary endpoint during follow-up were defined as events of ischaemic stroke. ischaemic stroke was defined as presentations of sudden-onset, focal, or global neurologic deficits that were not explained by other or known causes, with supporting evidence from the neurological imaging study. Although the ischaemic strokes were subtyped into either large-artery atherosclerosis or small-vessel occlusion, both were included as the primary outcome. Haemorrhagic stroke was excluded from the study.
The validation case population is those AF patients from the National Taiwan University Hospital Taipei (North Taiwan), Yun-Lin branch (Middle-South Taiwan), and Hsin-Chu Branch (North-Middle Taiwan). Genotyping these patients with a specific phenotype (stroke subgroup) in a diseased population (AF) is more likely to identify underlying genetic causes because of genetic predisposition, although the case number may be small due to the low prevalence of these patients.17 Thus, after a pioneer genome-wide scan, a validation study is needed. The control population is composed of those AF patients from our NTUAFR who have never experienced any thromboembolic event.
Detection of copy number variation regions by quantitative polymerase chain reaction and semi-quantitative conventional polymerase chain reaction
Quantitative polymerase chain reaction (qPCR) and semi-quantitative conventional PCR with densitometry were performed to verify the CNV and determine the copy number of the targeted gene (GSDMD) in the study samples. qPCR was performed with the TaqMan Copy Number Reference Assay RNase P following the TaqMan qPCR protocol. Each 15-μL reaction mixture comprised 50-ng genomic DNA and 1 TaqMan probe/primer mix in 1 TaqMan Master Mix, amplified on an Applied Biosystems QuanStudio 3D. qPCR data were collected and analysed by the ABI software (Life Technologies Co., CA, USA). Semi-quantitative conventional PCR with densitometry was performed for large samples with designed primers targeting the sequences within the postulated deletion segment. There was no PCR product from those samples with zero copy and the presence of PCR band from those with more or equal to one copy.18
Validation of association of the GSDMD gene with atrial fibrillation-related stroke in the Caucasian population
To confirm the role of GSDMD in AF-related stroke, we further validated the association of GSDMD gene with AF-related stroke in the United Kingdom (UK) population using the UK Biobank genotype data. In brief, UK Biobank is a prospective study of more than 500 000 people living in the UK with long-term follow-up. It was established to allow detailed investigations of the genetic and non-genetic determinants of the diseases of middle and old age. Participant DNA was genotyped on two arrays, UK BiLEVE and UK Biobank Axiom. Genotypes of ∼800 000 SNPs passing quality controls were imputed to the UK10K reference panel. Genotypes were called using Affymetrix Power Tools software. Finally, 1768 patients with AF and ischaemic stroke were used as the case population, and 18 456 patients with AF but without ischaemic stroke were used as the control population total.19
Statistical analysis
Continuous data were expressed as mean ± standard deviation. The comparison was conducted by independent two-sample Student’s t-test for the continuous variables and the χ2 test for the categorical variables.
Because this was a cross-sectional study, CNV association analyses were performed using the logistic regression model to adjust non-genetic covariates. The statistical significance level was set at P < 10−3 in the genome-wide discovery stage. We used a more liberal threshold of P < 10−3 in our stage I genome-wide discovery stage because of the small stage I sample size, and relied on subsequent validation in stage II with larger samples.
The statistical significance level was set at P < 0.05 in the validation study. Power estimation revealed that we had >95% power to replicate the association for an odds ratio of 2.0 at an alpha level of 0.05 with at least 100 cases and 300 controls in the replication population.
All statistical analyses were carried out using SPSS 25.0 (SPSS Inc. USA).
Results
Baseline characteristics
The study cohort comprised 400 participants, and the baseline characteristics of study patients is summarized in Table 1.
Table 1.
Comparisons of clinical characteristics of patients in the presence and absence of stroke during follow-up
| Variables | Stroke (n = 100) | non-stroke (n = 300) | P value |
|---|---|---|---|
| Age (years) | 76.3 ± 12.5 | 71.3 ± 13.0 | 0.701 |
| Male sex | 55 (55.0%) | 55 (55.0%) | 1.000 |
| VHD | 73 (73.0%) | 192 (64.0%) | 0.126 |
| Diabetes | 35 (35.0%) | 74 (24.7%) | 0.044 |
| Hypertension | 72 (72.0%) | 168 (56.0%) | 0.005 |
| Smoking | 37 (37.0%) | 75 (25.0%) | 0.021 |
| CAD | 39 (39.0%) | 90 (30.0%) | 0.083 |
| Heart failure | 13 (13.0%) | 31 (10.3%) | 0.460 |
| CHA₂DS₂-VASc | 3.54 ± 1.46 | 2.87 ± 1.53 | 0.636 |
CAD: coronary artery disease; VHD: valvular heart disease (at least moderate).
Patients with stroke were older (76.3 vs. 71.3-year-old, P = 0.701) and had a male predominance (55.0% vs. 55.0%, P = 1.000) and more co-morbidities such as diabetes mellitus (35.0% vs. 24.7%, P = 0.044), hypertension (72.0% vs. 56.0%, P = 0.005), smoking (37.0% vs. 25.0%, P = 0.021), but no difference in CAD (39.4% vs. 30.0%, P = 0.083), heart failure (13.0% vs. 10.3%, P = 0.460). The mean CHA2DS2-VASc score was higher in those with stroke than those without stroke without statistical significance. (3.54 ± 1.46 vs. 2.87 ± 1.53, P = 0.636).
Gasdermin D homogeneous deletion as a predictor of ischaemic stroke
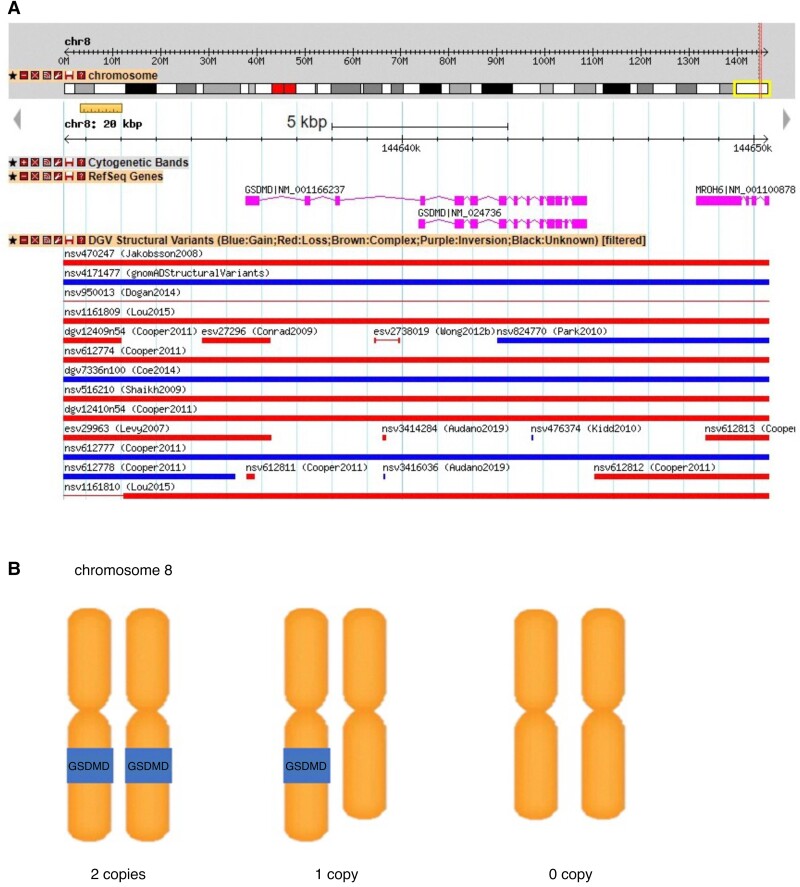
First, qPCR was performed and the result showed that GSDMD CNV was a diallelic polymorphism. For a larger number of samples, we performed semi-quantitative agarose gel electrophoresis to detect CNV. An illustrated figure was presented, demonstrating the variation in copy number deletions of the GSDMD gene located on the chromosome (Figure 1). PCR products of the candidate GSDMD gene showed a single band at the expected sizes in patients with either one or two copies and no band in those without any copy or homogeneous deletion. We then analysed whether the homozygous deletion was associated with stroke or not (recessive model).
Figure 1.
Graphical representation of the common copy number variation (CNV) region containing the GSDMD gene in chromosome 8q 24.3. A, This CNV region overlaps with previously reported CNV regions in the database of genomic variants (DGV) and 1000 genomes phase1 integrated variant calls. B, This CNV is a diallelic polymorphism. Individuals may have two copies, one copy or no copy of this CNV region containing GSDMD gene. CNV, copy number variation; DGV, database of genomic variants.
Table 2 presents the distribution of the GSDMD homozygous deletion genotypes in cases and controls, and the odds ratios for the association of GSDMD homozygous deletion, and ischaemic stroke. There was a significant association between the GSDMD homozygous deletion and the increasing risk of ischaemic stroke (odds ratio 2.048, P = 0.012).
Table 2.
Odds ratios for the association of GSDMD homozygous deletion with ischaemic stroke
| Variables | Stroke (n = 100) | non-stroke (n = 300) | Odds ratio (95% confidence interval) | P value |
|---|---|---|---|---|
| GSDMD deletion | 25.0% | 14.0% | 2.048 (1.172–3.577) | 0.012 |
Gasdermin D homogeneous deletion as a predictor of ischemic stroke, adjusted by CHA2DS2-VASc score
Previous studies have demonstrated that increasing the CHA2DS2-VASc score is associated with increased ischaemic stroke risk.
To eliminate the possible confounding effect of the CHA2DS2-VASc score on ischaemic stroke risk, we adjusted the risk by the CHA2DS2-VASc score.
Table 3 presents the results of the independent effect of GSDMD homozygous deletion on the risk of ischaemic stroke, adjusted by CHA2DS2-VASc score (model 1) or co-morbidities (model 2) in multivariable models. There was even a more significant association of the GSDMD homozygous deletion with an increased risk of ischaemic stroke (odds ratio 2.195, P = 0.007) after adjustment of CHA2DS2-VASc score or co-morbidities.
Table 3.
Odds ratios for the association of GSDMD homozygous deletion with ischaemic stroke in the multivariable analyses
| Variables | Odds ratio | P value |
|---|---|---|
| (95% confidence interval) | ||
| Model 1 | ||
| CHA2DS2-VASc score | 1.354 (1.159–1.581) | <0.001 |
| GSDMD deletion | 2.195 (1.237–3.896) | 0.007 |
| Model 2 | ||
| Age | 1.022 (0.571–1.827) | 0.942 |
| Male sex | 0.787 (0.444–1.391) | 0.409 |
| Diabetes | 1.318 (0.775–2.243) | 0.308 |
| Hypertension | 1.832 (1.110–3.025) | 0.018 |
| Coronary artery disease | 1.480 (0.862–2.540) | 0.155 |
| Heart failure | 1.033 (0.510–2.091) | 0.928 |
| GSDMD deletion | 2.017 (1.214–3.350) | 0.008 |
Validation of association of gasdermin D gene with atrial fibrillation-related to stroke in the Caucasian population
We further validated the association of the GSDMD gene with AF-related stroke in the UK Biobank. The results are shown in Table 4. The allele frequencies of SNPs (rs12541790, rs1349806, rs1377663, rs11551198, rs73375262, rs73375263, rs4874151, rs7840166, rs11551202) spanning the GSDMD gene were significantly different between those AF patients with stroke and those without stroke.
Table 4.
Validation of association of GSDMD gene with AF-related stroke in the Caucasian population
| SNP ID | SNP | MAF CASE | MAF CTL | OR | P value |
|---|---|---|---|---|---|
| rs12541790 | G/A | 0.190600 | 0.213580 | 0.869269 | 0.037841 |
| rs1349806 | C/G | 0.191850 | 0.214160 | 0.872873 | 0.043015 |
| rs1377663 | A/T | 0.191850 | 0.214070 | 0.873356 | 0.043866 |
| rs11551198 | G/A | 0.191850 | 0.213840 | 0.873884 | 0.044806 |
| rs73375262 | A/G | 0.206040 | 0.184740 | 1.141380 | 0.044858 |
| rs73375263 | G/A | 0.206040 | 0.184790 | 1.141050 | 0.045332 |
| rs4874151 | C/A | 0.206040 | 0.185020 | 1.140490 | 0.046122 |
| rs7840166 | G/A | 0.205760 | 0.184810 | 1.139060 | 0.048232 |
| rs11551202 | G/A | 0.205370 | 0.184430 | 1.138640 | 0.049234 |
CTL, control; MAF, minor allele frequency; OR, odds ratio; SNP, single-nucleotide polymorphism.
Discussion
The most serious morbidity of AF is thromboembolic stroke. With more clinical or genetic risk factors, the risk of developing stroke becomes higher. Currently, the major treatment for AF-related thromboembolic stroke is oral anti-coagulant,1 which could not reduce the risk to that of those without AF and is also associated with serious bleeding complications. When we better know the detailed molecular mechanism of AF-related thromboembolic stroke, we may have an opportunity to find a better treatment strategy for this serious complication.
In addition to the clinical risk factors such as CHA2DS2-VASC score, the impact of genetic factors on the risk of thromboembolic stroke in patients with AF has been largely unknown. Single-nucleotide polymorphisms (SNPs) in several genomic regions are associated with AF susceptibility.20
Regarding genes associated with thromboembolic stroke, we found several novel genes or variants by a genome-wide approach. We found deletions in GNB1, PRKCZ, and GNG7 genes that are related to the alpha-adrenergic receptor signalling pathway play a possible role in determining the risk of AF-related stroke.12 In this preliminary genome-wide study, we also found a novel gene gasdermin D (GSDMD), the deletion of which gene was also associated with AF-related stroke, but this was not confirmed in a larger validation population.
It has been reported autoimmunity serves as one of the novel risk factors or mechanisms for ischaemic stroke and transient ischaemic attacks.15 To the best of our knowledge, this is the first report to describe that GSDMD homozygous deletion is associated with ischaemic stroke among patients with AF, and we also validated this novel finding in the Causaaian population using the UK biobank data.
More deletions of the gene encoding GSDMD protein were found in AF patients with thromboembolic stroke compared to those AF patients without stroke. Therefore, we hypothesized that the impairment of GSDMD function and pore-forming ability and function may predispose AF patients to develop a thromboembolic stroke. However, how does impaired GSDMD function contribute to the susceptibility to thromboembolic stroke in patients with AF?
Gasdermin D (GSDMD) is a protein that in humans is encoded by the GSDMD gene on a chromosome. Several current studies have revealed that GSDMD serves as a specific substrate of inflammatory caspases (caspase-1, −4, −5, and −11) and as an effector molecule for the lytic and highly inflammatory form of programmed cell death known as pyroptosis. These caspases are localized on inflammasomes, large cytosolic protein complexes that serve as platforms upon which activated proinflammatory caspases cleave and activate their protein substrates. Cleaved GSDMD oligomerizes and forms pores in the plasma membrane, leading to disruption of local osmotic potential, formation of pyroptotic bodies, and eventual membrane rupture.14
Previous studies also showed that the N-terminal fragment of caspase-1-cleaved GSDMD rapidly targets the membrane fraction of macrophages which induces the formation of a plasma membrane pore in cells. These data suggest that GSDMD is the direct and final executor of pyroptotic cell death. The expression of the GSDMD gene is associated with other cardiovascular and neurological disorders.21
It has also been shown that NLRP3 inflammasome is a key driver of atherosclerosis and other vascular diseases.22 GSDMD is one of the downstream substrates of inflammasome and substrate of inflammatory caspases. Thus, GSDMD is one of the good therapeutic targets for alleviating cell ischemia/reperfusion injury in cardiovascular and neurological systems.23 Finally, activation of NLRPs inflammasome has been proposed as one of the mechanisms of AF, especially post-operative related AF.24–27
However, since activation of NLRPs inflammasome and GSDMD promotes AF, why deletion of the GSDMD gene and loss of GSDMD function is associated with AF-related thromboembolism?
We thought there may be several possible plausible explanations. First, it is possible that other inflammatory pathways also contribute to AF-related stroke or thromboembolism. For example, the interaction of Von Willebrand factor and Glycoprotein Ib receptor can activate thrombo-inflammatory pathways, which may lead to further cellular damage and potentially exacerbate AF and ischaemic stroke.28 Another study also discovered key genes and microRNAs related to inflammation including MEF2A, CAND1, PELI1, and PDCD4, and microRNAs (miR-1, miR-1-3p, miR-21, miR-21-5p, miR-192, miR-192-5p, miR-155, and miR-155-5p) in determining stroke risk for AF patients.29
Second, during AF, rapid depolarization may trigger cell death or injury, and dying cells or injured cells may trigger a prothrombotic state.30 Sustained AF was related to the structural change of atrial myocyte with loss of myofibrils, accumulation of glycogen, changes in mitochondrial shape and size, fragmentation of sarcoplasmic reticulum, and dispersion of nuclear chromatin. In fibrillating and dilated atria, self-cell death of myocytes with myolysis contributes to cellular remodelling. A previous study in electron microscopy from necropsy specimens of left atrial tissue showed a ‘rough endocardium’, with small areas of endothelial denudation related to thrombotic aggregation from patients with AF and cerebral embolism.31
It has been shown that pyroptosis may play a role in dying cell clearance.32 Therefore, we hypothesized that the impairment of GSDMD function resulted in the impairment of pyroptosis and might predispose AF patients to develop a thromboembolic stroke due to the impairment of clearance of injured atrial myocytes. This is a very novel idea and warrants further functional studies.
Finally, although the association of GSDMD with AF related was also validated in the UK population, our genetic finding should also be further validated in other studies.
Conclusion
We found a link between the homozygous deletion of the GSDMD gene and an increased risk of stroke in patients with AF. More studies are needed to substantiate these results.
Limitation
First, the case numbers are relatively small. Besides, this is a cross-sectional case-control genetic study, including the UK Biobank population. Therefore, we did not follow-up with the patients until the stroke happened in groups with different genotypes. Second, the study presents only an association and does not necessarily mean causality. We also did not measure the serum GSDMD level that could implicate causality. Third, we did not quantify the exact copy number and only dichotomized it into no copy and one or two copies by conventional PCR. Finally, GSDMD activation may also cause co-morbidities and may bias the results.24,26 Further studies are warranted to confirm the role of GSDMD deletion in AF-related thromboembolism.
Acknowledgements
The authors thank all the participating individuals for their contribution to this study.
Contributor Information
Pang-Shuo Huang, Division of Cardiology, Department of Internal Medicine, National Taiwan University Hospital Yun-Lin Branch, Yun-Lin, Taiwan; Graduate Institute of Clinical Medicine, College of Medicine, National Taiwan University, Taipei, Taiwan; Division of Cardiology, Department of Internal Medicine, National Taiwan University Hospital No. 7, Chung-Shan South Road, Taipei 100, Taiwan; Cardiovascular Center, National Taiwan University Hospital, No.7, Zhongshan S. Rd., Zhongzheng District, Taipei City 10002, Taiwan.
Jen-Fang Cheng, Division of Cardiology, Department of Internal Medicine, National Taiwan University Hospital No. 7, Chung-Shan South Road, Taipei 100, Taiwan; Cardiovascular Center, National Taiwan University Hospital, No.7, Zhongshan S. Rd., Zhongzheng District, Taipei City 10002, Taiwan; Division of Multidisciplinary medicine, Department of Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan.
Guan-Wei Li, Graduate Institute of Biomedical Electronics and Bioinformatics, National Taiwan University, Taipei, Taiwan.
Eric Y Chuang, Graduate Institute of Biomedical Electronics and Bioinformatics, National Taiwan University, Taipei, Taiwan; Bioinformatics and Biostatistics Core, Centers of Genomic and Precision Medicine, National Taiwan University, Taipei, Taiwan; Research and Development Center for Medical Devices, National Taiwan University, Taipei, Taiwan.
Jien-Jiun Chen, Division of Cardiology, Department of Internal Medicine, National Taiwan University Hospital Yun-Lin Branch, Yun-Lin, Taiwan.
Fu-Chun Chiu, Division of Cardiology, Department of Internal Medicine, National Taiwan University Hospital Yun-Lin Branch, Yun-Lin, Taiwan.
Cho-Kai Wu, Division of Cardiology, Department of Internal Medicine, National Taiwan University Hospital No. 7, Chung-Shan South Road, Taipei 100, Taiwan; Cardiovascular Center, National Taiwan University Hospital, No.7, Zhongshan S. Rd., Zhongzheng District, Taipei City 10002, Taiwan.
Yi-Chih Wang, Division of Cardiology, Department of Internal Medicine, National Taiwan University Hospital No. 7, Chung-Shan South Road, Taipei 100, Taiwan; Cardiovascular Center, National Taiwan University Hospital, No.7, Zhongshan S. Rd., Zhongzheng District, Taipei City 10002, Taiwan.
Juey-Jen Hwang, Division of Cardiology, Department of Internal Medicine, National Taiwan University Hospital No. 7, Chung-Shan South Road, Taipei 100, Taiwan; Cardiovascular Center, National Taiwan University Hospital, No.7, Zhongshan S. Rd., Zhongzheng District, Taipei City 10002, Taiwan.
Chia-Ti Tsai, Division of Cardiology, Department of Internal Medicine, National Taiwan University Hospital No. 7, Chung-Shan South Road, Taipei 100, Taiwan; Cardiovascular Center, National Taiwan University Hospital, No.7, Zhongshan S. Rd., Zhongzheng District, Taipei City 10002, Taiwan.
Author contributions
P.-S.H. and J.-F.C: study design, analyses, conducting, and manuscript preparation. G.-W.L. and E.-Y.C.: analyses. J.-J.C. and C.-K.W.: study design. Y.-C.W. and J.-J.H.: study design and planning. C.-T.T.: study design, analyses, and manuscript preparation. All authors contributed to the article and approved the submitted version.
Funding
The work was supported by grants from the Ministry of Science and Technology, Taiwan (MOST) in Taiwan [109-2314-B-002-244-MY3, 110-2314-B-002-198-MY3, and 110-2314-B-002-234] and grants from National Taiwan University Hospital Yunlin Branch [NTUHYL110.S012 and NTUHYL 111.S014]. Institutional Review Board (IRB) on National Taiwan University Hospital (200911002R).
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Hindricks G, Potpara T, Dagres N, Arbelo E, Bax JJ, Blomström-Lundqvist Cet al. 2020 ESC guidelines for the diagnosis and management of atrial fibrillation developed in collaboration with the European Association for Cardio-Thoracic Surgery (EACTS): the task force for the diagnosis and management of atrial fibrillation of the European Society of Cardiology (ESC) developed with the special contribution of the European Heart Rhythm Association (EHRA) of the ESC. Eur Heart J 2021;42:373–498. [DOI] [PubMed] [Google Scholar]
- 2. Ding WY, Proietti M, Boriani G, Fauchier L, Blomström-Lundqvist C, Marin Fet al. Clinical utility and prognostic implications of the novel 4S-AF scheme to characterize and evaluate patients with atrial fibrillation: a report from ESC-EHRA EORP-AF long-term general registry. Europace 2022;24:721–8. [DOI] [PubMed] [Google Scholar]
- 3. Ding WY, Blomström-Lundqvist C, Fauchier L, Marin F, Potpara TS, Boriani Get al. Contemporary management of atrial fibrillation and the predicted vs. absolute risk of ischaemic stroke despite treatment: a report from ESC-EHRA EORP-AF long-term general registry. Europace 2023;25:277–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van der Endt VHW, Milders J, Penning de Vries BBL, Trines SA, Groenwold RHH, Dekkers OMet al. Comprehensive comparison of stroke risk score performance: a systematic review and meta-analysis among 6 267 728 patients with atrial fibrillation. Europace 2022;24:1739–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Szymanski FM, Lip GY, Filipiak KJ, Platek AE, Hrynkiewicz-Szymanska A, Opolski G. Stroke risk factors beyond the CHA₂DS₂-VASc score: can we improve our identification of “high stroke risk” patients with atrial fibrillation? Am J Cardiol 2015;116:1781–8. [DOI] [PubMed] [Google Scholar]
- 6. Roberts JD, Davies RW, Lubitz SA, Thibodeau IL, Nery PB, Birnie DHet al. Evaluation of non-synonymous NPPA single nucleotide polymorphisms in atrial fibrillation. Europace 2010;12:1078–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kirchhof P, Breithardt G, Aliot E, Al Khatib S, Apostolakis S, Auricchio Aet al. Personalized management of atrial fibrillation: proceedings from the fourth atrial fibrillation competence NETwork/European heart rhythm association consensus conference. Europace 2013;15:1540–56. [DOI] [PubMed] [Google Scholar]
- 8. Tucker NR, Clauss S, Ellinor PT. Common variation in atrial fibrillation: navigating the path from genetic association to mechanism. Cardiovasc Res 2016;109:493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tsai CT, Chang SN, Chang SH, Lee JK, Lin LY, Wu CKet al. Renin-angiotensin system gene polymorphisms predict the risk of stroke in patients with atrial fibrillation: a 10-year prospective follow-up study. Heart rhythm 2014;11:1384–90. [DOI] [PubMed] [Google Scholar]
- 10. Chang SN, Lai LP, Chiang FT, Lin JL, Hwang JJ, Tsai CT. C-reactive protein gene polymorphism predicts the risk of thromboembolic stroke in patients with atrial fibrillation: a more than 10-year prospective follow-up study. J Thromb Haemost 2017;15:1541–6. [DOI] [PubMed] [Google Scholar]
- 11. Benjamin EJ, Rice KM, Arking DE, Pfeufer A, van Noord C, Smith AVet al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet 2009;41:879–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hsieh CS, Huang PS, Chang SN, Wu CK, Hwang JJ, Chuang EYet al. Genome-wide copy number variation association study of atrial fibrillation related thromboembolic stroke. J Clin Med 2019;8:332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Janardhan V, Wolf PA, Kase CS, Massaro JM, D'Agostino RB, Franzblau Cet al. Anticardiolipin antibodies and risk of ischemic stroke and transient ischemic attack: the Framingham cohort and offspring study. Stroke 2004;35:736–41. [DOI] [PubMed] [Google Scholar]
- 14. Ding J, Wang K, Liu W, She Y, Sun Q, Shi Jet al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 2016;535:111–6. [DOI] [PubMed] [Google Scholar]
- 15. Wu C, Lu W, Zhang Y, Zhang G, Shi X, Hisada Yet al. Inflammasome activation triggers blood clotting and host death through pyroptosis. Immunity 2019;50:1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tsai CT, Hsieh CS, Chang SN, Chuang EY, Ueng KC, Tsai CFet al. Genome-wide screening identifies a KCNIP1 copy number variant as a genetic predictor for atrial fibrillation. Nat Commun 2016;7:10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet 2010;11:415–25. [DOI] [PubMed] [Google Scholar]
- 18. Tsai CT, Wang DL, Chen WP, Hwang JJ, Hsieh CS, Hsu KLet al. Angiotensin II increases expression of alpha1C subunit of L-type calcium channel through a reactive oxygen species and cAMP response element-binding protein-dependent pathway in HL-1 myocytes. Circ Res 2007;100:1476–85. [DOI] [PubMed] [Google Scholar]
- 19. Lane JM, Vlasac I, Anderson SG, Kyle SD, Dixon WG, Bechtold DAet al. Genome-wide association analysis identifies novel loci for chronotype in 100,420 individuals from the UK biobank. Nat Commun 2016;7:10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roselli C, Rienstra M, Ellinor PT. Genetics of atrial fibrillation in 2020: GWAS, genome sequencing, polygenic risk, and beyond. Circ Res 2020;127:21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rui W, Xiao H, Fan Y, Ma Z, Xiao M, Li Set al. Systemic inflammasome activation and pyroptosis associate with the progression of amnestic mild cognitive impairment and Alzheimer’s disease. J Neuroinflammation 2021;18:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takahashi M. NLRP3 inflammasome as a key driver of vascular disease. Cardiovasc Res 2021;118:372–85. [DOI] [PubMed] [Google Scholar]
- 23. Wang K, Sun Z, Ru J, Wang S, Huang L, Ruan Let al. Ablation of GSDMD improves outcome of ischemic stroke through blocking canonical and non-canonical inflammasomes dependent pyroptosis in microglia. Front Neurol 2020;11:577927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heijman J, Muna AP, Veleva T, Molina CE, Sutanto H, Tekook Met al. Atrial myocyte NLRP3/CaMKII nexus forms a substrate for postoperative atrial fibrillation. Circ Res 2020;127:1036–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fender AC, Kleeschulte S, Stolte S, Leineweber K, Kamler M, Bode Jet al. Thrombin receptor PAR4 drives canonical NLRP3 inflammasome signaling in the heart. Basic Res Cardiol 2020;115:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scott L Jr, Fender AC, Saljic A, Li L, Chen X, Wang Xet al. NLRP3 inflammasome is a key driver of obesity-induced atrial arrhythmias. Cardiovasc Res 2021;117:1746–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dobrev D, Heijman J, Hiram R, Li N, Nattel S. Inflammatory signalling in atrial cardiomyocytes: a novel unifying principle in atrial fibrillation pathophysiology. Nat Rev Cardiol 2023;20:145–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guo Y, Lip GY, Apostolakis S. Inflammatory biomarkers and atrial fibrillation: potential role of inflammatory pathways in the pathogenesis of atrial fibrillation-induced thromboembolism. Curr Vasc Pharmacol 2015;13:192–201. [DOI] [PubMed] [Google Scholar]
- 29. Li Y, Tan W, Ye F, Wen S, Hu R, Cai Xet al. Inflammation as a risk factor for stroke in atrial fibrillation: data from a microarray data analysis. J Int Med Res 2020;48:300060520921671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grover SP, Mackman N. Tissue factor: an essential mediator of hemostasis and trigger of thrombosis. Arterioscler Thromb Vasc Biol 2018;38:709–25. [DOI] [PubMed] [Google Scholar]
- 31. Choudhury A, Lip GY. Atrial fibrillation and the hypercoagulable state: from basic science to clinical practice. Pathophysiol Haemost Thromb 2003;33:282–9. [DOI] [PubMed] [Google Scholar]
- 32. Atkin-Smith GK. Phagocytic clearance of apoptotic, necrotic, necroptotic and pyroptotic cells. Biochem Soc Trans 2021;49:793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Hindricks G, Potpara T, Dagres N, Arbelo E, Bax JJ, Blomström-Lundqvist Cet al. 2020 ESC guidelines for the diagnosis and management of atrial fibrillation developed in collaboration with the European Association for Cardio-Thoracic Surgery (EACTS): the task force for the diagnosis and management of atrial fibrillation of the European Society of Cardiology (ESC) developed with the special contribution of the European Heart Rhythm Association (EHRA) of the ESC. Eur Heart J 2021;42:373–498. [DOI] [PubMed] [Google Scholar]
- 2. Ding WY, Proietti M, Boriani G, Fauchier L, Blomström-Lundqvist C, Marin Fet al. Clinical utility and prognostic implications of the novel 4S-AF scheme to characterize and evaluate patients with atrial fibrillation: a report from ESC-EHRA EORP-AF long-term general registry. Europace 2022;24:721–8. [DOI] [PubMed] [Google Scholar]
- 3. Ding WY, Blomström-Lundqvist C, Fauchier L, Marin F, Potpara TS, Boriani Get al. Contemporary management of atrial fibrillation and the predicted vs. absolute risk of ischaemic stroke despite treatment: a report from ESC-EHRA EORP-AF long-term general registry. Europace 2023;25:277–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van der Endt VHW, Milders J, Penning de Vries BBL, Trines SA, Groenwold RHH, Dekkers OMet al. Comprehensive comparison of stroke risk score performance: a systematic review and meta-analysis among 6 267 728 patients with atrial fibrillation. Europace 2022;24:1739–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Szymanski FM, Lip GY, Filipiak KJ, Platek AE, Hrynkiewicz-Szymanska A, Opolski G. Stroke risk factors beyond the CHA₂DS₂-VASc score: can we improve our identification of “high stroke risk” patients with atrial fibrillation? Am J Cardiol 2015;116:1781–8. [DOI] [PubMed] [Google Scholar]
- 6. Roberts JD, Davies RW, Lubitz SA, Thibodeau IL, Nery PB, Birnie DHet al. Evaluation of non-synonymous NPPA single nucleotide polymorphisms in atrial fibrillation. Europace 2010;12:1078–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kirchhof P, Breithardt G, Aliot E, Al Khatib S, Apostolakis S, Auricchio Aet al. Personalized management of atrial fibrillation: proceedings from the fourth atrial fibrillation competence NETwork/European heart rhythm association consensus conference. Europace 2013;15:1540–56. [DOI] [PubMed] [Google Scholar]
- 8. Tucker NR, Clauss S, Ellinor PT. Common variation in atrial fibrillation: navigating the path from genetic association to mechanism. Cardiovasc Res 2016;109:493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tsai CT, Chang SN, Chang SH, Lee JK, Lin LY, Wu CKet al. Renin-angiotensin system gene polymorphisms predict the risk of stroke in patients with atrial fibrillation: a 10-year prospective follow-up study. Heart rhythm 2014;11:1384–90. [DOI] [PubMed] [Google Scholar]
- 10. Chang SN, Lai LP, Chiang FT, Lin JL, Hwang JJ, Tsai CT. C-reactive protein gene polymorphism predicts the risk of thromboembolic stroke in patients with atrial fibrillation: a more than 10-year prospective follow-up study. J Thromb Haemost 2017;15:1541–6. [DOI] [PubMed] [Google Scholar]
- 11. Benjamin EJ, Rice KM, Arking DE, Pfeufer A, van Noord C, Smith AVet al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet 2009;41:879–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hsieh CS, Huang PS, Chang SN, Wu CK, Hwang JJ, Chuang EYet al. Genome-wide copy number variation association study of atrial fibrillation related thromboembolic stroke. J Clin Med 2019;8:332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Janardhan V, Wolf PA, Kase CS, Massaro JM, D'Agostino RB, Franzblau Cet al. Anticardiolipin antibodies and risk of ischemic stroke and transient ischemic attack: the Framingham cohort and offspring study. Stroke 2004;35:736–41. [DOI] [PubMed] [Google Scholar]
- 14. Ding J, Wang K, Liu W, She Y, Sun Q, Shi Jet al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 2016;535:111–6. [DOI] [PubMed] [Google Scholar]
- 15. Wu C, Lu W, Zhang Y, Zhang G, Shi X, Hisada Yet al. Inflammasome activation triggers blood clotting and host death through pyroptosis. Immunity 2019;50:1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tsai CT, Hsieh CS, Chang SN, Chuang EY, Ueng KC, Tsai CFet al. Genome-wide screening identifies a KCNIP1 copy number variant as a genetic predictor for atrial fibrillation. Nat Commun 2016;7:10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet 2010;11:415–25. [DOI] [PubMed] [Google Scholar]
- 18. Tsai CT, Wang DL, Chen WP, Hwang JJ, Hsieh CS, Hsu KLet al. Angiotensin II increases expression of alpha1C subunit of L-type calcium channel through a reactive oxygen species and cAMP response element-binding protein-dependent pathway in HL-1 myocytes. Circ Res 2007;100:1476–85. [DOI] [PubMed] [Google Scholar]
- 19. Lane JM, Vlasac I, Anderson SG, Kyle SD, Dixon WG, Bechtold DAet al. Genome-wide association analysis identifies novel loci for chronotype in 100,420 individuals from the UK biobank. Nat Commun 2016;7:10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roselli C, Rienstra M, Ellinor PT. Genetics of atrial fibrillation in 2020: GWAS, genome sequencing, polygenic risk, and beyond. Circ Res 2020;127:21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rui W, Xiao H, Fan Y, Ma Z, Xiao M, Li Set al. Systemic inflammasome activation and pyroptosis associate with the progression of amnestic mild cognitive impairment and Alzheimer’s disease. J Neuroinflammation 2021;18:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takahashi M. NLRP3 inflammasome as a key driver of vascular disease. Cardiovasc Res 2021;118:372–85. [DOI] [PubMed] [Google Scholar]
- 23. Wang K, Sun Z, Ru J, Wang S, Huang L, Ruan Let al. Ablation of GSDMD improves outcome of ischemic stroke through blocking canonical and non-canonical inflammasomes dependent pyroptosis in microglia. Front Neurol 2020;11:577927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heijman J, Muna AP, Veleva T, Molina CE, Sutanto H, Tekook Met al. Atrial myocyte NLRP3/CaMKII nexus forms a substrate for postoperative atrial fibrillation. Circ Res 2020;127:1036–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fender AC, Kleeschulte S, Stolte S, Leineweber K, Kamler M, Bode Jet al. Thrombin receptor PAR4 drives canonical NLRP3 inflammasome signaling in the heart. Basic Res Cardiol 2020;115:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scott L Jr, Fender AC, Saljic A, Li L, Chen X, Wang Xet al. NLRP3 inflammasome is a key driver of obesity-induced atrial arrhythmias. Cardiovasc Res 2021;117:1746–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dobrev D, Heijman J, Hiram R, Li N, Nattel S. Inflammatory signalling in atrial cardiomyocytes: a novel unifying principle in atrial fibrillation pathophysiology. Nat Rev Cardiol 2023;20:145–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guo Y, Lip GY, Apostolakis S. Inflammatory biomarkers and atrial fibrillation: potential role of inflammatory pathways in the pathogenesis of atrial fibrillation-induced thromboembolism. Curr Vasc Pharmacol 2015;13:192–201. [DOI] [PubMed] [Google Scholar]
- 29. Li Y, Tan W, Ye F, Wen S, Hu R, Cai Xet al. Inflammation as a risk factor for stroke in atrial fibrillation: data from a microarray data analysis. J Int Med Res 2020;48:300060520921671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grover SP, Mackman N. Tissue factor: an essential mediator of hemostasis and trigger of thrombosis. Arterioscler Thromb Vasc Biol 2018;38:709–25. [DOI] [PubMed] [Google Scholar]
- 31. Choudhury A, Lip GY. Atrial fibrillation and the hypercoagulable state: from basic science to clinical practice. Pathophysiol Haemost Thromb 2003;33:282–9. [DOI] [PubMed] [Google Scholar]
- 32. Atkin-Smith GK. Phagocytic clearance of apoptotic, necrotic, necroptotic and pyroptotic cells. Biochem Soc Trans 2021;49:793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]