Abstract
Background:
BRCA2 alterations predict for a response to poly-ADP-ribose polymerase inhibition in metastatic castration-resistant prostate cancer (mCRPC). However, detection is hindered by insufficient tumor tissue and low sensitivity of cell-free DNA for detecting copy number loss.
Objective:
To evaluate the BRCA2 loss detection using single-cell, shallow whole-genome sequencing (sWGS) of circulating tumor cells (CTCs) in patients with mCRPC.
Design, setting, and participants:
We analyzed CTC samples collected concurrently with tumor biopsies intended for clinical sequencing in patients with progressing mCRPC.
Outcome measurements and statistical analysis:
Differences in proportions were evaluated using the chi-square test. Correlations between assays were analyzed in linear regression models. Associations between alterations and genomic instability were assessed on the single-cell level using mixed-effect negative binomial models.
Results and limitations:
We identified 138 patients with concurrent CTC and biopsy samples. CTC sWGS generated copy number profiles in a similar proportion of patients to biopsy samples (83% vs 78%,/) p = 0.23), but was more effective than bone biopsies (79% vs 50%; p = 0.009). CTC sWGS detected BRCA2 loss in more patients than tissue at the ≥1 (42% vs 16%; p < 0.001) and ≥2 (27% vs 16%; p = 0.028) CTC thresholds. The overall prevalence of BRCA2 loss was not increased in CTCs using sample-level composite z scores (p = 0.4), but was significantly increased compared with a lower-than-expected prevalence in bone samples (21% vs 3%, p =0.014). Positive/negative predictive values for CTC BRCA2 loss were 89%/96% using the ≥1 CTC threshold and 67%/92% using the composite z score. CTC BRCA2 loss was associated with higher genomic instability in univariate (1.4-fold large-scale transition difference, 95% confidence interval [CI]: 1.2–1.6;p < 0.001) and multivariable analysis (1.4-fold difference, 95% CI: 1.2–1.6; p < 0.001).
Conclusions:
Copy number profiles can reliably be generated using CTC sWGS, which detected a majority of tissue-confirmed BRCA2 loss and “CTC-only” losses. BRCA2 losses were supported by increases in genomic instability.
Patient summary:
Current testing strategies have limitations in their ability to detect BRCA2 loss, a relatively common alteration in prostate cancer that is used to identify patients who may benefit from targeted therapy. In this paper, we evaluated whether we could detect BRCA2 loss in individual tumor cells isolated from patient blood samples and found this method to be suitable for further analysis.
Keywords: BRCA2, Circulating tumor cells, Homologous recombination deficiency, Single-cell sequencing, Copy number loss
1. Introduction
Profiling for homologous recombination deficiency (HRD) is standard practice for men with metastatic castration-resistant prostate cancer (mCRPC) to guide treatment with poly-ADP-ribose polymerase (PARP) inhibitors [1-3]. A challenge is the difficulty obtaining sufficient tumor material for genetic profiling from bone, the most frequent site of spread [4,5]. Cell-free DNA (cfDNA) assays designed to detect mutations in BRCA2 and related genes are approved by the Food and Drug Administration [6,7], but tumor-derived DNA frequently represents only a small percentage of the genetic material [8]. The result is tumor fractions that are often too low (~<30% tumor vs normal) to detect copy number loss, which represent approximately one-third of BRCA2 alterations in mCRPC [9-11].
Recently, we showed that circulating tumor cells (CTCs) can be detected in >90% of patients with progressing mCRPC [12] and that shallow whole-genome sequencing (sWGS) of DNA from CTCs can detect losses and assess disease heterogeneity at the single-cell level [13,14]. Appropriately applied, CTC sWGS can potentially supplement tissue and/or cfDNA analyses to detect BRCA2 loss [15,16].
To initially evaluate this strategy, we profiled CTCs (Epic Sciences, San Diego, CA, USA) from men with progressing mCRPC undergoing a metastatic tumor biopsy for profiling using the Memorial Sloan Kettering Integrated Molecular Profiling of Actionable Cancer Targets (MSK-IMPACT) assay [17]. The objectives were to assess: (1) the feasibility of obtaining genetic information from CTCs, (2) the detection of BRCA2 loss in CTCs relative to tissue, and (3) the association between BRCA2 loss and chromosomal instability (CIN).
2. Patients and methods
2.1. Study cohort
Databases were queried in February 2018 to identify mCRPC patients who, as part of routine clinical management at MSKCC, underwent a biopsy within 30 d prior to starting a new treatment that was submitted for MSK-IMPACT sequencing (IRB #12-245, NCT01775072) [3,18]. Query results were matched to a database of banked CTC samples (IRB #06-107 and IRB #90-040) collected (when feasible) at therapeutic decision points within the same collection window (without intentional selection for specific patient populations) as part of a long-standing collaboration with Epic Sciences.
2.2. CTC sequencing
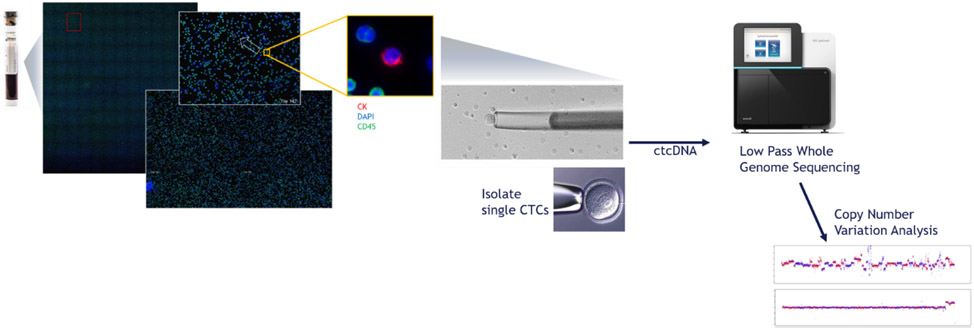
CTCs were identified and up to 25 individual CTCs were captured per patient for sWGS using a previously reported assay (Fig. 1) [15,19]. CTC sequencing was considered successful if copy number profiles were generated from one or more CTCs in a sample unless otherwise stated. For sequenced CTCs, the analysis pipeline aligns and bins the sequence reads using 1-Mb and 100-kb genomic bins, scales/normalizes the bin counts, and uses them for genome instability analysis and gene-centric copy number calls [15]. Composite (median) copy number ratios combining all sequenced CTCs in a sample were calculated for each 1-Mb bin. Gene-level z scores were calculated (using 100-kb bins) as the number of standard deviations above/below a panel of reference white blood cells (WBCs). Based on the results from prior validation studies using cell lines, z scores of ≥3 were considered amplifications and ≤−3 were considered losses [15]. Unless otherwise noted, CTC samples were considered positive for a gain/loss if that alteration was detected in one or more CTCs. The BRCA2 composite (median) z score of all CTCs in a given sample was calculated for sample-level comparisons. Large-scale transitions (LSTs), defined as copy number gains/losses of ≥10 Mb, were quantified independently for each CTC [20].
Fig. 1 –
Overview of Epic Sciences’ single-CTC sWGS methodology in which nucleated cells are extracted from whole blood, plated onto slides, stained (CK, DAPI, and CD45), and scanned for CTCs using automated imaging software. Individual CTCs are isolated from the slides for DNA extraction and sWGS. CTC = circulating tumor cell; ctDNA = circulating tumor DNA; sWGS = shallow whole-genome sequencing.
2.3. Tissue sequencing
Tissue samples were processed in MSKCC Department of Molecular Pathology for MSK-IMPACT and considered successful if a clinical sequencing report was generated. For successfully sequenced samples, BRCA2 deep deletions and mutations were called using an established variant-calling pipeline [17]. Additional analysis was performed to detect shallow deletions (any negative, significant fold-change), enabling more appropriate comparison.
2.4. Statistical analysis
Median values are accompanied by interquartile ranges (exclusionary method). Differences in proportions were analyzed with “N – 1” chi-square test [21,22]. Correlations between composite CTC and tissue copy number profiles were analyzed for each matched pair using Pearson’s correlation corrected for 91 replicate two-sided tests (Bonferroni’s method). Linear regression models were utilized to compare the number of CTCs sequenced and the correlation coefficient with tissue (Fisher transformed). Linear regression was also utilized to compare composite BRCA2 z scores and tissue fold change (log2 transformed). LST values were analyzed in mixed-effect negative-binomial models to determine the difference attributable to BRCA2 loss in univariate models and a multivariable model including AR, TP53, MYC, RB1, and PTEN. These models included random intercepts to account for intrapatient correlation.
3. Results
3.1. Cohort characteristics
A total of 138 matched CTC/tissue pairs met the study criteria (Supplementary Fig. 1). Patient demographics are shown in Table 1. The percentage of patients with metastasis to individual disease compartments (bone, nodal, liver, and lung) is consistent with prior literature [23]. Noted is that the biopsy targets were biased toward soft tissue sites because these lesions are more likely to provide sufficient material for sequencing.
Table 1 –
Overall cohort demographics broken down by assay success (≥1 CTC copy number profile generated for CTCs and clinical report generated for tissue)
| CTC success | Tissue success |
Concordance cohort |
All patients | ||
|---|---|---|---|---|---|
| Sample size (n) | 115(83) | 107 (78) | 91 (66) | 138 (100) | |
| Age (yr) | 68 (62–74) | 68(62–74) | 68 (62–74) | 67(62–74) | |
| Laboratory assessments | PSA (ng/dl) | 28.5 (6.5–166.5) | 34.2 (7.7–152.1) | 32.3 (7.3–165.5) | 28.2 (7.5–151.1) |
| ALP (U/l) | 121 (76–215) | 112 (75–182) | 112 (74–192) | 117 (76–190) | |
| LDH (U/l) | 246 (201–305) | 236 (201–324) | 247 (203–329) | 238 (194–305) | |
| Treatment exposures | Prior mCRPC Tx line | 1 (0–2) | 1 (0–2) | 1 (0–2) | 1 (0–2) |
| Prior ARSi (n) | 73 (64) | 73 (68) | 62 (68) | 85 (62) | |
| Prior taxane (n) | 33 (29) | 35 (33) | 30 (33) | 38 (28) | |
| Disease compartments with metastasis | Bone (n) | 93 (81) | 82 (77) | 69 (76) | 112 (81) |
| Bone only (n) | 17 (15) | 10 (9.3) | 4 (4.4) | 19 (14) | |
| LN (n) | 89 (77) | 86 (80) | 72 (79) | 107 (78) | |
| Liver (n) | 24 (21) | 26 (24) | 23 (25) | 29 (21) | |
| Lung (n) | 11 (9.6) | 11(10) | 9 (9.9) | 13 (9.4) | |
| Prostate/other (n) | 31 (27) | 30 (28) | 27 (289) | 35 (25) | |
| Biopsy site | Bone (n) | 30 (26) | 19 (18) | 15 (17) | 38 (28) |
| LN (n) | 55 (48) | 56 (52) | 48 (53) | 64 (46) | |
| Liver (n) | 17 (15) | 20 (19) | 17 (19) | 22 (16) | |
| Lung (n) | 5 (4.3) | 4 (3.7) | 4 (4.4) | 5 (3.6) | |
| Prostate/other (n) | 8 (7.0) | 8 (7.5) | 7 (7.7) | 9 (6.5) |
ALP = alkaline phosphatase; ARSi = androgen receptor signaling inhibitor; CTC = circulating tumor cell; LDH = lactate dehydrogenase; LN = lymph node; mCRPC = metastatic castration-resistant prostate cancer; PSA = prostate-specific antigen; Tx = treatment.
Statistics shown are median (interquartile range) for continuous variables and count (%) for discrete variables.
3.2. Success and concordance
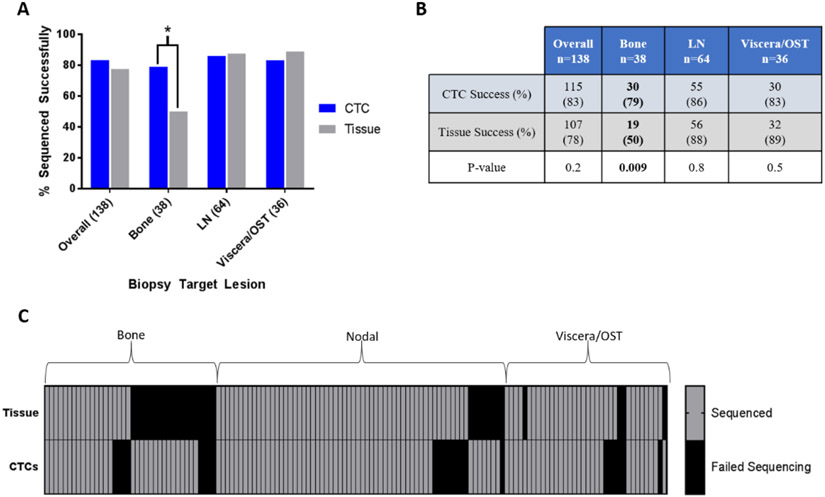
CTC sequencing was successful in 83% (115/138) of samples, similar to the 78% (107/138) success rate in the tissue samples (p = 0.2; Fig. 2A and 2B). Four (3%) CTC samples failed prior to staining (technical failures) and 19 (14%) had no detectable CTCs. From the 115 sequenced samples, a median of four CTCs (interquartile range [IQR] 2–10) were sequenced. CTC sequencing was successful in 77% (24/31) of the patients for whom tissue sequencing failed and in a significantly higher percentage of samples when profiling of bone lesions was attempted (n = 38; 79% vs 50%, p = 0.009; Fig. 2B and 2C).
Fig. 2 –
Ability of CTC and tissue sequencing to generate copy number profiles. (A) Proportion of successfully sequenced samples, by biopsy target (chi-square test). (B) Counts of sequenced samples, by biopsy target (p values from chi-square test). (C) Heat map of sequencing success rates (one or more CTCs) in matched tissue and CTC samples, showing individual patient data. CTC = circulating tumor cell; LN = lymph node; OST = other soft tissue site.
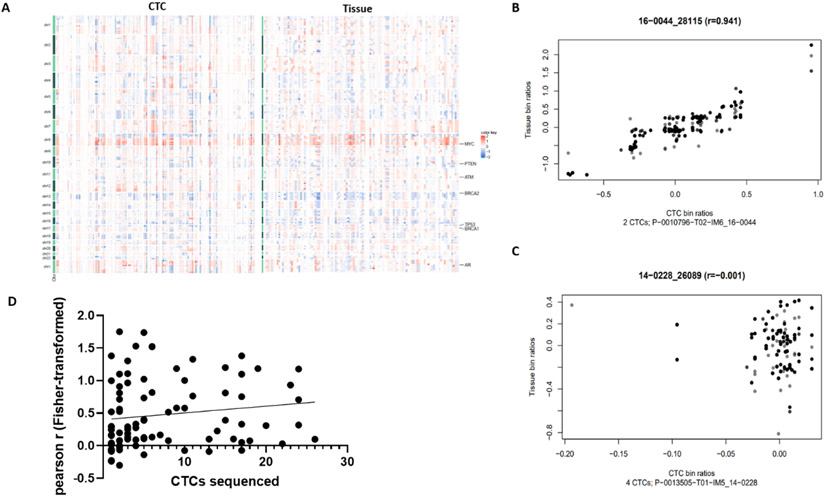
CTCs and tissue profiling identified recurring alterations associated with prostate cancer progression (del: 8p/13q/16q; gain: 7p/7q/8q; Fig. 3A) [24]. A significant positive correlation between composite CTC bin ratios and tissue bin ratios was observed in 77% (70/91) of matched sample pairs, with moderate-to-strong correlation (r > 0.5) in 37% (34/91; Fig. 3B and 3C). The median number of sequenced CTCs was higher in samples displaying a moderate-to-strong tissue correlation (6; IQR 2.5–15) than in samples with weak (4; IQR 2–11.75) and nonsignificant/negative (2; IQR 1–7.5) correlations. Although not significant, estimates obtained by linear regression suggest a possible association between the number of sequenced CTCs and Fisher-transformed correlation coefficient (estimated slope: 0.01, 95% confidence interval [CI] −0.005 to 0.026, R2 = 0.02,p = 0.19; Fig. 3D).
Fig. 3 –
Concordance of CTC copy number profiles. (A) Comparison of whole-genome copy number profiles, with each column representing copy number changes in an individual CTC (left panel) or a tissue (right panel) sample. (B) Scatter plot of bin ratios in composite CTCs (x axis) and tissue (y axis) in a representative patient demonstrating a strong correlation. Gray color is used to separate overlapping data points. (C) Scatter plot of bin ratios in composite CTCs (x axis) and tissue (y axis) in a representative patient demonstrating nonsignificant correlation. Gray color is used to separate overlapping data points. (D) Linear regression model demonstrating a nonsignificant trend towards the association between the number of CTCs sequenced and the CTC-tissue correlation coefficient (91 matched sample pairs plotted). CTC = circulating tumor cell.
3.3. BRCA2 loss detection
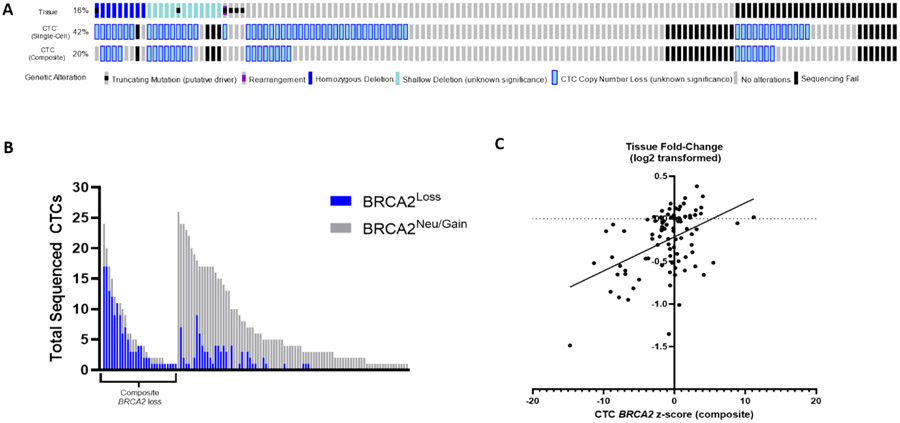
BRCA2 deep deletions were detected by MSK-IMPACT in 7% (9/138) of attempted samples and 8% (9/107) of successfully sequenced samples [25,26]. Including shallow loss, MSK-IMPACT detected BRCA2 loss in 16% (22/138) of patients overall and 21% (22/107) of those with successful sequencing. BRCA2 loss was detected on the single-CTC level in 42% (58/138) of attempted samples, a significantly higher frequency than tissue (p < 0.001; Fig. 4A). Given that BRCA2 loss was found in 7% of WBC controls, we applied a ≥2 CTC threshold to minimize false positives and the difference remained significant (27% vs 16%, p = 0.028). Notably, the difference remained significant only in bone biopsy cases at ≥2 (p = 0.014) and ≥3 (p = 0.049) CTC thresholds. The prevalence of composite BRCA2 loss in CTCs was similar to tissue (20% vs 16%, p = 0.4; Fig. 4A and 4B), but again significantly higher when the biopsy target was bone (21% vs 3%, p = 0.014).
Fig. 4 –
Comparison of BRCA2 detection in tissue and CTCs. (A) Oncoprint of BRCA2 alterations detected by MSK-IMPACT tumor testing (top), single-cell CTC calling (middle, ≥1 CTC threshold), and composite CTC calling (bottom). Shallow deletions and CTC losses have unknown clinical significance. (B) Number of successfully sequenced CTCs, by BRCA2 status (blue, loss; gray, neutral). Each bar represents an individual patient (left, patients with any composite BRCA2 loss; right, patients without composite BRCA2 loss in CTCs; n= 115). (C) Linear regression model demonstrating a significant association between BRCA2 fold-change (log2 transformed) in tissue and composite CTC z score (91 matched sample pairs plotted). CTC = circulating tumor cell; MSK-IMPACT = Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets.
BRCA2 loss was detected in 29% (220/765) of CTCs overall and in 39% (220/565) of CTCs from samples with detected loss. The median percentage of CTCs with BRCA2 loss within individual samples was 46% (IQR 23–77%; Fig. 4C) and was greater when the matched tissue sample had BRCA2 loss (58% vs 29%, p < 0.001).
These findings suggest that differences in BRCA2 loss prevalence may be explained by the challenges associated with profiling osseous disease, increased ability to detect subclonal losses in CTCs, and potential false positives.
Ninety-one successfully sequenced sample pairs were compared using tissue sequencing as “truth.” BRCA2 status was aligned in 66% (60/91) of matched pairs at the ≥1 CTC threshold and 75% (68/91) at the ≥2 CTC threshold. BRCA2 loss was detected in one or more CTCs in 16 of 18 samples (positive predictive value [PPV] = 89%) and two or more CTCs in 12 of 18 samples (PPV = 67%) matched to a tissue sample with BRCA2 loss. We identified one or more CTCs with BRCA2 loss in 40% (29/73), two or more CTCs in 23% (17/73), and three or more CTCs in 18% (13/73) of tissue-negative cases. When BRCA2 loss was not detected in CTCs (n = 46), it was present only in two tissue samples (negative predictive value [NPV] = 96%).
Using the composite z score, BRCA2 status was aligned in 85% (77/91) of sample pairs (PPV = 67% [12/18], NPV = 92% [65/71]). Additionally, there was a significant association between composite z score and tissue fold-change (log2-transformed; estimated slope: 0.04, 95% CI 0.024–0.057, R2 = 0.213, p < 0.001). In terms of discordance, eight (9%) samples had CTC-only BRCA2 loss and six (7%) had tissue-only loss. Tissue-only losses included four deep deletions, although only one of these samples had no CTCs with BRCA2 loss. These findings indicate that, compared with single-CTC calls, composite calls may reduce false positives and subclonal detection (potentially less/not responsive to PARP inhibition), but may have decreased sensitivity for deep deletions.
3.4. BRCA2 and CIN
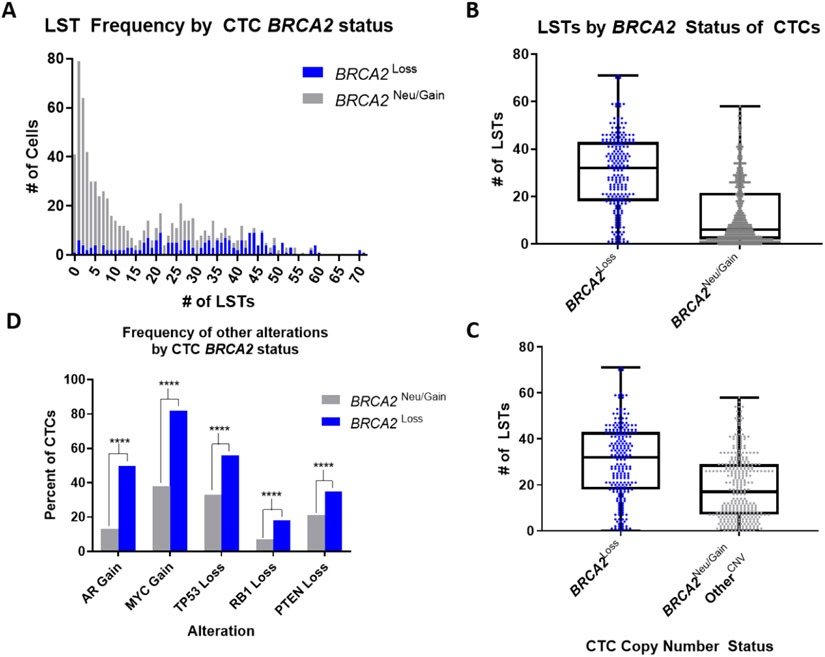
To evaluate the association between BRCA2 loss and CIN, we quantified genome-wide LSTs in all CTCs [27,28]. LST values by BRCA2 copy number status are plotted in Figure 5A. BRCA2 loss was associated with significantly higher LST values compared with BRCA2-neutral/amplified CTCs (2.2-fold LST difference, 95% CI 1.9–2.6, p < 0.001; Fig. 5B). BRCA2 loss was additionally associated with significantly higher LST values (1.4-fold LST difference, 95% CI 1.2–1.6, p < 0.001; Fig. 5C) compared with BRCA2-neutral/amplified CTCs, which harbor at least one copy number alteration in genes commonly implicated in prostate cancer progression (AR, TP53, MYC, PTEN, and/or RB1; n = 324). These results support that the observed increase in CIN is likely resulting from BRCA2 loss [29,30]. This analysis also controls for the possibility that WBCs and/or contaminating epithelial cells were misrepresented as BRCA2-neutral CTCs, as these cells would be less likely to have alterations in oncogenic driver genes.
Fig. 5 –
Comparison of genomic instability by CTC BRCA2 status. (A) Frequency of CTCs with (blue; n = 220) and without (gray; n = 565) BRCA2 CNL by LST number. (B) Average number of LSTs per CTC, by BRCA2 loss versus BRCA2 neutral (all n = 785 CTCs). Boxes span from the first to the third quartile, lines are medians, and whiskers are 1.5× IQR (Tukey); all data points are shown. (C) As in Figure 5B, but among CTCs with at least one other copy number variation in AR, TP53, ATYC, PTEN, and/or RBI (n = 324). (D) Prevalence of co-occurring AR, TP53, MYC, PTEN, and/or RB1 copy number variations in CTCs with and without BRCA2 CNL.
In a multivariable regression model, MYC gain (2.8-fold LST difference, 95% CI 2.5–3.3,p < 0.001), AR gain (1.8-fold LST difference, 95% CI 1.5–2.1, p < 0.001), and BRCA2 loss (1.4-fold difference, 95% CI 1.2–1.6, p < 0.001) demonstrated the strongest associations with LST number (Supplementary Table 1). MYC gain co-occurred in 82% (181/220) of CTCs with BRCA2 loss, consistent with prior studies [30,31]. Co-occurrence of TP53 loss (56%) and AR amplification (50%) was less frequent, followed by PTEN (35%) and RB1 (18%) loss. The prevalence of copy number alterations for each of the five genes was increased significantly in CTCs with BRCA2 loss (Fig. 5D). Additionally, since BRCA2 and RB1 are closely positioned on chromosome 13q, the modest rate of co-occurrence supports the ability of CTC sWGS to detect focal BRCA2 loss.
3.5. BRCA2 CNLs in CTCs from patients with other HRD alterations
In clinical trials evaluating PARP inhibitors, the highest response rates were seen in patients with BRCA2 alterations, with lower rates observed in patients with non-BRCA2 alterations predicted to result in HRD [2,32,33]. In this study, 116 CTCs were sequenced from 15 samples that had an oncogenic non-BRCA2 HRD alteration detected in matched tissue (Supplementary Fig. 2A and 3). CTCs from these samples had a range of LSTs from 0 to >70 (IQR = 36.5). Of CTCs with high CIN (nine or more LSTs), 64% (46/72) [20] harbored BRCA2 loss (Supplementary Fig. 2B), and the average LST number was higher in this CTC population (38.8 LSTs, n = 48) relative to those without loss (14.6 LSTs, n = 68; Supplementary Fig. 2C). These findings are strictly observational and require further investigation, but suggest that BRCA2 status may be an important factor in CIN in this patient population.
4. Discussion
Development of biomarker assays to detect BRCA2 alterations is increasingly important in mCRPC with the approval of multiple PARP inhibitors for this indication [1,2,6]. Copy number loss is estimated to account for about 30% of BRCA2 alterations in mCRPC patients [11], but bone biopsies often do not provide sufficient tumor for sequencing and/or detection of losses, and the detection of losses in cfDNA is feasible only with high tumor fractions [9,10]. Here, we report a cross-sectional study comparing single-CTC sWGS with concurrently collected tissue biopsies sequenced using MSK-IMPACT, focused specifically on BRCA2.
Our results showed that there was no significant difference in the overall likelihood of successfully profiling tissue and CTCs. A higher success rate (79%) was observed using CTC sWGS when attempting bone biopsies, with tissue sequencing being unsuccessful in half of cases. Although this preliminary observation requires further validation due to the relatively small number of bone biopsies represented in this cohort, it is an important finding given that osseous lesions are a hallmark of mCRPC and a substantial proportion of patients have exclusively osseous metastases. Additionally, since this study focused on patients with mCRPC, further investigation is needed to determine whether CTC sWGS is a reliable method for obtaining genomic profiles in earlier disease states where CTC detection rates are lower.
Next, we compared copy number profiles from CTCs and tissue to determine whether CTC sWGS produced accurate genomic information. Similarity in the most commonly altered chromosomal regions was observed across the cohort, and significant correlations were observed in a majority of cases on the patient level. Although not significant, our results suggested that sequencing more CTCs improves the accuracy of the assay, similar to the effect of tumor fraction on copy number detection in cfDNA [34,35]. Additionally, we found a significant association between composite BRCA2 z scores and tissue fold-change, supporting the BRCA2 calls. Going further, our data demonstrate that CTC sequencing detects the majority of tissue-detected BRCA2 loss using both the single CTC and the composite CTC calling method, although PPVs vary by method/threshold. This contrasts a recent cfDNA concordance study that reported a <40% PPV in respect to copy number alterations in concurrent sample pairs [35]. Conversely, a significant limitation of CTC sWGS is its inability to detect somatic mutations, a context in which cfDNA assays have demonstrated a clinical value.
Both single-cell and composite CTC calling methods detect losses not observed in matched tissue, but the frequency of this discordance is much higher when using the single-CTC threshold, potentially attributable to subclonal alterations and/or overcalling (false positives). Accordingly, the percentage of CTCs with BRCA2 loss (excluding patients without detected loss) was significantly lower in samples from patients without tissue-confirmed loss. These findings are similar to studies on somatic BRCA mutations showing high PPVs between cfDNA and tissue assays, with discordance resulting from low tumor fractions (analogous to low CTC number), subclonal heterogeneity, and clonal hematopoiesis [34,35]. Understanding that currently no single assay, including CTC sWGS, will be able to comprehensively profile the entirety of disease in a given patient for all classes of alterations, we hypothesize that multianalyte sequencing will be the most appropriate approach for profiling patients with mCRPC. Given the reciprocal strengths of cfDNA and CTC assays for the detection of mutations and copy number loss, respectively, a subsequent cfDNA analysis is ongoing to quantify the additive value of multianalyte profiling to detect actionable alterations.
To demonstrate that CTC BRCA2 loss is associated with CIN, we used the number of LSTs as a biomarker of CIN. CTC LSTs have previously been demonstrated to associate with BRCA2 loss and overall survival [20,36]. CTCs with BRCA2 loss had a significantly higher LST counts, with results indicating that CTCs BRCA2 loss results in increased CIN. This finding was supported in multivariable analysis including AR, PTEN, TP53, MYC, and RB1.
Our ability to determine the accuracy of BRCA2 calls was limited by the lack of true orthogonal comparison. Comparison with a clinically validated assay demonstrated a low frequency of false negatives, which increased when using composite CTC calls rather than single-cell calls. Conversely, inherent limitations of gross tissue sequencing and modest rates of BRCA2 loss observed in sequenced WBCs make it difficult to distinguish between false positives and true losses not detected in tissue. This was addressed in part by LST analyses, which mechanistically support the accuracy of single-cell BRCA2 calls in CTCs.
The cross-sectional design, sample size, alteration frequency, and patient population limited our ability to explore associations with clinical outcomes. Clinical validation studies remain as a critical next step to determine the predictive value and clinical relevance of CTC BRCA2 loss. To this end, the data from this analysis were used to inform the design of ongoing prospective biomarker validation studies attempting to replicate the observations from this study and determine the clinical relevance of CTC BRCA2 loss.
5. Conclusions
CTC sequencing has the potential to compliment tissue and/or cfDNA profiling to detect BRCA2 loss in mCRPC patients, especially those with predominantly/exclusively osseous disease. A further prospective study is needed to determine whether single-CTC and/or composite BRCA2 status are predictive of a response to PARP inhibitors.
Supplementary Material
Whole-genome sequencing of circulating tumor cells (CTCs) can detect BRCA2 loss in a majority of tissue-confirmed cases and in additional patients without tissue-confirmed loss. CTCs with BRCA2 loss have increased chromosomal instability, supporting the accuracy of the calls. CTCs can potentially address shortcomings of tissue/cell-free DNA for BRCA2 loss detection.
Funding/Support and role of the sponsor:
This work was funded in part by National Cancer Institute Cancer Center Support Grant P30 CA008748 to MSK, SPORE in Prostate Cancer grant P50 CA092629 to Howard I. Scher, the Department of Defense Early Investigator Research Award W81XWH-18-1-0330 to Konrad H. Stopsack, and Physician Research Award W81XWH-17-1-0124 to Wassim Abida, and by the Marie-Josée and Henry R. Kravis Center for Molecular Oncology at MSK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial disclosures: Howard I. Scher certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: E.S. Barnett, N. Schultz, and K.H. Stopsack: none. E.T. Lam: stock options and employment at Epic Sciences. A. Arfe: none. J. Lee: stock options and employment at Epic Sciences. J.L. Zhao: none. J.D. Schonhoft: stock options and employment at Epic Sciences. E.A. Carbone, N.M. Keegan, and A. Wibmer: none. Y. Wang: stock options and employment at Epic Sciences. D.B. Solit: has received honoraria/consulted for Pfizer, Loxo/Lilly Oncology, Vividion Therapeutics, Fore Therapeutics, Scorpion Therapeutics, and BridgeBio. W. Abida: consultant/advisory board member: Clovis Oncology, Janssen, ORIC Pharmaceuticals, and Daiichy Sankyo; institutional research funding: AstraZeneca, Zenith Epigenetics, Clovis Oncology, ORIC Pharmaceuticals, and Epizyme; honoraria: Roche, Medscape, Aptitude Health, Clinical Education Alliance, and OncLive/MJH Life Sciences. R. Wenstrup: stock options and employment at Epic Sciences. H.I. Scher: consultant/advisory board member: Ambry Genetics Corporation, Amgen, Bayer, ESSA Pharma, Janssen Biotech, Janssen Research & Development, Menarini Silicon Biosystems, Pfizer, Sun Pharmaceuticals Industries, Inc., and WCG Oncology; intellectual property rights: BioNTech, Elucida Oncology, MaBVAX, and Y-mAbs Therapeutics, Inc.; institutional research funding: Epic Sciences, Illumina, Janssen Diagnostics, Menarini Silicon Biosystems, and Thermo Fisher. Nikolaus Schultz, Konrad H. Stopsack, and Wassim Abida are Prostate Cancer Foundation Young Investigators.
References
- [1].Abida W, Patnaik A, Campbell D, et al. Rucaparib in men with metastatic castration-resistant prostate cancer harboring a BRCA1 or BRCA2 gene alteration. J Clin Oncol 2020;38:3763–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].de Bono J, Mateo J, Fizazi K, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med 2020;382:2091–102. [DOI] [PubMed] [Google Scholar]
- [3].Abida W, Armenia J, Gopalan A, et al. Prospective genomic profiling of prostate cancer across disease states reveals germline and somatic alterations that may affect clinical decision making. JCO Precis Oncol 2017;2017:PO.17.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ross RW, Halabi S, Ou SS, et al. Predictors of prostate cancer tissue acquisition by an undirected core bone marrow biopsy in metastatic castration-resistant prostate cancer—a Cancer and Leukemia Group B study. Clin Cancer Res 2005;11:8109–13. [DOI] [PubMed] [Google Scholar]
- [5].Sailer V, Schiffman MH, Kossai M, et al. Bone biopsy protocol for advanced prostate cancer in the era of precision medicine. Cancer 2018;124:1008–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Anscher MS, Chang E, Gao X, et al. FDA approval summary: rucaparib for the treatment of patients with deleterious BRCA-mutated metastatic castrate-resistant prostate cancer. Oncologist 2021;26:139–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Goodall J, Mateo J, Yuan W, et al. Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov 2017;7:1006–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Romanel A, Gasi Tandefelt D, Conteduca V, et al. Plasma AR and abiraterone-resistant prostate cancer. Sci Transl Med 2015;7:312re310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jayaram A, Wetterskog D, Attard G. Plasma DNA and metastatic castration-resistant prostate cancer: the odyssey to a clinical biomarker test. Cancer Discov 2018;8:392–4. [DOI] [PubMed] [Google Scholar]
- [10].Annala M, Struss WJ, Warner EW, et al. Treatment outcomes and tumor loss of heterozygosity in germline DNA repair-deficient prostate cancer. Eur Urol 2017;72:34–42. [DOI] [PubMed] [Google Scholar]
- [11].Chi KN, Barnicle A, Sibilla C, et al. Concordance of BRCA1, BRCA2 (BRCA), and ATM mutations identified in matched tumor tissue and circulating tumor DNA (ctDNA) in men with metastatic castration-resistant prostate cancer (mCRPC) screened in the PROfound study. J Clin Oncol 2021;39(6_suppl):26. [Google Scholar]
- [12].Scher HI, Armstrong AJ, Schonhoft JD, et al. Development and validation of circulating tumour cell enumeration (Epic Sciences) as a prognostic biomarker in men with metastatic castration-resistant prostate cancer. Eur J Cancer 2021;150:83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Scher HI, Graf RP, Schreiber NA, et al. Phenotypic heterogeneity of circulating tumor cells informs clinical decisions between AR signaling inhibitors and taxanes in metastatic prostate cancer. Cancer Res 2017;77:5687–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gupta S, Halabi S, Kemeny G, et al. Circulating tumor cell genomic evolution and hormone therapy outcomes in men with metastatic castration-resistant prostate cancer. Mol Cancer Res 2021;19:1040–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Greene SB, Dago AE, Leitz LJ, et al. Chromosomal instability estimation based on next generation sequencing and single cell genome wide copy number variation analysis. PLoS One 2016;11:e0165089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gupta S, Hovelson DH, Kemeny G, et al. Discordant and heterogeneous clinically relevant genomic alterations in circulating tumor cells vs plasma DNA from men with metastatic castration resistant prostate cancer. Genes Chromosomes Cancer 2020;59:225–39. [DOI] [PubMed] [Google Scholar]
- [17].Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 2015;17:251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Werner SL, Graf RP, Landers M, et al. Analytical validation and capabilities of the Epic CTC platform: enrichment-free circulating tumour cell detection and characterization. J Circ Biomark 2015;4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Schonhoft JD, Zhao JL, Jendrisak A, et al. Morphology-predicted large-scale transition number in circulating tumor cells identifies a chromosomal instability biomarker associated with poor outcome in castration-resistant prostate cancer. Cancer Res 2020;80:4892–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Campbell I. Chi-squared and Fisher-Irwin tests of two-by-two tables with small sample recommendations. Stat Med 2007;26:3661–75. [DOI] [PubMed] [Google Scholar]
- [22].Richardson JT. The analysis of 2 × 2 contingency tables—yet again. Stat Med 2011;30:890; author reply 891–2. [DOI] [PubMed] [Google Scholar]
- [23].Pezaro C, Omlin A, Lorente D, et al. Visceral disease in castration-resistant prostate cancer. Eur Urol 2014;65:270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Stopsack KH, Whittaker CA, Gerke TA, et al. Aneuploidy drives lethal progression in prostate cancer. Proc Natl Acad Sci U S A 2019;116:11390–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Marquard AM, Eklund AC, Joshi T, et al. Pan-cancer analysis of genomic scar signatures associated with homologous recombination deficiency suggests novel indications for existing cancer drugs. Biomark Res 2015;3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Popova T, Manie E, Rieunier G, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res 2012;72:5454–62. [DOI] [PubMed] [Google Scholar]
- [29].Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature 2012;481:287–94. [DOI] [PubMed] [Google Scholar]
- [30].Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer 2016;16:110–20. [DOI] [PubMed] [Google Scholar]
- [31].Castro E, Jugurnauth-Little S, Karlsson Q, et al. High burden of copy number alterations and c-MYC amplification in prostate cancer from BRCA2 germline mutation carriers. Ann Oncol 2015;26:2293–300. [DOI] [PubMed] [Google Scholar]
- [32].Abida W, Campbell D, Patnaik A, et al. Non-BRCA DNA damage repair gene alterations and response to the PARP inhibitor rucaparib in metastatic castration-resistant prostate cancer: analysis from the phase II TRITON2 study. Clin Cancer Res 2020;26:2487–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Stopsack KH. Efficacy of PARP inhibition in metastatic castration-resistant prostate cancer is very different with non-BRCA DNA repair alterations: reconstructing prespecified endpoints for cohort B from the phase 3 PROfound trial of olaparib. Eur Urol 2021;79:442–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wyatt AW, Annala M, Aggarwal R, et al. Concordance of circulating tumor DNA and matched metastatic tissue biopsy in prostate cancer. J Natl Cancer Inst 2017;109:djx118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tukachinsky H, Madison RW, Chung JH, et al. Genomic analysis of circulating tumor DNA in 3,334 patients with advanced prostate cancer identifies targetable BRCA alterations and AR resistance mechanisms. Clin Cancer Res 2021;27:3094–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jonsson P, Bandlamudi C, Cheng ML, et al. Tumour lineage shapes BRCA-mediated phenotypes. Nature 2019;571:576–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.