Abstract
Alcohol use among older adults is on the rise. This increase is clinically relevant as older adults are at risk for increased morbidity and mortality from many alcohol-related chronic diseases compared to younger patients. However, little is known regarding the synergistic effects of alcohol and age. There are intriguing data suggesting that aging may lead to impaired intestinal barrier integrity and dysbiosis of the intestinal microbiome, which could increase susceptibility to alcohol’s negative effects. To study the effects of alcohol in age we exposed aged and young mice to 3 days of moderate ethanol and evaluated changes in gut parameters. We found that these levels of drinking do not have obvious effects in young mice but cause significant alcohol-induced gut barrier dysfunction and expression of the pro-inflammatory cytokine TNFα in aged mice. Ethanol-induced downregulation of expression of the gutprotective antimicrobial peptides Defa-rs1, Reg3b, and Reg3g was observed in aged, but not young mice. Analysis of the fecal microbiome revealed age-associated shifts in microbial taxa, which correlated with intestinal and hepatic inflammatory gene expression. Taken together, these data demonstrate that age drives microbiome dysbiosis, while ethanol exposure in aged mice induces changes in the expression of antimicrobial genes important for separating these potentially damaging microbes from the intestinal lumen. These changes highlight potential mechanistic targets for prevention of the age-related exacerbation of effects of ethanol on the gut.
Keywords: aging, antimicrobial peptide, innate immunity, intestine, liver, microbiome
Introduction
Understanding how alcohol use may differentially effect those of advanced age is an urgent clinical issue. With the aging of the “baby boomer” generation the percent of the population over the age of 65 is growing rapidly and is expected to increase from 16% to 21.6% by 2040 (United States Census Bureau, 2017). This is accompanied by a concurrent rise in alcohol consumption among this age group over the last two decades (Breslow, Castle, Chen, & Graubard, 2017), with 10% of those over 65 reporting binge drinking in the last month (Substance Abuse and Mental Health Services Administration, 2019). This is of particular concern since older adults may be at increased risk for alcohol-related complications due to the existence of pre-existing chronic conditions and greater use of prescription and over-the-counter medications in this age group. In addition, the neurological effects of drinking have been shown to be exacerbated with age (Sullivan et al., 2018). However, alcohol can cause damage to multiple organs systems, including the gastrointestinal tract, and knowledge regarding how age alters these responses is lacking.
The gut plays a critical role in regulating the systemic effects of alcohol. In the healthy state, the intestinal epithelium acts as an important barrier between the microbiota residing in the lumen of the gastrointestinal tract and the systemic circulation. This intestinal barrier is maintained by the physical barrier formed by tight junction proteins connecting epithelial cells that line the lumen of the intestine (Chou et al., 2015). In addition, there is a chemical barrier that is generated by antimicrobial peptides (AMPs) on the luminal surface of the gut, which are produced by immune and epithelial cells (Mukherjee & Hooper, 2015). Multiple families of AMPs, including the C-type lectins (regenerating islet-derived protein 3-gamma and beta, or Reg3γ and Reg3β) and α-defensins, are produced by Paneth cells in the crypts of Leiberkuhn. These molecules are critical for preventing the microbiota in the lumen of the intestine from invading intestinal epithelial cells (Vaishnava et al., 2011). The resident microbiome also helps maintain epithelial barrier function. For example, short-chain fatty acids (SCFA), produced by microbial fermentation of dietary fiber and starch, act as an important energy source for epithelial cells and barrier function by increasing the expression of tight junction proteins (Kelly et al., 2015; Wang, Wang, Wang, Wan, & Liu, 2012). Therefore, decreased AMP production and/or changes in the intestinal microbiome and their metabolites can all have significant systemic effects resulting from loss of the integrity of the epithelial cell barrier.
In young populations, chronic exposure to high levels of ethanol results in disruption of gut homeostasis, promoting a range of pathologies both inside and outside the gastrointestinal tract. The breakdown of the intestinal epithelial cell barrier results in leakage of bacteria and bacterial products into multiple organs, including the liver, where they promote inflammation (Hartmann, Seebauer, & Schnabl, 2015; Seki et al., 2007). While the mechanisms for this barrier disruption are not fully understood, ethanol can have direct detrimental effects on intestinal epithelial cells (Rao, 2008). High levels of ethanol also induce dysbiosis of the intestinal microbiome in both mice (Bull-Otterson et al., 2013; Yan et al., 2011) and humans (Mutlu et al., 2012). Importantly, in chronic alcohol use, the observed changes in gut barrier function begin before there is evidence of liver inflammation and damage, suggesting they contribute to disease.
The gastrointestinal effects of alcohol may be a particularly critical area of study in the context of aging, as there is a growing body of evidence that the gut is altered in older individuals. Multiple studies have reported shifts in the human microbiome that are associated with advanced age (Claesson et al., 2011; Odamaki et al., 2016; O’Toole & Jeffery, 2015). While the microbiome itself does not age, it is likely that changes in the diet and environment of the aging host contribute to the dysbiosis. There is also evidence for impaired epithelial integrity in the aged (Mitchell et al., 2017), which could explain the increase in gut-related disorders in this population (Hall, Proctor, Fisher, & Rose, 2005). Importantly, these factors could have important implications for alcohol use in the elderly since alcohol-induced disruptions in the gut-liver axis contribute significantly to its harmful effects.
In this study, we employed a murine model of aging and acute ethanol exposure to test whether the gastrointestinal effects of ethanol were more severe in aged versus young mice. We characterized gut barrier function along with intestinal inflammatory cytokine and AMP expression. In addition, 16S rRNA gene sequencing was performed to profile the fecal microbiota in aged and young mice both before and after ethanol exposure. Our results show that aged mice are sensitized to the gastrointestinal effects of ethanol, with intestinal inflammation and barrier dysfunction occurring at ethanol levels that have no effects in young mice. Age alone was associated with a number of changes in the fecal microbiome that correlated with expression of inflammatory cytokines in the ileum and liver. Although 3 days of ethanol had minimal impact on the microbiota composition in either young or aged mice, it did induce a significant downregulation of AMPs in the ileum of aged mice alone, potentially exposing the lumen to the dysbiosis microbial populations expanded in aged mice.
Methods
Mice
BALB/cBy female mice were obtained from the National Institute on Aging (NIA) colony (Charles River Laboratories; Wilmington, Massachusetts). Mice were housed at the University of Colorado Anschutz Medical Campus for a minimum of 2 weeks prior to any experimentation. All animal experiments were performed humanely under a protocol approved by the Animal Care and Use Committee of the University of Colorado Anschutz Medical Campus. Young mice were 4–5 months of age (similar to humans 25–30 years) and aged mice were 21–22 months of age (similar to humans >65 years) (Flurkey, Currer, & Harrison, 2007). Mice were subjected to three sequential daily ethanol exposures by gavage of 1.5 g/kg and 1.25 g/kg, respectively. As the rate of ethanol metabolism differs between young and older subjects (Meier & Seitz, 2008), we adjusted ethanol dosages to elevate blood ethanol levels in both young and aged mice to an equivalent of ~80–100 mg/dL at 30 min post-gavage. Mice in the vehicle groups were gavaged with water. To test blood alcohol concentrations (BACs), tail-vein blood was collected 30 min after oral gavage of ethanol, and BAC was measured using the Ethanol Colormetric/Fluorometric Assay Kit from BioVision (McMahan et al., 2021). For all experiments, excluding those evaluating intestinal permeability (see below), mice were humanely euthanized using CO2 inhalation, and blood and organs were collected for analysis 1 h after the final ethanol or vehicle gavage.
Intestinal permeability assays
Intestinal permeability to fluorescein isothiocyanate (FITC)-conjugated dextran (FITC-dextran, 4kD) was measured using a previously described protocol (McMahan et al., 2021; Rendon, Li, Akhtar, & Choudhry, 2013). Briefly, 3 h prior to euthanasia, mice were gavaged with FITC-Dextran (600 mg/kg of body weight) simultaneously with the final dose of ethanol or vehicle. Serum was collected by cardiac puncture and samples were analyzed for levels of FITC-dextran with the Promega GloMax + fluorescence spectrophotometer plate reader.
Quantitative real-time PCR
Liver and ileal tissue were harvested and stored as previously described (McMahan et al., 2013; Zahs et al., 2012). RNA isolation and quantitative RT-PCR were also performed as previously described (McGettigan et al., 2019). Briefly, RNA was isolated using the RNeasy Mini kit and converted to cDNA using the QuantiTect RT kit (both from Qiagen) following manufacturer protocols. Real-time quantitative PCR was performed on the QuantStudio 3 Real-Time PCR System (Thermo Fisher) using TaqMan probes and reagents: Reg3g (Mm00441127-m1), Reg3b (Mm00440616_g1), Defa-rs1 (Mm00655850_m1), Tnfa (Mm00443258_m1), Il1b (Mm00434228_m1). Results were analyzed using the ΔΔCt algorithm (Livak & Schmittgen, 2001). GAPDH (4352339E) (Thermo-Fisher) was used as the endogenous control.
Microbiome analysis
Fecal bacterial profiles were determined by broad-range amplification and sequence analysis of 16S rRNA genes as previously described (Manuel et al., 2022; Wheatley et al., 2020). Briefly, DNA was extracted using the QIAamp PowerFecal kit (QIAGEN) and amplicons were generated using primers targeting the V3V4 variable region of the 16S rRNA gene. PCR products were then normalized using a SequalPrep kit (Invitrogen), purified and concentrated using a DNAClean and Concentrator Kit (Zymo), and quantified using Qubit Fluorometer 2.0 (Invitrogen). Illumina paired-end sequencing was performed on the MiSeq platform with versions v2.4 of the Miseq Control Software and of MiSeq Reporter, using a 600-cycle version three-reagent kit. Paired-end sequences were sorted by sample via barcodes in the paired reads with a Python script (Frank et al., 2021). The paired reads were assembled using phrap (Ewing & Green, 1998), and pairs that did not assemble were discarded. Assembled sequence ends were trimmed over a moving window of five nucleotides until average quality met or exceeded 20. Trimmed sequences with more than one ambiguity or shorter than 250 nt were discarded. Potential chimeras identified with Uchime (usearch6.0.203_i86linux32) (Edgar, Haas, Clemente, Quince, & Knight, 2011) using the Schloss (Schloss & Westcott, 2011) Silva reference sequences were removed from subsequent analyses. Assembled sequences were aligned and classified with SINA (1.3.0-r23838) (Pruesse, Peplies, & Glöckner, 2012) using the 418 497 bacterial sequences in Silva 115NR99 (Quast et al., 2013) as reference configured to yield the Silva taxonomy; taxonomic assignments used the lowest-common-ancestor approach with default SINA settings. Closed-reference, operational taxonomic units (OTUs) were produced by binning sequences with identical Silva/SINA LCA assignments. This process generated 3,667,872 quality-filtered 16S rRNA gene sequences for 28 samples (median of 130995 sequences per sample). All sequence libraries had a Good’s coverage score >98.5%, indicating excellent depth of sequence coverage.
The Explicet (v2.10.5) (Robertson et al., 2013) and MicrobiomeAnalyst software (Chong, Liu, Zhou, & Xia, 2020; Dhariwal et al., 2017) were used for data display and analysis. Overall differences in beta diversity were assessed by PERMANOVA using the Jensen-Shannon Distance. Differences in alpha diversity were assessed by ANOVA for both Chao1 richness (the number of different bacterial OTUs per sample) and Shannon diversity (number of OTUs and the evenness of their abundance within the sample) indexes. Differences in the relative abundances of individual OTUs between groups were calculated with the linear discriminant analysis (LDA) effect size (LEfSe) algorithm using Benjamini–Hochberg false discovery rate (FDR) adjusted p value cutoff value of 0.1 and the logarithmic LDA score cutoff of 2. A heat map correlating the abundance of bacterial genus-level taxa and ileal and liver gene expression was generated using nonparametric Spearman correlation tests, with significance denoted using raw p values.
Other statistical analysis
Prism 9 statistical analysis software (GraphPad) was used to perform all non-microbiome related statistical analysis. For comparison of two groups, an unpaired two-tailed Student’s t test was used. For comparison of more than two treatment groups, a two-way ANOVA with Tukey’s multiple comparisons statistical test was used. A p value of 0.05 or less was considered significant. Error bars indicate mean ± SEM.
Results
3-day moderate ethanol exposure results in intestinal barrier dysfunction and gastrointestinal inflammation in aged but not young mice
To evaluate whether the gastrointestinal effects of ethanol exposure are more dramatic in aged mice, we gavaged aged and young mice for three consecutive days with an ethanol dose that achieved a BAC of 0.08–0.10 g/dL (mean ethanol concentration [± SEM]; young mice given ethanol 98.56 g/dL ±13.02, aged mice given ethanol 98.13 g/dL ± 5.25) and examined intestinal inflammation and barrier parameters. Vehicle treatment of either young or aged mice had no demonstrable effect on intestinal barrier leakiness, as measured by evaluating serum levels of orally gavaged FITC-dextran (4Kd) in the serum (Fig. 1A). Similarly, barrier function remained intact in young mice given ethanol for three consecutive days. However, in aged mice, this ethanol exposure regimen led to a 2-fold increase (p = 0.005) in FITC-dextran levels in the serum compared to young vehicle-treated animals, demonstrating that 3 days of sequential ethanol exposure can result in impaired barrier function in aged mice.
Fig. 1. Impaired intestinal barrier function and increased intestinal and hepatic inflammation in aged, but not young, mice exposed to short-term ethanol.
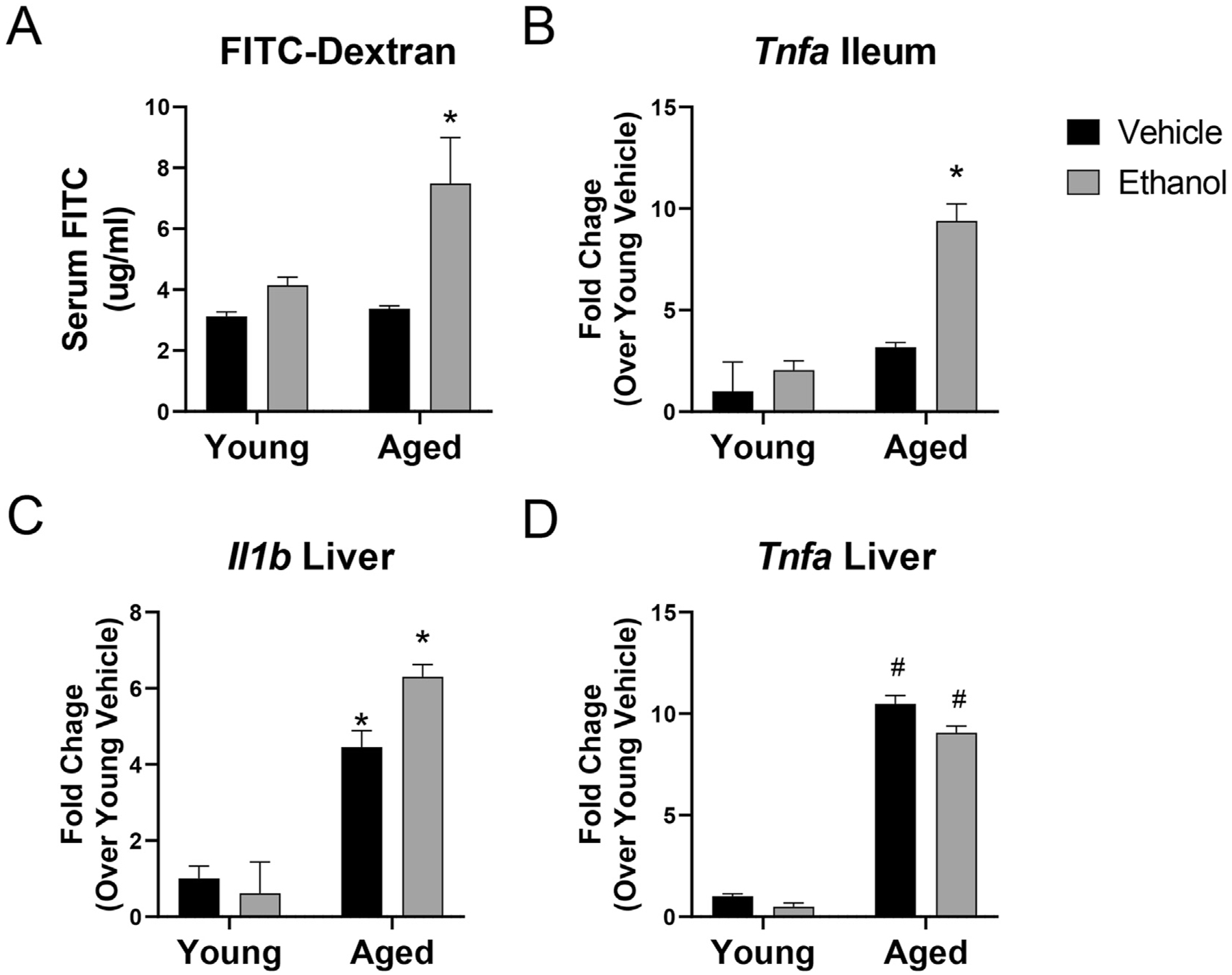
(A) Concentration of 4 kDa FITC-dextran in serum of young and aged mice given vehicle or ethanol. Data are shown as mean values ± SEM. n = 4–8 mice per group. (B–D) Quantitative RT-PCR pro-inflammatory gene expression in (B) whole ileum or (C & D) liver tissue. GAPDH was used as the endogenous control. Data are presented as mean fold change ±SEM relative to young vehicle group. n = 4–6 mice per group. *p < 0.05 from all other groups, #p < 0.05 from young groups by two-way ANOVA with Tukey’s multiple comparisons test.
The pro-inflammatory cytokine TNFα is a well-known mediator of inflammation in the gut and has been shown to impair barrier function (Mashukova, Wald, & Salas, 2011). Next, we evaluated transcriptional levels of Tnfa in the ileum of the mice from the four treatment groups. Intestinal Tnfa expression was unchanged in young mice given ethanol and in aged mice given vehicle, but there was a 10-fold increase in expression of this cytokine in the ileum of aged mice given ethanol (p < 0.001, compared to young vehicle-treated mice) (Fig. 1B). Breakdown of the intestinal barrier can also alter pro-inflammatory cytokine expression in the liver, as a result of increased passage of bacteria and bacterial products from the intestine to the liver via the gut-liver axis (Tripathi et al., 2018). Our group has previously shown that longer-term (4 week) exposure to ethanol leads to exacerbated hepatic inflammation in aged mice compared to young mice (McMahan et al., 2021). Therefore, we evaluated inflammatory gene expression in the livers of the young and aged mice in this shorter-term ethanol model. Interestingly, we saw increased expression of the pro-inflammatory cytokines Il1b and Tnfa in the livers of all aged mice, regardless of ethanol exposure, which is indicative of the baseline increase in levels of inflammation seen in “inflammaging” (Fig. 1C and D) (Franceschi et al., 2000). However, 3 days of ethanol led to an additional increase in Il1b expression in the liver of aged mice, suggesting it is further contributing to liver inflammation in this age group (Fig. 1C). We did not observe any significant changes in alanine transaminase (ALT), aspartate transaminase (AST), or liver triglycerides. Taken together, these results support the hypothesis that 3-day moderate ethanol consumption in age leads to a breakdown of the integrity of the intestinal epithelial barrier and induces increased expression of inflammatory markers.
Analysis of the fecal microbiome following ethanol exposure in young and aged mice
The maintenance of the intestinal barrier has been shown to be influenced by the intestinal microbiome in a number of murine disease models (reviewed in Ulluwishewa et al., 2011). Therefore, we performed microbiome bacterial profiling using 16S rRNA gene sequencing of fecal pellets from young and aged mice before and after 3-day ethanol exposure. Sequencing revealed differences in the fecal microbiota of aged mice both with and without ethanol exposure, compared to their younger counterparts (Fig. 2). Pairwise comparison of community beta diversity, based on Jensen-Shannon Distance, revealed a significant difference in microbial communities (p < 0.001 by PERMANOVA) in aged mice compared to young mice at baseline (Fig. 2A). Furthermore, differences in overall bacterial communities were observed between young ethanol-exposed and aged ethanol-exposed mice (p =0.05). Measurement of two common measures of alpha diversity revealed that neither age nor ethanol exposure influenced species richness (i.e., the number of different taxa within the sample; Fig. 2B). However, the Shannon diversity index, which takes into account both the number of different taxa along with their relative abundances, differed across the four groups (Fig. 2C, p = 0.009). Individual pairwise analysis revealed a significant difference between young and aged mice pre-treatment (p = 0.05) and young ethanol-treated mice compared to aged mice at baseline (p = 0.02).
Fig. 2. Fecal microbiome comparisons between young and aged mice.
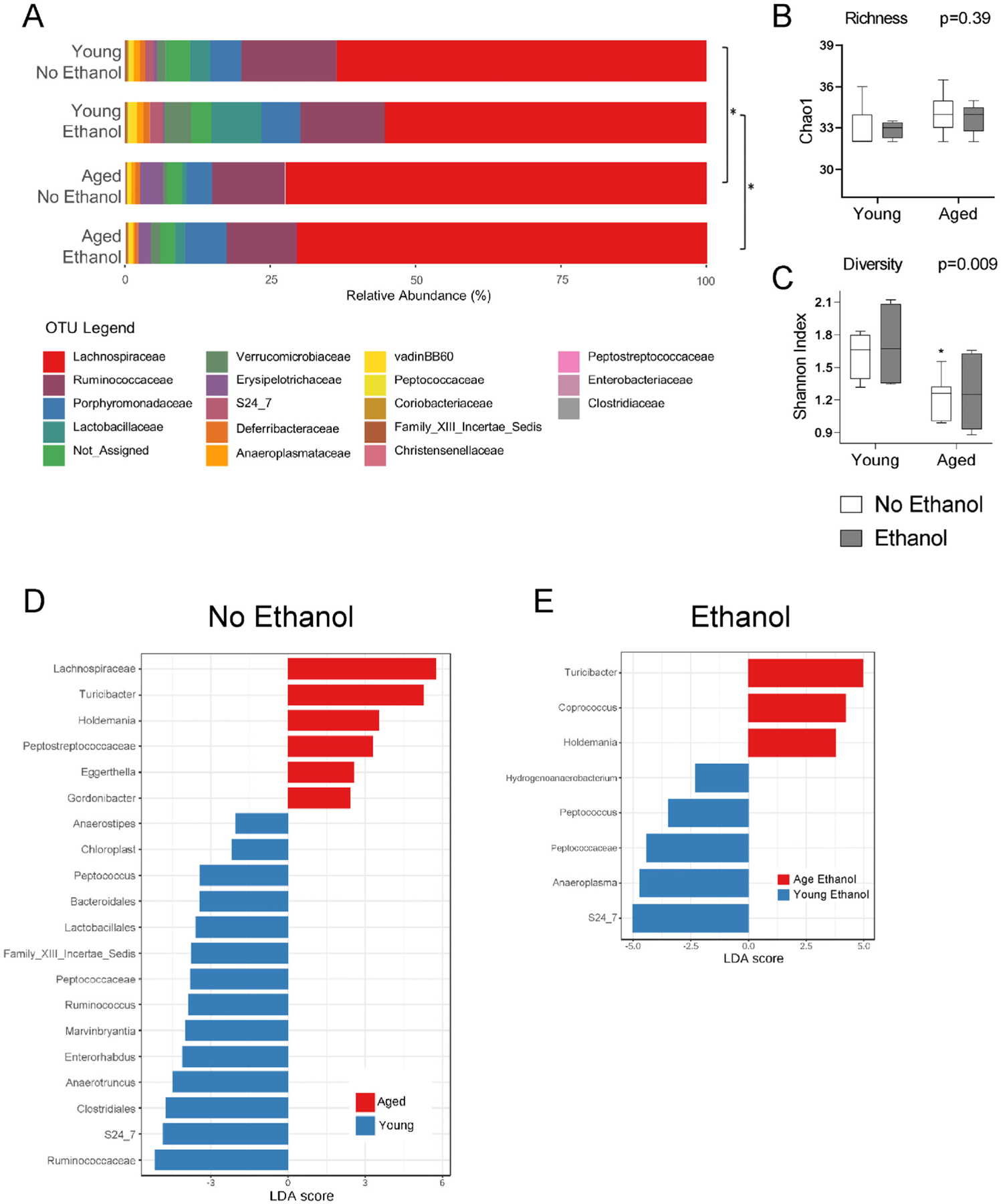
(A) Bar chart showing fecal microbiota family composition across young and aged mice with and without ethanol. Bars represent relative abundances (%). Pairwise analysis of bacterial taxa changes between treatment groups was performed and * = p < 0.05 by PERMANOVA test. (B & C) Measures of alpha biodiversity across all four treatment groups: (B) richness (Chao1) and (C) diversity (Shannon). *p < 0.05 compared to young groups by ANOVA with post hoc Tukey’s test. (D & E) Differences in the relative abundances of individual taxa between groups. The bar charts list the differentially abundant taxa based on effect size (LDA score [log10] > 2) with an FDR-adjusted p value cutoff of 0.1. (D) Enriched taxa in young (negative LDA score) and enriched taxa in aged (positive LDA score). (E) Enriched taxa in young ethanol (negative LDA score) and enriched taxa in aged ethanol (positive LDA score). n = 4–11 mice per group.
Next, we analyzed differences in individual taxa between each treatment group using the LDA effect size (LEfSe) algorithm with a false discovery rate (FDR) adjusted p-value cutoff value of 0.1 and the logarithmic LDA score cutoff of 2. Comparison of young and aged mice before ethanol treatment revealed 14 taxa enriched in young mice including beneficial bacteria such as Lactobacillales and Roseburia (Fig. 2D). Of the six taxa that were enriched in aged mice, two were members of the Actintobacteria phylum, Eggerthella and Gordonibacter, and three were members of the Firmicutes phylum, Holdemania, Turicibacter, and Peptostreptococcaceae (Fig. 2D). Comparison of microbial taxa between young ethanol-treated and aged-ethanol treated mice showed five taxa enriched in young ethanol-exposed mice and three taxa enriched in aged ethanol-exposed mice (Fig. 2E). There were no significant differences in individual taxa in young or aged mice before and after ethanol exposure. Taken together, the major differences in bacteria between groups could be attributed to age-specific changes in the microbiome.
Impaired intestinal antimicrobial peptide (AMP) responses in aged mice
While age alone led to bacterial changes that could contribute to increased inflammation and barrier dysfunction in the gut, we did not observe an increase in any of these parameters in aged mice in the absence of ethanol exposure. In addition, we detected minimal changes in the microbiome following 3 days of moderate ethanol exposure in both young and aged mice. Therefore, to further explore explanations for the increased intestinal permeability induced by ethanol in aged mice, we examined expression of AMPs in the intestines of young and aged mice after ethanol or vehicle exposure. The intestinal barrier is maintained, in part, by expression of AMPs, including the α-defensin Alpha-defensin-related sequence 1 precursor (Defa-rs1), and the C-type lectins Reg3γ and Reg3β. Interestingly, expression of Defars1 and Reg3g was significantly downregulated in aged mice in response to ethanol exposure (p = 0.04 and p=0.003, compared to aged vehicle mice) (Fig. 3). Expression of Reg3b was also decreased, although this did not reach significance (p = 0.07). In contrast, ethanol induced an increase in AMP expression in young mice with a 7-fold (p = 0.0003) and 3.5-fold (p = 0.0004) increase in ileal expression of Defars1 and Reg3g when compared to young mice given vehicle. There also was a baseline upregulation of AMPs with age, although only Reg3g was significantly increased compared to the young vehicle group (p = 0.004). Transcriptional analysis of levels of other AMPS, including lysozyme C-1 and C-2 (Lyz1, Lyz2) and α-defensins 1 and 2 (Defcr1, Defcr2), were unchanged in all treatment groups.
Fig. 3. Decreased expression of ileal AMPs in aged, but not young, mice in response to short-term ethanol exposure.
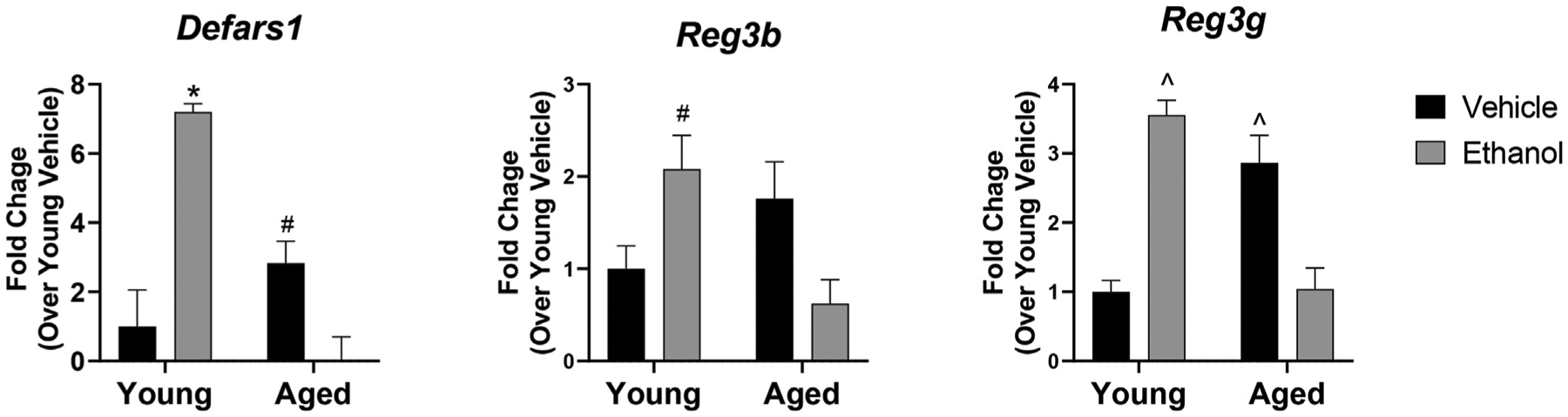
Quantitative RT-PCR of expression of the indicated AMP in whole ileum tissue. GAPDH was used as the endogenous control. n = 4–6 per group *p < 0.05 compared to all other groups, #p < 0.05 compared to age ethanol, ^p < 0.05 compared to young-vehicle mice and age-ethanol mice with Tukey’s multiple comparisons test. Data are presented as mean fold change ±SEM relative to young vehicle group.
As AMPs are important for maintaining a chemical barrier to prevent intestinal microbes from gaining access to the intestinal epithelial cells, the noted downregulation could contribute to increased inflammation in the ileum of aged mice following ethanol exposure. In addition, decreased AMP expression can also contribute to hepatic inflammation via the gut-liver axis. This led us to explore the relationship between AMPs, pro-inflammatory cytokine expression, and specific bacterial taxa in our model. The expression of all three AMPs, along with pro-inflammatory cytokine genes in the ileum and liver, was correlated with individual bacterial taxa across all four groups and visualized with a heat map (Fig. 4). Overall, 14 taxa were significantly correlated with expression of at least one of the intestinal AMPs (p < 0.05). Of the five taxa that correlated with downregulation of AMP genes, four were shown to be increased in relative abundance in aged mice with vehicle and ethanol exposure (Fig. 2D and E). Inversely, of the nine taxa that aligned with increased AMP expression, five were decreased in relative abundance in the aged mice regardless of ethanol exposure. Furthermore, bacterial taxa that correlated with decreased AMP expression were associated with increased pro-inflammatory cytokine expression, while those that were associated with increased AMP expression were significantly correlated with decreased expression of pro-inflammatory cytokines. Collectively, these observations reveal for the first time that there are age-specific ethanol-induced changes in AMP and pro-inflammatory gene expression, and these expression patterns correlate with distinct patterns of gut microbiota.
Fig. 4. Correlation of bacterial relative abundance with ileal and hepatic gene expression.
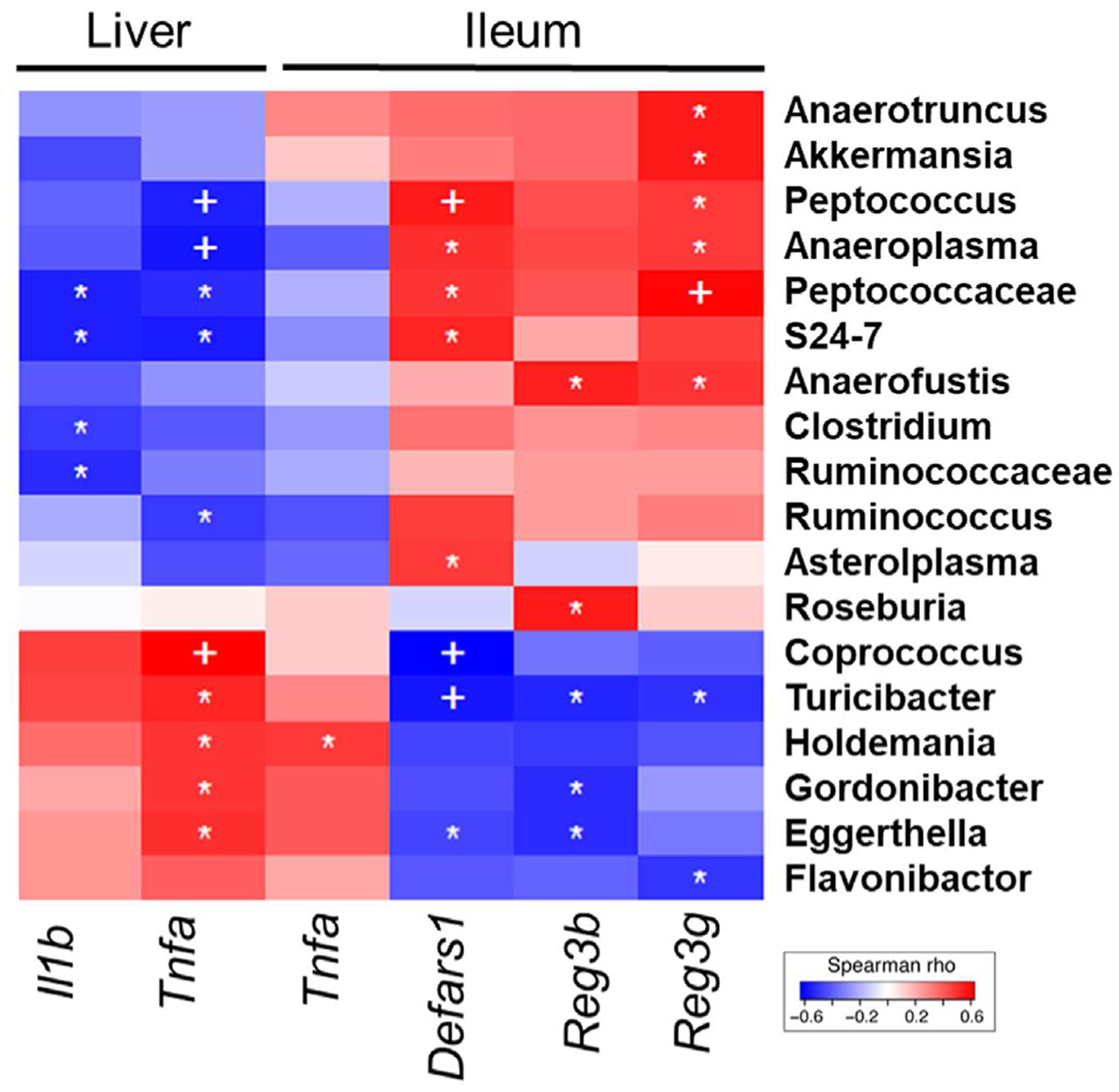
Heat map comparing relative abundance of bacterial taxa to gene expression in the ileum and liver across all treatment groups. Correlations were assessed by the non-parametric Spearman rank correlation test. Blue = negative correlation, red = positive correlation. n = 4–6 per group, +p < 0.01; *p < 0.05.
Discussion
As people are living longer, there is an increase in individuals of advanced age that participate in drinking, and understanding the disparate effects of alcohol in this age group is critical. A number of the detrimental effects of alcohol can be contributed to its effects on the gut, including a breakdown of the intestinal barrier and leakage of bacterial products out of the lumen and into other organs. In this study, we found that, when compared to young mice, a 3-day moderate exposure of ethanol to aged mice led to a significant breakdown of the intestinal barrier, showing that aged mice are sensitized to the effects of ethanol on the gut (Fig. 1). We also observed an upregulation of the gene encoding the cytokine TNFα in the ileum of aged, but not young mice, after ethanol gavage. TNFα is a pro-inflammatory cytokine that has been shown to directly induce a breakdown of the intestinal barrier (Ye, Ma, & Ma, 2006). Furthermore, blockade of this cytokine results in an improvement of barrier function in multiple inflammatory disorders, including Crohn’s disease (Suenaert et al., 2002).
There is a collection of literature suggesting that advanced age gives rise to changes in the intestinal environment, including microbial dysbiosis (Claesson et al., 2012; Langille et al., 2014), which could contribute to increased sensitivity to the effect of alcohol in patients over 65. Analysis of the fecal microbial populations in our model revealed an age-specific upregulation of a number of taxa, including multiple members of the Actinaobacteria and Firmicutes phyla (Fig. 2). Eggerthella, a member of the Actintobacteria phylum, which was increased in aged mice, can directly induce inflammatory responses in the gut (Alexander et al., 2022), and has been linked to a number of gastrointestinal disorders such as gastroenteritis, ulcerative colitis, and Crohn’s disease (Gardiner et al., 2015). We also observed higher relative abundance of the bacterial genus Holdemania in both aged groups when compared to their younger counterparts. This member of the Firmicutes phylum has been found to be enriched in the microbiome of heavy alcohol consumers (Bjørkhaug et al., 2019) and is decreased in response to alcohol withdrawal (Leclercq et al., 2014), suggesting it could play a mechanistic role in the gastrointestinal response to alcohol. Importantly, all the bacterial taxa that correlated with increased intestinal and hepatic inflammatory gene expression (Fig. 4) were enriched in the feces of aged mice (Fig. 2D and E).
Reciprocally, we found a reduction of beneficial bacteria in both aged mice compared to young mice and aged mice given ethanol compared to young mice given ethanol. These included multiple members of the Ruminococcaceae family (Ruminococcus, Anaerotruncus, Hydrogenoanaerobacterium) and the Lachnospiraceae family (Roseburia, Anaerostipes, Marvinbryantia). These taxa have beneficial effects on the intestinal environment, in part by production of SCFA, which has numerous beneficial effects in the gut, including maintenance of the intestinal barrier and regulation of inflammation. Although we did not observe any significant alterations in fecal SCFA in any of our treatment groups (data not shown), it should be noted that fecal levels of SCFA do not necessarily reflect the intestinal environment, as other confounding factors such as SFCA uptake can alter excreted levels (de la Cuesta-Zuluaga et al., 2018). However, this anti-inflammatory phenotype of microbial population in young mice is supported by the observed correlation of these taxa with decreased inflammatory gene expression (Fig. 4).
When comparing animals before and after ethanol exposure, there were no significantly altered taxa in either young or aged mice, suggesting that 3-day ethanol exposure does not appear to induce broad changes in the fecal microbiome. Instead, at baseline, the aged mice exhibit a pattern of bacterial dysbiosis that would predispose this age group to detrimental inflammatory consequences if other barrier functions were to be disrupted by ethanol. We did not observe increased barrier dysfunction or ileal inflammatory gene expression in aged mice in the absence of ethanol, indicating that microbial dysbiosis alone is not sufficient to induce the intestinal barrier disruption. Instead, ethanol is required to shift the intestinal environment toward a breakdown of barrier function. Future studies aimed at manipulation of the microbiome in aged mice, either through treatment with antibiotics or microbiome transfer from young mice, are warranted to further determine whether age-specific changes in the microbiome are drivers of the age-related exacerbation in the response to ethanol.
AMPs in the mucosal layer of the lumen are an important means of not only directly shaping microbial communities, via their ability to disrupt bacterial membranes, but also influencing how the microbiome interacts with luminal cells of the intestine (Mukherjee & Hooper, 2015). For example, Reg3γ is concentrated in the mucosal layer of the gut and is critical for maintaining a physical separation between the intestinal microbiome and host tissues, preventing induction of an inflammatory response (Loonen et al., 2014; Vaishnava et al., 2011). In young mice, ethanol exposure results in increased AMP expression (Fig. 3), suggesting that moderate ethanol levels lead to upregulation of barrier protective factors in young mice. Inversely, we noted a consistent downregulation of AMPs after ethanol exposure in the aged mice (Fig. 3). This downregulation would allow for increased exposure of the epithelial layer to bacterial components within the lumen in aged mice. The fact that AMP expression was inversely correlated with inflammatory gene expression in the ileum and liver (Fig. 4) would support this hypothesis. Interestingly, a baseline increase in AMP expression was seen in aged mice in the absence of ethanol, which could indicate a possible compensatory mechanism to prevent age-related changes in the microbiome from inducing an inflammatory response. The mechanisms by which ethanol induces a downregulation of AMPs in the aged gut remain to be determined. Both Reg3b and Reg3g are induced by intestinal immune cell production of IL-22 (Zheng et al., 2008), and it is possible that age-related defects in this immune pathway could contribute to the impaired response. Future studies investigating the effects of ethanol in aged mice overexpressing Reg3γ in the intestine (Wang et al., 2016) are underway to determine whether increased AMPs can protect aged mice from ethanol-induced barrier dysfunction.
Overall, this study demonstrates for the first time that 3 days of moderate ethanol exposure results in an age-specific decline in intestinal function and increased gastrointestinal inflammation. We also report the novel finding that these changes correlate with levels of specific bacterial taxa, providing insight into how age-related changes in the microbiome could contribute to intestinal dysfunction with age. Finally, the observed impaired antimicrobial response to ethanol within the aged gut identifies potential targets for future mechanistic studies in the context of aging and alcohol.
Funding
The work herein was supported in part by the National Institutes of Health R01 AG018859 (E.J.K.), R35 GM131831 (E.J.K.), R21 AA026295 (E.J.K.), F31AA027687 (H.J.H.), and the Veterans Administration 1 I01 BX004335 (E.J.K.).
References
- Alexander M, Ang QY, Nayak RR, Bustion AE, Sandy M, Zhang B, et al. (2022). Human gut bacterial metabolism drives Th17 activation and colitis. Cell Host & Microbe, 30(1), 17–30.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørkhaug ST, Aanes H, Neupane SP, Bramness JG, Malvik S, Henriksen C, et al. (2019). Characterization of gut microbiota composition and functions in patients with chronic alcohol overconsumption. Gut Microbes, 10(6), 663–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow RA, Castle IP, Chen CM, & Graubard BI (2017). Trends in alcohol consumption among older Americans: National health interview surveys, 1997 to 2014. Alcoholism: Clinical and Experimental Research, 41(5), 976–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull-Otterson L, Feng W, Kirpich I, Wang Y, Qin X, Liu Y, et al. (2013). Meta-genomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment. PLoS One, 8(1), Article e53028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Liu P, Zhou G, & Xia J (2020). Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nature Protocols, 15(3), 799–821. [DOI] [PubMed] [Google Scholar]
- Chou CH, Lai SL, Ho CM, Lin WH, Chen CN, Lee PH, et al. (2015). Lysophosphatidic acid alters the expression profiles of angiogenic factors, cytokines, and chemokines in mouse liver sinusoidal endothelial cells. PLoS One, 10(3), Article e0122060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Cusack S, O’Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, et al. (2011). Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proceedings of the National Academy of Sciences of the United States of America, 108(Suppl 1), 4586–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, et al. (2012). Gut microbiota composition correlates with diet and health in the elderly. Nature, 488(7410), 178–184. [DOI] [PubMed] [Google Scholar]
- de la Cuesta-Zuluaga J, Mueller NT, Álvarez-Quintero R, Velásquez-Mejía EP, Sierra JA, Corrales-Agudelo V, et al. (2018). Higher fecal short-chain fatty acid levels are associated with gut microbiome dysbiosis, obesity, hypertension and cardiometabolic disease risk factors. Nutrients, 11(1), 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhariwal A, Chong J, Habib S, King IL, Agellon LB, & Xia J (2017). MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Research, 45(W1), W180–W188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, & Knight R (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 27(16), 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, & Green P (1998). Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Research, 8(3), 186–194. [PubMed] [Google Scholar]
- Flurkey K, Currer JM, & Harrison DE (2007). Chapter 20 – mouse models in aging research. In Fox JG, Barthold SW, Davisson MT, Newcomer CE, Quimby FW, & Smith AL (Eds.), The mouse in biomedical research (2nd ed., pp. 637–672). Burlington: Academic Press. [Google Scholar]
- Franceschi C, Bonafè M, Valensin S, Olivieri F, De Luca M, Ottaviani E, et al. (2000). Inflamm-aging. An evolutionary perspective on immunosenescence. Annals of the New York Academy of Sciences, 908, 244–254. [DOI] [PubMed] [Google Scholar]
- Frank DN, Giese APJ, Hafren L, Bootpetch TC, Yarza TKL, Steritz MJ, et al. (2021). Otitis media susceptibility and shifts in the head and neck microbiome due to SPINK5 variants. Journal of Medical Genetics, 58(7), 442–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner BJ, Tai AY, Kotsanas D, Francis MJ, Roberts SA, Ballard SA, et al. (2015). Clinical and microbiological characteristics of Eggerthella lenta bacteremia. Journal of Clinical Microbiology, 53(2), 626–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall KE, Proctor DD, Fisher L, & Rose S (2005). American gastroenterological association future trends committee report: Effects of aging of the population on gastroenterology practice, education, and research. Gastroenterology, 129(4), 1305–1338. [DOI] [PubMed] [Google Scholar]
- Hartmann P, Seebauer CT, & Schnabl B (2015). Alcoholic liver disease: The gut microbiome and liver cross talk. Alcoholism: Clinical and Experimental Research, 39(5), 763–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, et al. (2015). Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host & Microbe, 17(5), 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MG, Meehan CJ, Koenig JE, Dhanani AS, Rose RA, Howlett SE, et al. (2014). Microbial shifts in the aging mouse gut. Microbiome, 2(1), 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S, Matamoros S, Cani PD, Neyrinck AM, Jamar F, Stärkel P, et al. (2014). Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proceedings of the National Academy of Sciences of the United States of America, 111(42), E4485–E4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, & Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 25(4), 402–408. [DOI] [PubMed] [Google Scholar]
- Loonen LM, Stolte EH, Jaklofsky MT, Meijerink M, Dekker J, van Baarlen P, et al. (2014). REG3γ-deficient mice have altered mucus distribution and increased mucosal inflammatory responses to the microbiota and enteric pathogens in the ileum. Mucosal Immunology, 7(4), 939–947. [DOI] [PubMed] [Google Scholar]
- Manuel CA, Johnson LK, Pugazhenthi U, Fong DL, Fink M, Habenicht LM, et al. (2022). Effect of antimicrobial prophylaxis on corynebacterium bovis infection and the skin microbiome of immunodeficient mice. Comparative Medicine, 72(2), 78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashukova A, Wald FA, & Salas PJ (2011). Tumor necrosis factor alpha and inflammation disrupt the polarity complex in intestinal epithelial cells by a posttranslational mechanism. Molecular and Cellular Biology, 31(4), 756–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGettigan B, McMahan R, Orlicky D, Burchill M, Danhorn T, Francis P, et al. (2019). Dietary lipids differentially shape nonalcoholic steatohepatitis progression and the transcriptome of kupffer cells and infiltrating macrophages. Hepatology, 70(1), 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan RH, Najarro KM, Mullen JE, Paul MT, Orlicky DJ, Hulsebus HJ, et al. (2021). A novel murine model of multi-day moderate ethanol exposure reveals increased intestinal dysfunction and liver inflammation with age. Immunity & Ageing, 18(1), 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan RH, Wang XX, Cheng LL, Krisko T, Smith M, El Kasmi K, et al. (2013). Bile acid receptor activation modulates hepatic monocyte activity and improves nonalcoholic fatty liver disease. Journal of Biological Chemistry, 288(17), 11761–11770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier P, & Seitz HK (2008). Age, alcohol metabolism and liver disease. Current Opinion in Clinical Nutrition and Metabolic Care, 11(1), 21–26. [DOI] [PubMed] [Google Scholar]
- Mitchell EL, Davis AT, Brass K, Dendinger M, Barner R, Gharaibeh R, et al. (2017). Reduced intestinal motility, mucosal barrier function, and inflammation in aged monkeys. The Journal of Nutrition, Health & Aging, 21(4), 354–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, & Hooper LV (2015). Antimicrobial defense of the intestine. Immunity, 42(1), 28–39. [DOI] [PubMed] [Google Scholar]
- Mutlu EA, Gillevet PM, Rangwala H, Sikaroodi M, Naqvi A, Engen PA, et al. (2012). Colonic microbiome is altered in alcoholism. American Journal of Physiology - Gastrointestinal and Liver Physiology, 302(9), G966–G978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole PW, & Jeffery IB (2015). Gut microbiota and aging. Science, 350(6265), 1214–1215. [DOI] [PubMed] [Google Scholar]
- Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao JZ, et al. (2016). Age-related changes in gut microbiota composition from newborn to centenarian: A cross-sectional study. BMC Microbiology, 16, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, & Glöckner FO (2012). SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics, 28(14), 1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Research, 41(Database issue), D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RK (2008). Acetaldehyde-induced barrier disruption and paracellular permeability in Caco-2 cell monolayer. Methods in Molecular Biology, 447, 171–183. [DOI] [PubMed] [Google Scholar]
- Rendon JL, Li X, Akhtar S, & Choudhry MA (2013). Interleukin-22 modulates gut epithelial and immune barrier functions following acute alcohol exposure and burn injury. Shock, 39(1), 11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson CE, Harris JK, Wagner BD, Granger D, Browne K, Tatem B, et al. (2013). Explicet: Graphical user interface software for metadata-driven management, analysis and visualization of microbiome data. Bioinformatics, 29(23), 3100–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, & Westcott SL (2011). Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Applied and Environmental Microbiology, 77(10), 3219–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki E, De Minicis S, Osterreicher CH, Kluwe J, Osawa Y, Brenner DA, et al. (2007). TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nature Medicine, 13(11), 1324–1332. [DOI] [PubMed] [Google Scholar]
- Substance Abuse and Mental Health Services Administration. (2019). National survey of drug use and Health (NSDUH) releases. Retrieved from: https://www.samhsa.gov/data/release/2019-national-survey-drug-use-and-health-nsduh-releases. [PubMed]
- Suenaert P, Bulteel V, Lemmens L, Noman M, Geypens B, Van Assche G, et al. (2002). Anti-tumor necrosis factor treatment restores the gut barrier in Crohn’s disease. American Journal of Gastroenterology, 97(8), 2000–2004. [DOI] [PubMed] [Google Scholar]
- Sullivan EV, Zahr NM, Sassoon SA, Thompson WK, Kwon D, Pohl KM, et al. (2018). The role of aging, drug dependence, and hepatitis C comorbidity in alcoholism cortical compromise. JAMA Psychiatry, 75(5), 474–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, et al. (2018). The gut-liver axis and the intersection with the microbiome. Nature Reviews Gastroenterology & Hepatology, 15(7), 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulluwishewa D, Anderson RC, McNabb WC, Moughan PJ, Wells JM, & Roy NC (2011). Regulation of tight junction permeability by intestinal bacteria and dietary components. Journal of Nutrition, 141(5), 769–776. [DOI] [PubMed] [Google Scholar]
- United States Census Bureau. (2017). Population estimates. July 1, 2017 (V2017) – Walla Walla City, WA: [Table 2]. Retrieved from: https://www.census.gov/data/tables/2017/demo/popproj/2017-summary-tables.html. [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, et al. (2011). The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science, 334(6053), 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Fouts DE, Stärkel P, Hartmann P, Chen P, Llorente C, et al. (2016). Intestinal REG3 lectins protect against alcoholic steatohepatitis by reducing mucosa-associated microbiota and preventing bacterial translocation. Cell Host & Microbe, 19(2), 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HB, Wang PY, Wang X, Wan YL, & Liu YC (2012). Butyrate enhances intestinal epithelial barrier function via up-regulation of tight junction protein Claudin-1 transcription. Digestive Diseases and Sciences, 57(12), 3126–3135. [DOI] [PubMed] [Google Scholar]
- Wheatley EG, Curtis BJ, Hulsebus HJ, Boe DM, Najarro K, Ir D, et al. (2020). Advanced age impairs intestinal antimicrobial peptide response and worsens fecal microbiome dysbiosis following burn injury in mice. Shock, 53(1), 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan AW, Fouts DE, Brandl J, Stäarkel P, Torralba M, Schott E, et al. (2011). Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology, 53(1), 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye D, Ma I, & Ma TY (2006). Molecular mechanism of tumor necrosis factor-alpha modulation of intestinal epithelial tight junction barrier. American Journal of Physiology. Gastrointestinal and Liver Physiology, 290(3), G496–G504 [DOI] [PubMed] [Google Scholar]
- Zahs A, Bird MD, Ramirez L, Turner JR, Choudhry MA, & Kovacs EJ (2012). Inhibition of long myosin light-chain kinase activation alleviates intestinal damage after binge ethanol exposure and burn injury. American Journal of Physiology - Gastrointestinal and Liver Physiology, 303(6), G705–G712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, et al. (2008). Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nature Medicine, 14(3), 282–289. [DOI] [PubMed] [Google Scholar]


