Abstract
It is currently assumed that the assimilation of sulfur into reduced forms occurs predominantly in the leaves of plants. However, developing seeds have a strong requirement for sulfur amino acids for storage protein synthesis. We have assessed the capacity of developing seeds of narrow-leaf lupin (Lupinus angustifolius) for sulfur assimilation. Cotyledons of developing lupin seeds were able to transfer the sulfur atom from 35S-labeled sulfate into seed proteins in vitro, demonstrating the ability of the developing cotyledons to perform all the steps of sulfur reduction and sulfur amino acid biosynthesis. Oxidized sulfur constituted approximately 30% of the sulfur in mature seeds of lupins grown in the field and almost all of the sulfur detected in phloem exuded from developing pods. The activities of three enzymes of the sulfur amino acid biosynthetic pathway were found in developing cotyledons in quantities theoretically sufficient to account for all of the sulfur amino acids that accumulate in the protein of mature lupin seeds. We conclude that sulfur assimilation by developing cotyledons is likely to be an important source of sulfur amino acids for the synthesis of storage proteins during lupin seed maturation.
The sulfur-containing amino acid Met is essential for animal nutrition, but is present in limiting amounts in many plants used for feed. This is particularly true of legume seeds (Waddell, 1958). Animal diets containing ingredients such as soybeans, peas, or lupins (Lupinus angustifolius) are supplemented with synthetic Met to correct the deficit in sulfur amino acids. There is some variation in the sulfur amino acid contents of different pulse cultivars, but plant breeders have met with only limited success in increasing seed Cys and Met (Schroeder, 1982). As an approach to understanding what limits the sulfur amino acid content of pulse seeds we investigated the capacity of developing lupin seeds for the reduction and assimilation of sulfur.
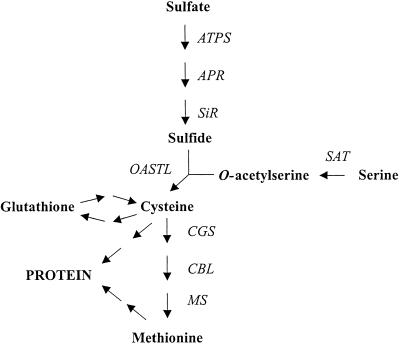
Plants take up sulfur from the soil, mainly in the form of sulfate. After transport into roots via trans-membrane transporters (Smith et al., 1995), sulfate moves into the vasculature and then to the leaves, which are presumed to be the major sites of sulfur assimilation in the plant (Hell, 1997; Leustek and Saito, 1999; Saito, 2000). The reduction of sulfur is thought to occur predominantly in leaf chloroplasts where reducing power from photosynthesis is harnessed to convert sulfate to sulfide. Sulfide is then combined with the amino acid skeleton of O-acetyl-Ser, producing Cys (Fig. 1). A subsequent series of reactions produces Met from Cys and O-phospho-homo-Ser (Ravanel et al., 1998). Although sulfur reduction and Cys biosynthesis are thought to take place mainly in plastids, enzymes of the sulfur metabolic pathway have been found in multiple subcellular compartments (Lunn et al., 1990; Ruffet et al., 1995) and in non-photosynthetic organs (Rolland et al., 1992).
Figure 1.
The pathway of sulfur assimilation in plants. ATPS, ATP sulfurylase; APR, adenosine 5′phosphosulfate reductase; SiR, sulfite reductase; SAT, Ser acetyltransferase; OASTL, O-acetyl-Ser (thiol) lyase; CGS, cystathionine γ-synthetase; CBL, cystathionine β-lyase; MS, Met synthase.
The re-distribution of sulfur within plants under conditions of sulfur nutritional deficiency has been extensively studied (Zhao et al., 1996; Sunarpi and Anderson, 1997; Blake-Kalff et al., 1998). However, the delivery of sulfur to developing seeds, particularly under conditions of adequate sulfur supply, has not been thoroughly investigated. The extent to which sulfur reduction, or sulfur amino acid biosynthesis, occurs in developing seeds in vivo has not been specifically established. Sulfate is abundant in the leaves and roots of plants (Bell et al., 1990; Zhao et al., 1996; Blake-Kalff et al., 1998) and has been shown to be phloem mobile (Rennenberg, 1984). If sulfate were the predominant form of sulfur being transported into the seed during maturation, it would have to undergo assimilation into organic forms in the seed itself. Sulfur is also stored and transported within the plant in organic forms, most notably as the tripeptide, glutathione (Lappartient and Touraine, 1996; Noctor et al., 1998). Glutathione can be metabolized to yield Cys, which can in turn be converted to Met, thereby supplying sulfur amino acids for protein synthesis. S-methyl-Met (SMM) is another abundant sulfur metabolite recently proposed to have an important role in sulfur storage and distribution in plants (Bourgis et al., 1999).
The aims of this study were to determine whether developing lupin seeds are capable of sulfur reduction and assimilation into amino acids, and to estimate the quantitative importance of sulfur assimilation in developing cotyledons to the accumulation of the stored, organic sulfur in the mature lupin seed. We demonstrate here that the sulfur arriving in developing lupin pods, via the phloem, is predominantly in an oxidized, rather than a reduced form. Developing cotyledons were able to incorporate sulfur atoms from sulfate into protein, demonstrating their capacity for sulfur reduction and sulfur amino acid biosynthesis in vitro. The activities of three enzymes of the sulfur amino acid biosynthetic pathway were found in developing cotyledons in quantities theoretically sufficient to account for all of the sulfur amino acids that accumulate in the protein of mature lupin seeds. We postulate that sulfur assimilation by developing cotyledons themselves is an important source of sulfur amino acids for the synthesis of lupin seed storage proteins.
RESULTS
Quantification of Sulfur in Organs of Lupin
X-ray fluorescence spectrometry (XRFS) was used to quantify oxidized and reduced sulfur in lupin seeds. XRFS is a fast and reliable technique for determining the content of sulfur in plant material. For S, the Kα transition shows dependence on chemical bonding of the atom, particularly on the oxidation number. Thus, there is a small SKα line shift between the valences of the two main forms of sulfur present in plant material (sulfate, oxidation no. +6, and sulfur amino acids, oxidation number < 0). Because the difference is small, there is considerable overlap of the peaks for oxidized and reduced S; however, they can be reliably quantified with a conventional XRFS using a germanium crystal and calibration with oxidized and reduced S standards. It has been demonstrated that the oxidized and reduced S measured in this way correspond mainly to sulfate and total sulfur amino acids, respectively, in plant material (Pinkerton et al., 1989).
Mature seeds from lupins grown in pots of soil with a gypsum supplement contained 116 μmol of atomic sulfur per gram of dry weight. Over 40% of this sulfur was in an oxidized form (Table I). Similar results were obtained for seed from lupins grown in sand and watered with nutrient solution containing 3 mm MgSO4. Sulfur was also measured in seed from lupins grown in the field over two seasons. The sulfur content of the field-grown seed was lower than that of the greenhouse-grown lupins, but oxidized sulfur still contributed a substantial proportion (over 25%) of the total (Table I). The reduced sulfur contents were similar in the lupin seeds grown in all three conditions.
Table I.
Sulfur in mature seed of soil-grown or nutrient-fed lupins
| Lupin Growth Conditions | Sulfur in Mature Seeds
|
||
|---|---|---|---|
| Reduced | Oxidized | Total | |
| μmol g−1 dry wt | |||
| Soil with sulfur supplement | 70.2 ± 1.4 | 48.6 ± 0.8 | 116.0 ± 1.2 |
| Sand plus nutrient | 73.3 ± 1.9 | 52.1 ± 2.5 | 123.2 ± 5.1 |
| Field | 74.8 ± 2.0 | 25.3 ± 2.0 | 97.6 ± 1.2 |
Each value is the mean (±sd) of the means of duplicate determinations on flour from pools of 6 g of mature seed from each of two plants, in the case of the lupins grown in soil or sand in the greenhouse. In the case of the field-grown seed, each value is the mean of the means of duplicate determinations on flour from 6-g samples of mature seed grown in the field in two different growing seasons. Total sulfur was determined separately; therefore, the total sulfur figures are not necessarily the perfect sum of the other two figures.
The sulfur contents of other lupin organs were quantified by analyzing freeze-dried samples of leaves, upper lateral stems, pods, and developing seeds of plants grown in sand and watered with 3 mm sulfate. All organs contained significant amounts of oxidized S: in most cases oxidized S constituted 30% to 50% of the total sulfur detected (Table II). In the upper lateral stems, oxidized S was extremely abundant, constituting 1.47% of the organ dry weight. Leaves and pooled, whole, developing seeds (representing a range of stages from early to late maturation) contained relatively high concentrations of reduced sulfur (Table II). In developing seeds that were dissected into cotyledon and testa, the higher concentrations of reduced and oxidized sulfur were found to be in the cotyledon. Testa, pod, and stem all contained relatively low concentrations of reduced sulfur, probably reflecting a lower protein content than the other organs.
Table II.
Sulfur in organs of nutrient-fed lupins
| Tissue | Sulfur
|
||
|---|---|---|---|
| Reduced | Oxidized | Total | |
| μmol g−1 dry wt | |||
| Whole seed | 72.0 | 32.4 | 102.6 |
| :- Cotyledon | 80.1 | 63.0 | 140.6 |
| :- Testa | 20.0 | 11.8 | 31.2 |
| Pod | 10.9 | 11.8 | 22.1 |
| Leaf | 63.0 | 55.2 | 116.0 |
| Stem | 11.2 | 451.5 | 459.0 |
Values shown are the results of single determinations on finely ground, pooled, freeze-dried plant material. For each sample, 10 to 20 g of fresh material was harvested from a single nutrient-fed plant (on the same day as the 40 d after flowering [DAF] seed sample was taken for enzyme analysis) and then freeze dried. Organs from seeds aged between early (18 DAF) and late (35 DAF) maturation were pooled to give the cotyledon, testa, and whole seed determinations. The concentration of sulfur in the cotyledon was higher than in the testa, and therefore, higher than in the whole seed, which included the cotyledon and the testa. Total sulfur was determined separately, therefore, the total sulfur figures are not necessarily the perfect sum of the other two figures.
Quantification of Sulfur in Lupin Phloem Exudates
Sulfur in phloem exudates from developing pods aged between 21 and 35 DAF was quantified by XRFS after drying onto cellulose powder. The concentration of sulfur in the original liquid sample was calculated and was corrected using a calibration curve derived from measuring the sulfur in standard solutions. Samples of the standards were dried onto cellulose, and then quantified using XRFS. The values obtained for the amount of sulfur in the solid sample (micrograms per gram of dry weight) were used to calculate the apparent concentration of sulfur in the original solution. These calculated figures were plotted against the known concentrations of sulfur in the standards. The regression equations were for Na2SO4, y (observed concentration) = 1.11x (known concentration) − 1.63, R2 = 0.99, and n = 2 to 10. For glutathione, y = 1.11x − 0.47, R2 = 0.99, and n = 2 to 4.
The XRFS method slightly under-estimated the concentrations of oxidized and reduced sulfur in the standard solutions. The detection limits were approximately 1.5 mm for oxidized sulfur and 0.5 mm for reduced sulfur. Millimolar concentrations of sulfur were found in the phloem of three cultivars of narrow-leaf lupin. All the sulfur detected was in the oxidized form (Table III). Reduced sulfur was below the detection limit of the XRFS method; therefore, more sensitive methods were used to quantify specific reduced sulfur metabolites in phloem. Glutathione was quantified using the GR-DTNB recycling method and was found to be present at concentrations of approximately 100 μm (Table III) in phloem from all three lupin cultivars. HPLC analysis after derivitization with O-phthaldialdehyde was used to quantify SMM in lupin phloem. The results showed an expected predominance of Asn and Gln, however, SMM was not detectable. SMM was reliably detected by XRFS and by HPLC of O-phthaldyal-dehyde adducts when added to phloem samples. The HPLC method would have detected SMM present at micromolar concentrations in the phloem. Met was present in concentrations similar to those of glutathione (results not shown). HPLC analysis after derivatization with monobromobimane was also used to confirm that significant quantities of other thiols such as Cys or inorganic reduced sulfur compounds were not present in lupin phloem. Glutathione (approximately 100 μm) was the most abundant thiol in the phloem (results not shown).
Table III.
Sulfur concentrations in lupin phloem
| Lupin Cultivar | Sulfur Content of Phloem from Developing Pods
|
||
|---|---|---|---|
| Oxidized Sa | Total reduced Sa | Glutathioneb | |
| mm | μm | ||
| Warrah | 6.0 ± 0.46 | <0.5 | 125 ± 16.3 |
| Ilyarrie | 6.1 ± 0.43 | <0.5 | 96.5 ± 7.8 |
| Merrit | 5.4 ± 0.15 | <0.5 | 90.5 ± 12.0 |
Phloem samples were from developing pods (aged between 18 and 35 DAF) of narrow-leaf lupin cv Warrah, cv Ilyarrie, and cv Merrit grown in soil in the greenhouse.
Quantification using XRFS; all values were corrected according to calibration curves derived from measurement of standards as described in “Materials and Methods.” For cv Warrah, the figures for oxidized S are means ± sd; n = 6 measurements on four independent phloem samples from two independent groups of plants; for cv Ilyarrie and cv Merrit, n = 3 independent measurements on phloem from two independent sets of plants.
Quantification by glutathione reductase-5,5′-dithiobis(2-nitrobenzoic acid) (GR-DTNB) recycling assay; the figures are means ± sd; n = 2 measurements on one phloem sample for each cultivar.
Incorporation of 35S from Sulfate into Seed Proteins in Developing Cotyledons
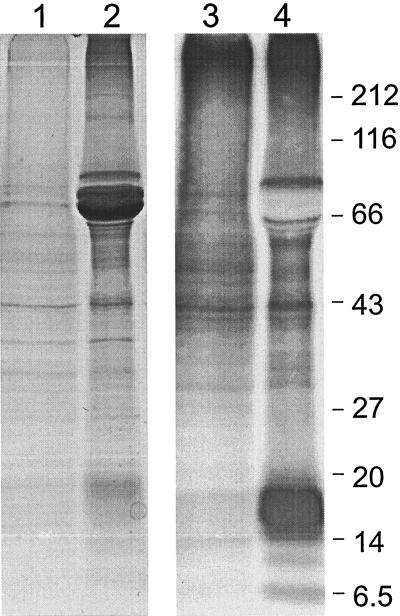
Cotyledons from developing lupin seeds in the early (18 DAF) or late (35 DAF) stages of seed storage protein accumulation were incubated with [35S]sulfate. The incorporation of 35S into proteins was demonstrated by extraction of total protein and analysis by SDS-PAGE and fluorography (Fig. 2). Protein bands of a molecular mass of approximately 14,000 to 20,000 were strongly labeled in the older seeds. These bands may correspond to lupin seed storage proteins that are relatively rich in Cys, for example conglutin δ (Lilley and Inglis, 1986). The most abundant conglutins, α and β, occur as precursors of approximately 70,000 molecular mass (Gayler et al., 1984) in Coomassie Blue-stained protein from developing seeds at 35 DAF.
Figure 2.
Lupin cotyledons transfer the sulfur atom from [35S]sulfate into protein. Cotyledons were dissected from developing narrow-leaf lupin seeds at an early (approximately 18 DAF, tracks 1 and 3) or late (approximately 35 DAF, tracks 2 and 4) stage of maturation. Total protein was visualized by Coomassie Blue staining (tracks 1 and 2) and proteins containing 35S were visualized by fluorography (tracks 3 and 4). Molecular mass marker sizes (×10−3) are shown at right.
Enzymes of the Sulfur Amino Acid Biosynthetic Pathway in Organs of Lupin
The activities of three enzymes of the pathway of sulfur amino acid biosynthesis were assayed in extracts from leaves, pods, and cotyledons or testa from developing seeds of lupins grown in soil. The plant organs were all harvested when the developing seeds were aged approximately 30 DAF, that is, during the phase of rapid accumulation of seed storage proteins. The three enzymes that were assayed were Ser acetyltransferase (SAT), which supplies the amino acid skeleton for Cys biosynthesis; O-acetyl-Ser (thiol) lyase (OASTL), which catalyzes the combination of O-acetyl-Ser and sulfide to form Cys; and cystathionine β-lyase (CBL), one of the three enzymes specific to Met biosynthesis.
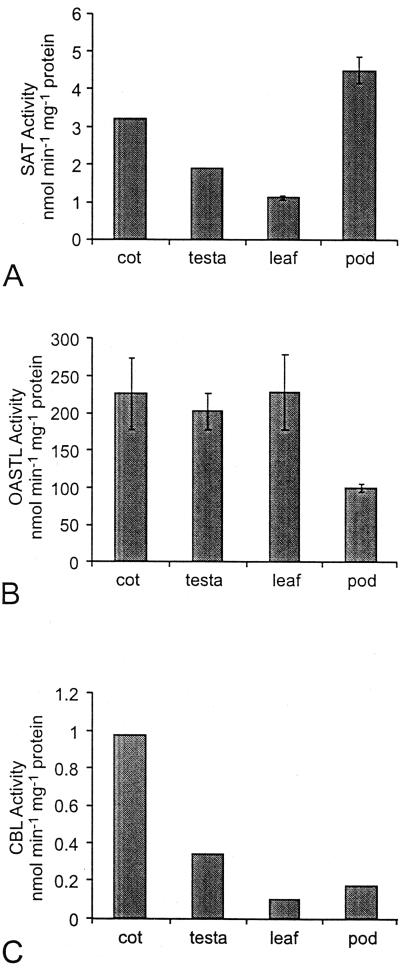
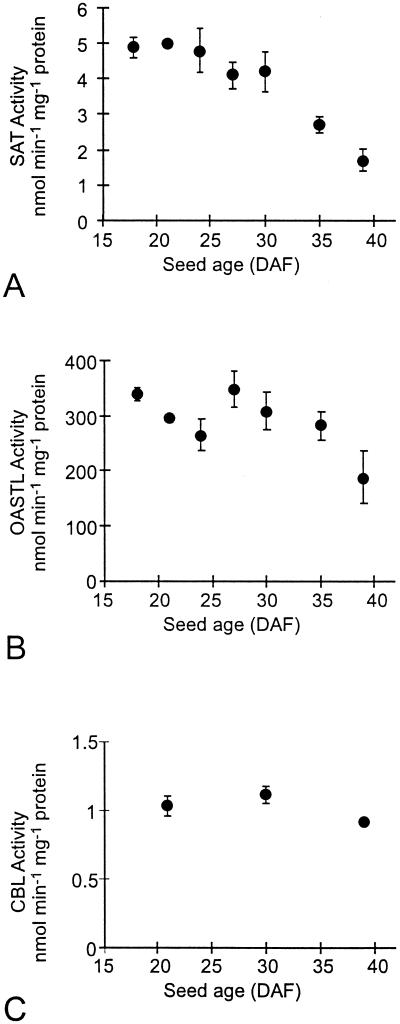
The three enzyme activities were detected in all organs tested. The specific activity of SAT was highest in pod, followed by cotyledon, testa, and fully expanded leaf (Fig. 3A). OASTL-specific activity was similar in cotyledon and testa of developing seed and in leaves, with the lowest specific activity in the pod (Fig. 3B). The specific activity of OASTL in lupin leaves and developing seeds was approximately two orders of magnitude higher than that of SAT. Similar absolute and relative levels of SAT and OASTL have been reported in crude extracts of pea leaf protoplasts (Ruffet et al., 1995). CBL-specific activity was highest in cotyledons of the developing seeds (Fig. 3C). The specific activities of the enzyme were lower and similar in testa of developing seed, leaves, and pods.
Figure 3.
SAT (A), OASTL (B), and CBL (C) in organs of narrow-leaf lupins grown in soil. For SAT and OASTL, means are plotted ±sd, n = 2 to 5 measurements on aliquots of a single extract from pooled samples representing approximately 50 seeds from a total of several plants (see “Materials and Methods”). The sds for SAT in the cotyledon and testa were too small for the error bars to show in the figure. CBL measurements were done only once (on the same pooled sample representing 50 seeds) for the soil-grown lupins; however, these figures are in agreement with the CBL measurements on organs from lupins grown in sand and watered with nutrient (see Fig. 5 and Table IV), demonstrating the reliability of the CBL assay.
Activities of Enzymes of the Sulfur Amino Acid Biosynthetic Pathway throughout Maturation of Lupin Seeds
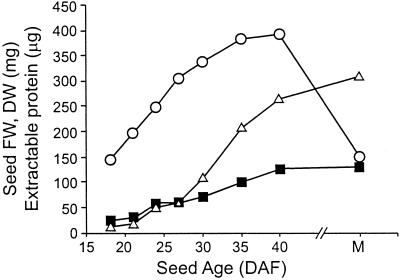
Developing seeds from lupins grown in conditions of defined mineral nutrition with adequate sulfur supply were harvested at a range of stages spanning the phase of storage protein accumulation (Fig. 4). The seed storage conglutins accumulated from 24 DAF onwards and dominated the total seed protein profiles in later stages (data not shown). The total protein content of the developing seeds increased with age, due to the accumulation of storage proteins (Fig. 4).
Figure 4.
Protein accumulation in developing narrow-leaf lupin seeds. Each point represents the mean of approximately 50 seeds pooled from six plants grown in sand watered with defined nutrient: fresh weight (○, milligrams) and dry weight (▪, milligrams). Total extractable protein (▵, micrograms) is the mean of duplicate extractions from a single pooled flour sample (representing approximately 50 seeds) for each seed age. M, Mature, approximately 60 DAF.
The activities of SAT, OASTL, and CBL were assayed in low-salt extracts from the developing seeds. To avoid distortion of specific activity estimates by the accumulation of storage proteins in older seeds we quantified enzyme activities in terms of low-salt-soluble protein that excluded the bulk of the seed storage proteins. Under the extraction and de-salting conditions used, the majority of the seed storage proteins were not soluble; therefore, the protein concentrations of the extracts were similar for all stages of developing seeds (protein concentrations for 18, 21, 24, 27, 30, 35, and 39 or 40 DAF extracts were 7.4, 8.8, 6.4, 7.5, 7.3, 8.6, and 5.8 mg protein mL−1, respectively).
The specific activity of SAT was highest in young seeds, but declined as development proceeded (Fig. 5A). The level of OASTL was fairly uniform throughout seed development, tailing off slightly at 40 DAF (Fig. 5B). The specific activity of CBL was relatively constant in developing seeds at the beginning, middle, and end of storage protein accumulation (Fig. 5C). The activities of the three enzymes in developing seeds were compared with the activities of the same enzymes in other organs from the same plants. The levels of all three enzymes were generally as high or higher in developing seeds at mid-maturation, as in fully expanded leaves or in roots. This was true whether enzyme activity was expressed in terms of low-salt-soluble protein, chlorophyll, dry weight, or fresh weight (Table IV).
Figure 5.
SAT (A), OASTL (B), and CBL (C) in developing seeds of narrow-leaf lupins grown in sand with high sulfur nutrient. Figures are means ± sd, n = 2 to 4 separate determinations on aliquots of a single extract from a pooled sample representing approximately 50 seeds.
Table IV.
Activities of three enzymes of the sulfur assimilation pathway in organs of nutrient-fed narrow-leaf lupin
| Enzyme/Lupin Organ | Enzyme Activity | |||
|---|---|---|---|---|
| nmol min−1 mg−1 proteina | nmol min−1 mg−1 chlorophyll | nmol min−1 g−1 dry wt | nmol min−1 g−1 fresh wt | |
| SAT | ||||
| Leaf | 1.7 ± 0.05 | 11.9 | 155 | 20.2 |
| Seed, 30 DAF | 4.2 ± 0.54 | 290 | 227 | 50 |
| Root | 3.9 ± 0.34 | n.a.b | 99 | 4.3 |
| OASTL | ||||
| Leaf | 320 ± 31.1 | 2,240 | 29,120 | 3,810 |
| Seed, 30 DAF | 309 ± 34.7 | 21,320 | 16,720 | 3,680 |
| Root | 377 ± 47.3 | n.a. | 9,610 | 420 |
| CBL | ||||
| Leaf | 0.18 ± 0 | 1.3 | 16.4 | 2.1 |
| Seed, 30 DAF | 1.1 ± 0.06 | 75.9 | 59.5 | 13.1 |
The same measurements were used to express the activities on the basis of chlorophyll or organ wt sds are quoted only for the original measurements.
Enzyme activities were quantified initially on the basis of low-salt soluble protein (means ± sd, n = 2 to 4 measurements on aliquots of a single extract from pooled samples representing approximately 50 leaves or seeds from a total of several plants. The root sample consisted of approximately 20 g fresh wt from a single nutrient-fed plant; see “Materials and Methods”).
n.a., Not applicable.
Organic Sulfur in Mature Lupin Seeds
The total amino acid content was determined in acid-hydrolyzed samples of mature lupin seed flour (Table V). We aimed to determine whether the observed activities of sulfur pathway enzymes in seeds were large enough to account for a significant proportion of the total Cys and Met accumulated by the mature lupin seed. We calculated the theoretical maximum amount of product that could be produced by each of the three enzymes examined in this study over the period of development during which seed storage proteins accumulated (Table VI). From these calculations it was deduced that, at least judging from maximal enzyme activities in vitro, the amounts of SAT, OASTL, and CBL in the developing lupin seeds were more than sufficient to account for all the sulfur amino acid stored in protein in the mature seeds.
Table V.
Sulfur amino acid content of mature narrow-leaf lupin seeds
| Amino Acid | μmol g−1 dry wt |
|---|---|
| Total Met | 14.2 ± 0.2 |
| Total Cys | 35.1 ± 4.1 |
| Total sulfur amino acids | 49.3 |
Total sulfur amino acids were determined, in duplicate, in flour from pooled seeds of one nutrient-fed lupin plant. An additional single determination was done on flour from pooled seeds from a second nutrient-fed plant. The results are the means ± sd of the single determination and the mean of the duplicate determinations on the other flour sample.
Table VI.
Integration of the maximum theoretical activities of SAT, OASTL, and CBL in lupin seeds throughout seed storage protein accumulation
| Enzyme | Average Enzyme Activity in Whole Seed during d 20 to 40 of Development | Total Maximum Activity during Storage Protein Accumulation |
|---|---|---|
| nmol min−1 mg−1 low-salt-soluble protein | μmol g−1 dry wt | |
| SAT | 3.8 | 1,475 |
| OASTL | 276 | 107,110 |
| CBL | 1.03 | 400 |
Storage protein accumulation occurred during the period between approximately 20 and 40 DAF in narrow-leaf lupin seeds grown in sand watered with defined nutrient (Fig. 4). A mean activity was calculated for each enzyme over this period, and then multiplied by time to give a total maximum enzyme activity per gram of dry wt. It was assumed that each enzyme operated at maximum activity for 12 h a day and that seed flours contained an average of 55 mg of low-salt extractable protein g−1 dry wt. There is 1 m sulfur in each mole of product for OASTL (product is Cys) and CBL (product is homo-Cys). One mole of the product of SAT (O-acetyl-Ser) gives rise, in the subsequent reaction, to 1 m of cysteine. Thus, moles of product calculated in this table can be compared directly with moles of sulfur amino acid in Table V.
DISCUSSION
The pathway of sulfur assimilation is now well characterized in plants (Fig. 1). It is generally accepted that the energy for sulfur reduction comes from coupling with photosynthetic electron transport. On a whole-plant level, it is, therefore, assumed that sulfur reduction and sulfur amino acid biosynthesis occur largely in the leaves (Hell, 1997; Saito 2000). Seeds, however, have a high requirement for amino acids, including Cys and Met, to support storage protein synthesis during maturation. Furthermore, developing seeds of many species contain appreciable levels of chlorophyll and some have been shown to be capable of electron transport and carbon fixation, presumably utilizing light transmitted through the pod wall in vivo (Atkins and Flinn, 1978; King et al., 1998). In this study we examined the capacity of developing narrow-leaf lupin seeds for sulfur reduction and sulfur amino acid biosynthesis.
Seeds accumulate reserves of carbon, nitrogen, and sulfur during maturation. Developing seeds import carbon and nitrogen that was assimilated into reduced forms in other parts of the plant, most notably the leaves. Less is known about the sulfur nutrition of developing seeds. It has been reported that soybean seeds could be cultured in vitro with sulfate as their sole sulfur source (Holowach et al., 1984). The fact that these seeds grew and accumulated storage proteins over periods of several days, albeit at non-physiological rates, can be taken as indirect evidence for the ability of developing seeds to perform sulfur reduction and sulfur amino acid biosynthesis, at least when supplied with sulfate in culture. In a similar manner, it has been reported that sulfur from sulfate appeared in seed storage proteins in a cotyledonary slice labeling system from broad bean (Bailey et al., 1970). Using a complementary approach, Macnicol (1977) reported that labeled, phloem-abundant amino acids were extensively metabolized to other amino acids, including Cys and Met, in isolated, developing cotyledons of pea.
In the present study we adopted a quantitative approach to assessing the importance of sulfur assimilation in the seeds of narrow-leaf lupin in vivo. It was demonstrated that oxidized S represents a significant pool of sulfur in lupin seeds under a range of growing conditions, and that phloem supplying developing lupin pods contained sulfur predominantly in the oxidized form. The phloem stream has been estimated to supply approximately 90% of the nitrogen that accumulates in fruits of white lupin (Pate et al., 1977). It would, therefore, be expected that, if significant sulfur were supplied to the developing fruits of narrow-leaf lupin in the form of sulfur amino acids or other nitrogenous sulfur metabolites such as glutathione or SMM, these reduced forms of sulfur should be detectable in the phloem. Although sulfate is known to be phloem mobile, the importance of glutathione as a means of transporting reduced sulfur to sinks such as roots has been emphasized (Rennenberg, 1984). It has been reported that sulfur in the phloem of canola is predominantly in the form of glutathione (Lappartient and Touraine, 1996), whereas glutathione, S-methyl-Met, and sulfate have all been detected in the phloem of cereals, with sulfate constituting a relatively minor proportion of the total sulfur (Bourgis et al., 1999; Kuzuhara et al., 2000). On the other hand, it was deduced from labeling studies in soybean that sulfate taken up in the xylem was rapidly transferred to the phloem, with glutathione being “quantitatively unimportant” in sulfur distribution to sinks within the plant (Smith and Lang, 1988). Thus, the three major transported forms of sulfur may have different importance in different plants, with sulfate being the most important in narrow-leaf lupin during seed maturation.
We report that developing lupin cotyledons are capable of utilizing the sulfur atom from sulfate in protein synthesis. It can, therefore, be deduced that developing lupin cotyledons are able to perform all the steps of sulfur reduction from sulfate to sulfide, as well as the subsequent steps of sulfur amino acid biosynthesis (see Fig. 1). We directly demonstrated the presence, in developing lupin seeds, of the two enzymes responsible for Cys biosynthesis. SAT catalyzes the initial reaction that activates Ser to produce O-acetyl-Ser, an important intermediate shown to have a key role in the control of the entire pathway of sulfur uptake and assimilation in plants (Saito, 2000). OASTL catalyzes the synthesis of Cys from sulfide and O-acetyl-Ser (Fig. 1). We also measured the activity of CBL as an indicator of the presence in seeds of the transulfuration pathway leading to Met.
We attempted to estimate the quantitative contribution of sulfur assimilation in developing seeds to the accumulated store of organic sulfur in the mature lupin seed. The specific activities of SAT, OASTL, and CBL were generally as high or higher in developing seeds as they were in photosynthetic source leaves from the same plants grown under sulfur-adequate conditions in soil (Fig. 3) or in sand watered with nutrient (Table IV). A similar conclusion was reached when enzyme activities were compared on the basis of chlorophyll content, dry weight, or fresh weight, all of which vary with time in developing seeds. However, the critical question is whether the enzyme activities measured in developing seeds are sufficient to account for the quantity of sulfur amino acids that accumulate during the course of maturation. To answer this question, the total activity of each enzyme was integrated over the period of seed maturation and storage protein accumulation (Table VI). The calculated capacity of the lupin seeds for sulfur amino acid synthesis can be compared with the actual quantities of organic sulfur found in the mature seeds (Table V).
The maximal activities of SAT, OASTL, and CBL measured in developing lupin seeds were theoretically more than sufficient to supply all of the sulfur amino acids that accumulate in storage proteins during maturation. These calculations obviously represent only approximate maxima for these activities since they are derived using maximal rates of enzyme activity measured in protein extracts in vitro. This ignores the in vivo regulation of enzyme activities mediated by endogenous regulators such as products and substrates. Furthermore, we have measured total enzyme activities in crude extracts, thereby pooling the activities of the multiple isoforms of each enzyme that are known to be present in different subcellular compartments.
In the case of SAT, the total enzyme activity found in crude extracts of pea leaf protoplasts was divided between the plastids (10% of total), the cytosol (14%), and the mitochondria (76%, Ruffet et al., 1995). OASTL activity was distributed fairly equally between the cytosol (44% of total) and the chloroplasts (42%) in extracts of spinach leaf protoplasts, with a minor proportion (14%) in the mitochondria (Lunn et al., 1990). It has recently been suggested that this mitochondrial activity may be attributable to β-cyano-Ala synthase (Warrilow and Hawkesford, 1998), although this is the subject of some debate (Berkowitz et al., 2000; Hatzfeld et al., 2000). CBL activity was found exclusively in plastids in Arabidopsis leaves (Ravanel et al., 1998). Assuming similar distributions of SAT, OASTL, and CBL activities between the different subcellular compartments in developing lupin seeds, as in leaves of other plants, we predict that there would be enough of any single isoform to supply sulfur amino acids for storage protein synthesis. It should also be noted that the three enzyme activities measured in extracts from freeze-dried lupin material underestimated the corresponding activities in fresh material by approximately 50% (results not shown). Despite the qualifications associated with in vitro studies, our results indicate that the activities of SAT, OASTL, and CBL in developing seeds have the potential to contribute significantly to the accumulation of sulfur amino acids in lupin seed storage proteins.
Within the lupin seed, higher activities of SAT, OASTL, and CBL were found in the cotyledon, compared with the testa (Fig. 3). The cotyledon is embryonic tissue and is the site of synthesis of the seed storage proteins. At mid-maturation (30 DAF), the testa contributed 40% of the fresh and dry weight of the whole seed, but less than 10% of the SAT or CBL activity, and approximately 14% of the total OASTL activity in the seed. In addition, almost all of the chlorophyll in developing seeds was found in the cotyledon (results not shown), so if photosynthetic electron transport does contribute reducing power for sulfur reduction in the seed, this process would be more likely to be occurring in the cotyledon than in the testa.
At 30 DAF, the pod wall contributed as much of the total weight of the fruit as did the seeds, in plants grown in soil. The average dry weight of each seed was 76.4 mg, whereas the average dry weight of pod, per seed, was 72.2 mg. Despite its relatively large mass, the pod wall contributed only 10% as much CBL activity and 30% as much OASTL activity as the cotyledons, on a per seed basis. The pod contained a relatively high SAT-specific activity (Fig. 3); however, protein concentration in the pod was relatively low (extractable protein = 2.9% of dry weight versus 7.3% of dry weight for the cotyledon), so on a per fruit basis, total SAT activity was actually higher in the cotyledons than in the pod.
Our work does not specifically address the role of the pod in the sulfur nutrition of developing seeds in lupin. In a study focussed on the distribution of sulfur in soybean during seed filling, it was argued that sulfate was assimilated in developing soybean pods and delivered to the expanding seeds as homoglutathione, under sulfur-limited growth conditions (Sunarpi and Anderson, 1997). Our finding that SAT-specific activity is relatively high in developing lupin pods is consistent with this organ having a role in assimilation of sulfur en route to the seeds in narrow-leaf lupin. However, the demonstration of sulfur assimilation in isolated lupin cotyledons and the predominance of the activities of OASTL and CBL, as well as high levels of SAT, in cotyledons seem more consistent with a major role for the embryo itself in sulfur assimilation. This interpretation is in agreement with the recently published finding that developing soybean seeds contain significant quantities of ATP sulfurylase, the first enzyme in the pathway of sulfur reduction (Sexton and Shibles, 1999). In this study the authors concluded that the developing seeds were the dominant sites of ATP sulfurylase activity in the plant during seed filling under field conditions. The soybean pods contained less than 10% as much ATP sulfurylase activity as the developing seeds.
In summary, we have demonstrated that developing lupin seeds contain large stores of oxidized S, that they are capable of utilizing the sulfur from sulfate in protein synthesis, and that they are therefore capable of all the steps of sulfur reduction and sulfur amino acid biosynthesis. Oxidized S was the dominant form of sulfur found in the phloem supplying pods during lupin seed development. Quantification of three enzymes central to the sulfur amino acid biosynthetic pathway indicated that developing lupin cotyledons have the potential to synthesize a significant part of the Cys and Met stored in mature lupin seeds. The accumulation of oxidized S in lupin seeds grown in sulfur-adequate conditions indicates that the rate of import of oxidized S exceeds the rate of reduction of sulfur in the cotyledon. This may be a limitation to the sulfur amino acid content of the seeds. Modification of the pathway of sulfur assimilation in developing seeds may, therefore, be a useful approach to improving the sulfur amino acid content, and hence the nutritional value of pulse seeds. Oxidized S accumulated in mature lupin seeds may act as a reserve for germination; however, it is not essential for successful germination, as demonstrated by the normal germination rate of lupin seeds lacking oxidized S after growth under conditions of sulfur limitation (results not shown). Our findings with narrow-leaf lupin might be expected to apply to other pulses with large, green seeds. However, the sulfur nutrition of cereal grains would probably be different, judging from the predominance of reduced sulfur in the phloem of rice and wheat (Bourgis et al., 1999; Kuzuhara et al., 2000).
MATERIALS AND METHODS
Plant Material
Narrow-leaf lupins (Lupinus angustifolius L. cv Warrah) were grown under several different conditions to generate material for this study.
Soil-Grown Plants
Lupins were grown in soil containing 0.6 g L−1 slow-release fertilizer (“Aboska,” containing 15.2% [w/w] nitrogen, 6.9% [w/w] phosphorus, and 5.2% [w/w] potassium sulfate) in 25-cm pots in a controlled temperature greenhouse at 23°C during the day (12 h) and 18°C during the night. Each pot, containing 9 L of soil, received a supplement of 2 g of solid calcium sulfate (gypsum), which was applied to the surface of the soil when the plants started to flower. The phloem samples and the seeds used for labeling with 35S (see below) were from lupin plants grown in soil as described, except that they did not receive a supplement of calcium sulfate. All these plants are referred to as soil-grown lupins. In addition, an analysis of sulfur content only was performed on mature seeds from lupins grown in the field at Wongan Hills in Western Australia.
Nutrient-Fed Plants
Six lupin (cv Warrah) plants were grown in a controlled-temperature greenhouse at 23°C during the day (12 h) and 18°C during the night, in separate 25-cm pots with a mixture of 50% (v/v) washed river sand and 50% (v/v) perlite, and were watered with defined nutrient solution. The plants were watered for 4 weeks after sowing with solution containing 0.3 mm MgSO4, 4 mm KNO3, 4 mm Ca(NO3)2, 1 mm Na(K) H2PO4, 0.1 mm ferric citrate/EDTA, 37 μm H3BO3, 10 μm MnCl2, 1.5 μm ZnCl2, 0.6 μm CuCl2, and 0.2 μm H2MoO4. The plants were subsequently watered with the same basal nutrient containing 3 mm instead of 0.3 mm MgSO4.
All pots were watered with 300 to 600 mL of nutrient once a day (until liquid started to drain from the pots) for 6 d a week. On the 7th d, plants received 600 mL of deionized water. In addition, during the second 3 months of growth, all pots were flushed with 600 mL of deionized water twice a week.
Sample Preparation
Phloem exudate samples were collected from developing pods aged between 21 and 35 DAF from soil-grown plants using the method of Pate et al. (1974). The tips of developing pods were cut off with a new razor blade, and the resulting droplet of exuded phloem (approximately 30 μL) was harvested within a few minutes using a disposable pipette tip. Phloem from several pods on the upper branches of several plants was pooled to produce samples of between 150 and 600 μL.
Pooled samples of leaves, pods, or developing seeds were collected from each group of soil-grown or nutrient-fed plants at various stages of development. Some of the seeds were dissected into cotyledons and testa. Each sample consisted of approximately 50 individual leaves or seeds representing several plants. Pod material represented approximately 12 to 15 individual pods at the same stage of development as the appropriate seed sample. The leaf samples used for enzyme analysis consisted of 10 to 15 g fresh weight of fully expanded leaves harvested on the same day as the 30 DAF seed samples. Between 5 and 23 g of leaf, stem, root, and developing pods with seeds were collected, for determination of sulfur content, from a single nutrient-fed plant on the same day as the 40 DAF seed sample. Enzyme analysis was also done on the root sample. All samples were frozen in liquid nitrogen and were then freeze-dried. The dried samples were conserved in a sealed container, with silica gel, at 4°C for between 6 and 10 weeks, and then at room temperature for 1 week, before analysis. Seeds were harvested fresh for labeling with [35S]sulfate.
Determination of Sulfur by XRFS
Solid Samples
Mature lupin seed samples weighing approximately 6 g (approximately 40 seeds) were milled to fine flour using a UDY Cyclone mill with a 0.5-mm screen. Other plant tissues were freeze-dried, and then pulverized using a puck mill. Powdered samples were compressed into aluminum planchettes or backed with solid boric acid. Total sulfur, reduced sulfur, and oxidized sulfur were determined using a spectrometer (PW 1404, Philips, Natick, MA) as described by Pinkerton et al. (1989).
Liquid Samples
Liquid samples were analyzed by dripping 150 to 300 μL of liquid onto approximately 350 mg of cellulose powder, drying it in a 60°C oven for 24 h, and then milling it to homogeneity in a puck mill. The concentration of sulfur in the original solution was calculated and compared with values calculated for the sulfur concentrations of standard solutions containing 300 mm Suc (to mimic the composition of phloem) and varying concentrations of sulfate or glutathione. Quantification of reduced sulfur gave the same results whether it was in the form of reduced glutathione (GSH), oxidized glutathione, or Met (results not shown). The solutions used for construction of a calibration curve were as follows: standard 1: 300 mm Suc, 5 mm GSH, and 5 mm Na2SO4; standard 2: 300 mm Suc, 2.5 mm GSH, and 7.5 mm Na2SO4; and standard 3: 300 mm Suc, 7.5 mm GSH, and 2.5 mm Na2SO4. Powdered samples were backed with solid boric acid and compressed in a die. Sulfur was quantified using XRFS as described above.
Determination of Glutathione by the GR-DTNB Recycling Method
Glutathione was measured using the spectrophotometric GR-DTNB recycling assay described by Noctor and Foyer (1998). Each set of assays included glutathione standards and was performed in a microtiter plate; each assay was in a volume of 350 μL. Samples were prepared in a volume of 30 μL of 0.1 m HCl, 1 mm EDTA in the microtiter plate wells. The reaction was started by adding 300 μL of assay buffer containing 120 mm NaH2PO4, pH 7.8, 0.6 mm DTNB, 0.1 mm NADPH, 6 mm EDTA, and 5 μg mL−1 GR. The change in A412 was monitored at 20- or 30-s intervals over a period of 3 to 5 min in a microtiter plate reader (SpectraMAX, Molecular Devices, Sunnyvale, CA). The rates of change in absorbance in the glutathione standards were used to create a standard curve that was used to calculate the glutathione concentrations in the unknowns.
Labeling Lupin Cotyledons with [35S]Sulfate
Developing seeds aged approximately 18 and 35 DAF were harvested from lupins grown in soil. These seed ages corresponded to early and late stages of seed storage protein accumulation, respectively (Gayler et al., 1984). Cotyledons were removed from the seeds within 30 to 60 min of the pods being harvested. The cotyledons from two seeds of each age were incubated, flat surface down, in a humidified Petri dish on a 20-μL droplet of carrier-free Na2[35S]O4 diluted 1:50 in sterile, distilled water, for 4 h at room temperature. After incubation, each group of four cotyledons was washed briefly in approximately 100 mL of water. The cotyledons were blotted dry and then a razor blade was used to slice off the part of the cotyledon that had been in direct contact with the 35S label (slices were approximately 1 mm thick). Total protein was extracted from the pooled slices from each of the four cotyledons by homogenizing them in 500 μL (approximately 4 volumes) of buffer containing 0.5 m NaCl, 1 mm EDTA, and 0.1 m TES [N-Tris(hydroxymethyl)-2-aminoethanesulfonic acid]-NaOH, pH 7.6. The homogenates were centrifuged at 10,000g for 10 min at room temperature. Four samples of 20 μL of each supernatant were spotted onto filters (GFA, Whatman, Clifton, NJ) and were allowed to dry. After drying, two of each set of filters were washed four times in 200 mL of ice-cold 5% (w/v) trichloroacetic acid (TCA). Washed filters were then rinsed briefly in ethanol then acetone, and then dried thoroughly. The 35S on the washed and unwashed filters was measured by scintillation counting using an external standard to correct for quenching, with a liquid scintillation counter (Minaxiβ, Packard Instrument Company, Meriden, CT).
Protein samples were electrophoresed on SDS-polyacrylamide gradient mini-gels (15%–30% [w/v] acrylamide) with equal numbers of TCA-insoluble counts loaded in each track. The gels were fluorographed after soaking in 20% (w/v) naphthalene, 0.5% (w/v) 2,5-diphenyloxazole in dimethylsulfoxide (Gill et al., 1981).
Protein Extraction
Dried samples were ground to a fine powder in liquid nitrogen. For enzyme activity measurements protein was extracted from 250 to 500 mg dry weight of powder into 3.5 mL of a buffer containing 50 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid]-NaOH, pH 7.5, 1 mm EDTA, 5 mm MgCl2, 1 mm dithiothreitol (DTT), and 100 μm pyridoxal phosphate. Each extract was desalted on a Sephadex G25 column (PD-10, Pharmacia, Uppsala), eluted in 50 mm HEPES-NaOH, pH 7.5, 1 mm EDTA, 5 mm MgCl2, and 0.1 mm DTT, and stored in aliquots at −80°C. Enzyme assays were performed on extracts thawed only once. The majority of the seed storage proteins precipitated during de-salting then thawing of the de-salted extracts. Thawed extracts were centrifuged at 10,000g for 10 min before quantification of protein (Bradford, 1976). The concentrations of low-salt-soluble protein in extracts from all stages of developing seed were similar (approximately 8 mg protein mL−1), which was equivalent to approximately 6% of dry weight of the starting material. The amount of low-salt-soluble protein recovered from the other organs varied from 1.5% of dry weight (testa) to 2.9% of dry weight (developing pods), 7.3% of dry weight (cotyledons), and 7.6% to 11% of dry weight for leaf.
Enzyme Assays
SAT activity was assayed in a coupled reaction with excess purified, recombinant OASTL as described by Droux et al. (1998). In addition to 20 μL of lupin extract (containing approximately 160 μg of protein), reactions contained 50 mm Tris-HCl, pH 7.5, 1 mm DTT, 10 mm Ser, 1 mm acetyl-coenzyme A, 2.5 mm Na2S, and 100 μg mL−1 recombinant spinach chloroplast OASTL (purified from Escherichia coli), in a final volume of 100 μL. Reactions were stopped by the addition of 50 μL of 20% (w/v) TCA. The product of this reaction was Cys, which was measured using an acidified ninhydrin reagent (Gaitonde, 1967).
OASTL activity was assayed as described (Droux et al., 1998) in a 100-μL reaction containing 50 mm Tris-HCl, pH 7.5, 1 mm DTT, 10 mm O-acetyl-Ser, 2.5 mm Na2S, and 2 μL (containing approximately 16 μg of protein) of lupin extract. The Cys produced was quantified by the method of Gaitonde (1967).
CBL activity was assayed by measuring the rate of hydrolysis of cystathionine to homo-Cys using HPLC (Droux et al., 1995). Reactions contained 42 mm HEPES-NaOH, pH 8, 0.8 mm DTT, 4.2 mm cystathionine, and 50 μL of lupin extract (approximately 400 μg of protein) in 120 μL of volume per time point. The reaction mix was incubated at 25°C, and at zero time, 10, 20, and 30 min, 120 μL was transferred to a fresh tube containing 5 μL of 100 mm monobromobimane (mBBr, Calbiochem, La Jolla, CA) in acetonitrile. This reaction was incubated at 25°C for a further 5 min before being stopped by addition of 50 μL of 1 m methane sulfonic acid. mBBr-conjugated homo-Cys was separated using HPLC and fluorescence detection (Droux et al., 1995). The homo-Cys was quantified by measuring peak areas using the Kromasystem 2000 software (Bio-Tek Instruments, Winooski, VT), and comparing with mBBr-homo-Cys standards.
Analysis of Amino Acid Composition
The amino acid composition of mature seeds was determined by complete hydrolysis of finely ground flour (with or without a prior oxidation step to convert Met residues to Met sulfone and Cys residues to cysteic acid). Oxidation was performed as follows. Oxidation mixture consisting of 0.5 mL of 30% (w/v) hydrogen peroxide and 4.5 mL of formic acid was prepared and kept for at least 1 h at room temperature in the dark. A sample of lupin flour weighing 100 mg was transferred to a 200-mL glass tube and the tube and the oxidation mixture were chilled in an ice bath. The oxidation mixture was added to the flour sample and was incubated on ice for 4 h, after which excess performic acid was destroyed by the addition of 0.8 g of sodium metabisulphite. After the oxidation step, the sample was hydrolyzed by adding 100 mL of 6 n HCl and boiling in a heating block at 120°C for 24 h. The sample was adjusted to a 200-mL volume, and then a 10-mL aliquot was evaporated to dryness and redissolved in 5 mL of water. The evaporation was repeated four times to eliminate any residual HCl. The sample was finally dissolved in 25 mL of lithium citrate buffer (9.4 g of tri-lithium citrate tetrahydrate, 7.4 g of citric acid in 1 L of 2% [w/v] thiodiglycol, pH 2.2, with HCl). The amino acids were separated and quantified using a autoanalyser (Beckman Instruments, Fullerton, CA) and post-column derivatization with ninhydrin.
ACKNOWLEDGMENTS
We gratefully acknowledge Dr. Francis Pierre and staff (Aventis Animal Nutrition, Commentry, France) for the analysis of total sulfur amino acid composition of lupin flour samples. Sincere thanks to Dr. Charles Hocart for his help with HPLC analysis and to Drs John Lunn, T.J. Higgins, and Don Spencer for helpful comments on the manuscript.
LITERATURE CITED
- Atkins CA, Flinn AM. Carbon dioxide fixation in the carbon economy of developing seeds of Lupinus albus (L.) Plant Physiol. 1978;62:486–490. doi: 10.1104/pp.62.4.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CJ, Cobb A, Boulter D. A cotyledon slice system for the electron autoradiographic study of the synthesis and intracellular transport of the seed storage protein of Vicia faba. Planta. 1970;95:103–118. doi: 10.1007/BF00387243. [DOI] [PubMed] [Google Scholar]
- Bell CI, Cram WJ, Clarkson DT. Turnover of sulfate in leaf vacuoles limits retranslocation under sulfur stress. In: Rennenberg H, Brunold C, Stulen I, DeKok L, editors. Sulfur Nutrition and Sulfur Assimilation in Higher Plants: Fundamental, Environmental and Agricultural Aspects. The Hague, The Netherlands: SPB Academic Publishing; 1990. pp. 163–165. [Google Scholar]
- Berkowitz JR, Wirtz M, Hopkins L, Hawkesford MJ, Hell R. Genomic and functional characterization of the oas gene family encoding O-acetylserine (thiol) lyases, enzymes catalyzing the final step in cysteine biosynthesis in Arabidopsis thaliana. Gene. 2000;253:237–247. doi: 10.1016/s0378-1119(00)00261-4. [DOI] [PubMed] [Google Scholar]
- Blake-Kalff MAA, Harrison KR, Hawkesford MJ, Zhao FJ, McGrath SP. Distribution of sulfur within oilseed rape leaves in response to sulfur deficiency during vegetative growth. Plant Physiol. 1998;118:1337–1344. doi: 10.1104/pp.118.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgis F, Roje S, Nuccio ML, Fisher DB, Tarczynski MC, Li C, Herschbach C, Rennenberg H, Pimenta MJ, Shen T. S-Methylmethionine plays a role in phloem sulfur transport and is synthesized by a novel type of methyltransferase. Plant Cell. 1999;11:1485–1497. doi: 10.1105/tpc.11.8.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principal of protein dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Droux M, Ravanel S, Douce R. Methionine biosynthesis in higher plants: II. Purification and characterization of cystathionine β-lyase from spinach chloroplasts. Arch Biochem Biophys. 1995;316:585–595. doi: 10.1006/abbi.1995.1078. [DOI] [PubMed] [Google Scholar]
- Droux M, Ruffet M, Douce R, Job D. Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants: structural and kinetic properties of the free and bound enzymes. Eur J Biochem. 1998;255:235–245. doi: 10.1046/j.1432-1327.1998.2550235.x. [DOI] [PubMed] [Google Scholar]
- Gaitonde MK. A spectrophotometric method for the direct determination of cysteine in the presence of other naturally occurring amino acids. Biochem J. 1967;104:627–633. doi: 10.1042/bj1040627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayler KR, Boadle BG, Snook M, Johnson ED. Precursors of storage proteins in Lupinus angustifolius. Biochem J. 1984;221:333–341. doi: 10.1042/bj2210333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill DS, Kumarasamy R, Symons RH. Cucumber mosaic virus-induced RNA replicase: solubilisation and partial purification of the particulate enzyme. Virology. 1981;113:1–8. doi: 10.1016/0042-6822(81)90131-8. [DOI] [PubMed] [Google Scholar]
- Hatzfeld Y, Maruyama A, Schmidt A, Noji M, Ishizawa K, Saito K. β-Cyanoalanine synthase is a mitochondrial cysteine synthase-like protein in spinach and Arabidopsis. Plant Physiol. 2000;123:1163–1171. doi: 10.1104/pp.123.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell R. Molecular physiology of plant sulfur metabolism. Planta. 1997;202:138–148. doi: 10.1007/s004250050112. [DOI] [PubMed] [Google Scholar]
- Holowach LP, Thompson JF, Madison JT. Effects of exogenous methionine on storage protein composition of soybean cotyledons cultured in vitro. Plant Physiol. 1984;74:576–583. doi: 10.1104/pp.74.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SP, Badger MR, Furbank RT. CO2 refixation characteristics of developing canola seeds and silique wall. Aust J Plant Physiol. 1998;25:377–386. [Google Scholar]
- Kuzuhara Y, Isobe A, Awazuhara M, Fujiwara T, Hayashi H. Glutathione levels in phloem sap of rice plants under sulfur deficient conditions. Soil Sci Plant Nutr. 2000;46:265–270. [Google Scholar]
- Lappartient AG, Touraine B. Demand-driven control of root ATP sulfurylase activity and SO42− uptake in intact canola. Plant Physiol. 1996;111:147–157. doi: 10.1104/pp.111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leustek T, Saito K. Sulfate transport and assimilation in plants. Plant Physiol. 1999;120:637–643. doi: 10.1104/pp.120.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley GG, Inglis AA. Amino acid sequence of conglutin δ, a sulfur-rich seed protein of Lupinus angustifolius L. FEBS Lett. 1986;195:235–241. [Google Scholar]
- Lunn JE, Droux M, Martin J, Douce R. Localization of ATP sulfurylase and O-acetylserine (thiol) lyase in spinach leaves. Plant Physiol. 1990;94:1345–1352. doi: 10.1104/pp.94.3.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnicol PK. Synthesis and interconversion of amino acids in developing cotyledons of pea (Pisum sativum L.) Plant Physiol. 1977;60:344–348. doi: 10.1104/pp.60.3.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Arisi AM, Jouanin L, Kunert KJ, Rennenberg H, Foyer CH. Glutathione: biosynthesis, metabolism and relationship to stress tolerance explored in transformed plants. J Exp Bot. 1998;49:623–647. [Google Scholar]
- Noctor G, Foyer C. Simultaneous measurements of foliar glutathione, γ-glutamylcysteine, and amino acids by high-performance liquid chromatography: comparison with two other assay methods for glutathione. Anal Biochem. 1998;264:98–110. doi: 10.1006/abio.1998.2794. [DOI] [PubMed] [Google Scholar]
- Pate JS, Sharkey PJ, Atkins CA. Nutrition of a developing legume fruit: functional economy in terms of carbon, nitrogen and water. Plant Physiol. 1977;59:506–510. doi: 10.1104/pp.59.3.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pate JS, Sharkey PJ, Lewis OAM. Phloem bleeding from legume fruits: a technique for study of fruit nutrition. Planta. 1974;120:229–243. doi: 10.1007/BF00390291. [DOI] [PubMed] [Google Scholar]
- Pinkerton A, Randall PJ, Norrish K. Estimation of sulfate and amino acid sulfur in plant material by X-ray spectrometry. Commun Soil Sci Plant Anal. 1989;20:1557–1574. [Google Scholar]
- Ravanel S, Gakiere B, Job D, Douce R. The specific features of methionine biosynthesis and metabolism in plants. Proc Natl Acad Sci USA. 1998;95:7805–7812. doi: 10.1073/pnas.95.13.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennenberg H. The fate of excess sulfur in higher plants. Annu Rev Plant Physiol. 1984;35:121–153. [Google Scholar]
- Rolland N, Droux M, Douce R. Subcellular distribution of O-acetylserine(thiol) lyase in cauliflower (Brassica oleracea L.) inflorescence. Plant Physiol. 1992;98:927–935. doi: 10.1104/pp.98.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffet M, Lebrun M, Droux M, Douce R. Subcellular distribution of serine acetyltransferase from Pisum sativum and characterization of an Arabidopsis thaliana putative cytosolic isoform. Eur J Biochem. 1995;227:500–509. doi: 10.1111/j.1432-1033.1995.tb20416.x. [DOI] [PubMed] [Google Scholar]
- Saito K. Regulation of sulfate transport and synthesis of sulfur-containing amino acids. Curr Opin Plant Biol. 2000;3:188–195. [PubMed] [Google Scholar]
- Schroeder HE. Quantitative studies on the cotyledonary proteins in the genus Pisum. J Sci Food Agric. 1982;33:623–633. doi: 10.1002/jsfa.2740330707. [DOI] [PubMed] [Google Scholar]
- Sexton PJ, Shibles RM. Activity of ATP sulfurylase in reproductive soybean. Crop Sci. 1999;39:131–135. [Google Scholar]
- Smith FW, Ealing PM, Hawkesford MJ, Clarkson DT. Plant members of a family of sulfate transporters reveal functional subtypes. Proc Natl Acad Sci USA. 1995;92:9373–9377. doi: 10.1073/pnas.92.20.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith IK, Lang AL. Translocation of sulfate in soybean (Glycine max L. Merr) Plant Physiol. 1988;86:798–802. doi: 10.1104/pp.86.3.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunarpi, Anderson JW. Allocation of S in generative growth of soybean. Plant Physiol. 1997;114:687–693. doi: 10.1104/pp.114.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell J. Supplementation of plant proteins with amino acids. In: Altschul AM, editor. Processed Plant Protein Foodstuffs. New York: Academic Press; 1958. pp. 307–351. [Google Scholar]
- Warrilow AGS, Hawkesford MJ. Separation, subcellular location and influence of sulfur nutrition on isoforms of cysteine synthase in spinach. J Exp Bot. 1998;49:1625–1636. [Google Scholar]
- Zhao FJ, Hawkesford MJ, Warrilow AGS, McGrath SP, Clarkson DT. Responses of two wheat varieties to sulfur addition and diagnosis of sulfur deficiency. Plant Soil. 1996;181:317–327. [Google Scholar]