Abstract
Rationale & Objective
Patients hospitalized with COVID-19 are at increased risk for major adverse kidney events (MAKE). We sought to identify plasma biomarkers predictive of MAKE in patients hospitalized with COVID-19.
Study Design
Prospective cohort study.
Setting and Participants
A total of 576 patients hospitalized with COVID-19 between March 2020 and January 2021 across 3 academic medical centers.
Exposures
26 plasma biomarkers of injury, inflammation, and repair from first available blood samples collected during hospitalization.
Outcomes
Major adverse kidney events (MAKE), defined as KDIGO stage 3 acute kidney injury (AKI), dialysis-requiring AKI, or mortality up to 60 days.
Analytic Approach
Cox proportional hazards regression to associate biomarker level with MAKE. We additionally applied the least absolute shrinkage and selection operator (LASSO) and random forest regression for prediction modeling and estimated model discrimination with time-varying C-index.
Results
The median length of stay for COVID-19 hospitalization was nine (IQR: 5-16) days. In total, 95 (16%) patients experienced MAKE. Each 1-SD increase in soluble tumor necrosis factor receptor 1 (sTNFR1) and sTNFR2 was significantly associated with an increased risk of MAKE (adjusted HR [aHR]: 2.30; 95% CI: 1.86-2.85 and aHR 2.26; 95% CI: 1.73-2.95, respectively). The C-index of sTNFR1 alone was 0.80 (95% CI: 0.78-0.84), while the C-index of sTNFR2 was 0.81 (95% CI: 0.77-0.84). LASSO and random forest regression modeling using all biomarkers yielded C-indices of 0.86 (95% CI: 0.83-0.89) and 0.84 (95% CI: 0.78-0.91) respectively.
Limitations
No control group of hospitalized patients without COVID-19.
Conclusions
TNFR1 and sTNFR2 are independently associated with MAKE in patients hospitalized with COVID-19, and can both also serve as predictors for adverse kidney outcomes.
Keywords: Index words: COVID-19, biomarkers, chronic kidney disease (CKD), acute kidney injury (AKI)
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to a global pandemic since the end of 2019.1 , 2 Acute kidney injury (AKI), defined using Kidney Disease: Improving Global Outcomes (KDIGO) serum creatinine criteria, occurs in 30%-50% of hospitalized patients with COVID-19. Among patients with AKI, nearly 20% require dialysis.3, 4, 5, 6 Furthermore, clinical AKI during hospital admission has been associated with a greater need for intensive care unit admission,7 worse short-term mortality, and adverse long-term outcomes.8 Moreover, compared with AKI due to other causes, COVID-19associated AKI may be associated with a greater long-term decline in kidney function.9
The pathogenesis of COVID-19–associated AKI is likely multifactorial with contributions by endothelial dysfunction, coagulopathy, complement activation, systemic inflammation, and immune cell activation affecting the kidney.10 Older age, male sex, diabetes mellitus, hypertension, obesity, and heart failure have all been associated with increased risk of AKI in the setting of COVID-19.7 , 10 We previously showed that even after adjusting for such risk factors, patients with COVID-19 have a more than 40% higher risk of AKI compared with patients without COVID-19.5 Further, the long-term impact of COVID-19 remains concerning, with numerous physiological and psychological effects reported.11 Thus, there remains a need to identify patients at the greatest risk of developing adverse kidney outcomes following COVID-19.
Biomarkers of kidney injury, inflammation, and repair may offer further insight beyond current standard methods of characterizing COVID-19–associated AKI.12, 13, 14, 15, 16 Plasma and urinary biomarkers have also been associated with COVID-19 disease severity.17 , 18 We recently showed that biomarkers of injury and inflammation measured in the urine are significantly associated with stage 3 AKI, dialysis, or death up to 60 days from hospital admission for COVID-19.19 Model discrimination using single biomarker values was moderate, with area under the receiver operating characteristic curve (AUC) levels approaching 0.80 at 60 days. A model combining the urine biomarkers EGF and neutrophil gelatinase-associated lipocalin (NGAL) yielded an AUC of 0.85.
Based on our previous work in other clinical settings of AKI, we identified 26 candidate plasma biomarkers representing different biological pathways of injury, inflammation, and repair.20, 21, 22, 23, 24 In this study, therefore, we evaluate the association of plasma biomarkers with major adverse kidney events (MAKE) and assess the predictive capability of top biomarkers. We hypothesized that plasma biomarkers would be strongly associated with MAKE in the setting of COVID-19, and have clinically significant predictive potential.
Methods
Study design, population, and data sources
We obtained patient data and biosamples for this study from three academic medical centers as part of the Translational Investigation of Kidney Disease in COVID-19 (TRIKIC) Consortium between March 2020 and January 2021: The Johns Hopkins Hospital in Baltimore, Maryland; Yale-New Haven Hospital in New Haven, Connecticut; and Mount Sinai Hospital in New York, New York. All three health care systems used similar institutional protocols to prospectively collect data and biosamples in hospitalized adult inpatients with COVID-19. At all three sites, adult patients with suspected COVID-19 were screened by PCR testing, and patients with confirmed COVID-19 approached for enrollment in institutional protocols to prospectively collect samples during hospitalization, linked with outpatient follow-up data. IRBs at each participating site of the TRIKIC Consortium approved the study, and written informed consent was obtained from all participants.
Baseline values for laboratory and clinical variables were defined as values present prior to hospital admission. Inpatients with baseline serum creatinine ≥4 mg/dL were excluded. We additionally limited our cohort to patients that were hospitalized for at least 2 days and did not have prior end stage kidney disease (ESKD) defined by ICD diagnosis codes for ESKD and kidney transplant and an admission serum creatinine ≥ 4mg/dL. Baseline comorbidities were defined by mapping ICD-10 codes to the Elixhauser comorbidity index.25 Laboratory values and vital signs on admission were defined as the first available values upon hospital presentation.
Sample collection and biomarker measurement
Plasma samples were collected after a patient’s admission to the hospital and with a confirmed COVID-19 test. Samples were collected following each institution’s informed consent protocol. After collection, all samples were centrifuged promptly at 3,000 × g at 4 ºC and stored in multiple aliquots at -80 ºC until biomarker measurement. For each participant, we used the first available plasma sample before the onset of stage 3 AKI if more than one plasma sample was collected. Patients who had stage 3 AKI present prior to the time of plasma sample collection, such as patients with AKI on presentation to the emergency department or admission to the hospital, were excluded from our analysis. We selected 26 plasma biomarkers as the primary exposure variables based on assay validity; previous work by our group and others have demonstrated significant associations between plasma biomarkers and AKI in various clinical settings.26, 27, 28, 29, 30 These 26 plasma biomarkers include the following: tumor necrosis factor-α (TNF-α); soluble TNF receptor 1 and 2 (sTNFR1 and sTNFR2); angiopoietin 1 and 2 (ANG1 and ANG2); chitinase-like protein-1 (YKL-40); monocyte chemoattractant protein-1 (MCP-1); kidney injury molecule-1 (KIM-1); neutrophil gelatinase-associated lipocalin (NGAL); IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12p70, IL-13, and IL-18; soluble forms-like tyrosine kinase 1 (sFlt-1); basic FGF (bFGF); placental growth factor (PIGF); VEGFA, VEGFC, and VEGFD; TIE2; and IFN-γ. All biomarkers were measured blinded, using the Meso Scale Discovery (MSD) platform after a single freeze-thaw cycle (Table S1).
Kidney function evaluation and outcome definitions
Estimated glomerular filtration rates (eGFRs) were calculated using the Chronic Kidney Disease Epidemiology Collaboration equation.31 Baseline kidney function was defined using the median of all outpatient serum creatinine measurements 7 to 365 days before hospitalization. In patients without any available outpatient serum creatinine measurements, the minimum serum creatinine value obtained during the COVID-19 hospitalization was used as the baseline creatinine (n = 369; 64%), as described previously.32 Clinical AKI was defined as an increase ≥50% in serum creatinine from baseline or dialysis at any time during the index hospitalization. AKI severity was classified by modified KDIGO staging criteria on the basis of the peak serum creatinine level during the index hospitalization. The 60-day time period for primary outcome determination was chosen as a relevant window of time to assess intermediate outcomes, aligning with our previous work.19
The primary outcome of the study was a composite of KDIGO stage 3 AKI, dialysis, or death within 60 days of hospital admission, which together defined MAKE. Additional details are outlined in Item S1.
Statistical Analysis
We limited the analytic cohort to the subset of participants who had all 26 biomarkers measured at least once during hospitalization. We summarized descriptive characteristics using the mean (SD) or median (IQR) for continuous variables and frequency (percentage) for categorical variables. All biomarkers were modeled continuously after log2 transformation. To explore the changes in biomarker levels over time, we used linear regression to calculate the slope of key biomarkers for each participant in the subset of participants with more than 1 plasma sample. World Health Organization (WHO) disease severity scale, as a marker of COVID-19 severity, was determined at the time of sample collection.34 Patients that did not experience the primary outcome were censored at 60 days.
Cox proportional hazards regression was used to examine the association between plasma biomarker level and MAKE using the first available biomarker measurement for each participant. Model 1 included each individual log2-transformed biomarker alone. Model 2 adjusted for demographic variables (age, sex, race), diabetes mellitus, obesity, hypertension, and baseline serum creatinine. Model 3 further adjusted for WHO disease severity scale. Kolmogorov-type supremum tests were used to evaluate proportional hazards assumptions for all models. HRs are reported per 1-SD increase for each log2-transformed biomarker.
To identify biomarker combinations, we used the endpoint of time to the development of MAKE within 60 days. We applied LASSO to model the risk of MAKE by biomarker level for the 26 candidate plasma biomarkers. Fivefold cross-validation was used for optimal shrinkage parameter selection. We performed random forest analysis as an alternative approach for dimensionality reduction in evaluating key biomarkers associated with MAKE to allow for more flexibility in accounting for interaction, with the data randomly partitioned: 80% (n = 461) into a training dataset and 20% (n = 115) into a test dataset.
We evaluated model discrimination by calculating the time-varying C-indices.35 We examined c-indices for each individual biomarker as well as several models including the LASSO and random forest model (on the test dataset). To identify the two highest-performing biomarkers, we examined biomarkers with the highest individual C-indices, with non-zero coefficients in LASSO models and the top biomarkers in variable importance plots from the random forest model. For the two highest-performing biomarkers , we created models with key clinical demographics to assess for any additional improvement in model performance. These clinical variables included age; sex; race/ethnicity; baseline diabetes, hypertension and obesity; baseline serum creatinine; and WHO disease severity scale at the time of sample collection. We additionally added in C-reactive protein to each model, as a clinically available and widely measured inflammatory biomarker in patients with COVID-19. We evaluated receiver operating characteristic (ROC) curves at 60 days, including sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). Based on these curves we determined optimal cutoff points for these biomarkers. Using the coefficients from the Cox model, we back-calculated the biomarker value for the optimal threshold.
In addition to analyses in the overall study population, we performed Cox proportional hazards regression and calculated C-indices in key subgroups, including in the subset of 499/576 (87%) patients with biosample collection occurring within the first week of admission, and in the 436/576 (76%) patients who did not receive steroids prior to biosample collection given the previously described association between steroid use and reduction in sTNFR1 and sTNFR2 levels.36 Furthermore, we developed Cox models and calculated model discrimination for the outcome of stage 3 AKI or dialysis. For the 38 patients that died without experiencing either kidney outcome, were censored at the time of death.
To determine if the association between biomarkers and outcomes was modified by patient comorbidities, we examined models with an interaction term between the biomarker and covariate of interest. P-values of the interaction term <0.1 were deemed as significant. Hazard ratios at each covariate value were calculated.
Since the timing of the plasma sample collection was not consistent across patients, we completed a supplementary analysis where time zero was defined as the time of sample collection. The imputation strategy for patients without a baseline outpatient serum creatinine measurement was modified to use the lowest serum creatinine prior to the time of sample collection as the baseline measure. Hazard ratios for individual biomarkers were calculated using the cox models as defined for the primary analysis.
All analyses were performed in SAS (version 9.4; SAS Institute, Cary, North Carolina)and R (version 3.1.2; R Foundation for Statistical Computing, Vienna, Austria). All tests of statistical significance were 2-sided, with P < 0.05 considered significant.
Results
Study Population
We included samples from a total of 576 patients during hospitalization for COVID-19 after applying inclusion and exclusion criteria (Figure S1). The mean patient age on admission was 60.3 ± 16.2 years, and 243 (42%) were women. The mean baseline serum creatinine was 0.87 ± 0.5 mg/dL and the mean admission serum creatinine was 1.23 ± 1.1 mg/dL (Table 1 ). Of the 576 patients, 203 (35%) had diabetes mellitus, 293 (51%) had hypertension, and 168 (29%) had obesity, with a BMI ≥30 kg/m2. A total of 62 patients (30%) had baseline chronic kidney disease (CKD), defined by an estimated glomerular filtration rate (eGFR) < 60 mL/min/1.73m2. The median length of a COVID-19 hospitalization in this study was 9 (IQR: 5-16) days. In total, 95 (16%) patients experienced the primary outcome of MAKE. Of these 95 patients, 57 (60%) developed stage 3 AKI, 31 (33%) required dialysis, and 69 (73%) died during the study period. A total of 125 (22%) patients received remdesivir and 140 (24%) received steroids prior to biomarker measurement. The majority (67%) of biosamples were collected within 72 hours of hospital admission with a smaller percentage collected between 4-7 days and later in admission.
Table 1.
Inpatient characteristics of patients with COVID-19
| Variable | Overall (N = 576) | Hopkins (N = 182) | Mt Sinai (N = 291) | Yale (N = 103) |
|---|---|---|---|---|
| Age, years | 60.34 ± 16.2 | 55.25 ±15.82 | 62.88 ±15.9 | 62.14 ±15.72 |
| Female | 243 (42) | 82 (45) | 116 (40) | 45 (44) |
| Black race | 171 (30) | 90 (49) | 48 (16) | 33 (32) |
| Diabetes | 203 (35) | 90 (49) | 70 (24) | 41 (40) |
| Obesity | 168 (29) | 104 (57) | 32 (11) | 32 (31) |
| Hypertension | 293 (51) | 121 (66) | 106 (36) | 66 (64) |
| Length of hospital stay (d) | 9 (5-16) | 9 (5-17) | 8 (5-15) | 11 (7-19) |
| WHO disease severity scale - maximum | ||||
| 3 | 84 (16) | 31 (17) | 52 (18) | 1 (2) |
| 4 | 264 (49) | 88 (48) | 135 (46) | 41 (64) |
| 5 | 64 (12) | 22 (12) | 34 (12) | 8 (13) |
| 6 | 3 (1) | 3 (1) | ||
| 7 | 75 (14) | 26 (14) | 39 (13) | 10 (16) |
| 8 | 47 (9) | 15 (8) | 28 (10) | 4 (6) |
| Admission C-reactive protein (mg/dL) | 83.9 (35.9-163.6) | 60.7 (25.3-111.3) | 101.7 (46.3-189.6) | 85 (39.8-146) |
| Number of serum creatinine values measured | 9 (5-16) | 8 (4-14) | 9 (5-15) | 10 (7-22) |
| Baseline serum creatinine (mg/dL) | 0.87 ± 0.45 | 0.83 ± 0.38 | 0.88 ± 0.49 | 0.90 ± 0.46 |
| Admission serum creatinine (mg/dL) | 1.23 ± 1.11 | 1.09 ± 0.53 | 1.34 ± 1.4 | 1.15 ± 0.85 |
| Discharge serum creatinine (mg/dL) | 1.14 ± 1.08 | 0.96 ± 0.51 | 1.28 ± 1.31 | 1.07 ± 1.02 |
| Baseline eGFR (mL/min per 1.73m2)31† | 77 (30) | 83 (31) | 78 (30) | 65 (25) |
| Baseline CKD† | 62 (30%) | 11 (21%) | 30 (26%) | 21 (54%) |
| AKI Stage 3, any dialysis, death within 60 d | 95 (16) | 15 (8) | 66 (23) | 14 (14) |
| Number of days to MAKE | 13 (9-24) | |||
| Dialysis requirement | 31 (5) | 4 (2) | 22 (8) | 5 (5) |
| Time to dialysis (d) | 14 (9-22) | 11 (8-24.5) | 15 (12-22) | 14 (13-14) |
| Death within 60 d | 69 (12) | 13 (7) | 46 (16) | 10 (10) |
| Time to death (d) | 18 (10-27) | 26 (15-32) | 13 (9-25) | 22.5 (15-35) |
| KDIGO AKI Stage | ||||
| 1 | 102 (18) | 45 (25) | 46 (16) | 11 (11) |
| 2 | 65 (11) | 18 (10) | 38 (13) | 9 (9) |
| 3 | 57 (10) | 7 (4) | 41 (14) | 9 (9) |
| Days to COVID-19-associated AKI | 5 (0-12) | 3 (1-10) | 5 (0-11) | 7 (1-15) |
| Biosample collection from time of admission | ||||
| Within 72 hours (3 days) | 385 (67%) | 99 (17) | 235 (41) | 51 (13) |
| 4-7 days | 114 (20%) | 40 (7) | 38 (7) | 36 (6) |
| After first week | 77 (13%) | 43 (7) | 18 (3) | 16 (3) |
| Medication use prior to biomarker measurement | ||||
| Tocilizumab | 61 (11%) | 7 (4) | 3 (1) | 51 (50) |
| Remdesivir | 125 (22%) | 40 (22) | 50 (17) | 35 (34) |
| Hydroxychloroquine | 196 (34%) | 25 (14) | 111 (38) | 60 (58) |
| Steroid* | 140 (24%) | 45 (25) | 65 (22) | 30 (29) |
Values are reported as mean ± SD, n (%), or median (IQR)
AKI = acute kidney injury; KDIGO = Kidney Disease: Improving Global outcomes; MAKE = major adverse kidney events; WHO = World Health Organization
* Steroid is any administration of dexamethasone, methylprednisolone, or hydrocortisone
†Baseline eGFR and CKD (defined as eGFR ≤60) is reported for the 206 patients with baseline serum creatinine available prior to the time of admission.
Associations of biomarkers with the primary outcome
Figure 1 (Table S2) highlights the risk of MAKE by individual biomarker level, using biomarkers measured from the first available plasma sample during admission. In the fully-adjusted analysis, 15 of 26 candidate biomarkers were significantly associated with MAKE, 11 with increased risk and 4 with decreased risk. Each 1-SD increase in log2-transformed ANG1, IL-13, VEGFA, and VEGFC was significantly associated with a lower risk of MAKE after adjustment for clinical covariates. Of the 11 biomarkers associated with increased risk of MAKE, each 1-SD increase in either sTNFR1 (adjusted HR [aHR] 2.30; 95% CI: 1.86-2.85) or sTNFR2 (aHR 2.26; 95% CI: 1.73-2.95) was associated with a greater than two-fold higher risk of 60-day MAKE.
Figure 1.
Risk of MAKE by 1-SD increase in plasma biomarker level in the fully adjusted analysis. MAKE is defined as having at least stage 3 AKI, dialysis, or death within 60 days of hospital admission with a confirmed COVID-19 test. Four biomarkers (ANG1, VEGFA, VEGFC, and IL-13) were associated with lower risk of MAKE, and 11 biomarkers were associated with higher risk of MAKE. The remaining 11 were not significantly associated with risk of MAKE.*Fully adjusted for the following: demographics (age, sex, race), diabetes, obesity, hypertension, baseline serum creatinine, World Health Organization (WHO) disease severity scale (reported as Model 3 in Table S2).
In sub-group analyses, we see largely similar results in the subset of 499 patients with biosample collection occurring within the first week of admission, with 73 events observed (Table S3). In the subset of 436 patients who did not receive steroids prior to biosample collection, we observed 73 events, with sTNFR1 and sTNFR2 still most strongly associated with risk of MAKE (Table S4).
In an additional secondary analysis limiting the outcome to stage 3 AKI or dialysis, sTNFR1 (aHR 2.98; 95% CI: 2.31-3.84), sTNFR2 (aHR 3.35; 95% CI: 2.35-4.77), and NGAL (aHR 2.34; 95% CI: 1.70-3.24) were still most strongly associated with greatest risk (Table S5). Figure S2 highlights sTNFR1 and sTNFR2 levels relative to the number of days from sample collection to the event for those that experienced MAKE. Similar trends were noted for other biomarkers based on time to MAKE (Figure S3). In a supplementary analysis on the subset of 70 participants with more than 1 plasma biomarker measurement before the onset of stage 3 AKI, both sTNFR1 and sTNFR2 increased in patients that went on to develop MAKE compared to those that did not (Table S6). Redefining time zero to the time of plasma sample collection resulted in a slightly lower rate of 91 MAKE events out of 575 patients included in this sensitivity analysis. Biomarker associations with MAKE remained largely unchanged (Table S7).
Discrimination of biomarkers with outcomes
Table 2 displays the results of discrimination for individual biomarkers using the first available measurement for each. Notably, sTNFR1 and sTNFR2 had the two highest C-index values of 0.80 (95% CI: 0.78-0.84) and 0.81 (95% CI: 0.77-0.84) respectively. Table S8 demonstrated individual biomarker discrimination for our key secondary analyses including the sub-groups of patients with biosample collection with 7 days of hospital admission, patients without steroid exposure prior to biosample collection, and in the overall patient population limiting the outcome to stage 3 AKI and dialysis.
Table 2.
Individual Biomarker Model Discrimination
| Direction of Association* | Biomarker | C-index (95% CI) |
|---|---|---|
| Increased risk | sTNFR2 | 0.81 (0.77-0.84) |
| sTNFR1 | 0.80 (0.78-0.84) | |
| YKL-40 | 0.78 (0.74-0.82) | |
| NGAL | 0.77 (0.72-0.81) | |
| ANG2 | 0.73 (0.69-0.79) | |
| KIM-1 | 0.73 (0.69-0.77) | |
| sFLT-1 | 0.69 (0.65-0.74) | |
| IL-18 | 0.68 (0.63-0.73) | |
| TNF-α | 0.67 (0.63-0.71) | |
| IL-10 | 0.63 (0.58-0.68) | |
| IL-2 | 0.63 (0.57-0.68) | |
| Decreased risk | ANG1 | 0.61 (0.56-0.66) |
| VEGFC | 0.56 (0.51-0.62) | |
| VEGFA | 0.55 (0.50-0.62) | |
| IL-13 | 0.52 (0.50-0.58) | |
| Not statistically significant | PIGF | 0.66 (0.60-0.72) |
| IL-6 | 0.64 (0.60-0.69) | |
| IL-8 | 0.63 (0.58-0.67) | |
| MCP-1 | 0.63 (0.59-0.68) | |
| VEGFD | 0.62 (0.56-0.68) | |
| IL-1β | 0.59 (0.53-0.64) | |
| IL-12p70 | 0.56 (0.51-0.61) | |
| IFN-γ | 0.55 (0.50-0.62) | |
| IL-4 | 0.55 (0.51-0.59) | |
| Tie2 | 0.51 (0.50-0.59) | |
| bFGF | 0.51 (0.50-0.57) |
†Taking into account time to event among 576 patients
*Direction of association is based on the HR of the biomarker adjusted for demographics (age, sex, race, diabetes, obesity, hypertension), baseline serum creatinine, and WHO disease severity scale, as presented in Figure 1
ANG1/ANG2 = angiopoietin 1/2; bFGF = basic fibroblast growth factor; KIM-1 = kidney injury molecule-1; MCP-1 = monocyte chemoattractant protein-1; NGAL = neutrophil gelatinase-associated lipocalin; PIGF = placental growth factor; sFlt-1 = soluble fms-like tyrosine kinase 1; sTNFR1/sTNFR2 = soluble tumor necrosis factor receptor-1 or -2; TIE2; YKL-40 = chitinase-like protein-1.
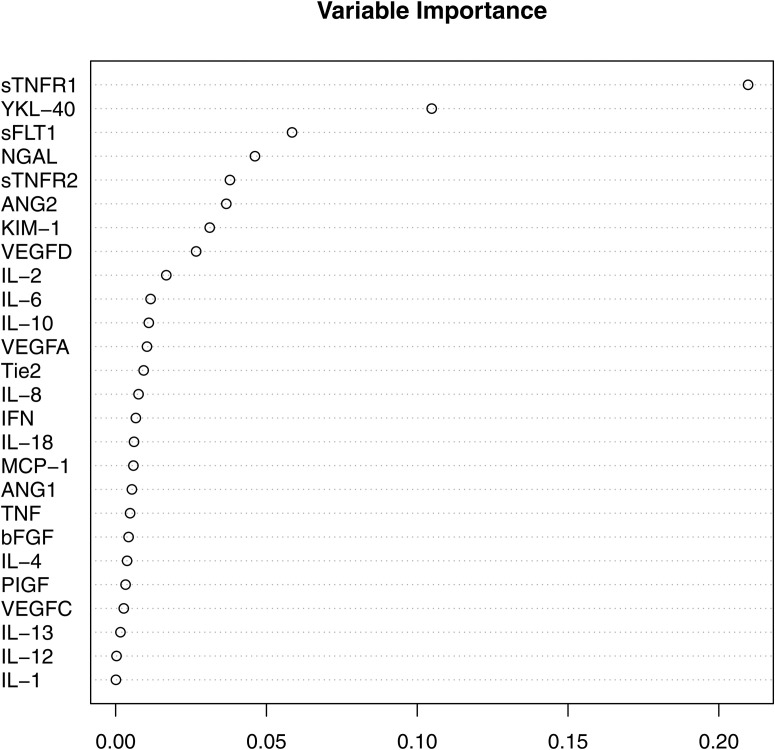
Using least absolute shrinkage and selection operator (LASSO), 20 of 26 biomarkers were found to have nonzero coefficients and were included in the final model: IFN-γ, IL-1β, IL-2, IL-4, IL-6, IL-8, IL-12p70, IL-13, IL-18, TNF-α, TIE2, PIGF, bFGF, sFLT1, NGAL, MCP-1, YKL-40, ANG1, ANG2, and sTNFR1. This combined model yielded a C-index of 0.86 (95% CI: 0.83-0.89; Table 3 ). As an alternative, complementary approach, we performed random forest regression using the first biomarker measurements of all 26 plasma biomarkers, which yielded a similar C-index of 0.84 (95% CI: 0.78-0.91) in the validation dataset. Several biomarkers were found to be significant predictors for MAKE, as determined through both LASSO and random forest analysis, with sTNFR1 and sTNFR2 being among the top predictors (Figure 2 ).
Table 3.
Combination Biomarker-Clinical Model Discrimination
| Model | C-index (95% CI) |
|---|---|
| LASSOa (20 biomarkers) | 0.86 (0.83-0.89) |
| Random forestb (26 biomarkers) | 0.84 (0.78-0.91) |
| Clinical modelc | 0.82 (0.76-0.88) |
| Clinical modelc + sTNFR1 | 0.86 (0.81-0.92) |
| Clinical modelc + sTNFR2 | 0.85 (0.79-0.91) |
| Clinical modelc + admission CRP | 0.82 (0.77-0.88) |
| Clinical modelc + sTNFR1 + admission CRP | 0.87 (0.81-0.92) |
| Clinical modelc + sTNFR2 + admission CRP | 0.85 (0.79-0.91) |
CRP = C-reactive protein; LASSO = least absolute shrinkage and selection operator
a: LASSO model had 20 biomarkers with nonzero coefficients
b: Random forest includes all 26 biomarkers
c: Clinical model comprises 8 variables: age; sex; race; baseline serum creatinine; history of diabetes, hypertension or obesity; WHO disease severity score at time of sample collection
Figure 2.
Variable importance in risk prediction using random forest regression, yielding an overall C-index of 0.84 (95% CI: 0.78-0.91).
A clinical prediction model that contained 8 variables based on known risk factors for adverse outcomes yielded a C-index of 0.82 (95% CI: 0.76-0.88), similar to the C-indices of sTNFR1 and sTNFR2 alone (Table 3).37 , 38 Addition of sTNFR1 to this 8-variable clinical model improved the C-index to 0.86 (95% CI: 0.81-0.92), similar to the LASSO and random forest regression models that utilized all 26 plasma biomarkers. Inclusion of C-reactive protein (CRP), as a marker of general inflammation and widely measured in patients with COVID-19, did not appreciably improve model discrimination of either the clinical model but did yield a higher C-index to the biomarker-enriched models, particularly to a model containing clinical variables, sTNFR1, and CRP (C-index 0.87; 95% CI: 0.81, 0.92).
A concentration of sTNFR1 at the optimal cutoff of 2,130 pg/mL, based on the ROC curve at 60 days, had a specificity of 65%, sensitivity of 70%, PPV of 30%, and most notably a NPV of 92% for the time to MAKE (Table S9). A concentration of sTNFR2 at the optimal cutoff of 14,670 pg/mL had a specificity of 74%, sensitivity of 70%, PPV of 35%, and NPV of 94%. There was no significant interaction on the association between sTNFR1 or sTNFR2 and MAKE by age, sex, race, diabetes, CKD (eGFR < 60 vs. ≥ 60), or obesity status (Figure S4).
Discussion
In this study, we demonstrated that 15 of 26 candidate plasma biomarkers were associated with MAKE up to 60 days from hospital admission, with each SD elevation in levels of sTNFR1 and sTNFR2 associated with an over twofold higher risk of MAKE. Through the complementary LASSO and random forest regression techniques using all 26 plasma biomarkers, we found that combination models yielded C-indices of 0.86 and 0.84. The biomarkers sTNFR1 or sTNFR2 were found to be the strongest individual biomarkers predictive of MAKE, and in combination with clinical variables yielded C-indices of 0.85 and 0.86.
TNFR1 and TNFR2 are cell-surface receptors that bind TNF-α and are important in mediating inflammatory responses. Downstream signaling through the TNF-α pathway is mediated through TNFR1 and TNFR2, and includes activation of the transcription factors NF-κB and JNK.40 Of note, measured levels of sTNFR1 and sTNFR2 reflect soluble forms of these receptors, in contrast to being bound to cell membrane.41 These soluble forms may result from either cleavage of the cell surface receptor or from the release of full-length sTNFR1 via exosome-like vesicles.42 , 43 However, the mechanistic relationship between elevated plasma sTNFR1 and adverse outcomes remains unclear. Xu and colleagues demonstrated that sTNFR1 plays a key role in TNF-mediated damage to glomerular epithelium in a mouse model of sepsis-associated AKI.44 Specifically, they showed that knockout mice deficient in sTNFR1 were resistant to lipopolysaccharide-induced AKI, whereas wild-type mice suffered significant kidney injury.
Elevations in sTNFR1 and sTNFR2 have been associated with progression of kidney disease in a number of clinical settings.42 , 46 In 2012 White and colleagues demonstrated that sTNFR1 is implicated in cellular apoptosis in the pulmonary microvasculature, noted on bronchoalveolar lavage analysis, in the setting of ischemic AKI.47 Both sTNFR1 and sTNFR2 predict CKD and end-stage kidney disease in the setting of types 1 and 2 diabetes mellitus.48 , 49 Furthermore, sTNFR1 was shown to be a strong prognostic factor for all-cause mortality in patients with CKD and type 2 diabetes mellitus.41
Our study adds to the growing body of literature evaluating the role of sTNFR1 and sTNFR2 as prognostic biomarkers in patients with COVID-19. Ferrando and colleagues demonstrated that in the acute setting, sTNFR1 and sTNFR2 are associated with AKI, with levels trending significantly higher by AKI stage. Additionally these authors showed that sTNFR1 was associated with 30-day mortality in critically ill hospitalized patients with COVID-19.50 Our study expands on these findings, evaluating the need for dialysis in our MAKE outcome with outcome ascertainment over a longer time period of 60 days. Notably, discrimination in a model containing CRP along with the 8 clinical variables was not as strong as a clinical model containing either sTNFR1 or sTNFR2. While CRP is a marker of general inflammation, sTNFR1 and sTNFR2 have been more strongly associated with inflammation in the kidney and may help explain this difference.51
While the focus of this investigation centered on hospitalized patients with COVID-19, our study adds to the existing literature of sTNFR1 as a marker of MAKE after hospitalized AKI, in the general setting. A recent investigation using data from the AKI Risk in Derby Study has shown that sTNFR1 and sTNFR2 measured post-discharge are strong predictors of CKD progression up to 3 years after hospitalization for AKI.52 Similarly, sTNFR1 and sTNFR2 were found to be significantly associated with long-term CKD and mortality in the Assessment, Serial Evaluation, and Subsequent Sequelae in Acute Kidney Injury Study.53 Furthermore, among patients with existing CKD, hospitalized AKI has been associated with increases in sTNFR1 and sTNFR2 even several months after hospital discharge.54 A number of experimental models have demonstrated that antibodies against sTNFR1 may in fact reduce chronic inflammation and tissue injury.55 , 56 Given their role in the body’s response to inflammation, elevations in sTNFR1 and sTNFR2 in the setting of COVID-19 may serve as markers of disease severity, with prolonged inflammation leading to worse kidney outcomes.
Our results have implications both for future research and clinical application. The ultimate goal of biomarker-based research is to improve the timely diagnosis and management of patients. Although a model containing 8 clinical variables did perform well in prediction of MAKE in our analysis, at the individual level, this may be difficult to implement given the lack of availability of all necessary variables, including baseline kidney function. On the other hand, the robust performance of sTNFR1 can be leveraged for early clinical prediction and timely management of patients who are likely to have poor outcomes. Our proposed cutoffs for sTNFR1 and sTNFR2 of 2,130 pg/mL and 14,670 pg/mL had high NPV’s of 92% and 94% for MAKE over 60 days respectively, largely excluding the outcome in our cohort at concentrations below the cutoff. Future studies can investigate sTNFR1 as a “rule-out” test with potential early discharge from ICU and hospital if the patient is improving clinically. After identification of high-risk patients, i.e. patients above the cutoff of 2,130 pg/mL, the goal of management would be to slow or prevent progression of CKD through meaningful post-discharge nephrology care. There has been increasing interest in evaluating the AKI-to-CKD transition, with interventions to help slow or prevent this process. The use of an sTNFR1 cutoff may offer significant contribution in such risk-stratification. While our current study was conducted exclusively in patients with COVID-19-associated AKI and in other clinical settings, exploring such an sTNFR1 cutoff for risk stratification should be explored more broadly.57 , 58
Study strengths include the use of a large and diverse patient population from 3 academic medical centers, allowing for improved generalizability of study findings. We also validated our discriminatory findings with high C-indices for a combined biomarker model generated through LASSO regression using the complementary approach of random forest. Given the predictive power of sTNFR1 alone, taking the first biomarker measurement available after admission may be a feasible approach to improve risk stratification of patients for future risk of adverse kidney outcomes. We have also identified a number of other plasma biomarkers including sTNFR2 that are strongly associated with MAKE and require further investigation. Our use of the agnostic LASSO approach for variable selection did not in fact identify sTNFR2 as having a non-zero coefficient, though this is likely due to its high collinearity with sTNFR1.
This study has several limitations. We previously showed that urinary biomarker measurements are robust to variations in sample collection, processing, and storage, though this is as yet unproven for plasma biomarkers.59 Despite standardized protocols, it is possible that the samples had differential handling across sites during the COVID-19 epidemic. Other biomarkers such as SuPAR and cystatin-C have been associated with COVID-19 associated AKI but were not clinically available in this cohort, and such associations were not known at the time of our biomarker measurement, and we did not have additional samples available at all sites to perform these additional biomarker measurements. Additionally, although this study includes data on over 500 patients with nearly 100 MAKE events, it is not sufficiently powered to separately evaluate the individual outcomes of KDIGO stage 3 AKI, dialysis, or death. Although we accounted for COVID-19 disease severity by adjusting for WHO disease severity score, this may not account for other unmeasured potentially confounding comorbidities. Similarly, while we explored patient cohorts across three academic medical centers, we did not externally validate our findings in a separate, larger patient cohort. The development of new therapies against SARS-CoV-2 infection may modify these associations and overall generalizability over time. Similarly, disease severity of COVID-19 varied over time, with the emergence of new variants that may differentially impact kidney outcomes, which we were not able to assess in this study.
In summary, increased plasma concentrations of sTNFR1 and sTNFR2 are each independently and strongly associated with MAKE in patients hospitalized with COVID-19. In particular, combining clinical variables with either sTNFR1 or sTNFR2 has very strong discrimination for predicting MAKE, and further studies should confirm these findings in COVID-19 and other hospitalized clinical settings to identify high risk patients. These results support that those with severe disease need post-discharge care and longer follow-up studies in a larger population are necessary to understand the full spectrum of health consequences from COVID-19.
Uncited reference
Acknowledgments
The specimens used for this publication were part of the Johns Hopkins Biospecimen Repository, which is based on the contribution of many patients, research teams, and clinicians.
Footnotes
Plain Language Summary
Patients hospitalized with COVID-19 are at increased risk for long-term adverse health outcomes but not all patients suffer long-term kidney dysfunction. Identification of patients with COVID-19 who are at high risk for adverse kidney events may have important implications in terms of nephrology follow-up and patient counseling. In this study, we found that the plasma biomarkers sTNFR1 and sTNFR2 measured in hospitalized patients with COVID-19 were associated with a greater risk of kidney outcomes. Along with clinical variables previously shown to predict adverse kidney events in patients with COVID-19, both sTNFR1 and sTNFR2 are also strong predictors of adverse kidney outcomes.
Article Information
Translational Research Investigating Kidney Outcomes in COVID-19 (TRIKIC) Consortium: Jie Deng, Mo Atta, Serena M. Bagnasco, and the Johns Hopkins COVID-19 and Data Research Evaluation Committee (Johns Hopkins University School of Medicine); Albert Ko, Akiko Iwasaki, Shelli Farhadian, Allison Nelson, Arnau Casanovas-Massana, Elizabeth B. White, Wade Schulz, Andreas Coppi, Patrick Young, Angela Nunez, Denise Shepard, Irene Matos, Yvette Strong, Kelly Anastasio, Kristina Brower, Maxine Kuang, Michael Chiorazzi, Santos Bermejo, Pavithra Vijayakumar, Bertie Geng, John Fournier, Maksym Minasyan, M. Catherine Muenker, Adam J. Moore (Yale University School of Medicine); and Girish Nadkarni (Icahn School of Medicine at Mount Sinai).
Authors’ Contributions: Research idea and study design: SM, HTP, CRP; specimen collection, processing: EUA (Mount Sinai); data analysis/interpretation: LC, UU, CJS, LJA, AZR; biomarker measurement: WO; development and principal investigator of the JH-CROWN Registry: BTG; statistical analysis: HTP; supervision/mentorship: CRP, DGM, HTP, FPW, SGC. Each author contributed important intellectual content during manuscript drafting or revision and agrees to be personally accountable for the individual’s own contributions and to ensure that questions pertaining to the accuracy or integrity of any portion of the work, even one in which the author was not directly involved, are appropriately investigated and resolved, including with documentation in the literature, if appropriate.
Support: This study was supported by NIDDK supplemental awards R01DK093770-09S1 (CRP) and R01 DK118222-03S1 (EUA) for COVID-19 research. These studies were also supported through the generosity of the collective community of donors to the Johns Hopkins University School of Medicine for COVID research as part of the JH-CROWN: The COVID PMAP Registry (IRB00253168, IRB00252317) and with funding from the JHU COVID-19 Research Response Program. Data presented here were also obtained through the Yale Department of Medicine’s COVID Explorer data repository, which is funded by the Department of Medicine, the George M. O'Brien Kidney Center at Yale (P30DK079310), and resources from the Clinical and Translational Research Accelerator (IMPACT Study IRB 2000027690) and the Mount Sinai data warehouse. FPW reports grant support from R01DK113191, R01HS027626 and P30DK079310. SM receives grant support from K23DK128538 and research support from RenalytixAI. DGM receives grant support from K23DK117065. This work was additionally supported by a JIT Award (to S.J.) from the Washington University Genome Technology Access Center at the McDonnell Genome Institute, which is partially supported by NCI Cancer Center Support (#P30 CA91842) to the Siteman Cancer Center, by ICTS/CTSA (# UL1TR002345) from the National Center for Research Resources (NCRR) and by the Washington University Kidney Translational Research Center (KTRC) supported by the Division of Nephrology. The KPMP is funded by the following grants from the NIDDK: U2C DK114886, UH3DK114861, UH3DK114866, UH3DK114870, UH3DK114908, UH3DK114915, UH3DK114926, UH3DK114907, UH3DK114920, UH3DK114923, UH3DK114933, and UH3DK114937. Funders did not have a role in study design, data collection, analysis, reporting, or the decision to submit for publication.
Financial Disclosure: SGC and CRP serve as members of the advisory board of and own equity in Renalytix AI. DGM and CRP are named co-inventors on a pending patent, “Methods and Systems for Diagnosis of Acute Interstitial Nephritis,” that is subject to an option for a license agreement with Renalytix AI, Inc. SGC has reported receiving consulting fees from Bayer, Boehringer-Ingelheim, Nuwellis, Axon Therapies, Reprieve Cardiovascular, 3ive, Renalytix AI, and Vifor in the past 3 years. BTG is a member of the Food and Drug Administration Pulmonary and Asthma Drug Advisory Committee, and has received consulting fees from Janssen Research and Development, LLC., Gilead Sciences, Inc. and Atea Pharmaceuticals, Inc. EUA reports research funding from Renalytix AI and Aurinia Pharmaceuticals. SM has received consulting fees from the Dedham Group and research funding from Renalytix AI. LC has received consulting fees from Vifor Pharma INC, and an honorarium from Fresenius Medical Care. The remaining authors declare that they have relevant financial interests.
Peer Review: Received August 29, 2022. Evaluated by 2 external peer reviewers, with direct editorial input from a Statistics/Methods Editor, an Associate Editor, and a Deputy Editor who served as Acting Editor-in-Chief. Accepted in revised form March 8, 2023. The involvement of an Acting Editor-in-Chief was to comply with AJKD’s procedures for potential conflicts of interest for editors, described in the Information for Authors & Journal Policies.
Contributor Information
on behalf of the TRIKIC Consortium:
Jie Deng, Mo Atta, Serena M. Bagnasco, Albert Ko, Akiko Iwasaki, Shelli Farhadian, Allison Nelson, Arnau Casanovas-Massana, Elizabeth B. White, Wade Schulz, Andreas Coppi, Patrick Young, Angela Nunez, Denise Shepard, Irene Matos, Yvette Strong, Kelly Anastasio, Kristina Brower, Maxine Kuang, Michael Chiorazzi, Santos Bermejo, Pavithra Vijayakumar, Bertie Geng, John Fournier, Maksym Minasyan, M. Catherine Muenker, Adam J. Moore, and Girish Nadkarni
References
- 1.Guan WJ, Ni ZY, Hu Y, et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. Feb 28 2020;382(18):1708 - 1720. doi:10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed]
- 2.Johns Hopkins University & Medicine Coronavirus Resource Center. COVID-19 Dashboard. Accessed January 21, 2021. https://coronavirus.jhu.edu/map.html
- 3.Chan L., Chaudhary K., Saha A., et al. AKI in Hospitalized Patients with COVID-19. J Am Soc Nephrol. Sep 3 2020;32(1):151–160. doi: 10.1681/asn.2020050615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirsch J.S., Ng J.H., Ross D.W., et al. Acute kidney injury in patients hospitalized with COVID-19. Kidney International. 2020/07/01/ 2020;98(1):209–218. doi: 10.1016/j.kint.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moledina D.G., Simonov M., Yamamoto Y., et al. The Association of COVID-19 With Acute Kidney Injury Independent of Severity of Illness: A Multicenter Cohort Study. American Journal of Kidney Diseases. 2021;77(4):490–499. doi: 10.1053/j.ajkd.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowe B., Cai M., Xie Y., Gibson A.K., Maddukuri G., Al-Aly Z. Acute Kidney Injury in a National Cohort of Hospitalized US Veterans with COVID-19. Clinical Journal of the American Society of Nephrology. 2021;16(1):14. doi: 10.2215/CJN.09610620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher M., Neugarten J., Bellin E., et al. AKI in Hospitalized Patients with and without COVID-19: A Comparison Study. J Am Soc Nephrol. Sep. 2020;31(9):2145–2157. doi: 10.1681/asn.2020040509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng JH, Hirsch JS, Hazzan A, et al. Outcomes Among Patients Hospitalized With COVID-19 and Acute Kidney Injury. Am J Kidney Dis. Feb 2021;77(2):204-215.e1. doi:10.1053/j.ajkd.2020.09.002 [DOI] [PMC free article] [PubMed]
- 9.Nugent J, Aklilu A, Yamamoto Y, et al. Assessment of Acute Kidney Injury and Longitudinal Kidney Function After Hospital Discharge Among Patients With and Without COVID-19. JAMA Network Open. 2021;4(3):e211095-e211095. doi:10.1001/jamanetworkopen.2021.1095 [DOI] [PMC free article] [PubMed]
- 10.Nadim MK, Forni LG, Mehta RL, et al. COVID-19-associated acute kidney injury: consensus report of the 25th Acute Disease Quality Initiative (ADQI) Workgroup. Nat Rev Nephrol. Dec 2020;16(12):747-764. doi:10.1038/s41581-020-00356-5 [DOI] [PMC free article] [PubMed]
- 11.del Rio C., Collins L.F., Malani P. Long-term Health Consequences of COVID-19. JAMA. 2020;324(17):1723–1724. doi: 10.1001/jama.2020.19719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delanaye P, Cavalier E, Pottel H. Serum Creatinine: Not So Simple! Nephron. 2017;136(4):302-308. doi:10.1159/000469669 [DOI] [PubMed]
- 13.Pode Shakked N., de Oliveira M.H.S., Cheruiyot I., et al. Early prediction of COVID-19-associated acute kidney injury: Are serum NGAL and serum Cystatin C levels better than serum creatinine? Clin Biochem. Apr. 2022;102:1–8. doi: 10.1016/j.clinbiochem.2022.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Temiz M.Z., Hacibey I., Yazar R.O., et al. Altered kidney function induced by SARS-CoV-2 infection and acute kidney damage markers predict survival outcomes of COVID-19 patients: a prospective pilot study. Ren Fail. Dec 2022;44(1):233–240. doi: 10.1080/0886022x.2022.2032743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azam TU, Shadid HR, Blakely P, et al. Soluble Urokinase Receptor (SuPAR) in COVID-19-Related AKI. J Am Soc Nephrol. Nov 2020;31(11):2725-2735. doi:10.1681/asn.2020060829 [DOI] [PMC free article] [PubMed]
- 16.Xu K., Shang N., Levitman A., et al. Elevated Neutrophil Gelatinase-Associated Lipocalin Is Associated With the Severity of Kidney Injury and Poor Prognosis of Patients With COVID-19. Kidney International Reports. 2021;6(12):2979–2992. doi: 10.1016/j.ekir.2021.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katagiri D, Ishikane M, Asai Y, et al. Evaluation of Coronavirus Disease 2019 Severity Using Urine Biomarkers. Crit Care Explor. Aug 2020;2(8):e0170. doi:10.1097/cce.0000000000000170 [DOI] [PMC free article] [PubMed]
- 18.Rodrigues T.S., de Sá K.S.G., Ishimoto A.Y., et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J Exp Med. Mar 1. 2021;218(3) doi: 10.1084/jem.20201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menez S, Moledina DG, Thiessen-Philbrook H, et al. Prognostic Significance of Urinary Biomarkers in Patients Hospitalized With COVID-19. American Journal of Kidney Diseases. 2022;79(2):257-267.e1. doi:10.1053/j.ajkd.2021.09.008 [DOI] [PMC free article] [PubMed]
- 20.Parikh C.R., Mishra J., Thiessen-Philbrook H., et al. Urinary IL-18 is an early predictive biomarker of acute kidney injury after cardiac surgery. Kidney Int. Jul. 2006;70(1):199–203. doi: 10.1038/sj.ki.5001527. [DOI] [PubMed] [Google Scholar]
- 21.Belcher J.M., Garcia-Tsao G., Sanyal A.J., et al. Urinary biomarkers and progression of AKI in patients with cirrhosis. Clin J Am Soc Nephrol. Nov 7. 2014;9(11):1857–1867. doi: 10.2215/cjn.09430913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coca SG, Garg AX, Thiessen-Philbrook H, et al. Urinary biomarkers of AKI and mortality 3 years after cardiac surgery. Journal of the American Society of Nephrology : JASN. 2014;25(5):1063-1071. doi:10.1681/ASN.2013070742 [doi] [DOI] [PMC free article] [PubMed]
- 23.Menez S., Moledina D.G., Garg A.X., et al. Results from the TRIBE-AKI Study found associations between post-operative blood biomarkers and risk of chronic kidney disease after cardiac surgery. Kidney International. 2021;99(3):716–724. doi: 10.1016/j.kint.2020.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puthumana J, Thiessen-Philbrook H, Xu L, et al. Biomarkers of inflammation and repair in kidney disease progression. J Clin Invest. 2021;131(3):e139927. doi:10.1172/jci139927 [DOI] [PMC free article] [PubMed]
- 25.Elixhauser A., Steiner C., Harris D.R., Coffey R.M. Comorbidity measures for use with administrative data. Med Care. Jan. 1998;36(1):8–27. doi: 10.1097/00005650-199801000-00004. [DOI] [PubMed] [Google Scholar]
- 26.Haase-Fielitz A., Haase M., Devarajan P. Neutrophil gelatinase-associated lipocalin as a biomarker of acute kidney injury: a critical evaluation of current status. Ann Clin Biochem. May. 2014;51(Pt 3):335–351. doi: 10.1177/0004563214521795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moledina D.G., Isguven S., McArthur E., et al. Plasma Monocyte Chemotactic Protein-1 Is Associated With Acute Kidney Injury and Death After Cardiac Operations. Ann Thorac Surg. Aug. 2017;104(2):613–620. doi: 10.1016/j.athoracsur.2016.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alge JL, Arthur JM. Biomarkers of AKI: a review of mechanistic relevance and potential therapeutic implications. Clinical journal of the American Society of Nephrology : CJASN. 2015;10(1):147-155. doi:10.2215/CJN.12191213 [doi] [DOI] [PMC free article] [PubMed]
- 29.Menez S, Parikh CR. Assessing the health of the nephron in acute kidney injury: biomarkers of kidney function and injury. Curr Opin Nephrol Hypertens. Nov 2019;28(6):560-566. doi:10.1097/mnh.0000000000000538 [DOI] [PMC free article] [PubMed]
- 30.Parikh C.R., Liu C., Mor M.K., et al. Kidney Biomarkers of Injury and Repair as Predictors of Contrast-Associated AKI: A Substudy of the PRESERVE Trial. Am J Kidney Dis. Feb. 2020;75(2):187–194. doi: 10.1053/j.ajkd.2019.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levey A.S., Stevens L.A., Schmid C.H., et al. A new equation to estimate glomerular filtration rate. Annals of Internal Medicine. 2009;150(9):604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siew E.D., Matheny M.E. Choice of Reference Serum Creatinine in Defining Acute Kidney Injury. Nephron. 2015;131(2):107–112. doi: 10.1159/000439144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kidney Precision Medicine Project. Kidney Tissue Atlas. Accessed December 18, 2021, https://atlas.kpmp.org/
- 34.World Health Organization. Living guidance for clinical management of COVID-19. Accessed 8/3/2021, https://www.who.int/publications/i/item/WHO-2019-nCoV-clinical-2021-2
- 35.Bansal A, Heagerty PJ. A Tutorial on Evaluating the Time-Varying Discrimination Accuracy of Survival Models Used in Dynamic Decision Making. Med Decis Making. Nov 2018;38(8):904-916. doi:10.1177/0272989x18801312 [DOI] [PMC free article] [PubMed]
- 36.Myśliwiec J, Kretowski A, Stepień A, Kinalska I. Serum levels of soluble TNFalpha receptors (sTNFR1 and sTNFR2) during corticosteroid treatment in patients with Graves' ophthalmopathy. Immunol Invest. Feb 2004;33(1):61-68. doi:10.1081/imm-120027685 [DOI] [PubMed]
- 37.Garibaldi B.T., Fiksel J., Muschelli J., et al. Patient Trajectories Among Persons Hospitalized for COVID-19. Annals of Internal Medicine. 2021;174(1):33–41. doi: 10.7326/M20-3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta S., Hayek S.S., Wang W., et al. Factors Associated With Death in Critically Ill Patients With Coronavirus Disease 2019 in the US. JAMA Internal Medicine. 2020;180(11):1436–1447. doi: 10.1001/jamainternmed.2020.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melms J.C., Biermann J., Huang H., et al. A molecular single-cell lung atlas of lethal COVID-19. Nature. Jul 2021;595(7865):114–119. doi: 10.1038/s41586-021-03569-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen G., Goeddel D.V. TNF-R1 signaling: a beautiful pathway. Science. May 31 2002;296(5573):1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 41.Saulnier P.J., Gand E., Ragot S., et al. Association of serum concentration of TNFR1 with all-cause mortality in patients with type 2 diabetes and chronic kidney disease: follow-up of the SURDIAGENE Cohort. Diabetes Care. May. 2014;37(5):1425–1431. doi: 10.2337/dc13-2580. [DOI] [PubMed] [Google Scholar]
- 42.Speeckaert M.M., Speeckaert R., Laute M., Vanholder R., Delanghe J.R. Tumor necrosis factor receptors: biology and therapeutic potential in kidney diseases. Am J Nephrol. 2012;36(3):261–270. doi: 10.1159/000342333. [DOI] [PubMed] [Google Scholar]
- 43.Hawari F.I., Rouhani F.N., Cui X., et al. Release of full-length 55-kDa TNF receptor 1 in exosome-like vesicles: a mechanism for generation of soluble cytokine receptors. Proc Natl Acad Sci U S A. Feb 3. 2004;101(5):1297–1302. doi: 10.1073/pnas.0307981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu C., Chang A., Hack B.K., Eadon M.T., Alper S.L., Cunningham P.N. TNF-mediated damage to glomerular endothelium is an important determinant of acute kidney injury in sepsis. Kidney Int. Jan. 2014;85(1):72–81. doi: 10.1038/ki.2013.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The Human Protein Atlas. Summary - TNFRSF1A. Accessed 1/17/2022, https://www.proteinatlas.org/ENSG00000067182-TNFRSF1A
- 46.Srivastava A., Schmidt I.M., Palsson R., et al. The Associations of Plasma Biomarkers of Inflammation With Histopathologic Lesions, Kidney Disease Progression, and Mortality-The Boston Kidney Biopsy Cohort Study. Kidney Int Rep. Mar. 2021;6(3):685–694. doi: 10.1016/j.ekir.2020.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.White L.E., Santora R.J., Cui Y., Moore F.A., Hassoun H.T. TNFR1-dependent pulmonary apoptosis during ischemic acute kidney injury. Am J Physiol Lung Cell Mol Physiol. Sep. 2012;303(5):L449–L459. doi: 10.1152/ajplung.00301.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gohda T., Niewczas M.A., Ficociello L.H., et al. Circulating TNF receptors 1 and 2 predict stage 3 CKD in type 1 diabetes. J Am Soc Nephrol. Mar. 2012;23(3):516–524. doi: 10.1681/asn.2011060628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niewczas M.A., Gohda T., Skupien J., et al. Circulating TNF receptors 1 and 2 predict ESRD in type 2 diabetes. J Am Soc Nephrol. Mar. 2012;23(3):507–515. doi: 10.1681/asn.2011060627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sancho Ferrando E, Hanslin K, Hultström M, Larsson A, Frithiof R, Lipcsey M. Soluble TNF receptors predict acute kidney injury and mortality in critically ill COVID-19 patients: A prospective observational study. Cytokine. 2022/01/01/ 2022;149:155727. doi: 10.1016/j.cyto.2021.155727 [DOI] [PMC free article] [PubMed]
- 51.Liu C., Debnath N., Mosoyan G., et al. Systematic Review and Meta-Analysis of Plasma and Urine Biomarkers for CKD Outcomes. Journal of the American Society of Nephrology. 2022;33(9):1657. doi: 10.1681/ASN.2022010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson M, Packington R, Sewell H, et al. Biomarkers During Recovery From AKI and Prediction of Long-term Reductions in Estimated GFR. Am J Kidney Dis. May 2022;79(5):646-656.e1. doi:10.1053/j.ajkd.2021.08.017 [DOI] [PubMed]
- 53.Coca S.G., Vasquez-Rios G., Mansour S.G., et al. Plasma Soluble Tumor Necrosis Factor Receptor Concentrations and Clinical Events After Hospitalization: Findings From the ASSESS-AKI and ARID Studies. American Journal of Kidney Diseases. 2023;81(2):190–200. doi: 10.1053/j.ajkd.2022.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCoy I.E., Hsu J.Y., Bonventre J.V., et al. Acute Kidney Injury Associates with Long-Term Increases in Plasma TNFR1, TNFR2, and KIM-1: Findings from the CRIC Study. J Am Soc Nephrol. Jun. 2022;33(6):1173–1181. doi: 10.1681/asn.2021111453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Müller P, Edelmann-Stephan B, Richter F, Pfizenmaier K, Fischer T. TNFR1 Blockade Rather then TNFR2 Blockade Reduces Chronic Inflammation in JAK2+/V617F Mice. Blood. 2019;134(Supplement_1):2966. doi: 10.1182/blood-2019-127997
- 56.Proudfoot A, Bayliffe A, O’Kane CM, et al. Novel anti-tumour necrosis factor receptor-1 (TNFR1) domain antibody prevents pulmonary inflammation in experimental acute lung injury. Thorax. 2018;73(8):723. doi:10.1136/thoraxjnl-2017-210305 [DOI] [PMC free article] [PubMed]
- 57.Silver SA, Adhikari NK, Bell CM, et al. Nephrologist Follow-Up versus Usual Care after an Acute Kidney Injury Hospitalization (FUSION): A Randomized Controlled Trial. Clin J Am Soc Nephrol. Jul 2021;16(7):1005-1014. doi:10.2215/cjn.17331120 [DOI] [PMC free article] [PubMed]
- 58.Singh G., Hu Y., Jacobs S., et al. Post-Discharge Mortality and Rehospitalization among Participants in a Comprehensive Acute Kidney Injury Rehabilitation Program. Kidney360. 2021;2(9):1424. doi: 10.34067/KID.0003672021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang C., Obeid W., Thiessen-Philbrook H., Parikh C.R. Sample Processing and Stability for Urine Biomarker Studies. J Appl Lab Med. Nov 1. 2021;6(6):1628–1634. doi: 10.1093/jalm/jfab082. [DOI] [PMC free article] [PubMed] [Google Scholar]