Abstract
Introduction
The objective of this study was to examine the evolution of carbapenem-resistant Klebsiella pneumoniae (CRKp) infections and their impact at a tertiary care hospital in South India.
Methods
A comparative analysis of clinical data from two prospective cohorts of patients with CRKp bacteremia (C1, 2014–2015; C2, 2021–2022) was carried out. Antimicrobial susceptibilities and whole genome sequencing (WGS) data of selected isolates were also analyzed.
Results
A total of 181 patients were enrolled in the study, 56 from C1 and 125 from C2. CRKp bacteremia shifted from critically ill patients with neutropenia to others (ICU stay: C1, 73%; C2, 54%; p = 0.02). The overall mortality rate was 50% and the introduction of ceftazidime-avibactam did not change mortality significantly (54% versus 48%; p = 0.49). Oxacillinases (OXA) 232 and 181 were the most common mechanisms of resistance. WGS showed the introduction of New Delhi metallo-β-lactamase-5 (NDM-5), higher genetic diversity, accessory genome content, and plasmid burden, as well as increased convergence of hypervirulence and carbapenem resistance in C2.
Conclusions
CRKp continues to pose a significant clinical threat, despite the introduction of new antibiotics. The study highlights the evolution of resistance and virulence in this pathogen and the impact on patient outcomes in South India, providing valuable information for clinicians and researchers.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40121-023-00803-3.
Keywords: Bacteremia, Carbapenem resistant, Klebsiella pneumoniae, Temporal evolution
Key Summary Points
| South Asia has the highest burden of CRKp infections globally with high fatality rates. The clinical and genomic evolution of CRKp between time periods is not studied so far from this region. |
| Introduction of ceftazidime-avibactam has improved outcomes in many areas of the globe. There is paucity of data evaluating its impact in this region. |
| CRKp bacteremia shifted from critically ill patients with neutropenia to others and pathogen genotype evolved continuously with the acquisition of NDM-5 and rise in hypervirulence between the two cohorts. |
| The overall mortality rate was 50% and the introduction of ceftazidime-avibactam did not result in a decrease in mortality. |
| Combination therapy may offer benefits, particularly in sicker patients. |
Introduction
Klebsiella pneumoniae is an opportunistic pathogen with profound clinical and public health impacts [1]. Its open genome enables continuous acquisition of antimicrobial resistance (AMR) and virulence genes [2]. In 2019, carbapenem-resistant Klebsiella pneumoniae (CRKp) infections contributed to more than 55,000 deaths and was only surpassed by Acinetobacter species as a cause of infection-related mortality [3]. This burden is highest among low- and middle-income countries (LMICs) in South Asia and Sub-Saharan Africa [3].
There are two specific knowledge gaps in current literature on CRKp from South Asia. It is unclear if novel agents such as CAZ-AVI have an advantage over repurposed agents such as polymyxin in the treatment of serious CRKp infections in this region. The newer β-lactam–β-lactamase combinations are associated with reduced mortality in other regions of the globe [4–6]. However, significant geographical differences exist in the genetic mechanisms of carbapenem resistance, the size of the pathogen resistomes, disease severity at presentation, and the outcomes in CRKp infections [7]. It is not clear whether CAZ-AVI improves clinical outcomes in South Asia. Analyzing region-specific prospective data across time periods prior to and after the introduction of CAZ-AVI may aid in clarifying the optimal therapy for such infections in that respective region.
Secondly, the dynamics of pathogen transmission and the genomic changes responsible for the disproportionately large clinical impact seen in South Asia are poorly understood. While the common oversimplified assumption is excessive antibiotic use resulting in increased resistance, emerging data point to a more complex scenario [8]. The acquisition of the ability to excessively thicken the capsule has increased the virulence of the hypermucoviscous pathotype of CRKp seen in South Asia [9]. Further, the spread in LMICs is driven by multiple closely related clones rather than a single dominant one [10]. It is unclear whether these small outbreaks are caused by infection from multiple persistent environmental point sources or by healthcare-worker-related transmissions rather than endogenous acquisition secondary to antibiotic pressure.
We analyzed two prospective cohorts of patients with CRKp bacteremia to examine the clinical and genomic evolution of CRKp over time to understand the impact of the introduction of CAZ-AVI on CRKp infections and the genetic changes that may have contributed to the increased mortality from CRKp infections in South Asia.
Methods
Study Design and Participants
We carried out a comparison of two prospective cohort studies among consecutive patients admitted at a 3500-bed academic center, with a bloodstream infection (BSI) confirmed in the laboratory as being caused by CRKp between June 2014 and June 2015 (clinical cohort 1 or C1) and March 2021 and March 2022 (clinical cohort 2 or C2). Patients with confirmed polymicrobial bacteremia and those under 15 years of age were excluded from the study. Only the first qualifying episode of CRKp bacteremia during the hospital stay of an individual patient was included. If CRKp bacteremia recurred within 30 days of follow-up, subsequent episodes were not included. This study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board and Ethics Committee of Christian Medical College, Vellore (IRB no. 9041/2014 and IRB no. 13464/2020). Before recruitment into the study, informed consent was obtained from all the patients or immediate kin for accessing clinical information and publishing data.
Procedures
Patients fulfilling the eligibility criteria were enrolled and followed up for 30 days. The investigators collected relevant demographic and infection-specific information. Using annual hospital data, the prevalence of CRKp was established among all patients with Klebsiella and Enterobacterales infections. Management decisions, including the antibiotics used and source control, were made by individual treating physicians. The infection control practices in the institution are maintained by the Hospital Infection Control Committee (HICC). All Carbapenem Resistant Gram Negative Blood Stream Infections (CRGNBSIs) are reported to the HICC on a real-time basis, and outbreak investigations are conducted wherever relevant. The HICC has protocols for management and infection control of drug-resistant pathogens and compliance to these protocols are regularly monitored and audited.
Definitions
Carbapenem resistance was defined as antibiotic resistance to either meropenem or imipenem as per the Clinical and Laboratory Standards Institute (CLSI) guidelines during the respective cohort periods. To be considered persistent bacteremia, a minimum of two blood cultures that were isolated during the same infectious episode on calendar days at least 48 h apart from each other were required to be positive.
Severe neutropenia was defined as an absolute neutrophil count of less than 500 cells/µL at the time of or within 48 h before blood culture positivity [11]. Serious bacterial infections, such as pneumonia, bacteremia, and meningitis, found 30 days before the day of CRKp isolation, were considered previous bacterial infections. Concomitant bacterial infections were defined as any serious bacterial infection other than the index CRKp bacteremia identified within the 30 days following CRKp isolation. We defined catheter-related bloodstream infection (CRBSI) in presence of an intravascular device with > 1 positive blood culture from a peripheral vein and no other reliable sources of infection with catheter blood culture positive at least 2 h earlier than the blood culture drawn peripherally at the same time. We concluded gut translocation as the source of bacteremia among patients with neutropenia with no clear other sources of infection and used the term “probable gut translocation” to label it.
The Pitt bacteremia score (PBS) was calculated on the basis of the highest values obtained for each parameter 48 h before or on the day of bacteremia. The INCREMENT score was calculated using the presence of septic shock or severe sepsis, a Pitt score of 6 or higher, a Charlson Comorbidity Index (CCI) of 2 or more, and a source of bacteremia other than urinary or biliary sources. INCREMENT scores ranged from 0 to 15; scores of less than 8 and 8 or higher were considered low and high risk for mortality, respectively [12–14].
All patients received antibiotics at standard recommended doses as defined in the antibiotic policy of the institution during the respective time periods, and the route of administration was intravenous. Patients were considered to have received appropriate therapy if the bacteremia was treated with drugs to which the isolate did not have reported resistance. Appropriateness of antibacterial treatment reported in the study is of definitive antibacterial treatment and not empirical therapy. Monotherapy was defined as treatment with a single agent with established in vitro susceptibility. The term combination therapy was used when two or more agents with documented in vitro susceptibility were used for treatment.
Clinical Outcomes
The primary outcome was 30-day all-cause mortality. Participants who were discharged within 30 days were telephoned on day 30 by the investigators to assess outcomes. Secondary outcomes included 14-day mortality, presence of septic shock, and requirement of intensive care or dialysis. We compared the antibiotic therapy received and its associations with 14-day and 30-day mortality.
Isolate Identification and Antimicrobial Susceptibility Testing
All CRKp isolates were identified from blood cultures using VITEK-MS (Biomerieux). The isolates were subjected to disk diffusion for a panel of antimicrobials consisting of ceftazidime, cefepime, piperacillin-tazobactam, meropenem, imipenem, gentamicin, amikacin, and ciprofloxacin. Results were interpreted according to the CLSI guidelines during the respective time period. Escherichia coli ATCC 25922, Enterococcus faecium ATCC 29212, and Pseudomonas aeruginosa ATCC 27853 were used as controls. Susceptibility to colistin, tigecycline, ceftazidime-avibactam, and aztreonam were determined using broth microdilution (BMD) and interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and Food and Drug Administration (FDA) guidelines, respectively. Due to issues in reviving some of the earlier cultures, we were not able to retest isolates to keep the methodologies uniform. The MICs of CAZ-AVI and aztreonam were independently determined by BMD. The susceptibility to combination of CAZ-AVI and aztreonam was determined using checkerboard assay as previously described [15]. To determine the synergy, CAZ-AVI concentration ranging from 0.12 to 8 μg/ml was tested with a fixed concentration of aztreonam of 8 μg/ml. Synergy was defined when the cumulative fractional inhibitory concentration (FIC) index of both agents was ≤ 0.5 [15].
Molecular Characterization
By convenience sampling, depending on the availability and viability of stored isolates, 90 bacteremic CRKp isolates that reflected the study periods were selected for genomic characterization; 43 isolates were from February 2014 to December 2016 (genomic cohort 1 or G1), 47 were from March 2020 to December 2021 (genomic cohort 2 or G2), and 41 isolates were from patients included in the clinical study. Genomic characterization of the isolates, including WGS, was performed using standard techniques as enumerated in the Supplementary Material.
Statistical Analysis
Clinical and microbiological data were compared between individuals enrolled in C1 and C2. Baseline characteristics and clinical events for individuals in both cohorts were presented as medians with interquartile ranges (IQR) for continuous variables and frequencies with proportions for categorical variables using appropriate statistical tests. A univariate analysis was performed for each variable, with 30-day mortality as the outcome. Hazard ratio (HR) with a 95% confidence interval (CI) was estimated, and a p-value of less than 0.05 was considered significant.
Multivariate analysis was done for factors that were significant in the overall estimate using Cox hazard regression analysis, with the Breslow method used for ties after checking for multicollinearity using the variance inflation factor (VIF). The adjusted hazard ratio (aHR) was estimated with a 95% CI. To visualize all-cause mortality within 30 days and 14 days of bacteremia among patients who received polymyxin or CAZ-AVI as monotherapy or both agents together as a combination, Kaplan–Meier survival estimates were calculated. A log-rank test was performed to determine the equality of survivor functions. Censoring was not required as all study participants were followed up until death or 30 days. We used STATA 16 (StataCorp. 2019, Stata Statistical Software: Release 16. College Station, TX: StataCorp LLC) for statistical analysis.
Results
Patient Population
We enrolled 181 patients with laboratory-confirmed CRKp bacteremia: 56 patients from 2014 to 2015 (C1) and 125 patients from 2021 to 2022 (C2). The median ages of participants in C1 and C2 were 43 years (IQR 20–67 years) and 44 years (IQR 18–69 years), respectively. In both the cohorts, 66% of patients were male. Location within the hospital where patients developed CRKp bacteremia was significantly different between cohorts. Almost three-quarters were in critical care units in C1, but in C2 the infections were distributed widely across the hospital, with just over half in the critical care unit (73% versus 54%; p = 0.02) (Table 1). Underlying hematological conditions were more frequently observed in C1 than in C2 (57% versus 32%, p = 0.001) (Table S1a). Consequently, severe neutropenia was also more common in C1 than in C2 (48% versus 28%, p = 0.01). CRBSI was the most common source of bacteremia in both cohorts (36% and 26%, respectively) followed by gut translocation (16% and 21%, respectively) (Table 1). In all patients, CRKp bacteremia occurred when hospitalized or after recent close hospital contact. Among all Gram-negative bloodstream infections, the prevalence of CRKp bacteremia significantly increased from 5% (98/1824) between January and December 2015 to 14% (591/2010) between January and December 2021 (p-value = < 0.001).
Table 1.
Demographic and clinical details of study participants
| Characteristics | Total n = 181 (%) |
Cohort C1 n = 56 (%) |
Cohort C2 n = 125 (%) |
p-value |
|---|---|---|---|---|
| Age, > 50 years age | 63 (35) | 19 (34) | 44 (35) | 0.87 |
| Gender, male | 120 (66) | 37 (66) | 83 (66) | 0.97 |
| Location at the time of bacteremia, ICU | 109 (60) | 41 (73) | 68 (54) | 0.02 |
| Presence of neutropenia | 62 (34) | 27 (48) | 35 (28) | 0.01 |
| Surgery within 30 days before bacteremia | 38 (21) | 5 (9) | 33 (26) | 0.01 |
| Previous exposure to colistin | 74 (41) | 27 (48) | 47 (38) | 0.18 |
| Previous exposure to carbapenem | 135 (75) | 44 (79) | 91 (73) | 0.41 |
| Infection details | ||||
| Previous bacterial infections | 90 (50) | 28 (50) | 62 (50) | 0.96 |
| Concomitant bacterial infections | 102 (56) | 32 (57) | 70 (56) | 0.89 |
| Source of infection | ||||
| CRBSI | 52 (29) | 20 (36) | 32 (26) | |
| Gut translocation | 35 (19) | 9 (16) | 26 (21) | 0.38 |
| Lung | 32 (18) | 7 (13) | 25 (20) | |
| Othersa | 62 (34) | 20 (36) | 42 (34) | |
| Presence of septic shock | 121 (67) | 40 (7) | 81 (65) | 0.38 |
| Dialysis | 41 (23) | 17 (30) | 24 (19) | 0.10 |
| ICU stay | 130 (72) | 53 (95) | 77 (62) | < 0.001 |
| Organism details | ||||
| Colistin resistance (n = 173 tested) | 27 (15.61) | 6 (10.71) | 21 (18) | 0.22 |
| Scores for prediction of mortality (median and IQR) | ||||
| Pitt bacteremia score | 2 (0–6) | 2 (0–6) | 2 (0–4) | 0.38 |
| Charlson Comorbidity Index | 2 (1–3) | 2 (0.5–3) | 2 (1–3) | 0.79 |
| INCREMENT score | 8 (6–12) | 11 (6–12) | 8 (6–11) | 0.01 |
| Increment score: high mortality (8–15) | 114 (62.9) | 41 (73.21) | 73 (58.4) | 0.06 |
| Treatment details | ||||
| Polymyxin or CAZ-AVI therapy (n = 128) | ||||
| Polymyxin or CAZ-AVI as monotherapy | 92 (72) | 37 (100) | 55 (60) | < 0.001 |
| Polymyxin and CAZ-AVI as combination | 36 (28) | 0 | 36 (40) | |
| Outcome assessment | ||||
| 30-day mortality | 90 (50) | 30 (54) | 60 (48) | 0.49 |
| 14-day mortality | 66 (37) | 22 (39) | 44 (35) | 0.59 |
ICU intensive care unit, CRBSI catheter-related bloodstream infection, CAZ-AVI ceftazidime-avibactam
aOther sources of infection included hepatobiliary, renal, skin and soft tissue, and peritoneal infections
Severity Scores
The median PBS and CCI score were similar in both time periods, while the median INCREMENT score was higher in C1 than in C2 (11 versus 8, p = 0.01). When stratified, among those in C1, 73% had an INCREMENT score greater than 8 (indicating a high risk of mortality), while only 58% had a score greater than 8 in C2 (Table 1).
Antimicrobial Susceptibilities
All 181 isolates (56 from C1 and 125 from C2) were resistant to ceftazidime, cefepime, meropenem, cefoperazone-sulbactam, and ciprofloxacin. Overall, 3 (2%) and 12 (7%) CRKp isolates retained susceptibility to gentamicin and amikacin, respectively, in C1 and C2. Colistin resistance, as determined by disk diffusion was 11% (6/56) in C1 and 18% (21/117) in C2, as determined by BMD. Tigecycline susceptibility fell from 88% (49/56) to 66% (49/74) between the cohorts (p = 0.007) (disk diffusion was performed for C1; BMD was performed for C2). In C2, 50% of the 68 isolates tested retained susceptibility to minocycline. Minocycline susceptibility was not available for isolates in C1. All tested isolates in C2 (100/100) were susceptible to CAZ-AVI (OXA48-like producers retain susceptibility to CAZ-AVI, while NDM producers were susceptible to CAZ-AVI in combination with aztreonam).
Details of Therapy
Combination therapy was used to manage CRKp bacteremia in 42% of patients in C2 and 18% of patients in C1. All patients in C1 received appropriate therapy, while 14% of patients in C2 did not receive appropriate therapy (p < 0.001). Patients in C1 were predominantly treated with colistin or polymyxin (75%), either as monotherapies or in combination with other adjunctive antibiotics. CAZ-AVI was introduced in the Indian setting in June 2019, after C1. In C2, 53% of patients received CAZ-AVI, while others (47%) received colistin with or without other adjunctive agents (n = 107; Table S1a). No patient in the study received monotherapy with aminoglycosides.
Primary Outcomes and Associations
Patients included in this study were very ill, with 60% of study participants in ICU at the time of bacterial isolation and a median INCREMENT score of 8 (IQR 6–12). Overall, 30-day mortality among the participants was 50% and not significantly (p = 0.49) different between cohort 1 (54%) and cohort 2 (48%). The 14-day mortality was 37% (39% in C1; 35% in C2; p = 0.59). The risk factors for 30-day mortality among the study population are summarized in Table 2 and Table S1b. Univariate analyses showed that age more than 50 years, presence of septic shock, admission to an intensive care unit at the time of bacterial isolation or during the 30-day follow-up period, requirement of dialysis, previous bacterial infections, pulmonary source of infection, mortality prediction/severity scores, and INCREMENT score of 8 or higher were all significantly associated with the risk of mortality (p < 0.05).
Table 2.
Risk factors for 30-day mortality among study participants
| Characteristics | Outcome | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|---|
| Alive n = 91 (%) |
Deceased n = 90 (%) |
Hazard ratio (95% CI) | p-value | Adjusted hazard ratio (95% CI) | p-value | |
| Demographic details | ||||||
| Age, > 50 years | 26 (29) | 37 (41) | 1.57 (1.03, 2.4) | 0.03 | 1.27 (0.80, 2.01) | 0.31 |
| Gender, male | 63 (69) | 57 (63) | 0.78 (0.51, 1.20) | 0.26 | – | |
| Location at the time of bacteremia, ICU | 41 (45) | 68 (76) | 2.69 (1.66, 4.36) | < 0.001 | – | |
| Presence of neutropenia | 33 (36.26) | 29 (32.22) | 0.93 (0.60, 1.44) | 0.73 | – | |
| Infection details | ||||||
| Previous bacterial infections | 37 (41) | 53 (59) | 1.58 (1.04, 2.40) | 0.03 | 1.47 (0.92, 2.36) | 0.11 |
| Source of infection | ||||||
| CRBSI | 30 (33) | 22 (24) | 0.83 (0.48, 1.43) | 0.50 | ||
| Lung | 11 (12) | 21 (23) | 1.78 (1.02, 3.11) | 0.04 | 1.24 (0.69, 2.23) | 0.47 |
| Gut translocation | 19 (21) | 16 (18) | 0.98 (0.54, 1.80) | 0.96 | ||
| Othersa | 31 (34) | 31 (34) | Ref | |||
| Presence of septic shock | 41 (45) | 80 (89) | 5.85 (3.02, 11.31) | < 0.001 | 4.63 (2.22, 9.66) | < 0.001 |
| Dialysis | 13 (14) | 28 (31) | 1.65 (1.06, 2.58) | 0.03 | 1.14 (0.69, 1.89) | 0.61 |
| ICU stay | 54 (59) | 76 (84) | 2.74 (1.55, 4.84) | 0.001 | 1.41 (0.70, 2.84) | 0.33 |
| Organism details | ||||||
| Colistin resistance | 7 (8) | 20 (24) | 2.18 (1.31, 3.63) | 0.003 | 1.43 (0.83, 2.46) | 0.19 |
| Scores for prediction of mortality (median and IQR) | ||||||
| Pitt bacteremia score | 1 (0–2) | 4 (1–6) | 1.31 (1.21, 1.41) | < 0.001 | – | |
| Charlson Comorbidity Index | 2 (1–2) | 2 (1–4) | 1.12 (1.01, 1.24) | 0.03 | – | |
| Increment score: high mortality (8–15) | 36 (40) | 78 (87) | 5.57 (3.02, 10.25) | < 0.001 | – | |
| Treatment details: polymyxin/CAZ-AVI therapy | ||||||
| Polymyxin and CAZ-AVI as combination | 23 (34) | 13 (22) | 0.63 (0.34, 1.17) | 0.146 | – | |
CI confidence interval, ICU intensive care unit, CRBSI catheter-related bloodstream infection, CAZ-AVI ceftazidime-avibactam
aOther sources include hepatobiliary, renal, skin and soft tissue infections and peritoneal infections
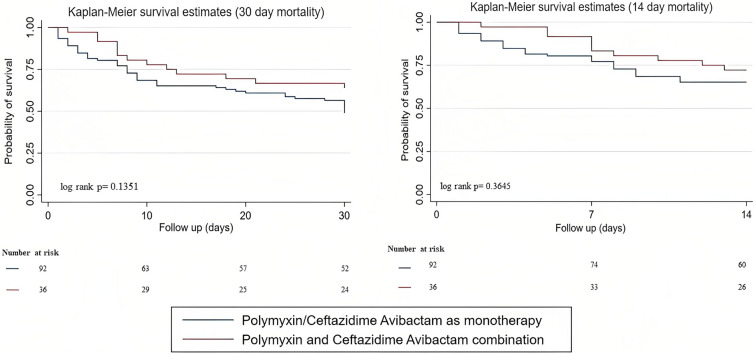
INCREMENT score greater than 8 and ICU stay at bacteremia were not included in multivariate analysis due to clinical correlation with septic shock and ICU stay during follow-up. The mean VIF for the chosen factors in the Cox multivariate model was 1.38 with no multicollinearity. The presence of septic shock was significantly associated with 30-day mortality on multivariate analysis (aHR 4.63; 95% CI 2.22–9.66; p < 0.001) (Table 2). Kaplan–Meier survival estimates, calculated for mortality at 14 days and 30 days, revealed a trend toward better survival among patients treated with a combination of polymyxin and CAZ-AVI than among those treated with either agent as monotherapy (72% versus 65% and 64% versus 49%, respectively), without statistical significance (p = 0.36 and p = 0.13, respectively) (Fig. 1).
Fig. 1.
Survival curves, among patients who received polymyxin or ceftazidime-avibactam as monotherapy or combination therapy, at 30 days and 14 days
Genomic Characteristics
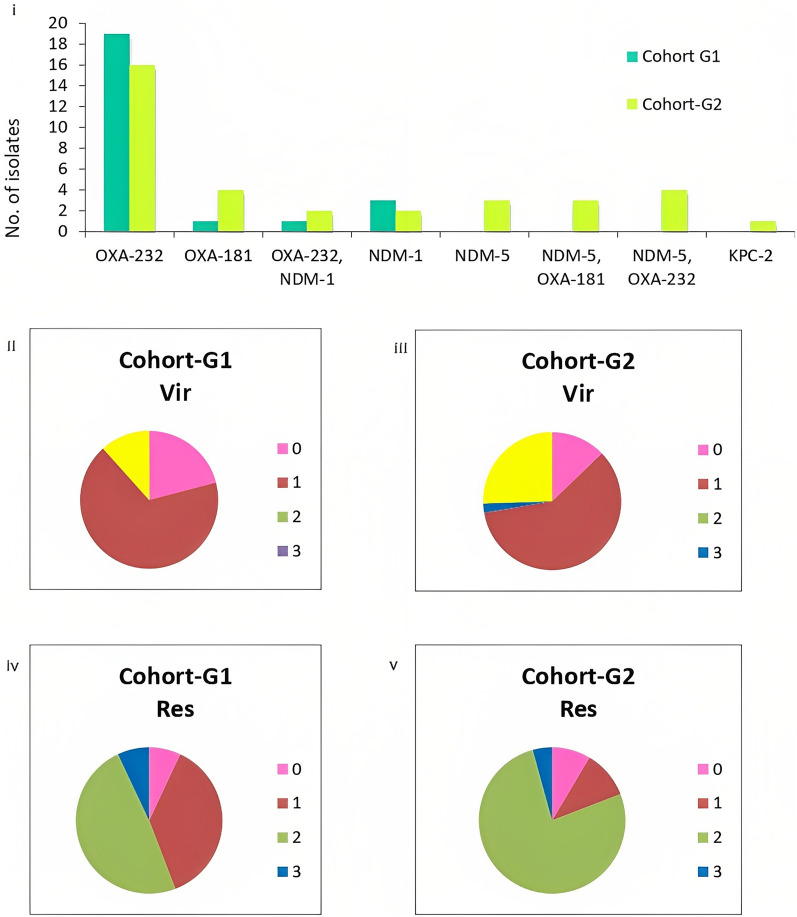
Of the 90 CRKp bacteremia isolates included for genomic characterization, 43 were collected between February 2014 and December 2016 (G1) and 47 between March 2020 and December 2021 (G2). A single isolate from G2 was identified as K. quasipneumoniae. Carbapenem resistance was mediated by carbapenemases in 69% of isolates (68/90) with OXA-232 being the commonest [39% (35/90)] (Fig. 2i). The number of CRKp isolates that had NDM-1 genes were similar in both datasets (G1, 4/43; G2, 4/47). Isolates with NDM-5 (10/47) were found in G2 (p = 0.001) either alone (3/47) or in combination with OXA-232 (4/47) and OXA-181 (3/47). Klebsiella pneumoniae carbapenamases was only present in one isolate of CRKp in G2. In addition to carbapenemases, other mechanisms of carbapenem resistance, including porins such as the Klebsiella pneumoniae outer membrane porin 35 (OmpK35) alterations, were observed among 72% of isolates (31/43) in G1 and 47% of isolates (22/47) in G2 (p = 0.015). OmpK36 alterations (truncation, OmpK36GD, and OmpK36TD) increased from 31% (17/43) in G1 to 53% (25/47) in G2 (p = 0.19).
Fig. 2.
Resistance and virulence profiles of carbapenem-resistant K. pneumoniae in the study. (i) Carbapenemase profile in genomic dataset 1 and 2; (ii–v) virulence and resistance scores of genomes as assigned by Kleborate. Vir virulence score, Res Resistance score. Virulence score: 0, no yersinabactin, colibactin, or aerobactin; 1, yersiniabactin only; 2, yersiniabactin and colibactin (or colibactin only); 3, aerobactin without yersiniabactin or colibactin; 4, aerobactin with yersiniabactin (no colibactin). Resistance score: 1, ESBL; 2, carbapenemase; 3, carbapenemase plus colistin resistance; 0, otherwise
Isolates were classified as carbapenem-resistant hypervirulent Klebsiella pneumoniae (CRhvKp) if they carried truncated rmpA2 gene and/or aerobactin with a virulence score of 4 on Kleborate. CRhvKp isolates were more frequent in G2 than in G1 (12/47 versus 5/43, p = 0.11) (Fig. 2ii–v). Notably, sequence type 2096 (ST2096), a single locus variant of ST14, emerged in G2 (n = 6) and was a predominant contributor to the convergence of resistance and virulence on a single large plasmid.
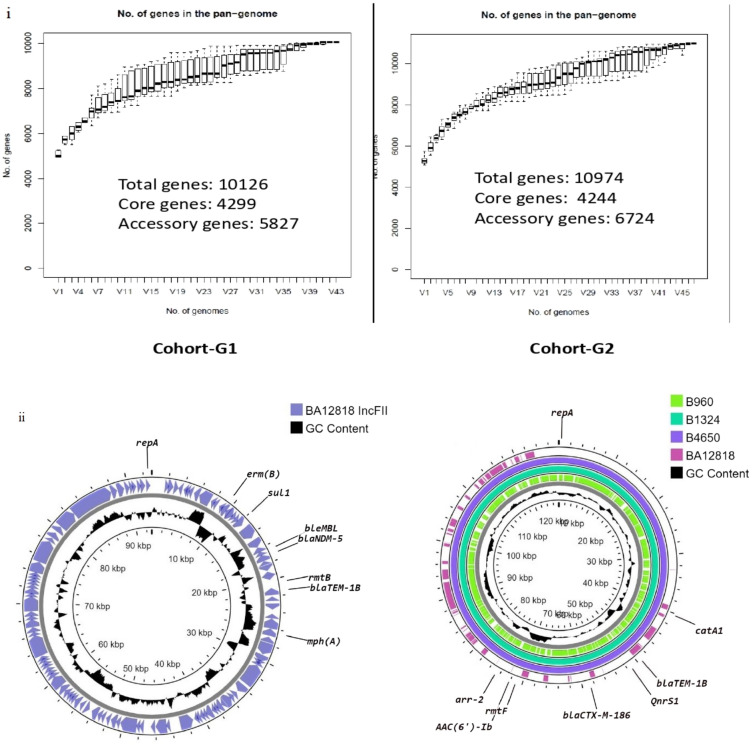
Genomic diversity of CRKp was marginally higher in G2, with 14 diverse clones, than in G1, which had 11 clones (p = 0.66). The Simpson’s Diversity Index was 0.91 and 0.81 in G2 and G1, respectively (Table 3). A significant increase in the accessory genome content (p < 0.0001, Fig. 3i) and higher plasmid burden (5 per genome versus 4 per genome) were also seen in G2 as compared with G1. The structure of IncFII plasmid carrying blaNDM-5 gene in G2 and the corresponding IncFII-IncFIB plasmid in G1 are shown in Fig. 3ii
Table 3.
Genome characteristics of similar genotypes of K. pneumoniae included in the study
| Sequence type (ST) | Cohorts | Capsular K antigen | O-antigen | Antimicrobial resistance (AMR) genes | Porin mutations | Plasmids | Virulence | Inference |
|---|---|---|---|---|---|---|---|---|
| ST15 | G1 (n = 7) | KL24 | O1/O2v1 | aac(6')-Ib-cr, aph3-Ia, blaOXA-1, blaTEM-1D, blaSHV-28, blaCTX-M-15, mphA, tetA, catB4 | None | IncFIIK, IncFIBK, ColpVC, Col156 | Absent | Diversity in K antigen, change in plasmid profile, addition of virulence genes and carbapenemases in G2 |
| G2 (n = 7) | KL2 (n = 1), KL19 (n = 1) KL24 (n = 1), KL112 (n = 5) | O1v1 | rmtF, strA, strB, blaOXA-1, blaTEM-1D, blaSHV-28, blaCTX-M-15, blaNDM-1, blaOXA-232, blaCMY, qnrB1, mphA, sul1, sul2, catA1, dfrA14 |
OmpK35, 17% OmpK36TD |
IncFIB(pQil), IncFIB, pVir, IncHI1B 7(pNDM_MAR), IncC, ColRNAI, ColKP3, Col440I | ybt 16; ICEKp12; single isolate carried aerobactin and rmpA2 | ||
| ST16 | G1 (n = 4) | KL51 | O3b |
aac(6')-Ib, aac(6')-Ib; aac(6')-Ib-cr, aadA, rmtF, mphA, blaDHA-1, blaOXA-1, blaSHV, blaOXA-9, blaTEM-1D, arr-2, arr-3, sul1, catB3, dfrA1 |
OmpK35, 22% OmpK36TD |
IncFIBK, IncR, IncFIIK, IncFII, Col440II | ybt 9; ICEKp3 | Diversity in K and O antigens, change in ybt, addition of IncX3 and FIA plasmids carrying OXA-181, NDM-5, and other ARGs |
| G2 (n = 6) | KL81 | OL101 | aac(3)-IIa; aac(6')-Ib-cr; strA, strB, rmtB, aadA2, blaSHV, blaOXA-1, blaTEM1D, blaCTX-M-15, blaNDM-5, blaOXA-181, sul1, sul2, qnrS1, mphA, catA, arr-2, dfrA1, dfrA12 |
OmpK35, 6% OmpK36TD |
IncFIBK, IncR, InFIIK, IncFII, IncX3, FIA, Col | ybt 16; ICEKp12 | ||
| ST231 | G1 (n = 4) | KL51 | O1/O2v2 | aac(6')-Ib, rmtF, blaSHV, blaTEM1D, blaCTX-M-15, blaOXA-232, qnrS1, catA, arr-2 |
ompK35, 30% ompK36TD |
IncFIB(pQil), IncFIIK, Col440I, ColKP3 | ybt 14; ICEKp5 | Diversity in K and O antigens, addition of IncFIA, NDM-5, and co-carried ARGs in G2; aerobactin acquired, diversity in ybt |
| G2 (n = 6) | KL51, KL64 | O1v1/v2 | aac(6')-Ib, aadA2, rmtF, blaSHV-1, blaTEM-1D, blaCTX-M-15, blaNDM-5, blaOXA-232, qnrS1, sul1, arr-2, ermB, mphA, catA1, dfrA12 | IncFIB(pQil), IncFIIK, Col440I, ColKP3, IncFIA, ColRNAI |
Aerobactin (n = 3); ybt 14, ICEKp5 (n = 3) ybt 9, ICEKp3 (n = 1) |
|||
| ST14 | G1 (n = 4) | KL2, KL64 | O1/O2v1 | aadA2;aph(3')-VI;armA;sat-2, strA;strB blaSHV-28, blaTEM-1D, blaCTX-M-15, blaNDM-1, blaOXA-232, blaOXA-1, blaOXA-9, mgrB truncated, ermB;mphA; mphE; msrE, sul1, sul2, catB4, catA1, tetD, dfrA1, dfrA12, dfrA14 |
OmpK35, 70% OmpK36, 45% OmpK36GD |
IncFIB(pNDM-Mar), IncFIB(K), IncHI1B(pNDM-MAR), ColKP3, IncFII, IncR, FIA |
ybt 14, ICEKp5 (truncated) | Change in AMEs, addition of OXA-181, shift from NDM-1 to NDM-5. Addition of IncFIIK carrying NDM-5 and other ARGs in G2 |
| G2 (n = 3) | O1v1 | aac(3)-IIa.;aac(3)-IId; aadA; sat-2; strA; strB, rmtB, blaSHV-28, blaTEM-1D, blaCTX-M-15, blaNDM-5, blaOXA-232, blaOXA-1, blaOXA-181, catA1;cmlA5, ereA; ermB; mphA; mphE; msrE, arr-2, sul-1, sul1, tetD, dfrA1;dfrA12;dfrA14 | OmpK36GD | IncFIB(pNDM-MAR), IncFIB(K), IncHI1B(pNDM-MAR), ColKP3, IncFII, IncR, IncFIIK, FIA | ybt 14, ICEKp5 |
ARGs antimicrobial resistance genes, AMEs aminoglycoside modifying enzyme
Fig. 3.
Structure of IncFII plasmids among carbapenem resistant K. pneumoniae. (i) Genome statistics of carbapenem-resistant K. pneumoniae genomic dataset 2 has a higher average number of genes per genome and higher accessory genome. Prevalence of core and accessory genes was inferred from Roary. (ii) IncFII plasmid of BA12818 carrying blaNDM-5 from genomic dataset 2 with accession number CP054171. Other antimicrobial resistance genes are co-carried on the same plasmid. IncFIIK-IncFIB (pQil) hybrid plasmid from cohort A carrying antimicrobial resistance genes. B960 (CP070411), B1324 (CP072403), and B4650 (CP072409) are from genomic dataset 1, which lack blaNDM, while BA12818, from genomic dataset 2, carrying blaNDM-5, is significantly different from the earlier plasmid
Over the 7-year study period, we found some persistent clones in each cohort that differed from each other by less than 25 single nucleotide polymorphisms (SNPs). CRKp ST15 showed two clusters: one was seen among three G-1 CRKp isolates from different wards (February–November 2016) and varied by 2–4 SNPs. The other was seen in four CRKp isolates in G2 that were distributed among hematology patients (January–October 2021) and varied from each other by 1–4 SNPs. CRKp ST231 was seen in ten isolates in G1 (February 2014–May 2015). They were closely related and varied by 0–20 SNPs, indicating that they were persistent within the hospital. The three CRKp ST16 isolates from G1 were identical (0 SNPs) and were distributed among diverse wards (December 2014–February 2015). The three CRKp ST16 isolates from G2 (May–October 2021) were closely related and scattered across different wards (Fig. S2).
Discussion
The proportion of CRKp among Gram-negative bacteremia has increased by nearly threefold (5–14%) between the two cohorts in 2015 and 2021. Despite the introduction of novel agents, mortality has remained unchanged in the two periods (54% versus 48%, p = 0.49). Increase in colistin resistance in C2 was associated with a precipitous reduction (p = 0.007) in susceptibility to tigecycline. This study highlights that the burden of CRKp is escalating, and that despite the introduction of novel agents, its clinical impact is sustained and the therapeutic options for clinicians in India are shrinking. We also show that the burden of CRKp bacteremia is mediated by varied clones with significant acquisition of AMR plasmids and emergence of convergence phenomenon among the isolates.
CRKp bacteremia was more common in the second period (n = 125 versus 56) in our study. This was associated with two significant shifts in the hospital epidemiology. Firstly, the number of patients with severe neutropenia significantly dropped from 48% in C1 to 28% in C2 (p = 0.01). Secondly, fewer patients were located in the critical care units in the second cohort (C1 74% versus C2 54%). This shows shift of CRKP bacteremia epidemiology from predominantly critically ill patients with neutropenia to widespread dissemination across the hospital wards. We propose that critically ill patients with neutropenia acted as reservoirs for these pathogens, enabling the subsequent spread. In addition, patients in C2 with CRKp had new risk factors, including chronic liver disease and recent surgery. Recent data support severe immunosuppression, including neutropenia, to promote antimicrobial resistance [16–18]. Significant antibiotic pressure in these patients and immunosuppression promotes the survival of persistent bacteria, enabling the development of multidrug resistance. This association is supported by observations in patient cohorts, animal data, and in other diseases, such as multidrug-resistant tuberculosis [16, 19, 20]. Infections in patients with neutropenia facilitated by high antibiotic pressure possibly contributed to the evolution and spread of CRKp to a more diverse group of patients in our hospital as well.
Unfortunately, the mortality associated with CRKp bacteremia remained very high, despite the introduction of ceftazidime-avibactam in our study. Patients in our study were sicker than those in other large cohorts such as PANORAMA, CRACKLE 2, and INCREMENT [7, 21, 22]. Patients in C1 more frequently required critical care support than those in C2. Ceftazidime-avibactam, with or without aztreonam, is associated with lower mortality than polymyxins across many cohorts with CRKp bacteremia [4–6, 23]. However, we did not observe this in our second cohort despite half of them receiving CAZ-AVI in their treatment. It is unclear whether this is due to the higher proportion of critically ill patients in our cohort, the newly acquired NDM-5, the presence of dual carbapenemase with coexistent porin alterations, the higher burden of colistin resistance, potential inoculum effect, or that a proportion of patients did not receive appropriate therapy in C2.
Though not statistically significant, combination therapy with polymyxin and CAZ-AVI was associated with improved survival, especially among those with high INCREMENT scores (42% versus 34%) (Fig. S3). Multiple observational studies have highlighted the beneficial role of combination therapy in the management of high-risk patients with CRKp infections [22, 24–26]. Polymyxins with carbapenems are the most common combination agent used, especially when the meropenem minimum inhibitory concentration is less than 16 mg/L [25, 27]. In vitro synergy testing with time kill assays have shown synergy with CAZ-AVI and polymyxin combination among Pseudomonas aeruginosa isolates with CAZ-AVI resistance [28]. While we want to be cautious in reporting this observation, as our study had small numbers and hence did not achieve statistical significance, this could be studied further in larger cohorts.
Finally, multiple factors coalesced to produce the large clinical impact in our population. The extremely sick, often immunosuppressed patient cohort with enormous antibiotic pressure sets the stage for endogenous acquisition of these pathogens. This is reflected by the varied pathogen serotypes emerging overtime. Small outbreaks, probably caused by the interplay between healthcare-worker-related and persistent point source transmissions, such as from water sinks, further adds to the complexity. The acquisition of novel carbapenemases, such as NDM-5, increase in porin-mediated changes, and constant increase in the genome size, content, and plasmids reflects the ability of the organism to constantly evolve in the hospital environment. Increased incidence of dual carbapenemases in the later cohort (NDM and OXA 48-like) further limits therapeutic choices, and the addition of aztreonam to CAZ-AVI also escalates the cost [4]. Hypervirulent Klebsiella spp. is endemic in the Indian subcontinent [29, 30]. Plasmid-mediated acquisition of hypervirulence in CRKp also enhances the virulence and invasiveness of the pathogen [31].
While this is the first large study from this region reporting the evolution of CRKp, there are several limitations. This was a single-center study with a relatively small number of patients. The methods of choice for testing sensitivity and the CLSI guidelines changed over the study time periods. Disc diffusion was used in cohort 1 and BMD was used in cohort 2 to determine colistin and tigecycline susceptibility. The above factors may have affected the definitions used in the study. Availability and revivability of isolates were another limitation, and hence molecular analysis was only performed for 41 isolates from the clinical cohort. While it would have been ideal to report CRKp data throughout the period, including years between C1 and C2, study approvals were limited for the above periods and we were not able to provide the same for this paper. To understand the transmission dynamics of CRKp, genomic analyses of all isolates causing infection and colonization would be required. In our study, genomic analyses were only done on some of the isolates.
Conclusions
This study calls attention to several clinical and genomic facets of CRKp in South India in two distinct time periods. There was a shift in patients who developed CRKp bacteremia from patients with neutropenia in the ICUs to ward inpatients with various comorbidities. Patients in both cohorts were critically ill and had high mortality rates. Combination therapy with CAZ-AVI and polymyxin offered the best probability for survival. The genomic analysis underscored unrelenting pathogen evolution both in terms of virulence and resistance. The resistome expanded by the continuous acquisition of plasmids with AMR genes, while the virulome expanded by increased hypervirulence in the later cohort.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful for the patients and medical personnel involved in this study.
Funding
This study is supported by the institutional research grant of Christian Medical College, Vellore, University of Dundee, and International Society for Infectious Diseases. The publication fee was waived.
Author contributions
George M Varghese, Balaji Veeraraghavan, Ian H. Gilbert, and Abi Manesh conceptualized the study and developed the study protocol. George M Varghese, Balaji Veeraraghavan, Abi Manesh, Chaitra Shankar, and Davinder Singh Jasrotia developed the methodology. George M Varghese and Abi Manesh acquired funding. George M Varghese, Balaji Veeraraghavan, Abi Manesh, Davinder Singh Jasrotia, Mithun Mohan George, Saranya Vijayakumar and Chaitra Shankar provided oversight and supervision. Mithun Mohan George did the statistical analysis. Davinder Singh Jasrotia and Mithun Mohan George did project administration, oversaw data collection and management. Chaitra Shankar, Abi Manesh, Balaji Veeraraghavan, George M Varghese, and Mithun Mohan George accessed and verified the data. George M Varghese, Abi Manesh, Chaitra Shankar, and Mithun Mohan George wrote the original draft of the manuscript. Balaji Veeraraghavan, Davinder Singh Jasrotia, Binesh Lal, Biju George, Vikram Mathews, C.E. Eapen, Philip Joseph, K Subramani, Shoma Rao, John Victor Peter, Binila Chacko, Anand Zachariah, Sowmya Sathyendra, Samuel George Hansdak, Ooriapadickal Cherian Abraham, Ramya Iyadurai, Saranya Vijayakumar, Rajiv Karthik, Charis A Marwick, Benjamin J Parcell, and Ian H. Gilbert provided critical review and edited the original draft. All authors had access to the raw data, reviewed and approved the final manuscript, and agreed to the submission for publication.
Disclosures
Abi Manesh, Chaitra Shankar, Mithun Mohan George, Davinder Singh Jasrotia, Binesh Lal, Biju George, Vikram Mathews, C.E. Eapen, Philip Joseph, K Subramani, Shoma Rao, John Victor Peter, Binila Chacko, Anand Zachariah, Sowmya Sathyendra, Samuel George Hansdak, Ooriapadickal Cherian Abraham, Ramya Iyadurai, Saranya Vijayakumar, Rajiv Karthik, Charis A Marwick, Benjamin J Parcell, Ian H. Gilbert, Balaji Veeraraghavan, and George M Varghese have nothing to disclose.
Compliance with ethics guidelines
The study was conducted according to the principles of the Helsinki Declaration and was approved by the Institutional Review Board and Ethics Committee of Christian Medical College, Vellore (IRB No.9041/2014 and IRB No.13464/2020). Before recruitment into the study, informed consent was obtained from all the patients or immediate kin for accessing clinical information and publishing data.
Data availability
The data sets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Balaji Veeraraghavan and George M. Varghese are co-corresponding authors and contributed equally to this manuscript.
Contributor Information
Balaji Veeraraghavan, Email: vbalaji@cmcvellore.ac.in.
George M. Varghese, Email: georgemvarghese@hotmail.com
References
- 1.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, et al. Multistate point-prevalence survey of health care-associated infections. N Engl J Med. 2014;370(13):1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wyres KL, Lam MMC, Holt KE. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol. 2020;18(6):344–359. doi: 10.1038/s41579-019-0315-1. [DOI] [PubMed] [Google Scholar]
- 3.Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Elsevier Enhanced Reader [Internet]. [cited 2022 Jul 8]. https://reader.elsevier.com/reader/sd/pii/S0140673621027240?token=1234BA86B083328F1CAD60E03C23B3B46819F0A4D0BAF7CD7875229243886E7AAA616CB5A5D035CAA2A01D3F55FD0611&originRegion=eu-west-1&originCreation=20220708060502.
- 4.Efficacy of ceftazidime-avibactam plus aztreonam in patients with bloodstream infections caused by metallo-β-lactamase—Producing Enterobacterales. Clinical Infectious Diseases. Oxford Academic. [cited 2022 Aug 7]. https://academic.oup.com/cid/article/72/11/1871/5840534?login=false. [DOI] [PubMed]
- 5.van Duin D, Lok JJ, Earley M, Cober E, Richter SS, Perez F, et al. Colistin versus ceftazidime-avibactam in the treatment of infections due to carbapenem-resistant enterobacteriaceae. Clin Infect Dis. 2018;66(2):163–171. doi: 10.1093/cid/cix783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karaiskos I, Daikos GL, Gkoufa A, Adamis G, Stefos A, Symbardi S, et al. Ceftazidime/avibactam in the era of carbapenemase-producing Klebsiella pneumoniae: experience from a national registry study. J Antimicrob Chemother. 2021;76(3):775–783. doi: 10.1093/jac/dkaa503. [DOI] [PubMed] [Google Scholar]
- 7.Wang M, Earley M, Chen L, Hanson BM, Yu Y, Liu Z, et al. Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): a prospective, multicentre, cohort study. Lancet Infect Dis. 2022;22(3):401–412. doi: 10.1016/S1473-3099(21)00399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laxminarayan R, Chaudhury RR. Antibiotic resistance in India: drivers and opportunities for action. PLoS Med. 2016;13(3):e1001974. doi: 10.1371/journal.pmed.1001974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Y, Wu C, Bao D, Jia H, Draz MS, He F, et al. Global evolution and geographic diversity of hypervirulent carbapenem-resistant Klebsiella pneumoniae. Lancet Infect Dis. 2022;22(6):761–762. doi: 10.1016/S1473-3099(22)00275-4. [DOI] [PubMed] [Google Scholar]
- 10.Lewis JM, Mphasa M, Banda R, Beale MA, Heinz E, Mallewa J, et al. Colonization dynamics of extended-spectrum beta-lactamase-producing Enterobacterales in the gut of Malawian adults. Nat Microbiol. 2022;7(10):1593–1604. doi: 10.1038/s41564-022-01216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wingard RJ. Diagnostic approach to the adult cancer patient with neutropenic fever. UpToDate; Dec 8 2020. [cited 2022 Aug 5]. https://www.uptodate.com/contents/diagnostic-approach-to-the-adult-cancer-patient-with-neutropenic-fever?search=Neutropenia&source=search_result&selectedTitle=8~150&usage_type=default&display_rank=8#H29410338.
- 12.Al-Hasan MN, Lahr BD, Eckel-Passow JE, Baddour LM. Predictive scoring model of mortality in Gram-negative bloodstream infection. Clin Microbiol Infect. 2013;19(10):948–954. doi: 10.1111/1469-0691.12085. [DOI] [PubMed] [Google Scholar]
- 13.A predictive model of mortality in patients with bloodstream infections due to carbapenemase-producing Enterobacteriaceae. Elsevier Enhanced Reader. [cited 2022 Aug 4]. https://reader.elsevier.com/reader/sd/pii/S0025619616303664?token=C190392B2B4FAC3A617D149578DDFB1B0A65712FC9FDFA40AC74CC3CD04407BB985A10629A7F930D11805A7C65A22447&originRegion=eu-west-1&originCreation=20220804102826. [DOI] [PubMed]
- 14.Jorgensen SCJ, Trinh TD, Zasowski EJ, Lagnf AM, Bhatia S, Melvin SM, et al. Evaluation of the INCREMENT-CPE, Pitt Bacteremia and qPitt scores in patients with carbapenem-resistant enterobacteriaceae infections treated with ceftazidime-avibactam. Infect Dis Ther. 2020;9(2):291–304. doi: 10.1007/s40121-020-00288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laishram S, Pragasam AK, Bakthavatchalam YD, Veeraraghavan B. An update on technical, interpretative and clinical relevance of antimicrobial synergy testing methodologies. Indian J Med Microbiol. 2017;35(4):445–468. doi: 10.4103/ijmm.IJMM_17_189. [DOI] [PubMed] [Google Scholar]
- 16.Huo W, Busch LM, Hernandez-Bird J, Hamami E, Marshall CW, Geisinger E, et al. Immunosuppression broadens evolutionary pathways to drug resistance and treatment failure during Acinetobacter baumannii pneumonia in mice. Nat Microbiol. 2022;7(6):796–809. doi: 10.1038/s41564-022-01126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honsa ES, Cooper VS, Mhaissen MN, Frank M, Shaker J, Iverson A, et al. RelA mutant Enterococcus faecium with multiantibiotic tolerance arising in an immunocompromised host. MBio. 2017;8(1):e02124-16. doi: 10.1128/mBio.02124-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ankomah P, Levin BR. Exploring the collaboration between antibiotics and the immune response in the treatment of acute, self-limiting infections. Proc Natl Acad Sci U S A. 2014;111(23):8331–8338. doi: 10.1073/pnas.1400352111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreau AS, Martin-Loeches I, Povoa P, Salluh J, Rodriguez A, Thille AW, et al. Impact of immunosuppression on incidence, aetiology and outcome of ventilator-associated lower respiratory tract infections. Eur Respir J. 2018;51(3):1701656. doi: 10.1183/13993003.01656-2017. [DOI] [PubMed] [Google Scholar]
- 20.Different immunosuppressive mechanisms in multi-drug-resistant tuberculosis and non-tuberculous mycobacteria patients. Clinical and Experimental Immunology. Oxford Academic. [cited 2022 Oct 15]. https://academic.oup.com/cei/article/171/2/210/6420943?login=true. [DOI] [PMC free article] [PubMed]
- 21.Stewardson AJ, Marimuthu K, Sengupta S, Allignol A, El-Bouseary M, Carvalho MJ, et al. Effect of carbapenem resistance on outcomes of bloodstream infection caused by Enterobacteriaceae in low-income and middle-income countries (PANORAMA): a multinational prospective cohort study. Lancet Infect Dis. 2019;19(6):601–610. doi: 10.1016/S1473-3099(18)30792-8. [DOI] [PubMed] [Google Scholar]
- 22.Effect of appropriate combination therapy on mortality of patients with bloodstream infections due to carbapenemase-producing Enterobacteriaceae (INCREMENT): a retrospective cohort study. Elsevier Enhanced Reader. https://reader.elsevier.com/reader/sd/pii/S1473309917302281?token=A380725BC568E13B449F907723F76DA246C038C79EAC97DFBF6EC399E1A628160A3910E3DF68A84FDF34F0BE20C860B7&originRegion=eu-west-1&originCreation=20220708070558. [DOI] [PubMed]
- 23.Efficacy and safety of ceftazidime-avibactam for the treatment of carbapenem-resistant enterobacterales bloodstream infection: a systematic review and meta-analysis. Microbiol Spectrum. 10.1128/spectrum.0260321. [DOI] [PMC free article] [PubMed]
- 24.Treatment outcome of bacteremia due to KPC-producing Klebsiella pneumoniae: superiority of combination antimicrobial regimens. Antimicrob Agents Chemother. 10.1128/AAC.06268-11. [DOI] [PMC free article] [PubMed]
- 25.Tumbarello M, Viale P, Viscoli C, Trecarichi EM, Tumietto F, Marchese A, et al. Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: importance of combination therapy. Clin Infect Dis. 2012;55(7):943–950. doi: 10.1093/cid/cis588. [DOI] [PubMed] [Google Scholar]
- 26.Antibiotic treatment of infections due to carbapenem-resistant enterobacteriaceae: systematic evaluation of the available evidence. Antimicrob Agents Chemother. 10.1128/AAC.01222-13. [DOI] [PMC free article] [PubMed]
- 27.Tumbarello M, Trecarichi EM, De Rosa FG, Giannella M, Giacobbe DR, Bassetti M, et al. Infections caused by KPC-producing Klebsiella pneumoniae: differences in therapy and mortality in a multicentre study. J Antimicrob Chemother. 2015;70(7):2133–2143. doi: 10.1093/jac/dkv086. [DOI] [PubMed] [Google Scholar]
- 28.Time-kill evaluation of antibiotic combinations containing ceftazidime-avibactam against extensively drug-resistant Pseudomonas aeruginosa and their potential role against ceftazidime-avibactam-resistant isolates. Microbiol Spectrum. 10.1128/Spectrum.00585-21. [DOI] [PMC free article] [PubMed]
- 29.Remya P, Shanthi M, Sekar U. Occurrence and characterization of hyperviscous K1 and K2 serotype in Klebsiella pneumoniael. J Lab Physicians. 2018;10(3):283–288. doi: 10.4103/JLP.JLP_48_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whole genome analysis of hypervirulent Klebsiella pneumoniae isolates from community and hospital acquired bloodstream infection. BMC Microbiol. 10.1186/s12866-017-1148-6. [DOI] [PMC free article] [PubMed]
- 31.Shankar C, Vasudevan K, Jacob JJ, Baker S, Isaac BJ, Neeravi AR, et al. Hybrid plasmids encoding antimicrobial resistance and virulence traits among hypervirulent Klebsiella pneumoniae ST2096 in India. Front Cell Infect Microbiol. 2022;27(12):875116. doi: 10.3389/fcimb.2022.875116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets generated and/or analysed during the current study are available from the corresponding author on reasonable request.