Abstract
Expansins comprise a multigene family of proteins in maize (Zea mays). We isolated and characterized 13 different maize expansin cDNAs, five of which are α-expansins and eight of which are β-expansins. This paper presents an analysis of these 13 expansins, as well as an expression analysis by northern blotting with materials from young and mature maize plants. Some expansins were expressed in restricted regions, such as the β-expansins ExpB1 (specifically expressed in maize pollen) and ExpB4 (expressed principally in young husks). Other expansins such as α-expansin Exp1 and β-expansin ExpB2 were expressed in several organs. The expression of yet a third group was not detected in the selected organs and tissues. An analysis of expansin sequences from the maize expressed sequence tag collection is also presented. Our results indicate that expansin genes may have general, overlapping expression in some instances, whereas in other cases the expression may be highly specific and limited to a single organ or cell type. In contrast to the situation in Arabidopsis, β-expansins in maize seem to be more numerous and more highly expressed than are α-expansins. The results support the concept that β-expansins multiplied and evolved special functions in the grasses.
Cell wall proteins are believed to play important roles in regulating cell wall extensibility (Taiz, 1984), which in turn controls cell enlargement (Green, 1968). Among cell wall proteins studied to date, expansins are unique in their ability to induce immediate cell wall extension in vitro and cell expansion in vivo (Cosgrove, 1999). Since they were first isolated from cucumber hypocotyls (McQueen-Mason et al., 1992), expansin proteins have been identified in many plant species and organs on the basis of activity assays and immunoblotting. Examples include tomato leaves (Keller and Cosgrove, 1995), oat coleoptiles (Li et al., 1993), maize roots (Wu et al., 1996), rice internodes (Cho and Kende, 1997a, 1997b), tobacco cell cultures (Link and Cosgrove, 1998), and various fruits (Civello et al., 1999; Rose et al., 2000).
The original sequencing of cucumber expansin cDNAs (Shcherban et al., 1995) has impacted our understanding of expansins in several respects. First, expansin genes have now been identified in many other plant species, and they appear to be restricted largely to the plant kingdom (Cosgrove, 2000). Second, expansins comprise a large multigene family in the plant species studied in detail. For example, in Arabidopsis, 31 expansins genes have been identified (D.J. Cosgrove, unpublished data). Third, studies of expression and localization of expansin mRNA are providing new insights and hypotheses concerning the developmental roles of specific expansin genes (Cho and Kende, 1997c, 1998; Rose et al., 1997; Shimizu et al., 1997; Reinhardt et al., 1998; Brummell et al., 1999; Civello et al., 1999; Hutchison et al., 1999; Catala et al., 2000; Cho and Cosgrove, 2000; Huang et al., 2000; Im et al., 2000; Vriezen et al., 2000). And fourth, sequence comparisons led to the discovery that another group of proteins, known previously as group-1 grass pollen allergens, have expansin activity (Cosgrove et al., 1997). These pollen-specific proteins are closely related to a group of sequences known, primarily from expressed sequence tag (EST) databases. These EST sequences, together with the group-1 pollen allergens, have now been classified as β-expansins, whereas the original group of expansins are now classified as α-expansins. Although these two expansin families have only about 20% amino acid identity, they are similar in size, they share a number of conserved motifs, and they have similar wall-loosening activities (Cosgrove et al., 1997).
To date, most studies have focused on α-expansins, and limited work has been done on β-expansins. A soybean cytokinin-induced gene known as CIM1 is now classified as a β-expansin, but the biological function of the CIM1 protein is uncertain (Crowell, 1994; Downes and Crowell, 1998). The maize group-1 pollen allergen, Zea m1, has wall-loosening activity with a high specificity for grass cell walls (Cosgrove et al., 1997). This β-expansin is hypothesized to aid fertilization by loosening the cell walls of the stigma and style, thereby facilitating penetration of the pollen tube. Many other β-expansin sequences are found in the rice EST databases, and most of these sequences come from cDNA libraries made from young seedlings and other plant materials that do not contain pollen. Thus, their biological functions clearly differ from those of the group-1 pollen allergens. These so-called vegetative β-expansins are hypothesized to function in cell enlargement and other processes where wall loosening is required. It is notable that the rice EST collection contains at least 75 entries representing at least 10 distinct β-expansin genes (Cosgrove, 2000; see also http://www.bio.psu.edu/expansins/). In contrast, only a single Arabidopsis EST is classified as a β-expansin (although a total of five β-expansin genes are found in the Arabidopsis genome). The disparity in the number of β-expansin entries in the rice and Arabidopsis EST collection, together with the specificity of Zea m1 activity for grass walls, leads to the proposal (Cosgrove et al., 1997) that β-expansins have evolved specialized functions in conjunction with the evolution of the grass cell wall, which has a distinctive set of matrix polysaccharides and structural proteins compared with other land plants (Carpita, 1996). If this is true, one would expect to find an abundance of β-expansin homologs in other grasses, with expression in many tissues beside pollen. In this paper we isolated and characterized expansin cDNAs from maize. Eight of these sequences fall into the β-expansin group, whereas five of them are α-expansins. Analysis of their expression patterns indicates large differences in expression among these genes, with most organs expressing α- and β-expansin genes.
RESULTS
Sequence Analysis
We isolated and sequenced 13 distinct expansin cDNAs in this study (Table I; Fig. 1). Ten of these maize cDNAs are predicted to encode the complete expansin protein and are probably full-length or nearly full-length cDNAs. Each of the 13 cDNAs has a unique 3′-untranslated region (UTR), so we conclude that they correspond to 13 distinct genes in the maize genome. This is also supported by Southern-blot analysis. Each of the encoded proteins is predicted to have a signal peptide. The longest and the shortest signal peptides are 29 and 18 amino acids for Exp3 and Exp5, respectively; the average length is 24 amino acids. The predicted mature protein (after cleavage of the signal peptide) varies from 208 (Exp5) to 284 amino acids (ExpB4). This is a larger variation in expansin protein structure than seen heretofore in other plants.
Table I.
A list of maize expansin cDNAs
| Name (α or β) | cDNA Length | 3′-UTR | 5′-UTR | Predicted Signal Peptide | Predicted Mature Protein |
|---|---|---|---|---|---|
| bp | amino acids | ||||
| Exp1, α | ∼1,160; full | 255 | 61 | 20 | 233 |
| Exp2, α | ∼1,285; full | 410 | 43 | 22 | 254 |
| Exp3, α | ∼1,040; full | 167 | 63 | 29 | 233 |
| Exp4, α | ∼1,029; partial | 321 | – | – | >235 |
| Exp5, α | ∼1,040; full | 324 | 9 | 18 | 208 |
| ExpB1, β | ∼1,145; full | 220 | 31 | 24 | 245 |
| ExpB2, β | ∼1,270; full | 380 | 45 | 24 | >285 |
| ExpB3, β | ∼800; partial | 138 | – | – | >216 |
| ExpB4, β | ∼1,279; full | 240 | 100 | 24 | 284 |
| ExpB5, β | ∼660; partial | 102 | – | – | >176 |
| ExpB6, β | ∼1,150; full | 211 | 83 | 26 | 250 |
| ExpB7, β | ∼1,170; full | 282 | 64 | 28 | 240 |
| ExpB8, β | ∼1,480; full | 520 | 80 | 26 | 260 |
Figure 1.
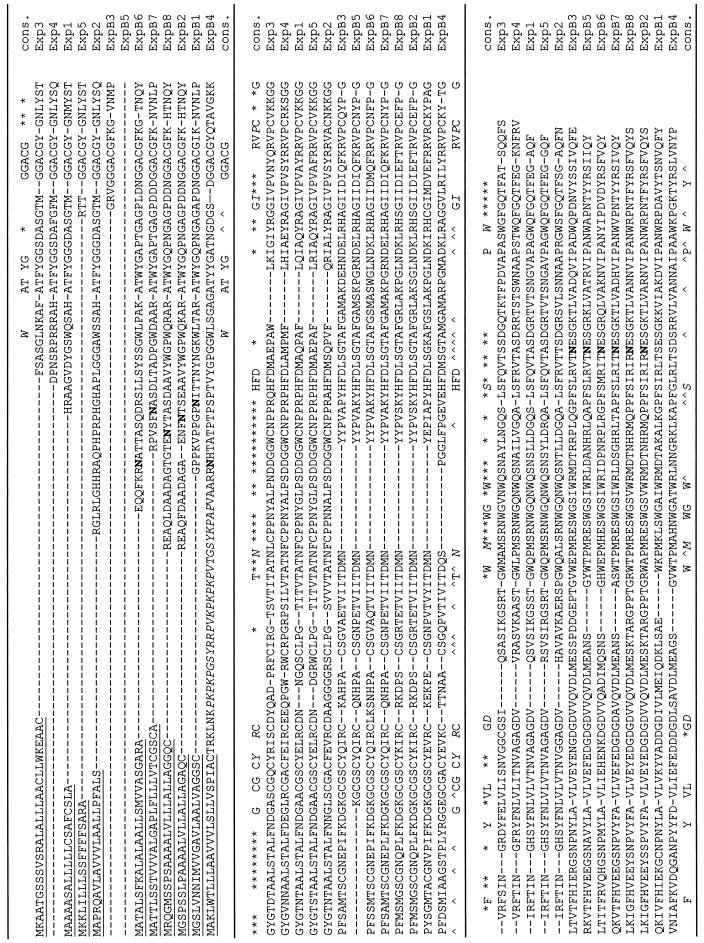
Alignment of amino acid sequences predicted from maize expansin cDNAs. Sequences were aligned by MEGALIGN program (DNAstar software) using the Clustal method with the PAM250 residue weight table and default parameters. The underlined sequences represent the signal peptides predicated by the SignalP program (www.cbs.dtu.dk/services/SignalP/). Potential N-linked glycosylation sites are shown in bold and were predicted using Prosite. Symbols in the consensus sequence, Capitalized letters indicate conserved amino acids for α- and β-expansins; italicized capital letters denote conserved amino acids for α- and β-expansins with occasional variance; an asterisk denotes amino acids conserved among α-expansins; and ∧ denotes amino acids conserved among β-expansins.
Upon sequence analysis we have divided the 13 cDNAs into two groups, α-expansins (five cDNAs, named Exp1–Exp5) and β-expansins (eight cDNAs, named ExpB1–ExpB8). Alignment of the predicted protein sequences shows that both groups of cDNAs contain the highly conserved regions that are diagnostic for expansins (Cosgrove, 1999), including a series of conserved C (Cys) residues, an HFD (His-Phe-Asp) motif, and four W (Trp) residues near the carboxy terminus (Fig. 1). For the conserved Cys residues, eight were very well conserved in α-expansins, and six of the eight Cys residues were conserved in the β-expansin group. This difference in Cys residues between α- and β-expansins corresponds to the absence of approximately 14 amino acids in the β-expansin group and appears to be a consistent difference between the two groups of expansins. Other conserved differences between α- and β-expansins are highlighted in Figure 1. It is notable that the β-expansins have a predicted N-gly-cosylation site near the amino terminus and in most, but not all, cases a second predicted glycosylation site near the carboxy terminus; in contrast, α-expansins lack glycosylation motifs.
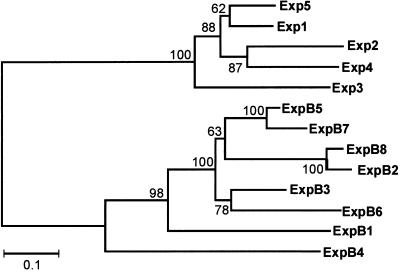
The distinction between α- and β-expansin proteins is also readily apparent in the corresponding sequence-based phylogenetic tree (Fig. 2), which shows the two families to be deeply divided from each other, with approximately 25% identity between the two families. In pair-wise comparisons within each family, high sequence similarity is typical of α-expansins (from 57%–83% identity), whereas the β-expansins tend to be more divergent from each other (28%–76% identity). The greater degree of sequence conservation in the α-expansin family can also be seen by inspection of the conserved residues marked in Figure 1.
Figure 2.
Phylogenetic analysis of maize expansin sequences. Phylogenetic analysis of 13 maize expansin sequences was performed using MEGA2 software (www.megasoftware.net). Sequences were aligned as indicated in Figure 1. Pairwise deletion was used to deal with gaps or missing data in sequences. The distance between sequences was estimated after Poisson correction and an unrooted tree was constructed using the neighbor-joining method. The bootstrap numbers indicated at each joining point were created from 500 samplings.
Although most of the predicted proteins appeared to be rather typical for α- and β-expansins, some unusual features of ExpB4, Exp2, and Exp5 are worth noting. The predicted mature ExpB4 protein contains an unusual extension of approximately 53 amino acids at its amino terminus (Fig. 1). This extension is highly basic (13 basic residues, no acidic residues; predicted pI of the extension is 11.6) and contains 13 Pro residues (25%). A BLAST search shows this short extension to be 40% to 50% identical with sequences from teosinte, rice, and maize encoding Hyp-rich glycoproteins (Raz et al., 1992). These are structural proteins of the cell wall. The predicted mature form of Exp2 likewise has a short extension (25 amino acids) at its amino terminus, and this extension is rich in His, Gly, Pro, and Arg residues. No significant sequence similarities were found in BLAST searches of GenBank when queried with this extension peptide. Exp5 is unusual in that a highly conserved motif (HATFYG or minor variations thereof) typically found at the amino terminus of α-expansins is missing.
Southern Analysis
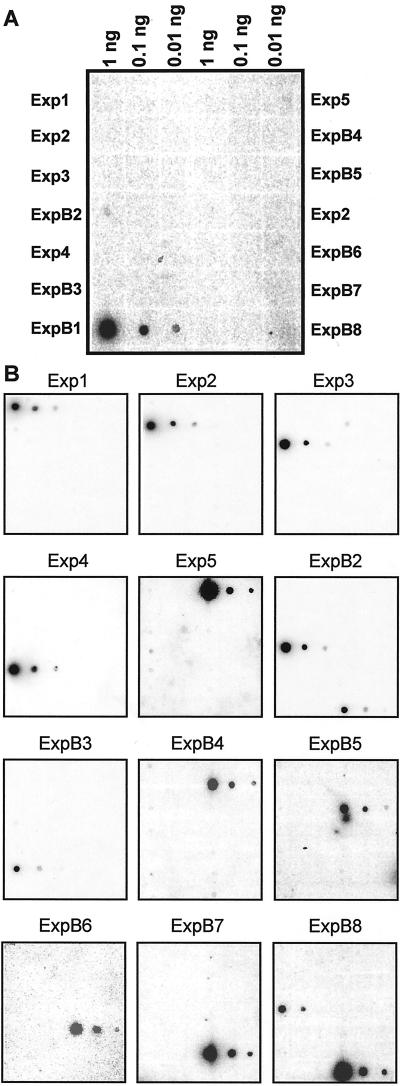
Because the 3′-UTR is the most divergent region of expansin cDNAs, we used them as gene-specific probes for Southern- and northern-blot analysis. A dot-blot analysis was first used to test the specificity of each probe. As an example, in Figure 3A the dot-blot probed with the ExpB1 probe was overexposed intentionally to show the arrangement of each cDNA on the dot blot. Except for probes from ExpB2 and ExpB8, all probes showed no cross reaction with any other cDNA sequences at a level of 0.1 ng of target DNA (Fig. 3B) Under the stringent conditions we used most probes could produce good signal even when the target DNA was only 0.01 ng. Probe for ExpB3 was an exception. It gave very week signal even when the target DNA was 0.1 ng. Probes from ExpB2 and ExpB8 showed cross reaction with each others cDNA at a level of 0.1 ng target DNA (Fig. 3B). These two cDNAs share very high sequence similarity in their coding region, with 97.5% identity in nucleotide sequence. The encoded proteins differ from each other at only 12 amino acids, excluding the signal peptide. Their 3′-UTRs are 74% identity over 127 bp, with a stretch of 65 bp having 96% identity. This degree of similarity for expansin cDNAs is unusual in our experience and we suspected they might be alleles for the same gene, but Southern analysis indicates they are distinct genes.
Figure 3.
Tests for cross hybridization of gene-specific probes using dot blots. A, Close up of dot blot for ExpB1, showing layout and DNA loadings. B, Dot blots for Exp1-5 and ExpB2-8. Each cDNA containing vector sequence was dotted onto a nylon membrane at three different loadings, as shown in A. Following denaturation and washing, the dot blot was then hybridized to a gene-specific probe (PCR-amplified from 3′-UTR) labeled with P32, washed at high stringency, and exposed to a phosphor screen overnight.
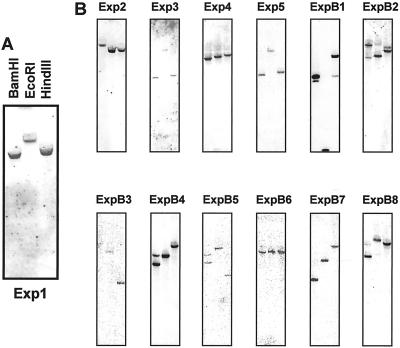
The specificity of these probes was also demonstrated in Southern blots, which indicated that most, but not all, of these genes are present as single-copy genes. The Southern blot for Exp1 is shown in Figure 4A as an example. For Exp1, Exp2, Exp3, Exp4, Exp5, ExpB3, ExpB6, and ExpB7, each probe detected a single band with different sizes in the three different enzyme digestions (Fig. 4B). The results indicate that each of these genes is probably present in the maize genome as a single copy.
Figure 4.
Southern-blot analysis for Exp1-5 and ExpB1-8. A, Close-up view of the Southern blot for Exp1, indicating the pattern of loadings. B, Southern blots for Exp2–Exp5 and ExpB1–8 in the same pattern as shown in A. Ten micrograms of genomic DNA was digested with BamHI, EcoRI, or HindIII, separated on a 0.8% (w/v) agarose gel, and transferred to a nylon membrane. The blot was hybridized with a P32-labeled gene-specific probe using the same conditions as in Figure 3.
The Southern blot also indicates that some of the probes hybridized with a second gene. There were double bands in the Southern blots for ExpB2 and ExpB8, which, as described above, are closely related in sequence. By comparing the two blots it is apparent that the major band in each lane corresponded to the probed gene itself and the fainter band corresponded to the other cross-reacting gene. Judged from Southern-blot results, the probe for ExpB8 is much more specific than the ExpB2 probe since the second band is hardly visible on the Southern blot for ExpB8. A second faint band was visible on the Southern blots for ExpB1, indicating that the probe might be interacting with another gene. A recent BLAST search against the maize EST database identified two additional sequences almost identical to ExpB1 in the coding region and with only a few nucleotide variations in the 3′-UTR. These two ESTs are listed as ExpB1b and ExpB1c in Table II, whereas ExpB1 is listed as ExpB1a. The Southern blots for ExpB4 and ExpB5 indicate there might be a second closely related and linked gene in the maize genome for each of these β-expansins.
Table II.
Analysis of expansin cDNA representation in maize EST libraries
| Name | Accession No. in Databasea | Library (Source) | No. of Hits in Library |
|---|---|---|---|
| Exp1 | gb: AI001321 | Shoots of green seedlings: shootsb | |
| Exp1 | gb: AF332169 | Rootsc | |
| Exp1 | CNDAH50 | Germinating maize seeds: 2 and 3 d, shootd | |
| Exp2 | AW062050 | Mixed stages of embryo development | 1 |
| Exp2 | gb: AF332170 | Rootsc | |
| Exp2 | CMAAR78 | Germinating maize seeds: 2 and 3 d, rootd | |
| Exp3 | CNDAA78 | Germinating maize seeds: 2 and 3 d, shootd | |
| Exp4 | gb: AF332172 | Rootsc | |
| CLDAC16 | 7 d after pollination, whole kernelsd | ||
| Exp5 | CODAM22 | 14-d Etiolated seedlings: coleoptiled | |
| Exp6 | TUC04-05-1482.1 | Ear tissuee | 2 |
| Exp7a | TUC04-05-8254.1 | Mixed stage of anther and pollenf | 7 |
| Exp7a | TUC04-05-8254.1 | Mixed adult tissuesg | 2 |
| Exp7b | TUC04-05-8082.1 | Mixed stage of anther and pollenf | 4 |
| Exp7b | TUC04-05-8082.1 | Mixed adult tissuesg | 4 |
| Exp8 | TUC04-05-8568.1 | Mixed stage of anther and pollenf | 11 |
| Exp9 | TUC04-05-3803.1 | Immature leafh | 1 |
| Exp9 | TUC04-05-3803.1 | Endospermi | 1 |
| Exp9 | TUC04-05-3803.1 | Mixed stages of embryo developmentj | 1 |
| ExpB1a | TUC04-05-9091.1 | Mixed stage of anther and pollenf | 18 |
| TC25561 | Mixed adult tissuesg | 11 | |
| ExpB1b | TUC04-05-8984.1 | Mixed stage of anther and pollenf | 17 |
| TC25560 | Mixed adult tissuesg | 3 | |
| ExpB1c | TUC04-05-8073.1 | Mixed stage of anther and pollenf | 3 |
| ExpB1c | TUC04-05-8073.1 | Mixed adult tissuesg | 5 |
| ExpB2 | gb: AF332175 | Rootsc | |
| ExpB3 | COMGF24 | 14-d Etiolated seedlings: mesocotyld | |
| ExpB4 | gb: AA979914 | Shoots of green seedlingsb | |
| ExpB5 | CRTAQ69 | 4-d Root tips shorter than 5 mm in lengthd | |
| ExpB6 | CMSAW71 | Scutellar node of 2-d seedlingsd | |
| ExpB7 | COMGX68 | 14-d Etiolated seedling: mesocotyld | |
| ExpB8 | CGEUY12 | Tassel: premeiotic to early uninucleated | |
| ExpB9 | TUC04-05-6266.1 | Mixed adult tissuesg | 4 |
| ExpB10 | TUC04-05-7808.1 | Mixed adult tissuesg | 7 |
| ExpB11 | TUC04-05-6295.1 | Mixed stages of anthers and pollenf | 1 |
| ExpB11 | TUC04-05-6295.1 | Mixed adult tissuesg | 3 |
| ExpB12 | TUC04-05-5656.1 | Endospermi | 2 |
| ExpB13 | AW066587 | 14-d Immature embryok | 1 |
| ExpB14 | gb: AI691979 | Ear tissuee | 1 |
| ExpB15 | gb: AI649785 | Stressed shootl | 1 |
| ExpB16 | TUC04-05-8939.1 | Mixed stages of anther and pollenf | 14 |
| Mixed adult tissueg | 4 | ||
| ExpB16 | TUC04-05-8706.1 | Mixed stages of anther and pollenf | 9 |
| Mixed adult tissueg | 4 | ||
| ExpB17 | TUC04-05-2135.1 | Rootm | 2 |
| ExpB18 | TUC04-05-9184.3 | Endospermi | 1 |
| ExpB18 | TUC04-05-9184.3 | Ear tissuee | 3 |
gb, A Genbank accession no.; TUC, a Zea mays Database assembly no.; TC, The Institute for Genomic Research assembly no.
ESTs deposited by Iowa State University.
Root library: gift from Dr. Terry R. Conley at Oklahoma City University.
Pioneer's maize library collection.
Ear tissue: Library 606 in ZmDB. Total ESTs sequenced = 5,523 at the date of survey.
Mixed stage of anther and pollen: Library 660 in ZmDB. Total EST sequenced = 4,651 at the date of survey.
Mixed adult tissues: Library 707 in ZmDB. Total EST sequenced = 5,436 at the date of survey.
Immature leaf: Library 486 in ZmDB. Total EST sequenced = 5,868 at the date of survey.
Endosperm: Library 605 in ZmDB. Total EST sequenced = 6,566 at the date of survey.
Mixed stages of embryo development: Library 687 in ZmDB. Total ESTs sequenced = 4,937 at the date of survey.
Fourteen-day immature embryo: Library 683 in ZmDB. Total EST sequenced = 1,138 at the date of survey.
Stressed shoot (NaCl): Library 496 in ZmDB. Total EST sequenced = 1,307 at the date of survey.
Root: Library 614 in ZmDB. Total EST sequenced = 10,652 at the date of survey.
Expression Analysis
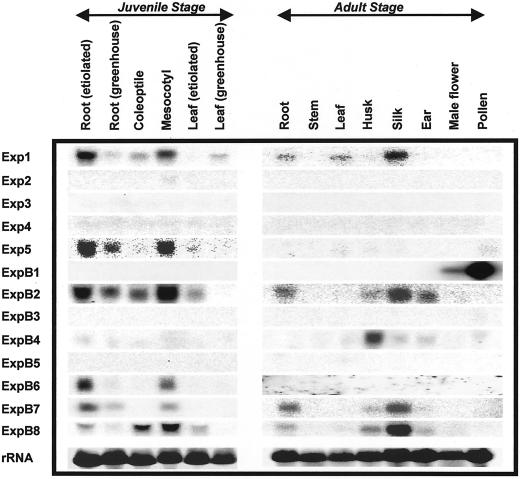
The expression patterns of these 13 expansins, studied by northern blotting with gene-specific probes, are complex (Fig. 5), with distinctive patterns for each gene. The strongest signals in the northern blots were found for Exp1, ExpB1, ExpB2, and ExpB8 (the Exp5 blot appears darker only because the phosporimage sensitivity was raised to bring out the exposed bands).
Figure 5.
Northern-blot analysis for Exp1–5 and ExpB1–8 using different tissues and organs. Twenty micrograms of total RNA extracted from the indicated organs and tissues of young or mature maize plants was separated on a 1% (w/v) denatured agarose gel and transferred to a nylon membrane. The blot was hybridized with P32-labeled gene-specific probes at the same condition as used for the dot blots in Figure 3.
The mRNA for α-expansin Exp1 was detected in several organs from juvenile and mature plants, whereas Exp5 mRNA was detected at low levels in juvenile roots and in the mesocotyl. The expression of the three other α-expansins (Exp2, Exp3, and Exp4) was not detected in any tissue studied here. This negative result could mean that these genes are expressed at very low levels, that they require particular conditions to be expressed, or that they are expressed in organs or tissue types not sampled here. We assume they are expressed genes because they were identified in cDNA libraries.
Six of the eight β-expansins were detected in northern blots. ExpB1 was detected with a very strong signal in pollen, as expected for the protein it encodes (Zea m1, a group-1 pollen allergen). The small signal seen in the male flower was probably due to the developing pollen in this organ. Another β-expansin that showed high specificity of expression is ExpB4, which gave a strong signal in young husks. In contrast to this high degree of specificity of expression, ExpB2 and ExpB8 were detected in many juvenile and mature organs. Because of high sequence similarity between these two genes, however, these northern-blot results may be a composite of signals from both β-expansin mRNAs. The northern-blot patterns are not identical for these two probes, however, so it is likely that these genes have distinct patterns of expression, but this point will need further work to resolve definitively. ExpB6 and ExpB7 were expressed in roots and mesocotyls. In addition, ExpB7 was expressed at relatively high levels in the silk.
Some tissues are notable for high levels of expansin mRNA expression. These include roots and mesocotyls in juvenile plants and silks in adult plants. These organs were sampled at stages of rapid grow, which likely accounts for their high degree of expansin expression. Pollen gave the strongest signal of all for ExpB1. Stem tissue is notable for its lack of detectable expansin gene expression; this is mostly likely due to sampling of the stem from a region lacking rapid growth.
We also analyzed the public maize EST databases for expansin gene expression and found an additional 17 sequence classes of expansins (named Exp6-9 and ExpB9-18) that are distinct from the 13 studied here. Table II summarizes all maize expansins we found from our screenings and from the EST database analysis. The source cDNA library and the frequency of each sequence class in the various libraries are also listed in the table as an indication of their expression pattern and relative transcript level. One of the interesting observations from the table is that several ESTs with the highest frequency are from anther and pollen (Exp8, ExpB1a, ExpB1b, and ExpB16). ExpB1a (which codes for the group-1 allergen Zea m1), for instance, was represented by 18 entries in 4,651 ESTs from the library of mixed stages of anther and pollen. This high frequency in pollen and anther libraries is consistent with the result of our northern-blot analysis in Figure 5, indicating that this expansin is highly expressed in pollen.
DISCUSSION
Our results show that expansins in maize make up a diverse family of at least 30 genes (32 if we count ExpB1b-c separately). Among the 13 cDNAs studied, eight encode β-expansin proteins and five encode α-expansin proteins. The β-expansin messages are expressed in a variety of organs, in some cases with high specificity and in other instances with wider expression. Analysis of the recently deposited maize ESTs confirms and extends this conclusion, bringing the total number of identified α-expansins in maize to nine and the number of identified maize β-expansins to 18 (20 if we count ExpB1b-c separately). This is undoubtedly an incomplete inventory, as the Southern blots indicate there are additional expansin genes yet to be found in maize and many of the expansins are represented in the EST databases with low counts.
Our results indicate that β-expansins are more numerous and more abundantly expressed in maize than are α-expansins. Analysis of the rice EST database indicates that this situation is likely the case in rice as well, and thus this may be a common characteristic of grasses. In comparison, only a single EST in Arabidopsis falls into the β-expansin family, which has a total of five β-expansin genes in its genome compared with 26 α-expansin genes (Cosgrove, 2000; D.J. Cosgrove, unpublished data). The large number of β-expansin genes in grasses is likely related to the fact that the cell wall matrix polysaccharides and structural proteins of grasses differ from those most other angiosperms (Carpita and Gibeaut, 1993). We suspect that β-expansins act on these unique wall components, such as the unique mixed-linked β-1,3:1,4-glucan or glucuronoarabinoxylans found in grass species. This is supported by the fact that Zea m1 (encoded by ExpB1) has high specificity of action for grass cell walls over dicot cell walls (Cosgrove et al., 1997). However, detailed biochemical tests of this hypothesis are needed, particularly for the β-expansins expressed in vegetative tissues.
Maize expansins are also notable in comparison with the expansin family in Arabidopsis because some of the β-expansins genes appear to be present in multiple copies of nearly identical genes, e.g. ExpB1a-c, ExpB2/8, and ExpB4. This is not the case in the Arabidopsis expansin gene family (D.J. Cosgrove, unpublished data).
Our new data allow us to identify particular sequence characteristics that are common to α- and β-expansins and that distinguish each group. The most notable features conserved in both groups of expansins include several sequence motifs, including the following: AT(F/W) YG, GGACG, CGSC, HFD, and RVPC (Fig. 1). In addition, a set of four tryptophans at the carboxy terminus with conserved spacing is common to α- and β-expansins, as is a set of six Cys residues (β-expansins lack an additional pair of cysteines that are characteristic of α-expansins).
Sequences that distinguish α- and β-expansins can be seen in the alignment of Figure 1. In β-expansins the absence of 14 consecutive residues in the middle of the protein, including a pair of cysteines that are conserved in α-expansins, suggests the absence of a loop that is present in α-expansins and is stabilized by a disulfide bridge. The sequence analysis also suggests that β-expansins are glycosylated, whereas α-expansins are not, and this conclusion is supported by the limited protein analysis published to date (see Cosgrove 2000, 1999). These conserved differences may play a role in the distinctive activities of α- and β-expansins, i.e. substrate preferences, binding characteristics, and biological roles (Cosgrove et al., 1997). Further biochemical characterization of β-expansins, particularly the proteins from vegetative tissues, is needed to test these ideas.
The expansin sequences predict mature proteins (after signal peptide removal) that vary from 208 to 284 amino acids. This is a larger variation in expansin protein size than has been observed up to now. At the short extreme (Exp5) the small size is due to the deletion of a region at the amino terminus that is normally conserved in the α-expansin group, whereas at the long extreme (ExpB4) the additional size is accomplished by an extension at the amino terminus of the mature protein. This extension is approximately 50% similar to regions of the Hyp-rich glycoproteins from maize and related species (Raz et al., 1992). These proteins are believed to play a structural role in the cell wall (Showalter, 1993; Cassab, 1998). Because of its high pI, this extension may bind to pectins and thus serve as an anchoring mechanism to locate the ExpB4 protein within a subregion of the cell wall. In contrast with the amino-terminus of the mature protein, which has extensions of varying length, the carboxy terminus of expansin appears to tolerate little variation in length. We note that the most divergent of the β-expansins (ExpB1 and ExpB4) are expressed principally in reproductive tissues (pollen and husk, respectively). This divergence in protein structure may be linked to specialized wall-loosening roles for these proteins, e.g. ExpB1 encodes the pollen allergen Zea m1 that aids pollen penetration of the stigma (Cosgrove et al., 1997).
The gene expression patterns of maize expansins indicate that expansins may be involved in many different developmental events. ExpB1 is expressed in pollen, indicating ExpB1 involvement in pollen development or in the pollination process (Cosgrove et al., 1997). Exp2 was not expressed in the tissues we examined. However, it is found in a cDNA library from endosperm, indicating that Exp2 may play a role in seed development. At this time it is difficult to attribute a specific function to other expansin genes since they are expressed in more than one organ or several genes were expressed in a same organ at the same time (Fig. 5). A similar situation was found in tomato fruit studies (Brummell et al., 1999). With gene-specific probes in hand, further work will be possible to examine the specific role of the numerous expansin genes, e.g. by inspecting in detail where each expansin is expressed (in situ hybridization) and how the expression of each expansin is associated with a specific developmental stage or an environmental stimulus.
MATERIALS AND METHODS
Plant Materials
Maize (Zea mays) plants (FR 697 inbred, Pioneer Hi-Bred International, Des Moines, IA) were used in this study. For analysis of gene expression we harvested tissues from maize plants at different growth stages. For etiolated plant materials a row of seeds was arranged on germination paper (Anchor Paper, St. Paul). The paper was rolled to form a tube and was placed vertically in a 2-L beaker containing 200 mL of distilled water. The beaker was loosely covered with another container to reduce water loss. After 96 h at 29°C in the dark, roots (apical 20 mm), coleoptiles, mesocotyls, and young leaves of maize seedlings were harvested under dim green light. For the greenhouse-grown plants, seeds were germinated in the greenhouse in soil (Pro-Mix, Geiger, PA). Young leaves and the whole primary roots of 1-week-old seedlings were collected. Several seedlings were transplanted into 6-L pots for further cultivation. Four days after silks appeared, the youngest leaf near the tassel, the stem (the internode below the tassel), several young adventitious roots (whole), male flowers, pollen, silk, husk (inner layers), and the whole unpollinated ear were harvested. Harvested tissues were immediately frozen in liquid nitrogen after collection and stored at −80°C before removal for RNA or genomic DNA extraction.
Library Screening and Sequencing
A maize root cDNA library (generously donated by Dr. Terry R. Conley, Oklahoma City University) was first screened by plaque hybridization using a radiolabeled probe (a mixture of nine rice expansin cDNAs). Nylon membranes with lifted plaques were pre-hybridized in hybridization buffer (Amersham, Buckinghamshire, UK) at 60°C overnight, and were then hybridized with the radioactive probe in the same buffer at 60°C for 20 h. Membranes were washed for 45 min once in 3× SSC, for 45 min once in 1× SSC, for 45 min twice in 0.1× SSC at 60°, and were then exposed to x-ray film (Fuji Photo Film, Tokyo) for 24 h at room temperature. From about 1.5 × 106 plaques screened a total of 255 positive plaques were picked for further screening. After the secondary and tertiary screening, 64 positive plaques were chosen for DNA isolation. Based on the restriction digestion pattern, 16 different inserts were subcloned into the pUC18 plasmid for subsequent sequencing. Five inserts turned out to be unique maize expansin cDNAs. Further sequencing work with positive clones from Pioneer's maize cDNA collection revealed another seven unique expansin cDNAs. The ExpB4 clone was a gift from Dr. P.S. Schnable (Iowa State University). Sequences have been deposited in GenBank under accession nos. AF332169 through AF332181.
Sequence Analysis
DNA sequences were analyzed with DNAstar software (DNAstar, Madison, WI). Predicted amino acid sequences were aligned by MEGALIGN (DNAstar) using the Clustal method with the PAM250 residue weight table and default parameters. The phylogenetic tree was based on this alignment, using the neighbor joining method with Poisson-corrected distances (MEGA2 software). Signal peptides were predicted with the SignalP program (Henrik et al., 1997).
Hybridization Analysis
A PCR-amplified 3′-UTR of each cDNA was used as a gene-specific probe (Table III) and P32-labeled probes were prepared with a Rediprime kit (Amersham). Plasmid DNA containing cDNA insert was dotted onto a nylon membrane to make an array of 1, 0.1, and 0.01 ng DNA for each expansin. The arrays were then hybridized to each gene-specific probe to test the specificity of the probe.
Table III.
Primer sets for gene-specific probe synthesis
| Name | Primer Sequences | Probe Length |
|---|---|---|
| bp | ||
| Exp1 | 5′CTACTACTACTCCATCGACG3′ | 259 |
| 5′AATAAGTTGCACGACACC3′ | ||
| Exp2 | 5′CGACGCAGTGGCACTAGT3′ | 300 |
| 5′CGAAGGAGCCATCACCAT3′ | ||
| Exp3 | 5′CGCAGCAGTTCTCTTAGT3′ | 182 |
| 5′GCACAAGTCAAAGCATATAT3′ | ||
| Exp4 | 5′GGGATGGGATGCGATGGG3′ | 315 |
| 5′GCTAGGGACCGAAGAGAC3′ | ||
| Exp5 | 5′AGGATGAACAGCTAGCGC3′ | 305 |
| 5′TAGTACTAGGAGCTGGATCG3′ | ||
| ExpB1 | 5′CCCTTCGATTCATCCGG3′ | 196 |
| 5′CCTCCAAAAATGGCATGT3′ | ||
| ExpB2 | 5′AGCTAGCTGGTTTGCGCC3′ | 296 |
| 5′AAGCAACAGTGGGCGGG3′ | ||
| ExpB3 | 5′CAGTTCGAGTAGCAGTATATG3′ | 144 |
| 5′CGTACATATATATCATCGATCC3′ | ||
| ExpB4 | 5′CCTGAAAAGAGAAATACCG3′ | 213 |
| 5′GTATCAAATTACCGGCATG3′ | ||
| ExpB5 | 5′TCCATCATTCAGTACTAGCTC′ | 118 |
| 5′CTAGTTAAGGCACCGTAATCC3′ | ||
| ExpB6 | 5′TAATGATCGAGCTAGCT3′ | 216 |
| 5′TTAGTATGAAATTATATAACC3′ | ||
| ExpB7 | 5′GGCCGGATAATAATATACA3′ | 240 |
| 5′ATCTTAATCGCACTGGTAC3′ | ||
| ExpB8 | 5′GGATCCGTGCCCTGCCCG3′ | 412 |
| 5′AGCAACAGTGGGCGGGAG3′ |
For Southern-blot analysis, genomic DNA was isolated from the shoots of etiolated maize seedlings with the cetyl-trimethyl-ammonium bromide method (Murray and Thompson, 1980). Ten micrograms of DNA was digested with EcoRI, BamHI, or HindIII at 37°C overnight, separated on a 0.8% (w/v) agarose gel, and vacuum-blotted to nylon membrane.
For northern-blot analysis, total RNA was extracted from plant tissues with TRIzol reagent (Gibco-BRL, Rockville, MD) using the manufacturer's instructions. Twenty micrograms of total RNA was separated on a 1% (w/v) denatured agarose gel (6.5% [v/v] formaldehyde) and vacuum-transferred to nylon membrane.
A set of blots consisting of one dot blot, one Southern blot, and two northern blots (one for young seedlings and one for adult stage) was processed together for hybridization to each of the 13 gene-specific probes. Blots were pre-hybridized in Ultrahyb solution (Ambion, Austin, TX) at 60°C overnight and hybridized to the probe in the same solution at 60°C for 20 h. The blots were washed at 65°C for 20 min twice in 5% SEN (5% [w/v] SDS, 1 mm EDTA, and 40 mm Na2HPO4), and for 20 min once in 1% SEN (1% [w/v] SDS, 1 mm EDTA, and 40 mm Na2HPO4). Blots were exposed to phosphor screens (Molecular Dynamics, Sunnyvale, CA) overnight at room temperature.
ACKNOWLEDGMENTS
We thank Daniel M. Durachko for technical assistance, Terry R. Conley for the maize root cDNA library, and Peter S. Schnable for the ExpB4 clone.
Footnotes
This work was supported by the U.S. Department of Energy (grant no. DE–FG02–84ER13179) and by the U.S. Department of Agriculture National Research Initiative (grant no. PENR–9601307).
LITERATURE CITED
- Brummell DA, Harpster MH, Dunsmuir P. Differential expression of expansin gene family members during growth and ripening of tomato fruit. Plant Mol Biol. 1999;39:161–169. doi: 10.1023/a:1006130018931. [DOI] [PubMed] [Google Scholar]
- Carpita NC. Structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:445–476. doi: 10.1146/annurev.arplant.47.1.445. [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Cassab GI. Plant cell wall proteins. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:281–309. doi: 10.1146/annurev.arplant.49.1.281. [DOI] [PubMed] [Google Scholar]
- Catala C, Rose JK, Bennett AB. Auxin-regulated genes encoding cell wall-modifying proteins are expressed during early tomato fruit growth. Plant Physiol. 2000;122:527–534. doi: 10.1104/pp.122.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ. Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2000;97:9783–9788. doi: 10.1073/pnas.160276997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Kende H. Expansins and internodal growth of deepwater rice. Plant Physiol. 1997a;113:1145–1151. doi: 10.1104/pp.113.4.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Kende H. Expansins in deepwater rice internodes. Plant Physiol. 1997b;113:1137–1143. doi: 10.1104/pp.113.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Kende H. Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell. 1997c;9:1661–1671. doi: 10.1105/tpc.9.9.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Kende H. Tissue localization of expansins in rice. Plant J. 1998;15:805–812. doi: 10.1046/j.1365-313x.1998.00258.x. [DOI] [PubMed] [Google Scholar]
- Civello PM, Powell ALT, Sabehat A, Bennett AB. An expansin gene expressed in ripening strawberry fruit. Plant Physiol. 1999;121:1273–1279. doi: 10.1104/pp.121.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Enzymes and other agents that enhance cell wall extensibility. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:391–417. doi: 10.1146/annurev.arplant.50.1.391. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Loosening of plant cell walls by expansins. Nature. 2000;407:321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Bedinger P, Durachko DM. Group I allergens of grass pollen as cell wall-loosening agents. Proc Natl Acad Sci USA. 1997;94:6559–6564. doi: 10.1073/pnas.94.12.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell DN. Cytokinin regulation of a soybean pollen allergen gene. Plant Mol Biol. 1994;25:829–835. doi: 10.1007/BF00028877. [DOI] [PubMed] [Google Scholar]
- Downes BP, Crowell DN. Cytokinin regulates the expression of a soybean β-expansin gene by a post-transcriptional mechanism. Plant Mol Biol. 1998;37:437–444. doi: 10.1023/a:1005920732211. [DOI] [PubMed] [Google Scholar]
- Green PB. Growth physics in Nitella: a method for continuous in vivo analysis of extensibility based on a micro-manometer technique for turgor pressure. Plant Physiol. 1968;43:1169–1184. doi: 10.1104/pp.43.8.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrik N, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Huang J, Takano T, Akita S. Expression of α-expansin genes in young seedlings of rice (Oryza sativa L.) Planta. 2000;211:467–473. doi: 10.1007/s004250000311. [DOI] [PubMed] [Google Scholar]
- Hutchison KW, Singer PB, Diaz-Sala C, Greenwood MS. Expansins are conserved in conifers and expressed in response to exogenous auxin. Plant Physiol. 1999;120:827–832. doi: 10.1104/pp.120.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im KH, Cosgrove DJ, Jones AM. Subcellular localization of expansin mRNA in xylem cells. Plant Physiol. 2000;123:463–470. doi: 10.1104/pp.123.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller E, Cosgrove DJ. Expansins in growing tomato leaves. Plant J. 1995;8:795–802. doi: 10.1046/j.1365-313x.1995.8060795.x. [DOI] [PubMed] [Google Scholar]
- Li Z-C, Durachko DM, Cosgrove DJ. An oat coleoptile wall protein that induces wall extension in vitro and that is antigenically related to a similar protein from cucumber hypocotyls. Planta. 1993;191:349–356. [Google Scholar]
- Link BM, Cosgrove DJ. Acid-growth response and α-expansins in suspension cultures of bright yellow 2 tobacco. Plant Physiol. 1998;118:907–916. doi: 10.1104/pp.118.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Durachko DM, Cosgrove DJ. Two endogenous proteins that induce cell wall expansion in plants. Plant Cell. 1992;4:1425–1433. doi: 10.1105/tpc.4.11.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M, Thompson W. Rapid isolation of high-molecular-weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz R, Jose M, Moya A, Martinez-Izquierdo JA, Puigdomenech P. Different mechanisms generating sequence variability are revealed in distinct regions of the hydroxyproline-rich glycoprotein gene from maize and related species. Mol Gen Genet. 1992;233:252–259. doi: 10.1007/BF00587586. [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Wittwer F, Mandel T, Kuhlemeier C. Localized up-regulation of a new expansin gene predicts the site of leaf formation in the tomato meristem. Plant Cell. 1998;10:1427–1437. doi: 10.1105/tpc.10.9.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Cosgrove DJ, Albersheim P, Darvill AG, Bennett AB. Detection of expansin proteins and activity during tomato fruit ontogeny. Plant Physiol. 2000;123:1583–1592. doi: 10.1104/pp.123.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Lee HH, Bennett AB. Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proc Natl Acad Sci USA. 1997;94:5955–5960. doi: 10.1073/pnas.94.11.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherban TY, Shi J, Durachko DM, Guiltinan MJ, McQueen-Mason S, Shieh M, Cosgrove DJ. Molecular cloning and sequence analysis of expansins: a highly conserved, multigene family of proteins that mediate cell wall extension in plants. Proc Natl Acad Sci USA. 1995;92:9245–9249. doi: 10.1073/pnas.92.20.9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, Aotsuka S, Hasegawa O, Kawada T, Sakuno T, Sakai F, Hayashi T. Changes in levels of mRNAs for cell wall-related enzymes in growing cotton fiber cells. Plant Cell Physiol. 1997;38:375–378. doi: 10.1093/oxfordjournals.pcp.a029178. [DOI] [PubMed] [Google Scholar]
- Showalter AM. Structure and function of plant cell wall proteins. Plant Cell. 1993;5:9–23. doi: 10.1105/tpc.5.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiz L. Plant cell expansion: regulation of cell wall mechanical properties. Annu Rev Plant Physiol. 1984;35:585–657. [Google Scholar]
- Vriezen WH, De Graaf B, Mariani C, Vosenek LACJ. Submergence induces expansin gene expressin in flooding tolerant Rumex palustris and not in flooding intolerant R. acetosa. Planta. 2000;210:956–963. doi: 10.1007/s004250050703. [DOI] [PubMed] [Google Scholar]
- Wu Y, Sharp RE, Durachko DM, Cosgrove DJ. Growth maintenance of the maize primary root at low water potentials involves increases in cell-wall extension properties, expansin activity, and wall susceptibility to expansins. Plant Physiol. 1996;111:765–772. doi: 10.1104/pp.111.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]