Abstract
Expansin proteins are essential components of acid-induced cell wall loosening in plants. β-Expansins, which constitute a subfamily of related expansin proteins, include the group I grass pollen allergens. To provide a better description of β-expansin expression, we have characterized a cytokinin-inducible β-expansin from soybean (Glycine max cv Mandarin) called Cim1. Our results demonstrate that the hormones cytokinin and auxin act synergistically to induce the accumulation and proteolytic processing of Cim1. Carboxyl terminal truncation of a 35-kD form of Cim1 is predicted to remove the putative cellulose binding domain from the amino terminal cysteine-rich domain, resulting in a 20-kD form of the protein. Furthermore, the identical amino termini of the 35- and 20-kD forms of Cim1 correspond to a position 11 amino acids downstream of the predicted signal sequence cleavage site, suggesting proteolysis of a short amino terminal propeptide after removal of the signal peptide. This propeptide fragment contains a consensus site for N-glycosylation and our data suggest that it is glycosylated by a tunicamycin-sensitive mechanism in cultured soybean cells. The onset of Cim1 expression correlates with increased growth of soybean cultures. Ultimately, Cim1 is rapidly and specifically proteolyzed as soybean cultures reach stationary phase. These findings are consistent with the hypothesis that β-expansin proteins are extensively modified by post-translational N-glycosylation and proteolysis.
At low pH, expansin proteins loosen cell walls and allow turgor-driven cell expansion. Thus, expansins irreversibly affect cell size and shape (Cosgrove, 1998). Expansin activity has been firmly established in monocots, dicots, and suspension-cultured cells (Link and Cosgrove, 1998; Cosgrove, 2000). However, the mechanism of expansin action remains elusive. Several findings suggest that expansins affect the stability of hydrogen bonds between cellulose and hemicellulose microfibrils and thus control the rigidity of the cell wall (Cosgrove, 1998). For example, expansins were found to associate with hemicellulose-coated cellulose microfibrils in vitro (McQueen-Mason and Cosgrove, 1995) and were able to induce loosening of pure cellulosic paper (McQueen-Mason and Cosgrove, 1994; Cosgrove, 2000).
In a recent report, Grobe et al. (1999) proposed that expansins possess a proteolytic activity that may influence plant cell wall loosening. The pollen allergen/β-expansin Phl pI from Timothy grass was expressed and secreted from the yeast strain, Pichia, which is capable of protein modifications typical of higher eukaryotes. The recombinant protein was found to catalyze the degradation of a peptide substrate containing a papain cleavage site. Furthermore, affinity-purified native protein exhibited autoproteolytic activity. This work suggested that the mechanism of expansin action may rely on digestion of proteinaceous structural elements of the cell wall (i.e. extensin proteins; Grobe et al., 1999). However, these results are not consistent with the finding that expansins loosen pure cellulosic paper, which is devoid of protein (McQueen-Mason and Cosgrove, 1994; Cosgrove, 2000).
Regulation of expansin activity by plant hormones is well documented. For example, the auxin-induced acid growth response relies on the activity of expansins for wall loosening and cell elongation (McQueen-Mason et al., 1992, 1993; Rayle and Cleland, 1992; Link and Cosgrove, 1998). In addition, plant hormones regulate expansin gene expression. Some examples include the tomato expansins LeExp1 and LeExp2, the deep water rice expansin OsExp4, and the soybean (Glycine max) β-expansin Cim1, which accumulate in response to ethylene, auxin, gibberellin, and cytokinin, respectively (Crowell et al., 1990; Cho and Kende, 1997; Rose and Bennett, 1999).
The cytokinin-induced Cim1 mRNA accumulates 20- to 60-fold after treatment of cytokinin-starved soybean suspension cultures with cytokinin (Crowell et al., 1990). Previous studies have focused on the cytokinin signal transduction pathway that leads to Cim1 message accumulation. These studies have shown that accumulation of Cim1 message occurs, in large part, by cytokinin-mediated changes in mRNA stability, and that protein dephosphorylation participates in propagating the cytokinin signal (Downes and Crowell, 1998). The Cim1 protein product is a member of the β-expansin subgroup of expansin proteins (Shcherban et al., 1995; Cosgrove et al., 1997), which includes the group I grass pollen allergens. However, unlike the pollen-specific pattern of expression typical of other β-expansins, Cim1 protein and mRNA are expressed in suspension cultures derived from soybean vegetative tissue and the mRNA has been detected in all soybean tissues analyzed (B.P. Downes and D.N. Crowell, unpublished data). Hence, Cim1 is classified as a vegetative homolog of the group I grass pollen allergens.
In this report, we present evidence that the β-expansin Cim1 is N-glycosylated and proteolyzed in several discrete steps in soybean suspension cultures. The expression pattern of multiple forms of Cim1 correlates with the application of cytokinin and/or auxin and with the expansive growth phase of soybean suspension cultures. Our results suggest that post-translational N-glycosylation and proteolysis are important for β-expansin processing and degradation.
RESULTS
Multiple Forms of the Cim1 Protein Are Specifically Detected in Soybean Suspension Cultures
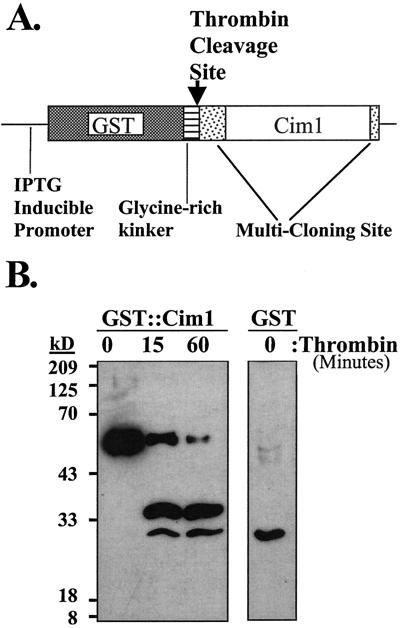
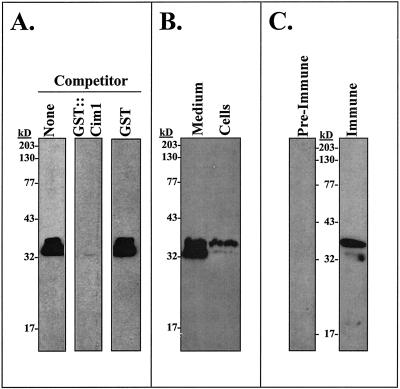
Antiserum was raised in rabbits against a recombinant fusion protein consisting of glutathione-S-transferase (GST) and Cim1. The fusion partners are separated by a kinker region, which contains a thrombin cleavage site. Recombinant protein cut with thrombin for various times was recognized by the antiserum (Fig. 1). The specificity of the Cim1 antibody for proteins in soybean suspension-cultured cells and medium is shown in Figure 2. In 4-d-old culture medium, 38- and 35-kD proteins were detected in the presence of purified GST, but were not detected in the presence of the specific competitor GST::Cim1 (Fig. 2A). The 38- and 35-kD forms of Cim1 were also present in 4-d-old cells (Fig. 2, B and C).
Figure 1.
Production of anti-GST::Cim1 serum. A, The fusion construct used for expression of GST::Cim1 protein is shown. B, GST::Cim1 fusion protein expressed in Escherichia coli and affinity purified on glutathione Sepharose beads was digested with thrombin for various times. A single thrombin site is predicted to cut within the kinker region, between GST and Cim1. Proteins were fractionated by 14% (w/v) SDS-PAGE and examined by immunoblot analysis using antibody raised against the fusion protein (1o antibody, anti-GST::Cim1, 1:6,000; 2o antibody, goat anti-rabbit-horseradish peroxidase, 1:7,500). GST protein expressed in E. coli from an empty pGEX-KG vector was also examined.
Figure 2.
Multiple forms of the Cim1 protein are detected in soybean suspension cultures. A, Four-d-old soybean suspension-culture medium was tested for the presence of Cim1 protein by immunoblot analysis as described in Figure 1 with no competitor (lane 1) or with pre-incubation of immune serum in the presence of 0.2 μm GST::Cim1 (lane 2) or 0.2 μm GST (lane 3). B, Four-d-old soybean suspension-culture medium and cells were analyzed as described in Figure 1. C, Four-d-old soybean suspension-cultured cells were analyzed as described in Figure 1 with preimmune and immune serum.
The Plant Hormones Cytokinin and Auxin Regulate Cim1 Accumulation and Processing
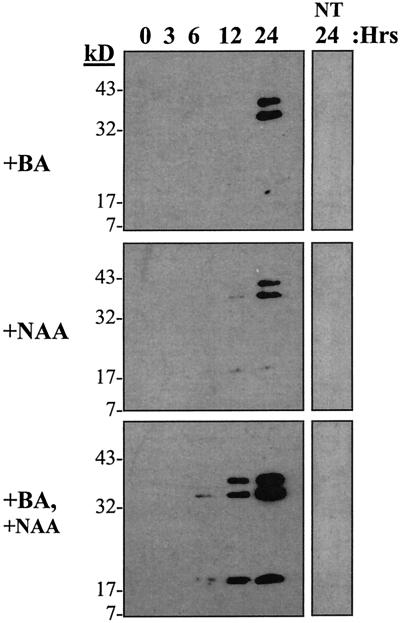
Regulation of Cim1 expression by cytokinin and auxin was analyzed in soybean cultures. Cim1 protein in the culture medium increased when cultures starved for cytokinin and auxin were treated with cytokinin, auxin, or both (Fig. 3). Cytokinin and auxin together had a cumulative effect on the abundance of the 38- and 35-kD forms of the protein described above. It is interesting that a 20-kD form of Cim1 also appeared in the medium of cultures treated with cytokinin and auxin.
Figure 3.
The plant hormones cytokinin and auxin regulate Cim1 accumulation and processing. Cytokinin and auxin regulate Cim1 expression and processing. Stationary phase soybean cells depleted of cytokinin and auxin received solvent (water, NT), or cytokinin (1 μm benzyladenine, +BA), or auxin (10 μm naphthalene acetic acid, +NAA), or cytokinin and auxin for 24 h. Medium samples taken at different times after hormone treatment were assayed for Cim1 protein by immunoblot analysis as described in Figure 1.
Characterization of Cim1 Glycosylation and Proteolysis
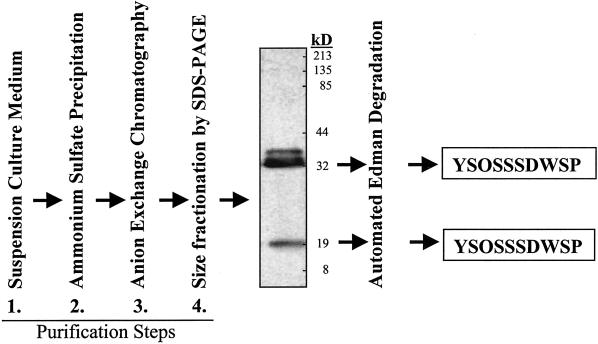
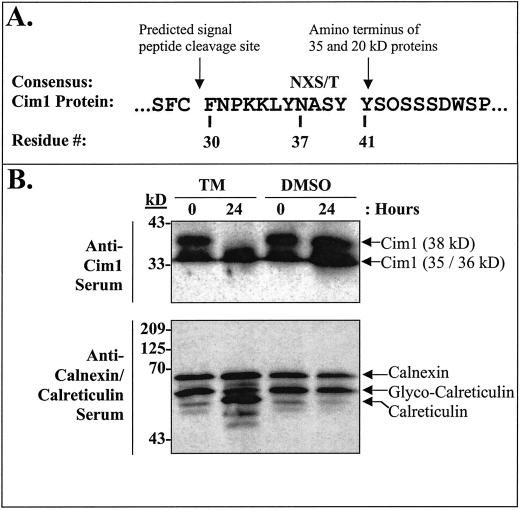
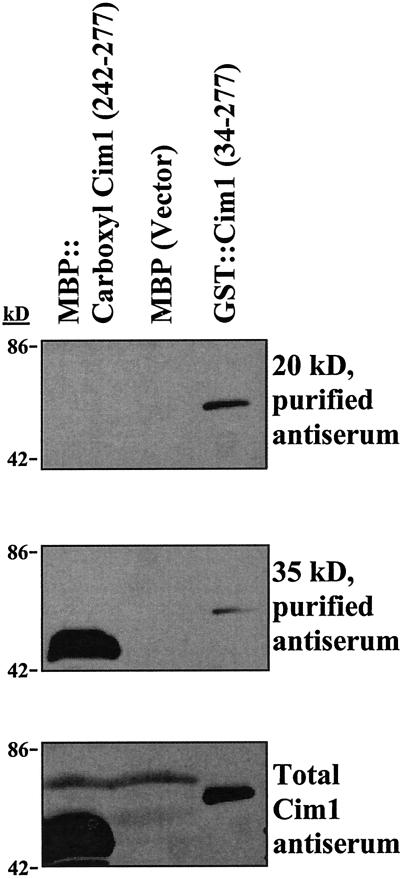
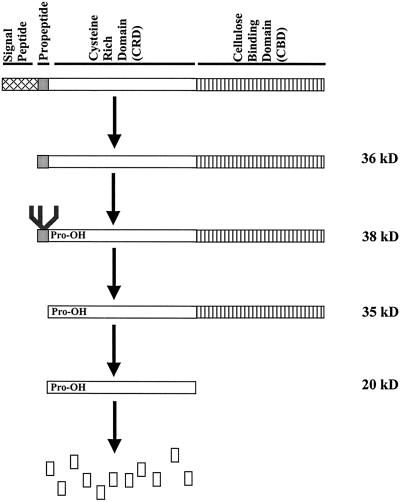
Purification and protein sequence analysis of the 35- and 20-kD forms of Cim1 (Fig. 4) from culture medium indicated that the amino terminus of both was at residue 41 (YSOSSSDWSP; O = Hyp) of the amino acid sequence deduced from the Cim1 cDNA (we were not successful at obtaining sufficient amounts of pure 38-kD protein from culture medium for amino terminal sequence analysis). The site of signal peptide cleavage was predicted using PROSITE (Bucher and Bairoch, 1994) to correspond to the peptide bond between amino acids 29 and 30, 11 amino acids upstream of the empirically determined amino terminus of the 35- and 20-kD forms of Cim1. This observation suggests the existence of a short 11-amino acid putative propeptide containing the consensus N-linked glycosylation motif characteristic of β-expansins (Fig. 5). Treatment of soybean cells with 10 μg mL−1 tunicamycin, an inhibitor of N-glycosylation, abolished the large 38-kD form of Cim1 and caused the expected shift in molecular mass for calreticulin (Denecke et al., 1995), a known N-glycosylated protein (tunicamycin treatment was expected to cause the accumulation of an unglycosylated Cim1 protein that comigrates with the 35-kD form of the protein and has an amino terminus corresponding to the signal sequence cleavage site). Thus, the 38-kD form of Cim1 appears to be a full-length glycoprotein lacking only the signal peptide. To test the hypothesis that the 20-kD form of Cim1 is truncated at the carboxyl terminus, antibodies eluted from the 35- and 20-kD forms of Cim1 were tested for binding to the carboxyl terminal 35 amino acids of Cim1 (Fig. 6). Only antibodies purified from the 35-kD form of Cim1 bound to an MBP::Cim1 fusion protein representing amino acids 242 through 277, whereas antibodies eluted from the 35- and 20-kD forms recognized a GST::Cim1 fusion protein corresponding to amino acids 34 through 277. In a reciprocal experiment, antibodies purified from the MBP::Cim1 fusion bound to the 35-kD, but not the 20-kD form of Cim1 (data not shown). These results are summarized in Figure 7.
Figure 4.
Protocol for purification and amino terminal sequence analysis of the 35- and 20-kD forms of Cim1. The image shown is a Cim1 immunoblot.
Figure 5.
The 38-kD form of Cim1 is N-glycosylated. A, Sequence of the Cim1 propeptide. The consensus sequence for N-glycosylation is NXS/T as indicated. B, Five-d-old soybean cells were treated for 0 or 24 h with 10 μg mL−1 tunicamycin in DMSO (TM) or an equivalent amount of DMSO and assayed for Cim1 and calreticulin/calnexin proteins by immunoblot analysis as described in Figure 1. The anti-calreticulin/calnexin serum, which was used at a dilution of 1:3,000, recognizes both proteins, but only calreticulin is N-glycosylated (Denecke et al., 1995; Coughlan et al., 1996).
Figure 6.
The carboxyl terminus of Cim1 is proteolytically processed. Maltose-binding protein (MBP) fused to a carboxyl terminal epitope of Cim1 (MBP::carboxyl Cim1), MBP, or GST::Cim1 were tested for reactivity to affinity purified anti-Cim1 antibodies by immunoblot analysis as described. Top, Antibodies were affinity purified from the 20-kD form of Cim1; middle, antibodies were affinity purified from the 35-kD form of Cim1; bottom, total Cim1 immune serum was used.
Figure 7.
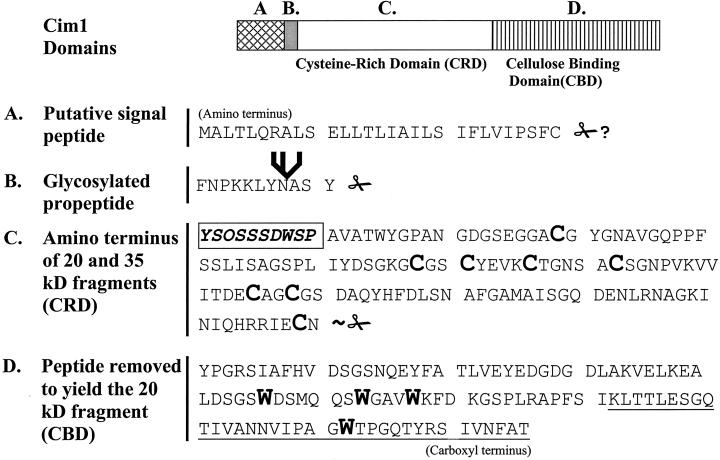
Cim1 primary structure. The amino terminal sequence of the 35- and 20-kD forms of Cim1 is shown boxed in bold type. Scissors indicate Cim1 proteolysis (✁, confirmed site of propeptide proteolysis; ∼✁, approximate site of carboxyl terminal proteolysis; ✁?, sequence-based prediction of signal peptide proteolysis). The branched structure represents an N-linked oligosaccharide chain. The conserved cysteines of the Cys-rich domain (CRD) and the conserved tryptophans of the cellulose-binding domain (CBD) are shown as large bold characters. The underlined region at the carboxyl terminus represents the region fused to MBP and analyzed in Figure 6. O, Hyp; all other letters represent standard amino acids.
The Expression of Cim1 Correlates with the Expansive Phase of Soybean Culture Growth
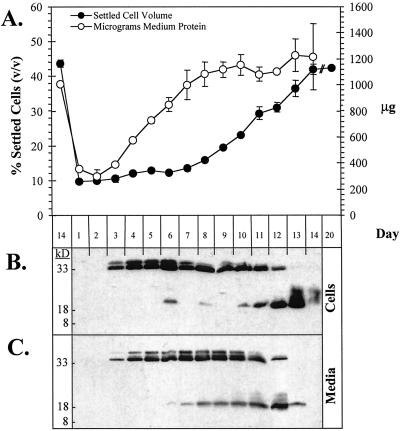
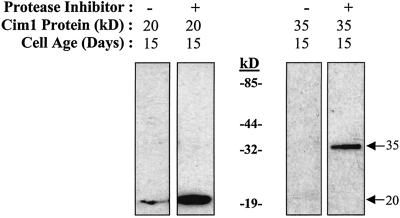
Soybean cell cultures exhibited a distinctive pattern of Cim1 expression (Fig. 8). The 38- and 35-kD forms of Cim1 were detected in soybean cells on the 3rd d of the culture period and increased until the 6th d, after which both declined in abundance. In contrast, the 20-kD form of Cim1 was observed transiently between the 6th and 8th d and accumulated again toward the end of the culture period. All forms of Cim1 disappeared completely in stationary phase cultures. Cim1 detected in the medium exhibited a similar pattern of accumulation. In cells and medium, Cim1 increased as cultures entered the expansive phase of growth and then declined as the cultures became stationary. The 20-kD form of Cim1 in the medium correlated especially well with the expansive phase of growth. A complete protease inhibitor mixture blocked the disappearance of Cim1 from medium samples added back to stationary phase cells for 16 h, suggesting that Cim1 is degraded, not taken up or cross-linked to cell walls at the end of the culture period (Fig. 9). The observation that Cim1 levels dropped sharply below detection at the end of the culture period without a corresponding decline in total extracellular protein suggests the presence of a specific mechanism for Cim1 degradation (Fig. 8).
Figure 8.
The expression of Cim1 correlates with the expansive phase of soybean culture growth. A, Settled cell volume was measured after 15 min of settling. Extracellular protein concentrations were determined by Bradford assay and then multiplied by the volume of medium to derive values for total extracellular protein. ses are shown (n = 2). B, Equal masses (7.5 μg) of cellular protein were assayed for Cim1 protein by immunoblot analysis as described in Figure 1. C, An equal volume of culture medium was sampled from each day of culture growth and analyzed as in B.
Figure 9.
Cim1 degradation occurs in the presence of stationary phase cells. Stationary phase cells were mixed with medium containing the 20-kD form of Cim1 or purified 35-kD Cim1 in the presence or absence of a complete protease inhibitor cocktail (Roche, Indianapolis) and analyzed as described in Figure 1.
DISCUSSION
Production of antibodies to the β-expansin Cim1 facilitated the specific detection of Cim1 processing intermediates in soybean suspension cultures (Fig. 10). The largest form of Cim1 had an apparent molecular mass of 38 kD and was present in cells and culture medium. The 38-kD form of Cim1 was abolished by tunicamycin treatment, suggesting that it is N-glycosylated, presumably at the consensus N-glycosylation sequence immediately downstream of the signal sequence cleavage site. Thirty-five- and 20-kD forms of Cim1 were also detected in cells and culture medium. These forms of Cim1 had identical amino termini that corresponded to amino acid 41 of the deduced Cim1 amino acid sequence, downstream of the site of N-glycosylation described above. Furthermore, the carboxyl terminal end of the 20-kD protein is truncated, because antibodies purified from the 35-kD form recognized epitopes at the carboxyl terminus of the amino acid sequence deduced from the Cim1 cDNA, whereas antibodies purified from the 20-kD form did not. Although the precise site of carboxyl terminal proteolysis is not known, it is reasonable to predict, based on the difference in size between the 35- and 20-kD forms, that the entire cellulose-binding domain is removed from the 20-kD form of Cim1. The fragment removed from Cim1 to create the 20-kD form was not detected in our experiments, despite the fact that this region of the protein is recognized by immune serum. One explanation for this is that the excised fragment is rapidly degraded. Alternatively, the carboxyl terminal fragment may be tightly associated with the cell wall and may thereby escape detection.
Figure 10.
Model for post-translational processing of Cim1.
A diagram of the predicted primary structure of the Cim1 protein is shown in Figure 7. Cim1 is predicted to have a signal peptide based on the -3, -1 rule (von Heijne, 1986), the hydrophobic profile of the first 30 amino acids deduced from the Cim1 cDNA, and alignment with other expansins, which exhibit amino acid divergence in the signal peptide, but significant similarity elsewhere. These predictions are consistent with a classic cotranslationally cleaved signal peptide ending at residue 29 of the deduced Cim1 amino acid sequence. However, as stated above, the amino terminus of the 35- and 20-kD forms of Cim1 corresponds to residue 41. The balance is an 11-amino acid peptide, which we propose to be a Cim1 propeptide because the N-linked glycosylation site conserved among β-expansins resides on this fragment. Indeed, our data suggest that a glycopropeptide accounts for the 38-kD form of Cim1. Glycosylation of the analogous site on Cim1 homologs from rye and timothy grass has been confirmed (Petersen et al., 1997). In addition, glycosylated propeptides have been described for proteins such as the p34 vacuolar storage protein from soybean and barley lectin (Wilkins et al., 1990; Kalinski et al., 1992). It is, therefore, conceivable that Cim1 propeptide glycosylation is a prerequisite for further processing.
A study of Cim1 regulation by cytokinin and auxin was designed to test the prediction that both hormones are required for Cim1 expression. When cytokinin and auxin were added back to cultures starved for both hormones, a striking accumulation of the 20-kD form of Cim1 was observed in the culture medium compared to that seen in cultures treated with cytokinin or auxin (Fig. 3). Apparently, both cytokinin and auxin must be present for maximal accumulation and carboxyl terminal truncation of Cim1. In contrast, the accumulation of the 38- and 35-kD forms of Cim1 in response to both hormones was only modestly increased over that seen in cultures treated with cytokinin or auxin.
Analyses of Cim1 protein forms secreted during the growth of soybean cultures revealed that the 20-kD form of Cim1 was present during the expansive phase of the culture (Fig. 8). In addition, all three Cim1 protein forms gradually accumulated, and then rapidly disappeared, at the end of the culture period. This disappearance was the result of proteolytic destruction, because the addition of stationary phase cells to Cim1 protein samples caused Cim1 to disappear in the absence, but not in the presence, of a protease inhibitor mixture (Fig. 9). Furthermore, the decline in Cim1 protein abundance in stationary phase cultures was not associated with a general decrease in extracellular protein (Fig. 8), suggesting the existence of a Cim1-specific protease that may function to deplete Cim1 protein when cell wall loosening is no longer required.
Despite the evidence shown in this paper for multiple processing intermediates of Cim1, we do not currently know which form(s) represent active expansin proteins. A rigorous purification and activity analysis of all forms of Cim1 will be necessary to assign function to the processing steps we have characterized. It is tempting to speculate that many of the effects of plant hormones on plant growth and development depend on the activities of specific expansin proteins. For example, cytokinin promotes cotyledon expansion, leaf expansion, shoot organogenesis, and release of lateral buds, all of which are potentially regulated by expansins. Furthermore, plant genomes contain a large number of expansin genes (i.e. the Arabidopsis genome contains 25 known expansin genes; Cosgrove, 2000), many of which may be regulated by plant hormones. Thus, understanding how plant hormones orchestrate the expression, processing, and activity of expansin proteins may provide insights into the mechanisms of action of phytohormones in plant growth and development.
MATERIALS AND METHODS
Plant Material and Chemical Treatments
Suspension cultures of soybean (Glycine max cv Mandarin) were grown in Murashige-Skoog liquid medium (pH 5.7; Murashige and Skoog, 1962) containing 10 μm α-naphthaleneacetic acid (NAA) and 4 μm benzyladenine (BA) at 26°C ± 1°C in continuous fluorescent light on a rotary shaker set to 100 rpm. These cultures were propagated at 14-d intervals by transferring 10 mL of stationary phase cells into 30 mL of fresh medium. Hormone starvations were performed by washing stationary phase cells with 1 L of medium (five washes of 200 mL each) lacking BA and NAA. Washed cells were collected by suction filtration on a Buchner funnel and cultured in 40 mL of fresh hormone-deficient medium for 6 d. Where indicated, BA and/or NAA were added to final concentrations of 1 and/or 10 μm, respectively. In a separate experiment, 10 μg mL−1 tunicamycin (Fluka, Milwaukee, WI) in dimethyl sulfoxide (DMSO) was added to 5-d-old soybean cells. Equivalent treatments with DMSO were performed to rule out solvent effects.
After various treatments, culture medium samples were filtered through a 10-μm nylon mesh, and the filtrates were microcentrifuged at 3,500 rpm for 1 min. Conditioned medium supernatants were then stored at 4°C. Cells used for Cim1 immunoblot analysis were washed extensively in Murashige-Skoog liquid medium (pH 5.7) containing no exogenous hormones (four washes of 4 volumes), frozen in liquid N2, and stored at −80°C.
Recombinant Protein Purification
HindIII-digested Cim1 cDNA was inserted into the HindIII site of pGEX-KG (Guan and Dixon, 1991), creating a GST::Cim1 fusion protein lacking the Cim1 signal peptide (i.e. 33 amino acids are encoded 5′ of the Cim1 HindIII site). Escherichia coli strain DH5α was used to express the GST::Cim1 fusion protein. Cultures were grown to an A595 of 0.85 and were then induced with 0.5 mm isopropyl-1-thio-β-d-galactopyranoside for 4 h. Cells were collected, washed once in cold ddH2O, resuspended in 150 mm NaCl, 10 mm Tris, pH 8.0, 1 mm EDTA, frozen in liquid N2, thawed, and sonicated for 2 min with a 0.8-cm probe. After centrifugation (10 min, 4°C, 10,000 rpm, Beckman JA-20, Beckman Instruments, Fullerton, CA), the pellet was resuspended in 2.5 m urea (the fusion protein formed inclusion bodies), centrifuged as before, and the insoluble material was dissolved completely in 8 m urea (Cardamone et al., 1995). The 8 m urea solution was then dialyzed to 0.1 μm urea and applied to reduced-glutathione Sepharose beads (Pharmacia, Uppsala). The beads were washed extensively and fusion protein was eluted with 10 mm glutathione, 0.5% (v/v) β-mercaptoethanol, 10 mm EDTA, 100 mm Tris, pH 8.0. Protein concentrations were determined by the method of Bradford (1976).
An MBP fusion to the 35 carboxyl terminal amino acids of Cim1 was constructed. The terminal 36 codons (35 amino acid codons plus the termination codon) from the 3′ end of the Cim1 gene were amplified by PCR (forward primer, 5′-GAA TTC GGA TCC AAG CTA ACC ACA CTT GA-3′; reverse primer, 5′-TCT AGA AAG CTT TTA GGT TGC AAA ATT CAC-3′) and subcloned into the BamHI and HindIII sites of the multicloning site downstream of MBP in the pMAL-c2X bacterial expression vector (New England Biolabs, Beverly, MA). E. coli (DH5α) carrying the construct was induced with 0.5 mm isopropyl-1-thio-β-d-galactopyranoside for 4 h, sedimented, resuspended in 2 volumes of 2× Laemmli sample buffer (25% [v/v] glycerol, 5% [w/v] SDS, 125 mm Tris, pH 6.8), boiled for 5 min, diluted to 1× Laemmli sample buffer with water, and boiled an additional 5 min. The sample was spun at 14,000 rpm in a microfuge for 10 min and the soluble material was stored at −80°C.
Antibody Preparation
Four milligrams of purified GST::Cim1 fusion protein was injected intraperitoneally into New Zealand White rabbits on a standard schedule at Cocalico Biolabs (Reamstown, PA). Ten weeks after the initial injection, serum was harvested and found to contain a high titer of anti-Cim1 IgG.
Protein Immunoblot Analysis
Proteins were separated by 14% (w/v) SDS-PAGE as described (Laemmli, 1970) and were electrophoretically transferred to nitrocellulose (Schleicher & Schuell, Keene, NH) in 25 mm Tris, pH 9.0, 192 mm Gly, 20% (v/v) methanol for 2 h at 400 mA with a Mini-Protean II transfer apparatus (Bio-Rad, Hercules, CA). Nitrocellulose membranes were blocked for 2 h in TBST (100 mm Tris, pH 7.5, 0.9% [w/v] sodium chloride, 0.1% [v/v] Tween 20) supplemented with 5% (w/v) non-fat dry milk and incubated with Cim1 antiserum (1:6,000) for 3 h at 25°C in TBST + 0.5% (w/v) non-fat dry milk. Membranes were then washed extensively in TBST. Cim1 immunoreactive proteins were detected using goat anti-rabbit IgG horseradish peroxidase conjugate (1:7, 500, Sigma, St. Louis), SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL), and X-Omat AR5 film (Eastman Kodak, Rochester, NY).
Antiserum Fractionation
Small scale antibody purification was performed essentially as described by Smith and Fisher (1984). Medium from 8-d-old soybean suspension cultures was separated by preparative 14% (w/v) SDS-PAGE and transferred to nitrocellulose as described above. The blot was then incubated in an excess of immune serum (50 mL, 1:6,000), the edges were visualized by luminescence following exposure to secondary antibody and chemiluminescence reagents, and the central regions corresponding to the 35- and 20-kD Cim1 proteins were excised. Antibodies were eluted with 0.5 mL of 100 mm Gly, pH 2.5, for 1 min and then again for 10 min. Each eluate was neutralized with 0.05 mL of 1 m Tris, pH 8.0. The eluates were then adjusted to 1× TBST in a final volume of 5 mL and used for subsequent immunoblot analyses.
Native Protein Purification and Sequencing
Analysis of the Cim1 protein secreted into the medium of soybean suspension cultures provided a substantial first purification step. Culture medium from 8-d-old cultures was filtered through a 10-μm nylon mesh and precipitated with 0.8 m ammonium sulfate. The supernatant was then adjusted to 3 m ammonium sulfate, and the resulting precipitate was dialyzed overnight against two changes of 25 mm sodium acetate, pH 6.3 (1,000 volumes), using a 12- to 14-kD filter (SPECTRA/POR, Fisher Scientific, Pittsburgh). The dialysate was bound to a DEAE column equilibrated with 25 mm sodium acetate, pH 6.3. Protein was eluted with a 0 to 1 m continuous sodium chloride gradient in 25 mm sodium acetate, pH 6.3. Fractions were tested for the presence of Cim1 protein by immunoblot analysis. Positive fractions were pooled, dialyzed, concentrated, and resolved by electrophoresis on thoroughly polymerized, pre-run 14% (w/v) SDS-polyacrylamide gels. Proteins were then electrophoretically transferred onto polyvinyldifluoride (Trans-Blot Transfer Medium, Bio-Rad) and Ponceau-stained bands corresponding to Cim1 immunoreactive material were excised and subjected to automated Edman degradation (Matsudaira, 1987) at the Indiana University Biochemistry and Biotechnology Facility (Indianapolis, IN).
ACKNOWLEDGMENTS
The authors wish to thank the Department of Biology at Indiana University-Purdue University at Indianapolis for generous financial support, which made possible the production of anti-Cim1 serum, and Dr. Stephen K. Randall for providing the anti-calreticulin/calnexin serum.
Footnotes
This work was supported by the U.S. Department of Agriculture (grant nos. 94–02330 and 00–03367 to D.N.C.) and by Indiana University-Purdue University at Indianapolis (Graduate Student Organization Educational Enhancement grant to B.P.D.).
LITERATURE CITED
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bucher P, Bairoch A. A generalized profile syntax for biomolecular sequence motifs and its function in automatic sequence interpretation. In: Altman R, Brutlag D, Karp P, Lathrop R, Searls D, editors. ISMB-94: Proceedings of the 2nd International Conference on Intelligent Systems for Molecular Biology. Menlo Park, CA: AAAI Press; 1994. pp. 53–61. [PubMed] [Google Scholar]
- Cardamone M, Puri NK, Brandon MR. Comparing the refolding and reoxidation of recombinant porcine growth hormone from a urea denatured state and from Escherichia coli inclusion bodies. Biochemistry. 1995;34:5773–5794. doi: 10.1021/bi00017a009. [DOI] [PubMed] [Google Scholar]
- Cho HT, Kende H. Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell. 1997;9:1661–1671. doi: 10.1105/tpc.9.9.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Cell wall loosening by expansins. Plant Physiol. 1998;118:333–339. doi: 10.1104/pp.118.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. New genes and new biological roles for expansins. Curr Opin Plant Biol. 2000;3:73–78. doi: 10.1016/s1369-5266(99)00039-4. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Bedinger PA, Durachko DM. Group I allergens of grass pollen as cell wall loosening agents. Proc Natl Acad Sci USA. 1997;94:6559–6564. doi: 10.1073/pnas.94.12.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlan SJ, Hastings C, Winfrey RJ. Molecular characterization of plant ER: identification of protein disulfide-isomerase as the major reticuloplasmin. Eur J Biochem. 1996;235:215–224. doi: 10.1111/j.1432-1033.1996.00215.x. [DOI] [PubMed] [Google Scholar]
- Crowell DN, Kadlecek AT, Amasino RM. Cytokinin-induced mRNAs in cultured soybean cells. Proc Natl Acad Sci USA. 1990;87:8815–8819. doi: 10.1073/pnas.87.22.8815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denecke J, Carlsson LE, Vidal S, Höglund A-S, Ek B, van Zeijl MJ, Sinjorgo KMC, Palva ET. The tobacco homolog of mammalian calreticulin is present in protein complexes in vivo. Plant Cell. 1995;7:391–406. doi: 10.1105/tpc.7.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes BP, Crowell DN. Cytokinin regulates the expression of a soybean β-expansin gene by a post-transcriptional mechanism. Plant Mol Biol. 1998;37:437–444. doi: 10.1023/a:1005920732211. [DOI] [PubMed] [Google Scholar]
- Grobe K, Becker WM, Schlaak M, Petersen A. Grass group I allergens (β-expansins) are novel, papain-related proteinases. Eur J Biochem. 1999;263:33–40. doi: 10.1046/j.1432-1327.1999.00462.x. [DOI] [PubMed] [Google Scholar]
- Guan K-L, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Kalinski A, Melroy DL, Dwivedi RS, Herman EM. A soybean vacuolar protein (p34) related to thiol proteases is synthesized as a glycoprotein precursor during seed maturation. J Biol Chem. 1992;267:12068–12076. [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Link BM, Cosgrove DJ. Acid-growth response and α-expansins in suspension cultures of bright yellow 2 tobacco. Plant Physiol. 1998;118:907–916. doi: 10.1104/pp.118.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- McQueen-Mason S, Cosgrove DJ. Disruption of hydrogen bonding between wall polymers by proteins that induce plant wall extension. Proc Natl Acad Sci USA. 1994;91:6574–6578. doi: 10.1073/pnas.91.14.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Cosgrove DJ. Expansin mode of action on cell walls. Plant Physiol. 1995;107:87–100. doi: 10.1104/pp.107.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Durachko DM, Cosgrove DJ. Two endogenous proteins that induce cell wall expansion in plants. Plant Cell. 1992;4:1425–1433. doi: 10.1105/tpc.4.11.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Fry SC, Durachko DM, Cosgrove DJ. The relationship between xyloglucan endotransglycosylase and in vitro cell wall extension in cucumber hypocotyls. Planta. 1993;190:327–331. doi: 10.1007/BF00196961. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Petersen A, Grobe K, Linder B, Schlaak M, Becker W-M. Comparison of natural and recombinant isoforms of grass pollen allergens. Electrophoresis. 1997;18:819–825. doi: 10.1002/elps.1150180528. [DOI] [PubMed] [Google Scholar]
- Rayle DL, Cleland RE. The acid growth theory of auxin-induced cell elongation is alive and well. Plant Physiol. 1992;99:1271–1274. doi: 10.1104/pp.99.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Bennett AB. Cooperative disassembly of the cellulose-xyloglucan network of plant cell walls: parallels between cell expansion and fruit ripening. Trends Plant Sci. 1999;4:176–183. doi: 10.1016/s1360-1385(99)01405-3. [DOI] [PubMed] [Google Scholar]
- Shcherban TY, Shi J, Durachko DM, Guiltinan MJ, McQueen-Mason S, Shieh M, Cosgrove DJ. Molecular cloning and sequence analysis of expansins: a highly conserved, multigene family of proteins that mediate cell wall extension in plants. Proc Natl Acad Sci USA. 1995;92:9245–9249. doi: 10.1073/pnas.92.20.9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DA, Fisher PA. Identification, developmental regulation, and response to heat shock of two antigenically related forms of a major nuclear envelope protein in Drosophila embryos: application of an improved method for affinity purification of antibodies using polypeptides immobilized on nitrocellulose blots. J Cell Biol. 1984;99:20–28. doi: 10.1083/jcb.99.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins TA, Bednarek SY, Raikhel NV. Role of propeptide glycan in post-translational processing and transport of barley lectin to vacuoles in transgenic tobacco. Plant Cell. 1990;2:301–313. doi: 10.1105/tpc.2.4.301. [DOI] [PMC free article] [PubMed] [Google Scholar]