Abstract
Applying 10 pmol of okadaic acid (OA), a specific inhibitor of type 1 or type 2A serine/threonine protein phosphatases, to the orchid (Phalaenopsis species) stigma induced a dramatic increase in ethylene production and an accelerated senescence of the whole flower. Aminoethoxyvinylglycine or silver thiosulfate, inhibitors of ethylene biosynthesis or action, respectively, effectively inhibited the OA-induced ethylene production and retarded flower senescence, suggesting that the protein phosphatase inhibitor induced orchid flower senescence through an ethylene-mediated signaling pathway. OA treatment induced a differential expression pattern for the 1-aminocyclopropane-1-carboxylic acid synthase multigene family. Accumulation of Phal-ACS1 transcript in the stigma, labelum, and ovary induced by OA were higher than those induced by pollination as determined by “semiquantitative” reverse transcriptase-polymerase chain reaction. In contrast, the transcript levels of Phal-ACS2 and Phal-ACS3 induced by OA were much lower than those induced by pollination. Staurosporine, a protein kinase inhibitor, on the other hand, inhibited the OA-induced Phal-ACS1 expression in the stigma and delayed flower senescence. Our results suggest that a hyper-phosphorylation status of an unidentified protein(s) is involved in up-regulating the expression of Phal-ACS1 gene resulting in increased ethylene production and accelerated the senescence process of orchid flower.
The gaseous plant hormone ethylene is involved in senescence of plant organs, such as fruits, leaves, and flowers. Application of inhibitors of ethylene biosynthesis or action delays the senescence process, indicating that ethylene plays an important signaling role in these processes (Yang and Hoffman, 1984).
Ethylene is synthesized in plant tissues via the conversions of S-adenosyl-Met to 1-aminocyclopropane-1-carboxylic acid (ACC), catalyzed by ACC synthase, and of ACC to ethylene, catalyzed by ACC oxidase. ACC synthase, which is encoded by a multiple gene family, is generally regarded as the rate-limiting enzyme in the ethylene biosynthetic pathway (Yang and Hoffman, 1984; Kende, 1993). The expression of the enzyme is controlled by a variety of environmental factors, such as flooding (Olson et al., 1995; Shiu et al., 1998) and chilling (Lelievre et al., 1997), and developmental factors, such as fruit ripening and flower senescence (Rottmann et al., 1991). However, the mechanism by which the expression of different ACC synthase genes is activated is not well understood at the signal transduction level. Senescence of the orchid (Phalaenopsis spp.) flower has been shown to serve as a good model system for addressing such a question.
Pollination of the orchid flower induces a dramatic increase in ethylene production, which subsequently causes a rapid petal wilting, whereas the longevity of intact un-pollinated flowers may reach as long as several months (O'Neill et al., 1993). Three ACC synthase genes, Phal-ACS1, Phal-ACS2, and Phal-ACS3, are differentially expressed in the pollinated orchid flower as recently shown by Bui and O' Neill (1998). They suggested that Phal-ACS2 and Phal-ACS3 are induced by primary, whereas Phal-ACS1 is induced by a secondary pollination signal. Porat et al. (1995) have shown that pollination induces significant increases in the level of protein phosphorylation and ethylene production in orchid flowers. Treating the pollinated flower with inhibitors of ethylene biosynthesis or action prevents the increase in protein phosphorylation (Porat et al., 1995). In an earlier paper by Porat et al. (1994), they briefly stated that treatment of un-pollinated orchid flowers with okadaic acid (OA), a specific inhibitor of type 1 and 2A protein phosphatase (Cohen et al., 1990), accelerated their senescence. However, whether ethylene was involved in OA-induced flower senescence was not investigated, nor was the possible relationships between protein phosphorylation/dephosphorylation and differential control of ACC synthase gene expression.
The use of protein kinase and protein phosphatase inhibitors provides a powerful approach for the initial assessment of the role of protein phosphorylation/dephosphorylation in controlling numerous cellular events (Smith and Walker, 1996). OA and staurosporine (STA), a potent inhibitor of different groups of protein kinase (Tamaoki, 1991), have been frequently used to study the regulation of signal transduction processes in plants. We have therefore chosen orchid flower as a model system and used OA and STA to study the possible role of protein phosphorylation/dephosphorylation in the regulatory mechanism of multiple ACC synthase gene expressions during the flower senescence.
RESULTS
OA-Induced Orchid Flower Senescence via an Ethylene-Mediated Pathway
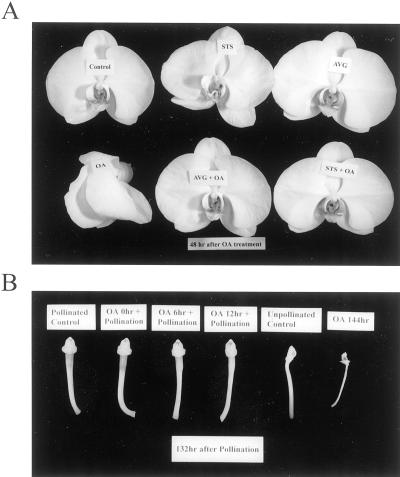
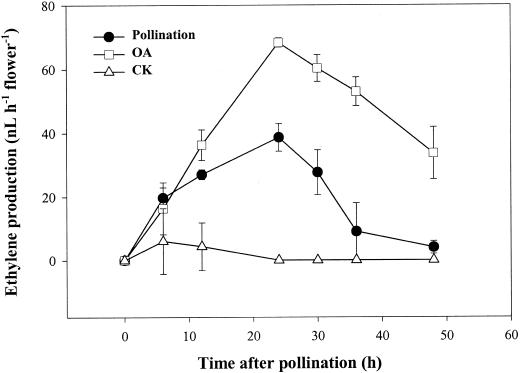
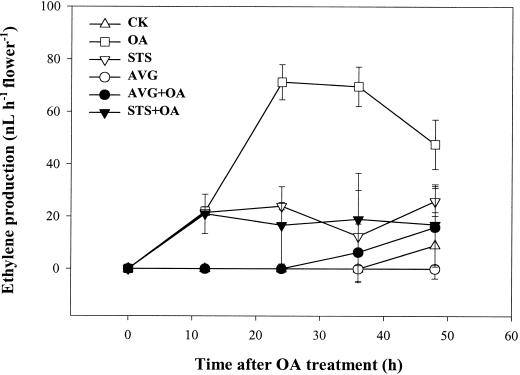
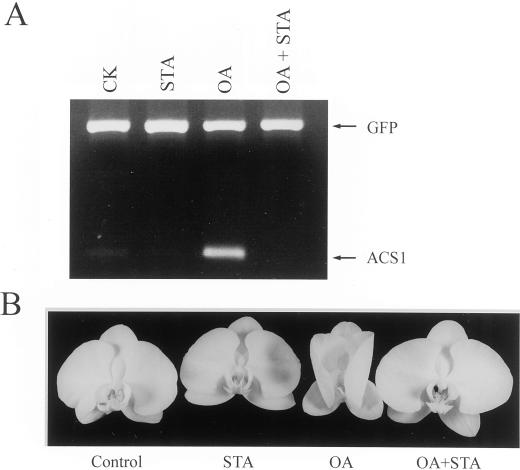
Applying 10 pmol of OA to the orchid stigma induced an accelerated senescence of all organs of the flower in 2 d (Fig. 1A), which was different from the phenotype of pollination-induced senescence by not exhibiting the swelling of stigma and ovary (Fig. 1B). A dramatic increase in ethylene production, which was consistently higher than that induced by pollination, was observed during the OA-induced flower senescence process (Fig. 2). Both the ethylene biosynthesis inhibitor aminoethoxyvinylglycine (AVG) and the ethylene action inhibitor silver thiosulfate (STS) effectively inhibited the OA-induced ethylene synthesis (Fig. 3) and retarded flower senescence (Fig. 1A), suggesting that OA induced orchid flower senescence through an ethylene-mediated signaling pathway. We also found that pollination after 0 to 12 h of OA treatment lowered OA-induced ethylene production to the pollination-induced level (data not shown) and sustained the post-pollination development of the stigma, ovary (Fig. 1B), and fruit (data not shown). These observations suggest that OA-induced senescence was not due to its toxic effect on the plant cells because this effect can be partially reversed by other signals, in this case, pollination.
Figure 1.
Effect of OA treatment or pollination on flower senescence (A) and stigma and ovary development (B). The flowers were detached and kept in distilled water overnight at room temperature followed by pollination, OA (10 pmol) or other chemical treatments as described in “Materials and Methods.”
Figure 2.
Effect of OA treatment or pollination on ethylene production in orchid flower. Flowers were detached and treated as described in Figure 1. At each indicated time point, the flowers were individually enclosed in a 1-L chamber for 1 h, and their ethylene production rates were analyzed by GC as described in “Materials and Methods.” Points and bars represent the mean of three replications ± se.
Figure 3.
Effects of AVG and STS treatments on the OA-induced ethylene production. Twenty microliters of solution containing 0.8 nmol AVG or 2 nmol STS, where indicated, was applied to the stigma 12 h prior to the OA treatment. Ethylene was measures as in Figure 2.
OA-Induced Differential Expression of ACC Synthase Genes in Orchid Flowers
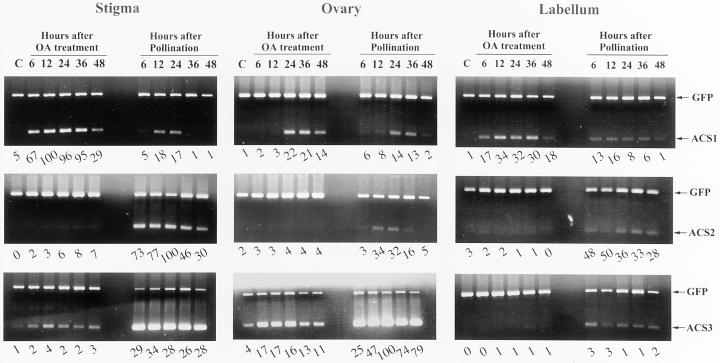
Since the OA-induced flower senescence was apparently caused by an increase in ethylene production, we have therefore investigated whether this compound exerts its effect on ACC synthase gene expression. With a “semiquantitative” reverse transcriptase (RT)-PCR method, we found that OA induced differential expression patterns of the multigene family in orchid flower organs, which was very different from that of the pollinated flower (Fig. 4).
Figure 4.
Time courses of the accumulation of ACC synthase mRNAs in OA-treated or pollinated flower organs analyzed by one-step RT-PCR. For each RT-PCR assay, 20 ng of total RNA and 200 pg of GFP mRNA (positive control) were used. RT-PCR products were resolved on agarose gel and quantified as described in “Materials and Methods.” Digitized intensities of Phal-ACS bands were normalized to that of the GFP band in each lane, and the relative expression levels in percentage for each gene were indicated in numeral at the bottom of each lane. C, Without OA or pollination treatment at time 48 h.
OA treatment induced a rapid accumulation of Phal-ACS1 mRNA in stigma and labelum (reaching its peak at 12 h) and then in ovary (detectable at 24 h after OA treatment) (Fig. 4). The OA-induced expression level of Phal-ACS1 was higher than that induced by pollination in all three organs. OA also induced the Phal-ACS3 gene expression in stigma and ovary, but the level was much lower than that induced by pollination (Fig. 4). In contrast with the results of pollinated flower, the transcript of Phal-ACS2 gene was hardly detected in OA-treated stigma, labelum, or ovary (Fig. 4).
Since Phal-ACS1 is positively regulated by ethylene, it is likely that OA promoted ethylene production by affecting ACC synthase activity in OA-treated flower, which subsequently up-regulated Phal-ACS1 expression. It should be noted that AVG, which inhibited the OA-induced ethylene production (Fig. 3), did not inhibit the OA-induced Phal-ACS1 expression in stigma (data not shown), suggesting that the initial induction of Phal-ACS1 was not associated with ethylene production. STA, a protein kinase inhibitor, on the other hand, inhibited the OA-induced Phal-ACS1 expression in stigma (Fig. 5A) and retarded OA-induced flower senescence (Fig. 5B). These results suggested that protein phosphorylation event(s) up-regulated, while protein dephosphorylation event(s) down-regulated, the Phal-ACS1 gene at the transcriptional level in orchid flower.
Figure 5.
Effect of STA on OA-induced Phal-ACS1 expression in orchid stigma (A) and flower senescence after 48 h of treatment (B). STA (0.3 nmol) or a combination of STA (0.3 nmol) and OA (10 pmol) were applied to the stigma. Total RNAs were isolated 12 h after chemical treatments. RT-PCR analysis was performed as described in Figure 4.
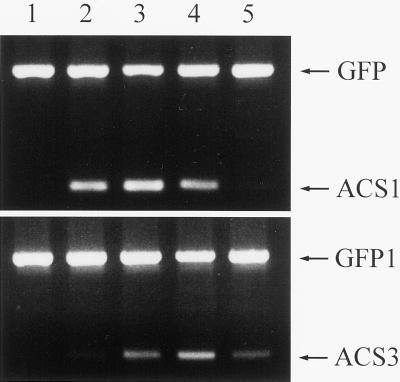
The semiquantitative RT-PCR method was also conducted to investigate the expression patterns of ACC synthase genes during natural senescence process in the stigma of orchid flower. Similar to the OA-induced senescence, naturally senescent flowers accumulated Phal-ACS1 and Phal-ACS3 transcripts in stigma at the onset of the senescence process (Fig. 6), whereas no detectable expression of Phal-ACS2 gene was observed (data not shown).
Figure 6.
Expression pattern of ACC synthase genes in stigma during natural senescence of orchid flower. Flowers were harvested from intact plants according to their senescence stages: lane 1, fully opened; lane 2, petals started to fold up; lane 3, petals fold one-fourth of the way up; lane 4, petals fold half way up; lane 5, petals fold all the way up. One-step RT-PCR analysis was performed as described in Figure 4.
DISCUSSION
Here, we report physiological and molecular changes during an accelerated senescence process of orchid flower induced by OA, a type 1 or type 2A Ser/Thr protein phosphatases inhibitor. OA treatment dramatically shortens the longevity of the flower from more than several months to just within 2 d, which is similar to the pollination effect (Porat et al., 1994; present study). OA apparently has little effect on the pollination and post-pollination process of the orchid flower, which is somehow unexpected. It has been shown that OA inhibits pollen tube growth during cross-pollination in Brassica flowers (Rundle et al., 1993). Since growing pollen tubes of orchid flowers are required to supply auxin to the ovary for a mature fruit development (Zhang and O'Neill, 1993), the normal fruit formation of the OA-treated flower followed by pollination indicates that OA treatment on stigma does not interfere with the orchid pollen tube growth.
In this study, we found that OA-induced flower senescence observed by Porat et al. (1994) was apparently mediated through the production of ethylene. Furthermore, transcription of ACC synthase genes were differentially influenced by OA treatment. Our result is the first report to show the possible relationship of ACC synthase gene expression and protein phosphorylation during flower senescence. Measurement of gene transcripts by RT-PCR revealed that expression patterns of ACC synthase genes in OA-treated flower organs differ notably from that of pollination (Fig. 4). The most obvious difference was the earlier and higher expression of Phal-ACS1 induced by OA (Fig. 4), indicating that the higher level of ethylene produced by OA-treated flower (Fig. 2) resulted from more abundance of its transcript.
It has been shown that protein phosphatase inhibtor calyculin A causes a rapid increase of ACC synthase activity in tomato suspension-cultured cells presumably by affecting its turnover (Spanu et al., 1994). Since Phal-ACS1 is known to be autocatalytically regulated by ethylene signal (Bui and O' Neill, 1998), it is possible that OA activated ACC synthase activity and promoted ethylene production, which in turn caused the induction of Phal-ACS1. The possible involvement of ethylene on OA-induced Phal-ACS1 expression was examined by using ethylene synthesis inhibitor AVG. AVG inhibited ethylene production by OA-treated flower to an undetectable level (Fig. 3) but did not affect Phal-ACS1 expression (data not shown). We found that detached orchid flower did not accumulate Phal-ACS1 transcript up to 12 h of treatment at exogenous ethylene concentration below 1 μL L−1. Therefore, the initial induction of Phal-ACS1 by OA was unlikely due to an increased ethylene production. However, we cannot exclude the possibility that OA might increase ethylene sensitivity or amplify its signal in the flower.
In the ethylene signal transduction pathway, the ethylene receptors resemble prokaryotic “two-component” regulators with a conserved His kinase domain (Chang et al., 1993). A putative Ser/Thr protein kinase, CTR1, located at the downstream of the receptor, is closely related to the animal Raf protein kinase family (Kieber et al., 1993). Both the ethylene receptors (Tieman et al., 2000) and CTR1 (Kieber et al., 1993) have been genetically shown to be negative regulators of the ethylene response pathway. So far, no evidence indicates that protein phosphatases are directly involved in the pathway. At this time, we are not clear how OA treatment could affect the expression of Phal-ACS1. One explanation for this phenomenon is that a delicate balance of the phosphorylation status of certain components of the ethylene signal transduction pathway is closely associated with the induction of the ethylene signal (Ecker, 1995).
The opposing effect exerted by STA on OA-induced Phal-ACS1 gene expression in un-pollinated flowers (Fig. 5) were consistent with the hypothesis that the proteins critical for signaling are the phosphorylated forms that undergo continual phosphorylation and dephosphorylation and that in the absence of the stimuli (e.g. ethylene) the phosphorylation status of the relevant proteins is kept low in the basal state because the protein phosphatases involved are more active than the corresponding protein kinases (Felix et al., 1994). In the presence of OA, the phosphorylation status becomes higher, and, hence, the signaling pathway is activated, while the protein kinase inhibitor STA antagonizes the effect of OA by depressing the transduction of the signal. Our results presented here inferred that hyperphosphorylation of certain unidentified signaling components was responsible for up-regulating Phal-ACS1 during the senescence of orchid flower, which is in agreement with the scheme explaining the induction of ethylene-dependent pathogenesis related genes by phosphorylation events (Raz and Fluhr, 1993).
Another notable difference between the effects of OA and pollination were the expression of Phal-ACS2 and Phal-ACS3 genes. Pollination signal elicited higher levels of both Phal-ACS2 and Phal-ACS3 transcription in stigma and ovary, whereas no expression of Phal-ACS2 gene and quite lower level of Phal-ACS3 expression were observed in OA-treated flower organs (Fig. 5). Phal-ACS2 and Phal-ACS3 have been shown to be regulated primarily by the signal of pollination (Bui and O' Neill, 1998). It is to be noted that the results of expression pattern of the three pollination-induced ACC synthase genes using the semiquantitative RT-PCR method in this study are similar to that reported previously done by northern blotting (Bui and O' Neill, 1998). In this study we observed that Phal-ACS3 gene, which has been suggested to be ovary-specific (Bui and O' Neill, 1998), was also detected in stigma in our experiments (Fig. 5). This may result from the higher sensitivity of RT-PCR to amplify low copy number of transcripts. Bui and O'Neill (1998) hypothesized that Phal-ACS3 in ovary is to synthesize ACC available for translocation to other floral organs. From the expression patterns in natural senescent orchid flower (Fig. 6), Phal-ACS2 seemed to be not required for the ethylene biosynthesis during this process. The transcript of Phal-ACS3 was detected later than that of Phal-ACS1 (Fig. 6), suggesting that, unlike pollination-induced senescence, Phal-ACS3 is not responsible for the induction of Phal-ACS1 in the natural senescence. It is likely that there exists another ACC synthase gene that is induced early during the natural senescence process and is responsible for triggering the subsequent induction of Phal-ACS1. Zhou et al. (1998) recently have shown that in Arabidopsis there exists a cross talk between Glc and ethylene signal transduction, which act antagonistically. Thus, a metabolic change, such as Glc level, during the onset of flower senescence also could induce the expression of Phal-ACS1, resulting in autocatalytic ethylene production and accelerated senescence process.
The involvement of protein phosphorylation events in regulating ACC synthase and hence ethylene biosynthesis has also been reported in other cases, but no definite conclusion have been reached. It is probable that ACC synthase gene could be regulated at multiple levels, i.e. transcriptional, translational, or post-translational, which in turn controls the production of this important hormone. Spanu et al. (1994) have shown that protein kinase inhibitor decreased, whereas protein phosphatase inhibitor increased ACC synthase activity in tomato suspension-cultured cells when pretreated with fungal elicitors. They suggested that protein phosphorylation and dephosphorylation were involved not by regulating the catalytic activity itself but by controlling the rate of turnover of the enzyme. Although Spanu et al. (1994) reported that the in vitro ACC synthase activity was not affected by protein kinase inhibitors, or by direct treatments with protein phosphatases, it is not known whether ACC synthase is phosphorylated or not. Tuomainen et al. (1997) also reported that ACC synthase activity was regulated by protein phosphorylation/dephosphorylation in ozone-exposed tomato plants. Mathooko et al. (1999) recently observed that inhibitors of protein kinase and type 1 and 2A protein phosphatases, which inhibited and stimulated, respectively, CO2 stress-induced ethylene production in cucumber fruit but had little effect on the expression of ethylene biosynthesis genes, suggesting a post-transcriptional control rather than a transcriptional control of ACC synthase. On the other hand, also by using inhibitors of protein kinase and phosphatase, Kim et al. (1997) postulated that both protein phosphorylation and protein dephosphorylation are involved in the ethylene-regulated induction of ACC oxidase transcripts and suppression of the ACC synthase transcript in mung bean hypocotyls. The differences found among these previous reports and our present study might be due to the tissue specificity of the regulatory mechanism.
MATERIALS AND METHODS
Plant Materials
Orchid (Phalaenopsis spp.) plants (cv KC1502) were obtained from a local supplier (KingCar Biotechnology, I-Lan, Taiwan). Fully opened flowers were harvested by excision at the pedicel abscission zone and placed immediately in tubes containing water and kept at room temperature (approximately 25°C) before and after various treatments. Following treatments, the stigma, ovary, and labelum parts were collected separately and frozen in liquid N2 for later use.
Experimental Treatments
For pollination experiments, the detached flowers were self-pollinated by placing the pollinia on the stigma. For chemical treatments, the detached flowers were treated by applying 20 μL of 0.01% (v/v) dimethyl sulfoxide in deionized water as a control or 20 μL of 0.01% (v/v) dimethyl sulfoxide containing 10 pmol OA, 0.3 nmol STA, 2 nmol STS, or 0.8 nmol AVG, or combinations thereof directly to the stigma. When STS or AVG were used, flowers were pretreated with these inhibitors overnight to ensure penetration of the inhibitors prior to treatment with other chemicals. At least four flowers were used for each treatment. Total RNA was isolated after a 12-h exposure to ethylene.
Ethylene Production Measurement
Individual, detached whole flowers in vials containing water were sealed in 1-L chambers for 1 h. One milliliter of gas sample was withdrawn with a syringe from the chamber, and its ethylene concentration was determined by gas chromatography.
RNA Isolation
Total RNA of orchid flower organs was isolated by using a commercial reagent according to the manufacturer's instructions (TRIZOL, Life Technologies/Gibco-BRL, Cleveland). To avoid amplification of Phal-ACS genomic DNA in RT-PCR analysis, isolated RNA was treated with RNase-free DNase I (Promega, Madison, WI) at 37°C for 1 h. The reaction mixture was extracted with phenol:chloroform:isoamyl alcohol (25:24:1, v/v) followed by ethanol precipitation. Phal-ACS1, 2, and 3 genes were not amplified by PCR with their specific primers when the DNase I-treated RNA was used as template, confirming that the RNA samples were apparently free from genomic DNA contamination. Each RNA sample was quantified with a spectrophotometeric method (Sambrook et al., 1989). The A260/A280 values of the RNA samples were all greater than 1.7. To ensure the quality of RNA for RT-PCR analysis, the RNA samples were also visualized on agarose gels following ethidium bromide staining (data not shown).
RT-PCR Analysis
For semiquantification of Phal-ACS transcripts in RNA samples, one-step RT-PCR experiments were performed according to Lee et al. (1997) with modification. In a final volume of 20 μL, the reaction mixtures contained 50 units of SuperScript II RNase H− reverse transcriptase (Life Technologies/Gibco-BRL), 1 unit of Taq DNA polymerase (Life Technologies/Gibco-BRL), 10 nmol of each dNTP, 10 units of RNase inhibitor (Promega), 1 μmol of KCl, 50 nmol MgCl2, 2 μg bovine serum albumin, 0.4 μmol Tris-HCl (pH 8.5), 10 pmol of each primer, and indicated amount of template RNA. To check the efficiency of individual RT-PCR, 200 pg of jellyfish green fluorescence protein (GFP) mRNA produced by in vitro transcription was spiked into the reaction mixtures as an internal control. The GFP mRNA was also found to be free from contamination of its cDNA after RNase-free DNase I treatment. A thermocycler was used to perform 1 cycle of 30 min at 50°C for reverse transcription followed by 35 cycles of 30 s at 94°C, 30 s at 57°C, and 45 s at 68°C, and then 1 cycle of 7 min at 68°C for PCR. Semiquantification of the RT-PCR products was performed by analyzing the digitized images of the DNA bands resolved on agarose gels using computer software (Bio-1D, version 97, Vilber Lourmat, France). Primers used for RT-PCR amplification of Phal-ACS cDNA were designed according to the sequences of orchid. ACC synthase cDNA published by Bui and O'Neill (1998) are as follows: 5′-TGGAGTCACCATCTTCCCAG-3′ and 5′-GGAGATCAGATGAATGGTTT-3′ for Phal-ACS1, 5′-GGATCTTAAGTGGAGAACTGGAGTCAAG-3′ and 5′-TGAGTAG ATTTCGTCGGAGATTAAATGG-3′ for Phal-ACS2, and 5′- GGATCTTAAATGGCGAACCGGAGC-3′ and 5′-GGAGTAGATTTCGTCGCAGATGAGATGAAG-3′ for Phal-ACS3. Plasmid harboring a fragment of ACS1, ACS2, or ACS3 cDNA was used as template to check the specificity of primers, and the results indicated that each primer set was so specific that they did not cross amplify other Phal-ACS cDNA under our PCR conditions. The sequences of the primers used for RT-PCR amplification of GFP cDNA were 5′-TGAGCAAGGGCGAGGAGCTGTTC-3′ and 5′-CTGCAGCCCGGGCGGCCGCTTTAC-3′. To find the amount of RNA suitable for linear amplification, we used 2, 5, 10, 20, and 50 ng of total RNA from pollinated or OA-treated orchid flower organs, and linear increase of RT-PCR products was observed in reactions with RNA up to 50 ng (data not shown). Therefore, we used 20 ng of total RNA for RT-PCR analysis in all of the experiments.
ACKNOWLEDGMENTS
We thank S.J. Chou and H.Y. Li for their helpful assistance in GC operation and RNA isolation, respectively.
Footnotes
This work was supported by the Academia Sinica and by the National Science Council of the Republic of China (grant nos. NSC 86–2311–B–001–031–A18 and NSC 88–2317–B–001–001).
LITERATURE CITED
- Bui AQ, O'Neill SD. Three 1-aminocyclopropane-1-carboxylate synthase genes regulated by primary and secondary pollination signals in orchid flowers. Plant Physiol. 1998;116:419–428. doi: 10.1104/pp.116.1.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Cohen P, Holmes CF, Tsukitani Y. Okadaic acid: a new probe for the study of cellular regulation. Trends Biochem Sci. 1990;15:98–102. doi: 10.1016/0968-0004(90)90192-e. [DOI] [PubMed] [Google Scholar]
- Ecker JR. The ethylene signal transduction pathway in plants. Science. 1995;268:667–675. doi: 10.1126/science.7732375. [DOI] [PubMed] [Google Scholar]
- Felix G, Regenass M, Spanu P, Boller T. The protein phosphatase inhibitor calyculin A mimics elicitor action in plant cells and induces rapid hyperphosphorylation of specific proteins as revealed by pulse labeling with [33P]phosphate. Proc Natl Acad Sci USA. 1994;91:952–956. doi: 10.1073/pnas.91.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende H. Ethylene biosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:283–307. [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Kim JH, Kim WT, Kang BG, Yang SF. Induction of 1-aminocyclopropane-1-carboxylate oxidase mRNA by ethylene in mung bean hypocotyls: involvement of both protein phosphorylation and dephosphorylation in ethylene signaling. Plant J. 1997;11:399–405. [Google Scholar]
- Lee EH, Sitaraman K, Schuster D, Rashtchian A. A highly sensitive method for one-step amplification of RNA by polymerase chain reaction. Focus. 1997;19:39–42. [Google Scholar]
- Lelievre JM, Tichit L, Dao P, Fillion L, Nam YW, Pech JC, Latche A. Effects of chilling on the expression of ethylene biosynthetic genes in Passe-Crassane pear (Pyrus communis L.) fruits. Plant Mol Biol. 1997;33:847–855. doi: 10.1023/a:1005750324531. [DOI] [PubMed] [Google Scholar]
- Mathooko FM, Mwaniki MW, Nakatsuka A, Shiomi S, Kubo Y, Inaba A, Nakamura R. Expression characteristics of CS-ACS1, CS-ACS2 and CS-ACS3, three members of the 1-aminocyclopropane-1-carboxylate synthase gene family in cucumber (Cucumis sativus L.) fruit under carbon dioxide stress. Plant Cell Physiol. 1999;40:164–172. doi: 10.1093/oxfordjournals.pcp.a029524. [DOI] [PubMed] [Google Scholar]
- Olson DC, Oetiker JH, Yang SF. Analysis of LE-ACS3, a 1-aminocyclopropane-1-carboxylic acid synthase gene expressed during flooding in the roots of tomato plants. J Biol Chem. 1995;270:14056–14061. doi: 10.1074/jbc.270.23.14056. [DOI] [PubMed] [Google Scholar]
- O'Neill SD, Nadeau JA, Zhang XS, Bui AQ, Halevy AH. Interorgan regulation of ethylene biosynthetic genes by pollination. Plant Cell. 1993;5:419–432. doi: 10.1105/tpc.5.4.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porat R, Borochov A, Halevy AH. Pollination-induced senescence in Phalaenopsis petals: relationship of ethylene sensitivity to activity of GTP-binding proteins and protein phosphorylation. Physiol Plant. 1994;90:679–684. [Google Scholar]
- Porat R, Halevy AH, Serek M, Borochov A. An increase in ethylene sensitivity following pollination is the initial event triggering an increase in ethylene production and enhanced senescence of Phalaenopsis orchid flowers. Physiol Plant. 1995;93:778–784. [Google Scholar]
- Raz V, Fluhr R. Ethylene signal is transduced via protein phosphorylation events in plants. Plant Cell. 1993;5:523–530. doi: 10.1105/tpc.5.5.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottmann WH, Peter GF, Oeller PW, Keller JA, Shen NF, Nagy BP, Taylor LP, Campbell AD, Theologis A. 1-Aminocyclopropane-1-carboxylate synthase in tomato is encoded by a multigene family whose transcription is induced during fruit and floral senescence. J Mol Biol. 1991;222:937–961. doi: 10.1016/0022-2836(91)90587-v. [DOI] [PubMed] [Google Scholar]
- Rundle SJ, Nasrallah ME, Nasrallah JB. Effects of inhibitors of protein serine/threonine phosphatases on pollination in Brassica. Plant Physiol. 1993;103:1165–1171. doi: 10.1104/pp.103.4.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shiu OY, Oetiker JH, Yip WK, Yang SF. The promoter of LE-ACS7, an early flooding-induced 1-amino- cyclopropane-1-carboxylate synthase gene of the tomato, is tagged by a Sol3 transposon. Proc Natl Acad Sci USA. 1998;95:10334–10339. doi: 10.1073/pnas.95.17.10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RD, Walker JC. Plant protein phosphatases. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:101–125. doi: 10.1146/annurev.arplant.47.1.101. [DOI] [PubMed] [Google Scholar]
- Spanu P, Grosskopf DG, Felix G, Boller T. The apparent turnover of 1-aminocyclopropane-1-carboxylate synthase in tomato cells is regulated by protein phosphorylation and dephosphorylation. Plant Physiol. 1994;106:529–535. doi: 10.1104/pp.106.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaoki T. Use and specificity of staurosporine, UCN-01, and calphostin C as protein kinase inhibitors. Methods Enzymol. 1991;201:340–347. doi: 10.1016/0076-6879(91)01030-6. [DOI] [PubMed] [Google Scholar]
- Tieman DM, Taylor MG, Ciardi JA, Klee HJ. The tomato ethylene receptors NR and LeETR4 are negative regulators of ethylene response and exhibit functional compensation within a multigene family. Proc Natl Acad Sci USA. 2000;97:5663–5668. doi: 10.1073/pnas.090550597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomainen J, Betz C, Kangasjarvi J, Ernst D, Yin ZH, Langebartels C, Sandermann H. Ozone induction of ethylene emission in tomato plants: regulation by differential accumulation of transcripts for the biosynthetic enzymes. Plant J. 1997;12:1151–1162. [Google Scholar]
- Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol. 1984;35:155–189. [Google Scholar]
- Zhang XS, O'Neill SD. Ovary and gametophyte development are coordinately regulated by auxin and ethylene following pollination. Plant Cell. 1993;5:403–418. doi: 10.1105/tpc.5.4.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Jang JC, Jones TL, Sheen J. Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc Natl Acad Sci USA. 1998;95:10294–10299. doi: 10.1073/pnas.95.17.10294. [DOI] [PMC free article] [PubMed] [Google Scholar]