Abstract
Background & Aims
Hepatocellular carcinoma (HCC) is a highly aggressive malignancy with dreadful clinical outcome. Tyrosine kinase inhibitors and immune checkpoint inhibitors are the only United States Food and Drug Administration-approved therapeutic options for patients with advanced HCC with limited therapeutic success. Ferroptosis is a form of immunogenic and regulated cell death caused by chain reaction of iron-dependent lipid peroxidation. Coenzyme Q10 (CoQ10)/ferroptosis suppressor protein 1 (FSP1) axis was recently identified as a novel protective mechanism against ferroptosis. We would like to explore whether FSP1 could be a potential therapeutic target for HCC.
Methods
FSP1 expression in human HCC and paired non-tumorous tissue samples were determined by reverse transcription-quantitative polymerase chain reaction, followed by clinicopathologic correlation and survival studies. Regulatory mechanism for FSP1 was determined using chromatin immunoprecipitation. The hydrodynamic tail vein injection model was used for HCC induction to evaluate the efficacy of FSP1 inhibitor (iFSP1) in vivo. Single-cell RNA sequencing revealed the immunomodulatory effects of iFSP1 treatment.
Results
We showed that HCC cells greatly rely on the CoQ10/FSP1 system to overcome ferroptosis. We found that FSP1 was significantly overexpressed in human HCC and is regulated by kelch-like ECH-associated protein 1/nuclear factor erythroid 2-related factor 2 pathway. FSP1 inhibitor iFSP1 effectively reduced HCC burden and profoundly increased immune infiltrates including dendritic cells, macrophages, and T cells. We also demonstrated that iFSP1 worked synergistically with immunotherapies to suppress HCC progression.
Conclusions
We identified FSP1 as a novel, vulnerable therapeutic target in HCC. The inhibition of FSP1 potently induced ferroptosis, which promoted innate and adaptive anti-tumor immune responses and effectively suppressed HCC tumor growth. FSP1 inhibition therefore represents a new therapeutic strategy for HCC.
Keywords: Hepatocellular Carcinoma, Immunogenic Cell Death, Lipid Peroxidation, Regulated Cell Death
Summary.
Inhibition of ferroptosis suppressor protein 1 suppresses hepatocellular carcinoma tumor growth via the induction of ferroptosis. Ferroptotic cell death also promotes innate and adaptive anti-tumor immunity. This represents a novel and potent therapeutic target for hepatocellular carcinoma with improved treatment outcome.
Hepatocellular carcinoma (HCC) is the most common form of primary liver cancer that originates from hepatocytes. Late symptom presentations result in most patients with HCC diagnosed at advanced stages, and curative treatments like surgical resection are unfeasible.1 Effective therapeutic options for patients with HCC are limited, as HCC is highly recurrent and highly resistant to conventional chemo- and radiotherapy. Currently, tyrosine kinase inhibitors (TKIs) (sorafenib, lenvatinib, regorafenib, cabozantinib) and immune checkpoint inhibitors (ICIs) (nivolumab [anti-PD-1] and atezolizumab [anti-PD-L1]) are the only United States Food and Drug Administration-approved drugs for patients with advanced HCC.2 Unfortunately, TKIs provide modest survival benefits, whereas ICIs are only effective in a small subset of patients with HCC. Drugs that can target HCC cells to cell death are worth investigation. Further investigation into drugs that can induce HCC cell death will benefit the development of effective HCC treatment, as these agents may be utilized as HCC therapeutics.
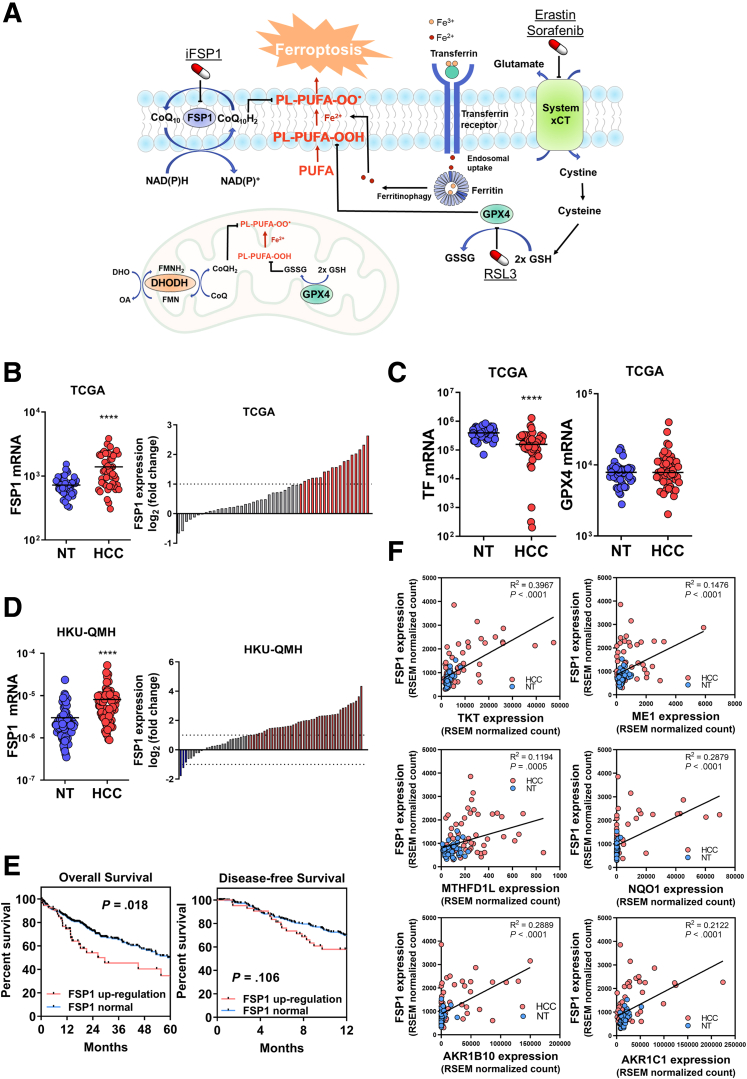
Ferroptosis is the most-recently discovered form of regulated cell death (RCD) that aggravates degenerative diseases and paradoxically suppresses cancer cell growth.3,4 Ferroptosis vastly differs from conventional forms of RCDs. Morphologically, ferroptosis displays outer mitochondrial membrane rupture, shrunken mitochondria with condensed membrane densities, and loss of cristae.5 Biochemically, ferroptosis is dependent on excessive iron-mediated lipid peroxidation.6 Iron reacts with hydrogen peroxides to generate hydroxyl radicals, highly toxic reactive oxygen species (ROS) that further react with polyunsaturated fatty acid (PUFA) tails on the cell membrane lipid bilayer to generate lipid peroxides, a process called lipid peroxidation. Lipid peroxides establish a chain reaction and produce more lipid peroxides. These further cause damage to the cell membrane, which eventually causes ferroptosis. Iron is essential for lipid peroxidation chain reaction propagation and also for catalyzing iron-dependent enzymes like lipoxygenases to generate lipid peroxides.7 Iron is transported from the blood into cells by transferrin (TF), stored in ferritin, and released upon ferritinophagy, an autophagic degradation process of ferritin (Figure 1A).8, 9, 10 Cells have 3 major protective mechanisms against ferroptosis: (1) the cysteine/glutathione (GSH)/glutathione peroxidase 4 (GPX4) pathway; (2) the mitochondrial GPX4-dependent coenzyme Q (CoQ)/dihydroorotate dehydrogenase (DHODH) pathway; and (3) the CoQ10/ferroptosis suppressor protein 1 (FSP1) pathway. The cysteine/GSH/GPX4 pathway was the first discovered and most well-known protective pathway against ferroptosis.11 In cysteine/GSH/GPX4 pathway, GPX4 removes phospholipid peroxides using reduced GSH. GSH synthesis depends on the xCT cystine-glutamate antiporter (encoded by SLC7A11 and SLC3A2), which pumps cystine into cells in exchange for glutamate.5 GPX4 reduces phospholipid hydroperoxides (PL-PUFA-OOH) to counteract ferroptosis. Dihydroorotate dehydrogenase is a mitochondrial enzyme localized on the outer face of the inner membrane. It converts dihydroorotate to orotate and generates FMNH2 to reduce ubiquinone (CoQ) into ubiquinol (CoQH2) to protect cells from ferroptosis at the mitochondrial membrane.12 The most common form of CoQ in human is CoQ10.13 In the CoQ10/FSP1 pathway, FSP1, a CoQ oxidoreductase, converts CoQ to CoQH2 to stabilize PL-PUFA-OOH.14,15 The N-terminal myristylation of FSP1 localizes FSP1 to cell membrane, which aids PL-PUFA-OOH removal.14,15 The CoQ10/FSP1 pathway acts independently of GPX4 as FSP1 does not affect the xCT antiporter and cysteine transport, GSH levels, and total PL-PUFA (Figure 1A). Although the GPX4 pathway is an attractive therapeutic target for cancer treatment through ferroptosis induction, the efficacy of current GPX4 inhibitors is limited to in vitro studies, and the responses vary across different cancer cell lines.16 Inhibitors, such as erastin and sorafenib, that target xCT antiporter are not highly specific. GPX4 is expressed in T cells, and these inhibitors could therefore inhibit T cells and cause T cell exhaustion in the cancer context.17 These limitations prompted us to explore new anti-ferroptotic mechanisms beyond the cysteine/GSH/GPX4 pathway to discover new therapeutic strategies against HCC. The CoQ10/FSP1 pathway is another protective pathway against ferroptosis that works independently of the cysteine/GSH/GPX4 pathway.14,15 A potent FSP1 inhibitor, iFSP1, was identified. iFSP1 induced ferroptosis and suppressed cancer cell growth in vitro with GPX4 inhibited. The in vivo efficacy of iFSP1 is yet to be investigated.15
Figure 1.
FSP1 is overexpressed in human HCC. (A) Schematic representation of pathways involved in ferroptosis regulation. (B) Left: TCGA data revealed upregulation of FSP1 in 49 human HCC tissues compared with corresponding NT liver tissues. Right: Waterfall plot showed upregulation of FSP1 in 37% (18/49) of patients with HCC retrieved from TCGA by at least 2-fold. (C) Left: TCGA data revealed downregulation of TF in human HCC tissues compared with corresponding NT liver tissues. Right: TCGA data revealed GPX4 is not upregulated in human HCC tissues compared with corresponding NT liver tissues. (D) Left: RT-qPCR results from an expanded cohort of 70 human HCC tissues from our institute demonstrated upregulation of FSP1 compared to corresponding NT liver tissues. Right: Waterfall plot showed upregulation of FSP1 in 63% (44/70) of patients with HCC by at least 2-fold. (E) Analysis of TCGA data revealed patients with HCC with FSP1 upregulation (Z-score > 1) are associated with poorer overall and disease-free survival. (F) TCGA data revealed positive correlations between expression of FSP1 with expressions of known NRF2-targeted genes including TKT, ME1, MTHFD1L, NQO1, AKR1B10, and AKR1C1 in human HCC and NT liver tissues. ∗∗∗∗P < .0001 vs NT. B‒D, F: Student t test. E: Kaplan-Meier followed by log-rank test. RSEM = RNA-Seq expression estimation by expectation maximization.
Immunogenic cell death (ICD) is characterized by its release or membrane exposure of damage-associated molecular patterns (DAMPs), which recruit and activate innate immune cells, subsequently eliciting adaptive immune responses.18,19 Innate immune cells play a key role in ICD-elicited anti-tumor immune responses. Dendritic cells (DCs) internalize dying cells or cellular fragments for presenting tumor-associated antigens to T cells via major histocompatibility complex molecules.20 Macrophages engulf dying cells and mediate the removal of cancer cells, accompanied by the production of pro-inflammatory cytokines.21 The immunomodulatory roles of ferroptotic cells in HCC are yet to be explored. Apart from activating innate immune cells through ICD induction, a new generation of immunotherapies that target innate immune cells has emerged as a promising therapeutic approach. Among these is CD40 agonist. CD40 is a co-stimulatory receptor expressed on antigen presenting cells (APCs), such as macrophages and DCs. Ligation of CD40 with CD40L expressed on activated T cells enhances antigen presentation by macrophages and DCs.22 CD40 agonist was shown to suppress tumor growth by enhancing major histocompatibility complex and co-stimulatory molecule expression on DCs23 and reprogramming tumor-associated macrophage (TAM).24,25 Here, we explore whether CD40 agonist can further activate innate immune cells and enhance iFSP1 efficacy in HCC.
We hypothesize that targeting FSP1 induces ferroptosis in cancer cells and promotes anti-tumor immune responses. Here, we showed that targeting FSP1 alone is sufficient to severely induce ferroptosis and suppress HCC cells proliferation as well as HCC tumor growth even with intact GPX4. Our work suggested that perturbation of the CoQ10/FSP1 pathway suffices to inhibit HCC development through ferroptosis induction. More excitingly, we unprecedentedly showed that iFSP1 treatment in mice profoundly increased the number of DCs, macrophages, and T cells in the tumors and suggested that iFSP1 works in synergy with multiple immunotherapies to improve the survival of mice with HCC.
Results
FSP1 Overexpression is Significantly Associated With Human HCC
To understand the clinical significance of ferroptosis regulation, especially FSP1 in human HCC, we made use of transcriptome sequencing results from 49 paired human HCC/non-tumorous (NT) liver tissue samples from The Cancer Genome Atlas (TCGA). Significant overexpression of FSP1 was found in human HCC tissues compared with NT tissues (Figure 1B). These transcriptome sequencing results also revealed significant downregulation of pro-ferroptotic gene TF (Figure 1C). Interestingly, genes from the classic cysteine/glutathione/GPX4 anti-ferroptosis pathway, including GPX4, a major anti-ferroptotic gene, were not upregulated in human HCC (Figure 1C). This suggested that HCC was protected from ferroptosis by limiting iron uptake into cancer cells and upregulation of FSP1 pathway but not GPX4 pathway, as both pathways work independently to counteract ferroptosis. An expanded cohort of 70 HCC cases from our institute (HKU-QMH) also demonstrated FSP1 overexpression in HCC tissues over NT liver tissues at the mRNA level determined by real-time quantitative polymerase chain reaction (RT-qPCR). FSP1 overexpression by at least 2-fold was found in 44 of 70 (62.9%) of the HCC tumor samples (Figure 1D). Clinicopathologically, FSP1 overexpression in HCC samples from our institute were significantly associated with patients with HCC with larger tumor size (>5 cm) (Table 1). Moreover, TCGA data indicated patients with HCC with higher FSP1 expression were significantly correlated with poorer overall survival (P = .018) and were associated with poorer disease-free survival (P = .106) (Figure 1E). Interestingly, we found positive correlation between mRNA expression of FSP1 and nuclear factor erythroid 2-related factor 2 (NRF2), transcriptional targets we previously identified, including transketolase (TKT), malic enzyme 1 (ME1), and methylenetetrahydrofolate dehydrogenase 1-like (MTHFD1L), as well as other well-known NRF2 targets NAD(P)H dehydrogenase, quinone 1 (NQO1), aldo-keto reductase family 1 member B10 (AKR1B10), and aldo-keto reductase family 1 member C1 (AKR1C1) in human HCC and NT liver tissues from TCGA (Figure 1F).26, 27, 28 Expression analyses from clinical HCC samples on TCGA and our institute converged to identify overexpression of FSP1 in human HCC is significantly correlated with dismal clinicopathological features of HCC.
Table 1.
Clinicopathological Correlation of FSP1 Overexpression in Human HCC
| Clinicopathological phenotype | Number of cases | Mean | P-value |
|---|---|---|---|
| Cellular differentiation by Edmonson grading | |||
| I‒III | 23 | 0.43 | .101 |
| IV‒VI | 40 | 0.23 | |
| Chronic liver disease | |||
| Normal | 12 | 0.25 | .638 |
| Chronic hepatitis and cirrhosis | 53 | 0.32 | |
| Direct liver invasion | |||
| Absent | 24 | 0.29 | .726 |
| Present | 36 | 0.25 | |
| Hepatitis B surface antigen by IHC | |||
| Absent | 16 | 0.25 | .511 |
| Present | 44 | 0.34 | |
| Hepatitis C surface antigen by IHC | |||
| Absent | 13 | 0.38 | .018a |
| Present | 21 | 0.00 | |
| Plasma hepatitis B surface antigen | |||
| Absent | 9 | 0.33 | .718 |
| Present | 40 | 0.40 | |
| pTNM stage | |||
| I/II | 30 | 0.30 | .718 |
| I/II/III/IV | 32 | 0.34 | |
| Tumor encapsulation | |||
| Absent | 27 | 0.41 | .440 |
| Present | 17 | 0.53 | |
| Tumor microsatellite formation | |||
| Absent | 25 | 0.36 | .550 |
| Present | 35 | 0.29 | |
| Tumor size | |||
| ≤ 5 cm | 33 | 0.18 | .021a |
| > 5 cm | 31 | 0.45 | |
| Venous invasion | |||
| Absent | 19 | 0.47 | .829 |
| Present | 25 | 0.44 |
Note: Mean: average values of FSP1 expression in human HCC samples relative to its paired NT tissues determined with the formula: ΔCtHCC(FSP1-18S) – ΔCtNT(FSP1-18S).
Note: Boldface P values indicate statistical significance.
FSP1, Ferroptosis suppressor protein 1; HCC, hepatocellular carcinoma; IHC, immunohistochemistry; NT, non-tumorous; pTNM, pathological tumor-node-metastasis.
T test: P < .05.
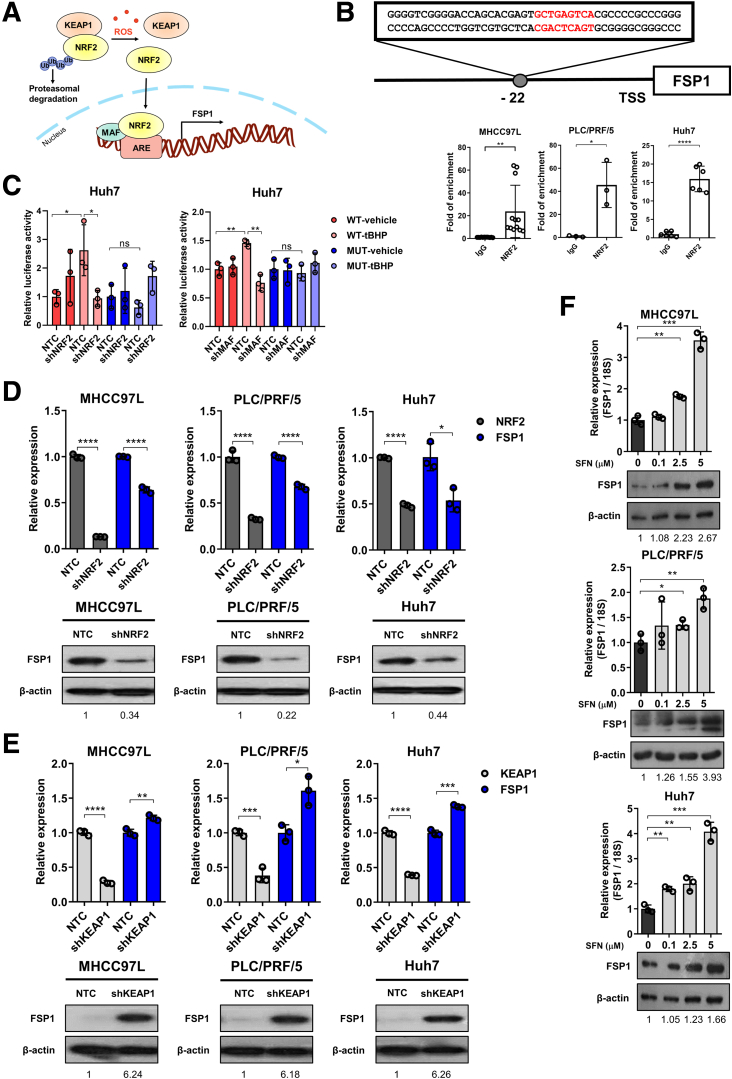
FSP1 is Regulated by the KEAP1/NRF2 Pathway
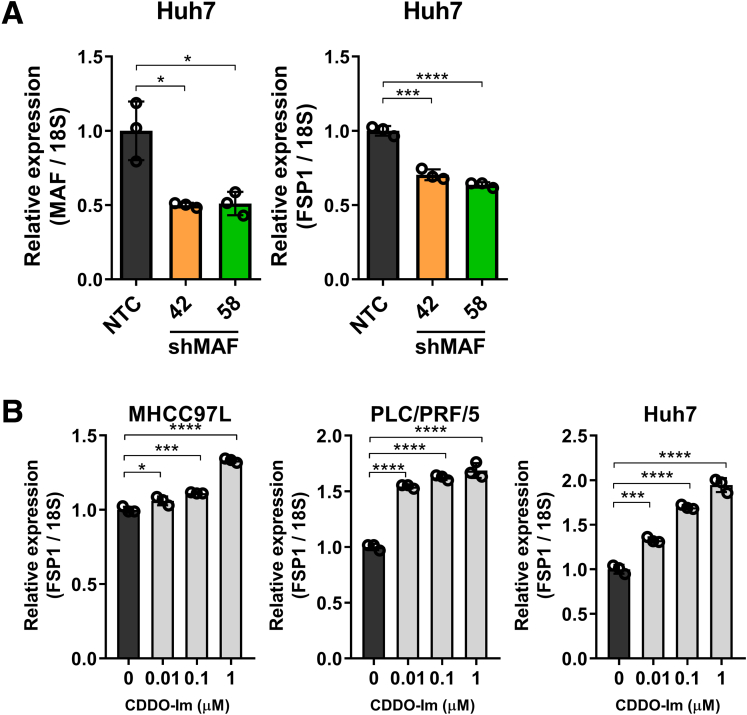
Next, we aimed to understand the regulatory mechanism that drives the overexpression of FSP1 in human HCC. Because FSP1 is essential for counteracting lipid peroxidation, it was postulated that FSP1 expression is regulated by the kelch-like ECH-associated protein 1 (KEAP1)/NRF2 pathway like other oxidative stress-related genes. KEAP1/NRF2 is the master regulatory pathway for an array of genes known for protection against oxidative stress.29 We found positive correlation between FSP1 expression with NRF2 transcriptional targets we previously found, including TKT, ME1, MTHFD1L, and known NRF2 targets NQO1, AKR1B10, and AKR1C1 (Figure 1F), strongly suggesting that FSP1 overexpression in human HCC is regulated by the KEAP1/NRF2 pathway (Figure 2A).26, 27, 28 Oxidative stress is a common feature in solid tumors and is believed to promote FSP1 upregulation in human HCC. In silico analysis identified a putative antioxidant response element (ARE) at −22 bp upstream of the transcription start site in FSP1. Furthermore, dramatic enrichment of NRF2 binding at the mapped putative ARE of FSP1 was confirmed by chromatin immunoprecipitation (ChIP) assay using NRF2 and IgG antibodies on multiple HCC cell lines, including MHCC97L, PLC/PRF/5, and Huh7 (Figure 2B). We next sought to confirm the regulatory role of KEAP1/NRF2 and small Maf proteins that dimerize with NRF2 to bind to ARE sequences on FSP1. Stable NRF2, KEAP1, and MAF BZIP transcription factor (MAF) knockdown clones were established in multiple HCC cell lines, including MHCC97L, PLC/PRF/5, and Huh7 using short hairpin RNAs (shRNAs) (shNRF2, shKEAP1, and shMAF). To show that the ARE we identified at FSP1 promoter region is functional and how NRF2 and MAF regulates FSP1, we performed luciferase reporter assay. Wild-type (WT) and mutated (MUT) AREs were first inserted into pGL4 luciferase reporter vector, and we transfected stable NRF2 and MAF knockdown clones of Huh7 with the WT-ARE or MUT-ARE containing luciferase plasmids. We then exposed the cells to treatment of tert-Butyl hydroperoxide (tBHP; Sigma-Aldrich, St. Louis, MO), an oxidant to induce NRF2, and determined the FSP1 promoter luciferase activities. We showed that tBHP treatment induced luciferase activities in cells expressing WT-ARE but not the MUT one (Figure 2C). The induction of promoter activities by tBHP in WT-ARE expressing cells could be abrogated by NRF2 or MAF knockdown (Figure 2C). We previously demonstrated the NRF2 stable knockdown clones down-regulated the expressions of known NRF2-target genes like TXNRD1 and ME1, whereas KEAP1 stable knockdown clones up-regulated the same targets.27,30 These results validate the validity of the NRF2 and KEAP1 inhibitions, and, consistent with our hypothesis, NRF2 inhibition effectively down-regulated FSP1 expressions (Figure 2D) and inhibition of KEAP1, the NRF2 suppressor, effectively up-regulated FSP1 expressions (Figure 2E) at both the mRNA and protein levels. We also confirmed that MAF contributes to FSP1 regulation in stable MAF knockdown clones. The inhibition of MAF effectively down-regulated FSP1 expression at the mRNA level (Figure 3A). Finally, we made use of a chemical NRF2 inducer L-sulforaphane (SFN; Sigma-Aldrich) and treated HCC cells (MHCC97L, PLC/PRF/5, Huh7) at various concentrations. Dose-dependent up-regulation of FSP1 were consistently observed at mRNA and protein levels upon SFN treatment across all 3 HCC cell lines (Figure 2F). We further confirmed our findings using another NRF2 inducer CDDO-Imidazolide (CDDO-Im; Tocris Bioscience, Bristol, UK), as dose-dependent up-regulation of FSP1 was also observed at the mRNA level across different HCC cells upon CDDO-Im treatment (Figure 3B). Taken together, in silico analysis, ChIP assay, luciferase reporter assay, and NRF2 inhibition strongly suggested that FSP1 is transcriptionally regulated by NRF2 in HCC.
Figure 2.
FSP1 is regulated by NRF2 in human HCC. (A) Schematic representation of FSP1 regulation mediated by the KEAP1/NRF2 pathway. (B) Top: ARE was located at -22 bp upstream of the transcription start site (TSS) of FSP1. Bottom: ChIP assay on HCC cells including MHCC97L, PLC/PRF/5, and Huh7 using NRF2, and IgG antibodies demonstrated specific and dramatic enrichment of NRF2 at the identified putative ARE of FSP1. (C) Left: Luciferase reporter assay showed that tert-Butyl Hydroperoxide (tBHP) treatment induced luciferase activities in Huh7 cells transfected with luciferase plasmid containing WT ARE we identified but not those with MUT ARE or knockdown of NRF2. Right: Luciferase reporter assay showed that tBHP-induced luciferase activities in Huh7 cells expressing WT ARE were abrogated upon knockdown of MAF. (D) Knockdown of NRF2 in MHCC97L, PLC/PRF/5, and Huh7 cells significantly downregulated FSP1 expressions at mRNA and protein levels confirmed by RT-qPCR and Western blotting, respectively. (E) Knockdown of KEAP1 in MHCC97L, PLC/PRF/5, and Huh7 cells significantly upregulated FSP1 expressions at mRNA and protein levels confirmed by RT-qPCR and Western blotting, respectively. (F) Activation of NRF2 upon treatments of 0.1, 2.5, 5 μM NRF2 activator SFN resulted in dose-dependent upregulation of FSP1 mRNA and protein expressions in MHCC97L, PLC/PRF/5, and Huh7 cells as confirmed by RT-qPCR and Western blotting, respectively. Error bars indicate mean ± SD. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001 vs IgG, NTC, or 0 μM as indicated. Student t test.
Figure 3.
FSP1 is regulated by NRF2 and MAF in human HCC. (A) Left: Knockdown of MAF in Huh7 cells is confirmed by RT-qPCR. Right: Knockdown of MAF in Huh7 cells significantly downregulated FSP1 expressions at mRNA level as confirmed by RT-qPCR. (B) Activation of NRF2 upon treatments of 0.01, 0.1, 1 μM NRF2 activator CDDO-Im resulted in dose-dependent upregulation of FSP1 mRNA expressions in MHCC97L, PLC/PRF/5, and Huh7 cells as confirmed by RT-qPCR. ∗P < .05, ∗∗∗P < .001, ∗∗∗∗P < .0001 vs NTC or 0 μM as indicated. Error bars indicate mean ± SD. Student t test.
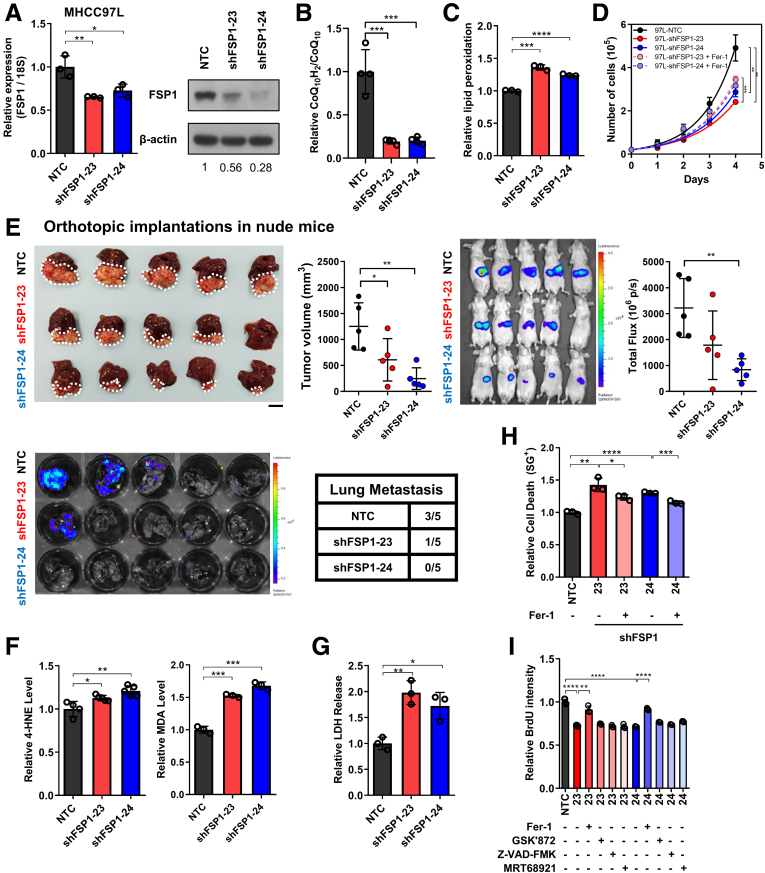
Knockdown of FSP1 Resulted in Ferroptosis-induced HCC Cell Death
We next studied the functional roles of FSP1 in human HCC, and we performed loss-of-function studies. First, stable FSP1 knockdown clones were established in MHCC97L cells with 2 independent shRNA sequences (shFSP1-23, -24). Significant inhibition of FSP1 expressions compared with non-targeting control (NTC) were evident in mRNA and protein levels (Figure 4A). FSP1 is an oxidoreductase, converting oxidized CoQ10 into reduced CoQ10H2. Liquid chromatography-mass spectrometry (LC-MS) confirmed that knockdown of FSP1 significantly reduced intracellular CoQ10H2/CoQ10 ratio (Figure 4B). Through the reduction of CoQ10, FSP1 is important for lipid peroxidation prevention, and we detected significant increase in intracellular lipid peroxidation in MHCC97L cells upon FSP1 knockdown (shFSP1-23, -24) compared with NTC control cells using BODIPY 581/591 C11 staining (Invitrogen, Carlsbad, CA) (Figure 4C). FSP1 knockdown obstructed HCC cell growth in vitro as cell proliferation rates of both knockdown clones (shFSP1-23, -24) were significantly lower than NTC cells. The reduced cell proliferation rates of the FSP1 knockdown clones were rescued upon treatment of ferroptosis inhibitor ferrostatin-1 (Fer-1; Sigma-Aldrich) (Figure 4D). This suggested that FSP1 knockdown adversely affects HCC growth through lipid peroxidation-mediated ferroptosis. Luciferase-labeled MHCC97L stable FSP1 knockdown clones and NTC were orthotopically injected into nude mice livers. Tumor volume and bioluminescent measurements consistently revealed that smaller HCC tumors were derived from FSP1 knockdown clones. Moreover, lung tissue imaging ex vivo revealed FSP1 knockdown suppressed the growth of lung metastases (Figure 4E). To confirm that FSP1 is essential for preventing ferroptosis-induced HCC cell death, we confirmed that FSP1 inhibition resulted in the accumulation of intracellular lipid peroxidation. A significant increase in intracellular levels of lipid peroxidation by-products 4-hydroxynonenal (4-HNE) and malondialdehyde (MDA) were detected from FSP1 knockdown stable clones relative to NTC (Figure 4F). We then examined whether cytotoxic effects were elicited because of lipid peroxidation. Quantification of lactate dehydrogenase (LDH) released from damaged cells, an indicator of cell death, revealed that FSP1 knockdown cells released significantly higher levels of LDH compared with NTC (Figure 4G). Furthermore, SYTOX Green dead cell staining (Invitrogen) also demonstrated similar results as LDH release. Fer-1 treatment to FSP1 knockdown clones (shFSP1-23, -24) significantly lowered the relative SYTOX Green staining compared with untreated cells (Figure 4H). Fer-1 treatment was also found to significantly increase relative bromodeoxyuridine (BrdU) intensities in MHCC97L-shFSP1-23 and -24 cells. Treatment of inhibitors of other forms of RCDs including necroptosis (GSK’872), apoptosis (Z-VAD-FMK), and autophagy (MRT68921), however, did not rescue the suppressed cell proliferation rates in FSP1 stable knockdown HCC cells (Figure 4I). The observations altogether demonstrated that only ferroptosis inhibitor (eg, Fer-1) was able to rescue FSP1 knockdown-induced cell death in HCC cells. This indicated the important role of FSP1 for the protection of HCC cells from ferroptosis and cell death.
Figure 4.
FSP1 knockdown induced ferroptotic HCC cell death. (A) Stable FSP1-knockdown subclones (shFSP1) were established using shRNAs. RT-qPCR and Western blotting confirmed the knockdown efficiencies at mRNA and protein levels, respectively. (B) LC-MS demonstrated decrease in relative CoQ10H2/CoQ10 ratio in MHCC97L-shFSP1 cells. (C) BODIPY 581/591 C11 staining in MHCC97L-shFSP1 cells revealed a significant increase in lipid peroxidation. (D) Cell proliferation rates of MHCC97L-NTC and -shFSP1 cells by cell counting daily revealed that FSP1 knockdown significantly suppressed HCC cell growth. Suppressed growth rates were rescued using ferroptosis inhibitor Fer-1. (E) Orthotopic implantation of luciferase-labeled MHCC97L-NTC and MHCC97L-shFSP1-23, and MHCC97L-shFSP1-24 cells into the livers of nude mice (n = 5 mice per experimental group) revealed that FSP1 knockdown suppressed HCC tumor formation in vivo. Top, left: Representative picture of orthotopic xenografts and quantification of tumor volume. Top, right: Bioluminescent images of mice implanted with different stable subclones. Bottom: Bioluminescent images of mice lung tissues with corresponding incidence of lung metastases. (F) Quantitative measurements of natural lipid peroxidation by-products Left: 4-HNE and Right: MDA in MHCC97L-NTC and MHCC97L-shFSP1 cells further suggest increased lipid peroxidation upon FSP1 knockdown. (G) Quantification of relative LDH release in MHCC97L-NTC and MHCC97L-shFSP1 cells demonstrated that intracellular lipid peroxidation induced by FSP1 knockdown significantly increased cell death. (H) SYTOX Green (SG+) staining in MHCC97L-NTC and MHCC97L-shFSP1 cells demonstrated increased cell death from FSP1 knockdown were rescued with Fer-1. (I) Cell proliferation rates of MHCC97L-NTC and MHCC97L-shFSP1 cells determined using BrdU assay indicated that Fer-1, not inhibitors of other modes of RCD GSK’872 (necroptosis), Z-VAD-FMK (apoptosis), and MRT68921 (autophagy), can rescue proliferation rates suppressed by FSP1 knockdown. Error bars indicate mean ± SD. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001 vs NTC or Fer-1 as indicated. Student t test. E: scale bar = 1 cm
iFSP1 Effectively Induced HCC Cell Death Via Immune Cell Recruitment
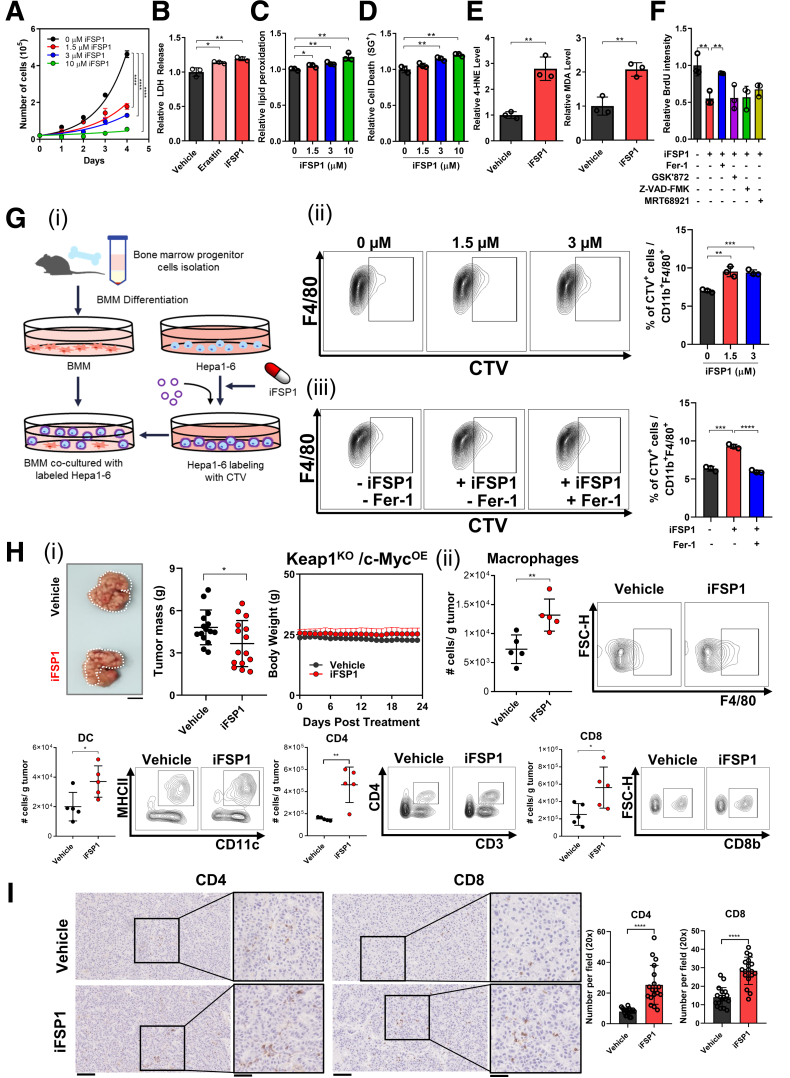
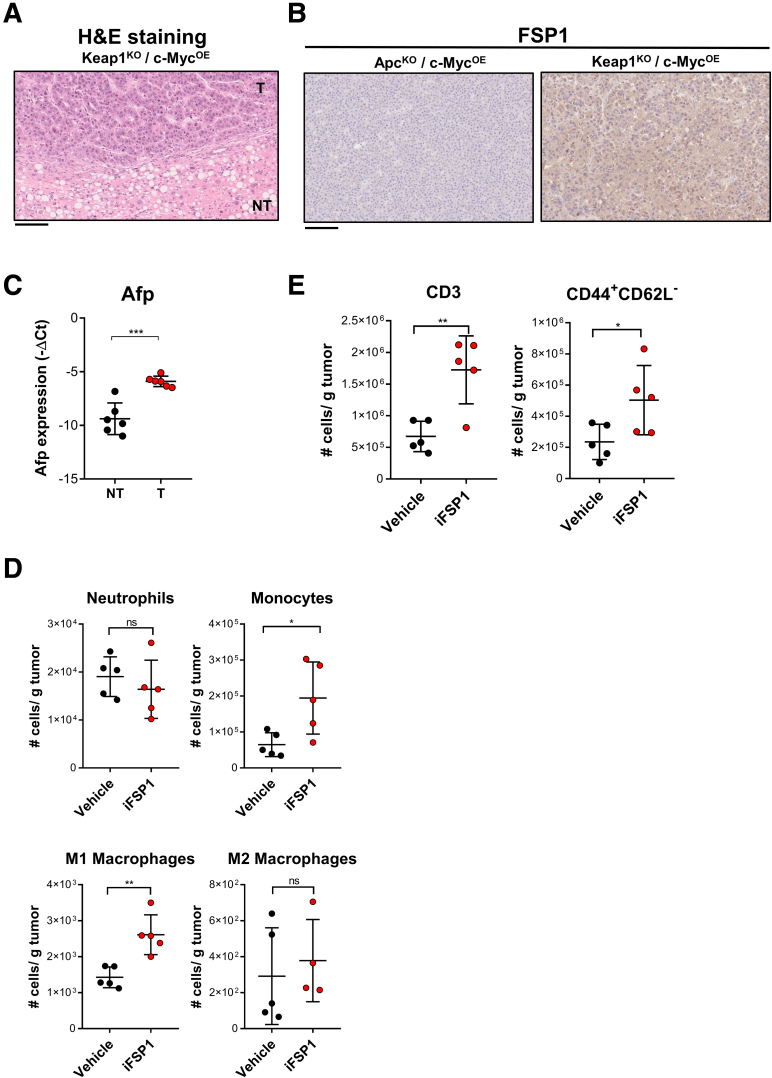
We demonstrated that FSP1 knockdown led to accumulation of intracellular lipid peroxidation, which ultimately resulted in ferroptosis-induced cell death in human HCC. As we now look to translate this concept for clinical application, we made use of a specific chemical FSP1 inhibitor, iFSP1.15 As a novel and sole FSP1 inhibitor, we tested the effects of iFSP1 on MHCC97L cells at multiple concentrations. Cell proliferation rates were significantly suppressed dose-dependently upon iFSP1 treatment (Figure 5A). Additionally, iFSP1 treatment, like erastin, which targets the xCT antiporter of the GPX4 system, also induced a greater extracellular release of LDH compared with vehicle control, further indicating that iFSP1 elicited cytotoxic effects on HCC cells (Figure 5B). Treatment of iFSP1 with the same concentrations as Figure 5A also displayed dose-dependent increase of intracellular lipid peroxidation with BODIPY 581/591 C11 staining, and cell death indicated by increased SYTOX Green dead cell staining (Figure 5C‒D). These results together suggested that pharmacological inhibition of FSP1 using iFSP1 effectively induces ferroptosis, which inhibits HCC growth. We then performed in vitro assays to detect the intracellular levels of lipid peroxidation by-products 4-HNE and MDA in HCC cells upon iFSP1 treatment. Both 4-HNE and MDA levels were significantly increased upon iFSP1 treatment relative to vehicle control cells (Figure 5E). Upon iFSP1 treatment in MHCC97L cells, we observed that co-treatment of Fer-1 reversed the effects as the relative BrdU intensities significantly increased. Co-treatment of necroptosis (GSK’872), apoptosis (Z-VAD-FMK), and autophagy (MRT68921) inhibitors had minimal rescue effect to the suppressed cell proliferation induced by iFSP1 in the BrdU assay (Figure 5F). Dying cells recruit APCs and phagocytes that mediate their clearance. We then asked whether iFSP1 treatment promotes HCC cells to be phagocytosed by macrophages. We first pre-treated mouse HCC cells Hepa1-6 with iFSP1 or vehicle control and labeled them with CellTrace Violet (CTV) dye (Invitrogen). Then, we co-cultured CTV-labeled Hepa1-6 cells with murine bone marrow-derived macrophages (BMMs). Phagocytic events were measured by CTV+ cells in CD11b+ F4/80+ macrophage population using flow cytometry (Figure 5Gi). We showed that iFSP1 treatment significantly enhanced phagocytosis of Hepa1-6 cells by BMMs (Figure 5Gii). Co-treatment with iFSP1 and Fer-1 abrogated the increase of phagocytosis of Hepa1-6 cells by BMMs (Figure 5Giii). We have demonstrated that ferroptotic HCC cells recruit macrophages and enhance the engulfment of ferroptotic cells, suggesting the potential immunomodulatory effect of iFSP1. To further study the therapeutic potential of iFSP1 and whether it affects the intra-tumoral immune composition, we induced HCC tumor with Keap1KO/c-MycOE genetic background by hydrodynamic tail vein injections (HDTVi) in immunocompetent C57BL/6N mice fed with high-fat diet (HFD) (60 kcal% fat, 0.1% methionine, no choline) (Research Diets, Inc, New Brunswick, NJ). In this model, HFD promoted lipid accumulation in liver, whereas Keap1KO resulted in Nrf2 activation, generating a highly aggressive murine HCC model (Figure 6). To characterize this model, we performed hematoxylin and eosin staining on the tumors, and they were confirmed to be HCC (Figure 7A). We showed that FSP1 expression was particularly induced in Keap1KO tumors (Figure 7B). Furthermore, conventional HCC marker alpha fetoprotein (Afp) was overexpressed in tumor tissues (Figure 7C). We showed that iFSP1 treatment significantly suppressed HCC tumor growth with no adverse effects on body weight (Figure 5Hi). Furthermore, flow cytometry analysis illustrated that iFSP1 greatly increased the number of macrophages, DCs, and CD4+ and CD8+ T cells in HCC tumor compared with vehicle control (Figure 5Hii). We observed no change in the number of neutrophils and a slight increase in monocytes (Figure 7D). Further characterizations showed that iFSP1 promoted tumor infiltration of M1 macrophages and effector T cells but no change in M2 macrophages (Figure 7D‒E). Tumor infiltration of CD4+ and CD8+ T cells was confirmed by immunohistochemical (IHC) staining (Figure 5I).
Figure 5.
iFSP1 induced ferroptosis and enhanced anti-tumor immune response. (A) Cell proliferation rates of MHCC97L cells determined by cell counting daily were significantly suppressed by treatments of 1.5, 3, and 10 μM FSP1 inhibitor iFSP1 in dose-dependent manner. (B) Quantification of relative LDH release from MHCC97L cells demonstrated that erastin and iFSP1 significantly induced cell death. (C) BODIPY 581/591 C11 staining in MHCC97L cells revealed significant increase of lipid peroxidation from treatments of 1.5, 3, and 10 μM iFSP1 in dose-dependent manner. (D) SYTOX Green (SG+) staining in MHCC97L cells revealed that treatments of 1.5, 3, and 10 μM iFSP1 resulted in significant increase in cell death dose-dependently. (E) Quantitative measurements of Left: 4-HNE and Right: MDA in MHCC97L cells with iFSP1 or vehicle treatment further suggested iFSP1 significantly increased lipid peroxidation. (F) Cell proliferation rates of MHCC97L cells treated with iFSP1 and vehicle determined using BrdU assay indicated that Fer-1, not inhibitors of other modes of RCD GSK’872 (necroptosis), Z-VAD-FMK (apoptosis), and MRT68921 (autophagy), can rescue proliferation rates suppressed by iFSP1 treatment. (G) Extent of phagocytosis by BMMs of iFSP1 pre-treated Hepa1-6 cells was determined by phagocytosis assay. (i) Schematic representation of the experimental design. (ii) Ferroptotic Hepa1-6 cells induced by iFSP1 pre-treatment were more frequently phagocytosed by BMMs. (iii) Fer-1 treatment abrogated the increase of phagocytosis by BMMs of iFSP1 pre-treated Hepa1-6 cells. (H) In vivo HCC tumors were induced via HDTVi of plasmids carrying Keap1KO/c-MycOE in C57BL/6N mice fed with HFD. (i) Administration of iFSP1 through intraperitoneal injection significantly reduced HCC tumor size compared with vehicle (n = 15 mice per experimental group). Left: Representative picture of harvested tumors. Middle & Right: Quantifications of tumor mass and body weight of mice. (ii) Administration of iFSP1 enhanced infiltration of macrophages, DCs, CD4+ T cells, and CD8+ T cells in HDTVi-induced HCC tumors. (I) IHC staining demonstrated the administration of iFSP1 enhanced the recruitment of CD4+ and CD8+ T cells in HDTVi-induced HCC tumors. Left: Representative staining of CD4+ and CD8+ T cells from harvested tumors treated with Vehicle or iFSP1. Right: Quantifications of number of CD4+ and CD8+ T cells stained per field. Error bars indicate mean ± SD. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001 vs vehicle or 0 μM as indicated. Student’s t test. H: Scale bar = 1 cm. I: original = 20× magnification, scale bar = 100 μm; Inset = 40× magnification, scale bar = 50 μm
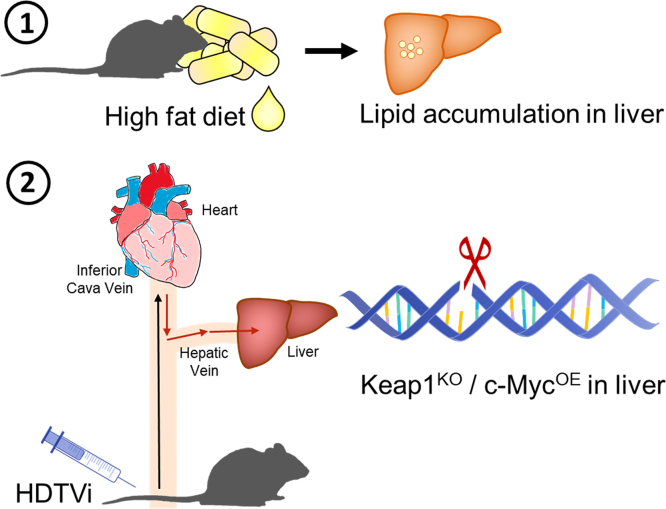
Figure 6.
Schematic diagram illustrating murine HCC model used for iFSP1 efficacy testing.In vivo HCC tumors were induced in HFD-fed C57BL/6N mice by HDTVi of plasmids carrying KeapKO/c-MycOE.
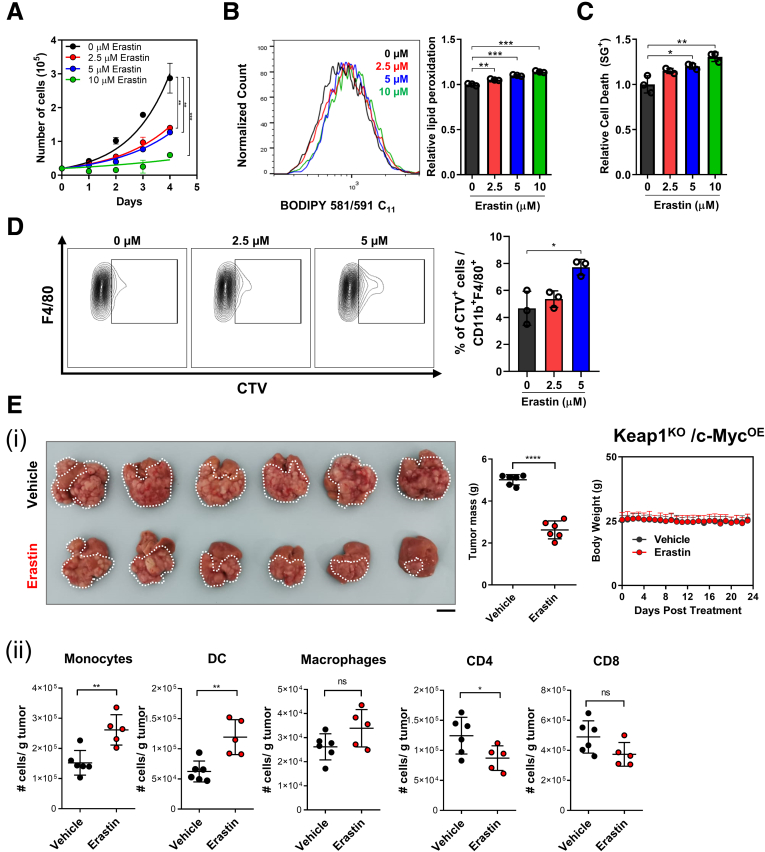
We further examined the effects of ferroptosis induction in human HCC using erastin (Selleck Chemicals, Houston, TX), another well-established ferroptosis inducer targeting the xCT antiporter and inhibiting the cysteine/GPX4 pathway.31 We found that erastin significantly induced ferroptosis, suppressed HCC cell proliferation, and promoted their uptake by BMMs in vitro (Figure 8A‒D). Furthermore, erastin effectively inhibited HCC tumor growth in vivo, but in stark contrast to iFSP1, the number of intratumoral CD4+ and CD8+ T cells was also reduced (Figure 8E). The discrepancy in T cell number upon iFSP1 (FSP1 inhibitor) and erastin (GPX4 inhibitor) treatments in HCC tumors may be due to T cells’ reliance on the GPX4 pathway to counteract ferroptosis.17,32 Our findings illustrated that ferroptosis induction via FSP1 inhibition using iFSP1 suppressed HCC tumor growth not only through mediating HCC cell death, but also by altering the immune landscape of HCC tumors. Our findings highlight FSP1 inhibition as a unique anti-tumor strategy in comparison to other ferroptosis inducers.
Figure 7.
Characterization of murine tumor model used and the immune landscape change upon iFSP1 treatment. (A‒C) In vivo HCC tumors were induced via HDTVi of plasmids carrying Keap1KO/c-MycOE (or ApcKO/c-MycOE) in C57BL/6N mice fed with HFD. (A) Representative hematoxylin and eosin staining of Keap1KO/c-MycOE HCC tumor. (B) Representative staining of FSP1 expression from ApcKO/c-MycOE or Keap1KO/c-MycOE HCC tumors at protein level by IHC staining. (C) RT-qPCR demonstrated the upregulation of Afp in tumorous (T) tissues when compared with NT liver tissues in Keap1KO/c-MycOE HCC tumor. (D) Administration of iFSP1 altered myeloid cells infiltration, including neutrophils (CD11b+ Ly6G+), monocytes (CD11b+ Ly6C+), M1 macrophages (CD11b+ F4/80+ CD86+ MHCII+), and M2 macrophages (CD11b+ F4/80+ CD206+) in HDTVi-induced HCC tumors. (E) Administration of iFSP1 increased number of T cells (CD3+) and T effector cells (CD44+ CD62L-) in HDTVi-induced HCC tumors. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 vs NT or vehicle as indicated. Student t test. A‒B: 20× magnification, scale bar = 100 μm.
Single-cell RNA Sequencing Revealed Immune Landscape Alteration Upon iFSP1 Treatment
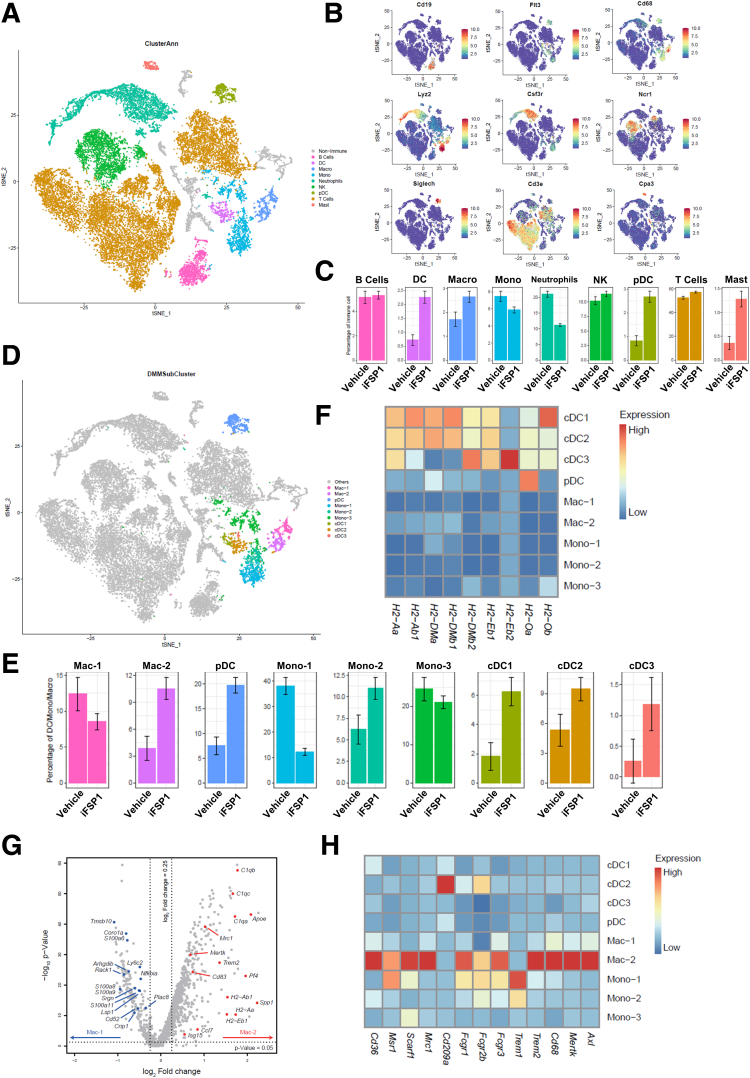
To further explore immune landscape changes in HCC tumors upon iFSP1 treatment, we performed single-cell RNA sequencing (scRNA-seq) on single cells isolated from murine HDTVi-induced HCC tumors treated with iFSP1 or vehicle control (Figure 9A). Using unsupervised clustering analysis, we identified major immune cell populations by their marker genes. The representative marker genes were visualized by t-distributed stochastic neighbor embedding (tSNE) plots. These cell populations included B cells (Cd19), DCs (Flt3), macrophages (Cd68), monocytes (Lyz2), neutrophils (Csf3r), NK cells (Ncr1), pDCs (Siglech), T cells (Cd3e), and mast cells (Cpa3) (Figure 10A‒B). Consistent with our observations from flow cytometry analysis, iFSP1 treatment resulted in increase of DCs, macrophages and T cells (Figure 10C).
Figure 8.
Erastin effectively induced ferroptotic HCC cell death. (A) Cell proliferation rates of MHCC97L cells determined by cell counting daily were significantly suppressed by treatments of 2.5, 5, and 10 μM ferroptosis inducer erastin in dose-dependent manner. (B) BODIPY 581/591 C11 staining in MHCC97L cells revealed significant increase of lipid peroxidation from treatments of 2.5, 5, and 10 μM erastin in dose-dependent manner. (C) SYTOX Green (SG+) staining in MHCC97L cells revealed that treatments of 2.5, 5, and 10 μM erastin resulted in significant increase in cell death dose-dependently. (D) Ferroptotic Hepa1-6 cells induced by erastin pre-treatment were more frequently phagocytosed by BMDMs. (E) In vivo HCC tumors were induced via HDTVi of plasmids carrying Keap1KO/c-MycOE in C57BL/6N mice fed with HFD. (i) Administration of erastin through intrperitoneal injection significantly reduced HCC tumor size compared with vehicle (n = 6 mice per experimental group). Left: Representative picture of harvested tumors. Middle & Right: Quantifications of tumor mass and body weight of mice. (ii) Administration of erastin increased the number of monocytes, DCs and macrophages, while it decreased the number of CD4+ T cells and CD8+ T cells in HDTVi-induced HCC tumors. Error bars indicate mean ± SD. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001 vs vehicle or 0 μM as indicated. Student t test. E: Scale bar = 1 cm.
Figure 9.
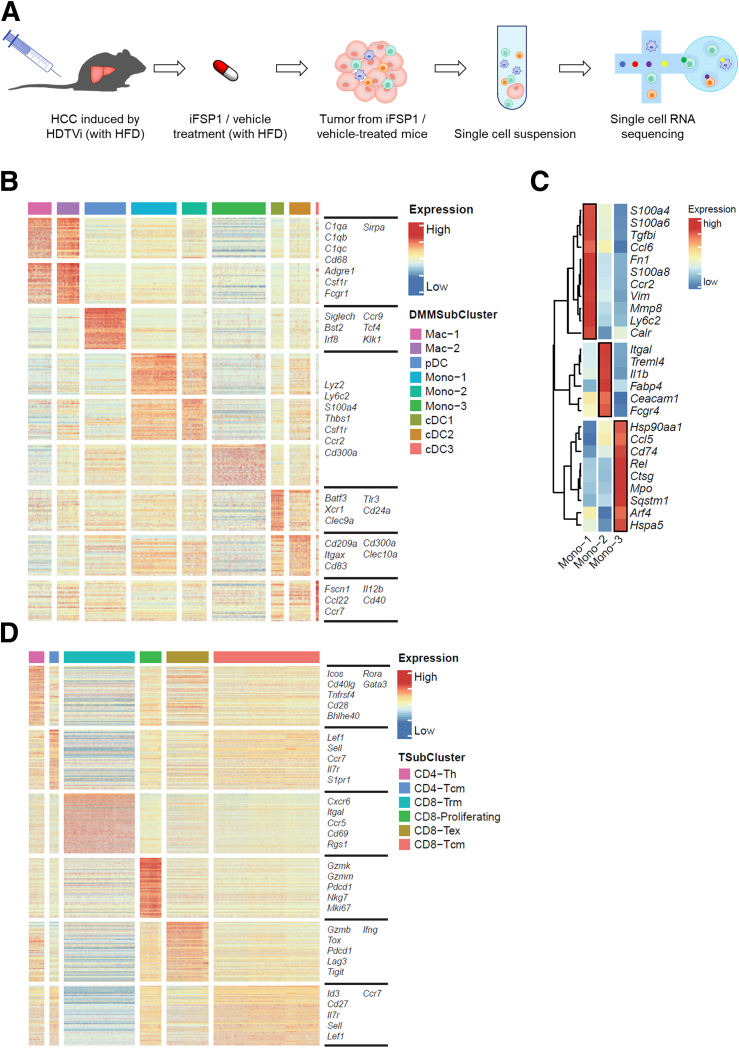
ScRNA-seq revealed iFSP1 altered immune landscape in mouse HCC. (A) Schematic diagram illustrating experimental design. (B) Gene expression heatmap of myeloid clusters showed the selected marker genes for each subset. (C) Gene expression heatmap of monocytes showed the selected marker genes for each subset. (D) Gene expression heatmap of T cells showed the selected marker genes for each subset. B, D: Expression: Z-score normalized expression. C: Expression: Z-score normalized mean expression.
We first investigated the myeloid compartment. Unsupervised clustering analysis further identified 4 DC subsets, 2 macrophage subsets, and 3 monocyte subsets (Figures 10D, 9B, 9C). All DC subsets increased upon iFSP1 treatment (Figure 10E). pDC represented plasmacytoid DCs expressing Siglech, Irf8, and Bst2. cDC1 expressed Xcr1, Clec9a, and Batf3, whereas cDC2 expressed Cd209a, Itgax, and Clec10a, corresponding to typical marker genes of conventional DC1 and conventional DC2, respectively (Figure 9B). cDC3 expressed Fscn1, Ccl22, and Ccr7 (Figure 9B). It resembled a mature DC subset that expressed high level of activation and regulatory markers and shown to be derived from both cDC1 and cDC2.33 This DC subset was also reported in multiple human cancer types as LAMP3+ cDCs.34 The 3 cDC subsets expressed a high level of major histocompatibility complex II molecules for antigen presentation (Figure 10F). For the 2 macrophage subsets, iFSP1 reduced Mac-1 and increased Mac-2 (Figure 10E). We further studied the differentially expressed genes among these 2 macrophage subsets. Interestingly, we found that Mac-2 was enriched in phagocytosis-related genes, such as C1qa, C1qb, C1qc, and Mertk (Figure 10G). Mac-2 also expressed a panel of receptors mediating phagocytosis (Figure 10H). These results illustrated that iFSP1 treatment recruit DCs with antigen presentation ability and phagocytic macrophages to tumor site.
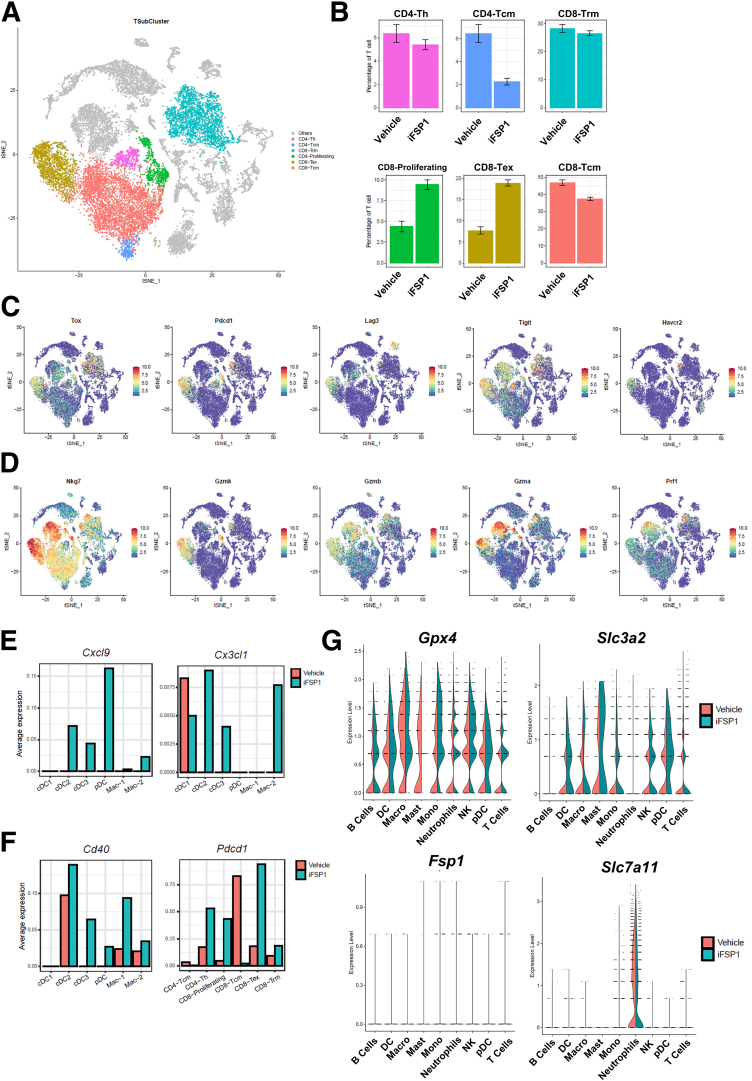
We next studied subsets in T cells. We identified 2 CD4+ and 4 CD8+ T cell clusters, and they were annotated based on their marker gene expression (Figures 11A, 9D). Proliferating and exhausted CD8+ T cells were particularly increased upon iFSP1 treatment (Figure 11B). Proliferating CD8+ T cells are characterized by their expression of Mki67. Exhausted T cells expressed exhaustion markers including Tox, Pdcd1, Lag3, Tigit, and Havcr2 (Figures 11C, 9D). Importantly, both T cell populations expressed multiple cytotoxic markers, including Nkg7, Gzmk, Gzmb, Gzma, and Prf1 (Figure 11D). This suggested that iFSP1 treatment promoted tumor infiltration of CD8+ T cells with cytotoxic functions.
Figure 10.
iFSP1 promoted anti-tumorigenic immune cells infiltration. (A) tSNE plot revealed clusters of major cell populations isolated from mouse HCC tumor treated with iFSP1 or vehicle. (B) tSNE plots showed the selected marker genes of major immune cell populations. (C) Percentage change of major immune cell clusters revealed the increase of DCs, macrophages, and T cells in tumor upon iFSP1 treatment. (D) tSNE plot illustrated further sub-clustering of selected myeloid cells. (E) Percentage change of myeloid cell subsets demonstrated iFSP1 treatment led to an increase in all DC subsets but differential effect on macrophages and monocytes. (F) Heatmap showing expression of major histocompatibility complex II genes in the myeloid subsets indicated that DCs expressed the highest level of antigen-presenting signature. (G) Volcano plot showing differentially expressed genes between Mac-1 and Mac-2 revealed that phagocytosis-related genes were enriched in Mac-2. P-value < .05 and log2 (fold change) > 0.25. (H) Heatmap showing expression of phagocytosis-related genes in the myeloid subsets indicated that Mac-2 expressed the highest level of phagocytic signature. B: Expression: normalized expression. C, E: Error bars indicate 95% confidence interval. F, H: Expression: Z-score normalized mean expression.
To understand how these immune cells were recruited to tumor site, we then explored the expression of chemokines and observed that Cxcl9 and Cx3cl1 were induced in multiple DC and macrophage populations upon iFSP1 treatment (Figure 11E). CXCL9 is a well-known chemoattractant for CXCR3+ T cells, whereas CX3CL1 recruit DCs and macrophages expressing receptor CX3CR1. Moreover, we observed induction of costimulatory receptor, Cd40 expression on DCs and macrophages and Pdcd1, encoding PD-1, on T cells (Figure 11F), pointing to potential targets for combination treatment.
As a systemic treatment, it is possible that iFSP1 also influences immune cell populations; therefore, we studied the expression of anti-ferroptotic genes in major immune cell populations. Interestingly, we found that immune cells expressed a high level of Gpx4 but a low level of Fsp1 (Figure 11G). Moreover, components of xCT antiporter were generally expressed in major immune cell populations (Figure 9E), suggesting that immune cells greatly rely on the GPX4 pathway to protect themselves from ferroptosis. This could also explain the reduction in T cells we observed in erastin-treated tumors.
iFSP1 in Combination With Immunotherapies Effectively Prolonged HCC Survival
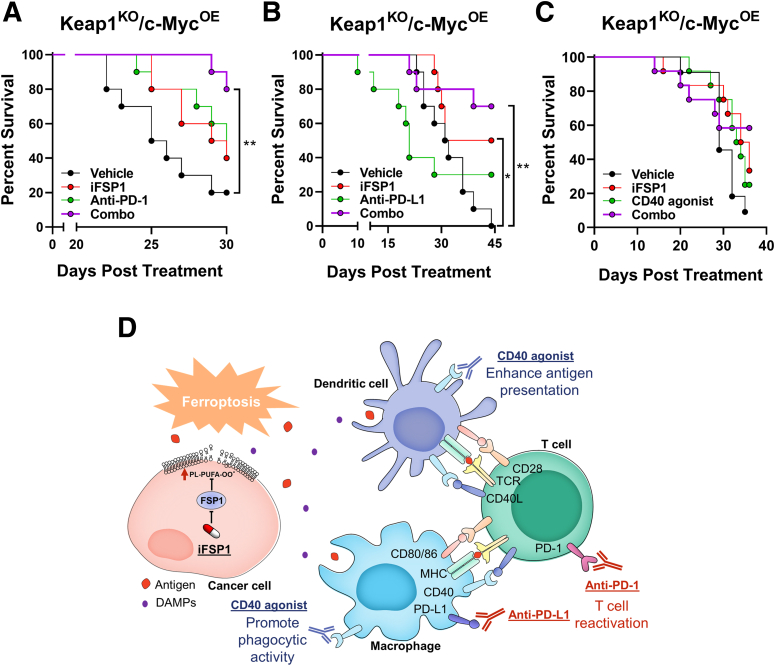
The upregulation of PD-1 on T cells and CD40 expression on DCs and macrophages prompted us to explore the synergistic effect of iFSP1 and immunotherapies. HCC tumors were induced by HDTVi in mice fed with HFD, and mice were treated with iFSP1 in combination with either ICIs (anti-PD-1 and anti-PD-L1; BioXcell, Lebanon, NH), or a new generation of immunotherapy, CD40 agonist (BioXcell). Single treatment of iFSP1 or anti-PD-1 improved survival of tumor-bearing mice similarly, whereas co-administration of them effectively prolonged survival of mice (Figure 12A). Interestingly, iFSP1 showed better efficacy in prolonging survival than anti-PD-L1 as a single treatment in our HCC model. Combining iFSP1 and anti-PD-L1 can further extend the survival of HCC-bearing mice (Figure 12B). Apart from the 2 ICIs currently used in patients with HCC, we also explored the effect of CD40 agonist in combination with iFSP1. CD40 agonist worked with iFSP1 to prolong the survival of tumor-bearing mice (Figure 12C). Here, we demonstrated that iFSP1 treatment can improve or potentiate the effect of multiple immunotherapies without adverse effects on body weight (Figure 13). Our results suggested that iFSP1 treatment recruited APCs and promoted antigen presentation by APCs to T cells. This is further enhanced by immunotherapies that either reinvigorate T cells (anti-PD-1 and anti-PD-L1) or activate APCs (CD40 agonist) (Figure 12D).
Figure 13.
Body weights of mice upon drug administration. Body weights of tumor-bearing mice co-treated with iFSP1 and anti-PD-1, anti-PD-L1, or CD40 agonist.
Figure 11.
ScRNA-seq revealed FSP1 as a potent therapeutic target in HCC. (A) tSNE plot showed further clustering of T cells into 6 subsets. (B) Percentage change of T cell subsets revealed an increase in proliferating and exhausted CD8+ T cells upon iFSP1 treatment. (C) tSNE plots showed the expression level of T cell exhaustion markers. (D) tSNE plots showed the expression level of cytotoxic markers. (E) Average expression level of Cxcl9 and Cx3cl1 were induced upon iFSP1 treatment in DC and macrophage subsets. (F) Average expression level of Cd40 in DC and macrophage subsets and Pdcd1 in T cell subsets were induced upon iFSP1 treatment. (G) Left: Violin plots showed the expression level of Gpx4 and Fsp1in major immune cell populations. Right: Violin plots showed the expression level of Slc3a2 and Slc7a11 in major immune cell populations. B: Error bars indicate 95% confidence interval. C‒G: Expression: normalized expression.
Figure 12.
Combination of iFSP1 and immunotherapies prolonged HCC survival. (A‒B) Co-administrations of iFSP1 with ICIs, (A) anti-PD-1 and (B) anti-PD-L1 antibodies, respectively, synergistically prolonged the survival of tumor-bearing C57BL/6N mice (n = 10 mice per experimental group). (C) Co-administrations of iFSP1 and CD40 agonist prolonged the survival of tumor-bearing C57BL/6N mice (n = 12 mice per experimental group). (D) Schematic summary of immunomodulatory effects and treatment strategies of iFSP1. A‒C: Log-rank test.
Discussion
Ferroptosis, as a distinct form of cell death, was first reported to suppress cancer in 2003; however, the implications of ferroptosis in cancers were only revealed in recent years.35 A CRISPR-Cas9 screening identified that peroxisome genes drove GPX4 inhibitor-induced ferroptosis in ovarian cancer cells by promoting polyunsaturated ether phospholipid synthesis.36 Another study revealed that therapy-resistant lung and pancreatic cancer cells were highly dependent on GPX4 and thus particularly susceptible to ferroptosis inducers.37 A recent study demonstrated that ferroptosis inhibits metastasis. Melanoma cells in the blood were shown to encounter a higher level of ferroptosis than melanoma cells in the lymph, resulting in more melanoma metastases formed from lymph node metastasis than vascular metastasis.38 Interestingly, triple-negative breast cancer cells express higher levels of pro-ferroptotic genes (TfR and ACSL4) and lower levels of anti-ferroptotic genes (ferritin heavy chain 1, ferroportin, GPX4, GSH synthetase) and are therefore more susceptible to ferroptosis.39 The roles of ferroptosis in HCC, especially FSP1-driven mechanisms, have not been fully explored. Our study unprecedentedly reported that HCC is reliant on FSP1 for protection against ferroptosis rather than GPX4, as we showed that FSP1 is upregulated in human HCC but not GPX4. FSP1 deficiency led to decreased ubiquinol-to-ubiquinone ratio, diminishing the ability of cancer cells to counteract lipid peroxidation. This induced ferroptosis in cancer cells and suppressed HCC tumor progression.
Multiple pathways are involved in the regulation of ferroptosis. P53 transcriptionally represses SLC7A11 to inhibit cysteine intake, which leads to ROS-induced ferroptosis in osteosarcoma, NSCLC, and breast cancer cells.40 Meanwhile, NRF2, the master regulator of antioxidant response, has been shown to protect HCC cells from ferroptosis.41,42 In HCC cells treated with ferroptosis inducers such as erastin and sorafenib, P62 releases NRF2 from KEAP1 binding, thereby stabilizing NRF2 so that NRF2 mediates the transcription of NQO1, heme oxygenase-1, and FTH1 to counteract ferroptosis.42 NRF2 inhibitor worked synergistically with these ferroptosis inducers to inhibit HCC growth in mice by eliciting severe ferroptosis in HCC cells.42 In this study, we showed for the first time that FSP1 is transcriptionally regulated by NRF2. NRF2 inhibitors such as clobetasol propionate, which we previously showed to have remarkable anti-tumor effects in HCC, are likely to induce ferroptosis through inhibiting GPX4 and FSP1 pathways.27 However, systemic administration of NRF2 inhibitors might also elicit ferroptosis of immune cells, affecting the combined effects with ICIs and CD40 agonist.
Ferroptosis is a form of ICD. Multiple studies illustrated that ferroptotic cancer cells released DAMPs such as ATP, HMGB1, and oxidized phospholipids, which promoted their uptake by DCs and macrophages.43,44 However, whether and how ferroptosis induction in cancer promotes anti-tumor immune responses are yet to be investigated. Here, we showed that induction of ferroptosis by FSP1 inhibition suppressed HCC tumor growth. This anti-tumor effect is not only due to the direct induction of cell death but also the activation of anti-tumor immune responses. We demonstrated that iFSP1 treatment suppressed HCC progression and promoted infiltration of antigen presenting DCs, phagocytic macrophages, and cytotoxic T cells into tumor. These immune cells could be recruited by chemokines that were upregulated upon iFSP1 treatment. CXCL9 is a well-studied chemokine that is important for CXCR3+ T cell trafficking and recruitment to tumor site.45,46 CX3CL1 could also mediate the recruitment of DCs and macrophages expressing CX3CR1.47,48 A recent study reported that ICIs (anti-CTLA4 and anti-PD-L1) could induce ferroptosis of cancer cells by unleashing the IFN-γ-producing capability of CD8+ T cells.49 IFN-γ produced by activated CD8+ T cells suppresses the protein expression of components in the xCT antiporter (SLC7A11 and SLC3A2), thereby inducing ferroptosis.49 This study mainly focuses on how T cells induce ferroptosis of cancer cells, whereas our study revealed that induction of ferroptosis in cancer cells could reciprocally promote T cell tumor infiltration. Together, these suggest a potential feedforward loop to suppress cancer progression.
Drugs that induce ICD are being hotly pursued because of their potential as combination treatments with ICIs in multiple cancer types.50 Suboptimal responses to ICIs could be due to the insufficient antigen presentation.51 Treatments inducing ICD lead to the release of tumor antigens and DAMPs by cancer cells and thus recruiting APCs to engulf the tumor antigens. This enhances antigen presentation by APCs to T cells, initiating anti-tumor immune response.52 Upon iFSP1 treatment, we observed an upregulation of CD40 on DCs and macrophages while PD-1 on T cells. Excitingly, we showed that combination treatment of iFSP1 and anti-PD-1, anti-PD-L1, or CD40 agonist can all prolong survival of tumor-bearing mice. Although anti-PD-1 and anti-PD-L1 aim at blocking PD-1 and PD-L1 ligation and thus reinvigorating T cells, CD40 agonist enhances antigen presentation ability of APCs and promotes the secretion of a plethora of pro-inflammatory cytokines and has been demonstrated to effectively suppress cholangiocarcinoma.53, 54, 55 CD40 agonist demonstrated clinical activity when used in combination with chemotherapies and nivolumab in treating metastatic pancreatic adenocarcinoma, whereas its effects in HCC remain to be investigated.56 Recently, it was reported that TYRO3 is also an anti-ferroptotic gene through inducing NRF2-mediated transcription to suppress lipid ROS accumulation. Cancers with TYRO3 overexpressed are resistant to anti-PD-1 treatment, indirectly suggesting that ferroptosis improves the efficiency of ICIs.57 Together with our findings, these show that ferroptosis induction could work synergistically with immunotherapies and shed light on a novel therapeutic approach for HCC.
The effects of ferroptosis inducers on immune cells require closer investigation. Interestingly, our flow cytometry data showed that total CD4 T cells were enriched in HCC from iFSP1-treated mice, whereas scRNA-seq data showed that the percentage of CD4 central memory T cells decreased. Although the detailed mechanisms explaining why CD4 central memory T cells are reduced remain unknown, studies showed that anti-PD-1/PD-L1 treatment was expected to promote the memory of T cells, further providing mechanistic explanation to combine iFSP1 with ICIs.58 A recent study showed that CD36 on CD8+ T cells increased their fatty acids uptake and lipid peroxidation.59 This contributed to CD8+ T cell ferroptosis and suppressed its cytotoxic functions, whereas CD36 blockade could enhance anti-tumor response.59 Most recently, it was further shown that the CD36-mediated CD8+ T cell dysfunction could be rescued by GPX4 overexpression.17 Another study performed a high throughput pharmacologic screening to identify specify metabolic vulnerabilities of CD8+ T cells and cancer cells and showed that CD8+ T cells are more susceptible to RSL3, a GPX4 inhibitor, than cancer cells.32 These indicated T cells highly rely on GPX4 pathway as a protective mechanism against ferroptosis. Our data showed that the dose of iFSP1 we used for in vivo treatment did not influence the T cell population, but we did observe a reduction in T cell population in GPX4 inhibitor (erastin)-treated tumors despite the suppression of tumor growth. This discrepancy could be due to the reliance of immune cells on the GPX4 pathway to protect themselves from ferroptosis because our scRNA-seq analysis demonstrated that immune cells in general only express GPX4 but not FSP1. Despite the tumor-suppressing effect of inhibitors targeting GPX4 pathway, immune cells also rely on GPX4 pathway to protect them from ferroptosis. Therefore, GPX4 inhibition may not fully demonstrate the potential of ferroptosis inducers as cancer treatment because it could dampen the immune responses. This highlights the importance of further investigation to identify a molecular target that can selectively induce ferroptosis in cancer cells but not immune cells. We showed that FSP1 is overexpressed in human HCC but not GPX4, whereas tumor-infiltrating immune cells express a high level of GPX4 but not FSP1. These suggest that cancer cells and immune cells rely on different mechanisms to protect themselves from ferroptosis in HCC. Therefore, FSP1 could be a superior candidate because its inhibition selectively induces ferroptosis in HCC cells and unleashes anti-tumor immune responses.
Materials and Methods
The Cancer Genome Atlas
Transcriptome sequencing results on 49 pairs of human HCC and corresponding NT liver tissues were retrieved from TCGA through cBioportal (http://www.cbioportal.org/). The mRNA expression levels of FSP1, TF, and GPX4 were examined. Correlation between expression levels of FSP1 and antioxidant genes TKT, ME1, MTHFD1L, NQO1, AKR1B10, and AKR1C1 in human HCC and NT liver tissues were also retrieved from TCGA.
Clinical Samples
Paired clinical HCC tumor and NT liver tissues were used in this study. Tissue samples were collected from patients with HCC admitted to Pamela Youde Nethersole Eastern Hospital, Queen Elizabeth Hospital, and/or Queen Mary Hospital. Resected tissue samples were snap-frozen with liquid nitrogen and stored at −80 °C for future use. Approval for the use of clinical HCC tumor samples and paired NT liver tissues was previously obtained from the Institutional Review Board of The University of Hong Kong and the Hospital Authority of Hong Kong (HKU/HA HKW IRB; ref. no. UW 09-158). Prior to the collection and use of resected clinical tissues, signed consents were obtained as acknowledgements from patients.
Immunohistochemical Staining
Clinical human and mouse HCC/NT tissues fixed in 4% formalin and 75% ethanol were embedded in paraffin. Before IHC staining, paraffin sections were dewaxed with xylene and rinsed with ethanol. Antigen retrieval was performed by boiling in EDTA buffer, and the liver tissue sections were stained with primary antibody (Table 2) at 4 °C overnight, followed by horseradish peroxidase-conjugated secondary antibody (Dako, Glostrup, Denmark) staining at room temperature for 30 minutes. Sections were developed using 3,3’-diaminobenzidine (Sigma-Aldrich) and counterstained with hematoxylin.
Table 2.
Antibodies Used
| Antibody | Source | Application | Clone | Cat no. | Company |
|---|---|---|---|---|---|
| NRF2 | Rabbit | ChIP | / | ab62352 | Abcam |
| IgG control | Rabbit | ChIP | / | sc-2027 | Santa Cruz |
| CD16/32 | Mouse | Flow cytometry | 93 | 101301 | Biolegend |
| CD45 | Mouse | Flow cytometry | 30-F11 | 103127 | Biolegend |
| CD8b | Mouse | Flow cytometry | YTS156.7.7 | 126619 | Biolegend |
| CD44 | Mouse | Flow cytometry | IM7 | 103005 | Biolegend |
| CD62L | Mouse | Flow cytometry | MEL-14 | 104431 | Biolegend |
| CD45 | Mouse | Flow cytometry | 30-F11 | 103115 | Biolegend |
| CD3 | Mouse | Flow cytometry | 17A2 | 100205 | Biolegend |
| CD4 | Mouse | Flow cytometry | GK1.5 | 100429 | Biolegend |
| CD11b | Mouse | Flow cytometry | M1/70 | 101205 | Biolegend |
| Ly6C | Mouse | Flow cytometry | HK1.4 | 128017 | Biolegend |
| Ly6G | Mouse | Flow cytometry | 1A8 | 127623 | Biolegend |
| CD11c | Mouse | Flow cytometry | N418 | 117307 | Biolegend |
| F4/80 | Mouse | Flow cytometry | BM8 | 123115 | Biolegend |
| I-A/I-E | Mouse | Flow cytometry | M5/114.15.2 | 107625 | Biolegend |
| CD8a | Mouse | IHC | D4W2Z | 98941 | Cell Signaling |
| CD4 | Mouse | IHC | D7D2Z | 25229 | Cell Signaling |
| FSP1 | Rabbit | IHC | / | 20886-1-AP | Proteintech |
| FSP1 | Rabbit | Western blot | / | PA5-28727 | Invitrogen |
| β-actin | Mouse | Western blot | / | A5316 | Sigma-Aldrich |
ChIP, Chromatin immunoprecipitation; FSP1, ferroptosis suppressor protein 1; IHC, immunohistochemistry; NRF2, nuclear factor erythroid 2-related factor 2.
Chromatin Immunoprecipitation
HCC cells were fixed in 3.7% formaldehyde for DNA crosslinking followed by sonication. Antibodies against NRF2 and IgG control (Table 2) were added to sheared DNA fragments. DNA-protein-antibody complexes were incubated with Protein A/Salmon Sperm DNA Agarose Beads (Merck Millipore, Burlington, MA), sequentially washed using gradients of salt buffer, and eluted in 1% SDS/NaHCO3. ChIP DNA samples were analyzed by RT-qPCR with primers flanking the putative ARE (Table 3).
Table 3.
Primer Sequences
| Primer | Sequence |
|---|---|
| Human FSP1 (ChIP) | Forward: TTCAAAGTTCAAGGCCGTTC Reverse: TGGTCGTCTTCCCGAGTTAC |
| Human FSP1 | Forward: GGAGACAGGGTTCGCCAAAA Reverse: CCCGTGGCCAGGATAAGATG |
| Human NRF2 | Forward: GGTTGCCCACATTCCCAAAT Reverse: AGCAATGAAGACTGGGCTCT |
| Human KEAP1 | Forward: CCTTCAGCTACACCCTGGAG Reverse: AACATGGCCTTGAAGACAGG |
| Human MAF | Forward: CCCGTCCTCTCCCGAGTTTT Reverse: ACACTGGTAAGTACACGATGCT |
| Human 18S | Forward: GAGGATGAGGTGGAACGTGT Reverse: AGAAGTGACGCAGCCCTCTA |
| Mouse Afp | Forward: AAGCAAAAGCCTGAACTGACA Reverse: TCAACTTTGGACCCTCTTCTG |
| Mouse Gapdh | Forward: GGGTGTGAACCACGAGAAAT Reverse: CCTTCCACAATGCCAAAGTT |
Afp, Alpha fetoprotein; ChIP, chromatin immunoprecipitation; FSP1, ferroptosis suppressor protein 1; KEAP1, kelch-like ECH-associated protein 1; MAF, MAF BZIP transcription factor; NRF2, nuclear factor erythroid 2-related factor 2.
Luciferase Reporter Assay
Oligonucleotides containing FSP1 WT or MUT AREs were inserted into the pGL4 firefly luciferase promoter reporter vector. Insert sequences were included in Table 4. These plasmids containing WT or MUT AREs were transfected into Huh7 cells (-NTC, -shNRF2 and -shMAF) together with pRL-CMV and treated with 10μM tBHP for 24 hours. Luciferase activities were determined using the Dual-Luciferase Reporter System (Promega) according to the manufacturer’s instructions.
Table 4.
shRNA Target Sequences and Insert Sequences for Luciferase Reporter Assay
| shRNA | Target sequence |
|---|---|
| shNRF2-55 | GCTCCTACTGTGATGTGAAAT |
| shNRF2-94 | AGTTTGGGAGGAGCTATTATCC |
| shKEAP1-57 | GCGAATGATCACAGCAATGAA |
| shKEAP1-76 | GTGGCGAATGATCACAGCAAT |
| shFSP1-23 | CGGGCAAGTTTAATGAGGTTT |
| shFSP1-24 | CATTTCTTACTCGGTGACTTT |
| shMAF-42 | TCAGTGGGATACGCCACATTT |
| shMAF-58 | TGGAAGACTACTACTGGATGA |
| Insert | Sequence |
| WT | GGGGTCGGGGACCAGCACGAGTGCTGAGTCACGCCCCGCCCGGG |
| MUT | GGGGTCGGGGACCAGCACGAGTAAAAAAAAACGCCCCGCCCGGG |
FSP1, ferroptosis suppressor protein 1; KEAP1, kelch-like ECH-associated protein 1; MAF, MAF BZIP transcription factor; MUT, mutated; NRF2, nuclear factor erythroid 2-related factor 2; shRNA, short hairpin RNA; WT, wild-type.
Establishment of Stable Cell Lines
As previously described, stable-knockdown HCC cell lines were established using lentiviral-mediated approach.60, 61, 62 Briefly, shRNAs targeting NRF2 (shNRF2), KEAP1 (shKEAP1), FSP1 (shFSP1), MAF (shMAF), or NTC contained within pLKO.1-puro vectors were stably transfected into HCC cells. Puromycin was used to select HCC cells that stably express the shRNA-containing vectors. Sequences of all shRNAs used are provided in Table 4.
HCC Cell Culture
Multiple human HCC cell lines including MHCC97L, PLC/PRF/5, Huh7, and a mouse HCC cell line (Hepa 1-6) were used in this study. MHCC97L and Huh7 were kind gifts from Dr Z.Y. Yang of Fudan University (Shanghai, China) and Professor H. Nakabayashi of Hokkaido University School of Medicine (Sapporo, Japan), respectively. PLC/PRF/5 and Hepa 1-6 were obtained from American Type Culture Collect (Manassas, VA). All cell lines were cultured in Dulbecco’s Modified Eagle Medium-high glucose media with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (P/S) supplementations (all reagents from Gibco by Life Technologies, Grand Island, NY). Cells were cultured in a 37 °C humidified incubator supplied with 5% CO2. Authentication of all cell lines within this study was performed using the AuthentiFiler PCR Amplification Kit (Applied Biosystems, Foster City, CA). All authenticated cell stocks were thawed and used within 4 passages.
RNA Extraction, Reverse Transcription, and RT-qPCR
Total RNA was extracted from paired clinical HCC/NT liver tissues, which were snap-frozen with liquid nitrogen and stored at −80 °C, and HCC cell lines with TRIzol reagent (Ambion by Life Technologies, Austin, TX). Purified RNA samples were synthesized into cDNA by reverse transcription using GeneAmp RNA PCR Core Kit (Applied Biosystems). From paired clinical HCC/NT liver tissue samples, RT-qPCR amplifications of FSP1 and internal control 18S were performed using TaqMan Gene Expression Assay (Applied Biosystems). RT-qPCR amplifications of NRF2, KEAP1, FSP1, internal control 18S from HCC cell lines, and Afp and Gapdh from mouse HCC/NT liver tissue samples were performed using SYBR Green PCR Master Mix (Applied Biosystems) with the respective RT-qPCR primers listed in Table 3. All RT-qPCR amplifications were performed using the StepOne Real-Time PCR System (Applied Biosystems).
Protein Extraction and Western Blotting
Total protein lysates from human HCC cell lines were extracted using RIPA lysis buffer with cOmplete protease and PhosSTOP phosphatase inhibitor supplementations (both from Roche Diagnostics, Mannheim, Germany). Equal volumes of protein lysates were separated by SDS-PAGE and transferred onto PVDF membranes (GE Healthcare, Little Chalfont, UK). Protein expression of targeted genes were determined with their respective antibodies listed in Table 2.
Liquid Chromatography-mass Spectrometry
Ubiquinol/ubiquinone were extracted from HCC cells plated on 10-cm culture dishes at 70% to 80% confluence using cold pure methanol. Supernatants from all pelleted samples were transferred to new tubes, dried using a vacuum concentrator, and reconstituted with 100 μL methanol. LC-MS analyses were performed on a ExionLC AC System coupled with X500R QTOF (Sciex Applied Biosystems, Framingham, MA) mass spectrometer. Samples were separated with isocratic elution of methanol containing 2 mM ammonium acetate at a flow rate of 0.4 mL/minute using an ACQUITY UPLC BEH C18 reverse-phase column (2.1 mm × 50 mm, 1.7 μm).63 The column temperature was kept at 40 °C. The mass spectrometer was operated in positive ionization mode. Quantification of CoQ10H2 and CoQ10 were performed using TOF-MS/MS as [M + NH4]+ with the following transitions from precursor to product ions: CoQ10H2 : m/z 883 >197, CoQ10: m/z 881 >197. Data acquisition and analyses were performed with SCIEX OS software (Sciex Applied Biosystems).
BODIPY 581/591 C11 Lipid Peroxidation Detection
Intracellular lipid peroxidation was detected in HCC cells plated on 6-well culture dishes at equal densities. Cells were incubated with 5 μmol/L lipid peroxidation sensor BODIPY 581/591 C11 (Invitrogen) at 37 °C for 30 minutes. Cells were washed twice with phosphate buffered saline (PBS) and collected by trypsinization. Pelleted cells were washed with PBS and re-suspended with 2% FBS in PBS. Relative cell staining was determined by the BD LSRFortessa Flow Cytometer (BD Biosciences, San Jose, CA), and results were analyzed using FlowJo Software (BD, Ashland, OR).
Cell Proliferation Assays
HCC cell proliferation rates were determined using 2 different methods: cell counting and Cell Proliferation enzyme-linked immunosorbent assay, BrdU (colorimetric) assay (Roche Diagnostics). For cell counting, HCC cells were plated onto 12-well culture dishes at the original density of 2 × 104 cells per well. Culture media were changed 24 hours after plating cells for pharmacological treatments. Cell counting began 24 hours after treatment. Cells were trypsinized and counted using the TC20 Automated Cell Counter (Bio-Rad Laboratories, Hercules, CA) daily for 4 consecutive days. Uncounted cells were replenished with fresh culture media with appropriate treatments every 24 hours. BrdU colorimetric assay was performed according to the manufacturer’s instructions. Briefly, HCC cells of equal density were seeded onto 96-well culture dishes. Pharmacological treatments and BrdU labeling were added 24 hours after cells were plated. Addition of conjugated anti-BrdU antibodies and substrate reactions were performed the next day, and absorbance for each sample was measured on a microplate reader to quantify the cell proliferation.
Animal Experiments
For orthotopic implantation, 1.5 × 106 luciferase-labeled MHCC97L cells were re-suspended in 100% Matrigel (BD Biosciences) and implanted into left lobes of livers of 6- to 8-week-old, male BALB/cAnN-nu (nude) mice. After 6 weeks (42 days) post implantation, tumor-bearing mice were subjected for bioluminescent imaging. The mice were injected intraperitoneally with 100 mg/kg D-Luciferin (Gold Biotechnology, St Louis, MO) then imaged using the PE IVIS Spectrum In Vivo Imaging System (PerkinElmer, Inc, Waltham, MA). Livers and lungs were also extracted for ex vivo imaging.
For HDTVi, plasmid DNA mixed with sterile saline (0.9% NaCl) at a total volume equivalent to 10% of body weights were injected into the lateral tail vein of 8- to 10-week-old, male C57BL/6N mice within 5 to 7 seconds. A total of 20 μg of CRISPR-Cas9 vector system carrying single guide RNA targeting Keap1 with transposon system carrying c-Myc vector were injected. For HDTVi model, fatty livers were induced by replacing standard diets with HFD 24 hours prior to HDTVi and for the duration of the experiment. FSP1 was inhibited in vivo by daily intraperitoneal injections of 10 mg/kg iFSP1 (Cayman Chemical). Administrations of anti-PD-1 mAb (clone RMP1.14, BioXcell) and anti-PD-L1 mAb (clone 10F.9G2, BioXCell) both began 1-week post HDTVi for 3 weeks, twice weekly by intraperitoneal injection at 10 mg/kg. CD40 agonist (clone FGK45, BioXCell) was administered 2 weeks post HDTVi by intraperitoneal injections at 2.5 mg/kg once per week. To study the effect of iFSP1 on tumor immune infiltrates, HDTVi-induced tumors were harvested 4 weeks post HDTVi for dissociation and histological analysis. To study the therapeutic benefits of combination treatment of iFSP1 and different immunotherapies, mice were sacrificed when they have reached humane endpoint.
All animal procedures performed throughout this study were approved by the Committee on the Use of Live Animals in Teaching and Research of The University of Hong Kong and adhered to the Animals (Control of Experiments) Ordinance of Hong Kong. All animal procedures were performed under the UK Co-ordinating Committee on Cancer Research (UKCCCR) PMID: 9459138 Guidelines for the Welfare of Animals in Experimental Neoplasia to minimize suffering on the animals throughout the experiments.
4-Hydroxynonenal Assay
Quantitation of relative 4-HNE in HCC cells were detected with the Lipid Peroxidation (4-HNE) Assay Kit (Abcam, Cambridge, UK) according to the manufacturer’s protocol. Briefly, samples and standards were incubated with anti-4-HNE antibody followed by secondary antibody-HRP conjugate on a 4-HNE conjugate pre-coated plate. After incubation with substrate, enzymatic reactions were stopped with a stop solution, and the absorbance for each sample mixture was measured on a microplate reader at OD 450 nm.
Malondialdehyde Colorimetric Assay
Relative productions of MDA in HCC cells were detected using the Lipid Peroxidation (MDA) Assay kit (Abcam) following the manufacturer’s instructions. Briefly, HCC cells were harvested and lysed by homogenization in lysis buffer. Homogenized samples and standards were incubated with thiobarbitoric acid. Absorbance measurement of each sample mixture was performed on a microplate reader at OD 532 nm.
Lactate Dehydrogenase Release Measurement
LDH released from damaged HCC cells were measured with Cytotoxicity Detection KitPLUS (LDH) (Roche Diagnostics) according to the manufacturer’s instructions. Briefly, equal densities of HCC cells were plated onto 6-well culture dishes for 24 hours followed by pharmacological treatments where applicable. Conditioned medium from HCC cells were collected, mixed with reaction mixture, and incubated at room temperature in dark for 30 minutes. Reactions in each sample mixture were stopped, and absorbance was measured with microplate reader at OD 490 nm.
Cell Death Analysis
The percentage of dead cells was determined in equal densities of HCC cells plated on 6-well culture dishes for 24 hours followed by pharmacological treatments where applicable. Conditioned medium and HCC cells were collected. The pelleted cells were washed with PBS and re-suspended in SYTOX Green dead cell stain (Invitrogen) at room temperature in the dark for 40 minutes. Dead cell percentages were determined by flow cytometry using the BD LSRFortessa Flow Cytometer (BD Biosciences).
Preparation of Bone Marrow-derived Macrophages
Femur and tibia were obtained from 5- to 7-week-old C57/BL6N male mice. Bones were cut at both ends, and bone marrow was flushed out with medium. Bone marrow cells were isolated using Ficoll Paque PLUS (GE Healthcare) according to manufacturer’s instruction. Cells were then cultured for 6 days in RPMI 1640 medium supplemented with 25 mM HEPES, 50 μM 2-mercaptoethanol (Sigma-Aldrich), 10% FBS, and 1% P/S in the presence of 20 ng/mL M-CSF (R&D Systems, Minneapolis, MN). More than 90% of the adherent cells were macrophages as identified by CD11b+ CD11c- F4/80+.
Phagocytosis Assay
BMMs were seeded at 1 × 105 per well in 12-well plate. Medium was changed the next day. Hepa1-6 cells were treated with iFSP1 at indicated concentration with or without Fer-1 for 24 hours. Hepa1-6 cells with or without treatment were labeled with 2.5μM CTV (Invitrogen) for 20 minutes at 37 °C with mixing at regular intervals. Labeled Hepa1-6 cells were co-cultured with macrophages in 5:1 ratio for 1 hour at 37 °C. Macrophages were harvested and stained for macrophage markers CD11b and F4/80. Phagocytosis events were analyzed by flow cytometry.
Preparation of Single-cell Suspension From Tumor Tissues
Single-cell suspensions from mouse tumors were prepared prior to flow cytometry analysis. Tumors harvested from HCC-bearing mice were cut into fine pieces and transferred to gentleMACS C tube (Milteyni Biotech, Bergisch Gladbach, Germany) containing 10 mL of DMEM/F12 medium (Gibco). Thirty μL of Liberase (2.5 mg/mL; Roche Diagnostics) and 60 μL of DNase I (10 mg/mL; Roche Diagnostics) were added. Samples were further dissociated with GentleMACS Dissociator (Miltenyi Biotech) using the installed programs “h_tumor_01” thrice, “h_tumor_02” twice, and “h_tumor_03” once. Samples were incubated at 37 °C for 15 minutes with regular mixing in between 2 programs. The cell suspensions were then passed through MACS SmartStrainer (Miltenyi Biotech) to remove undigested tissues and cell clumps, followed by centrifugation. The filtered cell suspensions were resuspended with 10 mL ACK lysing buffer (155 mM ammonium chloride, 10 mM potassium bicarbonate, and 0.1 mM EDTA in water) and incubated at room temperature for 5 minutes to lyse the red blood cells. The reaction was stopped by adding 2 volumes (20 mL) of cell staining buffer (0.5% w/v BSA and 2 mM EDTA in PBS). The cell suspensions were filtered again with MACS SmartStrainer and centrifuged. The cell pellets were then resuspended in proper amount of cell staining buffer and were ready for cell counting. All centrifugations were carried at 300 × g for 5 minutes.
Flow Cytometry Analysis
Cells were blocked with anti-mouse CD16/32 Antibody (Biolegend, San Diego, CA) at room temperature for 10 minutes and incubated with fluorochrome-conjugated primary antibody at 4 °C for 30 minutes. Antibodies were used in 1:100 dilution. The cells were washed with 500 μL cell staining buffer once. They were then resuspended in 300 μL cell staining buffer and ready for flow cytometry analysis with the analyzer BD LSRFortessa (BD Biosciences). Primary antibodies used were listed in Table 2. Software FlowJo was used for data analysis.
Single-cell RNA Sequencing
Library preparation
After obtaining single cell suspension from mouse tumors as described above, cells were purified with Dead Cell Removal Kit (Miltenyi Biotech). Purified cells were loaded on the 10× Chromium system and were encapsulated with Single Cell 5’ Kit (10× Genomics, Pleasanton, CA). Single-cell gene expression libraries were generated according to manufacturer’s protocol.
ScRNA-seq data processing and batch effect correction
The generated scRNA-seq FASTQ files was aligned to mm10 reference by Cell Ranger 3.0.2. First, the raw data files of barcodes, features, and matrix were imported and created as a Seurat object. Next, we counted the mitochondrial percentage of each cells, and each data of samples was normalized by the sctransform function64 with the glmGamPoi improvement.65 They were subsequently merged as a Seurat object. Then, the batch effect was corrected by Fast Mutual Nearest Neighbors (FastMNN) function. To confirm the correction and visualize the data, the non-linear dimension reduction method tSNE was applied using Seurat function RunTSNE with maximum 30 dimensions.
Major cell type annotation and cluster-specific marker gene identification
To annotate each cell with cell type, we first clustered the cells using Seurat function FindNeighbors and FindClusters with ‘resolution’ parameter from 0.1 to 1. We applied R-package Clustree to establish tree for visualizing the clustering results with different ‘resolution’ parameter. ‘Resolution’ of 0.8 was used in subsequent analysis. Next, we used another R-package SingleR to annotate the cell types based on the transcriptome of each cell with reference to ImmGen database. Then, we used the Seurat FindMarkers function to identify the marker genes of each cluster. We defined the marker genes by: (1) 0.25 increasing logfc and (2) P-value less than .05 based on the Wilcoxon rank sum test. After that, the ribosomal and mitochondrial genes were filtered out. We identified 9 major immune cell types (B cells, DCs, macrophages, monocytes, neutrophils, NK cells, pDCs, T cells, and mast cells). The dimensionality of the clustering result was reduced and visualized by tSNE.
Identification of T cell subsets and DC/monocyte/macrophage subsets
To identify the subsets of T cells and DC/monocyte/macrophage, raw data from T cells or monocytes, macrophages, DCs, and pDCs were extracted respectively. A second round of clustering was performed using the same procedures and parameters as the identification of major cell types described above. For T cell subsets, we identified 2 CD4+ T cell populations (CD4-Th and CD4-Tcm) and 4 CD8+ T cell subsets (CD8-Trm, CD8-Proliferating, CD8-Tex and CD8-Tcm). For DC/monocyte/macrophage subsets, we identified 2 macrophage subsets (Mac-1, Mac-2), 3 monocyte subsets (Mono-1, Mono-2, Mono-3), and 4 DC subsets (cDC1, cDC2, cDC3, and pDC). The subset clustering results were mapped to the tSNE plot of major cell types for visualization.
Comparison of cell type proportion in iFSP1 and vehicle-treated tumor
The percentage of cell types in iFSP1 and vehicle-treated tumor was calculated, respectively, based on the total cell numbers of each sample. The percentage of T cell subsets and DC/monocyte/macrophage subsets were normalized to the total number of cells extracted for the sub-clustering. The 95% confidence intervals were calculated by the freqCI function of R-package REdaS.
Software used in the scRNA-seq analysis are listed in Table 5.
Table 5.
Software Used in scRNA-seq Analysis
| Software and algorithms | Source | Identifier |
|---|---|---|
| Cell Ranger 3.0.2 | 10× Genomics | https://www.10xgenomics.com/ |
| Seurat 4.0.2 | 66 | https://satijalab.org/seurat/index.html |
| FastMNN | 67 | https://bioconductor.org/packages/release/bioc/html/scran.html |
| SingleR | 68 | https://github.com/LTLA/SingleR |
| Clustree | 69 | https://github.com/lazappi/clustree |
| REdaS | 70 | http://CRAN.R-project.org/package=REdaS |
| Complex heatmap | 71 | https://bioconductor.org/packages/ComplexHeatmap/ |
scRNA-seq, Single-cell RNA sequencing.
Statistical Analysis
All statistical analyses were performed using the GraphPad Prism 8.0 or above software (GraphPad Software, Inc, La Jolla, CA). Data were analyzed by the 2-tailed Student t test, the log-rank test, or as indicated otherwise. All functional assays are representations of at least 3 independent experiments and are expressed as mean ± standard deviation (SD). P values < .05 were considered statistically significant.
Acknowledgments
The authors thank the Centre for Comparative Medicine Research (CCMR) of The University of Hong Kong in support of our animal experiments, and the School of Biological Sciences, The University of Hong Kong for their technical support in our mass spectrometry experiment. We also thank the Genomics and Bioinformatics Cores, and the Imaging and Flow Cytometry Core of the Centre for PanorOmic Sciences (CPOS) from the Li Ka Shing Faculty of Medicine, The University of Hong Kong for their technical support in our single-cell RNA sequencing and in vivo imaging experiments, respectively.
CRediT Authorship Contributions
Jacinth Wing-Sum Cheu (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Derek Lee (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Qidong Li (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Validation: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Chi Ching Goh (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting)
Macus Hao-Ran Bao (Data curation: Supporting; Investigation: Supporting; Methodology: Supporting)
Vincent Wai-Hin Yuen (Data curation: Supporting; Investigation: Supporting)
Misty Shuo Zhang (Data curation: Supporting; Investigation: Supporting)
Chunxue Yang (Data curation: Supporting; Investigation: Supporting)
Cerise Yuen-Ki Chan (Data curation: Supporting; Investigation: Supporting)
Aki Pui-Wah Tse (Data curation: Supporting; Investigation: Supporting)
Grace Fu-Wan Sit (Data curation: Supporting; Investigation: Supporting)
Cindy Xinqi Liu (Data curation: Supporting; Investigation: Supporting)
Irene Oi-Lin Ng (Funding acquisition: Lead; Clinical sample provision: Lead)
Chun-Ming Wong (Conceptualization: Lead; Supervision: Supporting; Writing – review & editing: Lead)
Carmen Chak-Lui Wong (Conceptualization: Lead; Funding acquisition: Lead; Supervision: Lead; Validation: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding The study was supported by the Hong Kong Research Grant Council (RGC) General Research Fund (17112521), RGC Theme-Based Research Fund (T12-716/22-R), and Centre for Oncology and Immunology under the Health@InnoHK initiative funded by the Innovation and Technology Commission, The Government of Hong Kong, SAR, China. Carmen Chak-Lui Wong is a recipient of the Croucher Innovation Award, The University of Hong Kong Outstanding Young Researcher Award, and National Natural Science Foundation of China (NSFC) Excellent Young Scientists Fund (Hong Kong and Macau) (82022077).
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Llovet J.M., Kelley R.K., Villanueva A., et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6. doi: 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- 3.Dixon S.J., Lemberg K.M., Lamprecht M.R., et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang X., Stockwell B.R., Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22:266–282. doi: 10.1038/s41580-020-00324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J., Cao F., Yin H.L., et al. Ferroptosis: past, present and future. Cell Death Dis. 2020;11:88. doi: 10.1038/s41419-020-2298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao J.Y., Dixon S.J. Mechanisms of ferroptosis. Cell Mol Life Sci. 2016;73:2195–2209. doi: 10.1007/s00018-016-2194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah R., Shchepinov M.S., Pratt D.A. Resolving the role of lipoxygenases in the initiation and execution of ferroptosis. ACS Cent Sci. 2018;4:387–396. doi: 10.1021/acscentsci.7b00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng Y., Zak O., Aisen P., et al. Structure of the human transferrin receptor-transferrin complex. Cell. 2004;116:565–576. doi: 10.1016/s0092-8674(04)00130-8. [DOI] [PubMed] [Google Scholar]
- 9.Hou W., Xie Y., Song X., et al. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy. 2016;12:1425–1428. doi: 10.1080/15548627.2016.1187366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang W.S., Stockwell B.R. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem Biol. 2008;15:234–245. doi: 10.1016/j.chembiol.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seiler A., Schneider M., Forster H., et al. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. Cell Metab. 2008;8:237–248. doi: 10.1016/j.cmet.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Mao C., Liu X., Zhang Y., et al. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature. 2021;593:586–590. doi: 10.1038/s41586-021-03539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinzii C.M., DiMauro S., Hirano M. Human coenzyme Q 10 deficiency. Neurochem Res. 2007;32:723–727. doi: 10.1007/s11064-006-9190-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bersuker K., Hendricks J.M., Li Z., et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575:688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doll S., Freitas F.P., Shah R., et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575:693–698. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 16.Zou Y., Palte M.J., Deik A.A., et al. A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat Commun. 2019;10:1617. doi: 10.1038/s41467-019-09277-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu S., Chaudhary O., Rodríguez-Morales P., et al. Uptake of oxidized lipids by the scavenger receptor CD36 promotes lipid peroxidation and dysfunction in CD8+ T cells in tumors. Immunity. 2021;54:1561–1577.e7. doi: 10.1016/j.immuni.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green D.R., Ferguson T., Zitvogel L., et al. Immunogenic and tolerogenic cell death. Nat Rev Immunol. 2009;9:353–363. doi: 10.1038/nri2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garg A.D., Nowis D., Golab J., et al. Immunogenic cell death, DAMPs and anticancer therapeutics: an emerging amalgamation. Biochim Biophys Acta. 2010;1805:53–71. doi: 10.1016/j.bbcan.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Savina A., Amigorena S. Phagocytosis and antigen presentation in dendritic cells. Immunol Rev. 2007;219:143–156. doi: 10.1111/j.1600-065X.2007.00552.x. [DOI] [PubMed] [Google Scholar]
- 21.Acharya D., Li X.R.L., Heineman R.E.-S., et al. Complement receptor-mediated phagocytosis induces proinflammatory cytokine production in murine macrophages. Front Immunol. 2020;10:3049. doi: 10.3389/fimmu.2019.03049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quezada S.A., Jarvinen L.Z., Lind E.F., et al. CD40/CD154 interactions at the interface of tolerance and immunity. Annu Rev Immunol. 2004;22:307–328. doi: 10.1146/annurev.immunol.22.012703.104533. [DOI] [PubMed] [Google Scholar]
- 23.Murphy W.J., Welniak L., Back T., et al. Synergistic anti-tumor responses after administration of agonistic antibodies to CD40 and IL-2: coordination of dendritic and CD8+ cell responses. J Immunol. 2003;170:2727–2733. doi: 10.4049/jimmunol.170.5.2727. [DOI] [PubMed] [Google Scholar]
- 24.Beatty G.L., Chiorean E.G., Fishman M.P., et al. CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science. 2011;331:1612–1616. doi: 10.1126/science.1198443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoves S., Ooi C.-H., Wolter C., et al. Rapid activation of tumor-associated macrophages boosts preexisting tumor immunity. J Exp Med. 2018;215:859–876. doi: 10.1084/jem.20171440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu I.M., Lai R.K., Lin S.H., et al. Transketolase counteracts oxidative stress to drive cancer development. Proc Natl Acad Sci U S A. 2016;113:E725–E734. doi: 10.1073/pnas.1508779113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee D., Zhang M.S., Tsang F.H., et al. Adaptive and constitutive activations of malic enzymes confer liver cancer multi-layered protection against ROS. Hepatology. 2021;74:776–796. doi: 10.1002/hep.31761. [DOI] [PubMed] [Google Scholar]
- 28.Lee D., Xu I.M., Chiu D.K., et al. Folate cycle enzyme MTHFD1L confers metabolic advantages in hepatocellular carcinoma. J Clin Invest. 2017;127:1856–1872. doi: 10.1172/JCI90253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rojo de la Vega M., Chapman E., Zhang D.D. NRF2 and the hallmarks of cancer. Cancer Cell. 2018;34:21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee D., Xu I.M., Chiu D.K., et al. Induction of oxidative stress through inhibition of thioredoxin reductase 1 is an effective therapeutic approach for hepatocellular carcinoma. Hepatology. 2019;69:1768–1786. doi: 10.1002/hep.30467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conrad M., Pratt D.A. The chemical basis of ferroptosis. Nat Chem Biol. 2019;15:1137–1147. doi: 10.1038/s41589-019-0408-1. [DOI] [PubMed] [Google Scholar]
- 32.Drijvers J.M., Gillis J.E., Muijlwijk T., et al. Pharmacologic screening identifies metabolic vulnerabilities of CD8+ T cells. Cancer Immunol Res. 2021;9:184–199. doi: 10.1158/2326-6066.CIR-20-0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maier B., Leader A.M., Chen S.T., et al. A conserved dendritic-cell regulatory program limits antitumour immunity. Nature. 2020;580:257–262. doi: 10.1038/s41586-020-2134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng S., Li Z., Gao R., et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell. 2021;184:792–809. doi: 10.1016/j.cell.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 35.Dolma S., Lessnick S.L., Hahn W.C., et al. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285–296. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- 36.Zou Y., Henry W.S., Ricq E.L., et al. Plasticity of ether lipids promotes ferroptosis susceptibility and evasion. Nature. 2020;585:603–608. doi: 10.1038/s41586-020-2732-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viswanathan V.S., Ryan M.J., Dhruv H.D., et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547:453–457. doi: 10.1038/nature23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ubellacker J.M., Tasdogan A., Ramesh V., et al. Lymph protects metastasizing melanoma cells from ferroptosis. Nature. 2020;585:113–118. doi: 10.1038/s41586-020-2623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verma N., Vinik Y., Saroha A., et al. Synthetic lethal combination targeting BET uncovered intrinsic susceptibility of TNBC to ferroptosis. Sci Adv. 2020;6 doi: 10.1126/sciadv.aba8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang L., Kon N., Li T., et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun X., Niu X., Chen R., et al. Metallothionein-1G facilitates sorafenib resistance through inhibition of ferroptosis. Hepatology. 2016;64:488–500. doi: 10.1002/hep.28574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun X., Ou Z., Chen R., et al. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology. 2016;63:173–184. doi: 10.1002/hep.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Efimova I., Catanzaro E., Van der Meeren L., et al. Vaccination with early ferroptotic cancer cells induces efficient antitumor immunity. J Immunother Cancer. 2020;8 doi: 10.1136/jitc-2020-001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo X., Gong H.-B., Gao H.-Y., et al. Oxygenated phosphatidylethanolamine navigates phagocytosis of ferroptotic cells by interacting with TLR2. Cell Death Differ. 2021;28:1971–1989. doi: 10.1038/s41418-020-00719-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spranger S., Dai D., Horton B., et al. Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell. 2017;31:711–723. doi: 10.1016/j.ccell.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.House I.G., Savas P., Lai J., et al. Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin Cancer Res. 2020;26:487–504. doi: 10.1158/1078-0432.CCR-19-1868. [DOI] [PubMed] [Google Scholar]
- 47.Ozga A.J., Chow M.T., Luster A.D. Chemokines and the immune response to cancer. Immunity. 2021;54:859–874. doi: 10.1016/j.immuni.2021.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Truman L.A., Ford C.A., Pasikowska M., et al. CX3CL1/fractalkine is released from apoptotic lymphocytes to stimulate macrophage chemotaxis. Blood. 2008;112:5026–5036. doi: 10.1182/blood-2008-06-162404. [DOI] [PubMed] [Google Scholar]
- 49.Wang W., Green M., Choi J.E., et al. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569:270–274. doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vanmeerbeek I., Sprooten J., De Ruysscher D., et al. Trial watch: chemotherapy-induced immunogenic cell death in immuno-oncology. Oncoimmunology. 2020;9 doi: 10.1080/2162402X.2019.1703449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jenkins R.W., Barbie D.A., Flaherty K.T. Mechanisms of resistance to immune checkpoint inhibitors. Br J Cancer. 2018;118:9–16. doi: 10.1038/bjc.2017.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kroemer G., Galluzzi L., Kepp O., et al. Immunogenic cell death in cancer therapy. Annu Rev Immunol. 2013;31:51–72. doi: 10.1146/annurev-immunol-032712-100008. [DOI] [PubMed] [Google Scholar]
- 53.Chen L., Han X. Anti–PD-1/PD-L1 therapy of human cancer: past, present, and future. J Clin Invest. 2015;125:3384–3391. doi: 10.1172/JCI80011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vonderheide R.H. CD40 agonist antibodies in cancer immunotherapy. Annu Rev Med. 2020;71:47–58. doi: 10.1146/annurev-med-062518-045435. [DOI] [PubMed] [Google Scholar]
- 55.Diggs L.P., Ruf B., Ma C., et al. CD40-mediated immune cell activation enhances response to anti-PD-1 in murine intrahepatic cholangiocarcinoma. J Hepatol. 2021;74:1145–1154. doi: 10.1016/j.jhep.2020.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Hara M.H., O’Reilly E.M., Varadhachary G., et al. CD40 agonistic monoclonal antibody APX005M (sotigalimab) and chemotherapy, with or without nivolumab, for the treatment of metastatic pancreatic adenocarcinoma: an open-label, multicentre, phase 1b study. Lancet Oncol. 2021;22:118–131. doi: 10.1016/S1470-2045(20)30532-5. [DOI] [PubMed] [Google Scholar]
- 57.Jiang Z., Lim S.-O., Yan M., et al. TYRO3 induces anti–PD-1/PD-L1 therapy resistance by limiting innate immunity and tumoral ferroptosis. J Clin Invest. 2021;131 doi: 10.1172/JCI139434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ho W.J., Yarchoan M., Charmsaz S., et al. Multipanel mass cytometry reveals anti–PD-1 therapy–mediated B and T cell compartment remodeling in tumor-draining lymph nodes. JCI Insight. 2020;5 doi: 10.1172/jci.insight.132286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma X., Xiao L., Liu L., et al. CD36-mediated ferroptosis dampens intratumoral CD8+ T cell effector function and impairs their antitumor ability. Cell Metabolism. 2021;33:1001–1012.e5. doi: 10.1016/j.cmet.2021.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wong C.C., Gilkes D.M., Zhang H., et al. Hypoxia-inducible factor 1 is a master regulator of breast cancer metastatic niche formation. Proc Natl Acad Sci U S A. 2011;108:16369–16374. doi: 10.1073/pnas.1113483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wong C.C., Tse A.P., Huang Y.P., et al. Lysyl oxidase-like 2 is critical to tumor microenvironment and metastatic niche formation in hepatocellular carcinoma. Hepatology. 2014;60:1645–1658. doi: 10.1002/hep.27320. [DOI] [PubMed] [Google Scholar]
- 62.Wong C.C., Wong C.M., Tung E.K., et al. The microRNA miR-139 suppresses metastasis and progression of hepatocellular carcinoma by down-regulating Rho-kinase 2. Gastroenterology. 2011;140:322–331. doi: 10.1053/j.gastro.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 63.Tang Z., Li S., Guan X., et al. Rapid assessment of the coenzyme Q10 redox state using ultrahigh performance liquid chromatography tandem mass spectrometry. Analyst. 2014;139:5600–5604. doi: 10.1039/c4an00760c. [DOI] [PubMed] [Google Scholar]
- 64.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ahlmann-Eltze C., glmGamPoi Huber W. fitting Gamma-Poisson generalized linear models on single cell count data. Bioinformatics. 2021;36:5701–5702. doi: 10.1093/bioinformatics/btaa1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou B., Guo R. Integrative analysis of significant RNA-binding proteins in colorectal cancer metastasis. J Cell Biochem. 2018;119:9730–9741. doi: 10.1002/jcb.27290. [DOI] [PubMed] [Google Scholar]
- 67.Haghverdi L., Lun A.T.L., Morgan M.D., et al. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol. 2018;36:421–427. doi: 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aran D., Looney A.P., Liu L., et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zappia L., Oshlack A. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. Gigascience. 2018;7:giy083. doi: 10.1093/gigascience/giy083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hatzinger R., Hornik K., Nagel H.R. Pearson Deutschland GmbH; 2011. Einführung durch angewandte Statistik. [Google Scholar]
- 71.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]