Abstract
Background
Asthma is a common chronic respiratory disease worldwide. Recent studies have revealed the critical effects of the ceRNA network and ferroptosis on patients with asthma. Thus, this study aimed to explore the potential ferroptosis-related ceRNA network, investigate the immune cell infiltration level in asthma through integrated analysis of public asthma microarray datasets, and find suitable diagnostic biomarkers for asthma.
Methods
First, three asthma-related datasets which were downloaded from the Gene Expression Omnibus (GEO) database were integrated into one pooled dataset after correcting for batch effects. Next, we screened differentially expressed lncRNAs (DElncRNAs) between patients and healthy subjects, constructed a ceRNA network using the StarBase database and screened ferroptosis–related genes from the predicted target mRNAs for Disease Ontology (DO), Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses. We also performed Gene Set Enrichment Analysis (GSEA) and Gene Set Variation Analysis (GSVA) on the batch effect-corrected mRNA expression profile. Then, Least Absolute Shrinkage and Selection Operator (LASSO) regression was used to screen potential diagnostic biomarkers, and the diagnostic efficacy was assessed using a receiver operating characteristic (ROC) curve. Finally, we determined the proportion of 22 immune cells in patients with asthma using CIBERSORT and investigated the correlation between key RNAs and immune cells.
Results
We obtained 19 DElncRNAs, of which only LUCAT1 and MIR222HG had corresponding target miRNAs. The differentially expressed ferroptosis-related genes were involved in multiple programmed cell death-related pathways. We also found that the mRNA expression profile was primarily enriched in innate immune system responses. We screened seven candidate diagnostic biomarkers for asthma using LASSO regression (namely, BCL10, CD300E, IER2, MMP13, OAF, TBC1D3, and TMEM151A), among which the area under the curve (AUC) value for CD300E and IER2 were 0.722 and 0.856, respectively. Finally, we revealed the infiltration ratio of different immune cells in asthma and found a correlation between LUCAT1, MIR222HG, CD300E, and IER2 with some immune cells.
Conclusion
This study explored a potential lncRNA-miRNA-mRNA regulatory network and its underlying diagnostic biomarkers (CD300E and IER2) in asthma and identified the immune cells most associated with them, providing possible diagnostic markers and immunotherapeutic targets for asthma.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-023-09400-7.
Keywords: Asthma, Diagnostic biomarker, CeRNA network, Ferroptosis, Immune infiltration analysis
Introduction
Asthma is a chronic airway inflammatory disease, and the Global Asthma Network 1 (GAN1) study reported that the prevalence rates of asthma in children and adults were 10.5% and 4.4%, respectively [1, 2]. The overall global burden of asthma is still substantial. The main characteristics of asthma include airway inflammation, hyperresponsiveness, mucus hypersecretion, and airway remodeling [3]. However, the pathogenesis of asthma involves complex gene-environment interactions. Furthermore, current treatment options for asthma are primarily directed at symptom control rather than altering the prognostic endpoints of the disease, with a subset of patients with asthma whose symptoms cannot be controlled [4]. Therefore, identifying potential asthma biomarkers utilizing bioinformatic analysis might provide insights into the pathogenesis of asthma and determine new therapeutic targets.
The effect of competing endogenous RNAs (ceRNAs) on asthma has attracted the interest of researchers over recent years. Hence, further studies on ceRNAs might help to find new mechanisms for asthma treatment. CeRNAs, as a newly discovered mechanism of gene expression regulation, have an elaborate and complex regulatory network involving several RNA molecules, such as long non-coding RNAs (lncRNAs), pseudogenes, circular RNAs (circRNAs), viral RNAs, and mRNAs [5]. The present study aims to construct a lncRNA–miRNA–mRNA regulatory network. LncRNAs and mRNAs have miRNA response elements (MREs). LncRNAs indirectly regulate mRNA expression levels and cellular functions by competitively binding MREs. Furthermore, the trend in expression levels is consistent across ceRNAs [6]. A growing number of studies have revealed that lncRNAs could regulate different cellular processes (for example, inflammation [7], proliferation [8], apoptosis [9], migration, and epithelial–mesenchymal transition) through the lncRNA-miRNA-mRNA axis, thereby regulating asthma progression. Therefore, our study focuses on the lncRNA–miRNA–mRNA axis in asthma through bioinformatic analysis to investigate the molecular regulatory mechanism involved in peripheral blood mononuclear cells (PBMCs) among patients with asthma.
Ferroptosis has garnered interest of researchers over the past few years, and research on its role in the pathogenesis of asthma has gained momentum. The ferroptosis pathway, first proposed in 2012, is an iron-dependent novel form of programmed cell death, which varies from apoptosis, cell necrosis, and cell autophagy [10]. Ferroptosis could be triggered by iron-catalyzed lipid peroxidation, which is mediated by non-enzymatic (Fenton reaction) and enzymatic (lipoxygenases [LOXs]) mechanisms [11]. The excessive release of oxidized lipid mediators increases the activity of cyclooxygenase (COX) and LOX, accelerates the metabolism of arachidonic acid, promotes the secretion of inflammatory signaling molecules [12], and mediates inflammatory responses and immune cell chemotaxis. Studies have shown that the phosphatidylethanolamine-binding protein 1/15 LOX (PEBP1/15-LO) complex is a critical regulator of ferroptosis capable of stimulating IL-13/ IL-4-induced Th-cell inflammation. The colocalization level of PEBP1 and 15-LO in human airway epithelial cells (HAECs) of patients with asthma is higher than that in those of the normal population, indicating the likelihood of ferroptosis in HAECs of patients with asthma [13, 14]. Moreover, increased lipid peroxidation and ROS production levels as well as significantly reduced glutathione peroxidase 4 (GPX4) and solute carrier family 7 member 11 (SLC7A11) protein levels have been reported in lung tissues of a murine house dust mite–induced asthma model,, indicating an increase in ferroptosis in lung tissues of asthmatic mice [15]. Therefore, treatment of asthmatic mice with ferroptosis inhibitors such as deferoxamine (DFO) and ferrostatin-1 (Fer-1) significantly reduces airway inflammation [16], suggesting that ferroptosis plays a crucial role in airway epithelial cells of animal models with asthma.
Studies have revealed that Th1/Th2-mediated immune imbalance serves as the predominant mechanism of airway inflammatory response in asthma [17] and that various immune cells are involved in the pathogenesis of asthma, including eosinophils, mast cells, dendritic cells (DCs), macrophages, neutrophils, T lymphocytes, B lymphocytes, and innate lymphocytes [18]. Moreover, information on the ratio of different infiltrated immune cells can promote the classification and diagnosis of patients with asthma [19, 20]. For instance, asthma is classified as eosinophilic asthma, neutrophilic asthma, mixed granulocytic asthma, and paucigranulocytic asthma based on the analysis of inflammatory cell count in induced sputum. However, only few studies have explored whether other immune cells have diagnostic value. Therefore, identifying the ratios of different immune cells in asthma could help us understand asthma progression and establish an efficient diagnosis and personalized treatment strategy.
In this study, we obtained three microarray expression datasets of PBMC from patients with asthma and healthy individuals by screening the Gene Expression Omnibus (GEO) database, including two training sets (GSE143192/GSE165934) and one validation set (GSE117038). After removing the batch effect (nasthma = 14, nnormal = 13), the training sets were combined to obtain lncRNA and mRNA expression profiles. After screening the differentially expressed lncRNAs against the lncRNA expression profile, target mRNAs were predicted using the StarBase database and intersected with ferroptosis-related genes for Disease Ontology (DO), Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses to construct the ceRNA network. Then, we performed Gene Set Enrichment Analysis (GSEA) and Gene Set Variation Analysis (GSVA) analyses against the mRNA expression profile and screened key genes using the Least Absolute Shrinkage and Selection Operator (LASSO) machine learning algorithm. Afterward, receiver operating characteristic (ROC) curve analysis was applied to screen genes (as diagnostic markers) with high area under the curve (AUC) values. Finally, CIBERSORT was applied to perform immune infiltration analysis, while key lncRNAs and mRNAs were correlated with different immune cell infiltration levels. The experimental flow chart of this study is shown in Fig. 1, and the details of all data sets are shown in Table 1.
Fig. 1.

Complete flow chart of our research
Table 1.
Summary table of GEO dataset information
Materials and methods
Data downloading and processing
We downloaded three public asthma-related peripheral blood mononuclear cell gene expression dataset profiles from the Gene Expression Omnibus database managed by the National Center for Biotechnology Information [21] (https://www.ncbi.nlm.nih.gov). The profiles included training sets GSE143192 [7] (nasthma = 4, nnormal = 4) and GSE165934 (nasthma = 10, nnormal = 9) and validation set GSE117038 [22] (nasthma = 9, nnormal = 3). The species source of these three datasets is human (Homo sapiens) with platform numbers GPL22120, GPL23126, and GPL16791. Next, we downloaded human phenotype ontology (c5.hpo.v7.2.symbols.gmt), C2 (Curated gene sets, c2.all.v7.2.symbols.gmt), and C5 (c5.go.v7.2.symbols.gmt) gene sets from the MSigDB database [23] (https://www.gsea-msigdb.org/gsea/msigdb). Finally, we compiled 453 ferroptosis–related genes (Table S1) from the GeneCards database [24] (http://www.genecards.org), FerrDb database [25] (http://www.zhounan.org/ferrdb/), and ferroptosis–related literature in the journal CELL [26].
The datasets used in this study were derived from different study types and experimental conditions, generating the batch effect. Therefore, we first extracted common lncRNAs and mRNAs from GSE143192 and GSE165934, performed batch effect correction using the Combat function in the R package “sva” [27], and finally performed multiple analyses (as described below) on the obtained integrated lncRNA and mRNA expression profile (nasthma = 14, nnormal = 13). In addition, we evaluated data quality before and after the batch effect correction and then constructed box and Principal Components Analysis (PCA) plots of the lncRNA expression profile.
Differentially expressed lncRNA screening and ceRNA network construction
Differential analysis of the batch effect–corrected lncRNA expression profile was performed using the R package “limma” [28], using patients with asthma and healthy individuals as the grouping criterion, a difference threshold of Log2|FC|> 1, and p-value of < 0.05. Then, volcano plots and heat maps of differentially expressed lncRNAs were generated using the R package “ggplot2” [29] and “pheatmap” respectively.
Next, we constructed a ceRNA network using the obtained differentially expressed lncRNAs with StarBase [30] (http://StarBase.sysu.edu.cn/). Most lncRNAs showed no corresponding target miRNAs, with only lung cancer-associated transcript 1 (LUCAT1) and MIR222HG being identified as predictable target genes. Therefore, we constructed ceRNA networks using only these two lncRNAs. Furthermore, the ceRNA networks were visualized using Cytoscape [31].
Ferroptosis-related DEG enrichment analysis
The predicted target mRNAs from the ceRNA network constructed using differentially expressed lncRNAs were intersected with ferroptosis-related genes. Venn diagrams were plotted to obtain differentially expressed genes related to ferroptosis. These genes were subjected to DO [32], GO [33], and KEGG [34] enrichment analyses using the R package “clusterProfiler” [35].
GSEA and GSVA analyses of the mRNA expression profile
The mRNA expression profile corrected for batch effect was analyzed by GSEA [36] and GSVA [37]. GSEA uses genes in a predefined gene set to assess their distribution trend based on their correlation with the phenotype, thereby determining their contribution to this phenotype. For this analysis, “c2.all.v7.2.symbols.gmt” was selected as the reference gene set. GSVA is a non-parametric unsupervised analysis primarily used to assess the enrichment profile of gene sets in microarrays and transcriptomes. This analysis evaluates whether different metabolic pathways are enriched among samples by converting the expression matrix of genes into the expression matrix of gene sets. For GSVA, Hallmarker, KEGG, and GO-related gene sets were selected as reference gene sets. Then, the GSVA score of each gene set was quantified. Finally, we used the R package “limma” to perform a variance analysis based on the grouping information of patients with asthma and normal subjects and plotted histograms of upregulated and downregulated differentially expressed gene sets sorted by t-value.
Biomarker screening by LASSO regression
The integrated training set corrected for batch effect was first screened for key genes in asthma using the LASSO algorithm. We applied these candidate genes to the GSE117038 validation set to predict the asthma status in this cohort and validate the diagnostic validity of these candidate biomarkers. Then, ROC curves were used. The AUC values were calculated, and genes with strong ROC results (AUC value of > 0.7) were selected as diagnostic markers for asthma.
Immune infiltration analysis by CIBERSORT and the correlation between key genes and infiltrating immune cells
CIBERSORT [38] was used to analyze the proportion of various immune cells in PBMC in patients with asthma and healthy subjects based on the principle of linear support vector regression. The Wilcoxon Rank sum-test was used in the immune infiltration analysis by CIBERSORT, and p-value < 0.05 demonstrated that the immune cell infiltration matrix was acquired. Then, we investigated the association between the key genes screened in the previous analysis and the ratio of immune cell infiltration and calculated Pearson correlation coefficients along with p-values (p-value < 0.05 as the significance threshold). Finally, we plotted lollipop plots showing the correlation among each lncRNA, mRNA, and different immune cells using the R package “ggplot2”.
Statistical analysis
All the experiments were performed on R Software (https://www.r-project.org/ version 4.1.1). The correlation between genes and immune cells was measured by Spearman coefficient and corrected by Benjamini-Hochberg (BH) multiple test, FDR correction was performed on multiple tests to reduce false-positive rates. Due to the data species and characteristics, differences were analyzed by the Wilcoxon rank-sum test and Student’s t test for the categorical variables and the continuous variables, respectively. The two-sided test was considered statistically significant at p-value < 0.05.
Results
Batch effect correction of the data
After batch effect correction and integration of the GSE143192 and GSE165934 datasets, we obtained 12,125 and 3,376 common lncRNAs and mRNAs, respectively. We evaluated data quality before and after batch effect correction and generated box plots (Figs. 2A and C) and PCA plots (Figs. 2B and D) of the lncRNA expression profile before and after the batch effect correction. The results showed that the expression of genes in the two datasets before correction was unevenly distributed with significant principal component distinction (Figs. 2A and B). In contrast, the expression of genes in the two datasets after correction was evenly distributed with no significant principal component distinction (Figs. 2C and D), indicating that the batch effect was effectively reduced, which could improve subsequent data analysis.
Fig. 2.
Quality assessment of GSE143192 and GSE165934 before and after the batch effect correction. A Box plot before batch effect correction. B Principal Components Analysis (PCA) plot before batch effect correction. C Box plot after batch effect correction. D PCA plot after batch effect correction
Screening for differentially expressed lncRNAs and construction of ceRNA networks
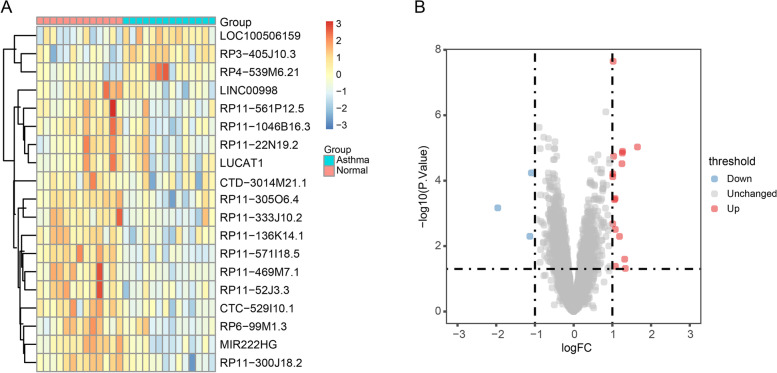
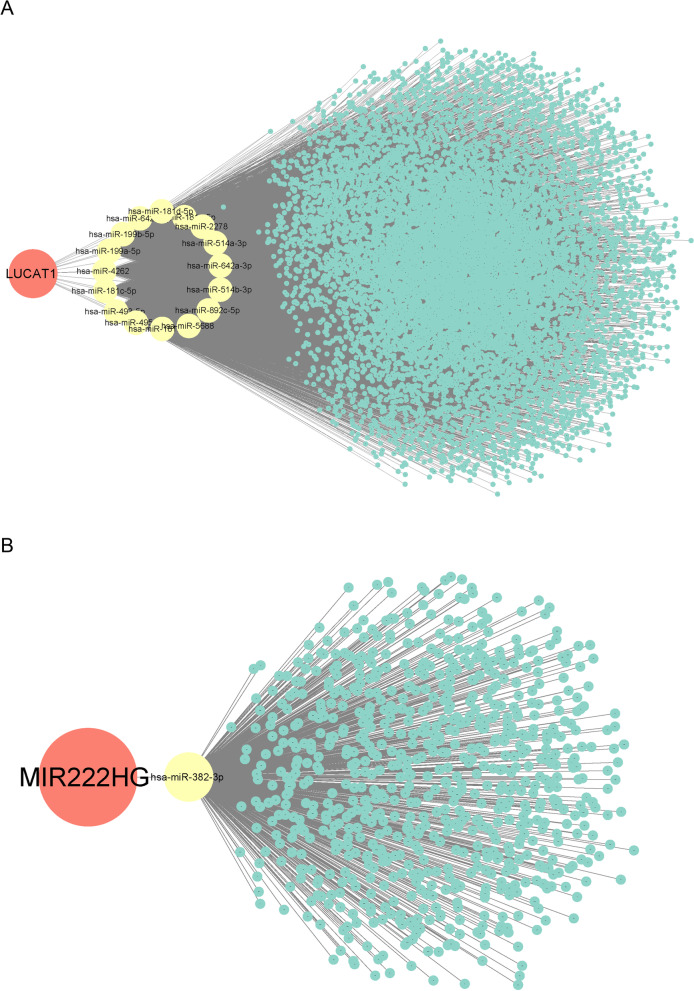
We performed variance analysis on the batch effect-corrected lncRNA expression profile data and obtained 19 DElncRNAs. The heat map (Fig. 3A) and volcano plot (Fig. 3B) allowed us to visualize the differentially expressed lncRNAs between patients with asthma and healthy individuals. Next, the results of the DElncRNA-miRNA-mRNA interaction ceRNA regulatory network showed that only two lncRNAs, namely, LUCAT1 and MIR222HG, had corresponding target miRNAs in the StarBase database. Therefore, we constructed lncRNA-miRNA-mRNA axes for LUCAT1 and MIR222HG, respectively. LUCAT1 had 16 target miRNAs, and 9136 mRNAs were targeted by these 16 miRNAs (Fig. 4A and Table S2); MIR222HG had only one target miRNA (has-miR-382-3p), and 756 mRNAs were targeted by this miRNA (Fig. 4B and Table S3).
Fig. 3.
Differential lncRNA analysis of the dataset after batch effect correction. A Heatmap showed the relative level of 19 upregulated and downregulated differentially expressed lncRNAs (DElncRNAs) in in the dataset after batch effect correction,with up-regulated lncRNAs (red) and down-regulated lncRNAs (blue). B Volcano plot of DElncRNAs in the dataset after batch effect correction, with high expression (red) and low expression (blue)
Fig. 4.
Competing endogenous RNA (CeRNA) networks for LUCAT1 and MIR222HG, with red dots for lncRNA, yellow dots for miRNA, and green dots for mRNA. A CeRNA network for LUCAT1. B CeRNA network for MIR222HG
Ferroptosis-related DEG enrichment analysis
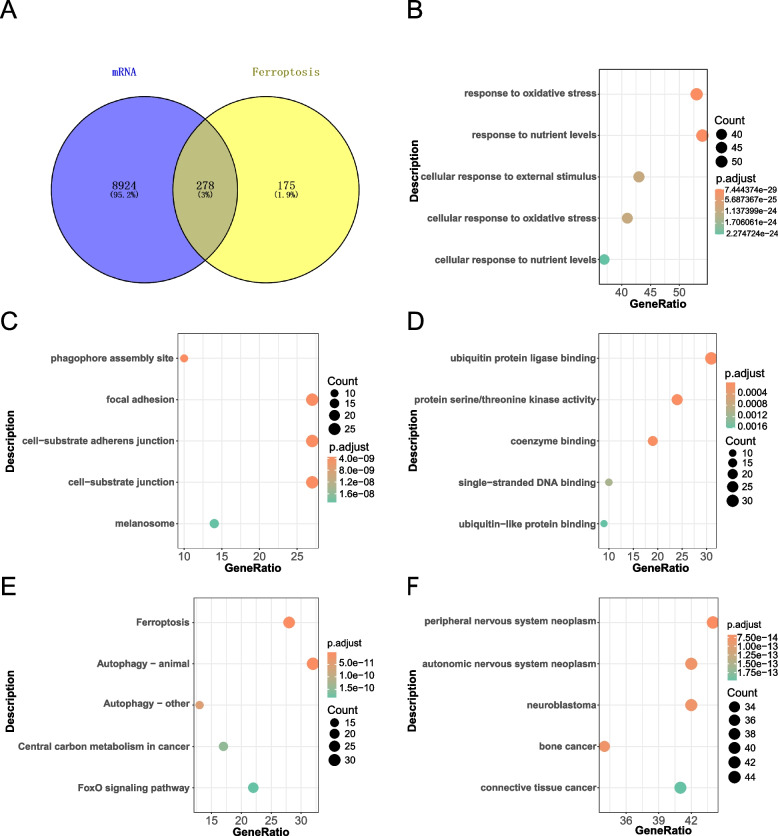
The target mRNAs of LUCAT1 and MIR222HG obtained in the previous procedure (a total of 9202 genes) were compared with ferroptosis-related genes (Fig. 5A) to obtain 278 differential ferroptosis-related genes. GO enrichment analysis revealed that these differential ferroptosis-related genes were significantly enriched during biological processes, including response to oxidative stress, response to nutrient levels, cellular response to external stimulus, cellular components (such as phagophore assembly sites, focal adhesion, cell-substrate junction, and melanosome), and molecular functions (such as ubiquitin protein ligase binding, protein serine/threonine kinase activity, coenzyme binding, and single-stranded DNA binding). KEGG pathway enrichment analysis showed that these differential genes were significantly enriched in signaling pathways, including ferroptosis, autophagy, central carbon metabolism in cancer, FoxO signaling pathway, and mitophagy. Moreover, DO enrichment analysis revealed that these differential genes were involved in peripheral nervous system neoplasm, autonomic nervous system neoplasm, neuroblastoma, and bone cancer. We selected the top five differentials ferroptosis-related genes from the enrichment analysis results to generate bubble plots for GO-BP, GO-CC, and GO-MF (Figs. 5B-D) and KEGG and DO analyses (Figs. 5E and F). The detailed GO enrichment analysis results are shown in Table 2, and the KEGG and DO enrichment analysis results are shown in Table 3.
Fig. 5.
GO, KEGG and DO enrichment analysis results of 278 differential ferroptosis–related genes. A Venn diagram showed 278 overlapping genes of mRNAs targeted by lncRNAs and ferroptosis–related genes. B Bubble plot of GO-BP enrichment results in differential ferroptosis–related genes. C Bubble plot of GO-CC enrichment results in differential ferroptosis–related genes. D Bubble plot of GO-MF enrichment results in differential ferroptosis–related genes. E Bubble plot of KEGG enrichment results in differential ferroptosis–related genes. F Bubble plot of DO enrichment results in differential ferroptosis–related genes
Table 2.
GO enrichment analysis of ferroptosis-related differentially expressed genes (DEGs)
| ID | Description | Count in gene set | p-value |
|---|---|---|---|
| GO-BP | |||
| GO:0006979 | response to oxidative stress | 53 | 1.66E-32 |
| GO:0031667 | response to nutrient levels | 54 | 2.78E-31 |
| GO:0071496 | cellular response to external stimulus | 43 | 9.36E-28 |
| GO:0034599 | cellular response to oxidative stress | 41 | 1.16E-27 |
| GO:0031669 | cellular response to nutrient levels | 37 | 3.05E-27 |
| GO:2001233 | regulation of apoptotic signaling pathway | 41 | 1.25E-22 |
| GO:0010038 | response to metal ion | 38 | 1.58E-21 |
| GO:0006914 | autophagy | 41 | 2.19E-19 |
| GO:0061919 | process utilizing autophagic mechanism | 41 | 2.19E-19 |
| GO:0035690 | cellular response to drug | 35 | 1.54E-18 |
| GO-CC | |||
| GO:0000407 | phagophore assembly site | 10 | 1.21E-11 |
| GO:0005925 | focal adhesion | 27 | 2.16E-11 |
| GO:0005924 | cell-substrate adherens junction | 27 | 2.55E-11 |
| GO:0030055 | cell-substrate junction | 27 | 3.19E-11 |
| GO:0042470 | melanosome | 14 | 2.67E-10 |
| GO:0048770 | pigment granule | 14 | 2.67E-10 |
| GO:0005776 | autophagosome | 12 | 6.78E-09 |
| GO:1902911 | protein kinase complex | 12 | 4.18E-08 |
| GO:0034045 | phagophore assembly site membrane | 6 | 5.07E-08 |
| GO:0031968 | organelle outer membrane | 15 | 1.64E-07 |
| GO-MF | |||
| GO:0031625 | ubiquitin protein ligase binding | 31 | 2.27E-17 |
| GO:0004674 | protein serine/threonine kinase activity | 24 | 1.03E-07 |
| GO:0050662 | coenzyme binding | 19 | 1.56E-07 |
| GO:0003697 | single-stranded DNA binding | 10 | 1.06E-05 |
| GO:0032182 | ubiquitin-like protein binding | 9 | 1.85E-05 |
| GO:0008198 | ferrous iron binding | 5 | 2.92E-05 |
| GO:0051219 | phosphoprotein binding | 8 | 4.42E-05 |
| GO:0003996 | acyl-CoA ligase activity | 4 | 8.96E-05 |
| GO:0016874 | ligase activity | 10 | 9.11E-05 |
| GO:0015175 | neutral amino acid transmembrane transporter activity | 5 | 0.000107 |
Table 3.
KEGG and DO enrichment analyses of ferroptosis-related DEGs
| ID | Description | Count in gene set | p-value |
|---|---|---|---|
| KEGG | |||
| hsa04216 | Ferroptosis | 28 | 1.91E-35 |
| hsa04140 | Autophagy—animal | 32 | 1.79E-21 |
| hsa04136 | Autophagy—other | 13 | 5.40E-13 |
| hsa05230 | Central carbon metabolism in cancer | 17 | 2.13E-12 |
| hsa04068 | FoxO signaling pathway | 22 | 3.34E-12 |
| hsa04137 | Mitophagy—animal | 16 | 4.18E-11 |
| hsa05167 | Kaposi sarcoma-associated herpesvirus infection | 25 | 4.45E-11 |
| hsa04066 | HIF-1 signaling pathway | 19 | 5.49E-11 |
| hsa04621 | NOD-like receptor signaling pathway | 24 | 8.65E-11 |
| hsa05161 | Hepatitis B | 22 | 2.51E-10 |
| DO | |||
| DOID:1192 | peripheral nervous system neoplasm | 44 | 8.98E-17 |
| DOID:2621 | autonomic nervous system neoplasm | 42 | 3.81E-16 |
| DOID:769 | neuroblastoma | 42 | 3.81E-16 |
| DOID:184 | bone cancer | 34 | 4.36E-16 |
| DOID:201 | connective tissue cancer | 41 | 1.33E-15 |
| DOID:0050736 | autosomal dominant disease | 43 | 1.65E-15 |
| DOID:3347 | osteosarcoma | 31 | 4.22E-15 |
| DOID:4450 | renal cell carcinoma | 38 | 1.57E-14 |
| DOID:3996 | urinary system cancer | 46 | 2.33E-14 |
| DOID:263 | kidney cancer | 42 | 8.72E-14 |
GSEA and GSVA analyses of the mRNA expression profile
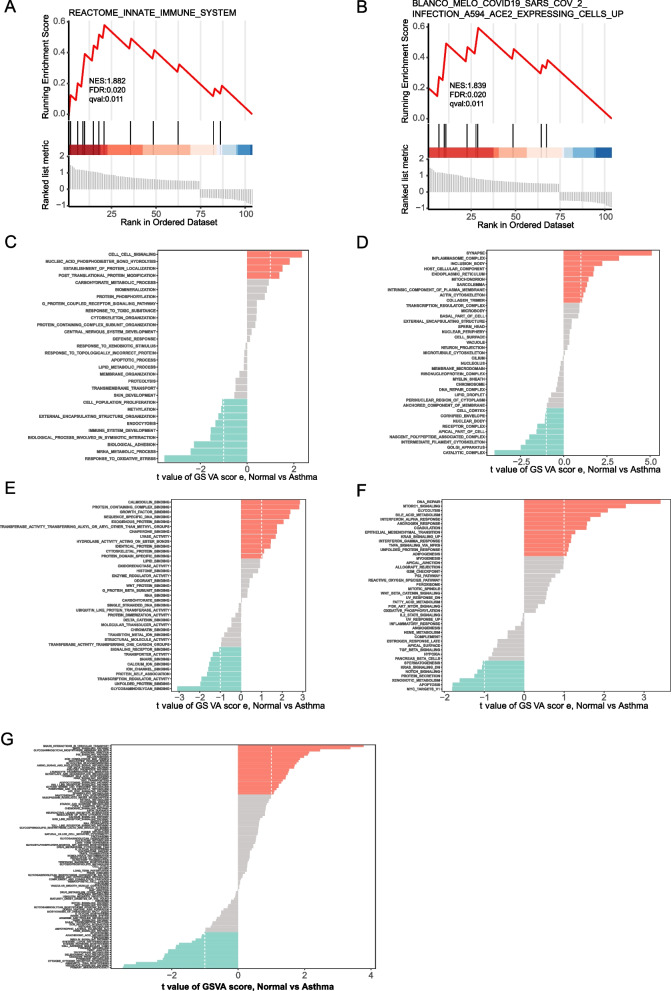
The GSEA result against the mRNA expression profile of asthma showed a significant correlation with gene sets, including “REACTOME_INNATE_IMMUNE_SYSTEM” and “BLANCO_MELO_COVID19_SARS_COV2_INFECTION_A594_ACE2_EXPRESSING_CELLS_UP” data sets (Figs. 6A and B). Meanwhile, the GSVA results primarily included the results of KEGG, GO (GO-BP, GO-CC, and GO-MF), and Hallmarker enrichment analyses. The top five highest GSVA values in GO-BP enrichment analysis results were associated with response to oxidative stress, mRNA metabolic process, cell–cell signaling, bio-adhesion, and nucleic acid phosphodiester bond hydrolysis. The GO-CC results showed that the relevant differential gene products primarily functioned in cellular components, including synapse, catalytic complex, inflammasome complex, Golgi apparatus, and inclusion body. The main results of GO-MF analysis include glycosaminoglycan binding, calmodulin binding, protein-containing complex binding, growth factor binding, and sequence-specific DNA binding. The results of the Hallmarker enrichment analysis included DNA repair, MTORC1 signaling, glycolysis, bile acid metabolism, and MYC-targeted v1. The results of the KEGG enrichment analysis primarily included SNARE interactions in vesicular transport, primary immunodeficiency, the Hedgehog signaling pathway, MAPK signaling pathway, and the aminoacyl tRNA biosynthesis pathway. We plotted histograms of upregulated and downregulated differential genes in gene sets based on the GSVA results (Figs. 6C-G). The GSEA results are shown in Table 4, and the GSVA results are shown in Table 5.
Fig. 6.
Plots of GSEA and GSVA results. A GSEA-enriched gene set REACTOME_INNATE_IMMUNE_SYSTEM. B GSEA-enriched gene set BLANCO_MELO_COVID19_SARS_COV_2_INFECTION_A594_ACE2_EXPRESSING_CELLS_UP. C Histogram of upregulated and downregulated gene sets in GSVA in the GO-BP reference set. D Histogram of GSVA enrichment results in the GO-CC reference set. E Histogram of GSVA enrichment results in the GO-MF reference set. F Histogram of GSVA enrichment results in the Hallmarker reference set. G Histogram of GSVA enrichment results in the KEGG reference set
Table 4.
GSEA results of the mRNA expression profile
| GSEA | |||
|---|---|---|---|
| ID | NES | p-adjust | q-value |
| REACTOME_INNATE_IMMUNE_SYSTEM | 1.882402 | 0.02038 | 0.010727 |
| BLANCO_MELO_COVID19_SARS_COV_2_INFECTION_A594_ACE2_EXPRESSING_CELLS_UP | 1.839356 | 0.02038 | 0.010727 |
Table 5.
GSVA results of the mRNA expression profile
| Gene Set | ID | Score |
|---|---|---|
| KEGG | SNARE_INTERACTIONS_IN_VESICULAR_TRANSPORT | 3.779257 |
| KEGG | PRIMARY_IMMUNODEFICIENCY | -3.4665 |
| KEGG | HEDGEHOG_SIGNALING_PATHWAY | -3.4328 |
| KEGG | MAPK_SIGNALING_PATHWAY | 3.381272 |
| KEGG | AMINOACYL_TRNA_BIOSYNTHESIS | -3.07139 |
| GO-BP | RESPONSE_TO_OXIDATIVE_STRESS | -3.52922 |
| GO-BP | MRNA_METABOLIC_PROCESS | -2.41036 |
| GO-BP | CELL_CELL_SIGNALING | 2.343272 |
| GO-BP | BIOLOGICAL_ADHESION | -2.26262 |
| GO-BP | NUCLEIC_ACID_PHOSPHODIESTER_BOND_HYDROLYSIS | 1.82118 |
| GO-CC | SYNAPSE | 5.093992 |
| GO-CC | CATALYTIC_COMPLEX | -3.99279 |
| GO-CC | INFLAMMASOME_COMPLEX | 3.192443 |
| GO-CC | GOLGI_APPARATUS | -2.481 |
| GO-CC | INCLUSION_BODY | 2.224472 |
| GO-MF | GLYCOSAMINOGLYCAN_BINDING | -3.08434 |
| GO-MF | CALMODULIN_BINDING | 2.854325 |
| GO-MF | PROTEIN_CONTAINING_COMPLEX_BINDING | 2.83028 |
| GO-MF | GROWTH_FACTOR_BINDING | 2.393979 |
| GO-MF | SEQUENCE_SPECIFIC_DNA_BINDING | 2.341345 |
| Hallmarker | DNA_REPAIR | 3.412268 |
| Hallmarker | MTORC1_SIGNALING | 2.539293 |
| Hallmarker | GLYCOLYSIS | 2.092539 |
| Hallmarker | BILE_ACID_METABOLISM | 1.911157 |
| Hallmarker | MYC_TARGETS_V1 | -1.80191 |
Biomarker screening by LASSO regression
Based on the constructed LASSO-penalized regression model (Fig. 7A), the number of variables was determined by finding λ with the lowest classification error (λ = 0.1583217) (Fig. 7B). Consequently, we screened seven potential marker genes (BCL10, CD300E, IER2, MMP13, OAF, TBC1D3, and TMEM151A). The ROC curve results plotted based on these seven candidate genes showed that only CD300E and IER2 featured good diagnostic effects in the validation set GSE117038, with AUC values of 0.722 and 0.856, respectively (Figs. 7C and D). These results indicate that CD300E and IER2 may serve as novel asthma biomarkers.
Fig. 7.
Biomarkers screened by the Least Absolute Shrinkage and Selection Operator (LASSO) regression algorithm. A The LASSO regression algorithm in training set. B The optimal value for penalization coefficient λ in training set. C ROC curve of CD300E in validation set. D ROC curve of IER2 in validation set
Immuno-infiltration analysis by CIBERSORT
We analyzed the changes in the proportion of various infiltrating immune cells in the GSE143192 and GSE165934 datasets using CIBERSORT (Fig. 8A). The results showed differences in immune cell composition among different groups. The correlation heat map of 20 types of immune cells (the algorithm identified a total of 22 types of immune cells, but the immune infiltration results of resting CD4 + memory T cells and naive B cells were null) showed (Fig. 8B) that naive CD4 + T cells were significantly and positively correlated with resting dendritic cells and resting mast cells. Meanwhile, such immune cells were significantly but negatively correlated with T follicular helper cells and activated CD4 + memory T cells. The heat map results of the variance analysis exhibited (Fig. 8C) that some immune cells differed in number between patients with asthma and normal groups. Meanwhile, the box plot results indicated (Fig. 8D) that the number of memory B cells, CD8 + T cells, naive CD4 + T cells, and M0 macrophages significantly differed between patients with asthma and normal groups.
Fig. 8.
Immune cell infiltration assessment and visualization results. A Bar chart of immune cell infiltration distribution across different samples in GSE143192 and GSE165934. B Correlation heat map of 20 types of infiltrating immune cells, with red representing positive correlation and blue representing negative correlation; the darker the color, the stronger the correlation. C Heat map of immune cells differentially expressed between asthma and normal cohorts, with red bands indicating high expression and blue representing low expression. D Box plot of the content of different immune cells between asthma and normal cohorts, with blue representing asthma samples and red representing normal samples,*p-value < 0.05
Correlation analysis of key genes and infiltrating immune cells
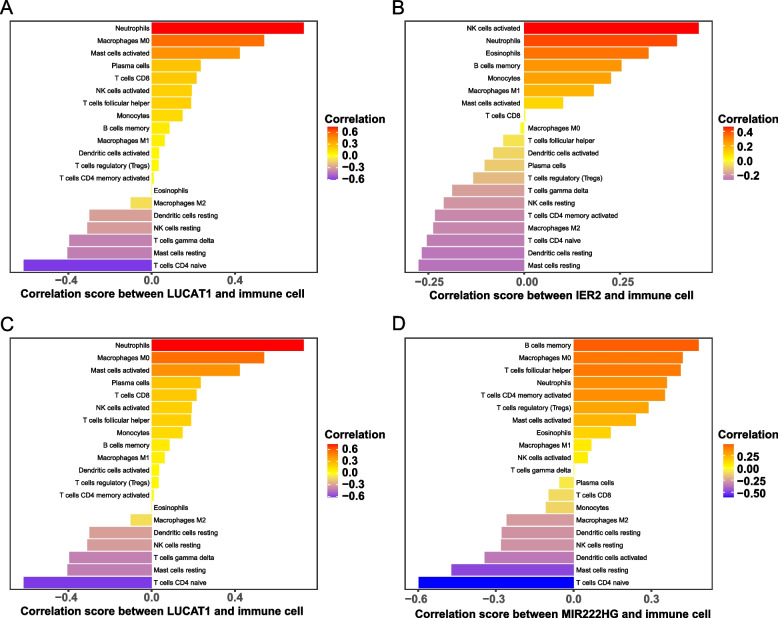
Finally, we investigated the association between the immune cell ratio and the expression of LUCAT1, MIR222HG, CD300E, and IER2 in patients with asthma to determine the biomarkers correlated with the immune cell ratio. The lollipop plots showed the correlation of LUCAT1, MIE222HG, CD300E, and IER2 with immune cells (Fig. 9). The results of immune cells with significant correlation (p-value < 0.05) showed that LUCAT1 expression was positively correlated with the ratios of M0 macrophages, activated mast cells, and neutrophils and negatively correlated with the ratios of resting mast cells, naive CD4 + T cells, and γ-δ T cells; MIR222HG expression was positively correlated with the ratios of memory B cells, M0 macrophages, and T follicular helper cells and negatively correlated with the ratios of resting mast cells and naive CD4 + T cells; CD300E expression was positively correlated with the ratios of M1 macrophages and activated NK cells; IER2 expression was positively correlated with the ratios of neutrophils and activated NK cells.
Fig. 9.
The correlation analysis results of key genes with immune cells. A Lollipop plot of the correlation between LUCAT1 in the mRNA expression profile and immune cells. B Lollipop plot of the correlation between MIR222HG in the mRNA expression profile and immune cells. C Lollipop plot of the correlation between CD300E in the mRNA expression profile and immune cells. D Lollipop plot of the correlation between IER2 in the mRNA expression profile and immune cells
Discussion
Asthma is a global problem affecting people of all ages. However, the current clinical diagnosis of asthma is usually based on clinical symptoms and variable and reversible airflow limitations, with no clear diagnostic gold standard. Although phenotypic analysis based on airway inflammatory biomarkers has elucidated the pathophysiological mechanism of asthma [39], the actual control of asthma remains suboptimal, with treatments mostly based on anti-inflammatory/bronchodilator regimens. Meanwhile, increasing studies have shown that lncRNAs serve as ceRNAs to regulate the onset and progression of asthma [40–42]. Therefore, we could make implications for achieving an early diagnosis and treatment of asthma by identifying novel biomarkers and targets of asthma and exploring the relationship between asthma and ferroptosis, as well as the diversity and complexity of the immune microenvironment.
In this study, we initially screened the differentially expressed lncRNAs using the StarBase database to obtain lncRNAs with targeted miRNAs (LUCAT1 and MIR222HG) and then constructed ceRNA networks by predicting targeting mRNAs from these miRNAs. LncRNAs are non-coding RNAs with a length of > 200 nucleotides, which account for nearly 70% of the human transcriptome. They are highly important in regulating almost all biological processes, and they serve as important regulators of histophysiology and disease processes [43]. LncRNAs also play a role in regulating the progression of asthma.
LUCAT1, which is also known as smoke and cancer-associated lncRNA1 [44], is highly expressed in various malignancies. It regulates the proliferation, migration, and invasion of malignant tumors through diverse mechanisms [45, 46]. Furthermore, studies have demonstrated that elevated LUCAT1 levels suppressed the expression of inflammatory genes and interferon-stimulated genes [47], whereas the overexpression of LUCAT1 in human lung cell lines cultured in high-glucose conditions resulted in reduced iNOS and NO levels [48]. In particular, LUCAT1 could regulate anti-inflammatory processes through different mechanisms. In the last two years, several studies that used bioinformatic analysis to screen key genes have shown that LUCAT1 is a differentially expressed lncRNA associated with ferroptosis [49, 50]. However, MIR222HG, which belongs to the lncRNA subclass of miRNA host genes and is a miR222/221 cluster host gene, remains poorly understood by researchers [51]. MIR222HG expression promotes the development of castration-resistant prostate cancer [52], whereas it was screened in an investigation of whether immune-related lncRNAs could be used as diagnostic markers for glioblastoma. In addition, the MIR222HG-ILF3 RNP complex regulates the RNA stability of DNM3OS, a cell cycle regulator [51]. However, neither LUCAT1 nor MIR222HG has been studied in the context of asthma remains unknown.
In recent years, there has been growing interest regarding the importance of ferroptosis in asthma, and lncRNAs are emerging as key mediators in ferroptosis studies [53]. For example, LINC00618 inhibits ferroptosis by attenuating the expression of lymphatic-like specific helicase and SLC7A11 [54]. LINC00336 also serves as a ceRNA to inhibit ferroptosis by binding to MIR6852 to regulate cystathionine-beta-synthase expression [55]. In this work, we obtained ferroptosis–related differential genes through ceRNA networks constructed by differentially expressed lncRNAs. The preliminary exploration of their enrichment analysis might provide insights into further research on the ferroptosis–related lncRNA-miRNA-mRNA regulatory axis. GO enrichment analysis revealed that these differential ferroptosis-related genes are closely associated with oxidative stress, which is consistent with the definition of ferroptosis (iron and oxidative stress-dependent programmed cell death), while the biochemical features of ferroptosis primarily include increased cellular unstable iron, massive ROS production, decreased GPX4, and lipid metabolite accumulation [56]. Meanwhile, recent studies have found that oxidative/antioxidative imbalance and increased oxidative stress products contribute to airway inflammation, mucus hypersecretion, and airway remodeling, thereby playing a role in asthma progression [57, 58]. Therefore, the role of oxidative stress in asthma has also received increasing attention from researchers. In this study, the KEGG analysis revealed that apart from being highly involved in the ferroptosis pathway, these differential ferroptosis–related genes were also significantly enriched in signaling pathways, including autophagy, FoxO signaling, and mitophagy. In addition, ferroptosis, autophagy, and mitophagy are closely related to each other [59, 60], affecting asthma progression under the regulation of different mechanisms. For example, the PEBP1/15-LO1 complex, which mediates the onset of ferroptosis, is elevated in patients with asthma, which induces airway redox imbalance, thereby causing T2 inflammation and asthma exacerbation [14]; MIR-335-5P targets and regulates ATG5, which reduces the onset of autophagy in the airways of patients with asthma, attenuating an inflammatory response [61]; estrogen receptor 2 transcriptionally suppresses the expression of miRNA-423, which increases the expression level of PINK1 in asthma and mitophagy mediated via PINK1, leading to the worsening of asthma [62]. A variety of programmed cell death pathways may interact with one another during asthma pathogenesis; however, this needs further exploration. Moreover, ferroptosis-related genes were enriched in the FoxO signaling pathway in a biomedical study on amyotrophic lateral sclerosis [63]. Christina et al. showed in their study using a Drosophila model that targeting the JNK/FoxO signaling pathway could regulate airway remodeling in chronic inflammatory lung diseases [64]. Furthermore, elevated FoxO1 expression in airway macrophages among patients with mild asthma induces macrophage polarization in the lungs, which is involved in airway inflammation and airway remodeling in asthma [65]. LUCAT1 and MIR222HG may modulate ferroptosis in asthma by various mechanisms, thereby influencing asthma progression. Analysis based on GSEA found that the innate immune system genes were significantly affected. Different form the GO and KEGG enrichment analyses of ferroptosis-related DEGs, GSVA was able to use whole-genome information and further distinguish the differences in biological behaviors between patients and healthy subjects.
In the present study, we screened seven key genes using the LASSO algorithm and identified differentially expressed genes, namely, CD300E and IER2, with AUC values greater than 0.7 as diagnostic markers for asthma using ROC curve analysis. CD300E, originally known as the immune receptor expressed by myeloid cells (IREM)-2, is a glycosylated surface receptor primarily expressed in human monocytes and myeloid DCs, which serves as an activating receptor that regulates inflammatory and immune responses [66, 67]. Immediate early response 2 (IER2) is a potential DNA-binding protein that serves as a transcription factor or transcriptional co-activator in regulating cellular biological processes [68]. Although CD300E and IER2 have not been reported in the context of asthma, CD300E serves as a biomarker for M2c macrophages [69]. Additionally, the level of M2c macrophages can be used as a marker for asthma to detect the severity of the disease and guide treatment strategies [70]. Therefore, subsequent studies must be conducted to validate the conjecture of CD300E as a diagnostic marker for asthma. Although we initially revealed several potential biomarkers using machine learning in this study that have a high diagnostic value for asthma, further examination of CD300E and IER2 expression levels in hospital-constructed asthma cohorts is necessary to verify the diagnostic power of these marker genes before they can be clinically used.
Finally, we applied the integrated genetic dataset to assess the differences in immune cell composition patterns between patients with asthma and healthy individuals by CIBERSORT. Among the 22 types of immune cells, CD4 + T cells, B cells, and mast cells showed significant differences between the two groups, which is consistent with previous reports on immune cell migration and infiltration in asthma [71–73]. In addition, we found a correlation between the infiltration of certain immune cells and the expression of key genes. We hypothesized that these genes may boost the abundance of immune cells through certain mechanisms, thereby influencing the progression of asthma. However, this correlation remains unclear, which requires validation by in vitro and in vivo experiments, which might be the endpoint of further studies. Meanwhile, we cannot exclude the possibility that there is no causal relationship between immune cell infiltration and the expression of these marker genes and that airway inflammation is responsible for the changes in immune cell ratios and marker gene expression during asthma progression.
This study also has some limitations: First, the small dataset collected from the GEO database and the relatively small sample size used for analysis and validation might lead to bias in the analysis of key genes and CIBERSORT. Next, the immune infiltration analysis based on PBMC samples may have some limitations due to potential interfering factors. In addition, our data analysis was based on open-source data sets, and the key genes screened by the analysis were not adequately supported by the literature, indicating that the credibility of the results should be verified in further experiments.
Conclusion
In this study, we screened key asthma-related genes (LUCAT1, MIR222HG, CD300E, and IER2) and immune cell infiltration profiles through bioinformatic analysis, which could provide insights into the pathogenesis of asthma and new perspectives and approaches for asthma diagnosis and treatment.
Supplementary Information
Additional file 1: Table S1. List of ferroptosis-related genes.
Additional file 2: Table S2. List of miRNAs and mRNAs targeted by LUCAT1.
Additional file 3: Table S3. List of miRNAs and mRNAs targeted by MIR222HG.
Acknowledgements
We are grateful to the GEO database for providing their platforms and contributors for uploading their meaningful datasets.
Abbreviations
- ceRNA
Competing endogenous RNA
- GEO
Gene Expression Omnibus
- DO
Disease Ontology
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GSEA
Gene Set Enrichment Analysis
- GSVA
Gene Set Variation Analysis
- LASSO
Least Absolute Shrinkage and Selection Operator
- ROC
Receiver operating characteristic
- AUC
Area under the curve
- GAN1
Global Asthma Network 1
- lncRNA
Long non-coding RNA
- circRNA
Circular RNA
- MRE
MiRNA response elements
- LOX
Lipoxygenase
- COX
Cyclooxygenase
- PEBP1/15-LO
Phosphatidylethanolamine-binding protein 1/15 LOX complex
- HAEC
Human airway epithelial cell
- GPX4
Glutathione peroxidase 4
- SLC7A11
Solute carrier family 7 member 11
- DFO
Deferoxamine
- Fer-1
Ferrostatin-1
- DC
Dendritic cell
- LUCAT1
Lung cancer–associated transcript 1
- IREM
Immune receptor expressed by myeloid cells
- IER2
Immediate early response 2
Authors’ contributions
NY conceived and designed the project. SC acquired and analyzed the data. All authors were involved in writing the manuscript and approved the submitted and published versions. All authors read, revised, and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81800027) and the Applied Basic Research Project of Liaoning province (2022JH2/101500004).
Availability of data and materials
Com Publicly available datasets were analyzed in this study, which can be found in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov), MSigDB database (https://www.gsea-msigdb.org/gsea/msigdb), GeneCards database (http://www.genecards.org), FerrDb database (http://www.zhounan.org/ferrdb/), and StarBase database (http://StarBase.sysu.edu.cn/).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.García-Marcos L, Asher MI, Pearce N, Ellwood E, Bissell K, Chiang CY, et al. The burden of asthma, hay fever and eczema in children in 25 countries: GAN Phase I study. Eur Respir J. 2022;60(3):2102866. [DOI] [PMC free article] [PubMed]
- 2.Mortimer K, Lesosky M, García-Marcos L, Asher MI, Pearce N, Ellwood E, et al. The burden of asthma, hay fever and eczema in adults in 17 countries: GAN Phase I study. Eur Respir J. 2022;60(3):2102865. [DOI] [PMC free article] [PubMed]
- 3.Barnes PJ. Immunology of asthma and chronic obstructive pulmonary disease. Nat Rev Immunol. 2008;8(3):183–192. doi: 10.1038/nri2254. [DOI] [PubMed] [Google Scholar]
- 4.Castillo JR, Peters SP, Busse WW. Asthma exacerbations: pathogenesis, prevention, and treatment. J Allergy Clin Immunol Pract. 2017;5(4):918–927. doi: 10.1016/j.jaip.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 6.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. [DOI] [PMC free article] [PubMed]
- 7.Xia L, Wang X, Liu L, Fu J, Xiao W, Liang Q, et al. lnc-BAZ2B promotes M2 macrophage activation and inflammation in children with asthma through stabilizing BAZ2B pre-mRNA. J Allergy Clin Immunol. 2021;147:921–932.e9. doi: 10.1016/j.jaci.2020.06.034. [DOI] [PubMed] [Google Scholar]
- 8.Wang WL, Luo XM, Zhang Q, Zhu HQ, Chen GQ, Zhou Q. The lncRNA PVT1/miR-590-5p/FSTL1 axis modulates the proliferation and migration of airway smooth muscle cells in asthma. Autoimmunity. 2021;54(3):138–147. doi: 10.1080/08916934.2021.1897977. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Chen J, Chen W, Liu L, Dong M, Ji J, et al. LINC00987 ameliorates COPD by regulating LPS-induced cell apoptosis, oxidative stress, inflammation and autophagy through Let-7b-5p/SIRT1 axis. Int J Chron Obstruct Pulm Dis. 2020;15:3213–3225. doi: 10.2147/COPD.S276429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirschhorn T, Stockwell BR. The Development of the concept of ferroptosis. Free Radic Biol Med. 2019;133:130–143. doi: 10.1016/j.freeradbiomed.2018.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang D, Kang R, Berghe TV, Vandenabeele P, Kroemer G. The molecular machinery of regulated cell death. Cell Res. 2019;29(5):347–364. doi: 10.1038/s41422-019-0164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Araújo AC, Wheelock CE, Haeggström JZ. The eicosanoids, redox-regulated lipid mediators in immunometabolic disorders. Antioxid Redox Signal. 2018;29(3):275–296. doi: 10.1089/ars.2017.7332. [DOI] [PubMed] [Google Scholar]
- 13.Wenzel SE, Tyurina YY, Zhao J, St Croix CM, Dar HH, Mao G, et al. PEBP1 Wardens Ferroptosis by Enabling Lipoxygenase Generation of Lipid Death Signals. Cell. 2017;171(3):628–641.e26. doi: 10.1016/j.cell.2017.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagasaki T, Schuyler AJ, Zhao J, Samovich SN, Yamada K, Deng Y, et al. 15LO1 dictates glutathione redox changes in the asthmatic airway epithelium to worsen type 2 inflammation. J Clin Invest. 2022;132(1):e151685. [DOI] [PMC free article] [PubMed]
- 15.Tang W, Dong M, Teng F, Cui J, Zhu X, Wang W, et al. Environmental allergens house dust mite-induced asthma is associated with ferroptosis in the lungs. Exp Ther Med. 2021;22(6):1483. doi: 10.3892/etm.2021.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng Z, Huang H, Zhang J, Liu Y, Zhong W, Chen W, et al. HDM induce airway epithelial cell ferroptosis and promote inflammation by activating ferritinophagy in asthma. FASEB J. 2022;36(6):e22359. doi: 10.1096/fj.202101977RR. [DOI] [PubMed] [Google Scholar]
- 17.Menzies-Gow A, Corren J, Bourdin A, Chupp G, Israel E, Wechsler ME, et al. Tezepelumab in Adults and Adolescents with Severe. Uncontrolled Asthma N Engl J Med. 2021;384(19):1800–1809. doi: 10.1056/NEJMoa2034975. [DOI] [PubMed] [Google Scholar]
- 18.Hammad H, Lambrecht BN. The basic immunology of asthma. Cell. 2021;184(6):1469–1485. doi: 10.1016/j.cell.2021.02.016. [DOI] [PubMed] [Google Scholar]
- 19.Agache I, Akdis CA. Endotypes of allergic diseases and asthma: An important step in building blocks for the future of precision medicine. Allergol Int. 2016;65(3):243–252. doi: 10.1016/j.alit.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 20.Terl M, Sedlák V, Cap P, Dvořáková R, Kašák V, Kočí T, et al. Asthma management: A new phenotype-based approach using presence of eosinophilia and allergy. Allergy. 2017;72(9):1279–1287. doi: 10.1111/all.13165. [DOI] [PubMed] [Google Scholar]
- 21.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991-D995. [DOI] [PMC free article] [PubMed]
- 22.Zhu Y, Mao D, Gao W, Han G, Hu H. Analysis of lncRNA Expression in Patients With Eosinophilic and Neutrophilic Asthma Focusing on LNC_000127. Front Genet. 2019;10:141. doi: 10.3389/fgene.2019.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, et al. GeneCards Version 3: the human gene integrator. Database (Oxford). 2010;2010:baq020. [DOI] [PMC free article] [PubMed]
- 25.Zhou N, Bao J. FerrDb: a manually curated resource for regulators and markers of ferroptosis and ferroptosis-disease associations. Database (Oxford). 2020;2020:baaa021. [DOI] [PMC free article] [PubMed]
- 26.Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell. 2017;171(2):273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito K, Murphy D. Application of ggplot2 to Pharmacometric Graphics. CPT Pharmacometrics Syst Pharmacol. 2013;2:e79. doi: 10.1038/psp.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92-D97. [DOI] [PMC free article] [PubMed]
- 31.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bello SM, Shimoyama M, Mitraka E, Laulederkind SJF, Smith CL, Eppig JT, et al. Disease Ontology: improving and unifying disease annotations across species. Dis Model Mech. 2018;11(3):dmm032839. [DOI] [PMC free article] [PubMed]
- 33.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–259. doi: 10.1007/978-1-4939-7493-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuruvilla ME, Lee FE-H, Lee GB. Understanding Asthma Phenotypes, Endotypes, and Mechanisms of Disease. Clin Rev Allergy Immunol. 2019;56(2):219–233. [DOI] [PMC free article] [PubMed]
- 40.Qiu YY, Wu Y, Lin MJ, Bian T, Xiao YL, Qin C. LncRNA-MEG3 functions as a competing endogenous RNA to regulate Treg/Th17 balance in patients with asthma by targeting microRNA-17/ RORγt. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie. 2019;111:386–394. [DOI] [PubMed]
- 41.Sun H, Wang T, Zhang W, Dong H, Gu W, Huang L, et al. LncRNATUG1 Facilitates Th2 Cell Differentiation by Targeting the miR-29c/B7-H3 Axis on Macrophages. Front Immunol. 2021;12:631450. doi: 10.3389/fimmu.2021.631450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu H, Qi N, Zhou Q. LncRNA H19 Inhibits Proliferation and Migration of Airway Smooth Muscle Cells Induced by PDGF-BB Through miR-21/PTEN/Akt Axis. J Asthma Allergy. 2021;14(71–80):H19. doi: 10.2147/JAA.S291333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thai P, Statt S, Chen CH, Liang E, Campbell C, Wu R. Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am J Respir Cell Mol Biol. 2013;49(2):204–211. doi: 10.1165/rcmb.2013-0159RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Xie Y, Wang J, Zhang Y, Liu S, Zhan Y, Zhao Y, et al. Characterization of a Novel LUCAT1/miR-4316/VEGF-A Axis in Metastasis and Glycolysis of Lung Adenocarcinoma. Front Cell Dev Biol. 2022;10:833579. doi: 10.3389/fcell.2022.833579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng A, Song X, Zhang L, Zhao L, Mao X, Wei M, et al. Long non-coding RNA LUCAT1/miR-5582-3p/TCF7L2 axis regulates breast cancer stemness via Wnt/β-catenin pathway. J Exp Clin Cancer Res. 2019;38(1):305. doi: 10.1186/s13046-019-1315-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agarwal S, Vierbuchen T, Ghosh S, Chan J, Jiang Z, Kandasamy RK, et al. The long non-coding RNA LUCAT1 is a negative feedback regulator of interferon responses in humans. Nat Commun. 2020;11(1):6348. doi: 10.1038/s41467-020-20165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li P, Zhang N, Ping F, Gao Y, Cao L. lncRNA SCAL1 inhibits inducible nitric oxide synthase in lung cells under high-glucose conditions. Exp Ther Med. 2019;18(3):1831–1836. doi: 10.3892/etm.2019.7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shu X, Zhang Z, Yao ZY, Xing XL. Identification of Five Ferroptosis-Related LncRNAs as Novel Prognosis and Diagnosis Signatures for Renal Cancer. Front Mol Biosci. 2021;8:763697. doi: 10.3389/fmolb.2021.763697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He Y, Ye Y, Tian W, Qiu H. A Novel lncRNA Panel Related to Ferroptosis, Tumor Progression, and Microenvironment is a Robust Prognostic Indicator for Glioma Patients. Front Cell Dev Biol. 2021;9:788451. doi: 10.3389/fcell.2021.788451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun Q, Hao Q, Lin YC, Song YJ, Bangru S, Arif W, et al. Antagonism between splicing and microprocessor complex dictates the serum-induced processing of lnc-MIGHT for efficient cell cycle reentry. RNA. 2020;26(11):1603–1620. doi: 10.1261/rna.075309.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun T, Du S-Y, Armenia J, Qu F, Fan J, Wang X, et al. Expression of lncRNA MIR222HG co-transcribed from the miR-221/222 gene promoter facilitates the development of castration-resistant prostate cancer. Oncogenesis. 2018;7(3):30. doi: 10.1038/s41389-018-0039-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mou Y, Wang J, Wu J, He D, Zhang C, Duan C, et al. Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J Hematol Oncol. 2019;12(1):34. doi: 10.1186/s13045-019-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Z, Chen X, Liu N, Shi Y, Liu Y, Ouyang L, et al. A Nuclear Long Non-Coding RNA LINC00618 Accelerates Ferroptosis in a Manner Dependent upon Apoptosis. Mol Ther. 2021;29(1):263–274. doi: 10.1016/j.ymthe.2020.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang M, Mao C, Ouyang L, Liu Y, Lai W, Liu N, et al. Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ. 2019;26(11):2329–2343. doi: 10.1038/s41418-019-0304-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao Y, Huang Z, Peng H. Molecular Mechanisms of Ferroptosis and Its Roles in Hematologic Malignancies. Front Oncol. 2021;11:743006. doi: 10.3389/fonc.2021.743006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mishra V, Banga J, Silveyra P. Oxidative stress and cellular pathways of asthma and inflammation: Therapeutic strategies and pharmacological targets. Pharmacol Ther. 2018;181:169–182. doi: 10.1016/j.pharmthera.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang MC, Huang WC, Chen LC, Yeh KW, Lin CF, Liou CJ. Sophoraflavanone G from Ameliorates Allergic Airway Inflammation by Suppressing Th2 Response and Oxidative Stress in a Murine Asthma Model. Int J Mol. Sci. 2022;23(11):6104. [DOI] [PMC free article] [PubMed]
- 59.Gao M, Monian P, Pan Q, Zhang W, Xiang J, Jiang X. Ferroptosis is an autophagic cell death process. Cell Res. 2016;26(9):1021–1032. doi: 10.1038/cr.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li W, Luo LX, Zhou QQ, Gong HB, Fu YY, Yan CY, et al. Phospholipid peroxidation inhibits autophagy via stimulating the delipidation of oxidized LC3-PE. Redox Biol. 2022;55:102421. doi: 10.1016/j.redox.2022.102421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang Q, He J, Yang Q, Zhang Q, Xu Y. MicroRNA-335-5p alleviates inflammatory response, airway fibrosis, and autophagy in childhood asthma through targeted regulation of autophagy related 5. Bioengineered. 2022;13(1):1791–1801. doi: 10.1080/21655979.2021.1996315. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Kong X, Chen R, Zhang L, Wu M, Wu J, Wei Y, et al. ESR2 regulates PINK1-mediated mitophagy via transcriptional repression of microRNA-423 expression to promote asthma development. Pharmacol Res. 2021;174:105956. doi: 10.1016/j.phrs.2021.105956. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Q, Zhao H, Luo M, Cheng X, Li Y, Li Q, et al. The Classification and Prediction of Ferroptosis-Related Genes in ALS: A Pilot Study. Front Genet. 2022;13:919188. doi: 10.3389/fgene.2022.919188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wagner C, Uliczka K, Bossen J, Niu X, Fink C, Thiedmann M, et al. Constitutive immune activity promotes JNK- and FoxO-dependent remodeling of Drosophila airways. Cell Rep. 2021;35(1):108956. doi: 10.1016/j.celrep.2021.108956. [DOI] [PubMed] [Google Scholar]
- 65.Chung S, Kim JY, Song MA, Park GY, Lee YG, Karpurapu M, et al. FoxO1 is a critical regulator of M2-like macrophage activation in allergic asthma. Allergy. 2019;74(3):535–548. doi: 10.1111/all.13626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brckalo T, Calzetti F, Pérez-Cabezas B, Borràs FE, Cassatella MA, López-Botet M. Functional analysis of the CD300e receptor in human monocytes and myeloid dendritic cells. Eur J Immunol. 2010;40(3):722–732. doi: 10.1002/eji.200939468. [DOI] [PubMed] [Google Scholar]
- 67.Isobe M, Izawa K, Sugiuchi M, Sakanishi T, Kaitani A, Takamori A, et al. The CD300e molecule in mice is an immune-activating receptor. J Biol Chem. 2018;293(10):3793–3805. doi: 10.1074/jbc.RA117.000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takaya T, Kasatani K, Noguchi S, Nikawa J-i. Functional analyses of immediate early gene ETR101 expressed in yeast. Biosci. Biotechnol. Biochem. 2009;73(7):1653–1660. [DOI] [PubMed]
- 69.Skronska-Wasek W, Durlanik S, Garnett JP, Pflanz S. Polarized cytokine release from airway epithelium differentially influences macrophage phenotype. Mol Immunol. 2021;132:142–149. doi: 10.1016/j.molimm.2021.01.029. [DOI] [PubMed] [Google Scholar]
- 70.Kuo CH, Tsai ML, Li CH, Hsiao HP, Chao MC, Lee MS, et al. Altered Pattern of Macrophage Polarization as a Biomarker for Severity of Childhood Asthma. J Inflamm Res. 2021;14:6011–6023. doi: 10.2147/JIR.S319754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Habener A, Grychtol R, Gaedcke S, DeLuca D, Dittrich AM, Happle C, et al. IgA+ memory B-cells are significantly increased in patients with asthma and small airways dysfunction. Eur Respir J. 2022:60(5):2102130. [DOI] [PMC free article] [PubMed]
- 72.Luo W, Hu J, Xu W, Dong J. Distinct spatial and temporal roles for Th1, Th2, and Th17 cells in asthma. Front Immunol. 2022;13:974066. doi: 10.3389/fimmu.2022.974066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu C, Li L, Wang C, Jiang J, Li L, Zhu L, Jin S, Jin Z, Lee JJ, Li G, Yan G. Effects of G-Rh2 on mast cell-mediated anaphylaxis via AKT-Nrf2/NF-κB and MAPK-Nrf2/NF-κB pathways. J Ginseng Res. 2022;46(4):550–560. doi: 10.1016/j.jgr.2021.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. List of ferroptosis-related genes.
Additional file 2: Table S2. List of miRNAs and mRNAs targeted by LUCAT1.
Additional file 3: Table S3. List of miRNAs and mRNAs targeted by MIR222HG.
Data Availability Statement
Com Publicly available datasets were analyzed in this study, which can be found in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov), MSigDB database (https://www.gsea-msigdb.org/gsea/msigdb), GeneCards database (http://www.genecards.org), FerrDb database (http://www.zhounan.org/ferrdb/), and StarBase database (http://StarBase.sysu.edu.cn/).