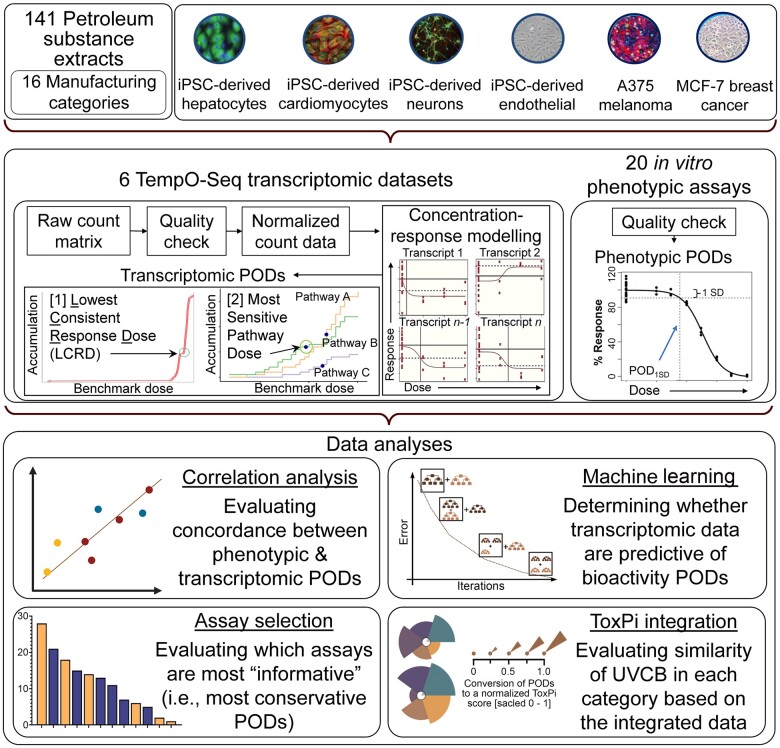
Abstract
Hazard evaluation of substances of “unknown or variable composition, complex reaction products and biological materials” (UVCBs) remains a major challenge in regulatory science because their chemical composition is difficult to ascertain. Petroleum substances are representative UVCBs and human cell-based data have been previously used to substantiate their groupings for regulatory submissions. We hypothesized that a combination of phenotypic and transcriptomic data could be integrated to make decisions as to selection of group-representative worst-case petroleum UVCBs for subsequent toxicity evaluation in vivo. We used data obtained from 141 substances from 16 manufacturing categories previously tested in 6 human cell types (induced pluripotent stem cell [iPSC]-derived hepatocytes, cardiomyocytes, neurons, and endothelial cells, and MCF7 and A375 cell lines). Benchmark doses for gene-substance combinations were calculated, and both transcriptomic and phenotype-derived points of departure (PODs) were obtained. Correlation analysis and machine learning were used to assess associations between phenotypic and transcriptional PODs and to determine the most informative cell types and assays, thus representing a cost-effective integrated testing strategy. We found that 2 cell types—iPSC-derived-hepatocytes and -cardiomyocytes—contributed the most informative and protective PODs and may be used to inform selection of representative petroleum UVCBs for further toxicity evaluation in vivo. Overall, although the use of new approach methodologies to prioritize UVCBs has not been widely adopted, our study proposes a tiered testing strategy based on iPSC-derived hepatocytes and cardiomyocytes to inform selection of representative worst-case petroleum UVCBs from each manufacturing category for further toxicity evaluation in vivo.
Keywords: transcriptomic, petroleum, grouping, dose-response, HTTr, NAMs
The study of changes in gene expression in response to chemical exposure, often referred to as transcriptomics, is now a common approach in mechanistic and predictive toxicology (National Research Council, 2007a; Nuwaysir et al., 1999) and in risk assessment (Buesen et al., 2017; Kavlock et al., 2018; National Toxicology Program, 2018). Because gene expression is reflective of the dynamic tissue and/or cell(s) states, transcriptional changes are considered to be both sensitive and early indicators of chemical-induced perturbations; they also may inform mode of action by providing gene- and pathway-level data (Chen et al., 2012; Cui and Paules, 2010). Over the last 2 decades, technologies used to query gene expression and associated data analysis methods have evolved from microarrays to next generation sequencing-based approaches (Kinaret et al., 2020). More recent toxicology studies have used high-throughput transcriptomics methods allowing for rapid evaluation of the effects of large numbers of chemicals in both time-course and concentration-response study designs (Harrill et al., 2019; House et al., 2017; Lamb et al., 2006; Yeakley et al., 2017). The data from high-throughput transcriptomic studies have been used not only to provide mechanistic underpinnings of the effects of chemicals on biological systems but also to derive quantitative estimates of chemical potency (ie, hazard) more broadly, without a narrow focus on the meaning of perturbed pathways or genes (Harrill et al., 2021).
With the availability of high-throughput transcriptomics methods, the use of these data in toxicology is rapidly evolving from hypothesis-driven observational studies to quantitative risk assessment with derivatio of points of departure (PODs). Several studies have demonstrated concordance among gene expression changes and the “apical” toxicity phenotypes (Geter et al., 2014; Lobenhofer et al., 2004; Rouquie et al., 2009; Zarbl et al., 2010). More recent work that included a larger number of chemicals confirmed that “apical” adverse effect-based PODs derived from sub-chronic (months) or chronic (years) studies in rodents are highly correlated with gene expression-based PODs derived from short-term (days) in vivo studies in the same species (Bhat et al., 2013; Bianchi et al., 2021; Gwinn et al., 2020; Johnson et al., 2020; Page-Lariviere et al., 2019; Thomas et al., 2011, 2012, 2013). For example, longer-term apical and shorter-term in vivo transcriptomic PODs are typically within one order of magnitude of each other across chemical categories and dose ranges; some studies showed that, transcriptomics PODs may be more protective (ie, the “effects” are detected at lower PODs) than the corresponding apical endpoints (Thomas et al., 2011, 2012). Based on these seminal studies, it was proposed that transcriptomic data from short-term dose-response in vivo studies may be used to refine the current regulatory toxicity testing paradigm that relies on the use of long-term animal studies (Farmahin et al., 2017; Johnson et al., 2022).
Although there is growing acceptance of in vivo-derived transcriptomic PODs, there is less evidence on whether gene expression data from in vitro assays has utility in decision-making on chemical safety. In response to the charge by the National Academies to accelerate toxicity testing (National Academies of Sciences Engineering and Medicine, 2017; National Research Council, 2007b), large-scale efforts to test thousands of chemicals in hundreds of cell-based and -free assays have generated a comprehensive compendium of data (Richard et al., 2016, 2021; Williams et al., 2017). Recent analyses of these data showed that in vitro-derived apical PODs are on average more sensitive than in vivo study-derived traditional PODs (Beal et al., 2022; Chen et al., 2020; Paul Friedman et al., 2020). High-throughput transcriptomic data from in vitro studies are the most recent type of information that may be used to advance the use of alternative methods for decision-making (De Abrew et al., 2016, 2019; Harrill et al., 2021; Yauk et al., 2020). Examples of applications of high-throughput transcriptomic data for decision-making range from supporting “biological similarity” among chemicals (De Abrew et al., 2019; Low et al., 2011), to ranking chemicals based on their “potency” to elicit transcriptional effects (Reardon et al., 2021; Rowan-Carroll et al., 2021), to exploring associations between high-throughput transcriptomic and high-throughput phenotypic profiling data (Harrill et al., 2021; Nyffeler et al., 2022). However, studies that evaluated effects of a large number of petroleum substances on a diverse set of human cell-based models showed that although high-throughput toxicogenomic data provided useful mechanistic information on the effects of substances in different manufacturing categories, they afforded only modest additional “value” for grouping (House et al., 2021, 2022). Therefore, the debate continues as to whether transcriptomic data is adding value to other in vitro data streams, especially because they add cost and complexity.
Although recent studies have made advances in demonstrating how combined use of in vitro phenotypes and transcriptomic data can support decision making regarding chemical safety, several challenges remain. First, the applicability of the data on the individual chemicals to the evaluation of multi-constituent substances and mixtures remains largely unexplored. Second, additional work on expanding both the chemical type and biological model (ie, cell lines) domains is needed to determine value added by each cell type and whether a representative set of models can suffice in being protective. Third, although many studies use transcriptomic data for grouping to inform read-across, an equally important potential outcome can be selection of representative (or worst-case) substances within an established group for animal testing if existing read-across data are deficient.
To address these challenges, we used a recently published comprehensive dataset consisting of human cell-based phenotypic and transcriptomic data on petroleum substances that are prototypical “unknown or variable composition, complex reaction products and biological materials” substances (UVCBs) (House et al., 2021, 2022). These studies were conducted to test a hypothesis that in vitro biological activity signatures, both phenotypic and gene expression, can be used to support grouping of UVCBs. Overall, 141 petroleum substances from 16 manufacturing categories (CONCAWE, 2020) were tested as representative UVCBs in a compendium of 15 human cell types representing a variety of tissues; of these, 6 cell types were also profiled for gene expression. Petroleum substances were assayed in dilution series to derive point of departure estimates for each cell type and phenotype. Some of these data were used in regulatory submissions to justify waiving of animal testing requirements, albeit the European Chemicals Agency did not accept the data as presented, in part because of the lack of clarity how these complex data can be integrated and interpreted (ECHA, 2020). Therefore, we aimed to re-analyze the data from (House et al., 2021, 2022) with a goal of informing the selection of worst-case sample(s) in each pre-existing manufacturing UVCB category for subsequent toxicity evaluation and read-across. The previously reported phenotypic PODs and newly modeled transcriptomic PODs were used to conduct concordance analyses, machine learning-based predictions, sensitivity testing for the selection of cell types and assays, and to propose a tiered testing strategy. The data were then integrated to identify substances that may best represent the range of potential hazards for each manufacturing category.
Materials and methods
Chemicals, cells, experimental design, and gene expression data
This study used data previously reported in (House et al., 2021, 2022). Briefly, dimethyl sulfoxide (0.25%–0.5% final concentration depending on a cell type) and cyclohexane extracts of 141 petroleum substances that were supplied by Concawe (Brussels, Belgium) (Figure 1, Table 1, and Supplementary Table 1) were used to expose (in a dilution-series) 6 human cell types representing diverse human organs/tissues (Figure 1 and Table 2). We used both induced pluripotent stem cell (iPSC)-derived cells as well as established cell lines. These in vitro models had to be reproducible (ie, a particular cell/donor can be obtained from a commercial source) and suitable for evaluation of both “functional” and “cytotoxicity” endpoints so that the specificity of the effects of tested compounds could be assessed. Four of these cell types (hepatocytes, endothelial cells, neurons, and cardiomyocytes) were human-iPSC-derived (FujiFilm-Cellular Dynamics International, Madison, Wisconsin). Two cell types (A375 and MCF7) were from ATCC (Manassas, Virginia). In total, 20 phenotype-based points of departure (pPODs) across all 6 cell types that passed the QC steps as described in (House et al., 2021) were included in the current study without additional processing (Table 2, Supplementary Tables 2 and 3). The method for pPOD derivation based on serial 1-log10 dilutions (4 dilutions and vehicle control) was detailed elsewhere (House et al., 2021). Briefly, vehicle control-scaled data for each substance and phenotype were fitted to a curve with a nonlinear logistic (Hill) function to determine POD values, defined as the concentrations at which the fitted curve exceeds 1 standard deviation (SD) above or below the mean of vehicle-treated controls (Sirenko et al., 2017). The choice of 1 SD “benchmark response” was based on the U.S. EPA guidance for dose-response modeling and determination of the point-of-departure values (U.S. EPA, 2012), as well as empirical testing of various thresholds as detailed in (Sirenko et al., 2017), which showed that a choice of 1 SD generates consistently high classification accuracy. The data on weight percentages of polycyclic aromatic compounds (PAC) in all tested samples were also from (House et al., 2021).
Figure 1.
Overview of the study design and data analyses of the in vitro effects of 141 petroleum UVCBs of the 6 cell types.
Table 1.
Petroleum substance categories (CONCAWE, 2020) examined in this study
| Category | Abbreviation | N of samples |
|---|---|---|
| Untreated distillate aromatic extracts | UDAE | 4 |
| Straight-run gas oils | SRGO | 6 |
| Vacuum gas oils, hydrocracked gas oils, and distillate fuels | VHGO | 10a |
| Cracked gas oils | CGO | 8a |
| Unrefined/acid treated oils | UATO | 4 |
| Heavy fuel oil components | HFO | 27a |
| Treated distillate aromatic extracts | TDAE | 2 |
| Residual aromatic extracts | RAE | 2 |
| Bitumens/oxidized asphalt | BIT | 5 |
| Other gas oils | OGO | 4 |
| Foots oils | FO | 3 |
| Kerosines/MK1 diesel fuel | KER | 10a |
| Other lubricant base oils/highly refined base oils | BO | 33a |
| Paraffin and hydrocarbon waxes/slack waxes | WAX | 10a |
| Low boiling point naphthas (gasolines) | NAPHTHA | 10a |
| Petrolatums | P.LAT | 3 |
Categories that contain 8 or more substances that were used for “worst case scenario” analysis.
Table 2.
Cell types and in vitro phenotypes used in this study
| Cell | Phenotype | Exposure duration | Abbreviation |
|---|---|---|---|
| iPSC-hepatocytes | Positive cytoplasmic staining | 48 h | CAM |
| Cellular mean area | 48 h | CMA | |
| Mitochondrial integrated intensity | 48 h | MII | |
| Positive mitochondrial staining | 48 h | posMT | |
| iPSC-cardiomyocytes | Total cells | 24 h | TC |
| Peak amplitude average | 90 min | amp | |
| Peak decay rise ratio | 90 min | decay.rise | |
| Peak spacing coefficient of variation | 90 min | spacing | |
| iPSC-endothelial cells | Branch points | 18 h | BrPo |
| Mean tube length | 18 h | MTL | |
| Nuclei mean intensity | 24 h | NI | |
| Positive mitochondrial staining | 24 h | posMT | |
| iPSC-neurons | Cells with significant growth | 72 h | CSG |
| Mean cell body area | 72 h | MCBA | |
| Total cells | 72 h | TC | |
| Total number of processes | 72 h | TotPr | |
| A375 | Alamar blue fluoresecnce | 24 h | AlmrB |
| Caspase-Glo® 3/7 luminescence | 24 h | CaspGlo | |
| MCF7 | Alamar Blue fluoresecnce | 24 h | AlmrB |
| ROS-Glo™ H2O2 assay | 24 h | Glo |
Transcriptional benchmark dose estimation
Gene expression analyses and data processing and filtering are described in detail elsewhere (House et al., 2022). All gene expression data (from human S1500+ TempOSeq assay, BioSpyder, San Diego, California) and experimental metadata are available from the public repository Gene Expression Omnibus (GSE186121). Normalized gene expression counts data analyzed herein did not include samples with <100K total counts and probes with <5% counts across all samples. The overall workflow for transcriptomic data analyses (3 dilutions and vehicle control) is shown in Figure 1. Specifically, normalized counts (counts + 1 to zero-protect the data for further analyses on the logarithmic scale) were processed using the BMDExpress (v.2.3) software (Phillips et al., 2019). First, data were pre-filtered with Williams trend test (p-value ≤.05 within each transcript and substance) and an absolute fold change ≥1.5 (compared with vehicle controls); probes that did not pass the criteria at any dose were removed from further analysis. Next, data were analyzed using Hill, power, linear, polynomial (2 and 3), and exponential (2, 3, 4, and 5) models. The models that were chosen as best fit varied slightly among the cell types, but the linear, exponential 2, and polynomial 2 models were selected over 90% of the time, indicating no concerns about overfitting (Supplementary Figure 1). A benchmark response of 1 SD was used. The best-fit model for each transcript/sample was selected based on the following settings: (1) maximum iterations of 250; (2) confidence level of 0.95; (3) constant variance; (4) Hill models with a k parameter <1/3 of the lowest positive dose were flagged and then the next best model with a p-value >.05 was used; and (5) a nested chi‐squared cutoff value of 0.05 to select the best polynomial models followed by minimum Akaike Information Criterion value and a goodness-of-fit p-value >.05. All benchmark dose (BMD) output data files from BMDExpress are available as Supplementary Files 1–12. The last data processing step using BMDExpress output detailed above was to cap the BMD values at the highest dose tested, and to remove transcripts that had BMD to BMD lower bound (BMDL) ratio >20. Vehicle control samples were assigned a “dose” one log10 unit below the lowest dose tested to allow plotting on the logarithmic scale, but for the analyses themselves, actual dose values were used.
Pathway analyses
Pathway analyses were performed using C2Reactome ontology gene sets and XGR R package (version 1.1.8) (Fang et al., 2016). For these analyses, the “background list” was composed of all transcripts that were retained after low-count removal for a given cell type as detailed in (House et al., 2022). Pathway enrichment was conducted with a false discovery rate (FDR) of 5% as a threshold for significantly enriched gene sets.
Derivation of transcriptomic data-based points of departure
Many statistical approaches for derivation of transcriptomic data-based points of departure (tPODs) are available (Farmahin et al., 2017) and this area of computational toxicology is rapidly evolving. It has been acknowledged that the specific methods used to derive the tPODs are of critical importance and need to be calibrated to be protective of toxicity while balancing sensitivity and specificity (Johnson et al., 2022). Overall, the most widely used approaches for tPOD derivation can be broadly summarized into 2 categories: tPOD derivation based on individual response genes, and tPOD derivation based on a subset of informative genes (eg, pathways or gene sets). Because the goal of this study was not to test all possible approaches, but to evaluate several most used, we selected one approach from each of these methods. For tPOD based on individual response genes, the lowest consistent response dose (LCRD) approach was used which identifies the most sensitive and consistent nonoutlier features (Crizer et al., 2021). The “lowest consistent response doses” (LCRD) were derived using a procedure detailed in (Crizer et al., 2021) by ranking the BMD values from the lowest to the highest, and identifying the lowest BMD values where all subsequent ratio values (calculated by rank n + 1/rank n) in down-rank BMDs are <1.66. For tPOD based on the choice of subsets of informative genes, the Most Sensitive Pathway Dose approach was used; it is one of the most commonly used approaches for tPOD derivation (National Toxicology Program, 2018; Thomas et al., 2013). Specifically, a median BMD value was derived from the individual BMDs for transcripts in each pathway; then, the lowest median value of all significant pathways was selected as the tPOD of the substance.
Our approaches to tPOD derivation were based on the inclusion criteria of BMD/BMDL < 20. Alternative suggestion was made that transcripts may be removed when a ratio of BMD/BMDL > 40 (National Toxicology Program, 2018). We explored whether such alternative consideration would be impactful. We found that the minimum tPOD selected using the current approach was highly concordant with that using a ratio of BMD/BMDL > 40 (Supplementary Figure 2); in a few instances where there were differences, our approach to tPOD selection resulted in a lower (ie, protective) tPOD.
Data analyses using pPOD, tPOD, and PACs (with 3–7 rings) content
The overall outline of various data analyses used in this study is shown in Figure 1. First, correlation analyses were performed using both Pearson’s and Spearman’s methods. Correlation significance was evaluated using adjusted p-values by each cell type using R v4.2 p.adjust function (method=“fdr”). Second, analyses of prediction of pPODs from transcriptomic data were conducted using tree learning algorithms of extreme gradient boosting (XGBoost) (Chen and Guestrin, 2016) using the R package xgboost (v 1.6.0.1). For this, BMD values for all transcripts and substances were used as predictors for a regression tree-based model training. The predicted values were obtained with leave-one-out cross-validation, and the performance of each prediction was evaluated by recording the Pearson correlation between the predicted results with the original pPOD values.
Third, we conducted data integration using the Toxicological Priority Index (ToxPi) approach (Marvel et al., 2018; Reif et al., 2013). The toxpiR R package (v 1.2.1) was used to integrate data from bioactivity, transcriptomics, and PAC for ranking petroleum substances within a category. The input data was inversely linearly scaled on a 0–1 scale, with 0 representing the lowest potency (highest POD or least PAC 3–7 ring content), and 1 representing the highest potency (lowest POD or most PAC 3–7 ring content).
Results
This study tested the hypothesis that a combination of human cell-based phenotypic and transcriptomic data can be integrated to make human health-protective decisions. Specifically, we evaluated this hypothesis in a case study of selecting worst-case petroleum UVCBs in a “manufacturing category” (ie, group of substances) for subsequent in vivo testing to fill in regulatory-required data gaps and enable read-across to other members of the category. The overall schematic of the study design and data analysis workflow is shown in Figure 1. The study included data on 141 petroleum UVCBs from 16 manufacturing categories/groups representing ∼75% of all petroleum substances that have been registered in the European Union (CONCAWE, 2020). We took advantage of the recently collected comprehensive “new approach methods” dataset comprising transcriptomic data and phenotypes in 6 human cell types (House et al., 2021, 2022). Although the previous studies focused on the use of such data to group petroleum substances, herein the in vitro, transcriptomic, and analytical data (ie, PAC content) were used to determine the most informative cell and data types, and to define a strategy to select worst-case substances within each category for further testing in animals.
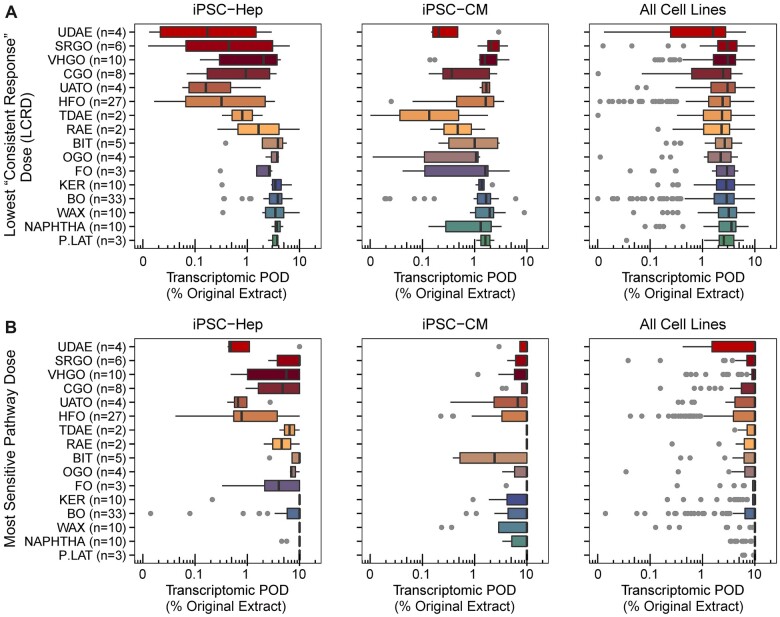
Our previous study (House et al., 2022) did not derive tPODs. A variety of approaches for extracting PODs from transcriptomic data have been proposed (Farmahin et al., 2017), yet no consensus exists as to what method is most appropriate for in vitro data. Here, we re-analyzed the in vitro transcriptomic data from (House et al., 2022) using two frequently used approaches (Figure 1). Figure 2 shows the representative tPOD values derived using each approach, data on the individual substances are grouped into manufacturing categories and the latter are sorted based on their overall bioactivity in vitro as detailed elsewhere (House et al., 2021). Overall, the tPOD values derived using LCRD (Figure 2A) appeared to be more protective than those generated from the Most Sensitive Pathway Dose approach (Figure 2B). Specifically, when comparing the tPOD values derived using the 2 approaches for each UVCB substance in each cell type, we found that 84%, 93%, 87%, 97%, 99%, and 93% of LCRD values were less than those derived using Most Sensitive Pathway Dose method in iPSC-Hep, iPSC-CM, MCF-7, iPSC-Endo, iPSC-Neu, and A375, respectively. Notably, median tPODs derived from iPSC-Hep using LCRD approach (for each category) showed a clear pattern that corresponds to the ranking of the categories with respect to their overall median in vitro bioactivity scores as detailed in House et al. (2021) (Spearman correlation coefficient = 0.79, p-value < .001). The categories with substances containing high amounts of PAC (ie, UDAE, SRGO, VHGO, and CGO) had lower LCRD-derived tPODs than those categories with substances low in PAC (ie, KER, BO, WAX, NAPHTHA, and P.LAT). The wide range of tPODs in the former was also concordant with greater within-category variation in both bioactivity and PAC content. These results suggest that tPODs derived using the LCRD approach are more protective (ie, lower) than those derived using the Most Sensitive Pathway Dose approach. The tPODs derived using the LCRD approach were also more concordant with both PAC content and bioactivity (Supplementary Figure 4 for correlation analysis between tPODs derived using the Most Sensitive Pathway Dose approach and PAC content, and for correlation analysis between tPODs derived using the Most Sensitive Pathway Dose approach and pPODs). Therefore, we used LCRD tPOD values in all subsequent analyses as quantitative transcriptomic data of choice.
Figure 2.
Box-and-whisker plots show manufacturing stream-based grouping of the transcriptomic points of departure (POD) for individual petroleum substances. Boxes represent the interquartile range, vertical line is the median value, and whiskers extend to the corresponding quartile plus 1.5× the interquartile range, or otherwise to the min–max range of values. See Supplementary Figure 3 for transcriptomic points of departure for all other cell types. A, Data derived using the Lowest Consistent Response Dose (LCRD) approach. B, Data derived using the Most Sensitive Pathway Dose approach. The data from iPSC-derived hepatocytes (Hep) are on the left, for iPSC-derived cardiomyocytes (CM) are in the middle, and for all 6 cell types combined are on the right.
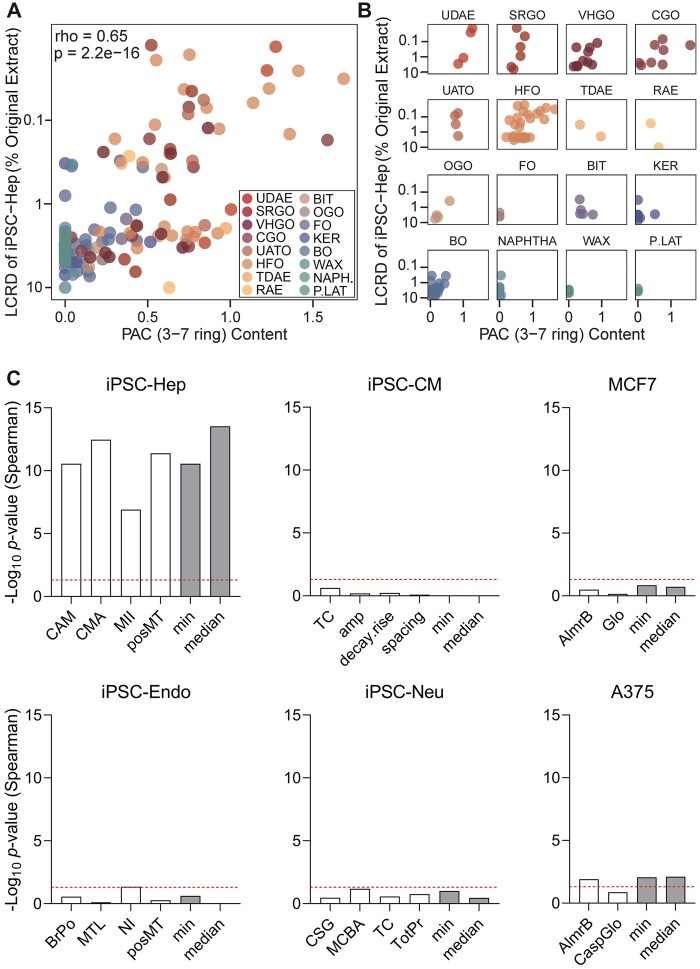
PAC content of each tested petroleum UVCB sample was previously found to be significantly correlated with the in vitro bioactivity data (House et al., 2021), as well as the number of differentially expressed and concentration-response genes in iPSC-Hep (House et al., 2022). Here, we compared the iPSC-Hep-derived tPOD values to PAC content (see Supplementary Table 4 in House et al. (2021)) and found them to be also highly correlated (Spearman correlation coefficient = 0.65, p-value = 2.2 × 10−16, Figure 3A). Specifically, the samples that contained higher levels of PAC elicited gene-level transcriptional effects at lower concentrations. Figure 3B shows these data sub-divided into each of 16 manufacturing categories; we found that positive correlations were discernable within those manufacturing categories that span a range of PAC content-containing samples.
Figure 3.
Correlations between transcriptomic point of departure (POD) values, polycyclic aromatic compound (PAC) content, and phenotypic PODs. A, A scatter plot of PAC score for 3–7 ring compounds (calculated by taking the weighed content of PAC [3–7 ring] and log10 transformed) in each UVCB sample and the transcriptomic POD values. Dots represent each substance and colors represent each manufacturing category (see inset). The rank-based correlation (rho) value and corresponding p-value are also shown. B, Same comparisons as in panel A but substances have been separated into each category. C, Bar plots show the -log10 transformed false discovery rate (FDR) adjusted p-values for rank-based (Spearman) correlation between transcriptomic and phenotypic POD values in each cell type. Individual phenotypes (white bars) and the minimum and median phenotypic (grey bars) PODs are shown. The horizontal red dotted lines show a significance threshold (adjusted p-value = .05). A color version of this figure appears in the online version of this article.
We also determined whether tPODs correlate with in vitro phenotypes-derived pPODs across tested cell types (Figure 3C). We conducted this analysis for each cell type and phenotype separately, as well as for the most conservative (min) and median pPOD values within each cell type regardless of the phenotype. We found that in iPSC-Hep, the correlations (Supplementary Table 4) were highly significant (after multiple testing correction) across all phenotypes. In other cell types, the correlations were lower (eg, A375 cell line), or not significant. These results indicate that when PODs are compared, only in the iPSC-Hep the phenotypic bioactivity was concordant with the transcriptomic data.
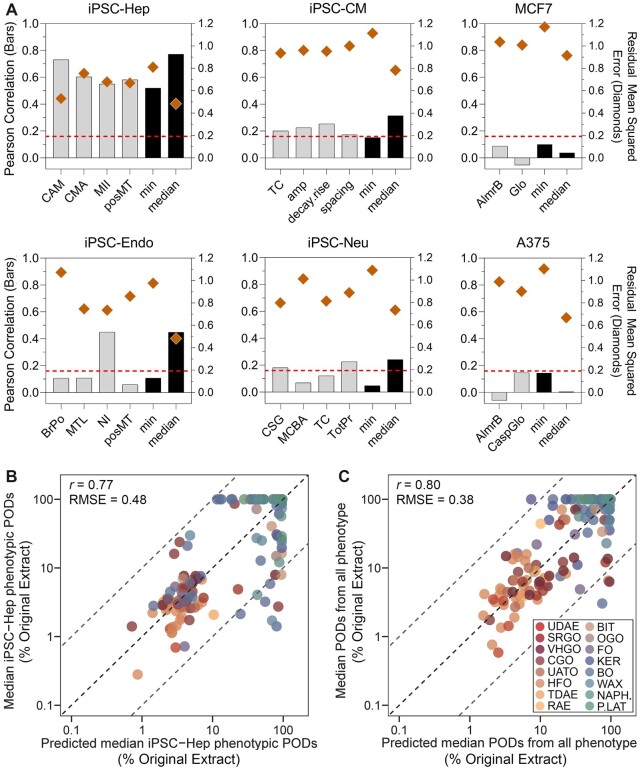
Another test of a relationship between transcriptomic and phenotypic data was to determine if transcriptomic data can be used to predict the bioactivity (ie, pPODs). For this, the transcript-level BMD values for all transcripts passing quality control criteria (see Materials and methods section) were used as input into a prediction model. We found that transcriptomic data from all iPSC-derived cells, but not MCF7 or A375 cell lines, could predict some or all pPODs (Figure 4A, bars). The strongest correlations were observed in iPSC-Hep where each phenotype could be predicted, a scatter plot example of the prediction for median pPODs in iPSC-Hep is shown in Figure 4B (r = 0.77). We also calculated the precision of such predictions (Figure 4A, diamonds) by calculating the residual mean square error (RMSE) of each prediction and found that they ranged from 0.48 to 1.17; the lowest RMSE overall were found in iPSC-Hep. In addition, when similar analysis was done across all data to predict the median pPOD across all cell types used, we found that transcriptomic data from iPSC-Hep were most informative (r = 0.80, RMSE = 0.38; Figure 4C).
Figure 4.
Cross-validated performance using transcriptomic benchmark dose values to predict phenotypic PODs. A, Bar plots show the cell-specific Pearson correlation coefficients (bars, left y-axis) and the residual mean square errors (diamonds, right y-axis). The horizontal red dotted lines represent a significance threshold for correlation values (adjusted p-value = .05). B, A scatter plot of benchmark dose values of all modeled probes in iPSC-Hep as the predictors versus the predicted PODs. C, Same as in panel B, but prediction made for the phenotypic data from all 6 cell types using transcriptomic data in iPSC-Hep. A color version of this figure appears in the online version of this article.
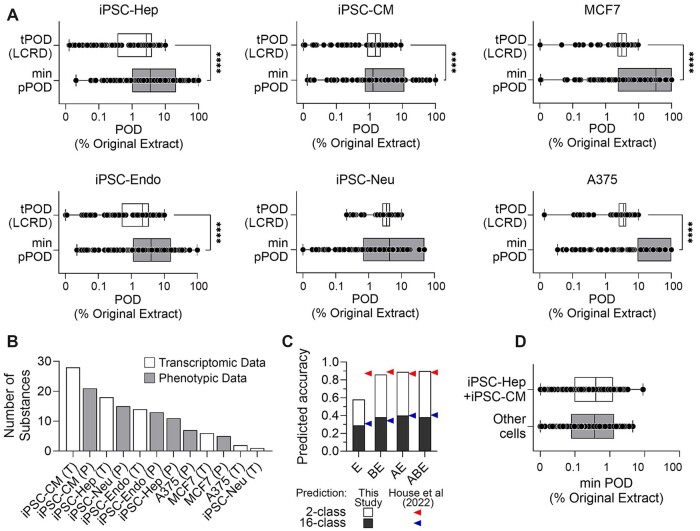
Because we observed that only some cell types provide informative data with respect to expected relationships between chemical composition of the samples and ensuing bioactivity, we tested whether a sub-set of the data (2 cell types) may be as informative as the overall dataset (all 6 cell types). This analysis was aimed at reducing the complexity and cost of future experiments with petroleum UVCBs whereby a more limited set of cells and assays may be used. To test this, we first compared tPODs to the lowest pPODs in each cell type (Figure 5A). We found that mean tPODs were almost always (except for iPSC-Neu) significantly (p < .001, t-test) lower than the means of the minimum pPODs. This result indicates that tPODs were more protective, on average, among all phenotypes collected. Second, we determined what cell type and assay (phenotypic or transcriptomic) was most sensitive in terms of the lowest POD for each of the tested substances. We found (Figure 5B) that tPOD or pPOD from iPSC-cardiomyocytes (iPSC-CM) and iPSC-Hep were most sensitive for the majority (55%) of tested substances, which demonstrates the ability of these 2 cell types to provide informative data across the wide range of petroleum UVCBs. Third, the ability of using tPODs and pPODs from only iPSC-CM and -Hep for grouping petroleum UVCBs into manufacturing categories was tested and compared to that when all in vitro, transcriptomic, and/or PAC content data were used (as reported previously in [House et al., 2022]). Figure 5C shows that tPOD data (column marked E) from iPSC-CM and -Hep was as informative as the whole transcriptomic data for 16-class (each manufacturing category separately) prediction, but less accurate for a 2-class (high- vs low-PAC compound classes) prediction. However, for multi-variable (tPODs combined with pPOD and/or PAC) predictions, the classification accuracy was indiscernible from that using all available data (columns marked BE, AE, and ABE). These results indicate that the predictions using data from iPSC-CM and -Hep are largely not less than those using a much larger dataset of 6 cell types. Fourth, we tested whether iPSC-CM and -Hep-derived minimum PODs (either transcriptomic or phenotypic) are equally sensitive to those derived from other cell types tested (Figure 5D). We found that the mean minimum PODs derived from iPSC-CM and -Hep were no greater than the mean minimum PODs from the rest of the cell types.
Figure 5.
Selection of most informative cell types and assays. A, Box and whiskers plots of cell type-specific transcriptomic POD values (boxes in white) and minimum phenotypic POD (boxes in grey). Boxes represent the interquartile range, vertical line is the median value, and whiskers extend to the min-max range of values. Individual substances are shown as black dots. The asterisks (****) denote a significant difference (p < .001) between conditions using a t-test. B, A bar plot showing the frequency of each cell type determining the lowest POD values for each tested substance. Transcriptomic (T, white bars) and phenotypic (P, grey bars) POD values are shown for each cell type. C, A stacked bar plot of the results for predicted accuracies in a supervised analysis in which the UVCB category was predicted from the transcriptomic PODs from iPSC-Hep and iPSC-CM only E, from both the phenotypic and transcriptomic PODs from iPSC-Hep and iPSC-CM (BE), from the pattern of PAC (3–7 rings) analytic data and transcriptomic PODs from iPSC-Hep and iPSC-CM (AE), and all data mentioned above (ABE). Each overall bar denotes the predicted accuracy of a binary prediction (see Results) with black section indicating the accuracy of predicting the exact manufacturing category (16-class prediction). Arrows (red is for a binary and blue for a 16-class prediction) denote previously reported accuracy of classification using all cell type data. D, A box and whiskers plot showing the sensitivity analysis for the means of lowest PODs of all samples obtaining from iPSC-Hep and iPSC-CM (white box) as compared to the data from all cell types (gray box). A color version of this figure appears in the online version of this article.
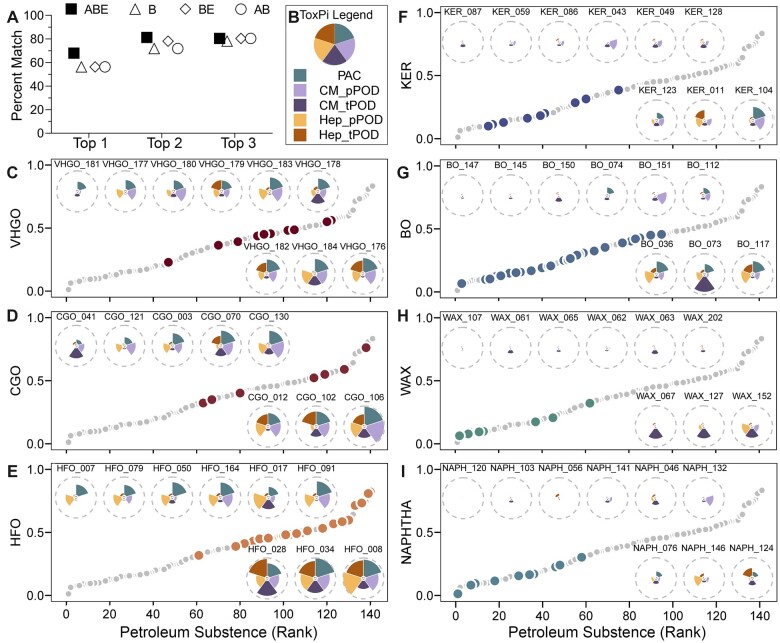
Overall, the results in Figure 5 demonstrated that both transcriptomic and phenotypic data from only 2 cell types, iPSC-CM and iPSC-Hep, when combined with the PAC content information, provide equivalently protective PODs and are sufficient for grouping. Therefore, we next determined if these data can be used as a tiered testing strategy for selecting worst-case petroleum UVCBs for toxicity studies in vivo. To accomplish this, we integrated tPOD and pPOD data with PAC content information using the ToxPi approach (Figure 6). A ToxPi score was calculated for each substance as the indicator of the overall hazard rank. To ensure that this approach was identifying the same substances of greatest potential concern regardless of whether the whole dataset from 6 cell types, or only the data from iPSC-CM and iPSC-Hep, was used, we compared the ranking between the 2 datasets (Figure 6A). We found that approximately 80% of the substances that were ranked in top 2 or 3 in their category based on ToxPi scores derived from the entire compendium of the data (all 6 cell types), remained in the top 2 or 3 when only iPSC-Hep and iPSC-CM data was used (Figure 6A). Notably, the best match was achieved when all 3 data streams (tPODs, pPODs, and PAC content) were used (Figure 6A, squares). Therefore, for subsequent data integration, we used ToxPi rankings based on this subset of data (Figure 6B). Figures 6C–I show the relative ranking of substances for 7 of 16 manufacturing categories in which there were at least 8 petroleum UVCBs. We reasoned that in a category containing a large number of substances (between 8 and 33), testing all of these substances in regulatory-required animal studies would not be commensurate with the 3R (Replacement, Reduction, and Refinement) principles and that a data-informed selection of the representative worst-case substance(s) can be made to reduce animal use. To demonstrate how data-informed selections could be made, ToxPi plots were created to summarize the content of PAC and t/pPOD information from iPSC-Hep and iPSC-CM. All 141 tested substances were ranked based on the overall ToxPi score and then substances within each category were identified by the colored dot on the overall ranking plot. We also display the actual ToxPi pie charts for the lowest, medium, and highest ranked substances in each category so it can be determined whether the rank is determined by the data from the same cell type and/or data type.
Figure 6.
Data integration and visualization using ToxPi to support selection of worst-case substance(s) in each category using PAC (3–7 ring) content and transcriptomic and phenotypic POD values from iPSC-Hep and iPSC-CM. A, A dot plot showing the percent match in ranking (top 1, 2, or 3 place) of the substances within their category using ToxPi scores using the data from all 6 cell types versus that from iPSC-Hep and iPSC-CM only. Hollow circles (AB): the pattern of PAC (3–7 rings) analytic data and transcriptomic PODs. Hollow diamonds (BE): the phenotypic and transcriptomic PODs. Hollow triangles (B): phenotypic PODs. Solid squares (ABE): all data mentioned above. B, ToxPi legend representing each included data type as a colored slice with equal weights. CM_pPOD: phenotypic POD data from iPSC-CM. CM_tPOD: transcriptomic POD data from iPSC-CM. Hep_pPOD: phenotypic POD data from iPSC-Hep. Hep_tPOD: transcriptomic POD data from iPSC-Hep. C–I, Scatter plots showing a rank order of all 141 petroleum UVCBs (gray dots) using their overall ToxPi score. Each panel represents the relative rank of the substances within each category (colored dots, see category labels on the y-axis and Table 1 for full description) for the categories with 8 or more substances. Insets show the actual ToxPi plots for selected individual substances in each category that are in the top ranked 3 (below the dot plot), middle 3, and bottom 3 (above the plot) substances.
Herein, we propose that an overall ToxPi rank based on the data from iPSC-Hep and iPSC-CM, together with a PAC score, can be used to select representative substance(s) for additional in vivo testing. For example, in the VHGO manufacturing category (Figure 6C), substances #184 and #176 are highest in rank among other VHGOs and are also among the highest ranked compounds of all tested. Their ToxPi profiles demonstrate that these can be deemed as representative of “worst cases” for this category, albeit there are some differences in their effects on transcriptional responses between iPSC-Hep and iPSC-CM. Another example is the HFO category where substances #028, #034, and #008 are all highly ranked but also show ToxPi profiles that are highly similar. Because other substances in this category (eg, other 6 substances shown in Figure 6E) show ToxPi profiles that are similar among them but somewhat different from the 3 top-ranked substances, some of them may be selected for additional in vivo testing as they would be representative of a sub-group based on their bioactivity. In other categories, similar choices can be made based on either overall ToxPi-based rank, or the ToxPi profiles to select representative worst-case substance(s).
Discussion
Previous studies (Grimm et al., 2016; House et al., 2021, 2022) used combined information of PAC content, cell-based phenotypic data, and/or in vitro transcriptional responses to determine whether grouping of petroleum UVCBs based on these new approach methods data are concordant with predefined categories established based on the manufacturing characteristics. These studies demonstrated that PAC content was (1) a strong determinant of the overall cell-based bioactivity, and (2) correlated with gene expression in iPSC-Hep (House et al., 2021, 2022). Although these previous results established that in vitro studies-derived gene expression and bioactivity are potentially useful for grouping of petroleum UVCBs, they also revealed considerable heterogeneity in bioactivity within each manufacturing category. Although it may be possible to use bioactivity data to group petroleum UVCBs, such an approach was questioned by regulators because “relationship between substances in in vitro test results has an unclear relationship to any in vivo toxicity assays on the test substances” (ECHA, 2020).
The existing categories of petroleum UVCBs are accepted by both regulators and the industry because they are based on the manufacturing process, physico-chemical characteristics, and product performance specifications (CONCAWE, 2017); however, the heterogeneity of substances within each category is also well acknowledged. Thus, there are disagreements as to how the data gaps in regulatory-required animal tests shall be filled. Although regulators demand either complete data package on every substance, or more substantive rationalization of proposed read-across within each category (ECHA, 2022), the registrants argue that “the biological activity profile of the substances [e.g., those from (House et al., 2021, 2022) studies] and other in vitro tests provides a basis for predicting the properties in relevant in vivo tests [ie, read-across]” (ECHA, 2020). Due to considerable gaps in the overall petroleum UVCB database of regulatory-required toxicological information, virtually no substance has data that would fully satisfy hazard characterization requirements under the European Union regulations; the regulators argue that some form of additional animal testing must be performed. Thus, the refinement of such additional required animal tests (ie, reducing the number of substances tested) is one of the primary drivers to strategies for selection of representative worst-case substances within each category for additional testing. Our study presents a series of arguments and case examples for how new approach methods data can be used to inform selection of worst-case substances within each category for such additional animal testing. By re-analyzing a very large dataset that contained data on various new approach methods (in vitro and transcriptomics data) specific to these substances, we reason that a tiered testing strategy based on select assays is a sensible path forward for choosing representative worst-case petroleum UVCBs for full-scale toxicity evaluation in vivo to meet regulatory requirements.
A recent opinion from a group of diverse stakeholders proposed a strategy for using tPODs in regulatory science (Johnson et al., 2022). They concluded that transcriptomic alterations and PODs derived from transcriptomic responses can serve as sensitive quantitative indicators of potential adverse human health outcomes. The authors found that although transcriptomic data are still fairly new with respect to its use in quantitative risk evaluations, there are a number of case studies that examined the ability of short-term animal exposure-derived tPODs to predict traditional sub-chronic and chronic apical endpoint-based PODs. The shift from using transcriptomics mainly in support of mechanistic considerations to quantitative risk assessment is an important advance toward the use of these data in decision-making. PODs are a critical information type in chemical safety assessments because they provide the basis for reference values and risk management. Because it is generally presumed that transcriptomic responses precede any apical effects, it follows that PODs derived from transcriptomic data constitute protective values, as supported by previous reports (Bhat et al., 2013; Bianchi et al., 2021; Gwinn et al., 2020; Johnson et al., 2020; Page-Lariviere et al., 2019; Thomas et al., 2011, 2012, 2013).
Still, there are very few examples of in vitro transcriptomic datasets that resulted in POD derivation, especially for complex substances and mixtures. Two recent studies demonstrated how in vitro high-throughput transcriptomics can be used for new approach methods-based hazard characterization of chemicals. In a study of 44 diverse chemicals that were tested in concentration-response in MCF7 cells, it was shown that in vitro tPODs were closely aligned with pPODs from other in vitro assays, and that gene expression signatures were associated with the known molecular targets of tested chemicals (Harrill et al., 2021). In a study of 24 chemicals in HepaRG cells, in vitro BMC modeling and pathway analysis were able to make qualitative and quantitative predictions between liver injury and non-liver injury compounds (Ramaiahgari et al., 2019). Based on these promising findings, the current study aimed to extend the evidence base to a greater number of cell types and a larger compendium of complex substances. Our proposed approach enables investigation of the potential value of transcriptomic data for data integration and decision-making with respect to “worst case” substances within a category, in a setting where detailed characterization of the chemical composition is difficult. In addition, we tested whether prioritization of assays can aid in reducing the complexity of new approach methods-based studies in the future. Overall, our work provides empirical evidence for the following broad considerations in how and where new approach methods-based data may be applied in regulatory decision-making of chemicals in general, but UVCBs and mixtures in particular.
First, although many studies of emerging new approach methods are evaluating effects of chemicals on a large number of cell types and endpoints, a strategy on how to use such data to focus on models fitting for a particular purpose is yet to emerge. For instance, many large-scale testing programs yield comprehensive datasets on hundreds to thousands of legacy chemicals (Richard et al., 2021; Williams et al., 2017), yet there are fewer examples of using these “big data” to narrow the choices of cell models and assays that can fit a certain regulatory purpose. Specific examples of such emerging efforts to “down-sample” are studies that demonstrated that a minimal set of in vitro assays can be selected to reliably determine estrogen agonist activity (Judson et al., 2017), or determined the minimal sample size needed for a regulatory context-specific precision and accuracy in estimating population variability using in vitro models (Blanchette et al., 2022; Chiu et al., 2017).
Previous studies of in vitro effects of petroleum UVCBs (House et al., 2021, 2022) were purposefully broad in terms of the number of cell-based models and phenotypes in an effort to extend the “biological space” and guard against underestimating potential hazard by neglecting a potentially “sensitive” cell type or phenotype. These studies already demonstrated that data derived from assays of biological activity in iPSC-derived models were highly informative, whereas data from cancer cell lines were less so (House et al., 2021). Accordingly, high-throughput transcriptomics data were obtained from a smaller set of cell types in a subsequent study, which also demonstrated that an even smaller set of cell types is most informative (House et al., 2022). Our study extends these observations by adding a quantitative argument with tPODs and demonstrating that iPSC-Hep and iPSC-CM are the most informative cell types by using multiple types of evidence, both in terms of their ability to yield the “most protective” t/p PODs, as well as to rank and prioritize substances. Although it was not surprising that hepatocyte-like cell type will be informative for PAC-containing substances because of their metabolic capacity to transform PACs to reactive intermediates, inclusion of cardiomyocytes was also found to be of great value for the same risk-based considerations. Our finding that tPODs from iPSC-CM were most protective for a majority of petroleum UVCBs is consistent with previous observations that this cell type can be used to distinguish between UVCBs with low to no PAC content (House et al., 2021). Although these studies demonstrated that iPSC-CM are a useful cell type for evaluation of hydrocarbon-containing substances, our previous finding that iPSC-CM yielded more protective phenotypic PODs across various chemical classes (Chen et al., 2020) argues for its utility beyond petroleum UVCBs. Combined, these results suggest that, in addition to in vitro pPODs, high-throughput transcriptomic-derived tPOD from iPSC-CM also provide unique value for quantitative risk evaluation using cell-based data. However, because iPSC-CM phenotypes were largely based on the ion channel activity, the lack of correlation of pPOD and tPOD in this cell type, and in other cells where cytotoxicity was the only phenotype queried, indicates that gene expression provides a far wider coverage of the potential biological effects and thus adds highly informative and complementary data. Still, an important limitation of our study and future consideration is the choice of tPOD derivation methods (Farmahin et al., 2017). Although we tested several commonly used approaches for tPOD derivation, inclusion of other methods and gene sets (Mubeen et al., 2022) could lead to lower (ie, more protective) tPODs, but also increase false discoveries. To ensure confidence that transcriptomics can be employed to establish a POD from both short-term in vivo and in vitro studies at a dose level below which a biological perturbation is not expected would require further methodological development and discussion to build an accepted weight of evidence around reproducible methods and appropriate study design (Johnson et al., 2022).
Second, we show that even though tPODs were, in general, more protective than pPODs for petroleum UVCBs tested herein, both types of data are informative for overall rank prioritization. The concordance between transcriptomic and phenotypic PODs has been evaluated recently in MCF7 cells (Harrill et al., 2021). It was found that although there was an overall agreement in the PODs of different type, depending on the compound, either one of those POD types may have been most protective. We found that concordance between tPOD and pPOD was highly dependent on the cell type. For example, in iPSC-Hep, correlations were highly significant, whereas in other cell types, this was not the case. On one hand, such a result may be interpreted as indicative of greater overall interpretability of the data from iPSC-Hep and enhance confidence in using these quantitative estimates of hazard for decision-making. For example, these cells exhibit metabolic competency comparable to that of many lots of primary human hepatocytes (Sirenko et al., 2014; Valdiviezo et al., 2022a,b), but questions remain about their fetal-like characteristics in terms of the metabolic enzyme expression (Yamaguchi et al., 2019). On the other hand, it is current practice to use any indication of the biological activity in vitro, across a range of assays and cell types, to provide a protective POD for risk assessment (Daston et al., 2022; Paul Friedman et al., 2020). Therefore, we conclude that once a targeted set of cell types is selected, in the case of our study of petroleum UVCBs this would be iPSC-Hep and iPSC-CM, both phenotypic and transcriptomic data shall be collected and included in determining worst-case substances for further evaluation in vivo. For other studies, the same or different cell types may be most informative depending on the expected toxicological responses of a particular chemical or a class of chemicals. For example, our previous work showed that a compendium of 5 cell types was sufficient to cover potential adverse effects of chemicals from different classes, defined mixtures, and environmental samples of unknown composition (Chen et al., 2020, 2021a,b; Hsieh et al., 2021).
Third, it is also increasingly clear that even though the aspiration to transition risk assessment of chemicals to a mechanistically based human-focused evaluation (U.S. EPA, 2021) may be informed by a desire to step away from the animal study as the gold standard (Piersma et al., 2018), most regulatory regimes still require testing in animals for hazard evaluations. Therefore, a full replacement of animal tests, or reliance only on in vitro data, may be an ambitious goal, but not a practical solution in the near future. Indeed, recent experience of submitting multi-dimensional in vitro data in support of grouping and read-across of petroleum UVCBs to reduce animal testing requirements was not successful (ECHA, 2020). Specifically, the European Chemicals Agency raised several concerns about the utility of using such data to waive animal testing requirements. Given the reality of unlikely elimination of animal tests for at least some endpoints in the near term, refinement appears to be a more sensible option where new approach methods data may contribute value. This study proposed a tiered integration of various data to arrive at selecting representative substance(s) for more scientifically justifiable and limited animal testing. The existing categories for petroleum UVCBs are already highly heterogeneous in terms of chemical composition of substances (eg, PAC content [House et al., 2021]), and there is paucity of data to determine representative compounds based solely on their chemical composition due to both complexity and variability (Roman-Hubers et al., 2023). Thus, additional data streams, eg, phenotypic and transcriptomic PODs, offer protective quantitative estimates across most informative cell types that, when combined with PAC content, yield actionable information that includes both overall hazard rank and can be also interpreted in terms of the similarity among compounds in a category for the ultimate purpose of selecting representative “worst case” substances for additional testing.
This tiered approach can be used not only for presumably hazardous substances, but also for those substances that are deemed to be without appreciable hazard. For example, substances in the base oils category are all classified as carcinogenic in the European Union (European Commission, 2008), despite the wide range of hazard profiles within the category. Additionally, KEROSINE category substances are classified as carcinogenic, whereas WAX category substances are not classified as hazardous, even though they all contain low levels of PAC substances. Furthermore, not all of the substances in these categories have been tested sufficiently to determine what substance(s) within a category may best serve as source for read-across. In this regard, the data integration approach proposed in our study may aid in determining what substances are more likely to pose human health hazard, regardless of how the entire category is classified. For example, using the ToxPi data integration strategy one can focus on a limited number of substances in base oils or WAX categories to conduct comprehensive animal testing and use the similarity in ToxPi profiles for data-informed selection of source-target choices for read-across.
Overall, this work provides an informative case study commensurate with a call (Price et al., 2022; Thomas et al., 2019) for improving the resource efficiency in chemical toxicity testing, both in vitro and in animals. We conclude that both phenotypic and transcriptomic data provide unique value and should be included as part of a tiered testing strategy to complete hazard evaluations of petroleum UVCBs. Among the cell types, iPSC-Hep and iPSC-CM, when coupled with information on PAC content, are the most informative for this purpose. Our specific strategy for selecting representative (or “worst case”) petroleum UVCBs in each manufacturing category for additional animal testing to fill-in data gaps includes: (1) narrowing the scope of any additional in vitro testing to a manageable set of cell models and assays that provide protective and informative PODs, (2) integrating both PAC, phenotypic, and transcriptomic data to visualize/evaluate the trends/patterns among substances in a category, (3) selecting a smaller set of representative substances for animal studies based on this integrative analysis, and (4) using the animal tests data from selected “source” substances to read-across to the remaining “target” data-poor substances in a category to complete regulatory evaluation and move to risk management.
Supplementary Material
Acknowledgments
The authors wish to thank Drs. Scott Auerbach (NIEHS), Alison Dickey (North Carolina State University), and George Daston (Procter&Gamble) for informative discussions about this study and assistance in using BMDExpress and other gene expression analysis software.
Contributor Information
Han-Hsuan Doris Tsai, Interdisciplinary Faculty of Toxicology, College Station, Texas 77843, USA; Department of Veterinary Physiology and Pharmacology, Texas A&M University, College Station, Texas 77843, USA.
John S House, National Institute of Environmental Health Sciences, Research Triangle Park, North Carolina 27709, USA.
Fred A Wright, Interdisciplinary Faculty of Toxicology, College Station, Texas 77843, USA; Department of Statistics and Bioinformatics Research Center, North Carolina State University, Raleigh, North Carolina 27603, USA; Department of Biological Sciences and Bioinformatics Research Center, North Carolina State University, Raleigh, North Carolina 27603, USA.
Weihsueh A Chiu, Interdisciplinary Faculty of Toxicology, College Station, Texas 77843, USA; Department of Veterinary Physiology and Pharmacology, Texas A&M University, College Station, Texas 77843, USA.
Ivan Rusyn, Interdisciplinary Faculty of Toxicology, College Station, Texas 77843, USA; Department of Veterinary Physiology and Pharmacology, Texas A&M University, College Station, Texas 77843, USA.
Supplementary data
Supplementary data are available at Toxicological Sciences online.
Declaration of conflicting interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
This work was funded by a contract with Concawe (Brussels, Belgium). The authors were also partially supported by the National Institute of Environmental Health Sciences (P42 ES027704). In addition, this research was supported in part by the Intramural Research Program of the National Institute of Environmental Health Sciences.
References
- Beal M. A., Gagne M., Kulkarni S. A., Patlewicz G., Thomas R. S., Barton-Maclaren T. S. (2022). Implementing in vitro bioactivity data to modernize priority setting of chemical inventories. Altex 39, 123–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat V. S., Hester S. D., Nesnow S., Eastmond D. A. (2013). Concordance of transcriptional and apical benchmark dose levels for conazole-induced liver effects in mice. Toxicol. Sci. 136, 205–215. [DOI] [PubMed] [Google Scholar]
- Bianchi E., Costa E., Yan Z. J., Murphy L., Howell J., Anderson D., Mukerji P., Venkatraman A., Terry C., Johnson K. J. (2021). A rat subchronic study transcriptional point of departure estimates a carcinogenicity study apical point of departure. Food Chem. Toxicol. 147, 111869. [DOI] [PubMed] [Google Scholar]
- Blanchette A. D., Burnett S. D., Rusyn I., Chiu W. A. (2022). A tiered approach to population-based in vitro testing for cardiotoxicity: Balancing estimates of potency and variability. J. Pharmacol. Toxicol. Methods. 114, 107154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buesen R., Chorley B. N., da Silva Lima B., Daston G., Deferme L., Ebbels T., Gant T. W., Goetz A., Greally J., Gribaldo L., et al. (2017). Applying 'omics technologies in chemicals risk assessment: Report of an ECETOC workshop. Regul. Toxicol. Pharmacol. 91Suppl 1, S3–S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Zhang M., Borlak J., Tong W. (2012). A decade of toxicogenomic research and its contribution to toxicological science. Toxicol. Sci. 130, 217–228. [DOI] [PubMed] [Google Scholar]
- Chen T., Guestrin C. (2016). XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, California, USA, pp. 785–794. 10.1145/2939672.2939785 [DOI]
- Chen Z., Jang S., Kaihatu J. M., Zhou Y. H., Wright F. A., Chiu W. A., Rusyn I. (2021a). Potential human health hazard of post-hurricane Harvey sediments in Galveston Bay and Houston Ship Channel: A case study of using in vitro bioactivity data to inform risk management decisions. Int J Environ Res Public Health 18, 13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Liu Y., Wright F. A., Chiu W. A., Rusyn I. (2020). Rapid hazard characterization of environmental chemicals using a compendium of human cell lines from different organs. Altex 37, 623–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Lloyd D., Zhou Y. H., Chiu W. A., Wright F. A., Rusyn I. (2021b). Risk characterization of environmental samples using in vitro bioactivity and polycyclic aromatic hydrocarbon concentrations data. Toxicol. Sci. 179, 108–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W. A., Wright F. A., Rusyn I. (2017). A tiered, Bayesian approach to estimating of population variability for regulatory decision-making. Altex 34, 377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONCAWE (2017). Hazard classification and labelling of petroleum substances in the European Economic Area—2017. Available at: https://www.concawe.eu/wp-content/uploads/2017/11/Rpt_17-13.pdf.
- CONCAWE (2020). Hazard Classification and Labelling of Petroleum substances in the European Economic Area—2020. Available at: https://www.concawe.eu/hazard-classification-and-labelling-of-petroleum-substances-in-the-european-economic-area-2020-2/.
- Crizer D. M., Ramaiahgari S. C., Ferguson S. S., Rice J. R., Dunlap P. E., Sipes N. S., Auerbach S. S., Merrick B. A., DeVito M. J. (2021). Benchmark concentrations for untargeted metabolomics versus transcriptomics for liver injury compounds in in vitro liver models. Toxicol. Sci. 181, 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Paules R. S. (2010). Use of transcriptomics in understanding mechanisms of drug-induced toxicity. Pharmacogenomics 11, 573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daston G. P., Mahony C., Thomas R. S., Vinken M. (2022). Assessing safety without animal testing: The road ahead. Toxicol. Sci. 187, 214–218. [DOI] [PubMed] [Google Scholar]
- De Abrew K. N., Kainkaryam R. M., Shan Y. K., Overmann G. J., Settivari R. S., Wang X., Xu J., Adams R. L., Tiesman J. P., Carney E. W., et al. (2016). Grouping 34 chemicals based on mode of action using connectivity mapping. Toxicol. Sci. 151, 447–461. [DOI] [PubMed] [Google Scholar]
- De Abrew K. N., Shan Y. K., Wang X., Krailler J. M., Kainkaryam R. M., Lester C. C., Settivari R. S., LeBaron M. J., Naciff J. M., Daston G. P. (2019). Use of connectivity mapping to support read across: a deeper dive using data from 186 chemicals, 19 cell lines and 2 case studies. Toxicology 423, 84–94. [DOI] [PubMed] [Google Scholar]
- ECHA (2022). Advice on Using Read-Across for UVCB Substances – Obligations Arising from Commission Regulation 2021/979, Amending REACH Annexes. European Chemicals Agency, Helsinki, Finland.
- ECHA (2020). Testing Proposal Decision on Substance EC 295-332-8 “Extracts (petroleum), deasphalted vacuum residue solvent”. European Chemicals Agency, Helsinki, Finland.
- European Commission (2008). Regulation (EC) No 1272/2008 of the European Parliament and of the Council of 16 December 2008 on classification, labelling and packaging of substances and mixtures, amending and repealing Directives 67/548/EEC and 1999/45/EC, and amencing Regulation (EC) nO 1907/2006. European Commission, Official Journal of the European Union.
- Fang H., Knezevic B., Burnham K. L., Knight J. C. (2016). XGR software for enhanced interpretation of genomic summary data, illustrated by application to immunological traits. Genome Med. 8, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmahin R., Williams A., Kuo B., Chepelev N. L., Thomas R. S., Barton-Maclaren T. S., Curran I. H., Nong A., Wade M. G., Yauk C. L. (2017). Recommended approaches in the application of toxicogenomics to derive points of departure for chemical risk assessment. Arch. Toxicol. 91, 2045–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geter D. R., Bhat V. S., Gollapudi B. B., Sura R., Hester S. D. (2014). Dose-response modeling of early molecular and cellular key events in the CAR-mediated hepatocarcinogenesis pathway. Toxicol. Sci. 138, 425–445. [DOI] [PubMed] [Google Scholar]
- Grimm F. A., Iwata Y., Sirenko O., Chappell G. A., Wright F. A., Reif D. M., Braisted J., Gerhold D. L., Yeakley J. M., Shepard P., et al. (2016). A chemical-biological similarity-based grouping of complex substances as a prototype approach for evaluating chemical alternatives. Green Chem. 18, 4407–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn W. M., Auerbach S. S., Parham F., Stout M. D., Waidyanatha S., Mutlu E., Collins B., Paules R. S., Merrick B. A., Ferguson S., et al. (2020). Evaluation of 5-day in vivo rat liver and kidney with high-throughput transcriptomics for estimating benchmark doses of apical outcomes. Toxicol. Sci. 176, 343–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill J., Shah I., Setzer R. W., Haggard D., Auerbach S., Judson R., Thomas R. S. (2019). Considerations for strategic use of high-throughput transcriptomics chemical screening data in regulatory decisions. Curr. Opin. Toxicol. 15, 64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill J. A., Everett L. J., Haggard D. E., Sheffield T., Bundy J. L., Willis C. M., Thomas R. S., Shah I., Judson R. S. (2021). High-throughput transcriptomics platform for screening environmental chemicals. Toxicol. Sci. 181, 68–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House J. S., Grimm F. A., Jima D. D., Zhou Y. H., Rusyn I., Wright F. A. (2017). A pipeline for high-throughput concentration response modeling of gene expression for toxicogenomics. Front. Genet. 8, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House J. S., Grimm F. A., Klaren W. D., Dalzell A., Kuchi S., Zhang S. D., Lenz K., Boogaard P. J., Ketelslegers H. B., Gant T. W., et al. (2022). Grouping of UVCB substances with dose-response transcriptomics data from human cell-based assays. Altex 39, 388–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House J. S., Grimm F. A., Klaren W. D., Dalzell A., Kuchi S., Zhang S. D., Lenz K., Boogaard P. J., Ketelslegers H. B., Gant T. W., et al. (2021). Grouping of UVCB substances with new approach methodologies (NAMs) data. Altex 38, 123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh N. H., Chen Z., Rusyn I., Chiu W. A. (2021). Risk characterization and probabilistic concentration-response modeling of complex environmental mixtures using new approach methodologies (NAMs) data from organotypic in vitro human stem cell assays. Environ. Health Perspect. 129, 17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. J., Auerbach S. S., Costa E. (2020). A rat liver transcriptomic point of departure predicts a prospective liver or non-liver apical point of departure. Toxicol. Sci. 176, 86–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. J., Auerbach S. S., Stevens T., Barton-Maclaren T. S., Costa E., Currie R. A., Dalmas Wilk D., Haq S., Rager J. E., Reardon A. J. F., et al. (2022). A transformative vision for an omics-based regulatory chemical testing paradigm. Toxicol. Sci. 190, 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R. S., Houck K. A., Watt E. D., Thomas R. S. (2017). On selecting a minimal set of in vitro assays to reliably determine estrogen agonist activity. Regul. Toxicol. Pharmacol. 91, 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavlock R. J., Bahadori T., Barton-Maclaren T. S., Gwinn M. R., Rasenberg M., Thomas R. S. (2018). Accelerating the pace of chemical risk assessment. Chem. Res. Toxicol. 31, 287–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinaret P. A. S., Serra A., Federico A., Kohonen P., Nymark P., Liampa I., Ha M. K., Choi J. S., Jagiello K., Sanabria N., et al. (2020). Transcriptomics in toxicogenomics, Part I: Experimental design, technologies, publicly available data, and regulatory aspects. Nanomaterials (Basel) 10, 750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J., Crawford E. D., Peck D., Modell J. W., Blat I. C., Wrobel M. J., Lerner J., Brunet J. P., Subramanian A., Ross K. N., et al. (2006). The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935. [DOI] [PubMed] [Google Scholar]
- Lobenhofer E. K., Cui X., Bennett L., Cable P. L., Merrick B. A., Churchill G. A., Afshari C. A. (2004). Exploration of low-dose estrogen effects: identification of No Observed Transcriptional Effect Level (NOTEL). Toxicol. Pathol. 32, 482–492. [DOI] [PubMed] [Google Scholar]
- Low Y., , UeharaT., , MinowaY., , YamadaH., , OhnoY., , UrushidaniT., , SedykhA., , MuratovE., , Kuz'minV., , Fourches D.,. et al. (2011). Predicting drug-induced hepatotoxicity using QSAR and toxicogenomics approaches. Chem. Res. Toxicol. 24, 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvel S. W., To K., Grimm F. A., Wright F. A., Rusyn I., Reif D. M. (2018). ToxPi Graphical User Interface 2.0: dynamic exploration, visualization, and sharing of integrated data models. BMC Bioinf 19, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mubeen S., Tom Kodamullil A., Hofmann-Apitius M., Domingo-Fernandez D. (2022). On the influence of several factors on pathway enrichment analysis. Brief Bioinform 23, bbac143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Academies of Sciences Engineering and Medicine (2017). Using 21st Century Science to Improve Risk-Related Evaluations. The National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- National Research Council (2007a). Applications of Toxicogenomic Technologies to Predictive Toxicology and Risk Assessment. The National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- National Research Council (2007b). Toxicity Testing in the 21st Century: A Vision and a Strategy. The National Academies Press, Washington, DC. [Google Scholar]
- National Toxicology Program (2018). NTP Research Report on National Toxicology Program Approach to Genomic Dose-Response Modeling: Research Report 5. [PubMed]
- Nuwaysir E. F., Bittner M., Trent J., Barrett J. C., Afshari C. A. (1999). Microarrays and toxicology: the advent of toxicogenomics. Mol. Carcinog. 24, 153–159. [DOI] [PubMed] [Google Scholar]
- Nyffeler J., Willis C., Harris F. R., Taylor L. W., Judson R., Everett L. J., Harrill J. A. (2022). Combining phenotypic profiling and targeted RNA-Seq reveals linkages between transcriptional perturbations and chemical effects on cell morphology: Retinoic acid as an example. Toxicol. Appl. Pharmacol. 444, 116032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page-Lariviere F., Crump D., O'Brien J. M. (2019). Transcriptomic points-of-departure from short-term exposure studies are protective of chronic effects for fish exposed to estrogenic chemicals. Toxicol. Appl. Pharmacol. 378, 114634. [DOI] [PubMed] [Google Scholar]
- Paul Friedman K., Gagne M., Loo L. H., Karamertzanis P., Netzeva T., Sobanski T., Franzosa J. A., Richard A. M., Lougee R. R., Gissi A., et al. (2020). Utility of in vitro bioactivity as a lower bound estimate of in vivo adverse effect levels and in risk-based prioritization. Toxicol. Sci. 173, 202–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J. R., Svoboda D. L., Tandon A., Patel S., Sedykh A., Mav D., Kuo B., Yauk C. L., Yang L., Thomas R. S., et al. (2019). BMDExpress 2: enhanced transcriptomic dose-response analysis workflow. Bioinformatics 35, 1780–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piersma A. H., Burgdorf T., Louekari K., Desprez B., Taalman R., Landsiedel R., Barroso J., Rogiers V., Eskes C., Oelgeschlager M., et al. (2018). Workshop on acceleration of the validation and regulatory acceptance of alternative methods and implementation of testing strategies. Toxicol. In Vitro 50, 62–74. [DOI] [PubMed] [Google Scholar]
- Price P. S., Hubbell B. J., Hagiwara S., Paoli G. M., Krewski D., Guiseppi-Elie A., Gwinn M. R., Adkins N. L., Thomas R. S. (2022). A framework that considers the impacts of time, cost, and uncertainty in the determination of the cost effectiveness of toxicity-testing methodologies. Risk Anal. 42, 707–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaiahgari S. C., Auerbach S. S., Saddler T. O., Rice J. R., Dunlap P. E., Sipes N. S., DeVito M. J., Shah R. R., Bushel P. R., Merrick B. A., et al. (2019). The power of resolution: Contextualized understanding of biological responses to liver injury chemicals using high-throughput transcriptomics and benchmark concentration modeling. Toxicol. Sci. 169, 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon A. J. F., Rowan-Carroll A., Ferguson S. S., Leingartner K., Gagne R., Kuo B., Williams A., Lorusso L., Bourdon-Lacombe J. A., Carrier R., et al. (2021). Potency ranking of per- and polyfluoroalkyl substances using high-throughput transcriptomic analysis of human liver spheroids. Toxicol. Sci. 184, 154–169. [DOI] [PubMed] [Google Scholar]
- Reif D. M., Sypa M., Lock E. F., Wright F. A., Wilson A., Cathey T., Judson R. R., Rusyn I. (2013). ToxPi GUI: an interactive visualization tool for transparent integration of data from diverse sources of evidence. Bioinformatics 29, 402–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard A. M., Huang R., Waidyanatha S., Shinn P., Collins B. J., Thillainadarajah I., Grulke C. M., Williams A. J., Lougee R. R., Judson R. S., et al. (2021). The Tox21 10K Compound Library: collaborative Chemistry Advancing Toxicology. Chem. Res. Toxicol. 34, 189–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard A. M., Judson R. S., Houck K. A., Grulke C. M., Volarath P., Thillainadarajah I., Yang C., Rathman J., Martin M. T., Wambaugh J. F., et al. (2016). ToxCast Chemical Landscape: paving the Road to 21st Century Toxicology. Chem. Res. Toxicol. 29, 1225–1251. [DOI] [PubMed] [Google Scholar]
- Roman-Hubers A. T., Cordova A. C., Barrow M. P., Rusyn I. (2023). Analytical chemistry solutions to hazard evaluation of petroleum refining products. Regul. Toxicol. Pharmacol. 137, 105310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquie D., Friry-Santini C., Schorsch F., Tinwell H., Bars R. (2009). Standard and molecular NOAELs for rat testicular toxicity induced by flutamide. Toxicol. Sci. 109, 59–65. [DOI] [PubMed] [Google Scholar]
- Rowan-Carroll A., Reardon A., Leingartner K., Gagne R., Williams A., Meier M. J., Kuo B., Bourdon-Lacombe J., Moffat I., Carrier R., et al. (2021). High-throughput transcriptomic analysis of human primary hepatocyte spheroids exposed to per- and polyfluoroalkyl substances as a platform for relative potency characterization. Toxicol. Sci. 181, 199–214. [DOI] [PubMed] [Google Scholar]
- Sirenko O., Grimm F. A., Ryan K. R., Iwata Y., Chiu W. A., Parham F., Wignall J. A., Anson B., Cromwell E. F., Behl M., et al. (2017). In vitro cardiotoxicity assessment of environmental chemicals using an organotypic human induced pluripotent stem cell-derived model. Toxicol. Appl. Pharmacol. 322, 60–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirenko O., Hesley J., Rusyn I., Cromwell E. F. (2014). High-content assays for hepatotoxicity using induced pluripotent stem cell-derived cells. Assay Drug Dev. Technol. 12, 43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R. S., Bahadori T., Buckley T. J., Cowden J., Deisenroth C., Dionisio K. L., Frithsen J. B., Grulke C. M., Gwinn M. R., Harrill J. A., et al. (2019). The next generation blueprint of computational toxicology at the U.S. environmental protection agency. Toxicol. Sci. 169, 317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R. S., Clewell H. J. 3rd, Allen B. C., Wesselkamper S. C., Wang N. C., Lambert J. C., Hess-Wilson J. K., Zhao Q. J., Andersen M. E. (2011). Application of transcriptional benchmark dose values in quantitative cancer and noncancer risk assessment. Toxicol. Sci. 120, 194–205. [DOI] [PubMed] [Google Scholar]
- Thomas R. S., Clewell H. J. 3rd, Allen B. C., Yang L., Healy E., Andersen M. E. (2012). Integrating pathway-based transcriptomic data into quantitative chemical risk assessment: a five chemical case study. Mutat. Res. 746, 135–143. [DOI] [PubMed] [Google Scholar]
- Thomas R. S., Wesselkamper S. C., Wang N. C., Zhao Q. J., Petersen D. D., Lambert J. C., Cote I., Yang L., Healy E., Black M. B., et al. (2013). Temporal concordance between apical and transcriptional points of departure for chemical risk assessment. Toxicol. Sci. 134, 180–194. [DOI] [PubMed] [Google Scholar]
- U.S. EPA (2012). Benchmark Dose Technical Guidance. Available at: https://www.epa.gov/risk/benchmark-dose-technical-guidance.
- U.S. EPA (2021). New approach methods work plan: Reducing use of vertebrate animals in chemical testing. US Environmental Protection Agency, Washington, DC. [Google Scholar]
- Valdiviezo A., Brown G. E., Michell A. R., Trinconi C. M., Bodke V. V., Khetani S. R., Luo Y. S., Chiu W. A., Rusyn I. (2022a). Reanalysis of trichloroethylene and tetrachloroethylene metabolism to glutathione conjugates using human, rat, and mouse liver in vitro models to improve precision in risk characterization. Environ. Health Perspect. 130, 117009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdiviezo A., Kato Y., Baker E. S., Chiu W. A., Rusyn I. (2022b). Evaluation of metabolism of a defined pesticide mixture through multiple in vitro liver models. Toxics 10, 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A. J., Grulke C. M., Edwards J., McEachran A. D., Mansouri K., Baker N. C., Patlewicz G., Shah I., Wambaugh J. F., Judson R. S., et al. (2017). The CompTox chemistry dashboard: A community data resource for environmental chemistry. J. Cheminform. 9, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Matsuzaki J., Katsuda T., Saito Y., Saito H., Ochiya T. (2019). Generation of functional human hepatocytes in vitro: current status and future prospects. Inflamm. Regen. 39, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauk C. L., Harrill A. H., Ellinger-Ziegelbauer H., van der Laan J. W., Moggs J., Froetschl R., Sistare F., Pettit S. (2020). A cross-sector call to improve carcinogenicity risk assessment through use of genomic methodologies. Regul. Toxicol. Pharmacol. 110, 104526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeakley J. M., Shepard P. J., Goyena D. E., VanSteenhouse H. C., McComb J. D., Seligmann B. E. (2017). A trichostatin A expression signature identified by TempO-Seq targeted whole transcriptome profiling. PLoS One. 12, e0178302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarbl H., Gallo M. A., Glick J., Yeung K. Y., Vouros P. (2010). The vanishing zero revisited: thresholds in the age of genomics. Chem. Biol. Interact. 184, 273–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.