Abstract
Germline GATA2 mutations predispose to myeloid malignancies resulting from the progressive acquisition of additional somatic mutations. Here we describe clinical and biological features of 78 GATA2-deficient patients. Hematopoietic stem and progenitor cell phenotypic characterization revealed an exhaustion of myeloid progenitors. Somatic mutations in STAG2, ASXL1 and SETBP1 genes along with cytogenetic abnormalities (monosomy 7, trisomy 8, der(1;7)) occurred frequently in patients with GATA2 germline mutations. Patients were classified into three hematopoietic spectra based on bone marrow cytomorphology. No somatic additional mutations were detected in patients with normal bone marrow (spectrum 0), whereas clonal hematopoiesis mediated by STAG2 mutations was frequent in those with a hypocellular and/or myelodys-plastic bone marrow without excess blasts (spectrum 1). Finally, SETBP1, RAS pathway and RUNX1 mutations were predominantly associated with leukemic transformation stage (spectrum 2), highlighting their implications in the transformation process. Specific somatic alterations, potentially providing distinct selective advantages to affected cells, are therefore associated with the clinical/hematological evolution of GATA2 syndrome. Our study not only suggests that somatic genetic profiling will help clinicians in their management of patients, but will also clarify the mechanism of leukemogenesis in the context of germline GATA2 mutations.
Introduction
During the last 15 years, with the development of next-generation sequencing (NGS), familial predisposition has emerged as an important issue in hematology, with the identification of recurrent mutated genes leading to myelodysplastic syndromes (MDS) or acute myeloid leukemia (AML). These germline mutations frequently encode master regulatory transcription factors such as RUNX1,1 CEBPA2 or GATA2.3-7 Myeloid neoplasms with germline predisposition became a separate entity of the World Health Organization (WHO) hematopoietic neoplasm classification in 2016.8 Germline heterozygous mutations of GATA2 account for heterogeneous clinical and hematological manifestations encompassing immunodeficiency (monocyte, B-cell, dendritic cell and natural killer [NK]-cell deficiencies) responsible for recurrent atypical mycobacterial, fungal, bacterial and viral infections,9 vascular disorders such as lymphedema (Emberger syndrome)4,6 or defects of alveolar macrophages leading to pulmonary alveolar proteinosis (PAP).10 Eighty percent of GATA2-deficient patients develop hematological disorders before the age of 40.11 Most patients display hypocellular bone marrows with or without myelodysplastic related changes.12,13
In hematological germline predisposition syndromes, additional somatic alterations could promote leukemic transformation,14-17 but few studies detailed the molecular landscape at different stages in patients with germline GATA2 mutations. Thanks to a series of 78 patients with a long-term follow-up, we now specify a correlation between the GATA2 genotype and the clinical phenotype. Furthermore, bone marrow cytological examination of each patient led to a stratification of hematological spectra, which correlates with somatic alterations.
Methods
Primary samples and diagnostic procedures
Sixty-two patients with heterozygous germline GATA2 mutations included in the GATA2 French-Belgium registry were enrolled in this survey, from 2011 to 2022. All participants gave written informed consent to participate in the study. This registry was labeled by the French health authorities in 2008 with a clinical database approved by the French national data protection agency (CNIL certificate 97.0). Sixteen patients were enrolled in the study from the UK with material obtained with informed consent from the Newcastle Biobank (Newcastle and North Tyneside 1 research Ethics Committee Reference 17/NE/0361.75). Thirty patients were previously reported.11,18-20 Data from 500 sporadic adult AML samples (2018-2022) were extracted from the AML database of Toulouse University Hospital registered at the Commission Nationale de l’Informatique et des Libertés (CNIL, #1778920), and sequenced with the same panel, coverage and sensibility of the technique.
Demographics, biological parameters and infectious status were recorded. Age at first symptom was defined as the age at the first pathological manifestation including cytopenia, MDS, chronic myelomonocytic leukemia (CMML) or AML, any infection (including recurrent bacterial infections, mycobacterial, fungal, human papillomavirus [HPV] and Eppstein Barr virus [EBV] infections), PAP, lymphedema and deafness. Peripheral blood counts were reported at the time of bone marrow evaluation. Bone marrow smears and karyotypes were evaluated by each center (n=76). Spectrum 0 was defined as a bone marrow with normal density without myelodysplastic related changes or normal blood counts, spectrum 1 as a hypocellular bone marrow and/or low-grade MDS (without excess blasts, <5%) and spectrum 2 includes patients with MDS with excess blasts (≥5%) or AML or CMML.
GATA2 germline analysis
GATA2 germline analyses were performed by Sanger or targeted NGS of exons 2 to 6 and the regulatory region in intron 4 using as reference sequence NM_032631.4. Large genomic deletions were investigated by quantitative polymerase chain reaction (PCR) and/or multiplex ligation-dependent probe amplification (MLPA) (MRC-Holland, SALSA MLPA Probemix P437 Familial MDS-AML). GATA2 germline mutation status was confirmed by analysis of non-hematopoietic tissue (cultured skin fibroblasts, hair follicles or nails). Mutations were classified as probably null (nonsense, frameshift, essential-splicing site mutations or large deletion), missense, intronic or synonymous, according to the potential consequence on protein function.
Somatic variant analysis
Mononuclear cells from bone marrow or blood samples were centrally collected from 76 patients and isolated by Ficoll centrifugation. Genomic DNA was extracted using standard procedures and sequenced using an Illumina NextSeq500 sequencer and Sureselect capture in-house panel (Agilent, Santa Clara, CA, USA) targeted on the complete coding regions (i.e., all exons were covered) and -5 to +5 splicing sites of 91 genes recurrently mutated in myeloid neoplasms (Online Supplementary Table S1). Deep sequencing was estimated with an average coverage of 4,859X, and specifically 3,379X for STAG2 exons (sensitivity 1%). Raw NGS data were analyzed using MuTect2, Haplo-typeCaller (both from the GATK suite developed by the Broad Institute) and SureCall (Agilent) algorithms for variant calling aggregated in the in-house remote pipeline (Institut Universitaire de Toulouse-Oncopôle) for data visualization, elimination of sequencing/mapping errors and retention of variants with high quality metrics. Variant interpretation was performed considering minor allele frequencies (MAF) in the public GnomAD database of polymorphisms (variants with MAF >0.02 in overall population/global ancestry or sub-continental ancestry are excluded), variant allele frequencies (VAF), prevalence and clinical interpretation (COSMIC, protein impact). All variants were checked manually on IGV and compared with other samples to check for possible sequencing artifacts, then named according to the Human Genome Variation Society, and compared with sequencing results generated by each local center. Cancer cell fractions (CCF) were calculated from VAF taking into account the chromosomal location of the genes and karyotype. All data are available on ENA_PRJEB55350.
Hematopoietic stem and progenitor cell phenotyping
Hemotopoietic stem and progenitor cell (HSPC) phenotyping was performed on fresh bone marrow samples using an antibody combination targeting CD34, CD38, CD133, CD135, CD45 and CD45RA (Online Supplementary Table S1) and analyzed on Navios instruments (Beckman-Coulter, Miami, FL) and compared to patients without hematological diseases (n=22) or aplasia (n=8) or AML (n=155). HSPC subpopulations were classified as multipotent progenitors (MPP), erythroid/myeloid progenitors (EMP), lymphoid/yeloid primed progenitors (LMPP), common myeloid progenitors (CMP), megakaryocyte erythroid progenitors (MEP), granulocyte macrophage progenitors (GMP) as described in the Online Supplementary Table S1.
Statistics
Data were summarized by frequency and percentage for categorical variables and median and range for continuous variables. Associations between variables were evaluated using Chi-square or Fisher’s exact test for qualitative variables and Kruskall-Wallis test for continuous variables. All survival times were calculated from the biological sampling date and survival are described using the graphical representation of Kaplan-Meier with death from any cause for overall survival (OS) as event. Patients who survived were censored at their last follow-up or for transplanted patients at their allograft day. Univariable analyses were performed using log-rank test. Tests were two-sided and P values <0.05 were considered significant. Statistical analyses were conducted using Stata, version 16. Statistical significance of differences between CBC counts, age or CCF data was determined using multiple unpaired t-test (Graph-Pad Prism 7.0) and P values ****P<0.001, ***P<0.005, **P<0.01, *P<0.05 were considered significant.
Results
GATA2-mutated patients display heterogeneous clinical disorders, depending of their genotype
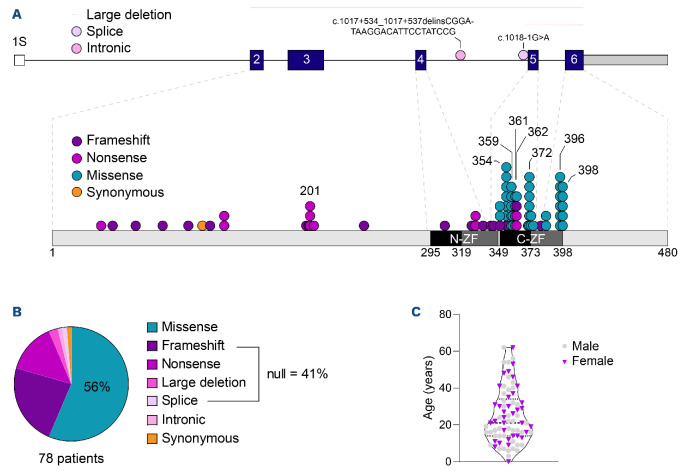
We investigated a series of 78 patients with heterozygous germline mutations of GATA2 (62 from the French-Belgian series and 16 British patients) from 61 kindreds bearing 46 distinct GATA2 germline mutations (Figure 1A, B; Online Supplementary Table S2). The mutations were either missense, located on or close to the second C-terminal zinc finger (44 patients; 56%) or predicted to be a null allele (32 patients; 41%) due to nonsense (11), frameshift (18) or splice defect mutations (1) or large deletions (2). In addition, one patient has an enhancer mutation in intron 4 and one a synonymous mutation (Thr117Thr) as described recently21 (Figure 1A, B).
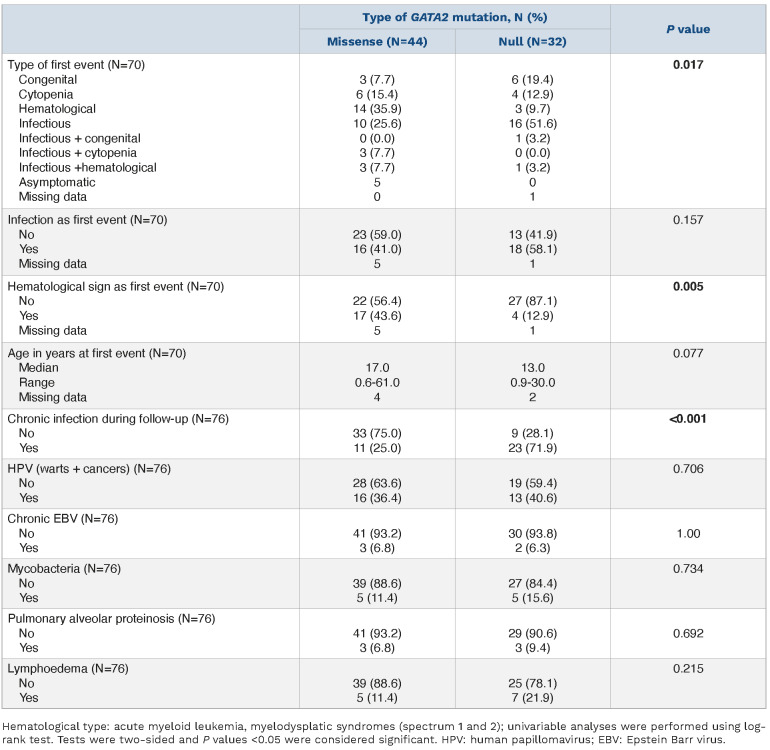
Age at the time of analysis ranged from 0 to 62 years with a median age of 21 years, a male/female ratio of 1.2 (Figure 1C). Seventy-three of 78 patients were symptomatic. The first symptoms occurred at a median age of 16 years, ranging from 7 months to 61 years. First symptoms were predominantly infections (27; 38%), hematological malignancies (17; 24%), cytopenias (11; 15%) or congenital abnormalities such as deafness or lymphedema (9; 13%). Eight patients combined infections with cytopenia (3; 4%), hematological malignancies (4; 6%) or lymphedema (1; 1%). There was a trend for first clinical symptoms occurring at a younger age in patients harboring null mutations, with a median age of detection of 13 years, compared to 17 years for patients with missense mutations (P=0.077). Chronic infectious complications reported before bone marrow transplantation (i.e., bacterial pneumonia, otitis, cellulitis, enteritis, arthritis) were more frequent in patients with null mutations (23 vs. 11; P<0.001). In contrast, hematological disorders were more frequent at the time of diagnosis in patients with missense mutations (17 vs. 4; P=0.005). Frequency of other infections (mycobacterial, EBV), PAP or lymphedema were similar regardless of the mutation type (Table 1).
Half of the patients (41; 53%) underwent bone marrow transplantation as curative treatment with no difference regarding the type of GATA2 mutations (missense vs. null; P=0.46). OS censored at allograft is 89% and 81% with a follow up of 1 and 2 years, respectively (Online Supplementary Figure S1). A total of 61 patients are still alive (78%).
GATA2-deficient patients acquire somatic mutations in a different patern than those with sporadic acute myeloid leukemia
Disease progression in sporadic MDS/AML is associated with karyotypic abnormalities and somatic genetic mutations.22 We compared GATA2-deficient patients to a series of 500 AML without germline GATA2 mutation (sporadic adult AML, 467 molecular samples sequenced with the same panel, coverage and threshold and 431 karyotypes). Karyotype was normal in 38 GATA2-deficient patients (50%) and 237 sporadic AML (55%; P=0.68). When compared to sporadic AML, GATA2-deficient patients more frequently have monosomy 7 (29% vs. 8%; P<0.0001), and der(1;7) (9% vs. 1%; P<0.0001), while rate of trisomy 8 was not different in the two groups (16% vs. 10%; P=0.48). Few other cytogenetic abnormalities were detected in GATA2-deficient patients (12% vs. 28% in sporadic AML; P=0.03) (Online Supplementary Figure S2A). Karyotypic abnormalities were more frequent in men (27 men vs. 11 women; P=0.006) especially chromosome 7 abnormalities (20 vs. 6; P=0.006) and trisomy 8 (10 vs. 2; P=0.033) despite a male/female ratio in the whole cohort of 1.2.
Figure 1.
Characterization of germline GATA2 mutations. (A) Distribution of coding germline GATA2 mutations across GATA2 protein. Predicted protein domains are indicated inside each bar (dark: link zinc finger domain, grey: enlarged zinc finger domain); each dot represents 1 single patient. Each color indicates a type of mutations (frameshift: purple; nonsense: magenta; missense: teal; synonymous: orange). (B) Proportion of germline GATA2 mutation types. (C) Distribution of patients (males, females) according to their age in years at the time of analysis.
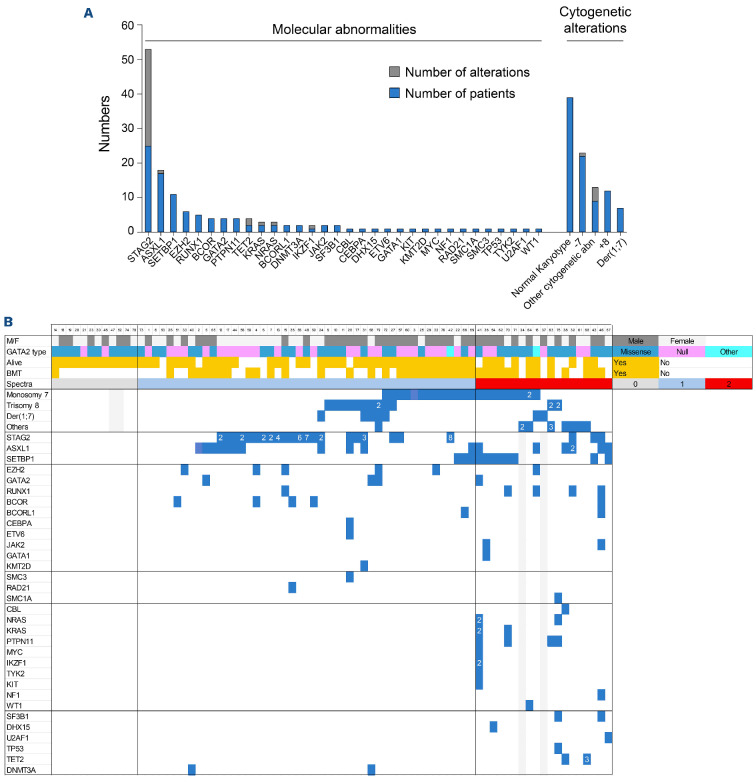
Molecular analysis was performed in 76 patients with germline GATA2 mutations at a median age of 21 years (range, 0-62). One hundred and forty-one somatic mutations were identified with an average of two mutations per patient (range, 1-13; Figure 2A, B; Online Supplementary Table S4). Fifty patients (66%) had at least one mutated gene with no significant difference between missense and null GATA2 mutations (P=0.13; Online Supplementary Table S1). The most frequently mutated genes were STAG2 (53 mutations in 25 patients, 33% vs. 4% in sporadic AML; P<0.0001), ASXL1 (18 mutations in 17 patients, 22% vs. 8%; P=0.0015), SETBP1 (11 patients, 15% vs. 1%; P<0.0001), EZH2 (6 patients, 8% vs. 3%; P=0.31), the RAS pathway (11 mutations combining PTPN11, NRAS, KRAS and CBL in 5 patients, 7% vs. 26%; P=0.031) and RUNX1 (5 patients, 7% vs. 11%; P=0.75) (Online Supplementary Figure S2A). Notably, somatic GATA2 mutations were identified in four patients including two missense mutations located in the first zinc finger domain and two in-frame mutations in the second zinc finger domain (Online Supplementary Table S3). The male/female ratio was not different across mutational identities.
In contrast, frequent mutations in sporadic AML were completely or almost absent in patients with germline GATA2 mutations. DNMT3A (3% vs. 28%; P<0.0001) and TET2 (3% vs. 17%; P=0.015) mutations were found in only two GATA2-deficient patients each. No mutations of NPM1 (37% in sporadic AML; P<0.0001), FLT3 (37% in sporadic AML; P<0.0001), IDH2 (10% in sporadic AML; P=0.03), IDH1 (9% in sporadic AML; P=0.06) or SRSF2 (6% in sporadic AML; P=0.16) were detected in GATA2-deficient patients.
The mutation profile was mainly C>T transitions (33%). A higher rate of deletions (23% vs. 8%; P<0.0001) and a lower rate of insertions (18% vs. 31%; P=0.012) compared to sporadic AML were a consequence of the absence of FLT3 and NPM1 mutations23, 24 (Online Supplementary Figure S2B).
GATA2-deficient patients have an exhaustion of common myeloid and granulocyte macrophage progenitor hematopoietic stem cells
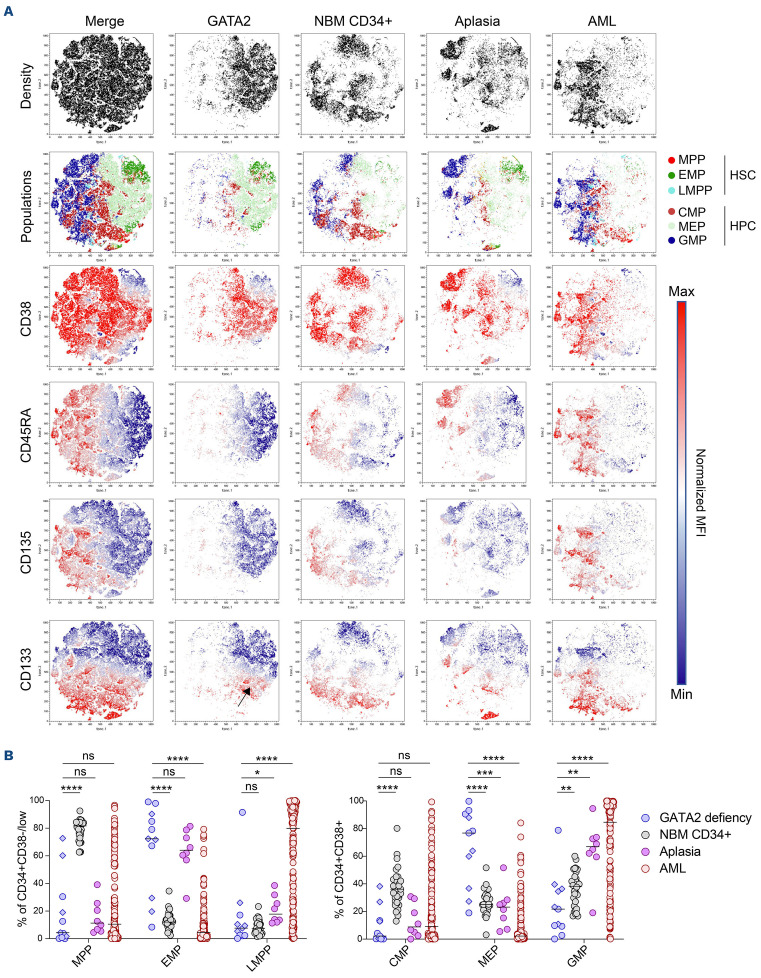
In order to further depict GATA2 syndrome at a cellular level, we analyzed the HSPC compartment of patients with germline GATA2 mutations (n=11 including 5 in spectrum 0 and 6 in spectrum 1). The majority had an EMP bias (63% vs. 13% in normal bone marrows; P<0.0001; Figure 3A, B; Online Supplementary Figure S3) with a loss of heterogeneity in contrast to normal bone marrow where MPP was the major population in the HSC compartment (79%; Figure 3A, B). Specifically, at the progenitor level, the MEP subpopulation was overrepresented compared to normal bone marrow and AML patients (66% vs. 27% and 10%, respectively; P<0.0001; Figure 3A, B; Online Supplementary Figure S3) characterizing a GATA2-specific profile. This pattern is close to aplastic patients with a decrease in MPP and CMP proportions. However, GATA2-deficient patients harbor a higher proportion of MEP and a lower proportion of GMP than aplastic anaemia patients (Figure 3B; Online Supplementary Figure S3). Moreover, it has been reported that the number of HSPC in GATA2-deficient patients is decreased in the same way as aplastic patients.25 Altogether, the differentiation bias in GATA2-deficient patients appears to be related rather to an exhaustion of CMP and GMP populations than to a proliferation of MEP cells. We note that part of the MEP population in GATA2 condition strongly express the CD133 marker which is usually little or not expressed on MEP surface26 (Figure 3A; Online Supplementary Figure S3). This observation may suggest that germline GATA2 mutations also lead to a phenotypic expression bias in the remaining majority population.
Table 1.
Genotype/phenotype correlation.
Figure 2.
GATA2 deficiency syndrome defines a distinct entity regarding molecular profiles. (A) Molecular and cytogenetic abnormalities in the cohort of 78 GATA2-mutated patients. (B) Somatic mutation occurrence. Summary of patients with GATA2 deficiency (n=78) organized by spectra (spectrum 0: grey, spectrum 1: blue and spectrum 2: red), germline GATA2 mutation type (missense: teal, null: fuchsia, other including synonymous and intronic mutations: turquoise), survival and bone marrow transplantation status and somatic mutation and cytogenetic status. Each vertical row represents 1 patient. Grey boxes indicate no data for that parameter. Cytogenetic abnormalities are grouped together in the same manner as the main molecular abnormalities (STAG2, ASXL1 and SETBP1). The number of abnormalities is indicated in each square. The other mutations are listed below including genes encoding transcription factors, splicing factors, chromatin modifiers, cohesin members, signaling pathway genes.
Figure 3.
Phenotypical characterization of hematopoietic stem and progenitor cells revealed loss of heterogeneity associated with common myeloid and granulocyte macrophage progenitors exhaustion. (A) Comparison of hematopoietic stem and progenitor cell (HSPC) phenotypic profiles visualized by a non-linear dimensionality reduction technique (t-SNE) between GATA2 deficiency patients (GATA2, blue, n=11) and control patients without hematological diseases (normal bone marrow [NBM] CD34+, grey, n=22) or acute myeloid leukemia (AML) (red, n=155) or aplasia (purple, n=6) patients without germline GATA2 mutations. Merged samples are localized at the left plots. The first plot line represents density of each condition, the second line the distribution of the different HSC and hematopoietic progenitor cells (HPC) subpopulations, the lines below show normalized mean fluorescent intensity (MFI) according the color scale of markers (CD38, CD45RA, CD135 and CD133). The black arrow on the CD133 plot of GATA2 condition identifies of megakaryocyte erythroid progenitors (MEP)-expressing CD133 marker. (B) Proportion of HSPC populations in CD34+ CD38- compartment (erythroid/myeloid progenitors [EMP]: CD135- CD45RA-; multipotent progenitors [MPP]: CD135+ CD45RA-; lymphoid/myeloid primed progenitors [LMPP]: CD135+ CD45RA+) and CD34+ CD38+ compartment (MEP: CD135-CD45RA-; common myeloid progenitors [CMP]: CD135+ CD45RA-; granulocyte macrophage progenitors [GMP]: CD135+ CD45RA+) of GATA2-deficient patients (blue) compared with NBM CD34+ (grey), AML (red) and aplastic (purple) patients. Spectrum 0 (diamond), spectrum 1 (round); median and P values are calculated using non-parametric unpaired Mann-Whitney test ****P<0.001,***P<0.005 **, P<0.01, *P<0.05.
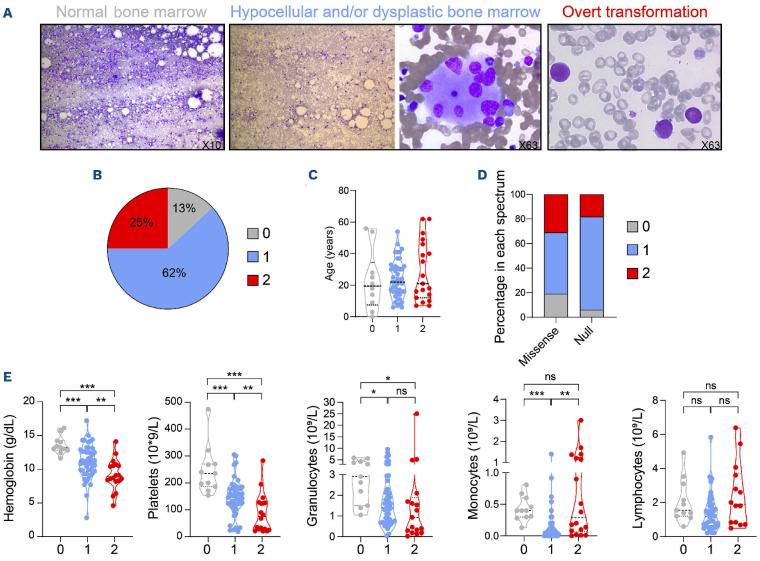
Hypocellular bone marrow - low-grade myelodysplastic syndromes represents the main hematological spectrum in GATA2-deficient patients
In order to further investigate the impact on hematopoiesis, we reviewed bone marrow smears of 76 patients and defined three distinct morphological categories: normal bone marrow (spectrum 0), hypoplastic and/or low-grade MDS (spectrum 1) or overt transformation (spectrum 2) (Figure 4A). The majority of GATA2-mutated patients had features of spectrum 1 (47; 62%), 19 had evidence of hematopoietic transformation (19; 25%) and only ten are in spectrum 0 (13%) (Figure 4B). Despite a different bone marrow pattern, there was no difference on survival, probably because most patients received an HSC transplantation before progression. Age had no impact (Figure 4C; P=0.83) but patients in spectrum 2 are at high risk of progression to MDS or AML in the year after diagnosis of the first event (P=0.036). As expected, null alleles are associated with spectrum 1 in contrast to missense mutations enriched in spectrum 2 (P=0.023; Figure 4D). Infections (mycobacterial, EBV), PAP or lymphoedema were not correlated with the spectra, with the exception of chronic HPV infections which were enriched in spectrum 1 (52 vs. 16; P=0.008).
Regarding the parameters of the peripheral blood count, absolute lymphocyte count showed no significant differences between spectra while hemoglobin concentration, platelet, absolute neutrophil and monocyte counts decreased progressively with advanced spectra. Six patients with spectrum 2 have an increased monocyte count (Figure 4E; Online Supplementary Table S5).
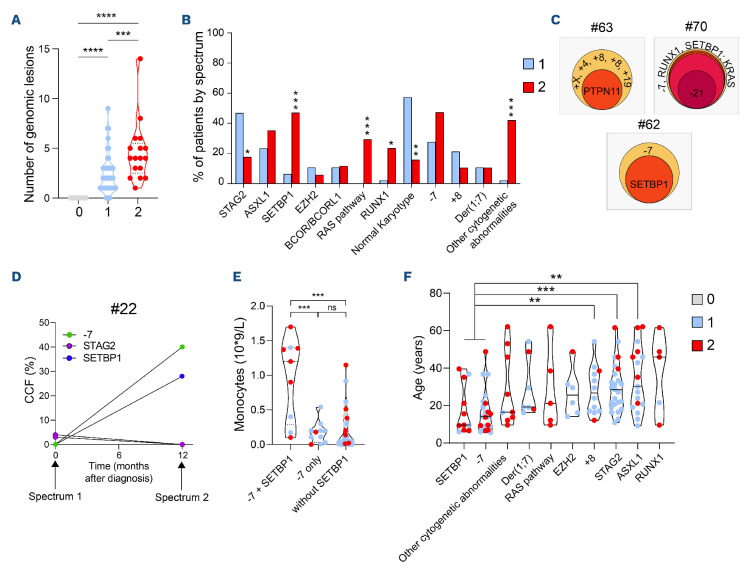
Somatic molecular and karyotypic abnormalities drive hematopoietic evolution
None of the patients at spectrum 0 have somatic mutations, or karyotypic abnormalities (Figure 5A). Mutation numbers increase at spectrum 2 (median of 3 vs. 1 at spectrum 1; P=0.022; Figure 2B) with an enrichment in SETBP1, RAS pathway and RUNX1 mutations (47% vs. 6% at spectrum 1; P<0.001; 29% vs. 0%; P<0.001; 23% vs. 2%; P=0.015, respectively). Karyotypic abnormalities increased from spectrum 0 to spectrum 2 (0% at spectrum 0, 47% at spectrum 1 and 84% at spectrum 2) and with an enrichment of the other cytogenetic abnormalities (42% vs. 2%; P<0.001). Notably, monosomy 7 increased through spectra (none at spectrum 0, 28% at spectrum 1 and 47% at spectrum 2; P=0.12; Figure 5B). Leukemic transformation is associated with a high clone size at spectrum 2 as demonstrated by patients #62, #63 and #70 (Figure 5C; Online Supplementary Tables S2 and S4). In patients #62 and #63, karyotypic abnormalities were the first event, followed by SETBP1 and PTPN11 mutations respectively. Patient #70 had a major driver clone with monosomy 7 and several mutated genes, with acquisition of monosomy 21. In patient #22, clonal-dynamics analysis showed a selective advantage of the major clone with monosomy 7 and SETBP1 mutation over STAG2-mutated clones detected at diagnosis (Figure 5D). These results suggest that the selective advantages of the clones were shaped by the acquisition of new abnormalities in distinct genes. According to these results, SETBP1 mutations may be critical for leukemic transformation in GATA2-deficient patients. Nine of 11 SETBP1-mutated patients also harbored a monosomy 7 (P<0.001), and their association was significantly enriched at spectrum 2 (31% vs. 6%; P=0.008). This co-occurrence was associated with a higher monocyte count (Figure 5E; P<0.001). Interestingly, SETBP1 mutations and monosomy 7 were the earliest oncogenic events to occur in patients (Figure 5F; median age 9.3 years). Patients with RUNX1 mutations had the highest median age (46 years) while being also enriched in spectrum 2. Patients with ASXL1 or EZH2 mutations or trisomy 8 were also older with a median age of 30 years, 26 and 27 years, respectively, without significant differences according to spectra. The only mutated gene significantly enriched in spectrum 1 was STAG2 (47% vs. 18%; P=0.035; Figure 5B) with a median age of 28 years (Figure 5F), and there was no difference across blood parameters, compared to patients in spectrum 1 without STAG2 mutation (Online Supplementary Figure S3). Our results strongly suggest that somatic acquired mutations in mutated GATA2 patients have a different impact on the leukemogenesis process.
Figure 4.
Classification in three hematological spectra based on cytological evaluation. (A) Representative pictures of normal density bone marrow defining spectrum 0 (left, objective 10x), hypocellular bone marrow (objective 10x) and an example of myelodysplastic-related changes: multinuclear megakaryocyte (objective 63x) reported in patients in spectrum 1 (middle), blast cells defining overt transformation for spectrum 2 including myelodysplastic syndromes (MDS) with excess of blasts, acute myeloid leukemia (AML) and chronic myelomonocytic leukemia (CMML) (objective 63x, right). (B) Proportion of patients in each spectrum (0, 1 and 2), P=not significant. (C) Distribution of patient age in each spectrum. (D) Percentage of patients with missense or null mutations in each spectrum. (E) Blood count parameters (hemoglobin level, platelet, granulocyte, monocyte and lymphocytes counts) of 78 patients in each spectrum. All data points represent each patient values according to spectra with median (wide dots) ± quartiles (small dots). P values are calculated using unpaired t-tests ***P<0.001, **P<0.01, *P<0.05.
Somatic mutations of STAG2 do not drive leukemic transformation in contrast to SETBP1 mutations
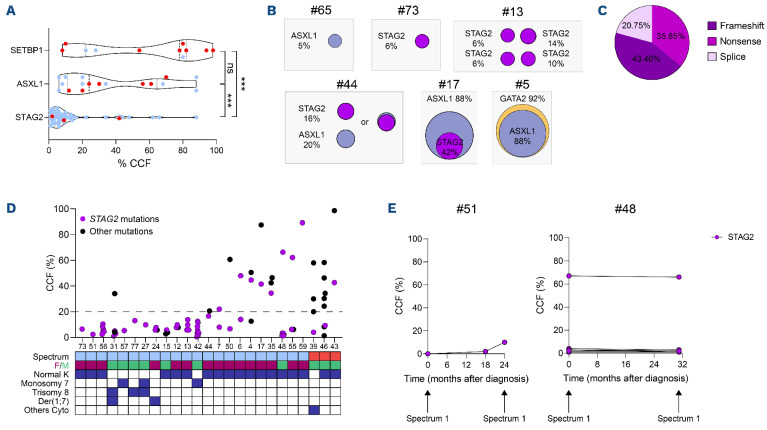
In order to further characterize the impact of somatic mutations in hematopoietic transformation, cancer cell fraction (CCF) was calculated for the most frequently mutated genes. STAG2 mutations exhibited a considerably lower CCF than those of ASXL1 and SETBP1 (median of 6% vs. 24% and 78%, respectively; Figure 6A). Some patients in spectrum 1 harbored a single mutation with low CCF (patient #65 for ASXL1 or #73 for STAG2), while others exhibited several mutations occurring in unique or probably multiple clones (Figure 6B, patients #13, #17, #5 and #43). Interestingly, despite a high proportion of mutated cells, patients #17 and #5 had no excess of blasts suggesting that not all abnormalities lead to spectrum 2.
STAG2 mutations were identified in 25 patients (10 males and 15 females) with up to eight different STAG2 mutations in the same individual (mean 2.1). STAG2 mutations are loss of function, mainly due to the introduction of premature stop codons leading to a destabilization of the cohesin complex27,28 (Figure 6C). Low CCF (≤20%) for STAG2 mutations were rarely associated with other molecular abnormalities in spectrum 1 patients (Figure 6D). Among the STAG2-mutated patients, only three were classified at spectrum 2. STAG2 mutations in patients #39 and #46 had low CCF suggesting that other mutations drove the leukemic transformation. Patient #43 exhibited a higher STAG2 CCF but in a leukemic clone probably driven by the primary SETBP1 mutation (Figure 6D). In order to gain insight in the potential of clonal growth linked to STAG2 mutations, we monitored longitudinal follow-up of two patients. After detection, STAG2-mutated clones could be little selected (patient #51) or with little CCF variations upon time (patient #48). Importantly, these two patients remained at spectrum 1 after several years (Figure 6E). Overall, our analysis demonstrate that STAG2 somatic mutations were recurrent in GATA2-deficient patients, indicating that they may provide a selective advantage to GATA2-deficient hematopoietic cells although insufficient for their transformation.
Discussion
While the majority of MDS and AML are sporadic, rare germline predisposition syndromes have been delineated.29 Myeloid transformations in the context of germline alterations have variable latency but usually occur in younger patients than sporadic malignancies, with the exception of germline DDX41 mutations.17 GATA2 deficiency syndrome, recognized as a major MDS/AML predisposition in the WHO classification, has a high although variable penetrance.11,30 This series highlights that patients with missense mutations may have a higher risk of transformation, and are thus over-represented in the spectrum 2 group, as opposed to patients with null mutations. The latter seems to correlate with an earlier age of diagnosis and chronic infections. Most patients have hypocellular and/or myelodysplastic bone marrows, including four patients with no somatic mutations, raising the interesting possibility that germline GATA2 mutations may intrinsically induce this condition. In line with this hypothesis, McReynolds and colleagues described in 2019 a GATA2 deficiency-related bone marrow and immunodeficiency disorder (G2BMID),13 with bone marrow hypocellularity, atypical megakaryocytes and minimal dysmyelopoiesis and dyserythropoiesis. As dysplastic threshold in GATA2 deficiency remains hard to define, we chose to group hypoplastic bone marrow and low-grade MDS in the same spectrum. In the recent report by West and colleagues,31 13 GATA2-deficient patients were asymptomatic with a normal bone marrow. Only three had somatic mutations including DNMT3A, that can be explained by the older median age of patients in link with age-related clonal hematopoiesis.32
Figure 5.
Stratification of genetic abnormalities according hematological spectra. (A) Distribution of genomic alterations including molecular and cytogenetic abnormalities in each spectrum. (B) Proportion of patients in each spectrum according to their mutational and cytogenetic abnormalities (blue: spectrum 1; red: spectrum 2). (C) Clonal hierarchy of 3 patients at spectrum 2, evaluated thanks to cancer cell fraction (CCF). (D) Clonal dynamics evaluated by molecular and cytogenetic follow-up of one patient with two STAG2 mutations at diagnosis (spectrum 1) which disappeared 1 year later (spectrum 2), concomitantly to the appearance of a clone with monosomy 7 and a SETBP1 mutation. CCF (%) was evaluated using variant allelic frequency for mutations and polymorphisms located on chromosome 7 for monosomy 7. (E) Monocyte count in patients with the association of SETBP1 mutation and monosomy 7 (n=9) or the monosomy 7 only (n=11) or patients without SETBP1 mutations (n=45). (F) Median age in years of patients according their genetic profiles. Patients harboring the association monosomy 7 and SETBP1 mutation were compared to STAG2-mutated patients, patients with trisomy 8 and with ASXL1 mutation. All data points represent each patient values according to hematological spectra with median ± quartiles and P values are calculated using unpaired t-tests ***P<0.001, **P<0.01, *P<0.05.
The molecular and cytogenetic profiles of GATA2-deficient patients differ from those of sporadic AML and adult MDS,33 and get closer to pediatric MDS.16,34 Indeed, no or very few mutations of NPM1, FLT3 or epigenetic-related genes such as IDH1/2, TET2 or DNMT3A or splicing genes (SRSF2, SF3B1, U2AF1 and SRSF2) have been identified in GATA2-deficient patients at spectrum 1 or 2, suggesting differences in the mechanism of clonal selection.33 In GATA2-deficient patients, we confirm in our study that the most frequent cytogenetic abnormalities involved chromosome 7 and somatic mutations target STAG2, ASXL1 and SETBP1. This profile more closely reflects pediatric hypocellular MDS also harbors monosomy 7 and SETBP1, ASXL1, RAS pathway and RUNX1 mutations.34,35 Sahoo et al. observed similar features in SAMD9/SAMD9L germline predisposition.16 However, GATA2-mutated patients harbored numerous STAG2 mutations, which are also reported in Down syndrome involving another GATA factor36 and in MDS with a poor OS,33 but not specifically in pediatric MDS, suggesting a specific mutational profile in GATA2-deficient patients.31,37
The analysis of HPC compartment38 reveals a loss of heterogeneity with exhaustion of CMP and GMP populations suggesting that hematopoiesis is less efficient in GATA2-deficient patients. Interestingly, some residual MEP showed aberrant strong expression of CD133 marker suggesting that GATA2 deficiency also alters the remaining progenitor compartment. Functional and quantitative defects in the Gata2+/- mouse model also suggest that germline GATA2 mutations could impair HSC fitness.39 Recently, the loss of fitness in HSC has been described in some inherited bone marrow failures also associated with a higher risk of myeloid transformation by acquisition of genetic alterations.40 Recent studies have highlighted the ability of some abnormalities to drive a leukemic clone while others preferentially define a state of clonal hematopoiesis.16,41 We found that SETBP1, RAS pathway, and RUNX1 mutations were enriched in spectrum 2, data which could be confirmed in a larger cohort. Monosomy 7 was present at spectrum 1 and 2, but the association with SETBP1 mutation was clearly enriched at spectrum 2. This co-occurrence suggests that the monosomy 7 clones can accumulate other genetic abnormalities and lead to leukemic transformation, corroborating recent findings in the context of other germline predisposition such as SAMD9 syndromes.16 Interestingly, patients with co-occurring monosomy 7 and SETBP1 mutations had a higher monocyte counts suggesting that it could improve monocytic differentiation in GATA2-deficient patients.
Figure 6.
Clonal hematopoiesis mediated by STAG2 mutations and clonal selection. (A) Comparison of cancer cell fraction (CCF) of the 3 main molecular abnormalities: SETBP1 (n=11), ASXL1 (n=17) and STAG2 (n=53) mutations. Blue and red dots correspond to spectra 1 and 2, respectively. (B) Representative examples of clonal hierarchy evaluated by CCF. (C) Proportion of the different STAG2 mutation types (n=53). (D) Percentage of STAG2 (purple) and the other (dark) mutation CCF from the 25 STAG2-mutated patients associated with the hematological spectrum (blue: spectrum 1; red: spectrum 2), sex (female: burgundy; male: green) and the cytogenetic profile (normal karyotype, monosomy 7, trisomy 8, der(1;7) and other cytogenetic abnormalities). The dotted line allows to visualize mutations with CCF less than or equal to 20%. (E) Clonal dynamic evaluated by longitudinal follow-up of 2 patients mutated only for STAG2.
As previously reported, STAG2 mutations are recurrent in GATA2 deficiency,31 we found that patients harboring STAG2 mutations are classified in spectrum 1, and were older than patients with SETBP1 mutations and monosomy 7. To go further, we showed that CCF of STAG2 mutations were lower than those of SETBP1 or ASXL1 mutations with up to eight STAG2 mutations per patient. Altogether, these observations suggest a different impact of STAG2 mutations, without obvious oncogenic potential but prone to induce a non-aggressive clonal hematopoiesis. Although our molecular analysis does not allow us to demonstrate that different STAG2 mutations were found in different clones, the presence of several low CCF mutations in distinct clones is a mechanism already reported in other IBMFS.41 Single- cell DNA sequencing analysis is required to confirm it in GATA2 deficiency. Moreover, mutations that modify the fitness of cells without association with transformation have been described in Shwachman-Diamond syndrome (SBDS) patients.41,42 The authors proposed that EIF6 mutations can counterbalance the initial deficiency induced by germline SBDS mutation and act as a somatic genetic rescue mechan ism. These mutations exhibit low VAF and up to eight different EIF6 mutations were present in the same individual, similar to what we found for STAG2 mutations. These mutations were found in multiple clones defining clonal hematopoiesis.42 STAG2 is a member of the cohesin complex, implicated in the three-dimensional folding of DNA and, thus in the regulation of numerous transcription factors28 such as GATA2 in HSPC. Indeed, its alteration can open chromatin at GATA and RUNX sites.27,43 This suggest that STAG2 mutations may increase the expression of GATA2 target genes by increasing GATA site opening. STAG2 mutations could act as a potential compensatory pathway, improving the fitness of clones with limited leukemic potential.
Figure 7.
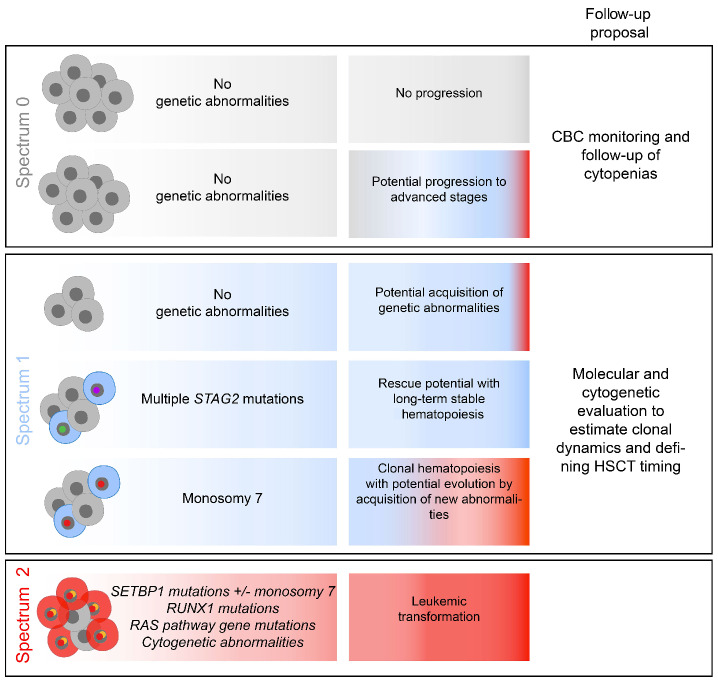
Distinct pathways of somatic clonal progression in GATA2 deficient patients. Summary of the different possibilities of evolution according to the spectra. Proposal of a personalized follow-up of patients from each spectrum.
Overall, monitoring these genetic abnormalities is consequently of importance in the context of GATA2 deficiency (Figure 7).
Supplementary Material
Acknowledgments
The team is also supported by the associations “Enfants Cancers et Santé”, “Capucine”, “Constance la Petite Guerrière Astronaute”. We acknowledge the contribution of the tumor bank HIMIP. We wish to thank all clinicians and biologists who participated in this study: Wadih Aboudchala, Pediatric Hematology, CHU Lille, Lille; Oana Balasanu, Department of Hematology, CHU Nancy, Nancy; Vincent Barlogis, Department of Pediatric Oncology and Hematology, CHU Marseille La Timone, Marseille; Sarah Beaussant Cohen, Pediatry Unit, CHU Besançon, Besançon; Nicolas Blein, Department of Hematology, CHU Nantes, Nantes; Anne Sophie Brunel, Department of Infectious Disease, CHU Montpellier, Montpellier; Benedicte Bruno, Pediatric Hematology, CHU Lille, Lille; Helene Cavé, APHP Robert-Debré, Paris; François Chabot, Department of Hematology, CHU Nancy, Nancy; Sophie Ducastelle Leprete, Department of Hematology, CHU Lyon, Hôpital Lyon SUD, Lyon; Bernard Drenou, Department of Hematology, CH Mulhouse, Mulhouse; Viviana Marin Esteban, UMR 996, University Paris 11, Clamart; Jean-Hugues Dalle Department of Pediatric Hematology and HSCT, APHP Robert Debré, Paris; Pierre-Yves Dumas, Department of Hematology, CHU Bordeaux, Bordeaux; Laurence Faivre, Department of Genetics, CHU Dijon, Dijon; Pascale Flandrin, Laboratory of Hematology, CHU Saint Etienne, Saint Etienne; Lionel Galicier, Department of Immunology, APHP St Louis, Paris; Virginie Gandemer, Pediatric Hematology, Rennes; Federica Giannotti, Department of Hematology, APHP Saint-Antoine, Paris; Jean Gutnecht, Department of Internal Medicine, CH Frejus Saint-Raphael, Fejus; Sébastien Héritier, Department of Pediatric Oncology and Hematology, Registre National des Neutropénies Chroniques, APHP Trousseau, Paris; Eric Jeziorski, Department of Pediatrics, CHU Montpellier, Montpellier; Thierry Lamy de la Chapelle, Department of Hematology, CHU Nantes, Nantes; Hélène Lapillone, Hematology Laboratory, APHP Trousseau, Paris; Yannick le Bris, Hematology Laboratory, Nantes; Guy Leverger, Department of Pediatric Oncology and Hematology, Registre National des Neutropénies Chroniques, APHP Trousseau, Paris; Ludovic Martin, Department of Dermatology, CHU Angers, Angers; Gérard Michel, Department of Pediatric Oncology and Hematology, CHU Marseille La Timone, Marseille; Fabrice Monpoux, Department of Pediatric Oncology and Hematology, CHU Nice, Nice; Despina Moushous, Department of Pediatric Immunology, APHP Necker-Enfants Malades, Paris; Benedicte Neven, Department of Pediatric Immunology, APHP Necker-Enfants Malades, Paris; Raphaele Nove Josserand, Department of Internal Medicine, CHU Lyon, Hôpital Lyon Sud, Lyon; Catherine Paillard, Department of Pediatric Oncology and Hematology, CHU Strasbourg, Strasbourg; Régis Pefault de la Tour, Department of Hematology, HSCT Unit, APHP Saint-Louis, Paris; Isabelle Pellier, Pediatric Hematology and Immunology, Angers; Arnaud Petit, Department of Pediatric Oncology and Hematology, Registre National des Neutropénies Chroniques, APHP Trousseau, Paris; Gaetan Sauvetre, Department of Internal Medicine, CHU Rouen, Rouen; Nicolas Schleinitz, Immunology and Internal Medicine, CHU Marseille La Timone, Marseille; Jean Soulier, APHP Saint Louis, Paris; Suzanne Tavitian, Department of Hematology, CHU Toulouse, Toulouse; Louis Terriou, Internal Medicine, CHU Lille, Lille; Pierre Van de Perre, ICR, Toulouse; Norbert Vey, Department of Hematology, HSCT Unit, Institut PaoliCalmette, Marseille and Stéphane Vignes, Department of Internal Medicine, Fondation Cognacq Jay, Paris, France.
Funding Statement
Funding: This project was supported by associations “111 des arts”, “Toulouse Recherche Enfant Cancer”, INCa and CONECT-AML project.
References
- 1.Song WJ, Sullivan MG, Legare RD, et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999;23(2):166-175. [DOI] [PubMed] [Google Scholar]
- 2.Smith ML, Cavenagh JD, Lister TA, Fitzgibbon J. Mutation of CEBPA in familial acute myeloid leukemia. N Engl J Med. 2004;351(23):2403-2407. [DOI] [PubMed] [Google Scholar]
- 3.Dickinson RE, Griffin H, Bigley V, et al. Exome sequencing identifies GATA-2 mutation as the cause of dendritic cell, monocyte, B and NK lymphoid deficiency. Blood. 2011;118(10):2656-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hahn CN, Chong CE, Carmichael CL, et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet. 2011;43(10):1012-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsu AP, Sampaio EP, Khan J, et al. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood. 2011;118(10):2653-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ostergaard P, Simpson MA, Connell FC, et al. Mutations in GATA2 cause primary lymphedema associated with a predisposition to acute myeloid leukemia (Emberger syndrome). Nat Genet. 2011;43(10):929-931. [DOI] [PubMed] [Google Scholar]
- 7.Pasquet M, Bellanne-Chantelot C, Tavitian S, et al. High frequency of GATA2 mutations in patients with mild chronic neutropenia evolving to MonoMac syndrome, myelodysplasia, and acute myeloid leukemia. Blood. 2013;121(5):822-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391-2405. [DOI] [PubMed] [Google Scholar]
- 9.Vinh DC, Patel SY, Uzel G, et al. Autosomal dominant and sporadic monocytopenia with susceptibility to mycobacteria, fungi, papillomaviruses, and myelodysplasia. Blood. 2010;115(8):1519-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spinner MA, Sanchez LA, Hsu AP, et al. GATA2 deficiency: a protean disorder of hematopoiesis, lymphatics, and immunity. Blood. 2014;123(6):809-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donadieu J, Lamant M, Fieschi C, et al. Natural history of GATA2 deficiency in a survey of 79 French and Belgian patients. Haematologica. 2018;103(8):1278-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wlodarski MW, Hirabayashi S, Pastor V, et al. Prevalence, clinical characteristics, and prognosis of GATA2-related myelodysplastic syndromes in children and adolescents. Blood. 2016;127(11):1387-1397. [DOI] [PubMed] [Google Scholar]
- 13.McReynolds LJ, Yang Y, Yuen Wong H, et al. MDS-associated mutations in germline GATA2 mutated patients with hematologic manifestations. Leuk Res. 2019;76:70-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Preudhomme C, Renneville A, Bourdon V, et al. High frequency of RUNX1 biallelic alteration in acute myeloid leukemia secondary to familial platelet disorder. Blood. 2009;113(22):5583-5587. [DOI] [PubMed] [Google Scholar]
- 15.Duployez N, Jamrog LA, Fregona V, et al. Germline PAX5 mutation predisposes to familial B-cell precursor acute lymphoblastic leukemia. Blood. 2021;137(10):1424-1428. [DOI] [PubMed] [Google Scholar]
- 16.Sahoo SS, Pastor VB, Goodings C, et al. Clinical evolution, genetic landscape and trajectories of clonal hematopoiesis in SAMD9/SAMD9L syndromes. Nat Med. 2021;27(10):1806-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duployez N, Largeaud L, Duchmann M, et al. Prognostic impact of DDX41 germline mutations in intensively treated acute myeloid leukemia patients: an ALFA-FILO study. Blood. 2022;140(7):756-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciullini Mannurita S, Vignoli M, Colarusso G, et al. Timely follow-up of a GATA2 deficiency patient allows successful treatment. J Allergy Clin Immunol. 2016;138(5):1480-1483. [DOI] [PubMed] [Google Scholar]
- 19.Tholouli E, Sturgess K, Dickinson RE, et al. In vivo T-depleted reduced-intensity transplantation for GATA2-related immune dysfunction. Blood. 2018;131(12):1383-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sekhar M, Pocock R, Lowe D, et al. Can somatic GATA2 mutation mimic germ line GATA2 mutation? Blood Adv. 2018;2(8):904-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozyra EJ, Pastor VB, Lefkopoulos S, et al. Synonymous GATA2 mutations result in selective loss of mutated RNA and are common in patients with GATA2 deficiency. Leukemia. 2020;34(10):2673-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osorio FG, Rosendahl Huber A, Oka R, et al. Somatic mutations reveal lineage relationships and age-related mutagenesis in human hematopoiesis. Cell Rep. 2018;25(9):2308-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee-Six H, Obro NF, Shepherd MS, et al. Population dynamics of normal human blood inferred from somatic mutations. Nature. 2018;561(7724):473-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganapathi KA, Townsley DM, Hsu AP, et al. GATA2 deficiency-associated bone marrow disorder differs from idiopathic aplastic anemia. Blood. 2015;125(1):56-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorgens A, Radtke S, Mollmann M, et al. Revision of the human hematopoietic tree: granulocyte subtypes derive from distinct hematopoietic lineages. Cell Rep. 2013;3(5):1539-1552. [DOI] [PubMed] [Google Scholar]
- 27.Ochi Y, Kon A, Sakata T, et al. Combined cohesin-RUNX1 deficiency synergistically perturbs chromatin looping and causes myelodysplastic syndromes. Cancer Discov. 2020;10(6):836-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viny AD, Bowman RL, Liu Y, et al. Cohesin members Stag1 and Stag2 display distinct roles in chromatin accessibility and topological control of HSC self-renewal and differentiation. Cell Stem Cell. 2019;25(5):682-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fenwarth L, Duployez N, Marceau-Renaut A, et al. Germline pathogenic variants in transcription factors predisposing to pediatric acute myeloid leukemia: results from the French ELAM02 trial. Haematologica. 2021;106(3):908-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wlodarski MW, Collin M, Horwitz MS. GATA2 deficiency and related myeloid neoplasms. Semin Hematol. 2017;54(2):81-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.West RR, Calvo KR, Embree LJ, et al. ASXL1 and STAG2 are common mutations in GATA2 deficiency patients with bone marrow disease and myelodysplastic syndrome. Blood Adv. 2022;6(3):793-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaiswal S, Natarajan P, Silver AJ, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377(2):111-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernard E, Tuechler H, Greenberg PL, et al. Molecular International Prognostic Scoring System for myelodysplastic syndromes. NEJM Evid. 2022;1(7). [DOI] [PubMed] [Google Scholar]
- 34.Schwartz JR, Ma J, Lamprecht T, et al. The genomic landscape of pediatric myelodysplastic syndromes. Nat Commun. 2017;8(1):1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pastor V, Hirabayashi S, Karow A, et al. Mutational landscape in children with myelodysplastic syndromes is distinct from adults: specific somatic drivers and novel germline variants. Leukemia. 2017;31(3):759-762. [DOI] [PubMed] [Google Scholar]
- 36.Yoshida K, Toki T, Okuno Y, et al. The landscape of somatic mutations in Down syndrome-related myeloid disorders. Nat Genet. 2013;45(11):1293-1299. [DOI] [PubMed] [Google Scholar]
- 37.Brown AL, Hahn CN, Scott HS. Secondary leukemia in patients with germline transcription factor mutations (RUNX1, GATA2, CEBPA). Blood. 2020;136(1):24-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vergez F, Green AS, Tamburini J, et al. High levels of CD34+CD38low/-CD123+ blasts are predictive of an adverse outcome in acute myeloid leukemia: a Groupe Ouest-Est des Leucemies Aigues et Maladies du Sang (GOELAMS) study. Haematologica. 2011;96(12):1792-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodrigues NP, Janzen V, Forkert R, et al. Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood. 2005;106(2):477-484. [DOI] [PubMed] [Google Scholar]
- 40.Tsai FD, Lindsley RC. Clonal hematopoiesis in the inherited bone marrow failure syndromes. Blood. 2020;136(14):1615-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kennedy AL, Myers KC, Bowman J, et al. Distinct genetic pathways define pre-malignant versus compensatory clonal hematopoiesis in Shwachman-Diamond syndrome. Nat Commun. 2021;12(1):1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan S, Kermasson L, Hilcenko C, et al. Somatic genetic rescue of a germline ribosome assembly defect. Nat Commun. 2021;12(1):5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mullenders J, Aranda-Orgilles B, Lhoumaud P, et al. Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J Exp Med. 2015;212(11):1833-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.