Abstract
PsaD subunit of Synechocystis sp PCC 6803 photosystem I (PSI) plays a critical role in the stability of the complex and is part of the docking site for ferredoxin (Fd). In the present study we describe major physiological and biochemical effects resulting from mutations in the accessible C-terminal end of the protein. Four basic residues were mutated: R111, K117, K131, and K135, and a large 36-amino acid deletion was generated at the C terminus. PSI from R111C mutant has a 5-fold decreased affinity for Fd, comparable with the effect of the C terminus deletion, and NADP+ is photoreduced with a 2-fold decreased rate, without consequence on cell growth. The K117A mutation has no effect on the affinity for Fd, but decreases the stability of PsaE subunit, a loss of stability also observed in R111C and the deletion mutants. The double mutation K131A/K135A does not change Fd binding and reduction, but decreases the overall stability of PSI and impairs the cell growth at temperatures above 30°C. Three mutants, R111C, K117A, and the C-terminal deleted exhibit a higher content of the trimeric form of PSI, in apparent relation to the removal of solvent accessible positive charges. Various regions in the C terminus of cyanobacterial PsaD thus are involved in Fd strong binding, PSI stability, and accumulation of trimeric PSI.
Photosystem I (PSI) is a membrane-embedded multisubunit complex capable of photoinduced electron transfer to soluble electron acceptors. In the cyanobacterium Synechocystis sp. PCC 6803 (Synechocystis), these acceptors can be ferredoxin (Fd) or flavodoxin, depending on the amount of iron present in the culture medium (Rogers, 1987; Laudenbach et al., 1988; Bottin and Lagoutte, 1992). PSI from most cyanobacteria is composed of 11 subunits (Golbeck, 1994), either integral like those binding the cofactors central to the electron transfer (PsaA and PsaB subunits; PsaX, psaX gene product), or peripheral and extractable by chaotropic agents such as PsaD, PsaE, or PsaC subunits (Golbeck and Cornelius, 1986; Tjus and Andersson, 1991; Lagoutte and Vallon, 1992; Zilber and Malkin, 1992). The functional reaction center, including associated pigments, has a Mr of about 320,000 and can be isolated in a monomeric as well as a trimeric form (Rögner et al., 1990; Kruip et al., 1994). Deletion mutagenesis studies revealed that Fd binding involves at least three PSI subunits, PsaC, PsaD, and PsaE, all of low molecular weight (Rousseau et al., 1993; Xu et al., 1994c; Hanley et al., 1996; Barth et al., 1998; Fischer et al., 1998). PsaD has been successfully cross-linked to Fd in a functional electron transfer complex, and the interacting sequences of both proteins were identified (Zanetti and Merati, 1987; Zilber and Malkin, 1988; Lelong et al., 1994). Various single site-directed mutagenesis results argue for a multi-site PsaD/Fd interaction. PsaD has also been proved to be a key protein in stabilizing the reducing side of PSI, mainly at the level of PsaC, the polypeptide bearing the two terminal [4Fe-4S] clusters (Li et al., 1991; Chitnis et al., 1996; Hanley et al., 1996). PsaD integration considerably decreases the turnover of PSI, a function partly shared with PsaE (Chitnis and Nelson, 1992a). When the psaD gene is deleted, the trimeric form of PSI is undetectable (Chitnis and Chitnis, 1993; Schluchter et al., 1996), probably due to the correlated destabilization of PsaL (Xu et al., 1994a). All these structural properties are in good agreement with the different topological views of this subunit on top of the PSI complex (Kruip et al., 1997; Kitmitto et al., 1998; Klukas et al., 1999). A large terminal end of PsaD extends over PsaC up to PsaE, and another part of the polypeptide is oriented toward the center of the PSI trimer, close to the region supposedly occupied by PsaL and PsaI. PsaD is a basic protein of 140 residues in Synechocystis, including several regions of clustered positive amino acids. Its structure is not yet well established. It seems to contain a β-sheet surrounding a short α-helix closely located to PsaC and to a membrane-embedded part of PSI (Klukas et al., 1999). Similar to the PSI-bound form, isolated PsaD shows only a low content of well-defined secondary structural elements, but it is still capable of Fd binding (Xia et al., 1998; Jin et al., 1999; Pandini et al., 1999). Lys 74 is part of a centrally located sequence including six basic amino acids out of 15 residues. It has been shown to be an important residue for the stable association of PsaD to the PSI core (Chitnis et al., 1997) and it is likely that the close surrounding sequence is buried inside the complex. More generally, the N-terminal one-half of the protein has been found to be poorly accessible in cyanobacteria, whereas different basic residues of the C terminus, starting at Lys 106, were found to be accessible to the external medium (Lelong et al., 1994; Xu et al., 1994b), thus appearing as possible candidates for interacting with soluble electron acceptors.
In the present work we describe the functional consequences of basic amino acid substitutions in this C-terminal region of PsaD, and of a 36-amino acid deletion starting at residue 105.
RESULTS
Description of the Mutations
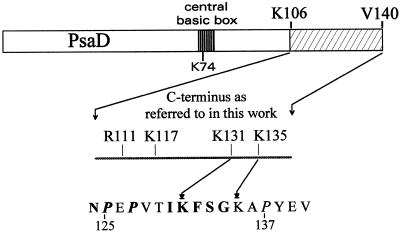
In the course of studying electrostatic interactions between PSI and Fd, numerous mutations of positively charged amino acids have been designed on the PsaD gene of the cyanobacterium Synechocystis (Hanley et al., 1996). Some of these mutations, located in the C terminus of this protein (Fig. 1), are more extensively described in the present work. Two are single-site changes, R111C (D10) and K117A (DE). One is a double mutant K131A/K135A (DF). The final one is a deletion of the complete C-terminal sequence starting at residue 105 (DΔCt). This latter drastic change deletes 36 amino acids, including four lysines, three arginines, and four glutamates, thus inducing a net loss of three positive charges. It resulted from a single base addition during the mutagenesis process, generating a frame shift and an early stop codon. All mutations were controlled by directly sequencing the gene from the corresponding Synechocystis strains after PCR amplification. The psaD gene in DΔCt appeared thus not only shortened, but also modified at residues 102 (Val to Gly) and 104 (Pro to Arg). The sequence of this truncated PsaD is now 104 amino acids long, ending with DGGFR. To control the absence of any further processing, this drastically modified protein was purified and cleaved with V8 protease (EC 3.4.21.19) as described previously (Rousseau et al., 1993). Peptide mapping followed by extensive sequencing revealed unprocessed N and C termini exactly fitting the DNA sequence.
Figure 1.
Location of PsaD residues from Synechocystis 6803 discussed in the present paper. The C terminus dashed area fits with the 36 amino acids deletion of the ΔCt mutation. Bold letters in the bottom sequence are for amino acids strictly conserved in cyanobacteria, or replaced homologous residues.
Physiology of the Mutant Strains
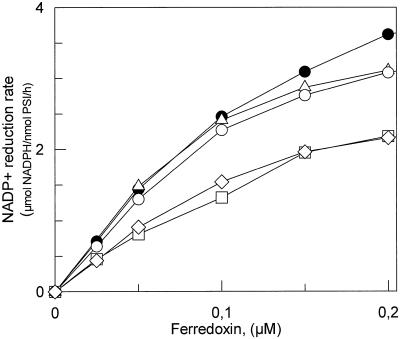
To study the physiological impact of mutations we measured the doubling time of the mutant strains under photoautotrophic growth conditions (Table I). Cells were grown in BG11 medium at 32°C and low light intensity (20 μmol m−2 s−1). Chloramphenicol, the selection marker for all mutants, was kept at a low level (5 μg/mL). Growth was monitored from absorption measurements at 730 nm. The double Lys mutant (DF) was the only strain to exhibit a considerably increased doubling time. This impaired growth almost disappeared when the temperature was decreased from 32°C to 28°C. NADP+ photoreduction mediated by Synechocystis Fd was also analyzed using heterologous Fd-NADP+ reductase (FNR, EC 1.18.1.2) from spinach. To quantify the effect of PSI mutations on the global electron transfer to NADP+ we first designed conditions where the limiting step was the electron transfer between PSI and Fd. This was achieved using artificial electron donors, PSI at 0.2 μm and Fd at 0.05 μm. At this low Fd concentration the rates of NADP+ photoreduction exhibited an almost linear relationship to the Fd concentration with all PSI studied (Fig. 2). The reduction rate catalyzed by D10 and DΔCt PSI showed a 40% decrease compared with wild type (WT) PSI (Table I). On the contrary, DE and DF PSI showed reduction rates identical to WT (Fig. 2). These rates closely paralleled the binding constants for Fd measured by flash absorption spectroscopy (Table II). In comparison, the PSI deleted of PsaD subunit (Hanley et al., 1996) still retained 20% of the WT rate using the same concentrations of proteins (data not shown). Increasing the Fd concentration to 0.4 μm increased the WT value from 1.4 to 6 to 7 μmol NADPH/nmol PSI/h. The use of the physiological electron donor cytochrome c6 (15 μm) allows a maximum value of 26 μmol NADPH/nmol PSI/h to be reached, very close to that found in a spinach homologous system (Aliverti et al., 1997). Variable NADP+ reduction rates ranging from 26 to 60 μmol NADPH/nmol PSI/h have been previously reported for Synechocystis thylakoids or purified PSI using heterologous spinach Fd (Xu et al., 1994c; Chitnis et al., 1996, 1997; van Thor et al., 1999).
Table I.
Description and physiological characteristics of the PsaD mutants
| Strain | Mutation | Doubling Time | Chlorophyll in Trimers | Fd-Mediated NADP+ Photoreduction Ratea |
|---|---|---|---|---|
| h | % | μmol NADPH/nmol PSI/h | ||
| WT | None | 40 | 46 | 1.44 |
| D10 | R111C | 38 | 72 | 0.81 |
| DE | K117A | 42 | 67 | 1.48 |
| DF | K131A/K135A | 58 | 41 | 1.30 |
| DΔCt | Δ105–140 | 36 | 65 | 0.91 |
Cell growth was followed by optical density measurements of the cell suspension at 730 nm.
Rates of NADP+ photoreduction were measured at a Fd concentration of 0.05 μm.
Figure 2.
Rate of NADP+ photoreduction by purified PSI reaction centers at varying Fd concentrations. Assay conditions were as described in “Material and Methods.” ●, WT PSI; □, D10 PSI; ▵, DE PSI; ○, DF PSI; ⋄, DΔCt PSI.
Table II.
Dissociation constants and kinetic parameters of PSI mutants
| PsaD Mutation | Kda | Kd mut /Kd wt | Submicrosecond Phase Amplitudeb | 20-μs Phase Amplitudeb |
|---|---|---|---|---|
| μm | ||||
| WT | 0.2 | 1 | 50% | 40% |
| D10 | 0.9–1.1 | 4.5–5.5 | 20% | 80% |
| DE | 0.2 | 1 | 35% | 50% |
| DF | 0.2–0.4 | 1–2 | 50% | 40% |
| DΔCt | 0.8–0.9 | 4.0–4.5 | 40% | 55% |
Average values from three complete Kd determinations.
Relative to the sum of the amplitudes of the three first-order Fd reduction phases.
Polypeptide Composition and Stability
Mutated PSI reaction centers have been isolated as monomers and trimers. The high-pressure method used for breaking the cells results in small membrane fragments. This could be impeding the in vitro reorganization between monomers and trimers at low salt concentration (Kruip et al., 1994). Under such conditions we found a proportion of 46% of the PSI chlorophyll in the trimers solubilized from WT membranes. The trimeric form was significantly increased in three mutants, D10, DE, and DΔCt (Table I), possibly due to the disappearance of accessible positive charge(s) at the surface of PSI.
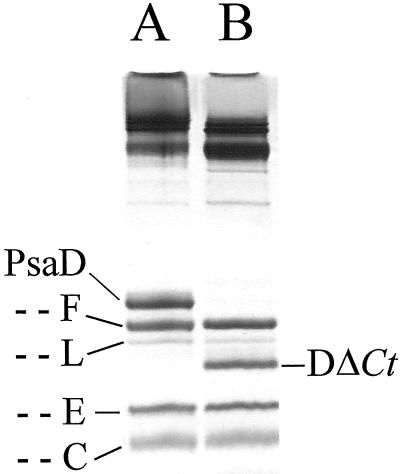
Pure monomeric PSI from the different mutants were subjected to denaturing urea/SDS gel electrophoresis. All except DΔCt exhibited a WT polypeptide pattern. The truncated PsaD of DΔCt was unambiguously identified by antibodies. It migrates as expected between PsaL and PsaE (Fig. 3), with an apparent Mr of 12,500, in good agreement with the calculated value of 12,699. In contrast, WT PsaD had a retarded migration with an apparent Mr of 18,000 compared with the predicted value of 15,502, as previously observed (Chitnis et al., 1995; Jin et al., 1999). It is likely that a highly stable conformation exists in the C terminus, preventing a complete unfolding of the polypeptide and a size-proportional migration. The decreased level of staining of this truncated PsaD is in agreement with what is expected from the size and positive charge decreases, arguing for a normal level of integration of the modified subunit. In addition, a low level of fast back-reaction of P700+ was observed at 820 nm (approximately 3% of the total amplitude; t1/2 = 1 ms). This rapid reaction has been shown to originate from charges recombination between P700+ and center Fx−, due to an alteration of the [4Fe-4S] centers of PsaC (Golbeck and Cornelius, 1986; Li et al., 1991). It is normally not observed when PsaC is protected from the solvent by a normal integration of PsaD subunit. These arguments are in favor of the truncated PsaD being incorporated in almost all reaction centers. DΔCt PSI most often exhibits a lower content of PsaL, at about 50% the WT amount. In addition, when the mannitol of the original preparation procedure is omitted during the washing steps (Rögner et al., 1990), this subunit becomes barely detectable.
Figure 3.
Comparative electrophoresis of denatured WT and DΔCt PSI. Equal amounts of 125 pmol of PSI reaction centers were loaded for each sample. A is WT PSI and B is DΔCt PSI. Gels were Coomassie Blue stained.
PSI from DF mutant was found to be quite unstable compared with WT PSI upon a long incubation (30 min) at 40°C in the gel loading buffer, exhibiting a progressive degradation of PsaF and PsaD. Denaturation for gel electrophoresis was thus performed at 95°C. This increased degradation rate was also observed in the absence of the denaturing buffer over a longer time scale (several hours at 37°C, data not shown).
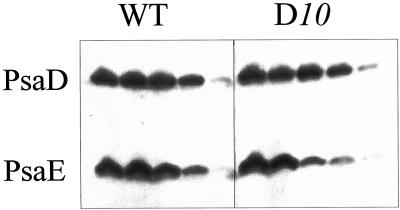
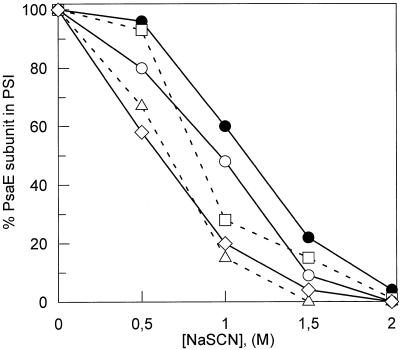
The stability of association of the mutated PsaD polypeptides has been analyzed by their resistance to removal by a chaotropic salt. Photosynthetic membranes were incubated in the presence of increasing concentrations of NaSCN (Lagoutte and Vallon, 1992), and the remaining fraction of the polypeptide still bound to the membranes after washing was probed and quantified by western blotting, as illustrated for WT and D10 membranes (Fig. 4). These values were corroborated by dot-blot quantifications of the polypeptides extracted in the supernatants. At a concentration of 1.5 m NaSCN, there is still 50% PsaD bound to the WT membranes, 60% in D10, and 40% for the two lysines mutants (DE and DF), but only 25% for DΔCt. The stability of truncated PsaD is largely impaired. A decreased stability of PsaE induced by PsaD mutations was also generally observed for all mutants and is illustrated in Figure 5 as a function of the chaotropic salt concentration. At 1 m NaSCN, 60% of PsaE was still bound to the WT membranes, whereas values of 28%, 10%, 48%, and 20% were observed for D10, DE, DF, and DΔCt, respectively. The more distant mutations in PsaD sequence (K131/K135) are less damaging for the stability of PsaE.
Figure 4.
Western blots of PSI polypeptides after extraction by increasing concentrations of NaSCN. For each sample, the five successive lanes from left to right are a membrane control, followed by comparable samples after a 30-min incubation at 0°C in the presence 0.5, 1.0, 1.5, and 2.0 m NaSCN. Blots have been probed using a mixture of anti-PsaD and anti-PsaE antibodies. A luminescent detection system was used to expose a light-sensitive film.
Figure 5.
Plots of the PsaE subunit bound to membrane-inserted PSI as a function of NaSCN concentration. Amounts of PsaE subunit remaining associated to PSI after NaSCN washing were quantified by western blotting followed by enhanced chemiluminescence detection and densitometric measurement of an exposed light-sensitive film. For each sample, remaining PsaE is expressed as the percent of the initial amount present after the control washing without chaotropic salt. ●, WT PSI; □, D10 PSI; ▵, DE PSI; ○, DF PSI; ⋄, DΔCt PSI.
Fd Photoreduction
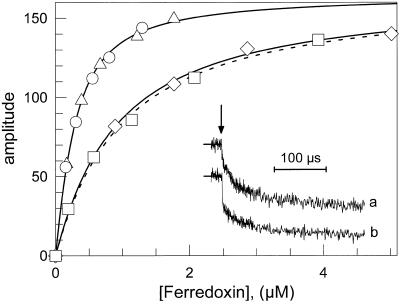
As already described in detail (Sétif and Bottin, 1994, 1995), the electron transfer between WT PSI and Fd (at pH 8) occurs mainly through three first-order kinetic phases, when measured by flash-induced absorption changes between 480 and 580 nm. The halftimes of these phases are 500 ns, 20 μs, and 100 μs. The total amplitude of these phases of intracomplex electron transfer (respectively, 50%, 40%, and 10% when measured at 580 nm) was directly related to the amount of PSI-Fd complex formed in the dark before light excitation (Fig. 6, inset). It varies according to the total Fd concentration, following a simple binding equilibrium:
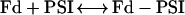 |
The amplitude plotted in Figure 6 is the sum of the sub-μs and 20-μs phases amplitudes resulting from kinetic analysis. The 100-μs halftime phase was not taken into account since it is of small amplitude (at 580 nm) and is difficult to differentiate from the second-order reaction, a process occurring between PSI and Fd unassociated before flash excitation. This is particularly true at high Fd concentration (needed for studying low affinity mutants). For all PSI described below, the halftimes of the two fastest first-order reduction phases were found unchanged (500 ns and 20 μs), whereas the respective amplitudes of these phases depend upon the nature of the mutation. To compare the effect of a mutation on KD, the dissociation constants for mutated PSI (KD mut) have been also expressed relative to KD for WT PSI (KD wt).
Figure 6.
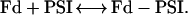 |
None of the mutated PSI described in the present work showed modification in the electron transfer rate to Fd. However, D10 mutant PSI exhibited a very reduced amplitude of the sub-μs phase (20%), compensated by a large 20-μs phase amplitude (80%; Fig. 6, inset). Mutants DΔCt and DE had also a decreased sub-μs phase amplitude, but to a smaller extent (40% and 35%, respectively; Table II).
Although DE and DF mutants show affinities for Fd identical to WT PSI, D10 and DΔCt mutants have affinities for Fd largely decreased (approximately 1 μm; Table II). This 5-fold increase of the KD indicates a strong effect if compared, for example, with results of most single mutations performed on PsaD or on Fd (Hanley et al., 1996; Guillouard et al., 2000). Such strong effects on KDs have been already described, but most often are the result of multiple mutations on Fd (Guillouard et al., 2000) or of complete subunit deletions on PSI (Barth et al., 1998).
DISCUSSION
The peripheral PsaD subunit is proposed to serve at least two distinct functions in the PSI complex. It was first attributed a key structural role in the completion of the PSI complex, being the first nuclear-encoded subunit of this membranous complex synthesized upon illumination (Nechustai and Nelson, 1985). This polypeptide is likely to be interacting with many different PSI subunits (Jansson et al., 1996). Well-established interactions are with the C-terminal part of the large PsaB core subunit (Xu and Chitnis, 1995), with PsaC, protecting FeS clusters FA and FB from the solvent (Oh-Oka et al., 1989; Li et al., 1991; Mannan et al., 1994; Naver et al., 1996), with PsaE (Andersen et al., 1992; Cohen et al., 1993), and also with PsaL, indirectly participating to the formation of trimers (Chitnis and Chitnis, 1993). The DΔCt PSI analyzed in the present work is missing the last 36 amino acids of PsaD, but still has a phenotype very close to WT regarding the general physiology and polypeptide composition. We assume that the structural functions of PsaD are supported to a large extent by the first 70% of the polypeptide chain. In agreement with this assumption, an α-helix region, previously predicted with more confidence in the middle or N-terminal part of PsaD sequence (Rousseau, 1992; Xia et al., 1998; Jin et al., 1999), has been recently internally located in the crystal structure (Klukas et al., 1999). Also in accordance with this result is a previous report that the highly basic sequence in the middle of the protein is important for the stable association of PsaD to the PSI core (Chitnis et al., 1997). In situ labeling of PSI by n-hydroxysuccinimidobiotin failed to target the Lys residues of this basic central region, whereas they become accessible in the isolated soluble form of PsaD (Xu et al., 1994b; von Leoprechting et al., 1998). Using fluorescein isothiocyanate, another primary amines reagent (Rousseau et al., 1993), it was also observed that the two residues predominantly labeled are K26 and K106, which are outside the central sequence (B. Lagoutte, unpublished data). This lack of accessibility of the central region of PsaD corroborates its internal location. Some subunits in the DΔCt PSI mutant have however a decreased stability compared with the WT PSI. In the absence of any treatment, PsaL appeared less stably associated in the monomer, with a mean 2-fold decreased level. In addition, PsaD and PsaE are easier to extract by chaotropic salts. A part of the deleted 36 amino acids in the C terminus could thus play the role of a structural lock, encompassing a large distance between PsaL and PsaE on the top of the cytoplasmic surface of PSI. Together with DΔCt, mutations at positions R111 and K117 are more destabilizing for PsaE. This could indicate that these two basic residues are in a close vicinity to PsaE subunit. Despite the loss of stability of some peripheral subunits in the DΔCt PSI, the presence of trimers is significantly increased. It is likely that the overall stability of the peripheral subunits is improved in the trimers, as reflected by the important decrease in the turnover of these subunits in this PSI form (Chitnis and Nelson, 1992b). Two other mutated PSIs described in the present work, R111C and K117A, have similarly higher levels of trimers. They have in common the loss of one basic accessible residue, which can decrease some electrostatic repulsive effect proposed as a limiting parameter in the trimer formation (Kruip et al., 1994). If this is the case, the sequence including R111 and K117, which we already suggested to be closely located to PsaE, should also be close to the center of the trimer. The very end of PsaD can be attributed to a more internal location, as lysines 131 and 135 are not accessible to chemical labeling (Xu et al., 1994b; Jin et al., 1999). In agreement with the proposed repulsive effect of charges in the formation of trimers, the suppression of lysines 131 and 135, which are apparently shielded from the external medium, does not lead to an increased presence of trimers (DF mutant).
Lys 131 is strictly conserved in cyanobacteria, as is also a part of the surrounding sequence between P125 and P137 (Fig. 1). This terminal sequence most often includes a second basic residue equivalent to Lys 135, and the first Pro is always in a P Z P motif, with Z being a hydrophilic residue (Golbeck, 1994). This Pro-rich sequence, predicted to generate β-turns, is likely to adopt a specific and stable conformation at the origin of the abnormal electrophoretic migration of WT PsaD. An impaired migration is no longer observed for DΔCt PsaD. Supporting this structural hypothesis, a C-terminal peptide obtained from Mastigocladus laminosus PsaD starting at R119 still migrates in denaturing gel electrophoresis with an apparent Mr of 5,600 instead of 3,600, whereas the remaining N-terminal fragment recovers a normal migration (Jin et al., 1999). Lys 131 and 135 in this structure would face internal acidic residues in PSI, thus providing an electrostatic stabilization of the C terminus. In the absence of these two lysines, DF PSI appeared far more unstable than WT PSI at moderate temperatures. This could explain the impaired growth of DF strain when incubated above 30°C as a result of a destabilization and increased turnover of PSI. A deletion of PsaD C terminus would then be less detrimental to the cell physiology than a disorganized structure in this part of the protein, as illustrated by the normal growth of the DΔCt strain.
Another important and well-established function of PsaD is to provide specific sequences for Fd electrostatic guiding and docking on PSI (Zanetti and Merati, 1987; Zilber and Malkin, 1988; Barth et al., 1998). This function is at least partly controlled by some residues in the C terminus. Lys 106 has been shown to be in electrostatic interaction with Glu 93 of Fd (Lelong et al., 1994). Its substitution for Ala was shown to induce a moderate 2-fold decrease of the affinity for Fd, whereas a cystein decreased this affinity 5-fold (Hanley et al., 1996). This important effect was interpreted as being due to the presence of the unprotonated form of this new Cys at pH 8.0. Arg 111, the next basic residue in PsaD sequence, is still in a region very accessible to labeling and proteases (Xu et al., 1994b; Jin et al., 1999). The replacement of this positive charge by a cystein induces also a similarly important 5-fold decrease of the affinity for Fd (Table II), together with an important loss of the sub-μs reduction phase. This Arg residue, as is Lys 106, is highly conserved and could play an equivalent and additive role in keeping Fd in the most favorable docking configuration on PSI. Suppression of the next positive charge on K117 does not significantly modify the KD. It is surprising that the large deletion of the last 36 amino acids does not induce a stronger effect on the KD than the R111C mutation, which could be partly due to the likely accessible Arg at the end of the new truncated C terminus. The kinetic analysis of Fd reduction by PSI shows that if the halftimes of the electron transfer reactions are not modified by the mutations on PsaD, the relative amplitude of the first-order phases vary greatly. This is illustrated by the D10 mutant in which the sub-μs phase represents only 20% of the absorption change, whereas it is 50% with DF mutant or WT PSI (Table II; Fig. 6, inset). It has been previously proposed that the complexity of the first-order kinetics originate from Fd molecules bound in different position within the docking site on PSI (Sétif and Bottin, 1995). In that respect, D10 mutant provides a good example of modifications in the equilibrium between subpopulations of the Fd-PSI complex, resulting from single-site mutation. Furthermore, unchanged halftimes of the electron transfer reactions indicate that the distance between the terminal [Fe-S] clusters of PSI (FA and FB) and Fd is not changed in the mutated PSI
The decreased Fd affinity of mutants R111C and DΔCt is also reflected by a decreased rate of NADP+ photoreduction (about 40%) measured in an in vitro reconstituted system. This almost 2-fold decrease does not impair normal cell growth. It can be due to the fact that even in WT cells, NADP+ reduction may not be the more limiting step in cell growth. An increased level of the FNR mRNA has also been reported in a PsaE-deleted strain. This was expected to compensate for the observed in vitro decrease of NADP+ photoreduction (van Thor et al., 1999). Using the same in vitro system, a PsaD-deleted PSI was still capable of NADP+ photoreduction at 20% of the WT rate, a somewhat higher value than the very low rate previously reported using heterologous Fd from spinach (Chitnis et al., 1996). This remaining low activity can be explained by the residual slow second-order Fd reduction observed by flash absorption spectroscopy (Barth et al., 1998). The growth rate of the corresponding PsaD deleted cells is severely affected unless Glc is added to the culture medium (Chitnis et al., 1989; Hanley et al., 1996). It has been briefly mentioned that Synechocystis Fd was 50% less efficient than spinach Fd in an in vitro NADP+ photoreduction experiment using spinach FNR (Xu et al., 1994c). This can explain the large range of values obtained when comparing experiments performed with spinach Fd, Synechocystis Fd, or Synechocystis flavodoxin. We conclude that a homologous Fd/FNR system is important for the maximum rate of NADP+ photoreduction, but that a homologous Fd/PSI is more adapted to the characterization PSI mutants.
Compared with cyanobacteria, PsaD subunits from plants and algae have an accessible extra N-terminal sequence of about 25 amino acids, in addition to the transit peptide. This additional sequence could be required for an efficient cleavage of the leader peptide (Cohen et al., 1992), a process that does not occur in cyanobacteria for this PSI subunit. The sequence of the C terminus is also somewhat different, and it has been proved to be a strict requirement for the stable integration of PsaD subunit in PSI (Cohen et al., 1995). An opposite situation seems to prevail in cyanobacteria where the N-terminal part is more important for integration than the dispensable C terminus (Chitnis and Nelson, 1992a; Cohen et al., 1995). Results obtained in the present work with DΔCt PSI are in good agreement with these previous studies. However, once PsaD is inserted in PSI, the terminal part of the polypeptide probably has a stabilizing function in cyanobacteria.
MATERIAL AND METHODS
Biological Samples
Recombinant Fd was overexpressed in Escherichia coli essentially as previously described (Guillouard et al., 2000). In brief, the fed1 gene of Synechocystis sp. PCC 6803 (Synechocystis) cloned into a pRSET5A vector was expressed in E. coli BL21 (DE3) pLysS. Cells were grown at 27°C in M9 minimum medium (Sambrook et al., 1989) supplemented with 100 μg/mL of ampicillin. Growth was maintained for 65 h in the presence of 2 mm Glc and 8 mm Gal used as inducer. Harvested cells were broken using a cell disrupter (Z model, Constant Systems, Warwick, UK) at a pressure of 1 kbar. Fd was purified from the whole-cell extract by a three-step procedure already described (Lelong et al., 1995), yielding 5 to 10 mg of purified recombinant Fd per liter of E. coli-induced cells.
Cytochrome c6 was purified from Synechocystis using stepwise ammonium sulfate precipitations followed by ion-exchange chromatography (Hervás et al., 1992). Spinach FNR was from Sigma (St. Louis).
Mutant and WT Synechocystis cells were grown in BG11 medium (Rippka et al., 1979) at 32°C and low light intensity (20 μmol m−2 s−1). Carbon dioxide was added to air at a concentration of 5% (v/v). The growth of liquid cultures was monitored by measuring the optical density at 730 nm.
Pelleted cells were resuspended in the following high osmolarity buffer containing 20 mm MES, pH 6.5, 10 mm MgCl2, and 1 m Suc (Rögner et al., 1990), and were broken in a cell disrupter at a pressure of 1.4 kbar (Z model, Constant Systems). For the washing steps, Suc was replaced by 0.5 m mannitol. Monomeric and trimeric PSI, solubilized by n-dodecyl β-d-maltoside (β-DM), were gradient purified according to the original method (Rögner et al., 1990). Quantification of chlorophyll (Chl) in the monomeric and trimeric forms was directly made on gradient fractions. Flash-absorption spectroscopy was performed using monomeric PSI further purified by anion-exchange chromatography on a mono Q column (Pharmacia Biotech, Piscataway, NJ; Rögner et al., 1990; Guillouard et al., 2000). This removed remaining carotenoids and a small PSII fraction, which never represented more than 5% of PSI and was thus neglected in the determination of the trimer to monomer ratio.
Site-Directed Mutagenesis
Site mutations in the isolated psaD gene were performed using the method of Kunkel (Kunkel, 1985) and the previously described M13 construct mpJCD (Hanley et al., 1996). The mutated gene was then reintroduced by direct transformation of the PsaD minus strain KDb3 (Hanley et al., 1996). Selection of transformants was carried out by serial dilution in the presence of increasing chloramphenicol concentrations (from 1–30 μg/mL). Once resistance to 30 μg/mL chloramphenicol was established, the selection procedure was continued through further eight to 10 rounds of liquid subculture in the absence of Glc.
Direct Sequencing of PCR-Amplified Genomic DNA
Genomic DNA from liquid cultures of Synechocystis variants was isolated and used for control sequencing. A 523-bp fragment encompassing the full psaD sequence was amplified using the PCR procedure, purified by electroelution (Hanley et al., 1996), and sequenced by the classical dideoxy termination method.
Protein Electrophoresis and Western Blotting
Protein electrophoresis were performed on mini-slab gels (Bio-Rad, Hercules, CA) using the Tris/Tricine [N-[2-hydroxy-1,1-Bis(hydroxymethyl)ethyl]glycine] buffer system (Schägger and von Jagow, 1987). The concentration of P700 was measured by flash-absorption spectroscopy at 820 nm in all PSI samples, and a constant amount of 125 pmol of P700 was loaded for each sample after a 3-min dissociation in the loading buffer at 95°C. Electroblotting of the proteins was made overnight at 4°C using Immobilon P membranes (Amersham, Buckinghamshire, UK) and a constant current of 10 mA. PsaD and PsaE subunits were probed with polyclonal antibodies (Rousseau et al., 1993) and revealed by an alkaline phosphatase anti-mouse IgG conjugate (Sigma) and a luminescent substrate (Immun-star, Bio-Rad, Hercules, CA). Densitometric analysis of the films (hyperfilm MP, Amersham) was made using a Gel Doc 1000 system (Bio-Rad).
Chaotropic Salt Extraction of Photosynthetic Membranes
For chaotropic salt extractions, crude membranes were suspended in a 100 mm Tris buffer, pH 8.0, at a Chl concentration of 0.1 mg/mL. Each sample containing a total amount of 0.2 mg of Chl was incubated for 30 min on ice in the presence of the appropriate concentration of NaSCN. Membranes were pelleted for 5 min at 5,000g and were quickly washed with the same volume of buffer in the absence of chaotropic salt. Pellets were solubilized in the electrophoretic loading buffer at a final concentration of 1 μg Chl/μL and were heat denatured for 3 min at 95°C. Supernatants were further diluted 10 times and bovine serum albumin (Sigma, fraction V) was added at a final concentration of 10 μg/mL to avoid non-specific adsorption. Quantitative dot blots using Bio-Dot apparatus (Bio-Rad) were on nitrocellulose membrane (Bio-Rad). Each well was loaded with 100 μL of sample of the soluble fractions. Specific detection and quantification of PsaD and PsaE were as described for western blotting.
NADP+ Photoreduction Assay
Reaction medium used for the reduction of NADP+ was the following: 50 mm Tricine, pH 8.0, 30 mm NaCl, 5 mm MgCl2, 4 mm Na-ascorbate (Asc), 100 μm 2, 6-dichlorophenolindophenol (DCPIP), and 1 mm NADP+. PSI was 0.2 μm, FNR was 0.5 μm, and Fd varied from 0.025 to 0.4 μm. Measurements were made on a total volume of 1.2 mL in a 1-cm cuvette thermostated at 20°C and constantly stirred. Illumination was provided at the top of the cuvette using an optical fiber system equipped with a yellow filter (band pass 500–650 nm) at a light intensity of 5 mmol m−2 s−1. The spectrophotometer detector was protected from actinic light by highly selective filters transmitting at 340 nm. Measurements were made on a 3-min time scale, during which the rate of NADPH formation remained constant for all experiments.
Flash-Absorption Spectroscopy
Measurements were made in a square 1-cm cuvette at 296 K as previously described (Sétif and Bottin, 1994, 1995). PSI was diluted to 0.15 to 0.2 μm in 20 mm Tricine/NaOH, pH 8.0, containing 0.03% (w/v) β-DM, 2 mm asc, 10 μm DCPIP, 30 mm NaCl, and 5 mm MgCl2. PSI excitation was provided by a frequency-doubled yttrium-aluminum-garnet laser pumping a dye laser (wavelength, 695 nm; pulse duration, 6 ns). Absorption changes were measured at 820 or 580 nm (repetition rate, 0.2 s−1; time resolution, 200 ns). The P700 content of a sample was calculated directly from the amplitude of the photoinduced absorption changes at 820 nm using an absorption coefficient of 6,500 m−1 cm−1 (Mathis and Sétif, 1981). Kinetics of Fd reduction were measured at 580 nm by subtracting the kinetics observed with and without Fd as described by Sétif and Bottin (1994). Each kinetic trace is the average of 64 measurements. Amplitudes and halftimes of the first-order phases were obtained by fitting data with exponential components using a Marquart algorithm. The total amplitude of the three first-order phases was estimated by measuring the amplitude of the absorption change 300 μs after the laser flash excitation as described by Sétif and Bottin (1994) and used to calculate the relative amplitudes of the first-order phases.
ACKNOWLEDGMENTS
We wish to thank C. Sigalat and F. Haraux for access to their spectrophotometer specifically designed for measuring absorption changes at 340 nm. We also thank Véronique Mary for skillful technical assistance.
LITERATURE CITED
- Aliverti A, Livraghi A, Piubelli L, Zanetti G. On the role of the acidic cluster Glu 92–94 of spinach ferredoxin I. Biochim Biophys Acta. 1997;1342:45–50. doi: 10.1016/s0167-4838(97)00079-4. [DOI] [PubMed] [Google Scholar]
- Andersen B, Koch B, Scheller HV. Structural and functional analysis of the reducing side of photosystem I. Physiol Plant. 1992;84:154–161. [Google Scholar]
- Barth P, Lagoutte B, Sétif P. Ferredoxin reduction by photosystem I from Synechocystis sp. PCC 6803: toward an understanding of the respective roles of subunits PsaD and PsaE in ferredoxin binding. Biochemistry. 1998;46:16233–16241. doi: 10.1021/bi981379t. [DOI] [PubMed] [Google Scholar]
- Bottin H, Lagoutte B. Ferredoxin and flavodoxin from the cyanobacterium Synechocystis sp PCC 6803. Biochim Biophys Acta. 1992;1101:48–56. doi: 10.1016/0167-4838(92)90465-p. [DOI] [PubMed] [Google Scholar]
- Chitnis PR, Chitnis VP, Xu Q, Jung Y-S, Yu L, Golbeck JH. Mutational analysis of photosystem I polypeptides. In: Mathis P, editor. Photosynthesis: From Light to Biosphere. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 17–22. [Google Scholar]
- Chitnis PR, Nelson N. Assembly of two subunits of the cyanobacterial photosystem I on the n-side of thylakoid membranes. Plant Physiol. 1992a;99:239–246. doi: 10.1104/pp.99.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis PR, Nelson N. Biogenesis of photosystem I: subunit PsaE is important for the stability. In: Akoyunoglou A, editor. Chloroplast Biogenesis. New York: Plenum Press; 1992b. pp. 285–290. [Google Scholar]
- Chitnis PR, Reilly PA, Nelson N. Insertional inactivation of the gene encoding subunit II of photosystem I from the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem. 1989;264:18381–18385. [PubMed] [Google Scholar]
- Chitnis VP, Chitnis PR. PsaL subunit is required for the formation of photosystem I trimers in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 1993;336:330–334. doi: 10.1016/0014-5793(93)80831-e. [DOI] [PubMed] [Google Scholar]
- Chitnis VP, Jung Y-S, Albee L, Golbeck JH, Chitnis PR. Mutational analysis of photosystem I polypeptides: role of PsaD and the lysyl 106 in the reductase activity of photosystem I. J Biol Chem. 1996;271:11772–11780. doi: 10.1074/jbc.271.20.11772. [DOI] [PubMed] [Google Scholar]
- Chitnis VP, Ke A, Chitnis PR. The PsaD subunit of photosystem I: mutations in the basic domain reduce the level of PsaD in the membranes. Plant Physiol. 1997;115:1699–1705. doi: 10.1104/pp.115.4.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Y, Chitnis VP, Nechustai R, Chitnis PR. Stable assembly of PsaE into cyanobacterial photosynthetic membranes is dependent on the presence of other accessory subunits of photosystem I. Plant Mol Biol. 1993;23:895–900. doi: 10.1007/BF00021544. [DOI] [PubMed] [Google Scholar]
- Cohen Y, Nelson N, Chitnis PR, Nechustai R. The carboxyl-terminal region of the spinach PsaD subunit contains information for its specific assembly into plant thylakoids. Photosynth Res. 1995;44:157–164. doi: 10.1007/BF00018306. [DOI] [PubMed] [Google Scholar]
- Cohen Y, Steppuhn J, Herrmann RG, Yalovsky S, Nechustai R. Insertion and assembly of the precursor of subunit II into the photosystem I complex may precede its processing. EMBO J. 1992;11:79–85. doi: 10.1002/j.1460-2075.1992.tb05030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Hippler M, Sétif P, Jacquot J-P, Rochaix J-D. The PsaC subunit of photosystem I provides an essential lysine residue for fast electron transfer to ferredoxin. EMBO J. 1998;17:849–858. doi: 10.1093/emboj/17.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golbeck JH. Photosystem I in cyanobacteria. In: Bryant DA, editor. Advances in Photosynthesis: The Molecular Biology of Cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 319–360. [Google Scholar]
- Golbeck JH, Cornelius JM. Photosystem I charge separation in the absence of centers A and B: I. Optical characterization of center A2 and evidence for its association with a 64-kDa protein. Biochim Biophys Acta. 1986;849:16–24. doi: 10.1016/0005-2728(86)90092-7. [DOI] [PubMed] [Google Scholar]
- Guillouard I, Lagoutte B, Moal G, Bottin H. Importance of the region including aspartates 57 and 60 of ferredoxin on the electron transfer complex with photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. Biochem Biophys Res Commun. 2000;271:647–653. doi: 10.1006/bbrc.2000.2687. [DOI] [PubMed] [Google Scholar]
- Hanley J, Sétif P, Bottin H, Lagoutte B. Mutagenesis of photosystem I in the region of the ferredoxin cross-linking site: modifications of the positively charged amino acids. Biochemistry. 1996;35:8563–8571. doi: 10.1021/bi960399x. [DOI] [PubMed] [Google Scholar]
- Hervás M, De la Rosa MA, Tollin G. A comparative laser-flash absorption spectroscopy study of algal plastocyanin and cytochrome c552 photooxidation by photosystem I particles from spinach. Eur J Biochem. 1992;203:115–120. doi: 10.1111/j.1432-1033.1992.tb19835.x. [DOI] [PubMed] [Google Scholar]
- Jansson S, Andersen B, Scheller HV. Nearest-neighbor analysis of higher-plant photosystem I holocomplex. Plant Physiol. 1996;112:409–420. doi: 10.1104/pp.112.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Sun J, Chitnis PR. Structural features and assembly of the soluble overexpressed PsaD subunit of photosystem I. Biochim Biophys Acta. 1999;1410:7–18. doi: 10.1016/s0005-2728(98)00169-8. [DOI] [PubMed] [Google Scholar]
- Kitmitto A, Mustafa AO, Holzenburg A, Ford RC. Three-dimensional structure of higher plant photosystem I determined by electron crystallography. J Biol Chem. 1998;273:29592–29599. doi: 10.1074/jbc.273.45.29592. [DOI] [PubMed] [Google Scholar]
- Klukas O, Schubert W-D, Jordan P, Krauβ N, Fromme P, Witt HT, Saenger W. Photosystem I, an improved model of the stromal subunits PsaC, PsaD, and PsaE. J Biol Chem. 1999;274:7351–7360. doi: 10.1074/jbc.274.11.7351. [DOI] [PubMed] [Google Scholar]
- Kruip J, Bald D, Boekema EJ, Rögner M. Evidence for the existence of trimeric photosystem I complexes in thylakoid membranes from cyanobacteria. Photosynth Res. 1994;40:279–286. doi: 10.1007/BF00034777. [DOI] [PubMed] [Google Scholar]
- Kruip J, Chitnis PR, Lagoutte B, Rögner M, Boekema EJ. Structural organization of the major subunits in cyanobacterial photosystem I: localization of subunits PsaC, -D, -E, -F and -J. J Biol Chem. 1997;272:17061–17069. doi: 10.1074/jbc.272.27.17061. [DOI] [PubMed] [Google Scholar]
- Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagoutte B, Vallon O. Purification and membrane topology of PSI-D and PSI-E, two subunits of the photosystem I reaction center. Eur J Biochem. 1992;205:1175–1185. doi: 10.1111/j.1432-1033.1992.tb16888.x. [DOI] [PubMed] [Google Scholar]
- Laudenbach DE, Reith ME, Strauss NA. Isolation, sequence analysis, and transcriptional studies of the flavodoxin gene from Anacystis nidulans R2. J Bacteriol. 1988;170:258–265. doi: 10.1128/jb.170.1.258-265.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelong C, Sétif P, Bottin H, André F, Neumann J-M. 1H and 15N NMR sequential assignment, secondary structure, and tertiary fold of [2Fe-2S] ferredoxin from Synechocystis sp. PCC 6803. Biochemistry. 1995;34:14462–14473. doi: 10.1021/bi00044a024. [DOI] [PubMed] [Google Scholar]
- Lelong C, Sétif P, Lagoutte B, Bottin H. Identification of the amino acids involved in the functional interaction between photosystem I and ferredoxin from Synechocystis sp. PCC 6803 by chemical cross-linking. J Biol Chem. 1994;269:10034–10039. [PubMed] [Google Scholar]
- Li N, Zhao J, Warren PV, Warden JT, Bryant DA, Golbeck JH. PsaD is required for the stable binding of PsaC to the photosystem I core protein of Synechococcus sp. PCC 6301. Biochemistry. 1991;30:7863–7872. doi: 10.1021/bi00245a028. [DOI] [PubMed] [Google Scholar]
- Mannan RM, Pakrasi HB, Sonoike K. The PsaC protein in necessary for stable association of the PsaD, PsaE, and PsaL proteins in the photosystem I complex: analysis of a cyanobacterial mutant strain. Arch Biochem Biophys. 1994;315:68–73. doi: 10.1006/abbi.1994.1472. [DOI] [PubMed] [Google Scholar]
- Mathis P, Sétif P. Near infra-red absorption spectra of the chlorophyll a cations and triplet state in vitro and in vivo. Isr J Chem. 1981;21:316–320. [Google Scholar]
- Naver H, Scott MP, Golbeck JH, Moller BL, Scheller HV. Reconstitution of barley photosystem I with modified PSI-C allows identification of domains interacting with PSI-D and PSI-A/B. J Biol Chem. 1996;271:8996–9001. doi: 10.1074/jbc.271.15.8996. [DOI] [PubMed] [Google Scholar]
- Nechustai R, Nelson N. Biogenesis of photosystem I reaction center during greening. Plant Mol Biol. 1985;4:377–384. doi: 10.1007/BF02418259. [DOI] [PubMed] [Google Scholar]
- Oh-Oka H, Takahashi Y, Matsubara H. Topological considerations on the 9-kDa polypeptide which contains centers A and B, associated with the 14- and 19-kDa polypeptides in the photosystem I complex of spinach. Plant Cell Physiol. 1989;30:869–875. [Google Scholar]
- Pandini V, Aliverti A, Zanetti G. Interaction of the soluble recombinant PsaD subunit of spinach photosystem I with ferredoxin I. Biochemistry. 1999;38:10707–10713. doi: 10.1021/bi990423h. [DOI] [PubMed] [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herdmann M, Stanier RY. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- Rogers LJ. Ferredoxins, flavodoxins and related proteins: structure, function and evolution. In: Fay P, Van Baalen C, editors. The Cyanobacteria. Amsterdam: Elsevier; 1987. pp. 35–67. [Google Scholar]
- Rögner M, Nixon PJ, Diner BA. Purification and characterization of photosystem I and photosystem II core complexes from wild-type and phycocyanin-deficient strains of the cyanobacterium Synechocystis PCC 6803. J Biol Chem. 1990;265:6189–6196. [PubMed] [Google Scholar]
- Rousseau F. Etude de deux sous-unités périphériques du photosystème I exposées du coté stromal de la membrane thylakoidale. PhD thesis. Orsay, France: University Paris XI; 1992. [Google Scholar]
- Rousseau F, Sétif P, Lagoutte B. Evidence for the involvement of PSI-E subunit in the reduction of ferredoxin by photosystem I. EMBO J. 1993;12:1755–1765. doi: 10.1002/j.1460-2075.1993.tb05823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Schluchter WM, Shen G, Zhao J, Bryant DA. Characterization of PsaI and PsaL mutants of Synechococcus sp. strain PCC 7002: a new model for state transitions in cyanobacteria. Photochem Photobiol. 1996;64:53–66. doi: 10.1111/j.1751-1097.1996.tb02421.x. [DOI] [PubMed] [Google Scholar]
- Sétif P, Bottin H. Laser flash absorption spectroscopy study of ferredoxin reduction by photosystem I in Synechocystis sp. PCC 6803: evidence for submicrosecond and microsecond kinetics. Biochemistry. 1994;33:8495–8504. doi: 10.1021/bi00194a014. [DOI] [PubMed] [Google Scholar]
- Sétif P, Bottin H. Laser flash absorption spectroscopy study of ferredoxin reduction by photosystem I: spectral and kinetic evidence for the existence of several photosystem I-ferredoxin complexes. Biochemistry. 1995;34:9059–9070. doi: 10.1021/bi00028a015. [DOI] [PubMed] [Google Scholar]
- Tjus SE, Andersson B. Extrinsic polypeptides of spinach photosystem I. Photosynth Res. 1991;27:209–219. doi: 10.1007/BF00035842. [DOI] [PubMed] [Google Scholar]
- van Thor JJ, Geerlings TH, Matthijs HCP, Hellingwerf KJ. Kinetic evidence for the PsaE-dependent transient ternary complex photosystem I/ferredoxin/ferredoxin: NADP(+) reductase in a cyanobacterium. Biochemistry. 1999;38:12735–12746. doi: 10.1021/bi9903502. [DOI] [PubMed] [Google Scholar]
- von Leoprechting A, Hörth P, Haehnel W, Schilz E, Mühlenhoff U. Identification of biotinylation sites on proteins by selective retrieval of 2-iminobiotinylated peptides from proteolytic peptide mixtures: localization of the accessible lysine residues on the photosystem I subunits PsaD and PsaE. Anal Biochem. 1998;262:110–121. doi: 10.1006/abio.1998.2751. [DOI] [PubMed] [Google Scholar]
- Xia Z, Broahurst RW, Laue ED, Bryant DA, Golbeck JH, Bendall DS. Structure and properties in solution of PsaD, an extrinsic polypeptide of photosystem I. Eur J Biochem. 1998;255:309–316. doi: 10.1046/j.1432-1327.1998.2550309.x. [DOI] [PubMed] [Google Scholar]
- Xu Q, Armbrust TS, Guikema JA, Chitnis PR. Organization of photosystem I polypeptides: a structural interaction between the PsaD and PsaL subunits. Plant Physiol. 1994a;106:1057–1063. doi: 10.1104/pp.106.3.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Chitnis PR. Organization of photosystem I polypeptides: identification of PsaB domains that may interact with PsaD. Plant Physiol. 1995;108:1067–1075. doi: 10.1104/pp.108.3.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Guikema JA, Chitnis PR. Identification of surface-exposed domains on the reducing side of photosystem I. Plant Physiol. 1994b;106:617–624. doi: 10.1104/pp.106.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Jung YS, Chitnis VP, Guikema JA, Golbeck JH, Chitnis PR. Mutational analysis of photosystem I polypeptides in Synechocystis sp. PCC 6803: subunit requirements for reduction of NADP+ mediated by ferredoxin and flavodoxin. J Biol Chem. 1994c;269:21512–21518. [PubMed] [Google Scholar]
- Zanetti G, Merati G. Interaction between photosystem I and ferredoxin: identification by chemical cross-linking of the polypeptide which binds ferredoxin. Eur J Biochem. 1987;169:143–146. doi: 10.1111/j.1432-1033.1987.tb13591.x. [DOI] [PubMed] [Google Scholar]
- Zilber AL, Malkin R. Ferredoxin cross-links to a 22-kDa subunit of photosystem I. Plant Physiol. 1988;88:810–814. doi: 10.1104/pp.88.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilber AL, Malkin R. Organization and topology of photosystem I subunits. Plant Physiol. 1992;99:901–911. doi: 10.1104/pp.99.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]