Abstract
The advent of high-throughput sequencing methods allowed researchers to fully characterize microbial community in environmental samples, which is crucial to better understand their health effects upon exposures. In our study, we investigated bacterial and fungal community in indoor and outdoor air of nine classrooms in three elementary schools in Seoul, Korea. The extracted bacterial 16s rRNA gene and fungal ITS regions were sequenced and their taxa were identified. Quantitative polymerase chain reaction for total bacteria DNA was also performed. The bacterial community was richer in outdoor air than classroom air whereas fungal diversity was similar indoors and outdoors. Bacteria such as Enhydrobacter, Micrococcus, and Staphylococcus that are generally found in human skin, mucous membrane, and intestine were found in great abundance. For fungi, Cladosporium, Clitocybe, and Daedaleopsis were the most abundant genera in classroom air and mostly related to outdoor plants. Bacterial community composition in classroom air was similar among all classrooms but differed from that in outdoor air. However, indoor and outdoor fungal community compositions were similar for the same school but different among schools. Our study indicated the main source of airborne bacteria in classrooms was likely human occupants; however, classroom airborne fungi most likely originated from outdoors.
Keywords: microbial communities, classroom air, elementary school classrooms, next-generation sequencing
1. Introduction
Most people in modern society spend more than 80 percent of their time indoors.1,2 Therefore, indoor environmental quality has a significant effect on health and quality of life, especially for groups disproportionately affected such as children, adolescents, the elderly, and people suffering from respiratory diseases.3 Children may also be exposed to airborne pollutants more than adults because their respiration rate per unit weight is higher.4 In addition, the high activity level of students in elementary schools may also contribute to increase classroom airborne concentrations of particulate matter (PM) through resuspension of settled surface dusts.5
Indoor PM consists of biological particles, organic compounds adsorbed onto particles, transition metals, ions, reactive gases, particle core of carbonaceous material, and minerals.6,7 Exposures to indoor bacteria and fungi have been associated with the occurrence of infectious diseases, and exacerbation of asthma and allergies.8 While reducing exposures to pathogens and bacterial toxins in the indoor environments is important,9 exposures to human-associated indoor microbiomes may also provide beneficial effects on occupants’ health.10–12 Hygiene hypothesis suggests that infants need to get sufficiently exposed to symbiotic microorganisms to properly develop the immune system that will prevent development of allergic diseases.13 Thus, exposure to indoor microbes might have both beneficial and adverse effects in occupants.14
It has been challenging to study the microbial community with the traditional culture method because most microorganisms existing in the environment are not easily cultured with selected media.15–18 Nonetheless, the microbial management standard for indoor air in Republic of Korea is based on the colony forming units using the culture method that is likely to neglect some important indoor microorganisms. As metagenome analysis using next generation sequencing becomes more readily available, studying microbial communities existing in the environment draws more attention from researchers.19 Nevertheless, only a limited number of studies exist that characterize microbial communities in school classroom air and examine their associations with classroom environments. Furthermore, the management of microbial community in indoor air in multi-user facilities has become a major issue during the pandemic of coronavirus disease 2019.20 In this study, we selected three elementary schools located near busy roads with high vehicular traffic in Seoul, Republic of Korea to: 1) characterize microbial communities in classroom air; and 2) compare microbiomes inside and outside classrooms by bacterial 16S rRNA gene and fungal ITS (internal transcribed spacer) region sequencing.
2. Materials and method
2.1. Site selection and sample collection
Three elementary schools adjacent to major roads with an average daily traffic volume of over 100,000 vehicles in Seoul, Korea were selected.21 The schools were, on average, 226 meters (standard deviation = 49) apart from the main roads. Air samples were collected from nine classrooms (one per classroom and two to four classrooms per school) with natural ventilation made through opening interior windows between classroom and corridor during October through December 2019. We selected 1st, 2nd, and 3rd graders’ classrooms considering their higher classroom activity levels compared to the higher grades.
Indoor and outdoor air filter samples for total suspended particulate matter were simultaneously collected during school hours on weekdays and during the absence of students on holidays or weekends. Three indoor sampling pumps (Sensidyne, St. Petersburg, FL USA) were placed on the lockers at the back of the classroom at a height of 130 cm, similar to the average height of elementary school students in Korea,22 and two outdoor sampling pumps at the school playground (Figure S1). The sampling pumps connected to 37-mm closed cassettes loaded with polyvinyl chloride filters (pore size: 5 μm, SKC, Inc., Eighty Four, PA USA) were operated at 2.5 L/min indoors and 4 L/min outdoors from 8:00 am to 3:30 pm. To secure enough genomic DNA in samples for the 16s rRNA gene and ITS region sequencing, three indoor filters were combined for each classroom and two outdoor filters for the corresponding outdoor sample. All equipment, materials, and supplies were sterilized before use. After completion of sampling, the filters were placed in a sterile filter container and sealed with parafilm using a sterile technique. The sealed filters were then placed in a cooler with ice packs to maintain cool temperature and minimize potential changes in the microbial community during transportation to the laboratory where all filter samples were stored at −80 °C until the analysis. Environmental conditions of the classrooms were recorded on the day when the air samples were collected and they included the number of students, temperature, relative humidity, natural ventilation time, and air cleaner operation time. Outdoor wind speed, precipitation, temperature, and relative humidity on the sampling day were also recorded.
2.2. Genomic DNA extraction
Genomic DNA (gDNA) in sampled filters was extracted using High Pure PCR Template Preparation Kit (Roche, Basel, Switzerland). The filter was put in a 2 mL screw-cap microcentrifuge tube along with 300 mg of 212–300 μm sterilized glass beads (Sigma, St. Louis, MO USA) and 650 μL of ultrapure water. It was pulverized twice for 30 s at 4,350 rpm using a homogenizer (Allsheng, Hangzhou, China).23,24 After centrifugation at 20,000 g for 1 min, the supernatant was collected and centrifuged again at 20,000 g for 1 min. A 400 μL aliquot of the supernatant was collected and used as a pretreatment sample. Then, the sample was eluted from the column as recommended by the kit manufacturer and stored at −20 °C.
2.3. Microbial diversity analysis
Each sample was prepared according to the Illumina 16S Metagenomic Sequencing Library protocols. The amount and purity of DNA were measured using the QuantiFluor dsDNA System (Promega, Madison, WI USA) and VICTOR Nivo (PerkinElmer, Waltham, MA USA). The samples that did not meet minimum cDNA concentration (10 ng/μL) were excluded from the 16s rRNA gene and ITS region sequencing (Table S1). ITS3-ITS4 region of fungal DNA25 and 16S V3-V4 region of bacterial DNA26 were used for gene amplification with the primer sequences shown in Table 1. The amplification step was performed to add multiplexing indices and Illumina sequencing adapters. The final products were normalized and pooled using the PicoGreen, and the size of libraries were verified using the TapeStation DNA screentape D1000 (Agilent, Santa Clara, CA USA). And then sequencing was performed using the Illumina MiSeq™ platform (Illumina, San Diego, CA USA).
Table 1.
The primer sequences used for gene sequencing and qPCR experiments.
| Primer | Sequence | ||
|---|---|---|---|
| Bacteria | V3-V4 | Forward | 5’ CCTACGGGNGGCWGCAG 3’ |
| Reverse | 5’ GACTACHVGGGTATCTAATCC 3’ | ||
| 518F | Forward | 5’ CCAGCAGCCGCGGTAATACG 3’ | |
| 800R | Reverse | 5’ TACCAGGGTATCTAATCC 3’ | |
| Fungi | ITS3-ITS4 | Forward | 5’ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGCATCGATGAAGAACGCAGC 3’ |
| Reverse | 5’ GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTCCTCCGCTTATTGATATGC 3’ | ||
Sequence analysis was performed using Quantitative Insights Into Microbial Ecology2 (QIIME2, v.2017.6.0), such as Caporaso et al. (2010).27 At the Demultiplex phase, each sequence was divided into samples using DADA2 (ver.1.1.1) and quality checked to eliminate low quality.28 The subsequent process also utilized DADA2 to filter out noisy sequences, correct errors in ambient sequences, and correct amplicon errors through the process of removing chimeric sequences and singletons. The clustering process used the q2-vsearch (ver.2019.7) tool and proceeded with an open reference OTU picking method. Sequences with similarity greater than 97% were clustered into Operational Taxonomic Units (OTUs), and OTUs were analyzed through SILVA and UNITE.29,30
2.4. Quantitative analysis of bacteria
For the quantitative analysis of the total bacterial DNA, qPCR (quantitative polymerase chain reaction) was performed by using 518F and 800R (Table 1) among 16S rRNA region.31 The reaction mixture was prepared with 4 μL of 5x HOT FIREPol EvaGreen qPCR Supermix (Solis BioDyne, Tartu, Estonia), 14.6 μL of Nuclease-Free Water (QIAGEN, Hilden, Germany), and forward and reverse primer that were mixed with 0.2 μL and 1 μL of DNA template, respectively. Initial denaturation was carried out at 95 °C for 12 min, and 40 cycles of three-step process were carried out- denaturation at 95 °C for 15 s, annealing at 55 °C for 20 s, and elongation at 72 °C for 30 s.
For qPCR, a LineGene 9600 (BIOER, Hangzhou, China) was used. Using the qPCR absolute quantification method,32 qPCR was performed with the diluted Escherichia coli (E. coli) sample. PCR efficiency (R2 = 0.993; PCR efficiency = 102.2%) was verified through a standard curve and specificity was confirmed through a melting curve. The amount of bacterial cDNA in sample was calculated according to the cycle threshold value (Ct value) of the diluted E. coli sample. To calculate the copy number in the bacterial sample, the Ct value of the diluted E. coli and the Ct value of the sample were compared. The OD600 value of the undiluted standard solution of E. coli sample used in the experiment was 0.7, and the corresponding DNA concentration measured after DNA extraction was 55.0 ng/μL. The E. coli standard sample solution was serially diluted to 10−5 times by a factor of 10.
2.5. Statistical analysis
Alpha and beta diversity were calculated and visualized using q2-diversity (ver.2019.7) in the QIIME2 plugin. Unidentified genera were excluded from the analysis. A rarefaction curve was created for each sample to estimate bacterial and fungal richness as a function of sequencing depth at genus level.33 To explore the abundance and uniformity of bacterial and fungal taxa in the samples, a Shannon-Wiener index (HI), which is a diversity index considering both abundance and diversity, and Pielou’s evenness index (EI), which represents the evenness of a community, were calculated. Using principal coordinates analysis (PCoA), we evaluated similarity of the microbial community among schools and between different sample types (inside versus outside classroom) with Bray-Curtis dissimilarity index. Spearman correlation coefficients among the diversity indices and environmental factors were calculated by sample location (indoors and outdoors). For bacteria, the DNA copy numbers from the qPCR analysis were also included in the correlation analysis. All statistical analyses were conducted in R version 4.0 (CRAN) and statistical significance set at p < 0.05 and marginal significance at p < 0.1.
3. Results and discussion
3.1. Classroom characteristics and outdoor weather conditions
All nine classrooms in three schools had wood flooring and the number of students per occupied classroom ranged from 21 to 26 with an average of 23. Classroom average temperature was 22.6 ± 1.6 °C (standard deviation), and the average relative humidity was 42.2 ± 3.2 % (Table 2). The average time of natural ventilation made through opening interior windows between classroom and corridor was 47.8 ± 80.5 minutes/day while the average operating time of a classroom air purifier was 151.7 ± 142.5 minutes/day. Outdoor temperature during the sampling campaign varied widely from −0.5 (in December) to 17.0 °C (in October), but was significantly lower than classroom temperature (p < 0.01). The outdoor average relative humidity was 62.5 ± 11.2 % and significantly higher than indoors (p < 0.01). Average wind speed during outdoor sampling was 7.7 ± 2.5 m/s. It rained during the sampling period for classrooms 1, 3, and 4 in school ‘b’, and the average precipitation was 3.3 ± 6.7 mm.
Table 2.
Indoor and outdoor environmental factors and number of students on the day when the air samples were collected.
| Sample type | Sampled month | Classroom/outdoor | Number of students | Time of Natural ventilation (minute) | Operation time of Air cleaner (minute) | Temp (°C) | RH (%) | Wind velocity (m/s) | Precipitation (mm) |
|---|---|---|---|---|---|---|---|---|---|
| OC-Sa1 | October | Classroom 1 | 26 | 0 | 160 | 21.4 | 46 | - | - |
| OUT-Sa1 | October | Outdoor | - | - | - | 15.2 | 61 | 4.7 | 0 |
| OC-Sa2 | October | Classroom 2 | 23 | 170 | 0 | 23.7 | 45 | - | - |
| OUT-Sa2 | October | Outdoor | - | - | - | 18.8 | 60 | 6.8 | 0 |
| OC-Sb1 | November | Classroom 3 | 21 | 220 | 290 | 23.1 | 45 | - | - |
| OUT-Sb1 | November | Outdoor | - | - | - | 11.2 | 72 | 8.6 | 1.1 |
| OC-Sb2 | November | Classroom 4 | 22 | 0 | 285 | 22.6 | 41 | - | - |
| OUT-Sb2 | November | Outdoor | - | - | - | 9.3 | 64 | 5 | 0 |
| OC-Sb3 | November | Classroom 5 | 24 | 40 | 0 | 23.7 | 43 | - | - |
| OUT-Sb3 | November | Outdoor | - | - | - | 7.2 | 72 | 10.4 | 7.5 |
| UOC-Sb4 | November | Classroom 3 | 0 | 0 | 0 | 21.5 | 45 | - | - |
| OUT-Sb4 | November | Outdoor | - | - | - | 8.3 | 79 | 11.2 | 21.1 |
| OC-Sc1 | December | Classroom 6 | 25 | 0 | 330 | 25.2 | 40 | - | - |
| OUT-Sc1 | December | Outdoor | - | - | - | -0.1 | 48 | 11.2 | 0 |
| OC-Sc2 | December | Classroom 7 | 23 | 0 | 300 | 22.4 | 37 | - | - |
| OUT-Sc2 | December | Outdoor | - | - | - | 2 | 42 | 6.5 | 0 |
| UOC-Sc3 | December | Classroom 7 | 0 | 0 | 0 | 19.4 | 38 | - | - |
| OUT-Sc3 | December | Outdoor | - | - | - | -0.9 | 65 | 5.0 | 0 |
Sample type: Occupied classroom sample (OC); Unoccupied classroom sample (UOC); outdoor sample (OUT); Schools a, b, and c (Sa, Sb, and Sc); and different days (sequential number within the same school). Classroom ID (number in the column for the classroom/outdoor); Temperature (Temp); Relative Humidity (RH).
3.2. Qualitative analysis of microbial community in classroom and outdoor air
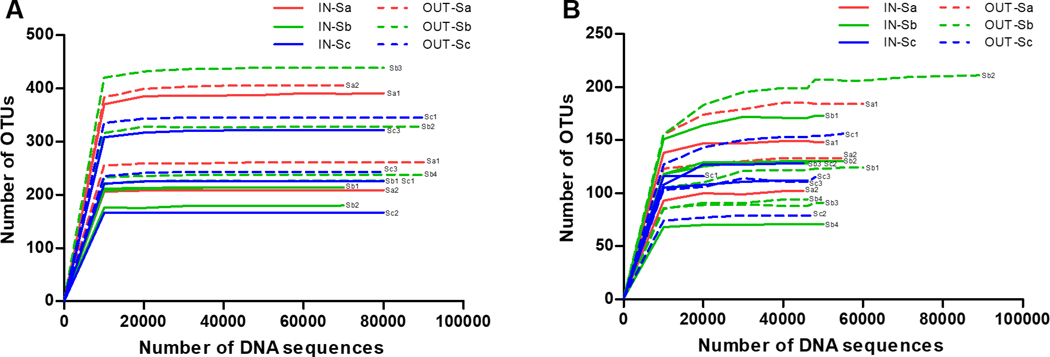
Richness of bacterial and fungal samples is presented using the rarefaction curves (Figure 1). As rarefaction curve reached the asymptote (represented by a straight line with a zero slope), sufficient sequencing depth was confirmed. For bacteria, the indoor average OTUs was 240.3 ± 70.7, which was lower than outdoors (310.9 ± 81.4). For fungi, the average OTUs of classroom and outdoor samples were similar− 120.7 ± 27.1 and 122.2 ± 37.7, respectively. This indicated that bacteria in outdoor air samples were more diverse and more variable in diversity than classroom air while fungal diversity was similar indoors and outdoors (similar in average richness) but more variable in diversity in outdoor air samples. The number of bacterial OTUs was significantly (p < 0.01) higher than fungal OTUs, indicating that bacterial taxa in both indoor and outdoor air were more diverse than fungal taxa. This result was similar to the studies that found more diverse bacteria than fungi in soil, sugarcane (roots, shoots and leaves) and floor dust of homes.34–36
Figure 1.
Rarefaction curves of (A) bacteria and (B) fungi in the samples. Operational taxonomic units (OTUs), Classroom sample (IN), outdoor sample (OUT), and School ID: Sa, Sb, Sc. Two to four classrooms per school were sampled.
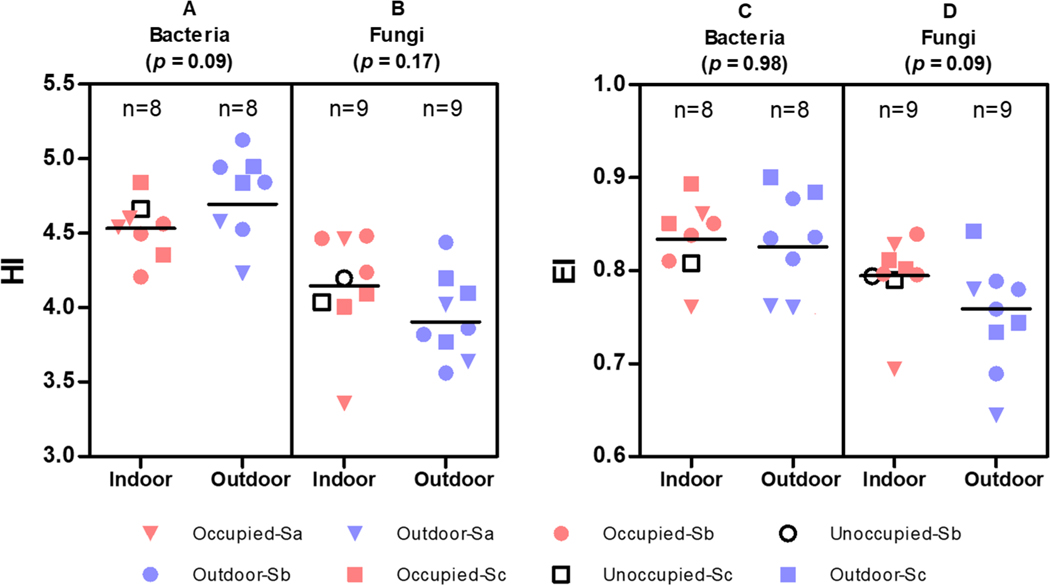
Diversity and evenness (uniformity) of bacteria and fungi in the classroom and outdoor air is presented in Figure 2. Shannon-Wiener index (HI) of classroom airborne bacteria was lower (marginally significant, p = 0.09) than outdoors, but Pielou’s evenness indices (EI) indoors and outdoors were similar while HI for fungi was not different between indoors and outdoors even if significance level was set at 0.1 although fungal taxa were more evenly distributed in classroom air than outdoor air (p = 0.09). Kembel et al. (2012)37 reported that diversity in the outdoor bacterial community was significantly greater than indoors, which was similar to our study finding. Comparison of bacterial and fungal diversities in our study indicated that both HI and EI for bacteria were significantly (p < 0.01) higher than fungi, which is consistent to the findings of Kettleson et al. (2015) report.34
Figure 2.
Shannon-Wiener index and Pielou’s evenness index by sample type. HI: Shannon-Wiener index; EI: Pielou’s evenness index; A: comparison of HI between indoor and outdoor bacterial samples; B: comparison of HI between indoor and outdoor fungal samples; C: comparison of EI between indoor and outdoor bacterial samples; D: comparison of EI between indoor and outdoor fungal samples. Occupied classroom: OC; Outdoor sample: OUT; School ID: Sa, Sb, or Sc; and classroom ID: 1, 2, or 3. The sample collected from ‘Unoccupied-Sa’ was not sequenced because the amount of cDNA was too low.
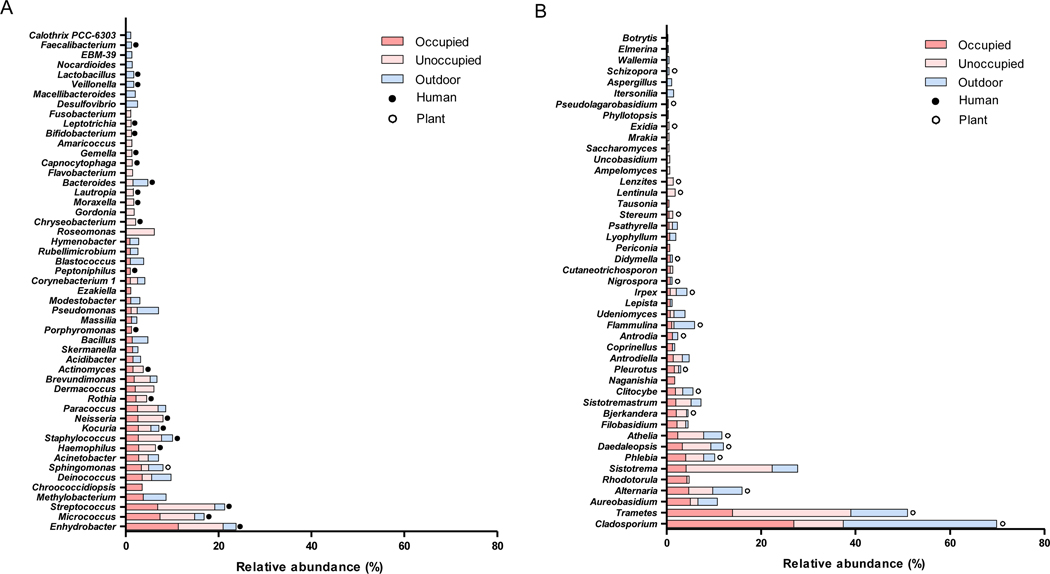
The top 30 bacterial and fungal genera in relative abundance are illustrated in the stacked bar graphs for the classroom and outdoor air samples (Figure 3). The average cumulative relative abundance of the top 15 genera accounted for 35.6 ± 9.0 % of total abundance for classroom bacterial samples, 22.3 ± 7.0 % for outdoor bacterial samples, 56.3 ± 7.8 % for classroom fungal samples, and 62.7 ± 8.0 % for outdoor fungal samples (Figure S2). For bacteria, Enhydrobacter, Streptococcus, and Micrococcus were the most abundant top three genera in classroom air while Methylobacterium, Pseudomonas, and Deinococcus were the most abundant top three genera in outdoor air (Figure 3A). The literature showed the main source of Enhydrobacter, Micrococcus, Staphylococcus, Streptococcus, Neisseria, and Haemophilus in indoor environments that were also found within the top 22 genera in our study was human occupants. These bacterial genera live in the human skin, mucous membrane, and the intestine.38–43 Methylobacterium, a genus living in soil and water and existing widely in the environment,44 was found in both indoor and outdoor environments as we observed in our study. Kocuria inhabits the human skin, mucous membrane, and larynx, and was also detected in indoor dust.45,46 Acinetobacter is widely distributed in the environment such as soils and water and also found in hospital settings because it can survive exposure to various disinfectants.47 For fungi, Cladosporium, Trametes, Aureobasidium, Sistotrema, and Alternaria were most abundant in both indoor and outdoor samples (Figure 3B). Cladosporium was the most commonly found genus inside and outside the classrooms.48 Fungal spores of some species of Alternaria are allergens that cause hypersensitivity reactions, allergic asthma, keratitis, and skin infection.49,50 Chronic exposure to Aureobasidium pullulans (a black and yeast-like fungus found in various environments such as soil, water, air, and limestone) from a contaminated humidifier or air conditioner may result in hypersensitivity pneumonitis (extrinsic allergic alveolitis).51,52 Of the top 30 fungal genera in our study, 20 were parasitic or symbiotic on plants. Cladosporium, Trametes, Phlebia, Daedaleopsis, Clitocybe, and Irpex that were found to be relatively abundant in our study were mostly related to trees.53–56 Our finding that more abundant bacterial genera indoors than outdoors were mostly related to humans is consistent to other study findings suggesting that students and teachers are likely to be a strong source of these airborne bacteria in classrooms; however, fungi in classroom air likely originate from outdoors.9,57,58
Figure 3.
Relative abundance of bacteria (A) and fungi (B) at the genus level in occupied and unoccupied classroom and outdoor air. (●: human-related genus; ○: plant-related genus)
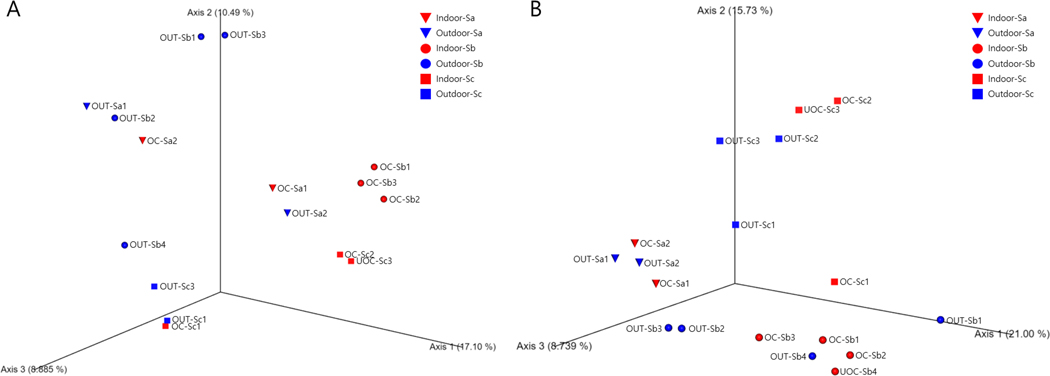
To confirm the main source of microbes in classroom air, we examined the similarity of bacterial and fungal community compositions among schools, classrooms, and outdoor air samples using principal coordinates analysis. Our results showed that bacterial community compositions in classroom air in all three schools were generally similar (clustered together) but different from those in outdoor air that also varied widely by school (Figure 4A). On the other hand, fungal community compositions in the samples were generally clustered by school (different location) regardless of indoor or outdoor samples, indicating that fungal communities in indoor and outdoor air were similar within the same school but different among schools (Figure 4B). However, we did not observe different community compositions among classrooms with or without operating air cleaner. These results of community composition confirmed that human occupants were likely to be the major factor determining bacterial community composition in classroom air whereas outdoor air was likely the main source contributing to fungal community composition in classroom air as described in Burge’s report.59
Figure 4.
Principal coordinates analysis plot using Bray-Curtis dissimilarity index at the genus level for bacteria (A) and fungi (B). Occupied classroom: OC; Outdoor sample: OUT; School ID: Sa, Sb, or Sc; and classroom ID: 1, 2, or 3.
3.3. Quantitative analysis of bacteria in classroom and outdoor air
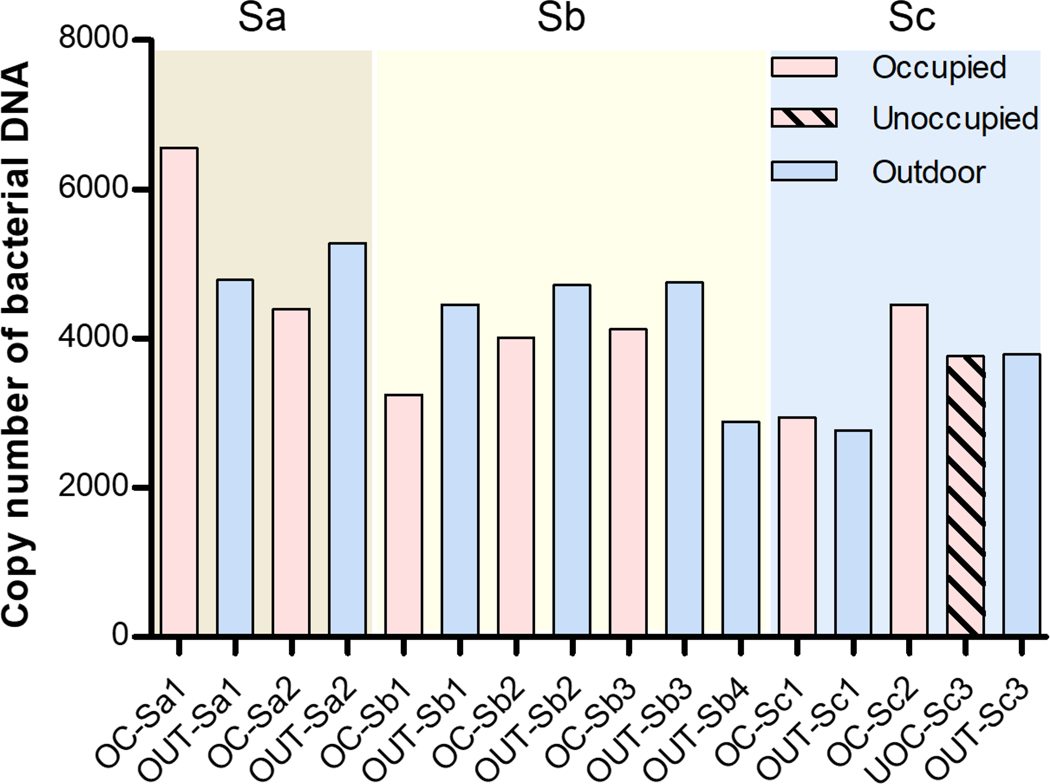
The copy number of bacterial DNA in the occupied classroom air was lower than that in outdoor air samples on the same day for the most samples, except for the classroom ‘1’ in school ‘a’ (Sa1) and classroom ‘1’ in school ‘c’ (Sc1) (Figure 5). For the classroom in the school ‘a’ (Sa1), the highest number of students (26) in the classroom might have influenced the concentration of airborne bacterial DNA in the classroom air. On the day of sampling at school ‘c’, outdoor temperature was below zero, and the sub-zero temperature might have influenced bacterial concentration in outdoor air. The copy number of bacterial DNA in the unoccupied classroom was similar to that in corresponding outdoor air. Our quantitative PCR results (amount of airborne bacterial DNA) were also consistent to the results of the semi-quantitative analyses of rarefaction curves and alpha diversity presented in the previous sections.
Figure 5.
Copy number of bacterial DNA by indoor and outdoor sample for the same school and classroom. (Occupied classroom: OC; Outdoor sample: OUT; School ID: Sa, Sb, or Sc; and classroom ID: 1, 2, or 3.)
3.4. Correlations between environmental factors and microbial community
Evenness index for bacteria in classroom air samples was significantly and positively correlated (correlation coefficient = 0.81, p < 0.05) with classroom temperature (Figure S3A). The number of students was positively correlated with the number of bacterial DNA copies but not statistically significant. For outdoor bacterial air samples, temperature was negatively correlated with evenness index (−0.83, p < 0.01) but positively correlated with DNA copy numbers (0.74, p < 0.05) (Figure S3B). For fungi, the number of students indoors was positively correlated with evenness index (0.70, p < 0.05) (Figure S3C) whereas outdoor wind speed and evenness index of outdoor fungi had a positive correlation (0.77, p < 0.05) (Figure S3D). However, relative humidity, natural ventilation time, air cleaner operation time, and precipitation were not correlated with the microbial diversity indices both indoors and outdoors.
One of the limitations in our study was the small sample size, which might limit generalizability of the study findings. The small sample size also did not allow us to statistically evaluate the effects of environmental conditions on microbial community in classroom air. Future studies with a large number of schools and classrooms in different areas and seasons might provide more generalizable results. Nonetheless, our study findings provide useful information to better understand airborne bacterial and fungal microbiome including their major sources in occupied classrooms.
4. Conclusions
From our study we found that bacterial microbiota in occupied classroom air was likely associated with humans and different from that in outdoor air, indicating that human occupants may be the major source of indoor airborne bacteria. However, fungal microbial community in classroom air is similar to those in outdoor air, indicating that outdoor airborne fungi may be the major contributor to indoor airborne fungi. Our findings from a study of Korean school classrooms were in line with those from studies conducted in other countries. However, health effects of human occupant-originated bacteria in school classrooms are largely unknown and thus epidemiologic studies of classroom microbiomes and health in schoolteachers and students are warranted. Our findings of higher number of bacterial DNA copies in outdoors than indoors, and outdoor airborne fungi as the main source of classroom fungi might indicate the importance of properly controlling infiltration of outdoor microbes into classrooms.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Practical implications.
We found that bacterial community composition in classroom air was similar among all classrooms but different from outdoor air. However, indoor and outdoor fungal community compositions were similar for the same school but different among the schools. Our study indicates that the main source of airborne bacteria in classrooms was likely human occupants whereas most fungi in classroom air likely originated from outdoors. Our finding that the amount of total bacterial DNA in outdoor air was higher than indoors also underscores the importance of controlling infiltration of outdoor microbes into classrooms.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT, MOE) and (NRF- 2019M3E7A1113076). Also, this study was supported by the Basic Science Research Program of NRF, which is funded by the Ministry of Education (NRF-2019R1A2C1004616).
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the National Institute for Occupational Safety and Health, Centers for Disease Control and Prevention.
Footnotes
Conflict of interest
The authors declare that they have no competing interest.
References
- 1.Klepeis NE, Nelson WC, Ott WR, et al. The National Human Activity Pattern Survey (NHAPS): a resource for assessing exposure to environmental pollutants. Journal of Exposure Science & Environmental Epidemiology. 2001;11:231–252. [DOI] [PubMed] [Google Scholar]
- 2.Yang J, Xu M, Zhang X, Hu Q, Sommerfeld M, Chen Y. Life-cycle analysis on biodiesel production from microalgae: Water footprint and nutrients balance. Bioresource Technology. 2011;102:159–165. [DOI] [PubMed] [Google Scholar]
- 3.Cincinelli A, Martellini T. Indoor Air Quality and Health. International Journal of Environmental Research and Public Health. 2017;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoon H, Seo J, Kim T, et al. Development of Korean Exposure Factors for Children in Korea. Korean Journal of Environmental Health Sciences. 2017;43:167–175. [Google Scholar]
- 5.Ferro AR, Kopperud RJ, Hildemann LM. Source Strengths for Indoor Human Activities that Resuspend Particulate Matter. Environmental Science & Technology. 2004;38:1759–1764. [DOI] [PubMed] [Google Scholar]
- 6.Jones AP. Indoor air quality and health. Atmospheric Environment. 1999;33:4535–4564. [Google Scholar]
- 7.Valavanidis A, Fiotakis K, Vlachogianni T. Airborne Particulate Matter and Human Health: Toxicological Assessment and Importance of Size and Composition of Particles for Oxidative Damage and Carcinogenic Mechanisms. Journal of Environmental Science and Health, Part C. 2008;26:339–362. [DOI] [PubMed] [Google Scholar]
- 8.Douwes J, Thorne P, Pearce N, Heederik D. Bioaerosol Health Effects and Exposure Assessment: Progress and Prospects. The Annals of Occupational Hygiene. 2003;47:187–200. [DOI] [PubMed] [Google Scholar]
- 9.Hospodsky D, Qian J, Nazaroff WW, et al. Human Occupancy as a Source of Indoor Airborne Bacteria. PLoS ONE. 2012;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nature Reviews Microbiology. 2009;7:887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nature Reviews Genetics. 2012;13:260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weyrich LS, Dixit S, Farrer AG, Cooper AJ, Cooper AJ. The skin microbiome: Associations between altered microbial communities and disease. Australasian Journal of Dermatology. 2015;56:268–274. [DOI] [PubMed] [Google Scholar]
- 13.Yazdanbakhsh M, Kremsner PG, Ree R van. Allergy, Parasites, and the Hygiene Hypothesis. Science. 2002;296:490–494. [DOI] [PubMed] [Google Scholar]
- 14.Liddicoat C, Waycott M, Weinstein P. Environmental Change and Human Health: Can Environmental Proxies Inform the Biodiversity Hypothesis for Protective Microbial–Human Contact? BioScience. 2016;66. [Google Scholar]
- 15.Amann RI, Ludwig W, Schleifer K-H. Phylogenetic Identification and In Situ Detection of Individual Microbial Cells without Cultivation. Microbiological reviews. 1995;59:143–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bloomfield SF, Stewart GSAB, Dodd CER, Booth IR, Power EGM The viable but non-culturable phenomenon explained? Microbiology. 1998;144:1–3. [DOI] [PubMed] [Google Scholar]
- 17.Streit WR, Schmitz RA. Metagenomics – the key to the uncultured microbes. Current Opinion in Microbiology. 2004;7:492–498. [DOI] [PubMed] [Google Scholar]
- 18.Jünemann S, Kleinbölting N, Jaenicke S, et al. Bioinformatics for NGS-based metagenomics and the application to biogas research. Journal of Biotechnology. 2017;261:10–23. [DOI] [PubMed] [Google Scholar]
- 19.Shokralla S, Spall JL, Gibson JF, Hajibabaei M. Next-generation sequencing technologies for environmental DNA research. Molecular Ecology. 2012;21:1794–1805. [DOI] [PubMed] [Google Scholar]
- 20.Morawska L, Tang JW, Bahnfleth W, et al. How can airborne transmission of COVID-19 indoors be minimised? Environment International. 2020;142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seoul. 2019 Seoul Metropolitan Government Traffic Survey Data. Seoul; 2019. [Google Scholar]
- 22.MOE. Results of 2018 Student Health Examination Sample Statistics Analysis. Sejong; 2019. [Google Scholar]
- 23.Rittenour WR, Park J-H, Cox-Ganser JM, Beezhold DH, Green BJ. Comparison of DNA extraction methodologies used for assessing fungal diversity via ITS sequencing. Journal of Environmental Monitoring. 2012;14:766–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemons AR, Hogan MB, Gault RA, et al. Microbial rRNA sequencing analysis of evaporative cooler indoor environments located in the Great Basin Desert region of the United States. Environmental Science: Processes & Impacts. 2017;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D, Kim H, Seo K. Microbial composition of Korean kefir and antimicrobial activity of Acetobacter fabarum DH1801. Journal of Food Safety. 2020;40:e12728. [Google Scholar]
- 26.Klindworth A, Pruesse E, Schweer T, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Research. 2013;41:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beloshapka AN, Forster GM. Fecal Microbial Communities of Overweight and Obese Client-Owned Dogs Fed Cooked Bean Powders as Assessed by 454-Pyrosequencing. Journal of Veterinary Science & Technology. 2016;7. [Google Scholar]
- 29.Abarenkov K, Henrik Nilsson R, Larsson K-H, et al. The UNITE database for molecular identification of fungi - recent updates and future perspectives. New Phytologist. 2010;186:281–285. [DOI] [PubMed] [Google Scholar]
- 30.Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research. 2012;41:D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Senthilraj R, Prasad GS, Janakiraman K. Sequence-based identification of microbial contaminants in non-parenteral products. Brazilian Journal of Pharmaceutical Sciences. 2016;52:329–336. [Google Scholar]
- 32.Larionov A, Krause A, Miller W. A standard curve based method for relative real time PCR data processing. BMC Bioinformatics. 2005;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaheer R, Noyes N, Ortega Polo R, et al. Impact of sequencing depth on the characterization of the microbiome and resistome. Scientific Reports. 2018;8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kettleson EM, Adhikari A, Vesper S, Coombs K, Indugula R, Reponen T. Key determinants of the fungal and bacterial microbiomes in homes. Environmental Research. 2015;138:130–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Souza RSC, Okura VK, Armanhi JSL, et al. Unlocking the bacterial and fungal communities assemblages of sugarcane microbiome. Scientific Reports. 2016;6:28774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagg C, Schlaeppi K, Banerjee S, Kuramae EE, van der Heijden MGA. Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nature Communications. 2019;10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kembel SW, Jones E, Kline J, et al. Architectural design influences the diversity and structure of the built environment microbiome. The ISME Journal. 2012;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kloos WE, Musselwhite MS. Distribution and Persistence of Staphylococcus and Micrococcus Species and Other Aerobic Bacteria on Human Skin1. Applied Microbiology. 1975;30:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rintala H, Pitkaranta M, Toivola M, Paulin L, Nevalainen A. Diversity and seasonal dynamics of bacterial community in indoor environment. BMC Microbiology. 2008;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson M. Bacteriology of Humans: An Ecological Perspective. 1st ed. John Wiley & Sons; 2009. [Google Scholar]
- 41.Tortora GJ, Funke BR, Case CL, Johnson TR. Microbiology : An Introduction. 11th ed. CA: Benjamin Cummings; 2012. [Google Scholar]
- 42.Liu G, Tang CM, Exley RM. Non-pathogenic Neisseria: members of an abundant, multi-habitat, diverse genus. Microbiology. 2015;161:1297–1312. [DOI] [PubMed] [Google Scholar]
- 43.Park J, Kim SJ, Lee J-A, Kim JW, Kim SB. Microbial forensic analysis of human-associated bacteria inhabiting hand surface. Forensic Science International: Genetics Supplement Series. 2017;6:e510–e512. [Google Scholar]
- 44.Green PN. Methylobacterium. In: Bergey’s Manual of Systematics of Archaea and Bacteria. Wiley; 2015:1–8. [Google Scholar]
- 45.McManus CJ, Kelley ST. Molecular survey of aeroplane bacterial contamination. Journal of Applied Microbiology. 2005;99:502–508. [DOI] [PubMed] [Google Scholar]
- 46.Grice EA, Kong HH, Renaud G, et al. A diversity profile of the human skin microbiota. Genome Research. 2008;18:1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doughari HJ, Ndakidemi PA, Human IS, Benade S. The Ecology, Biology and Pathogenesis of Acinetobacter spp.: An Overview. Microbes and Environments. 2011;26:101–112. [DOI] [PubMed] [Google Scholar]
- 48.Bensch K, Braun U, Groenewald JZ, Crous PW. The genus Cladosporium. Studies in Mycology. 2012;72:1–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rotem J. The Genus Alternaria: Biology, Epidemiology, and Pathogenicity. St Paul: American Phytopathological Society; 1994. [Google Scholar]
- 50.Meena M, Gupta SK, Swapnil P, Zehra A, Dubey MK, Upadhyay RS. Alternaria Toxins: Potential Virulence Factors and Genes Related to Pathogenesis. Frontiers in Microbiology. 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andrews JH, Spear RN, Nordheim E v. Population biology of Aureobasidium pullulans on apple leaf surfaces. Canadian Journal of Microbiology. 2002;48:500–513. [DOI] [PubMed] [Google Scholar]
- 52.Gostinčar C, Ohm RA, Kogej T, et al. Genome sequencing of four Aureobasidium pullulans varieties: biotechnological potential, stress tolerance, and description of new species. BMC Genomics. 2014;15:549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kirk PM, Cannon PF, David JC, Stalpers JA. Ainsworth and Bisby’s Dictionary of the Fungi. 9th ed. Egham: CABI publishing; 2001. [Google Scholar]
- 54.Zalar P, de Hoog GS, Schroers H-J, Crous PW, Groenewald JZ, Gunde-Cimerman N. Phylogeny and ecology of the ubiquitous saprobe Cladosporium sphaerospermum, with descriptions of seven new species from hypersaline environments. Studies in Mycology. 2007;58:157–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jakovlev J. Fungus gnats (Diptera: Sciaroidea) associated with dead wood and wood growing fungi: new rearing data from Finland and Russian Karelia and general analysis of known larval microhabitats in Europe. Entomologica Fennica. 2011;22:157–189. [Google Scholar]
- 56.LI H-J, SI J, HE S-H. Daedaleopsis hainanensis sp. nov. (Polyporaceae, Basidiomycota) from tropical China based on morphological and molecular evidence. Phytotaxa. 2016;275:294–300. [Google Scholar]
- 57.Shelton BG, Kirkland KH, Flanders WD, Morris GK. Profiles of Airborne Fungi in Buildings and Outdoor Environments in the United States. Applied and Environmental Microbiology. 2002;68:1743–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hewitt KM, Gerba CP, Maxwell SL, Kelley ST. Office space bacterial abundance and diversity in three metropolitan areas. PLoS ONE. 2012;7:e37849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burge HA. An update on pollen and fungal spore aerobiology. Journal of Allergy and Clinical Immunology. 2002;110:544–552. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.