SUMMARY
Understanding the mechanisms of blastocyst formation and implantation is critical for improving farm animal reproduction but is hampered by a limited supply of embryos. Here, we developed an efficient method to generate bovine blastocyst-like structures (termed blastoids) via assembling bovine trophoblast stem cells and expanded potential stem cells. Bovine blastoids resemble blastocysts in morphology, cell composition, single-cell transcriptomes, in vitro growth and the ability to elicit maternal recognition of pregnancy following transfer to recipient cows. Bovine blastoids represent an accessible in vitro model for studying embryogenesis and improving reproductive efficiency in livestock species.
Graphical Abstract
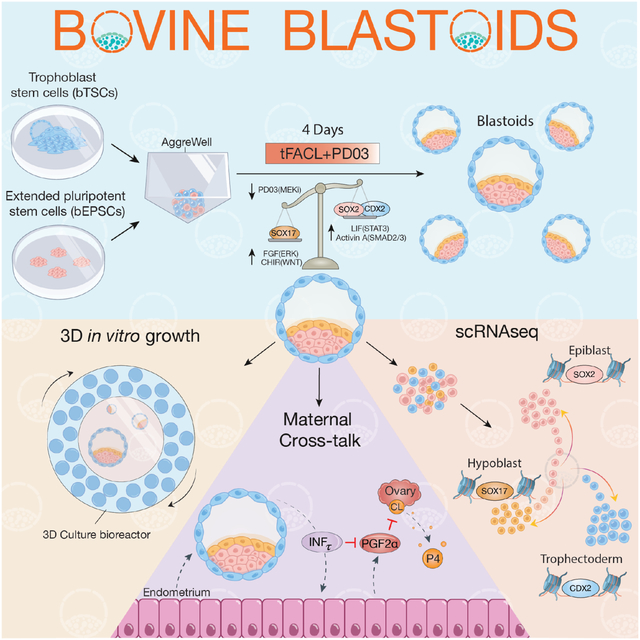
eTOC
Pinzon-Arteaga et al. develop a method for the efficient generation and in vitro growth of bovine blastocyst-like structures (termed blastoids) from stem cell cultures. They show via immunofluorescence and single-cell RNA-sequencing analyses that bovine blastoids resemble bovine blastocysts.
INTRODUCTION
Blastoids were initially developed in mice by assembling embryonic stem cells (ESCs)1 or extended pluripotent stem cells (EPSCs)2 with trophoblast stem cells (TSCs), or through EPSC differentiation and self-organization3, and have also been successfully generated in humans4–8. To date, however, blastoids from livestock species have not been reported. Several types of pluripotent stem cells (PSCs), including EPSCs, were recently derived from Bos taurus blastocysts9–15, which have great potential to advance animal agriculture16. Surprisingly, we found that LCDM medium previously used for EPSC culture13,17 could support de novo derivation and long-term culture of bovine trophoblast stem cells (TSCs) (CELL-REPORTS-D-23–01022). The availability of bovine EPSCs and TSCs (Figure S1A) prompted us to test whether bovine blastoids could be generated through 3D assembly.
RESULTS
To develop a condition that supports bovine blastoid formation, we adapted the FAC (FGF2, Activin-A, and CHIR99021) medium18, which supports the differentiation of hypoblast-like cells (HLCs) from naïve human PSCs4,19. We added the leukemia inhibitory factor (LIF) to the FAC medium that is known to improve preimplantation bovine embryo development20 (FACL). FGF signaling levels can bias the fate of inner cell mass (ICM), likely acting through the MEK-ERK pathway21,22, where high levels of FGF direct ICM cells towards the hypoblast (HYPO, or primitive endoderm [PE]) lineage23. To support both HYPO and epiblast (EPI) lineages, we optimized FGF signaling by lowering FGF2 concentration and including a low dose of a MEK inhibitor (PD0325901, 0.3μM), as MEK inhibition has been shown to suppress HYPO fate in bovine embryos in a dose dependent manner24. This optimized condition, termed titrated FACL+PD03 (tFACL+PD) (see Methods), supported the formation of bovine blastoids with high efficiency (64.2±7.6%) within four days (Figures 1A, 1B, S1B–S1H, S1O, and S1P).
Figure 1. Assembly of bovine blastoids from EPSCs and TSCs cultures.
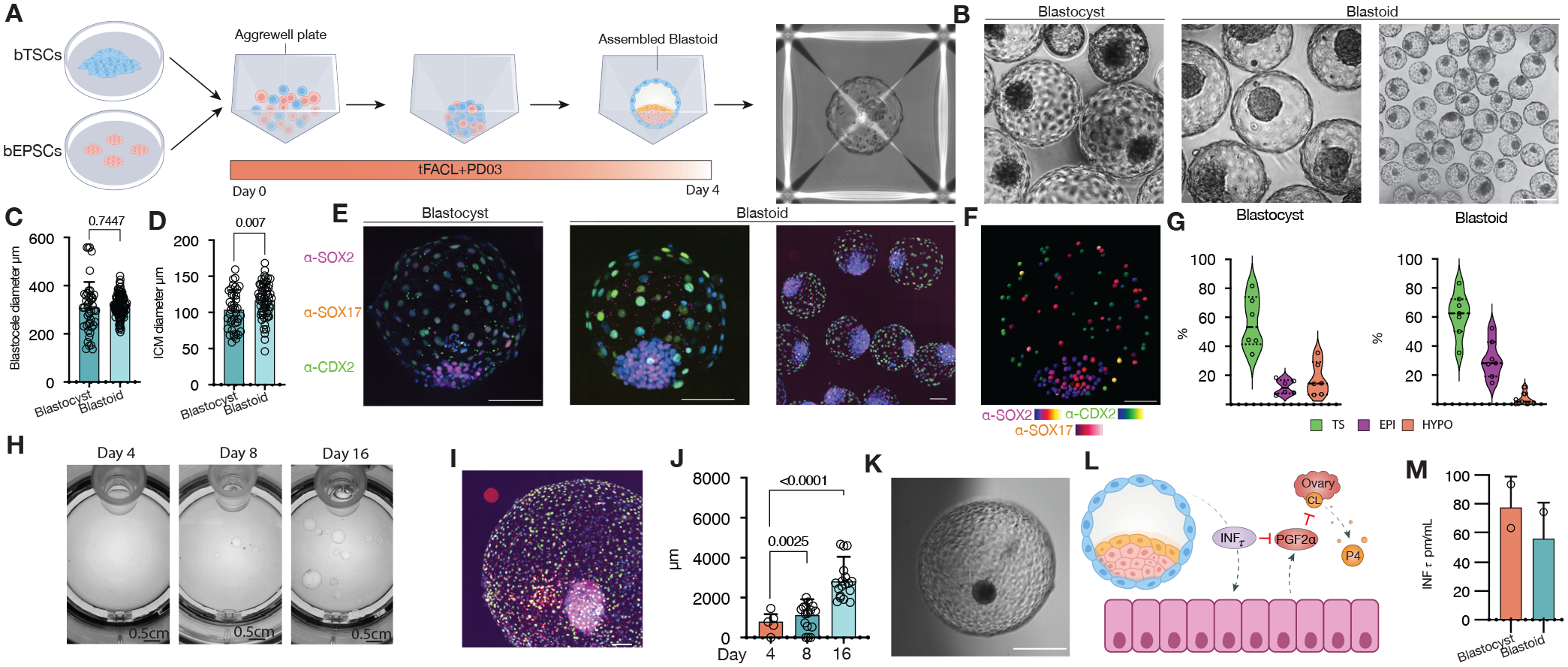
(A) Illustration of the assembly process via bovine EPSCs and TSCs aggregation. (B) Phase-contrast image comparing blastoids vs blastocysts. (C) Blastocele diameter measurement. (D) Inner cell mass (ICM) diameter measurement. (E) Immunostaining for epiblast marker SOX2 (magenta, EPI), hypoblast marker SOX17(red, HYPO) and trophectoderm marker CDX2(green, TS), individual markers in Figure S1. (F) Blastocyst and Blastoid lineage composition quantified via confocal microscopy 3D reconstruction and spots colocalization quantification using IMARIS. (G) Snapshots of in vitro growth of blastoids in a rotating culture system (Clinostar Incubator, Celvivo). (H) Representative image via immunostaining of all three lineages as in e, individual markers in Figure S4. (I) Blastoid diameter quantification. (J) representative micrographs of in vitro grown blastoid. (K) A schematic of the maternal recognition of the action of pregnancy signal interferon TAU (INFt). (L) Enzyme-linked immunosorbent assay (ELISA) measurement of (INFt) in surrogate recipients following embryo transfers. PGF2α: Prostaglandin F2α. CL: Corpus luteum. P4: Progesterone.
Morphologically each bovine blastoid contains a blastocele-like cavity, an outer trophectoderm (TE)-like layer, and an ICM-like compartment, which resembles bovine blastocysts produced by in vitro fertilization (IVF) (Figure 1B, and Video S1). Cavity and ICM sizes of day-4 bovine blastoids reached diameters equivalent to day-8 IVF blastocysts (Figures 1C and 1D). We performed immunofluorescence (IF) analysis and found that bovine blastoids contained cells that expressed markers characteristic of EPI (SOX2), HYPO (SOX17), and trophectoderm (TE) (GATA3, KRT18, and CDX2) lineages (Figures 1E and S1J), and stained positive for a tight junction marker ZO1(TJP1) and an apical marker F-actin (Phalloidin), comparable to blastocysts (Figures S1M and S1N). Despite the similarities, we found that the expression levels of lineage markers were different between IVF blastocysts and blastoids when quantified via IF, with blastoid trophoblast-like cells (TLCs) expressing higher levels of CDX2, HLCs expressing lower levels of SOX17 and epiblast-like cells (ELCs) expressing lower levels of SOX2 when compared to their corresponding cell types in IVF blastocysts (Figures S1I–S1K). While the proportion and cell number for TE were comparable between blastoids and blastocysts, as revealed by confocal microscopy, 3D reconstruction, and spots colocalization (Figure 1F), we found IVF blastocysts contained a higher number of HYP cells and cells co-expressing different lineage markers and a lower number of EPI cells than blastoids (blastoids: 30.87 ±13.11% [ELCs], 61.71±15.54% [TLCs], 3.61 ±4.41% [HLCs]; blastocysts: 9.7±5.02% [EPI], 54.18 ±18.34% [TE],15.27±11.59% [HYP]) (Figures 1F and S1L).
Next, we evaluated the in vitro growth of blastoids and blastocysts under a 3D suspension culture (ClinoStar, see Methods). We found trophoblast cells and cavities in both IVF blastocysts and blastoids continued to proliferate and expand for more than 2 weeks, which were also accompanied by an increase in the ICM size (Figures 1H–1K and S1S–S1W, and Video S2). We performed embryo transfer to synchronized surrogates to evaluate whether blastoids can establish maternal interaction and pregnancy (see Methods). Interestingly, we detected the anti-luteolytic hormone interferon-tau (INFτ) in surrogates’ blood25. INFτ is the signal for maternal recognition of pregnancy in ruminants, which acts by blocking prostaglandin release from the uterus and allowing the corpus luteum to persist and the pregnancy to be maintained26–28 (Figure 1L). INFτ was measured at concentrations of 56.53±25.13pm/ml in 2 out of 4 surrogates 7 days following blastoid transfer, which were comparable to those from IVF blastocyst transfers (78.36±21.54pm/ml) in 2 out of 5 surrogates (Figure 1M).
To determine the transcriptional states of bovine blastoid cells, we performed single-cell RNA-sequencing (scRNA-seq) using the 10x Genomics Chromium platform and carried out integrated analysis with Smart-seq2 single-cell transcriptomes derived from zygote29, 2-cell29, 8-cell30, 16-cell30, morula29 and two sets of day 7.5 blastocyst stage IVF bovine embryos29 as well as in vivo produced bovine blastocysts (see data availability). Joint uniform manifold approximation and projection (UMAP) embedding revealed blastoid-derived cells clustered with blastocyst-derived cells (Figures 2A and 2B). To further evaluate the temporal identity of blastoid cells, we performed two pseudo bulk analysis on the 10x blastoid data at a low and a high cluster resolution, to compensate for the differences in sequencing depth to Smart-seq2 data (Figures 2C, 2D and S2A–S2P). For the first analysis, we also included datasets from bovine early gastrulation-stage embryos31. We found that different embryo datasets were orderly arranged on the PCA plot according to their developmental time, and blastoid cells were mapped closer to blastocyst cells (Figures 2C, 2D and S2A–S2D).
Figure 2. Single-cell characterization of bovine assembled blastoids.
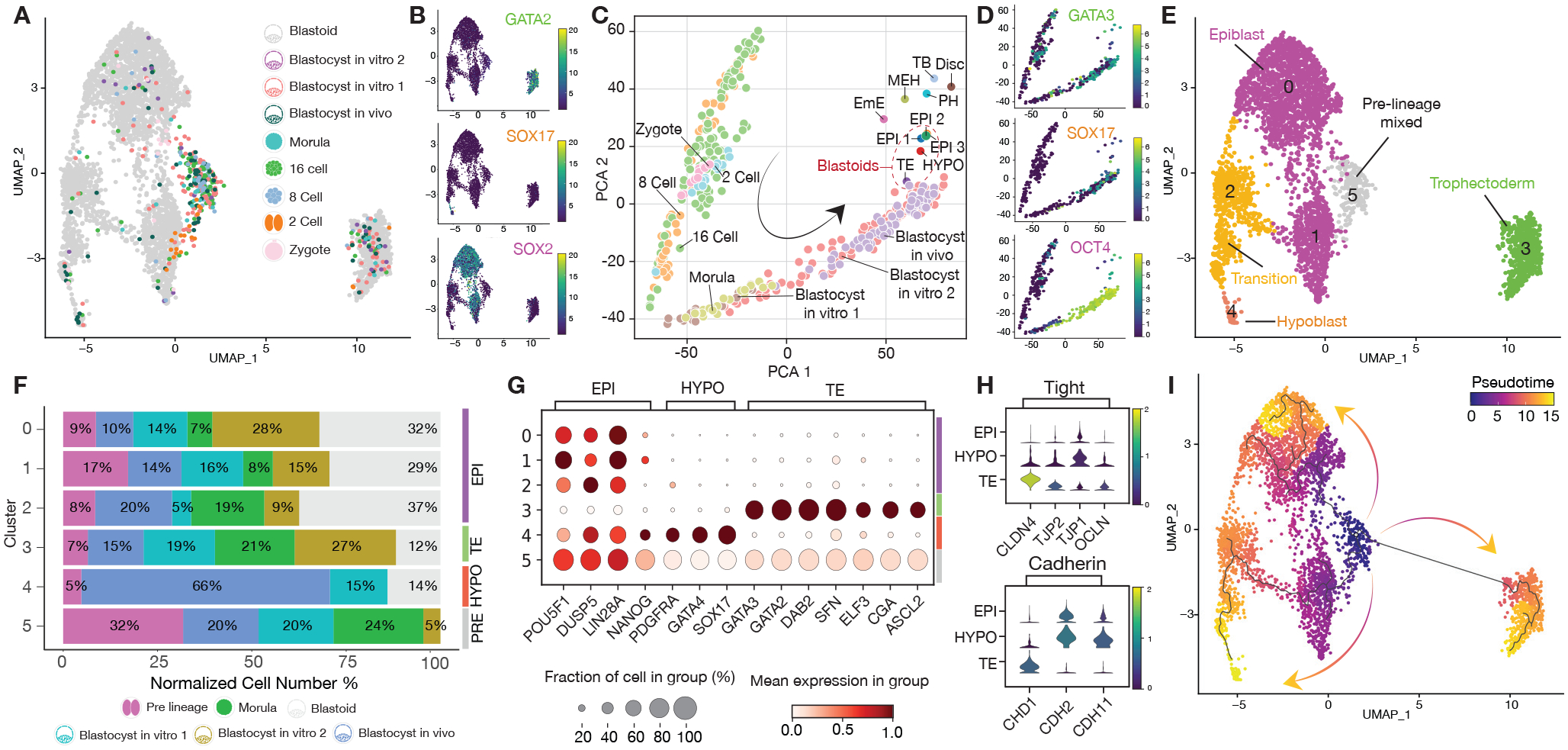
(A) Joint uniform manifold approximation and projection (UMAP) embedding of 10x Genomics single-cell transcriptomes of bovine blastoids (grey) and bovine zygote (pink), 2 cell (orange), 8 cell (blue), 16 cell (green), Morula (cyan) and in vivo and in vitro Blastocyst stage embryos (purple, dark green, light red). (B) UMAP Heatmap showing expression of Trophectoderm (TE), Hypoblast (HYPO), and epiblast (EPI) markers, GATA2, SOX17 and SOX2, respectively. (C) Principal component analysis (PCA) of pseudo bulk conversion of blastoid data. Gastrulation markers31: Disc: Embryonic disc (Day 14 Stage 4). EmE: Embryonic ectoderm (Day 14 Stage 5). MEH: Mesoderm, endoderm, and visceral hypoblast. (Day 14 Stage 5). PH: Parietal hypoblast. (Day 14 Stage 5). TB: Trophoblast. (Day 14 Stage 5). (D) PCA heatmaps showing expression of Trophectoderm (TE), Hypoblast (HYPO), and epiblast (EPI) markers, GATA3, SOX17 and OCT4 (also known as POU5F1), respectively. (E) Major cluster classification based on marker expression. (F) Normalized percentage of cells in each cluster. (G) Dot plot indicating the expression of markers of epiblast (EPI), trophectoderm (TE) and hypoblast (HYPO). (H) Violin plot of lineage-specific cell junction markers. (I) RNA velocity pseudotime analysis depicting the cell trajectories.
We annotated the six identified cell clusters based on marker gene expression and overlap with cells from bovine embryos (Figures 2E–2G and Table S1). Cluster 3 expresses TE markers, e.g., GATA2 and GATA3, and is annotated as TLCs; Cluster 4 expresses HYPO markers, e.g., GATA4 and SOX17, and thus represents HLCs; Three clusters (0, 1, 2) express EPI markers, e.g., SOX2 and LIN28a, and are designated as ELCs; Cluster 5 is mostly composed of cells from pre-blastocyst stage embryos (named pre-lineage), and each blastoid cluster expressed lineage-specific cadherin and tight junction markers (Figures 2E–2H). To evaluate the relationship between clusters, we performed pseudo-time analysis, which predicted the differentiation trajectories from pre-lineage cluster to blastocyst and blastoid lineages, showing markers of epiblast to hypoblast transitioning cells such as RSPO332,33(Figures 2E, 2I, S2H, S2R). Finally, cross-species comparison revealed similarities and differences of bovine blastoids with human blastoids and blastocysts (Figures S2T–S2V).
DISCUSSION
Here we report an efficient and robust protocol to generate bovine blastoids by assembling EPSCs and TSCs that can self-organize and faithfully recreate all blastocyst lineages. Bovine blastoids show a resemblance to bovine blastocysts in morphology, size, cell number, lineage composition, and could produce maternal recognition signal upon transfer to recipient cows. Bovine blastoids represent a valuable model to study early embryo development and understand the causes of early embryonic loss. Upon further optimization, bovine blastoid technology could lead to the development of new artificial reproductive technologies for cattle breeding, which may enable a paradigm shift in livestock reproduction.
Limitations of the study
Despite the similarities, we observed several differences between bovine blastoids and blastocysts, e.g., expression levels of lineage markers, and proportions of EPI and HYP lineages. Future comparisons with in vivo produced embryos should be made to provide a gold standard for lineage proportions. Also, the different culture conditions for IVF blastocysts and blastoids may account for some transcriptional differences observed. TE cells from mature bovine blastocysts are inherently difficult to be dissociated into single cells, and incompletely dissociated TE cells are lost during the filtering process, creating the observed underrepresentation of TE cells in the flow cytometry and scRNA-seq analyses. To better evaluate lineage proportions in blastocysts and blastoids we have performed quantification using confocal imaging and 3D reconstruction. Although pseudo bulk and batch correction allowed us to compare the transcriptomes of blastoid cells with reference blastocyst data, differences in scRNA-seq platforms might have generated bias in the comparative analysis. Processing and sequencing blastoid and blastocyst samples side-by-side to avoid batch differences will be needed in future studies for more proper comparisons. The lack of later stage (≥day 8.5) blastocysts transcriptomic datasets further complicated the analysis as many cells in early-stage bovine blastocyst co-expressed different lineage markers34 (Figures S2B–S2I). Finally, only INF τ measurements were included in the paper and assessment of the developmental potential of bovine blastoids warrants future studies.
STAR methods
RESOURCE AVAILABILITY
Lead contacts
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jun Wu (Jun2.Wu@UTSouthwestern.edu)
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
scRNA-seq data have been deposited at the Gene Expression Omnibus (GEO) and are publicly available as of the date of publication. Accession number is listed in the key resources table.
All original code has been deposited at GitHub and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLES
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CDX-2, Clone CDX2-88 antibody | BioGenex | AM392-5M, RRID: AB_2650531 |
| Human SOX17 Affinity Purified Polyclonal Ab antibody | R&D Systems | AF1924, RRID: AB_355060 |
| Sox-2 (E-4) antibody | Santa Cruz Biotechnology | sc-365823, RRID: AB_10842165 |
| Anti-Human SOX2 | BioGenex | AN833-5M, RRID: not available |
| GATA3-human antibody | Santa Cruz Biotechnology | sc-268, RRID: AB_2108591 |
| Keratin 18 | Sigma | SAB4501665, RRID: AB_10746153 |
| Anti-ZO-1 Monoclonal Antibody | Innovate Research | 33-9100, RRID: AB_87181 |
| Alexa Fluor® 594 Phalloidin antibody | Invitrogen | A12381, RRID: AB_2315633 |
| Phospho-Stat3 (Tyr705) (D3A7) | Cell Signaling Technology | 9145T, RRID: not available |
| Donkey anti-rabbit IgG (H+L) Antibody, Alexa Fluor™ 647 | Invitrogen | A31573, RRID: AB_2536183 |
| Donkey anti-mouse IgG (H+L) Antibody, Alexa Fluor™488 | Invitrogen | A21202, RRID: AB_141607 |
| Donkey anti-Goat IgG (H+L) Antibody, Alexa Fluor™ 555 | Invitrogen | A-21432, RRID: AB_2535853 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| N2 supplement (100X) | Gibco | Cat. No. 17502-048 |
| B27 supplement (50X) | Gibco | Cat. No. 17504-044 |
| Insulin-Transferrin-Selenium-Ethanolamine (ITS-X) | Gibco | Cat. No. 51500-056 |
| Recombinant Human LIF | Peprotech | Cat. No. 300-05 |
| CHIR-99021 | Selleckchem | Cat. No. S1263 |
| PD0325901 | Selleckchem | Cat. No. S1036 |
| Recombinant Human FGF-basic | Peprotech | Cat. No. 100-18B |
| Recombinant Human/Murine/Rat Activin A | Peprotech | Cat. No. 120-14E |
| Emricasan | Selleckchem | Cat. No. 50-136-5234 |
| Polyamine supplement 5ml | Sigma-Aldrich | Cat. No. P8483 |
| Antioxidant supplement, 5ml | Sigma-Aldrich | Cat. No. A1345 |
| trans-ISRIB,10mg | Tocris | Cat. No. 5284 |
| Chroman 1 (HY15392), 5mg | Medchemexpress | Cat. No. 502029121 |
| mTeSR™ Plus | STEMCELL Technologies | Cat. No. 100-1130 |
| WH-4-023 | Tocris | Cat. No. 5413 |
| endo-IWR 1 | Tocris | Cat. No. 3532 |
| XAV-939 | Tocris | Cat. No. 3748 |
| L-Ascorbic acid 2-phosphate | Sigma-Aldrich | Cat. No. A8960 |
| AlbumiNZ Free Fatty Acid | MP Biomedicals | Cat. No. 199899 |
| Dulbecco’s phosphate buffered saline (1X), no calcium, no magnesium (DPBS) | Corning | Cat. No. 354277 |
| DMEM/F12 | Gibco | Cat. No. 11320-033 |
| GlutaMAX (100X) | Gibco, Cat. No. 35050-061 | Cat. No. 35050-061 |
| MEM Non-Essential Amino Acids (100X) | Gibco | Cat. No. 11140-050 |
| 2-Mercaptoethanol (1000X) | Gibco | Cat. No. 21985-023 |
| Fetal Bovine Serum, heat inactivated | Gibco | Cat. No. A3840001 |
| Deposited Data | ||
| 8 cell and 16 cell | Single-cell RNA sequencing reveals developmental heterogeneity of blastomeres during major genome activation in bovine embryos30 | GSE99210 |
| Zygote, 2 cell, 8 cell, morula and blastocyst | Reprogramming barriers in bovine cells nuclear transfer revealed by single-cell RNA-seq analysis29 | PRJNA727165 |
| gastrulation embryos | Provided by Dr. Peter L. Pfeffer, Gene expression analysis of bovine embryonic disc, trophoblast and parietal hypoblast at the start of gastrulation31 | GSE215409 |
| In vivo blastocyst and in vitro blastocyst | Provided by Dr. Zongliang Jiang | GSE215409 |
| Blastoid | This paper | GSE221248 |
| Software and Algorithms | ||
| Cell Ranger (v 3.1.0) | www.10xgenomics.com | https://github.com/10XGenomics/cellranger; RRID:SCR_017344 |
| Seurat (v 3.1.4) | Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902. e1821. | https://satijalab.org/seurat/articles/get_started.html; RRID:SCR_016341 |
| Trim Galore (v 0.6.4) | www.bioinformatics.babraham.ac.uk/projects/trim_galore/ | http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/; RRID:SCR_011847 |
| HISAT2 (v 2.1.0) | Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature biotechnology. 2019;37(8):907–915. | http://daehwankimlab.github.io/hisat2/; RRID:SCR_015530 |
| HTSeq-count (v 2.0.2) | Putri GH, Anders S, Pyl PT, Pimanda JE, Zanini F. Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics. 2022;38(10):2943–2945. | https://htseq.readthedocs.io/en/master/htseqcount.html; RRID:SCR 011867 |
| clusterProfiler (v 3.14.3) | Wu T, Hu E, Xu S, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation. 2021;2(3):100141. | http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html; RRID:SCR 016884 |
| Monocle3 (v 0.2.3.0) | Cao J, Spielmann M, Qiu X, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566(7745):496–502. | https://cole-trapnell-lab.github.io/monocle3/; RRID:SCR 018685 |
| Scanpy (v 1.9.3) | Wolf, F., Angerer, P., Theis, F. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19(15). | https://github.com/scverse/scanpy |
| Scikit-learn (v 1.2.1) | Pedregosa, Fabian, et al. Scikit-learn: Machine learning in Python. the Journal of machine Learning research. 2011;12:2825–2830. | https://scikit-learn.org/stable/ |
| Imaris XT | Oxford Instruments | https://imaris.oxinst.com/products/imaris-for-cell-biologists |
| Code used in this study |
https://github.com/anaorsi/CSC_BovineBlastoids/tree/CSC
https://zenodo.org/record/7792885#.ZCnFqi-B2Ak |
DOI: 10.5281/zenodo.7792885 |
| Other | ||
| AggreWell 400 | STEMCELL Technologies | Cat. No. 34,415 |
| Anti-Adherence Rinsing | STEMCELL Technologies | Cat. No. 07,010 |
| Dispase | STEMCELL Technologies | Cat. No. 07913 |
| Accumax | Thermo Fisher | Cat. No. 00-4666-56 |
| TrypLE™ Express | Gibco | Cat. No. 12605036 |
| ProFreeze Freezing medium | Lonza, 12-769E | Cat. No. 12-769E |
| CoolCell | Corning | Cat. No. 432000 |
| PureCoat™ Amine 6 Well Plate | Corning | Cat. No. 354721 |
| BD LSR II Flow cytometer | BD Biosciences | N/A |
| Nikon CSU-W1 spinning-disk super resolution by optical pixel reassignment (SoRa) | Nikon | N/A |
| Chromium Next GEM Single Cell 3’ Kit v3.1 | 10X Genomics | Cat. No. 1000269 |
| Chromium Next GEM Chip G Single Cell Kit | 10X Genomics | Cat. No. 1000127 |
| Chromium Controller & Next GEM Accessory Kit | 10X Genomics | Cat. No. 000204 |
| Centrifuge HERMLE Z 326 K | HERMLE Labortechnik GmbH | Z 326 K |
| Stereo Microscope SMZ800N | Nikon | SMZ800N |
| Thermo Plate | TOKAI HIT | N/A |
| Micropipette Puller P-97 | Sutter Instruments | N/A |
| Micro Forge MF-900 | Narishige | N/A |
| ECHO Revolve | Echo | N/A |
| Nikon Eclipse Ts2 Inverted microscope | Nikon | N/A |
| Hypoxic humidified Incubator | NuAire | N/A |
| CO2 humidified Incubator | NuAire | N/A |
| 8 well μ-siles | Ibidi | Cat. No. 80821 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines and culture conditions
All cell lines used in this study were cultured at 37°C in a 5% CO2 humidified incubator. Bovine EPSCs and TSCs were cultured on 0.1% gelatin-coated dishes and a layer of inactivated mouse embryonic fibroblasts (iMEF) at 5×105 cells per cm2. All cell lines were periodically tested for mycoplasma contamination via PCR. Cell lines were authenticated by genomic PCR, RT-qPCRs, immunostaining, RNA-seq and/or in vitro differentiation.
Bovine EPSCs stem cells culture
Bovine female EPSCs35, generated via culture adaptation of bovine ESCs (derived and cultured in the FR1 [FGF+IWR1] condition9,10) in an bovine EPSC culture (3i+LAF)35: mTeSR base, 1% BSA,10ng/ml LIF, 20ng/ml Activin A, WH-4–023 0.3 μM, 1 μM Chir99021 20ng/ml FGF2, 5 μM IWRI and/or 5μM XAV-939, Ascorbic acid (Vitamin C) 50μg/ml. Bovine ESCs were adapted to the bEPSC (3i+LAF) condition for a minimum of 5 passages until doomed colony morphology was visible. N2B27 basal medium was prepared by adding 1× N2 supplement (Gibco), 1× B27 supplement (Gibco), 1× GlutaMAX, 1× NEAA(Gibco), and 2-mercaptonethanol (Gibco) (final concentration 0.1 mM) to 1:1 (vol/vol) mixture of DMEM/F12 (Gibco) and neurobasal medium (Gibco). Upon passaging, cells were washed with 1xPBS and dissociated with TrypLE (Thermo Fisher) for 3 minutes at 37°C; cells were then collected with 0.05% BSA in DMEM-F12 (Thermo Fisher) and centrifuged at 1000xg for 3 minutes and resuspended in 1ml of media per 9.6cm2. Each passage cells were counted using Countess II (Thermo Fisher) and plated at a density of 30,000 cells/cm2, at this plating ratio cells were passaged every 4 days. Upon plating, cells were treated with either (Y-27632) or the CEPT cocktail36 (50 nM chroman-1 [C, Tocris], 5 μM emricasan [E, Selleckchem], 0.7 μM trans-ISRIB [T, Tocris], and 1 x polyamine supplement [P, Thermo]) during the first 12h after passaging. Fresh culture media was added every day. Cells were cryopreserved in CoolCell freezing containers (Corning), in bEPSC media with 10% DMSO at 0.5×10^6 cells per ml and stored in liquid nitrogen the following day. Detailed descriptions of media are in the STAR methods Key Resource table.
Bovine TSCs stem cell culture
Bovine male TSCs were derived and cultured in LCDM media (CELL-REPORTS-D-23-01022) with slight modifications (N2B27 base, 1% BSA, 10ng/ml LIF, 3 μM Chir99021, 2μM Minocycline hydrochloride (M), 2μM (S)-(+)-Dimethindene maleate (D). During passaging cells were treated with Accumax (Thermo Fisher) or Dispase (STEMCELL Technologies) for 5 minutes at 37°C (no PBS wash), cells were collected with the same volume of bTSC medium and gently lifted of the plate using a wide opening p100 pipette tip and gentle force. Cells were split at a 1:3 ratio and plated on iMEFs with CEPT. Only 1ml of media was plated in a 6 well for the first 24h to facilitate bTSCs attachment. bTSCs do not survive well after single-cell dissociation and tend to form trophospheres if not plated correctly. These steps are critical for long term culture and expansion of bTSCs. Cells were cryopreserved in CoolCell freezing containers (Corning) in 45 % LCDM 45% FBS and 10% DMSO or ProFreeze Freezing medium (Lonza, 12–769E) at 2×10^6 cells per ml.
Animals
Cross breed (Bos taurus x Bos indicus) non-lactating female cows with an average age of 3 years were used as recipients. Cows were housed in open pasture, and under constant care of the farm staff at the Reproductive Biological Center (RBC) at the School of Animal Sciences, Louisiana State University Agriculture Center (LSU AgCenter). The animal protocol was approved by the Animal Care and Use Committee of LSU AgCenter (A2021–21).
Materials and Methods
Blastoid formation
For blastoid formation, EPSCs single-cells were collected as stated above. Bovine TSCs were washed with 1x PBS, dissociated with Trypsin for 10 minutes at 37 °C, with constant pipetting every 2–3 minutes and inactivated with DMEM-F12 containing 10% fetal bovine serum (FBS). Cells were washed twice and on final resuspension in their normal culture media with 1x CEPT and 10 UI per ml of DNase I (Thermo Fisher). To deplete iMEF cells, collected cells were placed in precoated 12 well plates (Corning) with 0.1% gelatin and incubated for 15 minutes at 37°C. Single-cell dissociation was made by gentle but constant pipetting and by passing the cells through a glass capillary pulled to an inner diameter of 50–100μm (micropipette puller, Sutter Instruments), hermetically attached to a p200 pipette tip. After single-cell dissociation, cells were collected and strained using a 70μm (TSCs) and then a 37μm cell strainers (EPSCs) (Corning). This same single-cell dissociation procedure was used for blastoids processing for 10x genomics. Cells were stained with 1x trypan blue and manually counted in a Neubauer chamber. Current protocol is optimized for 16 bEPSCs and 16 bTSCs per well in a ~1200 well Aggrewell 400 microwell culture plate (Stemcell technologies) for 19,200 of each cell type per well. Each well was precoated with 500μl of Anti-Adherence Rinsing Solution (Stemcell technologies) and spun for 5 minutes at 1500 rcf. Wells were rinsed with 1ml of PBS just before aggregation. An appropriate number of cells for the wells to be aggregated was centrifuged at 1000xg for 3 minutes and resuspended in 1ml of tFACL+PD media (N2B27 base, 1% BSA, 0.5x ITS-X, 20ng/ml LIF, 10ng/ml LIF, 10ng/ml FGF2, 0.3μM PD032590, 1μM Chir99021) per well, supplemented with 1x CEPT. To ensure even distribution of the cells within each microwell, cells were gently mixed by pipetting with a P200 pipette, then the plate was centrifuged at 1300 rcf for 2 minutes and put in a humidified incubator at 37°C with 5% CO2 and 5% Oxygen (NuAire). As MEK inhibition inhibits hypoblast differentiation a gradual decrease can be done if higher numbers of hypoblast cells are desired from 0.3 to 0.125μM. It is important to have viable, MEF free, cell debris free, and evenly distributed cells as any of these factors can negatively affect blastoid formation.
In vitro fertilization
Bovine IVF was performed as previously described37 with modifications. Briefly oocytes were collected at a commercial abattoir (DeSoto Biosciences) and shipped in an MOFA metal bead incubator (MOFA Global) at 38.5°C overnight in sealed sterile vials containing 5% CO2 in air-equilibrated Medium 199 with Earle’s salts (Thermo Fisher), supplemented with 10% fetal bovine serum (Hyclone), 1% penicillin–streptomycin (Invitrogen), 0.2-mM sodium pyruvate, 2-mM L-glutamine (Sigma), and 5.0 mg/mL of Folltropin (Vetoquinol). The oocytes were matured in this medium for 22 to 24 hours. Matured oocytes were washed twice in warm Tyrode lactate (TL) HEPES supplemented with 50 mg/mL of gentamicin (Invitrogen) while being handled on a stereomicroscope (Nikon) equipped with a 38.5°C stage warmer. In vitro fertilization was conducted using a 2-hour pre-equilibrated IVF medium modified TL medium supplemented with 250-mM sodium pyruvate, 1% penicillin–streptomycin, 6 mg/mL of fatty acid–free BSA (Sigma), 20-mM penicillamine, 10-mM hypotaurine, and 10 mg/mL of heparin (Sigma) at 38.5 C, 5% CO2 in a humidified air incubator. Frozen semen (Bovine-elite) was thawed at 35°C for 1 minute, then separated by centrifugation at 200xg for 20 minutes in a density gradient medium (Isolate, Irvine Scientific) 50% upper and 90% lower. Supernatant was removed; sperm pellet was resuspended in 2-mL modified Tyrode’s medium and centrifuged at 200 g for 10 minutes to wash. The sperm pellet was removed and placed into a warm 0.65-mL microtube before bulk fertilizing in Nunc four-well multidishes (VWR) containing up to 50 matured oocytes per well at a concentration of 1.0×10^6 sperm/mL. 18 hours after insemination, oocytes were cleaned of cumulus cells by constant pipetting for 3-minutes in vortex in 100μl drop of TL HEPES with 0.05% Hyaluronidase (Sigma), washed in TL HEPES, and then cultured in 500μl of IVC media (IVF-Biosciences) supplemented with 0.5xN2B27 (Thermo Fisher) and FLI20 under mineral oil (Irvine Scientific) cultured until the blastocyst stage. Cleavage rates were recorded on Day 2, and viable embryos were separated from nonviable embryos. Blastocyst rates were recorded on Day 8 after IVF.
Immunofluorescent staining
Samples (Cells, single-cells, blastoids and blastocysts) were fixed with 4% paraformaldehyde (PFA) in 1xDPBS with 0.1% PVA for 20 min at room temperature, washed in wash buffer (0.1% Triton X-100, 5% BSA in 1xDPBS) for 15 minutes and permeabilized with 0.1–1% Triton X-100 in PBS for 1 h. For phosphor-specific antibodies samples were treated with 0.5% SDS for 1h. Samples were then blocked with blocking buffer (PBS containing 5% Donkey serum, 5% BSA, and 0.1% Triton X-100) at room temperature for 1 h, or overnight at 4 °C. Because of the large number of blastoids, to facilitate processing blastoids were gently washed out of the aggrewell plate and separated from cell debris using a 100μM reversible strainer (Stem cells), blastoids were then placed in a 70μm strainer (Corning) in a 6 well plate containing wash buffer, and the strainer was moved from one well to another between steps. Primary antibodies were diluted in blocking buffer according to Key Resource table. Blastoids were incubated in primary antibodies in 96 wells for 2 h at room temperature or overnight at 4 °C. Samples were washed three times for 15 minutes with wash buffer, and incubated with fluorescent-dye conjugated secondary antibodies (AF-488, AF-555 or AF-647, Invitrogen) diluted in blocking buffer (1:300 dilution) for 2 h at room temperature or overnight at 4 °C. Samples were washed three times with PBS-T. Finally, cells were counterstained with 300 nM 4′,6-diamidino-2-phenylindole (DAPI) solution at room temperature for 20 min. Phalloidin was directly stained along with other secondary antibodies in the blocking buffer.
Imaging
Phase contrast images were taken using a hybrid microscope (Echo Laboratories, CA) equipped with objective x2/0.06 numerical aperture (NA) air, x4/0.13 NA air, x10/0.7 NA air and 20x/0.05 NA air. Fluorescence imaging was performed on 8 well μ-siles (Ibidi) on a Nikon CSU-W1 spinning-disk super-resolution by optical pixel reassignment (SoRa) confocal microscope with objectives x4/0.13 NA, a working distance (WD) of 17.1nm, air; ×20/0.45 NA, WD 8.9–6.9 nm, air; ×40/0.6 NA, WD 3.6–2.85 nm, air. In vitro growth blastocyst was imaged in a glass slide under a coverslip using a Keyence BZ-X810 scope.
Imaging analysis
Imaging experiments were repeated at least twice, with consistent results. In the figure captions, n denotes the number of biological repeats. Raw images were first processed in Fiji38 to create maximal intensity projection (MIP) and an export of representative images. Nuclear segmentation was performed in Ilastik. MIP images and segmentation masks were processed in MATLAB (R2022a) using custom code, available in a public repository. Nuclear localized fluorescence intensity was computed for each cell in each field, and the value was then normalized to the DAPI intensity of the same cell. Intensity values of all cells were plotted as mean ± s.d. Lineage cell number quantification was made using Imaris (v10, Oxford) XT module and spots colocalization tool. Total number of cells was calculated based on DAPI spots and spots for each channel were cleared if not overlapping DAPI spots. Hypoblast cells were calculated as SOX17 or SOX17 and SOX2 co-localized spots. Trophectoderm cells were calculated from CDX2 only, CDX2, and SOX17.
Flow Cytometry
Blastoids were collected under a stereo microscope and single-cell dissociated as stated above for the TSCs. Strained single-cells were processed as stated above for immunofluorescent staining performing wash steps in 1.5ml Eppendorf tubes on a 90° centrifuge. Flow cytometry was performed using the appropriate unstained and single stain controls in a DBiosciences LSR II flow cytometer and analyzed using Flow Jo. Gating Strategy is shown in Figure S1k–l.
In vitro growth
Prior to use for bovine blastoid culture, the water beads inside the humidity chamber of the ClinoReactor (CelVivo), were hydrated with sterile water (Corning) overnight at 4°C. Once hydrated and the growth chamber was filled with N2B27 basal media, and the reactor chamber was equilibrated for 1h at 37°C before exchanging for culture media. For rotating-culture blastoids were collected at day 4 post aggregation and placed in pre-equilibrated ClinoReactors in 10ml of tFACL+PD03 media and 1x CEPT (Key Resource table). ClinoReactors were placed in the ClinoStar incubator at 37 °C with a gas mix of 5%CO2, 5% O2 and air. The rotation speed was set between 10 and 12 rpm and was lowered progressively as the blastoids expanded. Optimal growth conditions were achieved by exchanging media every four days. Blastoid and blastocysts growth was also tested on N2B27 with rock inhibitor (Y27632) and activin A as reported in39. (Figure S1S–W). Blastoids could also grow in N2B27 with 20ng of FGF4, 20ng of FGF2 and Activin A (FFA), data not shown.
Embryo Transfer
Surrogate cows were synchronized with an intramuscular (IM) an injection of ovulation-inducing gonadotropin-release hormone (GnRH, Fertagyl), followed by a standard 7-day vaginal controlled drug internal release (CIDR) of progesterone. Upon CIDR removal, one dose of prostaglandin (Lutalyse) was administered. 48 hours after CIDR removal another dose of GnRH was administered via IM injection. A cohort of 15–20 bovine blastoids or 12–15 control IVF blastocysts were loaded into 0.5 mL straws in prewarmed Holding medium (ViGro) and transferred non-surgically to the uterine horn ipsilateral to the ovary with the corpus luteum (CL) as detected by transrectal ultrasound. 7 days after transfer, blastoids were be recovered by standard non-surgical flush with lactated ringers’ solution supplemented with 1% fetal bovine serum. All recipients were treated with prostaglandin (Lutalyse) after flushing.
Quantitative measurement of Bovine IFN-tau in blood
Blood samples from surrogate and controls were drawn from the coccygeal vein using serum separator tubes. The samples were immediately placed in refrigerator overnight before centrifugation for 15 minutes at 1000 ×g. IFN03c4 in the serum was determined by Bovine Interferon-Tau ELISA Kit (CSB-E 16948B) according to manufacturer’s protocol. Briefly, each well was added 100 μL standard or sample and incubated for 2 hours at 37 °C Then, liquid was removed and 100 μL Biotin-antibody (1X) was added to each well, incubating 1 hour at 37 °C. After aspirating the wells, 200 μL Wash Buffer was used to wash the wells for three times. After last wash, the plate was inverted and blotted against clean paper towels to remove any remaining Wash Buffer. 100 μL HRP-avidin (1X) was added to each well and incubated for 1 hour at 37 °C. 200 μL Wash Buffer was used to wash the wells for five times. 90 μL TMB Substrate was added and incubated for 20 minutes at 37 °C. Protect from light. 50 μL Stop Solution was added to each well, gently tapping plate to ensure thorough mixing. The plate was measured using microplate reader set to 450 nm.
Single-cell RNA-Seq library generation
Bovine blastoids were single-cell dissociated and strained cells were prepared as stated adobe. Cells were washed in PBS containing 0.04% BSA and centrifuged at 90° x500g for 5 min. Cell were resuspended in PBS containing 0.04% BSA at a single-cell suspension of 1,000 cells/μL. Cells were loaded into a 10x Genomics Chromium Chip following manufacturer instruction (10x Genomics, Pleasanton, CA, Chromium Next GEM Single Cell 3ʹ GEM, Library & Gel Bead Kit v3.1) and sequenced by Illumina NextSeq 500/550 sequencing systems (Illumina).
Published single-cell data collection
We collected single-cell sequencing data from published literature for comparative analysis. Two Bovine IVF single-cell sequencing raw FASTQ data were downloaded from the GEO database, including 179 IVF cells40 sequenced using Smart-seq2 and 98 IVF cells41 sequenced using STRT-seq. For the pseudo bulk analysis, bulk RNA sequencing data of gastrulation-stage embryos (Pfeffer et al., 2017) was also included.
Pre-processing single-cell data
For 10X Genomics single-cell data, we used the Cell Ranger pipeline (v.3.1.0) with default parameters to generate the expression count matrix. The bovine reference genome and gene annotation file were downloaded from Ensembl database (UMD3.1) and generated by Cell Ranger mkfastq with default parameters. Seurat42 (3.1.4) was used to single-cell quality control. To reduce multiplets and dead cells, we screened cells with expressed gene numbers between 2000 and 6000, unique molecular identifiers (UMIs) between 5000 and 30,000, and mitochondrial RNA genes counts below 15 percent.
For public Smart-seq2 and STRT-seq data, raw FASTQ reads were trimmed using Trim Galore (0.6.4, https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) with default parameters. In order to minimize processing differences, trimmed reads were aligned to the same genome reference (UMD3.1) by using HISAT243 (2.1.0) with default parameters. Read counts per gene were annotated by HTSeq-count44 software (2.0.2) using the same gene annotation files (UMD3.1). Then, transcripts per million (TPM) were calculated to reduce gene length differences. Also, dead cells were removed by filter mitochondrial gene counts content below 15%.
Normalization and dimensionality reduction
We used log-percentage value to normalize each single-cell expression matrix, which can reduce the bias of gene expression values caused by different sequencing depths and sequencing methods. In order to reducing the dimension of feature genes and improving the efficiency and accuracy of integration, the variance and mean of genes in each single-cell cohort were used to fit local polynomial regression and filter the top 2000 variable feature genes45.
Data integration and clustering
The Find Integration Anchors model in the Seurat package was used to find the similarity anchor structure between different single-cell data. Then, we completed the data integration according to the anchors information with 80 dimensions, 20 anchors, 40 candidate cells, and reciprocal PCA for dimensionality reduction (‘dims = 1:80, k.anchor = 20, k.filter = 40, reduction = “rpca”’). Single cells were clustered using the shared nearest neighbor (SNN) modularity optimization-based clustering algorithm in Seurat package, with 90 Principal Component (PC) and 0.6 resolution. Then, Uniform manifold approximation and projection (UMAP) was used to reduce the dimensions and show the visualize figure with non-default parameters: ‘dims = 1:90’.
Pseudo bulk analysis
Clusters of blastoid cells were annotated according to the expression of marker genes. To generate pseudo bulk counts, total counts for each gene were summed for cells sharing a cluster. Genes expressed in less than 5% of all samples were excluded. Cells were normalized by total counts and log-transformed. Data was scaled and the PCA was calculated by Python package Scikit-learn.
For single-cell RNA-seq pseudo bulk data integration, blastoid cells were processed with Python package Scanpy. Cells were divided into 52 clusters and raw counts per gene from all cells sharing a cluster were summed. After this process, blastoid pseudo bulk samples contained approximately the same number of counts of embryo cells. Samples containing more than 4% of counts from mitochondrial genes or with number of genes per counts less than 2000 were excluded. The data was normalized, log-transformed and the top 4000 variable genes were kept for further analysis. Samples were scaled and Principal Component Analysis was carried out with Harmony Integrate, using dataset as key.
Differentially expressed genes analysis
Differentially expressed genes between clusters and groups were determined using the Scanpy rank genes groups tool, using a Wilcoxon rank-sum test.
Matrix plot heatmap data graphical representation
Graphs were made using the Scanpy matrixplot function according to predetermined lists gathered from the MSigDB significant signatures or published stem cell signaling gene lists. Graphs were made by grouping pseudo bulk data by developmental stage and lines annotation.
Gene function annotation
Gene ontology (GO)46 terms and Kyoto encyclopedia of genes and genomes (KEGG)47 pathways enrichment were performed using clusterProfiler48 (3.14.3; org.Bt.eg.db v 3.10.0) with parameter: ‘pvalueCutoff = 0.05’.
Pseudotime construction
Monocle349 (0.2.3.0) was used for pseudotime analysis, with the UMI matrix and UMAP embedding matrix generated by Seurat as input. Cell pseudotime trend was learnt by using cells in all clusters to generate a single and acyclic structure graph (‘use_partition = F, close_loop = F’).
Datasets used
8 cell and 16 cell from GSE99210 (Single-cell RNA sequencing reveals developmental heterogeneity of blastomeres during major genome activation in bovine embryos)30. Zygote, 2 cell, 8 cell, morula and blastocyst from PRJNA727165 (Reprogramming barriers in bovine cells nuclear transfer revealed by single‐cell RNA‐seq analysis)29. Raw unprocessed data of gastrulation embryos was obtained from Dr. Peter L. Pfeffer31. In vivo blastocyst and in vitro blastocyst1 datasets were obtained from Dr. Zongliang Jiang (GSE215409). Bovine blastoid single-cell raw and processed data have been deposited in the Gene Expression Omnibus under accession code (GSE221248).
Supplementary Material
Table S1. Differentially expressed genes in blastoids and blastocysts, Related to Figure 2
Video S1. Bovine blastoid Z plane overview, Related to Figure 1
Video S2. Bovine blastoid 3D rotating culture on day 16, Related to Figure 1
Highlights.
Robust and efficient generation of bovine blastoids via assembly of EPSCs and TSCs
Bovine blastoids show molecular and cellular similarities to blastocysts
Bovine blastoids grow for more than two weeks under a 3D suspension culture
Bovine blastoids elicit maternal recognition of pregnancy following embryo transfer
Acknowledgements
We thank Dr. Joel Carter from J A Carter, CETS, LLC, for his assistance with embryo transfer and flushing. J.W. is a New York Stem Cell Foundation–Robertson Investigator and Virginia Murchison Linthicum Scholar in Medical Research and funded by CPRIT (RR170076), NIH (GM138565-01A1 and OD028763), Welch (854671) and UT Southwestern & Texas A&M clinical translation and translational award (CSTA) program supported by the NIH (1UL1TR003163-01A1). Z.J. is funded by the United States Department of Agriculture (2019–67016–29863), the National Institutes of Health (R01HD102533). A.E.R.O. is supported by the São Paulo Research Foundation (FAPESP) (grants #2020/16612-9 and #2022/02096-4). The Nikon SoRa spinning disk microscope was purchased by the UTSW quantitative light microscopy core with a Shared Instrumentation grant from NIH award: 1S10OD028630-01 to Katherine Luby-Phelps. We would like to thank the Lu Sun lab for providing access to the Keyence BZ-X810 scope. We would also like to thank the Celvivo team (Peter Willems-Alnøe, Reeham Motaher and Louis Lichten) for their support stabilizing the 3D culture system.
Inclusion and diversity
One of more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. We support inclusive, diverse, and equitable conduct of research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
To review GEO accession GSE221248, go to Enter token: ghwzaewsjjspdsr (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE221248)
To review GEO accession GSE215409, Enter token: cbevwsuilncpjcn (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE215409)
Declaration of interests
C.A.P-A., Y.Wang., Y.Wei., Z.J. and J.W., are co-inventors on US provisional patent application 63/370,068 relating to Bovine blastocysts like structures and uses thereof.
References
- 1.Rivron NC, Frias-Aldeguer J, Vrij EJ, Boisset J-C, Korving J, Vivié J, Truckenmüller RK, van Oudenaarden A, van Blitterswijk CA, and Geijsen N (2018). Blastocyst-like structures generated solely from stem cells. Nature 557, 106–111. 10.1038/s41586-018-0051-0. [DOI] [PubMed] [Google Scholar]
- 2.Sozen B, Cox AL, De Jonghe J, Bao M, Hollfelder F, Glover DM, and Zernicka-Goetz M (2019). Self-Organization of Mouse Stem Cells into an Extended Potential Blastoid. Developmental cell 51, 698–712.e698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li R, Zhong C, Yu Y, Liu H, Sakurai M, Yu L, Min Z, Shi L, Wei Y, Takahashi Y, et al. (2019). Generation of Blastocyst-like Structures from Mouse Embryonic and Adult Cell Cultures. Cell 179, 687–702.e618. 10.1016/j.cell.2019.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu L, Wei Y, Duan J, Schmitz DA, Sakurai M, Wang L, Wang K, Zhao S, Hon GC, and Wu J (2021). Blastocyst-like structures generated from human pluripotent stem cells. Nature 591, 620–626. 10.1038/s41586-021-03356-y. [DOI] [PubMed] [Google Scholar]
- 5.Yanagida A, Spindlow D, Nichols J, Dattani A, Smith A, and Guo G (2021). Naive stem cell blastocyst model captures human embryo lineage segregation. Cell Stem Cell 28, 1016–1022.e1014. 10.1016/j.stem.2021.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X, Tan JP, Schröder J, Aberkane A, Ouyang JF, Mohenska M, Lim SM, Sun YBY, Chen J, Sun G, et al. (2021). Modelling human blastocysts by reprogramming fibroblasts into iBlastoids. Nature 591, 627–632. 10.1038/s41586-021-03372-y. [DOI] [PubMed] [Google Scholar]
- 7.Kagawa H, Javali A, Khoei HH, Sommer TM, Sestini G, Novatchkova M, Scholte op Reimer Y, Castel G, Bruneau A, Maenhoudt N, et al. (2022). Human blastoids model blastocyst development and implantation. Nature 601, 600–605. 10.1038/s41586-021-04267-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu L, Ezashi T, Wei Y, Duan J, Logsdon D, Zhan L, Nahar A, Arteaga CAP, Liu L, Stobbe C, et al. (2022). Large scale production of human blastoids amenable to modeling blastocyst development and maternal-fetal crosstalk. bioRxiv, 2022.2009.2014.507946. 10.1101/2022.09.14.507946. [DOI] [PubMed] [Google Scholar]
- 9.Bogliotti YS, Wu J, Vilarino M, Okamura D, Soto DA, Zhong C, Sakurai M, Sampaio RV, Suzuki K, Izpisua Belmonte JC, and Ross PJ (2018). Efficient derivation of stable primed pluripotent embryonic stem cells from bovine blastocysts. Proc Natl Acad Sci U S A. 10.1073/pnas.1716161115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soto DA, Navarro M, Zheng C, Halstead MM, Zhou C, Guiltinan C, Wu J, and Ross PJ (2021). Simplification of culture conditions and feeder-free expansion of bovine embryonic stem cells. Sci Rep 11, 11045. 10.1038/s41598-021-90422-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinoshita M, Kobayashi T, Planells B, Klisch D, Spindlow D, Masaki H, Bornelöv S, Stirparo GG, Matsunari H, Uchikura A, et al. (2021). Pluripotent stem cells related to embryonic disc exhibit common self-renewal requirements in diverse livestock species. Development 148. 10.1242/dev.199901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang Y, Cai NN, An XL, Zhu WQ, Yang R, Tang B, Li ZY, and Zhang XM (2022). Naïve-like conversion of bovine induced pluripotent stem cells from Sertoli cells. Theriogenology 196, 68–78. 10.1016/j.theriogenology.2022.10.043. [DOI] [PubMed] [Google Scholar]
- 13.Xiang J, Wang H, Zhang Y, Wang J, Liu F, Han X, Lu Z, Li C, Li Z, Gao Y, et al. (2021). LCDM medium supports the derivation of bovine extended pluripotent stem cells with embryonic and extraembryonic potency in bovine-mouse chimeras from iPSCs and bovine fetal fibroblasts. The FEBS journal 288, 4394–4411. 10.1111/febs.15744. [DOI] [PubMed] [Google Scholar]
- 14.Su Y, Wang L, Fan Z, Liu Y, Zhu J, Kaback D, Oudiz J, Patrick T, Yee SP, Tian XC, et al. (2021). Establishment of Bovine-Induced Pluripotent Stem Cells. International journal of molecular sciences 22. 10.3390/ijms221910489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao L, Gao X, Zheng Y, Wang Z, Zhao G, Ren J, Zhang J, Wu J, Wu B, Chen Y, et al. (2021). Establishment of bovine expanded potential stem cells. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2018505118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Navarro M, Soto DA, Pinzon CA, Wu J, and Ross PJ (2020). Livestock pluripotency is finally captured in vitro. Reproduction, Fertility and Development 32, 11–39. [DOI] [PubMed] [Google Scholar]
- 17.Yang Y, Liu B, Xu J, Wang J, Wu J, Shi C, Xu Y, Dong J, Wang C, Lai W, et al. (2017). Derivation of Pluripotent Stem Cells with In Vivo Embryonic and Extraembryonic Potency. Cell 169, 243–257.e225. 10.1016/j.cell.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu L, Wei Y, Sun HX, Mahdi AK, Pinzon Arteaga CA, Sakurai M, Schmitz DA, Zheng C, Ballard ED, Li J, et al. (2021). Derivation of Intermediate Pluripotent Stem Cells Amenable to Primordial Germ Cell Specification. Cell Stem Cell 28, 550–567 e512. 10.1016/j.stem.2020.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Wei Y, Zhang E, Yu L, Ci B, Guo L, Sakurai M, Takii S, Liu J, Schmitz DA, Ding Y, et al. (2023). Dissecting embryonic and extra-embryonic lineage crosstalk with stem cell co-culture. bioRxiv, 2023.2003.2007.531525. 10.1101/2023.03.07.531525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stoecklein KS, Ortega MS, Spate LD, Murphy CN, and Prather RS (2021). Improved cryopreservation of in vitro produced bovine embryos using FGF2, LIF, and IGF1. PLoS One 16, e0243727. 10.1371/journal.pone.0243727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turner N, and Grose R (2010). Fibroblast growth factor signalling: from development to cancer. Nature reviews. Cancer 10, 116–129. 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 22.Lavoie H, Gagnon J, and Therrien M (2020). ERK signalling: a master regulator of cell behaviour, life and fate. Nat Rev Mol Cell Biol 21, 607–632. 10.1038/s41580-020-0255-7. [DOI] [PubMed] [Google Scholar]
- 23.Rossant J, and Tam PPL (2017). New Insights into Early Human Development: Lessons for Stem Cell Derivation and Differentiation. Cell Stem Cell 20, 18–28. [DOI] [PubMed] [Google Scholar]
- 24.Canizo JR, Ynsaurralde Rivolta AE, Vazquez Echegaray C, Suvá M, Alberio V, Aller JF, Guberman AS, Salamone DF, Alberio RH, and Alberio R (2019). A dose-dependent response to MEK inhibition determines hypoblast fate in bovine embryos. BMC Dev Biol 19, 13. 10.1186/s12861-019-0193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheikh AA, Hooda OK, Kalyan A, Kamboj A, Mohammed S, Alhussien M, Reddi S, Shimray PG, Rautela A, Pandita S, et al. (2018). Interferon-tau stimulated gene expression: A proxy to predict embryonic mortality in dairy cows. Theriogenology 120, 61–67. [DOI] [PubMed] [Google Scholar]
- 26.Hansen PJ, and Tríbulo P (2019). Regulation of present and future development by maternal regulatory signals acting on the embryo during the morula to blastocyst transition – insights from the cow. Biology of reproduction 101, 526–537. 10.1093/biolre/ioz030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brooks K, and Spencer TE (2015). Biological Roles of Interferon Tau (IFNT) and Type I IFN Receptors in Elongation of the Ovine Conceptus1. Biology of reproduction 92, 47, 41–10. 10.1095/biolreprod.114.124156. [DOI] [PubMed] [Google Scholar]
- 28.Bazer FW, and Thatcher WW (2017). Chronicling the discovery of interferon tau. Reproduction (Cambridge, England) 154, F11–f20. 10.1530/rep-17-0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao L, Long C, Zhao G, Su J, Ren J, Sun W, Wang Z, Zhang J, Liu M, Hao C, et al. (2022). Reprogramming barriers in bovine cells nuclear transfer revealed by single-cell RNA-seq analysis. J Cell Mol Med 26, 4792–4804. 10.1111/jcmm.17505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavagi I, Krebs S, Simmet K, Beck A, Zakhartchenko V, Wolf E, and Blum H (2018). Single-cell RNA sequencing reveals developmental heterogeneity of blastomeres during major genome activation in bovine embryos. Sci Rep 8, 4071. 10.1038/s41598-018-22248-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfeffer PL, Smith CS, Maclean P, and Berg DK (2017). Gene expression analysis of bovine embryonic disc, trophoblast and parietal hypoblast at the start of gastrulation. Zygote 25, 265–278. 10.1017/s0967199417000090. [DOI] [PubMed] [Google Scholar]
- 32.Corujo-Simon E, Radley AH, and Nichols J (2022). Evidence implicating sequential commitment of the founder lineages in the human blastocyst by order of hypoblast gene activation. bioRxiv, 2022.2012.2008.519626. 10.1101/2022.12.08.519626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boroviak T, Stirparo GG, Dietmann S, Hernando-Herraez I, Mohammed H, Reik W, Smith A, Sasaki E, Nichols J, and Bertone P (2018). Single cell transcriptome analysis of human, marmoset and mouse embryos reveals common and divergent features of preimplantation development. Development 145. 10.1242/dev.167833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo L, Shi Y, Wang H, Wang Z, Dang Y, Li S, Wang S, and Zhang K (2022). Base editing in bovine embryos reveals a species-specific role of SOX2 in regulation of pluripotency. PLOS Genetics 18, e1010307. 10.1371/journal.pgen.1010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao L, Gao X, Zheng Y, Wang Z, Zhao G, Ren J, Zhang J, Wu J, Wu B, Chen Y, et al. (2021). Establishment of bovine expanded potential stem cells. Proceedings of the National Academy of Sciences 118, e2018505118. 10.1073/pnas.2018505118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y, Tristan CA, Chen L, Jovanovic VM, Malley C, Chu PH, Ryu S, Deng T, Ormanoglu P, Tao D, et al. (2021). A versatile polypharmacology platform promotes cytoprotection and viability of human pluripotent and differentiated cells. Nat Methods 18, 528–541. 10.1038/s41592-021-01126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Golding MC, Snyder M, Williamson GL, Veazey KJ, Peoples M, Pryor JH, Westhusin ME, and Long CR (2015). Histone-lysine N-methyltransferase SETDB1 is required for development of the bovine blastocyst. Theriogenology 84, 1411–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramos-Ibeas P, Lamas-Toranzo I, Martínez-Moro Á, de Frutos C, Quiroga AC, Zurita E, and Bermejo-Álvarez P (2020). Embryonic disc formation following post-hatching bovine embryo development in vitro. Reproduction (Cambridge, England) 160, 579–589. 10.1530/rep-20-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lavagi I, Krebs S, Simmet K, Beck A, Zakhartchenko V, Wolf E, and Blum H (2018). Single-cell RNA sequencing reveals developmental heterogeneity of blastomeres during major genome activation in bovine embryos. Scientific reports 8, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao L, Long C, Zhao G, Su J, Ren J, Sun W, Wang Z, Zhang J, Liu M, and Hao C (2022). Reprogramming barriers in bovine cells nuclear transfer revealed by single-cell RNA-seq analysis. Journal of cellular and molecular medicine 26, 4792–4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck III WM, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive integration of single-cell data. Cell 177, 1888–1902. e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D, Paggi JM, Park C, Bennett C, and Salzberg SL (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature biotechnology 37, 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Putri GH, Anders S, Pyl PT, Pimanda JE, and Zanini F (2022). Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics 38, 2943–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hao Y, Hao S, Andersen-Nissen E, Mauck III WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, and Zager M (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587. e3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.The Gene Ontology resource: enriching a GOld mine. (2021). Nucleic acids research 49, D325–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanehisa M, Sato Y, and Kawashima M (2022). KEGG mapping tools for uncovering hidden features in biological data. Protein Science 31, 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, and Zhan L (2021). clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation 2, 100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, and Steemers FJ (2019). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Differentially expressed genes in blastoids and blastocysts, Related to Figure 2
Video S1. Bovine blastoid Z plane overview, Related to Figure 1
Video S2. Bovine blastoid 3D rotating culture on day 16, Related to Figure 1
Data Availability Statement
scRNA-seq data have been deposited at the Gene Expression Omnibus (GEO) and are publicly available as of the date of publication. Accession number is listed in the key resources table.
All original code has been deposited at GitHub and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLES
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CDX-2, Clone CDX2-88 antibody | BioGenex | AM392-5M, RRID: AB_2650531 |
| Human SOX17 Affinity Purified Polyclonal Ab antibody | R&D Systems | AF1924, RRID: AB_355060 |
| Sox-2 (E-4) antibody | Santa Cruz Biotechnology | sc-365823, RRID: AB_10842165 |
| Anti-Human SOX2 | BioGenex | AN833-5M, RRID: not available |
| GATA3-human antibody | Santa Cruz Biotechnology | sc-268, RRID: AB_2108591 |
| Keratin 18 | Sigma | SAB4501665, RRID: AB_10746153 |
| Anti-ZO-1 Monoclonal Antibody | Innovate Research | 33-9100, RRID: AB_87181 |
| Alexa Fluor® 594 Phalloidin antibody | Invitrogen | A12381, RRID: AB_2315633 |
| Phospho-Stat3 (Tyr705) (D3A7) | Cell Signaling Technology | 9145T, RRID: not available |
| Donkey anti-rabbit IgG (H+L) Antibody, Alexa Fluor™ 647 | Invitrogen | A31573, RRID: AB_2536183 |
| Donkey anti-mouse IgG (H+L) Antibody, Alexa Fluor™488 | Invitrogen | A21202, RRID: AB_141607 |
| Donkey anti-Goat IgG (H+L) Antibody, Alexa Fluor™ 555 | Invitrogen | A-21432, RRID: AB_2535853 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| N2 supplement (100X) | Gibco | Cat. No. 17502-048 |
| B27 supplement (50X) | Gibco | Cat. No. 17504-044 |
| Insulin-Transferrin-Selenium-Ethanolamine (ITS-X) | Gibco | Cat. No. 51500-056 |
| Recombinant Human LIF | Peprotech | Cat. No. 300-05 |
| CHIR-99021 | Selleckchem | Cat. No. S1263 |
| PD0325901 | Selleckchem | Cat. No. S1036 |
| Recombinant Human FGF-basic | Peprotech | Cat. No. 100-18B |
| Recombinant Human/Murine/Rat Activin A | Peprotech | Cat. No. 120-14E |
| Emricasan | Selleckchem | Cat. No. 50-136-5234 |
| Polyamine supplement 5ml | Sigma-Aldrich | Cat. No. P8483 |
| Antioxidant supplement, 5ml | Sigma-Aldrich | Cat. No. A1345 |
| trans-ISRIB,10mg | Tocris | Cat. No. 5284 |
| Chroman 1 (HY15392), 5mg | Medchemexpress | Cat. No. 502029121 |
| mTeSR™ Plus | STEMCELL Technologies | Cat. No. 100-1130 |
| WH-4-023 | Tocris | Cat. No. 5413 |
| endo-IWR 1 | Tocris | Cat. No. 3532 |
| XAV-939 | Tocris | Cat. No. 3748 |
| L-Ascorbic acid 2-phosphate | Sigma-Aldrich | Cat. No. A8960 |
| AlbumiNZ Free Fatty Acid | MP Biomedicals | Cat. No. 199899 |
| Dulbecco’s phosphate buffered saline (1X), no calcium, no magnesium (DPBS) | Corning | Cat. No. 354277 |
| DMEM/F12 | Gibco | Cat. No. 11320-033 |
| GlutaMAX (100X) | Gibco, Cat. No. 35050-061 | Cat. No. 35050-061 |
| MEM Non-Essential Amino Acids (100X) | Gibco | Cat. No. 11140-050 |
| 2-Mercaptoethanol (1000X) | Gibco | Cat. No. 21985-023 |
| Fetal Bovine Serum, heat inactivated | Gibco | Cat. No. A3840001 |
| Deposited Data | ||
| 8 cell and 16 cell | Single-cell RNA sequencing reveals developmental heterogeneity of blastomeres during major genome activation in bovine embryos30 | GSE99210 |
| Zygote, 2 cell, 8 cell, morula and blastocyst | Reprogramming barriers in bovine cells nuclear transfer revealed by single-cell RNA-seq analysis29 | PRJNA727165 |
| gastrulation embryos | Provided by Dr. Peter L. Pfeffer, Gene expression analysis of bovine embryonic disc, trophoblast and parietal hypoblast at the start of gastrulation31 | GSE215409 |
| In vivo blastocyst and in vitro blastocyst | Provided by Dr. Zongliang Jiang | GSE215409 |
| Blastoid | This paper | GSE221248 |
| Software and Algorithms | ||
| Cell Ranger (v 3.1.0) | www.10xgenomics.com | https://github.com/10XGenomics/cellranger; RRID:SCR_017344 |
| Seurat (v 3.1.4) | Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902. e1821. | https://satijalab.org/seurat/articles/get_started.html; RRID:SCR_016341 |
| Trim Galore (v 0.6.4) | www.bioinformatics.babraham.ac.uk/projects/trim_galore/ | http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/; RRID:SCR_011847 |
| HISAT2 (v 2.1.0) | Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature biotechnology. 2019;37(8):907–915. | http://daehwankimlab.github.io/hisat2/; RRID:SCR_015530 |
| HTSeq-count (v 2.0.2) | Putri GH, Anders S, Pyl PT, Pimanda JE, Zanini F. Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics. 2022;38(10):2943–2945. | https://htseq.readthedocs.io/en/master/htseqcount.html; RRID:SCR 011867 |
| clusterProfiler (v 3.14.3) | Wu T, Hu E, Xu S, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation. 2021;2(3):100141. | http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html; RRID:SCR 016884 |
| Monocle3 (v 0.2.3.0) | Cao J, Spielmann M, Qiu X, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566(7745):496–502. | https://cole-trapnell-lab.github.io/monocle3/; RRID:SCR 018685 |
| Scanpy (v 1.9.3) | Wolf, F., Angerer, P., Theis, F. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19(15). | https://github.com/scverse/scanpy |
| Scikit-learn (v 1.2.1) | Pedregosa, Fabian, et al. Scikit-learn: Machine learning in Python. the Journal of machine Learning research. 2011;12:2825–2830. | https://scikit-learn.org/stable/ |
| Imaris XT | Oxford Instruments | https://imaris.oxinst.com/products/imaris-for-cell-biologists |
| Code used in this study |
https://github.com/anaorsi/CSC_BovineBlastoids/tree/CSC
https://zenodo.org/record/7792885#.ZCnFqi-B2Ak |
DOI: 10.5281/zenodo.7792885 |
| Other | ||
| AggreWell 400 | STEMCELL Technologies | Cat. No. 34,415 |
| Anti-Adherence Rinsing | STEMCELL Technologies | Cat. No. 07,010 |
| Dispase | STEMCELL Technologies | Cat. No. 07913 |
| Accumax | Thermo Fisher | Cat. No. 00-4666-56 |
| TrypLE™ Express | Gibco | Cat. No. 12605036 |
| ProFreeze Freezing medium | Lonza, 12-769E | Cat. No. 12-769E |
| CoolCell | Corning | Cat. No. 432000 |
| PureCoat™ Amine 6 Well Plate | Corning | Cat. No. 354721 |
| BD LSR II Flow cytometer | BD Biosciences | N/A |
| Nikon CSU-W1 spinning-disk super resolution by optical pixel reassignment (SoRa) | Nikon | N/A |
| Chromium Next GEM Single Cell 3’ Kit v3.1 | 10X Genomics | Cat. No. 1000269 |
| Chromium Next GEM Chip G Single Cell Kit | 10X Genomics | Cat. No. 1000127 |
| Chromium Controller & Next GEM Accessory Kit | 10X Genomics | Cat. No. 000204 |
| Centrifuge HERMLE Z 326 K | HERMLE Labortechnik GmbH | Z 326 K |
| Stereo Microscope SMZ800N | Nikon | SMZ800N |
| Thermo Plate | TOKAI HIT | N/A |
| Micropipette Puller P-97 | Sutter Instruments | N/A |
| Micro Forge MF-900 | Narishige | N/A |
| ECHO Revolve | Echo | N/A |
| Nikon Eclipse Ts2 Inverted microscope | Nikon | N/A |
| Hypoxic humidified Incubator | NuAire | N/A |
| CO2 humidified Incubator | NuAire | N/A |
| 8 well μ-siles | Ibidi | Cat. No. 80821 |


