Abstract
Background
Antimicrobial resistance (AMR) is a severe public health problem that Bangladeshis are dealing with nowadays. However, we wanted to investigate the pooled prevalence of Salmonella and AMR in Salmonella strains isolated from livestock- and poultry-derived foods between 1 January 2000 and 31 August 2022.
Methods
The metafor and metareg packages in the R programming language were used to conduct all analyses. We used a random-effect or fixed-effect model for pooled prevalence of Salmonella and AMR to Salmonella, depending on the heterogeneity test for each antibiotic. The heterogeneity was examined using stratified analyses, the meta-regression approach and sensitivity analysis.
Results
The combined prevalence of Salmonella in livestock and poultry-derived food in Bangladesh is 37%, according to the 12-research considered (95% CI: 23%–52%). According to subgroup analysis, neomycin had the lowest prevalence of resistance (4%, 95% CI: 1%–13%), whereas tetracycline had the highest prevalence of resistance (81%, 95% CI: 53%–98%). According to univariate meta-analysis and correlation analysis, the prevalence of Salmonella increased with the study period (β = 0.0179; 95% CI: 0.0059–0.0298, P = 0.0034; R2 = 46.11%) and without this, none of aforementioned variables was significantly associated with the detected heterogeneity and there was a positive relationship (r = 0.692, P = 0.001) between the Salmonella prevalence and study period.
Conclusions
AMR is rising alarmingly in Bangladesh by livestock-derived food consumption. However, monitoring and evaluating antibiotic sensitivity trends and developing effective antibiotic regimens may improve Salmonella infection inhibition and control in Bangladesh. Policymakers should be concerned about food handling practices. Doctors should be concerned when using prescribing antibiotics.
Introduction
Salmonella is one of the most commonly recognized pathogens that cause gastroenteritis,1,2 which results in significant morbidity, mortality and economic loss.3,4 In 2010, the World Health Organization (WHO) reported 153 million cases of non-typhoidal Salmonella (NTS) enteric infections worldwide, of which 56 969 were fatal and 50% were foodborne.5Salmonella was the second foodborne epidemic in some regions’ illness monitoring reports from 2006 to 2010.6 Among the 2600 Salmonella serotypes discovered, NTS serovars such as Typhimurium and Enteritidis are the most common worldwide.7,8 Poultry has been identified as the one cause of human salmonellosis, and avian salmonellosis affects the poultry business and can infect humans when infected poultry meat and eggs are consumed.9 Eggs are the principal source of salmonellosis and other foodborne illnesses10–13Salmonella that grows in animal farms may contaminate eggs and meat during the slaughtering process before being transmitted to people via the food chain. Indeed, multiple earlier investigations have reported the isolation of Salmonella from animal and human diets.14–17 Human S. Enteritidis is commonly associated with the intake of infected eggs and chicken meat, whereas S. Typhimurium is commonly associated with the consumption of pork, poultry and beef.18,19 In addition, Salmonella enterica serovars have been found in varying concentrations in animal products and by-products worldwide.18,20,21 The most often-reported serovars connected with human foodborne diseases are Salmonella Typhimurium and Enteritidis.22 However, untyped Salmonella of animal origin is becoming more common in Bangladesh.23,24
However, domestic chickens in developing countries live close to humans in urban and rural communities and are frequently housed overnight in the family home.25 As soon as a chicken becomes infected, it sheds faeces into the environment. Additionally, interacting with employees in poultry farms and slaughterhouses, the main route of human Salmonella infections, involves contaminated meat and eggs.26 One of the primary reasons for animal management is the risk of antimicrobial resistance (AMR) in humans and animals.27
AMR is a worldwide public health issue.28,29 AMR can be caused by one of three fundamental processes: (i) antibiotic modification by lowering absorption or enhancing efflux of the antibiotic via their enzymes; (ii) alteration in the antibiotic's target site and (iii) gaining the capacity to break or change the antibiotic.30 Several lines of evidence showed that using antimicrobial agents in food animals contributes to the emergence and spread of AMR in foodborne Salmonella.31 AMR has recently been a significant issue in treating Salmonella infections.32,33Salmonella infections in food animals are essential in public health and, in particular, food safety because food products of animal origin are thought to be the most common source of human Salmonella infections.34 Contamination by healthy food handlers is also assessed during food processing. In recent years, it has been estimated that animals and their products can account for up to 96% of all Salmonella infections in humans.35,36 AMR is expected to increase by 70% in Asia, posing a national and global threat.37 The WHO estimates that Salmonella infections cause 93.8 million instances of gastroenteritis worldwide each year and that Salmonella infections cause 155 000 fatalities.38
Moreover, according to a recent Shanghai study, just 1.1% of strains were responsive to all 16 medications, and AMR rates for third and fourth-generation cephalosporins (cefotaxime and cefepime) were 10% and 8.1%, respectively.39 Furthermore, according to a study conducted in Guangzhou, annual resistance rates of ampicillin are reasonably consistent. However, resistance rates of NTS to ceftazidime in 2015 (31.43%) were significantly greater than in 2014 (16%). Furthermore, AMR to ampicillin was considerably higher in serotype Typhimurium and Enteritidis isolates than in other serotypes.40Salmonella drug resistance rates to cephalosporin and cefepime were 22.3% and 13.1%, respectively, in four hospitals in Shenzhen,41 which were all higher than the results of earlier Chinese investigations.42,43 This phenomenon demonstrates that the outlook for AMR is bleak. Regarding AMR mechanisms, the corresponding resistance genes are usually found on plasmids, transposons, gene cassettes or variants of the Salmonella genomic islands SGI1 and SGI2.44
However, this study revealed a prevalence of Salmonella in poultry and livestock-derived foods. In this regard, this meta-analytical study will be evidential for assessing the prevalence of Salmonella in livestock and poultry-derived food and AMR in livestock and poultry-derived foods and comprehensively investigate the whole scenario of Bangladesh. We hypothesize that Salmonella pooled prevalence and antibiotic resistance are increasing in Bangladesh.
Materials and methods
Data sources and systematic search strategy
We looked for research focused on investigating the prevalence of Salmonella and AMR in livestock and poultry-derived food in both English and Bangla. In English and Bangla, an attempt was made to locate grey and published scientific literature. From 2000 to August 2022, search engines such as Google Scholar, Scopus, ISI Web of Knowledge and PubMed were used to identify published literature, with reference lists of pertinent articles manually searched. We did not find any article on Bangla. The screening of titles and abstracts to find relevant publications was followed by full-text scanning of the relevant articles in a two-step approach.
Additionally, we were concerned with resolving any conflicts that developed during the data extraction to eliminate selection bias. ‘Salmonella’ AND ‘antibiotic resistance OR antimicrobial resistance OR drug resistance OR AMR’ AND ‘prevalence OR incidence OR morbidity OR odd ratio OR risk ratio OR confidence interval OR P value OR rate’ AND ‘antibiotic resistance OR antimicrobial resistance OR drug resistance’ AND ‘Bangladesh’. Studies conducted on livestock and poultry were chosen using the Species filters in PubMed and Google Scholar. Because there was no sorting filter for species in the Web of Science and Cochrane Library, new search phrases were added to select species. The articles found during the search were exported to EndNote to be checked for duplicates. The unique hits were then uploaded to the Rayyan QCRI website for data extraction and screening. The title and abstract were screened first, followed by the complete content of the article.
Data collection process and data items
As a result of our search approach, all items were exported to the EndNote program. Duplicated articles were removed from the database. One neutral reviewer (A.A.) examined the titles and abstracts of the discovered papers. The whole texts of qualifying papers were obtained and appropriately evaluated for eligibility. The records were examined for legitimacy by the second examiner (M.S.S.). In the event of a disagreement between the two reviewers (A.A. and M.S.S.), the reviewer was then consulted by F.A.M. and A.M.S. Then, the article information was included by the three reviewers (N.D., K.S.I. and R.B.G.).
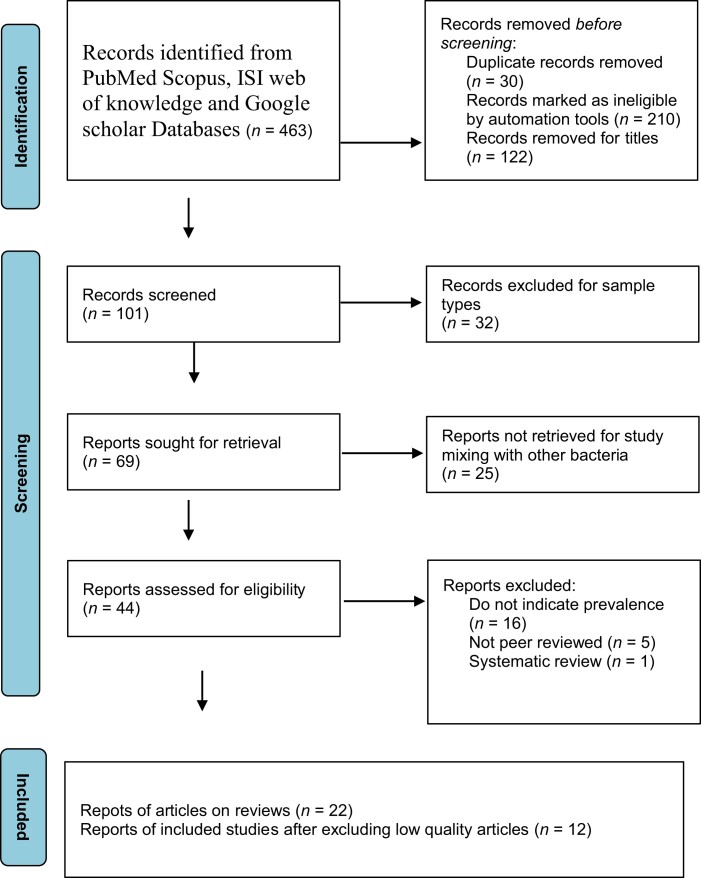
Furthermore, the Final was checked by one reviewer (R.K.R.). Next, statistical analysis was performed by R.K.R. and U.M. The search strategy was depicted in a PRISMA flow chart, which showed which studies were included and excluded and the reasons for exclusion (Figure 1).
Figure 1.
PRISMA flow diagram for this study.
Data extraction and quality assessment
We choose studies on the following criteria: (i) reported AMR in livestock and poultry-derived foods in Bangladesh; (ii) published between 2000 and August 2022; (iiii) samples indicated poultry and livestock-derived foods and (iv) were scientifically reported on location, number of samples and outcomes. The analysis did not cover the detection of Salmonella and the resistance pattern of sample types of faeces, faecal, cloacal, intestinal and rectal. In Bangladesh, there were no restrictions on when the study might be published.
Reporting bias assessment
Data quality was assessed according to the Joanna Briggs Institute.45–47 Appropriate sample frame, study participants sampled, sample size, description of study subjects and setting, sample size justification, power description or variance and effect estimates, valid methods for condition identification, a standard and reliable condition measured appropriate statistical analysis, and adequate response rate were among the nine factors used to assess the risk of bias. The phrases ‘yes’, ‘no’, ‘unclear’ and ‘not available’ were used to convey the risk assessment criteria. They received a one-point score, while the others received a zero. The total score was 0 to 9. The likelihood of bias was considered low when the overall score was greater than 70%, moderate when it was 50%–69% and high when it was 0%–49% (Table 1).45,47
Table 1.
Lists the explanatory factors for study regarding Salmonella that were retrieved from the investigations
| Author, year | Study year | Location | Host | Sample no. | Positive no. | Sample type | AST method | AST standard | Quality of articles | Resistance antibiotics |
|---|---|---|---|---|---|---|---|---|---|---|
| Fatema et al., 2014 | 2013 | Dhaka | Chicken | 20 | 10 | Meat | Disc diffusion | CLSI | 4 | Ampicillin, Tetracycline, Nalidixic acid, Ciprofloxacin Gentamicin, Erythromycin, Rifampicin, Streptomycin, Kanamycin, zithromycin |
| Mahmud et al., 2016 | 2013 | Chittagong | Chicken | 240 | 71 | egg shell surface, egg content |
Disc diffusion, Broth |
CLSI | 7 | Enrofloxacin |
| Khan et al., 2000 | 2004 | Mymensingh, Feni, Dhaka |
Chicken | 24 | 24 | Meat | Disc diffusion | CLSI | 4 | Ampicillin, Cloxacillin, Nalidixic acid, Erythromycin |
| Hosain et al., 2012 | 2012 | Mymensingh | pigeon | 112 | 40 | Clocal swab, Foot pad Faeces |
Disc diffusion | CLSI | 4 | Ampicillin, Amoxicillin, Tetracycline, Chloramphenicol, Nalidixic acid, Gentamicin, Erythromycin, Kanamycin |
| Akond et al., 2012 | 2010 | Dhaka | Chicken | 210 | 71 | Cloacal swab, Intestinal fluid, Egg surface |
Agar | CLSI | 4 | Ampicillin, Penicillin, Tetracycline, Chloramphenicol, Nalidixic acid, Ciprofloxacin, Erythromycin, Norfloxacin, Rifampicin, Cefixime, Cephalexin |
| Saifullah et al., 2016 | 2015 | Mymensingh | Pigeon | 50 | 17 | Cloacal swab, Pharyngeal swab |
Disc diffusion | CLSI | 4 | Ampicillin, Nalidixic acid, Gentamicin, Azithromycin, Cephalexin |
| Rahman et al., 2018 | 2017 | Mymensingh, Gazipur |
Chicken, Cow | 169 | 37 | Chicken meat, Milk, Beef |
Disc diffusion | CLSI | 6 | Amoxicillin, Oxytetracycline, Doxycycline, Sulpha/Trimetho/Cotri, Ciprofloxacin, Gentamicin, Erythromycin, Neomycin, Amikacin, Azithromycin |
| Islam et al., 2018 | 2017 | Dhaka | Chicken | 200 | 148 | Egg | Disc | NCCLS | 7 | Ampicillin, Amoxicillin, Tetracycline, Chloramphenicol, Nalidixic acid, Ciprofloxacin, Gentamicin, Kanamycin, Azithromycin, Ceftriaxone |
| Khan et al., 2019 | 2018 | Sylhet | Cattle | 200 | 24 | Milk | Agar | CLSI | 7 | Amoxicillin, Tetracycline, Sulpha/Trimetho/Cotri, Erythromycin, Streptomycin |
| Sobur et al., 2019 | 2018 | Mymensingh | Cattle | 100 | 60 | Cow dung, Milk | Disc | CLSI | 4 | Tetracycline, Oxytetracycline, Chloramphenicol, Ciprofloxacin, Gentamicin, Erythromycin, Neomycin, Kanamycin, Azithromycin, Ertapenem, Meropenem, Imipenem |
| Islam et al., 2018a | 2017 | Jamalpur, Netrokona, Tangail, Kishoreganj |
Chicken | 20 | 14 | Meat | Disc | CLSI | 8 | Ampicillin, Amoxicillin, Tetracycline, Ciprofloxacin, Erythromycin Azithromycin |
| Rahman et al., 2019 | 2017 | Dhaka | Chicken | 50 | 50 | Egg | Disc | CLSI | 4 | Ampicillin, Tetracycline, Gentamicin |
| Hassan et al., 2014 | 2012 | Chittagong | Chicken | 30 | 13 | Liver, Spleen | Disc | NCCLS | 9 | Amoxicillin, Tetracycline, Doxycycline, Colistin, Ciprofloxacin, Pefloxacin, Kanamycin, Enrofloxacin |
| Alam et al., 2020 | 2017 | Mymensingh | Chicken | 100 | 35 | Cloacal swab | Disc | 4 | Ampicillin, Tetracycline, Chloramphenicol, Ciprofloxacin, Streptomycin, Ertapenem |
|
| Jahan et al., 2018 | 2017 | Mymensingh | Quails | 75 | 10 | Cloacal swab | Disc | CLSI | 4 | Amoxicillin, Tetracycline, Colistin, Erythromycin, Neomycin |
| Joy et al., 2017 | 2017 | Sylhet | Chicken | 320 | 46 | Gut materials | Disc | CLSI | 4 | Ampicillin, Tetracycline, Sulpha/Trimetho/Cotri, Ciprofloxacin, Erythromycin, Neomycin |
| Islam et al., 2022 | 2019–2020 | Chattogram | Poultry Chickens | 16 | 8 | Chicken meat, Liver | Disc | CLSI | 9 | Not clear |
| Rabby et al., 2021 | 2020 | Dhaka | Poultry | 52 | 7 | Meat | Disc | CLSI | 7 | Erythromycin; Cloxacillin; Ampicillin; Trimethoprim; Nitrofurantoin; Ciprofloxacin; Cefotaxime; Ceftriaxone; Levofloxacin; Co-Trimoxazole |
| Rahman et al., 2022 | 2020–2021 | Barishal | Poultry | 40 | 13 | Chicken meat, Frozen milk | Disc | CLSI | 8 | Amikacin, Streptomycin, Gentamicin, Azithromycin, Oxytetracycline, Amoxicillin, Oxacillin, Sulfamethoxazole- Trimethoprim, Ceftriaxone, Ciprofloxacin |
| Matubber et al., 2021 | Not clear | Barishal, Pirojpur and Bhola | Chicken, Cattle, Goat | 205 | 19 | Chicken meat, Cattle meat, Buffalo meat and Goat meat | Disc | CLSI | 6 | Amoxicillin, Penicillin, Chloramphenicol, Erythromycin, Cefradine, Oxytetracycline, Enrofloxacin, Penicillin, Erythromycin, Cotrimoxazole, Cefradine, Amoxicillin, Penicillin, Cefradine |
| Rahman et al., 2018a | 2017 | Dhaka | Chicken | 189 | 82 | Meat | Disc | CLSI | 7 | Amoxicillin, Penicillin, Chloramphenicol, Erythromycin, Cefradine, Oxytetracycline, |
| Begum et al., 2007 | 2006 | Dhaka | Chicken | 50 | 35 | Meat | Disc | CLSI | 7 | Penicillin, Chloramphenicol, Erythromycin, Cefradine, Oxytetracycline |
Statistical analysis
After reviewing the entire article for quality, the authors independently extracted data using purpose-built forms that specified the pertinent factors. Disagreements were settled by debating the articles and coming to an agreement. The significant dependent variable was a binary categorization of study results on the basis of whether the article supported the prevalence of Salmonella and antibiotic resistance to Salmonella or not. The proportion for the study based on the standardized effect size was the dependent variable in the meta-analysis. The statistical analysis was carried out with the help of the R programming language. The metafor and metareg in the R programming language were used to determine the pooled prevalence after excluding low-quality articles.40,45,46 Metafor uses inverse-variance weights from a random-effects model to pool proportions and offers a weighted subgroup and overall pooled estimate. It entailed a meta-analysis of the prevalence values of each publication, weighted by sample size and allowing for potential heterogeneity across studies in this context. Before the random-effects model, the Freeman–Tukey transformation produced a proportional meta-analysis. Prevalence estimates were merged using a random-effects meta-analysis for accounting for between-study heterogeneity. The statistical heterogeneity was assessed using the chi-square test on the Q (H, I2) statistic, which was quantified by the I2 values under the assumption that I-square values of 25%, 50% and 75% were nominally assigned as low, moderate and high estimates, respectively.40,45 The following grouping variables were used in stratified analyses and meta-regression for covariates to examine potential sources of heterogeneity. To establish whether one or more studies had an impact on the outcomes by being excluded one at a time, a sensitivity analysis was also performed. The distribution of the observed studies was visually inspected on a funnel plot to determine publication bias. Egger's linear regression and Begg's rank correlation test were then used to measure the level of bias.40,45,46 The significance threshold was kept at <0.05.
Results
Search result and eligible studies
PRISMA was used to outline the specific steps of the systematic review and meta-analysis method, and Figure 1 outlines the process of selecting relevant papers. We identified 463 new studies via PubMed, Google Scholar, Scopus, ISI and Web of Knowledge databases. Owing to duplicate records, ineligible by automation tools, titles, sample types and study had mixing with other bacteria, 419 articles were removed. Twenty-two studies were removed for not indicating prevalence, being systematic and not being peer-reviewed. According to the quality assessment standards, two studies received a score of 9, two got an 8, six got a 7, two got a 6 and 10 got a 4. According to the quality scores, the studies were generally deemed to be of acceptable quality (scores more than or equal to 6)47,48. Ten articles were also removed for methodological quality. For that, we included 12 studies in this meta-analysis.
Characteristics of eligible studies
Table 1 included the characteristics of eligible studies The study sample included Chittagong, Mymensingh, Sylhet, Chittagong and Barisal divisions. The hosts were chickens, cattle and goats. These three are the most commonly used livestock for a portion of food in Bangladesh. According to the included study, 1411 livestock-derived food were used as a study sample, and the review found 471 samples to be Salmonella positive and resistant to some antibiotics. Most samples were identified by the disc diffusion method, and the CLSI method was used as an AST method. Tetracycline, oxytetracycline, doxycycline, sulpha/trimetho/cotri, ciprofloxacin, gentamycin and neomycin have frequently shown resistance against Salmonella in many reviewed studies.
Systematic review and meta-analysis results
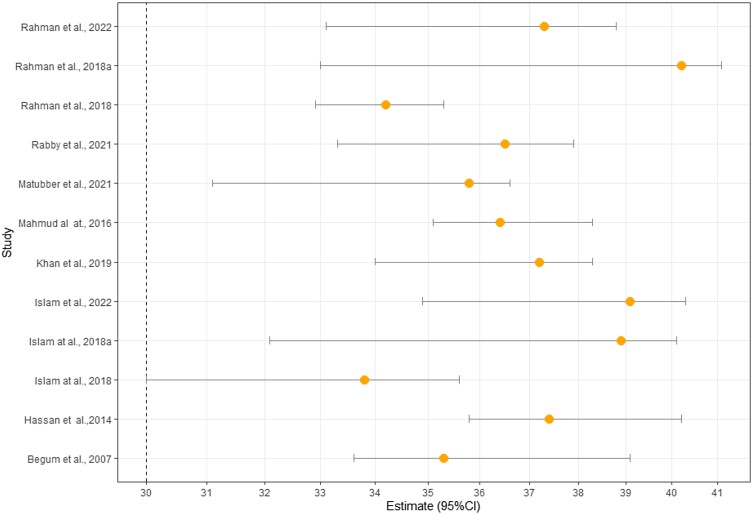
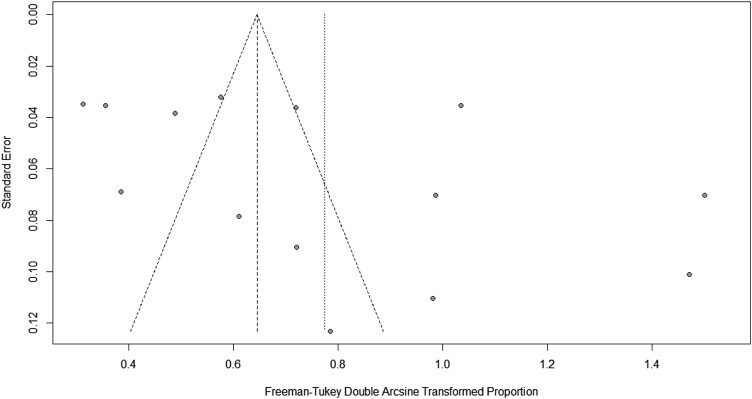
A higher level of multi-drug resistance was present in Bangladesh. In Bangladesh, the combined prevalence of Salmonella isolated from livestock and poultry-derived foods (sample types faeces, faecal, cloacal and rectal excluded) was 37% of the sample. Overall, 37% of the sample had Salmonella in livestock and poultry-derived foods [95% CI: (23%–52%), P ≤ 0.001, R2 = 97%]. A high prevalence was seen for some of the commonly prescribed antibiotics like tetracycline 81% [(95% CI:53–98), P ≤ 0.001, R2 = 93%], oxytetracycline 52% [(95% CI: 38%–66%), P = 0.06, R2 = 65%], doxycycline 51% [(95% CI: 12%–86%), P ≤ 0.001, R2 = 95%], sulpha/trimetho/cotri 42% [(95% CI: 11%–77%), P ≤ 0.001, R2 = 97%], ciprofloxacin 20% [(95% CI: 4–43), P ≤ 0.001, R2 = 95%], gentamycin 11% [(95% CI: 2%–25%), P ≤ 0.001, R2 = 90%] and neomycin 4% [(95% CI:1%–13%), P = 0.06, R2 = 60%] (Table 2 and Figures S1–S9 (available as Supplementary data at JAC-AMR Online)). Among the included covariate in this study, only the study period showed significance in the univariate meta-analysis (β = 0.0179; 95% CI: 0.0059–0.0298, P = 0.0034; R2 = 46.11%) (Table 3). Sensitivity analysis was carried out by calculating pooled Salmonella prevalence once more when any single study was eliminated to verify the meta-analysis's stability and liability. The related pooled Salmonella prevalence was shown range from 40.2% (33.0%–48.5%) to 33.8% (30.0%–35.6%) without significantly changing (Figure 2). The statistically identical findings indicated that no single study had an impact on the stability of the overall estimate of the prevalence of Salmonella in this meta-analysis. Even though the funnel plot's visual inspection exhibited asymmetry (Figure 3), Egg's test demonstrated substantial value, and there was no chance of publication bias (Table 2).
Table 2.
Overall result in meta-analysis
| Antibiotic name | Tau2 | I 2 | χ 2 | Random/fix effect of resistance prevalence, % [95% CI] | P for H | P for Egger |
|---|---|---|---|---|---|---|
| Erythromycin | 0.13 | 94% | 27.40 | 84[53–99] | <0.001 | 0.424 |
| Ciprofloxacin | 0.01 | 95% | 143.4 | 20[4–42] | <0.001 | 0.523 |
| Gentamicin | 0.05 | 90% | 71.82 | 11[2–24] | <0.001 | 0.242 |
| Oxytetracycline | 0.0097 | 65% | 5.57 | 52[39–66] | 0.06 | 0.654 |
| Sulpha/Trimetho/Cotri | 0.15 | 97% | 144.29 | 42[11–70] | <0.001 | 0.524 |
| Tetracycline | 0.1 | 93% | 60.48 | 80[53–98] | <0.001 | 0.525 |
| Neomycin | 0.1 | 60% | 7.45 | 4[1–13] | 0.06 | 0.131 |
| Doxycycline | 0.122 | 95% | 35.63 | 51[12–89] | <0.001 | 0.354 |
| Overall Salmonella prevalence | 0.105 | 97% | 558.13 | 37[23–52] | <0.001 | 0.636 |
Table 3.
Univariate meta-regression analysis
| Covariates | Beta [β] [95%CI] | P value | R 2 [%] |
|---|---|---|---|
| Study year | 0.0179 [0.0059 −0.0298] | 0.0034 | 46.11% |
| Location | −0.0073 [−0.0199 −0.0053] | 0.2557 | Nil |
| Host | −0.0085 [−0.0363 −0.0193] | 0.5497 | Nil |
| Sample number | 0.0024 [−0.0017 −0.0064] | 0.2623 | 20.98% |
| Positive sample | −0.0069 [−0.0230 −0.0093] | 0.4037 | Nil |
| AST method | −0.0106 [−0.0274 −0.0061] | 0.2125 | Nil |
| AST standard | 0.0028 [−0.0016 −0.0072] | 0.2140 | 15.22% |
| Quality of articles | −0.0384 [−0.1333 −0.0565] | 0.4280 | Nil |
| Resistance antibiotic | 0.0097 [−0.0313 −0.0119] | 0.3796 | Nil |
Figure 2.
Sensitivity analysis for individual studies.
Figure 3.
Funnel plot for study bias.
There was considerable variation between studies. To investigate potential sources of heterogeneity, meta-regression (univariate) was used. The meta-regression method was used to examine the study period, location, host, sample number, positive sample, AST method, AST standard, quality of articles and resistance antibiotic as potential sources of heterogeneity. Table 3 contains the meta-finding on regressions. None of those previously mentioned factors were found to be substantially correlated with the identified heterogeneity through the regression model, except for research study time (P = 0.004). Therefore, we conducted additional research to examine the relationship between Salmonella prevalence and the study period. The prevalence of Salmonella was found to be positively correlated with the research study period (r = 0.692, P = 0.001) (Table 4).
Table 4.
The correlation between the Salmonella prevalence and potential sources
| Covariate | Study year | Location | Host | Sample no. | Positive no. | Sample type | AST method | AST standard | Quality of articles | Resistance antibiotics |
|---|---|---|---|---|---|---|---|---|---|---|
| P | 0.001 | 0.744 | 0.563 | 0.785 | 0.325 | 0.325 | 0.563 | 0.566 | 0.743 | 0.774 |
| r | 0.692 | 0.764 | 0.346 | 0.633 | 0.764 | 0.763 | 0.567 | 0.568 | 0.366 | 0.852 |
Evidence-based antibiotic resistance drivers
AMR is an upcoming predicted pandemic for a low-income country such as Bangladesh.29–33 Bangladesh has limited information on antibiotic use and resistance.34 Antibiotics were considered able to treat both the physical and social elements of infection, which has severe implications for AMR.22 AMR has mainly spread throughout the country's hospitals and unregulated pharmacy stores for their widely available antibiotics, overprescribing and selling antibiotics without prescription.32 People in hospitals are predisposed to developing nosocomial and other infections due to the abuse of antibiotics and a lack of adherence to isolation techniques in these hospitals.27 In Bangladesh, most antibiotic prescriptions are written by unqualified practitioners.28,30 Owing to poor health systems, reported paucity of testing facilities, prescribing antibiotics without laboratory tests, using antibiotics for common infections and not finishing the entire course of antibiotics were causes of widespread resistance.33 Unsafe drinking water, inadequate sanitation, lack of diagnostic facilities growing private practice and excessive demand for antibiotics are all contributing causes of AMR.30,33 Antibiotic usage before seeking medical help may impair the sensitivity of blood cultures, which has ramifications for patients and doctors.29 Antibiotics are used in various food animal production and meat systems, such as commercial poultry and aquaculture.36 They have been discovered to be frequently used to boost food animal production, leading to the growth of antibiotic-resistant bacteria.33
Moreover, its use in aquaculture and aquatic ecology has been linked to AMR development.28 Antimicrobial drugs used in aquaculture have established AMR bacteria reservoirs in fish and other aquatic and non-aquatic animals.29–32 The long-term use of these antibiotics indiscriminately and unnecessarily increases the prevalence of AMR.28–31 This also leads to antibiotic residue. Antibiotic residues, other drugs and pollutants are found in high concentrations in Bangladesh's ponds, canals, lakes, rivers and other bodies of water.36 Higher residues are caused by poor sanitation, hygiene, antibiotic misuse on the farm and inappropriate use.33 As part of the food production cycle, food animals, seafood and vegetables are regarded significant reservoirs of AMR bacteria by these residues.26 Some antibiotics may have lost their effectiveness against specific microorganisms due to the misuse of those antibiotics.30 Antimicrobials are only available on prescription and to pharmacists who devote more time to patients. Antibiotics are sought and used during an emergency health problem.24 Policy measures such as restrictions on licensing specific antibiotics are vital to human health.25 In Bangladesh, specific and targeted measures to combat AMR should include teaching about the proper use of antibiotics.31 There is a significant frequency of AMR in Bangladesh, and major gaps in surveillance and information, and there has been a drop in the rate of new antibiotic development.23
Discussion
The present study found a significant prevalence of Salmonella from livestock and poultry-derived foods in Bangladesh (sample type faeces, faecal, collocal and rectal exclude). We found that 37% of livestock and poultry food had Salmonella. Consequently, livestock and poultry-derived foods appear to be one of the essential Salmonella reservoirs in Bangladesh. A meta-analysis of Salmonella in Ethiopia indicates that the prevalence of Salmonella in slaughterhouse may vary from 7% to 43%,46 and a worldwide meta-analysis of Salmonella prevalence in food indicate that it was less than 1%.45,49 Therefore, researchers have investigated Salmonella prevalence for many years, and different rates have been found in different studies. The reported prevalence of Salmonella found in this study matched and differed from the findings of other researchers studying the prevalence of Salmonella in poultry.50–57 Thus, it can be mentioned that the significant amount of Salmonella indicates a severe problem for both livestock and poultry-derived food and ensures public health.
Moreover, globally AMR is becoming a significant health issue.53 Resistant to many drugs, Salmonella has become a considerable public health concern worldwide.57 Our present study findings also revealed that a high percentage of the Salmonella isolates were resistant to routinely used antibiotics such as tetracycline, oxytetracycline, doxycycline, sulpha/trimetho/cotri, ciprofloxacin, gentamycin and neomycin and so on. Almost all the isolates tested in this investigation were determined to be multi-drug resistant, which is a concerning finding (Table 1).
However, among these antibiotics, the highest prevalence rate was seen for tetracycline (81%, 95% CI: 53–98). Some studies showed that isolated Salmonella was highly resistant to tetracycline in the antimicrobial investigation.50–52 The present study found the next high prevalence rate for resistance to oxytetracycline that shows 52% from the isolates (95% CI: 38–66), but some studies found a higher prevalence rate of oxytetracycline rather than our findings.45,46,48,49 According to a couple of studies, Salmonella resistance to tetracycline and oxytetracycline was found in layers and broilers in Bangladesh.58–60 Doxycycline is another antibiotic used in humans and animals to treat various diseases. Investigating multi-drug resistance due to Salmonella, our findings showed that a high prevalence for this commonly prescribed antibiotic is 51% (95% CI: 12–86). In addition, a recent study showed that Salmonella in poultry in Bangladesh has also been found to have a considerable number of isolates resistant to doxycycline.46,48,60 Moreover, according to univariate analysis and correlation in this meta-analysis, the prevalence of Salmonella increased with every year. Antibiotic resistance creates a severe problem in the treatment of Salmonella.
Furthermore, Salmonella has been proven to be a significant cause of creating resistance against several commonly prescribed antibiotics. Resistant to many drugs, Salmonella isolates were discovered in various food samples, and the gene responsible for multi-drug resistance could be passed on to consumers through food and unhealthy, unhygienic poultry handling systems, posing a severe public health risk. These findings also revealed that multi-drug resistance in Salmonella is rising due to the indiscriminate use of antibiotics in the dairy and poultry industries, pet animal usage and human practice in Bangladesh. In the future, rational usage of this antibiotic may help to prevent the formation of Salmonella-resistant isolates.
The main strength of this article was the pooled prevalence of Salmonella and antibiotic resistance to Salmonella from livestock and poultry-derived foods. We solely refer to the antibiotic subgroup. We looked through numerous databases and websites to discover all relevant and grey publications to eliminate database bias, but some databases or websites may be massing. The limitations mentioned before, as well as publication bias and heterogeneity for some of the pooled results, must be considered when interpreting the results.
Conclusions
Antimicrobial agents used in food animal production may lead to the emergence of multi-drug-resistant Salmonella strains. The irrational and excessive use of antibiotics in humans and food-producing animals raises the risk of AMR worldwide. To reduce the risk of pathogenic AMR bacteria originating from animal origin foods, raising awareness about the rational use of antibiotics in food animals, safe food handling and safe cooking practices is obligatory. Doctors should take concern when treating with an antibiotic.
Supplementary Material
Acknowledgements
We are thankful to Sojib Bhuiyan for his technical support, Department of Accounting, Government Bangla College, Bangladesh.
Contributor Information
Rezaul Karim Ripon, Department of Public Health and Informatics, Jahangirnagar University, Savar, Dhaka, Bangladesh.
Umma Motahara, Department of Public Health and Informatics, Jahangirnagar University, Savar, Dhaka, Bangladesh.
Ayesha Ahmed, Department of Public Health and Informatics, Jahangirnagar University, Savar, Dhaka, Bangladesh.
Nishrita Devnath, Department of Public Health and Informatics, Jahangirnagar University, Savar, Dhaka, Bangladesh.
Fatema Akter Mahua, Department of Public Health and Informatics, Jahangirnagar University, Savar, Dhaka, Bangladesh.
Rubaiya Binthe Hashem, Department of Public Health and Informatics, Jahangirnagar University, Savar, Dhaka, Bangladesh.
Kifayat Sadmam Ishadi, Department of Public Health and Informatics, Jahangirnagar University, Savar, Dhaka, Bangladesh.
Adiba Alam, Department of Public Health and Informatics, Jahangirnagar University, Savar, Dhaka, Bangladesh.
Md Safaet Hossain Sujan, Department of Public Health and Informatics, Jahangirnagar University, Savar, Dhaka, Bangladesh.
Md Samun Sarker, Antimicrobial Resistance Action Center (ARAC), Bangladesh Livestock Research Institute (BLRI), Savar, Dhaka, Bangladesh.
Funding
Self-funded.
Transparency declarations
None to declare.
Supplementary data
Figures S1 to S9 are available as Supplementary data at JAC-AMR Online.
References
- 1. Hawker J, Begg N, Reintjes Ret al. . Communicable Disease Control and Health Protection Handbook. John Wiley & Sons, 2018. [Google Scholar]
- 2. Jain P, Chowdhury G, Samajpati Set al. . Characterization of nontyphoidal Salmonella isolates from children with acute gastroenteritis, Kolkata, India, during 2000–2016. Braz J Microbiol 2020; 51: 613–27. 10.1007/s42770-019-00213-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lin D, Yan M, Lin Set al. . Increasing prevalence of hydrogen sulfide negative Salmonella in retail meats. Food Microbiol 2014; 43: 1–4. 10.1016/j.fm.2014.04.010 [DOI] [PubMed] [Google Scholar]
- 4. Sallam KI, Mohammed MA, Hassan MAet al. . Prevalence, molecular identification and antimicrobial resistance profile of Salmonella serovars isolated from retail beef products in Mansoura, Egypt. Food Control 2014; 38: 209–14. 10.1016/j.foodcont.2013.10.027 [DOI] [Google Scholar]
- 5. Kirk MD, Pires SM, Black REet al. . World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 2015; 12: e1001921. 10.1371/journal.pmed.1001921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pang L, Zhang Z, Xu J. Surveillance of foodborne disease outbreaks in China in 2006–2010. Chin J Food Hyg 2011; 23: 560–3. [Google Scholar]
- 7. Issenhuth-Jeanjean S, Roggentin P, Mikoleit Met al. . Supplement 2008–2010 (no. 48) to the White–Kauffmann–Le minor scheme. Res Microbiol 2014; 165: 526–30. 10.1016/j.resmic.2014.07.004 [DOI] [PubMed] [Google Scholar]
- 8. Takaya A, Yamamoto T, Tokoyoda K. Humoral immunity vs. Salmonella. Front Immunol 2020; 10: 3155. 10.3389/fimmu.2019.03155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Behravesh CB, Brinson D, Hopkins BAet al. . Backyard poultry flocks and salmonellosis: a recurring, yet preventable public health challenge. Clin Infect Dis 2014; 58: 1432–8. 10.1093/cid/ciu067 [DOI] [PubMed] [Google Scholar]
- 10. Gieraltowski L, Higa J, Peralta Vet al. . National outbreak of multidrug resistant Salmonella Heidelberg infections linked to a single poultry company. PLoS ONE 2016; 11: e0162369. 10.1371/journal.pone.0162369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Keerthirathne TP, Ross K, Fallowfield Het al. . Reducing risk of salmonellosis through egg decontamination processes. Int J Environ Res Public Health 2017; 14: 335. 10.3390/ijerph14030335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Biswas S, Li Y, Elbediwi Met al. . Emergence and dissemination of MCR-carrying clinically relevant Salmonella Typhimurium monophasic clone ST34. Microorganisms 2019; 7: 298. 10.3390/microorganisms7090298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu H, Elbediwi M, Zhou Xet al. . Epidemiological and genomic characterization of Campylobacter jejuni isolates from a foodborne outbreak at Hangzhou, China. Int J Mol Sci 2020; 21: 3001. 10.3390/ijms21083001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ed-Dra A, Karraouan B, El Allaoui Aet al. . Antimicrobial resistance and genetic diversity of Salmonella Infantis isolated from foods and human samples in Morocco. J Glob Antimicrob Resist 2018; 14: 297–301. 10.1016/j.jgar.2018.05.019 [DOI] [PubMed] [Google Scholar]
- 15. Paudyal N, Pan H, Wu Bet al. . Persistent asymptomatic human infections by Salmonella enterica serovar Newport in China. mSphere 2020; 5: e00163-20. 10.1128/mSphere.00163-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jiang Z, Paudyal N, Xu Yet al. . Antibiotic resistance profiles of Salmonella recovered from finishing pigs and slaughter facilities in Henan, China. Front Microbiol 2019; 10: 1513. 10.3389/fmicb.2019.01513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Elbediwi M, Pan H, Biswas Set al. . Emerging colistin resistance in Salmonella enterica serovar Newport isolates from human infections. Emerg Microbes Infect 2020; 9: 535–8. 10.1080/22221751.2020.1733439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Park HC, Baig IA, Lee SCet al. . Development of ssDNA aptamers for the sensitive detection of Salmonella Typhimurium and Salmonella Enteritidis. Appl Biochem Biotechnol 2014; 174: 793–802. 10.1007/s12010-014-1103-z [DOI] [PubMed] [Google Scholar]
- 19. Spector MP, Kenyon WJ. Resistance and survival strategies of Salmonella Enterica to environmental stresses. Food Res Int 2012; 45: 455–81. 10.1016/j.foodres.2011.06.056 [DOI] [Google Scholar]
- 20. de Freitas CG, Santana AP, da Silva PHet al. . PCR multiplex for detection of Salmonella Enteritidis, Typhi and Typhimurium and occurrence in poultry meat. Int J Food Microbiol 2010; 139: 15–22. 10.1016/j.ijfoodmicro.2010.02.007 [DOI] [PubMed] [Google Scholar]
- 21. Shah AH, Korejo NA. Antimicrobial resistance profile of Salmonella serovars isolated from chicken meat. J Vet Anim Sci 2012; 2: 40–6. [Google Scholar]
- 22. Suresh T, Hatha AA, Sreenivasan Det al. . Prevalence and antimicrobial resistance of Salmonella Enteritidis and other salmonellas in the eggs and egg-storing trays from retails markets of Coimbatore, South India. Food Microbiol 2006; 23: 294–9. 10.1016/j.fm.2005.04.001 [DOI] [PubMed] [Google Scholar]
- 23. Momtaz S, Saha O, Usha MKet al. . Occurrence of pathogenic and multidrug resistant Salmonella spp. in poultry slaughter-house in Bangladesh. Bioresearch Communications (BRC) 2018; 4: 506–15. https://www.bioresearchcommunications.com/index.php/brc/article/view/80 [Google Scholar]
- 24. Sultana M, Bilkis R, Diba Fet al. . Predominance of multidrug resistant zoonotic Salmonella Enteritidis genotypes in poultry of Bangladesh. J Poult Sci 2014; 51: 424–34. 10.2141/jpsa.0130222 [DOI] [Google Scholar]
- 25. Dana N, Van der Waaij LH, Dessie Tet al. . Production objectives and trait preferences of village poultry producers of Ethiopia: implications for designing breeding schemes utilizing indigenous chicken genetic resources. Trop Anim Health Prod 2010; 42: 1519–29. 10.1007/s11250-010-9602-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barrow PA, Jones MA, Smith ALet al. . The long view: Salmonella–the last forty years. Avian Pathol 2012; 41: 413–20. 10.1080/03079457.2012.718071 [DOI] [PubMed] [Google Scholar]
- 27. WHO . Drug Resistant Salmonella. 2014. http://www.who.int/mediacentre/factsheets/fs139/en/.
- 28. Su LH, Chu C, Cloeckaert Aet al. . An epidemic of plasmids? Dissemination of extended-spectrum cephalosporinases among Salmonella and other Enterobacteriaceae. FEMS Immunol Med Microbiol 2008; 52: 155–68. 10.1111/j.1574-695X.2007.00360.x [DOI] [PubMed] [Google Scholar]
- 29. García-Fernández A, Fortini D, Veldman Ket al. . Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J Antimicrob Chemother 2009; 63: 274–81. 10.1093/jac/dkn470 [DOI] [PubMed] [Google Scholar]
- 30. Strohl WA, Rouse H, Fisher BD. Microbiologia Ilustrada. Artmed, Porto Alegre, 2004. [Google Scholar]
- 31. Angulo FJ, Johnson KR, Tauxe RVet al. . Origins and consequences of antimicrobial-resistant nontyphoidal Salmonella: implications for the use of fluoroquinolones in food animals. Microb Drug Resist 2000; 6: 77–83. 10.1089/mdr.2000.6.77 [DOI] [PubMed] [Google Scholar]
- 32. Murugkar HV, Rahman H, Kumar Aet al. . Isolation, phage typing and antibiogram of Salmonella from man & animals in northeastern India. Indian J Med Res 2005; 122: 237–42. [PubMed] [Google Scholar]
- 33. Aragaw K, Molla B, Muckle Aet al. . The characterization of Salmonella serovars isolated from apparently healthy slaughtered pigs at Addis Ababa abattoir, Ethiopia. Prev Vet Med 2007; 82: 252–61. 10.1016/j.prevetmed.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 34. Zewdu E, Cornelius P. Antimicrobial resistance pattern of Salmonella serotypes isolated from food items and personnel in Addis Ababa, Ethiopia. Trop Anim Health Prod 2009; 41: 241–9. 10.1007/s11250-008-9181-y [DOI] [PubMed] [Google Scholar]
- 35. Bhan MK, Bahl R, Bhatnagar S. Typhoid and paratyphoid fever. Lancet 2005; 366: 749–62. 10.1016/S0140-6736(05)67181-4 [DOI] [PubMed] [Google Scholar]
- 36. WHO . Food Safety and Food Borne Illness. 2014. http://www.who.int/mediacentre/factsheets/fs237/edu.
- 37. Kang CI, Song JH. Antimicrobial resistance in Asia: current epidemiology and clinical implications. Infect Chemother 2013; 45: 22–31. 10.3947/ic.2013.45.1.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Majowicz SE, Musto J, Scallan Eet al. . The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 2010; 50: 882–9. 10.1086/650733 [DOI] [PubMed] [Google Scholar]
- 39. Wei ZQ, Chang HL, Li YFet al. . Clinical epidemiology and antimicrobial resistance of nontyphoidal Salmonella enteric infections in children: 2012–2014. Chin J Pediatr 2016; 54: 489–95. 10.3760/cma.j.issn.0578-1310.2016.07.003 [DOI] [PubMed] [Google Scholar]
- 40. Liang B, Xie Y, He Set al. . Prevalence, serotypes, and drug resistance of nontyphoidal Salmonella among paediatric patients in a tertiary hospital in Guangzhou, China, 2014–2016. J Infect Public Heal 2019; 12: 252–7. 10.1016/j.jiph.2018.10.012 [DOI] [PubMed] [Google Scholar]
- 41. Dong-Ling L, Jie-hong W, Jing-Song WUet al. . Characteristics of drug resistance and molecular typing for Salmonella in diarrhea patients from four hospitals in Shenzhen. Chin J Zoonoses 2017; 33: 897–902. [Google Scholar]
- 42. Hopkins KL, Kirchner M, Guerra Bet al. . Multi resistant Salmonella enterica serovar 4,[5],12:i:- in Europe: a new pandemic strain? Euro Surveill 2010; 15: 19580. 10.2807/ese.15.22.19580-en [DOI] [PubMed] [Google Scholar]
- 43. Liang DW, Lu JH, Jiang LX. Pattern of antimicrobial resistance and molecular typing for Salmonella isolated from diarrhea cases in Guangzhou. Mod Prev Med 2016; 43: 696–9. [Google Scholar]
- 44. Michael GB, Schwarz S. Antimicrobial resistance in zoonotic nontyphoidal Salmonella: an alarming trend? Clin Microbiol Infect 2016; 22: 968–74. 10.1016/j.cmi.2016.07.033 [DOI] [PubMed] [Google Scholar]
- 45. Lash TL, Fox MP, MacLehose RFet al. . Good practices for quantitative bias analysis. Int J Epidemiol 2014; 43: 1969–85. 10.1093/ije/dyu149 [DOI] [PubMed] [Google Scholar]
- 46. Nik Hazlina NH, Norhayati MN, Shaiful Bahari Iet al. . Worldwide prevalence, risk factors and psychological impact of infertility among women: a systematic review and meta-analysis. BMJ Open 2022; 12: e057132. 10.1136/bmjopen-2021-057132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Abd-Elghany SM, Sallam KI, Abd-Elkhalek Aet al. . Occurrence, genetic characterization and antimicrobial resistance of Salmonella isolated from chicken meat and giblets. Epidemiol Infect 2015; 143: 997–1003. 10.1017/S0950268814001708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ahmed MM, Rahman MM, Mahbub KRet al. . Characterization of antibiotic resistant Salmonella spp isolated from chicken eggs of Dhaka city. J Sci Res 2010; 3: 191–6. 10.3329/jsr.v3i1.6109 [DOI] [Google Scholar]
- 49. Tadesse G, Tessema TS. A meta-analysis of the prevalence of Salmonella in food animals in Ethiopia. BMC Microbiol 2014; 14: 270. 10.1186/s12866-014-0270-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Akond MA, Shirin M, Alam Set al. . Frequency of drug resistant Salmonella spp. isolated from poultry samples in Bangladesh. S J Microbiol 2013; 2: 15–9. 10.3329/sjm.v2i1.15207 [DOI] [Google Scholar]
- 51. Alam SB, Mahmud M, Akter Ret al. . Molecular detection of multidrug resistant Salmonella species isolated from broiler farm in Bangladesh. Pathogens 2020; 9: 201. 10.3390/pathogens9030201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nahar A, Islam MA, Sobur MAet al. . Detection of tetracycline resistant Salmonella and Salmonella spp. in sewage, river, pond and swimming pool in Mymensingh, Bangladesh. Afr J Microbiol Res 2019; 13: 382–7. 10.5897/AJMR2019.9156 [DOI] [Google Scholar]
- 53. Dahlén G, Blomqvist S, Almståhl Aet al. . Virulence factors and antibiotic susceptibility in enterococci isolated from oral mucosal and deep infections. J Oral Microbiol 2012; 4: 10855. 10.3402/jom.v4i0.10855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Iwabuchi E, Yamamoto S, Endo Yet al. . Prevalence of Salmonella isolates and antimicrobial resistance patterns in chicken meat throughout Japan. J Food Prot 2011; 74: 270–3. 10.4315/0362-028X.JFP-10-215 [DOI] [PubMed] [Google Scholar]
- 55. Karim MR, Giasuddin M, Samad MAet al. . Prevalence of Salmonella spp. in poultry and poultry products in Dhaka, Bangladesh. Int J Anim Biol 2017; 3: 18–22. [Google Scholar]
- 56. Mahmud MS, Kabir ML, Alam SSet al. . Prevalence of Salmonella spp. in poultry eggs from different retail markets at Savar area, Bangladesh. Am J Food Sci Health 2015; 1: 27–31. [Google Scholar]
- 57. Marshall BM, Levy SB. Food animals and antimicrobials: impacts on human health. Clin Microbiol Rev 2011; 24: 718–33. 10.1128/CMR.00002-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Paul P, Akther S, Zulfekar Ali Met al. . Isolation, identification and antibiogram study of Salmonella spp. from poultry farm environment. Int J Animal Biol 2017; 3: 5–11. [Google Scholar]
- 59. Talukder M, Islam MS, Ievy Set al. . Detection of multidrug resistant Salmonella spp. from healthy and diseased broilers having potential public health significance. J Adv Biotechnol Exp Ther 2021; 4: 248–55. 10.5455/jabet.2021.d125 [DOI] [Google Scholar]
- 60. Haque AKMZ, Akter MR, Islam SKSet al. . Salmonella gallinarum in small-scale commercial layer flocks: occurrence, molecular diversity and antibiogram. Vet Sci 2021; 8: 71. 10.3390/vetsci8050071 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.