Abstract
CD4+ T helper cells develop into subsets that are specialized in the secretion of particular cytokines to mediate restricted types of inflammation and immune responses. Among the subsets that promote development of allergic inflammatory responses, IL-9 producing TH9 are regulated by a number of transcription factors. We have previously shown that the Ets family members PU.1 and ETV5 function in parallel to regulate IL-9. In this study we identified a third member of the Ets-family of transcription factors, ERG, that mediates IL-9 production in TH9 cells in the absence of PU.1 and ETV5. Chromatin immunoprecipitation assays revealed that ERG interaction at the Il9 promoter region is restricted to the TH9 lineage and is sustained during TH9 polarization. Knock-down or knock-out of ERG during TH9 polarization in vitro led to a decrease in IL-9 production in TH9 cells. Deletion of ERG in vivo had modest effects on IL-9 production in vitro or in vivo. However, in the absence of PU.1 and ETV5, ERG was required for residual IL-9 production in vitro and for IL-9 production by lung-derived CD4-T cells in chronic allergic airway disease. Thus, ERG contributes to IL-9 regulation in TH9 cells.
Introduction:
Interleukin-9 (IL-9) is a pleotropic cytokine associated with type 2 immunity supporting the clearance of helminth infections and tumor cells (1-5). While appropriate production of IL-9 is important for human health, inappropriate or over production of IL-9 results in a myriad of diseases ranging from allergic to autoimmune disorders (6-8). Allergic asthma is a respiratory disease initiated by sensitization to inhaled aeroallergens and has been strongly correlated to IL-9 production (9). In chronic allergic airway disease, IL-9 producing CD4 T-helper (TH9) cells lead to accumulation of eosinophils and mast cells in the airway, increased type-2 cytokines, stimulated type 2 innate lymphoid cells (ILC2) cell proliferation, and induced mucus production from airway epithelium (2, 10-12). In response to antigen receptor activation in the presence of IL-4 and transforming growth factor beta (TGFβ), naïve CD4 T cells differentiate into TH9 cells by activating a complex network of transcription factors that can regulate the production of IL-9 (6, 13). Transcription factors including basic leucine zipper ATF-like transcription factor (BATF) and interferon regulatory factor 4 (IRF4) are induced by IL-4-mediated signal transducer and activator of transcription factor 6 (STAT6) signaling pathway (14-17). TGFβ signal activates SMAD proteins that lead to the induction of PU.1 and Ets translocation variant 5 (ETV5) (18, 19). While several transcription factors have been shown to affect IL-9 production in TH9 cells, the transcriptional network that governs the development of TH9 cells and their function during allergic responses is not entirely understood.
The E26 transformation specific (Ets) family of transcription factors play important roles in cellular proliferation, differentiation, angiogenesis and apoptosis, and are defined by the presence of a conserved ~85 amino acid long Ets domain, which facilitates DNA binding at the consensus binding site ‘GGAA/T’ (20, 21). In humans, the Ets family of transcription factors is comprised of 28 family members; there are 27 members in mice. The existence of a large family of closely related transcription factors suggests that individual Ets family members evolved with specific roles in the regulation of target genes. The Ets family member PU.1 is expressed in larger amounts in TH9 cells compared to TH2 cells and inhibits the production of TH2 cytokines (22). Interestingly, ectopic expression of PU.1 enhanced IL-9 production in TH2 cells. Moreover, deficiency in PU.1 expression impaired IL-9 production in vitro as well as TH9 function in allergic airway disease models (18). Additional studies also indicate that PU.1 binds at the Il9 promoter and facilitates recruitment of GCN5, a histone acetyltransferase (HAT) that induces IL-9 production in TH9 cells (23). PU.1 dependent TH9 function is associated with both allergic airway inflammation as well as inflammatory bowel disease (23).
ETV5, another member of the Ets-family of transcription factors, functions in parallel with PU.1 to modulate IL-9 secretion in TH9 cells. IL-9 production was decreased in the absence of ETV5, while overexpression of ETV5 in TH9 cultures lead to increased IL-9 production. Chromatin immunoprecipitation studies revealed that, although both PU.1 and ETV5 can regulate Il9 gene expression in a parallel manner, they bind to distinct conserved non-coding regions at the Il9 locus. In the absence of ETV5, lower binding of HAT p300 was observed at the Il9 locus, while GCN5 binding remained unaffected (19). Besides the similar roles of ETV5 and PU.1 in regulation of TH9 function, they had distinct effects on the overall development of allergic inflammation (19). In addition to PU.1 and ETV5, Koh et. al (19), showed additional members of the Ets-family of transcription factors that were expressed in the TH9 subset including ELK3 and ETV6 (19). However, ectopic expression of either of these transcription factors did not alter IL-9 production.
In a microarray analysis, Jabeen et. al. provided strong evidence that the TH9 subset has a unique transcription signature from other CD4 T helper subsets. In the analysis another member of the Ets-family of transcription factors Ets related gene (Erg) was preferentially expressed in TH9 cells compared to TH2 and T regulatory cell subset and expression was dependent on STAT6 and BATF (17). The function of ERG in TH9 cells has not been examined. ERG is primarily expressed in hematopoietic and vascular endothelial cells (24). Translocations associated with ERG are involved in the generation of oncogenic fusion proteins including TMPRSS2-ERG in prostate cancer and EWS-ERG in Ewing’s sarcoma (25, 26). Furthermore, aberrant expression of ERG has been strongly associated with poor prognosis of acute lymphoblastic leukemia (ALL) (27). Chromatin immuno-precipitation studies followed by sequencing (ChIP-seq) analysis revealed that ERG modulates gene expression patterns in prostate cancer by promoting chromatin rearrangements (28-31). Additionally, overexpression of ERG facilitates recruitment of transcription factors and co-activator proteins through the pointed (PNT) domain that can alter transcriptional activity of target genes (21). Some of the known Ets-transcription factor binding partners of ERG include PU.1, FLI-1, ETS-2, ER81, and ERG itself, (32). In the immune system, ERG controls B cell development and is induced during the initial stages of T cell lineage commitment and repressed once T cells are committed (20, 33). However, the function of ERG in mature T cells, T-helper subset differentiation and the development of effector function has not been studied. In this report, we demonstrate the function of ERG in the regulation of IL-9 and TH9 function in allergic airway inflammation.
Methods
Mice.
C57BL/6, dLck cre, CD4 cre and inducible CD4 (iCD4) cre mice (on C57BL/6 background) were purchased from The Jackson Laboratory. Ergfl/fl mice were obtained from Dr. Anna Randi (24) and crossed with dLck-cre, iCD4-Cre, or CD4-cre mice respectively. Spifl/flEtv5fl/flErgfl/fl CD4-cre mice were generated by crossing Spifl/flEtv5fl/fl CD4-cre mice (19, 34-37) with Ergfl/fl CD4-cre. Ergfl/fl Mx1-cre mice were maintained by Dr. Bo T. Porse and treated with poly(I:C) 2 weeks before harvesting for in vitro assays (33, 38). Erg exon 3-mutant mice were generated by Taconic as outlined in Supplementary Fig. 1. Mutation was confirmed by sequencing. In this study, 6-8 weeks old, male or female mice were utilized. All mice were used with the approval of the Indiana University Institutional Animal Care and Use Committee.
In vitro T-cell differentiation.
Naïve CD4 T cells were isolated from mouse spleens using Naïve CD4 T cell isolation kit provided by the supplier (Miltenyi Biotec). Cells were cultured in complete RPMI 1640 media on anti-CD3 (2 μg/ml) coated plates and soluble anti-CD28 (0.5 μg/ml) under T-helper subset polarizing conditions including TH9 (TGFβ1): TGF-β1 (2 ng/ml), IL-4 (20 ng/ml), hIL-2 (50 U/ml) and anti-IFN-γ (10 μg/ml); TH9 (Activin A): TGF-β1 (2 ng/ml), IL-4 (20 ng/ml), hIL-2 (50 U/ml) and anti-IFN-γ (10 μg/ml); TH2 polarizing condition: IL-4 (20 ng/ml), hIL-2 (50 U/ml) and anti-IFN-γ (10 μg/ml); TH1 polarizing condition: IL-12 (20 ng/ml), hIL-2 (50 U/ml), and anti-IL-4 (10 μg/ml); TH17 polarizing condition: IL-6 (100 ng/ml), TGF-β1 (2 ng/ml), IL-1β (10 ng/ml), IL-23 (10 ng/ml), anti–IFN-γ (10 μg/ml), anti–IL-4 (10 μg/ml), and anti–IL-2 (10 μg/ml); Treg polarizing condition: TGF-β1 (2 ng/ml), hIL-2 (50 U/ml), anti-IFNγ (10 μg/ml), and anti-IL-4(μg/ml). On day 3, cells were expanded into fresh media containing the original concentrations of cytokines in the absence of co-stimulatory signals for an additional 2 days. On day 5, mature TH9 or TH2 cells were harvested for further analysis.
In vitro human T-cell isolation and differentiation
PBMCs were isolated from de-identified buffy coat blood packs from healthy anonymous donors (Indiana Blood Center, IN) by density gradient centrifugation using Ficoll-Paque (GE Healthcare). Human naive CD4+ T cells were isolated from human PBMCs using magnetic separation (Miltenyi Biotec). Isolated naïve CD4 T cells were activated with an equal ratio of receptor crosslinking bead, Dynabead human T-activator CD3/CD28 (ThermoFisher Scientific) in cRPMI to generate TH9 cells in TH9 subset polarizing condition: hIL-4 (20 ng/ml), hTGF-β1 (2 ng/ml), and anti-IFNγ (10 μg/ml), and cultured at 37 °C under 5% CO2. Cells were harvested on day 5 for analysis.
Quantitative real-time PCR.
Total RNA was isolated from cells using Trizol (Life Technologies). RNA was reverse transcribed according to manufactures directions (Quantabio, Beverly, MA). Quantitative Reverse Transcriptase (qRT-PCR) was performed with commercially available primers (Life Technologies) with a 7500 Fast-PCR machine (Life Technologies). Gene expression was normalized to housekeeping gene expression (β2-microglobulin). In case of qPCR for ChIP assay, SYBR green master mix (Applied Biosystems) was used for gene expression analysis (39).
Retrovirus production.
Platinum E cells were grown in 10 ml of Dulbecco’s modified eagle medium 1640 (DMEM 1640) with 10% FBS and 1% antibiotics in a 100 mm tissue culture dish. When confluency reached 80~90%, cells were transfected with control vector or retroviral vector expressing PU.1-IRES-hCD4 (18), ETV5-IRES-Thy1.1 (19, 34), ERG-IRES-EGFP (ERG3 cDNA was cloned into the Mieg3-GFP plasmid)(40) or Cre-IRES-EGFP (41) using lipofectamine 3000 (ThermoFisher Scientific). For transfection, 18 μg of vector, 6 μg of pCL-Eco and 25 μl of P3000 were mixed in 500 μl of Opti-MEM®I reduced-serum medium (ThermoFisher Scientific), and 25 μl of lipofectamine 3000 was mixed in another 500 μl of Opti-MEM®I. After combining, this mixture was incubated for 10-15 mins at room temperature (RT). The mixture was gently pipetted into culture dish. After 16 hours, the media containing retrovirus was collected and changed with new fresh media. After 24 hours, the media was collected and centrifuged at 1500 rpm for 5 mins to remove cell debris. Supernatant containing retrovirus was used for retroviral transduction or stored at −80 °C for subsequent use.
Retroviral transductions.
Activated mouse CD4+ T cells were infected on day 1 with retrovirus containing control or expressing the interested gene by centrifugation at 2300 rpm at 32oC for 90 mins in the presence of 8 μg/ml polybrene (Sigma-Aldrich). After spin infection, the supernatant was replaced with the fresh T-helper subset cell condition media. Cells were expanded on day 3 and analyzed on day 4 or 5.
Erg small interfering RNA assay.
Mouse or human naïve CD4 T cells were cultured in TH9 polarizing condition as described in the in vitro T cell differentiation section. On day 1 of TH9 differentiation, the TH9 cell culture media was replaced with siRNA delivery media containing 1μM of SMARTpool Accell siRNA against mouse or human Erg (provided by Dharmacon) along with TH9 subset polarizing cytokines and growth factors for 48 hours. For control, non-targeting siRNA (provided by Dharmacon) was used at the same concentration as the targeting siRNA. On day 3, cells were expanded into fresh media containing the original concentrations of cytokines in the absence of co-stimulatory signals for an additional 2 days. On day 5, mature TH9 cells were harvested for further analysis.
Dual luciferase reporter assay
Jurkat cells were transfected with Il9 promoter renilla luciferase reporter construct in combination with the control or Erg overexpressing plasmid using FuGENE 6 roche reagent. After 24 hours of transfection, cells were stimulated with PMA and ionomycin for 6 hours and luciferase activities were measured using the dual luciferase reporter assay system (Promega) according to the manufacturer’s instructions. Firefly values were normalized to renilla values.
Chromatin immunoprecipitation assay.
In vitro-differentiated T-helper cells were activated with anti-CD3 for 3 hours and were crosslinked for 15 min with 1% formaldehyde at RT with rotation. The reaction was quenched by adding 0.125 M glycine and incubated at RT for 5 min. Fixed cells were lysed with cell lysis buffer, followed by nuclear lysis buffer. Nuclei were degraded and chromosomal DNA were fragmented to a size range of 200–500 bp through ultrasonic processor (Vibra-cell). After sonication, the supernatant was diluted 10-fold with ChIP dilution buffer. After pre-clearing, the supernatant was incubated with the ChIP antibodies against ERG ([EPR3864(2)], Rabbit mAb,Abcam), PU.1 (9G7, Rabbit mAb, Cell Signaling Technology), H3K27ac (Rabbit polyclonal, Abcam), p300 (N-15, Rabbit mAb, Santa Cruz Biotechnology) and IgG (Rabbit polyclonal, Abcam) at 4 °C overnight with rotation. The following day, immunocomplexes were precipitated with Protein Agarose A or G beads at 4 °C for 2–4 h. Immunocomplexes were washed with low salt, high salt, LiCl and two times with TE buffer. After elution followed by reverse crosslinks, DNA was purified and analyzed by qPCR. After normalization to the Input DNA, the amount of output DNA of each target protein was calculated by subtracting that of the IgG control. Quantification of ChIP assay by qPCR was performed using primers as previously described (39).
Aspergillus fumigatus (A.fumigatus) induced chronic allergic airway model and tissue processing.
In the chronic allergic airway model, mice were intranasally challenged with A. fumigatus protein extract (GREER, NC) in PBS (25μg/25μl PBS) 3 times per week for either 3 or 6 weeks to develop a chronic response. Bronchoalveolar fluid (BALF) and blood was collected prior to harvesting lung tissue samples. The mesenteric lymph node and spleen were collected manually dissociated to generate single cell suspensions. The lung vascular bed was perfused through the injection of 10mL of cold PBS via the right ventricle of the heart. Lungs were digested with 1mg/ml of collagenase A (Roche) for 1 hour at 37°C followed by pressing digested lungs through a wire mesh sieve (Bellco Glass) to generate single cell suspensions. Red blood cells (RBCs) were lysed using ammonium-chloride-potassium lysis buffer (LONZA). CD4 T cells were isolated via magnetic bead positive selection (clone L3T4; Miltenyi Biotec).
Flow-cytometric analysis.
For cytokine staining, CD4+ T cells were stimulated with Phorbol 12-myristate 13-acetate (PMA, 50ng/ml, Sigma) and ionomycin (1ug/ml, Sigma) for 3 hours followed by monensin (2μM, Biolegend) for total 6 hours at 37°C. After re-stimulation, cells were washed with FACS buffer (PBS with 0.5% BSA). CD4+ T were then stained with a fixable viability dye (eFluor780, eBioscience) and surface markers (CD4, GK1.5, Biolgend) for 30 min at 4 °C followed by washing and fixation with 4% formaldehyde for 10 min at room temperature. For cytokine staining, cells were then permeabilized with permeabilization buffer (eBioscience) for 30 min at 4 °C, and stained for cytokines including IL-9 (RM9A4, Biolegend), IL-4 (11B11, Biolegend), IL-5 (TRFK5, Biolegend), IL-13 (eBio13A, Invitrogen), IL-10 (JES6-5H4, Biolegend), IFNγ (XMG.1, Biolegend) for 30 min at 4 °C. To analyze cytokine production from CD4 T cells in retroviral transduction assays, cytokine expression was detected from viable CD4+ T cells that expressed respective reporters including: human-CD4 (PU.1 overexpression); Thy1.1 (ETV5 overexpression) and enhanced-GFP (ERG or Cre recombinase overexpression). For detection of cellular populations in the lung, single cell suspensions of lung cells were stained for different panels including: lymphocyte panel (CD4 T cells, CD8 T cells and B cells); Granulocyte panel (eosinophils, neutrophils and macrophages) and T-regulatory cells. CD4 T cells were identified by staining for CD4 and TCRβ (H57-597, Biolegend). CD8 T cells were identified as CD4−, CD8+ (53-6.7, Biolegend) and TCRβ+. B cells were identified as B220+ (RA3-6B2, Biolegend) and CD19+(6D5, Biolegend) cells. Amongst the granulocyte populations, eosinophils were identified as CD11b+ (M1/70, BD), CD11c− (N418, Biolegend), SiglecF+ (E50-2440, BD), F4/80+ (BM8, Biolegend), CD45+(104, Biolegend), Ly6G-(IA8, Biolegend) and neutrophils were detected as CD11b+, SiglecF−, F4/80−, CD45+, Ly6G+ cells. Alveolar macrophages were detected as CD11c+, CD11b−, SiglecF+, F4/80+, CD64+(X54-5/7.1, Biolegend), Mertk+(2B10C42, Biolegend) cells and interstitial macrophages were detected as CD11b+, CD11c−, SiglecF−, F4/80+, CD64+, Mertk+ cells. For transcription factor staining for the detection of T-regulatory cells, Foxp3 staining kit provided by eBiosciences was used. After surface staining cells were fixed using the Fix/Perm buffer provided in the kit overnight at 4°C in the dark. Samples were washed and incubated in the 1X permeabilization buffer along with the Foxp3 antibody (FJK-16s, Invitrogen) for 1 hour at 4°C. Cells were washed and resuspended in FACS buffer for further analysis.
Co-culture assay.
CD4+ T cells were sorted from the lungs of A. fumigatus challenged mice by magnetic CD4+ positive selection (Miltenyi). The 50,000 purified CD4+ T cells were cocultured with an equal number of CD11c+ antigen presenting cells isolated from splenocytes from naïve C57BL/6 mice in the presence of either BSA or A. fumigatus (100 mg/ml) for 72 hours in RPMI complete media in a 96-well U-bottom plate. In some experiments, additional controls and innate cytokines like IL-33 (50ng/ml) were added to the coculture. IL-9 was measured by ELISA MAX™ Deluxe Set Mouse IL-9 (Biolegend).
Statistics and data analysis
All statistics were done using Prism software version 9 (GraphPad). Unpaired student t test was used for the comparison of 2 samples. ANOVA with Tukey’s multiple comparison test was used for the comparison of 3 or more groups unless otherwise stated. Flow cytometry data was collected using a Nxt Attune flow cytometer (Life Technologies) and was analyzed using FlowJo version 10.8.0.
Results
ERG induced IL-9 production in TH9 cells
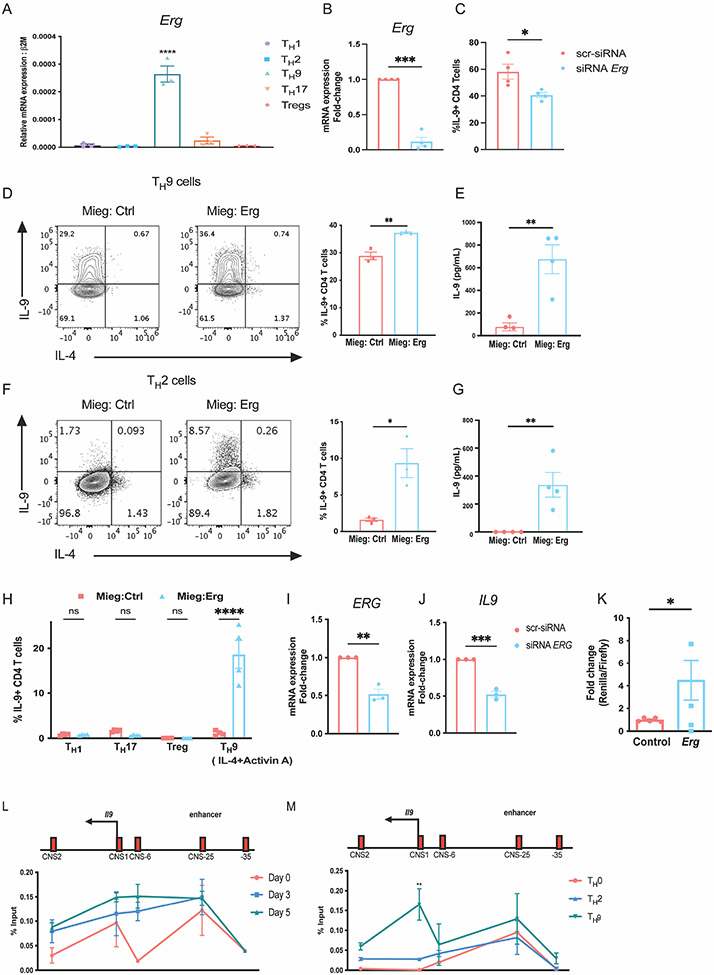
To investigate the function of ERG in the development of TH9 cells, we first assessed the expression of Erg in T-helper subset populations differentiated from naïve CD4 T cells. Amongst the subsets, Erg was preferentially expressed in the TH9 subset (Fig. 1A). This observation was consistent with previous findings reported by Jabeen et al.(17), where Erg expression was enriched in TH9 cells compared to TH2 and Treg populations in microarray and by qPCR analysis. To determine whether ERG contributes to the regulation of IL-9 production in TH9 cells, we performed in vitro siRNA mediated knockdown of Erg in TH9 cells. Erg knockdown in TH9 cells led to a decrease in IL-9 secretion by TH9 cells (Fig. 1 B and C). We then performed retroviral transduction assay to examine whether ectopic expression of Erg increased production of IL-9 by CD4 T helper subsets. Ectopic expression of Erg enhanced IL-9 production not only in TH9 cells but also in the TH2 subset (Fig. 1 D-G). Ectopic expression of Erg in cells cultured with Activin A, a member of the TGF-β superfamily that can also promote IL-9 production in combination with IL-4 (2) similarly enhanced IL-9 (Fig. 1 H). Ectopic expression of Erg in TH1, TH17 and Tregs failed to induce IL-9 production (Fig. 1 H). We examined whether ERG was required for IL-9 production in TH9 cells derived from human PBMCs (h-TH9 cells). Knockdown of ERG in h-TH9 cells, resulted in reduced expression of IL9 transcript (Fig. 1 I and J). To investigate whether ERG regulated the activity of Il9 promoter, we performed dual luciferase reporter assay. In comparison to the control plasmid, transfection of Erg expressing plasmid induced approximately 5-fold Il9 promoter activity in Jurkat cells cultured in vitro (Fig. 1 K). Overall, these findings indicate that Erg promotes IL-9 production in CD4 T-helper cells.
Figure 1: ERG promotes IL-9 production in murine and human CD4 T cells in vitro.
(A) Expression of Erg transcript in T-helper subsets. (B and C) SiRNA knock down of Erg in TH9 cells cultures in vitro showing, Erg expression and IL-9 production respectively. (D) IL-9 production in TH9 subsets after retroviral transductions of ERG overexpressing plasmid on day 1 of T cell differentiation in vitro. (E) Quantification of IL-9 protein secreted by TH9 cells overexpressing ERG. (F) IL-9 production in TH2 subsets after retroviral transductions of ERG overexpressing plasmid on day 1 of T cell differentiation in vitro. (G) Quantification of IL-9 protein secreted by TH2 cells overexpressing ERG. (H) IL-9 production in other T-helper subsets transduced with ERG overexpressing plasmid on day 1 of T cell differentiation in vitro. (I and J) ERG and IL9 transcript expression after siRNA-mediated knock-down of ERG in TH9 cells cultured in vitro from human PBMCs collected from healthy donors. (K) Il9 reporter activity measured as the ratio of Renilla/Luciferase with or without overexpression of Erg in Jurkat cells cultured in vitro. (L) ChIP-qPCR assay indicating ERG interactions at the Il9 regulatory regions in TH9 cells during differentiation through day 0, day 3 (expansion) and day 5. (M) ChIP-qPCR assay showing binding of ERG at Il9 regulatory regions on day 5 after re-stimulation with anti-CD3 antibody for 3 hours in TH9 cells compared TH0 and TH2 cells cultured in vitro. Data are mean of three-four mice or human PBMC donors per experiment and representative of two independent experiments. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
To begin to define the function of ERG, we investigated whether ERG binds the Il9 gene using chromatin immunoprecipitation (ChIP) from cells isolated during TH9 differentiation. We assessed the interaction of ERG at conserved non-coding sequence (CNS) regions across the Il9 locus including CNS1 (Il9 promoter region), CNS2 (located 2kb downstream of the Il9 promoter), CNS-6 (located 6kb upstream of the Il9 promoter), CNS-25 (located 25kb upstream of the Il9 promoter) and an adjacent site as a negative control (located 35kb upstream of the Il9 promoter)(39). ERG interaction at the CNS-25 region, recently identified as the Il9 enhancer region, was observed from day 0, through day 5 of TH9 polarization (Fig. 1 L). On day 5, ERG interaction was modestly enriched at the Il9 promoter region (CNS1) and CNS-6 region. ERG interaction at CNS1 was higher in TH9 cells compared to TH0 and TH2 subsets (Fig. 1 M). ERG binding at the Il9 enhancer region (CNS-25) also trended higher in TH9 cells compared to TH2 cells (Fig. 1 M). These observations indicate that ERG interacts with the Il9 regulatory regions in TH9 cells.
ERG regulates IL-9 production by TH9 cells during acute activation
Erg is expressed in multiple isoforms that include translational start sites in alternatively-spliced exons 3 or 4 (42). We assessed expression of the isoforms and observed that preferential expression in TH9 cells was predominantly from exon 3 (Supplementary Fig. 1A). We then generated mice using a CRISPR/Cas9 approach to mutate the initiation codon of exon 3 (Supplementary Figure 1B). Mutant mice carrying the mutant exon 3 allele were born in normal Mendelian frequencies and did not have any gross abnormalities. Mutation of exon 3 resulted in a decrease of exon 3-containing transcripts without obvious effects on levels of exon 4-containing transcripts and insignificant effects on overall Erg mRNA (Supplementary Fig. 1C-E). Culture of exon 3 mutant CD4 T cells in TH9 culture conditions resulted in IL-9 production that was similar to wild type cells (Supplementary Fig. 1F-H).
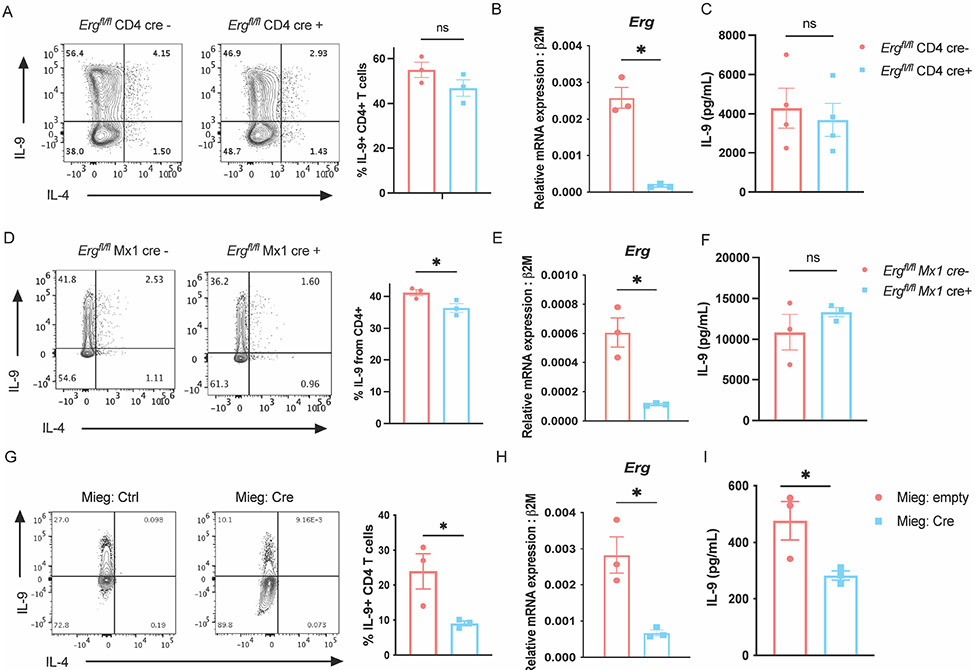
To further assess the role of ERG in the development of TH9 cells, Ergfl/fl mice were crossed with loxP sites flanking exon six (24), common to both isoforms, to multiple Cre-transgenic mice to generate Erg conditional knock-out mice. Multiple Cre-transgenic were used to test whether deletion at differing developmental time points resulted in distinct phenotypes. Deletion of Erg with CD4-Cre that deletes during thymic development was efficient but had negligible effects on IL-9 production in TH9 cells cultured in vitro (Fig. 2 A-C). The same Ergfl/fl mice crossed with dLck-cre mice allowed restricted deletion of Erg in peripheral T cells and resulted in a modest decrease in IL-9 production by cultured TH9 cells (Supplementary Fig. 2 A and B). We confirmed these results using Ergfl/fl Mx1-cre mice with loxP sites located on either side of exon 11 region of Erg (33, 38). Excision of Erg was achieved by treating Ergfl/fl Mx1-Cre mice with polyinosinic-polycytidylic acid (pIpC) (33). The deletion of Erg in peripheral cells using Mx1-Cre lead to a significant though modest reduction (approximately 5% decrease) in IL-9 producing CD4 T cells cultured in vitro. (Fig. 2 D and E), however we did not observe significant differences in IL-9 secretion (Fig. 2 F). We then tested whether deletion of Erg during in vitro differentiation of TH9 cells would affect IL-9 production. Ectopic expression of Cre recombinase in Ergfl/fl CD4 T cells during in vitro TH9 differentiation led to a greater decrease in IL-9 production in TH9 cells, similar to what was observed in siRNA-treated cells (Fig. 2 G-I). These results support the conclusion that ERG contributes to Il9 regulation, but that deletion of Erg early in T cell development or before activation has less of an effect on IL-9 production in TH9 cells. One possibility is that other factors, such as other ETS transcription factors, could compensate for loss of ERG.
Figure 2:
ERG regulates IL-9 production by TH9 cells during acute activation. Detection of IL-9 producing CD4 T cells, Erg gene expression, and secretion of IL-9 in TH9 cells derived from Ergfl/fl CD4 cre mice (A-C) and Ergfl/fl Mx1 cre mice (D-F). (G and H) Production of IL-9 and Erg transcript expression in Ergfl/fl TH9 cells with or without ectopic cre expression on day 3 of TH9 differentiation. (I) Quantification of IL-9 protein secreted. Data are mean of three mice donors per experiment and representative of two independent experiments. *p < 0.05; **p<0.01.
Cooperative function of Ets-transcription factors in the regulation of IL-9 production in TH9 cells.
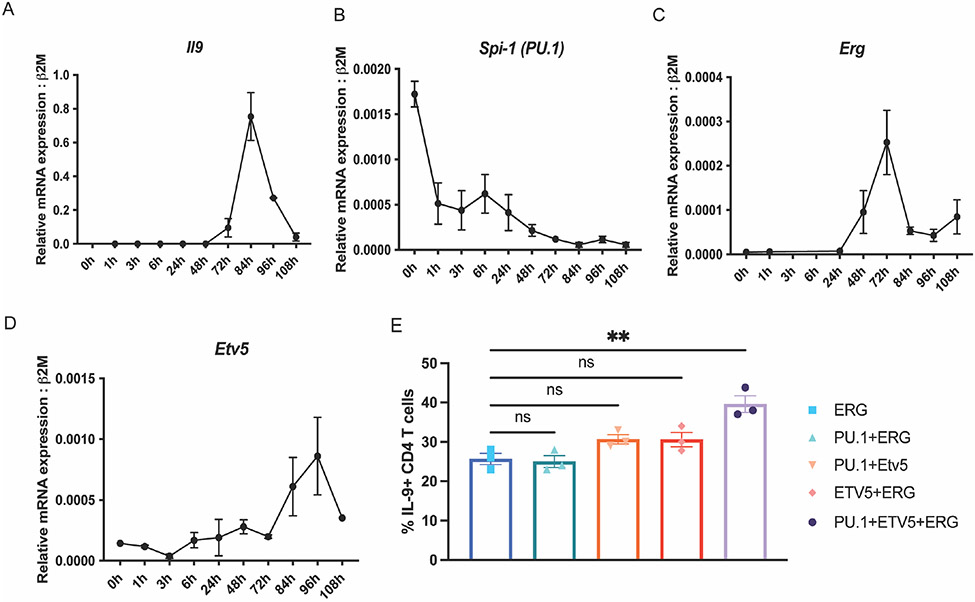
With increasing evidence for the involvement of multiple Ets-transcription factors (PU.1, ETV5 and ERG) in the development of TH9 cells, we profiled transcript expression of TH9 associated Ets-transcription factors during TH9 cell differentiation (Fig. 3 A-D). Previous studies have reported that PU.1-mediates TH9 differentiation during the early stages of TH9 development. Consistent with these findings, Spi-1(PU.1) transcript was observed to peak early during TH9 differentiation and declined during the later stages of differentiation (Fig. 3 B). Erg and Etv5 gene expression peaked on day 3 and day 4 of TH9 differentiation, along with Il9 expression (Fig. 3 A, C and D). Interestingly, Erg expression preceded Il9 transcription by 12 hours and a very similar pattern with a peak after 24 hours of transcription induction. These differing patterns of expression suggest distinct temporal but potentially overlapping roles for the factors in regulation of Il9.
Figure 3:
TH9 associated Ets-transcription factors in differentiating TH9 cells. Transcript expression of Il9 (A), Spi1 (B), Erg (C) and Etv5 (D) during TH9 polarization in vitro. (E) IL-9 production in TH9 subsets ectopically co-expressing ERG, PU.1 and ETV5. Data are mean of three mice per experiment and representative of two independent experiments. *p<0.05; **p<0.01.
As mentioned previously, overlapping roles of Ets-transcription factors in the regulation of Il9 gene expression may explain the minimal effects of Erg deletion on TH9 differentiation in some Cre models (Fig. 2). To begin to test if there was cooperativity between the Ets transcription factors PU.1, ERG and ETV5 on the regulation of IL-9 production during later stages of CD4 T cell differentiation, we performed retroviral transductions to ectopically express combinations of Ets-transcription factors on day 3 of T-helper cell differentiation in vitro. While transduction of ERG resulted in a significant increase of IL-9 (Fig. 1 D), paired transduction of ERG with PU.1 or ETV5 did not further increase IL-9 production (Fig. 3 E). However, combined ectopic expression of PU.1, ETV5, and ERG lead to a further significant increase of IL-9 production in TH9 cells. These data suggested that PU.1, ETV5, and ERG cooperatively regulate IL-9 production in TH9 cells.
ERG mediates IL-9 production in TH9 cells in the absence of PU.1 and ETV5.
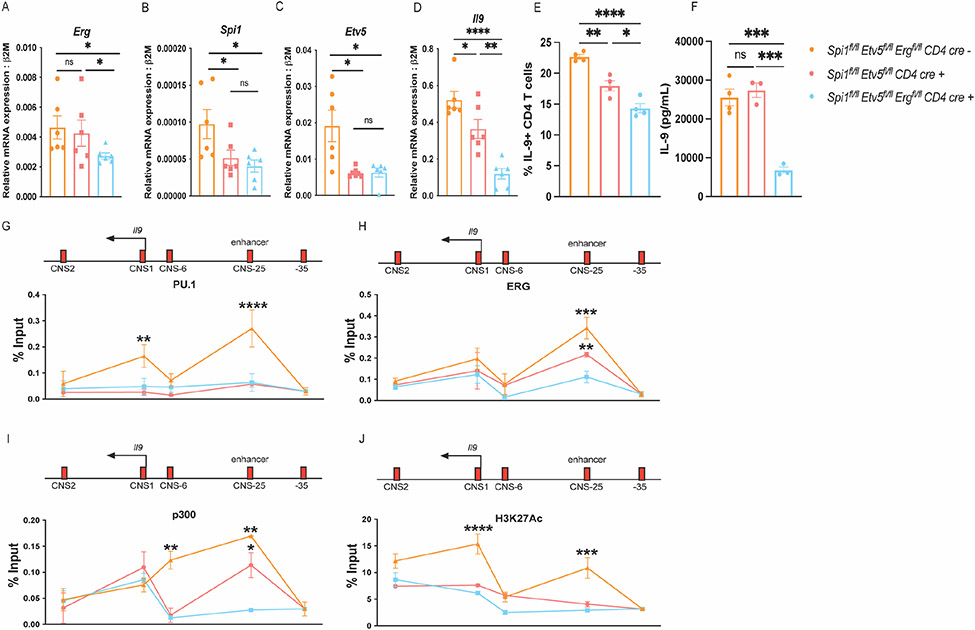
In order to dissect the function of ERG in TH9 development in the absence of other Ets-transcription factors including PU.1 and ETV5, we developed double- and triple knock out mice that lacked either PU.1 and ETV5 (Spifl/flEtv5fl/fl CD4-cre mice), or PU.1, ETV5 and ERG (Spifl/flEtv5fl/flErgfl/fl CD4-cre mice). The deletion of Ets-transcription factors in CD4 T cells was confirmed by assessing transcript expression of Erg, Spi1 (PU.1) and Etv5 in TH9 cultures derived from the knock-out mice (Fig. 4 A-C). Consistent with previous studies (19), deficiency in PU.1 and ETV5 resulted in reduced production of IL-9 production in TH9 cells cultured in vitro (Fig. 4 D and E). Deletion of ERG in the absence of PU.1 and ETV5 led to a further decrease in IL-9 production (Fig. 4 D-F).
Figure 4:
ERG regulated IL-9 production in TH9 cells in the absence of PU.1 and ETV5. These experiments were performed using transgenic conditional knock-out mice including: Spifl/flEtv5fl/flErgfl/fl CD4 cre−, Spifl/flEtv5fl/fl CD4 cre+ and Spifl/flEtv5fl/flErgfl/fl CD4 cre+ respectively (n=3 per group) (A-J). (A-D) Transcript expression of Erg, Spi1, Etv5, and Il9 respectively. (E) IL-9 production in TH9 cells cultured measured via flow-cytometry. (F) IL-9 secretion measured by performing ELISA. (G-J) ChIP qPCR assay to assess interactions of PU.1, ERG, p300 and H3K27Ac at the Il9 locus in resting TH9 cells. Data are mean of three-four mice per experiment and representative of two independent experiments. *p<.05; **p<.01; ***p<0.001; ****p<0.0001.
We further performed ChIP assays to define the binding of Ets factors at Il9 regulatory regions. As expected, there was reduced binding of PU.1 at both Il9 promoter and enhancer regions in cells lacking PU.1 and ETV5 (Fig. 4 G). ERG binding was observed at the Il9 enhancer region in PU.1-ETV5 double-deficient TH9 cells, albeit at lower abundance compared to the TH9 cells derived from Cre-negative mice. ERG binding was nearly absent from Spifl/flEtv5fl/flErgfl/fl CD4cre mice (Fig. 4 H). Binding of histone acetyl transferase (HAT) p300 was reduced at the Il9 enhancer and CNS-6 region in the absence of PU.1 and ETV5. Deletion of Erg further lead to a decrease in p300 binding at both CNS-6 and CNS-25 (enhancer) regions (Fig. 4 I). Both double and triple-deficiency of Ets proteins led to a decrease in H3K27 histone acetylation modifications across the Il9 locus in TH9 cells (Fig. 4 J). The deletion of Ets-transcription factors did not affect H3K27 histone acetylation or p300 interactions at the Il9 promoter regions (Fig. 4 I and J). Together these findings indicate that ERG interacts with the Il9 locus in the absence of PU.1 and ETV5 and may contribute to Il9 gene regulation in the absence of these Ets proteins. Deletion of Erg in the absence of PU.1 and ETV5 lead to a decrease in p300 interactions at the Il9 enhancer regions suggesting that at least one mechanism though which ERG regulates transcription of Il9 is by recruiting p300 to the Il9 regulatory regions.
Combined deletion of ERG in the absence of PU.1 and ETV5 reduced TH9 mediated lung inflammation in the chronic allergic airway disease model.
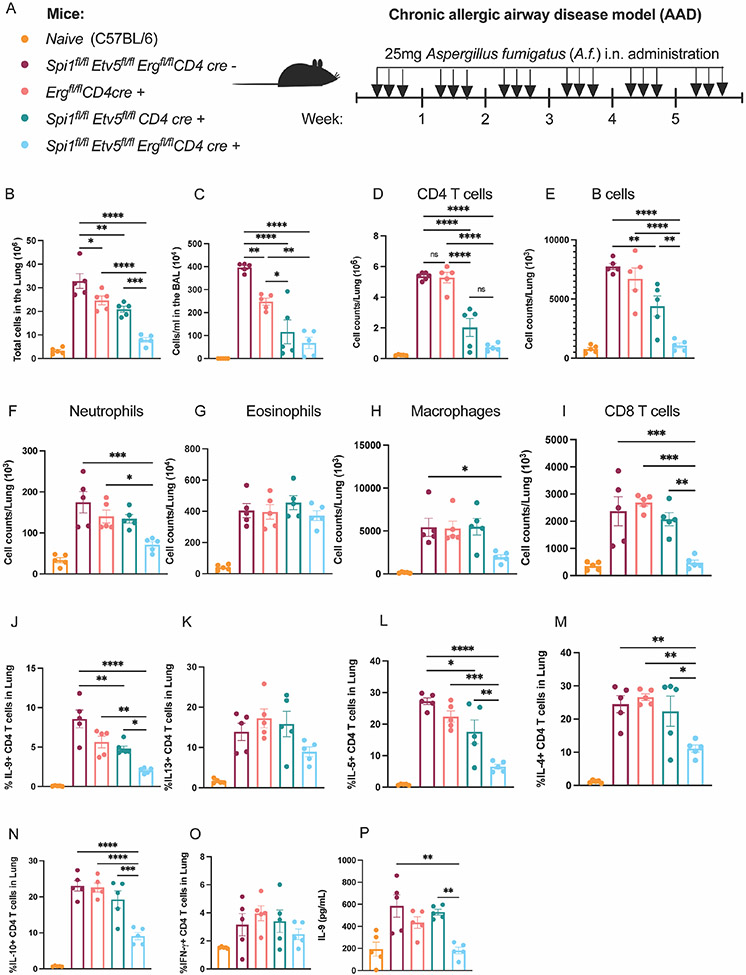
In the absence of PU.1 and ETV5, ERG facilitates IL-9 production in TH9 cells in vitro. We further wanted to determine whether ERG played a role in TH9 function in vivo in allergic airway diseases. To addresses this question, we measured TH9 function in Aspergillus fumigatus (A.f.)-induced chronic allergic airway disease (AAD) models in conditional knock out mice including Spifl/flEtv5fl/flErgfl/fl CD4 cre−, Ergfl/fl CD4 cre+, Spifl/flEtv5fl/fl CD4 cre+ and Spifl/flEtv5fl/flErgfl/fl CD4 cre+ mice (Fig. 5 A). Mice were intranasally challenged with A.f. over a period of 6 weeks and airway inflammation was examined at the end of 6 weeks. Deletion of Erg alone led to a modest decrease in total number of cells in the inflamed lungs as well as bronchoalveolar lavage (BAL) compared to CD4 cre− mice (Fig. 5 B and C). The absence of ERG alone failed to significantly reduce total CD4 T cell population or frequency of IL-9 producing CD4 T cells isolated from the lungs (Fig. 5 D and J).
Figure 5:
ERG regulates TH9 function in the absence of PU.1 and ETV5 in chronic AAD model. (A) Schematic of AAD model using transgenic mice of specified genotype. (B) Total number of cells in the lung. (C) Number of cells per ml in the BAL fluid. Number of CD4 T cells (D), B cells (E), neutrophils (F), eosinophils (G), macrophages (H), and CD8 T cell counts (I) in the lung. (J-O) Frequency of IL-9, IL-13, IL-5, IL-4, IL-10 and IFN-γ producing CD4 T cells in the lung. (P) IL-9 secretion from lung CD4 T cells co-cultured with naïve CD11c+ cells in the presence of IL-33 and A.f. (100mg/ml) for 72 hours ex vivo. Data are mean of 5 mice per experiment and representative of two independent experiments. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
In parallel experiments, we crossed Ergfl/fl mice with tamoxifen inducible CD4-cre mice to generate Ergfl/fl iCD4-cre transgenic mice. Mice were sensitized for 6 weeks with aspergillus fumigatus (A.f.) and CD4 specific cre expression was induced by administering 3 mg tamoxifen per mouse via oral gavage for 5 days during week 5 and week 6 of the chronic model (Supplementary Fig. 3 A). The deletion of Erg with tamoxifen treatment was confirmed by measuring Erg transcript in CD4 T cells isolated from the lung (Supplementary Fig. 3 B). Erg deletion in the last two weeks of the chronic AAD model led to a modest decrease in cellularity in the lung and BAL (Supplementary Fig. 3 C and D). ERG-deficient CD4 T cells had a significant decrease in total CD4 T cells and IL-9 producing CD4 T cell population in the lung (Supplementary Fig. 2 E-G). CD4 T cells from Ergfl/fl iCD4-Cre mice demonstrated reduced secretion of IL-9 by CD4 T cells in ex vivo, when cultured with naïve CD11c+ antigen presenting cells for 3 days in the presence of IL-33 and A.f. (Supplementary Fig. 3 H). Thus, as with in vitro results (Fig. 2 G-I), deletion of ERG in peripheral cells in vivo results in a greater decrease of IL-9 production, although this was still not sufficient to impact allergic inflammation.
We then determined if ERG cooperated with other Ets factors in the regulation of allergic airway inflammation. Deficiency in both PU.1 and ETV5 resulted in reduced cellularity in both lung and BAL compared to the CD4-cre- mice (Fig. 5 B and C). Spifl/flEtv5fl/fl CD4 cre+ mice also showed a decrease in CD4 T cells and the percentage of IL-9 producing CD4 T cell in the lung (Fig. 5 D and J). Deletion of Erg in the context of PU.1- and Etv5-double deficiency resulted in a further reduction in the total cellularity in both lung and BAL (Fig. 5 B and C). Spifl/flEtv5fl/flErgfl/fl CD4 cre+ mice had the greatest reduction in CD4 T cell populations and percentage of IL-9 secreting CD4 T cells in the lung (Fig. 5 D and J). Amongst the IL-9 responding immune cell populations, B cells and neutrophils showed a significant drop in cellularity in Spifl/flEtv5fl/flErgfl/fl CD4 cre+ mice (Fig. 5 E and F). In addition to IL-9+ CD4 T cell populations, we observed decreases in percentages of IL-4, IL-5 and IL-10 producing CD4 T cell in the lung with the deletion of Spi1, Etv5 and Erg (Fig. 5 L-O). Despite the reduced frequency of IL-5 secreting CD4 T cells in the Spifl/flEtv5fl/flErgfl/fl CD4 cre+ mice, the eosinophil population remained unaffected (Fig. 5 G). The populations of macrophages and CD8 T cells were only affected with the deletion of all three transcription factors: PU.1, ETV5 and ERG (Fig. 5 H and I). Moreover, deficiency of PU.1, ETV5 and ERG failed to affect IFNγ or IL-13 production by CD4 T cells, the latter which plays an important function in mucus production by airway epithelial cells (Fig. 5 K and O). Lastly, we also performed co-culture assays to measure IL-9 production by lung CD4 T cells in response to A.f. antigen. The combination of deficiency in PU.1, ETV5 and ERG leads to a significant decrease in IL-9 secretion by lung CD4 T cells specifically in response to A.f. (Fig. 5 P). Together these observations indicated that in the absence of PU.1 and ETV5, ERG regulated the development and function of TH9 cells in allergic airway diseases.
Discussion
Recent advances in TH9 biology have provided more evidence for the significance of IL-9 secreting CD4 T cells in mediating inflammatory responses by recruiting and activating innate and adaptive immune cells. Most T-helper cell subsets express a unique transcription factor that orchestrate the development of naïve CD4 T cells into specialized effector CD4 T-helper cells characterized by the production of signature cytokines. In the case of TH9 cells, a master regulator involved in the development of TH9 cells remains to be identified. Many transcription factors including PU.1, ETV5, STAT5, BATF, IRF4, IRF8 and more recently PPAR-γ have been reported to regulate IL-9 production in TH9 cells (1, 15-17, 19, 43-45). These findings suggest that TH9 differentiation is governed by a complex transcriptional network which may require recruitment of multiple transcription factors and co-activators to facilitate Il9 gene expression. Here we identify the Ets-transcription factor ERG that cooperates with PU.1 and ETV5 to enhance IL-9 production in TH9 cells.
Given the conserved Ets DNA binding domain and similar DNA target sequence amongst the members of Ets-family of transcription factors, it is not surprising that there are many examples of cooperativity. PU.1 and Spi-B exhibit redundant functions in the myeloid and lymphoid cell development, B cell function and the progression of leukemia (46, 47). Both ERG and Fli-1 have been implicated in the development of Ewing sarcoma due to the highly homologous Ets-DNA binding region in the C terminus (25, 26, 48). More relevant, PU.1 and ETV5 have also been reported to cooperatively regulate TH9 development in allergic airway disease model (19). That cooperation likely results in the modest decreases in IL-9 when ERG is deleted during T cell development or before activation. In vivo, deficiency of Erg expression alone leads to a modest decrease in IL-9 producing CD4 T cells in the lung and did not affect overall airway inflammation. However, when T cells in vivo are already deficient in PU.1 and ETV5, the additional deficiency in ERG results in greater decreases in IL-9 and allergic airway inflammation. The cooperativity among the Ets factors undoubtedly contributes to the differential effects of ERG deletion in vivo and during differentiation in vitro. The cooperativity, which is further supported by retroviral transduction and ChIP assays, allows for other factors to compensate for ERG deficiency, at least in some circumstances. However, it is important to note that in the absence of all three Ets factors we could still observe IL-9 producing TH9 cells, albeit at much lower frequency, suggesting that other factors could be facilitating IL-9 production by CD4 T cells.
Erg splice forms add additional complexity to understanding the contribution to IL-9 regulation. Mutation of the TH9-enriched exon 3 resulted in diminished expression of that isoform without changes in the exon 4 transcript or the overall Erg mRNA expression. This suggests that while enriched, exon 3 containing Erg transcripts represent a minor component of total Erg transcripts. It also suggests, in combination with other data presented, that either exon 4-containing Erg transcripts are sufficient to contribute to IL-9 production, or that there are additional cryptic splice forms with translational start sites that are not yet characterized. Importantly, utilizing two conditional mutant mouse strains that targeted distinct exons common to both exon 3- and exon 4-containing transcripts and deleted at the mature T cell stage we observed modest phenotypes suggesting that it is redundant function with other ETS proteins and not other splice forms that are responsible for the phenotype.
Mechanistically, ERG promotes Il9 expression by impacting binding of other transcription factors and histone modifying enzymes. Deletion of ERG results in decreased PU.1 binding, and deletion of ERG in the absence of PU.1 and ETV5 significantly reduced HAT p300 binding at the Il9 enhancer region. HAT p300 is a histone acetyltransferase that has been previously shown to bind at the Il9 locus to catalyze H3K27 acetylation in TH9 cells co-stimulated with OX40 (49). Studies on angiogenesis in endothelial cells show that vascular endothelial growth factor (VEGF) signaling induces activation of VEGF/ERK signaling pathway that facilitates ERG-dependent recruitment of p300 to promote transcription of genes associated with VEGF-dependent angiogenesis (50). In prostate cancer, ERG has been reported to interact with BAF chromatin remodeling complexes to promote expression of oncogenes (30). These findings suggest that ERG could enhance Il9 gene expression in TH9 cells by recruiting transcription co-activators and chromatin remodeling proteins to facilitate chromatin-chromatin interactions amongst cis-regulatory regions at the Il9 locus.
As noted, there is also evidence of ERG-dependent TH9 function in allergic airway diseases. Although deletion of Erg alone by either a constitutive or inducible CD4-Cre has modest effects on overall inflammation in the lung, in the absence of PU.1 and Etv5, Erg deficiency results in significant decreases in cellularity in the lung, including decreased CD4 T cells, CD8 T cells and B cell populations. The combined deficiency of ERG, PU.1 and ETV5 results in a significant decrease in IL-9 production by antigen specific CD4 T cells in the lung. Erg deletion also leads to a decrease in production of some of the other proinflammatory cytokines including IL-5, IL-10 and IL-4. Together these findings show that while Ets-transcription factors PU.1, ETV5 and ERG each contribute to IL-9 regulation, they can partially compensate for the absence of one another. Our data suggest that Ets-proteins function most effectively in concert to regulate IL-9 production and TH9-mediated inflammation in allergic airway diseases and that the redundancy suggests the importance of maintaining IL-9 expression in the organism.
Supplementary Material
Key Points:
ERG binds to and regulates Il9
ERG functionally overlaps with other ETS family members
ERG-dependent IL-9 production is important for allergic airway inflammation
Acknowledgments
Research presented here was supported by NIH grant AI057459. B.J.U. was supported by NIH grants T32 AI060519 and F30 HL147515. M.C. was supported by T32 HL091816 and F30AI174762. M.M.H. was supported by NIH grant T32 AI060519. Core facility usage was also supported by IU Simon Cancer Center Support Grant P30 CA082709 and U54 DK106846. Support provided by the Herman B Wells Center was in part from the Riley Children’s Foundation. Work in the Porse lab was supported through center grant from the Novo Nordisk Foundation (Novo Nordisk Foundation Center for Stem Cell Biology, DanStem; Grant Number NNF17CC0027852).
References:
- 1.Micosse C, von Meyenn L, Steck O, Kipfer E, Adam C, Simillion C, Seyed Jafari SM, Olah P, Yawlkar N, Simon D, Borradori L, Kuchen S, Yerly D, Homey B, Conrad C, Snijder B, Schmidt M, and Schlapbach C. 2019. Human "TH9" cells are a subpopulation of PPAR-gamma(+) TH2 cells. Sci Immunol 4. [DOI] [PubMed] [Google Scholar]
- 2.Jones CP, Gregory LG, Causton B, Campbell GA, and Lloyd CM. 2012. Activin A and TGF-beta promote T(H)9 cell-mediated pulmonary allergic pathology. J Allergy Clin Immunol 129: 1000–1010 e1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jia L, Wang Y, Li J, Li S, Zhang Y, Shen J, Tan W, and Wu C. 2017. Detection of IL-9 producing T cells in the PBMCs of allergic asthmatic patients. BMC Immunol 18: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerlach K, Hwang Y, Nikolaev A, Atreya R, Dornhoff H, Steiner S, Lehr HA, Wirtz S, Vieth M, Waisman A, Rosenbauer F, McKenzie AN, Weigmann B, and Neurath MF. 2014. TH9 cells that express the transcription factor PU.1 drive T cell-mediated colitis via IL-9 receptor signaling in intestinal epithelial cells. Nat Immunol 15: 676–686. [DOI] [PubMed] [Google Scholar]
- 5.Licona-Limon P, Henao-Mejia J, Temann AU, Gagliani N, Licona-Limon I, Ishigame H, Hao L, Herbert DR, and Flavell RA. 2013. Th9 Cells Drive Host Immunity against Gastrointestinal Worm Infection. Immunity 39: 744–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaplan MH, Hufford MM, and Olson MR. 2015. The development and in vivo function of T helper 9 cells. Nat Rev Immunol 15: 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaplan MH 2013. Th9 cells: differentiation and disease. Immunol Rev 252: 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angkasekwinai P, and Dong C. 2021. IL-9-producing T cells: potential players in allergy and cancer. Nat Rev Immunol 21: 37–48. [DOI] [PubMed] [Google Scholar]
- 9.Yao W, Tepper RS, and Kaplan MH. 2011. Predisposition to the development of IL-9-secreting T cells in atopic infants. J Allergy Clin Immunol 128: 1357–1360 e1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Longphre M, Li D, Gallup M, Drori E, Ordonez CL, Redman T, Wenzel S, Bice DE, Fahy JV, and Basbaum C. 1999. Allergen-induced IL-9 directly stimulates mucin transcription in respiratory epithelial cells. J Clin Invest 104: 1375–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louahed J, Toda M, Jen J, Hamid Q, Renauld JC, Levitt RC, and Nicolaides NC. 2000. Interleukin-9 upregulates mucus expression in the airways. Am J Respir Cell Mol Biol 22: 649–656. [DOI] [PubMed] [Google Scholar]
- 12.Stassen M, Arnold M, Hultner L, Muller C, Neudorfl C, Reineke T, and Schmitt E. 2000. Murine bone marrow-derived mast cells as potent producers of IL-9: costimulatory function of IL-10 and kit ligand in the presence of IL-1. J Immunol 164: 5549–5555. [DOI] [PubMed] [Google Scholar]
- 13.Schmitt E, Germann T, Goedert S, Hoehn P, Huels C, Koelsch S, Kuhn R, Muller W, Palm N, and Rude E. 1994. IL-9 production of naive CD4+ T cells depends on IL-2, is synergistically enhanced by a combination of TGF-beta and IL-4, and is inhibited by IFN-gamma. J Immunol 153: 3989–3996. [PubMed] [Google Scholar]
- 14.Staudt V, Bothur E, Klein M, Lingnau K, Reuter S, Grebe N, Gerlitzki B, Hoffmann M, Ulges A, Taube C, Dehzad N, Becker M, Stassen M, Steinborn A, Lohoff M, Schild H, Schmitt E, and Bopp T. 2010. Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells. Immunity 33: 192–202. [DOI] [PubMed] [Google Scholar]
- 15.Fu Y, Koh B, Kuwahara M, Ulrich BJ, Kharwadkar R, Yamashita M, and Kaplan MH. 2019. BATF-Interacting Proteins Dictate Specificity in Th Subset Activity. J Immunol 203: 1989–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu Y, Wang J, Panangipalli G, Ulrich BJ, Koh B, Xu C, Kharwadkar R, Chu X, Wang Y, Gao H, Wu W, Sun J, Tepper RS, Zhou B, Janga SC, Yang K, and Kaplan MH. 2020. STAT5 promotes accessibility and is required for BATF-mediated plasticity at the Il9 locus. Nat Commun 11: 4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jabeen R, Goswami R, Awe O, Kulkarni A, Nguyen ET, Attenasio A, Walsh D, Olson MR, Kim MH, Tepper RS, Sun J, Kim CH, Taparowsky EJ, Zhou B, and Kaplan MH. 2013. Th9 cell development requires a BATF-regulated transcriptional network. J Clin Invest 123: 4641–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang HC, Sehra S, Goswami R, Yao W, Yu Q, Stritesky GL, Jabeen R, McKinley C, Ahyi AN, Han L, Nguyen ET, Robertson MJ, Perumal NB, Tepper RS, Nutt SL, and Kaplan MH. 2010. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat Immunol 11: 527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koh B, Hufford MM, Pham D, Olson MR, Wu T, Jabeen R, Sun X, and Kaplan MH. 2016. The ETS Family Transcription Factors Etv5 and PU.1 Function in Parallel To Promote Th9 Cell Development. J Immunol 197: 2465–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson MK, Hernandez-Hoyos G, Diamond RA, and Rothenberg EV. 1999. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development 126: 3131–3148. [DOI] [PubMed] [Google Scholar]
- 21.Hollenhorst PC, McIntosh LP, and Graves BJ. 2011. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem 80: 437–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang HC, Zhang S, Thieu VT, Slee RB, Bruns HA, Laribee RN, Klemsz MJ, and Kaplan MH. 2005. PU.1 expression delineates heterogeneity in primary Th2 cells. Immunity 22: 693–703. [DOI] [PubMed] [Google Scholar]
- 23.Goswami R, and Kaplan MH. 2012. Gcn5 is required for PU.1-dependent IL-9 induction in Th9 cells. J Immunol 189: 3026–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birdsey GM, Shah AV, Dufton N, Reynolds LE, Osuna Almagro L, Yang Y, Aspalter IM, Khan ST, Mason JC, Dejana E, Gottgens B, Hodivala-Dilke K, Gerhardt H, Adams RH, and Randi AM. 2015. The endothelial transcription factor ERG promotes vascular stability and growth through Wnt/beta-catenin signaling. Dev Cell 32: 82–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kruse EA, Loughran SJ, Baldwin TM, Josefsson EC, Ellis S, Watson DK, Nurden P, Metcalf D, Hilton DJ, Alexander WS, and Kile BT. 2009. Dual requirement for the ETS transcription factors Fli-1 and Erg in hematopoietic stem cells and the megakaryocyte lineage. Proc Natl Acad Sci U S A 106: 13814–13819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ida K, Kobayashi S, Taki T, Hanada R, Bessho F, Yamamori S, Sugimoto T, Ohki M, and Hayashi Y. 1995. EWS-FLI-1 and EWS-ERG chimeric mRNAs in Ewing's sarcoma and primitive neuroectodermal tumor. Int J Cancer 63: 500–504. [DOI] [PubMed] [Google Scholar]
- 27.Yi H, Fujimura Y, Ouchida M, Prasad DD, Rao VN, and Reddy ES. 1997. Inhibition of apoptosis by normal and aberrant Fli-1 and erg proteins involved in human solid tumors and leukemias. Oncogene 14: 1259–1268. [DOI] [PubMed] [Google Scholar]
- 28.Selvaraj N, Kedage V, and Hollenhorst PC. 2015. Comparison of MAPK specificity across the ETS transcription factor family identifies a high-affinity ERK interaction required for ERG function in prostate cells. Cell Commun Signal 13: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hollenhorst PC, Shah AA, Hopkins C, and Graves BJ. 2007. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev 21: 1882–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandoval GJ, Pulice JL, Pakula H, Schenone M, Takeda DY, Pop M, Boulay G, Williamson KE, McBride MJ, Pan J, St Pierre R, Hartman E, Garraway LA, Carr SA, Rivera MN, Li Z, Ronco L, Hahn WC, and Kadoch C. 2018. Binding of TMPRSS2-ERG to BAF Chromatin Remodeling Complexes Mediates Prostate Oncogenesis. Mol Cell 71: 554–566 e557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kedage V, Selvaraj N, Nicholas TR, Budka JA, Plotnik JP, Jerde TJ, and Hollenhorst PC. 2016. An Interaction with Ewing's Sarcoma Breakpoint Protein EWS Defines a Specific Oncogenic Mechanism of ETS Factors Rearranged in Prostate Cancer. Cell Rep 17: 1289–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooper CD, Newman JA, and Gileadi O. 2014. Recent advances in the structural molecular biology of Ets transcription factors: interactions, interfaces and inhibition. Biochem Soc Trans 42: 130–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sondergaard E, Rauch A, Michaut M, Rapin N, Rehn M, Wilhelmson AS, Camponeschi A, Hasemann MS, Bagger FO, Jendholm J, Knudsen KJ, Mandrup S, Martensson IL, and Porse BT. 2019. ERG Controls B Cell Development by Promoting Igh V-to-DJ Recombination. Cell Rep 29: 2756–2769 e2756. [DOI] [PubMed] [Google Scholar]
- 34.Pham D, Sehra S, Sun X, and Kaplan MH. 2014. The transcription factor Etv5 controls TH17 cell development and allergic airway inflammation. J Allergy Clin Immunol 134: 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schweitzer BL, and DeKoter RP. 2004. Analysis of gene expression and Ig transcription in PU.1/Spi-B-deficient progenitor B cell lines. J Immunol 172: 144–154. [DOI] [PubMed] [Google Scholar]
- 36.Dakic A, Metcalf D, Di Rago L, Mifsud S, Wu L, and Nutt SL. 2005. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J Exp Med 201: 1487–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, Sui P, Dong A, Hassell J, Cserjesi P, Chen YT, Behringer RR, and Sun X. 2010. Preaxial polydactyly: interactions among ETV, TWIST1 and HAND2 control anterior-posterior patterning of the limb. Development 137: 3417–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knudsen KJ, Rehn M, Hasemann MS, Rapin N, Bagger FO, Ohlsson E, Willer A, Frank AK, Sondergaard E, Jendholm J, Thoren L, Lee J, Rak J, Theilgaard-Monch K, and Porse BT. 2015. ERG promotes the maintenance of hematopoietic stem cells by restricting their differentiation. Genes Dev 29: 1915–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koh B, Abdul Qayum A, Srivastava R, Fu Y, Ulrich BJ, Janga SC, and Kaplan MH. 2018. A conserved enhancer regulates Il9 expression in multiple lineages. Nat Commun 9: 4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campbell TB, Basu S, Hangoc G, Tao W, and Broxmeyer HE. 2009. Overexpression of Rheb2 enhances mouse hematopoietic progenitor cell growth while impairing stem cell repopulation. Blood 114: 3392–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao C, Teng EM, Summers RG Jr., Ming GL, and Gage FH. 2006. Distinct morphological stages of dentate granule neuron maturation in the adult mouse hippocampus. J Neurosci 26: 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vijayaraj P, Le Bras A, Mitchell N, Kondo M, Juliao S, Wasserman M, Beeler D, Spokes K, Aird WC, Baldwin HS, and Oettgen P. 2012. Erg is a crucial regulator of endocardial-mesenchymal transformation during cardiac valve morphogenesis. Development 139: 3973–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olson MR, Ulrich BJ, Hummel SA, Khan I, Meuris B, Cherukuri Y, Dent AL, Janga SC, and Kaplan MH. 2017. Paracrine IL-2 Is Required for Optimal Type 2 Effector Cytokine Production. J Immunol 198: 4352–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ulrich BJ, Verdan FF, McKenzie AN, Kaplan MH, and Olson MR. 2017. STAT3 Activation Impairs the Stability of Th9 Cells. J Immunol 198: 2302–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Humblin E, Thibaudin M, Chalmin F, Derangere V, Limagne E, Richard C, Flavell RA, Chevrier S, Ladoire S, Berger H, Boidot R, Apetoh L, Vegran F, and Ghiringhelli F. 2017. IRF8-dependent molecular complexes control the Th9 transcriptional program. Nat Commun 8: 2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solomon LA, Li SK, Piskorz J, Xu LS, and DeKoter RP. 2015. Genome-wide comparison of PU.1 and Spi-B binding sites in a mouse B lymphoma cell line. BMC Genomics 16: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeKoter RP, Geadah M, Khoosal S, Xu LS, Thillainadesan G, Torchia J, Chin SS, and Garrett-Sinha LA. 2010. Regulation of follicular B cell differentiation by the related E26 transformation-specific transcription factors PU.1, Spi-B, and Spi-C. J Immunol 185: 7374–7384. [DOI] [PubMed] [Google Scholar]
- 48.Wang ZB, An XJ, Deng JF, Liu JH, and Shi HY. 2017. [Characteristics of ERG, Fli-1, CD34, CD31 and FRAg expression in hepatic malignant vascular tumors]. Zhonghua Bing Li Xue Za Zhi 46: 760–763. [DOI] [PubMed] [Google Scholar]
- 49.Xiao X, Fan Y, Li J, Zhang X, Lou X, Dou Y, Shi X, Lan P, Xiao Y, Minze L, and Li XC. 2018. Guidance of super-enhancers in regulation of IL-9 induction and airway inflammation. J Exp Med 215: 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fish JE, Cantu Gutierrez M, Dang LT, Khyzha N, Chen Z, Veitch S, Cheng HS, Khor M, Antounians L, Njock MS, Boudreau E, Herman AM, Rhyner AM, Ruiz OE, Eisenhoffer GT, Medina-Rivera A, Wilson MD, and Wythe JD. 2017. Dynamic regulation of VEGF-inducible genes by an ERK/ERG/p300 transcriptional network. Development 144: 2428–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.