Abstract
Background:
Accurate interpretation of hip MRI is time-intensive and difficult, prone to inter- and intrareviewer variability, and lacks a universally accepted grading scale to evaluate morphological abnormalities.
Purpose:
To 1) develop and evaluate a deep-learning-based model for binary classification of hip osteoarthritis (OA) morphological abnormalities on MR images, and 2) develop an artificial intelligence (AI)-based assist tool to find if using the model predictions improves interreader agreement in hip grading.
Study Type:
Retrospective study aimed to evaluate a technical development.
Population:
A total of 764 MRI volumes (364 patients) obtained from two studies (242 patients from LASEM [FORCe] and 122 patients from UCSF), split into a 65–25–10% train, validation, test set for network training.
Field Strength/Sequence:
3T MRI, 2D T2 FSE, PD SPAIR.
Assessment:
Automatic binary classification of cartilage lesions, bone marrow edema-like lesions, and subchondral cyst-like lesions using the MRNet, interreader agreement before and after using network predictions.
Statistical Tests:
Receiver operating characteristic (ROC) curve, area under curve (AUC), specificity and sensitivity, and balanced accuracy.
Results:
For cartilage lesions, bone marrow edema-like lesions and subchondral cyst-like lesions the AUCs were: 0.80 (95% confidence interval [CI] 0.65, 0.95), 0.84 (95% CI 0.67, 1.00), and 0.77 (95% CI 0.66, 0.85), respectively. The sensitivity and specificity of the radiologist for binary classification were: 0.79 (95% CI 0.65, 0.93) and 0.80 (95% CI 0.59, 1.02), 0.40 (95% CI −0.02, 0.83) and 0.72 (95% CI 0.59, 0.86), 0.75 (95% CI 0.45, 1.05) and 0.88 (95% CI 0.77, 0.98). The interreader balanced accuracy increased from 53%, 71% and 56% to 60%, 73% and 68% after using the network predictions and saliency maps.
Data Conclusion:
We have shown that a deep-learning approach achieved high performance in clinical classification tasks on hip MR images, and that using the predictions from the deep-learning model improved the interreader agreement in all pathologies.
Level of Evidence:
3
Technical Efficacy Stage:
1
HIP OSTEOARTHRITIS (OA) is a debilitating joint disease that affects over 27 million Americans annually.1 Even though the reported radiographic and symptomatic hip OA prevalence is 28% and 10% in subjects over age 45,2 the exact primary hip OA etiology remains unknown, and there is no unanimous criteria for its diagnosis.3 Currently, radiography is the most common imaging modality used to diagnose joint space narrowing and osteophyte formation in the hip and is graded using the Kellgren–Lawrence or Tönnis classification systems.3-5
Magnetic resonance imaging (MRI) has emerged as an important tool to evaluate morphological abnormalities in joints such as the knee and hip.6 This is due to its ability to evaluate anomalies in the cartilage, bone marrow, and labrum and other pathologies, making it more robust to assess early morphological abnormalities in the hip, which are characterized by internal derangement of the joint structures often not seen with standard radiographs. MRI of hip OA remains challenging, given the thin articular cartilage in the hip, its distance from the surface of the body, and the complexity of assessing the labrum.4 Due to the quantity of images in each hip MR exam, accurate interpretation of hip MRI is time-intensive and difficult, being prone to inter- and intrareviewer variability, compounded by the absence of a universally accepted grading scale to evaluate the morphological abnormalities in hip MRI.7-9
To address these current limitations and standardize image interpretation, various hip OA grading scales were developed. Semiquantitative MRI-based classification systems such as HOAMS (Hip Osteoarthritis MRI Scoring System) and SHOMRI (Scoring Hip Osteoarthritis with MRI) have been proposed,4,10 showing associations with radiographic OA severity and patient reported outcomes, but still limited by low kappa values (weighted kappa between 0.18–0.85 in HOAMS and 0.55–0.79 in SHOMRI), but with moderate to good percent agreement (83.1–83.8% in HOAMS and 66–99% in SHOMRI) for interreader reliability. The low kappa values were attributed to low frequencies of some features in the dataset.10 A classification system for hip morphological abnormalities that is reliable, agreed upon by radiologists, and can achieve widespread use for grading commonly occurring hip pathologies is still required. Even with the multiple grading systems discussed above, clinical translation of these systems has been difficult, considering the time and expertise required to grade hip MR images along with issues of inter- and intrareader variability.
Recently, deep-learning-based models have shown promise in several musculoskeletal imaging diagnostic tasks due to their ability to detect features automatically, making them suitable to find relationships between features in medical images and their interpretations. Deep learning can classify joint abnormalities in the knee, including detection of cartilage lesion, OA prediction, fractures, meniscal tears, and chondrocyte patterns.11-16 These studies were facilitated by the availability of large and well-curated knee datasets such as the Osteoarthritis Initiative, MOST dataset, and MRNet dataset, but these data are not publicly available for hip OA classification by deep learning. For the hip, deep learning has been used in detecting OA on radiographs.17
While deep learning has shown promising results when adopted in a controlled setting, it is well understood that generalization beyond the statistical distribution of the training set is still an unmet challenge. Deep convolutional networks can learn representations that are generically used across a variety of tasks and visual domains. However, due to a phenomenon known as dataset bias or domain shift,18 models trained on one large dataset do not generalize well to novel datasets and tasks. In MRI this translates into poor performance when trained models are tested on different imaging protocols or images acquired on different MRI systems. The typical solution is to fine-tune these networks on specific datasets; however, it is often prohibitive to obtain enough labeled data to properly fine-tune the large number of parameters employed by a deep model on the new datasets.
In this study we aimed to address these challenges and fill this gap. Specifically, we aimed 1) to develop a deep-learning-based model for binary classification of cartilage lesions, bone marrow edema-like lesions, and subchondral cyst-like lesions in coronal hip MR images acquired from two centers without any initial calibration or standardization of the sequence, 2) to evaluate the performance of the models by comparing the performance to an experienced musculoskeletal radiologist, and 3) to develop an artificial intelligence (AI)-based assist tool to investigate whether using the model predictions improves interreader agreement in assessing hip cartilage lesions, bone marrow edema-like lesions, and subchondral cyst-like lesions.
Materials and Methods
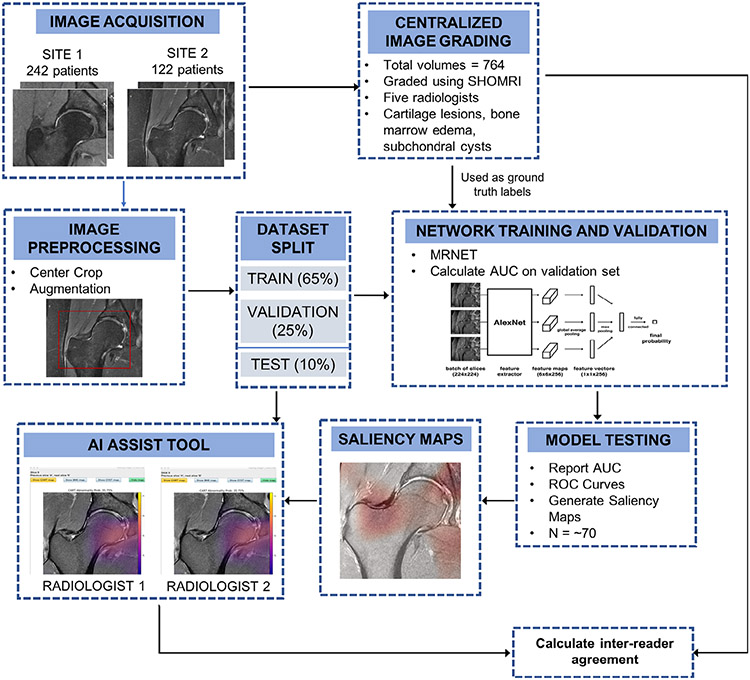
An overview of the entire process is shown in Fig. 1. In this study data were obtained from the femoroacetabular impingement and hip osteoarthritis cohort (FORCe) study19 conducted at LASEM (La Trobe Sport and Exercise Medicine Research Centre) in Melbourne, Australia, and a hip osteoarthritis study20 conducted at the UCSF (University of California San Francisco) in San Francisco, United States.
FIGURE 1:
Method and analysis pipeline. Images were acquired at two different sites, and a centralized image grading was performed using the SHOMRI grading system for each pathology. These grades were used as ground truth labels in network training, validation, and testing. All the images underwent image preprocessing and augmentation if needed. The dataset was split into training, validation, and testing groups. Three networks were trained separately for each pathology, using the MRNET architecture. Models that had an AUC greater than 0.7 were selected and used for testing the network performance on the test dataset, while also generating saliency maps. These maps were loaded into the AI assist tool, used by two radiologists. The interreader agreement was calculated between the two radiologists for each pathology.
The UCSF study was Health Insurance Portability and Accountability Act (HIPAA)-compliant, approved by the Institutional Review Board (IRB). Written informed consent was obtained from each patient at the time of image acquisition and the authors from UCSF retained control of all study information and statistical analysis. Data acquired from the FORCe cohort were collected in accordance with the Declaration of Helsinki, with ethics approval obtained from the La Trobe University Human Ethics Committee (HEC 15-019 and HEC16-045) and the University of Queensland Human Ethics Committee (2015000916 & 2016001694).
Image Dataset
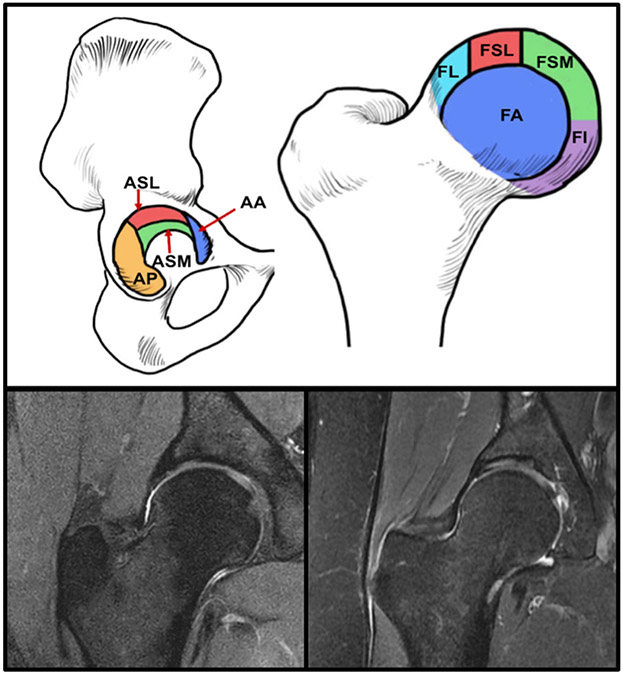
A total of 764 coronal hip volumes were used in this project, including bilateral scans and multiple timepoints of 364 unique patients. Of these, 242 patient data were obtained from the LASEM FORCe dataset and 122 were part of the UCSF dataset. Images were acquired on a 3.0T Scanner (Philips, Eindhoven, The Netherlands) scanner at LASEM and a 3.0T Scanner (GE Healthcare, Milwaukee, WI) scanner at UCSF. The coronal sequences acquired that were used in this study are described in Table 1, and examples of both study site images are shown in Fig. 2.
TABLE 1.
MRI Sequence Parameters Shown by Study Sites (UCSF and LASEM)
| UCSF | LASEM | |
|---|---|---|
| Sequence type |
Fat-saturated T2- weighted |
PD-weighted SPAIR |
| TE/TR | 60/3090 msec | 25/2574 msec |
| FOV | 20 cm | 17 cm |
| Matrix size | 288 × 224 | 244 × 234 |
| Slice thickness | 4 mm | 2.5 mm |
| Resolution | 0.69 × 0.89 mm | 0.69 × 0.72 mm |
| Scanner | 3.0 T (GE) | 3.0 T (Philips) |
TR = repetition time; TE = echo Time; FOV = field of view; PD = proton density; SPAIR = Spectral Attenuated Inversion Recovery; GE = General Electric.
FIGURE 2:
(Top) Subregions of the hip used for SHOMRI grading: acetabular superolateral (ASL), acetabular superomedial (ASM), and femoral lateral (FL), femoral superolateral (FSL), femoral superomedial (FSM), and femoral inferior (FI). (Bottom-left) Example coronal hip image from the UCSF. (Bottom-right) Example hip image from LASEM.
Image Grading
The training set (496 studies) was annotated between 2013 and 2019 by five board-certified radiologists, all with 5+ years of experience. All images were read by an attending and a fellow to describe the presence or absence of cartilage lesions, bone marrow edema type patterns, and subchondral cysts. These annotations served as ground truth data to train the deep-learning models. The grading was done using the SHOMRI scoring system. All radiologists performing the SHOMRI gradings were trained by a senior radiologist (T.M.L.), who was involved in developing the scoring system and reviewed at least 20 imaging studies with each of the radiologists at two different timepoints. The senior radiologist also adjudicated any cases that had discrepancies. The test set (78 studies) was independently graded by three radiologists for the absence or presence of cartilage lesions, bone marrow edema-type patterns, and subchondral cysts, and a majority vote was taken to serve as the final ground truth. All the model performances reported in this study were computed in comparison to this ground truth obtained by three independent readers. As part of an interreader experiment, these test data were further graded by two radiologists with and without the aid of the automatic system to evaluate efficacy and use of the developed model in practice (see section AI Assist Tool Development and Experiment). In the coronal hip images SHOMRI differentiates six anatomic compartments: acetabular superolateral (ASL), acetabular superomedial (ASM), and femoral lateral (FL), femoral superolateral (FSL), femoral superomedial (FSM), and femoral inferior (FI). Each compartment was graded on a 3-point scale for cartilage lesions, a 4-point scale for bone marrow edema-like lesions, and a 3-point scale for subchondral cyst-like abnormalities. The total score was calculated for each abnormality (cartilage, bone marrow edema-like lesions, subchondral cyst-like lesions) by adding up the scores of each compartment (Fig. 2).4
Network Architecture
MRNet architecture was used for this study. This architecture has been shown to achieve an area under the curve (AUC) of 0.937 (95% confidence interval [CI] 0.895, 0.980) in detecting abnormalities in the knee, with performance comparable to that of radiologists. Briefly described, the input to the MRNet has dimensions s × 3 × 256 × 256, where s is the number of slices in the series acquisition. Each slice is passed through a feature extractor: the AlexNet (which consists of five convolutional layers, three fully connected layers, and an ReLU activation function)21 pretrained on ImageNet to obtain features for that slice of dimensions s × 256 × 7 × 7. After going through a global average pooling layer to reduce these features to s × 256, a max pooling was applied across the slices to obtain a 256-dimensional vector, which was then passed through a fully connected layer and a sigmoid activation function to obtain a probability between 0 and 1.11 In our study we used a threshold of 0.5 to dichotomize the model’s predictions into abnormal and normal.
Data Processing for Input
All input volumes underwent identical preparation regardless of study site. Each input volume consisted of all slices from each patient, with each slice cropped at the center to 150 × 150 mm, by using the image resolution from the different sites. This way, assuming a well-centered scan, the same hip area was covered in patients from both sites. If an image underwent augmentation, any one, two, or all of the following were used: 1) rotation along superior–inferior and medial-lateral axes of a random integer between −9 and 9 degrees; 2) an affine transformation with spline interpolation of order 3, and diagonal values of the inverse transform matrix =1, 0.78, 0.87; and 3) an intensity rescaling and histogram equalization with values between the 2 and 98 percentiles. All image processing and augmentations were performed using Python’s (Wilmington, DE) Pydicom and Scipy packages. All images were visually inspected after cropping and augmentation to ensure no important structures were being omitted or distorted. In each network implementation, the label for each image corresponded to the maximum cartilage lesion, bone marrow edema-like lesion, or subchondral cyst-like lesion grade of all six compartments (ASL, ASM, FSL, FSM, FI, FL), binarized as 0 or 1, with the label 1 corresponding to a maximum lesion grade greater than 0. Thus, the network was being trained to identify the presence or absence of lesions.
Data Split
We trained three separate networks for binary detection of the cartilage lesions, bone marrow edema-like lesions, and subchondral cyst-like lesions. All three networks consisted of the same training, validation, and test set so that their performance could be compared. The 764 volumes were split into 65–25–10% sets used for train, validation, and test. The ratio of images between the two study sites (1.98, 1 for LASEM:UCSF) was maintained in training, validation, and test sets. It was also ensured that all images corresponding to one patient (different timepoints, laterality) were contained within the same split.
Network Training and Model Selection
Since there was no previous knowledge regarding efficient network hyperparameters to train for the hip abnormalities, an experimental approach was used to find an optimal combination of parameters that would result in optimal models for detecting the cartilage, bone marrow edema-like lesions, and subchondral cyst-like lesions. Thus, a grid search including different values for the learning rate and probability of dropout, combined with the presence of augmentation and weight decay, was implemented to find the best combination of these parameters for training the networks. The learning rates ranged between 1E-06 and 4.02E-05, the probability of dropout varied from 0.3 to 0.6, augmentation was turned on or off, and weight decay was turned off or set at 0.01. Therefore, for each pathology, 100 networks were trained, each with a different combination of parameters.
The network used a weighted cross-entropy loss to account for class imbalance by assigning weights that were inverse to the number of labels present in that class. An Adam optimizer with a scheduler with patience = 5 was used. The network was implemented in Pytorch, v. 0.3.0, trained for 80 epochs (3 hours on a Nvidia Titan X GPU).22 Using Python’s Sklearn package, receiver operating characteristic (ROC) curves and AUC values were plotted and saved for each epoch in each model, for training and validation data. Among the different parameter combinations, the combinations that resulted in the highest validation AUC over all epochs were retained. For those selected combinations models, the epoch with the highest AUC was selected before the model started to overfit the dataset. Hyperparameter optimization and model performance tracking was done to optimize performance on the validation set. The test set was not used in any of the model development phases.
Model Interpretation Using Class Activation Maps
To check if the networks were learning relevant features, class activation maps were generated for the training, validation, and test model predictions. For each slice, a weighted average across the 256 convolutional neural network (CNN) feature maps using weights from the classification layer was obtained and then upsampled to 256 × 256 pixels, and overlaid with the original input image. Since the final layer of the network was used for weighting the features, more predictive feature maps appear in different intensities of color.11 These maps were visually inspected to see if the network was looking within the hip regions, and used in the development of the AI Assist tool described below.
Network Performance
After selecting one model for each pathology, the test set was identically processed (without augmentations) and run through this model. The model probabilities corresponding to each volume were saved for the cartilage lesions, bone marrow edema-like lesions, and subchondral cyst-like lesions. The same test set was given to the radiologist for binary grading of the three pathologies and their responses were recorded. Using R on this test set only, ROC curves were created for the models’ performance for each pathology against the ground truth. AUC values were calculated by finding the AUC for these ROC curves. On the same ROC space, the sensitivity and specificity of the senior radiologist performance on the test set against the ground truth was plotted to compare the performance of the radiologist and model.
AI Assist Tool Development and Experiment
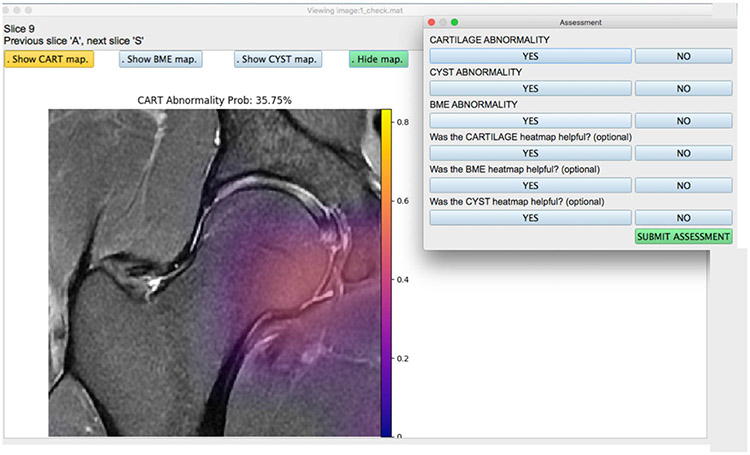
Once the test set was run through each model for all three pathologies, an application (Fig. 3) was developed in Python v. 3.1 with an interface that consisted of the following key features: 1) visualize hip image and scroll through slices; 2) toggle on or off the saliency map for any one of the pathologies at a time; 3) visualize the networks predicted lesion probability; 4) input binary assessment from the radiologist; 5) select from the list of patient images; and 6) ensure that there was no metadata or patient health information data visible anywhere. The application was loaded onto computers for the radiologists and ran locally without needing any Internet access.
FIGURE 3:
View of the user-friendly AI-based assist tool for radiologists. Images were completely stripped of their metadata and any patient information and loaded onto the tool. The radiologist has the option of turning on or off the cartilage lesions, bone marrow edema-like lesions, and subchondral cyst-like lesion saliency maps generated on the test set using the model. Once the assessment was submitted, the responses were recorded and stored for each patient image. The radiologist can select from a list of patient images, where each file is simply labeled with a number, and corresponding grades are mapped onto the Excel sheet.
The application was used by two radiologists at two different trial periods. For the first trial, they were given the application without the option for the saliency maps or the network prediction probability, where they provided a binary assessment on the test set patients for the presence of cartilage lesions, bone marrow edema-like lesions, and subchondral cyst-like lesions. After a 1-month washout period, the radiologists repeated the assessment, this time with the saliency maps and network prediction probability present. The responses of both the radiologists from both trials were recorded and used to calculate balanced accuracy (calculated in R [v. 1.1.456] using the caret library), which was chosen as a metric of quantification, since it avoids inflated performance metrics on imbalanced datasets, which our study contained. For binary classifications, the balanced accuracy is the arithmetic mean of the sensitivity and specificity, and is equal to conventional accuracy if the classifier performs equally well on both classes.23
Results
Image Dataset and Grading
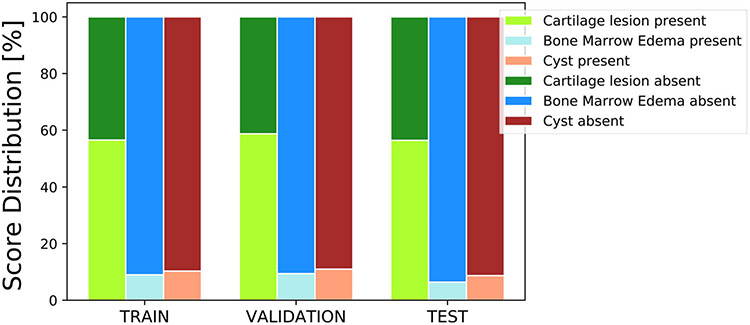
From the UCSF cohort, the patients had a mean age of 43 (22–72) years, a mean body mass index (BMI) of 23 (16–32) kg/m2, and were 56% female. From the LASEM cohort, the patients had a mean age of 27 (18–49) years, a mean BMI of 24 (17–35) kg/m2, and were 22% female. The distribution of the binary cartilage lesion, bone marrow edema-like lesion, and subchondral cyst-like lesion scores in the training, validation, and test sets are shown in Fig. 4. As seen, there is a strong class imbalance in the subchondral cyst-like lesion and bone marrow edema-like lesion scores, as very few patients presented with these abnormalities in the whole dataset.
FIGURE 4:
Distribution of binary (present or absent) grades in the training, validation, and testing splits used in this study. The bone marrow edemas and cysts have a low representation of positive lesions in this study; however, they are uniform in all three training, validation, and test sets.
Grid Search and Network Training
From all the parameter combinations run, for the cartilage the following parameters were selected based on their performance: learning rate = 3.08E-05, dropout = 0.36, for the bone marrow edema-like lesions: learning rate = 3.08E-05, dropout = 0.42, for the subchondral cyst-like lesions: learning rate = 2.12E-05, dropout = 0.30 were selected and their performance was tested. No models selected used augmentation or weight decay, as these were found to have lower performance on the validation than their counterparts. By dividing the job on different GPUs and training simultaneous networks, the grid search took ~11 days to run through all its combinations.
Class Activation Maps
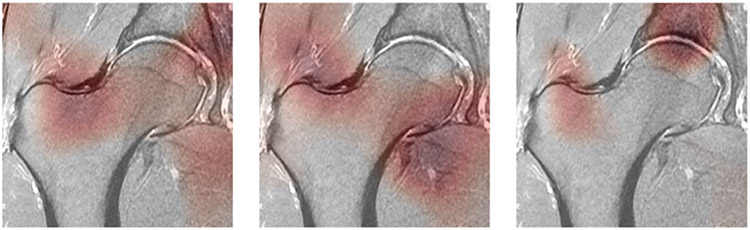
Examples of class activation maps on the selected models on the test set can be seen in Fig. 5. All three images shown in Fig. 5 are the same slice from the same patient from the test set, and show cartilage lesion, bone marrow edema-like lesion, and subchondral cyst-like lesion saliency maps (left to right). The maps appear darker in regions where the network has paid more attention in making its decision.
FIGURE 5:
Class activation maps (CAMs) highlight which pixels in the images are important for the model to make its classification probability. The darker areas of the map show where the network focused its decision-making on. The three images are the same slice from one patient in the test set. (Left) Map from the cartilage lesion detection network. (Middle) Map from the bone marrow edema-like lesion detection edema detection network. (Right) Map from the subchondral cyst-like lesion detection network.
Network Performance
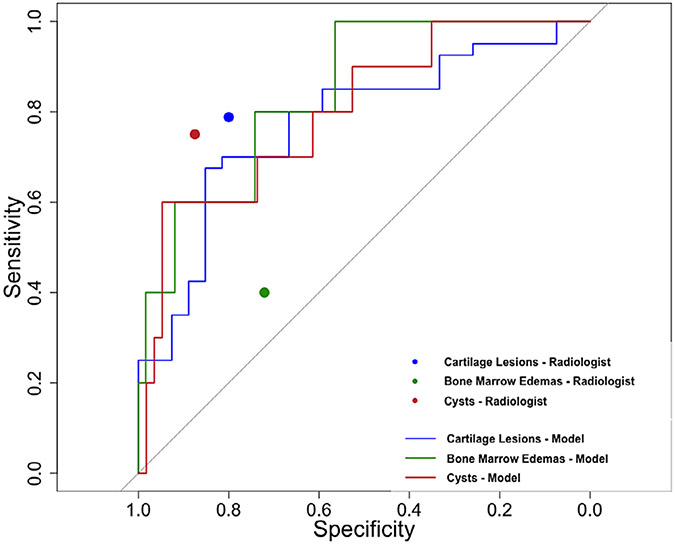
Figure 6 shows the ROC curves (color coded) for the models selected for cartilage lesions, bone marrow edema-like lesions, and subchondral cyst-like lesions for binary classification as they performed against the ground truth data. The AUC for the network performance was as follows: 0.80 (95% CI 0.65, 0.95) for cartilage lesions, 0.84 (95% CI 0.67, 1.00) for bone marrow edema-like lesions, and 0.77 (95% CI 0.66, 0.85) for subchondral cyst-like lesions. When stratified by study site, the network performance AUC on the UCSF images was as follows: 0.79 (95% CI 0.57, 1.00) for cartilage lesions, 0.67 (95% CI 0.24, 1.00) for bone marrow edema-like lesions, and 0.67 (95% CI 0.41, 0.92) for subchondral cyst-like lesions. When stratified by study site, the network performance AUC on the LASEM images was as follows: 0.75 (95% CI 0.60, 0.90) for cartilage lesions, 0.71 for bone marrow edema-like lesions, and 0.91 (95% CI 0.81, 1.00) for subchondral cyst-like lesions. The sensitivity and specificity of the radiologist on the same test set for binary classification are as follows: 0.79 (95% CI 0.65, 0.93) and 0.80 (95% CI 0.59, 1.02) for cartilage lesions, 0.40 (95% CI −0.02, 0.83) and 0.72 (95% CI 0.59, 0.86) for bone marrow edemas-like lesions, 0.75 (95% CI 0.45, 1.05) and 0.88 (95% CI 0.77, 0.98) for subchondral cyst-like lesion. The sensitivity and specificity values for the radiologist on the same test set are also plotted on the same curve in Fig. 6.
FIGURE 6:
Receiver operating characteristic curves of the three binary classification models (cartilage lesion detection model: blue line, bone marrow edema-like lesion detection model: green line, subchondral cyst-like lesion detection model: red line) along with performance of an experienced musculoskeletal radiologist in terms of sensitivity and specificity for binary detection of hip pathology (cartilage lesion: blue point, bone marrow edema-like lesion: green point, subchondral cyst-like lesion: red point).
AI Assist Tool and Interreader Agreement
Before using the aided detection application, the interreader balanced accuracy for binary classification on cartilage lesions, bone marrow edema-like lesions, and subchondral cyst-like lesions was 53%, 71%, and 56%, respectively. After a month-long washout period and using the saliency maps and network prediction probabilities, the interreader balanced accuracy for binary classification improved for all three abnormalities: cartilage lesions, bone marrow edema-like lesions, and subchondral cyst-like lesions was 60%, 73%, and 68%, respectively.
Predicting Disease Severity Using Total SHOMRI Score
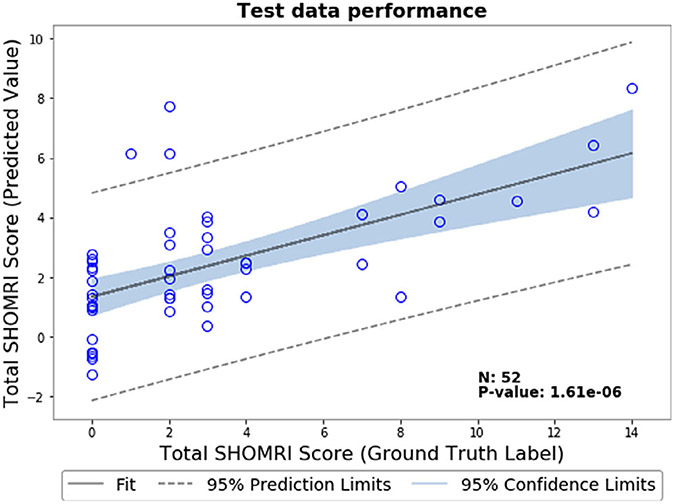
As part of a preliminary study, we evaluated a deep-learning based model for predicting disease severity using the total SHOMRI score of each patient instead of binary classification. Briefly described, this score is calculated as the total of the cartilage lesion scores, bone marrow edema scores, and cyst scores in the coronal and sagittal images. We used the same dataset and split as used in the binary classification experiment, with the difference of adding the sagittal images of each patient in addition to the coronal images in the input for the MRNet. The details of the sequences acquired and used in this study are in Table 1 (both coronal and sagittal sequences had the same acquisition parameters). Since we now only included patients that had both coronal and sagittal image acquisitions, this reduced our dataset to a total of 558 volumes (a 26.9% reduction in dataset size). The distribution of the total SHOMRI scores in this dataset is skewed; more than 50% of the patients had scores between 0 and 5. We changed the network loss from a binary cross-entropy to an Mean Square Error-based loss function and used a linear activation function instead of sigmoidal to have a continuous output. We used a learning rate of 1e-05 and a weight decay of 0.01. We did not use dropout and augmentation as part of this initial study. To evaluate the results, we calculated the Pearson correlation coefficient between the input score and network predicted score on the test set, which was R = 0.61 (P = 1.61E-06). A correlation plot between the input score and network predicted score on the test set is shown in Fig. 7.
FIGURE 7:
Correlation plot between input and predicted total SHOMRI scores on the test dataset for the severity staging model. Along with the line of best fit, 95% confidence limits and 95% prediction limits for this dataset are shown in this plot.
Discussion
The purpose of this study was to develop a deep-learning model to classify pathologies on hip MRI and to compare its performance with radiologists considered clinical experts in the field, and to develop an AI-based assist tool to see if it improves interreader reliability in classifying these abnormalities. Our results demonstrate that a deep-learning approach achieved high performance in clinical classification tasks on hip MR images, and that using the predictions from the deep-learning model improved the interreader agreement quantified as balanced accuracy in all pathologies.
Besides its promising performance in classifying abnormalities, the MRNet was chosen for a number of reasons for this study. First, for each patient volume the label stated the presence or absence of lesion; however, there was no information regarding the slice location of this lesion, making this a weakly supervised problem. Thus, a 2D model was not suitable for this problem, since it would require ground truth label annotations on each slice, and not just each volume. While a 3D architecture could have been chosen for this problem, there were no suitable pretrained model weights for a 3D architecture that could be transferred to this problem. With a relatively small sample size, a lack of pretrained weights would lead to overfitting. Additionally, the MR sequences used in this study were not isotropic, and a 3D convolution would have a receptive field that is different inplane and out-of-plane. Therefore, the MRNet was chosen since it retains information from all the slices without using a 3D convolution, and allows pretraining on the ImageNet for the filters and trains a fully connected layer after globally pooling the features from all the slices.
The network was able to generalize its performance over two study sites, both acquired with different imaging parameters on scanners produced by different manufacturers, without any additional harmonization of the images, showing the network’s strength in generalizing performance over two different datasets and its potential to work on different datasets. With this approach we successfully created a model that performs well while accounting for such differences. To the best of our knowledge, this was the first musculoskeletal deep-learning-based study that included a mixed-training dataset from multiple study sites. The datasets used for training and validation were annotated by five different radiologists, all of whom underwent the same training. Since the images were not annotated by just one radiologist, this potentially reduced bias in the labels, and helped the network adapt to different radiologists’ performance, making it more robust. When the performance was stratified by sites, it was better for the bone marrow edema and subchondral cysts on the images from the site that had more data, showing that this network would improve further if it was trained on more images. Since the final AUC values were obtained for each model after using the grid search, it suggested that for a problem like this with a relatively smaller dataset, an experimental approach was useful in finding the right set of parameters to tune the network.
The grid search results also indicated that augmentation did not help in generalizing the network or prevent overfitting. This could have been for a few different reasons. First, since the hip lesions are hard to detect by a radiologist, it is possible that any slight modifications done by even the mild augmentations could throw the network off. Second, it is possible that since this is already a binary classification, the network does not need to learn on any more augmented data. The results suggest that augmentation may help in a three-class classification or when the dataset is bigger, suggesting that the network needs to learn more 7class activation maps show that the network is looking within the hip anatomy; however, further experiments would have to be performed with the assistance of a radiologist to determine if the network is focusing on areas where actual lesions are present.24 To improve on the results, it is possible that these images could be cropped even further to focus on parts of the acetabulum and femur head where most cartilage lesions, bone marrow edema-like lesions, and subchondral cyst-like lesions are present.
There are numerous potential applications of this study. These results suggest that if incorporated in a radiology workflow, the network could detect patients with higher lesion probability and triage abnormal cases for review by the radiologist, therefore freeing up the radiologist to work on more complicated cases.25,26 Additionally, the results of the all the networks could be combined in order to find which patients have cartilage lesions, bone marrow edema-like lesions, and subchondral cyst-like lesions, classifying those as patients of higher priority. Furthermore, these trained networks could be used as an assist tool when training incoming radiologists, assuming all the network predictions are vetted by an experienced radiologist, therefore reducing the workload on the experienced radiologist.27,28
Limitations
First, this network is currently able to do a binary classification and not trained to do a multiclass (mild, moderate, severe lesion) classification. A three-class classification would be useful in further stratifying lesion types, but was not possible with the limited data size available for this study. Second, the training dataset was highly imbalanced in the bone marrow edema-like lesions and subchondral cyst-like lesions containing volumes. While we did use a weighted cross-entropy loss function to account for this imbalance, a well-balanced dataset may further improve performance, especially if used for a three-class detection. While we found moderate to good balanced accuracy in classifying the cartilage, bone marrow edema-like lesions, and subchondral cyst-like lesions between the two radiologists, there was still scope for obtaining an improvement of balanced accuracy with an enhanced version of the AI assist tool. The current version of the tool was built to incorporate the single, coronal view of the patient volume. While this did help our readers in this study, radiologists in general often use multiple views (sagittal, axial, coronal, oblique axial) of the same patient volume when annotating images for lesions, especially when the cartilage is as thin as it is in the hip. A previous study has shown that using the combination of the axial and coronal planes of T2-weighted fast spin echo sequence with fat saturation instead of using only the coronal planes increased the sensitivity of cartilage abnormality detection in the knee from 61% to 94%, suggesting that multiplanar images are important when assessing cartilage lesions.29 For future experiments, we aim to combine the predictions from models trained on all three views (axial, coronal, and sagittal). Providing radiologists to assess with more than one view at a time along with the model predictions and saliency maps should further improve the inter- and intrareader agreement along with closely mimicking the actual clinical radiologist practice. Furthermore, this dataset did not contain multiparametric data, which could have possibly improved the network performance, by providing the network with different contrasts for training. This study used images graded by a single radiologist for training and images graded by multiple radiologists for model evaluation and testing. Previous studies11,12 have used similar weakly supervised designs where the training data have one ground truth but the test data have multiple readers to annotate the dataset—creating a more robust testing design. Having a weakly supervised network design enabled us to have a larger dataset to train on, where the variability within the data resulted in a more generalizable model. However, the weak supervision could have possibly introduced more noise in the data, which we will analyze and address in a future study. On the other hand, the test set, which is typically of a smaller size, was graded by independent readers where the ground truth was established as majority voting, making the final evaluation of the model and reported performance values accurate.
As a preliminary study, we incorporated the sagittal images along with the coronal images to train the same network to predict a total severity score instead of binary classification. The results of this preliminary study show that while there is scope in training this network to predict disease severity instead of binary classification, more data are needed, especially images containing more severe lesions to teach the network how to recognize patients with more severe disease. In future studies, we will continue to train this regression network to predict total score, using more data and incorporating more networking tuning to improve the results.
Conclusion
We developed a deep-learning based network that achieves high performance on clinical tasks of detecting hip abnormalities related to OA on MR images. With further experiments and additional data, we will continue to improve the network generalizability and performance, with the goal of assisting radiologists in the detection and grading of hip lesions on MRI.
Contract grant sponsor:
GE Healthcare and National Institutes of Health; Contract grant numbers: R01AR069006 and P50AR060752; Contract grant sponsor: Australian National Health and Medical Research Council (NHMRC) project grant; Contract grant number: 1088683. The funding bodies did not have a role in the study design, collection, analysis, and interpretation of data, writing of the article, or decision to submit the article for publication.
References
- 1.Barbour KE, Helmick CG, Boring M, Brady TJ. Vital signs: Prevalence of doctor-diagnosed arthritis and arthritis-attributable activity limitation — United States, 2013-2015. MMWR Morb Mortal Wkly Rep 2017;66(9):246–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jordan JM, Helmick CG, Renner JB, et al. Prevalence of hip symptoms and radiographic and symptomatic hip osteoarthritis in African Americans and Caucasians: The Johnston County Osteoarthritis Project. J Rheumatol 2009;36(4):809–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lespasio MJ, Sultan AA, Piuzzi NS, et al. Hip osteoarthritis: A primer. Perm J 2018;22:17–084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee S, Nardo L, Kumar D, et al. Scoring hip osteoarthritis with MRI (SHOMRI): A whole joint osteoarthritis evaluation system. J Magn Reson Imaging 2015;41(6):1549–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kellgren JH, Lawrence JS. Radiological assessment of osteo-arthrosis. Ann Rheum Dis 1957;16(4):494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menashe L, Hirko K, Losina E, et al. The diagnostic performance of MRI in osteoarthritis: A systematic review and meta-analysis. Osteoarthr Cartil 2012;20(1):13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmid MR, Notzli HP, Zanetti M, Wyss TF, Hodler J. Cartilage lesions in the hip: Diagnostic effectiveness of MR arthrography. Radiology 2003;226(2):382–386. [DOI] [PubMed] [Google Scholar]
- 8.Keeney JA, Peelle MW, Jackson J, Rubin D, Maloney WJ, Clohisy JC. Magnetic resonance arthrography versus arthroscopy in the evaluation of articular hip pathology. Clin Orthop Relat Res 2004;429:163–169. [DOI] [PubMed] [Google Scholar]
- 9.Zazgyva A, Gurzu S, Gergely I, Jung I, Roman CO, Pop TS. Clinicoradiological diagnosis and grading of rapidly progressive osteoarthritis of the hip. Medicine (Baltimore) 2017;96(12):e6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roemer FW, Hunter DJ, Winterstein A, et al. Hip Osteoarthritis MRI Scoring System (HOAMS): Reliability and associations with radiographic and clinical findings. Osteoarthr Cartil 2011;19(8):946–962. [DOI] [PubMed] [Google Scholar]
- 11.Bien N, Rajpurkar P, Ball RL, et al. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: Development and retrospective validation of MRNet. PLoS Med 2018;15(11):e1002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu F, Zhou Z, Samsonov A, et al. Deep learning approach for evaluating knee MR images: Achieving high diagnostic performance for cartilage lesion detection. Radiology 2018;289(1):160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Couteaux V, Si-Mohamed S, Nempont O, et al. Automatic knee meniscus tear detection and orientation classification with Mask-RCNN. Diagn Interv Imaging 2019;100(4):235–242. [DOI] [PubMed] [Google Scholar]
- 14.Roblot V, Giret Y, Bou Antoun M, et al. Artificial intelligence to diagnose meniscus tears on MRI. Diagn Interv Imaging 2019;100(4):243–249. [DOI] [PubMed] [Google Scholar]
- 15.Abidin AZ, Deng B, AM DS, Nagarajan MB, Coan P, Wismüller A. Deep transfer learning for characterizing chondrocyte patterns in phase contrast x-ray computed tomography images of the human patellar cartilage. Comput Biol Med 2018;95:24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pedoia V, Lee J, Norman B, Link TM, Majumdar S. Diagnosing osteoarthritis from T2 maps using deep learning: an analysis of the entire osteoarthritis initiative baseline cohort. Osteoarthr Cartil 2019;27(7):1002–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue Y, Zhang R, Deng Y, Chen K, Jiang T. A preliminary examination of the diagnostic value of deep learning in hip osteoarthritis. PLoS One 2017;12(6):e0178992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jason Y, Jeff C, Yoshua B, Hod L. How transferable are features in deep neural networks? In Neural Information Processing Systems (NIPS) 2014:3320–3328. https://arxiv.org/abs/1411.1792 [Google Scholar]
- 19.Crossley KM, Pandy MG, Majumdar S, et al. Femoroacetabular impingement and hip OsteoaRthritis Cohort (FORCe): Protocol for a prospective study. J Physiother 2018;64(1):55. [DOI] [PubMed] [Google Scholar]
- 20.Wyatt C, Kumar D, Subburaj K, et al. Cartilage T1rho and T2 relaxation times in patients with mild-to-moderate radiographic hip osteoarthritis. Arthritis Rheumatol 2015;67(6):1548–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krizhevsky ASI, Hinton G. ImageNet classification with deep convolutional neural networks. Adv Neur Inform Process Syst 2012. https://dl.acm.org/doi/10.1145/3065386 [Google Scholar]
- 22.Paszke A, Gross S, Chintala S, et al. Automatic differentiation in Pytorch. 2017. http://papers.nips.cc/paper/9015-pytorch-an-imperative-style-high-performance-deep-learning-library [Google Scholar]
- 23.Urbanowicz RJ, Moore JH. ExSTraCS 2.0: Description and evaluation of a scalable learning classifier system. Evol Intell 2015;8(2):89–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larson DB, Chen MC, Lungren MP, Halabi SS, Stence NV, Langlotz CP. Performance of a deep-learning neural network model in assessing skeletal maturity on pediatric hand radiographs. Radiology 2018;287(1):313–322. [DOI] [PubMed] [Google Scholar]
- 25.Annarumma M, Withey SJ, Bakewell RJ, Pesce E, Goh V, Montana G. Automated triaging of adult chest radiographs with deep artificial neural networks. Radiology 2019;291(1):272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yala A, Schuster T, Miles R, Barzilay R, Lehman C. A deep learning model to triage screening mammograms: A simulation study. Radiology 2019;293(1):38–46. [DOI] [PubMed] [Google Scholar]
- 27.Kolachalama VB, Garg PS. Machine learning and medical education. NPJ Digit Med 2018;1:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choy G, Khalilzadeh O, Michalski M, et al. Current applications and future impact of machine learning in radiology. Radiology 2018;288(2):318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bredella MA, Tirman PF, Peterfy CG, et al. Accuracy of T2-weighted fast spin-echo MR imaging with fat saturation in detecting cartilage defects in the knee: Comparison with arthroscopy in 130 patients. AJR Am J Roentgenol 1999;172(4):1073–1080. [DOI] [PubMed] [Google Scholar]