Abstract
Repetitive rounds of differential subtraction screening, followed by nucleotide sequence determination and northern-blot analysis, identified 84 salt-regulated (160 mm NaCl for 4 h) genes in Arabidopsis wild-type (Col-0 gl1) seedlings. Probes corresponding to these 84 genes and ACP1, RD22BP1, MYB2, STZ, and PAL were included in an analysis of salt responsive gene expression profiles in gl1 and the salt-hypersensitive mutant sos3. Six of 89 genes were expressed differentially in wild-type and sos3 seedlings; steady-state mRNA abundance of five genes (AD06C08/unknown, AD05E05/vegetative storage protein 2 [VSP2], AD05B11/S-adenosyl-l-Met:salicylic acid carboxyl methyltransferase [SAMT], AD03D05/cold regulated 6.6/inducible2 [COR6.6/KIN2], and salt tolerance zinc finger [STZ]) was induced and the abundance of one gene (AD05C10/circadian rhythm-RNA binding1 [CCR1]) was reduced in wild-type plants after salt treatment. The expression of CCR1, SAMT, COR6.6/KIN2, and STZ was higher in sos3 than in wild type, and VSP2 and AD06C08/unknown was lower in the mutant. Salt-induced expression of VSP2 in sos1 was similar to wild type, and AD06C08/unknown, CCR1, SAMT, COR6.6/KIN2, and STZ were similar to sos3. VSP2 is regulated presumably by SOS2/3 independent of SOS1, whereas the expression of the others is SOS1 dependent. AD06C08/unknown and VSP2 are postulated to be effectors of salt tolerance whereas CCR1, SAMT, COR6.6/KIN2, and STZ are determinants that must be negatively regulated during salt adaptation. The pivotal function of the SOS signal pathway to mediate ion homeostasis and salt tolerance implicates AD06C08/unknown, VSP2, SAMT, 6.6/KIN2, STZ, and CCR1 as determinates that are involved in salt adaptation.
High soil salinity, which is caused typically by NaCl, results in ion toxicity and hyperosmotic stress leading to numerous pathologies including generation of reactive oxygen species (ROS) and programmed cell death (Niu et al., 1995; Zhu et al., 1997; Hasegawa et al., 2000b). Salt tolerance determinants are categorized either as effectors that directly modulate stress etiology or attenuate stress effects, or as regulatory molecules that are involved in stress perception, signal transduction, or modulation of effector function. Enzymes that catalyze rate-limiting steps in the biosynthesis of compatible solutes or osmoprotectants, e.g. sugar alcohol, quaternary ammonium, and tertiary sulfonium compounds, are categorical examples of osmotic stress tolerance effectors (Hanson et al., 1994; Ishitani et al., 1997; Yoshiba et al., 1997; Nelson et al., 1998; Hasegawa et al., 2000b). Other effectors include proteins that protect membrane integrity, control water or ion homeostasis, and ROS scavenging (Bray, 1994; Ingram and Bartels, 1996; Hasegawa et al., 2000b). Determinant function of some effectors has been confirmed because expression in transgenic plants enhances salt tolerance sufficiency (Hasegawa et al., 2000b).
Regulatory determinants include transcription factors and signal pathway intermediates that posttranscriptionally activate effectors (Hasegawa et al., 2000b). Basic Leu zipper motif, MYB and MYC, and zinc finger transcription factors, including rd22BP1 (MYC), AtMYB2 (MYB), DREB1A, and DREB2A (AP2 domain), and ALFIN1 (zinc finger), interact with promoters of osmotic-regulated genes (Abe et al., 1997; Liu et al., 1998; Hasegawa et al., 2000b). The osmotic stress tolerance function of DREB1A in Arabidopsis (Kasuga et al., 1999) and ALFIN1 in alfalfa (Medicago sativa; Winicov, 2000) has been confirmed by ectopic expression in transgenic plants. Regulatory intermediates that modulate plant salt stress responses include SOS3 (Ca2+-binding protein), SOS2 (Suc non-fermenting-like) kinase, Ca2+-dependent protein kinases, and mitogen-activated protein kinases (Sheen, 1996; Halfter et al., 2000; Kovtun et al., 2000). Additional signal intermediates have been implicated in the plant response to salt (Hwang and Goodman, 1995; Mizoguchi et al., 1996; Mikami et al., 1998; Piao et al., 1999; Hasegawa et al., 2000b).
The Zhu laboratory recently has pioneered identification of salt tolerance determinants using forward genetics in the plant model Arabidopsis (Hasegawa et al., 2000a; Sanders, 2000; Zhu, 2000). This effort has identified three complementation groups of ion hypersensitive (salt overly sensitive [sos1-sos3]) mutants. Genetic and physiological data indicate that SOS3, SOS2, and SOS1 are components of a signal pathway that regulates ion homeostasis and salt tolerance and their functions are Ca2+ dependent. Positional cloning revealed that SOS1 encodes a putative plasma membrane Na+/H+ antiporter, SOS2 encodes a Suc non-fermenting-like (SNF) kinase, and SOS3 encodes a Ca2+-binding protein with sequence similarity to the regulatory subunit of calcineurin and neuronal Ca2+ sensors (Liu and Zhu, 1998; Liu et al., 2000; Shi et al., 2000). Molecular interaction and complementation analyses indicate that SOS3 is required for activation of SOS2 that regulates SOS1 transcription (Halfter et al., 2000; Shi et al., 2000), further confirming that the order of the signal pathway is SOS3 → SOS2 → SOS1 (Hasegawa et al., 2000a; Sanders, 2000; Zhu, 2000).
Herein is described a functional dissection of the plant salt stress response by analysis of gene expression controlled by the SOS signal pathway. Differential subtraction and northern analysis identified 84 salt-regulated genes, the majority of which have not been annotated to be salt responsive in Arabidopsis. Comparison of salt regulation in wild type and sos3 identified six genes that are controlled by the SOS signal pathway. Transcription of one gene (VSP2) is controlled as an output from the SOS3/SOS2 pathway, similar to SOS1 (Shi et al., 2000), whereas regulation of five other genes (AD06C08, CCR1, SAMT, COR6.6/KIN2, and STZ) is dependent on both SOS3 and SOS2 as well. Because SAMT, COR6.6/KIN2, and STZ are induced by NaCl, negative control by SOS3 indicates that the SOS pathway functions to reduce the numerous signals induced by salt to those that function more specifically, to mediate processes like ion homeostasis.
RESULTS
Identification of Expressed Sequence Tags (ESTs) Corresponding to Salt-Regulated Genes
A combination of approaches was used to identify stress-regulated transcriptional outputs from the SOS pathway on the premise that these are salt tolerance determinants. A population of ESTs representing salt-regulated transcripts was identified by screening, through three rounds, a subtracted cDNA library prepared from wild-type (Col-0 gl-1) seedlings treated for 4 h without or with 160 mm NaCl. Differential dot-blot hybridization, and sequence and northern-blot analyses identified unique ESTs (salt-regulated ESTs [SREs]) that detected salt-regulated transcripts (Table I). The second and third rounds of screening eliminated highly repetitive clones from the initial subtracted library, including those ESTs identified during the first round of screening. The differential subtractive chain selection protocol was used in the third round (Luo et al., 1999) to further enrich for less abundant cDNAs in the library. The different rounds of screening led to identification of 84 nonredundant ESTs that hybridize to salt-regulated transcripts based on northern-blot analysis (Table II). The SREs represent the greatest number that has been used in one study to define the transcriptional response of plants to salt stress. These results identify genes whose expression is most likely controlled by transcriptional activation, although other factors such as salt stress-dependent mRNA stability might contribute also to steady-state transcript abundance.
Table I.
Arabidopsis (Col-1 gl1) SREs identified after successive rounds of differential subtraction
| Round | Dot Blot | Sequenced | Unique Clones | Salt Regulated |
|---|---|---|---|---|
| First | 1,000 | 137 | 34 | 25 |
| Second | 6,000 | 157 | 91 | 42 |
| Third | 3,000 | 320 | 80 | 17 |
| Total | 10,000 | 614 | 205 | 84 |
Salt regulation was based on detection of altered steady-state transcript abundance after NaCl treatment.
Table II.
Arabidopsis (Col-0 and gl1) ESTs corresponding to salt-responsive genes determined by northern-blot analysis
| GenBank Accession No. | EST No. | Chromosome | BAC Clone No. | geneID | proteinID | Annotation on Arabidopsis Gene/Homologa | Score | E Value | Comments | gl-1 Control | gl-1 Salt |
|---|---|---|---|---|---|---|---|---|---|---|---|
| bits | |||||||||||
| Primary metabolism | |||||||||||
| Photosynthesis | |||||||||||
| BE844959 | AD04H11 | 3 | F5N5 | ATU05218 | AAA21570.1 | ATP sulfurylase | 492 | e-138 | ++++ | +++++ | |
| BE845179 | AD08A06 | 2 | 25 | At2g05100 | AAD31358.1 | Putative chlorophyll a/b-binding protein | 789 | 0.0 | – | ++ | |
| BE845351 | AD10C12 | 4 | 69 | AT4g28750 | CAB81463.1 | Photosystem I subunit PSI-E-like protein | 513 | e-144 | ++++ | +++++ | |
| BE845356 | AD10D06 | 1 | F12A21 | F12A21.11 | AAF26934.1 | Putative PSBY emblCAA11248 | 728 | 0.0 | ++ | +++ | |
| Carbohydrate | |||||||||||
| BE844849 | AD03C01 | 4 | 7 | dl4575c | CAB46051.1 | Putative β-amylase | 1078 | 0.0 | Gene duplication? | – | + |
| BE844998 | AD05E02 | 3 | 45 T22E16 | AT4g17090 T22E16.70 | CAB80980.1 CAB75899.1 | 2-Oxoglutarate dehydrogenase, E1subunit-like protein | 242 | 2e-63 | + | ++ | |
| Amino acid | |||||||||||
| BE844834 | AD03A02 | 4 | 59 | AT4g23600 | CAB79315.1 | Tyrosine transaminase-like protein | 605 | e-172 | + | +++++> | |
| BE844976 | AD05B09 | 1 | F15I1 | F15I1.19 | AAD25783.1 | Strong similarity to gblS77096 aldehyde dehydrogenase homolog from Brassica napus and is a member of PFI00171 aldehyde dehydrogenase family | 137 | 5e-31 | Proline degradation? | + | +++ |
| BE845063 | AD06C01 | 3 | T21L8 | T21L8.90 | CAB51206.1 | Glutamine-dependent asparagine synthetase | 210 | 4e-53 | + | +++ | |
| BE845111 | AD06G04 | 4 | 82 | AT4g34710 | CAB80188.1 | Arginine decarboxylase SPE2 | 809 | 0.0 | Polyamine synthesis | – | ++++ |
| Others | |||||||||||
| BE844989 | AD05D03 | 5 | K24M7 | – | – | Not annotated | 55.4 | 6e-08 | Cytochrome p450 | – | ++ |
| pirllA29368 Prostaglandin omega-hydroxylase (EC 1.14.15.-) cytochrome P450 4A4, rabbita | 48 | e-05 | |||||||||
| Cellular function | |||||||||||
| Cell wall | |||||||||||
| BE844904 | AD04B12 | 5 | MMI9 | AB019235.1 | BAA97199.1 | Ripening related, protein like; contains similarity to pectinesterase | 712 | 0.0 | + | +++ | |
| BE845087 | AD06E01 | 5 | F7K24 | – | – | Not annotated | 339 | 3e-92 | + | ++ | |
| pirllT11610 Probable cinnamyl-alcohol dehydrogenase (EC 1.1.1.195) CPRD14 (Vigna unguiculata)a | 191 | 2e-48 | |||||||||
| bits | |||||||||||
| BE845121 | AD06H02 | 4 | 6 | AT4g02330 | CAB80726.1 | Strong similarity to similar to pectinesterase, contains pectinesterase signatures AA407-414 | 424 | e-118 | + | +++ | |
| Vesicle/protein trafficing | |||||||||||
| BE845046 | AD06A05 | 5 | F17I14 | F17I14_270 | CAB89377.1 | Periaxin-like protein | 856 | 0.0 | PDZ domain | + | +++ |
| BE845048 | AD06A07 | 1 | F23N19 | F23N19.7 | AAF19550.1 | Similar to vacuolar processing enzyme | 385 | e-106 | Protease | – | + |
| BE845064 | AD06C02 | 2 | 189 | At2g34250 | AAC27401.1 | Putative protein transport protein SEC61 alpha subunit | 216 | 6e-55 | Vesicle transport | – | +++ |
| BE845086 | AD06D12 | 1 | T25N20 | T25N20.17 | AAF79733.1 | Putative transport protein | 155 | 2e-36 | Vesicle transport | + | ++++ |
| refINP_062761.1I SEC23B (Saccharomyces cerevisiae)a | 692 | 0.0 | |||||||||
| Other cellular functions | |||||||||||
| BE844934 | AD04F04 | 4 | 67 | AT4g27400 | CAB81391.1 | Putative protein: similarity to Arabidopsis nap gene, PID:e1234813 | 749 | 0.0 | Cell division and expansion | – | +++ |
| BE844985 | AD05C10 | 4 | 90 | AT4g39260 | CAB80589.1 | Gly-rich protein (clone AtGRP8): Contains eukaryotic putative RNA-binding region RNP-1 signature AA47-54 | 59.7 | e-08 | Ccr1 | +++ | + |
| BE845050 | AD06A09 | 1 | F1P2 | F1P2.100 | CAB61981.1 | Putative protein: similarity to PIT1, Arabidopsis, GB: AF130849 | 480 | e-134 | Plant growth | – | +++++ |
| BE845059 | AD06B07 | 4 | 63 | AT4g25630 | CAB81373.1 | Fibrillarin-like protein: strong similarity to probable fibrillarin (Sb21) mRNA, Picea mariana, AF051216 | 412 | e-114 | Pre-rRNA processing | – | ++ |
| BE845095 | AD06E12 | ? | ? | ATHATJ | AAB86799.1 | Chaperone protein (atj) | 509 | e-143 | Chaperon | – | + |
| BE845137 | AD07C05 | 2 | 210 | At2g38860 | AAC79625.1 | Unknown protein, splO59413IPFPI_PYRHO PROTEASE I (Pyrococcus horikoshii)a | 15764 | 4e-373e-09 | Protease | + | +++ |
| BE845172 | AD07H10 | 2 | 185 | At2g33210 | AAC04902.1 | Mitochondrial chaperonin (HSP60) | 186 | 4e-46 | Chaperon | + | +++ |
| BE845190 | AD08B06 | 2 | 21 | At2g04460 | AAD25832.1 | Putative retroelement pol polyprotein | 638 | 0.0 | Transposon | +++ | + |
| BE845392 | AD10H03 | 4 | 14 | AT4g05050 | CAB81047.1 | Contains similarity to Pfam family PF00240, ubiquitin family | 1086 | 0.0 | ++ | +++ | |
| Transporters/nutrient uptake | |||||||||||
| BE844897 | AD04B05 | 5 | F9D12 | F9D12.17 | AAC26243.1 | Contains similarity to sugar transporters | 424 | 4e-32 | Transporter | – | ++++ |
| BE844907 | AD04C05 | 5 | F14F18 | F14F18_180 | CAB87674.1 | Putative protein, splQ96250IATP3_ARATH ATP SYNTHASE GAMMA CHAIN, MITOCHONDRIAL PRECURSORa | 99535.2 | 0.01.6 | Transporter? | – | + |
| BE844924 | AD04E04 | 2 | 91 | At2g15620 | AAD17406.1 | Ferredoxin-nitrite reductase | 406 | e-112 | Nitrogen | + | ++ |
| BE845007 | AD05E11 | 1 | T17F3 | T17F3.10 | AAF07386.1 | Putative peptide transporter | 609 | e-173 | Transporter | – | +++ |
| BE845058 | AD06B06 | 5 | F8M21 | F8M21_130 | CAB89334.1 | Putative protein: similarity to amino acid transport protein, Arabidopsis, EMBL U39783 | 527 | e-148 | Transporter | – | + |
| BE845182 | AD08A09 | 1 | F5I6 | F5I6.6 | AAF27688.1 | Putative sulfate transporter | 234 | e-60 | Transporter | – | +++ |
| bits | |||||||||||
| BE845347 | AD10C07 | 5 | MUG13 | AB021934 | BAA74589.1 | Nicotianamine synthase | 541 | e-152 | Iron uptake | – | +++ |
| BE845350 | AD10C11 | 3 | F4P12 | F4P12_210 | CAB67658.1 | ABC transporter-like protein | 5866 | 0.0 | Transporter | – | + |
| Signaling | |||||||||||
| General signal components | |||||||||||
| BE844894 | AD04B01 | 2 | 106 | At2g18190 | AAD31347.1 | Putative AAA-type ATPase | 884 | 0.0 | + | +++ | |
| BE844899 | AD04B07 | 1 | T10O24 | T10O24.2 | AAD39582.1 | Hypothetical protein, dbjIBAB11554.1I (AB011479), contains similarity to bHLH DNA-binding protein: gene_id:MNA5.5 (Arabidopsis)a | 108536.7 | 2.9e-1850.16 | Transcription | – | +++ |
| BE844973 | AD05B05 | 5 | MJC20 | MJC20.11 | BAB08434.1 | Contains similarity to unknown protein (AF117897), rab11-binding protein (Bos taurus)a | 507187 | e-1424e-46 | WD repeat | + | +++ |
| BE845012 | AD05F04 | 3 | MMM17 | MMM17.7 | BAB01914.1 | Casein kinase-like protein | 353 | 4e-96 | +++ | ++++ | |
| BE845028 | AD05G10 | 3 | T5P19 | T5P19_160 | CAB88054.1 | Putative protein: similarity to TATA-binding protein-binding protein, ABT1: Mus musculus, EMBL AB021860 | 291 | e-77 | Transcription | + | ++ |
| BE845052 | AD06A11 | 5 | MQN23 | MQN23.23 | BAB11664.1 | G protein-coupled receptor-like protein | 377 | e-103 | +++ | +++++ | |
| BE845077 | AD06D03 | 3 | F24M12 | F24M12.170 | CAB62635.1 | Putative protein: similarity to lin-10 protein: Rattus norwegicus, PIR:JE0239 | 268 | 2e-70 | Receptor targeting | + | ++ |
| BE845082 | AD06D08 | 5 | MAC9 | MAC9.10 | BAB10078.1 | Transcription factor-like protein | 317 | 2e-85 | Transcription | + | +++ |
| BE845106 | AD06F11 | 1 | F24B9 | F24B9.4 | AAF75068.1 | Contains similarity to a protein kinase gblD88207 | 139 | 9e-33 | + | +++ | |
| BE845234 | AD08G06 | 3 | F26F24 | F26F24.8 | AAF86997.1 | Hypothetical protein, emblCAB92072.1I (AL121575), dJ914N13.2.1 (cofactor required for Sp1 transcriptional activation, subunit 3; 130kd; CRSP130, DRIP130, SUR2, and KIAA1216; isoform 1) (Homo sapiens) | 27484.3 | 2e-729e-15 | – | + | |
| BE845243 | AD08H07 | 2 | 77 | At2g13790 | AAD28318.1 | Putative receptor-like protein kinase | 184 | 7e-46 | – | ++ | |
| BE845394 | AD10H07 | 3 | MGL6 | MGL6.10 | BAB00069.1 | Translationally controlled tumor protein like | 559 | e-157 | Ca2+ binding | +++ | +++++ |
| Lipid signaling/response | |||||||||||
| BE844846 | AD03B08 | 3 | T8P19 | T8P19.200 | CAB62358.1 | Putative protein, splP24484ILIP2_MORSP LIPASE 2 (TRIACYLGLYCEROL LIPASE)a | 117658.6 | 0.09e-08 | Lipase | + | +++ |
| BE844917 | AD04D07 | 3 | K13N2 | K13N2.11 | BAA95764.1 | Unknown, emblCAB95731.1I (AJ272026) allene oxide cyclase (Lycopersicon esculentum)a | 472249 | e-1323e-65 | JA synthesis | + | ++++ |
| BE844950 | AD04G12 | 3 | T14D7 | T14D3.80 | CAB72152.1 | Lipoxygenase AtLOX2 | 432 | e-120 | + | +++++> | |
| BE845001 | AD05E05 | 5 | ? | AB006778 | BAA33447.1 | Vsp2 gene for vegetative storage protein | 509 | e-143 | – | +++ | |
| bits | |||||||||||
| BE845091 | AD06E08 | 5 | MTI20 | MTI20.3 | BAB08850.1 | Lipid transfer protein; glossy1 homolog | 414 | e-114 | – | +++ | |
| BE845100 | AD06F05 | 3 | T02O04 | T02O04.11 | AAB63638.1 | Jasmonate-inducible protein isolog, myrosinase binding protein like | 466 | e-130 | ++ | ++++ | |
| Hormone related | |||||||||||
| BE844978 | AD05B11 | 3 | F28D10 | F28D10_50 | CAC03536.1 | AtPP-like protein: prokaryotic membrane lipoprotein lipid attachment site AA199-209 (AF133053), S-adenosyl-l-methionine:salicylic acid carboxyl methyltransferase (Clarkia breweri)a | 706129 | 0.06e-29 | SA | – | ++ |
| BE845401 | AD05C05 | ? | ? | AF183827 | AAF22295.1 | Beta-glucosidase homolog (BG1) | 3515 | 0.0 | – | ++++ | |
| BE845084 | AD06D10 | 3 | T10D17 | T10D17_90 | CAB88998.1 | Nitrilase 2 | 202 | e-50 | + | +++ | |
| Cell death | |||||||||||
| BE844851 | AD03C06 | 5 | MQL5 | MQL5.19 | BAA97167.1 | Palmitoyl-protein thioesterase precursor like | 422 | e-117 | Anti-apoptosis? | + | ++ |
| BE844958 | AD04H10 | 3 | F28D10 | F28D10_70 | CAC03538.1 | Lethal leaf-spot 1 homolog Lls1: contains prokaryotic membrane lipoprotein lipid attachment site AA150-160 | 472 | e-132 | Dioxygenase | + | ++ |
| Defense response | |||||||||||
| Defense proteins | |||||||||||
| BE844875 | AD03G09 | 1 | F22K20 | F22K20.19 | AAC00625.1 | Alcohol dehydrogenase | 486 | e-136 | + | +++ | |
| BE844896 | AD04B03 | 5 | MSG15 | MSG15.3 | BAB11043.1 | Mandelonitrile lyase-like protein | 519 | e-146 | Cyanogenesis | – | ++ |
| BE844926 | AD04E06 | 3 | MDB19 | MDB19.5 | BAB02775.1 | Contains similarity to endo-1,3-1,4-beta-d-glucanase gene_id:MDB19.5 | 414 | e-115 | + | +++ | |
| BE844972 | AD05B03 | 1 | F5F19 | F5F19.6 | AAD12691.1 | Similar to gblY09437 myrosinase binding protein from B. napus | 194 | 8e-49 | – | + | |
| BE844990 | AD05D04 | 1 | F24B9 | F24B9.34 | AAF75098.1 | Metallothionein | 605 | e-172 | ++++ | +++++ | |
| BE845403 | AD06E03 | 1 | F15K9 | F15K9.17 | AAC72119.1 | Strong similarity to gblD14550 extracellular dermal glycoprotein (EDGP) precursor from Daucus carota | 745 | 0.0 | + | ++ | |
| BE845117 | AD06G10 | 1 | F14N23 | F14N23.25 | AAD32887.1 | Similar to glutathione S-transferase TSI-1 (gil2190992) | 763 | 0.0 | Cyanogenesis | + | +++++ |
| BE845152 | AD07F07 | 2 | 236 | At2g43620 | AAB64044.1 | Putative endochitinase | 549 | e-154 | + | ++++ | |
| BE845153 | AD07F08 | 5 | MOJ9 | MOJ9.4 | BAB11145.1 | Polygalacturonase-inhibiting protein | 509 | e-142 | – | ++++ | |
| Cor/RD proteins | |||||||||||
| BE844850 | AD03C02 | 4 | 73 | AT4g30650 | CAB79783.1 | Strong similarity to low temperature and salt-responsive protein LTI6A, Arabidopsis | 383 | e-105 | + | +++ | |
| BE844854 | AD03D05 | 5 | F1N13 | F1N13_110 | CAC01796.1 | Cold-regulated protein COR6.6 (KIN2) | 341 | 2e-92 | – | +++++> | |
| BE844855 | AD03D11 | 2 | F14N22 | At2g42540 | AAD22999.1 | Cold-regulated protein cor15a precursor | 519 | e-14 | – | +++++> | |
| BE844865 | AD03F02 | 5 | K24M7 | D13044 | BAA02376.1 | Desiccation-responsive rd29A | 371 | e-101 | – | +++++> | |
| BE845022 | AD05G04 | 5 | T14C9 | ATHRD22 | BAA01546.1 | rd22 Gene | 448 | e-124 | – | +++++ | |
| BE845049 | AD06A08 | 1 | F15M15 | F5M15.21 | AAF79613.1 | Similar to cold-regulated protein cor47 | 127 | 4e-28 | ? | ? | |
| bits | |||||||||||
| Anthocyanin synthesis | |||||||||||
| BE844845 | AD03B06 | 5 | MOP10 | MOP10.14 | BAB11549.1 | Leucoanthocyanidin dioxygenase-like protein | 664 | 0.0 | – | +++++> | |
| BE844892 | AD04A11 | 2 | 206 | At2g38240 | AAC27173.1 | Putative anthocyanidin synthase | 203 | 4e-87 | – | ++++ | |
| BE845101 | AD06F06 | 4 | T29A15 | AT4g27560 | CAB38268.1 | Strong similarity to UDP rhamnose-anthocyanidin-3-glucoside rhamnosyltransferase, Petunia hybrida | 837 | 0.0 | + | ++ | |
| Antioxidation | |||||||||||
| BE844884 | AD04A02 | 5 | F5O24 | AB035137 | BAA86999.1 | Blue copper-binding protein | 494 | e-138 | Anti-oxidation | + | ++++ |
| Unknown | |||||||||||
| BE844860 | AD03E08 | 3 | T19D11 | – | – | Not annotated, dbjlBAB02819.1I (AB024036), dbjlBAA87936.1: gene_id:MQC12.15, similar to unknown | 13546.1 | 4e-315e-05 | – | +++ | |
| BE844992 | AD05D06 | 5 | K3M16 | K3M16_30 | CAC01890.1 | Hypothetical protein | 777 | 0.0 | – | ++++ | |
| BE845016 | AD05F09 | 3 | F14O13 | F14O13.28 | BAB03026.1 | Similar to unknown protein | 476 | e-133 | + | ++ | |
| BE845069 | AD06C07 | 1 | F22O13 | F22O13.29 | AAF99773.1 | Unknown protein | 690 | 0.0 | + | ++ | |
| BE845070 | AD06C08 | 3 | F16J14 | – | – | Not annotated | 910 | 0.0 | + | ++ | |
| emblCAB86422.1I (AL138648) putative protein (Arabidopsis)a | 78.8 | 4e-14 | |||||||||
| BE845097 | AD06F02 | ? | ? | – | – | Not annotated | + | ++++ | |||
| BE845355 | AD10D05 | 1 | F24J8 | – | – | Not annotated | 775 | 0.0 | Transporter | – | ++++ |
| gblAAF28474.1IAF173553_1 (AF173553) V-ATPase 110-kD integral membrane subunit (Manduca sexta)a | 29.7 | 16 | |||||||||
| BE845376 | AD10F07 | 1 | T27G7 | – | – | Not annotated | 579 | e-164 | + | +++ | |
| splQ08180IICCR_DROME IRREGULAR CHIASM C-ROUGHEST PROTEIN PRECURSOR (IRREC PROTEIN)a | 32.1 | 3.7 | |||||||||
Total RNA was isolated from seedlings transferred to medium without or
with 160 mm NaCl for 4 h SREs were subjected to Blastn
analysis and illustrated are annotations based on the information from
the Arabidopsis genome sequence project: chromosome no., section on
chromosomes 2 and 4, or BAC clone identification no. (for
chromosomes 1, 3, and 5), and gene/protein accession nos. Highlighted
in bold are SREs that detected transcripts differentially regulated in
gl1 and sos3. Transcript abundance is rated based
on the scale illustrated.
Blastp analysis was performed on any Arabidopsis open reading frame (ORF) that corresponded to an SRE that is not functionally annotated. Any SRE sequence that did not match an Arabidopsis ORF was subjected to Blastx/Blastp analysis.
Database comparisons of SREs using Blast programs determined that the corresponding encoded proteins included those involved in primary metabolism, cell wall synthesis or degradation, other cellular functions, transport or nutrient assimilation, signaling, and defensive responses (Table II). The SREs were compared with Arabidopsis genome data using Blastn. Blastp analysis was performed on any SRE ORF, without predicted function, that was identified in an Arabidopsis database. Blastx/Blastp analysis was performed on SRE sequences that were unannotated as an ORF.
Several of the salt-responsive genes identified in this evaluation encode components of octadecanoid signaling through jasmonic acid (Table II, lipid signaling responses). The plant hormone appears to be derived from hydrolysis of membrane phospholipids (Koiwa et al., 1997). Triacyl glycerol lipase (AD03B08) can release free linolenic acid from phospholipids that is then oxidized by lipoxygenase (AD04G12) and cyclized by allene oxide synthase and allene oxide cyclase (AD04D07) to 12-oxo-phytodienoic acid. Two SREs were annotated previously as jasmonic acid regulated, VSP2 (AD05E05) and AD06F05. Furthermore, the genes of a number of SREs are involved in plant defense and may be regulated by the octadecanoid signal pathway. Some of these genes have been shown to express upon dehydration in tomato (Lycopersicon esculentum; Reymond et al., 2000). Several abscisic acid (ABA)-responsive SREs are included in Table II and the plant hormone is a potentiator of octadecanoid signaling.
Genes Differentially Regulated by Salt in Wild Type and sos3
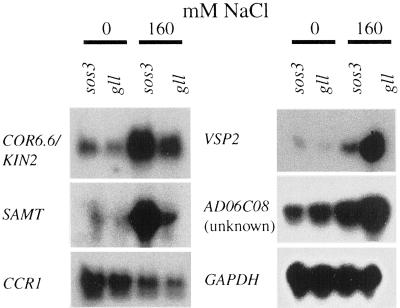
The salt regulated expression profile of SRE transcripts, as well as that of previously characterized stress-regulated genes (ACP1, RD22BP1, MYB2, STZ, and PAL), revealed that most are controlled similarly in wild type and sos3. Six of the salt-responsive genes, including STZ (Lippuner et al., 1996), were differentially regulated in wild type and sos3 (Fig. 1). Transcript abundance of two was lower (AD06C08/unknown and AD05E05/vegetative storage protein2 [VSP2]) and of four was higher (encoding AD05C10/cold-circadian rhythm-RNA binding1 [CCR1], STZ/salt tolerance zinc finger [not shown], AD05B11/S-adenosyl-l-Met: salicyclic acid carboxyl methyltransferase [SAMT], AD03D05/cold regulated/cold inducible [COR6.6/KIN2]) in the salt-sensitive mutant. SAMT and COR6.6/KIN2 transcript abundance was slightly elevated in sos3 but the steady-state mRNA levels were hyper-induced by salt treatment. CCR1 was the only gene for which transcript abundance is reduced by salt treatment.
Figure 1.
Salt-responsive gene expression that is dependent on the SOS pathway in Arabidopsis. Genes that are differentially regulated in wild-type (Col-0 gl1) and sos3. Total RNA (40 μg) from seedlings grown in liquid culture (14 d) and treated without or with 160 mm NaCl for 4 h. The northern blot was hybridized with 32P-labeled probe corresponding to: COR6.6/KIN2, SAMT, CCR1, VSP2, and AD06C08 (unknown). AtGAPDH (glyceraldehyde-3-phosphate-dehydrogenase) is the control.
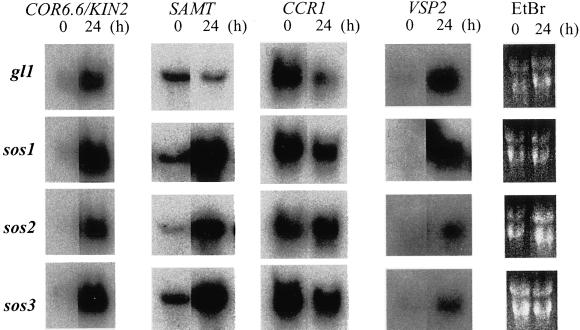
Salt regulation of VSP2 is similar in wild type and sos1 indicating that transcriptional activation is not dependent on SOS1 (Fig. 2). Methyl jasmonate (MeJA) induces VSP2 transcript abundance in wild type and sos3 (not shown). The SOS3/2 pathway and the hormone seem to regulate VSP2 independently. Signal pathways often converge to regulate transcription of key effectors involved in cellular adaptation to environmental perturbation (Rep et al., 2000). VSP2 is a member of a two-gene family (87% nucleotide sequence identity over the coding region) that encodes a protein with similarity to soybean VSPs (Berger et al., 1995; Utsugi et al., 1998), which are vacuolar-localized glycoproteins with acid phosphatase activity (Mason and Mullet, 1990). These proteins are presumed to be amino acid sinks during water deficit but are important reduced nitrogen sources after stress relief (Mason and Mullet, 1990).
Figure 2.
Comparative expression of genes dependent on the SOS pathway in wild type (Col-0 and gll) and sos1, sos2, and sos3. Illustrated is the northern blot of steady-state mRNA levels of COR6.6/KIN2, SAMT, CCR1, and VSP2 in plants without (0 h) or 160 mm NaCl (24 h). Ethidium bromide staining was used to monitor RNA loading.
CCR1, STZ, SAMT, COR6.6/KIN2, and AD06C08/unknown have similar expression profiles in sos1, sos2, and sos3, implicating these as transcriptional outputs requiring all components of the SOS pathway. CCR1 encodes a Gly-rich RNA-binding protein implicated in posttranscriptional regulation. CCR1 and CCR2 comprise a two-gene family, and their expression is regulated by a diurnal circadian clock (Carpenter et al., 1994; Heintzen et al., 1997; Kreps and Simon, 1997). CCR1 or CCR2 expression negatively regulates either gene and this feedback loop presumably facilitates diurnal oscillation controlled by the master circadian clock (Heintzen et al., 1997). CCR1 and CCR2 steady-state mRNA levels are induced by cold but CCR1 transcript is negatively regulated by ABA and dehydration, whereas CCR2 expression is induced by dehydration (Carpenter et al., 1994). CCR1 transcript abundance was down-regulated in sos3 indicating that the SOS pathway is at least another negative regulator that controls CCR1 expression downstream of the circadian rhythm clock (Heintzen et al., 1997; Kreps and Simon, 1997). These results indicate that control of circadian oscillations may be required during the salt stress response.
COR6.6//KIN2 and SAMT are both implicated in plant stress responses. COR6.6/KIN2 is linked in tandem to its homolog KIN1 (95% nucleotide sequence identity in the coding region) and both encode hydrophilic peptides that are boiling soluble but of unknown function (Thomashow, 1999). KIN1 and 2 transcript abundance is cold and ABA induced (Kurkela and Franck, 1990; Kurkela and Borg-Franck, 1992; Thomashow, 1999) but KIN2 and not KIN1 expression is modulated positively by drought and salt (Kurkela and Borg-Franck, 1992). SAMT catalyzes the formation of methylsalicylate from salicylic acid using S-adenosyl-l-Met as the methyl donor (Ross et al., 1999). The volatile ester is implicated as a pollinator attractant and a signal in plant defense mediated by salicylic acid (Ross et al., 1999; Dudareva et al., 2000).
DISCUSSION
Many salt-regulated genes are responsive also to other biotic or abiotic perturbations, indicating that these stresses have common etiologies, e.g. water deficit, and cold are both osmotic stresses, occur simultaneous in the environment or elicit similar pathologies (Bohnert et al., 1995; Shinozaki and Yamaguchi-Shinozaki, 1997; Zhu et al., 1997; Hasegawa et al., 2000b). Furthermore, osmotic and ionic stresses induce secondary cellular perturbations that arise from ROS, elicitors from degradation of cell wall and plasma membrane macromolecules, or wounding, which initiate signal transduction pathways that modulate other plant defensive processes (Hasegawa et al., 2000b). In fact, many of the proteins encoded by the 84 salt-responsive genes identified in this study can be categorized as functional outputs from these different signal cascades (Table II). Most of these genes were not known previously to be modulated as a part of the Arabidopsis salt response. It is interesting that even after subtractive hybridization only approximately 13% of the ESTs (84/614) were unique and detected salt-regulated transcripts.
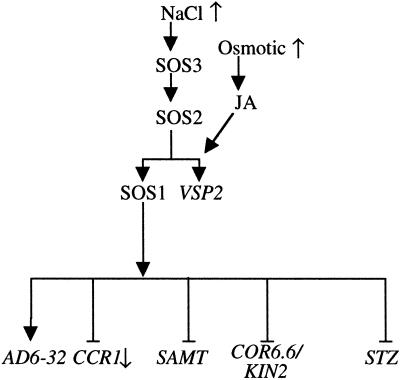
Six of 89 genes examined were differentially responsive to NaCl in wild type and sos3, implicating the SOS pathway in their transcriptional regulation. AD06C08/unknown and VSP2 are induced and CCR1, STZ, SAMT, and COR6.6/KIN2 are controlled negatively by SOS3. CCR1 expression was lower in wild type after NaCl treatment, whereas the message abundance of the others was salt induced. From these results, a model for the SOS pathway regulation of these genes is illustrated in Figure 3 (Zhu, 2000). Salt regulated expression of VSP2 is the same in wild type and sos1 defining this gene as a transcriptional output from the SOS pathway that does not require SOS1. This supports the premise that SOS3 and SOS2 are signal intermediates and SOS1 is an effector of Na+ homeostasis (Shi et al., 2000). However, salt regulation of AD06C08/unknown, CCR1, STZ, SAMT, and COR6.6/KIN2 is dependent on SOS2 and SOS1, perhaps implicating a signaling function for SOS1. Genes encoding an enzyme catalyzing the penultimate step in Pro biosynthesis (P5CS) and a putative transcription factor (AtMYB) are hyper-induced by salt in sos1 compared with wild type (Liu and Zhu, 1997). Furthermore, some transport proteins include sensor domains or function in association with sensors (Ozcan et al., 1998; Heinisch et al., 1999; Sabirov et al., 1999) as could the putative Na+/H+ antiporter SOS1 (Shi et al., 2000).
Figure 3.
Illustrated is a model depicting the SOS pathway regulation of salt responsive genes. Hypersaline conditions activate the SOS (SOS3 → SOS2 → SOS1) signal pathway (Zhu, 2000) and transcript abundance of AD0608 (unknown), VSP2 (vegetative storage protein 2, AD05E05), SAMT (S-adenosyl-l-Met:salicylic acid carboxyl methyltransferase, AD05B11), COR6.6/KIN2 (cold regulated 6.6/inducible 2, AD03D05 [COR6.6/KIN2], and STZ [salt tolerance zinc finger], Lippuner et al., 1996) increase and of CCR1 (circadian rhythm-RNA binding1, AD05C10) decreases in Arabidopsis seedlings. Positive (↓) or negative (⊥) regulation by the SOS pathway is indicated.
The experimental evidence presented here indicates that the SOS pathway controls expression of only a few salt stress-specific tolerance determinant genes among the numerous genes (six of 89 in this study) that are regulated in the plant response to NaCl treatment (Zhu et al., 1997). This is similar to the paradigm that has been established recently for the salt stress response of the unicellular eukaryote yeast (Saccharomyces cerevisiae). Genome-wide array analysis determined that osmotic upshock causes a rapid and multi-fold increase in mRNA of between 186 and 1,359 genes and reduced transcript abundance of more than 100 genes depending on the severity of osmotic shock, the osmotic agent (NaCl or sorbitol), and time after treatment (Posas et al., 2000; Rep et al., 2000; J. Yale and H.J. Bohnert, unpublished data). Salt-induced expression of most is either partially or completely controlled by the high osmolarity glycerol and mitogen-activated protein kinase pathway. The yeast calcineurin pathway is analogous to the Arabidopsis SOS pathway, controls ion homeostasis and is essential for salt tolerance in yeast (Mendoza et al., 1994, 1996), and affects expression of many fewer osmotic responsive genes (T.K. Matsumoto, unpublished data). Like the SOS pathway, calcineurin regulates expression of genes that encode salt tolerance effectors such as Na+ efflux transporters (Mendoza et al., 1994; Shi et al., 2000; Matusmoto et al., unpublished data).
It is conceivable that signal transduction through the SOS pathway that mediates salt tolerance may have a substantial component that involves posttranscriptional activation of salt tolerance effectors, particularly over the time span (minimum of 4 h) of the salt treatment used in experiments reported here. Plant survival in severe stress likely requires very immediate cellular responses, whereas transcriptional regulation may be sufficient for stress recovery and adaptation. Notwithstanding, salt induces transcriptional activation of genes in yeast within minutes (Posas et al., 2000; Rep et al., 2000). Genes that are transiently induced or weakly expressed further complicate inference of function from expression profile analysis. The majority of yeast genes induced by mild salt shock exhibit transient expression (Posas et al., 2000). Determinant gene transcript abundance differences in wild type and sos3 may be insufficient for the resolution limits of the subtraction protocols, yet are biologically meaningful to salt stress adaptation.
It is interesting that the SOS pathway negatively controls the expression of four salt-regulated genes (SAMT, C0R 6.6/KIN2, STZ, and CCR1), three (SAMT, C0R6.6/KIN2, and STZ) of which are induced by NaCl treatment. So, salt tolerance determinants include genes that must be repressed, at least temporally, during the plant stress response. Negatively regulated genes may include those that contribute to growth arrest necessary during the period of adjustment or ameliorate other etiologies that occur coincidentally with salt in the native environment of the organism. CCR1 and COR6.6/KIN2 are cold induced, whereas SAMT is implicated in plant defense against pathogens indicating their principal function is not in salt adaptation. Together, this suggests that a function of the SOS pathway is to discriminate against the myriad of stress signals that are elicited by salt and to focus the capacity of the plant to cope with the principal etiology; in this instance, ion dis-equilibrium. Furthermore, the SOS pathway may coordinate temporal gene expression to focus the availability of effectors, as required, during stress perception, amelioration, or adaptation. Confirmation that the genes identified in this study encode tolerance determinants awaits molecular genetic confirmation by loss- or gain-of-function experimentation.
MATERIALS AND METHODS
Plant Material
Arabidopsis (ecotype Columbia-0 gl1) and sos1, sos2, and sos3 were wild-type and salt-hypersensitive genotypes, respectively (provided by Dr. Jian-Kang Zhu, University of Arizona). Seeds were surface disinfected and stratified for 2 d at 4°C. Seeds were germinated in liquid medium (Murashige and Skoog salts [Murashige and Skoog, 1962] and 3% [w/v] Suc [pH 5.8]) in 250-mL flasks on a gyratory shaker (80–100 rpm) under low-intensity Cool White fluorescent illumination (light/dark:16/8 h daily) at 22°C to 24°C. After 14 d, seedlings were transferred to fresh medium without or with 160 mm NaCl for the time interval indicated. Seedlings were harvested, frozen in liquid nitrogen, and stored at −80°C.
Construction of Subtraction Libraries
Total RNA was isolated as described by Gong et al. (1997). mRNA was isolated using the Poly(A)+ RNA purification kit (CLONTECH, Palo Alto, CA). The PCR-Select cDNA subtraction kit (K1804-1, CLONTECH) was used to obtain subtracted cDNA libraries.
Subtraction of cDNAs Obtained from mRNA of Salt-Treated sos3 and gl1
Subtractive hybridization was used to identify cDNAs corresponding to salt regulated genes differentially expressed in sos3. Both forward and reverse subtractive hybridizations were performed with salt-treated (160 mm NaCl, 4 h) seedlings. The forward subtraction used tester cDNA obtained from mRNA of gl1 and driver cDNA from sos3. In the reverse subtraction, the tester cDNA was obtained from sos3 and driver cDNA from gl1. Driver cDNAs are the reference and are targets for elimination during subtraction leaving unique tester cDNAs. Forward and reverse subtractive hybridization was meant to identify salt-responsive genes that are specifically regulated in sos3.
Subtraction of cDNAs Obtained from gl1 after Treatment without or with NaCl
This subtraction was intended for identification of genes differentially regulated by salt in gl1. The forward subtraction tester cDNA was obtained from mRNA of gl1 seedlings 4 h after transfer to medium with 160 mm NaCl and driver cDNA from gl1 plants grown in medium without salt. In the reverse subtraction, tester and driver cDNA was obtained from gl1 grown without and with 160 mm NaCl, respectively.
The subtracted libraries were subjected to two rounds of PCR amplification, the second using nested primers for adaptors 1 and 2R (CLONTECH). The PCR products were ligated into pT-Adv (CLONTECH), and transformed into Escherichia coli. The white colonies were isolated and inserts amplified by PCR.
Enrichment of Unique Salt-Regulated cDNAs in the Subtraction Library
A modification of the differential subtraction chain method (Luo et al., 1999) was used to enrich the subtracted library for unique salt-regulated cDNAs. The driver for this subtraction (driver 2) is a mixture of second PCR product from reverse subtraction (100 μL; tester DNA:gl1 without salt, driver DNA:gl1 with salt) and PCR products amplified from highly repetitive salt-induced cDNAs isolated from previous rounds of screening (0.5 μL of each). To remove adaptor sequences from driver 2, the mixture was digested at the restriction sites of adaptors 1 (SmaI and RsaI) and 2R (EagI and RsaI) by consecutive restriction digestion using SmaI, and then EagI + RsaI. The digested DNA was extracted with phenol and phenol/chloroform, and then precipitated with ethanol. The precipitate was redissolved in 100 μL of water and passed through the Microcon YM-30 column (Amicon, Beverly, MA) to separate the DNA from Adaptor fragments. Driver 2 DNA was recovered in 20 μL of water and used as driver in the following PCR subtraction. About 10 times excess amount of driver 2 DNA (2 μL) was mixed with 1μL of forward subtraction library (driver and tester = gl1 grown without and with 160 mm NaCl, respectively) in hybridization buffer {5 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid]-HCl (pH 8.3), 12 mm NaCl, and 0.05 mm EDTA} in a total volume of 12 μL. The hybridization solution was denatured at 94°C for 5 min, and incubated at 72°C for 12 h.
After hybridization, the mixture was precipitated with ethanol, and digested with mung bean nuclease (Promega, Madison, WI) to remove adaptor sequence from products of the driver-tester hybridization. Adaptor sequences in tester-tester hybridization products are aligned with matching ends and these are not digested by mung bean nuclease. The mung bean nuclease digested mixture was passed through the Microcon column and purified DNA was rehybridized at 72°C. The mung bean nuclease digestion and DNA purification procedures were repeated. The DNA was amplified by PCR using nested primers for 1 and 2R, 11 cycles. The cloning of PCR products was as described as above.
Dot-Blot Analysis
For dot-blot analysis, cDNA inserts of forward subtraction library clones were individually amplified by PCR and 2 μL of PCR product was mixed with 2 μL of 0.6 m NaOH. Two microliters of the mixture was blotted onto each of two duplicate nylon membrane filters. Probes for dot-blot analysis were either forward or reverse subtraction products or cDNA of RNA from plants, as indicated. The products of forward and reverse subtraction were digested with SmaI, RsaI, and EagI to remove the adapter sequences and labeled with 32P using the Ready-To-Go kit (Amersham Pharmacia Biotech, Piscataway, NJ). The labeled forward probes were hybridized to one membrane and the reverse probes to the duplicate.
Template Preparation, DNA Sequencing, and Data Analysis
Plasmid templates were prepared from selected bacterial colonies by 96-well alkaline lysis minipreps according to the manufacturer's instructions (Edge BioSystems, Inc., Gaithersburg, MD).
DNA sequencing reactions were conducted using DyeDeoxy “Terminator PRISM” mix (Perkin-Elmer-ABI, Foster City, CA) according to the manufacturer's instructions in a multiplate thin-wall 96-well microplate on an MJ Research PTC-100-96 (MJ Research, Watertown, MA) programmable thermal controller using the following profile: 96°C for 30 s, 45°C for 15 s, and 60°C for 4 min for 49 cycles. Unincorporated dye terminators were removed by passing reactions over a 96-well gel filtration block (Edge BioSystems). Recovered sequencing reaction products were analyzed on either an ABI 373A-XL Stretch or an ABI 3700 capillary array automated DNA sequencing system (Perkin-Elmer Applied BioSystems). Raw sequence data was analyzed using PHRED (Ewing and Green, 1998; Ewing et al., 1998) and Cross match to removal vector sequences. Additional vector sequence removal and editing was done manually using FACTURA software (Perkin-Elmer Applied BioSystems). Polished EST sequence files were assembled into singleton and contig files using PHRAP (P. Green, unpublished data). EST identities were determined by sequence comparison to the nonredundant GenBank database using BLASTN (BLAST 2.0) using default parameters (Altschul et al., 1997). In instances where an unannotated match was obtained, BLASTX searchers were conducted and sequence homology information was used to assign putative identities. All EST sequences reported here have been deposited in dbEST and can be browsed and retrieved from the NCBI website (http://www.ncbi.nlm.nih.gov) under accession numbers BE844684 through BE845405.
Northern-Blot Analysis of Putative Clones
Total RNA was isolated from seedlings and 40 μg from each sample was separated on 1.2% (w/v) agarose formaldehyde gels and transferred to Hybond-N nylon membranes (Amersham) as previously described (Gong et al., 1997). The cDNA insert of each clone was amplified by PCR using nested primers that hybridize to adaptors 1 and 2R, and purified from agarose gels using the Qiagen Gel Purification kit. The probes for RD22BP1, MYB2, PAL, STZ, and ACP1 were obtained by PCR amplification using cDNA obtained from mRNA of salt-treated gl1 as template; RD22BP1 (AB000875), forward primer: 5′-ATGACGCTGT-TGATGAGGAG-3′ and reverse primer: 5′-TTTCGGATT-CTGGGTCTGAG-3′ (0.56 kb); MYB2 (D14712), forward primer: 5′-GAAATGGAAGATTACGAGCG-3′ and reverse primer: 5′-TTAATTATACGAATACGATGTC-3′ (1.0 kb); PAL (L33677), forward primer: 5′-ATGGAGATT-AACG-GGGCACAC-3′ and reverse primer: 5′-ACGT-TCACCG-TTGGGACCAG-3′ (1.1 kb); STZ (X95573), ORF; and ACP1 (AF009228), forward primer: 5′-CAA-AAGCCATTTTT-CAAATTTCAAACTCAG and reverse primer 5′-GTTTT-CAATGATAGTGAAGAAAGATG-TAC-AAC (0.83 kb). The purified PCR products were labeled using 32P dCTP using the Ready-To-Go kit. Blot-blot hybridization and washes were as described (Gong et al., 1997). The blots were stripped by boiling in 0.5% (w/v) SDS solution for 3 min and were rehybridized with another probe.
ACKNOWLEDGMENTS
The authors would like to thank Sue Ann Hudiburg and Janet Rogers of the Oklahoma State University Recombinant DNA/Protein Resource Facility for providing oligonucleotide synthesis and automated DNA sequencing analysis services.
Footnotes
This work was supported by a National Science Foundation Plant Genome award (no. DBI–9813360). This is Purdue University Agricultural Experiment Station paper no. 16427.
LITERATURE CITED
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipma DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S, Bell E, Sadka A, Mullet JE. Arabidopsis thaliana Atvsp is homologous to soybean VspA and VspB, genes encoding vegetative storage protein acid phosphatases, and is regulated similarly by methyl jasmonate, wounding, sugars, light and phosphate. Plant Mol Biol. 1995;27:933–942. doi: 10.1007/BF00037021. [DOI] [PubMed] [Google Scholar]
- Bohnert HJ, Nelson DE, Jensen RG. Adaptation to environmental stresses. Plant Cell. 1995;7:1099–1111. doi: 10.1105/tpc.7.7.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray EA. Molecular responses to water deficit. Plant Physiol. 1994;103:1035–1040. doi: 10.1104/pp.103.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter CD, Kreps JA, Simon AE. Genes encoding glycine-rich Arabidopsis thalianaproteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol. 1994;104:1015–25. doi: 10.1104/pp.104.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Murfitt LM, Mann CJ, Gorenstein N, Kolosova N, Kish CM, Bonham C, Wood K. Developmental regulation of methyl benzoate biosynthesis and emission in snapdragon flowers. Plant Cell. 2000;12:949–961. doi: 10.1105/tpc.12.6.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using Phred: II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using Phred: I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Gong Z, Yamazaki M, Sugiyama M, Tanaka Y, Saito K. Cloning and molecular analysis of structural genes involved in anthocyanin biosynthesis and expressed in a forma-specific manner in Perilla frutescens. Plant Mol Biol. 1997;35:915–927. doi: 10.1023/a:1005959203396. [DOI] [PubMed] [Google Scholar]
- Halfter U, Ishitani M, Zhu J-K. The ArabidopsisSOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA. 2000;97:3735–3740. doi: 10.1073/pnas.040577697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Rathinasabapathi B, Rivoal J, Burnet M, Dillon MO, Gage DA. Osmoprotective compounds in the Plumbaginaceae: a natural experiment inmetabolic engineering of stress tolerance. Proc Natl Acad Sci USA. 1994;91:306–310. doi: 10.1073/pnas.91.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Pardo JM. The dawn of plant salt tolerance genetics. Trends Plant Sci. 2000a;5:317–319. doi: 10.1016/s1360-1385(00)01692-7. [DOI] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu J-K, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol. 2000b;51:463–499. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- Heinisch JJ, Lorberg A, Schmitz H-P, Jacoby JJ. The protein kinase C-mediated MAP kinase pathway involved in the maintenance of cellular integrity in Saccharomyces cerevisiae. Mol Microbiol. 1999;32:671–680. doi: 10.1046/j.1365-2958.1999.01375.x. [DOI] [PubMed] [Google Scholar]
- Heintzen C, Nater M, Apel K, Staiger D. AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1997;94:8515–8520. doi: 10.1073/pnas.94.16.8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Goodman H. An Arabidopsis thalianaroot-specific kinase homolog is induced by dehydration, ABA and NaCl. Plant J. 1995;8:37–43. doi: 10.1046/j.1365-313x.1995.08010037.x. [DOI] [PubMed] [Google Scholar]
- Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Stevenson B, Zhu J-K. Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell. 1997;9:1935–1949. doi: 10.1105/tpc.9.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt and freezing tolerance by gene transfer of a single stress-inducible transcriptional factor. Nature Biotech. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Koiwa H, Bressan RA, Hasegawa PM. Regulation of protease inhibitors and plant defense. Trends Plant Sci. 1997;2:379–384. [Google Scholar]
- Kovtun Y, Chiu W-L, Tena G, Sheen J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA. 2000;97:2940–2945. doi: 10.1073/pnas.97.6.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps JA, Simon AE. Environmental and genetic effects on circadian clock-reggulated gene expression in Arabidopsis. Plant Cell. 1997;9:297–304. doi: 10.1105/tpc.9.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkela S, Borg-Franck M. Structure and expression of KIN2, one of two cold- and ABA-induced genes of Arabidopsis thaliana. Plant Mol Biol. 1992;19:689–692. doi: 10.1007/BF00026794. [DOI] [PubMed] [Google Scholar]
- Kurkela S, Franck M. Cloning and characterization of a cold- and ABA-inducible Arabidopsisgene. Plant Mol Biol. 1990;15:137–144. doi: 10.1007/BF00017731. [DOI] [PubMed] [Google Scholar]
- Lippuner V, Cyert MS, Gasser CS. Two classes of plant cDNAs differentially complement yeast calcineurin mutants and increase salt tolerance of wild-type yeast. J Biol Chem. 1996;271:12859–12866. doi: 10.1074/jbc.271.22.12859. [DOI] [PubMed] [Google Scholar]
- Liu J, Ishitani M, Halfter U, Kim C-S, Zhu J-K. The Arabidopsis thaliana SOS2gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA. 2000;79:3730–3734. doi: 10.1073/pnas.060034197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu J-K. Proline accumulation and salt-stress-induced gene expression in a salt-hypersensitive mutant of Arabidopsis. Plant Physiol. 1997;114:591–596. doi: 10.1104/pp.114.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu J-K. A calcium sensor homolog required for plant salt tolerance. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- Luo J-H, Puc JA, Slosberg ED, Yao Y, Bruce JN, Wright TC, Becich MJ, Parsons R. Differential subtraction chain, a method for identifying differences in genomic DNA and mRNA. Nucleic Acids Res. 1999;27:24e. doi: 10.1093/nar/27.19.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason HS, Mullet JE. Expression of two soybean vegetative storage protein genes during development and in response to water deficit, wounding, and jasmonic acid. Plant Cell. 1990;2:569–579. doi: 10.1105/tpc.2.6.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza I, Quintero FJ, Bressan RA, Hasegawa PM, Pardo JM. Activated calcineurin confers high tolerance to ion stress and alters the budding pattern and cell morphology of yeast cells. J Biol Chem. 1996;271:23061–23067. doi: 10.1074/jbc.271.38.23061. [DOI] [PubMed] [Google Scholar]
- Mendoza I, Rubio F, Rodriguez-Navarro A, Pardo JM. The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae. J Biol Chem. 1994;269:8792–8796. [PubMed] [Google Scholar]
- Mikami K, Katagiri T, Iuchi S, Yamaguchi-Shinozaki K, Shinozaki K. A gene encoding phosphatidylinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J. 1998;15:563–568. doi: 10.1046/j.1365-313x.1998.00227.x. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Irie K, Hirayama T, Hayashida N, Yamaguchi-Shinozaki K, Matsumoto K, Shinozaki K. A gene encoding a mitogen-activated protein kinase kinase kinase is induced simultaneously with genes for a mitogen-activated protein kinase and an S6 ribosomal protein kinase by touch, cold, and water stress in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;84:765–769. doi: 10.1073/pnas.93.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant. 1962;15:473–479. [Google Scholar]
- Nelson DE, Shen B, Bohnert HJ. The regulation of cell-specific inositol metabolism and transport in plant salinity tolerance. Plant Cell. 1998;10:753–764. doi: 10.1105/tpc.10.5.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X, Bressan RA, Hasegawa PM, Pardo JM. Ion homeostasis in NaCl stress environments. Plant Physiol. 1995;109:735–742. doi: 10.1104/pp.109.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S, Dover J, Johnston M. Glucose sensing and signalling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:2566–2573. doi: 10.1093/emboj/17.9.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao HL, Pih KT, Lim JH, Kang SG, Jin JB, Kim SH, Hwang I. An Arabidopsis GSK3/shaggy-likegene that complements yeast salt stress-sensitive mutant is induced by NaCl and abscisic acid. Plant Physiol. 1999;119:1527–1534. doi: 10.1104/pp.119.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Chambers JR, Heyman JA, Hoeffler JP, Nadal ED, Arino J. The transcriptional response of yeast to saline stress. J Biol Chem. 2000;275:17249–17255. doi: 10.1074/jbc.M910016199. [DOI] [PubMed] [Google Scholar]
- Rep M, Krantz M, Thevelein JM, Hohmann S. The transcriptional response of Saccharomyces cerevisiaeto osmotic shock. J Biol Chem. 2000;275:8290–8300. doi: 10.1074/jbc.275.12.8290. [DOI] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell, 2000;12:707–719. doi: 10.1105/tpc.12.5.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JR, Nam KH, D'Auria JC, Pichersky E. S-adenosyl-L-methionine: salicylic acid carboxyl methyltransferase, an enzyme involved in floral scent production and plant defense, represents a new class of plant methyltransferases. Arch Biochem Biophys. 1999;367:9–16. doi: 10.1006/abbi.1999.1255. [DOI] [PubMed] [Google Scholar]
- Sabirov RZ, Azimov RR, Ando-Akatsuka, Miyoshi T, Okada Y. Na+ sensitivity of ROMK1 K+ channel: role of the Na+/H+antiporter. J Membr Biol. 1999;172:67–76. doi: 10.1007/s002329900584. [DOI] [PubMed] [Google Scholar]
- Sanders D. Plant biology: the salty tale of Arabidopsis. Curr Biol. 2000;10:R-486–R-488. doi: 10.1016/s0960-9822(00)00554-6. [DOI] [PubMed] [Google Scholar]
- Sheen J. Ca2+-dependent protein kinases and stress signal transduction in plants. Science. 1996;274:1900–1902. doi: 10.1126/science.274.5294.1900. [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu J-K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+antiporter. Proc Natl Acad Sci USA. 2000;97:6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene expression and signal transduction in water-stress response. Plant Physiol. 1997;115:327–334. doi: 10.1104/pp.115.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. Plant cold Acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Utsugi S, Sakamoto W, Murata M, Motoyoshi F. Arabidopsis thaliana vegetative storage protein (VSP) genes: gene organization and tissue-specific expression. Plant Mol Biol. 1998;38:565–576. doi: 10.1023/a:1006072014605. [DOI] [PubMed] [Google Scholar]
- Winicov I. Alfin1 transcription factor overespression enhances plant root growth under normal and saline conditions and improves salt tolerance in alfalfa. Planta. 2000;210:416–22. doi: 10.1007/PL00008150. [DOI] [PubMed] [Google Scholar]
- Yoshiba Y, Kiyosue T, Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. Regulation of levels of proline as an osmolyte in plants under water stress. Plant Cell Physiol. 1997;38:1095–1102. doi: 10.1093/oxfordjournals.pcp.a029093. [DOI] [PubMed] [Google Scholar]
- Zhu J-K. Genetic analysis of plant salt tolerance using Arabidopsis thaliana. Plant Physiol. 2000;124:941–948. doi: 10.1104/pp.124.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J-K, Hasegawa PM, Bressan RA. Molecular aspects of osmotic stress in plants. Crit Rev Plant Sci. 1997;16:253–277. [Google Scholar]