ABSTRACT
Measles virus (MeV), the causative agent of measles, is an enveloped RNA virus of the family Paramyxoviridae, which remains an important cause of childhood morbidity and mortality. MeV has two envelope glycoproteins, the hemagglutinin (H) and fusion (F) proteins. During viral entry or virus-mediated fusion between infected cells and neighboring susceptible cells, the head domain of the H protein initially binds to its receptors, signaling lymphocytic activation molecule family member 1 (SLAM) and nectin-4, and then the stalk region of the H protein transmits the fusion-triggering signal to the F protein. MeV may persist in the human brain and cause a fatal neurodegenerative disease, subacute sclerosing panencephalitis (SSPE). Recently, we showed, using in vitro cell culture, that cell adhesion molecule (CADM) 1 and CADM2 are host factors that trigger hyperfusogenic mutant F proteins, causing cell-to-cell fusion and the transfer of the MeV genome between neurons. Unlike conventional receptors, CADM1 and CADM2 interact in cis (on the same membrane) with the H protein and then trigger membrane fusion. Here, we show that alanine substitutions in part of the stalk region (positions 171-175) abolish the ability of the H protein to mediate membrane fusion triggered by CADM1 and CADM2, but not by SLAM. The recombinant hyperfusogenic MeV carrying this mutant H protein loses its ability to spread in primary mouse neurons as well as its neurovirulence in experimentally infected suckling hamsters. These results indicate that CADM1 and CADM2 are key molecules for MeV propagation in the brain and its neurovirulence in vivo.
IMPORTANCE Measles is an acute febrile illness with skin rash. Despite the availability of highly effective vaccines, measles is still an important cause of childhood morbidity and mortality in many countries. The World Health Organization estimates that more than 120,000 people died from measles worldwide in 2021. Measles virus (MeV), the causative agent of measles, can also cause a fatal progressive neurological disorder, subacute sclerosing panencephalitis (SSPE), several years after acute infection. There is currently no effective treatment for this disease. In this study, using recombinant MeVs with altered receptor usage patterns, we show that cell adhesion molecule (CADM) 1 and CADM2 are host factors critical for MeV spread in neurons and its neurovirulence. These findings further our understanding of the molecular mechanism of MeV neuropathogenicity.
KEYWORDS: CADM1, CADM2, SSPE, cis-acting receptor, encephalitis, hemagglutinin stalk, measles virus, membrane fusion, neuropathogenicity, neurovirulence
INTRODUCTION
Measles is a highly contagious disease characterized by high fever, respiratory symptoms, conjunctivitis, and maculopapular rash (1). Despite the availability of effective live-attenuated vaccines, measles remains an important cause of childhood morbidity and mortality worldwide, particularly in developing countries (2). Measles virus (MeV), the causative agent of measles, is an enveloped RNA virus belonging to the genus Morbillivirus in the family Paramyxoviridae. MeV has two envelope glycoproteins, the hemagglutinin (H), and the fusion (F) proteins. These proteins cooperatively mediate membrane fusion required for viral entry and direct cell-to-cell spread (1). The signaling lymphocytic activation molecule family member 1 (SLAM; also known as SLAMF1 and CD150) on immune cells and nectin-4 on epithelial cells act as receptors for MeV (3–6).
MeV persists, albeit rarely, in the central nervous system (CNS), causing a progressive neurological disorder, subacute sclerosing panencephalitis (SSPE), several years after acute infection (7, 8). Although wild-type (WT) MeV isolates from acute measles patients are not neurotropic because of the lack of SLAM and nectin-4 in neurons (9, 10), neurons are mainly affected cells in the brains of SSPE patients, where MeV is thought to spread presumably by transsynaptic cell-to-cell transmission (11–13). Recently, many studies have shown that hyperfusogenic mutations found in the MeV F proteins derived from SSPE patients (e.g., T461I) enable MeV spread in primary neurons in vitro and in the brains of experimentally infected mice and hamsters, leading to death (14–23). These mutations destabilize the prefusion form of the F protein, rendering it hyperfusogenic (17). In addition, structurally unstable F proteins are triggered even by the weak interaction between the so-called “receptor-blind” H proteins having substitutions within the receptor binding sites and the corresponding receptors (24). Recently, we have reported that cell adhesion molecule 1 (CADM1, also known as IGSF4A, Necl-2, and SynCAM1) and CADM2 (also known as IGSF4D, Necl-3, and SynCAM2) are “cis-acting receptors” (host molecules that weakly interact with the H protein on the same membrane [in cis] and trigger membrane fusion caused by the hyperfusogenic mutant F proteins) in neurons, although they do not function as the entry receptor (22, 25). Interestingly, CADM1 and CADM2 can interact with the H protein even when it lacks the head domain which binds to ordinary receptors SLAM and nectin-4, triggering membrane fusion (25).
In this study, we identified the segment of the H protein stalk essential for mediating membrane fusion triggered by CADM1 and CADM2. Using recombinant MeVs possessing mutant H proteins with altered receptor usage patterns, we demonstrated that CADM1 and CADM2 are indeed critical for MeV spread in neurons and its neurovirulence.
RESULTS
The H protein lacking the head domain and a part of the stalk region loses the ability to support CADM1/2-dependent membrane fusion.
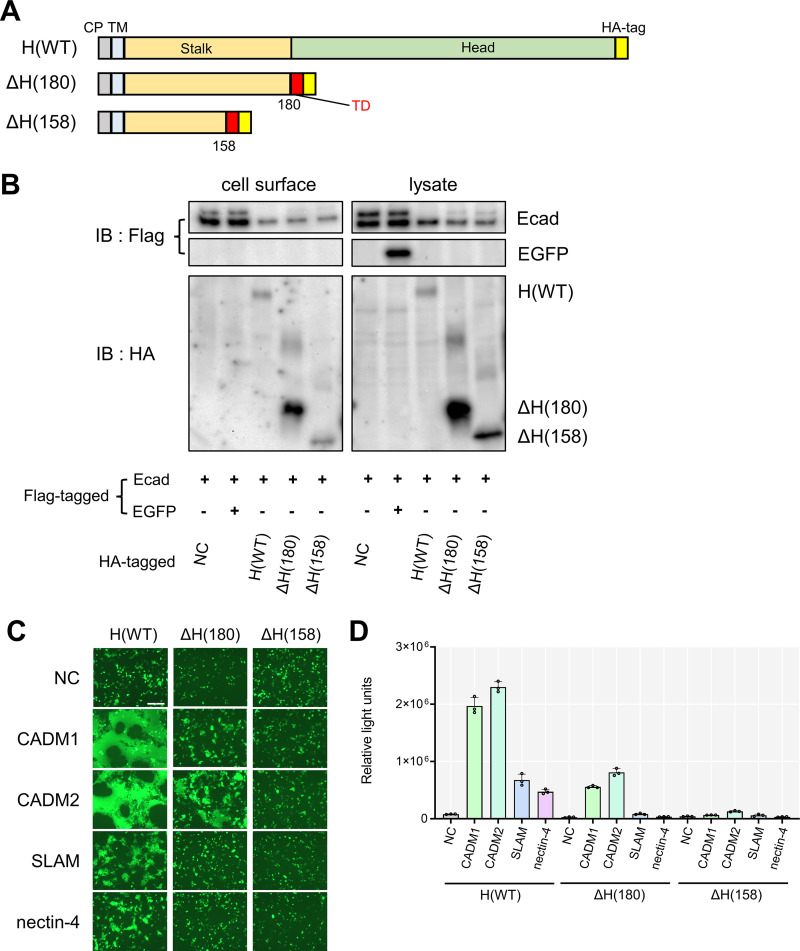
We have previously shown that the cis-acting receptors CADM1 and CADM2, but not the trans-acting receptors SLAM and nectin-4, can interact with the H protein lacking its head domain, presumably through its stalk region, and trigger hyperfusogenic F protein-mediated membrane fusion (25). To investigate which part in the H stalk is involved in the interaction with CADM1 and CADM2, we examined the full-length WT H protein (H[WT]), the tetramerized H protein lacking its head domain [designated as ΔH(180)] and a further truncated H protein (ΔH[158]) for their functions (Fig. 1A). ΔH(158) was reported previously as the truncated H stalk containing the H-F interaction domain (26).
FIG 1.
The truncated headless H protein ΔH(158) does not mediate CADM1/2-dependent membrane fusion. (A) Schematic diagrams of the HA-tagged full-length H protein (H[WT]), H protein lacking its head domain (residues 1 to 180) (ΔH[180]), and H protein lacking its head and a part of the stalk region (resides 1 to 158) (ΔH[158]). A tetramerization domain (TD) was fused to the C-terminus of the ΔH(180) and ΔH(158). CP, cytoplasmic domain; TM, transmembrane domain. (B) The cell surface biotinylation assay with H(WT), ΔH(180), and ΔH(158). The pCA7 expression plasmids respectively encoding E-cadherin (a cell surface control) and one of HA-tagged H [H(WT), ΔH(180), or ΔH(158)] or none (NC, plasmid alone as a negative control) were transfected into 293FT cells. That encoding Flag-tagged EGFP (an intracellular protein control) was also transfected. Precipitate of biotinylated cell surface proteins and cell lysates were detected by Western blotting using anti-Flag (upper and middle) or anti-HA Ab (lower). (C) The fusion assay with H protein mutants. The pCA7 plasmids respectively encoding one of the H constructs (H[WT], ΔH[180], and ΔH[158]), the F(T461I) protein, EGFP, and one of the host proteins (CADM1, CADM2, SLAM, and nectin-4) or none were transfected into 293FT cells. Cells were observed under a fluorescence microscope 48 h after transfection. Scale bar = 200 μm. (D) The DSP assay with various H constructs. The pCA7 plasmids respectively encoding one of the H protein constructs (H[WT], ΔH[180], and ΔH[158]), the F(T461I) protein, and one of the host proteins (CADM1, CADM2, SLAM, and nectin-4) or none were transfected into cocultured 293FT/DSP1 and 293FT/DSP2 cells. Renilla luciferase activity was measured 48 h after transfection (n = 3, means ±SD).
The pCA7 expression plasmids respectively encoding H(WT), ΔH(180), and ΔH(158) proteins modified by the influenza virus hemagglutinin (HA) tag were generated (Fig. 1A). ΔH(180) and ΔH(158) proteins were designed such that they have a tetramerization domain at their C-terminus (26, 27). The cell surface expression levels of these H proteins were evaluated by the cell surface biotinylation (Fig. 1B). The results showed that all of the H(WT), ΔH(180), and ΔH(158) proteins were expressed on the cell surface 24 h after transfection into 293FT cells.
The fusion assay was then performed to evaluate the levels of cell-to-cell membrane fusion mediated by these H proteins, as previously reported (22). Briefly, expression plasmids respectively encoding one of the H proteins [H(WT), ΔH(180), and ΔH(158)], the hyperfusogenic F(T461I) protein, the enhanced green fluorescent protein (EGFP), and one of the host proteins (CADM1, CADM2, SLAM, and nectin-4), were transfected into 293FT cells, and the cells were observed under a fluorescence microscope 48 h after transfection (Fig. 1C). When the H(WT) protein was expressed, all the host molecules induced syncytium formation (CADM1 and CADM2 induced larger syncytia than SLAM and nectin-4, probably because the cells that received the plasmids encoding cis-acting receptors, but not trans-acting receptors, could fuze even with neighboring untransfected cells). On the other hand, the ΔH(180) protein supported syncytium formation induced by CADM1 and CADM2, but not by SLAM and nectin-4, confirming our previous study (25). Notably, when the ΔH(158) protein was expressed, all the host molecules including CADM1 and CADM2 did not induce syncytium formation.
The levels of membrane fusion supported by these H proteins were also quantified by the dual split protein (DSP) assay (24, 28–30). In this assay, a pair of chimeric reporter proteins, DSP1 and DSP2, each comprised of the split Renilla luciferase and split green fluorescent protein (GFP), are stably expressed in 293FT cells, respectively (293FT/DSP1 and 293FT/DSP2 cells). When cell-to-cell fusion is induced between 293FT/DSP1 and 293FT/DSP2 cells, Renilla luciferase and GFP activities are restored by the association of DSP1 and DSP2. The 293FT/DSP1 and 293FT/DSP2 cells were cocultured, and then transfected with plasmids respectively encoding one of the H proteins [H(WT), ΔH(180) and ΔH(158)], the F(T461I) protein, and one of the host proteins (CADM1, CADM2, SLAM, and nectin-4). The results with the DSP assay completely paralleled those with the fusion assay (Fig. 1D). Thus, the deleted part (resides at positions 159 to 180) of the stalk region appears to contain the residue(s) essential for the fusion-triggering function of the H protein in conjunction with CADM1 and CADM2.
Alanine scanning reveals a segment in the H stalk essential for membrane fusion induced by CADM1 and CADM2.
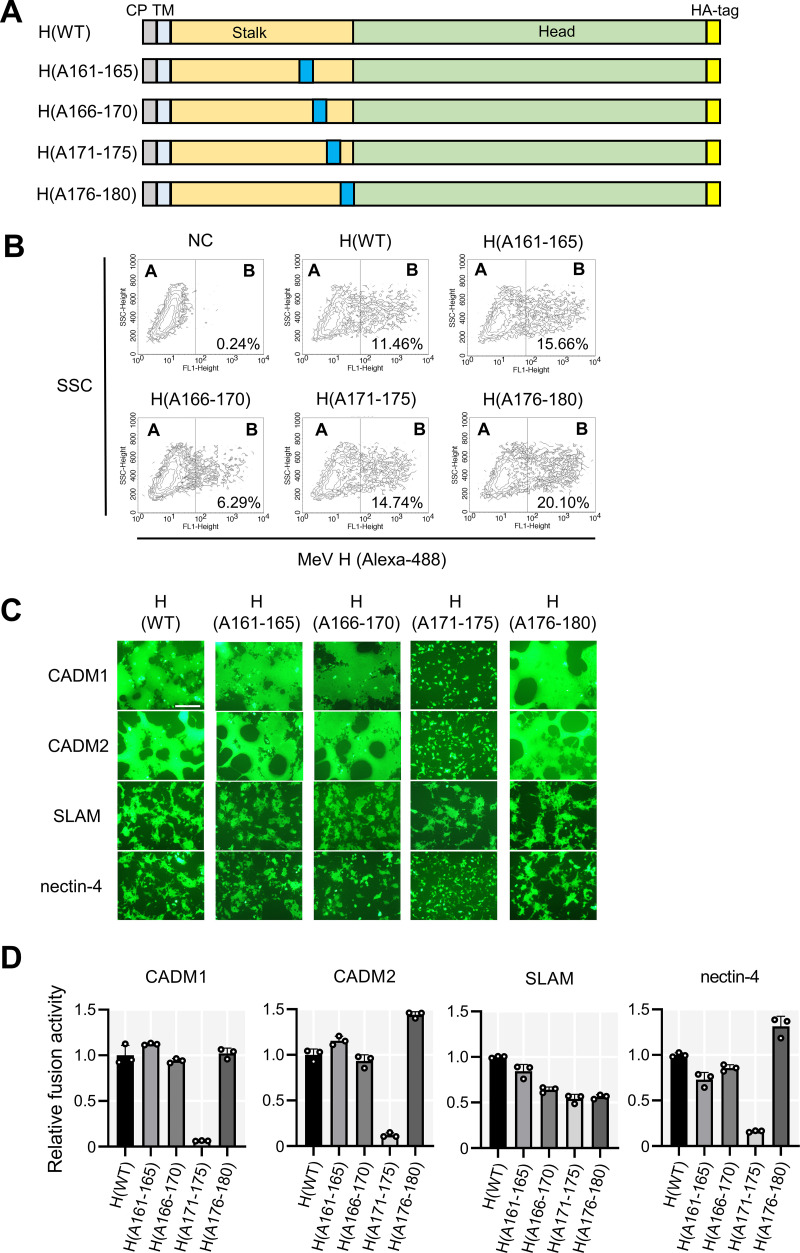
We then attempted to identify the residues in the H stalk region that mediate membrane fusion induced by CADM1 and CADM2. The pCA7 expression plasmids encoding H mutants with alanine substitutions at residues 161 to 165, 166 to 170, 171 to 175, and 176 to 180 [designated H(A161-165), H(A166-170), H(A171-175), and H(A176-180), respectively] were generated (Fig. 2A). Flow cytometry analysis confirmed that these mutant proteins were expressed comparably to the H(WT) protein on the cell surface 24 h after transfection of these expression plasmids into 293FT cells (Fig. 2B).
FIG 2.
Alanine scanning mutagenesis of the H stalk region. (A) Schematic diagrams of the alanine scanning mutants of the H protein. Residues 161 to 165, 166 to 170, 171 to 175, and 176 to 180 were changed to alanines [designated H(A161-165), H(A166-170), H(A171-175), and H(A176-180), respectively]. The area of alanine substitution is indicated by the blue rectangle. CP, cytoplasmic domain; TM, transmembrane domain. (B) The cell surface expression of mutant H proteins. The pCA7 encoding the H(WT) protein, one of the mutant H proteins [H(A161-165), H(A166-170), H(A171-165), and H(A176-180)], or none (NC) was transfected into 293FT cells. Surface expression of the H protein was analyzed by flow cytometry. Percentages of the positive populations in compartment B are shown. SSC, Side-scattered light. (C) The fusion assay with the WT and mutant H proteins. The pCA7 plasmids encoding one of the H proteins [H(WT), H(A161-165), H(A166-170), H(A171-175), and H(A176-180)], the F(T461I) protein, EGFP, and one of the host proteins (CADM1, CADM2, SLAM, and nectin-4) were transfected into 293FT cells. Cells were observed under a fluorescence microscope 30 h after transfection. Scale bar = 200 μm. (D) The DSP assay with the WT and mutant H proteins. The pCA7 plasmids respectively encoding one of the H proteins [H(WT), H(A161-165), H (A166-170), H(A171-175), and H(A176-180)], the F(T461I) protein, and one of the host proteins (CADM1, CADM2, SLAM, and nectin-4) were transfected into cocultured 293FT/DSP1 and 293FT/DSP2 cells. Renilla luciferase activity was measured 48 h after transfection. Luciferase activity of the sample expressing the H(WT) protein and the corresponding host molecule was set to 1.0 (n = 3, means ±SD).
We then performed the fusion assay with the mutant H proteins (Fig. 2C). All the mutant H proteins supported SLAM-induced syncytium formation, indicating that these mutations did not greatly disrupt the overall structure of the H protein. CADM1, CADM2, and nectin-4 also induced membrane fusion supported by the H(WT), H(A161-165), H(A166-170), and H(A176-180) proteins, but not by the H(A171-175) protein. These findings were confirmed by the DSP assay. CADM1, CADM2, and nectin-4, but not SLAM, were unable to induce membrane fusion with the H(171-175) protein (Fig. 2D). The results indicate that the resides at positions 171 to 175 of the H protein stalk region are important for supporting cell-to-cell fusion dependent on CADM1, CADM2, and nectin-4.
CADM1 and CADM2 play an essential role in the MeV spread in neuronal cells.
To investigate whether CADM1- and CADM2-induced membrane fusion is critically involved in MeV spread in neurons and its neurovirulence, we generated the recombinant MeVs possessing the H(WT) or H(A171-175) protein in addition to the hyperfusogenic F(T461I) protein and a fluorescent protein Venus [designated hyperfusogenic(hf)-MeV-H(WT) and hf-MeV-H(A171-175), respectively] by the reverse genetics system as previously reported (31) (Fig. 3A). Since hf-MeV-H(A171-175) is expected to fail to induce CADM1/2-dependent cell-to-cell fusion as well as nectin-4-dependent cell-to-cell fusion, we also prepared the recombinant MeV possessing the H(Y543S) protein, which interacts with nectin-4 only with a low affinity [designated hf-MeV-H(Y543S)] (32) (Fig. 3A). Although human neurons do not express nectin-4 (10), neurons of dogs and mice do (10, 33, 34). Therefore, hf-MeV-H(Y543S) is necessary as a control to evaluate the role of nectin-4 in MeV spread in neurons and its neurovirulence. These recombinant MeVs propagated efficiently in Vero cells stably expressing human SLAM (Vero/hSLAM), ascertaining that all the viruses retain the ability to use SLAM as a receptor. We then examined nectin-4 usage of these viruses for their entry into cells. The numbers of Venus-expressing cells were counted 48 h after infection of 293FT cells transiently expressing human, mouse, or hamster nectin-4 (designated h-, m- and ham-nectin-4, respectively) with the same titers of the viruses. The results showed that hf-MeV-H(WT) and hf-MeV-H(A171-175) could use all types of nectin-4 as entry receptors, while hf-MeV-H(Y543S) could not (Fig. 3B). We then evaluated the ability of these recombinant MeVs to induce syncytium formation in cell lines expressing different MeV receptors. Syncytium formation was observed in Vero/hSLAM cells, nectin-4-positive II-18 cells, and CADM1-positive IMR32 cells infected with hf-MeV-H(WT) (Fig. 3C). In contrast, hf-MeV-H(Y543S) infection induced syncytium formation efficiently in Vero/hSLAM and IMR32 cells, but not in II-18 cells (Fig. 3C). When the cells were infected with hf-MeV-H(A171-175), syncytium formation occurred only in Vero/hSLAM cells, not in II-18 and IMR32 cells (Fig. 3C). These results indicate that whereas the Y543S substitution affects the H protein’s ability to support nectin-4-dependent entry and syncytium formation, alanine substitutions at positions 171 to 175 suppress CADM1/2-dependent and nectin-4-dependent syncytium formation, but not nectin-4-dependent cell entry.
FIG 3.
The Recombinant MeV with the F(T461I) and H(A171-175) proteins does not spread in neurons. (A) Schematic diagrams of recombinant MeVs possessing F (T461I) and one of the H proteins [H(WT), H(A171-175), and H(Y543S)] [designated as hf-MeV-H (WT), hf-MeV-H (A171-75), and hf-MeV-H (Y543S), respectively]. (B) Virus entry assay. The 293FT cells were transfected with pCA7 encoding one of the human, mouse, and hamster nectin-4, or vector plasmid alone (NC), and subsequently infected with respective recombinant MeVs. The number of infected cells was counted 48 h after infection (when syncytium was rarely formed) under a fluorescence microscope, and entry efficiency was expressed as infectious unit (IU)/mL. (C) Syncytium formation in Vero/hSLAM, II-18 (a nectin-4-postiive human lung cancer cell line), and IMR32 (a CADM1-positive human neuroblastoma cell line) cells infected with the recombinant MeVs at an MOI of 0.1. At 72 h after infection, the cells were observed under a fluorescence microscope. Scale bar = 200 μm. (D) Mouse primary neurons were infected with the recombinant MeVs at an MOI of 0.1. At 72 h after infection, the cells were observed under a fluorescence microscope. (E) The percentages of the “single” and “spread” types of infected spots when mouse primary neurons were infected with the recombinant MeVs. Scale bar = 200 μm.
We also examined whether these viruses spread in mouse primary neurons isolated from the cerebral cortex of fetal C57BL/6 mice (E17). hf-MeV-H(WT) and hf-MeV-H(Y543S) spread efficiently in neurons, but hf-MeV-H(A171-175) did not (Fig. 3D). The ability of these viruses to spread in mouse primary neurons was quantitatively expressed as follows. After MeV infection, the “single” type (singly infected cells) and the “spread” type (clusters of multiple infected cells) of infected spots were observed on neurons. When neurons were infected with hf-MeV-H(WT) or hf-MeV-H(Y543S), the percentage of the spread type in infected spots was more than 90%. In contrast, when they were infected with hf-MeV-H(A171-175), the percentage of the spread type in infected spots was less than 10% (Fig. 3E). Taken together, these results clearly indicate that CADM1 and CADM2, but not nectin-4, are involved in MeV spread in mouse primary neurons.
hf-MeV-H(A171-175) lacks neurovirulence.
To evaluate the neurovirulence of these recombinant MeVs, we used a hamster model as previously reported (16). Nine 10-day-old suckling hamsters each were intracerebrally inoculated with hf-MeV-H(WT), hf-MeV-H(A171-175), and hf-MeV-MeV-H(Y543S). All hamsters inoculated with hf-MeV-H(WT) or hf-MeV-H(Y543S) died 5 to 7 days after inoculation (Fig. 4A). In contrast, no hamsters inoculated with hf-MeV-H(A171-175) showed any symptoms for 2 weeks after inoculation (Fig. 4A). In another experiment, the virus-infected brains were collected 1, 3, and 5 days after inoculation and examined under a fluorescence stereomicroscope (Fig. 4B). While hamsters inoculated with hf-MeV-H(WT) or hf-MeV-H(Y543S) showed strong Venus expression throughout their brains at 5 days after inoculation, Venus was not detected in the brains of hamsters inoculated with hf-MeV-H(A171-175). These data indicate that CADM1 and CADM2, but not SLAM and nectin-4, are important for MeV spread in neurons and its neurovirulence in vivo.
FIG 4.
hf-MeV-H(A171-175) lacks neurovirulence. (A) Mortality of hamsters after intracerebral inoculation with the recombinant MeVs. Nine 10-day-old hamsters were infected with 1,500 PFU of hf-MeV-H(WT), hf-MeV-H(A171-175), or hf-MeV-H(Y543S). The hamsters were monitored for 14 days. (B) Spread of the recombinant MeVs in the brains of infected hamsters. The brains of euthanized hamsters at 1, 3, and 5 days after infection were observed under a fluorescence stereomicroscope.
DISCUSSION
The MeV H protein is a type II glycoprotein consisting of an N-terminal cytoplasmic tail, a transmembrane region, a membrane-proximal stalk region, and a C-terminal head domain. The head domain binds to MeV entry receptors, SLAM and nectin-4, and the crystal structures of the H head domain in complex with these receptors are already available (35, 36). On the other hand, the function of the H stalk (residues 59 to 180) has remained unclear, and its crystal structure has not been determined. Recent studies have shown that different sites on the H stalk have different roles in supporting membrane fusion. The central segments of the H stalk (residues 84 to 117) are important for triggering the conformational change of the F protein (37), and the residues 111 to 118 are also involved in the physical association of the H protein with the F protein (38). In addition, Navaratnarajah et al. reported that the deletion of residues 167 to 183 in the H protein abolishes its function in supporting membrane fusion (39). The authors suggested that this segment probably acts as a leash to provide the flexible head movement necessary to induce the fusion-triggering conformational change of the H protein (39). However, the detailed functions of these and other parts of the stalk region are largely unknown. We recently reported that CADM1 and CADM2, cis-acting receptors that enable neuropathogenic MeV spread in neurons (22), interact with the H protein lacking the head domain, presumably through the stalk region (25).
In this study, we show that the residues 171 to 175 of the H protein play an important role in membrane fusion caused by hyperfusogenic F proteins. Unexpectedly, the mutant H protein with alanine substitutions at positions 171 to 175 failed to support membrane fusion induced not only by CADM1 and CADM2, but also by nectin-4 (Fig. 2). At present, we have not been able to further define the critical residue(s) within this segment. What is the function of these five residues? First, they may be directly involved in the interaction with CADM1, CADM2 and nectin-4. Nectin-4 is known to interact with the head domain of the H protein, but it may additionally interact with the stalk region. The substitutions of these five residues could also indirectly affect the interaction of the H protein with CADM1, CADM2, and nectin-4 by altering its conformation. Second, they may be involved in the transfer of the fusion-triggering signal to the F protein. In this case, the property of the fusion-triggering signal must be dependent on the receptors since the H(A171-175) protein can still support SLAM-dependent membrane fusion. Importantly, hf-MeV-H(A171-175) does not efficiently induce nectin-4-dependent syncytium formation but retains the ability to use nectin-4 as an entry receptor. The results suggest that nectin-4-dependent virus-to-cell fusion (entry) and cell-to-cell fusion (syncytium formation) may be regulated differently, as found in other studies (40–43). To fully understand how the H stalk region is involved in the membrane fusion process, we need further functional, biochemical, and structural studies.
Although our previous study suggested using knockdown experiments that CADM1 and CADM2 are necessary to cause MeV transmission in neurons (22), it is unknown whether these molecules are also essential for MeV neurovirulence in vivo. We show here that hf-MeV-H(A171-175) neither spreads in neurons, nor does it exhibit neurovirulence in experimentally infected suckling hamsters. In contrast, hf-MeV-H(WT) and nectin-4 blind hf-MeV-H(Y543S) spread in neurons and exhibit neurovirulence (Fig. 3 and 4). Thus, the ability to cause CADM1- and CADM2-dependent syncytium formation parallels the neurotropism and neurovirulence in vivo of these recombinant MeVs (Table 1). The results indicate that CADM1 and CAMD2, which interact with the H stalk region, are essential for MeV neurovirulence. These findings further our understanding of the molecular mechanism of MeV neuropathogenicity and present the interaction between the H stalk region and CADM1/2 as a potential target for antiviral therapy against SSPE.
TABLE 1.
Properties of the recombinant MeVs used in this study
| Virus | Syncytium formation (cell-to-cell fusion) dependent on |
Spread in neurons | Neurovirulence in hamsters | ||
|---|---|---|---|---|---|
| SLAM | Nectin-4 | CADM1/2 | |||
| hf-MeV-H(WT) | Yes | Yes | Yes | Yes | Yes |
| hf-MeV-H(A171-175) | Yes | Noa | No | No | No |
| hf-MeV-H(Y543S) | Yes | No | Yes | Yes | Yes |
No indicates that nectin-4-dependent syncytium formation, but not entry, is suppressed.
MATERIALS AND METHODS
Cells.
The 293FT cells, Vero cells stably expressing human SLAM (Vero/hSLAM cells) (44), II-18 cells, and IMR-32 cells were maintained in Dulbecco’s modified Eagle medium (DMEM) (FUJIFILM Wako Pure Chemical Corporation) supplemented with 10% fetal bovine serum (FBS). The 293FT cell line is a derivative of 293T and was obtained from Invitrogen. The 293FT cells stably expressing DSP1 and DSP2 (28–30), kindly provided by Z. Matsuda, the University of Tokyo, were maintained in DMEM supplemented with 10% FBS and 1 μg/mL puromycin (InvivoGen). Mouse primary neurons were isolated from the cortex of the C57BL/6 mouse at embryonic day 17 and cultured according to the protocol described previously (22).
Plasmids.
The eukaryotic expression vector pCA7 (45) is a derivative of pCAGGS (46). The pCA7 plasmids respectively encoding H(WT) (the IC-B strain), ΔH(180) [residues 1 to 180 of the H protein with a tetramerization domain (GRMKQIEDKLEEILSKLYHIENELARIKKLLGER) derived from a derivative of the leucine-zipper domain of the yeast GCN4 transcriptional activator (26, 27) at the C-terminus], F(T461I), SLAM (accession no. NM_003037.5), nectin-4 (NM_030916.3), human CADM1 (NM_001098517.2), mouse CADM1 (NM_001025600.1), hamster CADM1 (XM_005069479.4), human CADM2 (NM_001167675.2), mouse CADM2 (NM_178721.4), hamster CADM2 (XM_005082055.4), E-cadherin (NM_004360.5), and EGFP were described previously (16, 22, 24, 47). The synthesized DNAs encoding mouse SLAM (NM_013730.4), hamster SLAM (XM_005078217.3), mouse nectin-4 (NM_027893.3), and hamster nectin-4 (XM_021223926.2), were purchased from Eurofins Genomics K.K. DNA fragments respectively encoding ΔH(158), H(A161-165), H(A166-170), H(A171-175), and H(A176-180) were inserted into pCA7 predigested with EcoRI and NotI. For cell surface expression analysis, the Flag tag (DYKDDDDK) sequences were fused to the C termini of the E-cadherin and EGFP, and the HA-tag (YPYDVPDYA) sequences were fused to the C termini of the H(WT), ΔH(180), and ΔH(158), respectively.
Virus preparation.
Recombinant MeVs were recovered as described previously (31). The viruses were propagated in Vero/hSLAM cells. Cells were then harvested and lysed by three freeze-thaw cycles. Lysates were centrifuged at 2,500 × g for 10 min at 4°C. Supernatants were collected and stored at −80°C. The titer of each recombinant virus (plaque forming unit [PFU] per mL) was determined in Vero/hSLAM cells as described previously (44). Titers of these recombinant viruses were all comparable (2.5-5.0 × 105 PFU/mL).
Fusion assay.
The 293FT cells cultured in 24-well plates were transfected with different combinations of pCA7 plasmids respectively encoding one of the H proteins [H(WT), ΔH(180), ΔH(158), H(A161-165), H(A166-170), H(A171-175), and H(A176-180)] (0.1 μg), the F(T461I) protein (0.1 μg), and EGFP (0.1 μg) with pCA7 encoding one of host molecules (CADM1, CADM2, SLAMF1, and nectin-4) or a vector plasmid alone (0.3 μg) using Lipofectamine LTX (Thermo Fisher Scientific). The cells were observed 30 to 48 h after transfection by fluorescence microscopy.
DSP assay.
DSP1 and DSP2 are split proteins of the Renilla luciferase and GFP which become functional when reassociated with each other after 293FT/DSP1 and 293FT/DSP2 cells are fused. The protocol of the DSP assay was previously described (24). Briefly, pCA7 expression plasmids respectively encoding one of the H proteins [H(WT), ΔH(180), ΔH(158), H(A161-165), H(A166-170), H(A171-175), and H(A176-180)] (0.1 μg) and the F(T461I) proteins (0.1 μg) with pCA7 encoding one of the host molecules (CADM1, CADM2, SLAM, and nectin-4) or pCA7 alone (0.3 μg), were transfected into cocultured 293FT/DSP1 and 293FT/DSP2 cells in 24-well plates using Lipofectamine LTX (Thermo Fisher Scientific). The Renilla luciferase activity was analyzed 24 h after transfection using the Renilla luciferase assay system (Promega).
Virus entry assay.
The 293FT cells cultured in 12-well plates were transfected with pCA7 plasmids encoding human, mouse, or hamster nectin-4 (1 μg) using Lipofectamine LTX (2 μL). The cells were subsequently infected with the hf-MeV-H(WT), the hf-MeV-H(A171-175), or the hf-MeV-H(Y543S) 24 h after transfection. At 48 h after infection, infectious units of the recombinant viruses were determined by the number of Venus-expressing cells under a fluorescence microscope.
Infection of the hamster model with recombinant viruses.
Following procedures previously described in (16), 10-day-old Syrian golden hamsters (SLC-Japan, Shizuoka, Japan) were anesthetized with sevoflurane. Then, 25 μL of diluted viruses were inoculated into the left hemisphere of the brains of hamsters. After the inoculation, clinical symptoms were observed every day and moribund hamsters were euthanized.
Two hamsters inoculated with the hf-MeV-H(WT), the hf-MeV-H(A171-175), or the hf-MeV-H(Y543S) were euthanized at the point of 1 day, 3 days, or 5 days after inoculation, and their brains were then collected. The MeV-infected brains were observed under a fluorescence stereomicroscope. All animal experiments were reviewed by the Institutional Committee of Ethics on Animal Experiments and carried out according to the Guidelines for Animal Experiments of the Faculty of Medicine, Kyushu University, Fukuoka, Japan.
Cell surface biotinylation assay.
Following procedures previously described in (25), subconfluent monolayers of 293FT cells cultured on 12-well plates were transfected with 0.5 μg of pCA7 encoding the HA-tagged H [H(WT), ΔH(180), and ΔH(158)], the Flagged EGFP (an intracellular protein control), or a pCA7 vector alone (a negative control) with 0.5 μg of pCA7 encoding the Flag-tagged E-cadherin (a cell surface protein control) using Lipofectamine LTX. At 24 h after transfection, cells were washed with phosphate-buffered saline (PBS), and then incubated with 200 μL of the biotin reagent solution (2 mM EZ-Link Sulfo-NHS-Biotin [Thermo Scientific] in PBS) for 30 min (min) at 4°C. After washing three times with PBS containing 100 mM glycine for quenching, the cells were lysed in 200 μL of the immunoprecipitation (IP) lysis buffer (Thermo Fisher Scientific) containing the protease inhibitor cocktail (Sigma). The lysates were cleared by centrifugation for 30 min at 17,360 × g and 4°C. Then, 50 μL of each supernatant was mixed with an equal volume of the 2× sodium dodecyl sulfate (SDS) loading buffer [125 mM Tris-HCl (pH 6.8), 10% 2-mercaptoethanol, 4% SDS, 0.1% bromophenol blue, and 20% glycerol], boiled for 5 min, and stocked at −20°C as the cell lysate samples. The rest of the supernatant was incubated with avidin-agarose beads (A-9207, Sigma) for 3 h at 4°C. The samples were centrifuged and washed three times with the IP lysis buffer. Pellets were mixed with 30 μL of the 2× SDS loading buffer, boiled for 5 min, and stocked at −20°C as the biotinylated cell surface protein samples.
Western blotting.
Following procedures previously described in (25), proteins in samples were separated by SDS-polyacrylamide gel electrophoresis, and then blotted onto polyvinylidene difluoride membranes (Hybond-P, Amersham Biosciences). The membranes were incubated with primary Abs (abs) for 1 h. Rabbit polyclonal abs against HA-tag (GTX115044, GeneTex), His-tag (PM 032, MBL), and mouse monoclonal abs against E-cadherin (Clone SHE78-7, TaKaRa Bio), Flag-tag (F1804, Sigma-Aldrich), and β-actin (clone BA3R, BioVision) were used, respectively. The membranes were washed with Tris-buffered saline containing 0.05% Tween 20 (TBS-T) and incubated with horseradish peroxidase-conjugated goat anti-mouse (Bio-Rad) or anti-rabbit IgG ab (Zymed) for 1 h at room temperature. After washing with TBS-T, the membranes were treated with the ECL Plus reagent (Amersham Biosciences), and chemiluminescent signals were detected and imaged using a VersaDoc 5000 imager (Bio-Rad).
Flow cytometry.
Following procedures previously described in (24), pCA7 expression plasmids respectively encoding one of the H proteins [H(WT), H(A161-165), H (A166-170), H(A171-175), and H(A176-180)] (1.0 μg) or pCA7 alone (1.0 μg), were transfected into 293FT cells in 6-well plates using Lipofectamine LTX (Thermo Fisher Scientific). The cells were incubated with human polyclonal antibody against MeV 48 h after transfection, followed by Alexa Fluor 488-conjugated anti-human IgG (Molecular Probes, Inc.), as described (24). The cells were then analyzed on a FACSCalibur HD flow cytometer using BD CellQuest Pro, version 5.2.1, software. Target cell populations were first distinguished from cell debris by forward scatter (FSC) and side scatter (SSC) gating, followed by detection of the MeV H-positive populations.
ACKNOWLEDGMENTS
We thank Z. Matsuda for providing the DSP assay system. We also appreciate the technical assistance from The Research Support Center, Research Center for Human Disease Modeling, Kyushu University Graduate School of Medical Sciences.
This work was supported by JSPS KAKENHI Grant Numbers JP20K07527 (to Y.S.), JP22K06016 (to S.W.), JP20H00507 (to Y.Y.), Takeda Science Foundation (to Y.S.), The Chemo-Sero-Therapeutic Research Institute (to Y.S.), The Uehara Memorial Foundation (to Y.S.), JSPS Core-to-Core Program A grant number JPJSCCA20190008 (to T.H.), and AMED grant number JP22wm0325002h (to T.H.).
Contributor Information
Yuta Shirogane, Email: shirogane.yuta.528@m.kyushu-u.ac.jp.
Rebecca Ellis Dutch, University of Kentucky College of Medicine.
REFERENCES
- 1.Griffin DE. 2013. Measles virus, p 1042–1069. In Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Coughlin MM, Beck AS, Bankamp B, Rota PA. 2017. Perspective on global measles epidemiology and control and the role of novel vaccination strategies. Viruses 9:11. doi: 10.3390/v9010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tatsuo H, Ono N, Tanaka K, Yanagi Y. 2000. Slam (CDw150) is a cellular receptor for measles virus. Nature 406:893–897. doi: 10.1038/35022579. [DOI] [PubMed] [Google Scholar]
- 4.Tahara M, Takeda M, Shirogane Y, Hashiguchi T, Ohno S, Yanagi Y. 2008. Measles virus infects both polarized epithelial and immune cells by using distinctive receptor-binding sites on its hemagglutinin. J Virol 82:4630–4637. doi: 10.1128/JVI.02691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noyce RS, Bondre DG, Ha MN, Lin LT, Sisson G, Tsao MS, Richardson CD. 2011. Tumor cell marker pvrl4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog 7:e1002240. doi: 10.1371/journal.ppat.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mühlebach MD, Mateo M, Sinn PL, Prüfer S, Uhlig KM, Leonard VHJ, Navaratnarajah CK, Frenzke M, Wong XX, Sawatsky B, Ramachandran S, McCray PB, Cichutek K, Von Messling V, Lopez M, Cattaneo R. 2011. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature 480:530–533. doi: 10.1038/nature10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellini WJ, Rota JS, Lowe LE, Katz RS, Dyken PR, Zaki SR, Shieh WJ, Rota PA. 2005. Subacute sclerosing panencephalitis: more cases of this fatal disease are prevented by measles immunization than was previously recognized. J Infect Dis 192:1686–1693. doi: 10.1086/497169. [DOI] [PubMed] [Google Scholar]
- 8.Mekki M, Eley B, Hardie D, Wilmshurst J. 2019. Subacute sclerosing panencephalitis: clinical phenotype, epidemiology, and preventive interventions. Dev Med Child Neurol 61:1139–1144. doi: 10.1111/dmcn.14166. [DOI] [PubMed] [Google Scholar]
- 9.McQuaid S, Cosby SL. 2002. An immunohistochemical study of the distribution of the measles virus receptors, CD46 and SLAM, in normal human tissues and subacute sclerosing panencephalitis. Lab Invest 82:403–409. doi: 10.1038/labinvest.3780434. [DOI] [PubMed] [Google Scholar]
- 10.Reymond N, Fabre S, Lecocq E, Adelaide J, Dubreuil P, Lopez M. 2001. Nectin4/PRR4, a new afadin-associated member of the nectin family that trans-interacts with Nectin1/PRR1 through V domain interaction. J Biol Chem 276:43205–43215. doi: 10.1074/jbc.M103810200. [DOI] [PubMed] [Google Scholar]
- 11.Iwasaki Y, Koprowski H. 1974. Cell to cell transmission of virus in the central nervous system. I. Subacute sclerosing panencephalitis. Lab Invest 31:187–196. [PubMed] [Google Scholar]
- 12.Allen IV, McQuaid S, McMahon J, Kirk J, McConnell R. 1996. The significance of measles virus antigen and genome distribution in the CNS in SSPE for mechanisms of viral spread and demyelination. J Neuropathol Exp Neurol 55:471–480. doi: 10.1097/00005072-199604000-00010. [DOI] [PubMed] [Google Scholar]
- 13.Lawrence DM, Patterson CE, Gales TL, D'Orazio JL, Vaughn MM, Rall GF. 2000. Measles virus spread between neurons requires cell contact but not CD46 expression, syncytium formation, or extracellular virus production. J Virol 74:1908–1918. doi: 10.1128/jvi.74.4.1908-1918.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ayata M, Takeuchi K, Takeda M, Ohgimoto S, Kato S, Sharma LB, Tanaka M, Kuwamura M, Ishida H, Ogura H. 2010. The F gene of the Osaka-2 strain of measles virus derived from a case of subacute sclerosing panencephalitis is a major determinant of neurovirulence. J Virol 84:11189–11199. doi: 10.1128/JVI.01075-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shirogane Y, Watanabe S, Yanagi Y. 2012. Cooperation between different RNA virus genomes produces a new phenotype. Nat Commun 3:1235. doi: 10.1038/ncomms2252. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe S, Shirogane Y, Suzuki SO, Ikegame S, Koga R, Yanagi Y. 2013. Mutant fusion proteins with enhanced fusion activity promote measles virus spread in human neuronal cells and brains of suckling hamsters. J Virol 87:2648–2659. doi: 10.1128/JVI.02632-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe S, Ohno S, Shirogane Y, Suzuki SO, Koga R, Yanagi Y. 2015. Measles virus mutants possessing the fusion protein with enhanced fusion activity spread effectively in neuronal cells, but not in other cells, without causing strong cytopathology. J Virol 89:2710–2717. doi: 10.1128/JVI.03346-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jurgens EM, Mathieu C, Palermo LM, Hardie D, Horvat B, Moscona A, Porotto M. 2015. Measles fusion machinery is dysregulated in neuropathogenic variants. mBio 6:e02528-14. doi: 10.1128/mBio.02528-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato Y, Watanabe S, Fukuda Y, Hashiguchi T, Yanagi Y, Ohno S. 2018. Cell-to-cell measles virus spread between human neurons is dependent on hemagglutinin and hyperfusogenic fusion protein. J Virol 92:e02166-17. doi: 10.1128/JVI.02166-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watanabe S, Shirogane Y, Sato Y, Hashiguchi T, Yanagi Y. 2019. New Insights into measles virus brain infections. Trends Microbiol 27:164–175. doi: 10.1016/j.tim.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 21.Angius F, Smuts H, Rybkina K, Stelitano D, Eley B, Wilmshurst J, Ferren M, Lalande A, Mathieu C, Moscona A, Horvat B, Hashiguchi T, Porotto M, Hardie D. 2019. Analysis of a subacute sclerosing panencephalitis genotype B3 virus from the 2009–2010 South African measles epidemic shows that hyperfusogenic F proteins contribute to measles virus infection in the brain. J Virol 93:e01700-18. doi: 10.1128/JVI.01700-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shirogane Y, Takemoto R, Suzuki T, Kameda T, Nakashima K, Hashiguchi T, Yanagi Y. 2021. CADM1 and CADM2 triggers neuropathogenic measles virus-mediated membrane fusion by acting in cis. J Virol 95:e0052821. doi: 10.1128/JVI.00528-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shirogane Y, Harada H, Hirai Y, Takemoto R, Suzuki T, Hashiguchi T, Yanagi Y. 2023. Collective fusion activity determines neurotropism of an en bloc transmitted enveloped virus. Sci Adv 9:eadf3731. doi: 10.1126/sciadv.adf3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shirogane Y, Hashiguchi T, Yanagi Y. 2020. Weak cis and trans interactions of the hemagglutinin with receptors trigger fusion proteins of neuropathogenic measles virus isolates. J Virol 94:e01727-19. doi: 10.1128/JVI.01727-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takemoto R, Suzuki T, Hashiguchi T, Yanagi Y, Shirogane Y. 2022. Short-stalk isoforms of CADM1 and CADM2 trigger neuropathogenic measles virus-mediated membrane fusion by interacting with the viral hemagglutinin. J Virol 96:e0194921. doi: 10.1128/JVI.01949-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brindley MA, Suter R, Schestak I, Kiss G, Wright ER, Plemper RK. 2013. A stabilized headless measles virus attachment protein stalk efficiently triggers membrane fusion. J Virol 87:11693–11703. doi: 10.1128/JVI.01945-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harbury PB, Zhang T, Kim PS, Alber T. 1993. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 28.Kondo N, Miyauchi K, Meng F, Iwamoto A, Matsuda Z. 2010. Conformational changes of the HIV-1 envelope protein during membrane fusion are inhibited by the replacement of its membrane-spanning domain. J Biol Chem 285:14681–14688. doi: 10.1074/jbc.M109.067090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishikawa H, Meng F, Kondo N, Iwamoto A, Matsuda Z. 2012. Generation of a dual-functional split-reporter protein for monitoring membrane fusion using self-associating split GFP. Protein Eng Des Sel 25:813–820. doi: 10.1093/protein/gzs051. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, Li X, Nakane S, Liu S, Ishikawa H, Iwamoto A, Matsuda Z. 2014. Co-expression of foreign proteins tethered to HIV-1 envelope glycoprotein on the cell surface by introducing an intervening second membrane-spanning domain. PLoS One 9:e96790. doi: 10.1371/journal.pone.0096790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seki F, Yamada K, Nakatsu Y, Okamura K, Yanagi Y, Nakayama T, Komase K, Takeda M. 2011. The SI strain of measles virus derived from a patient with subacute sclerosing panencephalitis possesses typical genome alterations and unique amino acid changes that modulate receptor specificity and reduce membrane fusion activity. J Virol 85:11871–11882. doi: 10.1128/JVI.05067-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mateo M, Navaratnarajah CK, Syed S, Cattaneo R. 2013. The measles virus hemagglutinin β-propeller head β4-β5 hydrophobic groove governs functional interactions with nectin-4 and CD46 but not those with the signaling lymphocytic activation molecule. J Virol 87:9208–9216. doi: 10.1128/JVI.01210-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pratakpiriya W, Seki F, Otsuki N, Sakai K, Fukuhara H, Katamoto H, Hirai T, Maenaka K, Techangamsuwan S, Lan NT, Takeda M, Yamaguchi R. 2012. Nectin4 is an epithelial cell receptor for canine distemper virus and involved in neurovirulence. J Virol 86:10207–10210. doi: 10.1128/JVI.00824-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pratakpiriya W, Ping Teh AP, Radtanakatikanon A, Pirarat N, Thi Lan N, Takeda M, Techangamsuwan S, Yamaguchi R. 2017. Expression of canine distemper virus receptor nectin-4 in the central nervous system of dogs. Sci Rep 7:349. doi: 10.1038/s41598-017-00375-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hashiguchi T, Ose T, Kubota M, Maita N, Kamishikiryo J, Maenaka K, Yanagi Y. 2011. Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat Struct Mol Biol 18:135–141. doi: 10.1038/nsmb.1969. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, Lu G, Qi J, Li Y, He Y, Xu X, Shi J, Zhang CWH, Yan J, Gao GF. 2013. Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4. Nat Struct Mol Biol 20:67–72. doi: 10.1038/nsmb.2432. [DOI] [PubMed] [Google Scholar]
- 37.Navaratnarajah CK, Kumar S, Generous A, Apte-Sengupta S, Mateo M, Cattaneo R. 2014. The measles virus hemagglutinin stalk: structures and functions of the central fusion activation and membrane-proximal segments. J Virol 88:6158–6167. doi: 10.1128/JVI.02846-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brindley MA, Takeda M, Plattet P, Plemper RK. 2012. Triggering the measles virus membrane fusion machinery. Proc Natl Acad Sci USA 109:E3018–E3027. doi: 10.1073/pnas.1210925109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Navaratnarajah CK, Rosemarie Q, Cattaneo R. 2016. A structurally unresolved head segment of defined length favors proper measles virus hemagglutinin tetramerization and efficient membrane fusion triggering. J Virol 90:68–75. doi: 10.1128/JVI.02253-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hasegawa K, Hu C, Nakamura T, Marks JD, Russell SJ, Peng K-W. 2007. Affinity thresholds for membrane fusion triggering by viral glycoproteins. J Virol 81:13149–13157. doi: 10.1128/JVI.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kondo N, Marin M, Kim JH, Desai TM, Melikyan GB. 2015. Distinct requirements for HIV-cell fusion and HIV-mediated cell-cell fusion. J Biol Chem 290:6558–6573. doi: 10.1074/jbc.M114.623181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto M, Du Q, Song J, Wang H, Watanabe A, Tanaka Y, Kawaguchi Y, Inoue JI, Matsuda Z. 2019. Cell-cell and virus-cell fusion assay-based analyses of alanine insertion mutants in the distal α9 portion of the JRFL gp41 subunit from HIV-1. J Biol Chem 294:5677–5687. doi: 10.1074/jbc.RA118.004579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fukui A, Maruzuru Y, Takeshima K, Koyanagi N, Kato A, Kawaguchi Y. 2023. Establishment of a system to quantify wild-type herpes simplex virus-induced cell-cell fusion reveals a role of N-glycosylation of HSV-1 envelope glycoprotein B in cell-cell fusion. Microbiol Immunol 67:114–119. doi: 10.1111/1348-0421.13050. [DOI] [PubMed] [Google Scholar]
- 44.Ono N, Tatsuo H, Hidaka Y, Aoki T, Minagawa H, Yanagi Y. 2001. Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J Virol 75:4399–4401. doi: 10.1128/JVI.75.9.4399-4401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeda M, Ohno S, Seki F, Nakatsu Y, Tahara M, Yanagi Y. 2005. Long untranslated regions of the measles virus M and F genes control virus replication and cytopathogenicity. J Virol 79:14346–14354. doi: 10.1128/JVI.79.22.14346-14354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 47.Tahara M, Takeda M, Yanagi Y. 2007. Altered interaction of the matrix protein with the cytoplasmic tail of hemagglutinin modulates measles virus growth by affecting virus assembly and cell-cell fusion. J Virol 81:6827–6836. doi: 10.1128/JVI.00248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]