ABSTRACT
Several recent studies indicate that mutations in the human immunodeficiency virus type 1 (HIV-1) 3′polypurine tract (3′PPT) motif can reduce sensitivity to the integrase inhibitor dolutegravir (DTG). Using an in vivo systematic evolution of ligands by exponential enrichment (SELEX) approach, we discovered that multiple different mutations in this viral RNA element can confer DTG resistance, suggesting that the inactivation of this critical reverse transcription element causes resistance. An analysis of the viral DNA products formed upon infection by these 3′PPT mutants revealed that they replicate without integration into the host cell genome, concomitant with an increased production of 1-LTR circles. As the replication of these virus variants is activated by the human T-lymphotropic virus 1 (HTLV-1) Tax protein, a factor that reverses epigenetic silencing of episomal HIV DNA, these data indicate that the 3′PPT-mutated viruses escape from the integrase inhibitor DTG by switching to an integration-independent replication mechanism.
IMPORTANCE The integrase inhibitor DTG is a potent inhibitor of HIV replication and is currently recommended in drug regimens for people living with HIV. Whereas HIV normally escapes from antiviral drugs by the acquisition of specific mutations in the gene that encodes the targeted enzyme, mutational inactivation of the viral 3′PPT sequence, an RNA element that has a crucial role in the viral reverse transcription process, was found to allow HIV replication in the presence of DTG in cell culture experiments. While the integration of the viral DNA into the cellular genome is considered one of the hallmarks of retroviruses, including HIV, 3′PPT inactivation caused integration-independent replication, which can explain the reduced DTG sensitivity. Whether this exotic escape route can also contribute to viral escape in HIV-infected persons remains to be determined, but our results indicate that screening for 3′PPT mutations in patients that fail on DTG therapy should be considered.
KEYWORDS: antiretroviral agents, dolutegravir, drug resistance mechanisms, human immunodeficiency virus, retroviruses
INTRODUCTION
Integration of the reverse-transcribed viral DNA in the cellular DNA is considered an essential step in the replication cycle of all retroviruses, including human immunodeficiency virus type 1 (HIV-1). This step is executed by the virus-encoded integrase protein, and several antiretroviral drugs targeting integrase are potent inhibitors of virus replication. The integrase strand-transfer inhibitor (INSTI) dolutegravir (DTG) is currently recommended in first-line and second-line drug regimens for people living with HIV (1, 2). Although DTG has a high genetic barrier for resistance, integrase mutations have been linked to DTG resistance (3). More recently, in vitro and in vivo studies linked mutations in the viral 3′-polypurine tract (3′PPT) to DTG resistance (4, 5). Malet et al. (4) selected a DTG-resistant HIV variant with mutations in the 3′PPT region by long-term virus passage in the presence of DTG. Wijting et al. (5) observed 3′PPT mutations in a patient that failed on DTG monotherapy, although others reported that these 3′PPT mutations are not sufficient to confer DTG resistance in a phenotypic drug-resistance assay (6, 7). We and others confirmed that mutations in the 3′PPT region can indeed reduce DTG sensitivity (8, 9). However, these mutations also significantly reduce viral fitness, and we demonstrated that replication of 3′PPT-mutated HIV variants can be activated by the HTLV-1 Tax protein that was previously shown to reverse epigenetic silencing of unintegrated HIV DNA (8, 10, 11).
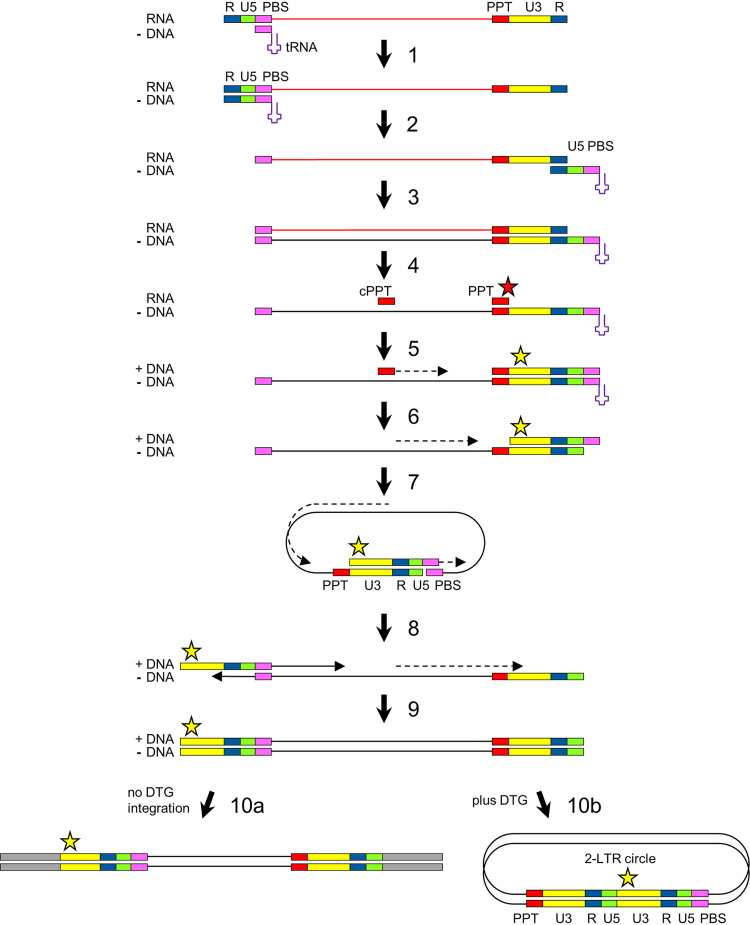
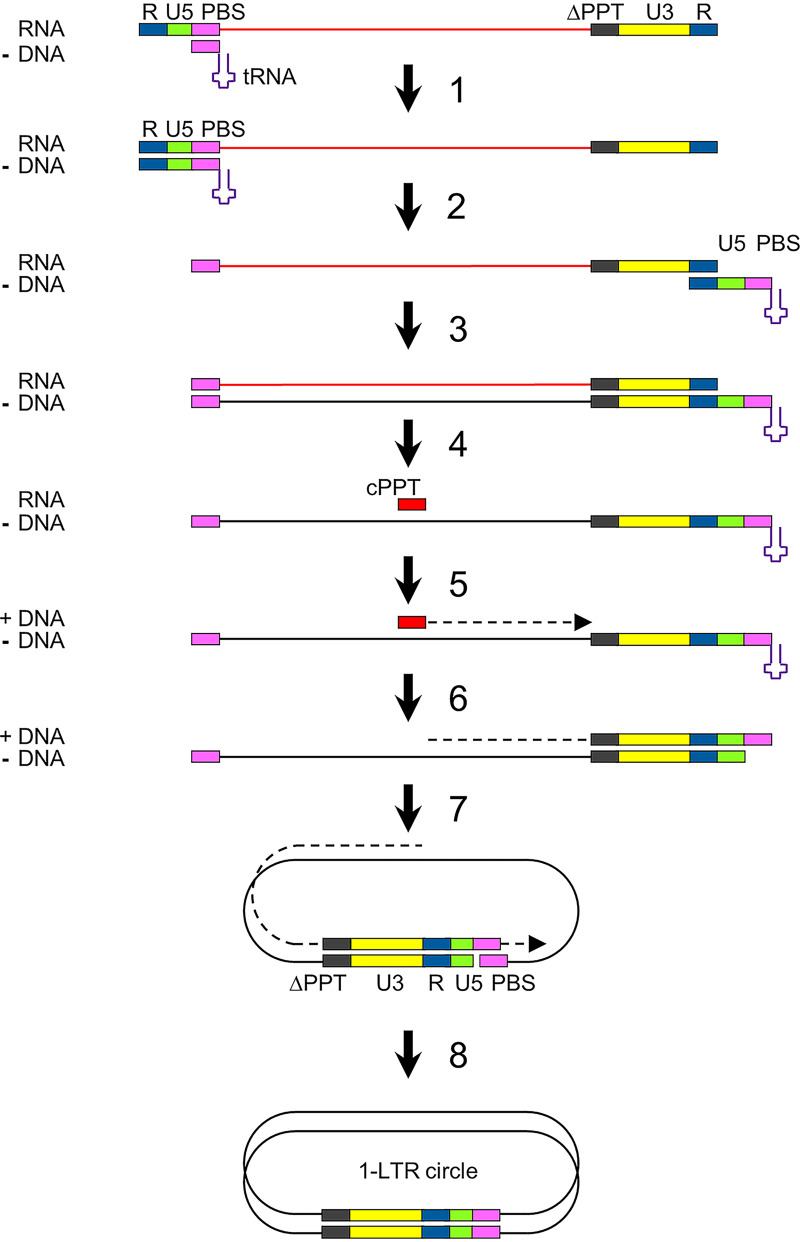
Upon infection of a cell by HIV, the viral RNA genome is copied into double-stranded DNA (dsDNA) by the viral reverse transcriptase (Fig. 1). Following minus-strand DNA synthesis, the viral RNA template is degraded by the RNase H activity of reverse transcriptase, but the 3′PPT and central PPT (cPPT) motifs (5′-AAAAGAAAAGGGGGG-3′) resist cleavage (step 4) and the short leftover RNA molecules function as primers for plus-strand DNA synthesis (step 5) (12–14). Complementarity of the sequences at the termini of the partially double-stranded linear HIV DNA induces circularization to form 1-LTR circles (step 7). The subsequent continuation of second-strand DNA synthesis and linearization, which involves strand displacement of the LTR sequences, results in a linear dsDNA molecule with complete LTR motifs at both termini (steps 8 to 9) (15). The 3′ terminus of the 3′PPT primer (indicated with a red star in Fig. 1) thereby determines the 5′ end of the dsDNA product (indicated with yellow star), which—together with the 3′ end of the dsDNA—forms the substrate for integration into the host cell DNA by the HIV integrase (step 10a) (16). Binding of DTG to the dsDNA-integrase complex prevents integration, which results in an increased production of 2-LTR circles (step 10b) (17). The 3′PPT mutations linked to DTG resistance are mostly located in the 3′ terminal G6-motif (Fig. 2A), and mutations in this motif have been described to affect RNase H cleavage and thereby the formation of the 5′ end of the HIV dsDNA (18, 19).
FIG 1.
HIV reverse transcription. Upon infection, the viral positive-strand RNA genome is copied into double-stranded DNA (dsDNA) by the viral reverse transcriptase. The cellular tRNALys3 binding to the primer binding site (PBS) functions as a primer for minus-strand DNA synthesis (step 1). After being reverse transcribed, the viral RNA template is degraded by the RNase H activity of reverse transcriptase, but the 3′PPT (indicated with PPT) and central PPT (cPPT) motifs resist cleavage and function as a primer for plus-strand DNA synthesis (steps 4 and 5). Complementarity of the sequences at the termini of the partially double-stranded linear HIV DNA induces circularization to form 1-LTR circles (step 7). Subsequent continuation of second-strand DNA synthesis and linearization, which involves strand displacement of the LTR sequences, results in a linear dsDNA molecule with complete LTR motifs at both termini (steps 8 and 9). This full-length 2-LTR dsDNA is integrated into the cellular DNA by the viral integrase protein (step 10a; no DTG). When integration is blocked, for example in the presence of DTG, 2-LTR circles accumulate (step 10b). The red star indicates the 3′ end of the 3′PPT RNA fragment, and the yellow star indicates the 5′ end of the 5’LTR in the 2-LTR dsDNA products.
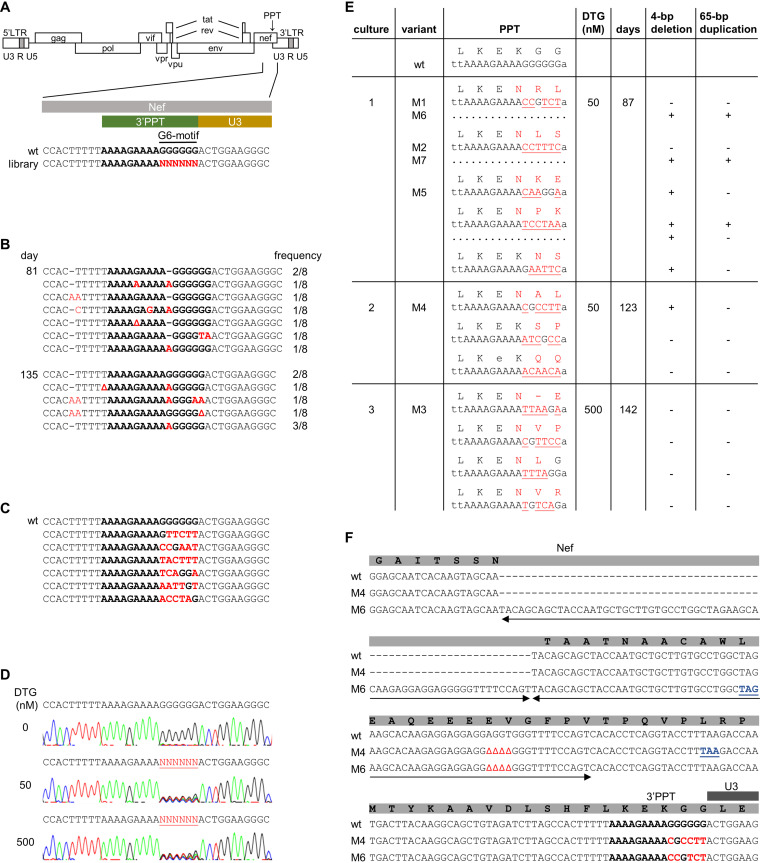
FIG 2.
Selection of DTG-resistance mutations in the HIV 3’PPT. (A) Schematic of the HIV genome and nucleotide sequence of the 3′PPT region. The 3′PPT nucleotide sequence is indicated in bold. The G6-motif that is randomized in the SELEX library is indicated in red. (B) 3′PPT-mutations identified upon long-term culturing of HIV with DTG on SupT1 T cells. After 81 and 135 days of virus passage, intracellular DNA was isolated and the HIV 3′PPT region was amplified by PCR, followed by sequencing of 8 individual PCR clones. The frequency by which each 3′PPT sequence was found is indicated (Δ, nucleotide deletion; dashes are used to align sequences). The 3′PPT nucleotide sequence is indicated in bold. Nucleotide substitutions are in red. (C) The 3′PPT sequence in individual plasmid clones from the full-length HIV-1 library in which the G6-motif was randomized. The wt sequence is shown on top for comparison. The 3′PPT nucleotide sequence is indicated in bold. Nucleotide substitutions are in red. (D) C8166 T cells were infected with the HIV-1 library and cultured with 0, 50, or 500 nM DTG. Upon syncytium formation (at day 7 without DTG and day 14 with DTG), the 3′PPT region of the virus population was PCR amplified and analyzed by sequencing. The randomized G6-motif is indicated in red and underlined. (E) Sequence analysis of the 3′PPT/nef region upon long-term culturing of the HIV-1 library with 50 (cultures 1 and 2) or 500 nM DTG (culture 3). Intracellular viral DNA was isolated from the infected C8166 cells at the indicated day, followed by PCR amplification of the 3′PPT/nef region and ligation of the PCR product into a TOPO TA cloning vector. Several TA clones were sequenced. The observed 3'PPT nucleotide sequences with their amino acid translation of the overlapping nef open reading frame are shown. Nucleotide substitutions are in red and underlined. Amino acid substitutions are in red. When indicated, a 4-bp deletion and 65-bp duplication (sequence shown in F) was detected in the upstream nef region. The variants indicated with M1-M7 were cloned into the full-length HIV-1 LAI construct for further analysis. (F) Sequence alignment of the 3′PPT/nef region in wt HIV and the M4 and M6 variants. The 3′PPT nucleotide sequence is indicated in bold. Nucleotide substitutions are indicated in red, deletions are indicated by Δ, and the duplicated sequence is indicated with two-sided arrows. The deletion and duplication result in a premature stop codon in the nef open reading frame (in blue and underlined).
Here, we describe the use of the powerful in vivo systematic evolution of ligands by exponential enrichment (SELEX) method to identify novel mutations in the 3′PPT that confer DTG resistance and investigated the resistance mechanism by analyzing the viral DNA products formed upon the infection of cells. Our data suggest that the DTG-resistant viruses switched to an integration-independent, episomal replication mode, thus elegantly explaining the resistance phenotype.
RESULTS
To identify mutations that can confer DTG resistance, we initially cultured the HIV-1 subtype B LAI strain, a primary CXCR4-using virus isolate (20), on SupT1 T cells in the presence of high DTG levels. When massive virus-induced syncytia were observed, the virus-containing culture medium was passaged onto fresh SupT1 cells. We repeated this procedure to extend the culturing time and thus the time for virus evolution. Successful passaging to fresh cells required a relatively large culture volume (100 μL) as we frequently lost the replicating virus with smaller volumes, indicating no or poor DTG resistance development. After 81 days of virus passage, intracellular DNA was isolated and the HIV 3′PPT region was amplified by PCR, followed by sequencing of several individual PCR clones (Fig. 2B). This analysis revealed the frequent presence of point mutations in the 3′PPT and occasionally in the upstream T-stretch, but different PCR clones carried distinct mutations and the wild-type (wt) sequence was usually still present at a low frequency. Similar results were obtained at day 135 of the HIV evolution experiment. An analysis of the HIV integrase coding region did not reveal any known DTG resistance-associated mutations (RAMs) or new candidates (results not shown). Overall, we were underwhelmed by these initial results. Although fluctuation of the 3′PPT was readily observed, we did not see the outgrowth of a particular mutant clone that outcompeted the wt virus.
As an alternative approach to identify 3′PPT mutations that confer DTG resistance, we used in vivo systematic evolution of ligands by exponential enrichment (SELEX), a method that we developed to probe the sequence requirements of several HIV RNA domains (21, 22). A full-length HIV-1 DNA genome library based on the pLAI plasmid was constructed in which the 3′PPT G6-motif was randomized (Fig. 2A), theoretically yielding a library of 4,096 virus variants. To obtain a large and diverse collection, bacterial transformation of the plasmids was scaled up until approximately 7,400 transformants were obtained. Multiple individual clones were sequenced to confirm sequence diversity in the G6-motif and the absence of a predominant sequence (Fig. 2C). All transformed bacteria were pooled for DNA isolation. The resulting HIV DNA library was transfected into 293T cells to produce a virus stock that was used to infect C8166 cells. The C8166 cell line is an HTLV-1-transformed T cell line that is frequently used in HIV replication studies because it supports efficient virus replication (23, 24). Infected cells were cultured in the absence or presence of DTG. When massive syncytia were observed, intracellular DNA was isolated and the 3′PPT region was amplified by PCR and analyzed by sequencing (population sequence). Analysis of the 3′PPT after culturing without DTG for 8 days indicated the rapid outgrowth of the wt virus (Fig. 2D) (5′-AAAAGAAAAGGGGGG-3′), which unequivocally demonstrates the importance of the 3′PPT motif for efficient HIV replication, consistent with previous studies (13, 18). The analysis upon culturing with 50 or 500 nM DTG for 14 days revealed a mixed 3′PPT sequence with variable nucleotides in the randomized part (Fig. 2D) (5′-AAAAGAAAANNNNNN-3′). The 3′PPT sequence remained mixed upon prolonged culturing for up to 142 days (results not shown). The subsequent analysis of individual PCR fragments confirmed the presence of multiple sequence variants (Fig. 2E). As the 3′PPT sequence overlaps with the nef open reading frame, most 3′PPT mutations cause amino acid substitutions in the Nef protein (Fig. 2E). Furthermore, we observed a 4-bp deletion and a 65-bp duplication in the upstream nef region in the 50 nM DTG culture. These modifications create a premature stop codon in the nef open reading frame (Fig. 2F). Nef is an accessory viral factor that is important for in vivo HIV replication but not for in vitro replication on transformed T cell lines, such as the C8166 cells used in this experiment (25). For a control, we PCR amplified and sequenced the HIV integrase coding region, which did not reveal any additional changes.
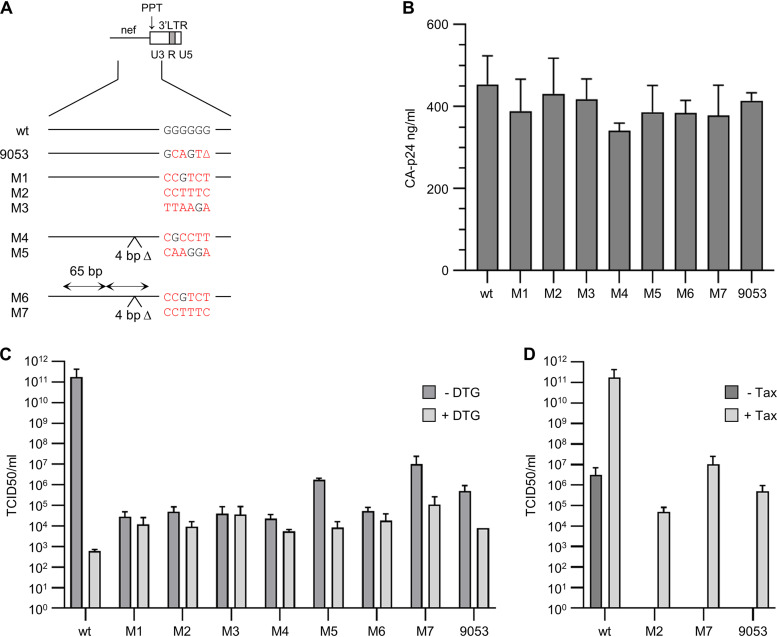
To demonstrate that the 3′PPT/nef mutations confer DTG resistance, HIV LAI molecular clones were generated for some of the selected variants without (M1 to M3) or with the additional 4-bp deletion (M4 and M5) plus 65-bp duplication (M6 and M7) (Fig. 3A). We reported previously that the M1, M2, and M3 mutations do indeed reduce DTG sensitivity in a single-cycle infection assay but also significantly reduce viral fitness (8). These variants were found to replicate in C8166 cells and in genetically modified HTLV-1 Tax-expressing CEM-SS T cells but not in CEM-SS cells that did not express Tax (8, 10). Here, we set out to compare the replication capacity of the M1 to M3 variants and the newly generated M4 to M7 variants. The molecular clones were transfected into 293T cells to produce virus stocks. All variants showed a similar level of viral CA-p24 production in the culture medium as the wt construct, which demonstrates that the 3′PPT/nef changes do not affect viral gene expression and virus production (Fig. 3B). Tax-expressing CEM-SS cells were infected with serial dilutions of the virus stocks. For comparison, we included an HIV LAI variant with the 3′PPT mutations identified by Malet et al. (4) (9053 mutant). After culturing the infected cells without or with 500 nM DTG for up to 10 days, we calculated the median tissue culture infectious dose (TCID50) for each variant (Fig. 3C). In the absence of DTG, all 3′PPT variants demonstrated significantly reduced viral infectivity compared with the wt virus (4 to 7 log reduction). Whereas DTG administration reduced the infectivity of the wt virus by more than 8 log, the infectivity of the 3′PPT variants was not or was significantly less reduced (up to 2 log). In fact, the infectivity of the mutants was 1 or 2 log higher than that of the wt virus in the presence of DTG. Although the infectivity and DTG sensitivity varied for the different variants, no obvious differences could be observed for the HIV variants with or without the 4-bp deletion and 65-bp duplication. For some virus variants (wt, M2, M7, and 9053), the TCID50 was also determined upon infection of CEM-SS cells that did not express Tax (Fig. 3D). Whereas the wt virus did replicate in the absence of Tax, none of the 3′PPT mutants showed a detectable level of replication (no virus-induced syncytium formation or cell death), which is in agreement with our previous observation that Tax activates the replication of the 3′PPT mutants (8).
FIG 3.
Production and replication of 3′PPT-mutated viruses. (A) Mutations in the 3′PPT/nef region of different HIV-1 pLAI molecular clones. The mutations M1 to M7 were selected in the SELEX experiment (Fig. 2). Nucleotide substitutions in the G6-motif of the different variants are in red. Nucleotide substitutions in the G6-motif of the different variants are in red. The 4-bp deletion in variants M4-M7 (4 bp Δ) and 65-bp duplication in the upstream nef region in variants M6-M7 (65 bp) are indicated. The 9053 variant corresponds to the 3′PPT mutant that was earlier described by Malet et al. (4). In addition to the indicated mutations in the G6-motif, this variant has a C-to-T substitution immediately upstream of the 3’PPT. (B) HEK293T cells were transfected with the HIV molecular clones, and the CA-p24 level in the culture supernatant was measured at 2 days after transfection. Average values obtained in 2 to 4 experiments are shown, with the error bars indicating the standard deviation. Statistical analysis using one-way analysis of variance (ANOVA) demonstrated that the CA-p24 production of the different variants did not differ significantly (P > 0.05). (C and D) Equal amounts of the different viruses, based on the viral CA-p24 protein, were serially diluted and used to infect CEM-SS cells with a doxycycline-inducible HTLV-1 tax gene (10). Viral replication was scored based on the virus-induced syncytium formation at 10 days after infection. Average TCID50 values obtained in two experiments are shown, with the error bars indicating the standard deviation. (C) The infected cells were cultured in the presence of 0.5 μg/mL doxycycline to induce Tax expression, and either in the absence or presence of 500 nM DTG to analyze the DTG sensitivity of the different variants. (D) The infected cells were cultured without doxycycline (−Tax) or with 0.5 μg/mL doxycycline (+Tax) to analyze the effect of Tax on virus replication (+Tax values in D are identical to the −DTG values in C). Statistical analysis using one-way ANOVA demonstrated that the TCID50 values obtained for the different mutants did not differ significantly (P > 0.05).
These results demonstrate that all tested 3’PPT mutations reduce HIV replication in the absence of DTG and improve replication in the presence of DTG, but we did not observe an obvious effect of the additional mutations in the nef region. We therefore set up very sensitive virus competition assays that are ideally suited to detect small replication differences in an internally controlled manner (26). We compared variants with an identical 3′PPT variation but without (M1 and M2) and with (M6 and M7) the upstream modifications in nef (Fig. 4A). C8166 cells were infected with a virus mixture (M1+M6 or M2+M7) and cultured without or with 500 nM DTG. Total intracellular DNA was isolated at 1 and 8 days after infection, and the produced viral DNA was analyzed by PCR amplification of the nef-3′PPT-U3 region (PPT PCR, Fig. 4A) and gel electrophoresis. Two differently sized PCR products, corresponding to the two input viruses, were detectable at 1 day after infection, but the smaller fragment, corresponding to the M1 or M2 variant, was slightly more abundant than the larger fragment, corresponding to the M6 or M7 variant, both without and with DTG (Fig. 4B). This difference in intensity is possibly due to the more efficient PCR amplification of smaller DNA fragments. Importantly, at 8 days postinfection, the ratio between the fragments had changed and the larger M6 and M7 products were more abundant than the smaller M1 and M2 products. This shift in ratio indicates that the M6 and M7 variants replicate more efficiently than the M1 and M2 variants, respectively. These results thus suggest that the additional nef modifications improve replication of the 3′PPT mutants, both in the absence and presence of DTG. Variants M6 and M7 were therefore used for further analysis of the viral DNA products formed upon infection.
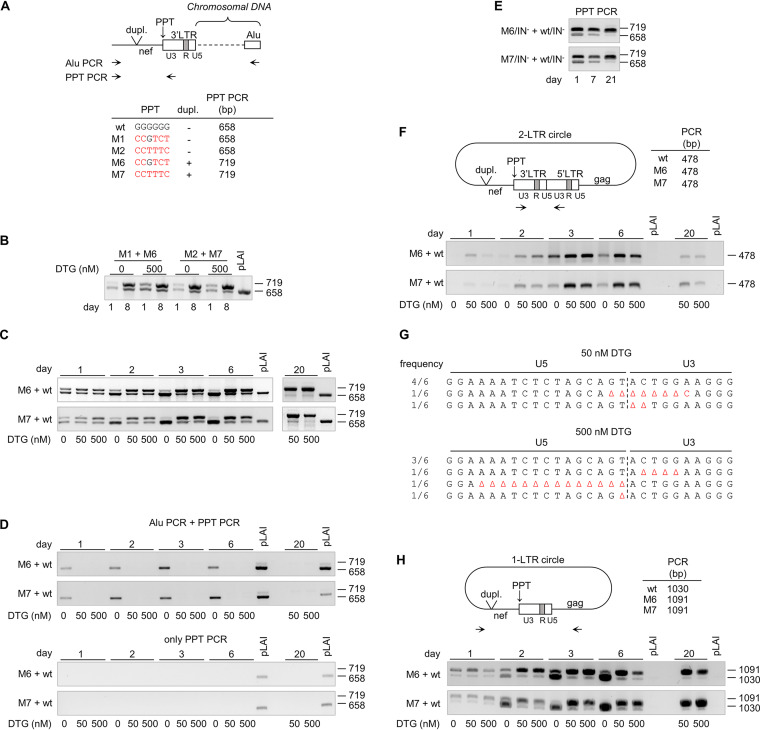
FIG 4.
Viral DNA products formed during the replication of 3′PPT-mutated viruses. (A) Schematic of the 3′ end of integrated proviral DNA with the primers used for Alu PCR and PPT PCR indicated. The PPT PCR amplicon size for the different 3′PPT/nef variants is indicated. The nucleotide substitutions in the G6-motif of the different variants are in red. (B) C8166 cells were coinfected with M1+M6 or M2+M7 virus and cultured without or with 500 nM DTG. Intracellular DNA was isolated at 1 and 8 days after infection, and viral DNA production was analyzed by PPT PCR. pLAI, control wt plasmid. (C and D) Upon coinfection of C8166 cells with M6+wt or M7+wt virus, cells were cultured with 0, 50, or 500 nM DTG. (C) Viral DNA production was analyzed at several times after infection by PPT PCR. (D) Integrated HIV DNA was analyzed by Alu PCR followed by PPT PCR (top). As a control, the same DNA input and PCR conditions (limited number of PCR cycles) were used in the PPT PCR (bottom). (E) C8166 cells were coinfected with integrase-deficient HIV variants with either a mutated or wt 3′PPT (M6/IN−+wt/IN− or M7/IN−+wt/IN−). Viral DNA production was analyzed at several times after infection by PPT PCR. (F to H) 2-LTR and 1-LTR circle formation in M6+wt- and M7+wt-infected cells (cultures as used in C and D) was analyzed by PCR. (F) The 2-LTR PCR amplifies the 2-LTR circle junction and results in a 478-bp product for both the wt and 3′PPT-mutated virus. (G) PCR products obtained for M6+wt-infected cells after culturing for 6 days in the presence of 50 or 500 nM DTG (as shown in F) were cloned into a Topo TA cloning vector, and individual clones were sequenced. The frequency of each observed sequence is indicated at the left side. (H) The 1-LTR circle PCR results in a 1,030-bp product for the wt virus and a 1,091-bp fragment for the 3′PPT variants.
Similar internally controlled virus competition assays were used to analyze the effect of the 3′PPT/nef mutations on viral DNA production. C8166 cells were coinfected with the wt virus and either the M6 or M7 mutant (M6+wt and M7+wt cultures) and cultured with 0, 50, or 500 nM DTG. Without DTG, virus-induced syncytium formation was rapidly observed and the culture was stopped after 6 days when massive syncytia were present. In the presence of DTG, syncytium formation was much slower. At day 11, syncytia were observed and the virus-containing culture medium was used to infect fresh cells, which were subsequently cultured for up to day 20. At several times after infection, total cellular DNA was isolated and used as the template for the PPT PCR to detect viral DNA (Fig. 4C). While the virus input for the different DTG conditions was identical, the ratio between the shorter (wt) and the larger (mutant) PCR products varied for the different conditions. Noticeably, the wt product rapidly became more dominant over time in cultures without DTG, whereas the mutant product became more dominant in cultures with 50 and 500 nM DTG. These ratio changes are in agreement with the more efficient replication of the wt virus in the absence of DTG and the more efficient replication of the 3′PPT/nef-mutated viruses with DTG.
We next analyzed the effect of the M6 and M7 mutations on viral DNA integration. We analyzed the DNA isolated from the M6+wt- and M7+wt-infected cells in an Alu-PCR assay in which the size variation in nef can again be used to distinguish wt and mutant products. In a first PCR step, integrated viral DNA was amplified with primers annealing to nef and to Alu repeat sequences, which are likely present in the flanking cellular DNA (Fig. 4A). As integration will occur at different chromosomal positions, this PCR will not result in a single product with a specific size but in a mixture of differently sized products. These products were used as the template in the PPT PCR that creates discrete products for wt (658 bp) and mutant viruses (719 bp). The number of PCR cycles in this second step was limited such that no PCR product was detected when the first PCR step was omitted (Fig. 4D, bottom). In both the M6+wt and M7+wt cultures without DTG, predominantly the wt PCR product was detected after 1 day of culturing and later (Fig. 4D, top), reflecting efficient integration of the wt virus. In cultures with DTG, neither the wt nor the mutant PCR product was detected, indicating that the 3′PPT/nef mutations do not restore viral integration in the presence of DTG.
The combined data suggest that the 3′PPT variants can replicate in the presence of DTG but without detectable HIV DNA integration. Although this surprising finding would provide an easy explanation of the DTG resistance phenotype, it is remarkable as integration is considered one of the hallmarks of the retroviral replication cycle (27, 28). To confirm that the 3′PPT/nef-mutated viruses can indeed replicate without the viral integrase function, we inactivated the integrase function by mutation of the crucial enzymatic DDE motif into NNQ (29, 30). C8166 cells were coinfected with the integrase-deficient viruses with a wt and a M6 -or M7-mutated 3′PPT/nef region. At 7 days after infection, massive virus-induced syncytia were observed and the virus-containing culture supernatant was used to infect fresh cells. Viral DNA production was analyzed by PPT PCR at several times after infection. Both the wt and mutant DNA products (658 and 719 bp, respectively) were observed at day 1 (Fig. 4E). The ratio between these products changed over time, and a strongly reduced level of the wt product and an increased level of the mutant product were observed at day 21. These results confirm that the 3′PPT/nef-mutated viruses can replicate in C8166 cells without the viral integrase function.
Nakajima et al. (30) previously reported low-level replication of integrase-defective HIV variants in C8166 cells. Irwan et al. (10, 11) demonstrated more recently that this replication was due to the expression of Tax protein in these HTLV-1-transformed T cells (23). Tax was shown to prevent epigenetic silencing of unintegrated HIV DNA, in particular the 2-LTR DNA circles that accumulate when integration is blocked (Fig. 1). We used C8166 cells for the selection of the DTG-resistant 3′PPT-mutated HIV variants and demonstrated that the replication of these 3′PPT mutants is activated by Tax (Fig. 3D) (8). We therefore analyzed the effect of the 3′PPT-mutations on 2-LTR circle production. Unfortunately, the size variation in nef cannot be used to distinguish wt and mutant 2-LTR circles by PCR, as combining a primer annealing upstream of the nef modifications with a primer annealing in the LTR region will result in a short PCR product that does not include the U5-U3 junction, while combining the upstream primer with a primer annealing in gag will result in the preferential amplification of a 1-LTR circle fragment (see below). Previous studies demonstrated that mutations in the 3′PPT, like single- and dual-nucleotide substitutions in the G6-motif, can cause aberrant RNase H cleavage and lead to the formation of 2-LTR circle U5-U3 junctions with an extended U3 region due to the retention of 3′PPT sequences (19, 31, 32). We therefore analyzed the 2-LTR circles present in the M6+wt- and M7+wt-infected cultures by PCR with forward and reverse primers binding to U3 sequences upstream and downstream of the 2-LTR junction, followed by sequencing of the PCR product (Fig. 4F and G). For both cultures, we observed a single DNA product on the agarose gel, both in the absence and presence of DTG. Sequencing of this product obtained from the cultures with DTG at day 6 revealed only regular U5-U3 junctions and small U5 and U3 deletions, as expected for the wt virus (19, 31, 32), but we did not detect any U5-U3 junctions in which the U3 region was extended (Fig. 4G). The level of 2-LTR circles was higher in the presence of DTG than that without DTG (Fig. 4F), which is consistent with an increase in 2-LTR circle formation when the integration of wt HIV is blocked (17). Moreover, the level of 2-LTR circles in the cultures with DTG was relatively high at days 3 and 6, and much lower at day 20, which corresponds with the total viral DNA level that was observed for the wt virus in the PPT PCR (Fig. 4C). These results indicate that the 2-LTR circles are formed by the wt virus and not by the M6 and M7 mutant viruses.
Our experiments indicate that 3′PPT-mutated viruses can replicate in the presence of DTG without detectable 2-LTR circle formation and chromosomal integration. We therefore analyzed the production of 1-LTR circles, which are normally formed as an intermediate DNA product during reverse transcription (Fig. 1, step 7) (33). For this analysis, the intracellular DNA isolated from the M6+wt- and M7+wt-infected cells was analyzed by PCR with primers in nef and gag (Fig. 4H). This 1-LTR PCR results in a 1,030-bp product for the wt virus and a 1,091-bp product for the mutant viruses. At 1 day after infection, both products were detected, but the 1,091-bp product was more abundant, both with and without DTG, which indicates that the 3′PPT/nef mutations increase 1-LTR circle production upon infection. In the cultures with DTG, the mutant product became clearly more abundant over time. In cultures without DTG, the wt product became more dominant at later times, which can be explained by the efficient replication of the wt virus resulting in a high level of the reverse transcription intermediate. The 1-LTR circle production by the wt and mutant viruses thus resembles total viral DNA production at later times (compare Fig. 4H and C). Taken together, our results indicate that 3′PPT/nef mutations do not restore integration nor increase 2-LTR circle formation in the presence of DTG but instead increase 1-LTR circle formation, which we argue is the source of the observed low-level replication of these 3′PPT-mutated viruses.
DISCUSSION
We used the in vivo SELEX approach to identify novel mutations in the HIV 3′PPT motif that confer DTG resistance. Surprisingly, many different virus variants with diverse 3′PPT mutations were identified. This information adds considerable sequence diversity to the few 3′PPT mutations that were already reported to provide DTG resistance in other in vitro studies (4, 8, 9). What these mutants have in common is that the 3′PPT function is likely affected or even abolished, as this motif acts in a very sequence-specific manner (13, 14). All of this evidence suggests that mutational inactivation of the 3′PPT motif is what is really needed to achieve DTG resistance. This evolutionary route is quite exotic, as resistance to antiviral drugs is usually caused by one or multiple mutations in the targeted enzyme (3). This DTG resistance mechanism is thus exceptional in at least two ways. First, the resistance mutations are positioned in a regulatory 3′PPT signal that controls second-strand reverse transcription and not in the HIV gene that encodes the integrase enzyme. Second, many different mutations instead of a specific mutation can cause resistance, which led to the idea that 3′PPT-inactivation is the critical event in the acquisition of DTG resistance.
An analysis of the viral DNA products formed upon infection revealed that these 3′PPT mutants replicate without integration in the cellular genome but with increased formation of 1-LTR circle products. During reverse transcription of the HIV RNA genome, second-strand DNA synthesis normally initiates from the central PPT (cPPT) and 3′PPT RNA fragments that resist RNase H cleavage (Fig. 1, steps 4 and 5) (12, 34). Sequence complementarity at the termini of the partially double-stranded linear HIV DNA induces formation of 1-LTR circles (step 7). Subsequent continuation of second-strand DNA synthesis and strand displacement of the LTR sequences results in a linear 2-LTR dsDNA molecule (steps 8 and 9) (15), which is integrated into the cellular DNA (step 10a). It is likely that mutation of the 3′PPT sequence affects the special RNA-DNA structure that provides protection against RNase H cleavage (18, 35). Upon degradation of the 3′PPT RNA fragment, second-strand DNA synthesis may initiate only from the cPPT RNA (Fig. 5, steps 4 and 5), which will result in the formation of completely double-stranded 1-LTR circles (step 8). Our results are in accordance with the previous observation that 3′PPT deletion in a lentiviral vector induces the formation of 1-LTR circles (36).
FIG 5.
Reverse transcription of 3′PPT-mutated HIV variants. Upon infection, the viral RNA genome is copied into double-stranded DNA (dsDNA) by the viral reverse transcriptase. We hypothesize that the mutation of the 3′PPT (indicated with ΔPPT) causes RNase H cleavage of this RNA fragment. As a consequence, second-strand DNA synthesis may initiate only at the cPPT (steps 4 and 5), which results in the production of 1-LTR circles that cannot linearize because of the extended double-stranded LTR region (steps 7 and 8).
Unintegrated HIV DNA, which includes linear and circularized 2-LTR DNA and 1-LTR DNA circles, is normally silenced at the transcriptional level by cellular factors (37). We demonstrated that the replication of 3′PPT-mutated virus variants is activated by the HTLV-1 Tax protein (Fig. 3D) (8), a factor that reverses epigenetic silencing of unintegrated HIV DNA (10, 11). Accordingly, the 3′PPT variants replicate and cause cytopathogenic effects in C8166 cells, which are HTLV transformed and express Tax. Tax activation can explain why our initial in vitro evolution experiment in SupT1 cells, which lack HTLV-1 sequences, did not result in the selection of 3′PPT-mutated DTG-resistant virus variants. The combined data suggest that the 3′PPT-mutated viruses escape from DTG inhibition by switching to an integration-independent replication mechanism in which 1-LTR circles are produced. The presence of Tax likely prevents epigenetic silencing of this episomal DNA and stimulates the production of all HIV transcripts and proteins needed to support virus replication.
Irwan et al. (10, 11) recently described Tax-dependent and integration-independent replication of an HIV variant lacking a functional integrase but with a wt 3′PPT. It was suggested that this HIV variant replicates via episomal 2-LTR DNA circles that are formed when integration is blocked (Fig. 1). Here, we show that the replication of an integrase-deficient HIV variant in Tax-expressing cells is improved by the 3′PPT mutations (Fig. 4E). Superior replication of the 3′PPT-mutated variant is probably due to the increased production of episomal 1-LTR DNA circles (Fig. 4H).
This and previous studies demonstrate that the 3′PPT-mutated viruses replicate more efficient than wt HIV with DTG but worse than wt virus without DTG (6–8). 3′PPT mutations thus reduce the sensitivity toward DTG (resistance) but at the same time reduce the viral replication capacity (fitness). Notably, we describe 3′PPT-mutated viruses with additional upstream frameshift mutations in the nef gene (4-bp deletion and 65-bp duplication) that result in a premature stop codon in the nef open reading frame (Fig. 2F). Nef is an accessory viral protein that is not essential for viral replication in vitro, and mutations in the nef gene have been observed frequently upon long-term HIV culturing in vitro, in particular in transformed T cell lines that support a high level of virus replication (25, 38, 39). We demonstrated that the nef mutations improved replication of 3′PPT-mutated viruses in the C8166 T cell line. However, these additional mutations are not essential for the observed DTG-resistance phenotype (8).
Reverse transcription of the HIV RNA genome into a double-stranded DNA copy and integration of this linear DNA into the cellular DNA are considered hallmarks of the replication cycle of HIV and in fact all retroviruses. We describe 3′PPT-mutated HIV variants that produce 1-LTR DNA circles and do not integrate, and yet, these variants can replicate at a low level. We propose that 3′PPT-mutated viruses replicate via the episomal 1-LTR circle DNA intermediate. We suggest that transcription occurs on these 1-LTR DNA circles to produce the HIV RNA transcripts for protein production and to drive a low level of virus replication. Indeed, there is literature showing that this exotic scenario is feasible (40–42). The replication cycle of these HIV variants thus mimics that of the Hepadnaviridae-like hepatitis B virus (HBV) that reverse transcribes the full-length RNA into a circular DNA product that does not integrate but is used as the template for transcription of all viral transcripts. Thus, HIV may change its basic replication strategy upon DTG pressure.
Whether 3′PPT mutations contribute to virus escape from DTG in vivo remains to be determined. In our in vitro assays, replication of the 3′PPT-mutated viruses was found to be activated by the HTLV-1 Tax protein, likely to prevent epigenetic silencing of the 1-LTR DNA circles. Such an HTLV-1 Tax effect on HIV-1 replication in vivo may seem a remote possibility, but both HIV-1 and HTLV-1 infect T lymphocytes and coinfections are frequently observed in some patient populations. For example, up to 10% of HIV-1-infected persons in parts of Brazil are coinfected with HTLV-1 (43–45). Moreover, other viral proteins may similarly stimulate the replication of nonintegrating HIV variants. For example, the HIV Vpr, herpes simplex virus ICP0, and HBV HBx proteins can induce transcription from unintegrated viral DNA (46–48). Additionally, although 3′PPT-mutated viruses replicate poorly, they may acquire additional mutations that improve virus replication upon continued evolution (49). During preparation of this paper, Richetta et al. (50) reported that the 3′PPT-mutated HIV variant selected by Malet et al. (4) in HTLV-1-transformed MT-4 cells replicates without integration or increased formation of 2-LTR circles and with an increased production of 1-LTR circles, suggesting the involvement of this episomal DNA in virus replication, like we describe here for our 3′PPT variants. Interestingly, these authors describe that the mutated virus can also replicate in the absence of Tax. However, their results do not exclude a stimulatory effect of this protein. Therefore, the role of Tax in the proposed episomal DNA replication pathway, or another protein or compensatory mutation in the HIV genome that may have a similar effect, deserves further investigation. This exotic drug-escape route will be missed by regular resistance assays that are based on integrase genotyping. Screening for 3′PPT mutations in patients that fail on DTG therapy, in particular when no resistance-associated mutations are detected in integrase, will reveal the clinical importance of this drug resistance pathway. However, routine 3′PPT screening for patients on DTG therapy is complicated by the fact that many different 3′PPT mutations can cause the resistance phenotype, unlike the specific resistance mutations in targeted gene products regularly observed with antiretroviral drugs.
MATERIALS AND METHODS
Cell culture.
Human embryonic kidney 293T (HEK293T; ATCC CRL-11268) cells were cultured in Dulbecco’s modified Eagle medium (DMEM) (Gibco, Life Technologies) supplemented with 10% fetal calf serum (FCS), nonessential amino acids (0.1 mM; Invitrogen), and penicillin and streptomycin (both at 100 U/mL). SupT1 (51) and C8166 (23) T cell lines were obtained through the NIH HIV Reagent Program (AIDS Research and Reference Reagents Program, Division of AIDS, ARP-100 and ARP-404). CEM-SS T cells carrying a doxycycline-inducible HTLV-1 tax gene were kindly provided by Bryan Cullen and Hal Bogard (Duke University Medical Center, Durham, NC). T cell lines were cultured in advanced RPMI 1640 medium (Gibco, Life Technologies) with 1% l-glutamine, 1% FCS, penicillin (15 U/mL), and streptomycin (15 U/mL). All cells were maintained in a humidified chamber at 37°C and 5% CO2. When indicated, cells were cultured in the presence of 50 or 500 nM DTG (Tivicay, ViiV Healthcare).
DNA constructs.
All HIV-1 constructs are based on the pLAI plasmid encoding the full-length HIV-1 LAI genome of a subtype B primary isolate (20). The duplicated 3′PPT/nef sequence present upstream of the 5′LTR in this plasmid was removed by digestion with XbaI and PteI, followed by ligation of a gBlock gene fragment without these 3′PPT/nef sequences (Integrated DNA Technologies) into these sites (pLAI-5’exPPT). A gBlock gene fragment encoding the 3′PPT/nef region with a randomized G6-motif was digested with XhoI and Kpn2I and ligated into the corresponding sites in the pBlue3′LTR plasmid (40). The ligation mix was transformed in max efficiency Stbl2 competent cells (ThermoFisher). Multiple individual bacterial clones were isolated and analyzed by sequencing of the randomized segment to confirm that all sequenced clones carry a different 3′PPT sequence. The pooled Blue3′LTR-3′PPT randomized library was digested with XhoI and Kpn2I, and the fragments with the randomized 3′PPT were isolated and ligated into the corresponding sites in pLAI-5′exPPT to create the pLAI-5′exPPT-3′PPT randomized library. The ligation mix was transformed in max efficiency Stbl2 competent cells, and multiple individual clones were isolated and sequenced.
Topo TA vectors carrying the 3′PPT regions that were selected in the in vivo SELEX assay (described below) were digested with XhoI and Kpn2I, and the purified 3′PPT/nef sequences were ligated into the corresponding sites in pLAI-5′exPPT, creating variants M1 to M7. A gBlock gene fragment encoding the 3′PPT/nef region with the 3′PPT mutations as described by Malet et al. (4) was similarly digested with XhoI and Kpn2I and ligated into pLAI-5′exPPT, which yielded the 9053 variant.
The pNL4-3-NNQ molecular clone (30) was digested with restriction enzymes PshAI and NcoI. The integrase fragment was ligated into the corresponding sites of pLAI-5′exPPT, pLAI-5′exPPT-M6, and pLAI-5′exPPT-M7. Ligation products were transformed in max efficiency Stbl2 competent cells (ThermoFisher), and all sequences were confirmed by sequencing with the BigDye terminator kit (ThermoFisher).
Production of virus stocks.
Virus stocks were produced by transient transfection of HEK293T cells (5 × 105 cells per 10 cm2 well) with 4 μg of the pLAI-based molecular clones using Lipofectamine 2000 (ThermoFisher). The culture supernatant was collected at 48 h after transfection and centrifuged at 180 × g for 5 min to remove cells, and aliquots were stored at −80°C. The CA-p24 concentration of the virus stocks was determined by ELISA as described previously (52).
HIV-1 evolution in SupT1 T cells.
A total of 1 × 106 SupT1 T cells in 5 mL culture medium were infected with HIV-1 LAI (corresponding to 100 ng CA-p24) and cultured with 50 or 500 nM DTG. When cells formed massive syncytia, 100 μL of the culture supernatant was passaged onto fresh cells to continue virus replication. To analyze the intracellular viral DNA, cells were pelleted from 250 μL culture medium by centrifugation at 180 × g for 5 min. Intracellular DNA was solubilized by resuspending the cells in 75 μL 10 mM Tris-HCl (pH 8.0), 0.1 mM EDTA, and 0.5% Tween 20, followed by incubation with 200 μg/mL proteinase K at 56°C for 60 min and 95°C for 10 min. For PCR amplification of the 3’PPT/nef region, 1.5 μL of the lysed cells was mixed with 2 μL DreamTaq green PCR master mix (ThermoFisher), 500 nM primer End-env-s (5′-TAGAAGAATAAGACAGGGCTTGG-3′), and 500 nM primer Circh (5′-CAGTTCTTGAAGTACTCCGG-3′), and the volume was completed to 20 μL with water. The PCR mix was incubated at 95°C for 1 min; followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and a final incubation at 72°C for 5 min. The PCR products were cloned into a Topo TA cloning vector (ThermoFisher) and sequenced (53).
In vivo SELEX in C8166 cells.
A total of 2 × 105 C8166 T cells in 1 mL culture medium was infected with LAI-randomized-3′PPT virus (corresponding to 50 ng CA-p24) and cultured without or with DTG (50 or 500 nM). When cells showed massive syncytia, the intracellular viral DNA was isolated and the 3′PPT/nef region was amplified by PCR, as described above. The PCR product was directly sequenced (population sequencing) or cloned into a Topo TA cloning vector followed by sequencing of individual clones. The virus-containing culture supernatant was used to infect fresh C8166 cells. Initially, 100 μL of the supernatant was used to infect 2× 105 cells in 1 mL culture medium, but the volume of this inoculum was gradually reduced to 5 μL after 10 weeks.
TCID assay.
CEM-SS cells with a doxycycline-inducible tax gene (2 × 104 cells in 100 μL culture medium) were infected with serial dilutions (dilution factor 3) of wt or 3′PPT-mutated virus stocks (starting at 10 ng CA-p24 per 100-μL culture). When indicated, cells were cultured with 0.5 μg/mL doxycycline (Sigma D-9891) and 500 nM DTG. Syncytium formation was scored at 10 days after infection and the TCID50 was calculated (54).
Virus competition and viral DNA analysis.
A premix of two HIV variants was prepared by mixing equal amounts (based on CA-p24) of two viruses. A total of 2 × 105 C8166 cells in 1 mL culture medium was infected with an aliquot of this premix (corresponding to 5 ng CA-p24 of each virus) and cultured with 0, 50, or 500 nM DTG. The intracellular viral DNA was isolated at several times after infection, and the 3′PPT/nef region was amplified by PCR, as described above. The same PCR protocol with different primers was used for an analysis of the 2-LTR circle production (primers TA019 [5′-AAGTGTTAGAGTGGAGGTTTG-3′] and 3’envN [5′-CTGCCAATCAGGGAAGTAGCCTTGTGT-3′]) and 1-LTR circle production (primers End-env-S [5′-TAGAAGAATAAGACAGGGCTTGG-3′] and Circ4 [5′-CTTAACCGAATTTTTTCCCA-3′]). For the first step of the Alu-PCR analysis, the same PCR protocol, but with 35 (instead of 30) PCR cycles, was used with primers 5′NEF-1 (5′-GCAGTAGCTGAGGGGACAGATAGG-3′) and Alu_reverse (5′-TGCTGGGATTACAGGCGTGAG-3′). For the second step of the Alu-PCR analysis, 2 μL of the PCR product from the first step was used as input DNA for 3′PPT/nef PCR analysis as described above, but with only 16 (instead of 30) PCR cycles.
ACKNOWLEDGMENTS
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R01AI147330. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We thank Bryan Cullen and Hal Bogard (Duke University Medical Center, Durham, North Carolina) for kindly providing us the CEM-SS cell line with doxycycline-controlled HTLV-1 Tax expression.
Contributor Information
Atze T. Das, Email: a.t.das@amsterdamumc.nl.
Frank Kirchhoff, Ulm University Medical Center.
REFERENCES
- 1.Smith SJ, Zhao XZ, Passos DO, Lyumkis D, Burke TR, Jr, Hughes SH. 2021. Integrase strand transfer inhibitors are effective anti-HIV drugs. Viruses 13:205. doi: 10.3390/v13020205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. 2019. Policy brief: update of recommendations on first- and second-line antiretroviral regimens. WHO, Geneva, Switzerland. https://apps.who.int/iris/handle/10665/325892. [Google Scholar]
- 3.Anstett K, Brenner B, Mesplede T, Wainberg MA. 2017. HIV drug resistance against strand transfer integrase inhibitors. Retrovirology 14:36. doi: 10.1186/s12977-017-0360-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malet I, Subra F, Charpentier C, Collin G, Descamps D, Calvez V, Marcelin AG, Delelis O. 2017. Mutations located outside the integrase gene can confer resistance to HIV-1 integrase strand transfer inhibitors. mBio 8:e00922-17. doi: 10.1128/mBio.00922-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wijting IEA, Lungu C, Rijnders BJA, Van Der Ende ME, Pham HT, Mesplede T, Pas SD, Voermans JJC, Schuurman R, Van De Vijver DAMC, Boers PHM, Gruters RA, Boucher CAB, Van Kampen JJA. 2018. HIV-1 resistance dynamics in patients with virologic failure to dolutegravir maintenance monotherapy. J Infect Dis 218:688–697. doi: 10.1093/infdis/jiy176. [DOI] [PubMed] [Google Scholar]
- 6.Wei Y, Sluis-Cremer N. 2021. Mutations in the HIV-1 3′-polypurine tract and integrase strand transfer inhibitor resistance. Antimicrob Agents Chemother 65:e02432-20. doi: 10.1128/AAC.02432-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith SJ, Ferris A, Zhao X, Pauly G, Schneider JP, Burke TR, Jr, Hughes SH. 2021. INSTIs and NNRTIs potently inhibit HIV-1 polypurine tract mutants in a single round infection assay. Viruses 13:2501. doi: 10.3390/v13122501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dekker JG, Klaver B, Berkhout B, Das AT. 2022. Mutations in the HIV-1 3′-polypurine tract can confer dolutegravir resistance. Antimicrob Agents Chemother 66:e01027-21. doi: 10.1128/AAC.01027-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hachiya A, Kubota M, Shigemi U, Ode H, Yokomaku Y, Kirby KA, Sarafianos SG, Iwatani Y. 2022. Specific mutations in the HIV-1 G-tract of the 3′-polypurine tract cause resistance to integrase strand transfer inhibitors. J Antimicrob Chemother 77:574–577. doi: 10.1093/jac/dkab448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Irwan ID, Karnowski HL, Bogerd HP, Tsai K, Cullen BR, Griffin DE. 2020. Reversal of epigenetic silencing allows robust HIV-1 replication in the absence of integrase function. mBio 11:e01038-20. doi: 10.1128/mBio.01038-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irwan ID, Cullen BR. 2021. Tax induces the recruitment of NF-κB to unintegrated HIV-1 DNA to rescue viral gene expression and replication. J Virol 95:e0028521. doi: 10.1128/JVI.00285-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charneau P, Alizon M, Clavel F. 1992. A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J Virol 66:2814–2820. doi: 10.1128/JVI.66.5.2814-2820.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rausch JW, Le Grice SFJ. 2004. “Binding, bending and bonding”: polypurine tract-primed initiation of plus-strand DNA synthesis in human immunodeficiency virus. Int J Biochem Cell Biol 36:1752–1766. doi: 10.1016/j.biocel.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 14.Figiel M, Krepl M, Park S, Poznański J, Skowronek K, Gołąb A, Ha T, Šponer J, Nowotny M. 2018. Mechanism of polypurine tract primer generation by HIV-1 reverse transcriptase. J Biol Chem 293:191–202. doi: 10.1074/jbc.M117.798256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuentes GM, Rodríguez-Rodríguez L, Palaniappan C, Fay PJ, Bambara RA. 1996. Strand displacement synthesis of the long terminal repeats by HIV reverse transcriptase. J Biol Chem 271:1966–1971. doi: 10.1074/jbc.271.4.1966. [DOI] [PubMed] [Google Scholar]
- 16.Ellison V, Brown PO. 1994. A stable complex between integrase and viral DNA ends mediates human immunodeficiency virus integration in vitro. Proc Natl Acad Sci USA 91:7316–7320. doi: 10.1073/pnas.91.15.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hazuda DJ, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler JA, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller MD. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 18.Rausch JW, Tian M, Li Y, Angelova L, Bagaya BS, Krebs KC, Qian F, Zhu C, Arts EJ, Le Grice SF, Gao Y. 2015. siRNA-induced mutation in HIV-1 polypurine tract region and its influence on viral fitness. PLoS One 10:e0122953. doi: 10.1371/journal.pone.0122953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Julias JG, McWilliams MJ, Sarafianos SG, Alvord WG, Arnold E, Hughes SH. 2004. Effects of mutations in the G tract of the human immunodeficiency virus type 1 polypurine tract on virus replication and RNase H cleavage. J Virol 78:13315–13324. doi: 10.1128/JVI.78.23.13315-13324.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peden K, Emerman M, Montagnier L. 1991. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology 185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 21.Berkhout B, Klaver B. 1993. In vivo selection of randomly mutated retroviral genomes. Nucleic Acids Res 21:5020–5024. doi: 10.1093/nar/21.22.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Bel N, Das AT, Berkhout B. 2014. In vivo SELEX of single-stranded domains in the HIV-1 leader RNA. J Virol 88:1870–1880. doi: 10.1128/JVI.02942-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salahuddin SZ, Markham PD, Wong-Staal F, Franchini G, Kalyanaraman VS, Gallo RC. 1983. Restricted expression of human T-cell leukemia–lymphoma virus (HTLV) in transformed human umbilical cord blood lymphocytes. Virology 129:51–64. doi: 10.1016/0042-6822(83)90395-1. [DOI] [PubMed] [Google Scholar]
- 24.Ongrádi J, Laird HM, Szilágyi JF, Horváth A, Bendinelli M. 2000. Unique morphological alterations of the HTLV-I transformed C8166 cells by infection with HIV-1. Pathol Oncol Res 6:27–37. doi: 10.1007/BF03032655. [DOI] [PubMed] [Google Scholar]
- 25.Jere A, Fujita M, Adachi A, Nomaguchi M. 2010. Role of HIV-1 Nef protein for virus replication in vitro. Microbes Infect 12:65–70. doi: 10.1016/j.micinf.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 26.Koken SE, van Wamel JL, Goudsmit J, Berkhout B, Geelen JL. 1992. Natural variants of the HIV-1 long terminal repeat: analysis of promoters with duplicated DNA regulatory motifs. Virology 191:968–972. doi: 10.1016/0042-6822(92)90274-s. [DOI] [PubMed] [Google Scholar]
- 27.Temin HM. 1976. The DNA provirus hypothesis. Science 192:1075–1080. doi: 10.1126/science.58444. [DOI] [PubMed] [Google Scholar]
- 28.Varmus H. 1988. Retroviruses. Science 240:1427–1435. doi: 10.1126/science.3287617. [DOI] [PubMed] [Google Scholar]
- 29.Engelman AN, Craigie R. 1992. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol 66:6361–6369. doi: 10.1128/JVI.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakajima N, Lu R, Engelman AN. 2001. Human immunodeficiency virus type 1 replication in the absence of integrase-mediated DNA recombination: definition of permissive and nonpermissive T-cell lines. J Virol 75:7944–7955. doi: 10.1128/jvi.75.17.7944-7955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McWilliams MJ, Julias JG, Sarafianos SG, Alvord WG, Arnold E, Hughes SH. 2003. Mutations in the 5′ end of the human immunodeficiency virus type 1 polypurine tract affect RNase H cleavage specificity and virus titer. J Virol 77:11150–11157. doi: 10.1128/jvi.77.20.11150-11157.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McWilliams MJ, Julias JG, Sarafianos SG, Alvord WG, Arnold E, Hughes SH. 2006. Combining mutations in HIV-1 reverse transcriptase with mutations in the HIV-1 polypurine tract affects RNase H cleavages involved in PPT utilization. Virology 348:378–388. doi: 10.1016/j.virol.2005.12.042. [DOI] [PubMed] [Google Scholar]
- 33.Munir S, Thierry S, Subra F, Deprez E, Delelis O. 2013. Quantitative analysis of the time-course of viral DNA forms during the HIV-1 life cycle. Retrovirology 10:87. doi: 10.1186/1742-4690-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu WS, Hughes SHJ. 2012. HIV-1 reverse transcription. Cold Spring Harb Perspect Med 2:a006882–a006882. doi: 10.1101/cshperspect.a006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rausch JW, Le Grice SFJ. 2007. Purine analog substitution of the HIV-1 polypurine tract primer defines regions controlling initiation of plus-strand DNA synthesis. Nucleic Acids Res 35:256–268. doi: 10.1093/nar/gkl909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kantor B, Bayer M, Ma H, Samulski J, Li C, McCown T, Kafri T. 2011. Notable reduction in illegitimate integration mediated by a PPT-deleted, nonintegrating lentiviral vector. Mol Ther 19:547–556. doi: 10.1038/mt.2010.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geis FK, Goff SP. 2019. Unintegrated HIV-1 DNAs are loaded with core and linker histones and transcriptionally silenced. Proc Natl Acad Sci USA 116:23735–23742. doi: 10.1073/pnas.1912638116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertels F, Leemann C, Metzner KJ, Regoes R. 2019. Parallel evolution of HIV-1 in a long-term experiment. Mol Biol Evol 36:2400–2414. doi: 10.1093/molbev/msz155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pawlak EN, Dikeakos JD. 2015. HIV-1 Nef: a master manipulator of the membrane trafficking machinery mediating immune evasion. Biochim Biophys Acta 1850:733–741. doi: 10.1016/j.bbagen.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Klaver B, Berkhout B. 1994. Comparison of 5′ and 3′ long terminal repeat promoter function in human immunodeficiency virus. J Virol 68:3830–3840. doi: 10.1128/JVI.68.6.3830-3840.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thierry S, Thierry E, Subra F, Deprez E, Leh H, Bury-Moné S, Delelis O. 2016. Opposite transcriptional regulation of integrated vs unintegrated HIV genomes by the NF-κB pathway. Sci Rep 6:25678. doi: 10.1038/srep25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Y, Marsh JW. 2003. Early transcription from nonintegrated DNA in human immunodeficiency virus infection. J Virol 77:10376–10382. doi: 10.1128/jvi.77.19.10376-10382.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silva MT, Neves ES, Grinsztejn B, de Melo Espíndola O, Schor D, Araújo A. 2012. Neurological manifestations of coinfection with HIV and human T-lymphotropic virus type 1. AIDS 26:521–523. doi: 10.1097/QAD.0b013e32834c4a3e. [DOI] [PubMed] [Google Scholar]
- 44.Galetto LR, Lunge VR, Béria JU, Tietzmann DC, Stein AT, Simon D. 2014. Short communication: prevalence and risk factors for human T cell lymphotropic virus infection in Southern Brazilian HIV-positive patients. AIDS Res Hum Retroviruses 30:907–911. doi: 10.1089/AID.2013.0210. [DOI] [PubMed] [Google Scholar]
- 45.Ribeiro ML, Gonçales JP, Morais VMS, Moura L, Coêlho M. 2019. HTLV 1/2 prevalence and risk factors in individuals with HIV/AIDS in Pernambuco, Brazil. Rev Soc Bras Med Trop 52:e20180244. doi: 10.1590/0037-8682-0244-2018. [DOI] [PubMed] [Google Scholar]
- 46.Dupont L, Bloor S, Williamson JC, Cuesta SM, Shah R, Teixeira-Silva A, Naamati A, Greenwood EJD, Sarafianos SG, Matheson NJ, Lehner PJ. 2021. The SMC5/6 complex compacts and silences unintegrated HIV-1 DNA and is antagonized by Vpr. Cell Host Microbe 29:792–805.e6. doi: 10.1016/j.chom.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murphy CM, Xu Y, Li F, Nio K, Reszka-Blanco N, Li X, Wu Y, Yu Y, Xiong Y, Su L. 2016. Hepatitis B virus X protein promotes degradation of SMC5/6 to enhance HBV replication. Cell Rep 16:2846–2854. doi: 10.1016/j.celrep.2016.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poon B, Chen IS. 2003. Human immunodeficiency virus type 1 (HIV-1) Vpr enhances expression from unintegrated HIV-1 DNA. J Virol 77:3962–3972. doi: 10.1128/jvi.77.7.3962-3972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Duyne R, Kuo LS, Pham P, Fujii K, Freed EO. 2019. Mutations in the HIV-1 envelope glycoprotein can broadly rescue blocks at multiple steps in the virus replication cycle. Proc Natl Acad Sci USA 116:9040–9049. doi: 10.1073/pnas.1820333116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richetta C, Subra F, Malet I, Leh H, Charpentier C, Corona A, Collin G, Descamps D, Deprez E, Parissi V, Calvez V, Tramontano E, Marcelin AG, Delelis O. 2022. Mutations in the 3′-PPT lead to HIV-1 replication without integration. J Virol 96:e0067622. doi: 10.1128/jvi.00676-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith SD, Shatsky M, Cohen PS, Warnke R, Link MP, Glader BE. 1984. Monoclonal antibody and enzymatic profiles of human malignant T-lymphoid cells and derived cell lines. Cancer Res 44:5657–5660. [PubMed] [Google Scholar]
- 52.Jeeninga RE, Jan B, van den Berg H, Berkhout B. 2006. Construction of doxycyline-dependent mini-HIV-1 variants for the development of a virotherapy against leukemias. Retrovirology 3:64. doi: 10.1186/1742-4690-3-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou M-Y, Gomez-Sanchez CE. 2000. Universal TA cloning. Curr Issues Mol Biol 2:1–7. [PubMed] [Google Scholar]
- 54.Lei C, Yang J, Hu J, Sun X. 2021. On the calculation of TCID(50) for quantitation of virus infectivity. Virol Sin 36:141–144. doi: 10.1007/s12250-020-00230-5. [DOI] [PMC free article] [PubMed] [Google Scholar]