ABSTRACT
Perenniporia fraxinea can colonize living trees and cause severe damage to standing hardwoods by secreting a number of carbohydrate-activate enzymes (CAZymes), unlike other well-studied Polyporales. However, significant knowledge gaps exist in understanding the detailed mechanisms for this hardwood-pathogenic fungus. To address this issue, five monokaryotic P. fraxinea strains, SS1 to SS5, were isolated from the tree species Robinia pseudoacacia, and high polysaccharide-degrading activities and the fastest growth were found for P. fraxinea SS3 among the isolates. The whole genome of P. fraxinea SS3 was sequenced, and its unique CAZyme potential for tree pathogenicity was determined in comparison to the genomes of other nonpathogenic Polyporales. These CAZyme features are well conserved in a distantly related tree pathogen, Heterobasidion annosum. Furthermore, the carbon source-dependent CAZyme secretions of P. fraxinea SS3 and a nonpathogenic and strong white-rot Polyporales member, Phanerochaete chrysosporium RP78, were compared by activity measurements and proteomic analyses. As seen in the genome comparisons, P. fraxinea SS3 exhibited higher pectin-degrading activities and higher laccase activities than P. chrysosporium RP78, which were attributed to the secretion of abundant glycoside hydrolase family 28 (GH28) pectinases and auxiliary activity family 1_1 (AA1_1) laccases, respectively. These enzymes are possibly related to fungal invasion into the tree lumens and the detoxification of tree defense substances. Additionally, P. fraxinea SS3 showed secondary cell wall degradation capabilities at the same level as that of P. chrysosporium RP78. Overall, this study suggested mechanisms for how this fungus can attack the cell walls of living trees as a serious pathogen and differs from other nonpathogenic white-rot fungi.
IMPORTANCE Many studies have been done to understand the mechanisms underlying the degradation of plant cell walls of dead trees by wood decay fungi. However, little is known about how some of these fungi weaken living trees as pathogens. P. fraxinea belongs to the Polyporales, a group of strong wood decayers, and is known to aggressively attack and fell standing hardwood trees all over the world. Here, we report CAZymes potentially related to plant cell wall degradation and pathogenesis factors in a newly isolated fungus, P. fraxinea SS3, by genome sequencing in conjunction with comparative genomic and secretomic analyses. The present study provides insights into the mechanisms of the degradation of standing hardwood trees by the tree pathogen, which will contribute to the prevention of this serious tree disease.
KEYWORDS: tree pathogen, wood decay fungi, CAZy, Basidiomycetes, genomics
INTRODUCTION
The vast majority of organic carbon in a terrestrial ecosystem is first accumulated in plant cell walls via photosynthesis. Wood decay fungi are well known to be primarily responsible for the depolymerization of complex plant cell wall polymers, including various polysaccharides and recalcitrant lignin (1), and recent meta-omics analyses of decayed woods provided evidence of their roles in natural environments (2). Some wood decay fungi have also been reported to aggressively invade standing trees as serious pathogens and to damage and fell living trees by structurally weakening their cell wall, which consists of a thin primary cell wall layer and a thick secondary cell wall layer, in turn causing the necrosis of living cells (3). Therefore, such tree pathogens represent pioneer degraders in a terrestrial carbon cycle. However, the molecular mechanisms of how these tree pathogens infect living trees, overcome tree defense systems, weaken the cell walls, and, finally, kill the cells in living trees remain largely unknown, except for those of a softwood pathogen, Heterobasidion annosum (4).
Many white-rot Polyporales, including Phanerochaete chrysosporium, Ceriporiopsis subvermispora, Trametes versicolor, and Phlebiopsis gigantea, possess an impressive ability to degrade dead trees, such as stumps, fallen trees, and woody debris; hence, they are called wood decay fungi (5–8). A combination of genomic, transcriptomic, and proteomic analyses of these fungi showed that a series of carbohydrate-active enzymes (CAZymes) was secreted to degrade plant cell wall components such as cellulose, hemicellulose, and lignin in dead trees (9). Unlike the above-mentioned representative wood decay Polyporales, Perenniporia fraxinea is capable of colonizing living hardwood trees and causes mortality and severe mechanical damage by exhibiting the white-rot type of wood decay (10). In urban environments, fungus-related diseases have led to hazardous situations for a broad range of hardwoods, including downed trees or limbs of planted black locusts, oaks, ashes, and flowering cherries, throughout North America, Europe, and Asia (10–14). However, knowledge of the genetic information and the molecular mechanisms possessed by this hardwood pathogen is limited to date.
To explore the mechanism of the ability of P. fraxinea to degrade living trees, five monokaryotic strains of P. fraxinea were isolated from natural environments. Among them, P. fraxinea strain SS3 showed fast growth and a high polysaccharide-degrading capacity compared to the other four strains; hence, genomic and proteomic analyses were performed on this strain. A draft genome sequence of P. fraxinea SS3 was obtained by whole-genome sequencing, and a set of genes that encode CAZymes was annotated. The CAZyme compositions of the P. fraxinea genome were compared to those of the genomes of phylogenetically related nonpathogenic Polyporales and another tree pathogen, H. annosum, which belongs to a different order, Russulales (8). In addition, carbon source-dependent CAZyme secretion by P. fraxinea SS3 was monitored using polysaccharide-degrading activity measurements along with a well-studied model wood degrader of the Polyporales, Phanerochaete chrysosporium, which does not infect living trees. Finally, a proteome secreted by P. fraxinea was analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) and compared with that of P. chrysosporium to elucidate CAZymes related to the degradation of the plant cell wall by the hardwood tree pathogen.
RESULTS
Isolation and selection of P. fraxinea monokaryotic strains.
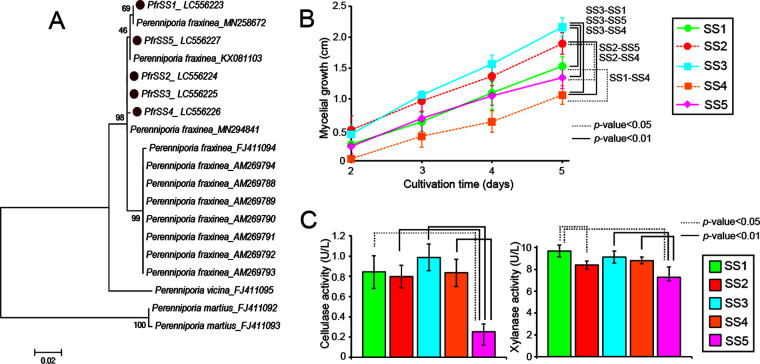
We isolated five monokaryotic strains, SS1 to SS5, from a P. fraxinea fruiting body grown on a Robinia pseudoacacia tree. The internal transcribed spacer (ITS) regions of the strains were analyzed, and the best hits from a BLASTn search showed 99.2 to 100% identity to previously reported P. fraxinea strains (see Table S1 in the supplemental material). A phylogenetic tree of the ITS sequences showed that all strains are closely related to P. fraxinea, compared to the outgroups of Perenniporia vicina and P. martius (11, 15), indicating that they belong to P. fraxinea (Fig. 1A). The five isolates were cultivated on a potato dextrose agar (PDA) plate, and the growth of the mycelium of P. fraxinea SS3 was found to be relatively faster than that of the other isolates (Fig. 1B and Fig. S1). To evaluate the ability of the pathogen to degrade plant components, five strains, SS1 to SS5, were grown in liquid medium containing wood powder as the sole carbon source in biological triplicates, and the plant polysaccharide-degrading activities in each culture supernatant were measured for cellulose and xylan (Fig. 1C). As a result, both cellulase and xylanase activities were similar among strains SS1 to -4, while strain SS5 displayed significantly low activities. Overall, strain SS3 seemed to be a monokaryotic strain with robust growth and high wood degradation activities; therefore, this strain was selected for further analysis.
FIG 1.
(A) Phylogenetic tree of P. fraxinea species, including five strains, SS1 to SS5, isolated from a P. fraxinea fruiting body, using the ITS region sequences. (B) Time course of mycelial growth of the five P. fraxinea strains SS1 to SS5 grown on agar plates. (C) Major wood polysaccharide-degrading activities in the culture supernatants of the five P. fraxinea strains grown in liquid culture containing poplar wood powder as a sole carbon source for 7 days. PASC cellulase activities and xylanase activities are shown on the left and right, respectively. The averages and standard deviations were calculated from three independent biological replicates. Student’s t test was performed.
Genomic analysis of P. fraxinea SS3.
The genomic DNA of monokaryotic P. fraxinea strain SS3 was extracted, the whole genome was sequenced, and the total output data comprised 6.15 Gbp. The total assembled size of the draft genome was found to be 32.6 Mbp (GC content, 52.5%), as shown in Table 1. The genome includes 571 scaffolds with an N50 length of 255,461 bp, 7,796 annotated genes, and an average mRNA size of 1,540 nucleotides (nt). Its genome features are similar to those of the previously disclosed genomes of other wood decay fungi deposited in the MycoCosm portal (16). The putative functions of these genes were annotated based on a BLAST homology search (17) and a dbCAN database search (18) (Data Set S1).
TABLE 1.
Genome summary of P. fraxinea strain SS3
| Parametera | Value for P. fraxinea SS3 |
|---|---|
| Assembled genome size (Mbp) | 32.6 |
| GC content (%) | 52.5 |
| No. of assembled scaffolds | 571 |
| Scaffold N50 length (bp) | 255,461 |
| No. of annotated gene models | 7,798 |
| Avg CDS length (bp) | 1,543 |
CDS, coding DNA sequence.
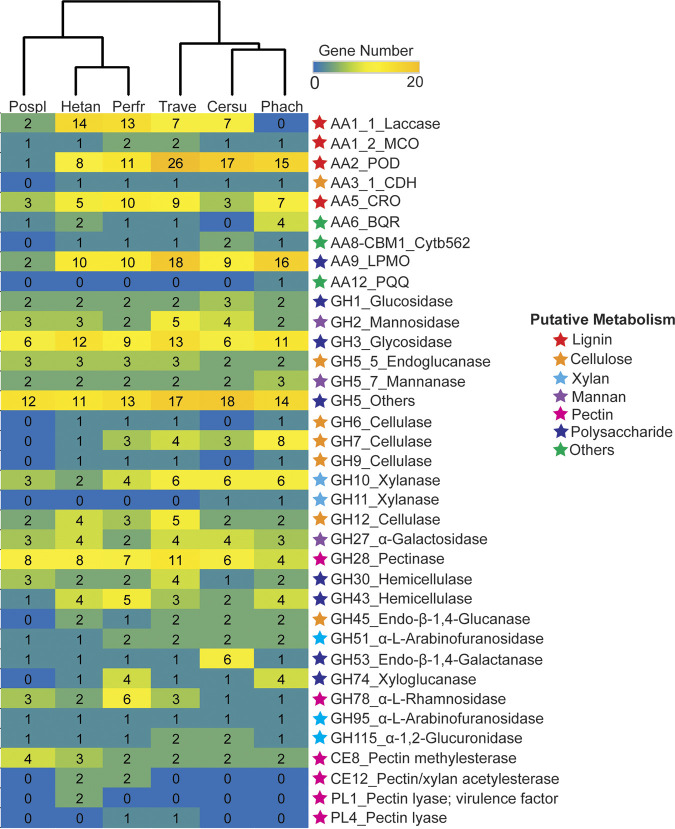
Four hundred seventy-five CAZyme genes were annotated in the genome of P. fraxinea SS3, and the number of CAZy families was compared to those of other reported Polyporales, a brown-rot fungus, Postia placenta, and the white-rot fungi P. chrysosporium, C. subvermispora, and T. versicolor, together with a plant-pathogenic Russulales member, H. annosum (Fig. 2). Clustering analysis of the numbers of genes of representative CAZymes for plant cell wall degradation clearly showed that P. fraxinea SS3 possesses characteristic CAZymes different from those of other white-rot and brown-rot Polyporales and rather similar to those of H. annosum, which strongly indicates that tree pathogens utilize common specialized CAZymes. When we observed the details of the glycoside hydrolase (GH) families, there was no significant difference with respect to the number of genes encoding conventional fungal cellulases (19) such as cellobiohydrolase (CBH) and endoglucanase (EG) classified into GH7, GH6, GH5_5, GH12, and GH45 between P. fraxinea SS3 and other white-rot fungi (20). Interestingly, the GH7 genes in the genome of P. fraxinea SS3 (mRNA6940, mRNA3133, and mRNA5626) have unique domain structures that lack carbohydrate-binding modules (CBMs) of family 1 (CBM1) (Fig. S2), the accessory domain of which is known to introduce a catalytic domain to the substrate surface (19). It has been known that nonpathogenic white-rot fungi have a GH7 domain with CBM1 at the C-terminal region; therefore, all of the GH7 sequences from P. fraxinea SS3, P. chrysosporium, C. subvermispora, T. versicolor, and H. annosum were analyzed, and it was found that a GH7 cellulase from T. versicolor and H. annosum also lacks CBM1 (Fig. S2). These results indicated that the cellulase system of P. fraxinea SS3 is unique among those of other Polyporales but is conserved in tree pathogens.
FIG 2.
Heatmap and hierarchical tree of CAZyme genes predicted in the genome of P. fraxinea SS3 (Perfr) compared to other reported fungal genomes, including Phanerochaete chrysosporium RP78 ver 2.2 (Phach), Ceriporiopsis subvermispora (Cersu), Trametes versicolor (Trave), Heterobasidion annosum (Hetan), and Postia placenta (Pospl). The CAZy family and the putative function of the genes are shown, and the corresponding possibly related metabolism is indicated as a colored star on the right of the heatmap. All of the predicted genes are described in Data Set S1 in the supplemental material.
For hemicellulose degradation, there was also no difference in the numbers of xylan-degrading enzymes (e.g., GH10 and GH11 endoxylanases, GH51 and GH95 arabinofuranosidases, and GH115 glucuronidase) (21, 22) and mannan-degrading enzymes (e.g., GH2 mannosidase, GH5_7 mannanase, and GH27 α-galactosidase) (21) between P. fraxinea SS3 and other white-rot fungi. However, the number of the major hemicellulases, which are thought to be involved in the degradation of the primary cell wall hemicelluloses, was unique in P. fraxinea SS3. Four GH74 xyloglucanase genes were located in the genomes of P. fraxinea SS3 and P. chrysosporium, while no or only one GH74-encoding gene was found in the genomes of other wood decay fungi (Fig. 2). Regarding pectin-degrading enzymes, the numbers of genes encoding GH28 pectinase and carbohydrate esterase (CE) family 8 (CE8) pectin methylesterase were not different, while two CE12 pectin/xylan acetylesterases were predicted only in the genomes of P. fraxinea SS3 and H. annosum. In the case of oxidoreductases involved in plant degradation categorized into auxiliary activity (AA) families, P. fraxinea SS3 and H. annosum had a larger number of AA1 laccases than other Polyporales. In contrast, the numbers of genes in other AA families, such as the AA2 fungal peroxidase (POD) playing central roles in lignin degradation (23), AA3 and AA5 H2O2 supply enzymes for the POD (23), and the AA9 lytic polysaccharide monooxygenase (LPMO) involved in cellulose and hemicellulose degradation (24), were almost the same as those in other nonpathogenic white-rot fungi. Overall, the CAZyme potential of P. fraxinea SS3 seems to support a greater pectin degradation ability and more laccases than a common nonpathogenic white-rot Polyporales fungus and was similar to that of the distantly related tree pathogen H. annosum.
Comparison of the CAZyme activities of P. fraxinea SS3 to those of a model white-rot Polyporales fungus.
To assess the features of the CAZyme-encoding genes predicted in the P. fraxinea SS3 genome, P. fraxinea SS3 and P. chrysosporium RP78 were cultivated in synthetic medium containing glucose, cellulose, or wood powder as the sole carbon source for 5 days, and the CAZyme activities in the culture supernatants were measured using purified polysaccharides as the substrates in biological triplicates (Fig. 3). The protein concentrations in six culture supernatants were measured, and P. fraxinea SS3 secreted relatively higher concentrations proteins than P. chrysosporium RP78 (Fig. S3). The highest protein concentration was detected in cellulose medium for both fungi, followed by wood- and glucose-containing media. The mycelium grew well in glucose medium, but the levels of secreted proteins were very low, probably due to carbon catabolite repression, which is often observed in wood decay fungi (25, 26).
FIG 3.
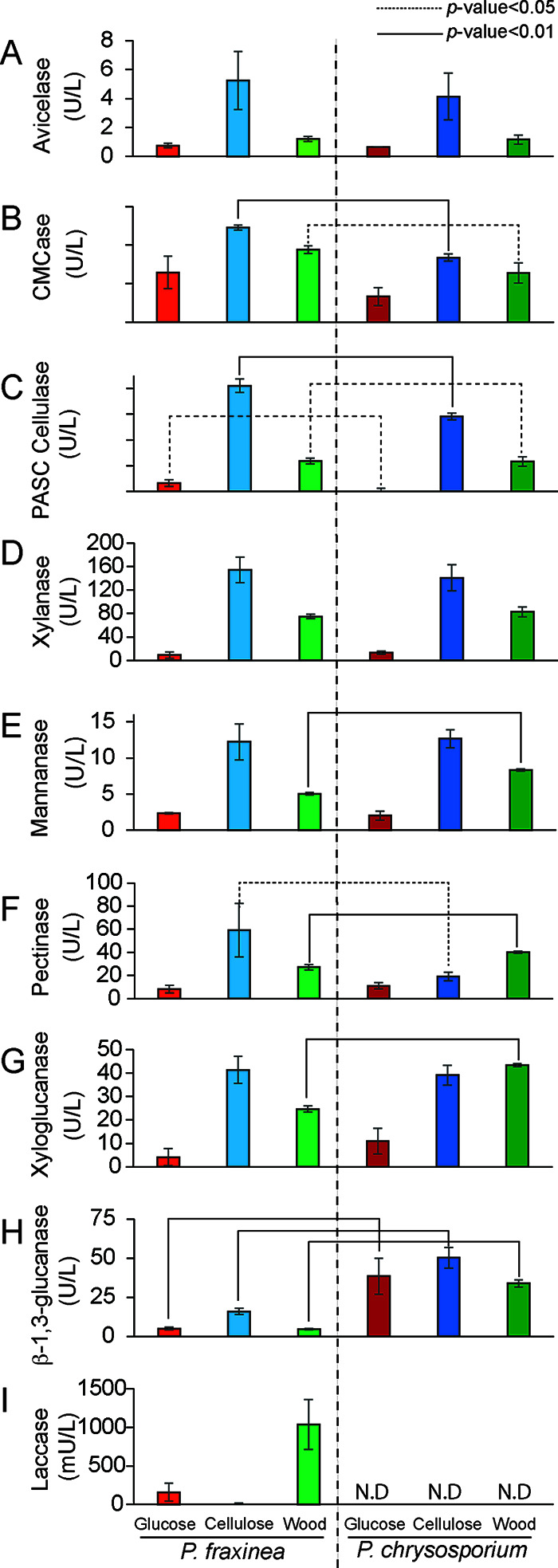
CAZyme activities in the culture supernatants of P. fraxinea SS3 or P. chrysosporium RP78 grown in liquid culture containing glucose, cellulose, or poplar wood powder as the sole carbon source for 5 days. Microcrystalline cellulose-degrading activities (Avicelase) (A), carboxymethyl cellulose-degrading activities (CMCase) (B), phosphoric acid-swollen cellulose-degrading activities (PASC cellulase) (C), glucuronoxylanase (D), mannanase (E), pectinase (F), xyloglucanase (G), β-1,3-glucanase (H), and laccase (I) activities are shown. The averages and standard deviations were calculated from three independent biological replicates. Student’s t test was performed for comparison of P. fraxinea SS3 and P. chrysosporium RP78 under the same culture conditions. N.D, not determined.
For cellulose degradation measurements, microcrystalline cellulose (Avicel), carboxymethyl cellulose (CMC), and phosphoric acid-swollen cellulose (PASC) were used as the substrates for determining cellobiohydrolase, endoglucanase, and whole-cellulase activities, respectively, and the amount of released reducing end products was measured by a dinitrosalicylic acid (DNS) method (Fig. 3A to C) (27). As expected, cellulose in the culture medium strongly induced cellulase activities in both fungi compared to medium containing complex wood or glucose. Interestingly, P. fraxinea SS3 showed significantly higher CMC- and PASC-degrading activities than those of P. chrysosporium RP78, while the Avicelase activities were almost the same between the two fungi. These results could be attributed to the enriched endo-acting cellulases of P. fraxinea SS3 (Fig. S2). Glucuronoxylan and glucomannan were used as the substrates to measure representative wood hemicellulose degradation (Fig. 3D and E). P. fraxinea SS3 showed the same levels of xylan- and mannan-degrading activities as those of P. chrysosporium RP78 when the fungi were grown on cellulose medium. Among other hemicellulose degradation potentials (Fig. 3F and G), the pectinase activities in the cellulose medium of P. fraxinea SS3 were significantly higher than those of P. chrysosporium RP78, consistent with the comparison of their genomes. On the other hand, the mannan-, pectin-, and xyloglucan-degrading activities of P. fraxinea SS3 in wood medium were relatively lower than those of P. chrysosporium RP78. Additionally, β-1,3-glucanase activities toward β-1,3/1,6-glucan, which is often found in the fungal cell wall, were significantly weaker in the culture supernatant of P. fraxinea SS3 than in that of P. chrysosporium RP78 (Fig. 3H). Moreover, laccase activities were detected only in P. fraxinea SS3 (Fig. 3I), and these activities were strongly induced in the wood-containing medium, followed by the glucose and cellulose media, respectively, which were not detected in the P. chrysosporium RP78 culture supernatant, as expected by the genome analysis. Overall, the CAZyme potential found in the P. fraxinea SS3 genome and CAZyme activities were well correlated.
Proteomic analysis of P. fraxinea SS3.
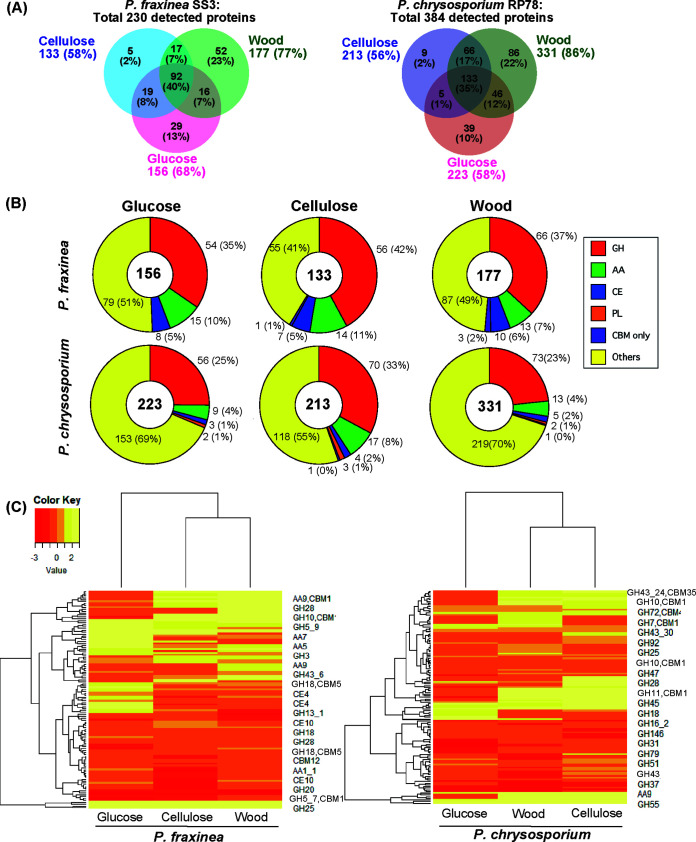
To determine the compositions of CAZymes of P. fraxinea SS3 responsible for the significantly different CAZyme activities from those of P. chrysosporium RP78, the culture supernatants in biological triplicates were analyzed by LC-MS/MS. Two hundred thirty proteins were detected overall, and 156, 133, and 177 proteins were identified in the glucose-, cellulose-, and wood-containing culture media, respectively (Fig. 4A). The identified proteins met our criteria, requiring a minimum of two unique peptides per detected protein. Exponentially modified protein abundance index (emPAI) values (28) were calculated accordingly for each protein in the data set (Data Set S2). Proteomics analysis of the culture supernatant of P. chrysosporium RP78 grown on glucose-, cellulose-, and wood-containing media was also performed; the numbers of secreted proteins in each sample were determined to be 223 (glucose), 213 (cellulose), and 331 (wood), and the total number of detected proteins was 384 (Fig. 4A). Overall, the total number of proteins detected in P. chrysosporium RP78 was higher than that in P. fraxinea SS3. Next, the predicted functions of the detected proteins were categorized into CAZy families such as GHs, AAs, CEs, polysaccharide lyases (PLs), and CBMs (Fig. 4B). As a result, P. fraxinea SS3 produced more abundant CAZymes than P. chrysosporium RP78 under all of the culture conditions. In addition to CAZymes, P. fraxinea SS3 produced 25, 12, and 13 hypothetical proteins in the glucose-, cellulose-, and wood-containing culture media, respectively (Table S2), and they are thought to be potential candidates for plant cell wall degradation or tree pathogenesis. Clustering analysis of the expression of all of the detected CAZymes clearly indicated that differences in the carbon-dependent secretion of CAZymes were similar in both fungi (Fig. 4C); i.e., the expression patterns in cellulose and wood media are categorized as being closer than those under glucose conditions for both fungi. However, CAZymes that were induced in the presence of cellulose and wood in the two fungi seemed highly different. To gain further insight into the detailed CAZyme secretion profiles of P. fraxinea SS3 and P. chrysosporium RP78, the 10 most abundant proteins were compared (Table S3). Regarding the cellulose-degrading system, GH7 CBH/EG and AA9 LPMO were abundantly secreted by both fungi. In the culture supernatant of P fraxinea SS3, GH6, GH12, and GH45 were also abundantly secreted in cellulose and wood cultures. All of the GH7 cellulases lacking CBM1 (mRNA6940, mRNA3133, and mRNA5626) were produced by P. fraxinea SS3, while P. chrysosporium RP78 abundantly produced the GH7 cellulases associated with the CBM1 domain (Protein ID 2971601 and 137372). Since GH7 cellulases are often determined to be dominant proteins in cellulose cultures of wood-decaying fungi (29, 30), the secretion patterns of the two fungi in cellulose-containing medium were compared by SDS-PAGE (Fig. S4). The major protein bands detected in the cellulose medium of P. fraxinea SS3 and P. chrysosporium RP78, which were thought to be cellulases, showed different apparent molecular weights of approximately 40 kDa and 60 kDa, respectively. These results confirmed that the two fungi utilize different GH7-involved cellulase systems and that P. fraxinea SS3 has a greater cellulose-degrading ability than other white-rot Polyporales fungi.
FIG 4.
Venn diagrams of proteomes (unique peptide counts of ≥2) secreted by P. fraxinea SS3 (A) and P. chrysosporium RP78 (B) cultivated in glucose-, cellulose-, or poplar wood powder-containing medium for 5 days. Percentages represent ratios to the total number of detected proteins, 230 and 384, in all of the culture supernatants of P. fraxinea SS3 and P. chrysosporium RP78, respectively. (C) Clustering analysis of CAZymes secreted by P. fraxinea SS3 and P. chrysosporium RP78 grown on glucose, cellulose, and poplar wood powder media by using the emPAI values from biological triplicates as the expression levels. All of the detected proteomes are referred to in Data Set S3 in the supplemental material.
For hemicellulases, GH10 xylanases were highly produced by both fungi in the cellulose- and wood-containing media (Table S3), consistent with the measured CAZyme activities (Fig. 3D). Notably, two GH28 pectinases (mRNA5111 and mRNA3447) were abundant in the cellulose medium of P. fraxinea SS3, supporting the observed significantly high pectin-degrading activity of this fungus (Fig. 3F). A CE12 pectin/xylan acetylesterase (mRNA3979), found in the genomes of pathogenic fungi (Fig. 2), was detected in the glucose and cellulose media of P. fraxinea SS3 and was not detected in any medium of P. chrysosporium RP78 (Data Set S2). Moreover, a large proportion of AA1 laccase (mRNA6925) was detected in the wood medium of P. fraxinea SS3, which explained the observed high laccase activity in the culture supernatant (Fig. 3I). In the case of lignin degradation, an AA2 manganese peroxidase (MnP) (mRNA3095) was secreted by P. fraxinea SS3 in glucose and cellulose media, and H2O2 supply enzymes such as AA3_2 aryl alcohol oxidase/glucose oxidase and AA5_1 glyoxal oxidase (GLOX) were also secreted into the same media (Data Set S2). Meanwhile, an MnP (protein ID 3589) was secreted by P. chrysosporium RP78 in wood medium along with AA3_2 aryl alcohol oxidase/glucose oxidase, AA3_3 alcohol oxidase, AA3_4 pyranose oxidase, and AA5_1 GLOX. Additionally, the high β-1,3-glucanase activity in P. chrysosporium RP78 was thought to be due to a large portion of secreted GH152 β-1,3-glucanases (thaumatin-like proteins [Protein ID 2983557 and 3003339]) (Fig. 3H). These results indicated that we successfully identified important CAZymes for the degradation of plant cell wall substrates by P. fraxinea SS3.
DISCUSSION
The detailed mechanisms underlying most plant pathogens that damage plant leaves have been comprehensively analyzed (31), while little is known about tree pathogens that seriously damage tree xylem, which contains more recalcitrant plant cell walls than leaves. Few studies have described the CAZyme potential encoded in the genomes of plant pathogens (32). Several comparative genome analyses of tree pathogens have been reported for serious conifer pathogens of the species Heterobasidion (4, 33) and tree pathogens of the species Armillaria belonging to the Agaricales (34). Here, we report the genome-wide identification of CAZymes potentially related to plant cell wall degradation and pathogenesis factors in P. fraxinea. P. fraxinea belongs to the Polyporales, a group of strong wood decayers, and this fungus is known to aggressively attack and fell standing hardwood trees all over the world (10–14).
The novel P. fraxinea strains SS1 to SS5 were identified in this study, and the most robust P. fraxinea strain, SS3, was selected for further investigation using a combination of genomic, biochemical, and proteomic analyses. Overall, a complete set of CAZymes for the degradation of plant cell walls is encoded in the P. fraxinea SS3 genome as well as those of several well-studied white-rot Polyporales, and key features of CAZyme potential in the genome of P. fraxinea SS3 as a pathogen were determined (Fig. 2). For example, P. fraxinea SS3 possesses genes with higher pectin-degrading potentials in its genome, unlike other Polyporales (Fig. 2), as reported in previous studies describing other tree-pathogenic white-rot fungi (4, 33, 34). Consistent with this, P. fraxinea SS3 showed stronger pectin-degrading activities than the nonpathogenic wood decay fungus P. chrysosporium RP78 (Fig. 3), which was thought to be due to the high level of secretion of the GH28 pectinases mRNA5111 and mRNA3447 (see Table S3 in the supplemental material). In addition, our results suggested that a CE12 pectin/xylan acetylesterase is involved in the high pectin degradation activity and pathogenesis since only P. fraxinea SS3 and H. annosum possess CE12 pectin/xylan acetylesterases in their genomes among the wood decay fungi (Fig. 2). CE12 mRNA3979 was secreted by P. fraxinea SS3 during wood component degradation (Data Set S1). Pectic polysaccharide is known to be abundant in the primary cell wall; hence, the observed pectin degradation ability is thought to be important for microbial invasion (31). Why a tree pathogen like P. fraxinea SS3 possesses high pectic polysaccharide-degrading potential is an intriguing question since tree xylem consists mainly of a secondary cell wall, with a negligible amount of primary cell wall. Our explanation is that pectic polysaccharides are distributed in cell wall adhesion region and the pit membrane as well as the primary cell wall in the xylem (35), which must be removed during fungal invasion within the lumens of the xylem. Therefore, the strong pectic polysaccharide-degrading capacity of these tree-pathogenic fungi may enable fungal invasion through the cell wall lumens in the xylem in a living tree, although further genetic studies will be needed to clarify the contributions of these CAZymes to the above-described fungal pathogenesis.
In contrast to the series of CAZymes for pectin degradation, the numbers of xylanases and mannanases encoded in the genomes of pathogenic and nonpathogenic fungi were comparable (Fig. 2), and the xylanase activities of P. fraxinea SS3 were comparable to those of P. chrysosporium RP78 (Fig. 3). Xylan and mannan are abundant in the secondary cell wall, and therefore, both fungi are capable of degrading secondary cell wall hemicelluloses. Regarding the degradation of cellulose, enriched in the secondary cell wall, there was also no significant difference in the numbers of conventional cellulases encoded in the genomes of P. fraxinea SS3 and other white-rot Polyporales. However, P. fraxinea SS3 possesses a unique domain structure of GH7 CBH genes that lacked CBM1 (Fig. S2), yet other cellulases and hemicellulases such as GH6, GH5_5, and GH10 that had a CBM1 domain were the same as those of other white-rot wood decay fungi. It is known that a CBM-lacking cellulase can readily access amorphous cellulose rather than crystalline cellulose (29), and indeed, the endoglucanase activities in the culture supernatants of P. fraxinea SS3 grown in all of the cultures including glucose, cellulose, or wood as the sole carbon source were significantly higher than those of P. chrysosporium RP78, a strong white-rot fungus (Fig. 3). High endoglucanase activities might be important for tree pathogens, yet how they function in tree pathogenesis processes such as fungal invasion and xylem cell wall degradation is still elusive. Regarding lignin degradation, one AA2 MnP and several H2O2 supply enzymes were detected in the secretomes of both fungi (Data Set S2). These results suggested that P. fraxinea SS3 is able to decompose all of the major components of the secondary cell wall dominantly existing in tree xylem as efficiently as other strong wood decayers in the Polyporales group.
In addition to the above-mentioned GHs, many AA1_1 laccases, which oxidize a broad range of phenolic compounds, were encoded in the genome of P. fraxinea SS3, and mRNA6925 and mRNA5962 were abundantly secreted by this fungus during the degradation of plant cell wall components (Fig. 2 and 3 and Table S3). Although it has been suggested that a higher number of laccase genes present in the genomes of tree-pathogenic fungi is correlated with tree pathogenesis, their function is still elusive (4, 33). It is known that defense substrates of polyphenols are filled in the xylem cells, called a reaction zone, which functions in response to microbial attack (36), and pathogenic fungi need to detoxify the polyphenols. Indeed, pathogenic fungi, i.e., Ganoderma adspersum, move through the lumens filled with phenols in hardwoods (37, 38), yet the details of the defense substances in trees and the detoxification mechanisms in these fungi need further studies.
In conclusion, we isolated P. fraxinea SS3 from a natural environment and successfully determined that the hardwood-pathogenic Polyporales have strong CAZyme potential, equivalent to those reported for strong wood decayers at the genomic and secretomic levels. Notably, this pathogenic fungus has substantial degradation activities toward pectin distributed in the primary cell wall, membrane pit, and cell wall adhesions, while there was no significant difference found in the degradation of the secondary cell wall components, which are also abundant in dead trees. In addition to plant cell wall degradation, we successfully identified AA1_1 laccases, which were highly produced from the tree-pathogenic fungi, potentially contributing to a system to detoxify defense substances produced by living trees. Altogether, the present study provides insights into the mechanisms of the degradation of standing hardwood trees by this tree pathogen and suggests the potential of CAZymes to differentiate tree-pathogenic fungi from nonpathogenic fungi. These results will contribute to a fundamental understanding of interactions between fungi and trees, which is important for the carbon cycle in nature and applications for preventing fungus-related diseases in the future.
MATERIALS AND METHODS
Isolation and identification of monokaryotic strains of P. fraxinea.
To isolate monokaryotic strains of P. fraxinea, basidiospores of P. fraxinea were collected from a fruiting body grown on a Robinia pseudoacacia tree on the campus of Hokkaido University, Sapporo, Japan, and spread onto potato dextrose agar (PDA; Nihon Pharmaceutical Co., Ltd., Tokyo, Japan). The colonies were separated and grown several times until five single filamentous fungi were isolated (P. fraxinea strains SS1 to SS5). The internal transcribed spacer (ITS) region, including 5.8 rRNA, was amplified by a colony PCR method using Kod Fx polymerase (Toyobo Co., Ltd., Osaka, Japan). Primer set ITS1 and ITS4 was used as previously described (39). The ITS sequences were searched using NCBI BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=tblastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome), and all of the best hits showed >99% identity to Vanderbylia fraxinea (synonym of P. fraxinea) (see Table S1 in the supplemental material). Phylogenetic tree analysis (1,000 bootstraps) was performed with the BLASTn best-hit ITS sequences of five identified single spores (Table S1) and the previously reported ITS sequences of P. fraxinea (11, 15) by using MEGA7 software (40).
Fungal culture conditions for analysis of secreted proteins.
To select a strain for the following genomic and proteomic analyses from the 5 monokaryotic strains of P. fraxinea, P. fraxinea SS1 to SS5 were grown in 100 mL of modified Highley medium (41) containing 1 wt% poplar wood ground powder (<0.5 mM) as a carbon source in a 500-mL Erlenmeyer flask for 5 days at 26.5°C at 150 rpm. The growth of the five strains was within the exponential growth phase under these cultivation conditions. To compare the selected monokaryotic strain of P. fraxinea SS3 with a model wood-decaying fungus, P. chrysosporium RP78 (42), both strains were grown in 100 mL of modified Highley medium (41) containing 0.5 wt% glucose, 1 wt% cellulose, and 1 wt% poplar wood ground powder (<0.5 mm) as a carbon source in a 500-mL Erlenmeyer flask for 5 days at 26.5°C at 150 rpm.
The culture supernatant was collected by filtration with a glass filter (Advantec GA-100; Toyo Roshi Kaisha, Ltd., Japan), and the protein concentration was estimated by the Bradford method (Bio-Rad Laboratories, Inc., CA). The protein patterns of the supernatants were visualized by SDS-PAGE (Bio-Rad Laboratories, Inc., CA) and Coomassie brilliant blue (CBB) R-250 staining (Quick CBB; Fujifilm Wako Pure Chemical Corporation, Osaka, Japan). The collected supernatants were used for plant polysaccharide degradation assays and proteomic analyses.
Plant polysaccharide degradation activity assays.
For enzyme activity assays of the culture supernatants, 45 μL of the crude protein sample was mixed with 50 μL of the substrates described below and 5 μL of 1 M sodium acetate (pH 5.0) in a total reaction volume of 100 μL. Both soluble and insoluble polysaccharides were adjusted to contain 2 wt% microcrystalline cellulose (Avicel PH-101 Fluka; Sigma-Aldrich, Inc., St. Louis, MO), 2 wt% carboxymethyl cellulose (CMC 4M; Megazyme, Ireland), 2 wt% glucuronoxylan (xylan from beech; Sigma-Aldrich, St. Louis, MO), 2 wt% mannan (Megazyme, Ireland), 2 wt% pectin (pectin from citrus; Nacalai Tesque, Kyoto, Japan), and 2 wt% β-1,3/1,6-glucan (laminarin, Sigma-Aldrich, MO and Sigma-Aldrich). Additionally, 1 wt% xyloglucan (tamarind gum; Tokyo Chemical Industry Co., Ltd., Tokyo, Japan) and 1 wt% phosphoric acid-swollen cellulose (PASC) were used due to high viscosity. PASC was prepared by using Avicel and 85% phosphoric acid (Nacalai Tesque, Kyoto, Japan) as described in a previous report (43). The reaction mixture was incubated for 16 h at 37°C, and the reducing end product was measured by a dinitrosalicylic acid (DNS; Fujifilm Wako Pure Chemical Corporation, Osaka, Japan) assay at 540 nm (27). The molar equivalent of reducing sugar was estimated by using the glucose standard. One unit of enzyme activity was defined as 1 μM glucose released per min. For laccase activity assays, 2,6-dimethoxyphenol (DMP) was used as a substrate (44). The slope of the absorbance change at 470 nm over time was determined by using a UV-visible (UV-Vis) spectrophotometer (V-730; Jasco Corporation, Tokyo, Japan). One unit of laccase activity was defined as the oxidation of 1 μmol of the substrate per min. The molar extinction coefficient (ε) of oxidized DMP at 470 nm is 14,800. The average and standard deviation were calculated based on three biological replicates, and Student’s t tests were performed to compare samples of P. fraxinea SS3 and P. chrysosporium RP78 under the same culture conditions.
DNA preparation and sequencing of P. fraxinea SS3.
The selected strain, P. fraxinea SS3, was grown in potato dextrose broth. The collected mycelium was crushed by using a crushing device (μT-12; Taitec, Saitama, Japan), and the genomic DNA was extracted by using IsoplantII (Nippon gene Co., Ltd., Japan). The obtained DNA was treated with RNase A (Nippon Gene Co., Ltd., Japan). The quality and quantity of the DNA were confirmed by spectrometry and agarose gel electrophoresis. The qualified DNA was cut into fragments by using a restriction enzyme. The construction of the DNA libraries included processes of end repair, phosphorylation, the addition of A to 3′ tails, the ligation of index adaptors, purification, and PCR amplification according to the manufacturer’s instructions (Illumina). The library was sequenced by using the Hiseq4000 system (Illumina, Inc., CA) with a 150-bp paired-end sequencing strategy.
Genome assembly and gene annotations.
The acquired DNA sequences were assembled by using Platanus v1.2.4 (45) after trimming the adaptor sequences. Only scaffolds of >1 kbp were used for the genome assembly of P. fraxinea SS3. The protein-encoding genes were predicted by a homology search. Known proteins from the UniProt database (46) were aligned against the genome assembly using GhostX (47) to roughly identify the gene locations. The detailed gene structures were then determined using Spaln (48). Among the predicted gene candidates, those with in-frame stop codons and those with low coverage (<60%) for the reference proteins were removed to construct the final gene set. For CAZyme analysis, the obtained amino acid sequences predicted in the genome sequence of P. fraxinea SS3 were applied for dbCAN analysis (18). Some CAZyme genes were manually curated. All detected protein sequences were searched using BLASTp with an E value of 10−15 using the default parameters of blast2go version 5.2 software (17) to annotate putative functions (2). Signal peptide secretion was predicted using SignalP version 5.0 (http://www.cbs.dtu.dk/services/SignalP/) (49). The theoretical Mw and pI of the corresponding proteins were calculated by using the ExPASy server (https://web.expasy.org/compute_pi/). Heatmap analysis was performed by using Heatplus in the Bioconductor package.
Proteomic analysis of secreted proteins.
One-hundred-microgram equivalents of crude secreted proteins of the three fungi under three culture conditions from three independent biological replicates were prepared for proteomic analyses by using a previously described method (41). Tryptic digestion was performed by using 1 mg/mL trypsin (proteomics grade; Sigma-Aldrich, Inc., St. Louis, MO) overnight at 37°C, and trypsin-digested peptides were desalted by using Ziptip (Merck KGaA, Darmstadt, Germany). The peptides dissolved in a 0.1% formic acid solution were applied to the nanoLC1000 system (Thermo Fisher Scientific, IL) equipped with a C18 column (catalog no. NTCC-360/75-3-125; Nikkyo Technos, Tokyo, Japan) connected to a hybrid linear ion trap-orbitrap mass spectrometer (Q-Exactive plus; Thermo Fisher Scientific, IL) and then monitored at a scan range of 300.0 to 2,000.0 m/z. Raw MS/MS data were analyzed by using Proteome Discoverer software version 2.1 (Thermo Fisher Scientific, IL) with the following settings: the peptide mass tolerance was set at 10 ppm, and the fragment mass tolerance was set at 0.8 Da. Fixed modifications of carbamidomethyl at Cys and dynamic modifications of oxidation at Met were used for the database search. The annotated genes of P. fraxinea SS3 were used for a database search. We omitted the proteins with unique peptides of <2 fragments for the stringent screening of proteins determined by proteomics in each secretome. Based on the acquired LC-MS/MS spectra, the abundance of each protein was semiquantitatively calculated using the emPAI value (28). The averages and standard deviations of the area values were estimated from 3 biological replicates obtained by LC-MS/MS, and the emPAIs represent the values from 3 biological replicates.
Data availability.
The ITS sequences of P. fraxinea SS1 to SS5 were deposited in the NCBI database (DDBJ/EMBL/GenBank accession no. LC556223 to LC556227).
ACKNOWLEDGMENTS
We thank Ayane Yamamoto for technical assistance in DNA extraction.
This study was partly supported by JSPS grants-in-aid for scientific research 16K18727, 21H02251, and 22K19195; JST-ACTX PJ2519A059; and the Asahi Glass Foundation (to C.H.).
R.M. performed experiments. J.J.M. analyzed the data and wrote the draft. H.N. analyzed genome data and wrote the draft. T.M. collected monokaryotic spores. T.E.T. investigated proteome analysis and wrote the draft. C.H. designed the research, investigated experiments, analyzed the data, and wrote the draft. All authors reviewed and approved the paper.
We have no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Chiaki Hori, Email: chori@ees.hokudai.ac.jp.
Yvonne Nygård, Chalmers University of Technology.
REFERENCES
- 1.Eriksson K-E, Blanchette RA, Ander P. 1990. Microbial and enzymatic degradation of wood and wood components. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 2.Hori C, Gaskell J, Cullen D, Sabat G, Stewart PE, Lail K, Peng Y, Barry K, Grigoriev IV, Kohler A, Fauchery L, Martin F, Zeiner CA, Bhatnagar JM. 2018. Multi-omic analyses of extensively decayed Pinus contorta reveal expression of a diverse array of lignocellulose-degrading enzymes. Appl Environ Microbiol 84:e01133-18. doi: 10.1128/AEM.01133-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmidt O. 2006. Wood and tree fungi: biology, damage, protection, and use. Springer, Berlin, Germany. [Google Scholar]
- 4.Olson Å, Aerts A, Asiegbu F, Belbahri L, Bouzid O, Broberg A, Canbäck B, Coutinho PM, Cullen D, Dalman K, Deflorio G, van Diepen LTA, Dunand C, Duplessis S, Durling M, Gonthier P, Grimwood J, Fossdal CG, Hansson D, Henrissat B, Hietala A, Himmelstrand K, Hoffmeister D, Högberg N, James TY, Karlsson M, Kohler A, Kües U, Lee Y-H, Lin Y-C, Lind M, Lindquist E, Lombard V, Lucas S, Lundén K, Morin E, Murat C, Park J, Raffaello T, Rouzé P, Salamov A, Schmutz J, Solheim H, Ståhlberg J, Vélëz H, de Vries RP, Wiebenga A, Woodward S, Yakovlev I, Garbelotto M, et al. 2012. Insight into trade-off between wood decay and parasitism from the genome of a fungal forest pathogen. New Phytol 194:1001–1013. doi: 10.1111/j.1469-8137.2012.04128.x. [DOI] [PubMed] [Google Scholar]
- 5.Hori C, Ishida T, Igarashi K, Samejima M, Suzuki H, Master E, Ferreira P, Ruiz-Dueñas FJ, Held B, Canessa P, Larrondo LF, Schmoll M, Druzhinina IS, Kubicek CP, Gaskell JA, Kersten P, St John F, Glasner J, Sabat G, Splinter BonDurant S, Syed K, Yadav J, Mgbeahuruike AC, Kovalchuk A, Asiegbu FO, Lackner G, Hoffmeister D, Rencoret J, Gutiérrez A, Sun H, Lindquist E, Barry K, Riley R, Grigoriev IV, Henrissat B, Kües U, Berka RM, Martínez AT, Covert SF, Blanchette RA, Cullen D. 2014. Analysis of the Phlebiopsis gigantea genome, transcriptome and secretome provides insight into its pioneer colonization strategies of wood. PLoS Genet 10:e1004759. doi: 10.1371/journal.pgen.1004759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez-Fueyo E, Ruiz-Dueñas FJ, Ferreira P, Floudas D, Hibbett DS, Canessa P, Larrondo LF, James TY, Seelenfreund D, Lobos S, Polanco R, Tello M, Honda Y, Watanabe T, Watanabe T, Ryu JS, Kubicek CP, Schmoll M, Gaskell J, Hammel KE, St John FJ, Vanden Wymelenberg A, Sabat G, Splinter BonDurant S, Syed K, Yadav JS, Doddapaneni H, Subramanian V, Lavín JL, Oguiza JA, Perez G, Pisabarro AG, Ramirez L, Santoyo F, Master E, Coutinho PM, Henrissat B, Lombard V, Magnuson JK, Kües U, Hori C, Igarashi K, Samejima M, Held BW, Barry KW, LaButti KM, Lapidus A, Lindquist EA, Lucas SM, Riley R, et al. 2012. Comparative genomics of Ceriporiopsis subvermispora and Phanerochaete chrysosporium provide insight into selective ligninolysis. Proc Natl Acad Sci USA 109:5458–5463. doi: 10.1073/pnas.1119912109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martínez AT, Otillar R, Spatafora JW, Yadav JS, Aerts A, Benoit I, Boyd A, Carlson A, Copeland A, Coutinho PM, de Vries RP, Ferreira P, Findley K, Foster B, Gaskell J, Glotzer D, Górecki P, Heitman J, Hesse C, Hori C, Igarashi K, Jurgens JA, Kallen N, Kersten P, Kohler A, Kües U, Kumar TKA, Kuo A, LaButti K, Larrondo LF, Lindquist E, Ling A, Lombard V, Lucas S, Lundell T, Martin R, McLaughlin DJ, Morgenstern I, Morin E, Murat C, Nagy LG, Nolan M, Ohm RA, Patyshakuliyeva A, et al. 2012. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336:1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- 8.Binder M, Justo A, Riley R, Salamov A, Lopez-Giraldez F, Sjökvist E, Copeland A, Foster B, Sun H, Larsson E, Larsson KH, Townsend J, Grigoriev IV, Hibbett DS. 2013. Phylogenetic and phylogenomic overview of the Polyporales. Mycologia 105:1350–1373. doi: 10.3852/13-003. [DOI] [PubMed] [Google Scholar]
- 9.Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. 2014. The Carbohydrate-Active Enzymes Database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kehr R, Dujesiefken D, Wohlers A, Wulf A. 2000. Root and butt decay of Robinia pseudoacacia caused by Perenniporia fraxinea. Mitt Biol Bundesanst Land Forstwirtsch 370:92–96. [Google Scholar]
- 11.Guglielmo F, Bergemann SE, Gonthier P, Nicolotti G, Garbelotto M. 2007. A multiplex PCR-based method for the detection and early identification of wood rotting fungi in standing trees. J Appl Microbiol 103:1490–1507. doi: 10.1111/j.1365-2672.2007.03378.x. [DOI] [PubMed] [Google Scholar]
- 12.Szczepkowski A. 2004. Perenniporia fraxinea (fungi, Polyporales), a new species for Poland. Pol Bot J 49:73–77. [Google Scholar]
- 13.Zhao C-L, Cui B-K. 2012. A new species of Perenniporia (Polyporales, Basidiomycota) described from southern China based on morphological and molecular characters. Mycol Prog 11:555–560. doi: 10.1007/s11557-011-0770-1. [DOI] [Google Scholar]
- 14.Sillo F, Savino E, Giordano L, Girometta C, Astegiano D, Picco AM, Gonthier P. 2016. Analysis of genotypic diversity provides a first glimpse on the patterns of spread of the wood decay fungus Perenniporia fraxinea in an urban park in northern Italy. J Plant Pathol 98:617–624. [Google Scholar]
- 15.Robledo GL, Amalfi M, Castillo G, Rajchenberg M, Decock C. 2009. Perenniporiella chaquenia sp. nov. and further notes on Perenniporiella and its relationships with Perenniporia (Poriales, Basidiomycota). Mycologia 101:657–673. doi: 10.3852/08-040. [DOI] [PubMed] [Google Scholar]
- 16.Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F, Smirnova T, Nordberg H, Dubchak I, Shabalov I. 2014. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res 42:D699–D704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. 2012. DbCAN: a Web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Payne CM, Knott BC, Mayes HB, Hansson H, Himmel ME, Sandgren M, Ståhlberg J, Beckham GT. 2015. Fungal cellulases. Chem Rev 115:1308–1448. doi: 10.1021/cr500351c. [DOI] [PubMed] [Google Scholar]
- 20.Igarashi K, Ishida T, Hori C, Samejima M. 2008. Characterization of an endoglucanase belonging to a new subfamily of glycoside hydrolase family 45 of the basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol 74:5628–5634. doi: 10.1128/AEM.00812-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van den Brink J, de Vries RP. 2011. Fungal enzyme sets for plant polysaccharide degradation. Appl Microbiol Biotechnol 91:1477–1492. doi: 10.1007/s00253-011-3473-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Dilokpimol A, Kabel MA, de Vries RP. 2022. Fungal xylanolytic enzymes: diversity and applications. Bioresour Technol 344:126290. doi: 10.1016/j.biortech.2021.126290. [DOI] [PubMed] [Google Scholar]
- 23.Kersten P, Cullen D. 2007. Extracellular oxidative systems of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Fungal Genet Biol 44:77–87. doi: 10.1016/j.fgb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Forsberg Z, Sørlie M, Petrović D, Courtade G, Aachmann FL, Vaaje-Kolstad G, Bissaro B, Røhr ÅK, Eijsink VG. 2019. Polysaccharide degradation by lytic polysaccharide monooxygenases. Curr Opin Struct Biol 59:54–64. doi: 10.1016/j.sbi.2019.02.015. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki H, Igarashi K, Samejima M. 2008. Real-time quantitative analysis of carbon catabolite derepression of cellulolytic genes expressed in the basidiomycete Phanerochaete chrysosporium. Appl Microbiol Biotechnol 80:99–106. doi: 10.1007/s00253-008-1539-6. [DOI] [PubMed] [Google Scholar]
- 26.Daly P, Peng M, Falco M, Lipzen A, Wang M, Ng V, Grigoriev I, Tsang A, Mäkelä MR, de Vries RP. 2019. Glucose-mediated repression of plant biomass utilization in the white-rot fungus Dichomitus squalens. Appl Environ Microbiol 85:e01828-19. doi: 10.1128/AEM.01828-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller GL. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 28.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. 2005. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Momeni MH, Payne CM, Hansson H, Mikkelsen NE, Svedberg J, Engström A, Sandgren M, Beckham GT, Stahlberg J. 2013. Structural, biochemical, and computational characterization of the glycoside hydrolase family 7 cellobiohydrolase of the tree-killing fungus Heterobasidion irregulare. J Biol Chem 288:5861–5872. doi: 10.1074/jbc.M112.440891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hori C, Igarashi K, Katayama A, Samejima M. 2011. Effects of xylan and starch on secretome of the basidiomycete Phanerochaete chrysosporium grown on cellulose. FEMS Microbiol Lett 321:14–23. doi: 10.1111/j.1574-6968.2011.02307.x. [DOI] [PubMed] [Google Scholar]
- 31.Giraldo MC, Valent B. 2013. Filamentous plant pathogen effectors in action. Nat Rev Microbiol 11:800–814. doi: 10.1038/nrmicro3119. [DOI] [PubMed] [Google Scholar]
- 32.Kubicek CP, Starr TL, Glass NL. 2014. Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu Rev Phytopathol 52:427–451. doi: 10.1146/annurev-phyto-102313-045831. [DOI] [PubMed] [Google Scholar]
- 33.Zeng Z, Sun H, Vainio EJ, Raffaello T, Kovalchuk A, Morin E, Duplessis S, Asiegbu FO. 2018. Intraspecific comparative genomics of isolates of the Norway spruce pathogen (Heterobasidion parviporum) and identification of its potential virulence factors. BMC Genomics 19:220. doi: 10.1186/s12864-018-4610-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sipos G, Prasanna AN, Walter MC, O’Connor E, Bálint B, Krizsán K, Kiss B, Hess J, Varga T, Slot J, Riley R, Bóka B, Rigling D, Barry K, Lee J, Mihaltcheva S, LaButti K, Lipzen A, Waldron R, Moloney NM, Sperisen C, Kredics L, Vágvölgyi C, Patrignani A, Fitzpatrick D, Nagy I, Doyle S, Anderson JB, Grigoriev IV, Güldener U, Münsterkötter M, Nagy LG. 2017. Genome expansion and lineage-specific genetic innovations in the forest pathogenic fungi Armillaria. Nat Ecol Evol 1:1931–1941. doi: 10.1038/s41559-017-0347-8. [DOI] [PubMed] [Google Scholar]
- 35.Jonas H, Geoffrey D, Ulla W. 2000. The distribution of acidic and esterified pectin in cambium, developing xylem and mature xylem of Pinus sylvestris. IAWA J 21:157–168. [Google Scholar]
- 36.Morris H, Brodersen C, Schwarze FWMR, Jansen S. 2016. The parenchyma of secondary xylem and its critical role in tree defense against fungal decay in relation to the CODIT model. Front Plant Sci 7:1665. doi: 10.3389/fpls.2016.01665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwarze FWMR, Baum S. 2000. Mechanisms of reaction zone penetration by decay fungi in wood of beech (Fagus sylvatica). New Phytol 146:129–140. doi: 10.1046/j.1469-8137.2000.00624.x. [DOI] [Google Scholar]
- 38.Ueta M, Hori C, Tamai Y, Yamagishi Y, Miyamoto T, Sano Y. 2018. Changes in the secondary xylem of the living stem of four tree species in response to inoculation with Perenniporia fraxinea. Mokuzai Gakkaishi 64:1–9. doi: 10.2488/jwrs.64.1. [DOI] [Google Scholar]
- 39.Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 40.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hori C, Song R, Matsumoto K, Matsumoto R, Minkoff BB, Oita S, Hara H, Takasuka TE. 2020. Proteomic characterization of lignocellulolytic enzymes secreted by the insect-associated fungus Daldinia decipiens oita, isolated from a forest in northern Japan. Appl Environ Microbiol 86:e02350-19. doi: 10.1128/AEM.02350-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez D, Larrondo LF, Putnam N, Gelpke MDS, Huang K, Chapman J, Helfenbein KG, Ramaiya P, Detter JC, Larimer F, Coutinho PM, Henrissat B, Berka R, Cullen D, Rokhsar D. 2004. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol 22:695–700. doi: 10.1038/nbt967. [DOI] [PubMed] [Google Scholar]
- 43.Schülein M. 1997. Enzymatic properties of cellulases from Humicola insolens. J Biotechnol 57:71–81. doi: 10.1016/s0168-1656(97)00090-4. [DOI] [PubMed] [Google Scholar]
- 44.Slomczynski D, Nakas J, Tanenbaum S. 1995. Production and characterization of laccase from Botrytis cinerea 61-34. Appl Environ Microbiol 61:907–912. doi: 10.1128/aem.61.3.907-912.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M, Yabana M, Harada M, Nagayasu E, Maruyama H, Kohara Y, Fujiyama A, Hayashi T, Itoh T. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res 24:1384–1395. doi: 10.1101/gr.170720.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.The UniProt Consortium. 2017. UniProt: the universal protein knowledgebase. Nucleic Acids Res 45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki S, Kakuta M, Ishida T, Akiyama Y. 2014. GHOSTX: an improved sequence homology search algorithm using a query suffix array and a database suffix array. PLoS One 9:e103833. doi: 10.1371/journal.pone.0103833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iwata H, Gotoh O. 2012. Benchmarking spliced alignment programs including Spaln2, an extended version of Spaln that incorporates additional species-specific features. Nucleic Acids Res 40:e161. doi: 10.1093/nar/gks708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aem.00272-23-s0001.pdf, PDF file, 7.4 MB (7.4MB, pdf)
Supplemental material. Download aem.00272-23-s0002.xlsx, XLSX file, 4.2 MB (4.2MB, xlsx)
Data Availability Statement
The ITS sequences of P. fraxinea SS1 to SS5 were deposited in the NCBI database (DDBJ/EMBL/GenBank accession no. LC556223 to LC556227).