ABSTRACT
Porcine deltacoronavirus (PDCoV) is an emerging swine enteropathogenic coronavirus that has the potential to infect humans. Histone deacetylase 6 (HDAC6) is a unique type IIb cytoplasmic deacetylase with both deacetylase activity and ubiquitin E3 ligase activity, which mediates a variety of cellular processes by deacetylating histone and nonhistone substrates. In this study, we found that ectopic expression of HDAC6 significantly inhibited PDCoV replication, while the reverse effects could be observed after treatment with an HDAC6-specific inhibitor (tubacin) or knockdown of HDAC6 expression by specific small interfering RNA. Furthermore, we demonstrated that HDAC6 interacted with viral nonstructural protein 8 (nsp8) in the context of PDCoV infection, resulting in its proteasomal degradation, which was dependent on the deacetylation activity of HDAC6. We further identified the key amino acid residues lysine 46 (K46) and K58 of nsp8 as acetylation and ubiquitination sites, respectively, which were required for HDAC6-mediated degradation. Through a PDCoV reverse genetics system, we confirmed that recombinant PDCoV with a mutation at either K46 or K58 exhibited resistance to the antiviral activity of HDAC6, thereby exhibiting higher replication compared with wild-type PDCoV. Collectively, these findings contribute to a better understanding of the function of HDAC6 in regulating PDCoV infection and provide new strategies for the development of anti-PDCoV drugs.
IMPORTANCE As an emerging enteropathogenic coronavirus with zoonotic potential, porcine deltacoronavirus (PDCoV) has sparked tremendous attention. Histone deacetylase 6 (HDAC6) is a critical deacetylase with both deacetylase activity and ubiquitin E3 ligase activity and is extensively involved in many important physiological processes. However, little is known about the role of HDAC6 in the infection and pathogenesis of coronaviruses. Our present study demonstrates that HDAC6 targets PDCoV-encoded nonstructural protein 8 (nsp8) for proteasomal degradation through the deacetylation at the lysine 46 (K46) and the ubiquitination at K58, suppressing viral replication. Recombinant PDCoV with a mutation at K46 and/or K58 of nsp8 displayed resistance to the antiviral activity of HDAC6. Our work provides significant insights into the role of HDAC6 in regulating PDCoV infection, opening avenues for the development of novel anti-PDCoV drugs.
KEYWORDS: porcine deltacoronavirus, nsp8, HDAC6, deacetylation, ubiquitination
INTRODUCTION
Porcine deltacoronavirus (PDCoV) is an emerging enteropathogenic coronavirus that can cause acute watery diarrhea, vomiting, dehydration, and even death in nursing piglets (1, 2). PDCoV was first detected from pig rectal swab samples for virological surveillance in Hong Kong in 2012 (3); the initial outbreak of PDCoV was reported in several pig farms with symptoms of acute diarrhea in Ohio in the United States in 2014. PDCoV spread rapidly to many countries and regions, including China, South Korea, Thailand, Japan, Lao People’s Democratic Republic, Vietnam, Mexico, and Peru, leading to significant economic losses (4–8). Accumulating evidence has shown that in addition to pigs, PDCoV can infect calves, turkeys, chickens, mice, and even humans, suggesting that it possesses cross-species transmission and zoonotic potential (1, 9–12). It was recently proposed that PDCoV can be considered the eighth coronavirus infecting humans after human coronavirus 229E, human coronavirus NL63, human coronavirus OC43, human coronavirus HKU1, severe acute respiratory syndrome coronavirus (SARS-CoV), SARS-CoV-2, and Middle Eastern respiratory syndrome coronavirus (MERS-CoV) (13). The significant threat posed by PDCoV to human and animal health has sparked tremendous attention.
PDCoV is a single-stranded, positive-sense RNA virus that belongs to the newly identified genus Deltacoronavirus in the family Coronaviridae. Its genome is approximately 25.4 kb in length (3, 14), encoding two large polyproteins, polyprotein1a (pp1a) and polyprotein 1ab (pp1ab), four structural proteins (spike [S], envelope [E], membrane [M], and nucleocapsid [N]), and three accessory proteins (NS6, NS7, and NS7a). The two polyproteins pp1a and pp1ab are cleaved proteolytically into 15 mature nonstructural proteins (nsp2 to nsp16) by the papain-like protease (PLpros) nsp3 and the 3C-like protease nsp5 (15, 16). Of them, nsp8 is a 189-amino-acid polypeptide that is relatively conserved within the family Coronaviridae. It combines with nsp7 and nsp12 to form a viral replication and transcription complex (RTC) that is indispensable for viral replication (17, 18). However, the detailed role of nsp8 in the infection and pathogenesis of coronaviruses remains largely unknown.
Lysine acetylation is a reversible posttranslational modification that affects many cellular processes and modulates viral replication (19–22), which is competitively regulated by two classes of enzymes, histone acetyltransferases (HATs) and histone deacetylases (HDACs) (23). To date, at least 18 HDACs have been identified in mammals. Among them, HDAC6 is a unique type IIb cytoplasmic deacetylase with two deacetylase structural domains and a C-terminal zinc finger ubiquitin binding domain (BUZ) (24), which mediates a variety of cellular processes by deacetylating nonhistone substrates, including heat shock protein 90, microtubulin, cortical agonist protein, and retinoic-acid-inducible gene-I (RIG-I) (25–27). Increasing evidence has demonstrated that HDAC6 is involved in the regulation of viral replication. For example, overexpression of HDAC6 inhibits the acetylation of α-tubulin (Ac-α-tubulin) and remarkably suppresses human immunodeficiency virus type 1 (HIV-1) envelope-dependent cell fusion and infection (28). HDAC6 downregulates the trafficking of viral components to the plasma membrane, resulting in the suppression of influenza A virus (IAV) release (15, 29). HDAC6 enzyme inhibitor treatment promotes the gene expression of human T-lymphocytic leukemia virus type 1 (HTLV-1) (30). HDAC6 depletion enhances the replication of vesicular stomatitis virus (VSV) by negatively regulating the production of type I interferon (31). Overexpression of HDAC6 enhances resistance to porcine reproductive and respiratory syndrome virus (PRRSV) both in vitro and in vivo (32). Collectively, most studies have shown that HDAC6 overexpression enhances the resistance of cells and transgenic mice/pigs to viral infection. However, whether HDAC6 is involved in the regulation of PDCoV replication is unknown.
In this study, we identified HDAC6 as a novel negative regulator of PDCoV infection. Mechanistically, HDAC6 interacted with nsp8, resulting in its deacetylation and ubiquitination modification and subsequent proteasomal degradation. Furthermore, we demonstrated that lysine 46 (K46) and K58 of nsp8 were required for its degradation by HDAC6. Importantly, recombinant PDCoV, mutated at K46 and/or K58, displayed obvious resistance to the antiviral activity of HDAC6.
RESULTS
PDCoV infection downregulates HDAC6 expression.
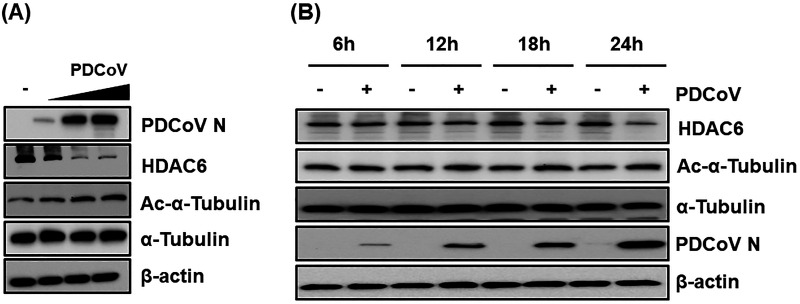
To investigate the role of HDAC6 in PDCoV infection, we first examined the dynamic changes in HDAC6 protein expression after PDCoV infection in LLC-PK1 cells. As shown in Fig. 1A, PDCoV infection significantly downregulated HDAC6 expression in a dose-dependent manner. A previous study demonstrated that cellular microtubulin, α-tubulin, is a substrate of HDAC6 and that the acetylation of α-tubulin, which represents the polymerization of microtubules, is favorable for viral proliferation (26); thus, we also detected cellular acetylation level after PDCoV infection and found that the acetylation of α-tubulin was gradually increased in PDCoV-infected cells (Fig. 1A). In addition, we detected the expression kinetics of HDAC6 at different time points after PDCoV infection. As shown in Fig. 1B, HDAC6 expression was reduced in a time-dependent manner, especially in the late stages of PDCoV infection. These results suggest that HDAC6 may play a regulatory role in PDCoV infection.
FIG 1.
The dynamic changes in HDAC6 expression during PDCoV infection. (A) LLC-PK1 cells were infected with PDCoV at an increasing multiplicity of infection (MOI = 0.1, 0.5, or 1). At 12 hpi, cells were collected for Western blotting using antibodies against HDAC6, α-tubulin, Ac-α-tubulin, and PDCoV N protein. (B) LLC-PK1 cells were infected with PDCoV (MOI = 0.5). At 6, 12, 18, and 24 hpi, cells were collected for Western blotting assay to detect the expression of HDAC6, α-tubulin, Ac-α-tubulin, PDCoV N, and β-actin.
HDAC6 inhibits PDCoV infection.
To determine the effect of HDAC6 on PDCoV infection, experiments based on three strategies, namely, ectopic expression, chemical inhibition, and small interfering RNA (siRNA), were performed. First, LLC-PK1 cells were transfected with increasing amounts of expression plasmids encoding Flag-HDAC6, which were then infected with PDCoV (multiplicity of infection [MOI] of 0.5) for 12 h. The results showed that overexpression of HDAC6 inhibited PDCoV infection, as evidenced by decreased N protein expression (Fig. 2A), viral mRNA expression (Fig. 2B), and viral titers (Fig. 2C). To clarify which stages of PDCoV infection are affected by HDAC6 most, the experiments were performed to investigate the viral kinetics in the context of HDAC6 overexpression. As shown in Fig. 2D, HDAC6 overexpression significantly downregulated the acetylation of α-tubulin and PDCoV N protein expression, but the inhibitory effects were weakened in a time-dependent manner, suggesting that the early stages of PDCoV infection are most affected by HDAC6.
FIG 2.
HDAC6 inhibits PDCoV infection. (A to C) The effect of HDAC6 overexpression on PDCoV infection. LLC-PK1 cells were transfected with increasing amounts of pCAGGS-Flag-HDAC6 or empty vector for 12 h and then infected with PDCoV (MOI = 0.5). At 12 hpi, the cells were collected and subjected to Western blotting (A), RT-qPCR (B), and a TCID50 assay (C). (D) LLC-PK1 cells were transfected with pCAGGS-Flag-HDAC6 or empty vector and then infected with PDCoV (MOI = 0.5). Cells were collected at 6, 12, and 18 hpi and subjected to Western blotting assay for detecting the expressions of Flag-HDAC6, α-tubulin, Ac-α-tubulin, PDCoV N, and β-actin. Density analysis for targeted proteins in Western blotting pictures was performed via ImageJ software. The relative expression levels of Ac-α-tubulin and PDCoV N compared to those of the control vector group were shown on the right side. (E and F) LLC-PK1 cells were pretreated with tubacin and then untreated (E) or infected with PDCoV (MOI = 0.5) (F). At 12 hpi, cells were collected and subjected to Western blotting assay for detecting the expressions of HDAC6, Ac-α-tubulin, α-tubulin, PDCoV N, and β-actin. (G and H) The effect of an HDAC6-specific inhibitor tubacin on PDCoV infection. LLC-PK1 cells were pretreated with tubacin for 2 h and then infected with PDCoV (MOI = 0.5). At 12 hpi, cells were collected and subjected to RT-qPCR (G) and a TCID50 assay (H). (I to K) The effect of HDAC6 knockdown on PDCoV infection. LLC-PK1 cells were transfected with siRNA against HDAC6 or with control siRNA and were then infected with PDCoV (MOI = 0.5) for 12 h. Cell samples were harvested and subjected to Western blotting (I), RT-qPCR (J), and a TCID50 assay (K) to determine relative HDAC6 protein expression, viral mRNA levels, and viral titers, respectively. The presented results represent the means and standard deviations of the data from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Tubacin, an HDAC6-specific selective inhibitor, was extensively used in previous studies (33). We first tested the HDAC6 protein expression after tubacin treatment. The results showed that tubacin treatment alone increased the Ac-α-tubulin expression in a dose-dependent manner but had little effect on HDAC6 and α-tubulin expression (Fig. 2E), in agreement with a previous study that tubacin affects the activity of HDAC6 but not its protein expression level (34). To test the effect of tubacin on PDCoV infection, LLC-PK1 cells were pretreated with different doses of tubacin for 2 h with no detectable cytotoxicity and then infected with PDCoV. The results showed that tubacin treatment significantly reduced the HDAC6 expression while increasing the acetylation of α-tubulin, N protein expression (Fig. 2F), viral mRNA expression (Fig. 2G), and viral titers (Fig. 2H) in a dose-dependent manner, suggesting that the inhibition of HDAC6 activity promotes PDCoV infection. To further confirm the anti-PDCoV function of HDAC6, LLC-PK1 cells were transfected with specific siRNA to knock down HDAC6 expression and then infected with PDCoV. Compared with the control siRNA, HDAC6-specific siRNA notably knocked down HDAC6 expression and promoted the acetylation of α-tubulin and PDCoV infection, as demonstrated by the results of Western blotting, quantitative real-time RT-PCR (RT-qPCR), and a 50% tissue culture infective dose (TCID50) assay (Fig. 2I to K). Collectively, these findings indicate that HDAC6 exerts anti-PDCoV effects.
HDAC6 interacts with nsp5 and nsp8 of PDCoV.
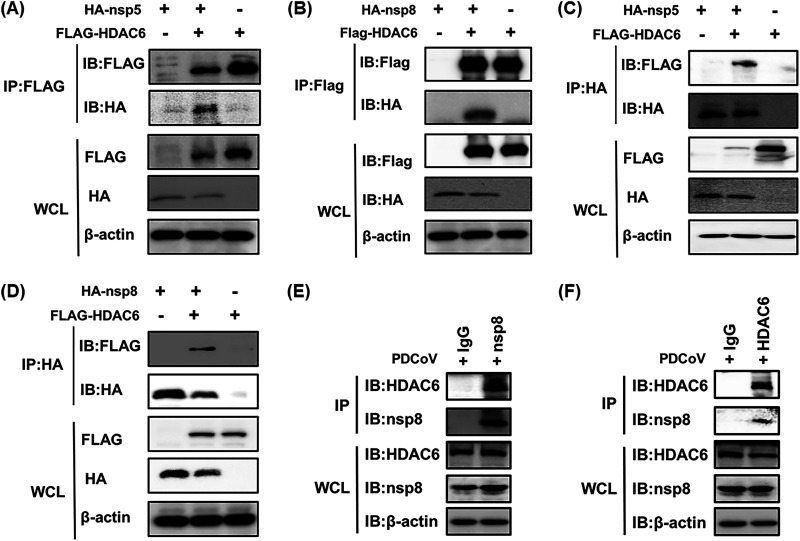
Since HDAC6 negatively regulates PDCoV infection, we next explored the mechanism by which this occurs. We speculated that HDAC6 could function by interacting with viral protein(s) encoded by PDCoV. To confirm this hypothesis, HEK-293T cells were cotransfected with pCAGGS-Flag-HDAC6 and HA-tagged expression plasmids encoding each individual PDCoV protein, followed by a coimmunoprecipitation (Co-IP) assay with anti-Flag antibodies. The results showed that HDAC6 interacted with nsp5, nsp8, NS7, NS7a, and N protein (Fig. 3A and B; Fig. S1A in the supplemental material). To further confirm the interactions between HDAC6 and these viral proteins, a reverse Co-IP experiment was performed with anti-HA antibodies. The results showed that only nsp5 and nsp8 could be efficiently coimmunoprecipitated with HDAC6, not NS7, NS7a, and N protein (Fig. 3C and D; Fig. S1B). Combining the results of forward and reverse Co-IP experiments, we concluded that HDAC6 interacts with nsp5 and nsp8 of PDCoV in an overexpression system. To further confirm the endogenous interaction in the context of PDCoV infection, we expressed and purified the recombinant nsp5 and nsp8 proteins in Escherichia coli to generate the corresponding rabbit polyclonal antibodies, respectively. However, we only successfully obtained the antibody against nsp8. As shown in Fig. 3E and F, the results from Co-IP assays confirmed that an interaction between PDCoV nsp8 and endogenous HDAC6 was detected in PDCoV-infected cells. Interestingly, from the results in Fig. 3B and D, we found that the expression level of nsp8 significantly decreased on coexpression with HDAC6. A previous study showed that HDAC6 possesses both deacetylase activity and ubiquitin E3 ligase activity (35), which has the potential to affect protein stability. We speculated that HDAC6 inhibited PDCoV replication by affecting the stability of nsp8. Thus, we focused on the HDAC6-nsp8 interaction in subsequent studies.
FIG 3.
HDAC6 interacts with nsp5 and nsp8 of PDCoV. (A to D) HEK-293T cells were cotransfected with pCAGGS-Flag-HDAC6 and pCAGGS-HA-nsp5 (A and C) or pCAGGS-HA-nsp8 (B and D) for 28 h. The cells were lysed and immunoprecipitated with anti-Flag (A and B) or anti-HA antibodies (C and D). (E and F) LLC-PK1 cells were infected with PDCoV (MOI = 0.5) for 12 h. The cells were lysed and then subjected to Co-IP assay using anti-nsp8 (E) or anti-HDAC6 (F) antibodies. Whole-cell lysates (WCL) and immunoprecipitation (IP) complexes were analyzed by Western blotting with antibodies against Flag, HA, nsp8, HDAC6, or β-actin.
HDAC6 degrades nsp8 via the ubiquitin-proteasome pathway.
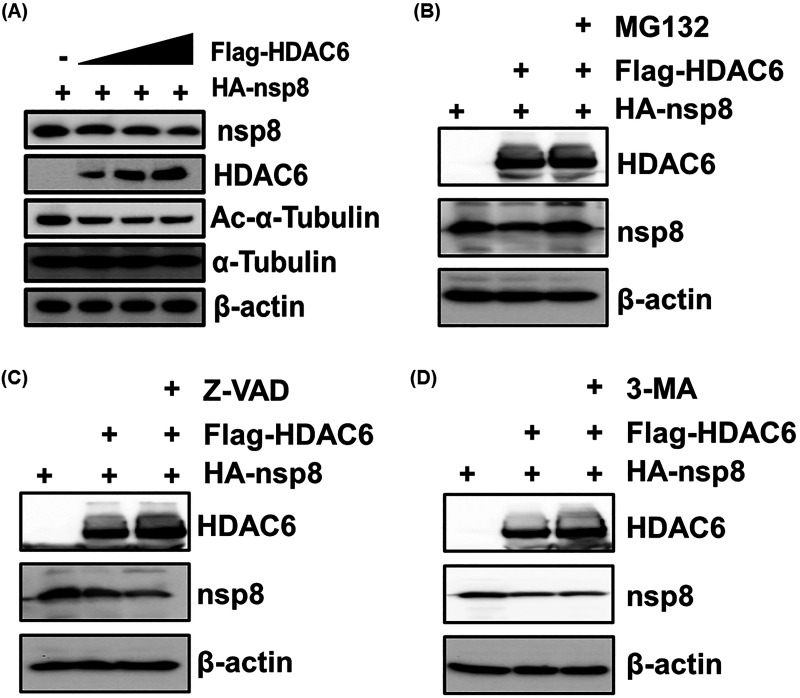
To further determine whether HDAC6 degrades nsp8, LLC-PK1 cells were cotransfected with fixed amounts of pCAGGS-HA-nsp8 and increasing quantities of pCAGGS-Flag-HDAC6 for 28 h, followed by a Western blotting assay. The results showed that the levels of both Ac-α-tubulin and nsp8 expression gradually decreased with increasing HDAC6 expression (Fig. 4A), demonstrating that HDAC6 mediated the degradation of nsp8. We next investigated the degradation pathways by which this event occurred. As shown in Fig. 4, the results revealed that the HDAC6-mediated degradation of nsp8 was reversed by treatment with the proteasome pathway inhibitor MG132 but not the apoptosis pathway inhibitor Z-VAD or the autophagy pathway inhibitor 3-MA (Fig. 4B to D). Taken together, these results suggest that HDAC6 mediates the degradation of nsp8 via the ubiquitin proteasomal pathway.
FIG 4.
HDAC6-mediated degradation of nsp8 is dependent on the ubiquitin proteasome pathway. (A) LLC-PK1 cells were cotransfected with fixed amounts of pCAGGS-HA-nsp8 and increasing quantities of pCAGGS-Flag-HDAC6 for 28 h, and then the cell lysates were analyzed by Western blotting with the indicated antibodies. (B to D) HEK-293T cells were cotransfected with pCAGGS-HA-nsp8 and pCAGGS-Flag-HDAC6 or empty vector and then treated by MG132 (20 μM) (B), Z-VAD (20 μM) (C), or 3-MA (5 mM) (D) for 8 h before collecting cell samples were collected and Western blotting assay was performed.
HDAC6-mediated degradation of nsp8 is dependent on its deacetylase activity.
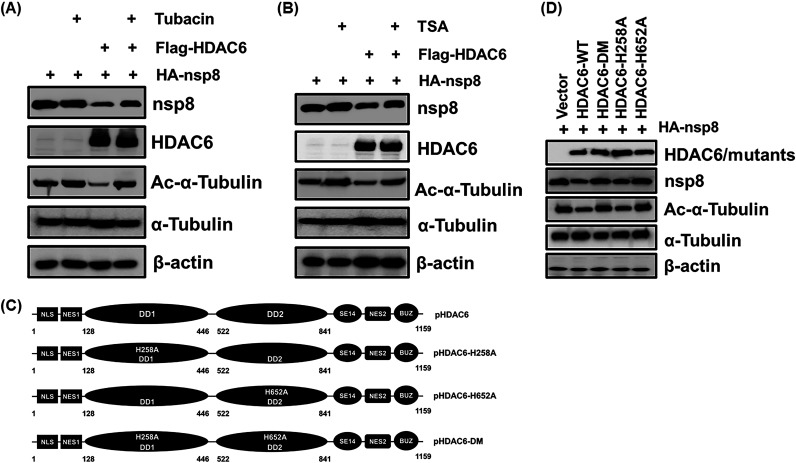
A previous study showed that N-terminal acetylated proteins can be stabilized by avoiding the proteasome-dependent degradation pathways (36). To explore whether HDAC6-mediated degradation of nsp8 is associated with its deacetylase activity, LLC-PK1 cells were cotransfected with pCAGGS-HA-nsp8 and pCAGGS-Flag-HDAC6 and then, respectively, treated with tubacin and the pan-HDAC inhibitor TSA, followed by a Western blotting assay. The results showed that HDAC6 significantly suppressed the acetylation of α-tubulin and nsp8 expression; however, the inhibition was obviously restored by treatment with both tubacin and TSA (Fig. 5A and B). To further confirm this result, three HDAC6 deacetylase dead mutants, namely, HDAC6-H258A, HDAC6-H652A, and HDAC6-DM (-H258/652A), were constructed according to a previous study (Fig. 5C) (37). As shown in Fig. 5D, HDAC6-WT notably reduced nsp8 expression; however, the inhibition, to a large extent, was attenuated in cells expressing HDAC6-DM and HDAC6-H652A, but not in cells expressing HDAC6-H258A, in agreement with a previous report that HDAC6 H258 plays a minor role in its deacetylase activity (38, 39). These results demonstrate that HDAC6 mediates the degradation of nsp8 and that this is dependent on its deacetylase activity.
FIG 5.
HDAC6-mediated degradation of PDCoV nsp8 is dependent on its deacetylase activity. (A and B) LLC-PK1 cells were cotransfected with pCAGGS-HA-nsp8 and pCAGGS-Flag-HDAC6 or empty vector, along with or without treatment with tubacin (5 μM) (A) or TSA (1 μM) (B) for 8 h. The cells were collected and subjected to Western blotting for detecting HA-nsp8, Flag-HDAC6, Ac-α-tubulin, α-tubulin, and β-actin. (C) Schematic diagram of porcine HDAC6 (pHDAC6) and pHDAC6 deacetylase dead mutants. (D) LLC-PK1 cells were cotransfected with pCAGGS-HA-nsp8 and expression plasmids encoding HDAC6-WT, HDAC6-DM, HDAC6-H258A, HDAC6-H652A, or empty vector for 28 h. The cell lysates were subjected to a Western blotting assay.
Identification of nsp8 acetylation sites at K37 and K46 and its ubiquitination site at K58.
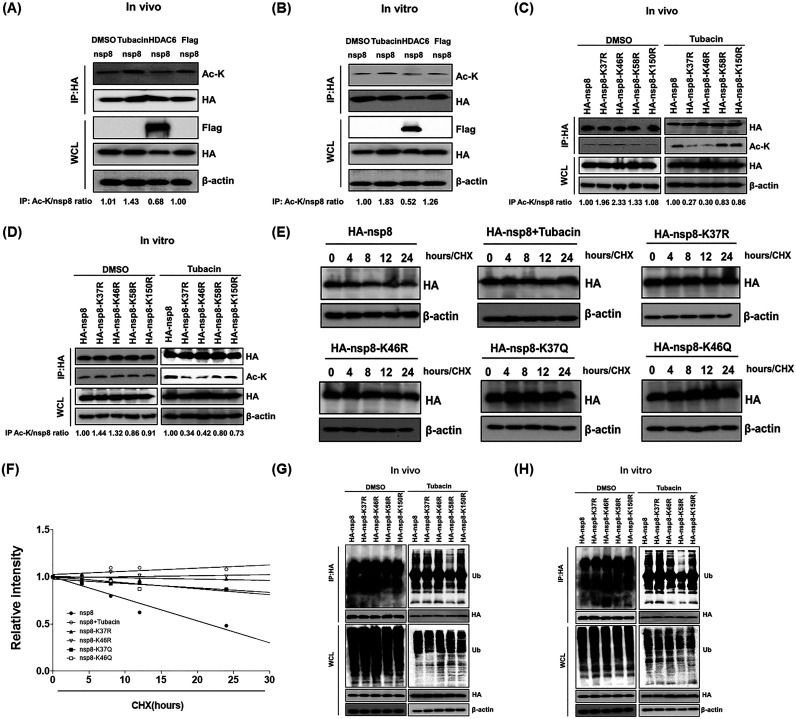
To determine whether HDAC6-mediated deacetylation of nsp8 occurs, we detected the acetylation level of nsp8 using two different methods defined as in vitro and in vivo, respectively, as described in previous studies (40, 41). As shown in Fig. 6A and B, the acetylation level of nsp8 was significantly higher after treatment with tubacin compared with coexpression or coincubation with HDAC6, demonstrating that HDAC6 was able to mediate deacetylation of nsp8. Because HDAC6 also mediated nsp8 degradation via the ubiquitin-proteasome pathway (Fig. 4B) and HDAC6 possesses both deacetylase activity and ubiquitin E3 ligase activity (35), we predicted the potential acetylation and ubiquitination sites of nsp8 using online resources (www.biocuckoo.org and www.abcepta.com.cn). Based on the obtained scores, lysine 37 (K37) and K150 in nsp8 were the most probable acetylation sites, and ubiquitination sites were possibly located at K46 and K58. To confirm these possible acetylation and ubiquitination sites, four nsp8 mutants with a single point mutation of lysine (K) to arginine (R) (nsp8-K37R, nsp8-K46R, nsp8-K58R, and nsp8-K150R) were constructed and overexpressed in HEK-293T cells, followed by in vitro and in vivo acetylation assays. The results showed that the acetylation level of nsp8-K37R and -K46R mutants had no reduction without tubacin treatment compared with that of wild-type HA-nsp8, which inversely had a slight increase. However, the acetylation levels of mutants nsp8-K37R and nsp8-K46R were significantly lower than that of wild-type nsp8 after tubacin treatment (Fig. 6C and D), demonstrating that HDAC6 mediates the deacetylation of nsp8 at the K37 and K46 sites.
FIG 6.
Determination of nsp8 acetylation sites at K37 and K46, and its ubiquitination at K58. (A) HEK-293T cells were transfected with pCAGGS-HA-nsp8 and pCAGGS-Flag-HDAC6 or empty vector, and then treated with DMSO or tubacin. The cell samples were collected and subjected to a Co-IP assay with anti-HA antibodies and Western blotting for detecting the acetylation level of nsp8 in vivo. (B) The cell lysates with nsp8 expression were immunoprecipitated with anti-HA antibodies and then the immunoprecipitation complexes were incubated with DMSO, tubacin, or the lysates of cell with HDAC6 expression or empty vector transfection for 8 h in vitro, followed by treatment as described in panel A and Western blotting assay. (C) HEK-293T cells were transfected with pCAGGS-HA-nsp8 or expression plasmids encoding nsp8 mutants and then treated with DMSO or tubacin for 8 h in vivo, followed by a Co-IP assay with anti-HA antibodies and Western blotting as described in panel A. (D) The cell lysates with the expression of HA-nsp8 or HA-nsp8 mutants were immunoprecipitated with anti-HA antibodies, and then the immunoprecipitation complexes were incubated with DMSO or tubacin for 8 h in vitro, followed by treatment as described in panel A and Western blotting assay. (E) HEK-293T cells were transfected with pCAGGS-HA-nsp8 or expression plasmids encoding each of nsp8 mutants and then left untreated or treated with tubacin, followed by treatment with CHX (10 μg/mL). The cells were collected at different time points (0, 4, 8, 12, and 24 h) and subjected to a Western blotting assay. (F) Calculated relative half-lives of HA-nsp8 or HA-nsp8 mutants from panel E. The relative intensity was plotted versus time. (G) HEK-293T cells were transfected with expression plasmids encoding HA-nsp8 or HA-nsp8 mutants and then treated with DMSO or tubacin for 8 h in vivo as described in panel A. The cell lysates were subjected to a Western blotting assay for detecting the ubiquitination level of HA-nsp8 and its mutants. (H) The cell lysates with the expression of HA-nsp8 or HA-nsp8 mutants were immunoprecipitated with anti-HA antibodies and then the immunoprecipitation complexes were incubated with DMSO or tubacin for 8 h in vitro, followed by detection of the ubiquitination level of HA-nsp8 and its mutants via Western blotting as described in panel G.
We further investigated whether the acetylation status of nsp8 affects its stability. To address this, two acetylation-mimicking nsp8 mutants (nsp8-K37Q and nsp8-K46Q) and two acetylation-dead nsp8 mutants (nsp8-K37R and nsp8-K46R) were overexpressed in HEK-293T cells and then treated by CHX, followed by a Western blotting assay. Tubacin treatment served as a control. The results showed that the protein levels of wild-type nsp8 gradually reduced in a time-dependent manner; however, the relative half-life of nsp8 was significantly increased by tubacin treatment or acetylation mimicry and death mutations (Fig. 6E and F). These results supported the notion that HDAC6-mediated deacetylation of nsp8 promoted its proteasomal degradation. We further identified the potential ubiquitination sites of nsp8 by in vitro and in vivo ubiquitination assays according to the similar description in Fig. 6C and D. As shown in Fig. 6G and H, the ubiquitination level of HA-nsp8 mutants had no reduction without tubacin treatment compared with that of wild-type HA-nsp8. However, the ubiquitination level of the nsp8-K58R mutant was significantly reduced compared with that of wild-type nsp8 after tubacin treatment. Taken together, we conclude that the acetylation sites of nsp8 are located at K37 and K46, and its ubiquitination site is located at K58.
Sites K46 and K58 of nsp8 are required for its degradation by HDAC6.
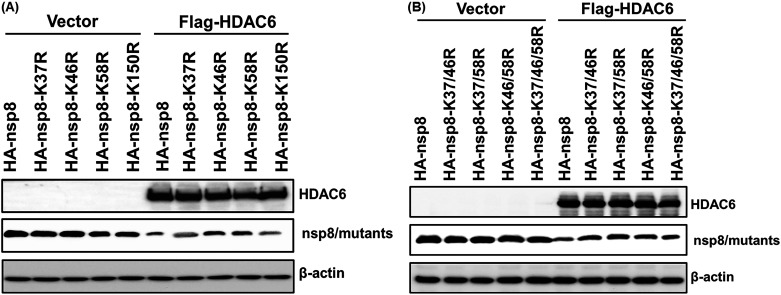
To determine whether HA-nsp8-K37R, HA-nsp8-K46R, and HA-nsp8-K58R have resistance to the degradation mediated by HDAC6, Flag-HDAC6 or the empty vector were, respectively, coexpressed with HA-nsp8-K37R, HA-nsp8-K46R, and HA-nsp8-K58R in HEK-293T cells. HA-nsp8 and HA-nsp8-K150R served as positive controls. The results showed that HA-nsp8 and HA-nsp8-K150R were significantly degraded by HDAC6 compared with the control group; however, HA-nsp8-K46R and HA-nsp8-K58R displayed obvious resistance to HDAC6-mediated degradation (Fig. 7A). We further constructed expression plasmids encoding nsp8 mutants with double mutations (HA-nsp8-K37/46R, HA-K37/58R, and HA-K46/58R) and triple mutations (HA-nsp8-K37/46/58R) and found that the four nsp8 mutants tested, with either K46 or K58 mutations, exhibited distinct resistance to HDAC6-mediated degradation (Fig. 7B). Taken together, we conclude that the sites at both K46 and K58 of nsp8 are required for HDAC6-mediated degradation.
FIG 7.
The sites at both K46 and K58 of nsp8 are required for HDAC6-mediated degradation. (A) HEK-293T cells were transfected with expression plasmids encoding HA-nsp8 or its mutants with a single point mutation, along with or without pCAGGS-Flag-HDAC6 for 28 h. The cells were collected and subjected to Western blotting with antibodies against Flag and HA. (B) HEK-293T cells were transfected with expression plasmids encoding HA-nsp8 or its mutants with double or triple mutations, along with or without pCAGGS-Flag-HDAC6 for 28 h, followed by Western blotting with antibodies against Flag and HA.
Recombinant PDCoV with a mutation at K46 and/or K58 of nsp8 displays resistance to the antiviral activity of HDAC6.
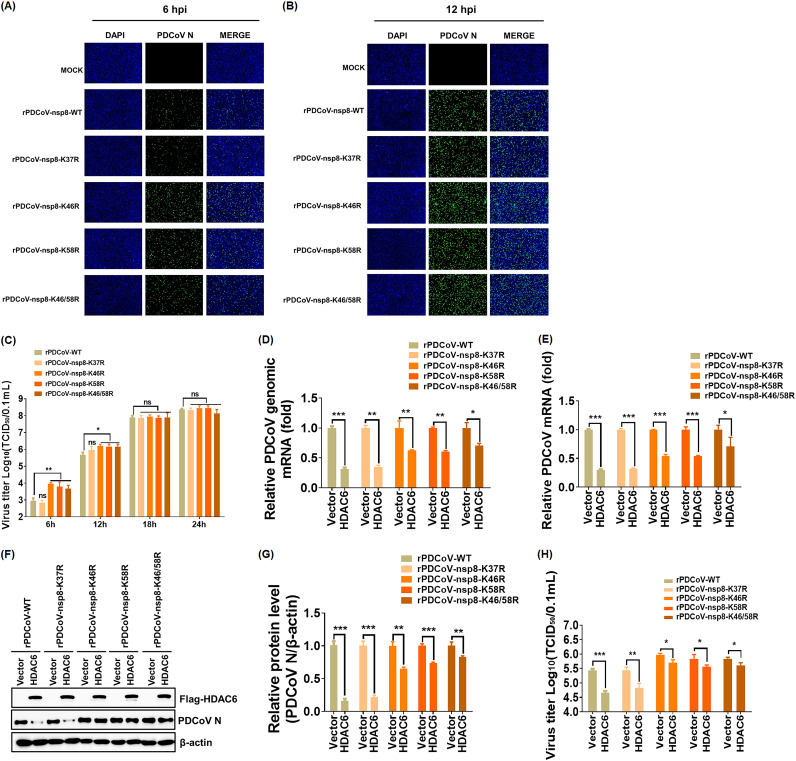
To further confirm the contribution of K46 and K58 of nsp8 to resistance to the antiviral activity of HDAC6, we successfully constructed recombinant PDCoV with a single point mutation of nsp8 at K37, K46, or K58 and with double mutations at both K46 and K58, designated rPDCoV-nsp8-K37R, rPDCoV-nsp8-K46R, rPDCoV-nsp8-K58R, and rPDCoV-nsp8-K46/58R, respectively. rPDCoV-nsp8-K37R served as a positive control. As expected, the indirect immunofluorescence assay (IFA) results showed that rPDCoV-WT and rPDCoV-nsp8-K37R exhibited similar fluorescence intensity, which was significantly lower than that of the other three rPDCoVs tested, especially in the early stages of viral infection (6 h postinfection [6 hpi]) (Fig. 8A and B). Consistent with the results from the IFA, a TCID50 assay demonstrated that the titers of rPDCoV-WT or rPDCoV-nsp8-K37R were significantly lower than those of the other three rPDCoVs tested at 6 and 12 hpi; however, no obvious difference was observed in the titers of all tested rPDCoVs at 18 and 24 hpi (Fig. 8C). These results suggest that the mutation of K46 or K58 of nsp8 results in the promotion of viral replication in the early stage of replication but not in the mid and late stages.
FIG 8.
Recombinant PDCoV with mutation at K46 and/or K58 of nsp8 displays resistance to the antiviral activity of HDAC6. (A and B) IFA of LLC-PK1 cells infected with rPDCoV-WT, rPDCoV-nsp8-K37R, rPDCoV-nsp8-K46R, rPDCoV-nsp8-K58R, and rPDCoV-nsp8-K46/58R at 6 hpi (A) or 12 hpi (B) using an anti-PDCoV N monoclonal antibody. Bar, 100 μm. (C) Multiple-step growth curves of rPDCoVs on LLC-PK cells. Cells were infected with rPDCoVs (MOI = 0.5) and then collected at the different time points postinfection (6, 12, 18, 24 hpi) and subjected to a TCID50 assay. (D to H) LLC-PK1 cells were transfected with pCAGGS-Flag-HDAC6 or empty vector for 12 h and then infected with rPDCoVs (MOI = 0.5) for 12 h. Cell samples were harvested and subjected to RT-qPCR for the detection of viral genomic mRNA (D) and N subgenomic mRNA (E), Western blotting (F), including density analysis of panel F by ImageJ software (G), and a TCID50 assay (H). The presented results represent the means and standard deviations of data from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonsignificant difference.
To further verify this result, LLC-PK1 cells were transfected with pCAGGS-Flag-HDAC6 or empty vector for 12 h and then infected with rPDCoV, followed by RT-qPCR, Western blotting, and a TCID50 assay. As shown in Fig. 8D to H, HDAC6 significantly suppressed the replication of rPDCoV-WT and rPDCoV-nsp8-K37R; however, the inhibitory effects were obviously attenuated in rPDCoV-nsp8-K46R-, rPDCoV-nsp8-K58R-, and rPDCoV-nsp8-K46/58R-infected cells. Taken together, these results confirm that rPDCoV mutated at either K46 or K58 of nsp8 displays obvious resistance to the antiviral activity of HDAC6.
DISCUSSION
Accumulating evidence has established HDAC6 as a critical regulator of viral infection either directly, via orchestrating different stages of the viral life cycle, or indirectly, via modulating the cytokine production of host cells, suggesting that this protein may represent an antiviral target. However, little is known about the role of HDAC6 in the infection and pathogenesis of coronaviruses. In this study, for the first time, we found that HDAC6 directly targets nsp8 for degradation through deacetylation and ubiquitination, thereby inhibiting PDCoV proliferation. Importantly, we confirmed that the K46 and K58 sites of nsp8 were, respectively, targeted for acetylation and ubiquitination, which were indispensable for the HDAC6-mediated degradation of nsp8. Recombinant PDCoV, with a mutation at either K46 or K58 of nsp8, showed obvious resistance to the antiviral activity of HDAC6.
Many studies have shown that HDAC6 plays important roles in a variety of biological processes through the regulation of posttranslational modifications. For example, HDAC6 deacetylates the transcriptional activator Tat protein, thereby preventing transcriptional activation of the HIV-1 promoter and inhibiting the formation of ribonucleoprotein complexes (42). HDAC6-dependent regulation of Hsp90 deacetylation affects its affinity for most clients and certain cochaperones, which in turn modulates viral replication (43). Of 18 identified deacetylases, HDAC6 is the one most associated with ubiquitin. Previous studies indicated that HDAC6 harbors a unique BUZ domain with the ability to bind to ubiquitin and ubiquitinated proteins, as well as misfolded proteins, for proteasomal or autophagic degradation (37, 44). This suggested that HDAC6 is involved in the ubiquitin-proteasome pathway and lysosome-mediated degradation pathway. In this study, we confirmed that HDAC6 degraded nsp8 via the ubiquitin-proteasome pathway and that the deacetylase activity of HDAC6 was required for the degradation of nsp8. We further detected the effects of the ubiquitin-proteasome pathway, the apoptotic pathway, or the autophagic pathway on the deacetylation of nsp8 by HDAC6, respectively, and found that the three host degradation pathways had no significant effect on HDAC6-mediated deacetylation of nsp8 (data not shown). These results further demonstrated that the ubiquitin-proteasome pathway is involved in HDAC6-mediated nsp8 degradation, but does not affect its deacetylation.
Direct competition between lysine acetylation and ubiquitination is thought to be an important regulator for the degradation of ubiquitinated protein. For example, MDM2 could recruit HDAC1 to mediate the deacetylation of the tumor suppressor p53, followed by MDM2-dependent ubiquitylation at the same sites as deacetylation and subsequent protein degradation (45). However, in this study, we found that the identified acetylation site at K46 of nsp8 was not the same as its ubiquitination site at K58, unlike previous studies reporting that the same lysine of target proteins acts as both an acetylation and ubiquitination site (35, 46). A previous study showed that E3 ubiquitin ligase E6AP promotes HDAC6-mediated deacetylation of tumor suppressor FHL1 at K157, followed by its ubiquitination at K102, resulting in the decreased expression of FHL1 (47). Thus, the mechanism of degradation of different proteins mediated by deacetylation and ubiquitination is complex and may be distinct. Our results showed that K37 is also an acetylation site of nsp8 but is not required for HDAC6 degradation. We speculated that the acetylation site at K37 of nsp8 may be regulated by other components of the deacetylation family to function. In addition, one of them may be the primary target of action in the presence of multiple acetylation sites. Indeed, a previous study indicated that the effect of enzyme activity depends specifically on site-specific acetylation when multiple acetylation sites are present (48). We experimentally verified that K46 and K37 are the acetylation sites and K58 is the ubiquitination site. A total of 18 lysine residues are present in the PDCoV nsp8. Thus, we could not rule out the possibility that other sites are involved in HDAC6-mediated modifications and degradations. In addition, our results showed that nsp8-K46R, nsp8-K58R, and nsp8-K46/58R exhibited similar resistance to HDAC6-mediated degradation. We speculated that HDAC6 interacted with nsp8 and regulated the turnover of nsp8 via sequential deacetylation and ubiquitination. A previous study indicated that HDAC6 enhances the polyubiquitination of Cdc20, dependent on its BUZ domain, rather than its deacetylase activity (49). Another study showed that the deacetylase activity and E3 ligase activity of HDAC6 function in concert to ubiquitinate MSH2, leading to the degradation of MSH2 (35). Our results from in vitro and in vivo ubiquitination assays suggested that the ubiquitination of nsp8 may be dependent on the deacetylase activity of HDAC6 (Fig. 6). However, whether HDAC6 directly ubiquitinates nsp8, whether its deacetylase activity and E3 ligase activity work collaboratively, and whether HDAC6 recruits other ubiquitin ligases to degrade nsp8 are interesting topics that deserve further investigation.
The cytoskeleton consists mainly of a protein fibril meshwork structure consisting of microtubules, microfilaments, and intermediate fibers. A previous study showed that the acetylation of lysine at position 40 of α-tubulin mediated by HDAC6 stabilizes microtubules and facilitates transport and signaling (50). Viruses always hijack the transport system of microtubule proteins to facilitate the intracellular transport, assembly, and release of viral particles or viral genomes. Thus, HDAC6 has been considered a crossroad for viral fusion, entry, nuclear shuttling, replication, assembly, and export, playing crucial roles in regulating viral infection, as demonstrated in the well-studied IAV (51, 52). Our results also showed that the acetylation level of α-tubulin significantly increased after PDCoV infection. We further used the inhibitor nocodazole to disrupt microtubule aggregation and demonstrated that nocodazole treatment with no detectable cytotoxicity significantly suppressed the acetylation level of α-tubulin and reduced N protein, viral mRNA expression, and viral titers in a dose-dependent manner (Fig. S2). These results suggest that HDAC6 may also inhibit PDCoV proliferation by reducing the acetylation of microtubulin and microtubule polymerization.
Previous studies suggested that HDAC6 can link the microtubule cytoskeleton to immune synaptic tissues and limit viral transmission and pathogenicity by inducing an immune defense response in host cells through its deacetylase activity (31, 53). In addition, HDAC6 is able to deacetylate β-linked protein to induce the expression of the transcription factor IRF3 (54) and directly deacetylate RIG-I at lysine 909, promoting RIG-I oligomerization and resulting in the activation of downstream signals (MAVS-IRF3-NF-κB). Thus, HDAC6 knockout mice, similar to IRF3 knockout mice, were highly susceptible to RNA viruses, highlighting the important role of HDAC6 in triggering innate antiviral immune signaling (25, 55, 56). In this study, we found that the inhibition of recombinant PDCoV with a mutation at either K46 or K58 by HDAC6 was not completely eliminated compared with the control group. We speculated that in addition to directly targeting viral protein nsp8 for degradation, HDAC6 may modulate innate antiviral immune signaling to inhibit PDCoV infection, such as enhancing the RIG-I-mediated pathway or other antiviral signaling pathways. However, this hypothesis needs to be further verified via additional experiments.
In this study, we found that in addition to nsp8, HDAC6 also interacted with the PDCoV nsp5. This interaction of HDAC6 with nsp5 did not result in the significant degradation of nsp5 but inhibited HDAC6 expression (Fig. 3). We raised several possibilities: (i) HDAC6 may regulate the acetylation modification of nsp5, thereby inhibiting its enzymatic activity. Indeed, a previous study showed that HATs and HDACs could effectively modulate the function of p65 but did not result in its degradation (57); (ii) PDCoV utilizes nsp5 to occupy part of the HDAC6 protein, reducing the degradation of nsp8 by HDAC6 and antagonizing the antiviral effect of HDAC6; and (iii) coronavirus nsp5, also called 3C-like protease, is responsible for processing viral polyprotein precursors in coronavirus replication. Some studies reported that coronaviruses nsp5 could inhibit the host’s antiviral innate immunity through the cleavage of key host factors (58–61). PDCoV nsp5 may employ similar or unknown mechanisms to reduce HDAC6 expression and impair the antiviral effects of HDAC6. In addition, our results showed that rPDCoV with a mutation at either K46 or K58 possessed significantly higher replication ability than rPDCoV-WT during early infection (Fig. 8C). Furthermore, PDCoV downregulated HDAC6 expression and exhibited an increased resistance to HDAC6-mediated antiviral activity, especially at the late stage of infection (Fig. 1B and Fig. 2D). Thus, it is possible that endogenous HDAC6 contributes to the lower replication level of rPDCoV-WT compared with rPDCoV with a mutation at either K46 or K58, prior to the expression of virus-encoded HDAC6 antagonists, such as PDCoV nsp5 during early infection; however, the increased expression of nsp5 protein during the mid and late stages of infection effectively inhibits the antiviral activity of HDAC6, resulting in insignificant differences in the replication level of all rPDCoVs (Fig. 8C).
Nsp8 is well conserved among the Coronaviridae family and is considered to be a second RdRp encoded uniquely by coronaviruses (62). Coronavirus nsp8 possesses polymerizing activity toward short RNA oligonucleotides, proposed to be used as primers for primer-dependent nsp12 RdRp during RNA synthesis (17, 18, 62). It was previously shown that numerous RNA viruses used primers for RNA synthesis through different strategies, such as the recruitment of cellular mRNA and tRNA by influenza viruses and retroviruses (63, 64). However, coronavirus may have evolved another strategy to produce primers by using nsp8 RdRp as the primase (62). As a primase, nsp8 is involved in the formation of RTC, playing a relatively conserved role in coronavirus replication. Because HDAC6 could directly target PDCoV nsp8, and the amino acid homology of the enzymatic activity region of HDAC6 among different species was relatively conserved, we speculated that HDAC6 has a broad-spectrum antiviral effect on PDCoV infection in different host cells. We further detected the effect of human HDAC6 on PDCoV infection in HeLa cells. As expected, overexpression of human HDAC6 notably inhibited PDCoV infection in HeLa cells (data not shown). In addition, we examined whether HDAC6 also inhibits other swine coronaviruses, like transmissible gastroenteritis virus (TGEV) and porcine epidemic diarrhea virus (PEDV). The results showed that HDAC6 overexpression significantly inhibited the infection of TGEV and PEDV (data not shown). A recent study showed that HDAC6 inhibitors have high therapeutic efficacy because of their potential to reduce the morbidity associated with severe COVID-19 by acting on the innate and adaptive immune system (65), but the detailed inhibitory mechanism has not been further investigated. Whether HDAC6-mediated nsp8 degradation through deacetylation and ubiquitination is conserved across coronaviruses deserves further study.
In summary, our present study demonstrated that HDAC6 interacted with and degraded nsp8 through deacetylation and ubiquitination at K46 and K58 of nsp8, respectively, thereby inhibiting PDCoV replication. Our findings reveal a novel regulatory function of HDAC6 on PDCoV infection, which deepens our understanding of the function of HDACs and may direct us toward novel therapeutic targets.
MATERIALS AND METHODS
Cells, virus, and reagents.
HEK-293T cells were obtained from the China Center for Type Culture Collection (Wuhan, China). LLC-PK1 cells were purchased from the American Type Culture Collection (ATCC CL-101; Manassas, VA, USA). All cells were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen, Madison, WI, USA) supplemented with 10% fetal bovine serum at 37°C in 5% CO2. PDCoV strain CHN-HN-2014 (GenBank accession number KT336560) used in this study was isolated from a piglet with severe diarrhea in China in 2014 (66). Anti-Flag antibody and anti-HA antibody were purchased from Medical and Biological Laboratories (MBL, Nagoya, Japan). Rabbit anti-HDAC6 monoclonal antibody was purchased from Huabio Technology (Hangzhou, China). Mouse anti-Ac-α-tubulin and anti-ac-lysine were purchased from Santa Cruz (Dallas, TX, USA). Rabbit anti-β-actin antibody was purchased from ABclonal (Wuhan, China). Rabbit anti-α-tubulin antibody was purchased from PTM BioLab (Hangzhou, China). The mouse anti-PDCoV N protein monoclonal antibody used herein was described previously (22). The rabbit polyclonal antibody against PDCoV nsp8 was prepared by DIA-AN (Wuhan, China) using the purified recombinant fusion protein His-nsp8. 3-MA and cycloheximide (CHX) were purchased from MedChemExpress (MCE, NJ, USA). MG132 and Z-VAD were purchased from Beyotime (Shanghai, China). Tubacin and TSA were purchased from Selleck (Radnor, PA, USA).
Plasmid construction and RNA interference.
The full-length cDNA of porcine HDAC6 (GenBank accession number XM_005673600.3) was amplified from LLC-PK1 cells and then cloned into pCAGGS-Flag vector containing an N-terminal Flag tag, generating the expression plasmid pCAGGS-Flag-HDAC6. The porcine HDAC6 deacetylase dead mutants, including the substitution mutants HDAC6-H258A, -H652A, and -H258/652A (DM), were also cloned into pCAGGS-Flag vector. The nsp5 and nsp8-coding sequences of PDCoV strain CHN-HN-2014 and mutants, including the substitution mutations nsp8-K37R, -K37Q, -K46R, -K46Q, -K58R, and K150R, were all amplified and cloned into pCAGGS-HA-C with a HA tag at the C terminus. All primers used are listed in Table S1. For RNA interference experiments, the siRNA sequences against HDAC6 (si-HDAC6-1: 5′-GGAGGAGCUUAUGUUGGUUTT-3′; si-HDAC6-2: 5′-GCUACGAUCAUGGCACCUUTT-3′) and a nontarget control siRNA (si-Ctrl: 5′-ACGUGACACGUUCGGAGAATT-3′) were designed by Genepharma Co. Ltd. (Shanghai, China). These siRNAs were transfected into LLC-PK1 cells using jetPRIME (Polyplus, Illkirch, France) according to the manufacturer’s instructions.
RNA extraction and quantitative real-time RT-PCR.
LLC-PK1 cells grown in six-well plates were treated under various experimental conditions and then infected with PDCoV for the indicated time points. Cellular total RNA was extracted using TRIzol reagent (Invitrogen) and then reverse transcribed into cDNA by avian myeloblastosis virus reverse transcriptase (TaKaRa, Kusatsu, Japan). The above cDNA was used as the template to perform RT-qPCR, repeated at least three times. Relative mRNA expression levels were normalized to the expression of GAPDH. Absolute mRNA levels were calculated using standard curves. The primers used in these experiments are listed in Table S1.
Coimmunoprecipitation and Western blotting assay.
HEK-293T cells were transfected with plasmids for the indicated time and then lysed with a lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 10% glycerin, 0.1% SDS, and 2 mM Na2EDTA). Cellular lysates were collected and centrifuged at 12,000 rpm at 4°C for 10 min. A portion of each supernatant from the lysed cells was used in the whole-cell extract assay. The remaining portions of the supernatants from the lysed cells were immunoprecipitated with the indicated antibodies overnight at 4°C and then added with protein A+G agarose beads (Beyotime, Shanghai, China) for 4 h at 4°C. The beads containing the immunoprecipitation complex were washed three times with 1 mL of lysis buffer. Whole-cell lysates and the immunoprecipitation complex were resuspended in SDS-PAGE loading buffer, boiled for 10 min, and then subjected to Western blotting.
For Western blotting, the protein samples separated by SDS-PAGE were transferred onto polyvinylidene difluoride membranes (Millipore, Darmstadt, Germany). The membranes were blocked with 5% nonfat milk in phosphate-buffered solution with 0.1% polysorbate-20 for 3 h and then incubated with the indicated primary antibodies. After three washes, the membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit antibody or goat anti-mouse antibody (Beyotime, Shanghai, China) for 45 min. Finally, the membranes were washed three times and then visualized using enhanced chemiluminescence reagents (Bio-Rad, CA, USA).
Indirect immunofluorescence assay.
LLC-PK1 cells seeded in 24-well plates were infected with PDCoV for the indicated time. Cells were fixed with 4% paraformaldehyde and then permeabilized with cold methyl alcohol. The cells were then blocked with 5% bovine serum albumin and incubated with primary and secondary antibodies, consecutively. Subsequently, cells were incubated with 4′,6-diamidino-2-phenylindole for 15 min and then visualized by confocal laser scanning microscopy (Fluoviewver.3.1; Olympus, Tokyo, Japan).
Generation of recombinant viruses.
Recombinant PDCoV with a single point mutation (K37R, K46R, or K58R) or double mutations (K46/58R) of nsp8 was constructed based on CRISPR/Cas9 technology, as described previously (67). Briefly, two specific primers (sgPDCoV-1 and sgPDCoV-2) targeting the upstream and downstream sequences of the fragment of interest were designed and synthesized for generating sgRNA-1 and sgRNA-2. pBAC-CHN-HN-2014 was then cleaved into the linearized BAC vector by the in vitro addition of Cas9, sgRNA-1, and sgRNA-2. At the same time, fragments containing each of the nsp8 gene sequences with the indicated mutation were generated through overlapping PCR using the indicated primers (Table S1) and then were, respectively, ligated into the purified linearized BAC vector via homologous recombination, generating the recombinant BAC plasmids (pBAC-CHN-HN-2014-nsp8-K37R, -K46R, -K58R, or -K46/58R). These recombinant plasmids were, respectively, transfected into LLC-PK1 cells seeded in 6-well plates with 80% confluence using jetPRIME for 4 h. The cells were washed twice and then added to DMEM containing 7.5 μg/mL of trypsin (Sigma) and incubated at 37°C and 5% CO2. Daily observations were made.
Cell viability assay.
The cytotoxic effects of the drugs on cells were measured using a CCK-8-based cell viability assay (Beyotime) according to the manufacturer’s instructions.
Statistical analysis.
Statistical differences were determined by one-way ANOVA or a Student’s t test performed using GraphPad Prism 5.0 software (GraphPad Software, CA, USA). Differences were considered to be statistically significant when the corresponding P values were <0.05.
ACKNOWLEDGMENTS
This work is supported by the National Key Research and Development Program of China (2021YFD1801104) and the National Natural Science Foundation of China (32072846, 32272983).
Footnotes
Supplemental material is available online only.
Contributor Information
Puxian Fang, Email: pxfang@mail.hzau.edu.cn.
Shaobo Xiao, Email: vet@mail.hzau.edu.cn.
Tom Gallagher, Loyola University Chicago-Health Sciences Campus.
REFERENCES
- 1.Li W, Hulswit RJG, Kenney SP, Widjaja I, Jung K, Alhamo MA, van Dieren B, van Kuppeveld FJM, Saif LJ, Bosch BJ. 2018. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc Natl Acad Sci USA 115:E5135–E5143. doi: 10.1073/pnas.1802879115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jung K, Hu H, Saif LJ. 2016. Porcine deltacoronavirus infection: etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res 226:50–59. doi: 10.1016/j.virusres.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY. 2012. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol 86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong N, Fang L, Zeng S, Sun Q, Chen H, Xiao S. 2015. Porcine deltacoronavirus in mainland China. Emerg Infect Dis 21:2254–2255. doi: 10.3201/eid2112.150283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jang G, Lee KK, Kim SH, Lee C. 2017. Prevalence, complete genome sequencing and phylogenetic analysis of porcine deltacoronavirus in South Korea, 2014–2016. Transbound Emerg Dis 64:1364–1370. doi: 10.1111/tbed.12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saeng-Chuto K, Lorsirigool A, Temeeyasen G, Vui DT, Stott CJ, Madapong A, Tripipat T, Wegner M, Intrakamhaeng M, Chongcharoen W, Tantituvanont A, Kaewprommal P, Piriyapongsa J, Nilubol D. 2017. Different lineage of porcine deltacoronavirus in Thailand, Vietnam and Lao PDR in 2015. Transbound Emerg Dis 64:3–10. doi: 10.1111/tbed.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Z, Zhong H, Zhou Q, Du Y, Chen L, Zhang Y, Xue C, Cao Y. 2018. A highly pathogenic strain of porcine deltacoronavirus caused watery diarrhea in newborn piglets. Virol Sin 33:131–141. doi: 10.1007/s12250-018-0003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turlewicz-Podbielska H, Pomorska-Mol M. 2021. Porcine coronaviruses: overview of the state of the art. Virol Sin 36:833–851. doi: 10.1007/s12250-021-00364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boley PA, Alhamo MA, Lossie G, Yadav KK, Vasquez-Lee M, Saif LJ, Kenney SP. 2020. Porcine deltacoronavirus infection and transmission in poultry, United States. Emerg Infect Dis 26:255–265. doi: 10.3201/eid2602.190346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jung K, Hu H, Saif LJ. 2017. Calves are susceptible to infection with the newly emerged porcine deltacoronavirus, but not with the swine enteric alphacoronavirus, porcine epidemic diarrhea virus. Arch Virol 162:2357–2362. doi: 10.1007/s00705-017-3351-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang Q, Zhang H, Li B, Ding Q, Wang Y, Gao W, Guo D, Wei Z, Hu H. 2019. Susceptibility of chickens to porcine deltacoronavirus infection. Viruses 11:573. doi: 10.3390/v11060573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Wang B, Liang QZ, Shi FS, Ji CM, Yang XL, Yang YL, Qin P, Chen R, Huang YW. 2021. Roles of two major domains of the porcine deltacoronavirus s1 subunit in receptor binding and neutralization. J Virol 95:e0111821. doi: 10.1128/JVI.01118-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhai SL, Sun MF, Zhang JF, Zheng C, Liao M. 2022. Spillover infection of common animal coronaviruses to humans. Lancet Microbe 3:e808. doi: 10.1016/S2666-5247(22)00198-7. [DOI] [PubMed] [Google Scholar]
- 14.Mai K, Li D, Wu J, Wu Z, Cheng J, He L, Tang X, Zhou Z, Sun Y, Ma J. 2018. Complete genome sequences of two porcine deltacoronavirus strains, CHN-GD16-03 and CHN-GD16-05, isolated in Southern China, 2016. Genome Announc 6:e01545-17. doi: 10.1128/genomeA.01545-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sawicki SG, Sawicki DL, Younker D, Meyer Y, Thiel V, Stokes H, Siddell SG. 2005. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog 1:e39. doi: 10.1371/journal.ppat.0010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang P, Fang L, Liu X, Hong Y, Wang Y, Dong N, Ma P, Bi J, Wang D, Xiao S. 2016. Identification and subcellular localization of porcine deltacoronavirus accessory protein NS6. Virology 499:170–177. doi: 10.1016/j.virol.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, Wang T, Sun Q, Ming Z, Zhang L, Ge J, Zheng L, Zhang Y, Wang H, Zhu Y, Zhu C, Hu T, Hua T, Zhang B, Yang X, Li J, Yang H, Liu Z, Xu W, Guddat LW, Wang Q, Lou Z, Rao Z. 2020. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirchdoerfer RN, Ward AB. 2019. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun 10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hai T, Hao J, Wang L, Jouneau A, Zhou Q. 2011. Pluripotency maintenance in mouse somatic cell nuclear transfer embryos and its improvement by treatment with the histone deacetylase inhibitor TSA. Cell Reprogram 13:47–56. doi: 10.1089/cell.2010.0042. [DOI] [PubMed] [Google Scholar]
- 20.Valenzuela-Fernandez A, Cabrero JR, Serrador JM, Sanchez-Madrid F. 2008. HDAC6: a key regulator of cytoskeleton, cell migration and cell-cell interactions. Trends Cell Biol 18:291–297. doi: 10.1016/j.tcb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Sprung R, Pei J, Tan X, Kim S, Zhu H, Liu CF, Grishin NV, Zhao Y. 2009. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol Cell Proteomics 8:215–225. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Fang P, Duan P, Chen J, Fang L, Xiao S. 2022. Porcine deltacoronavirus infection cleaves HDAC2 to attenuate its antiviral activity. J Virol 96:e0102722. doi: 10.1128/jvi.01027-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falkenberg KJ, Johnstone RW. 2014. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov 13:673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Gilquin B, Khochbin S, Matthias P. 2006. Two catalytic domains are required for protein deacetylation. J Biol Chem 281:2401–2404. doi: 10.1074/jbc.C500241200. [DOI] [PubMed] [Google Scholar]
- 25.Choi SJ, Lee HC, Kim JH, Park SY, Kim TH, Lee WK, Jang DJ, Yoon JE, Choi YI, Kim S, Ma J, Kim CJ, Yao TP, Jung JU, Lee JY, Lee JS. 2016. HDAC6 regulates cellular viral RNA sensing by deacetylation of RIG-I. EMBO J 35:429–442. doi: 10.15252/embj.201592586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. 2002. HDAC6 is a microtubule-associated deacetylase. Nature 417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 27.Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. 2005. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell 18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 28.Valenzuela-Fernandez A, Alvarez S, Gordon-Alonso M, Barrero M, Ursa A, Cabrero JR, Fernandez G, Naranjo-Suarez S, Yanez-Mo M, Serrador JM, Munoz-Fernandez MA, Sanchez-Madrid F. 2005. Histone deacetylase 6 regulates human immunodeficiency virus type 1 infection. Mol Biol Cell 16:5445–5454. doi: 10.1091/mbc.e05-04-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D, Meng Q, Huo L, Yang M, Wang L, Chen X, Wang J, Li Z, Ye X, Liu N, Li Q, Dai Z, Ouyang H, Li N, Zhou J, Chen L, Liu L. 2015. Overexpression of Hdac6 enhances resistance to virus infection in embryonic stem cells and in mice. Protein Cell 6:152–156. doi: 10.1007/s13238-014-0120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mosley AJ, Meekings KN, McCarthy C, Shepherd D, Cerundolo V, Mazitschek R, Tanaka Y, Taylor GP, Bangham CR. 2006. Histone deacetylase inhibitors increase virus gene expression but decrease CD8+ cell antiviral function in HTLV-1 infection. Blood 108:3801–3807. doi: 10.1182/blood-2006-03-013235. [DOI] [PubMed] [Google Scholar]
- 31.Nusinzon I, Horvath CM. 2006. Positive and negative regulation of the innate antiviral response and beta interferon gene expression by deacetylation. Mol Cell Biol 26:3106–3113. doi: 10.1128/MCB.26.8.3106-3113.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu T, Song Z, Li Q, Li Z, Wang M, Liu L, Tian K, Li N. 2017. Overexpression of histone deacetylase 6 enhances resistance to porcine reproductive and respiratory syndrome virus in pigs. PLoS One 12:e0169317. doi: 10.1371/journal.pone.0169317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scholz C, Weinert BT, Wagner SA, Beli P, Miyake Y, Qi J, Jensen LJ, Streicher W, McCarthy AR, Westwood NJ, Lain S, Cox J, Matthias P, Mann M, Bradner JE, Choudhary C. 2015. Acetylation site specificities of lysine deacetylase inhibitors in human cells. Nat Biotechnol 33:415–423. doi: 10.1038/nbt.3130. [DOI] [PubMed] [Google Scholar]
- 34.Evans CA, Kim HR, Macfarlane SC, Nowicki PIA, Baltes C, Xu L, Widengren J, Lautenschlager F, Corfe BM, Gad AKB. 2022. Metastasising fibroblasts show an HDAC6-dependent increase in migration speed and loss of directionality linked to major changes in the vimentin interactome. Int J Mol Sci 23:1961. doi: 10.3390/ijms23041961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang M, Xiang S, Joo HY, Wang L, Williams KA, Liu W, Hu C, Tong D, Haakenson J, Wang C, Zhang S, Pavlovicz RE, Jones A, Schmidt KH, Tang J, Dong H, Shan B, Fang B, Radhakrishnan R, Glazer PM, Matthias P, Koomen J, Seto E, Bepler G, Nicosia SV, Chen J, Li C, Gu L, Li GM, Bai W, Wang H, Zhang X. 2014. HDAC6 deacetylates and ubiquitinates MSH2 to maintain proper levels of MutSalpha. Mol Cell 55:31–46. doi: 10.1016/j.molcel.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hershko A, Heller H, Eytan E, Kaklij G, Rose IA. 1984. Role of the alpha-amino group of protein in ubiquitin-mediated protein breakdown. Proc Natl Acad Sci USA 81:7021–7025. doi: 10.1073/pnas.81.22.7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. 2003. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 38.Hai Y, Christianson DW. 2016. Histone deacetylase 6 structure and molecular basis of catalysis and inhibition. Nat Chem Biol 12:741–747. doi: 10.1038/nchembio.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyake Y, Keusch JJ, Wang L, Saito M, Hess D, Wang X, Melancon BJ, Helquist P, Gut H, Matthias P. 2016. Structural insights into HDAC6 tubulin deacetylation and its selective inhibition. Nat Chem Biol 12:748–754. doi: 10.1038/nchembio.2140. [DOI] [PubMed] [Google Scholar]
- 40.Lu YF, Xu XP, Lu XP, Zhu Q, Liu G, Bao YT, Wen H, Li YL, Gu W, Zhu WG. 2020. SIRT7 activates p53 by enhancing PCAF-mediated MDM2 degradation to arrest the cell cycle. Oncogene 39:4650–4665. doi: 10.1038/s41388-020-1305-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, Jacobson MP, Verdin E. 2010. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab 12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huo L, Li D, Sun X, Shi X, Karna P, Yang W, Liu M, Qiao W, Aneja R, Zhou J. 2011. Regulation of Tat acetylation and transactivation activity by the microtubule-associated deacetylase HDAC6. J Biol Chem 286:9280–9286. doi: 10.1074/jbc.M110.208884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scroggins BT, Robzyk K, Wang D, Marcu MG, Tsutsumi S, Beebe K, Cotter RJ, Felts S, Toft D, Karnitz L, Rosen N, Neckers L. 2007. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol Cell 25:151–159. doi: 10.1016/j.molcel.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ouyang H, Ali YO, Ravichandran M, Dong A, Qiu W, MacKenzie F, Dhe-Paganon S, Arrowsmith CH, Zhai RG. 2012. Protein aggregates are recruited to aggresome by histone deacetylase 6 via unanchored ubiquitin C termini. J Biol Chem 287:2317–2327. doi: 10.1074/jbc.M111.273730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ito A, Kawaguchi Y, Lai CH, Kovacs JJ, Higashimoto Y, Appella E, Yao TP. 2002. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J 21:6236–6245. doi: 10.1093/emboj/cdf616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu Y, Wang X, Xu F, Zhang L, Wang T, Fu X, Jin T, Zhang W, Ye L. 2020. The regulation of acetylation and stability of HMGA2 via the HBXIP-activated Akt-PCAF pathway in promotion of esophageal squamous cell carcinoma growth. Nucleic Acids Res 48:4858–4876. doi: 10.1093/nar/gkaa232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Chen Z, Lin J, Liu J, Lin Y, Li H, Xi Y, Wei B, Ding L, Ye Q. 2020. The ubiquitin ligase E6AP facilitates HDAC6-mediated deacetylation and degradation of tumor suppressors. Signal Transduct Target Ther 5:243. doi: 10.1038/s41392-020-00330-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jew KM, Le VTB, Amaral K, Ta A, Nguyen May NM, Law M, Adelstein N, Kuhn ML. 2021. Investigation of the importance of protein 3D structure for assessing conservation of lysine acetylation sites in protein homologs. Front Microbiol 12:805181. doi: 10.3389/fmicb.2021.805181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim AH, Puram SV, Bilimoria PM, Ikeuchi Y, Keough S, Wong M, Rowitch D, Bonni A. 2009. A centrosomal Cdc20-APC pathway controls dendrite morphogenesis in postmitotic neurons. Cell 136:322–336. doi: 10.1016/j.cell.2008.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perdiz D, Mackeh R, Pous C, Baillet A. 2011. The ins and outs of tubulin acetylation: more than just a post-translational modification? Cell Signal 23:763–771. doi: 10.1016/j.cellsig.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 51.Husain M, Harrod KS. 2011. Enhanced acetylation of alpha-tubulin in influenza A virus infected epithelial cells. FEBS Lett 585:128–132. doi: 10.1016/j.febslet.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L, Ogden A, Aneja R, Zhou J. 2016. Diverse roles of HDAC6 in viral infection: implications for antiviral therapy. Pharmacol Ther 164:120–125. doi: 10.1016/j.pharmthera.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 53.Serrador JM, Cabrero JR, Sancho D, Mittelbrunn M, Urzainqui A, Sanchez-Madrid F. 2004. HDAC6 deacetylase activity links the tubulin cytoskeleton with immune synapse organization. Immunity 20:417–428. doi: 10.1016/s1074-7613(04)00078-0. [DOI] [PubMed] [Google Scholar]
- 54.Zhu J, Coyne CB, Sarkar SN. 2011. PKC alpha regulates Sendai virus-mediated interferon induction through HDAC6 and beta-catenin. EMBO J 30:4838–4849. doi: 10.1038/emboj.2011.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chattopadhyay S, Fensterl V, Zhang Y, Veleeparambil M, Wetzel JL, Sen GC. 2013. Inhibition of viral pathogenesis and promotion of the septic shock response to bacterial infection by IRF-3 are regulated by the acetylation and phosphorylation of its coactivators. mBio 4:e00636-12. doi: 10.1128/mBio.00636-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu HM, Jiang F, Loo YM, Hsu S, Hsiang TY, Marcotrigiano J, Gale M. Jr, 2016. Regulation of retinoic acid inducible gene-I (RIG-I) activation by the histone deacetylase 6. EBioMedicine 9:195–206. doi: 10.1016/j.ebiom.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang B, Yang XD, Lamb A, Chen LF. 2010. Posttranslational modifications of NF-kappaB: another layer of regulation for NF-kappaB signaling pathway. Cell Signal 22:1282–1290. doi: 10.1016/j.cellsig.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen J, Li Z, Guo J, Xu S, Zhou J, Chen Q, Tong X, Wang D, Peng G, Fang L, Xiao S. 2022. SARS-CoV-2 nsp5 exhibits stronger catalytic activity and interferon antagonism than its SARS-CoV ortholog. J Virol 96:e0003722. doi: 10.1128/jvi.00037-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hameedi MAETP, Garvin MR, Mathews II, Amos BK, Demerdash O, Bechthold M, Iyer M, Rahighi S, Kneller DW, Kovalevsky A, Irle S, Vuong VQ, Mitchell JC, Labbe A, Galanie S, Wakatsuki S, Jacobson D. 2022. Structural and functional characterization of NEMO cleavage by SARS-CoV-2 3CLpro. Nat Commun 13:5285. doi: 10.1038/s41467-022-32922-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang D, Fang L, Shi Y, Zhang H, Gao L, Peng G, Chen H, Li K, Xiao S. 2016. Porcine epidemic diarrhea virus 3C-like protease regulates its interferon antagonism by Cleaving NEMO. J Virol 90:2090–2101. doi: 10.1128/JVI.02514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu X, Wang D, Zhou J, Pan T, Chen J, Yang Y, Lv M, Ye X, Peng G, Fang L, Xiao S. 2017. Porcine deltacoronavirus nsp5 antagonizes type I interferon signaling by cleaving STAT2. J Virol 91:e00003-17. doi: 10.1128/JVI.00003-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Imbert I, Guillemot JC, Bourhis JM, Bussetta C, Coutard B, Egloff MP, Ferron F, Gorbalenya AE, Canard B. 2006. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J 25:4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mak J, Kleiman L. 1997. Primer tRNAs for reverse transcription. J Virol 71:8087–8095. doi: 10.1128/JVI.71.11.8087-8095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Plotch SJ, Bouloy M, Ulmanen I, Krug RM. 1981. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- 65.Ripamonti C, Spadotto V, Pozzi P, Stevenazzi A, Vergani B, Marchini M, Sandrone G, Bonetti E, Mazzarella L, Minucci S, Steinkuhler C, Fossati G. 2022. HDAC inhibition as potential therapeutic strategy to restore the deregulated immune response in severe COVID-19. Front Immunol 13:841716. doi: 10.3389/fimmu.2022.841716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dong N, Fang L, Yang H, Liu H, Du T, Fang P, Wang D, Chen H, Xiao S. 2016. Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN-HN-2014. Vet Microbiol 196:98–106. doi: 10.1016/j.vetmic.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fang P, Zhang H, Sun H, Wang G, Xia S, Ren J, Zhang J, Tian L, Fang L, Xiao S. 2021. Construction, characterization and application of recombinant porcine deltacoronavirus expressing nanoluciferase. Viruses 13:1991. doi: 10.3390/v13101991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 and Fig. S1 and S2. Download jvi.00375-23-s0001.docx, DOCX file, 0.7 MB (759.8KB, docx)