ABSTRACT
The porcine reproductive and respiratory syndrome viruses (PRRSV) led to a global panzootic and huge economical losses to the pork industry. PRRSV targets the scavenger receptor CD163 for productive infection. However, currently no effective treatment is available to control the spread of this disease. Using bimolecular fluorescence complementation (BiFC) assays, we screened a set of small molecules potentially targeting the scavenger receptor cysteine-rich domain 5 (SRCR5) of CD163. We found that the assay examining protein-protein interactions (PPI) between PRRSV glycoprotein 4 (GP4) and the CD163-SRCR5 domain mainly identifies compounds that potently inhibit PRRSV infection, while examining the PPI between PRRSV-GP2a and the SRCR5 domain maximized the identification of positive compounds, including additional ones with various antiviral capabilities. These positive compounds significantly inhibited both types 1 and 2 PRRSV infection of porcine alveolar macrophages. We confirmed that the highly active compounds physically bind to the CD163-SRCR5 protein, with dissociation constant (KD) values ranging from 28 to 39 μM. Structure-activity-relationship (SAR) analysis revealed that although both the 3-(morpholinosulfonyl)anilino and benzenesulfonamide moieties in these compounds are critical for the potency to inhibit PRRSV infection, the morpholinosulfonyl group can be replaced by chlorine substituents without significant loss of antiviral potency. Our study established a system for throughput screening of natural or synthetic compounds highly effective on blocking of PRRSV infection and shed light on further SAR modification of these compounds.
IMPORTANCE Porcine reproductive and respiratory syndrome virus (PRRSV) causes significant economic losses to the swine industry worldwide. Current vaccines cannot provide cross protection against different strains, and there are no effective treatments available to hamper the spread of this disease. In this study, we identified a group of new small molecules that can inhibit the PRRSV interaction with its specific receptor CD163 and dramatically block the infection of both types 1 and type 2 PRRSVs to host cells. We also demonstrated the physical association of these compounds with the SRCR5 domain of CD163. In addition, molecular docking and structure-activity relationship analyses provided new insights for the CD163/PRRSV glycoprotein interaction and further improvement of these compounds against PRRSV infection.
KEYWORDS: PRRSV, CD163, SRCR5, small molecule compound, antiviral treatment
INTRODUCTION
Porcine reproductive and respiratory syndrome (PRRS) is one of the most economically significant swine diseases, leading to an annual loss of $664 million to the United States pork industry (1), a similar loss in China (2), and over €1.5 billion in Europe (3). PRRS results in severe reproductive failure in sows and respiratory diseases in growing pigs (4) and is frequently associated with secondary infections resulting in severe clinical manifestations and mortality (5–7). The causative agent, PRRS virus (PRRSV) is an enveloped, positive-sense, single-stranded RNA virus of the family Arteriviridae within the order Nidovirales (8, 9). PRRS was initially detected in central Europe and North America in the late 1980s (10–12), with two genotypes (types 1 and 2) of PRRSV isolated (9, 11, 13), which shared 60% genetic similarity (14). Unfortunately, due to the high genetic, antigenic, and pathogenic heterogeneities, broadly effective vaccines against PRRSV are still lacking (15–17). Novel antiviral approaches are needed to help mitigate its devastating consequences.
PRRSV preferentially targets and replicates in well-differentiated cells of the monocyte/macrophage lineages (4, 18–21). While productive viral infection by oronasal exposure primarily occurs through porcine alveolar macrophages (PAMs) residing in pig lungs (22), there are many other potential routes of viral transmission via bodily fluids of infected pigs (23). Multiple studies including gene knockout pigs identified CD163, a macrophage-specific membrane scavenger receptor as the key receptor for PRRSV infection (24–30). The interactions between PRRSV minor envelope glycoproteins GP2 and GP4 with CD163 are necessary for successful infection (31, 32), while the scavenger receptor cysteine-rich domain 5 (SRCR5) in CD163 is needed for the PRRSV-receptor recognition (33–35). The evidence, thus, suggested that identification of drug compounds that interfere with the PRRSV-CD163 recognition may have therapeutic potential in controlling PRRS.
Bimolecular fluorescence complementation (BiFC) assay recognizes protein-protein interactions (PPI) via the reconstitution of two fragments of Venus yellow fluorescence protein (YFP) (36). We previously used BiFC to screen for compounds interfering with the interaction between CD163-SRCR5 and PRRSV glycoproteins GP2a or GP4 (37) and identified new compound B7 and its derivatives (B7-A1 to B7-A4) that can inhibit PRRSV infection by blocking its binding to CD163 (37). However, the structure-activity relationships (SAR) for this scaffold are still unclear, and physical evidence is still lacking for specific ligand-receptor association. Here, we conducted BiFC screening of a defined small molecule library based on the B7 chemical structure. We report the identification of new compounds with potent anti-PRRSV infection activity and confirmed their physical binding to CD163-SRCR5 domain. We also compared the compound antiviral potency-predicting power by BiFC assays utilizing different PRRSV glycoproteins-SRCR5 domain interactions and characterized the SAR based on the positive hits identified.
RESULTS
Screening of new compounds that block PRRSV/CD163 interaction.
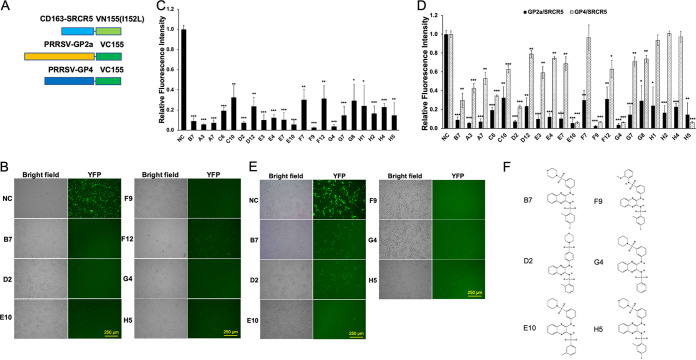
In order to identify SAR with respect to the anti-PRRSV activity of our scaffold, we utilized the artificial intelligence-based AtomNet platform (38) to design a chemically defined library containing 94 compounds (see Table S1 in the supplemental material), within which 28 compounds were from a set of compounds with structural similarity to B7 and B7-A4 identified previously (37), while the remaining 65 represented varying scaffolds and were predicted to bind to CD163. We first screened the library using a BiFC assay investigating the PPI between PRRSV glycoprotein GP2a and CD163-SRCR5, with B7 and dimethyl sulfoxide (DMSO) as the positive and negative inhibitory control (PC and NC), respectively (Fig. 1A). Overall, 20 positive hits were identified from the library, which exerted at least 70% reduction of the YFP fluorescence signal compared with the NC (Fig. 1B and C; see also Table S1). As expected, positive control B7 significantly inhibited the PPI between PRRSV GP2a and CD163-SRCR5 (Fig. 1B and C). All hits identified were analogs of B7, and no additional scaffolds were identified from the 65 additional compounds screened.
FIG 1.
Screen of new compounds that block PRRSV GP2a/GP4-CD163 interaction. (A) Scheme diagram for the BiFC assay constructs between CD163-SRCR5 or PRRSV envelope proteins GP2a/GP4 and the fragments of Venus protein VN155(I152L) or VC155, respectively. (B) BiFC screening images for select positive compounds that inhibit the PPI between SRCR5-VN and GP2a-VC proteins. NC, DMSO Ctrl. Bar = 250 μm. (C) Relative fluorescence intensity of the SRCR5/GP2a BiFC screening results for all 20 positive compounds. Mean ± SD; n = 3. (D) Comparison for the relative fluorescence intensity of the SRCR5/GP2a and SRCR5/GP4 BiFC screening results for the 20 positive compounds. Mean ± SD; n = 3. (E) BiFC screening images for select positive compounds that inhibit the PPI between SRCR5-VN and GP4-VC proteins. NC, DMSO Ctrl. Bar = 250 μm. (F) Chemical structure of the 5 identified strong positive compounds by both BiFC assay.
Successful PRRSV infection requires the PPI between CD163 with both glycoproteins GP2a and GP4 (31, 32). To determine if the candidate compounds identified above could inhibit both interactions, we conducted another BiFC assay investigating the PPI between PRRSV GP4 and CD163-SRCR5 domain (Fig. 1A). Out of the 20 positive hits identified above, only 5 (D2, E10, F9, G4, and H5) exhibited more than 70% reduction of the YFP fluorescence signal compared with NC (Fig. 1D to F). Interestingly, these 5 compounds all exerted more than 85% and 90% inhibition in GP2a/SRCR5 and GP4/SRCR5 BiFC assays, respectively (Fig. 1D). Among them, compounds D2 and H5 (Fig. 1F) represented B7-A2 and B7, respectively, as reported by us previously (37). The remaining three new positive hits—E10, F9, and G4—all exerted more than 90% inhibition in both assays (Fig. 1D and E). To investigate if the compounds with less than 70% inhibition in GP2a/SRCR5 BiFC assay may exhibit significant potency in GP4/SRCR5 BiFC analysis, we screened the remaining 78 chemicals in the library using GP4/SRCR5 BiFC assay. No additional compound was identified with more than 70% reduction of the fluorescence signal compared to NC (Table S1). Therefore, both GP2a/SRCR5 and GP4/SRCR5 BiFC screening of the library demonstrated a similar trend for compound activities. However, assay screening using the PPI between GP4 and SRCR5 is more productive for identifying compounds with high inhibitory activity.
The combined GP2a/SRCR5 and GP4/SRCR5 BiFC assays accurately predict potent PRRSV-inhibitory compounds.
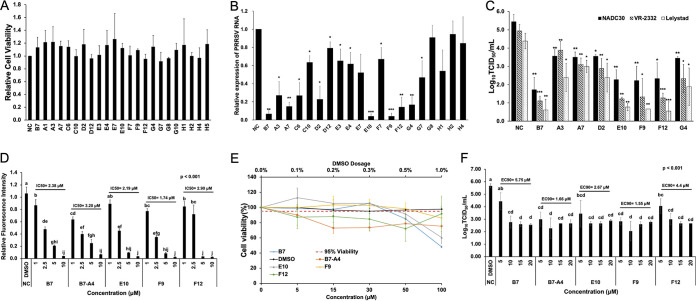
MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] analysis of the inhibitory compounds identified above revealed no obvious cytotoxicity in 48 h for PAMs incubated with the 20 small chemicals at a 15 μM level (Fig. 2A). We then inoculated fresh PAMs with different PRRSV strains including NADC30, VR-2332 (type 2), and Lelystad (type 1) followed by the addition of each compound at 15 μM in culture medium immediately after infection (H5 was excluded, as it is structurally identical to B7). At 24 h postinfection, viral RNAs were extracted from PAMs for the evaluation of viral loads, and cell supernatants were collected to measure the viral titer. Quantitative reverse transcription-PCR (qRT-PCR) revealed that 8 out of the 19 compounds tested showed more than 70% inhibition of viral RNA loads for the average of three viral strains (Fig. 2B). Among these, five compounds, A7, E10, F9, F12, and G4, appeared to have the strongest inhibitory effect against infection with more than 85% deduction and seconded by three other compounds, A3, C6, and D2, with a 70% to 80% decrease (Fig. 2B). This included all of the highly positive compounds identified by the GP4/SRCR5 BiFC analysis—D2, E10, F9, and G4 (Fig. 2B). The remaining four compounds (A3, A7, C6, and F12) that exhibited over 70% inhibition of GP2a/SRCR5 but not GP4/SRCR5 BiFC screening (Fig. 1D) also showed mild (A3 and C6) or strong (A7 and F12) inhibitory effect against PRRSV infection, respectively (Fig. 2B).
FIG 2.
The Combined GP2a/SRCR5 and GP4/SRCR5 BiFC assays accurately predict potent PRRSV-inhibitory compounds. (A) MTT analysis to the 20 positive compounds in PAMs. NC, DMSO Ctrl. Mean ± SD; n = 3. (B) qRT-PCR for PRRSV in total RNAs extracted from infected PAMs treated with various compounds. Values are normalized with GAPDH of PAMs, and the average of three strains (NADC30, VR-2332, and Lelystad) are plotted. Bars = mean ± SD; n = 3. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) Titration assay results for PRRSV in the culture media of PAMs treated as described in panel B. Bars = mean ± SD; n = 3. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (D) Relative fluorescence intensity of the SRCR5/GP2a BiFC assay for B7, B7-A4, E10, F9, and F12 at different concentrations. Mean ± SD; n = 3. P values are calculated by one-way ANOVA and letters on the top of bars indicate significant differences in Tukey post hoc test. (E) MTT analysis to B7, B7-A4, E10, F9, and F12 at different concentrations in PAMs. NC, DMSO Ctrl. Mean ± SD; n = 3. (F) Titration for PRRSV in the culture media from infected PAMs treated with B7, B7-A4, E10, F9, and F12 at different concentrations. Bars = mean ± SD; n = 3. P values are calculated by one-way ANOVA and letters on the top of bars indicate significant differences in Tukey post hoc test.
The antiviral effects of selected positive compounds were further confirmed by viral titration assay of culture medium harvested from infected PAMs. Similar to the qRT-PCR analysis, treatment by compounds E10, F9, and F12 exhibited strong reduction in the viral titer across different PRRSV strains (3.1 to 3.3 logs for NADC30, 3.6 to 3.8 logs for VR-2332, and 3.6 to 3.9 logs for Lelystad), while compounds A3, A7, D2, and G4 also reduced the viral titer of three strains (1.8 to 2 logs for NADC30, 1 to 2.6 logs for VR-2332, and 1.4 to 2.5 logs for Lelystad) (Fig. 2C). Therefore, compounds that effectively interfered with the GP4/SRCR5 PPI all exerted strong inhibitory potency against PRRSV infection. Of note, although not all compounds effectively interfering with the GP2a/SRCR5 PPI exhibited high potency against viral infection, this BiFC screening identified additional compounds not detected by the GP4/SRCR5 BiFC assay, including F12. Taken together, a combined interrogation to the PPIs of GP4/SRCR5 and GP2a/SRCR5 can facilitate the identification of highly effective compounds against both types 1 and 2 PRRSV strains.
To further evaluate these highly positive compounds, we compared the 50% inhibitory concentration (IC50) values for B7, B7-A4, E10, F9, and F12. The GP2a/SRCR5 BiFC assay was performed for each compound with concentrations ranging from 1 to 10 μM. The results showed that the IC50 values of compounds B7, B7-A4, E10, F9 and F12 against the GP2a/SRCR5 PPI are 2.38 μM, 3.2 μM, 2.19 μM, 1.74 μM, and 2.90 μM, respectively (Fig. 2D). We further performed toxicity analysis for these compounds from 5 to 100 μM in PAMs. MTT assay revealed that treatment with compound F9 for 48 h still maintained 86% PAM cell viability at 100 μM, and treatment with B7 and E10 maintained 85% and 92% cell viability at 50 μM, while B7-A4 and F12 slightly reduced cell viability to 73% and 88% at 15 μM, respectively (Fig. 2E). We then examined the 90% effective concentration (EC90) values of these compounds against PRRSV infection. PAMs were inoculated with VR-2332 at a multiplicity of infection (MOI) of 0.1 and then treated by each compound with concentrations ranging from 5 to 20 μM. Viral titration for supernatants at 24 h revealed that the EC90 values for B7, B7-A4, E10, F9, and F12 are 5.75 μM, 1.66 μM, 2.67 μM, 1.55 μM, and 4.4 μM, respectively (Fig. 2F). The IC50 and EC90 values of these compounds correlated well with each other, and among them compound F9 exhibited the lowest combination of IC50/EC90 values with minimum cytotoxicity, indicating the identification of a new compound with superior property over B7.
Validation of the positive compounds—SRCR5 molecular interaction.
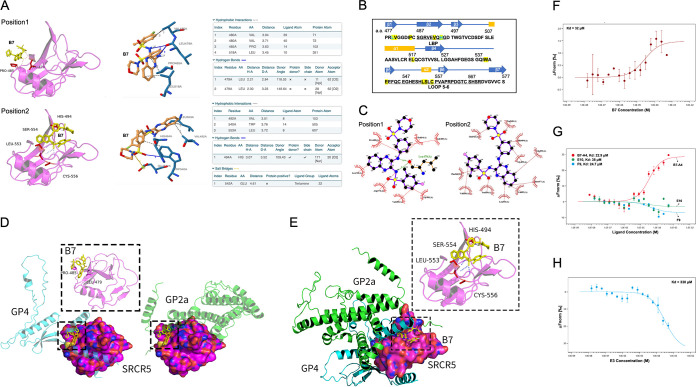
In order to confirm the ligand-receptor physical binding between the small chemicals and CD163, we first performed molecular docking analysis using the prototypical inhibitory compound B7 and the resolved CD163-SRCR5 protein crystal structure (39). The docking and protein-ligand interaction profiler (PLIP) (40) analyses indicate that B7 potentially binds to two positions of SRCR5 (Fig. 3A). At position 1, B7 forms a hydrogen bond with residue L479 located in the first β-turn, and hydrophobic interactions with V480, P485, and L518 of SRCR5 (Fig. 3A, top, and Fig. 3B). At position 2, B7 forms a hydrogen bond with residue H494, and hydrophobic interactions with V492 within the ligand-binding pocket (LBP) (41), W540, and L553 within the conserved loop 5–6 region (42) as well as a salt bridge with E542 of SRCR5 (Fig. 3A, bottom, and Fig. 3B). Similar association analysis results were obtained by using LigPlot+ (43) for the B7 ligand-SRCR5 docking (Fig. 3C). As no crystal structure information is available for PRRSV GP2a and GP4, we applied ColabFold (44) to estimate their 3-dimerntional structures and performed protein-protein docking between SRCR5 and GP2a or GP4, respectively, using the ZDOCK platform (45). The results indicate that both GP2a and GP4 could interact with SRCR5 (Fig. 3D), consistent with our BiFC data. Furthermore, we found that the B7-SRCR5 interaction predicted at position 1 disturbs both protein-protein docking conformations by interfering with the association of SRCR5 with the second α-helix of GP2a or the fifth α-helix and its surrounding region of GP4, respectively (Fig. 3D). Similarly, multiprotein docking analysis revealed that SRCR5, GP4, and GP2a could form a protein complex (Fig. 3E). However, the B7-SRCR5 interaction predicted at position 2 disturbs this docking conformation by interfering with the association between SRCR5 and the sixth α-helix of GP4 (Fig. 3E).
FIG 3.
Validating of physical interactions between the compounds and SRCR5. (A) Molecular docking (left) and PLIP (right) analyses depicting two potential interaction sites between compounds B7 and CD163-SRCR5 domain. (B) Residues in CD163-SRCR5 with potential interactions with B7 (yellow highlighted), residues forming potential hydrogen bonds with B7 are marked with green. The amino acids in the LBP and loop 5–6 regions are underlined. (C) Two-dimensional ligand-protein interaction diagram by LigPlot+ showing the hydrogen bond and hydrophobic interactions of B7 with SRCR5. (D) Molecular docking analyses by ZDOCK depicting the potential conformation of PPIs between SRCR5 with GP4 (left) or GP2a (right). B7 was placed in position 1 (boxes) to show the potential disturbing of both conformations. (E) Molecular docking analyses by ZDOCK depicting the PPI conformation between SRCR5 with both GP4 and GP2a. B7 was placed in position 2 (box) to show the potential disturbing of this conformation. (F) MST analysis of GFP-fused SRCR5 thermal dynamic association with ligand B7. Values represent Mean ± SD; n = 3. (G) MST analysis of GFP-fused SRCR5 thermal dynamic association with ligand B7-A4, E10, and F9. Values represent Mean ± SD; n = 3. (H) MST analysis of GFP-fused SRCR5 thermal dynamic association with ligand E3. Values represent Mean ± SD; n = 3.
To determine whether there was physical association between the chemicals and CD163, we expressed green fluorescent protein (GFP)-fusion CD163-SRCR5 recombinant protein, with GFP and GFP-fusion CD163-SRCR2 as a control because they do not interact with GP2a and GP4 (37) or bind to B7 (see Fig. S1 in the supplemental material). We then used the microscale thermophoresis (MST) assay (46) to evaluate the direct association between B7 and the GFP-fusion proteins. The MST analysis demonstrated a physical binding between B7 and the GFP-SRCR5 with a dissociation constant (KD) value of 32 μM (Fig. 3F). However, little or no association was detected between B7 and GFP-SRCR2 or GFP (see Fig. S2 in the supplemental material). Similar ligand-protein interactions were identified between GFP-SRCR5 and compounds B7-A4, E10, and F9, with KD values similar as B7 (Fig. 3G). The KD values of these small molecules correlate well with their demonstrated potency against PRRSV infection (Fig. 2B and C; see also the previous report [37]). On the contrary, compound E3 which showed low activity against PRRSV infection (Fig. 2B) exhibited a KD value 10 times greater than B7 (Fig. 3H). Therefore, these data indicate that the positive compounds including B7 and its analogs physically target the CD163-SRCR5 domain to exert their function against PRRSV infection.
As PRRSV escapes the endosome upon cell entry through a low pH-dependent endocytic pathway (47), the inhibitory effect of B7 analogues at pH = 6.0 was assessed using the GP2a/SRCR5 BiFC assay. Although HEK293T cells grew slow at this low pH and could not survive after treatment with compounds B7, B7-A4, E10, or F9 at the 5 μM level, the cells did survive upon treatment with 5 μM F12, and the YFP fluorescence intensity was reduced to 25% compared with the DMSO control (see Fig. S3A and B in the supplemental material). This indicates that the B7 analogue can inhibit the GP2a/CD163 PPI under an acidic environment similar as the endosomes.
SAR evaluation of the compounds for anti-PRRSV function.
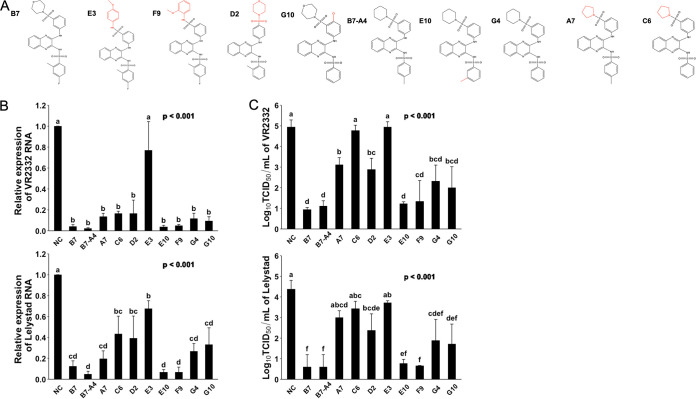
It is important to understand the structure-activity relationship for better design of ligands with improved potency. We therefore asked how the structural changes in different moieties of these compounds may affect their property of anti-PRRSV infection. We organized these chemicals into 2 different groups based on the chemical structure similarities and compared their potency on inhibiting the infection by different strains of PRRSV. In the first group (Fig. 4A), a near complete loss of antiviral potency was observed for compound E3 (Fig. 4B and C), with the 3-morpholin in B7 replaced by the 4-methoxyphenyl group. Interestingly, however, the antiviral potency did not change at all for compound F9 (Fig. 4B and C), which has the 3-morpholin replaced by the 2-methoxyphenyl group (Fig. 4A). Other changes, including the reposition of 3-morpholinosulfonyl in B7 to 4-morpholinosulfonyl group in compound D2 (Fig. 4A) and the removal of 4-fluoro and 2-methyl groups from the benzenesulfonamide moiety in G10 (Fig. 4A) also mildly but significantly decreased the PRRSV infection inhibitory effect for these compounds (Fig. 4B and C). Therefore, in the B7 molecule, a moiety topologically similar to the 3-(morpholinosulfonyl)anilino is critical, while side group modifications to the benzenesulfonamide moiety also exert some effects on the antiviral potency.
FIG 4.
Evaluating compound structure-activity relationship for anti-PRRSV infection. (A) Molecular structures of B7 and B7-A4 analogues. (B) qRT-PCR for PRRSV in total RNAs extracted from infected PAMs treated with B7 and B7-A4 analogues. Values are normalized with GAPDH of PAMs. Bars = mean ± SD; n = 3. P values are calculated by one-way ANOVA, and letters on the top of bars indicate significant differences in Tukey post hoc test. (C) Titration assay results for PRRSV in the culture media of PAMs treated as described in panel B. Bars = mean ± SD; n = 3. P values are calculated by one-way ANOVA, and letters on the top of bars indicate significant differences in Tukey post hoc test.
The analog B7-A4 has the replacement of the 3-morpholinosulfonyl group in B7 by the 3-piperidinylsulfonyl group and shared similar anti-PRRSV potency as B7 (37) (Fig. 4A to C). We used it as a control to evaluate the impact to antiviral effect by modification of the benzenesulfonamide moiety. Compared with B7-A4, compound E10 with the reposition of 4-methyl group to 3-methyl on the benzenesulfonamide moiety (Fig. 4A) showed no change in antiviral potency (Fig. 4B and C). However, complete removal of the methyl group in compound G4 (Fig. 4A) significantly decreased its antiviral potency (Fig. 4B and C). Therefore, these data confirmed that 2- or 3-methyl modification of the benzenesulfonamide moiety is also important for the antiviral effect of B7. Furthermore, similar to that observed above for the importance of 3-(morpholinosulfonyl)anilino moiety, the replacement of piperidin by the pyrrolidine group in compound A7 (Fig. 4A) caused a significant drop of antiviral potency (Fig. 4B and C).
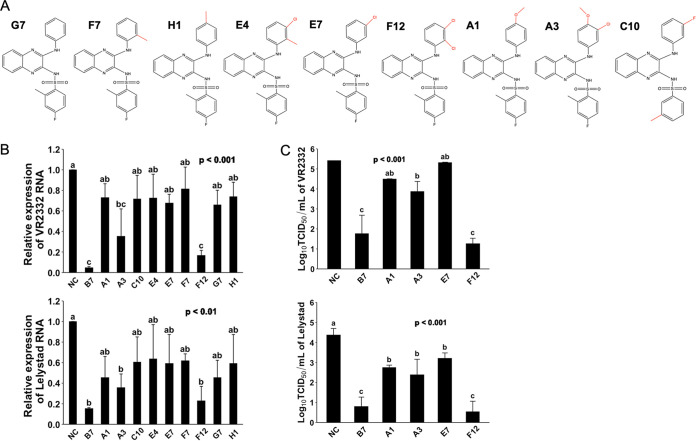
We further asked whether in these compounds, the 3-morpholinosulfonyl group could be replaced by other modifications without a significant decline in the anti-PRRSV potency. Compound G7, which does not have a 3-morpholinosulfonyl group, showed a much lower antiviral effect compared with B7 (Fig. 5B and C). Compounds F7, H1, E4, E7, F12, C10, A1, and A3 have either an additional methyl, chloro, fluoro, or methoxymethyl group in the place of 3-morpholinosulfonyl group (Fig. 5A). Among these compounds, only F12 with two chloro group substitutions and A3 with one chloro and one methoxymethane group substitutions exhibited a significantly increased antiviral effect, while all nonpolar modifications, single chloro, or single methoxymethyl substitution showed no effect on improving the antiviral potency compared with G7 (Fig. 5B and C). Noticeably, compound F12 exerted an antiviral potency nearly identical to B7 (Fig. 5B and C). Therefore, these data demonstrate that the 3-morpholinosulfonyl group of the B7 compound can be replaced by other chemical modifications, including multiple chloro substituents without affecting its inhibitory potency.
FIG 5.
Evaluating compound structure-activity relationship for anti-PRRSV infection (2). (A) Molecular structures of additional compounds with similarity to B7. (B) qRT-PCR for PRRSV in total RNAs extracted from infected PAMs treated with B7 analogues. Values are normalized with GAPDH of PAMs. Bars = mean ± SD; n = 3. P values are calculated by one-way ANOVA, and letters on the top of bars indicate significant differences in Tukey post hoc test. (C) Titration assay results for PRRSV treated by selected compounds as described in panel B. Bars = mean ± SD; n = 3. P values are calculated by one-way ANOVA, and letters on the top of bars indicate significant differences in Tukey post hoc test.
DISCUSSION
PRRS is one of the most economically significant porcine diseases affecting the swine industry globally. While there are vaccines to control the disease, these are not widely protective due to the constant evolution of the virus. There are no other effective antivirals that can be utilized to aid in the control of PRRS. The interactions between PRRSV minor envelope glycoproteins with CD163 are necessary for successful infection (31, 32), while the CD163-SRCR5 domain is needed for receptor recognition by PRRSV (33–35). However, it is unclear how each of the glycoprotein-CD163 interactions contributes to efficient PRRSV infection. Combining the BiFC and viral infection assays, we found here that compounds that inhibit the PPI between PRRSV-GP4 and CD163-SRCR5 can efficiently block the PRRSV infection. Also, compared with the GP4/SRCR5 BiFC, which is highly selective on compound with potent activity, the GP2a/SRCR5 BiFC analysis identified a wider range of compounds with various efficiencies against PRRSV infection. This includes compound F12 that does not effectively interfere with the PPI between GP4/SRCR5 (Fig. 1D). These data thus confirm that GP4 and GP2a are both critical for their respective interaction with CD163 in order to establish productive viral infection to host cells. Further investigation of the PPIs between PRRSV glycoproteins and CD163 would be highly relevant to better understand the mechanism and importance of these PPIs for PRRSV infection and to identify targeting sites or develop more effective assays for drug screening.
Although we previously reported the interference of the PPI between PRRSV glycoproteins and porcine CD163-SRCR5 domain by B7 and B7-A4 small molecules, no direct evidence was provided for the specific ligand-protein interaction by those compounds. In this study, we demonstrated that the CD163-SRCR5 domain can interact directly with B7, B7-A4, and the other newly identified compounds that potently inhibit PRRSV infection. We also found that B7 did not interact with SRCR2, highlighting the SRCR5-targeting specificity of B7. Molecular docking further indicates that the B7 molecule interacts directly with SRCR5 and may form hydrogen bonds with either L479 or H494, residues next to or within the LBP region. The LBP is a potential PRRSV binding site based on the ligand-binding role of homologous regions in SRCR superfamily members (41). Recently, it was found that LBP-deletion pigs confer resistance to type 2 PRRSV infection (48), which was also confirmed by cell infection assay with CD163 mutants disturbing this region (49). Docking analysis also found that B7 could form hydrophobic interactions with residues located in other regions of SRCR5, including the loop 5–6 region. The loop 5–6 region in CD163 SRCR5 remains highly conserved across species from humans, monkeys, swine, and dogs to mice (42), consistent with the fact that the CD163 of multiple species could mediate PRRSV infection (24). Expressing a mutant pig CD163 with R561 changed to alanine in the loop 5–6 region of SRCR5 significantly inhibited PRRSV infection compared with wild-type CD163 (39). Further investigation of the small molecule ligand- SRCR5 associations, and how they may disturb the PPIs between SRCR5 and PRRSV glycoproteins would be critical to refine the protein targeting area for development of highly specific drugs, and for complete elucidation of the molecular mechanism of PRRSV-CD163 receptor recognition.
Our SAR study provided evidence that the inhibitory function of B7 and its analogues is correlated with the topology of 3-(morpholinosulfonyl)anilino moiety and additional side group modifications to the benzenesulfonamide moiety in B7. More interestingly, we found that although complete removal of the 3-morpholinosulfonyl group in compound G7 resulted in obvious loss of the antiviral activity compared with B7, this can be rescued by two chloro substituents as demonstrated by compound F12. The addition of chloro groups may rescue the antiviral potency of the compound by changing the topology of anilino moiety or simply by improving its polarity. Overall, our SAR analysis provided useful functional information and clues for downstream modification of these compounds to further improve the antiviral potency and bioavailability.
To find out other possible mechanisms for the inhibitory effect of B7 analogues, we tested CD163 expression in PAMs with treatment of B7. The result showed that B7 treatment from 5 to 20 μM for 24 h could reduce the transcription of CD163, which might be caused by an undefined negative-feedback signal stimulated by B7-CD163 ligand-protein interaction (see Fig. S4 in the supplemental material). However, unlike the dosage-response curves observed for the inhibition of GP2a/SRCR5 PPI and PRRSV infection by B7 (Fig. 2D and F), the CD163 transcription inhibition by B7 exhibited no dosage-response across the same concentration range (Fig. S4). Since all B7 antiviral treatments reported here were performed after 1 h viral inoculation when the CD163-PRRSV association shall not be affected by later change of CD163 transcription upon B7 administration, and our previous study demonstrated that pretreatment of PAMs with B7 exerted no significant inhibition on PRRSV infection, and that B7 does not affect viral-host cell binding and cell internalization (37), we conclude that the main inhibitory function of B7 is mediated through direct interruption of the PPIs between SRCR5-GP2a/GP4 proteins that may be the key for PRRSV uncoating as previously reported (50), while the inhibition of CD163 transcription may provide some additional benefits.
Currently, there is no broadly effective vaccine developed to prevent PRRS and control the huge economic losses for the global pork industry (17). This study, together with a better understanding of the PPIs in PRRSV glycoprotein-CD163 recognition, will facilitate the identification of effective antivirals against PRRS. The B7 compound and the potent new derivatives identified here have the potential to serve as veterinary antiviral drugs if they pass in vivo efficacy tests against PRRSV infection and other safety measures. These antivirals have promising activities with the potential to be applied as means to control the disease. In conclusion, our study provided powerful screening tools and structure-functional information for the development and optimization of veterinary antivirals to prevent/treat PRRSV infection in pigs.
MATERIALS AND METHODS
Chemicals, cells, plasmids, and virus.
All chemicals (dissolved in 10 mM in DMSO) were designed and provided by Atomwise Inc. (CA, USA). Compounds were further diluted in phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, and 2 mM KH2PO4, pH 7.4) (Gibco, MA, USA) to 200 μM as the working solution. PAMs were harvested from healthy 4- to 6-month Landrace/Yorkshire pigs. Briefly, pig lungs were removed from euthanized pigs during necropsy and transferred on ice to the lab. Two hundred milliliters warm PBS with 200 U/mL penicillin and 200 μg/mL streptomycin was injected into the trachea and reached major bronchi of both sides of the lungs. Lungs were then massaged, and bronchoalveolar lavage fluid (BALF) was collected. PAMs were harvested from the BALF by centrifugation at 400 × g for 15 min and washed in PBS twice. Fresh PAMs were counted and frozen in 90% fetal bovine serum (FBS) and 10% DMSO (Sigma-Aldrich, MO) in liquid nitrogen. PAMs were cultivated in RPMI 1640 (Gibco, MA) supplemented with 10% FBS, 2 mM GlutaMAX, 0.1 mM minimal essential medium (MEM) nonessential amino acids, 1 mM sodium pyruvate, 100 U/mL penicillin and 100 μg/mL streptomycin, and 0.5 μg/mL amphotericin B. SRCR5 domain of porcine CD163 receptor was amplified from cDNA of pig cells and inserted into pBiFC-VN155 (I152L). GP2a and GP4 were amplified from the reverse-transcribed genome of the PRRSV VR-2332 strain (GenBank accession number EF536003.1) and cloned into the pBiFC-VC155 vector as previously described (37). Porcine CD163 was cloned into pMXs vector (Cell Biolabs, CA, USA). For protein purification, PRRSV GP2a and GP4 were cloned into pET28b-GFP (Addgene, MA, USA).
For CD163-expressing retroviral packaging, pMXs-CD163 was cotransfected with pUMVC and pCMV-VSV-G (Addgene, MA, USA) packaging plasmids into HEK293T cells using Fugene 6 (Promega, WI, USA). Supernatants containing viruses were collected at 48 h and 72 h after transfection. To established the CD163 expression Marc145 cell line (CD163-Marc145), Marc145 cells were cultivated in FP medium (Dulbecco modified Eagle medium [DMEM] containing 10% FBS, 2 mM GlutaMAX supplement, 0.1 mM MEM nonessential amino acids, 50 U/mL and 50 μg/mL penicillin-streptomycin) and were incubated with CD163-expressing retrovirus at 32°C while centrifuging at 650 × g for 45 min. The infection was repeated after 24 h. Cells were stained with CD163 monoclonal antibody (2A10/11) conjugated with Phycoerythrin (PE) (1:10; Thermo Fisher Scientific, MA, USA) at 24 h after the second infection, and fluorescence-positive cells were sorted using the FACSAria II cell sorter (BD, NJ, USA). Genotype I PRRSV strain Lelystad and genotype II PRRSV strain NADC30 were propagated in PAMs and titrated in CD163-Marc145 cells.
AIMS screen.
Virtual screening was performed using the AtomNet model, the first deep neural network for structure-based drug design trained to predict protein-ligand binding affinity (38). For targeting the interaction between the porcine CD163 and PRRSV glycoprotein (GP2a or GP4), the X-ray structure of CD163-SRCR5 domain (PDB accession number 5HRJ) was used to define a screening site centered around R561 comprising residues C502, S503, D505, W540, A541, E543, A559, P560, R561, P562, D563, G564, and C566. The Mcule library of commercially available organic small molecule compounds (~5 M; v20171018) was prepared and screened, as described previously (51), using an ensemble of protein-ligand conformations. Similar to that previously described (37), a set of 69 chemically diverse compounds was obtained for experimental testing to identify new scaffolds targeting the CD163-SRCR5 interaction. In addition, analogs of B7 sampled from the Mcule library were screened using the AtomNet models, and 29 compounds were selected for testing.
Bimolecular fluorescence complementation assay.
HEK293T cells were cultivated in 6-well plates in FP medium and transfected with 1 μg pBiFC-VN155 (I152L)-SRCR5 and 1 μg pBiFC-VC155-GP2/GP4 using Fugene 6. After 4 h, cells were digested with 0.05% Typsin-EDTA (Thermo Fisher Scientific, MA, USA) and passaged to 48-well plates, which were coated with EmbryoMax 0.1% gelatin solution (Sigma-Aldrich, MO, USA) and preloaded with 1 to 10 μM B7 analogues. Fluorescence images were taken at 24 h after transfection, and fluorescence intensity was measured by ImageJ (52).
Cytotoxicity assay.
PAMs were cultivated in 48-well plates and incubated with 5 to 100 μM B7 analogues for 48 h. PAMs treated with 20% DMSO in PBS served as controls. The viability of cells was measured using an in vitro toxicology assay kit (MTT based; Sigma-Aldrich, MO, USA) following the manufacturer’s instruction. Briefly, 20 μL labeling reagent was added to the cells and incubated at 37°C for 4 h. And then, 200 μL solubilization solution was added and incubated at 37°C overnight. Absorbance at 550 nm was measured using the CLARIOstar Plus plate reader (BMG LABTECH, NC, USA). The cell viability was calculated from the percentage of absorbance of experimental groups to the control groups.
PRRSV infection assay.
PAMs were cultivated in 12-well plates and infected with the PRRSV NADC30 strain, VR-2332 strain, or Lelystad strain at an MOI of 0.05 or 0.1 for 1 h. Cells were then incubated with 5 to 20 μM B7 analogues for 24 h at 37°C. RNAs were extracted from the PAMs for quantitative reverse transcription-PCR (qRT-PCR) using RNeasy minikits (Qiagen, MD, USA), and supernatants of cells were collected with PRRSV titration assay.
Quantitative reverse transcription-PCR.
RNAs isolated from infected PAMs were reverse transcribed to cDNA using iScript cDNA synthesis kit (Bio-Rad Laboratories, CA, USA). Specific qRT-PCR primers for ORF7 gene of PRRSV NADC30, VR-2332, and Lelystad strains and for porcine glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are shown in Table S2 in the supplemental material. qRT-PCR was conducted with SYBR green supermix (Bimake, TX, USA) using the ABI 7500 fast platform (Thermo Fisher Scientific, MA, USA). GAPDH served as the housekeeping gene for normalization.
PRRSV titration assay.
CD163-Marc145 cells were cultivated in 48-well plates and were inoculated with 10-fold serially diluted supernatants from PRRSV infection assay (6 wells for each dilution) at 37°C for 2 h. The inoculum was replaced by DMEM supplemented with 2% FBS, 2 mM GlutaMAX supplement, 0.1 mM MEM nonessential amino acids, 50 U/mL and 50 μg/mL penicillin-streptomycin. Cells with cytopathic effect were recorded, and the median tissue culture infectious does (TCID50/mL) was calculated using the Reed and Muench method.
Protein expression and purification.
The recombinant pET28b-GFP/GFP-SRCR2/SRCR5 constructs were transformed into Escherichia coli strain BL21(DE3) (New England Biolabs, MA, USA). The transformed clones were cultured at 37°C in LB broth with 50 mg/mL kanamycin, induced by adding 1 mM isopropyl-β-d-1-thiogalactopyranoside at an optical density of 0.6 to 0.8 and then incubated at 37°C for 3 h. For protein purification, cells were pelleted and resuspended in xTractor buffer containing DNase I, lysozyme solution, and protease inhibitor cocktail (TaKaRa Bio USA, Inc., CA). The suspension was sonicated and centrifuged. The supernatant was incubated with equilibrated TALON metal affinity resin (TaKaRa Bio USA, Inc., CA, USA) and then eluted from resin with the elution buffer (pH 7.0, 150 mM imidazole, 50 mM NaH2PO4, and 300 mM NaCl). The eluted proteins were concentrated and with buffer exchanged to phosphate-buffered saline (pH 7.4) containing 0.05% Tween 20 (PBST) using protein concentrator PES, 10K MWCO (Pierce Biotechnology, PA, USA).
Microscale thermophoresis and docking analyses.
The purified GFP, GFP-fused SRCR2, or SRCR5 proteins were mixed with serially diluted compounds. The mixtures were loaded to capillaries, and the affinity was measured by Monolith NT.115 (NanoTemper Technologies, Inc., CA, USA) at corresponding MST power with the blue channel. Molecular docking was performed via AutoDock MGL tools version 1.5.6. (53) for preparation of CD163-SRCR5 PDB structure 5JFB (39) and small molecule compounds for docking. AutoDock Vina version 1.2.3 (54) was used for docking analysis with comparative analysis of the obtained binding energy (kilocalorie per mole). PyMol version 2.5.2 was used to visualize the top-rated ligand-protein docking conformation. LigPlot+ version 2.2.5 and PLIP 2021 were used to visualize the two-dimensional structure of ligand-protein docking results (40, 43). The three-dimensional structures of GP4 or GP2a were resolved by the ColabFold, which offers accelerated prediction of protein structures and complexes by combining the fast homology search of MMseqs2 with AlphaFold2 or RoseTTAFold (44). ZDOCK version 3.0.4 was used for docking analysis between CD163-SRCR5 and GP4 or GP2a (45).
Statistics analysis.
Data were analyzed by one-way analysis of variance (ANOVA) with Tukey’s post hoc comparison or 2 sample Student's t tests. The figures were presented as mean ± standard deviation (SD) and a P value of <0.05 was considered statistically significant.
Ethics statement.
Porcine alveolar macrophages (PAMs) used in this study were harvested from pigs based on a protocol (A17-004) approved by the institutional animal care and use committee (IACUC) at the University of Connecticut.
Availability of data and materials.
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
ACKNOWLEDGMENTS
We are grateful for M. Kyle Hadden at UConn School of Pharmacy for his review and comments on this manuscript.
This study was supported by the USDA/National Institution for Food and Agriculture (NIFA) grants 2016-09339 to Y.T. and 2022-67016-37126 to Y.T., X.T., A.G., and N.M.; the Atomwise Inc. Artificial Intelligence Molecular Screen (AIMS) award to Y.T.; the Program in Innovative Therapeutics for Connecticut’s Health (PITCH) Promising Project Award to Y.T. and A.G.; and the University of Connecticut Research Excellence Program to Y.T. and A.G.
Footnotes
[This article was published on 3 May 2023 with Andrew Wolek’s name misspelled in the byline. The spelling was corrected in the current version, posted on 9 May 2023.]
Supplemental material is available online only.
Contributor Information
Xiuchun Tian, Email: xiuchun.tian@uconn.edu.
Antonio Garmendia, Email: antonio.garmendia@uconn.edu.
Young Tang, Email: young.tang@nwafu.edu.cn.
Tom Gallagher, Loyola University Chicago, Health Sciences Campus.
REFERENCES
- 1.Pork-Checkoff. 2012. Checkoff research helps wage war on PRRS. Pork Checkoff Special Edition Report 8. https://www.nationalhogfarmer.com/health/checkoff-research-helps-wage-war-prrs. [Google Scholar]
- 2.Zhou L, Yang X, Tian Y, Yin S, Geng G, Ge X, Guo X, Yang H. 2014. Genetic diversity analysis of genotype 2 porcine reproductive and respiratory syndrome viruses emerging in recent years in China. Biomed Res Int 2014:748068. doi: 10.1155/2014/748068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paz XD. 17 August 2015. PRRS cost for the European swine industry. Pig333, Ripoll, Spain. https://www.pig333.com/articles/prrs-cost-for-the-european-swine-industry_10069/.Pig333.com. [Google Scholar]
- 4.Music N, Gagnon CA. 2010. The role of porcine reproductive and respiratory syndrome (PRRS) virus structural and non-structural proteins in virus pathogenesis. Anim Health Res Rev 11:135–163. doi: 10.1017/S1466252310000034. [DOI] [PubMed] [Google Scholar]
- 5.Halbur PG. 1998. “PRRS Plus”–PRRS virus infection in combination with other agents. Hypothesis 1997:1998b–2000. [Google Scholar]
- 6.Zimmerman JJ, Yoon KJ, Wills RW, Swenson SL. 1997. General overview of PRRSV: a perspective from the United States. Vet Microbiol 55:187–196. doi: 10.1016/s0378-1135(96)01330-2. [DOI] [PubMed] [Google Scholar]
- 7.Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, Hu Y, Chen X, Hu D, Tian X, Liu D, Zhang S, Deng X, Ding Y, Yang L, Zhang Y, Xiao H, Qiao M, Wang B, Hou L, Wang X, Yang X, Kang L, Sun M, Jin P, Wang S, Kitamura Y, Yan J, Gao GF. 2007. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2:e526. doi: 10.1371/journal.pone.0000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavanagh D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol 142:629–633. [PubMed] [Google Scholar]
- 9.Benfield DA, Nelson E, Collins JE, Harris L, Goyal SM, Robison D, Christianson WT, Morrison RB, Gorcyca D, Chladek D. 1992. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J Vet Diagn Invest 4:127–133. doi: 10.1177/104063879200400202. [DOI] [PubMed] [Google Scholar]
- 10.Paton DJ, Brown IH, Edwards S, Wensvoort G. 1991. ‘Blue ear’ disease of pigs. Vet Rec 128:617. https://pubmed.ncbi.nlm.nih.gov/1897099/. [DOI] [PubMed] [Google Scholar]
- 11.Hill H. 1990. Overview and history of mystery swine disease (swine infertility respiratory syndrome), p 29–30. In Proceedings of the Mystery Swine Disease Committee Meeting. Livestock Conservation Institute, Bowling Green, KY. [Google Scholar]
- 12.Keffaber KK. 1989. Reproductive failure of unknown etiology. Am Assoc Swine Pract Newsletter 1:1–9. [Google Scholar]
- 13.Wensvoort G, Terpstra C, Pol JM, ter Laak EA, Bloemraad M, de Kluyver EP, Kragten C, van Buiten L, den Besten A, Wagenaar F. 1991. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet Q 13:121–130. doi: 10.1080/01652176.1991.9694296. [DOI] [PubMed] [Google Scholar]
- 14.An TQ, Li JN, Su CM, Yoo D. 2020. Molecular and cellular mechanisms for PRRSV pathogenesis and host response to infection. Virus Res 286:197980. doi: 10.1016/j.virusres.2020.197980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng XJ. 2000. Heterogeneity of porcine reproductive and respiratory syndrome virus: implications for current vaccine efficacy and future vaccine development. Vet Microbiol 74:309–329. doi: 10.1016/s0378-1135(00)00196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murtaugh MP, Stadejek T, Abrahante JE, Lam TT, Leung FC. 2010. The ever-expanding diversity of porcine reproductive and respiratory syndrome virus. Virus Res 154:18–30. doi: 10.1016/j.virusres.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 17.Nan Y, Wu C, Gu G, Sun W, Zhang YJ, Zhou EM. 2017. Improved vaccine against PRRSV: current progress and future perspective. Front Microbiol 8:1635. doi: 10.3389/fmicb.2017.01635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duan X, Nauwynck HJ, Pensaert MB. 1997. Effects of origin and state of differentiation and activation of monocytes/macrophages on their susceptibility to porcine reproductive and respiratory syndrome virus (PRRSV). Arch Virol 142:2483–2497. doi: 10.1007/s007050050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossow KD. 1998. Porcine reproductive and respiratory syndrome. Vet Pathol 35:1–20. doi: 10.1177/030098589803500101. [DOI] [PubMed] [Google Scholar]
- 20.Teifke JP, Dauber M, Fichtner D, Lenk M, Polster U, Weiland E, Beyer J. 2001. Detection of European porcine reproductive and respiratory syndrome virus in porcine alveolar macrophages by two-colour immunofluorescence and in-situ hybridization-immunohistochemistry double labelling. J Comp Pathol 124:238–245. doi: 10.1053/jcpa.2000.0458. [DOI] [PubMed] [Google Scholar]
- 21.Thanawongnuwech R, Thacker EL, Halbur PG. 1997. Effect of porcine reproductive and respiratory syndrome virus (PRRSV) (isolate ATCC VR-2385) infection on bactericidal activity of porcine pulmonary intravascular macrophages (PIMs): in vitro comparisons with pulmonary alveolar macrophages (PAMs). Vet Immunol Immunopathol 59:323–335. doi: 10.1016/s0165-2427(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 22.Murtaugh MP, Xiao Z, Zuckermann F. 2002. Immunological responses of swine to porcine reproductive and respiratory syndrome virus infection. Viral Immunol 15:533–547. doi: 10.1089/088282402320914485. [DOI] [PubMed] [Google Scholar]
- 23.Pileri E, Mateu E. 2016. Review on the transmission porcine reproductive and respiratory syndrome virus between pigs and farms and impact on vaccination. Vet Res 47:108. doi: 10.1186/s13567-016-0391-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calvert JG, Slade DE, Shields SL, Jolie R, Mannan RM, Ankenbauer RG, Welch SK. 2007. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J Virol 81:7371–7379. doi: 10.1128/JVI.00513-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delrue I, Van Gorp H, Van Doorsselaere J, Delputte PL, Nauwynck HJ. 2010. Susceptible cell lines for the production of porcine reproductive and respiratory syndrome virus by stable transfection of sialoadhesin and CD163. BMC Biotechnol 10:48. doi: 10.1186/1472-6750-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weingartl HM, Sabara M, Pasick J, van Moorlehem E, Babiuk L. 2002. Continuous porcine cell lines developed from alveolar macrophages: partial characterization and virus susceptibility. J Virol Methods 104:203–216. doi: 10.1016/s0166-0934(02)00085-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee YJ, Park CK, Nam E, Kim SH, Lee OS, Lee Du S, Lee C. 2010. Generation of a porcine alveolar macrophage cell line for the growth of porcine reproductive and respiratory syndrome virus. J Virol Methods 163:410–415. doi: 10.1016/j.jviromet.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 28.Whitworth KM, Rowland RR, Ewen CL, Trible BR, Kerrigan MA, Cino-Ozuna AG, Samuel MS, Lightner JE, McLaren DG, Mileham AJ, Wells KD, Prather RS. 2016. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat Biotechnol 34:20–22. doi: 10.1038/nbt.3434. [DOI] [PubMed] [Google Scholar]
- 29.Prather RS, Rowland RR, Ewen C, Trible B, Kerrigan M, Bawa B, Teson JM, Mao J, Lee K, Samuel MS, Whitworth KM, Murphy CN, Egen T, Green JA. 2013. An intact sialoadhesin (Sn/SIGLEC1/CD169) is not required for attachment/internalization of the porcine reproductive and respiratory syndrome virus. J Virol 87:9538–9546. doi: 10.1128/JVI.00177-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wells KD, Bardot R, Whitworth KM, Trible BR, Fang Y, Mileham A, Kerrigan MA, Samuel MS, Prather RS, Rowland RR. 2017. Replacement of porcine CD163 scavenger receptor cysteine-rich domain 5 with a CD163-like homolog confers resistance of pigs to genotype 1 but not genotype 2 porcine reproductive and respiratory syndrome virus. J Virol 91:e01521-16. doi: 10.1128/JVI.01521-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das PB, Dinh PX, Ansari IH, de Lima M, Osorio FA, Pattnaik AK. 2010. The minor envelope glycoproteins GP2a and GP4 of porcine reproductive and respiratory syndrome virus interact with the receptor CD163. J Virol 84:1731–1740. doi: 10.1128/JVI.01774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wissink EH, Kroese MV, van Wijk HA, Rijsewijk FA, Meulenberg JJ, Rottier PJ. 2005. Envelope protein requirements for the assembly of infectious virions of porcine reproductive and respiratory syndrome virus. J Virol 79:12495–12506. doi: 10.1128/JVI.79.19.12495-12506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Gorp H, Van Breedam W, Van Doorsselaere J, Delputte PL, Nauwynck HJ. 2010. Identification of the CD163 protein domains involved in infection of the porcine reproductive and respiratory syndrome virus. J Virol 84:3101–3105. doi: 10.1128/JVI.02093-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burkard C, Opriessnig T, Mileham AJ, Stadejek T, Ait-Ali T, Lillico SG, Whitelaw CBA, Archibald AL. 2018. Pigs lacking the scavenger receptor cysteine-rich domain 5 of CD163 are resistant to porcine reproductive and respiratory syndrome virus 1 infection. J Virol 92:e00415-18. doi: 10.1128/JVI.00415-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burkard C, Lillico SG, Reid E, Jackson B, Mileham AJ, Ait-Ali T, Whitelaw CB, Archibald AL. 2017. Precision engineering for PRRSV resistance in pigs: macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLoS Pathog 13:e1006206. doi: 10.1371/journal.ppat.1006206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kodama Y, Hu CD. 2010. An improved bimolecular fluorescence complementation assay with a high signal-to-noise ratio. Biotechniques 49:793–805. doi: 10.2144/000113519. [DOI] [PubMed] [Google Scholar]
- 37.Huang C, Bernard D, Zhu J, Dash RC, Chu A, Knupp A, Hakey A, Hadden MK, Garmendia A, Tang Y. 2020. Small molecules block the interaction between porcine reproductive and respiratory syndrome virus and CD163 receptor and the infection of pig cells. Virol J 17:116. doi: 10.1186/s12985-020-01361-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wallach I, Dzamba M, Heifets A. 2015. AtomNet: a deep convolutional neural network for bioactivity prediction in structure-based drug discovery. arXiv doi: 10.48550/arXiv.1510.02855. [DOI]
- 39.Ma H, Jiang L, Qiao S, Zhi Y, Chen XX, Yang Y, Huang X, Huang M, Li R, Zhang GP. 2017. The crystal structure of the fifth scavenger receptor cysteine-rich domain of porcine CD163 reveals an important residue involved in porcine reproductive and respiratory syndrome virus infection. J Virol 91:e01897-16. doi: 10.1128/JVI.01897-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adasme MF, Linnemann KL, Bolz SN, Kaiser F, Salentin S, Haupt VJ, Schroeder M. 2021. PLIP 2021: expanding the scope of the protein-ligand interaction profiler to DNA and RNA. Nucleic Acids Res 49:W530–W534. doi: 10.1093/nar/gkab294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Gorp H, Delputte PL, Nauwynck HJ. 2010. Scavenger receptor CD163, a Jack-of-all-trades and potential target for cell-directed therapy. Mol Immunol 47:1650–1660. doi: 10.1016/j.molimm.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 42.Welch SK, Calvert JG. 2010. A brief review of CD163 and its role in PRRSV infection. Virus Res 154:98–103. doi: 10.1016/j.virusres.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 43.Laskowski RA, Swindells MB. 2011. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 44.Mirdita M, Schutze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. 2022. ColabFold: making protein folding accessible to all. Nat Methods 19:679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, Weng Z. 2014. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 30:1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seidel SA, Dijkman PM, Lea WA, van den Bogaart G, Jerabek-Willemsen M, Lazic A, Joseph JS, Srinivasan P, Baaske P, Simeonov A, Katritch I, Melo FA, Ladbury JE, Schreiber G, Watts A, Braun D, Duhr S. 2013. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 59:301–315. doi: 10.1016/j.ymeth.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kreutz LC, Ackermann MR. 1996. Porcine reproductive and respiratory syndrome virus enters cells through a low pH-dependent endocytic pathway. Virus Res 42:137–147. doi: 10.1016/0168-1702(96)01313-5. [DOI] [PubMed] [Google Scholar]
- 48.Guo C, Wang M, Zhu Z, He S, Liu H, Liu X, Shi X, Tang T, Yu P, Zeng J, Yang L, Cao Y, Chen Y, Liu X, He Z. 2019. Highly efficient generation of pigs harboring a partial deletion of the CD163 SRCR5 domain, which are fully resistant to porcine reproductive and respiratory syndrome virus 2 infection. Front Immunol 10:1846. doi: 10.3389/fimmu.2019.01846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoian AMM, Rowland RRR, Brandariz-Nunez A. 2022. Mutations within scavenger receptor cysteine-rich (SRCR) protein domain 5 of porcine CD163 involved in infection with porcine reproductive and respiratory syndrome virus (PRRS). J Gen Virol 103. doi: 10.1099/jgv.0.001740. [DOI] [PubMed] [Google Scholar]
- 50.Van Breedam W, Delputte PL, Van Gorp H, Misinzo G, Vanderheijden N, Duan X, Nauwynck HJ. 2010. Porcine reproductive and respiratory syndrome virus entry into the porcine macrophage. J Gen Virol 91:1659–1667. doi: 10.1099/vir.0.020503-0. [DOI] [PubMed] [Google Scholar]
- 51.Hsieh CH, Li L, Vanhauwaert R, Nguyen KT, Davis MD, Bu G, Wszolek ZK, Wang X. 2019. Miro1 marks Parkinson's disease subset and Miro1 reducer rescues neuron loss in Parkinson's models. Cell Metab 30:1131–1140. doi: 10.1016/j.cmet.2019.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ. 2016. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc 11:905–919. doi: 10.1038/nprot.2016.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trott O, Olson AJ. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4 and Table S2. Download jvi.00054-23-s0001.pdf, PDF file, 1.3 MB (1.3MB, pdf)
Table S1. Download jvi.00054-23-s0002.xlsx, XLSX file, 0.06 MB (62.4KB, xlsx)
Data Availability Statement
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.