ABSTRACT
The availability of fixed nitrogen is a limiting factor in the net primary production of all ecosystems. Diazotrophs overcome this limit through the conversion of atmospheric dinitrogen to ammonia. Diazotrophs are phylogenetically diverse bacteria and archaea that exhibit a wide range of lifestyles and metabolisms, including obligate anaerobes and aerobes that generate energy through heterotrophic or autotrophic metabolisms. Despite the diversity of metabolisms, all diazotrophs use the same enzyme, nitrogenase, to reduce N2. Nitrogenase is an O2-sensitive enzyme that requires a high amount of energy in the form of ATP and low potential electrons carried by ferredoxin (Fd) or flavodoxin (Fld). This review summarizes how the diverse metabolisms of diazotrophs utilize different enzymes to generate low potential reducing equivalents for nitrogenase catalysis. These enzymes include substrate-level Fd oxidoreductases, hydrogenases, photosystem I or other light-driven reaction centers, electron bifurcating Fix complexes, proton motive force-driven Rnf complexes, and Fd:NAD(P)H oxidoreductases. Each of these enzymes is critical for generating low potential electrons while simultaneously integrating the native metabolism to balance nitrogenase’s overall energy needs. Understanding the diversity of electron transport systems to nitrogenase in various diazotrophs will be essential to guide future engineering strategies aimed at expanding the contributions of biological nitrogen fixation in agriculture.
KEYWORDS: diazotrophs, electron transport, nitrogen fixation, physiology
INTRODUCTION
Nitrogen in the form of ammonia, nitrate, or urea is required for primary production in terrestrial and marine ecosystems (1). Most terrestrial nitrogen is stored in the atmosphere as dinitrogen (N2) gas and is unavailable to the majority of organisms. However, some bacteria and archaea, called diazotrophs, can convert N2 into bioavailable ammonia (NH3) through biological nitrogen fixation (BNF). Diazotrophs account for 60% of the fixed N2 input into the biogeochemical nitrogen cycle (2). Diazotrophs fix N2 via the activity of nitrogenase, which catalyzes the cleavage of the N2 triple bond and its reduction to ammonia. BNF can be extraordinarily challenging for diazotrophs, as nitrogenase requires energy from ATP hydrolysis and low potential electrons and is sensitive to oxygen inactivation (3). Despite the aforementioned limitations of nitrogenase, diazotrophs are found in most ecological niches having a primary role in a diversity of microbial communities. Diazotrophs reside in oligotrophic marine ecosystems, the extreme environments of hot springs, the guts of termites, and in symbiotic relationships with crops (4–8). Diazotrophs can occupy these diverse niches through various physiological adaptations that shape native metabolisms to protect nitrogenase from oxygen while supplying enough ATP and reducing equivalents. This review details how a diversity of diazotrophs use electron transport enzymes in concert with physiological adaptations to support the energetic needs of nitrogenase-dependent reduction of N2.
The enzymatic reduction of N2 has one of the highest energy barriers in biology, requiring large amounts of reducing chemical energy and intricate metal cofactors to perform the reaction (9). Nitrogenase is the only enzyme that can catalyze the reduction of N2 to NH3 and exists in 3 related forms with different metal dependencies: Mo-dependent, V-dependent, and heterometal-independent forms. These 3 forms of nitrogenase have been demonstrated to share the same overall mechanism but have slightly different reaction stoichiometries (10). Mo-dependent nitrogenase is the most commonly occurring, extensively studied, and most efficient operating with the optimal reaction stoichiometry shown in (Equation 1) (9, 11).
| (1) |
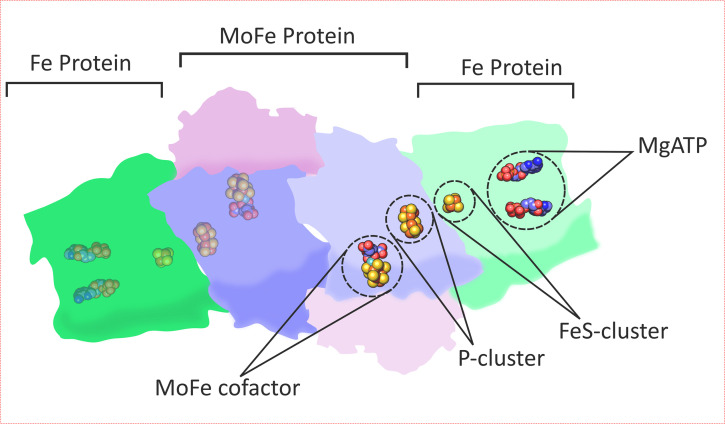
Mo-dependent nitrogenase consists of 2 separable component proteins termed the Fe protein and the MoFe protein to reflect the composition of their respective metal-containing cofactors (9, 12). The Fe protein is a homodimer containing a single [4Fe-4S] cluster and 2 binding sites for ATP (13). The MoFe protein is an α2β2 tetramer containing two [8Fe-7S] clusters (P-clusters) and two [7Fe-Mo-9S-C-citrate] FeMo-cofactors (FeMo-cos) that are located at the sites of substrate binding and reduction (Fig. 1) (14, 15). The metal cofactors are labile to oxygen, and organisms have evolved various strategies to protect nitrogenase from oxygen inactivation (16, 17).
FIG 1.
The nitrogenase enzyme is a heterodimer consisting of 3 subunits per dimer, the Fe protein (NifH), and the 2 subunits of the MoFe protein (NifDK). The Fe-protein contains 1 FeS cluster that accepts electrons from Fd or Fld and reduces the P-cluster in the MoFe protein in an ATP-dependent electron transfer. The P-cluster delivers electrons to the MoFe cofactor allowing electrons to be loaded for the reduction of N2. For complete N2 reduction, the Fe protein must deliver 8 electrons to the MoFe protein, requiring 2 ATP per electron transfer. Each metal cofactor is very sensitive to oxygen damage.
The Fe protein is reduced by ferredoxin (Fd) or flavodoxin (Fld) in vivo (18–20). Fd and Fld are small electron transfer proteins containing either FeS cluster(s) or a flavin mononucleotide (FMN), respectively (21–23). Diazotrophs typically express several Fds and Flds, and both can serve as electron donors to the Fe protein in an individual organism (24–27). During catalysis, the Fe protein switches between an ADP-bound and ATP-bound conformational state, modulating the reduction potential of the [4Fe-4S] cluster (28, 29). Fd/Fld most likely reduces the ADP-bound Fe protein preferentially, requiring Fd/Fld to have a reduction potential of at least -415 to -430 mV (Fig. 2) (30, 31) (Table 1).
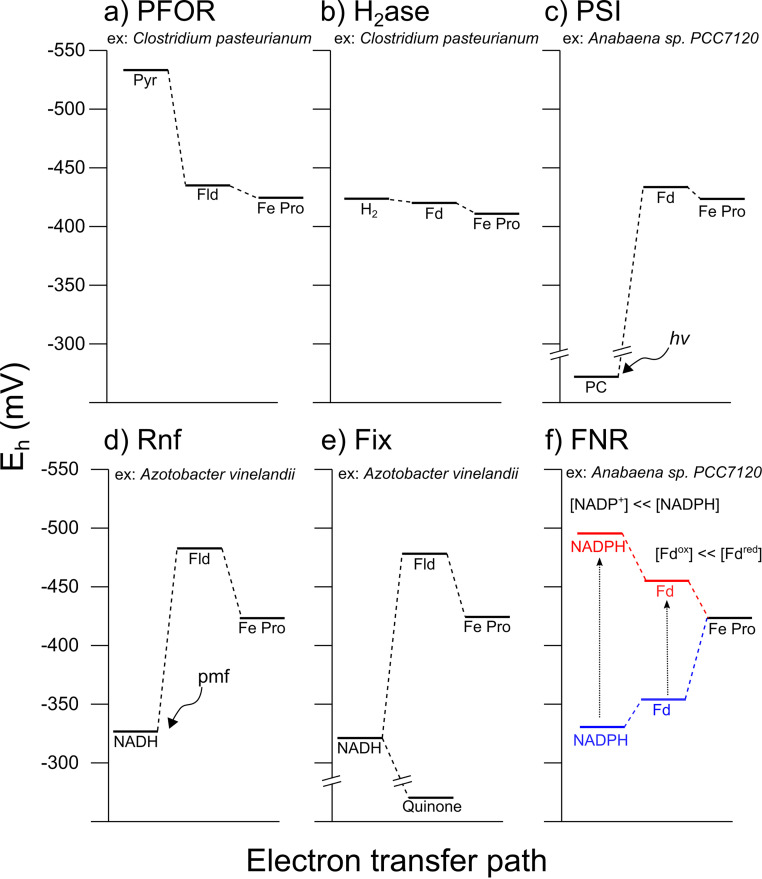
FIG 2.
Electrochemical potential landscapes of electron transport mechanism to Fd/Fld and the Fe protein of nitrogenase (Fe Pro) (values in Table 1). (a) Pyruvate ferredoxin oxidoreductase (PFOR) catalyzes the reduction of Fd from pyruvate to form acetyl-CoA and CO2. (b) Bi-directional hydrogenase (H2ase) in C. pasteurianum catalyzes the reduction of Fd directly from H2. Fd and the Fe protein of nitrogenase are at a higher potential in C. pasteurianum than other diazotrophs, facilitating the electron transfer. (c) Light-coupled Fd reduction in photosystem I or other reaction centers catalyzes the reduction of Fd from plastocyanin or other cytochromes. (d) Rnf catalyzes the reduction of Fd through the coupling of the proton motive force (pmf). (e) Fix couples the reduction of quinone with the reduction of Fd using electron bifurcation. (f) Ferredoxin:NADPH oxidoreductase (FNR) catalyzes Fd reduction by using the redox ratios of the NADPH and Fd. Heterocyst Fd only has a reduction potential of −351mV, which is not low enough for Fe protein reduction. An increase in the concentration of NADPH and reduced Fd will decrease the Eh (blue to red color), allowing the FNR reaction to be thermodynamically feasible.
TABLE 1.
Reduction potential values and references from Fig. 2
| Species | Type | Potential (mV) | Figure 2 | Reference |
|---|---|---|---|---|
| Azotobacter vinelandii | Nitrogenase ATP bound | −430 | d and e | 31 |
| Azotobacter vinelandii | Nitrogenase ADP bound | −470 | d and e | 31 |
| Clostridium pasteurianum | Nitrogenase ATP bound | −415 | a and b | 31 |
| Clostridium pasteurianum | Nitrogenase ADP bound | −415 | a and b | 31 |
| Azotobacter vinelandii | Flavodoxin | −483 | d and e | 21 |
| Azotobacter vinelandii | Ferredoxin | −619 | d and e | 207 |
| Anabaena 7120 | Ferredoxin heterocyst | −351 | f | 208 |
| Anabaena 7120 | Ferredoxin vegetative | −384 | f | 208 |
| Synechocystis PCC 6803 | Cytochrome c6 | +320 | c | 176 |
| General | Quinone | 4 | e | 32 |
| General | NAD+/ NADH (1:1) | −320 | e, d, and f | 32 |
| General | NAD+/ NADH (10:1) | −290 | e, d, and f | 32 |
| General | NADP+/ NADPH (1:1) | −320 | e, d, and f | 32 |
| General | NADP+/ NADPH (1:10) | −350 | e, d, and f | 32 |
| General | NADP+/ NADPH (1:100) | −380 | e, d, and f | 32 |
| General | H2 | −420 | b | 32 |
Maintaining a low reduction potential is essential for efficient electron transport to nitrogenase. When discussing electron transfer mechanisms, we use standard reduction potentials (E0’), which is the potential at pH 7, where the concentrations of the oxidized and reduced forms are equal. In reality, biological processes occur far from equilibrium, allowing reactions to become thermodynamically favorable, meaning a redox couple with E0’ of -300 mV might be maintained at -350 mV within the cell (32). Diazotrophs utilize the disequilibrium of redox couples to sustain the production of ATP and reduce Fd for nitrogenase.
Diazotroph metabolism differs mainly in the intercellular oxidation-reduction potential and nature of primary electron carriers. For example, low potential Fd is one of the central electron carriers in anaerobic bacteria, making Fd readily available for nitrogen fixation. In contrast, aerobic diazotrophs carry most electrons on the NAD(P)+/NAD(P)H reduction potentials at ~−320 mV and need to input energy or maintain favorable redox ratios to reduce Fd/Fld at ~−400 to −500 mV. There are 6 known or proposed mechanisms that generate low potential electrons in the form of Fd or Fld in order to donate to nitrogenase: (i) substrate-level Fd oxidoreductases, (ii) hydrogenases, (iii) photosystem I or other light-driven reaction centers, (iv) electron bifurcating Fix complexes, (v) proton motive force (pmf) driven Rnf complexes, and (vi) Fd:NAD(P)H oxidoreductases (Fig. 2).
OBLIGATE OR FACULTATIVE ANAEROBIC METABOLISM AND SUBSTRATE-LEVEL FD REDUCTION
During anaerobic metabolism, low potential electrons are generated through substrate-level Fd reduction. For example, obligate anaerobes Clostridium pasteurianum and Desulfovibrio africanus, and facultative anaerobes like Klebsiella pneumoniae and Bacillus polymyxa produce Fd in CoA-dependent reactions (33–40). The half-reaction of pyruvate oxidation to acetyl-CoA and CO2 by pyruvate-ferredoxin oxidoreductases (PFORs) has a reduction potential of −540 mV, allowing low potential Fds to act as the electron acceptor (Fig. 2a) - (Equation 2) (41–43). PFORs contain FeS clusters sensitive to oxygen, restricting PFORs from being expressed and functioning only in anaerobic conditions (44).
| (2) |
PFORs are phylogenetically diverse, found throughout all kingdoms, and represent key steps in the Wood-Ljungdahl and reductive TCA pathways (45–47). Many PFORs are found within the genomes of diazotrophs, but only a few have been shown to support nitrogenase activity. In K. pneumoniae, a facultative anaerobe that only fixes nitrogen under anaerobic or microaerobic conditions, the first characterized electron transport mechanism to nitrogenase was determined to be Fd produced by PFOR (38, 48). The gene for PFOR was designated nifJ, and an associated flavodoxin gene was termed nifF. Genes homologous to nifJ have been found in multiple diazotrophs, but not all are associated with nitrogen fixation. In the cyanobacterium Anabaena 7120, its PFOR is not required for diazotrophic growth under standard conditions but is essential under Fe-limitation (49, 50). In the purple nonsulfur bacterium (PNSB) Rhodospirillum rubrum, there is a significant amount of PFOR activity, and purified PFOR supports pyruvate-dependent in vitro nitrogenase activity with purified R. rubrum Fd (51, 52). PFOR in Rhodobacter capsulatus is upregulated under nitrogen-fixing conditions, but decreased levels are found in acetate and dark aerobically grown cells (45). However, PFOR’s overall contribution to nitrogen fixation in the PNSB may be low since other mechanisms, discussed later in the review, have been found to provide the primary source of low potential electrons (53–56). Overall, PFORs tend to be essential to general anaerobic metabolism and can support nitrogen fixation in obligate or facultative anaerobes.
In other anaerobic metabolisms, hydrogenases have been implicated in substrate-level Fd reduction for nitrogen fixation. Two classes of hydrogenase, [FeFe]- and [NiFe]-, catalyze the reversible oxidation of H2 (57–59). Under standard conditions, the H+/H2 reduction potential is −414 mV; therefore, reducing Fd (~ −400 to −500 mV) from H2 is a favorable or a slightly endergonic reaction (60). Nitrogen-fixing C. pasteurianum produces 3 non-bifurcating [FeFe]-hydrogenases that exhibit different catalytic properties. C. pasteurianum hydrogenase II (CpII) is upregulated 7.5-fold under nitrogen-fixing conditions and is proposed to recapture electrons that are produced as H2 during nitrogenase catalysis (59, 61). In C. pasteurianum, the standard reduction potential of Fd is −412 mV, and the ADP-bound Fe protein standard reduction potential has been measured at −415 mV (62, 63). Thus, the reduction potentials of H2, Fd, and Fe protein are within the thermodynamically favorable range allowing for the forward reaction and the reduction of the Fe protein from H2 (Fig. 2b).
While hydrogenase may contribute to Fd reduction, many other reactions in anaerobic metabolisms result in Fd reduction (64). For example, during nitrogen-fixing conditions in C. pasteurianum, 3 distinct PFOR genes are all expressed. Under these conditions, Fld is also expressed 14-fold higher, suggesting the ability of PFORs in C. pasteurianum to reduce both Fd and Fld (65). Other reactions also produce reduced Fd through formate oxidation and the reduction of crotonyl-CoA to butyryl-CoA, allowing multiple pathways to deliver electrons to nitrogenase (66, 67).
Anaerobic metabolisms, either through fermentation or anaerobic respiration, yield far less ATP compared to aerobic metabolism. For example, the facultative anaerobe K. pneumoniae fixes nitrogen under anaerobic conditions by fermenting glucose (68). Glucose catabolism and pyruvate degradation through PFOR to acetyl-P via phosphotransacetylase, then to the final fermentation product acetate, through acetate kinase, creates 2 reduced Fds and 1 ATP (69, 70). As nitrogenase requires a 2:1 ATP to reduced Fd ratio, anaerobic nitrogen fixation is limited by ATP synthesis (71). Reductant excess is balanced by shifting to ethanol or H2 as a final fermentation product which consumes the extra reductant but lowers the overall biomass yields (68). So, while anaerobic organisms and their reducing environment are ideal for protecting nitrogenase from oxygen and supplying electrons, overall energy expenditure is limited for optimizing growth.
ANAEROBIC LIGHT-DRIVEN FD REDUCTION
In phototrophic metabolism, reduced Fd can be generated from inputs of light energy. Bacterial FeS-type I reaction centers (RCI) found in anaerobic green sulfur bacteria and heliobacteria, as well as type I photosystems (PSI) found in cyanobacteria, reduce Fd (72) (Fig. 2c). Direct electron transfer has been proposed from Fd produced by RCI/PSI to nitrogenase. Heliobacterium modesticaldum is an active diazotroph with a minimal photosynthetic RC, containing just 2 subunits PshA and PshB (73). The terminal electron acceptor in PshB contains 2 Fd-like [4Fe-4S] clusters that donate electrons to Fd and is upregulated under diazotrophic conditions (74, 75). H. modesticaldum does have PFOR and Ferredoxin:NAD(P)H oxidoreductase (FNR) activity, but reduced diazotrophic growth rates observed under dark chemotrophic conditions suggest that RCI’s have a role in providing reduced Fd for nitrogenase catalysis (76). Reduction of Fd through FeS-type RC also occurs in nitrogen-fixing green sulfur bacteria Chlorobium tepidum, where Fd is primarily used in the reductive TCA cycle but could also be used for nitrogen fixation (77, 78). The Fd produced by PSI in cyanobacteria can also transport electrons to nitrogenase, but other electron transport mechanisms in cyanobacteria might be favored (see section on heterocyst metabolism).
NADH OXIDATION IN AEROBIC METABOLISM
Aerobic metabolism avoids using Fd as the primary electron carrier for most redox reactions due to its sensitivity to oxygen and therefore utilizes nucleotide electron carriers NAD(P)+/NAD(P)H as the primary electron source. Since nitrogenase can only accept electrons for Fd or Fld, various mechanisms are required to maintain reduced Fd/Fld production from the NAD(P)+/NAD(P)H pool. Primarily, aerobic diazotrophs couple the endergonic reduction of Fd from NAD(P)H with an exergonic reaction, such as electron bifurcation or the pmf. Alternatively, some aerobic diazotrophs can maintain a low reduction potential by decreasing the NAD(P)+/NAD(P)H ratio to overcome the thermodynamic barrier of Fd reduction in FNR.
There are multiple exergonic reactions to couple with the endergonic reduction of NAD(P)H and Fd in aerobic metabolism. One of the most studied is the flavin-based electron bifurcating enzyme Fix. The Fix enzyme was initially discovered in a genetic locus on the Rhizobium meliloti 2011 pSym megaplasmid that was essential for nitrogen fixation (79–81). Some genes in this region have homology to known K. pneumoniae nif genes, but other genes also required for nitrogen fixation have an unknown function. These genes were given the name fix (82). Some of the fix genes, fixABCX, have been implicated in nitrogenase biosynthesis in Azorhizobium caulinodans ORS571, but others have been suggested to have a role in electron transport as FixX has amino acid sequence homology with Fd (83, 84). The FixABCX complex was finally determined to be involved in electron transfer to nitrogenase in R. rubrum, in which the ΔfixABCX strains showed an ~ 80% decrease of in vivo nitrogenase activity while maintaining full activity in vitro (54, 55).
FixABCX is a membrane-bound oxidoreductase enzyme that generates low potential electrons Fd or Fld (Fig. 2e). FixAB shares similarities to the family of electron transfer flavoproteins (ETF) found in all life, including the ETF ubiquinone oxidoreductase, which transfers electrons to the quinone pool in the membrane (85). FixABCX employs flavin-based electron bifurcation (FBEB) to overcome the energy barrier of the endergonic reduction of Fd/Fld via NADH oxidation (86, 87). FBEB couples a thermodynamically favorable exergonic reaction to drive a thermodynamically unfavorable endergonic reaction (88). Using FBEB, FixABCX couples the exergonic reduction of quinone (10 mV) and the endergonic reduction of Fd (−460 mV) to the oxidation of 2 NADH (−320 mV) in an overall thermodynamically feasible reaction (Fig. 2e) (Equation 3) (89). Recently the structure of FixABCX homolog termed EtfABCX from Thermotoga maritima was determined by cryo-electron microscopy providing insights into the energy landscape of this class of flavin-based electron bifurcating enzymes (90).
| (3) |
Another mechanism to drive the endergonic reduction of Fd by NADH is harnessing the proton motive force. Seven genes in R. capsulatus were discovered to be required for nitrogen fixation called Rhodobacter nitrogen fixation or rnf (91, 92). The R. capsulatus genes rnfABCDGEH are similar to the respiratory Na+-dependent NADH:ubiquinone oxidoreductase (Na+-NQR) (93, 94). While Rnf has been implicated in Fd reduction for nitrogen fixation in R. capsulatus (91, 92), Pseudomonas stutzeri (95), and A. vinelandii (89, 96, 97), the biochemical characterization of Rnf has been in the directionality of Fd oxidation and H+/Na+ pumping. In Acetobacterium woodii, purification and characterization of Rnf has shown Na+-dependent reversible reduction of NAD+ with Fd (Fig. 2d) (Equation 4) (98–101). In general, rnf genes are widely distributed in bacteria and required in some methanogenic archaea (102).
| (4) |
Recently, cryo-EM structures of Rnf from A. vinelandii and Clostridium tetanomorphum implicate strict electron and proton transfer coupling through multiple flavins and FeS clusters (103, 104). This coupling is controlled by significant conformational changes to combine the endergonic reduction of Fd with the energy available in the H+/Na+ motive force.
Many aerobic heterotrophs store electrons in the NAD+/NADH electron couple with a reduction potential of -320 mV and funnel these electrons to oxidative phosphorylation in the electron transport chain (ETC). In order to produce low potential electrons in Fd, they must balance the use of Fix or Rnf with the production of ATP while maintaining a low oxygen environment. One of the most extensively studied diazotrophs is the obligate aerobic gamma-proteobacteria, A. vinelandii (105). The ability of A. vinelandii to fix nitrogen in fully saturated oxygen media is due to multiple mechanisms; from the creation of an extracellular alginate barrier to limit O2 diffusion, production of superoxide dismutase and catalase to consume oxygen radicals, and as a final barrier of protection, reversible inactivation, and protection of nitrogenase via the FeSII/Shethna protein (106–109). While the above mechanism provides some level of oxygen protection, the key mechanism that supports nitrogen fixation under high oxygen tensions is respiratory protection, where a terminal oxidase consumes oxygen at a high rate at the cell surface, preventing the accumulation of oxygen in the cytoplasm (110).
Respiratory protection is facilitated by a unique branch of the ETC, including a decoupled NADH-dehydrogenase II (NDHII) and a quinol terminal oxidase called cytochrome bd (111–113). This abbreviated pathway is partially coupled to the membrane, only translocating 1 H+/e-, allowing for a high respiration rate (113, 114). As a result, accumulation of cytochrome bd during high aeration and nitrogen fixation is observed, and knockouts of bd oxidase can no longer grow under diazotrophic conditions (115). A. vinelandii simultaneously expresses a fully proton coupled ETC under nitrogen-fixing conditions with an NDH-I, cytochrome bc1-oxidase, and cytochrome-o oxidases similar to complex I-IV seen in other proteobacteria and mitochondria (Fig. 3a) (115–118). The implementation of respiratory protection occurs at an energetic cost reducing the biomass yield of A. vinelandii and is sometimes termed a “wasting” mechanism. It has recently been proposed that this respiratory protection mechanism may be elicited in response to a high carbon and oxygen environment (119–121).
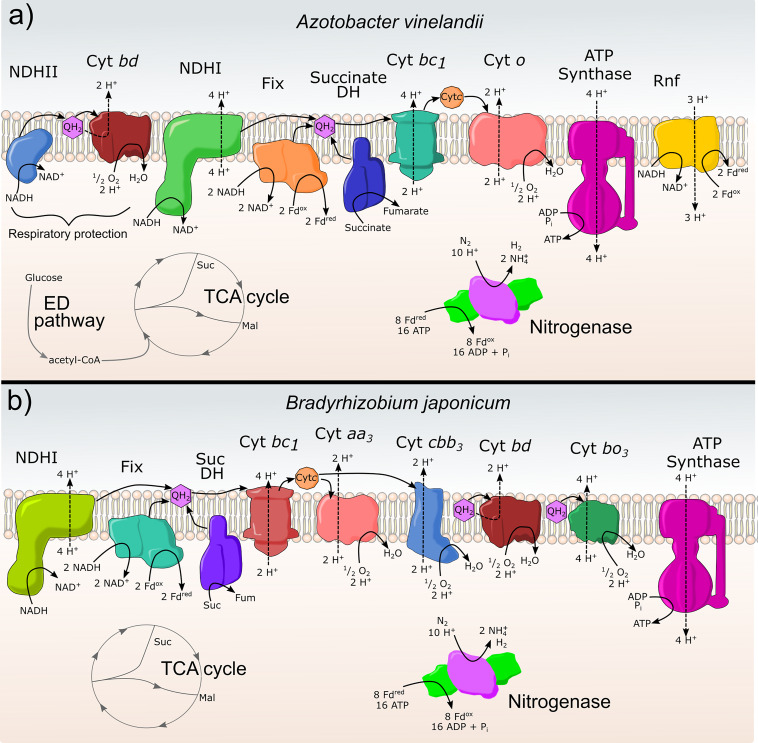
FIG 3.
The electron transport systems (ETS) of free-living and symbiotic aerobic diazotrophs. (a) The ETS in A. vinelandii can use 2 pathways. The first (left side) is a respiratory protection pathway consisting of an uncoupled type II NADH dehydrogenase (NDHII) and terminal oxidase cytochrome bd (Cyt bd). The second path is a traditional proteobacterial electron transport chain with full proton coupled complexes: NADH dehydrogenase (NDHI), succinate dehydrogenase (Succinate DH), cytochromes bc1 (Cyt bc1), and terminal oxidase cytochrome o (cyt o). The production of reduced ferredoxin (Fd) for nitrogenase is also part of the ETS with Fix and Rnf complexes. (b) Electron transport of Rhizobia. A generic look at the electron transport chain of rhizobia primarily based on data from Bradyrhizobium japonicum. Carbon enters the bacteroid mostly as organic acids malate (Mal) and succinate (Suc). NADH produced from the TCA cycle is oxidized by NADH dehydrogenase NDHI and produces quinol (QH2). Succinate dehydrogenase (SucDH) also reduces quinone to QH2. Electrons for nitrogenase are produced through the enzyme complex Fix which bifurcates electrons from NADH to quinone and ferredoxin (Fd). QH2 is oxidized by cytochrome bc1 and reduces cytochrome c (Cytc). There is a diversity of terminal oxidases in rhizobia: under higher oxygen concentrations, cytochrome aa3 is used, while under lower oxygen conditions (like those present for nitrogen fixation), cytochrome cbb3 is used. Two proton translocating quinol oxidases are also present as cytochrome bd and cytochrome bo3.
A. vinelandii possesses both Fix and Rnf mechanisms for generating low potential electrons for nitrogen fixation. The rnf gene cluster is located upstream from the small nif cluster and is regulated by nifA, the nif gene transcriptional activator. The fix gene cluster is not a part of the major or minor nif gene clusters in A. vinelandii but is upregulated under nitrogen-fixing conditions (122, 123). The Rnf and Fix complexes create partial redundancy for electron transport to nitrogenase, and deletion of either rnf or fix has little effect on diazotrophic growth, but the double mutant, Δrnf1Δfix is unable to grow diazotrophically (89). Other diazotrophs that encode for Rnf rarely have genes encoding for Fix and vice versa, making A. vinelandii unusual in having 2 ways to reduce Fd from NAD+/NADH (17, 124). Recently our lab has investigated the roles of both Rnf and Fix in differing conditions and has shown that while they are redundant in standard conditions of high carbon and oxygen, Fix is required for efficient growth under low oxygen conditions (125).
Other aerobic heterotrophic diazotrophs form symbiotic relationships with plants that provide carbon and a microaerobic environment in exchange for fixed nitrogen. These organisms, which belong to the alpha- and betaproteobacteria, are generally called rhizobia and are critical for agriculture (126, 127). Root nodules of leguminous plants house specialized symbiotic rhizobia (bacteroids) within infected plant cells enclosed in plant-derived membranes called the symbiosome membrane, allowing for the exchange of nutrients and protection from oxygen diffusion (128). Rhizobia are a diverse category of bacteria with various hosts with specialized metabolism to support nitrogen fixation. Here, we generalize the production of energy and electron transport to nitrogenase in a nodule environment.
The carbon source for rhizobia is predominately C4-dicarboxylic acids, primarily succinate and malate (129). Dicarboxylates are consumed by an NAD+-dependent malic enzyme paired with PEP-carboxykinase to produce acetyl-CoA, which can be used for carbon storage or consumption in the TCA cycle (130–132). Rhizobia express FixABCX encoded on the Sym plasmid and immediately upstream and regulated by nifA (133, 134). Metabolic changes are required to maintain redox balance during nitrogen fixation, including carbon storage in polyhydroxy-3-butyrate (PHB), lipids, or glycogen (135, 136). Amino acid export from the rhizobium and carbon sources from the plant also regulate redox balance in the cell with dicarboxylates creating a highly reduced environment requiring a higher oxygen demand but increasing the supply of reducing equivalents to nitrogenase (137). This balance between carbon storage, amino acid export, nitrogenase demand, and oxygen demand is critical for efficient ammonia export to the plant. Recent studies have shown that R. leguminosarum ΔfixAB mutants accumulated an order of magnitude more PHB and glycogen, suggesting balancing the redox homeostasis in the rhizobia by shuttling unused electrons to PHB or glycogen (133).
Rhizobia also have complex respiratory chains with multiple terminal oxidases (Fig. 3b). Oxygen levels in the nodules are maintained at 10 to 40 nM, protecting nitrogenase from oxygen damage (138). Bradyrhizobium japonicum encodes 8 terminal oxidases, including 2 bd-type oxidases and 6 heme-copper oxidases, which can be further divided into 2 quinol oxidases and 4 cytochrome c oxidases (139). Of these, 2 are predominant, the cbb3-type oxidase is encoded by the fixNOQP genes, and the caa3-type oxidase is encoded by coxBACF. The cbb3-type oxidases have a high affinity for oxygen (KM = 7 nM) and are essential in symbiosis and transport 0.5 H+/e- protons across the membrane (140–142). The cytochrome caa3 seems to not be essential under low oxygen concentrations in B. japonicum and R. leguminosarum but is the most prominent terminal oxidase in aerobic conditions (139, 143, 144).
Recent work has led to the proposal of combining the co-catabolism of succinate and arginine and FixABCX’s electron bifurcation mechanism to maintain flux through the electron transport chain (145). The acidification of the symbiosomes may make the driving force of NADH dehydrogenase no longer sufficient, requiring FixABCX as the major NADH regenerating pathway. This proposed path, called CATCH-N, suggests a co-catabolism of 2 equivalents of succinates and 2 arginine leads to the export of 2 alanine and 6 ammonia. This pathway, which nets ~ 2 to 3 ATPs per cycle, depends on using FixABCX as the primary NADH consumer (145). The intricacy of the symbiotic metabolism of plants and rhizobia is still being investigated, but the energy-saving bifurcation of the Fix mechanism allows for maximal ammonia production in highly acidic and low oxygen environments of the symbiosome (146).
ANOXYGENIC PHOTOTROPHS
Purple nonsulfur bacteria (PNSB) are metabolically versatile, growing in aerobic or anaerobic environments as heterotrophs or autotrophs. They can also generate energy through respiration, photosynthesis, and in some cases, fermentation (147, 148). However, PNSB can only fix nitrogen anaerobically in the light, creating ATP through anoxygenic photosynthetic cyclic electron transport and maintaining carbon and electrons supply from organic substrates, such as butyrate (149). To produce ATP, PNSB use light energy absorbed by reaction center 1 (RC1) to reduce quinone to quinol using electrons donated from cytochrome c. Cytochrome bc1 oxidizes quinol and reduces cytochrome c while translocating protons to complete the cyclic electron transport. The generated pmf can be used to produce ATP and NADH. Generation of NADH is achieved through the reverse reaction of NADH dehydrogenase, and consumption of pmf for NADH production is shown to be required for photoautotrophic growth in R. capsulatus and Rhodobacter sphaeroides (150, 151). During anaerobic phototrophic growth, PNSB must use the Calvin-Benson-Bassham (CBB) cycle as an electron sink, with the benefit of increasing biomass yield (152, 153). However, when cells become nitrogen-limited, nitrogenase becomes the electron sink, and the CBB cycle is no longer needed (154). Therefore, the cyclic electron transport of anoxygenic photosynthesis and reducing environment is ideal for nitrogen fixation.
PNSB have been shown to possess Rnf or Fix as mechanisms for transporting electrons to nitrogenase. In Rhodopseudomonas palustris and R. rubrum, FixABCX is located directly downstream of the nif genes, regulated by nifA, and essential for nitrogen fixation (27, 54, 155) (Fig. 4a). In contrast, R. capsulatus and R. sphaeroides only possess Rnf (92, 149, 156–158) (Fig. 4b). The contrast of 2 different mechanisms for Fd reduction within the same metabolic model of photoheterotrophy offers a potential comparison between Fix and Rnf. Different routes to generate biosynthetic precursors from acetate assimilation also determine the balance of electrons in PNSB. R. sphaeroids uses the reductive ethylmalonyl-CoA pathway, while R. palustris uses the oxidative glyoxylate shunt (154, 159). In R. sphaeroids which relies on Rnf for Fd production, the reductive ethylmalonyl-CoA pathway consumes reductant in the initial assimilation of acetate to malate and negates the use of the CBB cycle as an electron sink (159). In contrast, R. palustris, which relies on the Fix complex for Fd production, uses the glyoxylate shunt to produce net NADH and requires an electron sink (160). The contrasting acetate assimilation and Fd production pathways might suggest the co-evolution of Fix and Rnf within corresponding PNSB metabolism. In R. palustris the use of Fix means for every Fd produced and utilized as an electron sink in nitrogenase, another pair of electrons is recycled to quinol and, eventually, NADH. The less-reducing environment of R. sphaeroids uses Rnf to consume pmf, and there is no corresponding excess reductant. Each enzyme may play a role in balancing electron flow in certain PNSB bacteria and depend on the broader redox homeostasis of the specific metabolism.
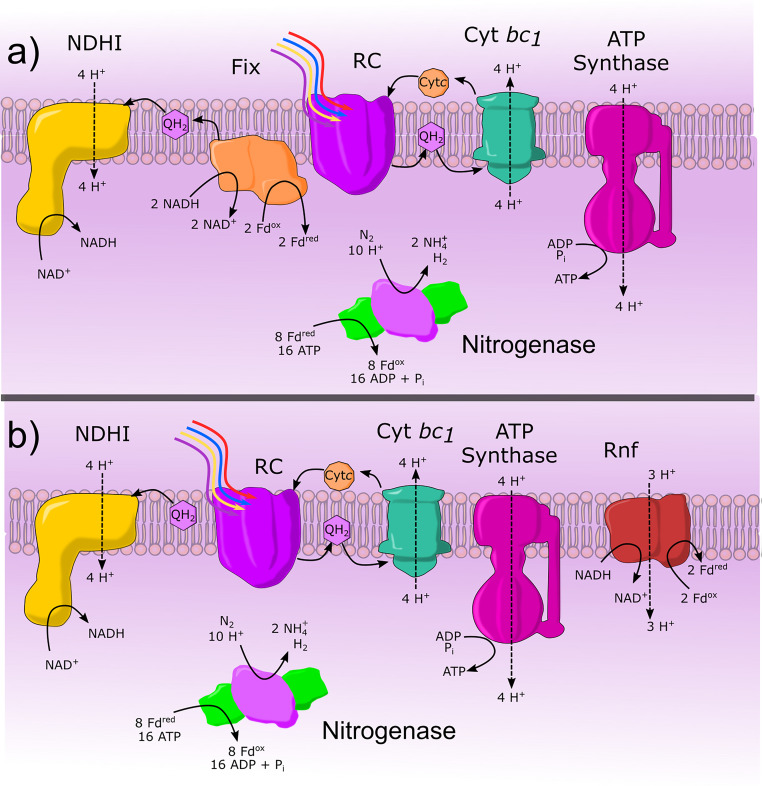
FIG 4.
Electron transport to nitrogenase in PNSB. a) A generalization of Fd and ATP production in R. palustris and R. rubrum with cyclic electron transport between the rection center 1 (RC), which reduces quinone to quinol (QH2) and oxidation of QH2 by cytochrome bc1 and while reducing cytochrome c (Cytc) and translocating protons, Cytc is then oxidized by RC to complete the cycle. NADH is produced by the reverse reaction of NADH dehydrogenase (NDHI). Fix catalyzes the production of Fd in R. palustris and R. rubrum through electron bifurcation where half of the electrons are directed to the reduction of quinone, either contributing to cyclic electron transport of reduction of NAD+. b) Fd and ATP production in R. capsulatus and R. sphaeroides is very similar to the ETC of R. palustris and R. rubrum but does not contain Fix but only contains Rnf. Rnf consumes pmf and competes with ATP synthase and NDH-I for pmf.
OXYGENIC PHOTOTROPHS
While substrate-level reduction of Fd, such as that carried out by PFORs in anaerobic metabolism, is thermodynamically favorable, reducing Fd from NADH is thermodynamically unfavorable. As previously covered, enzymes overcome this by coupling Fd reduction with a thermodynamically favorable reaction, such as using the pmf in Rnfs, redistributing exothermic reactions with electron bifurcation in Fix, or the input of light energy in RC/PSI. Reducing Fd directly from NAD(P)H, however, is possible via FNR under high NADPH/NADP+ ratios (Equation 5) (Fig. 2f).
| (5) |
FNR was first isolated from pea thylakoids and was shown to be the final step in photosynthetic electron transport from Fd to NADP+ (161, 162). FNRs ubiquity among living organisms and its diversity of cellular roles highlight the requirement for this simple electron transfer mechanism. While thermodynamically favorable in the direction of Fd oxidation and NAD(P)+ reduction, many FNRs serve primarily in the reverse Fd reduction direction (163). So how does FNR overcome the energy barrier of Fd reduction and avoid added input energy mechanisms seen in Rnf, Fix, or PSI?
While there is a diversity of FNRs and well characterized FNRs in photosynthetic electron transfer, we will focus on FNRs involved in nitrogen fixation (see reviews [164, 165] for more on other FNRs). The involvement of FNR in nitrogen fixation is well characterized in the filamentous cyanobacteria Anabaena sp. PCC 7120, which forms specialized heterocyst cells that create a microaerobic environment to protect nitrogenase from oxygen. Heterocysts depend on vegetative cells for electrons and carbon, and vegetative cells depend on fixed nitrogen from the heterocysts, making them interdependent (166, 167).
In Anabaena, 2 separate transcriptional start sites produce 2 isoforms of FNR from a single gene (petH). There is an ~ 34 kDa short FNR (FNRS) and a longer ~ 46 kDa FNR (FNRL), which contains an extra N-terminal domain (168, 169). This N-terminal domain allows interaction with cyanobacteria light-harvesting complexes called phycobilisomes (PBS), where interaction in the FNRL-PBS complex increases the activity toward NADP+ reduction (170–172). In contrast, the FNRS isoform accumulates in the heterocyst under nitrogen and Fe-deficient conditions and is not associated with the thylakoid membrane (173). The cellular localization suggests specific roles for the isoforms in either the forward reaction to reduce NADP+ for FNRL or the reverse reaction of Fd reduction for FNRS. While subcellular localization and protein interaction may help facilitate the reversibility of the reaction, other studies have shown that cofactor environment and protein surface residues tune the reduction potential of the FMN cofactor (reviewed in reference [165]). While tuning the cofactors allows for reversibility, reaction rates still play a significant role in the overall directionality of the reactions. Within the heterocyst, the NADP+/NADPH ratio is usually found in a reductive state of ~ 0.01, powered by the fast glucose-6-phosphate dehydrogenase and oxidative pentose phosphate pathway (OPP) (32). In vitro studies with NADPH, FNR, oxidized Fd, and nitrogenase showed maximal nitrogenase activity with a ratio of ~ 0.01 NADP+/NADPH (174, 175). The reduction potential of Fd also affects the overall favorability and kinetics of the reaction. Heterocysts have a specific Fd (HtFd), expressed during nitrogen starvation, that has a higher reduction potential at −351 mV than its vegetative counterpart (VFd) at −384 mV (Fig. 2f) (176). The HtFd is important for nitrogen fixation, but other redundant electron carriers supplement electron transport when deleting the HtFd gene (177). Overall, at equilibrium, the reduction of Fd directly from NADPH is thermodynamically unfavorable; however, cell physiology, cofactor environment, and cellular localization can modulate the reaction parameters to overcome the thermodynamic barriers.
Heterocyst metabolism specializes in protecting nitrogenase from oxygen while maintaining ATP and Fd for nitrogen fixation. First, photosystem II and its water oxidizing activity is limited, while PSI is upregulated, and cyclic electron transport is used to create pmf for ATP synthesis (178). Carbon fixation through rubisco is not active, and heterocysts do not contain carboxysomes (179, 180). Nitrogen metabolism also shifts as the glutamine oxoglutarate aminotransferase enzymes are not expressed in heterocysts, leaving glutamine synthetase to incorporate ammonia from nitrogenase and transport glutamine back to the vegetative cells (181). An increase in respiration also occurs through 2 specialized cytochrome c-type terminal oxidases (182).
Energy in the form of carbohydrates, in particular sucrose, is imported to the heterocysts from the photosynthetic vegetative cells and metabolized through the OPP (183). Deriving the right balance of low potential electrons and ATP for nitrogen fixation from the OPP pathway requires delivering electrons from NADPH to Fd, as well as to linear electron transport in the thylakoid membrane to produce ATP. Original experiments in crude extracts showed that nitrogenase activity could be measured with the addition of NADP+, glucose-6-phosphate, and a renewable ATP source, demonstrating the use of the OPP pathway to produce Fd (184). In addition, nitrogenase was active both in the light and the dark but had higher activity in the light, indicating the possibility of directly reducing nitrogenase through NADPH by FNR in the dark and the thylakoid membrane supporting nitrogen fixation under light conditions (184, 185). Within the ocean environment, nitrogen fixation can occur at night, even if at a much lower rate (185, 186). Thylakoid membrane reduction of Fd also contributes to nitrogenase as dibromothymoquinone (DBMIB) which blocks the plastoquinone reduction of Cytb6/f, inhibits nitrogen fixation (187). DBMIB inhibition suggests that NADPH electrons enter linear electron transport at plastoquinone, and that PSI can produce Fd for nitrogenase.
Thylakoid membranes of cyanobacteria contain an NAD(P)H:quinone oxidoreductase named NDH-1, similar to the complex I found in mitochondria (188, 189). The NDH-1 complex, along with ATP synthase and PSI complexes, are more abundant in heterocysts cells than in vegetative cells (190, 191). Recently, Fd has been implicated as the electron donor to NDH-1 as cyanobacterial NDH-1 lacks NADH binding subunits and should be considered a ferredoxin:plastoquinone oxidoreductases (192–194). This new insight places Fd as the central electron carrier in heterocysts where Fd produced either by FNR or PSI is responsible for nitrogen fixation, linear electron transport, and terminal oxidase activity (Fig. 5).
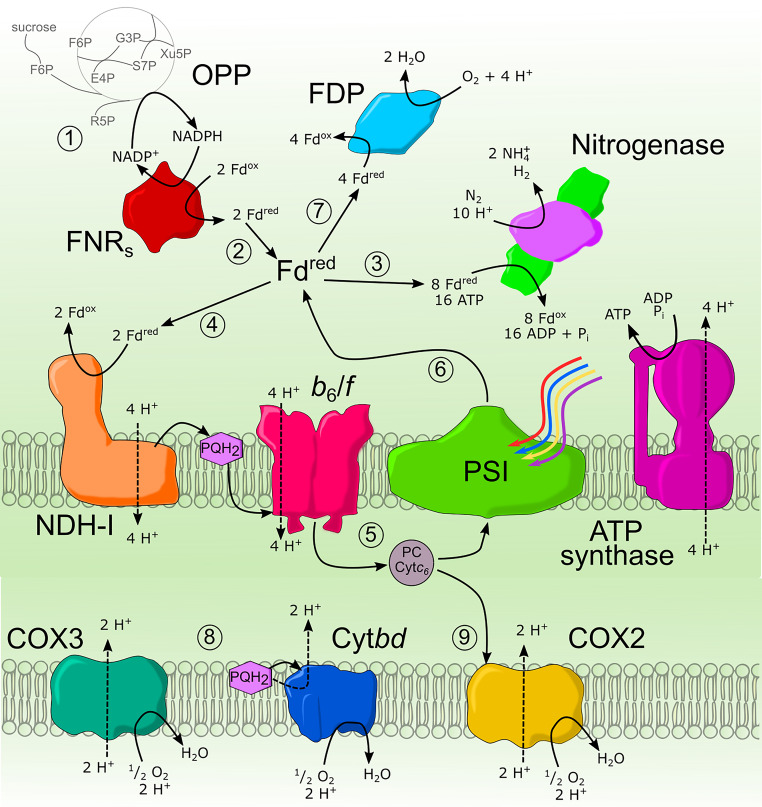
FIG 5.
Electron transport in heterocyst of Anabaena sp PCC 7120. (1) Carbon enters the heterocyst from the vegetative cells, usually as sucrose, entering the oxidative pentose phosphate (OPP) pathway through fructose-6-phosphate (F6P) to produce NADPH. (2) NADPH is oxidized by the short isoform of ferredoxin:NADPH oxidoreductase (FNRS). Reduced Fd then has multiple paths; the main pathway (3) would be the reduction of the Fe protein in nitrogenase for nitrogen fixation. Another path (4) for Fd is the cyclic electron transport starting by reducing plastoquinone to plastiquinol (PQH2) in the ferredoxin:plastoquinone oxidoreductase (NDH-1). Plastiquinone is oxidized by cytochrome b6/f translocating protons across the membrane, and plastocyanin (PC) or cytochrome c6 (CytC6) is reduced (5). Finally, photosystem 1 (PSI) oxidizes PC and uses energy from photons to reduce Fd (6), completing the cyclic electron transport and producing pmf for ATP synthase to produce ATP. Heterocysts have a diversity of terminal oxidases capable of reducing O2 from multiple electron sources. First, flavodiiron proteins (FDP) can reduce O2 directly from Fd (7). Second, 3 terminal oxidases are found in the thylakoid membrane of heterocysts. First, ubiquinol-oxidase bd (Cytbd) and Cox3 (8), while less characterized, are not essential for nitrogen fixation but contribute to respiration. Cox2 (9) exclusively accepts electrons from Cytc6 and translocate protons for ATP synthase.
While heterocysts maintain a microaerobic environment through morphological adaptations, consumption of excess O2 within the heterocysts is required for nitrogenase protection. Flavodiiron protein (FDP), which reduces O2 directly to H2O using Fd as an electron source, is responsible for light-induced O2 uptake in heterocysts of Anabaena sp. PCC 7120 (195). Multiple terminal oxidases contribute to oxygen consumption, including the heme-copper-type cox1 and cox2 genes encoding 2 aa3-type terminal oxidases and a cox3 gene cluster encoding a quinol oxidase. During nitrogen fixation, only cox2 and cox3 are expressed in heterocysts along with a cytochrome c6 that delivers electrons specifically to the caa3-type Cox2 terminal oxidase and is required for nitrogen fixation (196). Uptake hydrogenase (Hup) also plays a crucial role in maintaining redox status in the cell. Although they do not directly produce Fd in the heterocyst, Hups recycle H2 from nitrogenase back into quinone (197). Heterocysts must maintain the flux of electrons to nitrogenase and through the respiratory chain to produce ATP. Then, electrons also must be prioritized in terminal oxidases and FDPs to protect nitrogenase from any excess oxygen. The total balance of these reactions has not been fully investigated.
More work is needed to elucidate the electron transport mechanism to nitrogenase in heterocysts, but a main electron transport network can be proposed. First, carbon from vegetative cells enters as sucrose and is hydrolyzed to hexoses or hexose phosphates, then metabolized through the OPP pathway to create NADPH, and this NADPH reduces Fd using the FNRS isoform. From here, reduced Fd can be delivered directly to nitrogenase or used in linear electron flow to produce pmf and ATP. Electrons in reduced Fd can also be used to consume O2 and protect nitrogenase through multiple terminal oxidases and FDPs, which limits O2 in the cytoplasm (Fig. 5) (195, 196). As stated in the section above, FNRS activity primarily depends on the NADPH/NADP+ and Fdred/Fdox ratios. Maintaining high NADPH and low reduced Fd concentrations could also be controlled by the thylakoid membranes and varying terminal electron acceptors (198).
TRANSFER OF NITROGENASE TO NON-DIAZOTROPHIC ORGANISMS
As shown above, bacterial metabolisms have been able to adapt their metabolisms to support the strict criteria of nitrogenase. These complex adaptations require specific electron transport enzymes but also a total rebalance of metabolism and redox state. Efforts to transfer nitrogenase into non-diazotrophic organisms have become a promising strategy to overcome the disadvantages of industrial nitrogen fertilizers in agriculture (199–201). Significant advances have been made in synthesizing nitrogenase subunits or its biosynthetic enzymes in yeast, plant mitochondria, and non-diazotrophic cyanobacteria (202–205). In addition to the challenges of synthesizing a working nitrogenase in non-diazotrophic organisms, the metabolic context around nitrogenase must be considered. Yang et al. tested plant FNRs and Fd’s ability to reduce nitrogenase in an E. coli construct. The authors first showed that Klebsiella oxytoca PFOR reduced chloroplast and root-plastid Fds, and these reduced plant Fds could support nitrogenase activity in vivo (206). Further, they tested the ability of plant FNRs and plant Fd to supply nitrogenase with electrons and saw a restoration of ~ 45% of nitrogenase activity compared to K. oxytoca PFOR and Fld. This review highlights that many mechanisms can provide low potential reducing equivalents for nitrogen fixation, but each mechanism is adapted for a specific role in each diazotroph. Total physiology, such as maintenance of NADP+/NADPH ratios, Fd reduction potentials, and oxygen protection mechanisms, must be considered if the goal is to support nitrogen fixation with plant FNRs in plant organelles such as root plastids.
Conclusion.
The reduction of N2 by nitrogenase is one of the most energy-intensive and complex reactions in biology. A large amount of energy is required, and nitrogenase’s susceptibility to oxygen damage limits its potential in aerobic environments. Even with the intense evolutionary pressure of nitrogen limitation, life has not been able to reduce N2 through any other enzymatic mechanism, nor has it made the requirements for the nitrogenase reaction conditions less stringent. Diazotrophs utilize specific electron transport enzymes depending on the mode of metabolism to supply low potential electrons for nitrogenase catalysis such as substrate-level Fd oxidoreductases, hydrogenases, photosystem I or other light-driven reaction centers, electron bifurcating Fix complexes, proton motive force-driven Rnf complexes, and Fd:NAD(P)H oxidoreductases. Despite the diversity in pathways and enzymes, each diazotroph has the same criteria of maintaining a microaerobic environment, producing low potential electrons, and providing a high amount of ATP to maximize flux to nitrogenase.
ACKNOWLEDGMENTS
This work was supported by the U.S. DOE, Office of Science, Office of Basic Energy Sciences, under award DE-SC0018143 to J.W.P. Partial salary for J.W.P. was supported by the United States Department of Agriculture National Institute of Food and Agriculture, Hatch umbrella project #1015621.
Biographies

Alexander B. Alleman received his bachelor’s in biochemistry at Western State Colorado University in 2014 and Ph.D. in molecular plant science from Washington State University in 2021. He is currently a postdoctoral fellow at the University of Idaho in the biological sciences department. His main interests are physiology and energy metabolism in bacteria that form beneficial plant interactions.

John W. Peters obtained his PhD from Virginia Tech in 1995 and was an NIH postdoctoral fellow at Caltech. He is currently a Presidential Professor and Chair of the Department of Chemistry and Biochemistry at his alma mater, The University of Oklahoma. He is appointed a joint appointee and laboratory fellow at the Pacific Northwest National Laboratory and has been elected fellow of the American Association for the Advancement of Science and the American Academy of Microbiology. His main interests are in electron transfer processes focusing mainly on aspects of hydrogen and nitrogen metabolism and electron bifurcation.
Contributor Information
John W. Peters, Email: jw.peters@ou.edu.
Jennifer B. Glass, Georgia Institute of Technology
REFERENCES
- 1.Gruber N, Galloway JN. 2008. An Earth-system perspective of the global nitrogen cycle. Nature 451:293–296. doi: 10.1038/nature06592. [DOI] [PubMed] [Google Scholar]
- 2.Fowler D, Coyle M, Skiba U, Sutton MA, Cape JN, Reis S, Sheppard LJ, Jenkins A, Grizzetti B, Galloway JN, Vitousek P, Leach A, Bouwman AF, Butterbach-Bahl K, Dentener F, Stevenson D, Amann M, Voss M. 2013. The global nitrogen cycle in the twenty- first century. Philos Trans R Soc Lond B Biol Sci 368:1–13. doi: 10.1098/rstb.2013.0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robson RL, Postgate JR. 1980. Oxygen and hydrogen in biological nitrogen fixation. Annu Rev Microbiol 34:183–207. doi: 10.1146/annurev.mi.34.100180.001151. [DOI] [PubMed] [Google Scholar]
- 4.Hamilton TL, Boyd ES, Peters JW. 2011. Environmental constraints underpin the distribution and phylogenetic diversity of nifH in the Yellowstone geothermal complex. Microb Ecol 61:860–870. doi: 10.1007/s00248-011-9824-9. [DOI] [PubMed] [Google Scholar]
- 5.Zehr JP, Capone DG. 2020. Changing perspectives in marine nitrogen fixation. Science 368:1–9. doi: 10.1126/science.aay9514. [DOI] [PubMed] [Google Scholar]
- 6.Noda S, Ohkuma M, Usami R, Horikoshi K, Kudo T. 1999. Culture-independent characterization of a gene responsible for nitrogen fixation in the symbiotic microbial community in the gut of the termite Neotermes koshunensis. Appl Environ Microbiol 65:4935–4942. doi: 10.1128/AEM.65.11.4935-4942.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohkuma M, Kudo T. 1996. Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus. Appl Environ Microbiol 62:461–468. doi: 10.1128/aem.62.2.461-468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poole P, Ramachandran V, Terpolilli J. 2018. Rhizobia: from saprophytes to endosymbionts. Nat Rev Microbiol 16:291–303. doi: 10.1038/nrmicro.2017.171. [DOI] [PubMed] [Google Scholar]
- 9.Seefeldt LC, Hoffman BM, Dean DR. 2009. Mechanism of Mo-dependent nitrogenase. Annu Rev Biochem 78:701–722. doi: 10.1146/annurev.biochem.78.070907.103812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mus F, Alleman AB, Pence N, Seefeldt LC, Peters JW. 2018. Exploring the alternatives of biological nitrogen fixation. Metallomics 10:523–538. doi: 10.1039/c8mt00038g. [DOI] [PubMed] [Google Scholar]
- 11.Seefeldt LC, Yang ZY, Duval S, Dean DR. 2013. Nitrogenase reduction of carbon-containing compounds. Biochim Biophys Acta 1827:1102–1111. doi: 10.1016/j.bbabio.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peters JW, Fisher K, Dean DR. 1995. Nitrogenase structure and function: a biochemical-genetic perspective. Annu Rev Microbiol 49:335–366. doi: 10.1146/annurev.mi.49.100195.002003. [DOI] [PubMed] [Google Scholar]
- 13.Georgiadis MM, Komiya H, Chakrabarti P, Woo D, Kornuc JJ, Rees DC. 1992. Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science 257:1653–1659. doi: 10.1126/science.1529353. [DOI] [PubMed] [Google Scholar]
- 14.Peters JW, Stowell MHB, Soltis SM, Finnegan MG, Johnson MK, Rees DC. 1997. Redox-dependent structural changes in the nitrogenase P-cluster. Biochemistry 36:1181–1187. doi: 10.1021/bi9626665. [DOI] [PubMed] [Google Scholar]
- 15.Chan MK, Kim J, Rees DC. 1993. The nitrogenase FeMo-cofactor and P-cluster pair: 2.2 A resolution structures. Science 260:792–794. doi: 10.1126/science.8484118. [DOI] [PubMed] [Google Scholar]
- 16.Wang ZC, Burns A, Watt GD. 1985. Complex formation and O2 sensitivity of Azotobacter vinelandii nitrogenase and its component proteins. Biochemistry 24:214–221. doi: 10.1021/bi00322a031. [DOI] [PubMed] [Google Scholar]
- 17.Boyd ES, Garcia Costas AM, Hamilton TL, Mus F, Peters JW. 2015. Evolution of molybdenum nitrogenase during the transition from anaerobic to aerobic metabolism. J Bacteriol 197:1690–1699. doi: 10.1128/JB.02611-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klugkist J, Haaker H, Veeger C. 1986. Studies on the mechanism of electron transport to nitrogenase in Azotobacter vinelandii. Eur J Biochem 155:41–46. doi: 10.1111/j.1432-1033.1986.tb09456.x. [DOI] [PubMed] [Google Scholar]
- 19.Lowery TJ, Wilson PE, Zhang B, Bunker J, Harrison RG, Nyborg AC, Thiriot D, Watt GD. 2006. Flavodoxin hydroquinone reduces Azotobacter vinelandii Fe protein to the all-ferrous redox state with a S = 0 spin state. Proc Natl Acad Sci USA 103:17131–17136. doi: 10.1073/pnas.0603223103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Z-Y, Ledbetter R, Shaw S, Pence N, Tokmina-Lukaszewska M, Eilers B, Guo Q, Pokhrel N, Cash VL, Dean DR, Antony E, Bothner B, Peters JW, Seefeldt LC. 2016. Evidence that the P i release event is the rate-limiting step in the nitrogenase catalytic cycle. Biochemistry 55:3625–3635. doi: 10.1021/acs.biochem.6b00421. [DOI] [PubMed] [Google Scholar]
- 21.Segal HM, Spatzal T, Hill MG, Udit AK, Rees DC. 2017. Electrochemical and structural characterization of Azotobacter vinelandii flavodoxin II. Protein Sci 26:1984–1993. doi: 10.1002/pro.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen K, Jung YS, Bonagura CA, Tilley GJ, Prasad GS, Sridhar V, Armstrong FA, Stout CD, Burgess BK. 2002. Azotobacter vinelandii ferredoxin I. A sequence and structure comparison approach to alteration of [4Fe-4S]2+/3+ reduction potential. J Biol Chem 277:5603–5610. doi: 10.1074/jbc.M108916200. [DOI] [PubMed] [Google Scholar]
- 23.Pérez-Dorado I, Bortolotti A, Cortez N, Hermoso JA. 2013. Structural and phylogenetic analysis of Rhodobacter capsulatus NifF: uncovering general features of nitrogen-fixation (nif)-flavodoxins. Int J Mol Sci 14:1152–1163. doi: 10.3390/ijms14011152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin AE, Burgess BK, Iismaa SE, Smartt CT, Jacobson MR, Dean DR. 1989. Construction and characterization of an Azotobacter vinelandii strain with mutations in the genes encoding flavodoxin and ferredoxin I. J Bacteriol 171:3162–3167. doi: 10.1128/jb.171.6.3162-3167.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yakunin AF, Gennaro G, Hallenbeck PC. 1993. Purification and properties of a nif-specific flavodoxin from the photosynthetic bacterium Rhodobacter capsulatus. J Bacteriol 175:6775–6780. doi: 10.1128/jb.175.21.6775-6780.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gennaro G, Hübner P, Sandmeier U, Yakunin AF, Hallenbeck PC. 1996. Cloning, characterization, and regulation of nifF from Rhodobacter capsulatus. J Bacteriol 178:3949–3952. doi: 10.1128/jb.178.13.3949-3952.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fixen KR, Pal Chowdhury N, Martinez-Perez M, Poudel S, Boyd ES, Harwood CS. 2018. The path of electron transfer to nitrogenase in a phototrophic alpha-proteobacterium. Environ Microbiol 20:2500–2508. doi: 10.1111/1462-2920.14262. [DOI] [PubMed] [Google Scholar]
- 28.Braaksma A, Haaker H, Grande HJ, Veeger C. 1982. The effect of the redox potential on the activity of the nitrogenase and on the Fe-protein of Azotobacter vinelandii. Eur J Biochem 121:483–491. doi: 10.1111/j.1432-1033.1982.tb05813.x. [DOI] [PubMed] [Google Scholar]
- 29.Watt GD, Wang ZC, Knotts RR. 1986. Redox reactions of and nucleotide binding to the iron protein of Azotobacter vinelandii. Biochemistry 25:8156–8162. doi: 10.1021/bi00373a005. [DOI] [Google Scholar]
- 30.Pence N, Tokmina-Lukaszewska M, Yang Z-Y, Ledbetter RN, Seefeldt LC, Bothner B, Peters JW. 2017. Unraveling the interactions of the physiological reductant flavodoxin with the different conformations of the Fe protein in the nitrogenase cycle. J Biol Chem 292:15661–15669. doi: 10.1074/jbc.M117.801548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rutledge HL, Tezcan FA. 2020. Electron transfer in nitrogenase. Chem Rev 120:5158–5193. doi: 10.1021/acs.chemrev.9b00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicholls DG, Ferguson SJ. 2013. Bioenergetics 4 p. 27–51, 4th ed Elsiver, London, England. [Google Scholar]
- 33.Mortenson LE. 1964. Ferredoxin requirement for nitrogen fixation by extracts of Clostridium pasteurianum. Biochim Biophys Acta 81:473–478. doi: 10.1016/0926-6569(64)90132-4. [DOI] [PubMed] [Google Scholar]
- 34.Carpenter CE, Reddy DS, Cornforth DP. 1987. Inactivation of clostridial ferredoxin and pyruvate-ferredoxin oxidoreductase by sodium nitrite. Appl Environ Microbiol 53:549–552. doi: 10.1128/aem.53.3.549-552.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wahl RC, Orme-Johnson WH. 1987. Clostridial pyruvate oxidoreductase and the pyruvate-oxidizing enzyme specific to nitrogen fixation in Klebsiella pneumoniae are similar enzymes. J Biol Chem 262:10489–10496. doi: 10.1016/S0021-9258(18)60987-1. [DOI] [PubMed] [Google Scholar]
- 36.Pieulle L, Magro V, Hatchikian EC. 1997. Isolation and analysis of the gene encoding the pyruvate-ferredoxin oxidoreductase of Desulfovibrio africanus, production of the recombinant enzyme in Escherichia coli, and effect of carboxy-terminal deletions on its stability. J Bacteriol 179:5684–5692. doi: 10.1128/jb.179.18.5684-5692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nieva-Gómez D, Roberts GP, Klevickis S, Brill WJ. 1980. Electron transport to nitrogenase in Klebsiella pneumoniae. Proc Natl Acad Sci USA 77:2555–2558. doi: 10.1073/pnas.77.5.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hill S, Kavanagh EP. 1980. Roles of nifF and nifJ gene products in electron transport to nitrogenase in Klebsiella pneumoniae. J Bacteriol 141:470–475. doi: 10.1128/jb.141.2.470-475.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grau FH, Wilson PW. 1962. Physiology of nitrogen fixation by Bacillus polymyxa. J Bacteriol 83:490–496. doi: 10.1128/jb.83.3.490-496.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoch DC. 1973. Purification and properties of two ferredoxins from the nitrogen-fixing bacterium Bacillus polymyxa. Arch Biochem Biophys 158:633–640. doi: 10.1016/0003-9861(73)90555-9. [DOI] [PubMed] [Google Scholar]
- 41.Ragsdale SW. 2003. Pyruvate ferredoxin oxidoreductase and its radical intermediate. Chem Rev 103:2333–2346. doi: 10.1021/cr020423e. [DOI] [PubMed] [Google Scholar]
- 42.Chen PY-T, Aman H, Can M, Ragsdale SW, Drennan CL. 2018. Binding site for coenzyme A revealed in the structure of pyruvate:ferredoxin oxidoreductase from Moorella thermoacetica. Proc Natl Acad Sci USA 115:3846–3851. doi: 10.1073/pnas.1722329115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saeki K. 2004. Electron transport to nitrogenase: diverse routes for a common destination, p 257–290. In Genetics and Regulation of Nitrogen Fixation in Free-living Bacteria. Kluwer Academic Publishers. Dordreht, Netherlands. [Google Scholar]
- 44.Vita N, Hatchikian EC, Nouailler M, Dolla A, Pieulle L. 2008. Disulfide bond-dependent mechanism of protection against oxidative stress in pyruvate-ferredoxin oxidoreductase of anaerobic Desulfovibrio bacteria. Biochemistry 47:957–964. doi: 10.1021/bi7014713. [DOI] [PubMed] [Google Scholar]
- 45.Yakunin AF, Hallenbeck PC. 1998. Purification and characterization of pyruvate oxidoreductase from the photosynthetic bacterium Rhodobacter capsulatus. Biochim Biophys Acta 1409:39–49. doi: 10.1016/s0005-2728(98)00145-5. [DOI] [PubMed] [Google Scholar]
- 46.Charon M-H, Volbeda A, Chabriere E, Pieulle L, Fontecilla-Camps JC. 1999. Structure and electron transfer mechanism of pyruvate:ferredoxin oxidoreductase. Curr Opin Struct Biol 9:663–669. doi: 10.1016/s0959-440x(99)00027-5. [DOI] [PubMed] [Google Scholar]
- 47.Moulis J-M, Davasse V, Meyer J, Gaillard J. 1996. Molecular mechanism of pyruvate-ferredoxin oxidoreductases based on data obtained with the Clostridium pasteurianum enzyme. FEBS Lett 380:287–290. doi: 10.1016/0014-5793(96)00062-2. [DOI] [PubMed] [Google Scholar]
- 48.Smith A, Hill S, Anthony C. 1990. The purification, characterization and role of the d-type cytochrome oxidase of Klebsiella pneumoniae during nitrogen fixation. J Gen Microbiol 136:171–180. doi: 10.1099/00221287-136-1-171. [DOI] [PubMed] [Google Scholar]
- 49.Bauer CC, Scappino L, Haselkorn R. 1993. Growth of the cyanobacterium Anabaena on molecular nitrogen: NifJ is required when iron is limited. Proc Natl Acad Sci USA 90:8812–8816. doi: 10.1073/pnas.90.19.8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmitz O, Kentemich T, Zimmer W, Hundeshagen B, Bothe H. 1993. Identification of the nifJ gene coding for pyruvate: ferredoxin oxidoreductase in dinitrogen-fixing cyanobacteria. Arch Microbiol 160:62–67. doi: 10.1007/BF00258146. [DOI] [PubMed] [Google Scholar]
- 51.Brostedt E, Nordlund S. 1991. Purification and partial characterization of a pyruvate oxidoreductase from the photosynthetic bacterium Rhodospirillum rubrum grown under nitrogen-fixing conditions. Biochem J 279:155–158. doi: 10.1042/bj2790155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ludden PW, Burris RH. 1981. In vivo and in vitro studies on ATP and electron donors to nitrogenase in Rhodospirillum rubrum. Arch Microbiol 130:155–158. doi: 10.1007/BF00411070. [DOI] [Google Scholar]
- 53.Lindblad A, Jansson J, Brostedt E, Johansson M, Hellman U, Nordlund S. 1996. Identification and sequence of a nifJ-like gene in Rhodospirillum rubrum: partial characterization of a mutant unaffected in nitrogen fixation. Mol Microbiol 20:559–568. doi: 10.1046/j.1365-2958.1996.5311054.x. [DOI] [PubMed] [Google Scholar]
- 54.Edgren T, Nordlund S. 2004. The fixABCX genes in Rhodospirillum rubrum encode a putative membrane complex participating in electron transfer to nitrogenase. J Bacteriol 186:2052–2060. doi: 10.1128/JB.186.7.2052-2060.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgren T, Nordlund S. 2005. Electron transport to nitrogenase in Rhodospirillum rubrum: identification of a new fdxN gene encoding the primary electron donor to nitrogenase. FEMS Microbiol Lett 245:345–351. doi: 10.1016/j.femsle.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 56.Saeki K, Kumagai H. 1998. The rnf gene products in Rhodobacter capsulatus play an essential role in nitrogen fixation during anaerobic DMSO-dependent growth in the dark. Arch Microbiol 169:464–467. doi: 10.1007/s002030050598. [DOI] [PubMed] [Google Scholar]
- 57.Peters JW, Lanzilotta WN, Lemon BJ, Seefeldt LC. 1998. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution. Science 282:1853–1858. doi: 10.1126/science.282.5395.1853. [DOI] [PubMed] [Google Scholar]
- 58.Volbeda A, Charon M-H, Piras C, Hatchikian EC, Frey M, Fontecilla-Camps JC. 1995. Crystal structure of the nickel–iron hydrogenase from Desulfovibrio gigas. Nature 373:580–587. doi: 10.1038/373580a0. [DOI] [PubMed] [Google Scholar]
- 59.Therien JB, Artz JH, Poudel S, Hamilton TL, Liu Z, Noone SM, Adams MWW, King PW, Bryant DA, Boyd ES, Peters JW. 2017. The physiological functions and structural determinants of catalytic bias in the [FeFe]-hydrogenases CpI and CpII of Clostridium pasteurianum strain W5. Front Microbiol 8:1–11. doi: 10.3389/fmicb.2017.01305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schuchmann K, Chowdhury NP, Müller V. 2018. Complex multimeric [FeFe] hydrogenases: biochemistry, physiology and new opportunities for the hydrogen economy. Front Microbiol 9:1–22. doi: 10.3389/fmicb.2018.02911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Artz JH, Zadvornyy OA, Mulder DW, Keable SM, Cohen AE, Ratzloff MW, Williams SG, Ginovska B, Kumar N, Song J, McPhillips SE, Davidson CM, Lyubimov AY, Pence N, Schut GJ, Jones AK, Soltis SM, Adams MWW, Raugei S, King PW, Peters JW. 2020. Tuning catalytic bias of hydrogen gas producing hydrogenases. J Am Chem Soc 142:1227–1235. doi: 10.1021/jacs.9b08756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prince RC, Adams MW. 1987. Oxidation-reduction properties of the two Fe4S4 clusters in Clostridium pasteurianum ferredoxin. J Biol Chem 262:5125–5128. doi: 10.1016/S0021-9258(18)61163-9. [DOI] [PubMed] [Google Scholar]
- 63.Morgan TV, Prince RC, Mortenson LE. 1986. Electrochemical titration of the S = 3/2 and S = ½ states of the iron protein of nitrogenase. FEBS Lett 206:4–8. doi: 10.1016/0014-5793(86)81329-1. [DOI] [Google Scholar]
- 64.Horner DS, Hirt RP, Embley TM. 1999. A single eubacterial origin of eukaryotic pyruvate: ferredoxin oxidoreductase genes: implications for the evolution of anaerobic eukaryotes. Mol Biol Evol 16:1280–1291. doi: 10.1093/oxfordjournals.molbev.a026218. [DOI] [PubMed] [Google Scholar]
- 65.Groeger C, Wang W, Sabra W, Utesch T, Zeng AP. 2017. Metabolic and proteomic analyses of product selectivity and redox regulation in Clostridium pasteurianum grown on glycerol under varied iron availability. Microb Cell Fact 16:1–16. doi: 10.1186/s12934-017-0678-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scherer PA, Thauer RK. 1978. Purification and properties of reduced ferredoxin: CO2 oxidoreductase from Clostridium pasteurianum, a molybdenum iron-sulfur-protein. Eur J Biochem 85:125–135. doi: 10.1111/j.1432-1033.1978.tb12220.x. [DOI] [PubMed] [Google Scholar]
- 67.Buckel W, Thauer RK. 2013. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim Biophys Acta 1827:94–113. doi: 10.1016/j.bbabio.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 68.Hill S. 1976. The apparent ATP requirement for nitrogen fixation in growing Klebsiella pneumoniae. J Gen Microbiol 96:297–312. doi: 10.1099/00221287-95-2-297. [DOI] [PubMed] [Google Scholar]
- 69.Köpke M, Held C, Hujer S, Liesegang H, Wiezer A, Wollherr A, Ehrenreich A, Liebl W, Gottschalk G, Dürre P. 2010. Clostridium ljungdahlii represents a microbial production platform based on syngas. Proc Natl Acad Sci USA 107:13087–13092. doi: 10.1073/pnas.1004716107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schuchmann K, Müller V. 2014. Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nat Rev Microbiol 12:809–821. doi: 10.1038/nrmicro3365. [DOI] [PubMed] [Google Scholar]
- 71.Haaker H, Klugkist J. 1987. The bioenergetics of electron transport to nitrogenase. FEMS Microbiol Lett 46:57–71. doi: 10.1111/j.1574-6968.1987.tb02452.x. [DOI] [Google Scholar]
- 72.Hohmann-Marriott MF, Blankenship RE. 2011. Evolution of photosynthesis. Annu Rev Plant Biol 62:515–548. doi: 10.1146/annurev-arplant-042110-103811. [DOI] [PubMed] [Google Scholar]
- 73.Sattley WM, Madigan MT, Swingley WD, Cheung PC, Clocksin KM, Conrad AL, Dejesa LC, Honchak BM, Jung DO, Karbach LE, Kurdoglu A, Lahiri S, Mastrian SD, Page LE, Taylor HL, Wang ZT, Raymond J, Chen M, Blankenship RE, Touchman JW. 2008. The genome of Heliobacterium modesticaldum, a phototrophic representative of the Firmicutes containing the simplest photosynthetic apparatus. J Bacteriol 190:4687–4696. doi: 10.1128/JB.00299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heinnickel M, Shen G, Golbeck JH. 2007. Identification and characterization of PshB, the dicluster ferredoxin that harbors the terminal electron acceptors FA and FB in Heliobacterium modesticaldum. Biochemistry 46:2530–2536. doi: 10.1021/bi0622165. [DOI] [PubMed] [Google Scholar]
- 75.Sheehy D, Lu Y, Osman F, Alattar Z, Flores C, Sussman H, Zaare S, Dooling M, Meraban A, Baker P, Touchman J, Redding K. 2018. Genome-wide transcriptional response during the shift to N2-fixing conditions in Heliobacterium modesticaldum. J Proteom Bioinform 11:143–160. doi: 10.4172/jpb.1000481. [DOI] [Google Scholar]
- 76.Tang K-H, Yue H, Blankenship RE. 2010. Energy metabolism of Heliobacterium modesticaldum during phototrophic and chemotrophic growth. BMC Micro 10:1–14. doi: 10.1186/1471-2180-10-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seo D, Tomioka A, Kusumoto N, Kamo M, Enami I, Sakurai H. 2001. Purification of ferredoxins and their reaction with purified reaction center complex from the green sulfur bacterium Chlorobium tepidum. Biochim Biophys Acta 1503:377–384. doi: 10.1016/s0005-2728(00)00245-0. [DOI] [PubMed] [Google Scholar]
- 78.Eisen JA, Nelson KE, Paulsen IT, Heidelberg JF, Wu M, Dodson RJ, Deboy R, Gwinn ML, Nelson WC, Haft DH, Hickey EK, Peterson JD, Durkin AS, Kolonay JL, Yang F, Holt I, Umayam LA, Mason T, Brenner M, Shea TP, Parksey D, Nierman WC, Feldblyum TV, Hansen CL, Craven MB, Radune D, Vamathevan J, Khouri H, White O, Gruber TM, Ketchum KA, Venter JC, Tettelin H, Bryant DA, Fraser CM. 2002. The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc Natl Acad Sci USA 99:9509–9514. doi: 10.1073/pnas.132181499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Batut J, Terzaghi B, Ghérardi M, Huguet M, Terzaghi E, Garnerone AM, Boistard P, Huguet T. 1985. Localization of a symbiotic fix region on Rhizobium meliloti pSym megaplasmid more than 200 kilobases from the nod-nif region. Mol Gen Genet 199:232–239. doi: 10.1007/BF00330264. [DOI] [Google Scholar]
- 80.Corbin D, Barran L, Ditta G. 1983. Organization and expression of Rhizobium meliloti nitrogen fixation genes. Proc Natl Acad Sci USA 80:3005–3009. doi: 10.1073/pnas.80.10.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Renalier MH, Batut J, Ghai J, Terzaghi B, Gherardi M, David M, Garnerone AM, Vasse J, Truchet G, Huguet T. 1987. A new symbiotic cluster on the pSym megaplasmid of Rhizobium meliloti 2011 carries a functional fix gene repeat and a nod locus. J Bacteriol 169:2231–2238. doi: 10.1128/jb.169.5.2231-2238.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pühler A, Aguilar MO, Hynes M, Müller P, Klipp W, Priefer U, Simon R, Weber G. 1984. Advances in the genetics of free-living and symbiotic nitrogen fixing bacteria, p 609–619. In Veeger C, Newton WE (ed), Advances in nitrogen fixation research: proceedings of the 5th international symposium on nitrogen fixation, Noordwijkerhout, The Netherlands, August 28 – September 3, 1983. Springer; Dordrecht, Netherlands. [Google Scholar]
- 83.Earl CD, Ronson CW, Ausubel FM. 1987. Genetic and structural analysis of the Rhizobium meliloti fixA, fixB, fixC, and fixX genes. J Bacteriol 169:1127–1136. doi: 10.1128/jb.169.3.1127-1136.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kaminski PA, Norel F, Desnoues N, Kush A, Salzano G, Elmerich C. 1988. Characterization of the fixABC region of Azorhizobium caulinodans ORS571 and identification of a new nitrogen fixation gene. Mol Gen Genet 214:496–502. doi: 10.1007/BF00330486. [DOI] [PubMed] [Google Scholar]
- 85.Sperotto RA, Gross J, Vedoy C, Passaglia LMP, Schrank IS. 2004. The electron transfer flavoprotein fixABCX gene products from Azospirillum brasilense show a NifA-dependent promoter regulation. Curr Microbiol 49:267–273. doi: 10.1007/s00284-004-4318-3. [DOI] [PubMed] [Google Scholar]
- 86.Buckel W, Thauer RK. 2018. Flavin-based electron bifurcation, ferredoxin, flavodoxin, and anaerobic respiration with protons (Ech) or NAD+ (Rnf) as electron acceptors: a historical review. Front Microbiol 9:1–24. doi: 10.3389/fmicb.2018.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Peters JW, Miller A-F, Jones AK, King PW, Adams MW. 2016. Electron bifurcation. Curr Opin Chem Biol 31:146–152. doi: 10.1016/j.cbpa.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 88.Peters JW, Beratan DN, Bothner B, Dyer RB, Harwood CS, Heiden ZM, Hille R, Jones AK, King PW, Lu Y, Lubner CE, Minteer SD, Mulder DW, Raugei S, Schut GJ, Seefeldt LC, Tokmina-Lukaszewska M, Zadvornyy OA, Zhang P, Adams MW. 2018. A new era for electron bifurcation. Curr Opin Chem Biol 47:32–38. doi: 10.1016/j.cbpa.2018.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ledbetter RN, Garcia Costas AM, Lubner CE, Mulder DW, Tokmina-Lukaszewska M, Artz JH, Patterson A, Magnuson TS, Jay ZJ, Duan HD, Miller J, Plunkett MH, Hoben JP, Barney BM, Carlson RP, Miller AF, Bothner B, King PW, Peters JW, Seefeldt LC. 2017. The electron bifurcating FixABCX protein complex from Azotobacter vinelandii: generation of low-potential reducing equivalents for nitrogenase catalysis. Biochemistry 56:4177–4190. doi: 10.1021/acs.biochem.7b00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Feng X, Schut GJ, Lipscomb GL, Li H, Adams MWW. 2021. Cryoelectron microscopy structure and mechanism of the membrane-associated electron-bifurcating flavoprotein Fix/EtfABCX. Proc Natl Acad Sci USA 118:e2016978118. doi: 10.1073/pnas.2016978118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Saeki K, Tokuda K, Fujiwara T, Matsubara H. 1993. Nucleotide sequence and genetic analysis of the region essential for functional expression of the gene for ferredoxin I, fdxN, in Rhodobacter capsulatus: sharing of one upstream activator sequence in opposite directions by two operons related to nitrogen fixation. Plant Cell Physiol 34:185–199. doi: 10.1093/oxfordjournals.pcp.a078406. [DOI] [PubMed] [Google Scholar]
- 92.Schmehl M, Jahn A, Meyer zu Vilsendorf A, Hennecke S, Masepohl B, Schuppler M, Marxer M, Oelze J, Klipp W. 1993. Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol Gen Genet 241:602–615. doi: 10.1007/BF00279903. [DOI] [PubMed] [Google Scholar]
- 93.Jouanneau Y, Jeong HS, Hugo N, Meyer C, Willison JC. 1998. Overexpression in Escherichia coli of the rnf genes from Rhodobacter capsulatus–characterization of two membrane-bound iron-sulfur proteins. Eur J Biochem 251:54–64. doi: 10.1046/j.1432-1327.1998.2510054.x. [DOI] [PubMed] [Google Scholar]
- 94.Reyes-Prieto A, Barquera B, Juárez O. 2014. Origin and evolution of the sodium -pumping NADH: ubiquinone oxidoreductase. PLoS One 9:e96696–14. doi: 10.1371/journal.pone.0096696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lu W, Yan Y, Ping S, Peng J, Han Y, Li L, Yang J, Dou Y, Li Y, Fan H. 2010. Global transcriptional analysis of nitrogen fixation and ammonium repression in root- associated Pseudomonas stutzeri A1501. BMC Genet 11:1–13. doi: 10.1186/1471-2164-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bertsova YV, Serebryakova MV, Baykov AA, Bogachev AV. 2021. The flavin transferase ApbE flavinylates the ferredoxin:NAD+-oxidoreductase Rnf required for N2 fixation in Azotobacter vinelandii. FEMS Microbiol Lett 368:fnab130. doi: 10.1093/femsle/fnab130. [DOI] [PubMed] [Google Scholar]
- 97.Alleman AB, Mus F, Peters JW. 2021. Low potential electron generating enzymes serve different functions during aerobic nitrogen fixation in Azotobacter vinelandii. bioRxiv. doi: 10.1101/2021.10.06.463449. [DOI]
- 98.Hess V, Schuchmann K, Müller V. 2013. The ferredoxin: NAD+ Oxidoreductase (Rnf) from the acetogen Acetobacterium woodii requires Na+ and is reversibly coupled to the membrane potential. J Biol Chem 288:31496–31502. doi: 10.1074/jbc.M113.510255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Biegel E, Muller V. 2010. Bacterial Na+-translocating ferredoxin:NAD+ oxidoreductase. Proc Natl Acad Sci USA 107:18138–18142. doi: 10.1073/pnas.1010318107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wiechmann A, Trifunović D, Klein S, Müller V. 2020. Homologous production, one-step purification, and proof of Na+ transport by the Rnf complex from Acetobacterium woodii, a model for acetogenic conversion of C1 substrates to biofuels. Biotechnol Biofuels 13:1–9. doi: 10.1186/s13068-020-01851-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Westphal L, Wiechmann A, Baker J, Minton NP, Müller V. 2018. The Rnf complex is an energy-coupled transhydrogenase essential to reversibly link cellular NADH and ferredoxin pools in the acetogen Acetobacterium woodii. J Bacteriol 200:e00357-18. doi: 10.1128/JB.00357-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Biegel E, Schmidt S, González JM, Müller V. 2011. Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell Mol Life Sci 68:613–634. doi: 10.1007/s00018-010-0555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang L, Einsle O. 2022. Architecture of the NADH:ferredoxin oxidoreductase RNF that drives biological nitrogen fixation. bioRxiv. doi: 10.1101/2022.07.08.499327. [DOI]
- 104.Vitt S, Prinz S, Eisinger M, Ermler U, Buckel W. 2022. Purification and structural characterization of the Na+-translocating ferredoxin: NAD+ reductase (Rnf) complex of Clostridium tetanomorphum. Nat Commun 13:1–11. doi: 10.1038/s41467-022-34007-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Noar JD, Bruno-Bárcena JM. 2018. Azotobacter vinelandii: the source of 100 years of discoveries and many more to come. Microbiology (Reading) 164:421–436. doi: 10.1099/mic.0.000643. [DOI] [PubMed] [Google Scholar]
- 106.Sabra W, Zeng AP, Lünsdorf H, Deckwer WD. 2000. Effect of oxygen on formation and structure of Azotobacter vinelandii alginate and its role in protecting nitrogenase. Appl Environ Microbiol 66:4037–4044. doi: 10.1128/AEM.66.9.4037-4044.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schlesier J, Rohde M, Gerhardt S, Einsle O. 2016. A conformational switch triggers nitrogenase protection from oxygen damage by Shethna protein II (FeSII). J Am Chem Soc 138:239–247. doi: 10.1021/jacs.5b10341. [DOI] [PubMed] [Google Scholar]
- 108.Dingler C, Oelze J. 1987. Superoxide dismutase and catalase in Azotobacter vinelandii grown in continuous culture at different dissolved oxygen concentrations. Arch Microbiol 147:291–294. doi: 10.1007/BF00463490. [DOI] [Google Scholar]
- 109.Kabasakal BV, Cotton CaR, Murray JW. 2021. Crystal structure of the [2Fe–2S] protein I (Shethna protein I) from Azotobacter vinelandii. Acta Crystallogr F Struct Biol Commun 77:407–411. doi: 10.1107/S2053230X21009936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Poole RK, Hill S. 1997. Respiratory protection of nitrogenase activity in Azotobacter vinelandii: roles of the terminal oxidases. Biosci Rep 17:303–317. doi: 10.1023/a:1027336712748. [DOI] [PubMed] [Google Scholar]
- 111.Bertsova YV, Bogachev AV, Skulachev VP. 2001. Noncoupled NADH : ubiquinone oxidoreductase of Azotobacter vinelandii is required for diazotrophic growth at high oxygen concentrations. J Bacteriol 183:6869–6874. doi: 10.1128/JB.183.23.6869-6874.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wu G, Cruz-Ramos H, Hill S, Green J, Sawers G, Poole RK. 2000. Regulation of cytochrome bd expression in the obligate aerobe Azotobacter vinelandii by CydR (Fnr). Sensitivity to oxygen, reactive oxygen species, and nitric oxide. J Biol Chem 275:4679–4686. doi: 10.1074/jbc.275.7.4679. [DOI] [PubMed] [Google Scholar]
- 113.Bertsova YV, Bogachev AV, Skulachev VP. 1997. Generation of protonic potential by the bd-type quinol oxidase of Azotobacter vinelandii. FEBS Lett 414:369–372. doi: 10.1016/s0014-5793(97)01047-8. [DOI] [PubMed] [Google Scholar]
- 114.Bertsova YV, Bogachev AV, Skulachev VP. 1998. Two NADH:ubiquinone oxidoreductases of Azotobacter vinelandii and their role in the respiratory protection. Biochim Biophys Acta 1363:125–133. doi: 10.1016/s0005-2728(97)00094-7. [DOI] [PubMed] [Google Scholar]
- 115.D'mello R, Purchase D, Poole RK, Hill S. 1997. Expression and content of terminal oxidases in Azotobacter vinelandii grown with excess NH4+ are modulated by O2 supply. Microbiology (Reading) 143:231–237. doi: 10.1099/00221287-143-1-231. [DOI] [PubMed] [Google Scholar]
- 116.D'Mello R, Hill S, Poole RK. 1994. Determination of the oxygen affinities of terminal oxidases in Azotobacter vinelandii using the deoxygenation of oxyleghaemoglobin and oxymyoglobin: Cytochrome bd is a low-affinity oxidase. Microbiology 140:1395–1402. doi: 10.1099/00221287-140-6-1395. [DOI] [Google Scholar]
- 117.Rey L, Maier RJ. 1997. Cytochrome c terminal oxidase pathways of Azotobacter vinelandii: analysis of cytochrome c4 and c5 mutants and up-regulation of cytochrome c- dependent pathways with N2 fixation. J Bacteriol 179:7191–7196. doi: 10.1128/jb.179.22.7191-7196.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Moshiri F, Chawla A, Maier RJ. 1991. Cloning, characterization, and expression in Escherichia coli of the genes encoding the cytochrome d oxidase complex from Azotobacter vinelandii. J Bacteriol 173:6230–6241. doi: 10.1128/jb.173.19.6230-6241.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Oelze J. 2000. Respiratory protection of nitrogenase in Azotobacter species: is a widely held hypothesis unequivocally supported by experimental evidence? FEMS Microbiol Rev 24:321–333. doi: 10.1111/j.1574-6976.2000.tb00545.x. [DOI] [PubMed] [Google Scholar]
- 120.Inomura K, Bragg J, Riemann L, Follows MJ. 2018. A quantitative model of nitrogen fixation in the presence of ammonium. PLoS One 13:e0208282. doi: 10.1371/journal.pone.0208282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Alleman AB, Mus F, Peters JW. 2021. Metabolic model of the nitrogen-fixing obligate aerobe Azotobacter vinelandii predicts its adaptation to oxygen concentration and metal availability. mBio 12:e02593-21. doi: 10.1128/mBio.02593-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hamilton TL, Ludwig M, Dixon R, Boyd ES, Dos Santos PC, Setubal JC, Bryant DA, Dean DR, Peters JW. 2011. Transcriptional profiling of nitrogen fixation in Azotobacter vinelandii. J Bacteriol 193:4477–4486. doi: 10.1128/JB.05099-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Barney BM, Plunkett MH, Natarajan V, Mus F, Knutson CM, Peters JW. 2017. Transcriptional analysis of an ammonium excreting strain of Azotobacter vinelandii deregulated for nitrogen fixation. Appl Environ Microbiol 83:e01534-17. doi: 10.1128/AEM.01534-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Poudel S, Colman DR, Fixen KR, Ledbetter RN, Zheng Y, Pence N, Seefeldt LC, Peters JW, Harwood CS, Boyd ES. 2018. Electron transfer to nitrogenase in different genomic and metabolic backgrounds. J Bacteriol 200:1–19. doi: 10.1128/JB.00757-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Alleman AB, Garcia Costas A, Mus F, Peters JW. 2022. Rnf and fix have specific roles during aerobic nitrogen fixation in Azotobacter vinelandii. Appl Environ Microbiol 88:e01049-22. doi: 10.1128/aem.01049-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dixon R, Kahn D. 2004. Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol 2:621–631. doi: 10.1038/nrmicro954. [DOI] [PubMed] [Google Scholar]
- 127.Hirsch PR, Mauchline TH. 2015. The importance of the microbial N cycle in soil for crop plant nutrition. Adv Appl Microbiol 93:45–71. doi: 10.1016/bs.aambs.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 128.Haag AF, Arnold MFF, Myka KK, Kerscher B, Dall'Angelo S, Zanda M, Mergaert P, Ferguson GP. 2013. Molecular insights into bacteroid development during Rhizobium-legume symbiosis. FEMS Microbiol Rev 37:364–383. doi: 10.1111/1574-6976.12003. [DOI] [PubMed] [Google Scholar]
- 129.Zhang Y, Aono T, Poole P, Finan TM. 2012. NAD(P)+-malic enzyme mutants of Sinorhizobium sp. strain NGR234, but not Azorhizobium caulinodans ORS571, maintain symbiotic N2 fixation capabilities. Appl Environ Microbiol 78:2803–2812. doi: 10.1128/AEM.06412-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mulley G, Lopez-Gomez M, Zhang Y, Terpolilli J, Prell J, Finan T, Poole P. 2010. Pyruvate is synthesized by two pathways in pea bacteroids with different efficiencies for nitrogen fixation. J Bacteriol 192:4944–4953. doi: 10.1128/JB.00294-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Finan TM, McWhinnie E, Driscoll B, Watson RJ. 1991. Complex symbiotic phenotypes result from gluconeogenic mutations in Rhizobium meliloti. MPMI 4:386–392. doi: 10.1094/MPMI-4-386. [DOI] [Google Scholar]
- 132.Taté R, Cermola M, Riccio A, Diez-Roux G, Patriarca EJ. 2012. Glutathione is required by Rhizobium etli for glutamine utilization and symbiotic effectiveness. Mol Plant Microbe Interact 25:331–340. doi: 10.1094/MPMI-06-11-0163. [DOI] [PubMed] [Google Scholar]
- 133.Webb I, Xu J, Sanchez-Cañizares C, Karunakaran R, Ramachandran V, Rutten P, East A, Huang W, Watmough N, Poole P. 2021. Regulation and characterization of mutants of fixABCX in Rhizobium leguminosarum. Mol Plant Microbe Interact 34:1167–1180. doi: 10.1094/MPMI-02-21-0037-R. [DOI] [PubMed] [Google Scholar]
- 134.Degli EM, Martinez Romero E. 2016. A survey of the energy metabolism of nodulating symbionts reveals a new form of respiratory complex I. FEMS Microbiol Ecol 92:1–13. doi: 10.1093/femsec/fiw084. [DOI] [PubMed] [Google Scholar]
- 135.Lodwig EM, Leonard M, Marroqui S, Wheeler TR, Findlay K, Downie JA, Poole PS. 2005. Role of polyhydroxybutyrate and glycogen as carbon storage compounds in pea and bean bacteroids. Mol Plant Microbe Interact 18:67–74. doi: 10.1094/MPMI-18-0067. [DOI] [PubMed] [Google Scholar]
- 136.Terpolilli JJ, Masakapalli SK, Karunakaran R, Webb IUC, Green R, Watmough NJ, Kruger NJ, Ratcliffe RG, Poole PS. 2016. Lipogenesis and redox balance in nitrogen-fixing pea bacteroids. J Bacteriol 198:2864–2875. doi: 10.1128/JB.00451-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Schulte CCM, Borah K, Wheatley RM, Terpolilli JJ, Saalbach G, Crang N, de Groot DH, Ratcliffe RG, Kruger NJ, Papachristodoulou A, Poole PS. 2021. Metabolic control of nitrogen fixation in rhizobium-legume symbioses. Sci Adv 7:eabh2433. doi: 10.1126/sciadv.abh2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Vance CP, Heichel GH. 1991. Carbon in N2 fixation: limitation or exquisite adaptation. Annu Rev Plant Physiol Plant Mol Biol 42:373–390. doi: 10.1146/annurev.pp.42.060191.002105. [DOI] [Google Scholar]
- 139.Bühler D, Rossmann R, Landolt S, Balsiger S, Fischer H-M, Hennecke H. 2010. Disparate pathways for the biogenesis of cytochrome oxidases in Bradyrhizobium japonicum. J Biol Chem 285:15704–15713. doi: 10.1074/jbc.M109.085217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Preisig O, Zufferey R, Thöny-Meyer L, Appleby CA, Hennecke H. 1996. A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J Bacteriol 178:1532–1538. doi: 10.1128/jb.178.6.1532-1538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Preisig O, Zufferey R, Hennecke H. 1996. The Bradyrhizobium japonicum fixGHIS genes are required for the formation of the high-affinity cbb3-type cytochrome oxidase. Arch Microbiol 165:297–305. doi: 10.1007/s002030050330. [DOI] [PubMed] [Google Scholar]
- 142.Arslan E, Kannt A, Thöny-Meyer L, Hennecke H. 2000. The symbiotically essential cbb3-type oxidase of Bradyrhizobium japonicum is a proton pump. FEBS Lett 470:7–10. doi: 10.1016/s0014-5793(00)01277-1. [DOI] [PubMed] [Google Scholar]
- 143.Wheatley RM, Ramachandran VK, Geddes BA, Perry BJ, Yost CK, Poole PS. 2017. Role of O2 in the growth of Rhizobium leguminosarum bv. viciae 3841 on glucose and succinate. J Bacteriol 199:e00572-16. doi: 10.1128/JB.00572-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bott M, Preisig O, Hennecke H. 1992. Genes for a second terminal oxidase in Bradyrhizobium japonicum. Arch Microbiol 158:335–343. doi: 10.1007/BF00245362. [DOI] [PubMed] [Google Scholar]
- 145.Flores-Tinoco CE, Tschan F, Fuhrer T, Margot C, Sauer U, Christen M, Christen B. 2020. Co-catabolism of arginine and succinate drives symbiotic nitrogen fixation. Mol Syst Biol 16:e9419. doi: 10.15252/msb.20199419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ledermann R, Schulte CCM, Poole PS. 2021. How rhizobia adapt to the nodule environment. J Bacteriol 203:e00539-20. doi: 10.1128/JB.00539-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hädicke O, Grammel H, Klamt S. 2011. Metabolic network modeling of redox balancing and biohydrogen production in purple nonsulfur bacteria. BMC Syst Biol 5, 150:1–18. doi: 10.1186/1752-0509-5-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Klamt S, Grammel H, Straube R, Ghosh R, Gilles ED. 2008. Modeling the electron transport chain of purple non-sulfur bacteria. Mol Syst Biol 4:156–119. doi: 10.1038/msb4100191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ryu MH, Hull NC, Gomelsky M. 2014. Metabolic engineering of Rhodobacter sphaeroides for improved hydrogen production. Int J Hydrog Energy 39:6384–6390. doi: 10.1016/j.ijhydene.2014.02.021. [DOI] [Google Scholar]
- 150.Spero MA, Brickner JR, Mollet JT, Pisithkul T, Amador-Noguez D, Donohue TJ. 2016. Different functions of phylogenetically distinct bacterial complex I isozymes. J Bacteriol 198:1268–1280. doi: 10.1128/JB.01025-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tichi MA, Meijer WG, Tabita FR. 2001. Complex I and its involvement in redox homeostasis and carbon and nitrogen metabolism in Rhodobacter capsulatus. J Bacteriol 183:7285–7294. doi: 10.1128/JB.183.24.7285-7294.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hallenbeck PL, Lerchen R, Hessler P, Kaplan S. 1990. Roles of CfxA, CfxB, and external electron acceptors in regulation of ribulose 1,5-bisphosphate carboxylase/oxygenase expression in Rhodobacter sphaeroides. J Bacteriol 172:1736–1748. doi: 10.1128/jb.172.4.1736-1748.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Falcone DL, Tabita FR. 1991. Expression of endogenous and foreign ribulose 1,5-bisphosphate carboxylase-oxygenase (RubisCO) genes in a RubisCO deletion mutant of Rhodobacter sphaeroides. J Bacteriol 173:2099–2108. doi: 10.1128/jb.173.6.2099-2108.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.McKinlay JB, Harwood CS. 2011. Calvin cycle flux, pathway constraints, and substrate oxidation state together determine the H2 biofuel yield in photoheterotrophic bacteria. mBio 2:e00323-10. doi: 10.1128/mBio.00323-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Huang JJ, Heiniger EK, McKinlay JB, Harwood CS. 2010. Production of hydrogen gas from light and the inorganic electron donor thiosulfate by Rhodopseudomonas palustris. Appl Environ Microbiol 76:7717–7722. doi: 10.1128/AEM.01143-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jeong HS, Jouanneau Y. 2000. Enhanced nitrogenase activity in strains of Rhodobacter capsulatus that overexpress the rnf genes. J Bacteriol 182:1208–1214. doi: 10.1128/JB.182.5.1208-1214.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ma H, Yang H, Zheng X, Lie T, Yan W. 2021. Promoting photo-fermentative hydrogen production performance by substituting the rnf promoter in Rhodobacter capsulatus. Int J Hydrog Energy 46:3742–3752. doi: 10.1016/j.ijhydene.2020.10.270. [DOI] [Google Scholar]
- 158.Kontur WS, Ziegelhoffer EC, Spero MA, Imam S, Noguera DR, Donohue TJ. 2011. Pathways involved in reductant distribution during photobiological H2 production by Rhodobacter sphaeroides. Appl Environ Microbiol 77:7425–7429. doi: 10.1128/AEM.05273-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Erb TJ, Berg IA, Brecht V, Müller M, Fuchs G, Alber BE. 2007. Synthesis of C5-dicarboxylic acids from C2-units involving crotonyl-CoA carboxylase/reductase: the ethylmalonyl-CoA pathway. Proc Natl Acad Sci USA 104:10631–10636. doi: 10.1073/pnas.0702791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Laguna R, Tabita FR, Alber BE. 2011. Acetate-dependent photoheterotrophic growth and the differential requirement for the Calvin–Benson–Bassham reductive pentose phosphate cycle in Rhodobacter sphaeroides and Rhodopseudomonas palustris. Arch Microbiol 193:151–154. doi: 10.1007/s00203-010-0652-y. [DOI] [PubMed] [Google Scholar]
- 161.Avron M, Jagendorf AT. 1956. A TPNH diaphorase from chloroplasts. Arch Biochem Biophys 65:475–490. doi: 10.1016/0003-9861(56)90207-7. [DOI] [PubMed] [Google Scholar]
- 162.Shin M, Arnon DI. 1965. Enzymic mechanisms of pyridine nucleotide reduction in chloroplasts. J Biol Chem 240:1405–1411. doi: 10.1016/S0021-9258(18)97591-5. [DOI] [PubMed] [Google Scholar]
- 163.Carrillo N, Ceccarelli EA. 2003. Open questions in ferredoxin-NADP+ reductase catalytic mechanism. Eur J Biochem 270:1900–1915. doi: 10.1046/j.1432-1033.2003.03566.x. [DOI] [PubMed] [Google Scholar]
- 164.Aliverti A, Pandini V, Pennati A, de Rosa M, Zanetti G. 2008. Structural and functional diversity of ferredoxin-NADP+ reductases. Arch Biochem Biophys 474:283–291. doi: 10.1016/j.abb.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 165.Mulo P, Medina M. 2017. Interaction and electron transfer between ferredoxin–NADP+ oxidoreductase and its partners: structural, functional, and physiological implications. Photosynth Res 134:265–280. doi: 10.1007/s11120-017-0372-0. [DOI] [PubMed] [Google Scholar]
- 166.Fay P. 1992. Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol Rev 56:340–373. doi: 10.1128/mr.56.2.340-373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kumar K, Mella-Herrera RA, Golden JW. 2010. Cyanobacterial heterocysts. Cold Spring Harb Perspect Biol 2:1–19. doi: 10.1101/cshperspect.a000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Thomas J-C, Ughy B, Lagoutte B, Ajlani G. 2006. A second isoform of the ferredoxin:NADP oxidoreductase generated by an in-frame initiation of translation. Proc Natl Acad Sci USA 103:18368–18373. doi: 10.1073/pnas.0607718103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Valladares A, Muro-Pastor AM, Fillat MF, Herrero A, Flores E. 1999. Constitutive and nitrogen-regulated promoters of the petH gene encoding ferredoxin:NADP+ reductase in the heterocyst-forming cyanobacterium Anabaena sp. FEBS Lett 449:159–164. doi: 10.1016/s0014-5793(99)00404-4. [DOI] [PubMed] [Google Scholar]
- 170.Korn A, Ajlani G, Lagoutte B, Gall A, Sétif P. 2009. Ferredoxin:NADP+ oxidoreductase association with phycocyanin modulates its properties. J Biol Chem 284:31789–31797. doi: 10.1074/jbc.M109.024638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Grzyb J, Gieczewska K, Łabuz J, Sztatelman O. 2018. Detailed characterization of Synechocystis PCC 6803 ferredoxin:NADP+ oxidoreductase interaction with model membranes. Biochim Biophys Acta Biomembr 1860:281–291. doi: 10.1016/j.bbamem.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 172.Liu H, Weisz DA, Zhang MM, Cheng M, Zhang B, Zhang H, Gerstenecker GS, Pakrasi HB, Gross ML, Blankenship RE. 2019. Phycobilisomes harbor FNRL in cyanobacteria. mBio 10:e00669-19. doi: 10.1128/mBio.00669-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Alcántara-Sánchez F, Leyva-Castillo LE, Chagolla-López A, González de la Vara L, Gómez-Lojero C. 2017. Distribution of isoforms of ferredoxin-NADP+ reductase (FNR) in cyanobacteria in two growth conditions. Int J Biochem Cell Biol 85:123–134. doi: 10.1016/j.biocel.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 174.Apte S, Rowell P, Duncan W, Stewart P. 1978. Electron donation to ferredoxin in heterocysts of the N2-fixing alga Anabaena cylindrica. Proc Royal Soc B P ROY SOC B-BIOL SCI 200:1–25. doi: 10.1098/rspb.1978.0001. [DOI] [Google Scholar]
- 175.Schrautemeier B, Böhme H. 1984. Different functions assigned to NAD(H) and NADP(H) in light-dependent nitrogen fixation by heterocysts of Anabaena variabilis. FEMS Microbiol Lett 25:215–218. doi: 10.1111/j.1574-6968.1984.tb01459.x. [DOI] [Google Scholar]
- 176.Hurley JK, Weber-Main AM, Stankovich MT, Benning MM, Thoden JB, Vanhooke JL, Holden HM, Chae YK, Xia B, Cheng H, Markley JL, Martinez-Júlvez M, Gómez-Moreno C, Schmeits JL, Tollin G. 1997. Structure−function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants. Biochemistry 36:11100–11117. doi: 10.1021/bi9709001. [DOI] [PubMed] [Google Scholar]
- 177.Masepohl B, Schölisch K, Görlitz K, Kutzki C, Bohme H. 1997. The heterocyst-specific fdxH gene product of the cyanobacterium Anabaena sp. PCC 7120 is important but not essential for nitrogen fixation. Mol Gen Genet 253:770–776. doi: 10.1007/s004380050383. [DOI] [PubMed] [Google Scholar]
- 178.Thomas J. 1970. Absence of the pigments of Photosystem II of photosynthesis in heterocysts of a blue–green alga. Nature 228:181–183. doi: 10.1038/228181b0. [DOI] [PubMed] [Google Scholar]
- 179.Madan AP, Nierzwicki-Bauer SA. 1993. In situ detection of transcripts for ribulose-1,5-bisphosphate carboxylase in cyanobacterial heterocysts. J Bacteriol 175:7301–7306. doi: 10.1128/jb.175.22.7301-7306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Valladares A, Maldener I, Muro-Pastor AM, Flores E, Herrero A. 2007. Heterocyst development and diazotrophic metabolism in terminal respiratory oxidase mutants of the cyanobacterium Anabaena sp. Strain PCC 7120. J Bacteriol 189:4425–4430. doi: 10.1128/JB.00220-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Martín-Figueroa E, Navarro F, Florencio FJ. 2000. The GS-GOGAT pathway is not operative in the heterocysts. Cloning and expression of glsF gene from the cyanobacterium Anabaena sp. PCC 7120. FEBS Lett 476:282–286. doi: 10.1016/s0014-5793(00)01722-1. [DOI] [PubMed] [Google Scholar]
- 182.Valladares A, Herrero A, Pils D, Schmetterer G, Flores E. 2003. Cytochrome c oxidase genes required for nitrogenase activity and diazotrophic growth in Anabaena sp. PCC 7120. Mol Microbiol 47:1239–1249. doi: 10.1046/j.1365-2958.2003.03372.x. [DOI] [PubMed] [Google Scholar]
- 183.Wolk CP, Ernst A, Elhai J. 1994. Heterocyst metabolism and development, p 769–823. In Bryant DA (ed), The molecular biology of cyanobacteria. Springer; Netherlands, Dordrecht. [Google Scholar]
- 184.Winkenbach F, Wolk CP. 1973. Activities of enzymes of the oxidative and the reductive pentose phosphate pathways in heterocysts of a blue-green alga. Plant Physiol 52:480–483. doi: 10.1104/pp.52.5.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lockau W, Peterson RB, Wolk CP, Burris RH. 1978. Modes of reduction of nitrogen in heterocysts isolated from Anabaena species. Biochim Biophys Acta 502:298–308. doi: 10.1016/0005-2728(78)90051-8. [DOI] [PubMed] [Google Scholar]
- 186.Fay P. 1976. Factors influencing dark nitrogen fixation in a blue-green alga. Appl Environ Microbiol 31:376–379. doi: 10.1128/aem.31.3.376-379.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Schrautemeier B, BöHme H, BöGer P. 1984. In vitro studies on pathways and regulation of electron transport to nitrogenase with a cell-free extract from heterocysts of Anabaena variabilis. Arch Microbiol 137:14–20. doi: 10.1007/BF00425801. [DOI] [Google Scholar]
- 188.Battchikova N, Eisenhut M, Aro EM. 2011. Cyanobacterial NDH-1 complexes: novel insights and remaining puzzles. Biochim Biophys Acta 1807:935–944. doi: 10.1016/j.bbabio.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 189.Berger S, Ellersiek U, Steinmüller K. 1991. Cyanobacteria contain a mitochondrial complex I-homologous NADH-dehydrogenase. FEBS Lett 286:129–132. doi: 10.1016/0014-5793(91)80957-5. [DOI] [PubMed] [Google Scholar]
- 190.Christman HD, Campbell EL, Meeks JC. 2011. Global transcription profiles of the nitrogen stress response resulting in heterocyst or hormogonium development in Nostoc punctiforme. J Bacteriol 193:6874–6886. doi: 10.1128/JB.05999-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sandh G, Ramström M, Stensjö K. 2014. Analysis of the early heterocyst Cys-proteome in the multicellular cyanobacterium Nostoc punctiforme reveals novel insights into the division of labor within diazotrophic filaments. BMC Genomics 15:1–16. doi: 10.1186/1471-2164-15-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhang C, Shuai J, Ran Z, Zhao J, Wu Z, Liao R, Wu J, Ma W, Lei M. 2020. Structural insights into NDH-1 mediated cyclic electron transfer. Nat Commun 11:1–13. doi: 10.1038/s41467-020-14732-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Peltier G, Aro E-M, Shikanai T. 2016. NDH-1 and NDH-2 plastoquinone reductases in oxygenic photosynthesis. Annu Rev Plant Biol 67:55–80. doi: 10.1146/annurev-arplant-043014-114752. [DOI] [PubMed] [Google Scholar]
- 194.He Z, Zheng F, Wu Y, Li Q, Lv J, Fu P, Mi H. 2015. NDH-1L interacts with ferredoxin via the subunit NdhS in Thermosynechococcus elongatus. Photosynth Res 126:341–349. doi: 10.1007/s11120-015-0090-4. [DOI] [PubMed] [Google Scholar]
- 195.Ermakova M, Battchikova N, Richaud P, Leino H, Kosourov S, Isojärvi J, Peltier G, Flores E, Cournac L, Allahverdiyeva Y, Aro E-M. 2014. Heterocyst-specific flavodiiron protein Flv3B enables oxic diazotrophic growth of the filamentous cyanobacterium Anabaena sp. PCC 7120. Proc Natl Acad Sci USA 111:11205–11210. doi: 10.1073/pnas.1407327111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Torrado A, Ramírez-Moncayo C, Navarro JA, Mariscal V, Molina-Heredia FP. 2019. Cytochrome c6 is the main respiratory and photosynthetic soluble electron donor in heterocysts of the cyanobacterium Anabaena sp. PCC 7120. Biochim Biophys Acta Bioenerg 1860:60–68. doi: 10.1016/j.bbabio.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 197.Khanna N, Lindblad P. 2015. Cyanobacterial hydrogenases and hydrogen metabolism revisited: recent progress and future prospects. Int J Mol Sci 16:10537–10561. doi: 10.3390/ijms160510537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Magnuson A. 2019. Heterocyst thylakoid bioenergetics. Life 9:1–13. doi: 10.3390/life9010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Burén S, Rubio LM. 2018. State of the art in eukaryotic nitrogenase engineering. FEMS Microbiol Lett 365:1–9. doi: 10.1093/femsle/fnx274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ryu M-H, Zhang J, Toth T, Khokhani D, Geddes BA, Mus F, Garcia-Costas A, Peters JW, Poole PS, Ané J-M, Voigt CA. 2020. Control of nitrogen fixation in bacteria that associate with cereals. Nat Microbiol 5:314–330. doi: 10.1038/s41564-019-0631-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Mus F, Crook MB, Garcia K, Garcia Costas A, Geddes BA, Kouri E-DD, Paramasivan P, Ryu M-H, Oldroyd GED, Poole PS, Udvardi MK, Voigt CA, Ané J-M, Peters JW. 2016. Symbiotic nitrogen fixation and challenges to extending it to non-legumes. Appl Environ Microbiol 82:3698–3710. doi: 10.1128/AEM.01055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Burén S, Young EM, Sweeny EA, Lopez-Torrejón G, Veldhuizen M, Voigt CA, Rubio LM. 2017. Formation of nitrogenase NifDK tetramers in the mitochondria of Saccharomyces cerevisiae. ACS Synth Biol 6:1043–1055. doi: 10.1021/acssynbio.6b00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Burén S, Jiang X, López-Torrejón G, Echavarri-Erasun C, Rubio LM. 2017. Purification and in vitro activity of mitochondria targeted nitrogenase cofactor maturase NifB. Front Plant Sci 8:1–16. doi: 10.3389/fpls.2017.01567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Okada S, Gregg CM, Allen RS, Menon A, Hussain D, Gillespie V, Johnston E, Byrne K, Colgrave ML, Wood CC. 2020. A synthetic biology workflow reveals variation in processing and solubility of nitrogenase proteins targeted to plant mitochondria, and differing tolerance of targeting sequences in a bacterial nitrogenase assay. Front Plant Sci 11:1–12. doi: 10.3389/fpls.2020.552160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Liu D, Liberton M, Yu J, Pakrasi HB, Bhattacharyya-Pakrasi M. 2018. Engineering nitrogen fixation activity in an oygenic phototroph. mBio 9:e01029-18. doi: 10.1128/mBio.01029-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Yang J, Xie X, Yang M, Dixon R, Wang Y-P. 2017. Modular electron-transport chains from eukaryotic organelles function to support nitrogenase activity. Proc Natl Acad Sci USA 114:E2460–E2465. doi: 10.1073/pnas.1620058114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chen K, Tilley GJ, Sridhar V, Prasad GS, Stout CD, Armstrong FA, Burgess BK. 1999. Alteration of the reduction potential of the [4Fe-4S]2+/+ cluster of Azotobacter vinelandii ferredoxin I. J Biol Chem 274:36479–36487. doi: 10.1074/jbc.274.51.36479. [DOI] [PubMed] [Google Scholar]
- 208.Cho YS, Wang QJ, Krogmann D, Whitmarsh J. Extinction coe¤cients and midpoint potentials of cytochrome c6 from the cyanobacteria Arthrospira maxima, Microcystis aeruginosa, and Synechocystis 6803. Biochimica et Biophysica Acta 6:92–97. doi: 10.1016/S0005-2736(99)00124-8. [DOI] [PubMed] [Google Scholar]