ABSTRACT
The 16S rRNA gene has been extensively used as a molecular marker to explore evolutionary relationships and profile microbial composition throughout various environments. Despite its convenience and prevalence, limitations are inevitable. Variable copy numbers, intragenomic heterogeneity, and low taxonomic resolution have caused biases in estimating microbial diversity. Here, analysis of 24,248 complete prokaryotic genomes indicated that the 16S rRNA gene copy number ranged from 1 to 37 in bacteria and 1 to 5 in archaea, and intragenomic heterogeneity was observed in 60% of prokaryotic genomes, most of which were below 1%. The overestimation of microbial diversity caused by intragenomic variation and the underestimation introduced by interspecific conservation were calculated when using full-length or partial 16S rRNA genes. Results showed that, at the 100% threshold, microbial diversity could be overestimated by as much as 156.5% when using the full-length gene. The V4 to V5 region-based analyses introduced the lowest overestimation rate (4.4%) but exhibited slightly lower species resolution than other variable regions under the 97% threshold. For different variable regions, appropriate thresholds rather than the canonical value 97% were proposed for minimizing the risk of splitting a single genome into multiple clusters and lumping together different species into the same cluster. This study has not only updated the 16S rRNA gene copy number and intragenomic variation information for the currently available prokaryotic genomes, but also elucidated the biases in estimating prokaryotic diversity with quantitative data, providing references for choosing amplified regions and clustering thresholds in microbial community surveys.
IMPORTANCE Microbial diversity is typically analyzed using marker gene-based methods, of which 16S rRNA gene sequencing is the most widely used approach. However, obtaining an accurate estimation of microbial diversity remains a challenge, due to the intragenomic variation and low taxonomic resolution of 16S rRNA genes. Comprehensive examination of the bias in estimating such prokaryotic diversity using 16S rRNA genes within ever-increasing prokaryotic genomes highlights the importance of the choice of sequencing regions and clustering thresholds based on the specific research objectives.
KEYWORDS: 16S rRNA gene, interspecific conservation, intragenomic heterogeneity, microbial diversity
INTRODUCTION
The 16S rRNA gene, with a length of ~1,500 bp, includes nine hypervariable regions (V1 to V9) interspersed by highly conserved sequences (1). The conserved regions can be used for binding universal primers targeting 16S rRNA genes for a wide range of prokaryotes, while the variable regions are taxon-specific and are commonly used to distinguish between microbial taxa (2). Next-generation sequencing (NGS) of marker genes, notably the 16S rRNA gene, has provided the means to profile microbial composition in different environmental samples, ranging from human body sites (e.g., gut, oral cavity) (3, 4) to various natural habitats, including soil, ocean, and glacier (5–7). One of the most popular and widely used NGS platforms is Illumina MiSeq (8), which is capable of generating, at most, 300-bp paired-end reads. Consequently, only partial fragments, rather than the complete 16S rRNA genes, were amplified and sequenced in the majority of microbial ecology studies, which generally covered single to three variable regions, such as the V4 (used in the Earth Microbiome Project) (9), V3-V4, and V1-V3 regions.
Despite the amplicon analysis based on 16S rRNA gene becoming the principal methodology in surveys of microbial communities, there are inevitable with limitations and challenges (10, 11). Estimation of the microbial diversity of any specific environment via amplicon analysis could be biased due to errors introduced by PCR and sequencing, intragenomic divergence, and insufficient intergenomic variation. It is now generally accepted that most individual bacterial genomes harbor multiple 16S rRNA genes, either identical or frequently distinct, thus resulting in the intragenomic heterogeneity of prokaryotes’ 16S rRNA genes (12–15). Consequently, taxonomic misclassification and overestimation of microbial diversity are likely to occur when conducting bioinformatic analysis. Throughout the 16S rRNA gene, the V4 to V5 region was proposed as the optimal region for 16S rRNA gene-based microbial analyses due to its least intragenomic variation (12). This result was drawn via the analysis of 2,013 complete genomes available during the study, which is remarkably fewer than the amount that can be retrieved nowadays, thus calling into question the tenability of this conclusion. In addition, highly similar and even identical rRNA gene sequences exist in different species, and thus they are likely to be classified into the same cluster based on the commonly used 97% identity threshold regardless of their differentia, resulting in the underestimation of the extant prokaryotic diversity (16). Different subregions of rRNA genes could introduce various degrees of bias in the estimation of community diversity, and this attaches vital significance to assessing the subregions of the 16S rRNA gene for selecting appropriate variable regions and minimizing the risk of misestimating microbial diversity.
Sequence clustering is most commonly used in data analysis after 16S rRNA gene partial amplicon reads are generated by next-generation sequencing, which assigns reads to specific sets of operational taxonomic units (OTUs). The process can be performed based on a given identity threshold (e.g., 97%) using algorithms such as OptiClust, nearest neighbor, or furthest neighbor. With the rapid development of whole-genome sequencing technology and the rapidly growing number of small subunit rRNA genes available, higher thresholds for full-length 16S rRNA gene have been recommended as the boundary for species delineation, such as 98.5% (17), 98.65% (18), and 98.7 to 99.0% (19), revealing more stringent standards than the canonical value, 97% (20). These studies generally focused on the full-length 16S rRNA genes, but appropriate identity thresholds for different variable regions, largely used in ecological studies, have not been thoroughly investigated. In addition to the aforementioned clustering methods, several denoising methods are increasingly prevalent in processing amplicon reads. A denoising algorithm infers sample sequences exactly by correcting amplicon errors, and each unique sequence obtained is defined as an amplicon sequence variant (ASV), approximately equal to 100% OTU (the identity threshold 100% is used). ASV offers higher taxonomic resolution but increases the possibility of splitting single strain into multiple clusters. For example, Escherichia coli K-12 MG1655 harbors seven copies of the 16S rRNA gene that can be divided into five types based on sequence differences, and each type represents an ASV. However, quantitative information about the risk of diversity-biased estimation by applying ASVs for different variable regions remains limited. This study aims to quantify the risk of splitting a single genome into multiple clusters and lumping together different species into the same cluster when using different identity thresholds (ASV or OTU).
In this study, we analyzed the 16S rRNA gene copy number and the intragenomic variation among divergent copies in 24,248 prokaryotic genomes (as of November 2021) retrieved from the RefSeq database. The bias in estimating microbial diversity introduced by intragenomic heterogeneity or interspecific conservation was quantified under a range of identity thresholds using full-length or subregions of the 16S rRNA gene. In addition, different clustering thresholds for 11 amplified regions were evaluated based on three metrics: oversplitting, overmerging, and total error rate. Results indicate that the clustering threshold can be adjusted according to the selected amplified regions in specific circumstances for minimizing the risk of splitting one strain into multiple clusters or merging distinct species into the same cluster.
RESULTS
In this study, 24,248 complete genomes (399 archaea, 23,849 bacteria) belonging to 6,889 unique species were obtained from the National Center for Biotechnology Information (NCBI) complete genome database in November 2021 (see detailed information for genomes in File S1 in the supplemental material). The 16S rRNA gene sequences were successfully retrieved from each genome. These 6,889 species were from 46 different phyla (Table 1), of which the Proteobacteria, with 3,198 unique species, were the most abundant, followed by Actinobacteria (1,172 species), Firmicutes (1,039 species), Bacteroidetes (518 species), Euryarchaeota (217 species), Cyanobacteria (159 species), and Tenericutes (137 species). The remaining 39 phyla were represented by fewer than 100 species per phylum. When performing Genome Taxonomy Database (GTDB) classification, 24,037 of the 24,248 genomes were taxonomically identified to the species level (6,265 GTDB species), and the remaining genomes were not classified into specific species. Because both NCBI taxonomy and GTDB taxonomy were used, taxonomic names mentioned in this study refer to the NCBI taxonomy unless otherwise specified.
TABLE 1.
Overview of archaeal and bacterial genomes at the NCBI phylum level
| Superkingdom | Phylum | No. of genera | No. of species | No. of genomes | Avg 16S rRNA gene copy (mean ± SD) |
|---|---|---|---|---|---|
| Archaea | “Candidatus Korarchaeota” | 1 | 1 | 1 | 1 |
| “Candidatus Lokiarchaeota” | 1 | 1 | 1 | 1 | |
| “Candidatus Micrarchaeota” | 2 | 2 | 2 | 1 | |
| “Candidatus Nanohaloarchaeota” | 1 | 1 | 1 | 1 | |
| “Candidatus Thermoplasmatota” | 8 | 10 | 14 | 1 | |
| Crenarchaeota | 26 | 56 | 92 | 1 | |
| Euryarchaeota | 72 | 217 | 263 | 2.0 ± 0.9 | |
| Thaumarchaeota | 9 | 25 | 25 | 1.2 ± 0.5 | |
| Bacteria | Unclassified | 3 | 4 | 4 | 1 |
| Acidobacteria | 10 | 20 | 26 | 1.1 ± 0.3 | |
| Actinobacteria | 200 | 1,172 | 2,372 | 3.2 ± 1.9 | |
| Aquificae | 9 | 13 | 14 | 1.7 ± 0.6 | |
| Armatimonadetes | 1 | 1 | 1 | 2 | |
| Atribacterota | 1 | 1 | 1 | 2 | |
| Bacteroidetes | 145 | 518 | 879 | 4.1 ± 2.3 | |
| Balneolaeota | 1 | 1 | 1 | 3 | |
| Caldiserica | 1 | 1 | 1 | 1 | |
| Calditrichaeota | 1 | 1 | 1 | 1 | |
| “Candidatus Bipolaricaulota” | 1 | 1 | 1 | 1 | |
| “Candidatus Cloacimonetes” | 1 | 1 | 1 | 2 | |
| “Candidatus Omnitrophica” | 1 | 1 | 1 | 1 | |
| “Candidatus Saccharibacteria” | 2 | 2 | 2 | 1 | |
| Chlamydiae | 8 | 25 | 188 | 1.7 ± 0.5 | |
| Chlorobi | 5 | 13 | 13 | 2.0 ± 0.6 | |
| Chloroflexi | 14 | 20 | 42 | 1.4 ± 0.7 | |
| Chrysiogenetes | 1 | 1 | 2 | 3 | |
| Coprothermobacterota | 1 | 1 | 1 | 2 | |
| Cyanobacteria | 50 | 159 | 188 | 2.5 ± 1.1 | |
| Deferribacteres | 5 | 5 | 5 | 2 | |
| Deinococcus-Thermus | 6 | 32 | 61 | 2.6 ± 0.8 | |
| Dictyoglomi | 1 | 2 | 2 | 2 | |
| Elusimicrobia | 2 | 3 | 4 | 1 | |
| Fibrobacteres | 1 | 1 | 2 | 3 | |
| Firmicutes | 257 | 1,039 | 5,266 | 6.9 ± 2.8 | |
| Fusobacteria | 7 | 27 | 76 | 5.2 ± 1.1 | |
| Gemmatimonadetes | 2 | 4 | 4 | 1.8 ± 0.5 | |
| Ignavibacteriae | 2 | 2 | 2 | 1 | |
| Kiritimatiellaeota | 1 | 1 | 1 | 1 | |
| Nitrospirae | 4 | 9 | 10 | 1.6 ± 0.7 | |
| Planctomycetes | 40 | 49 | 55 | 2.1 ± 1.3 | |
| Proteobacteria | 706 | 3,198 | 13,871 | 5.5 ± 2.5 | |
| Spirochaetes | 13 | 59 | 160 | 1.7 ± 0.8 | |
| Synergistetes | 5 | 5 | 5 | 3.2 ± 1.1 | |
| Tenericutes | 10 | 137 | 426 | 1.7 ± 0.7 | |
| Thermodesulfobacteria | 5 | 7 | 7 | 1.1 ± 0.4 | |
| Thermotogae | 11 | 32 | 41 | 1.9 ± 1.2 | |
| Verrucomicrobia | 12 | 18 | 112 | 2.8 ± 0.6 |
16S rRNA gene copy number in archaea and bacteria.
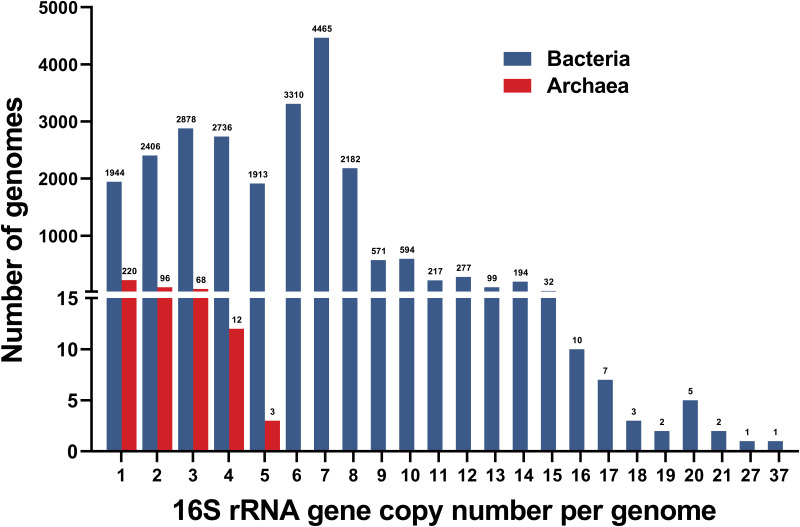
The 16S rRNA gene copy number ranged from 1 to 5 in archaea and 1 to 37 in bacteria (Fig. 1). The maximum copy number of 37 was observed in Tumebacillus avium AR23208, which was considerably larger than the previously published value (15 copies) obtained using several smaller databases (12, 13, 15). Only 8% of the bacterial genomes contained a single 16S rRNA gene, while more than half of the archaeal genomes contained one copy. Bacteria with seven copies were the most abundant (4,465 genomes), and copy numbers greater than 15 were relatively rare, with a total of 31 genomes. The average copy number was 5.3 ± 2.8 for bacteria, which is apparently higher than that for archaea (1.7 ± 0.9). In total, 127,003 copies of the 16S rRNA gene were identified, with an average of 5.2 copies per prokaryotic genome.
FIG 1.
Distribution of 16S rRNA gene copy numbers within bacterial and archaeal genomes. Values above columns represent the numbers of genomes containing the corresponding copies of 16S rRNA genes.
The copy number of 16S rRNA genes within individual genomes was taxon specific at several taxonomic levels. The mean copy number per phylum ranged between 1 and 6.9 ± 2.8 (Table 1, Fig. S1). Among bacterial phyla, the Firmicutes had an average copy number of 6.9 ± 2.8, which was the highest of all phyla, followed by the Proteobacteria (5.5 ± 2.5) and Fusobacteria (5.2 ± 1.1). Low mean copy numbers were observed in the Acidobacteria (1.1 ± 0.3), Thermodesulfobacteria (1.1 ± 0.4), and Chloroflexi (1.4 ± 0.7). For archaeal phyla, the average copy number of 16S rRNA genes was 2.0 ± 0.9 in Euryarchaeota and 1.2 ± 0.5 in Thaumarchaeota, while the rest of the archaeal phyla contained only one copy of the 16S rRNA gene on average.
Intragenomic heterogeneity of 16S rRNA genes in prokaryotic genomes.
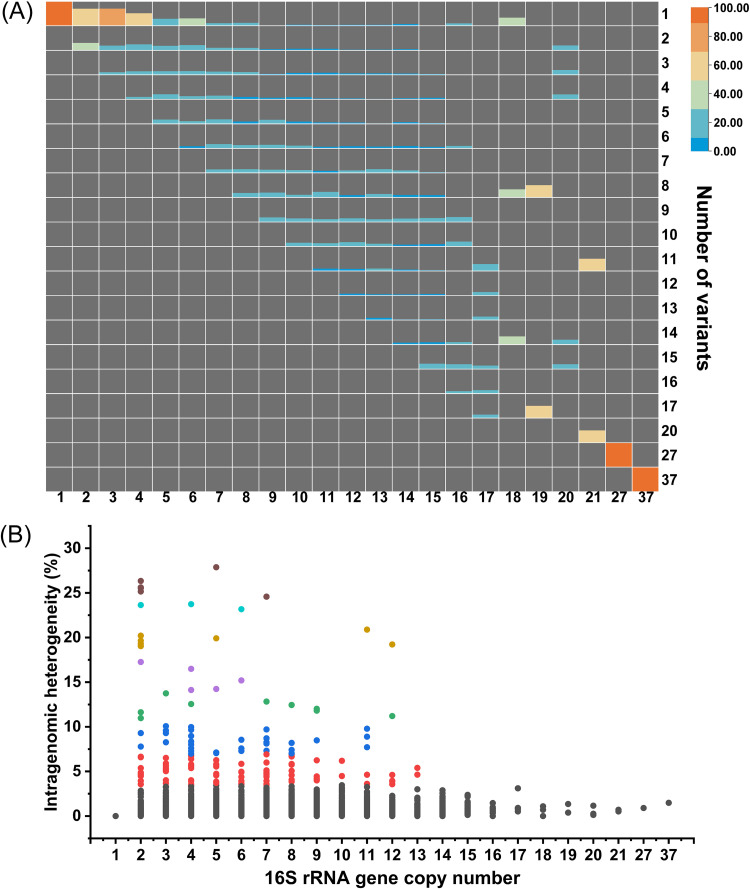
As described above, many prokaryotic genomes harbored multiple 16S rRNA genes, and we further investigated whether different copies are identical and the extent of possible differences. The number of 16S rRNA gene sequence variants showed an upward trend as the copy number within individual genomes increased (Fig. 2A). On average, there were three 16S rRNA gene variants in a genome. The intragenomic variation of 16S rRNA gene sequences was observed in about 60% of genomes (14,502 out of 24,248 genomes) (Fig. S2), in which the number of variants ranged between 2 and 37. The detailed information about intragenomic variation detected within each genome is available in File S1. The majority of the heterogeneity detected was below 1% in DNA sequences (12,642 out of 14,502 genomes), but the remaining 1,860 genomes had intragenomic heterogeneity greater than 1%, which may result in false taxonomic classification using 16S rRNA-based methods. There were as many as 37 variants in Tumebacillus avium AR23208, and the distances of variants varied from 0.06% to 1.49%. The greatest heterogeneity calculated was found in Listeria monocytogenes 10-092876-1155 LM6 (27.9%) (Fig. 2B), which contained 5 distinct copies of the 16S rRNA gene (Fig. S3). It is also noteworthy that although archaea generally contain fewer copies of the 16S rRNA gene, the intragenomic divergence of the 16S gene existed in 119 out of 179 archaeal strains with multiple copies. Moreover, we also found that all strains in some species with two or more sequenced genomes exhibited high intragenomic variation, as shown in Table S1, including pathogenic bacteria Borreliella afzelii and Streptococcus iniae and extremophiles Thermobispora bispora, Haloarcula marismortui, Haloarcula hispanica, and Halomicrobium mukohataei. The stable presence of distinct rRNA variants in these species suggested that variants may have specific physiological functions.
FIG 2.
The numbers of variants and sequence differences between variants in prokaryotic genomes with variable 16S rRNA gene copy numbers. (A) The percentile distribution of sequence variant counts in prokaryotic genomes with different 16S rRNA gene copy numbers. The colored areas in each grid indicate the percentage of the number of genomes containing corresponding 16S gene variant counts (vertical coordinate) in all genomes harboring corresponding 16S gene copy numbers (horizontal coordinate). The background color of the grid is gray, indicating that the proportion is 0. (B) Intragenomic heterogeneity in prokaryotic genomes with different 16S rRNA gene copy numbers; each dot represents a genome.
Bias in estimating microbial diversity using different hypervariable regions.
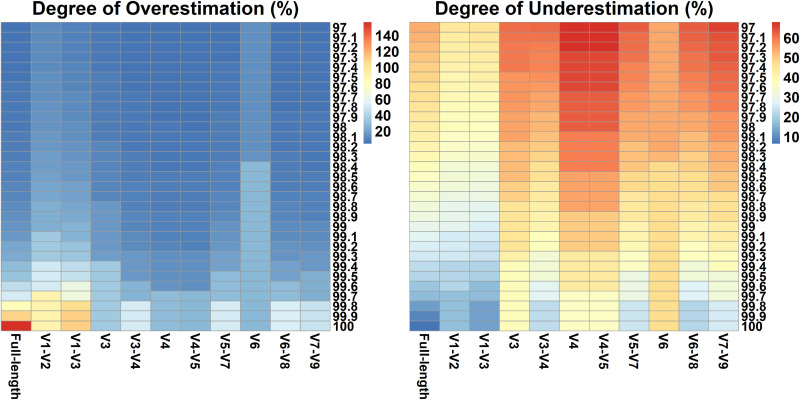
Given the intragenomic divergence of 16S RNA genes, multiple variants were likely to be classified into different sequence clusters, resulting in an overestimation of microbial diversity when using 16S rRNA gene-based approaches. The overestimation rates were drawn in a previous study via the analysis of 2,013 complete genomes (12), yet the number of sequenced genomes in the RefSeq database has already reached over 20,000, and an updated analysis seems necessary. Therefore, the degree of overestimation was quantified using currently available larger data sets, and the results of this analysis are shown in Table 2. At the ASV level, i.e., identity threshold 100%, an overestimation of 156.5% was measured when using full-length 16S rRNA genes, which was significantly higher than values (27.3% to 110.5%) obtained when focusing on partial regions. Intragenomic variation of the V6 region led to the lowest overestimation (27.3%), followed by the V4 to V5 (28.3%) and V4 regions (28.8%). At the commonly used threshold of 97%, the degree of overestimation was the lowest for the V4 to V5 (4.4%) and V7 to V9 (4.4%) regions, while it was highest for the V6 (14.0%) and V1 to V2 (11.7%) regions. Similar results were obtained when using other clustering thresholds between 97% and 100% (Fig. 3). It should be noted that, for full-length 16S genes, the intragenomic heterogeneity-caused overestimation of species richness was significantly decreased at the threshold level of 97% compared with 100%, indicating that a specific threshold should be chosen to avoid biased estimations of species diversity.
TABLE 2.
Degree of overestimation due to intragenomic heterogeneity and underestimation caused by insufficient interspecific variation for different 16S rRNA gene regions under the ASV and 97%-OTU levels
| Identity threshold | 16S gene region | HIQ-T-NCBIa |
HIQ-C-NCBIa |
Overestimation (%)b | Underestimation (%)b | ||
|---|---|---|---|---|---|---|---|
| No. of sequences | No. of OTUs | No. of sequences | No. of OTUs | ||||
| 100% | Full-length | 29,416 | 15,727 | 6,550 | 6,131 | 156.5 | 6.4 |
| V1–V2 | 29,459 | 10,287 | 6,562 | 5,433 | 89.3 | 17.2 | |
| V1–V3 | 28,883 | 11,623 | 6,339 | 5,523 | 110.5 | 12.9 | |
| V3 | 29,246 | 5,467 | 6,554 | 4,060 | 34.6 | 38.0 | |
| V3–V4 | 29,994 | 8,079 | 6,866 | 5,325 | 51.7 | 22.4 | |
| V4 | 29,903 | 5,593 | 6,829 | 4,341 | 28.8 | 36.4 | |
| V4–V5 | 30,020 | 5,636 | 6,890 | 4,392 | 28.3 | 36.2 | |
| V5–V7 | 29,058 | 7,333 | 6,402 | 4,823 | 52.0 | 24.7 | |
| V6 | 26,669 | 3,816 | 5,713 | 2,998 | 27.3 | 47.5 | |
| V6–V8 | 30,027 | 8,310 | 6,890 | 5,407 | 53.7 | 21.5 | |
| V7–V9 | 26,179 | 6,474 | 5,875 | 4,374 | 48.0 | 25.6 | |
| 97% | Full-length | 29,416 | 3,181 | 6,550 | 3,035 | 4.8 | 53.7 |
| V1–V2 | 29,459 | 4,074 | 6,562 | 3,647 | 11.7 | 44.4 | |
| V1–V3 | 28,883 | 3,788 | 6,339 | 3,478 | 8.9 | 45.1 | |
| V3 | 29,246 | 2,715 | 6,554 | 2,556 | 6.2 | 61.0 | |
| V3–V4 | 29,994 | 2,794 | 6,866 | 2,663 | 4.9 | 61.2 | |
| V4 | 29,903 | 2,284 | 6,829 | 2,186 | 4.5 | 68.0 | |
| V4–V5 | 30,020 | 2,314 | 6,890 | 2,217 | 4.4 | 67.8 | |
| V5–V7 | 29,058 | 2,511 | 6,402 | 2,384 | 5.3 | 62.8 | |
| V6 | 26,669 | 2,989 | 5,713 | 2,623 | 14.0 | 54.1 | |
| V6–V8 | 30,027 | 2,692 | 6,890 | 2,566 | 4.9 | 62.8 | |
| V7–V9 | 26,179 | 2,114 | 5,875 | 2,025 | 4.4 | 65.5 | |
HIQ-T-NCBI, the data set constructed considering intragenomic heterogeneity; HIQ-C-NCBI, the data set constructed ruling out intragenomic heterogeneity.
The overestimation rate was calculated as (A – B)/B · 100%, where A represents the number of OTUs from HIQ-T-NCBI and B represents the number of OTUs from HIQ-C-NCBI. The underestimation rate was calculated as (C – B)/C · 100%, where C is the number of sequences in HIQ-C-NCBI.
FIG 3.
The degree of the overestimation introduced by intragenomic heterogeneity (left panel) and the underestimation caused by the insufficient interspecific variation (right panel) for full-length and partial 16S rRNA genes (horizontal coordinates) at an identity threshold of 97% to 100% (vertical coordinates). The above-described overestimation and underestimation rates were calculated using data sets constructed based on NCBI taxonomy. As shown in the legends, the colors range from blue to red represent the over/underestimation rates ranging from low to high, respectively.
On the other hand, some closely related but distinct taxa may share the same or part of the same 16S rRNA gene, namely, insufficient interspecific variation, resulting in the underestimation of microbial diversity. The degree of underestimation was calculated by comparing the number of species and OTUs generated by clustering. For longer amplicons such as fragments targeting the entire gene or the V1 to V3 region, their diversities were generally underestimated by less than 15% under the 100% threshold, whereas the number of species was greatly underestimated by shorter amplicons, such as the V3, V4, and V6 regions (Table 2, Fig. 3). These results demonstrated that some closely related species were indistinguishable when using partial 16S genes due to the conservative part of variable regions in certain taxa.
Considering that GTDB taxonomy is a phylogenetically consistent and rank-normalized genome-based taxonomy (21), the GTDB taxonomic ranks of each genome were obtained using GTDB-Tk (v2) (22), and the 16S rRNA gene sequence sets were constructed to calculate the bias in estimating microbial diversity based on GTDB taxonomy. Apparently, similar results were obtained using GTDB data sets with respect to NCBI data sets (Fig. S4; the detailed data for the bias are shown in File S1). Intragenomic heterogeneity of the V4 to V5 region of the 16S rRNA gene resulted in the lowest overestimation (3.7% to 26.4%) of species diversity at the threshold of 97% to 100% (Fig. S4A). As indicated in Fig. S4B, the full-length amplicon yielded a diversity estimation which is close to the number of species at the identity threshold of 100%, with only 1.7% fewer OTUs. In contrast, the partial gene amplicons resulted in over 5% fewer OTUs. These results suggested that partial 16S rRNA genes from different GTDB species tended to be clustered together and thus provided limited implications for evolutionary relationship.
Intragenomic and intergenomic variation per base along the 16S rRNA gene.
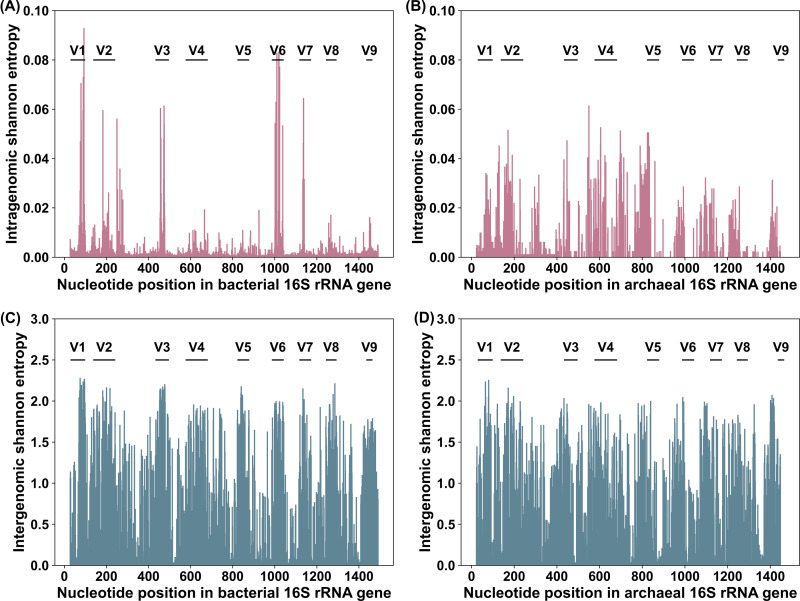
To further investigate potential nucleotide positions that may lead to the overestimation of species diversity, the average Shannon entropy of each base across the 16S rRNA gene was calculated for archaea and bacteria. The entropy is used to represent intragenomic variation per base position, and a high entropy value means that the nucleotide position contains rich information, also referred to as high heterogeneity. It is apparent from Fig. 4A that nucleotide positions with high Shannon entropy values (>0.04) tended to focus on specific regions, such as the V1, V3, and V6 regions, whereas the nucleotide sites in the V4, V5, V8, and V9 regions exhibited low Shannon entropy (<0.02), consistent with the observation of a smaller degree of overestimation introduced by targeting the V4 to V5 region. In contrast, no significant differences were found between the entropy values of different variable regions in archaea (Fig. 4B). This could be attributed to a smaller number of archaeal genomes available compared to those of bacteria, and another possible explanation for this is that archaea typically contain fewer 16S rRNA genes per genome. In terms of the intergenomic variation per nucleotide position along the 16S gene, high Shannon entropy was observed for nine hypervariable regions, as shown in Fig. 4C and D. The presence of sufficient intergenomic variation likely enables 16S gene fragment-based analyses to be employed for distinguishing different taxa at the genus and higher levels.
FIG 4.
Intragenomic and intergenomic variation across 16S rRNA genes for bacteria and archaea. (A) The mean intragenomic variation was calculated by averaging the Shannon entropy at each position of the 16S rRNA gene in all 23,899 bacterial genomes studied. Sequence coordinates refer to the 16S gene of Escherichia coli K-12 MG1655 (gene ID 948332). (B) The mean intragenomic variation per base across 16S rRNA genes in all 399 archaeal genomes studied. Sequence coordinates refer to the 16S gene of Saccharolobus solfataricus IFO 15331 (accession number NR_029127.1). (C and D) The intergenomic variation across 16S rRNA genes based on the alignment of all bacterial (C) and archaeal (D) sequences present in the SILVA Ref NR99 database.
Oversplitting and overmerging rates under different clustering thresholds.
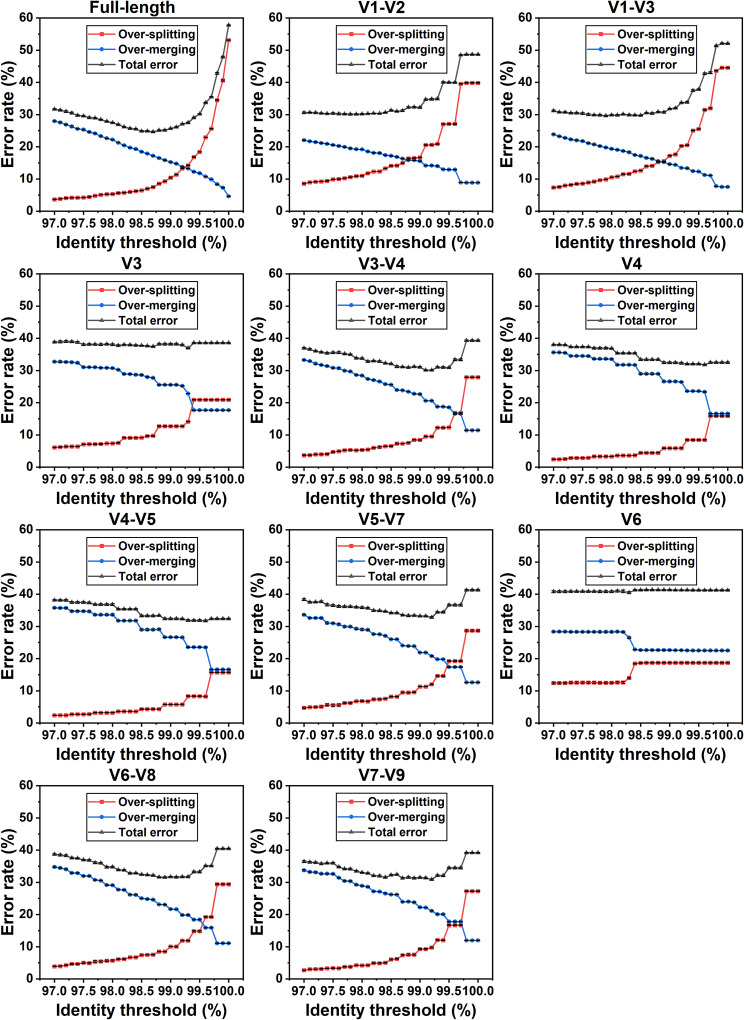
For the 11 amplified fragments, as shown in Fig. 5, the percentage of species assigned to multiple clusters (oversplitting), the percentage of OTUs (ASVs) containing sequences from different species (overmerging), and the sum of these two values (total error) were calculated under serial thresholds. In general, the oversplitting rate rose as the identity thresholds increased, reaching 53% when considering the full-length rRNA gene with a threshold of 100%. When using thresholds of <99%, the oversplitting rates were below 20% (Fig. 5), indicating that lower thresholds could be used to reduce the risk of splitting multiple rRNA gene variants within the same species into different OTUs.
FIG 5.
Oversplitting, overmerging, and total error rate for full-length or partial 16S genes at clustering thresholds of 97% to 100%. The three metrics were calculated using data sets constructed based on NCBI taxonomy. The oversplitting rate was defined as the ratio of species which were assigned into multiple clusters (OTUs or ASVs). The overmerging rate was calculated as the ratio of clusters containing sequences from distinct species. The total error rate represented the sum of the two values above.
In contrast to oversplitting, the overmerging rate showed a significant decline as the clustering threshold increased. For longer amplified fragments containing at least three variable regions, such as full-length regions or the V7 to V9 region, as few as ~5% of the overmerging rates were shown at high thresholds. However, for shorter fragments comprising one or two adjacent variable regions, the percentage tended to be higher, for example, up to 22% for the V6 region. This suggested that sequences from different species were more likely to be combined into the same cluster when using shorter amplicons.
As shown in Fig. 5, there are three major changing trends for the total error rates with the thresholds ranging from 97% to 100%. One is a clear trend of falling followed by rising with increasing identity thresholds, such as full-length regions and the V3 to V4 region, and the minimum can be found at specific thresholds: 98.5% to 99.0% for the full-length 16S rRNA gene, 97.5% to 98.0% for the V1 to V2 region, 98.0% to 98.5% for the V1 to V3 region, 99.0% to 99.5% for the V3 to V4 region, 98.8% to 99.3% for the V6 to V8 region, and 98.7% to 99.2% for the V5 to V7/V7 to V9 regions. Another pattern is displayed in the V3 and V6 regions; the total error remained nearly constant at different thresholds from 97% to 100%. The third trend is as shown in the V4 and V4 to V5 regions; the total error declines monotonically with increasing identity thresholds, and the clustering thresholds from 99% to 100% were recommended for the two variable regions. Furthermore, the optimal identity thresholds obtained using data sets constructed based on GTDB taxonomy (Fig. S5), i.e., the thresholds corresponding to the minimum total error rates, were consistent with those based on NCBI taxonomy.
DISCUSSION
The copy number of 16S genes per genome ranged from 1 to 4 in archaea and 1 to 15 in bacteria (12, 13); these results were obtained a decade ago, so an updated analysis is warranted, as the database has been amplified by almost 10-fold. The present analysis revealed that, of the 24,248 prokaryotic genomes now available, the 16S rRNA gene copy number of 31 genomes falls outside the boundaries reported previously (up to 15 copies), and 3 of the 399 archaeal genomes fall outside the previous reported analysis (up to 4 copies) (Fig. 1). Variable copies of the 16S gene within different strains have substantially affected the estimation of the relative abundance per taxon (23). For example, a low-abundance species with high copy number of 16S genes was likely to be observed more frequently than a high-abundance species with a low copy number, which prompted the development of tools attempting to overcome such biases, including rrnDB (24), CopyRighter (25), and PICRUSt (26). There has been an ongoing debate about whether relative abundance should be corrected via the mean 16S rRNA gene copy number (GCN). Even though an improvement in the prediction of microbial community profiles by GCN was reported in the literature (27), a recent study showed that 16S GCN failed to more reliably uncover the community structure in microbial ecology studies (28), which might be limited by inadequate records of copy numbers in the reference database. In this study, we have provided a more comprehensive catalogue of copy numbers at the strain level or other taxonomic levels, which could be taken as a reference for 16S GCN to allow more accurate estimation of microbial community structure.
Consistent with previous studies, the intragenomic variation of 16S rRNA genes was widely observed in bacteria and archaea; 1,860 strains of these possessed multiple copies of 16S rRNA genes that differed by more than 1%. Additionally, it is worth noting that some species in which all strains exhibited high intragenomic heterogeneity (>1%) (Table S1), including several bacterial species such as Thermobispora bispora, Streptococcus iniae, and Vibrio natriegens, as well as a few previously reported halophilic archaea such as Haloarcula hispanica, Haloarcula marismortui, and Halomicrobium mukohataei (29). A possible explanation for the presence of intragenomic heterogeneity was horizontal gene transfer (HGT) of the 16S rRNA gene, which has been supported by several studies. Yap et al. provided evidence that the rrnB operon of Thermomonospora chromogena was acquired from Thermobispora bispora or other closely related species via HGT (30). Divergence of rRNA genes within a single strain has been proposed as an adaptation to the changing natural environment, with distinct variants being functional under different environmental conditions. For example, Haloarcula marismortui harbored three rRNA operons, of which operon B was markedly different from the other two operons in nucleotide sequence and GC content, displaying a higher expression level at high temperatures (e.g., 50°C) than at low temperatures (e.g., 15°C) (29). The similar phenomenon that different rRNA genes exhibited temperature-dependent expression also took place in other halophilic archaea, such as Haloarcula hispanica, Haloarcula japonica, and Haloarcula amylolytica (31, 32). In addition, divergent rRNA variants regulated gene expression and cell phenotype at the ribosome level (33, 34). In Vibrio vulnificus, the ribosomes carrying the most variable rRNAs, namely, I-ribosomes, preferentially translated specific mRNAs associated with stress response and carbon metabolism, generating the ability to adapt rapidly to temperature and nutrient shifts (35). Our analysis of intragenomic heterogeneity for a larger number of sequenced genomes can provide the reference for further exploring the origin and function of distinct rRNA gene variants within the same strain.
Intragenomic heterogeneity is an interference factor for estimating microbial diversity using 16S rRNA gene-based analysis (1), which may overestimate the number of observed OTUs. Overestimation rates of 27.3% to 156.5% were generated for full-length or partial 16S genes at the ASV level (threshold, 100%) (Table 2), generally higher than previously reported values (12). This was probably due to the utilization of a larger number of sequenced genomes containing higher copy numbers of 16S rRNA genes. The V3 to V4 region is a widely used sequencing target in microbial ecology studies because of its high overall coverage and broad phylum spectrum (36); however, the amplicon analysis based on the V3 to V4 region may cause an overestimation of 51.7% for microbial diversity. At the most commonly used threshold, 97%, microbial diversity was overestimated by 4.4% when using the V4 to V5 region for such analysis, which was the lowest among all fragments used in this study. Meanwhile, microbial diversity may be underestimated by the interspecific conservation of 16S rRNA genes, as highly conservative sequences from closely related but distinct species were prone to be lumped together (16). We also found that the V4 to V5 region exhibited a relatively high underestimation rate, with the OTU count being 36.2% lower than the number of species, even using the most stringent threshold, 100%, indicating lower species resolution. Obviously, if one expects to achieve taxonomic resolution at the species level, the V4 to V5 region is not recommended as a target for sequencing, and full-length sequencing seems to be more appropriate.
Sequence clustering has been routinely used for bioinformatics analysis of amplicon reads retrieved from high-throughput sequencing platforms, and this assigned highly similar sequences to multiple clusters based on a given identity threshold, which appreciably improved the efficiency of the downstream analysis. In fact, the choice of clustering threshold significantly affects the calculation of community alpha diversity and should be carefully considered. A low identity threshold (e.g., 97%) may cluster sequences from different species or even genera into the same OTU (overmerging), as the 16S rRNA genes belonging to different species within a genus (e.g., Escherichia) are highly similar in sequence identity, whereas a too high threshold is likely to split a species or even a strain into multiple distinct clusters (oversplitting) due to intragenomic variation (14, 37). Ideally, the clustering process produces a set of OTUs or ASVs, each of which corresponds to a specific taxon, with all taxa at the same taxonomic level for convenient comparison. Moreover, because different variable regions provided different taxonomic resolutions, and the appropriate thresholds for different amplified fragments should be determined separately rather than depending on general experience. Hence, we also attempted to find the optimal identity thresholds of species delimitation for full-length and 10 partial 16S gene segments to minimize the total error rate. For full-length 16S genes, the total error rate was comparatively low (25% to 26%) with thresholds of 98.5% to 99.0% (Fig. 5), whereas it rapidly increased with thresholds exceeding 99%. For the V4 or V4 to V5 region, thresholds between 99% and 100% led to the minimum total error; this means that ASVs were more suitable to be adopted than OTUs with a threshold of 97%.
It has to be conceded that, regardless of which threshold is employed, it was impossible to yield an actual microbial structure profile when only the 16S rRNA gene was used as the sequencing target. Hence, what we can do is minimize bias in the estimation of microbial diversity. Our results provide quantitative data on the degree of overestimation due to intragenomic heterogeneity and the extent of underestimation caused by the insufficient interspecific variation of 16S rRNA genes in prokaryotic genomes. We also propose the optimal identity thresholds for full-length and multiple partial 16S genes to minimize the risk of overmerging and oversplitting. Intriguingly, when NCBI taxonomy was replaced by GTDB taxonomy for the aforementioned analysis, conclusions about the bias in estimating microbial diversity as well as the appropriate identity thresholds for different sequencing regions remain largely unchanged. This study will contribute to understanding how 16S rRNA gene redundancy and heterogeneity within prokaryotic genomes affect the estimation of microbial diversity and will provide general guidance on the choice of amplified regions and clustering thresholds when using 16S rRNA gene-based analysis in microbial ecology studies.
MATERIALS AND METHODS
Genomes and taxonomic information retrieval.
A total of 24,248 complete prokaryotic genome assemblies were retrieved from the National Center for Biotechnology Information (NCBI) nonredundant RefSeq database (https://ftp.ncbi.nlm.nih.gov/genomes/refseq/). All of the 16S rRNA gene sequences of each genome were extracted from *_rna_from_genomic.fna.gz file using SeqKit (v2.1.0) (38). In addition, the taxonomy file containing the names and phylogenetic lineages of all the organisms in NCBI was downloaded from their FTP site (https://ftp.ncbi.nih.gov/pub/taxonomy/new_taxdump/); mapping of each genome to NCBI taxonomy was subsequently performed to obtain the standard ranks (superkingdom, phylum, class, order, family, genus, and species). The GTDB taxonomy of each genome was obtained using GTDB-Tk (v2) (22). The taxonomic information was used to further analyze the 16S rRNA gene copy number per genome and the intragenomic variation of multiple copies at various taxon ranks.
Analysis of intragenomic heterogeneity of 16S rRNA genes in prokaryotes.
The number of 16S rRNA gene copies and variants and the intragenomic heterogeneity per genome were calculated as previously described (12). Specifically, for each genome, dereplication of 16S rRNA genes was performed using the fastx_uniques command in Usearch (v11.0.667) (39), which resulted in a file with unique sequences (variants). Multiple 16S rRNA gene variants belonging to the same genome were aligned using MUSCLE (v3.8.1551) (40), and pairwise distances between aligned sequences were then calculated using the dist.seqs command from the mothur package (v1.46.0) (41) with default options (calc=onegap, only counts a string of gaps as a single gap). The command will generate a distance matrix for each genome, the maximum value of which is defined as the intragenomic heterogeneity index (%).
Calculation of the bias in estimating microbial diversity.
Because of the uneven distribution of publicly available genomes across species, we randomly selected one representative for each species to avoid overrepresentation. All 16S rRNA gene sequences of the selected genome were merged to construct a data set (HIQ-T). Ideally, each species harbors one specific 16S rRNA gene sequence, so the sole representatives were randomly selected to construct the reference sequence data set (HIQ-C). The random selection was executed 10 times using a Python script (https://github.com/sjtu-piaopiao/16S_analysis.git). Due to the differences in NCBI species and GTDB species, data sets based on NCBI taxonomy (HIQ-T-NCBI and HIQ-C-NCBI) and GTDB taxonomy (HIQ-T-GTDB and HIQ-C-GTDB) were constructed, respectively. For each data set, in silico PCR was performed by running the pcr.seqs program in the mothur toolset (v1.46.0) using commonly used primers targeting different variable regions (Table 3), such as the V1 to V9 (full-length), V4, and V4 to V5 regions, in which three mismatches per primer were allowed. The amplicon sequences obtained were aligned to the SILVA reference database (v138.1) (42), and pairwise distances between aligned sequences were calculated using the mothur package. Based on the distance, each amplicon was assigned to a specific OTU or ASV using the OptiClust algorithm, with the identity thresholds set at 97% to 100%. Finally, the degrees of overestimation caused by full-length or partial 16S rRNA genes were calculated by comparing the numbers of OTUs generated from HIQ-T and HIQ-C, whereas the underestimation rates were measured by the comparison of the numbers of sequences and OTUs counts in HIQ-C.
TABLE 3.
Primers used in this study to extract different regions of 16S rRNA genes
| Region | Primer name | Sequence (5′–3′) | Reference |
|---|---|---|---|
| V1–V2 | 27F | AGAGTTTGATYMTGGCTCAG | 44 |
| 338R | TGCTGCCTCCCGTAGGAGT | 44 | |
| V1–V3 | 27F | AGAGTTTGATYMTGGCTCAG | 3 |
| 534R | ATTACCGCGGCTGCTGG | 3 | |
| V1–V9 | 27F | AGAGTTTGATCMTGGCTCAG | 1 |
| 1492R | TACGGYTACCTTGTTAYGACTT | 1 | |
| V3 | 341F | CCTACGGGAGGCAGCAG | 12 |
| 518R | ATTACCGCGGCTGCTGG | 12 | |
| V3–V4 | 341F | CCTACGGGNGGCWGCAG | 36 |
| 785R | GACTACNNGGGTATCTAATCC | 36 | |
| V4 | 515F | GTGYCAGCMGCCGCGGTAA | 45 |
| 806R | GGACTACNVGGGTWTCTAAT | 46 | |
| V4–V5 | 515F | GTGYCAGCMGCCGCGGTAA | 45 |
| 926R | CCGTCAATTCMTTTRAGT | 45 | |
| V5–V7 | 799F | AACMGGATTAGATACCCKG | 47 |
| 1193R | ACGTCATCCCCACCTTCC | 47 | |
| V6 | 967F | CAACGCGAAGAACCTTACC | 12 |
| 1046R | CGACAGCCATGCANCACCT | 12 | |
| V6–V8 | 926F | AAACTYAAAKGAATTGACGG | 48 |
| 1392R | ACGGGCGGTGTGTRC | 48 | |
| V7–V9 | 1115F | CAACGAGCGCAACCCT | 2 |
| 1492R | TACGGYTACCTTGTTACGACTT | 2 |
Shannon entropy analysis of nucleotide positions across the 16S rRNA gene.
All 16S rRNA genes per genome were aligned to the SILVA data set, and Shannon entropy was calculated at each nucleotide position along a specific reference 16S gene sequence using the open-source software Oligotyping (v3.1) (43) based on the equation , where pi is the probability of nucleotide base i (A, T, C, G) or the gap character. There was a considerable difference in the 16S rRNA genes between bacteria and the archaea domain. Thus, the Escherichia coli sequence (gene ID 948332) was used as the reference for bacteria, and the Saccharolobus solfataricus sequence (accession number NR_029127.1) for archaea to determine the nucleotide position throughout the 16S rRNA gene. As the variable site is genome specific, the average information entropy per base was then calculated to quantify the overall intragenomic heterogeneity at each position of the 16S rRNA gene. Analogously, the intergenomic variation per base was measured by the Shannon entropy analysis of each column in an alignment of sequences present in the SILVA Ref NR99 database.
Calculation of oversplitting, overmerging, and total error rates.
In order to minimize the risk of splitting a single genome into multiple clusters and lumping together different species to the same cluster, a range of thresholds between 97% and 100% in increments of 0.1% were evaluated using full-length or partial 16S rRNA genes based on three metrics: oversplitting, overmerging, and total error rate, as previously reported (37). The oversplitting rate was defined as the percentage of species which were split into separate clusters (OTUs or ASVs), and the overmerging rate, as the percentage of clusters containing amplicons from multiple species. Total error rates were computed by summing the above two values. Python scripts that we wrote were executed to perform this calculation.
ACKNOWLEDGMENT
This work was supported by a grant from the National Natural Science Foundation of China (NSFC) (92051104).
Footnotes
Supplemental material is available online only.
Contributor Information
Ning-Yi Zhou, Email: ningyi.zhou@sjtu.edu.cn.
Isaac Cann, University of Illinois Urbana-Champaign.
REFERENCES
- 1.Johnson JS, Spakowicz DJ, Hong BY, Petersen LM, Demkowicz P, Chen L, Leopold SR, Hanson BM, Agresta HO, Gerstein M, Sodergren E, Weinstock GM. 2019. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun 10:5029. doi: 10.1038/s41467-019-13036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abellan-Schneyder I, Matchado MS, Reitmeier S, Sommer A, Sewald Z, Baumbach J, List M, Neuhaus K. 2021. Primer, pipelines, parameters: issues in 16S rRNA gene sequencing. mSphere 6:e01202-20. doi: 10.1128/mSphere.01202-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker AW, Martin JC, Scott P, Parkhill J, Flint HJ, Scott KP. 2015. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome 3:26. doi: 10.1186/s40168-015-0087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eren AM, Borisy GG, Huse SM, Mark Welch JL. 2014. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci USA 111:E2875–E2884. doi: 10.1073/pnas.1409644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Segawa T, Yonezawa T, Edwards A, Akiyoshi A, Tanaka S, Uetake J, Irvine-Fynn T, Fukui K, Li Z, Takeuchi N. 2017. Biogeography of cryoconite forming cyanobacteria on polar and Asian glaciers. J Biogeogr 44:2849–2861. doi: 10.1111/jbi.13089. [DOI] [Google Scholar]
- 6.Faoro H, Alves AC, Souza EM, Rigo LU, Cruz LM, Al-Janabi SM, Monteiro RA, Baura VA, Pedrosa FO. 2010. Influence of soil characteristics on the diversity of bacteria in the southern Brazilian Atlantic Forest. Appl Environ Microbiol 76:4744–4749. doi: 10.1128/AEM.03025-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang N, Tian C, Lv Y, Hou J, Yang Z, Xiao X, Zhang Y. 2022. Novel primers for 16S rRNA gene-based archaeal and bacterial community analysis in oceanic trench sediments. Appl Microbiol Biotechnol 106:2795–2809. doi: 10.1007/s00253-022-11893-3. [DOI] [PubMed] [Google Scholar]
- 8.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert JA, Meyer F, Jansson J, Gordon J, Pace N, Tiedje J, Ley R, Fierer N, Field D, Kyrpides N, Glöckner F-O, Klenk H-P, Wommack KE, Glass E, Docherty K, Gallery R, Stevens R, Knight R. 2010. The Earth Microbiome Project: meeting report of the “1st EMP meeting on sample selection and acquisition” at Argonne National Laboratory October 6th 2010. Stand Genomic Sci 3:249–253. doi: 10.4056/aigs.1443528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vos M, Quince C, Pijl AS, De Hollander M, Kowalchuk GA. 2012. A comparison of rpoB and 16S rRNA as markers in pyrosequencing studies of bacterial diversity. PLoS One 7:e30600. doi: 10.1371/journal.pone.0030600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bajagai YS, Trotter M, Williams TM, Costa DFA, Whitton MM, Ren X, Wilson CS, Stanley D. 2022. The role of microbiota in animal health and productivity: misinterpretations and limitations. Anim Prod Sci 62:944–956. doi: 10.1071/AN21515. [DOI] [Google Scholar]
- 12.Sun DL, Jiang X, Wu QL, Zhou NY. 2013. Intragenomic heterogeneity of 16S rRNA genes causes overestimation of prokaryotic diversity. Appl Environ Microbiol 79:5962–5969. doi: 10.1128/AEM.01282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Větrovský T, Baldrian P. 2013. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One 8:e57923. doi: 10.1371/journal.pone.0057923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schloss PD. 2021. Amplicon sequence variants artificially split bacterial genomes into separate clusters. mSphere 6:e00191-21. doi: 10.1128/mSphere.00191-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pei AY, Oberdorf WE, Nossa CW, Agarwal A, Chokshi P, Gerz EA, Jin Z, Lee P, Yang L, Poles M, Brown SM, Sotero S, Desantis T, Brodie E, Nelson K, Pei Z. 2010. Diversity of 16S rRNA genes within individual prokaryotic genomes. Appl Environ Microbiol 76:3886–3897. doi: 10.1128/AEM.02953-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez RL, Castro JC, Kyrpides NC, Cole JR, Tiedje JM, Konstantinidis KT. 2018. How much do rRNA gene surveys underestimate extant bacterial diversity? Appl Environ Microbiol 84:e00014-18. doi: 10.1128/AEM.00014-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole JR, Konstantinidis K, Farris RJ, Tiedje JM. 2010. Microbial diversity and phylogeny: extending from rRNAs to genomes. In Liu WT, Jansson JK, Environmental molecular microbiology. Caister Academic Press, Poole, UK. [Google Scholar]
- 18.Kim M, Oh H-S, Park S-C, Chun J. 2014. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 19.Stackebrandt E, Ebers J. 2006. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 4:6–9. [Google Scholar]
- 20.Stackebrandt E, Goebel BM. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849. doi: 10.1099/00207713-44-4-846. [DOI] [Google Scholar]
- 21.Parks D, Chuvochina M, Rinke C, Mussig A, Chaumeil P-A, Hugenholtz P. 2021. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res 50:D785–D794. doi: 10.1093/nar/gkab776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. 2022. GTDB-Tk v2: memory friendly classification with the genome taxonomy database. Bioinformatics 38:5315–5316. doi: 10.1093/bioinformatics/btac672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Louca S, Doebeli M, Parfrey LW. 2018. Correcting for 16S rRNA gene copy numbers in microbiome surveys remains an unsolved problem. Microbiome 6:41. doi: 10.1186/s40168-018-0420-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoddard SF, Smith BJ, Hein R, Roller BRK, Schmidt TM. 2015. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res 43:D593–D598. doi: 10.1093/nar/gku1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Angly FE, Dennis PG, Skarshewski A, Vanwonterghem I, Hugenholtz P, Tyson GW. 2014. CopyRighter: a rapid tool for improving the accuracy of microbial community profiles through lineage-specific gene copy number correction. Microbiome 2:11. doi: 10.1186/2049-2618-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langille MGI, Zaneveld J, Caporaso JG, Mcdonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kembel SW, Wu M, Eisen JA, Green JL. 2012. Incorporating 16S gene copy number information improves estimates of microbial diversity and abundance. PLoS Comput Biol 8:e1002743. doi: 10.1371/journal.pcbi.1002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Starke R, Pylro VS, Morais DK. 2021. 16S rRNA gene copy number normalization does not provide more reliable conclusions in metataxonomic surveys. Microb Ecol 81:535–539. doi: 10.1007/s00248-020-01586-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.López-López A, Benlloch S, Bonfá M, Rodríguez-Valera F, Mira A. 2007. Intragenomic 16S rDNA divergence in Haloarcula marismortui is an adaptation to different temperatures. J Mol Evol 65:687–696. doi: 10.1007/s00239-007-9047-3. [DOI] [PubMed] [Google Scholar]
- 30.Yap WH, Zhang ZS, Wang Y. 1999. Distinct types of rRNA operons exist in the genome of the actinomycete Thermomonospora chromogena and evidence for horizontal transfer of an entire rRNA operon. J Bacteriol 181:5201–5209. doi: 10.1128/Jb.181.17.5201-5209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato Y, Fujiwara T, Kimura H. 2017. Expression and function of different guanine-plus-cytosine content 16S rRNA genes in Haloarcula hispanica at different temperatures. Front Microbiol 8:482. doi: 10.3389/fmicb.2017.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato Y, Kimura H. 2019. Temperature-dependent expression of different guanine-plus-cytosine content 16S rRNA genes in Haloarcula strains of the class Halobacteria. Antonie Van Leeuwenhoek 112:187–201. doi: 10.1007/s10482-018-1144-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leppek K, Barna M. 2019. An rRNA variant to deal with stress. Nat Microbiol 4:382–383. doi: 10.1038/s41564-019-0396-7. [DOI] [PubMed] [Google Scholar]
- 34.Kurylo CM, Parks MM, Juette MF, Zinshteyn B, Altman RB, Thibado JK, Vincent CT, Blanchard SC. 2018. Endogenous rRNA sequence variation can regulate stress response gene expression and phenotype. Cell Rep 25:236–248.e6. doi: 10.1016/j.celrep.2018.08.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song W, Joo M, Yeom JH, Shin E, Lee M, Choi HK, Hwang J, Kim YI, Seo R, Lee JE, Moore CJ, Kim YH, Eyun SI, Hahn Y, Bae J, Lee K. 2019. Divergent rRNAs as regulators of gene expression at the ribosome level. Nat Microbiol 4:515–526. doi: 10.1038/s41564-018-0341-1. [DOI] [PubMed] [Google Scholar]
- 36.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonin A, Guerrieri A, Ficetola GF. 2023. Optimal sequence similarity thresholds for clustering of molecular operational taxonomic units in DNA metabarcoding studies. Molecular Ecology Resources 23:368–381. doi: 10.1111/1755-0998.13709. [DOI] [PubMed] [Google Scholar]
- 38.Shen W, Le S, Li Y, Hu F. 2016. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS One 11:e0163962. doi: 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 40.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glöckner FO. 2014. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res 42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG, Sogin ML. 2013. Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol 4:1111–1119. doi: 10.1111/2041-210X.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Z, Hui PC, Hui M, Yeoh YK, Wong PY, Chan MCW, Wong MCS, Ng SC, Chan FKL, Chan PKS. 2019. Impact of preservation method and 16S rRNA hypervariable region on gut microbiota profiling. mSystems 4:e00271-18. doi: 10.1128/mSystems.00271-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parada AE, Needham DM, Fuhrman JA. 2016. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 46.Apprill A, Mcnally S, Parsons R, Weber L. 2015. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol 75:129–137. doi: 10.3354/ame01753. [DOI] [Google Scholar]
- 47.Wang F, Men X, Zhang G, Liang K, Xin Y, Wang J, Li A, Zhang H, Liu H, Wu L. 2018. Assessment of 16S rRNA gene primers for studying bacterial community structure and function of aging flue-cured tobaccos. AMB Expr 8:182. doi: 10.1186/s13568-018-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, Ochman H, Hugenholtz P. 2010. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J 4:642–647. doi: 10.1038/ismej.2009.153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aem.02108-22-s0001.xlsx, XLSX file, 2.8 MB (2.8MB, xlsx)
Supplemental material. Download aem.02108-22-s0002.docx, DOCX file, 1.5 MB (1.5MB, docx)