ABSTRACT
Foot-and-mouth disease virus (FMDV) is a picornavirus, which infects cloven-hoofed animals to cause foot-and-mouth disease (FMD). The positive-sense RNA genome contains a single open reading frame, which is translated as a polyprotein that is cleaved by viral proteases to produce the viral structural and nonstructural proteins. Initial processing occurs at three main junctions to generate four primary precursors; Lpro and P1, P2, and P3 (also termed 1ABCD, 2BC, and 3AB1,2,3CD). The 2BC and 3AB1,2,3CD precursors undergo subsequent proteolysis to generate the proteins required for viral replication, including the enzymes 2C, 3Cpro, and 3Dpol. These precursors can be processed through both cis and trans (i.e., intra- and intermolecular proteolysis) pathways, which are thought to be important for controlling virus replication. Our previous studies suggested that a single residue in the 3B3-3C junction has an important role in controlling 3AB1,2,3CD processing. Here, we use in vitro based assays to show that a single amino acid substitution at the 3B3-3C boundary increases the rate of proteolysis to generate a novel 2C-containing precursor. Complementation assays showed that while this amino acid substitution enhanced production of some nonenzymatic nonstructural proteins, those with enzymatic functions were inhibited. Interestingly, replication could only be supported by complementation with mutations in cis acting RNA elements, providing genetic evidence for a functional interaction between replication enzymes and RNA elements.
IMPORTANCE Foot-and-mouth disease virus (FMDV) is responsible for foot-and-mouth disease (FMD), an important disease of farmed animals, which is endemic in many parts of the world and can results in major economic losses. Replication of the virus occurs within membrane-associated compartments in infected cells and requires highly coordinated processing events to produce an array of nonstructural proteins. These are initially produced as a polyprotein that undergoes proteolysis likely through both cis and trans alternative pathways (i.e., intra- and intermolecular proteolysis). The role of alternative processing pathways may help coordination of viral replication by providing temporal control of protein production and here we analyze the consequences of amino acid substitutions that change these pathways in FMDV. Our data suggest that correct processing is required to produce key enzymes for replication in an environment in which they can interact with essential viral RNA elements. These data further the understanding of RNA genome replication.
KEYWORDS: FMDV, picornavirus, cleavage, replication, replication complex, foot-and-mouth disease virus
INTRODUCTION
Small RNA viruses such as members of the Picornaviridae have limited genome size and therefore minimal coding capacity. These viruses have evolved several strategies to overcome this limitation, including the use of protein precursors that can perform different functions to the mature proteins. Furthermore, individual proteins and their precursors can also sometimes perform more than one function (1–4).
The Picornaviridae family includes several important human and animal pathogens, including but not limited to poliovirus (PV) and foot-and-mouth disease virus (FMDV). PV is responsible for the incapacitating (and potentially fatal) human disease poliomyelitis, while FMDV is the causative agent of the economically damaging foot-and-mouth disease, an acute vesicular disease of livestock, including cloven-hoofed ruminants and pigs. The FMDV genome contains one large open reading frame that encodes a ~250 kDa polyprotein (5). Initial processing of the FMDV polyprotein occurs at three positions to produce four primary products: Lpro, the capsid precursor P1-2A and two nonstructural protein precursors 2BC (also termed P2) and 3AB1,2,3CD (also termed P3) (6, 7). Lpro is autocatalytically released from the N-terminal region of the polyprotein (6, 8). The P1-2A precursor is released from the polyprotein via a co-translational 2A-driven ribosome skipping mechanism (9) before the 2BC-3AB1,2,3CD polyprotein is processed by 3Cpro. For FMDV, initial processing is believed to be predominantly at the 2C-3A junction, generating 2BC and 3AB1,2,3CD precursors. Further 3Cpro-mediated proteolysis releases the final proteins via a succession of intermediate precursors (7, 10, 11). Processing of the 2BC precursor ultimately generates the 2B and 2C proteins, both of which have multiple roles in replication. The 3AB1,2,3CD precursor is composed of the transmembrane protein 3A, three 3B peptides (individually referred to as 3B1, 3B2, and 3B3,), the protease 3Cpro, and the polymerase 3Dpol (12, 13).
Processing of the nonstructural polyprotein by 3Cpro is thought to occur through at least two separate pathways to generate mutually exclusive sets of precursors (14). For example, for FMDV, the 3AB1,2,3CD precursor is processed to generate the precursors 3AB1,2,3C and 3CD, which must be derived from alternative processing strategies. Likewise, for PV, the 3ABCD precursor (the equivalent of 3AB1,2,3CD in FMDV) can be processed to generate 3ABC and 3CD. Furthermore, it appears that this alternative processing may be temporally controlled and used to regulate virus replication. For example, previous studies with PV have demonstrated that later production of 3AB and 3CD can delay the initiation of viral RNA replication (15). For FMDV, reducing cleavage of 3CD inhibits replication by limiting the supply of 3Dpol (16). Processing through alternative pathways is likely to be driven (in part) through a switch between intramolecular versus intermolecular proteolysis (i.e., cis versus trans cleavage events). However, methods to differentiate between these cis versus trans cleavage events are challenging, and as a result, the mechanism(s) that controls this switch are not completely understood.
Like all positive-sense RNA viruses, picornavirus genome replication is associated with virus-induced cytoplasmic membranous structures, sometimes referred to as “replication complexes” or “replication organelles” (17). In these assemblies, multiple new viral positive-strand RNAs are synthesized via a complementary negative-sense template. For FMDV, the full composition of these assemblies is unknown, but they are likely composed of multiple viral and cellular factors, including the nonstructural proteins 3B, 3Dpol, and 3CD. Some of the viral nonstructural proteins and precursors associate with RNA elements located in the 5′ and 3′ untranslated regions (UTRs) that flank the open reading frame (5). The 5′ UTR of FMDV is uncharacteristically long for a picornavirus and contains several distinct structural elements, including an internal ribosome entry site (IRES), a cis-acting replicative element (cre), a large stem-loop (termed the S-fragment) and a tandem series of pseudoknots (18–21). The IRES has been well studied and is used to initiate protein translation in a cap-independent manner (19, 20). The cre is essential for replication and acts as the template for uridylation of 3B (also known as VPg) to generate the replicative primer, VPg-pUpU (22–27). The role of the S-fragment in FMDV replication is less well understood but may be involved in both replication and modulating the innate immune response (28). In other picornaviruses, such as PV, an RNA structure termed the cloverleaf (or oriL) is located at the 5′ terminus of the genome at the site occupied by the S-fragment stem-loop in FMDV (29). This interacts with the precursor protein 3CD as well as host proteins and is involved in initiating negative-strand RNA synthesis (30–35). Furthermore, for PV, other precursor proteins have also been implicated in 5′ UTR interactions, including 3AB (the equivalent to 3AB1,2,3 in FMDV), 3BCD, and 3ABCD, and have been suggested to be important for controlling replication (15, 31, 36, 37). The role of precursors in FMDV replication is less well established.
In a previous study, we used an FMDV replicon system based on serotype O1K, where replication is monitored by fluorescent protein (e.g., GFP/RFP) expression over time, to investigate FMDV replication by mutation of the 3B proteins (16). We reported a series of amino acid substitutions that increased the efficiency of processing at the 3B3-3C junction but inhibited replication by abrogating the release of free 3Dpol. Simultaneously, we observed that these substitutions caused an overall shift in 3AB1,2,3CD processing and channeled precursor synthesis mainly down one pathway generating 3AB1,2,3 and 3CD precursors. This series of mutations has enabled us to separate alternative cleavage pathways and study the function of different precursor sets. Here, we investigated the mechanism by which these substitutions result in increased processing between 3B3 and 3C. Our data suggest that a single amino acid substitution increases sensitivity to trans-mediated proteolysis at this boundary. Furthermore, when placed into the context of a full-length polyprotein, this single substitution resulted in accumulation of a novel precursor. Interestingly, it also prevented reciprocal complementation of replicons in trans, which we demonstrate is due to a deficiency in the functions of essential viral enzymes.
RESULTS
A single amino acid substitution in 3B3 prevents replicon replication.
In a previous study, we reported that amino acid substitutions within 3B3, at the boundary with 3Cpro, dramatically changed processing of the FMDV 3AB1,2,3CD polyprotein and prevented release of active 3Dpol. However, through blind passage, a compensatory mutation was selected which restored replication and wild-type (WT) 3AB1,2,3CD processing. This was identified as a reversion of a lysine at the P2 residue of the 3B3-3C junction to the WT threonine (16). These data suggested that the amino acid in the P2 position of the 3B3-3C junction alone can be a major determinant of altered polyprotein processing.
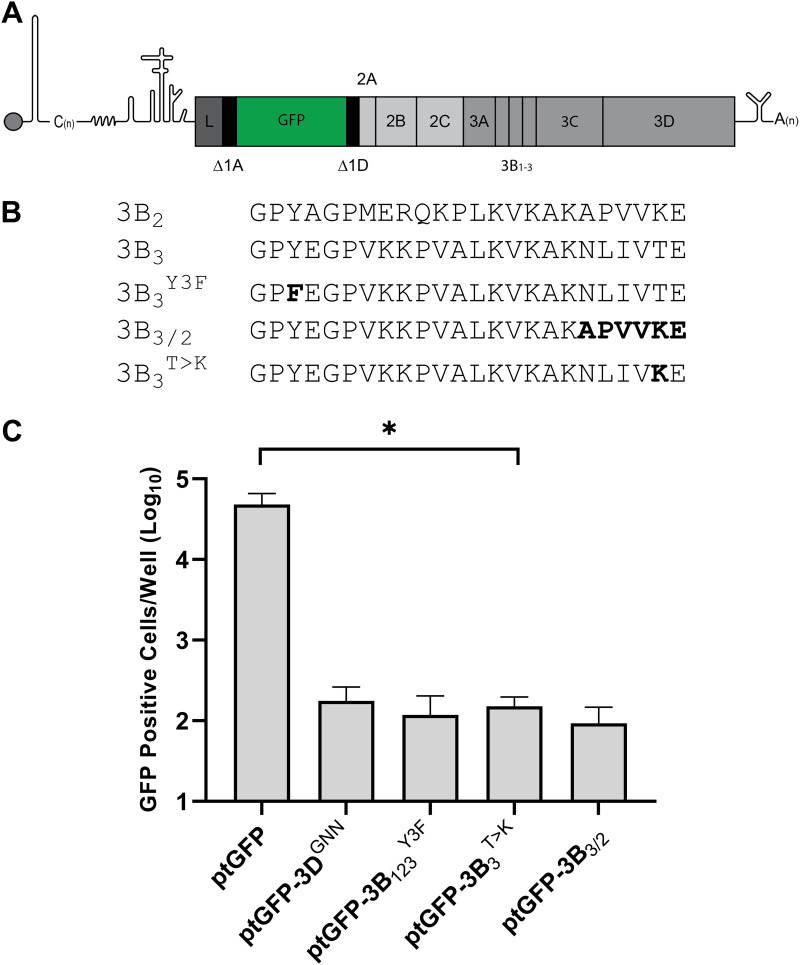
Before investigating the mechanism by which substitutions at this junction increased processing, we first sought to establish that this residue alone was sufficient to change FMDV polyprotein processing and prevent replicon replication. To this end, we generated an FMDV replicon with a threonine to lysine substitution at the P2 residue of the 3B3-3C cleavage junction (Fig. 1A and B). In this replicon (termed ptGFP-3B3T>K) the reporter protein ptGFP replaced the structural proteins, allowing ptGFP expression to be used as an indicator of replicon replication (Fig. 1A). This replicon RNA was transfected into BHK-21 cells alongside the previously published mutant replicons ptGFP-3B1,2,3Y3F (contains inactivating point mutations to the triptych of 3B genes) and ptGFP-3B3/2 (the six C-terminal residues of 3B3 replaced by those of 3B2) (Fig. 1B). A WT ptGFP expressing replicon (ptGFP) and a replicon containing an inactivating double point mutation in 3Dpol (ptGFP-3DGNN) were included as controls. The latter replicon serves as a negative control for ptGFP production from translation of the input transfected RNA, as we have previously described (38). RNAs from these replicons were transfected into BHK-21 cells and replication monitored by ptGFP fluorescence using an Incucyte real-time imaging system (Fig. 1C).
FIG 1.
A single amino acid substitution at the 3B3-3C junction prevents FMDV replicon replication. (A) Schematic diagram of the FMDV replicon. (B) Sequence alignments of the 3B cleavage junctions with the 3B1,2,3Y3F, 3B3/2, and 3B3T>K mutants. (C) Replication of replicons containing 3B3 mutations as well as the WT ptGFP replicon (ptGFP) and replication-defective controls containing inactivating mutations in 3Dpol (3DGNN), or 3B proteins (3B1,2,3Y3F). GFP expression was monitored hourly for 24 h. The graph shows GFP positive cells per well at 8 h posttransfection when replication is maximal. Significance compared to WT control (n = 3 ± SEM; * = P ≤ 0.05).
As anticipated, the WT ptGFP replicon produced ptGFP >100-fold greater than the ptGFP-3DGNN control replicon, as previously reported (39). In comparison, the ptGFP-3B3T>K replicon showed GFP expression equivalent to the replication-defective ptGFP-3DGNN control. These data were in agreement with those obtained with the ptGFP-3B3/2 and ptGFP-3B1,2,3Y3F replication-defective replicons we previously reported (16). Thus, the 3B3T>K substitution alone is sufficient to prevent replicon replication.
A single amino acid substitution at the 3B3-3C boundary increases the rate of proteolysis.
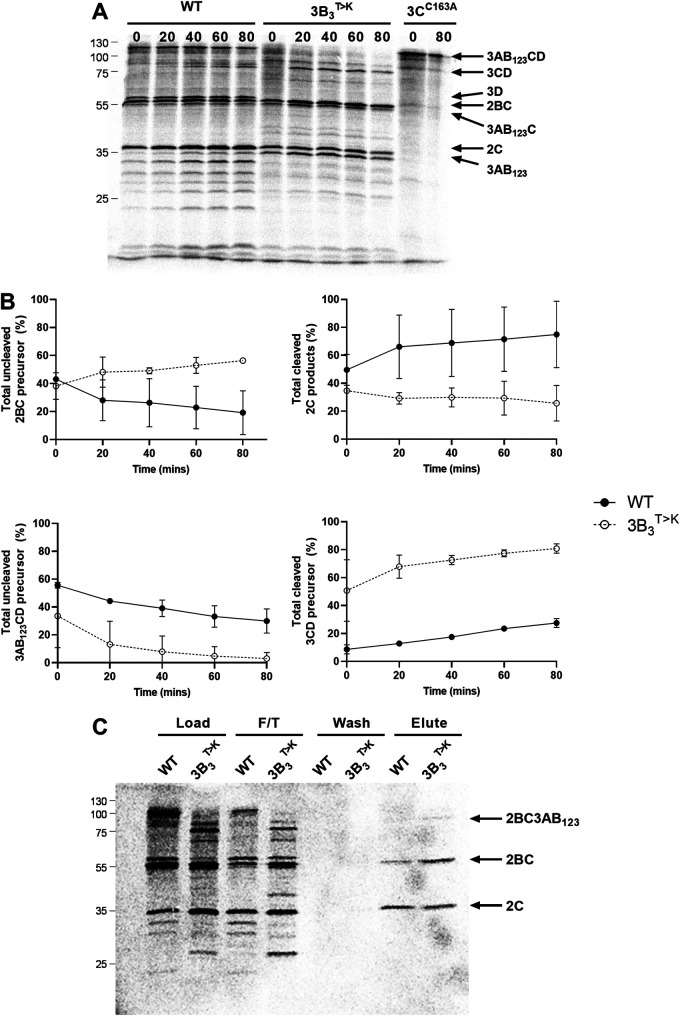
We previously demonstrated that the ptGFP-3B3/2 mutation inhibited replication and changed processing of the 3AB1,2,3CD polyprotein. To confirm that the 3B3T>K substitution alone was sufficient to induce the same changes, we employed the previously described in vitro coupled transcription/translation assay (16). T7 expression constructs were generated to express either the WT FMDV 2BC-3AB1,2,3CD polyprotein or a polyprotein containing the 3B3T>K amino acid substitution. For simplicity, in these experiments Lpro and GFP were not included in the polyprotein. The polyprotein used in these experiments included 2BC as well as the 3AB1,2,3CD region to determine changes to the entire NS polyprotein. These experiments also included a control polyprotein containing an inactivating mutation in 3Cpro (3CC163A) predicted to prevent its proteolytic activity (40). Processing was investigated by [35S] methionine/cysteine pulse/chase labeling in in vitro coupled transcription/translation reactions, harvesting samples at regular time points and analyzing protein products by SDS-PAGE (Fig. 2A and B).
FIG 2.
A single amino acid substitution at the 3B3-3C boundary increases the rate of proteolysis and drives the production of novel precursors. (A) Plasmids expressing the WT or mutant 3B3T>K FMDV polyprotein precursors were used to prime [35S] labeled pulse-chase in vitro coupled transcription/translation reactions. At regular intervals, samples were taken and stopped by the addition of 2× Laemmli buffer. Proteins were separated by SDS-PAGE and visualized by autoradiography. The identity of some FMDV proteins is shown. (B) The percentage of protein or protein precursor was quantified as a total percentage of 2C or 3Dpol containing products, as appropriate (n = 2 ± SD). (C) Duplicate reactions were incubated for 90 min before immunoprecipitation of 2C containing precursors with anti-2C antibodies. The pre- and post precipitation samples were separated by SDS-PAGE and visualized by autoradiography. Arrows show the identity of 2C containing proteins, based on predicted molecular weights. F/T refers to the flow through samples.
For the WT construct, the full-length 3AB1,2,3CD precursor was detected at early time points, and was steadily processed over time primarily into 3AB1,2,3C and 3Dpol, with a small amount of 3CD derived from an alternative processing pathway. At the later time points (40 and 60 min), 3AB1,2,3 was also detected. Both 2B and 2C were present, in addition to a limited amount of the precursor 2BC at earlier time points. Compared to WT, the construct containing the 3B3T>K substitution resulted in greater amounts of the 3CD and 3AB1,2,3 precursors and less of the 3AB1,2,3CD precursor. There were also increased levels of the 2BC precursor in addition to a high molecular weight precursor, possibly 2BC3AB1,2,3, which was detected at early time points and gradually decreased over time. These data extended our previous observations demonstrating that the 3B3T>K substitution alone is sufficient to accelerate proteolysis at the 3B3-3C junction and so increases the relative amounts of 2BC, 3CD, and 3AB1,2,3.
Increasing polyprotein proteolysis generates a novel 2BC containing precursor.
The previous in vitro translation experiments suggested that the 3B3T>K substitution increased the production of the 2BC precursor in addition to a larger molecular weight precursor not observed in the WT control. To confirm the identity of the 2C-containing precursors, the in vitro translation samples were immunoprecipitated using an anti-2C antibody. T7 constructs expressing the WT polyprotein or polyprotein containing the 3B3T>K substitution were used in in vitro coupled transcription/translation reactions with [35S] methionine/cysteine pulse/chase labeling. Samples were taken after 90 min and immunoprecipitation performed on half of the sample. Both pre- and post-immunoprecipitation protein samples were analyzed by SDS-PAGE (Fig. 2C).
In comparison to WT, a smaller proportion of mature 2C (but more unprocessed 2BC precursor) were immunoprecipitated with an anti-2C antibody following expression from the polyprotein containing the 3B3T>K substitution. Furthermore, the additional higher molecular weight band which was only present following expression of the 3B3T>K precursor was also immunoprecipitated with the anti-2C antibody. Based on these observations and the estimated molecular weight, this product is mostly likely a 2BC3AB1,2,3 precursor. These results agree with previous data and suggest that the 3B3T>K substitution preferentially increases the rate of proteolysis at the 3B3-3C junction, compared to the 2C-3A junction, resulting in the accumulation of 2BC3AB1,2,3, which is not normally detected.
The 3B3T>K substitution stimulates trans-mediated processing.
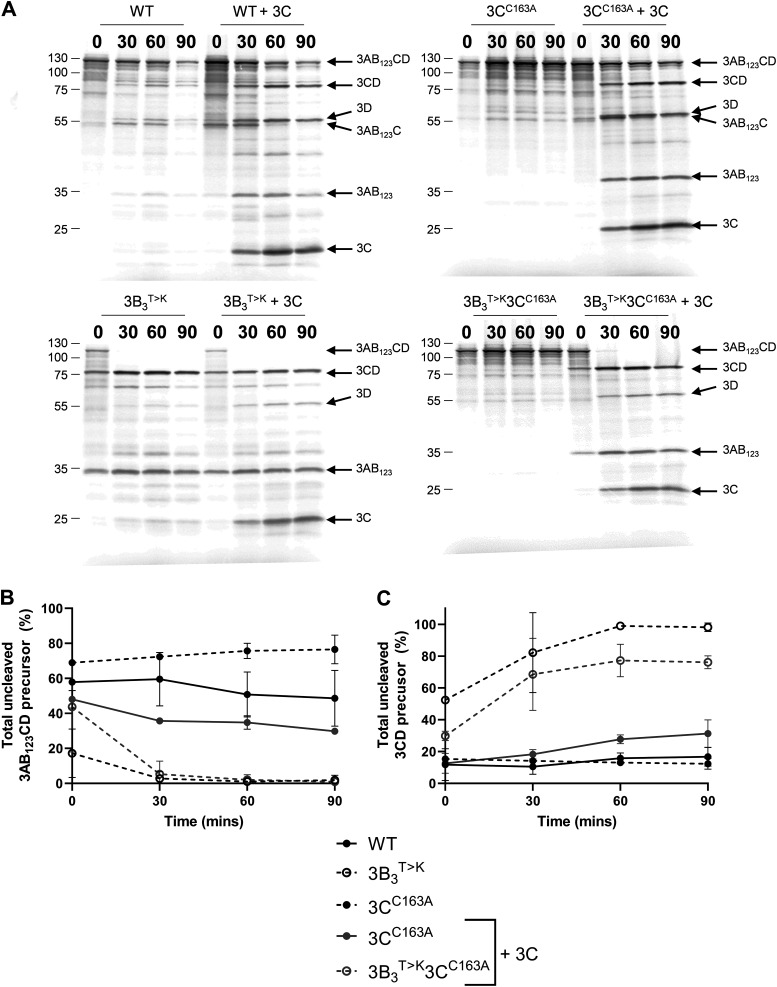
We speculated that the most likely mechanism by which the order of polyprotein processing was altered was that the substitution generated a boundary sequence that was preferentially recognized in trans by 3Cpro and/or a 3Cpro-containing precursor. To explore the latter possibility, we adapted our in vitro assay to investigate trans-mediated cleavage. For simplicity, we adapted both the WT 3AB1,2,3CD precursor or precursor containing the 3B3T>K substitution to also contain the inactivating mutation in 3Cpro (3CC163A) to prevent self-proteolysis (40). These precursor substrates (termed 3CC163A and 3B3T>K-3CC163A, respectively) were translated in vitro with [35S] methionine/cysteine before adding excess unlabeled methionine/cysteine and purified active 3Cpro to a duplicate set of reactions (plus 3Cpro). Samples were harvested at regular time points and processing of the [35S] labeled precursor was analyzed by SDS-PAGE (Fig. 3). As controls, the experiment was conducted with the WT and 3B3T>K constructs that did not contain the 3CC163A point mutation (Fig. 3). To aid interpretation, the relative amounts of 3AB1,2,3CD and 3CD products were quantified by phosphorimaging (Fig. 3).
FIG 3.
The 3B3T>K amino acid substitution drives trans-mediated precursor proteolysis. Plasmids expressing proteolytically inactive polyproteins with or without the 3B3T>K substitution (termed 3B3T>K, 3CC163A, and 3CC163A, respectively) were used to prime coupled [35S] labeled transcription/translation assays, followed by unlabeled amino acid chase. To a duplicate set of reactions 10 μM purified 3Cpro was added (+3C) immediately after chase. As controls, reactions were set up alongside the WT or 3B3T>K polyproteins without an inactivated 3Cpro. At regular intervals, samples were taken and reactions stopped by the addition of 2× Laemmli buffer. (A) Proteins were separated by SDS-PAGE and visualized by autoradiography. The relative proportion of uncleaved 3AB1,2,3CD (B) or 3CD (C) was quantified as a percentage of 3Dpol containing products (n = 2 ± SD).
Both WT and 3B3T>K precursors carrying the 3CC163A mutation produced only uncleaved full-length 3AB1,2,3CD in the absence of 3Cpro provided in trans. The addition of 3Cpro resulted in the production of smaller proteins due to trans-mediated proteolysis of 3AB1,2,3CD. For the WT precursor, these were predominantly 3AB1,2,3, 3CD, and 3Dpol, in addition to a cluster of 3B1,2,3CD-derived products, including some 3CD, indicative of alternative cleavage pathways. In comparison, the precursor containing the 3B3T>K substitution was processed to 3AB1,2,3 and a greater proportion of 3CD compared to WT over the duration of the experiment, as observed previously with the active precursor molecule (Fig. 2A) (16).
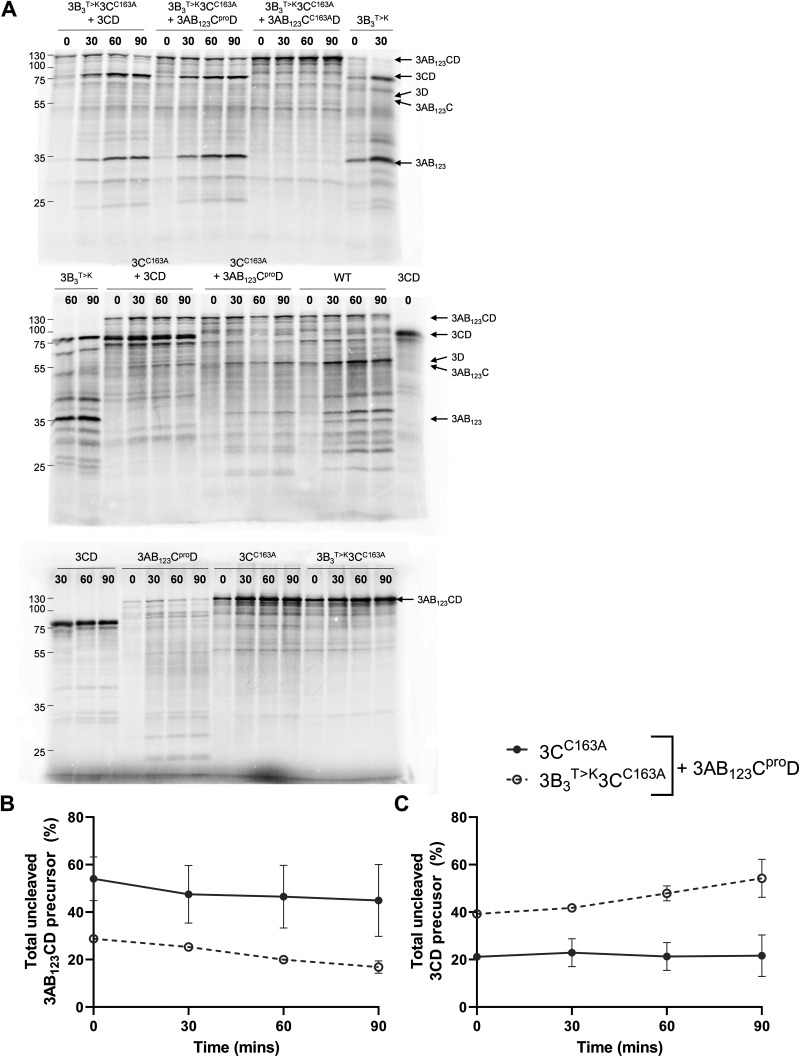
To investigate whether the 3B3T>K precursor was also sensitive to cleavage by 3Cpro when this is present as part of a larger precursor molecule, we generated a 3AB1,2,3CD expression construct in which all the cleavage boundaries had been mutated. Thus, the protease activity of this precursor was retained but only in the context of a full-length 3AB1,2,3CD polyprotein. Our trans-cleavage assay was repeated using this new construct, termed 3AB1,2,3CproD, in place of the purified 3Cpro enzyme used above (Fig. 4).
FIG 4.
The 3B3T>K amino acid substitution drives trans-mediated precursor proteolysis. Plasmids expressing proteolytically inactive versions of WT or 3B3T>K polyproteins (3CC163A and 3B3T>K3CC163A, respectively) were used to prime coupled [35S] labeled transcription/translation assays in the presence of 3AB1,2,3CproD, a proteolytically active precursor with all cleavage boundaries mutated to prevent self-proteolysis (+3AB123CproD). At regular intervals, samples were taken and reactions stopped by the addition of 2× Laemmli buffer. (A) Proteins were separated by SDS-PAGE and visualized by autoradiography. The relative proportion of uncleaved 3AB1,2,3CD (B) or 3CD (C) was quantified as a percentage of 3Dpol containing products (n = 2 ± SD).
As before, the 3CC163A mutation prevented self-proteolysis when present within the WT precursor or the precursor containing the 3B3T>K substitution, as expected. Addition of the proteolytically active 3AB1,2,3CproD construct resulted in processing of the proteolytically inactive 3AB1,2,3CD precursor bearing the 3B3T>K substitution to generate 3AB1,2,3 and 3CD. This pattern of processing was similar to that observed following addition of active 3Cpro, as observed above (Fig. 3). This contrasts with the WT proteolytically inactive 3AB1,2,3CD precursor, which was not significantly processed in trans by 3AB1,2,3CproD. Taken together, these data suggest that the 3B3T>K substitution at the 3B3-3C junction generates a cleavage boundary that is preferentially recognized by 3Cpro (even when delivered as part of a larger precursor). Thus, driving rapid trans-mediated proteolysis at this junction results in overproduction of a specific set of viral precursor proteins.
Increasing the rate of 3B3-3C cleavage prevents production of trans-functional 2C and 3Dpol but not 3B.
The in vitro polyprotein processing data above implies that the 3B3T>K substitution increases the rate of proteolysis at the 3B3-3C junction by stimulating trans-mediated proteolysis. In doing so, it drives the formation of 2BC, 3AB1,2,3, and 3CD precursors to the detriment of other products such as the enzymes 2C and 3Dpol. In our previous studies, we used trans-complementation assays to investigate protein function. These assays involve the co-transfection of two replication defective replicon constructs that express different fluorescent reporter genes, allowing replication to be differentially monitored. Co-transfection of the two replicons allows exchange of viral nonstructural proteins within replication complexes to permit replication of one (or both) of the input genomes (39) (Fig. 5A). Here, we used this approach to investigate whether stimulating trans-mediated proteolysis at the 3B3-3C junction prevented the production of functional 2C. To do this, inactivating substitutions introduced into 2C were investigated to determine if these could be compensated by a replicon harboring a 3B3T>K substitution. This possibility would indicate that changing the temporal order of polyprotein processing does not prevent 2C function.
FIG 5.
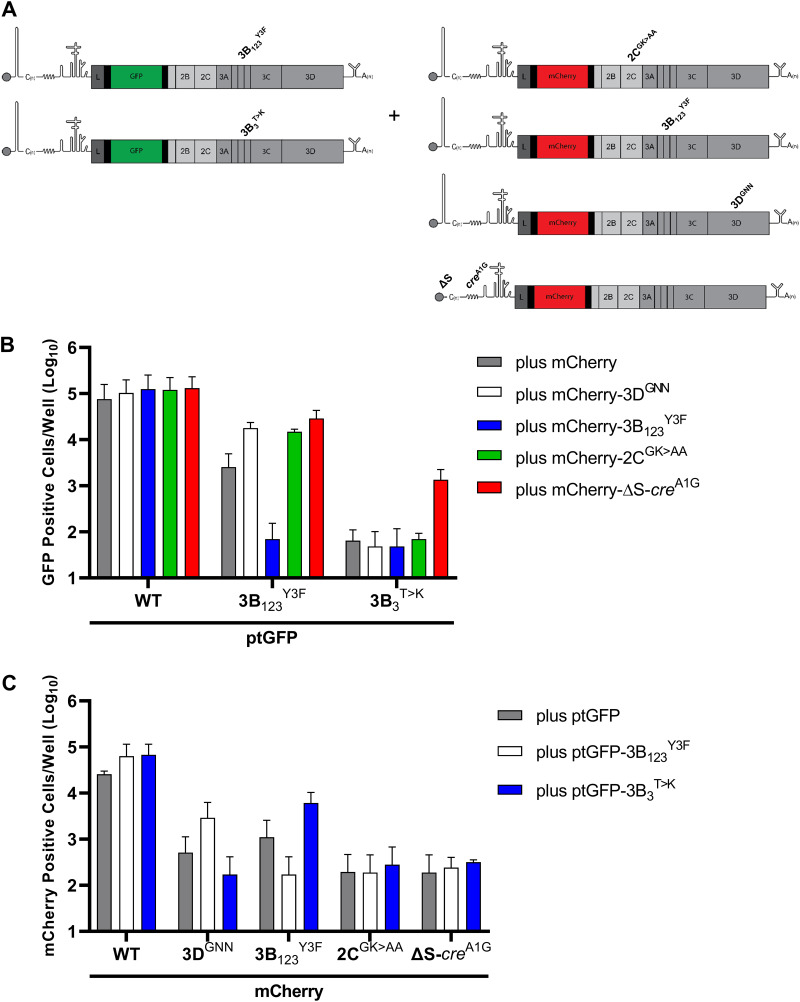
The 3B3T>K substitution prevents complementation of 2C mutants in trans. (A) Schematic of the trans-complementation experiment which involved co-transfecting BHK-21 cells with mCherry replicons containing replication-defective 2C or 3B mutations together with a WT ptGFP, ptGFP-3B3T>K or ptGFP-3DGNN replicon. Fluorescent protein expression was monitored hourly for 24 h. The data show (B) ptGFP positive cells per well or (C) mCherry positive cells per well at 8 h posttransfection (n = 2 ± SD).
To this end, replication-defective mCherry constructs were generated which contained inactivating substitutions at catalytic 2C residues (termed, mCherry-2CGK>AA) (4). This construct was co-transfected with the ptGFP-3B3T>K replicon (as used in Fig. 1C), or a WT ptGFP replicon control. As a positive control, co-transfections were also performed with an mCherry-3B1,2,3Y3F replicon (which contains inactivating point mutations to the triptych of 3B genes), which we have shown can be complemented in trans (16). Co-transfections were also performed with WT mCherry or ptGFP replicons to eliminate the possibility of any dominant negative effects and yeast tRNA to act as a negative control for no complementation (Fig. 5A). Replication was monitored by both ptGFP and mCherry expression and the number of fluorescent positive cells quantified at 8 h posttransfection, as documented previously (39). For brevity, the key data sets and controls are shown (Fig. 5B and C) with the complete data set shown in Fig. S1.
Replication of the WT mCherry or ptGFP replicon did not significantly change upon co-transfection with any of the RNAs tested, suggestive of no dominant negative effects. Replication of the mCherry-3B1,2,3Y3F replicon was significantly enhanced by the ptGFP-3B3T>K construct, as anticipated. The mCherry-2CGK>AA replicon was not recovered by any of the helper replicons, suggesting the functions of 2C cannot be provided in trans (Fig. 5C). We also noticed that no functional complementation was provided to the ptGFP-3B3T>K replicon by co-transfection with mCherry-3B1,2,3Y3F. To investigate this further, we extended our complementation experiments to include mCherry replicons containing mutations to cis-acting RNA replication elements such as the S-fragment or cre. The rationale here was that previous studies have demonstrated that deleting cis-acting replication elements can improve or allow recovery of replicons in trans (39), presumably by increasing the free pool of proteins which would otherwise be sequestered by cis interactions. When the ptGFP-3B3T>K replicon was co-transfected with a mCherry-ΔS or mCherry-creA1G replicon, we observed a significance increase in ptGFP expression, indicating that inactivation of these cis-acting replication elements permitted complementation of the ptGFP-3B3T>K replicon in trans (Fig. 5B). Together, these data suggest that the ptGFP-3B3T>K replicon can supply material in trans to recover replicons defective in 3B but not 2C or 3Dpol and can only receive complementation in trans from replicons with inactivating mutations or deletions to cis-acting RNA elements.
Complementation of replicons with the 3B3T>K substitution reveals a requirement for the nonstructural polyprotein.
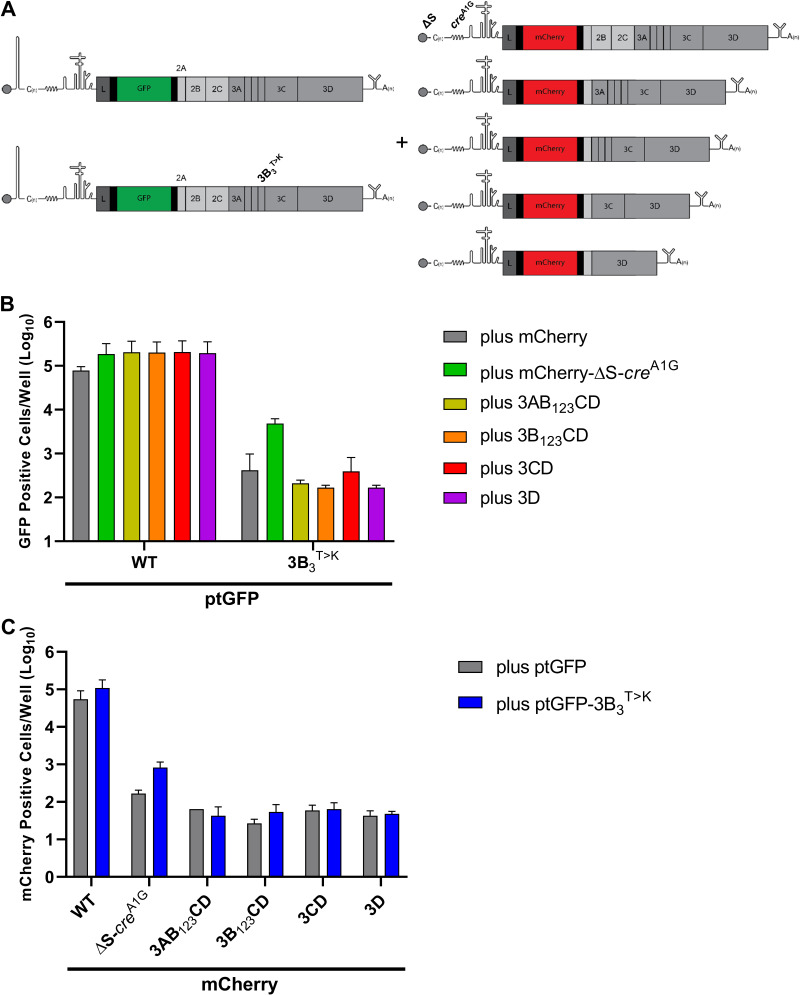
Having shown that deletion of cis-acting RNA elements allowed complementation of the ptGFP-3B3T>K replicon, we took advantage of this system to identify which protein component was required for complementation. To this end, we generated a new panel of mCherry replicons in which both the S-fragment and cre were inactivated but which expressed just a subset of the polyprotein, namely, 3AB1,2,3CD, 3B1,2,3CD, 3CD, or 3Dpol (Fig. 6A). With these constructs, we were able to probe whether the ptGFP-3B3T>K replicon is missing the ability to generate one of these protein components to initiate replication. This new panel of replicons was used in complementation assays with the same controls as described above, with replication monitored by both ptGFP and mCherry expression, and the number of fluorescent positive cells quantified at 8 h posttransfection. Again, for clarity, the key data sets and controls are shown (Fig. 6B and C) with the complete data set shown in Fig. S2.
FIG 6.
S-fragment deletions allow trans-complementation of cis-acting replication components. (A) Schematic of the trans-complementation experiment which involved co-transfecting BHK-21 cells with mCherry replicons containing S-fragment deletions together with a WT ptGFP, ptGFP-3B3T>K or ptGFP-3DGNN replicon. Fluorescent protein expression was monitored hourly for 24 h. The data show (B) ptGFP positive cells per well or (C) mCherry positive cells per well at 8 h posttransfection (n = 2 ± SD).
As described above, we found that replicons lacking a functional S-fragment and/or cre were able to significantly increase the replication of the ptGFP-3B3T>K replicon, with ptGFP expression increasing >10-fold compared to the controls. In contrast, no complementation of the ptGFP-3B3T>K replicon was observed when co-transfected with RNAs expressing just 3AB1,2,3CD, 3B1,2,3CD, 3CD, or 3Dpol. Hence, it would appear that 3AB1,2,3CD is not sufficient to support replication of a 3B3T>K replicon and provision of 2BC containing proteins is also required.
The 3B3T>K substitution does not prevent RNA-protein interactions.
A key observation from our trans-complementation work is that the 3B3T>K substitution prevents complementation of this nonfunctional replicon unless the cis-acting S-fragment or cre elements are deleted from the helper RNA. A possible explanation is that the altered cleavage pattern induced by the 3B3T>K substitution prevents replication components (e.g., the 2C helicase or 3Dpol) from interacting with the template RNA, thus preventing its replication.
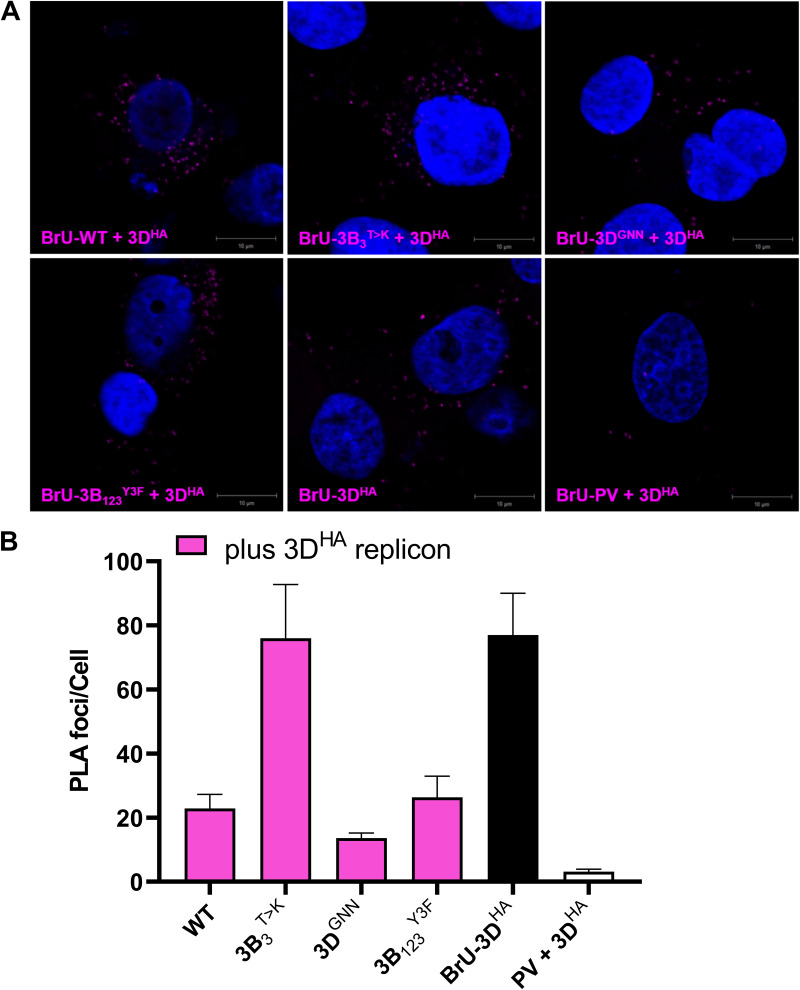
To investigate this possibility, we adapted a proximity ligation assay (PLA) to study interactions between replicon RNA and components of the replication complex. First, ptGFP-3B3T>K replicon RNA (or ptGFP-3DGNN and ptGFP-3B1,2,3Y3F controls) was in vitro transcribed in the presence of BrUTP to generate BrU-labeled replicon RNA. Alongside, a WT ptGFP was transcribed with BrUTP and used to confirm that BrU labeling had no significant inhibitory effect on replicon replication (data not shown). The ptGFP-3B3T>K BrU replicon was co-transfected into BHK-21 cells seeded on coverslips together with a WT replicon in which 3Dpol had been labeled with a HA epitope (termed 3DHA). HA epitopes are derived from the human influenza virus hemagglutinin (HA) protein and we have previously demonstrated that tagging 3Dpol with HA in this manner does not affect replication (39). The coverslips were fixed and the interaction between the BrU-3B3T>K RNA and 3DHA probed by PLA using anti-BrU and anti-HA antibodies. This allows RNA-protein interactions between the ptGFP-BrU-3B3T>K RNA and 3DHA protein to be measured in situ. A positive PLA signal indicates that the mutant replicon RNA can associate with enzymes of the replication complex provided in trans. No signal indicates a lack of association (Fig. 7A). The number of PLA foci per GFP positive cell was quantified from at least 20 cells per sample (Fig. 7B).
FIG 7.
Detection of FMDV RNA-protein complexes by PLA. (A) BHK-21 cells were co-transfected with BrU-labeled ptGFP replicon RNA together with 3DHA-labeled replicon RNA. At 4 h posttransfection cells were fixed and 3DHA-BrU RNA complexes detected by proximity ligation assay (PLA) using anti-HA and anti-BrU primary antibodies together with PLA-labeled secondary antibodies. The in-situ PLA signal is detected as foci in the cell cytoplasm (pseudo-colored magenta). Cell nuclei were stained with DAPI (pseudo-colored blue). Images were captured on a Zeiss LSM-880 confocal microscope (bar 10 μm). (B) The mean number of individual PLA foci per cell were quantified from an average of at least 20 GFP positive cells per condition spread across two biological replicate experiments (n = 2 ± SD).
Co-transfection of the WT ptGFP BrU labeled replicon with the WT 3DHA replicon generated PLA signals that were easily detectable, indicative of interactions between the ptGFP RNA and 3DHA protein as predicted from mixing of replication complex components. In contrast, little or no PLA signal was detected when a WT BrU labeled poliovirus replicon (BrU-PV) was co-transfected with the 3DHA helper replicon, with a ~40-fold decrease in the number of PLA foci/cell compared to the positive control. This suggests the PLA signal is generated through specific mixing of replication complexes. Co-transfection of the ptGFP-3B3T>K, ptGFP-3DGNN, or ptGFP-3B1,2,3Y3F BrU labeled replicon RNAs with the WT 3DHA replicon also generated PLA signals that were easily detectable, with ~40-, ~10-, and ~15-fold increase in PLA foci/cell compared to the BrU-PV control. This would suggest the 3B3T>K substitution does not prevent recruitment of its cognate RNA with a functional RNA polymerase.
DISCUSSION
All well-studied positive-sense RNA viruses produce polyproteins that help compensate for a relatively limited coding capacity. These polyproteins are processed by viral protease(s) to generate mature proteins via functional intermediates in a highly regulated manner that modulates viral replication. However, establishing how polyprotein processing regulates viral replication can be challenging due to the intricate nature of these interactions. In a previous study of the FMDV polyprotein, we showed that changing six amino acids at the 3B3-3C cleavage junction prevents viral replication by disrupting polyprotein processing (16). We postulated that this observation provided an opportunity to investigate the role of FMDV polyproteins in viral genome replication. First, we investigated whether consequences of substituting the six amino acids could be replicated by a single change. To do this, we substituted the P2 residue at the 3B3-3C cleavage junction, as our previous investigation showed this to be the most important for dictating cleavage efficiency. Using a GFP encoding replicon, we showed that a single substitution to lysine (3B3T>K) prevented viral replication and in in vitro translation assays changed polyprotein processing, as predicted.
To build a more complete picture of how the 3B3T>K construct changed polyprotein processing and understand the mechanism that underpinned this change, we employed a combination of in vitro translation, immunoprecipitation, and trans-cleavage assays in the context of the FMDV nonstructural polyprotein. We found that a 3B3-3C cleavage junction bearing this substitution was more efficiently cleaved by 3Cpro in trans compared to WT. Furthermore, the 3B3T>K substitution also rendered this cleavage junction sensitive to proteolysis by 3Cpro when the protease was present as part of a larger precursor (i.e., as part of another molecule of 3AB1,2,3CD). Thus, the consequence of introducing the 3B3T>K substitution was to generate a substrate that was more efficiently cleaved (potentially equal to or greater than either 2B-2C or 2C-3A cleavage sites). This changed the subset of proteins produced; generating 2BC3AB1,2,3 (not typically observed during infection [7, 41]), significantly increasing levels of 3CD and 2BC and reducing levels of 3AB1,2,3CD. The identity of these products was based on both molecular weight and immunoprecipitation experiments. The 3B3T>K substitution construct did generate fully cleaved 2B and 2C at a reduced rate, demonstrating that 3Cpro as part of 3CD is proteolytically active, but processing of 3AB1,2,3 and 3CD was severely impaired as observed previously (16). Further experiments to confirm the identity of these proteins could be conducted in replicon transfected cells, dependent on sufficient translation of input RNA for replication-defective replicons. It would also be interesting to investigate processing of the 3B3T>K substitute when placed in the context of polyprotein containing the capsid precursor (P1-2A). This would introduce another two cleavage sites that may subtly change the hierarchy of processing of the entire polyprotein.
Our data are consistent with biochemical investigations using purified 3Cpro which showed that charged residues in the P2 position of the cleavage junction are more efficiently recognized (40, 42–44). They are also consistent with the suggestion that 3Cpro retains activity as part of a larger precursor, albeit at lower efficiency (45–48). Our data therefore agree with a mechanism of polyprotein processing (suggested from studies with other picornaviruses) in which 3Cpro as part of a larger precursor can process the WT 2B-2C and/or 2C-3A junctions most efficiently in trans, hence, providing a level of regulation to the processing cascade. After these initial cleavage events, 3AB1,2,3CD is processed more slowly to allow intermediate and mature cleavage products to fulfil their roles in viral replication (36). Processing of the 3AB1,2,3CD precursor can thus proceed potentially through both cis and trans mechanisms (i.e., intra- and intermolecular cleavage) and can give rise to alternative precursors, for example, 3AB1,2,3C and 3CD. It is clearly suggested from our data that trans cleavage of one 3AB1,2,3CD molecule by another polyprotein is possible. However, the WT precursor was not processed efficiently by 3AB1,2,3CD in trans, and the observation that 3Cpro in the context of 3CD is proteolytically active, but only able to cleave efficiently at 2B-2C and 2C-3A junctions, could suggest that other factors (e.g., a cis mediated mechanism), are also involved. Picornavirus 3Cpro proteins are also implicated in RNA binding and lipid biogenesis (potentially as part of a precursor), and therefore may act as co-factors to dictate processing pathways (29, 49, 50). A study by Escarmís et al. previously demonstrated that a single amino acid substitution in FMDV VP1 had the ability to affect a protein processing between VP1 and VP3 by 3Cpro, despite the substitution being located 54 amino acids away from the cleavage site. This is another example of distal effects amino acid substitutions can have on proteolytic processing patterns (51).
Using trans-complementation assays, we investigated the function of these different sets of precursors in FMDV replication. A replicon containing the 3B3T>K substitution (i.e., producing increased levels of 2BC, 3AB1,2,3, and 3CD) was able to complement defective mutations in the 3B proteins but not 2C or 3Dpol, suggesting that this replicon can produce active primers for replication, 3B1, 3B2, and/or 3B3 but inactive replication enzymes, 2C and 3Dpol. Interestingly, the 3B3T>K replicon was only complemented by a replicon lacking cis-acting RNA replication elements (cre, S-fragment or both) and when the protein components were provided as part of an entire polyprotein. However, the PLA data suggested that the 3B3T>K substitution does not prevent recruitment of its cognate RNA with 3Dpol in trans but instead potentially increases the association. The increased association between the 3B3T>K RNA and 3Dpol provided by a WT replicon in trans may suggest the formation of more stable RNA-protein complexes that are detrimental to replication.
One interpretation of our data combined is that a correct functional interaction between the nonstructural polyprotein and viral RNA elements is required to generate functional enzymes for replication (e.g., 2C and 3Dpol but not 3B). Hence, if processing of the precursor occurs too rapidly, it cannot associate correctly with these RNA elements required for replication, or generates a complex that is too stable (Fig. 8). These mechanisms could provide a level of temporal control of replication. This could also explain the greater number of foci seen in the PLA experiments when ptGFP-3B3T>K BrU-labeled replicons were co-transfected with 3DHA replicons (compared to co-transfections of ptGFP-3DGNN or ptGFP-3B1,2,3Y3F BrU-labeled replicon RNAs with the WT 3DHA replicons). A similar mechanism has been suggested for PV where two molecules of 3ABCD are required for replication, one which produces 3CD that interacts with viral RNA structures while one produces enzymatically active 3Dpol (32, 36, 52, 53). This two-molecule model of processing of 3ABCD would also be compatible with data that suggest that a larger precursor (such as 3BC) is required to deliver 3B for RNA replication (15, 54, 55). This work extends the growing body of evidence suggesting that processing intermediates are essential for controlling temporally and structurally the organization of the picornavirus replication organelle.
FIG 8.
Model for regulation of FMDV replication by polyprotein processing. The 3B3T>K substitution drives rapid processing preferentially down one pathway, as demonstrated by the reduced levels of of 2C and 3Dpol with increased formation of 3AB1,2,3 and 3CD. To support replication of the 3B3T>K replicon in trans, the entire non structural polyprotein is required to deliver a complex of functional components including 2C and 3Dpol, as demonstrated by the wild-type processing schematic. This likely provides products from alternative cleavage pathways such as 3B1,2,3 and 3CD.
MATERIALS AND METHODS
Cell lines and plasmids.
BHK-21 cells obtained from the ATCC (LGC Standard) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with glutamine (Sigma-Aldrich) supplemented with 10% FCS, 50 U/mL penicillin, and 50 μg/mL streptomycin.
Plasmids carrying wild-type FMDV replicons, pRep-mCherry and pRep-ptGFP, have already been described (38, 56), along with equivalent plasmids containing 3DGNN, 3B3/2 and ΔS-fragment mutations (16, 38, 39). Mutations within these plasmids were performed by standard two-step overlapping PCR mutagenesis. For coupled in vitro transcription and translation experiments pcDNA3.1(+) based expression plasmids were generated by PCR. Briefly, the relevant FMDV sequence was amplified to include flanking NotI restriction enzymes and upstream Kozak modified translational start site. The NotI digested PCR products were cloned into NotI digested pcDNA3.1(+) (Thermo Fisher Scientific). The sequence of all plasmids used in this study was confirmed by Sanger sequencing. The sequences of all primers and plasmids are available on request.
Coupled transcription and translation reactions.
Coupled in vitro transcription and translation assays were performed using the TNT Quick Coupled Transcription/Translation system (Promega) as described previously (16). Reactions contained 10 μL lysate with 250 ng of pcDNA T7 expression plasmid and 0.5 μL [35S] methionine/cysteine (PerkinElmer). Reactions were incubated at 30°C for 40 min chasing with 2 μL of 50 mg/mL unlabeled methionine/cysteine. Reactions were stopped at 20-min or hourly intervals by the addition of 2× Laemmli buffer. Samples were separated by SDS-PAGE before visualization of radiolabeled products by autoradiography.
For the trans-cleavage assays, the TNT reactions were supplemented with purified FMDV 3Cpro (a kind gift from Dr. Tobias Tuthill [57]) to the indicated final concentration from dilution of a 1 mM stock, simultaneous to the addition of unlabeled methionine/cysteine. Reactions were stopped at 20-min or hourly intervals by the addition of 2× Laemmli buffer and the 3Cpro-mediated proteolysis of radiolabeled precursor monitored by SDS-PAGE.
In vitro transcription.
Plasmids containing cDNA copies of FMDV replicons were linearized with AscI before being used to generate T7 in vitro transcribed RNA as previously described (38, 39). The reaction was incubated at 32°C for 4 h before being treated with DNase for 20 min at 37°C then purified using an RNA clean and concentrate kit (Zymo Research). The RNA quality was checked using a MOPS/formaldehyde agarose gel electrophoresis.
Replication and complementation assays.
BHK-21 cells were seeded into 24-well tissue cultures vessels, allowed to adhere overnight for 16 h, before duplicate wells were transfected with 1 μg of each in vitro transcribed RNA using Lipofectin (Thermo Fisher Scientific) as previously described (38). For co-transfection complementation assays, 500 ng of each RNA molecules were mixed prior to the addition of Lipofectin reagent as previously described (39). Fluorescent reporter expression was monitored using an IncuCyte Zoom Dual Color FLR (Essen BioSciences) live-cell imaging system housed within a humidified incubator scanning hourly up to 24 h posttransfection. Images were captured and analyzed using the associated software for fluorescent protein expression, as previously described (39). Control transfections (untransfected and the 3DGNN transfection for input translation) were used to determine fluorescent thresholds and identify positive objects from background fluorescence. A positive object was determined as having an average fluorescent intensity of >8 green calibration units (GCU; an arbitrary fluorescent unit) and >2 RCU (red calibration units), which were kept constant throughout the experiments. The number of positive cells per well was determined from the average of up to nine nonoverlapping images per well. Unless stated otherwise, data are presented as mean fluorescent positive cells per well at 8 h posttransfection when replication was approximately maximal. For each experiment, the data were analyzed as both fluorescent cell counts per well and total fluorescent intensity per well. There was no difference observed when the data were analyzed in either way. Unless otherwise stated, statistical analysis was performed using a two-tailed unpaired t test.
Immunofluorescence and proximity ligation assays.
BHK-21 cells seeded onto coverslips were co-transfected with replicon RNA before fixing in 4% paraformaldehyde and washing with PBS. Immunofluorescence was conducted as previously described (39). Primary antibodies used were sheep anti-BrdU (Sigma-Aldrich), rabbit anti-FMDV 3D (a kind gift from Francisco Sobrino), and mouse anti-HA (Sigma-Aldrich). Proximity ligation assays (PLA) were conducted using the Duolink In Situ Red Kit (Sigma-Aldrich), following manufacturer’s instructions. Images from more than 20 GFP positive cells per condition were analyzed using the “Find maxima” function on ImageJ to count PLA foci across conditions with prominence set to strictly >25.
Immunoprecipitation.
Immunoprecipitation reactions were performed using Dynabeads Protein G (Invitrogen). To bind the antibody to magnetic beads, 5 μL of the FMDV 2C antibody (a kind gift from Francisco Sobrino) was mixed with 195 μL PBS and incubated at room temperature with 50 μL magnetic beads, shaken for 1 h, after which the supernatant was removed from the beads. Transcription and translation reaction samples were mixed with 200 μL PBS and incubated shaking at room temperature with 25 μL of Dynabeads as a preclear step. The tube was placed on the magnet and the supernatant removed. This was added to the 50 μL of Dynabeads with the 2C antibody bound and incubated at room temperature shaking for 1 h. The flow through was removed and added to 2× Laemmli buffer. The beads were washed three times with PBS pH 7.4 with 0.02% Tween 20 and each wash supernatant retained. Proteins were eluted from the beads by adding 50 μL of 2× Laemmli buffer and heating to 100°C.
ACKNOWLEDGMENTS
We thank Francisco Sobrino (Centro De Biologia Molecular Severo Ochoa, Madrid) for the gift of FMDV primary antibodies and Toby Tuthill (The Pirbright Institute, Pirbright) for the gift of purified 3C.
M.R.H., N.J.S., and D.J.R. designed the study and wrote the manuscript; D.M.P. conducted the in vitro translation experiments; D.M.P. and M.R.H. conducted the replication assays; M.R.H. conducted the immunofluorescence assays; D.M.P., C.H., and M.R.H. analyzed the data; M.R.H., N.J.S., and D.J.R. provided supervision.
This work was supported by BBSRC grant (BB/T015748/1) awarded to M.R.H., D.J.R., and N.J.S. D.M.P. and C.H. were funded by BBSRC DTP studentships (BB/M011151/1). M.R.H. was also supported by the MRC (MR/S007229/1). The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Correspondence and materials requests should be directed to M.R.H.
Footnotes
Supplemental material is available online only.
Contributor Information
Nicola J. Stonehouse, Email: n.j.stonehouse@leeds.ac.uk.
Morgan R. Herod, Email: m.r.herod@leeds.ac.uk.
Susana Lopez, Instituto de Biotecnologia/UNAM.
REFERENCES
- 1.Yost SA, Marcotrigiano J. 2013. Viral precursor polyproteins: keys of regulation from replication to maturation. Curr Opin Virol 3:137–142. doi: 10.1016/j.coviro.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moffat K, Knox C, Howell G, Clark SJ, Yang H, Belsham GJ, Ryan M, Wileman T. 2007. Inhibition of the secretory pathway by foot-and-mouth disease virus 2BC protein is reproduced by coexpression of 2B with 2C, and the site of inhibition is determined by the subcellular location of 2C. J Virol 81:1129–1139. doi: 10.1128/JVI.00393-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu Z, Wang G, Yang F, Cao W, Mao R, Du X, Zhang X, Li C, Li D, Zhang K, Shu H, Liu X, Zheng H. 2016. Foot-and-mouth disease virus Viroporin 2B antagonizes RIG-I-mediated antiviral effects by inhibition of its protein expression. J Virol 90:11106–11121. doi: 10.1128/JVI.01310-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sweeney TR, Cisnetto V, Bose D, Bailey M, Wilson JR, Zhang X, Belsham GJ, Curry S. 2010. Foot-and-mouth disease virus 2C is a hexameric AAA+ protein with a coordinated ATP hydrolysis mechanism. J Biol Chem 285:24347–24359. doi: 10.1074/jbc.M110.129940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrillo C, Tulman ER, Delhon G, Lu Z, Carreno A, Vagnozzi A, Kutish GF, Rock DL. 2005. Comparative genomics of foot-and-mouth disease virus. J Virol 79:6487–6504. doi: 10.1128/JVI.79.10.6487-6504.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts PJ, Belsham GJ. 1995. Identification of critical amino acids within the foot-and-mouth disease virus leader protein, a cysteine protease. Virology 213:140–146. doi: 10.1006/viro.1995.1554. [DOI] [PubMed] [Google Scholar]
- 7.Vakharia VN, Devaney MA, Moore DM, Dunn JJ, Grubman MJ. 1987. Proteolytic processing of foot-and-mouth disease virus polyproteins expressed in a cell-free system from clone-derived transcripts. J Virol 61:3199–3207. doi: 10.1128/JVI.61.10.3199-3207.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strebel K, Beck E. 1986. A second protease of foot-and-mouth disease virus. J Virol 58:893–899. doi: 10.1128/JVI.58.3.893-899.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donnelly MLL, Luke G, Mehrotra A, Li X, Hughes LE, Gani D, Ryan MD. 2001. Analysis of the aphthovirus 2A/2B polyprotein 'cleavage' mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal 'skip'. J Gen Virol 82:1013–1025. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- 10.Kitamura N, Semler BL, Rothberg PG, Larsen GR, Adler CJ, Dorner AJ, Emini EA, Hanecak R, Lee JJ, van der Werf S, Anderson CW, Wimmer E. 1981. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature 291:547–553. doi: 10.1038/291547a0. [DOI] [PubMed] [Google Scholar]
- 11.Pallansch MA, Kew OM, Semler BL, Omilianowski DR, Anderson CW, Wimmer E, Rueckert RR. 1984. Protein processing map of poliovirus. J Virol 49:873–880. doi: 10.1128/JVI.49.3.873-880.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Y, Sun SQ, Guo HC. 2016. Biological function of foot-and-mouth disease virus non-structural proteins and non-coding elements. Virol J 13:107. doi: 10.1186/s12985-016-0561-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grubman MJ, Baxt B. 2004. Foot-and-mouth disease. Clin Microbiol Rev 17:465–493. doi: 10.1128/CMR.17.2.465-493.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bedard KM, Semler BL. 2004. Regulation of picornavirus gene expression. Microbes Infect 6:702–713. doi: 10.1016/j.micinf.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Oh HS, Pathak HB, Goodfellow IG, Arnold JJ, Cameron CE. 2009. Insight into poliovirus genome replication and encapsidation obtained from studies of 3B-3C cleavage site mutants. J Virol 83:9370–9387. doi: 10.1128/JVI.02076-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herod MR, Gold S, Lasecka-Dykes L, Wright C, Ward JC, McLean TC, Forrest S, Jackson T, Tuthill TJ, Rowlands DJ, Stonehouse NJ. 2017. Genetic economy in picornaviruses: foot-and-mouth disease virus replication exploits alternative precursor cleavage pathways. PLoS Pathog 13:e1006666. doi: 10.1371/journal.ppat.1006666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.den Boon JA, Ahlquist P. 2010. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol 64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- 18.Belsham GJ, Brangwyn JK. 1990. A region of the 5' noncoding region of foot-and-mouth disease virus RNA directs efficient internal initiation of protein synthesis within cells: involvement with the role of L protease in translational control. J Virol 64:5389–5395. doi: 10.1128/JVI.64.11.5389-5395.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez de Quinto S, Martinez-Salas E. 1997. Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J Virol 71:4171–4175. doi: 10.1128/jvi.71.5.4171-4175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez de Quinto S, Saiz M, de la Morena D, Sobrino F, Martinez-Salas E. 2002. IRES-driven translation is stimulated separately by the FMDV 3'-NCR and poly(A) sequences. Nucleic Acids Res 30:4398–4405. doi: 10.1093/nar/gkf569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mason PW, Bezborodova SV, Henry TM. 2002. Identification and characterization of a cis-acting replication element (cre) adjacent to the internal ribosome entry site of foot-and-mouth disease virus. J Virol 76:9686–9694. doi: 10.1128/jvi.76.19.9686-9694.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forss S, Schaller H. 1982. A tandem repeat gene in a picornavirus. Nucleic Acids Res 10:6441–6450. doi: 10.1093/nar/10.20.6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul AV, Peters J, Mugavero J, Yin J, van Boom JH, Wimmer E. 2003. Biochemical and genetic studies of the VPg uridylylation reaction catalyzed by the RNA polymerase of poliovirus. J Virol 77:891–904. doi: 10.1128/jvi.77.2.891-904.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul AV, van Boom JH, Filippov D, Wimmer E. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 25.Falk MM, Sobrino F, Beck E. 1992. VPg gene amplification correlates with infective particle formation in foot-and-mouth disease virus. J Virol 66:2251–2260. doi: 10.1128/JVI.66.4.2251-2260.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paul AV, Rieder E, Kim DW, van Boom JH, Wimmer E. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J Virol 74:10359–10370. doi: 10.1128/jvi.74.22.10359-10370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nayak A, Goodfellow IG, Belsham GJ. 2005. Factors required for the Uridylylation of the foot-and-mouth disease virus 3B1, 3B2, and 3B3 peptides by the RNA-dependent RNA polymerase (3Dpol) in vitro. J Virol 79:7698–7706. doi: 10.1128/JVI.79.12.7698-7706.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kloc A, Diaz-San Segundo F, Schafer EA, Rai DK, Kenney M, de Los Santos T, Rieder E. 2017. Foot-and-mouth disease virus 5'-terminal S fragment is required for replication and modulation of the innate immune response in host cells. Virology 512:132–143. doi: 10.1016/j.virol.2017.08.036. [DOI] [PubMed] [Google Scholar]
- 29.Amero CD, Arnold JJ, Moustafa IM, Cameron CE, Foster MP. 2008. Identification of the oriI-binding site of poliovirus 3C protein by nuclear magnetic resonance spectroscopy. J Virol 82:4363–4370. doi: 10.1128/JVI.02087-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcotte LL, Wass AB, Gohara DW, Pathak HB, Arnold JJ, Filman DJ, Cameron CE, Hogle JM. 2007. Crystal structure of poliovirus 3CD protein: virally encoded protease and precursor to the RNA-dependent RNA polymerase. J Virol 81:3583–3596. doi: 10.1128/JVI.02306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiang W, Harris KS, Alexander L, Wimmer E. 1995. Interaction between the 5'-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J Virol 69:3658–3667. doi: 10.1128/JVI.69.6.3658-3667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gamarnik AV, Andino R. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5' untranslated region of the poliovirus genome. J Virol 74:2219–2226. doi: 10.1128/jvi.74.5.2219-2226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris KS, Xiang W, Alexander L, Lane WS, Paul AV, Wimmer E. 1994. Interaction of poliovirus polypeptide 3CDpro with the 5' and 3' termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J Biol Chem 269:27004–27014. doi: 10.1016/S0021-9258(18)47118-9. [DOI] [PubMed] [Google Scholar]
- 34.Parsley TB, Towner JS, Blyn LB, Ehrenfeld E, Semler BL. 1997. Poly (rC) binding protein 2 forms a ternary complex with the 5'-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y, Rijnbrand R, Watowich S, Lemon SM. 2004. Genetic evidence for an interaction between a picornaviral cis-acting RNA replication element and 3CD protein. J Biol Chem 279:12659–12667. doi: 10.1074/jbc.M312992200. [DOI] [PubMed] [Google Scholar]
- 36.Spear A, Ogram SA, Morasco BJ, Smerage LE, Flanegan JB. 2015. Viral precursor protein P3 and its processed products perform discrete and essential functions in the poliovirus RNA replication complex. Virology 485:492–501. doi: 10.1016/j.virol.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Towner JS, Mazanet MM, Semler BL. 1998. Rescue of defective poliovirus RNA replication by 3AB-containing precursor polyproteins. J Virol 72:7191–7200. doi: 10.1128/JVI.72.9.7191-7200.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herod MR, Loundras EA, Ward JC, Tulloch F, Rowlands DJ, Stonehouse NJ. 2015. Employing transposon mutagenesis to investigate foot-and-mouth disease virus replication. J Gen Virol 96:3507–3518. doi: 10.1099/jgv.0.000306. [DOI] [PubMed] [Google Scholar]
- 39.Herod MR, Ferrer-Orta C, Loundras EA, Ward JC, Verdaguer N, Rowlands DJ, Stonehouse NJ. 2016. Both cis and trans activities of foot-and-mouth disease virus 3D polymerase are essential for viral RNA replication. J Virol 90:6864–6883. doi: 10.1128/JVI.00469-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sweeney TR, Roque-Rosell N, Birtley JR, Leatherbarrow RJ, Curry S. 2007. Structural and mutagenic analysis of foot-and-mouth disease virus 3C protease reveals the role of the beta-ribbon in proteolysis. J Virol 81:115–124. doi: 10.1128/JVI.01587-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clarke BE, Sangar DV. 1988. Processing and assembly of foot-and-mouth disease virus proteins using subgenomic RNA. J Gen Virol 69:2313–2325. doi: 10.1099/0022-1317-69-9-2313. [DOI] [PubMed] [Google Scholar]
- 42.Birtley JR, Curry S. 2005. Crystallization of foot-and-mouth disease virus 3C protease: surface mutagenesis and a novel crystal-optimization strategy. Acta Crystallogr D Biol Crystallogr 61:646–650. doi: 10.1107/S0907444905007924. [DOI] [PubMed] [Google Scholar]
- 43.Birtley JR, Knox SR, Jaulent AM, Brick P, Leatherbarrow RJ, Curry S. 2005. Crystal structure of foot-and-mouth disease virus 3C protease. New insights into catalytic mechanism and cleavage specificity. J Biol Chem 280:11520–11527. doi: 10.1074/jbc.M413254200. [DOI] [PubMed] [Google Scholar]
- 44.Zunszain PA, Knox SR, Sweeney TR, Yang J, Roque-Rosell N, Belsham GJ, Leatherbarrow RJ, Curry S. 2010. Insights into cleavage specificity from the crystal structure of foot-and-mouth disease virus 3C protease complexed with a peptide substrate. J Mol Biol 395:375–389. doi: 10.1016/j.jmb.2009.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis GJ, Wang QM, Cox GA, Johnson RB, Wakulchik M, Dotson CA, Villarreal EC. 1997. Expression and purification of recombinant rhinovirus 14 3CD proteinase and its comparison to the 3C proteinase. Arch Biochem Biophys 346:125–130. doi: 10.1006/abbi.1997.0291. [DOI] [PubMed] [Google Scholar]
- 46.Jurgensen D, Kusov YY, Facke M, Krausslich HG, Gauss-Muller V. 1993. Cell-free translation and proteolytic processing of the hepatitis A virus polyprotein. J Gen Virol 74:677–683. doi: 10.1099/0022-1317-74-4-677. [DOI] [PubMed] [Google Scholar]
- 47.Probst C, Jecht M, Gauss-Muller V. 1998. Processing of proteinase precursors and their effect on hepatitis A virus particle formation. J Virol 72:8013–8020. doi: 10.1128/JVI.72.10.8013-8020.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ypma-Wong MF, Dewalt PG, Johnson VH, Lamb JG, Semler BL. 1988. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology 166:265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]
- 49.Banerjee S, Aponte-Diaz D, Yeager C, Sharma SD, Ning G, Oh HS, Han Q, Umeda M, Hara Y, Wang RYL, Cameron CE. 2018. Hijacking of multiple phospholipid biosynthetic pathways and induction of membrane biogenesis by a picornaviral 3CD protein. PLoS Pathog 14:e1007086. doi: 10.1371/journal.ppat.1007086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shengjuler D, Chan YM, Sun S, Moustafa IM, Li ZL, Gohara DW, Buck M, Cremer PS, Boehr DD, Cameron CE. 2017. The RNA-binding site of poliovirus 3C protein doubles as a phosphoinositide-binding domain. Structure 25:1875–1886. doi: 10.1016/j.str.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Escarmis C, Perales C, Domingo E. 2009. Biological effect of Muller's Ratchet: distant capsid site can affect picornavirus protein processing. J Virol 83:6748–6756. doi: 10.1128/JVI.00538-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toyoda H, Franco D, Fujita K, Paul AV, Wimmer E. 2007. Replication of poliovirus requires binding of the poly(rC) binding protein to the cloverleaf as well as to the adjacent C-rich spacer sequence between the cloverleaf and the internal ribosomal entry site. J Virol 81:10017–10028. doi: 10.1128/JVI.00516-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen M, Reitman ZJ, Zhao Y, Moustafa I, Wang Q, Arnold JJ, Pathak HB, Cameron CE. 2008. Picornavirus genome replication. Identification of the surface of the poliovirus (PV) 3C dimer that interacts with PV 3Dpol during VPg uridylylation and construction of a structural model for the PV 3C2-3Dpol complex. J Biol Chem 283:875–888. doi: 10.1074/jbc.M707907200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pathak HB, Oh HS, Goodfellow IG, Arnold JJ, Cameron CE. 2008. Picornavirus genome replication: roles of precursor proteins and rate-limiting steps in oriI-dependent VPg uridylylation. J Biol Chem 283:30677–30688. doi: 10.1074/jbc.M806101200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nayak A, Goodfellow IG, Woolaway KE, Birtley J, Curry S, Belsham GJ. 2006. Role of RNA structure and RNA binding activity of foot-and-mouth disease virus 3C protein in VPg uridylylation and virus replication. J Virol 80:9865–9875. doi: 10.1128/JVI.00561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tulloch F, Pathania U, Luke GA, Nicholson J, Stonehouse NJ, Rowlands DJ, Jackson T, Tuthill T, Haas J, Lamond AI, Ryan MD. 2014. FMDV replicons encoding green fluorescent protein are replication competent. J Virol Methods 209:35–40. doi: 10.1016/j.jviromet.2014.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Newman J, Asfor AS, Berryman S, Jackson T, Curry S, Tuthill TJ. 2018. The cellular chaperone heat shock protein 90 is required for foot-and-mouth disease virus capsid precursor processing and assembly of capsid pentamers. J Virol 92. doi: 10.1128/JVI.01415-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Download jvi.00171-23-s0001.docx, DOCX file, 0.06 MB (60.4KB, docx)
Fig. S2. Download jvi.00171-23-s0002.docx, DOCX file, 0.06 MB (62.6KB, docx)