Abstract
Development and regeneration are orchestrated by gene regulatory networks that operate in part through transcriptional enhancers. Although many enhancers are pleiotropic and are active in multiple tissues, little is known about whether enhancer pleiotropy is due to 1) site pleiotropy, in which individual transcription factor binding sites (TFBS) are required for activity in multiple tissues, or 2) multiple distinct sites that regulate expression in different tissues. Here, we investigated the pleiotropy of an intronic enhancer of the stickleback Bone morphogenetic protein 6 (Bmp6) gene. This enhancer was previously shown to regulate evolved changes in tooth number and tooth regeneration, and is highly pleiotropic, with robust activity in both fins and teeth throughout embryonic, larval, and adult life, and in the heart and kidney in adult fish. We tested the hypothesis that the pleiotropy of this enhancer is due to site pleiotropy of an evolutionarily conserved predicted Foxc1 TFBS. Transgenic analysis and site-directed mutagenesis experiments both deleting and scrambling this predicted Foxc1 TFBS revealed that the binding site is required for enhancer activity in both teeth and fins throughout embryonic, larval, and adult development, and in the heart and kidney in adult fish. Collectively these data support a model where the pleiotropy of this Bmp6 enhancer is due to site pleiotropy and this putative binding site is required for enhancer activity in multiple anatomical sites from the embryo to the adult.
Keywords: Enhancer, Enhancer pleiotropy, Site pleiotropy, Transgenesis, Tooth, Stickleback
1. Introduction
Eukaryotic gene expression is governed by enhancers, non-coding cis-regulatory elements that positively regulate transcription. Enhancers bind transcription factors that promote cell-type-specific gene expression programs throughout development and drive transcription in response to stimuli (Levine et al., 2014). Decades of genetic studies have revealed that most human disease phenotypes (Farh et al., 2015; Maurano et al., 2012), and much of natural phenotypic variation in animals are associated with sequence changes in non-coding DNA (Rebeiz and Tsiantis, 2017). These findings suggest that enhancers play a critical role in both evolution and disease, thereby motivating further study of basic enhancer biology.
Many enhancers are known to function in multiple distinct tissues, suggesting that these elements are pleiotropic. A study on chromatin from a subset of human tissues found that at least 1% of identified cis-regulatory elements were active in at least two spatially distinct domains (Singh and Yi, 2021). In vivo studies in fish and mice have additionally identified enhancer sequences that drive gene expression in more than one tissue type (Cleves et al., 2018; Erickson et al., 2015; Jackman and Stock, 2006; Jumlongras et al., 2012; Stepaniak et al., 2021). Two different described mechanisms of enhancer pleiotropy are (1) tightly clustered, but different, transcription factor binding sites (TFBSs) that drive expression in different tissues and (2) site pleiotropy, where a single TFBS drives expression in multiple tissues (Preger-Ben Noon et al., 2018). These two mechanisms of enhancer pleiotropy have different implications for evolution: site pleiotropy would constrain evolution to preserve the integrity of critical regulatory sequences, as a single mutation in the TFBS could have a significant impact on gene expression in multiple tissues (Boffelli et al., 2004; Fish et al., 2017; Infante et al., 2015; Sabarís et al., 2019). In contrast, tightly clustered TFBSs could allow for more modular evolution of gene expression in different tissues or at different developmental stages.
Threespine stickleback fish (Gasterosteus aculeatus) are a powerful system to study enhancer biology, as abundant natural variation throughout their adaptive radiation occurs largely due to non-coding variation, pointing to the importance of enhancers in phenotypic evolution (Jones et al., 2012). For example, changes in tooth number have evolved repeatedly in sticklebacks, with derived freshwater populations having increases in tooth number and tooth regeneration rates relative to ancestral marine populations (Cleves et al., 2014; Ellis et al., 2015). Quantitative trait loci mapping revealed a large effect locus that contains the gene Bone Morphogenetic Protein 6 (Bmp6), which is dynamically expressed in both epithelial and mesenchymal cells within developing teeth (Cleves et al., 2014). Further high-resolution genetic mapping of this large effect locus linked the evolved phenotypic effects to an enhancer in the fourth intron of Bmp6 (Cleves et al., 2018). This intronic enhancer is highly pleiotropic and drives expression in all developing and regenerating teeth, as well as the distal edges of the pectoral, median, and caudal fins (Cleves et al., 2018). Comparing the marine and freshwater versions of this intronic enhancer in doubly transgenic lines revealed evolved spatial shifts in enhancer activity in both tooth epithelium and mesenchyme, consistent with an evolved change in enhancer activity driving phenotypic evolution of tooth number (Stepaniak et al., 2021). In addition, although both enhancers drive strong expression in the distal edges of embryonic fins, only the freshwater enhancer was detected in fin ray joints in pectoral and caudal fins (Stepaniak et al., 2021). However, whether the enhancer utilizes common inputs for tooth and fin activity, and which sites are required for this activity are unknown.
Previous studies have found that fish teeth and mammalian hair share many aspects of early development: both are derived from placodes (Pispa and Thesleff, 2003), continuously regenerate in adults, are regulated by BMP signaling during replacement (Jia et al., 2013; Vainio et al., 1993; Wang et al., 2012), and express similar batteries of genes during regeneration (Square et al., 2021). In mice, BMP signaling has been proposed as a major regulator of maintaining stem cell quiescence. Conditional ablation of the Bmpr1a gene in skin epithelium activated the stem cell niche, causing quiescent hair follicle stem cells to proliferate (Kandyba et al., 2013; Kobielak et al., 2007). Further, Foxc1 regulates Bmp6 expression in regenerating hair. Foxc1 binds to a regulatory region adjacent to Bmp6 in mice and inhibits hair regeneration, as conditional knockout of Foxc1 in skin results in accelerated hair regeneration (Wang et al., 2016). Moreover, Foxc1 was proposed to be a candidate regulator of the Bmp6 intron 4 enhancer, as a putative Foxc1 TFBS was identified within this enhancer (Cleves et al., 2018). Given that this enhancer is expressed in regenerating teeth and has been linked to evolved changes in tooth regeneration, and the similarities between hair and tooth regeneration, a parsimonious model is that BMP signaling negatively regulates mammalian hair and fish tooth regeneration using homologous gene regulatory networks. Here we test the hypotheses that (1) a predicted Foxc1 TFBS is required for Bmp6 enhancer activity in developing and regenerating teeth and (2) this predicted Foxc1 TFBS has site pleiotropy and is required for enhancer activity in teeth and fins.
2. Methods
2.1. Animal husbandry
All animal work was approved by UC Berkeley IACUC protocol AUP-2015–01–7117. Sticklebacks were raised as previously described (Cleves et al., 2014; Square et al., 2021). All experiments used lab-reared marine (Rabbit Slough, Alaska) sticklebacks.
2.2. Generation of transgenic GFP enhancer stickleback lines
Tol2 plasmid transgenesis was performed using pT2HE plasmid backbone as previously described (Erickson et al., 2016; O’Brown et al., 2015). To generate GFP reporter constructs, PCR-based site-directed mutagenesis was done on plasmids containing the ~1300 bp Paxton benthic (PAXB) freshwater high-tooth-associated intron four enhancer allele (Cleves et al., 2018; Stepaniak et al., 2021). Primers for site-directed mutagenesis were designed using the Quickchange tool (https://www.agilent.com/store/primerDesignProgram.jsp) and PCR was carried out according to Erickson et al. (2015). Primer sequences used for the deleted construct were CTAATTACCCCGACGAGGTCG GGTGGGGAG and CTCCCCACCCGACCTCGTCGGGGTAATTAG and for the scrambled construct CCCCACCCGACCTTTTGAATACCGTCGGGG TAATTA and TAATTACCCCGACGGTATTCAAAAGGTCGGGTGGGG. Transposase messenger RNA was synthesized as described previously (Kawakami and Shima, 1999). Stickleback embryos at the one-cell stage were microinjected as described (Erickson et al., 2016). Injected embryos rarely (~<5%) display atypical patterns of GFP fluorescence, possibly the result of position effects. To raise founders, typical injected embryos were grown up for germline transmission. Two stable transgenic lines were generated with each of the intact, scrambled, and deleted reporter constructs.
2.3. Imaging fluorescent transgenic sticklebacks
Transgenic GFP reporter lines were imaged using a Leica M165FC stereoscope with a DFC340 FX camera, using the same exposure time for each series of images being compared within each figure. Transgenic embryos were imaged live. Hearts and kidneys were imaged in unfixed adults. Transgenic juvenile and adult fish were fixed at 4 °C in 4% paraformaldehyde for 4 h for 9–19 mm standard length (SL) or overnight (>20 mm SL). Teeth were imaged using Montage z-stack projections on a Leica M165FC dissecting microscope with a GFP2 filter.
2.4. Bioinformatics
The following genome assemblies were used to compare Bmp6 sequences: stickleback Gasterosteus aculeatus, “Gac”: Broad/gasAcu, medaka Oryzias latipes, “Ola”: NIG/UT MEDAKA1/oryLat2, zebrafish Danio rerio, “Dre”: GRCz10/danRer10, gar Lepisosteus oculatus “Loc”: LepOcu1 (GCA_000242695.1). A ~25 kb window centered on stickleback Bmp6 was aligned to orthologous sequences from these three other species using mVISTA and LAGAN at https://genome.lbl.gov/vista/index.shtml (Frazer et al., 2004) using default visualization parameters of 100 bp windows showing minimum y-axis of 50% and 70% sequence identity to color windows as conserved. The entire fourth intron sequence from all four species was aligned as above, and the first 5.5 kb of the ~6.5 kb intron shown in Fig. 1C. Sequences orthologous to the minimally sufficient ~500 bp Bmp6 intron 4 enhancer (Cleves et al., 2018) were aligned using Clustal Omega at https://www.ebi.ac.uk/Tools/msa/clustalo/(Sievers et al., 2011) and default parameters. The resulting text alignment was opened in MS Word and conserved bases in all four species highlighted manually.
Fig. 1.
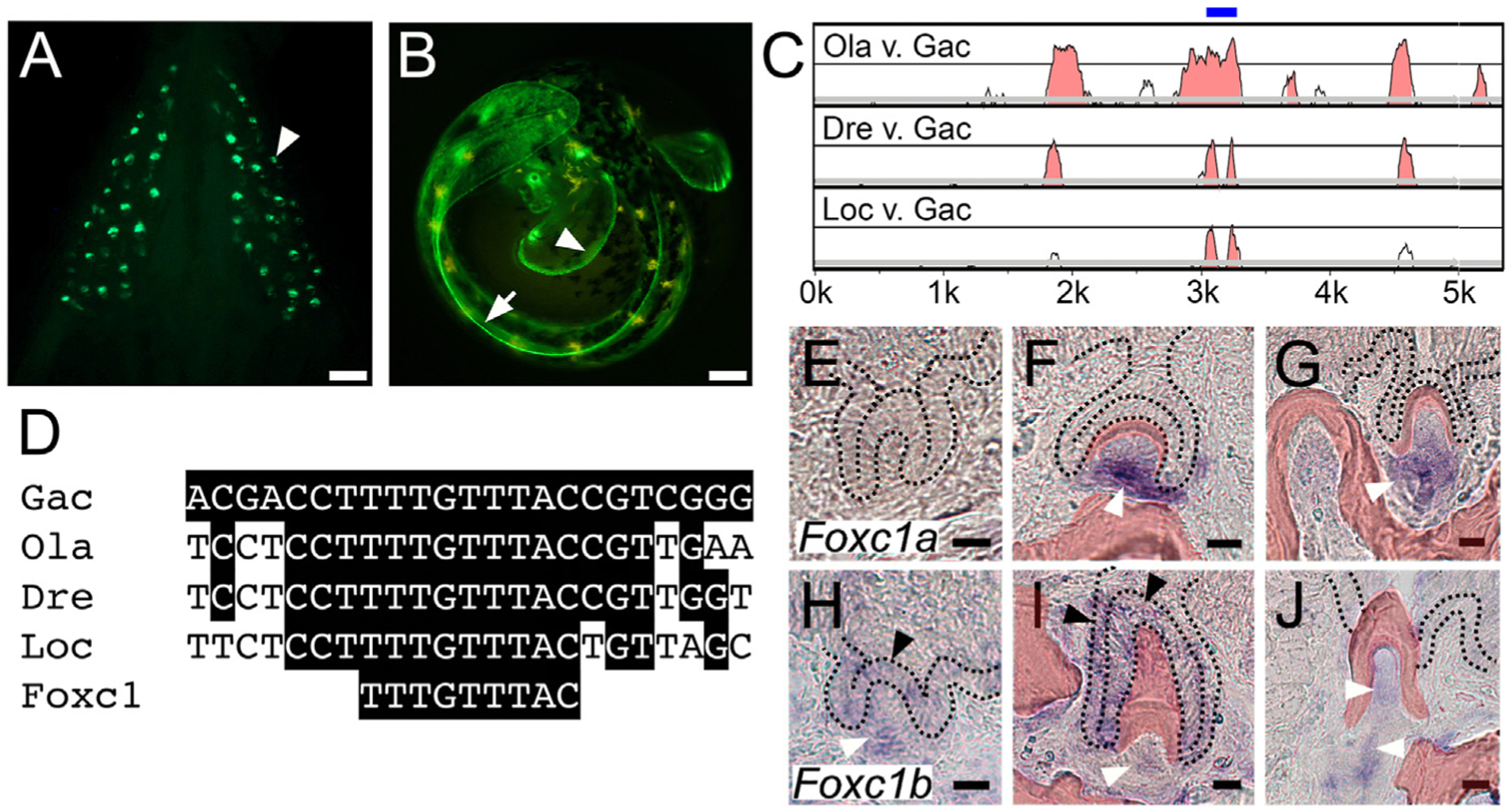
A pleiotropic tooth and fin enhancer includes an evolutionarily conserved predicted Foxc1 binding site and Foxc1 genes are expressed in teeth. GFP reporter gene driven by a 1.3 kb Bmp6 intronic enhancer (Cleves et al., 2018; Stepaniak et al., 2021) reveals expression in (A) ventral pharyngeal teeth (arrowhead) of juvenile stickleback fish and (B) distal edge of pectoral (arrowhead) and median (arrow) fin fold of unhatched 7 dpf developing stickleback embryo. (C) mVISTA LAGAN sequence alignment showing central region of stickleback Bmp6 intron four is conserved to gar. Intron four of stickleback Bmp6 (x-axis) is aligned to medaka (top), zebrafish (middle) and gar (bottom). Graph shows a sliding window of 100 base pairs covering the first ~5.5 kb of this ~7 kb intron, with y-axis starting at 50% identity and windows shaded at a threshold of 70% nucleotide identity. The blue bar above the peaks indicates the central ~200 bp island of conservation (see Fig. S1). Gac = Gasterosteus aculeatus (stickleback), Ola = Oryzias latipes (medaka), Dre = Danio rerio (zebrafish), Loc = Lepisosteus oculatus (gar). (D) Predicted Foxc1 binding site TTTGTTTAC (Berry et al., 2008) within the central island of conservation is conserved from sticklebacks to gar. In situ hybridization of Foxc1a (E–G) and Foxc1b (H–J) expression on sections of adult stickleback pharyngeal teeth at three different stages: mid bell (E,H), late bell (F,I), and erupted (G,J). Foxc1a expression was detected in dental mesenchyme at late bell and erupted stages (white arrowheads), but not at mid bell. Foxc1b expression was detected in tooth germ epithelium at mid bell (black arrowhead in H) and in both the inner (left black arrowhead) and outer dental epithelium (top black arrowhead) at late bell (I). Foxc1b was also detected in tooth mesenchyme at all three stages (white arrowheads, H-J). Black dotted lines demarcate the dental epithelium, and bone and dentine is pseudocolored red (see Fig. S2 for unlabeled panels and DAPI staining). Scale bars = 10 μm.
2.5. In situ hybridizations on sections
Sample preparation, sectioning, in situ hybridization, and riboprobe syntheses were carried out as previously described (Square et al., 2021). Riboprobes were designed as described previously (Ellis et al., 2016) against Foxc1a and Foxc1b. Riboprobe template plasmids were created by PCR cloning gene fragments from genomic DNA using primers to Foxc1a: 5′-GCCGctcgagGGGACAGGTCTAGCCACTTG-3’; 5′-GCCGtctagaACGGCGATATACACGTTCCT-’3 and Foxc1b: 5’-GCCGctcgagCCTGCCCGACTATTGCATCA-3’; 5′- GCCGactagtAGACGGCACTTTATTAAACAAACA-3.
3. Results
Previous research mapped evolved increases in tooth number in freshwater sticklebacks to an enhancer located in the fourth intron of Bmp6 (Cleves et al., 2014, 2018). This enhancer drives robust expression in all developing pharyngeal and oral teeth, as well as in the distal margins of the pectoral and median fins (Fig. 1A and B) (Cleves et al., 2018; Stepaniak et al., 2021). The sequence of this intronic enhancer is evolutionarily conserved in teleosts, with clear homology detectable in medaka and zebrafish, as well as to gar, an outgroup to teleosts (Fig. 1C and D). Comparison of enhancer sequences within fish and outgroups suggests different levels of constraint within this enhancer, possibly representing functionally required TFBSs. Of those regions with 100% sequence conservation across the aforementioned fish species, one includes a nine base pair motif TTTGTTTAC that perfectly matches the binding site consensus previously reported for human FOXC1 (Fig. 1D, Fig. S1) (Berry et al., 2008).
Teleosts underwent a whole genome duplication (Amores et al., 1998), resulting in pairs of co-orthologs of many teleost genes relative to outgroups. In sticklebacks, both Foxc1a and Foxc1b have been maintained. Expression of both Foxc1 genes was detected in developing adult teeth, with Foxc1a largely restricted to the dental mesenchyme at late bell and eruption stages but not at mid bell (Fig. 1E–G). Foxc1b expression was detected in dental epithelium of germs at mid bell stages (Fig. 1H), throughout the inner and outer dental epithelium at late bell stages (Fig. 1I), and in dental mesenchyme at all three stages examined (Fig. 1H–J). Thus, Foxc1 gene expression appears to overlap with all dental domains of the Bmp6 intronic enhancer in teeth (Stepaniak et al., 2021).
To test the hypothesis that the predicted Foxc1 TFBS is required for enhancer activity, we introduced mutations in a Bmp6 reporter construct previously described (Cleves et al., 2018; Stepaniak et al., 2021). The unaltered construct drove GFP expression in several tissues including the pectoral and median fins, and oral and pharyngeal teeth. Site directed mutagenesis was used to make two mutant enhancer variants: (1) a scrambled enhancer, where two thymine nucleotides in the predicted Foxc1 TFBS were mutated to adenine, and (2) a deleted enhancer, where the entire nine base pair Foxc1 predicted TFBS was deleted (Fig. 2A). We generated two stable transgenic lines each for three different enhancer transgenes: intact wild-type, scrambled, and deleted (Fig. 2A). For all three genotypes, we imaged transgenic larvae, juveniles, and adults to test for possible effects on enhancer activity in different tissues. All scrambled and deleted transgenes dramatically reduced GFP expression in the pectoral and median fins of the developing embryo (Fig. 2B).
Fig. 2.
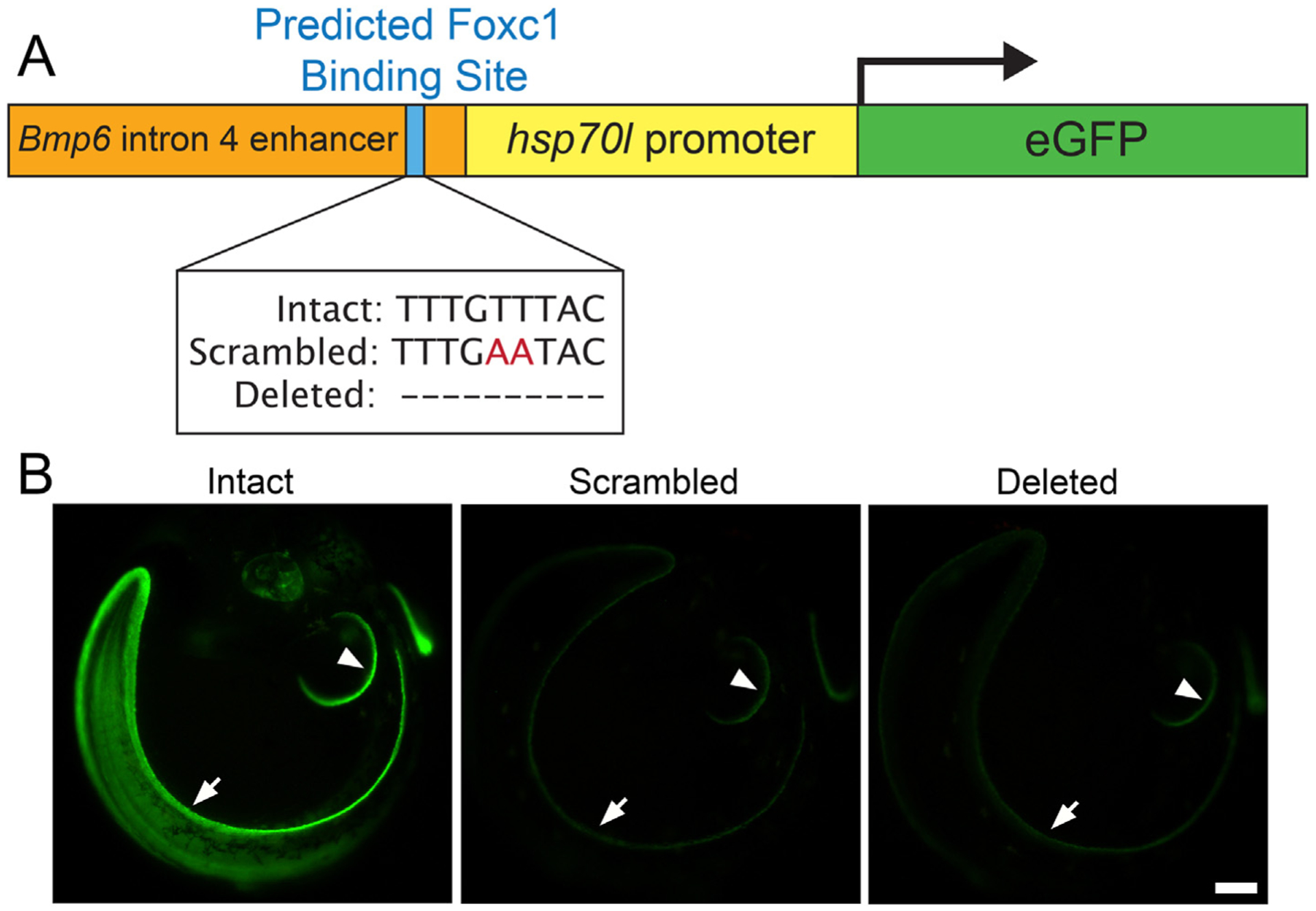
Predicted Foxc1 binding site is required for embryonic enhancer activity. (A) Schematic of GFP reporter construct with Bmp6 intronic enhancer with intact, scrambled, and deleted sequences for the predicted Foxc1 binding site upstream of the hsp70l promoter driving eGFP. (B) GFP reporter expression pattern driven by the Bmp6 intronic enhancer with intact, scrambled, or deleted sequence for the Foxc1 binding site in 7 dpf (stage 23, (Swarup, 1958)) stickleback embryos. Strong expression was seen in the distal edge of the developing pectoral fin (arrowhead) and median fin (arrow) for the enhancer with an intact sequence of the predicted Foxc1 binding site (left). Severely reduced GFP expression in all domains of enhancer activity was detected for enhancers with the scrambled (middle) or deleted (right) sequences of the predicted Foxc1 binding site. Scale bar = 100 μm.
While the stable transgenic reporter lines containing the intact binding site drove robust expression in every detectable tooth in the pharyngeal and oral jaw, neither the scrambled nor deleted reporter lines drove detectable GFP expression in pharyngeal teeth (Fig. 3). As an internal positive control, all transgenic constructs to test enhancer activity used the zebrafish hsp70l promoter, which is strongly active in the lens of the eye in sticklebacks (Erickson et al., 2015). This lens domain serves as a positive control for the promoter, GFP fluorophore, and permissive integration site of the transgenes, and was comparably bright in the intact, scrambled, and deleted transgenic lines (insets in Fig. 3A). Within developing primary oral and pharyngeal teeth, transgene GFP expression was observed in the dental epithelium and mesenchyme for the intact enhancer, but in neither epithelium nor mesenchyme for the scrambled or deleted enhancer in pharyngeal teeth (Fig. 3). GFP expression was also seen in the epithelium and mesenchyme of adult teeth for the intact enhancer. Faint GFP expression was detected in some oral teeth in the premaxilla at adult stages for the scrambled enhancer (Fig. 3C). Overall, adult tooth expression was largely abolished in the scrambled and deleted enhancer reporter lines, suggesting that the predicted Foxc1 TFBS is required for enhancer activity in replacement as well as primary teeth (Fig. 3).
Fig. 3.
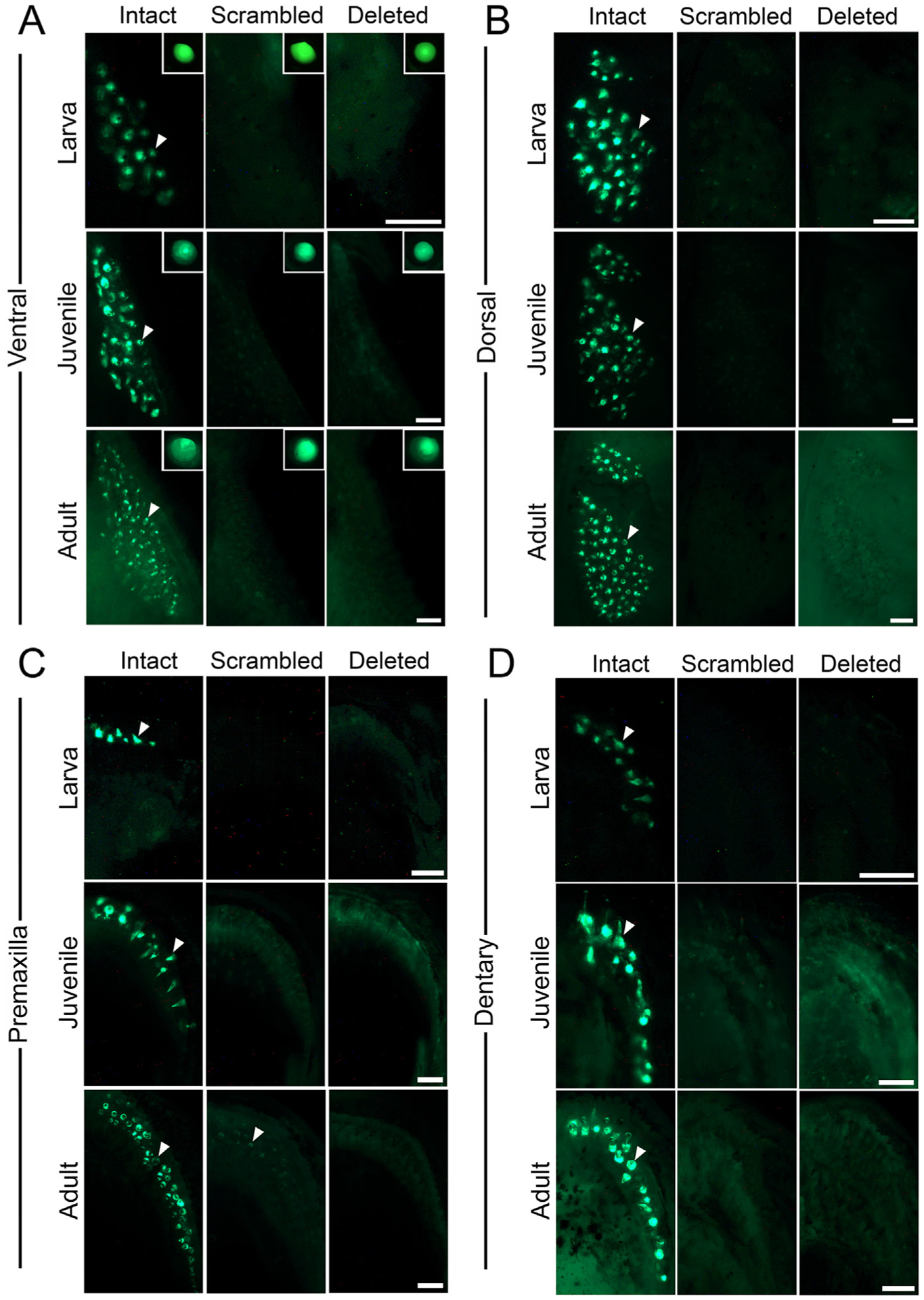
Predicted Foxc1 binding site is required for pharyngeal and oral tooth expression in larva, juveniles, and adults. GFP reporter gene expression in ventral (A,D) and dorsal (B,C) pharyngeal (A,B) and oral (C,D) jaws in larva (7–11 mm SL), juvenile (17–21 mm SL), and adult (37–55 mm SL) sticklebacks. GFP expression was detected in the epithelium and mesenchyme of developing teeth (arrowheads) in larva, juvenile, and adult stages with the intact sequence of the predicted Foxc1 binding site (left columns). No expression was detected in any developing teeth of larva, juvenile, and adults with the scrambled or deleted sequence of the predicted Foxc1 binding site (middle and right columns, respectively), except for faint expression in some adult premaxilla teeth (arrow in adult premaxilla panel in C). (A insets) Lens of eye with GFP expression as an internal control driven by the hsp70l promoter used in the transgenic construct. Scale bars = 100 μm (larva and juvenile), 250 μm (adult).
Additionally, we found that the predicted Foxc1 TFBS is required for enhancer function in fins at all developmental time points examined (Fig. 4). The intact enhancer drove robust reporter gene expression in the distal edges of the pectoral and caudal fins at early larval stages, in intersegmental joints of the juvenile and adult pectoral and caudal fins, and also at the base of the caudal fin throughout the caudal peduncle at all stages examined, including in cells adjacent to the fused hypural fan and the parhypural (Fig. 4) (Bowne, 1994), All fin domains were nearly abolished in both the scrambled and deleted reporter lines at all developmental stages (Fig. 4). GFP expression was detected in rare intersegmental joints of the pectoral fins in juveniles for the scrambled enhancer, and in the base of the pectoral fin rays for both the scrambled and deleted construct (Fig. 4A). Some GFP expression persisted in the caudal peduncle for the deleted enhancer (Fig. 4B). In general, all sites of fin expression, like tooth expression, were severely reduced in both the scrambled and deleted enhancer lines relative to the intact enhancer lines. These data support a crucial role for this single putative Foxc1 binding site in positively regulating both tooth and fin expression across development from embryos to adults.
Fig. 4.
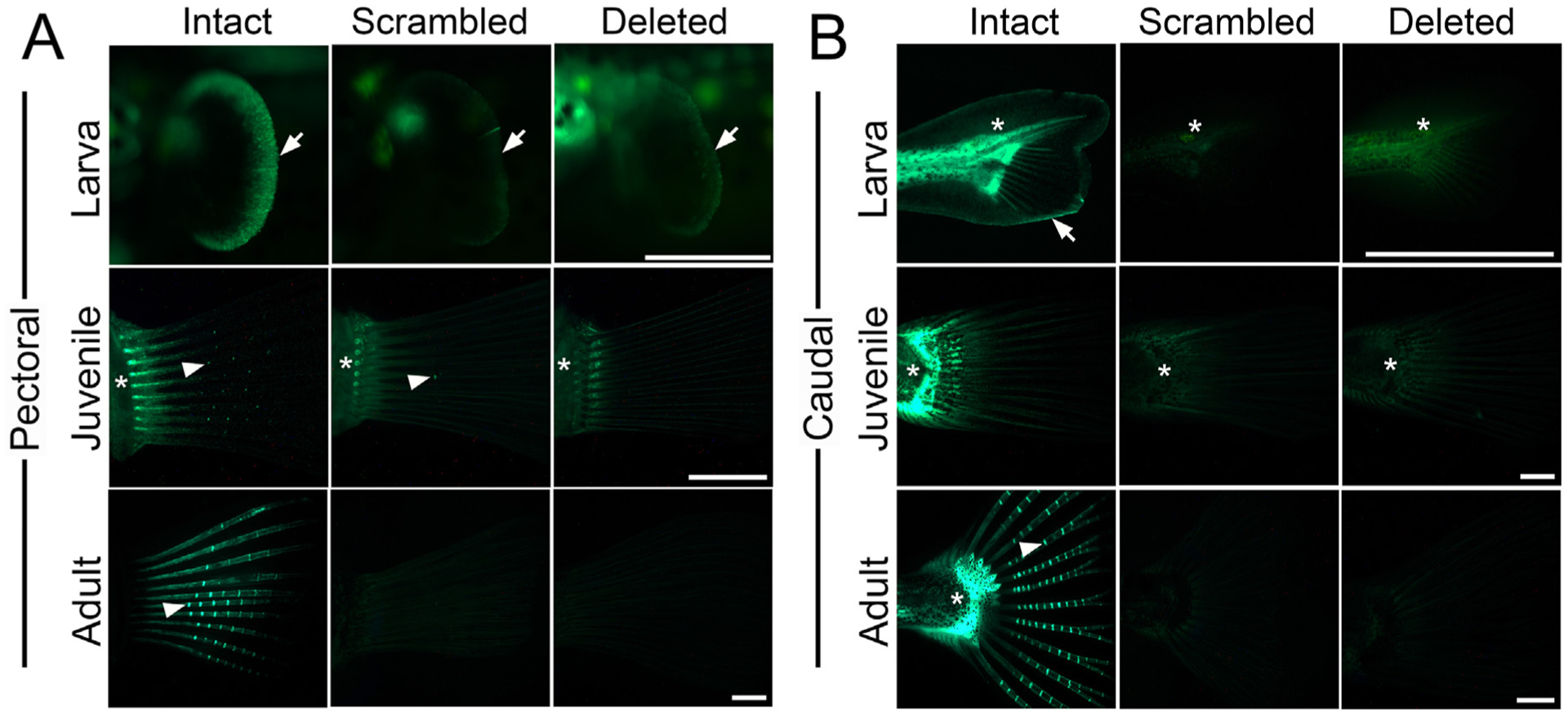
Predicted Foxc1 binding site is required for fin expression in larvae, juveniles, and adults. GFP reporter gene expression in the left pectoral fin (A) and caudal fin (B) of larvae, juveniles and adults. (A) Expression was detected in the distal edges of the pectoral fin (arrows) in larvae, the base of the pectoral fin rays (asterisk), and the intersegmental joints (arrowheads) of juveniles and adults with the intact sequence of the predicted Foxc1 binding site (left column). Severely reduced expression was seen in the distal edges of the pectoral fin in larvae and the intersegmental joints of juveniles and adults with the scrambled or deleted sequence of the predicted Foxc1 binding site (middle and right columns, respectively), while some expression at the base of the fin rays remained. (B) GFP reporter expression in the caudal fin. In larvae, expression was observed in the distal edge of the median fin fold (arrow) and caudal peduncle (asterisk) with the intact enhancer, but was severely reduced with the scrambled and deleted enhancers. Some peduncle expression remained with the deleted enhancer (right). In juveniles and adults, expression was detected in the peduncle (asterisks) and the intersegmental joints of the caudal fin (arrowheads) for the intact enhancer, but not for the scrambled or deleted enhancer. Scale bars = 500 μm (larva), 1 mm (juvenile), 2 mm (adult).
To test for possible additional internal sites of enhancer expression in adult fish, we dissected three adult fish from both lines of the intact, scrambled, and deleted constructs. We discovered robust GFP expression driven by the intact enhancer in the adult heart and kidney (Fig. 5). In the adult heart, brighter GFP expression appeared to be present in the atrium than the ventricle (Fig. 5A). In contrast, using the same exposure time (139 ms), expression was not detected in the heart in either of the scrambled lines or either of the deleted lines (Fig. 5A–C). However, we noticed that increasing exposure time to 2 seconds revealed that the first scrambled line had faint expression throughout the heart, again with the atrium appearing brighter than the ventricle. In contrast, in the second scrambled line, the remaining GFP expression was fainter, with only the atrium having detectable GFP expression (Fig. S3). In adults, and in larval and juvenile fish, no additional sites of expression were detected for either of the deleted or scrambled lines.
Fig. 5.
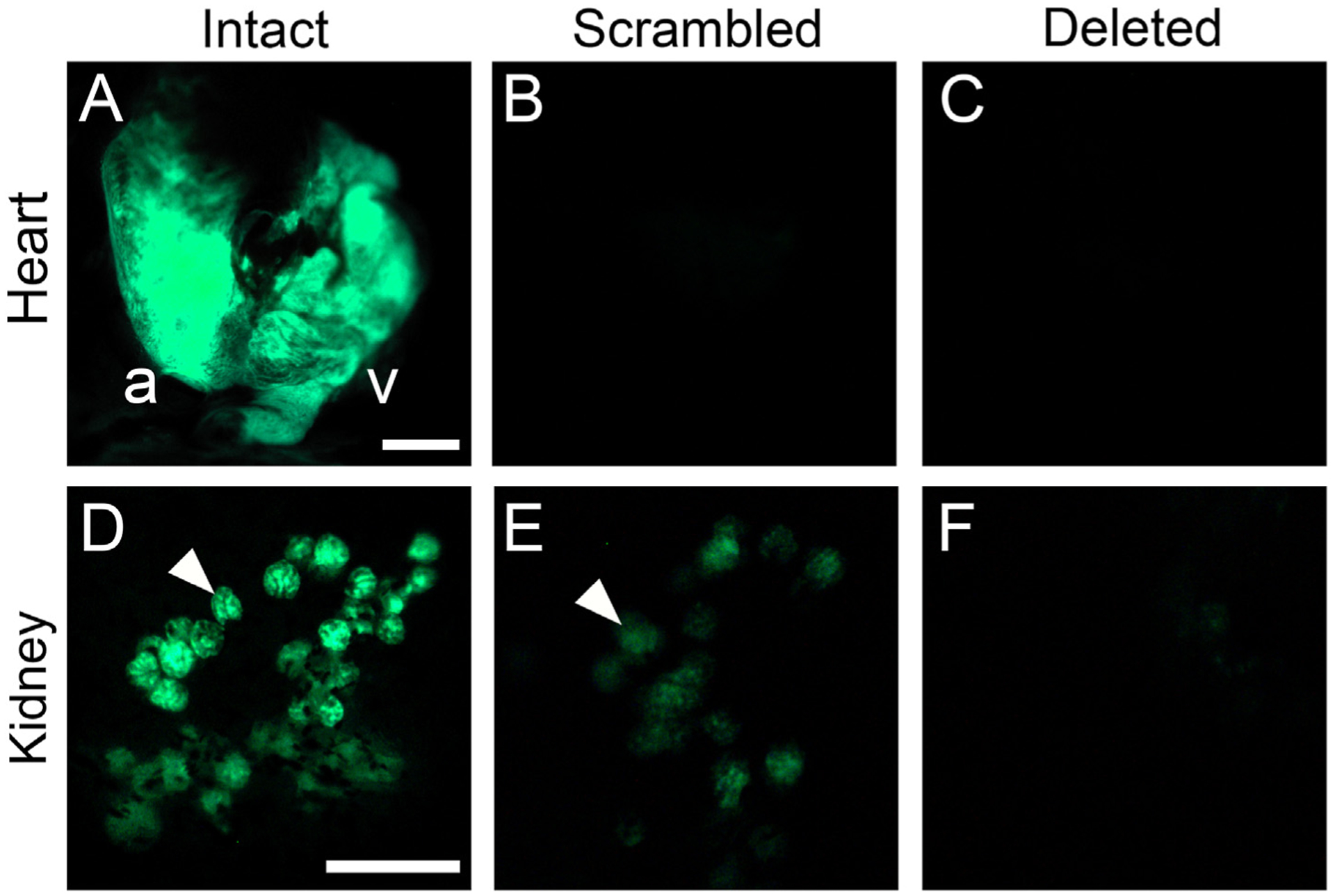
Predicted Foxc1 binding site is required for heart and kidney expression in adults. The intact enhancer drove robust GFP reporter gene expression in the adult heart (A), with overall stronger expression present in the atrium than ventricle. At the same exposure time (139 ms) used for the intact transgene, no heart expression was detected for either the scrambled (B) or deleted (C) constructs (but see Fig. S3). The intact enhancer also drove robust GFP expression in renal corpuscles (arrowhead) of the kidney (D). This domain was severely reduced for the scrambled construct (E) and not detectable for the deleted construct (F). Abbreviations: a = atrium, v = ventricle. Scale bars = 1 mm (A–C), 200 μm (D–F).
4. Discussion
4.1. A short, conserved, pleiotropic binding site is required for enhancer activity in teeth, fins, heart, and kidney
Here we show a predicted Foxc1 TFBS is required for Bmp6 intron four enhancer activity in teeth and fins at all stages examined of stickleback development, and in the heart and kidney of adult fish. Altering the predicted Foxc1 TFBS by scrambling two nucleotides or deleting nine nucleotides via site-directed mutagenesis severely downregulated enhancer activity in teeth, fins, heart, and kidney, suggesting that this predicted TFBS is critical for enhancer function in multiple expression domains. This site pleiotropy is reminiscent of a previously described enhancer 5′ to the stickleback Bmp6 gene, where a conserved predicted SMAD3 binding site is required for enhancer activity in teeth and fins (Erickson et al., 2015). We previously showed some spatial differences between the 5′ Bmp6 enhancer and the intronic Bmp6 enhancer studied here, including expression in early tooth germ epithelium and gill rakers driven by the 5′ enhancer but not the intronic enhancer (Erickson et al., 2015; Cleves et al., 2018). Deletion of the 5′ enhancer resulted in severe reductions in endogenous expression of Bmp6 in both fins and teeth (Erickson et al., 2015), suggesting this intronic enhancer cannot fully compensate for loss of the 5′ enhancer. Thus, while the 5’ and intronic enhancers drive overlapping expression in many tissues, they do not seem redundant as proposed for shadow enhancers (Kvon et al., 2021).
While both marine and freshwater intron four Bmp6 enhancers drive strong expression in the distal edges of embryonic fins, only the freshwater enhancer was detected in distal intersegmental joints in pectoral and caudal fins (Stepaniak et al., 2021). The intact enhancer tested here was the freshwater allele and drove robust fin ray joints as previously reported. Both early distal fin fold and later fin joint expression domains driven by the intact enhancer were largely absent in the scrambled and deleted Foxc1 TFBS reporter lines. In addition, no unique domains of expression were detected in any of the scrambled or deleted lines, suggesting no repressive roles for the Foxc1 predicted TFBS and also suggesting minimal position effects within these transgenic lines. However, the quantitative differences in adult heart expression levels between the two scrambled lines suggest position effects can contribute to enhancer activity and need to be controlled for by generating multiple stable transgenic lines.
Enhancers have displayed both site pleiotropy and tightly linked binding sites driving expression in different tissues at different developmental stages. A study investigating the shavenbaby enhancers in Drosophila found that while one enhancer used the same TFBS to regulate embryonic and pupal expression (site pleiotropy), another enhancer required distinct tightly linked binding sites for expression in different tissues (Preger-Ben Noon et al., 2018). One interesting but unanswered question is whether enhancers with site pleiotropy are more likely to be evolutionarily conserved, as site pleiotropy likely constrains enhancer sequence evolution. We note that we discovered this intronic enhancer not by using sequence conservation, but instead by using a combined QTL mapping and genome resequencing approach (Cleves et al., 2018).
While we hypothesize that this predicted TFBS studied here is bound by Foxc1, it may serve as a binding site for other Fox or non-Fox transcription factors with similar binding site affinity. It is possible that in generating scrambled or deleted TFBSs, one or more new TFBSs were generated. However, it would be unlikely that both deleted and scrambled nucleotides would create the same TFBS, given the different primary sequence of these two mutations. Once stickleback Foxc1 antibodies are available, biochemical experiments could test whether stickleback Foxc1a and/or Foxc1b binds this predicted TFBS. Genetic experiments, crossing this enhancer into Foxc1a and Foxc1b single and double mutants could directly test whether either or both of these duplicate Foxc1 genes regulate this pleiotropic enhancer. In addition, these experiments will test for functional requirements for Foxc1 genes in regulating tooth development and/or regeneration.
4.2. Roles for Foxc1 in BMP regulation
Previous studies have revealed Foxc1 as a molecular switch for regulating regeneration in mouse hair follicles. As a transcription factor, Foxc1 activates BMP signaling in self-renewing hair follicle stem cells (HFSCs) (Wang et al., 2016). The conditional knockout of Foxc1 in mouse results in reduced quiescence in HFSCs and an increase in hair regeneration rates. RNA sequencing (RNA-Seq) and quantitative polymerase chain reaction (qPCR) revealed that genes associated with quiescence of HFSCs, like Bmp6, were down-regulated. The direct regulation of Bmp6 by Foxc1 was supported by chromatin immunoprecipitation PCR (Wang et al., 2016). Here we suggest Foxc1 regulates the BMP pathway for tooth, and possibly fin, heart, and kidney development and regeneration through this TFBS in Bmp6.
4.3. Foxc1 in organogenesis and chromatin remodeling
Previous studies of enhancer evolution have suggested that enhancers are born as “proto-enhancers” with an initial low-information content but containing a TFBS that serves as a nucleation point around which other TFBSs then evolve (Emera et al., 2016). The winged-helix structure of the forkhead DNA-binding domain, which is highly conserved across all members of the Fox family, resembles the structure of the linker histone H1, a key modulator of chromatin structure. Because of its structure and chromatin binding interactions, FoxA proteins have been proposed to act as pioneer transcription factors, opening compacted chromatin to allow the binding of other transcription factors (Cirillo et al., 2002; Lee et al., 2005). Foxd3 has been shown to act as a pioneer factor by re-arranging the chromatin landscape and opening cis-regulatory elements to maintain multipotency in the early neural crest lineage (Lukoseviciute et al., 2018). Together, these studies suggest Fox family transcription factors promote chromatin accessibility within promoter and enhancer regions.
Mutations in Foxc1 co-orthologs (foxc1a and foxc1b) in zebrafish results in severe reductions of upper facial cartilages as well as missing trabecular cartilage of the neurocranium. The zebrafish foxc1a−/− mutants died by 7 dpf, and displayed facial cartilage defects and foxc1b−/− mutants displayed a truncation in the symplectic cartilage (Xu et al., 2018). In double homozygous foxc1a; foxc1b mutants, severe reductions of upper facial cartilages occurred, suggesting that Foxc1a and Foxc1b act redundantly in upper facial cartilage development (Xu et al., 2018). In zebrafish embryos doubly homozygous foxc1a and foxc1b mutations, the cartilaginous skeleton did not form properly, likely because many of the regulatory sequences in cartilage regulatory genes were not accessible (Xu et al., 2021). These results support a model in which zebrafish Foxc1 proteins serve as pioneer transcription factors to open chromatin, making it accessible for other transcription factors to bind and regulate genomic regions responsible for cartilage development. It is possible that stickleback Foxc1 proteins may serve similar roles by opening chromatin regions specific to tooth, fin, heart, and kidney enhancers so other transcription factors can regulate development or regeneration.
Supplementary Material
Acknowledgements
We thank Mark Stepaniak and Josh Tworig for assistance and advice on developing and injecting transgenic constructs, and Sophie Arch-ambeault, Naama Weksler, and Hernan Garcia for suggestions on the manuscript.
Funding
This work was supported by the National Institutes of Health (DE021475 and DE027871).
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ydbio.2022.09.012.
Data availability
Data will be made available on request.
References
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Postlethwait JH, 1998. Zebrafish hox clusters and vertebrate genome evolution. Science 282 (5394), 1711–1714. 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Berry FB, Skarie JM, Mirzayans F, Fortin Y, Hudson TJ, Raymond V, et al. , 2008. FOXC1 is required for cell viability and resistance to oxidative stress in the eye through the transcriptional regulation of FOXO1A. Hum. Mol. Genet 17 (4), 490–505. 10.1093/hmg/ddm326. [DOI] [PubMed] [Google Scholar]
- Boffelli D, Nobrega MA, Rubin EM, 2004. Comparative genomics at the vertebrate extremes. , June Nat. Rev. Genet. 10.1038/nrg1350. [DOI] [PubMed] [Google Scholar]
- Bowne P, 1994. Systematics and morphology of the gasterosteiformes. In: Bell MA, Foster SA (Eds.), The Evolutionary Biology of the Threespine Stickleback. Oxford Science, Oxford, pp. 28–60. [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS, 2002. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 9 (2), 279–289. 10.1016/S1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Cleves PA, Ellis NA, Jimenez MT, Nunez SM, Schluter D, Kingsley DM, Miller CT, 2014. Evolved tooth gain in sticklebacks is associated with a cis-regulatory allele of Bmp6. Proc. Natl. Acad. Sci. U.S.A 111 (38), 13912–13917. 10.1073/pnas.1407567111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves PA, Hart JC, Agoglia RM, Jimenez MT, Erickson PA, Gai L, Miller CT, 2018. An intronic enhancer of Bmp6 underlies evolved tooth gain in sticklebacks. PLoS Genet. 14 (6), e1007449. 10.1371/journal.pgen.1007449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis NA, Donde NN, Miller CT, 2016. Early development and replacement of the stickleback dentition. J. Morphol 277 (8), 1072–1083. 10.1002/jmor.20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis NA, Glazer AM, Donde NN, Cleves PA, Agoglia RM, Miller CT, 2015. Distinct developmental and genetic mechanisms underlie convergently evolved tooth gain in sticklebacks (June). Development 2442–2451. 10.1242/dev.124248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emera D, Yin J, Reilly SK, Gockley J, Noonan JP, 2016. Origin and evolution of developmental enhancers in the mammalian neocortex. Proc. Natl. Acad. Sci. U.S.A 113 (19), E2617–E2626. 10.1073/pnas.1603718113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson PA, Cleves PA, Ellis NA, Schwalbach KT, Hart JC, Miller CT, 2015. A 190 base pair, TGFB responsive tooth and fin enhancer is required for stickleback Bmp6 expression. Dev. Biol 401 (2), 310–323. 10.1016/j.ydbio.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson PA, Ellis NA, Miller CT, 2016. Microinjection for transgenesis and genome editing in threespine sticklebacks. JoVE (111), e54055. 10.3791/54055e54055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KKH, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, et al. , 2015. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518 (7539), 337–343. 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish A, Chen L, Capra JA, 2017. Gene regulatory enhancers with evolutionarily conserved activity aremore pleiotropic than those with species-specific activity. Genome Biology and Evolution 9 (10), 2615–2625. 10.1093/gbe/evx194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I, 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279. 10.1093/nar/gkh458. WEB SERVER ISS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante CR, Mihala AG, Park S, Wang JS, Johnson KK, Lauderdale JD, Menke DB, 2015. Shared enhancer activity in the limbs and phallus and functional divergence of a limb-genital cis-regulatory element in snakes. Dev. Cell 35 (1), 107–119. 10.1016/j.devcel.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman WR, Stock DW, 2006. Transgenic analysis of Dlx regulation in fish tooth development reveals evolutionary retention of enhancer function despite organ loss. Proc. Natl. Acad. Sci. U.S.A 103 (51), 19390–19395. 10.1073/pnas.0609575103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S, Zhou J, Gao Y, Baek J-A, Martin JF, Lan Y, Sucov HM, 2013. Roles of Bmp4 during tooth morphogenesis and sequential tooth formation. Development (Cambridge, U. K.) 140 (2), 423–432. 10.1242/dev.081927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, et al. , 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484 (7392), 55–61. 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumlongras D, Lachke SA, O’Connell DJ, Aboukhalil A, Li X, Choe SE, Ho JWK, Turbe-Doan A, Robertson EA, Olsen BR, et al. , 2012. An evolutionarily conserved enhancer regulates Bmp4 expression in developing incisor and limb bud. PLoS ONE 7, e38568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandyba E, Leung Y, Chen Y-B, Widelitz R, Chuong C-M, Kobielak K, 2013. Competitive balance of intrabulge BMP/Wnt signaling reveals a robust gene network ruling stem cell homeostasis and cyclic activation. Proc. Natl. Acad. Sci. USA 110 (4), 1351–1356. 10.1073/pnas.1121312110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Shima A, 1999. Identification of the Tol2 transposase of the medaka fish Oryzias latipes that catalyzes excision of a nonautonomous Tol2 element in zebrafish Danio rerio. Gene 240 (1), 239–244. 10.1016/S0378-1119(99)00444-8. [DOI] [PubMed] [Google Scholar]
- Kobielak K, Stokes N, de la Cruz J, Polak L, Fuchs E, 2007. Loss of a quiescent niche but not follicle stem cells in the absence of bone morphogenetic protein signaling. Proc. Natl. Acad. Sci. USA 104 (24), 10063–10068. 10.1073/pnas.0703004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon EZ, Waymack R, Gad M, Wunderlich Z, 2021. Enhancer redundancy in development and disease. Nat. Rev. Genet 22, 324–336. 10.1038/s41576-020-00311-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Friedman JR, Fulmer JT, Kaestner KH, 2005. The initiation of liver development is dependent on Foxa transcription factors. Nature 435 (7044), 944–947. 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- Levine M, Cattoglio C, Tjian R, 2014. Looping back to leap forward: transcription enters a new era. Cell. 10.1016/j.cell.2014.02.009. Elsevier B.V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukoseviciute M, Gavriouchkina D, Williams RM, Hochgreb-Hagele T, Senanayake U, Chong-Morrison V, et al. , 2018. From pioneer to repressor: bimodal foxd3 activity dynamically remodels neural crest regulatory landscape in vivo. Dev. Cell 47 (5), 608–628. 10.1016/j.devcel.2018.11.009 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. , 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science 337 (6099), 1190–1195. 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brown NM, Summers BR, Jones FC, Brady SD, Kingsley DM, 2015. A recurrent regulatory change underlying altered expression and Wnt response of the stickleback armor plates gene EDA, 2015 Elife (4). 10.7554/eLife.05290. [DOI] [PMC free article] [PubMed]
- Pispa J, Thesleff I, 2003. Mechanisms of ectodermal organogenesis. Dev. Biol 262 (2), 195–205. 10.1016/S0012-1606(03)00325-7. [DOI] [PubMed] [Google Scholar]
- Preger-Ben Noon E, Sabarís G, Ortiz DM, Sager J, Liebowitz A, Stern DL, Frankel N, 2018. Comprehensive analysis of a cis-regulatory region reveals pleiotropy in enhancer function. Cell Rep. 22 (11), 3021–3031. 10.1016/j.celrep.2018.02.073. [DOI] [PubMed] [Google Scholar]
- Rebeiz M, Tsiantis M, 2017. Enhancer evolution and the origins of morphological novelty. , August 1 Curr. Opin. Genet. Dev. 10.1016/j.gde.2017.04.006. Elsevier Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabarís G, Laiker I, Preger-Ben Noon E, Frankel N, 2019. Actors with multiple roles: pleiotropic enhancers and the paradigm of enhancer modularity. Trends Genet. 10.1016/j.tig.2019.03.006. Elsevier Ltd. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. , 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol 7 (1), 539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D, Yi SV, 2021. Enhancer pleiotropy, gene expression, and the architecture of human enhancer–gene interactions. Mol. Biol. Evol 38 (9), 3898–3909. 10.1093/molbev/msab085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Square TA, Sundaram S, Mackey EJ, Miller CT, 2021. Distinct tooth regeneration systems deploy a conserved battery of genes. EvoDevo 12 (1), 1–17. 10.1186/s13227-021-00172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepaniak MD, Square TA, Miller CT, 2021. Evolved Bmp6 enhancer alleles drive spatial shifts in gene expression during tooth development in sticklebacks. Genetics. 10.1093/genetics/iyab151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup H, 1958. Stages in the development of the stickleback Gasterosteus aculeatus (L.). J. Embryol. Exp. Morphol 6, 373–383. [PubMed] [Google Scholar]
- Vainio S, Karavanova I, Jowett A, Thesleff I, 1993. Identification of BMP-4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell 75 (1), 45–58. 10.1016/S0092-8674(05)80083-2. [DOI] [PubMed] [Google Scholar]
- Wang L, Siegenthaler JA, Dowell RD, Yi R, 2016. Foxc1 reinforces quiescence in self-renewing hair follicle stem cells. Science (New York, N.Y.) 351 (6273), 613–617. 10.1126/science.aad5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li L, Zheng Y, Yuan G, Yang G, He F, Chen Y, 2012. BMP activity is required for tooth development from the lamina to bud stage. J. Dent. Res 91 (7), 690–695. 10.1177/0022034512448660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Balczerski B, Ciozda A, Louie K, Oralova V, Huysseune A, Gage Crump J, 2018. Fox proteins are modular competency factors for facial cartilage and tooth specification. Development 145 (12). 10.1242/dev.165498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Yu HV, Tseng KC, Flath M, Fabian P, Segil N, Crump JG, 2021. Foxc1 establishes 1 enhancer accessibility for craniofacial cartilage differentiation. Elife 10, 1–50. 10.7554/eLife.63595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.


