Abstract
Sucrose and auxin are well-known determinants of root system architecture (RSA). However, the factors that connect the signaling pathways evoked by these two critical factors during root development are poorly understood. In this study, we report the role of MEDIATOR SUBUNIT17 (MED17) in RSA and its involvement in the transcriptional integration of sugar and auxin signaling pathways in Arabidopsis (Arabidopsis thaliana). Sucrose regulates root meristem activation through the TARGET OF RAPAMYCIN-E2 PROMOTER BINDING FACTOR A (TOR-E2FA) pathway, and auxin regulates lateral root (LR) development through AUXIN RESPONSE FACTOR-LATERAL ORGAN BOUNDARIES DOMAIN (ARF-LBDs). Both sucrose and auxin play a vital role during primary and LR development. However, there is no clarity on how sucrose is involved in the ARF-dependent regulation of auxin-responsive genes. This study establishes MED17 as a nodal point to connect sucrose and auxin signaling. Transcription of MED17 was induced by sucrose in an E2FA/B-dependent manner. Moreover, E2FA/B interacted with MED17, which can aid in the recruitment of the Mediator complex on the target promoters. Interestingly, E2FA/B and MED17 also occupied the promoter of ARF7, but not ARF19, leading to ARF7 expression, which then activates auxin signaling and thus initiates LR development. MED17 also activated cell division in the root meristem by occupying the promoters of cell–cycle genes, thus regulating their transcription. Thus, MED17 plays an important role in relaying the transcriptional signal from sucrose to auxin-responsive and cell–cycle genes to regulate primary and lateral root development, highlighting the role of the Mediator as the transcriptional processor for optimal root system architecture in Arabidopsis.
Mediator subunit MED17 regulates root system architecture by establishing a link between auxin and sucrose signaling pathways.
Graphical Abstract
Graphical Abstract.
Introduction
Plants have an amazing capacity to grow and produce organs throughout their life cycle. This ability is due to the presence of stem cells in the meristematic tissues. The root apical meristem (RAM) is a highly organized structure that allows for the elaboration of the root. In Arabidopsis (Arabidopsis thaliana), continuous proliferation and transition of cells in RAM lead to the development of primary root (PR) (Ivanov and Dubrovsky, 2013). Lateral roots (LR) emerge from the pericycle of PR (Malamy and Benfey, 1997; Dubrovsky et al., 2008). From the epidermis of PR and LR, root hairs emerge and increase the surface area of the root system (Ketelaar et al., 2002). This entire root system architecture (RSA) helps the plant in anchorage, water/nutrient acquisition, and sensing environmental conditions (Ingram and Malamy, 2010).
The development of roots is a complex process that involves different hormones such as auxin, brassinosteroid (BR), gibberellic acid (GA), cytokinin, etc. (Petricka et al., 2012). Among all, auxin plays a decisive role (De Smet et al., 2007). Auxin biosynthesis, transport, and response are essential for proper root patterning and development (Posner and Peterson, 2008; Gallavotti, 2013). An auxin gradient is formed at the root tip and LR primordia by local auxin biosynthesis and its long-distance transport (Blilou et al., 2005; Petricka et al., 2012). Mechanistically, auxin works in a module where it activates AUXIN RESPONSE FACTORs (ARFs) by causing the degradation of their negative regulators AUXIN/INDOLE-3 ACETIC ACID (AUX/IAA) (Korasick et al., 2014; Nanao et al., 2014). The ARF transcription factors (TFs) bind to the auxin response elements (AuxREs) present in the promoters of target genes (Korasick et al., 2014). ARF7 and ARF19 have been established as the main regulators of LR development. These two TFs bind to the promoters of LATERAL ORGAN BOUNDARIES DOMAIN (LBD) genes such as LBD16, LBD18, LBD29, and LBD33 (Okushima et al., 2007). LBDs are lateral organ boundaries (LOB) domain-containing proteins. LOB is a plant-specific domain indicating the functional importance of LBDs in plants (Iwakawa et al., 2002; Shuai et al., 2002). LBDs regulate the expression of E2 PROMOTER BINDING FACTOR A (E2FA) for the maintenance of asymmetric cell division during LR initiation (Berckmans et al., 2011).
E2F TFs play an important role in deciding between cell proliferation and cell differentiation (Wildwater et al., 2005; Wyrzykowska et al., 2006). In Arabidopsis, three E2F TFs (E2FA, E2FB, and E2FC) form a complex with RETINOBLASTOMA RELATED (RBR) protein to regulate various developmental programs (Magyar et al., 2016). E2Fs require two dimerization proteins (DPA or DPB) for their binding to the target DNA elements (De Veylder et al., 2007; Magyar et al., 2012). The overexpression studies reveal that E2FA and E2FB function as activators, while E2FC has been found to work as a repressor (Magyar et al., 2016). E2FA and E2FB are phosphorylated by TARGET OF RAPAMYCIN (TOR) kinase and have been hypothesized to work redundantly for cell cycle activation in SAM and RAM (Xiong et al., 2013; Li et al., 2017). TOR kinase is an evolutionarily conserved protein that regulates signaling pathways evoked by nutrients and energy (Poss et al., 2013; Xiong et al., 2013). In response to sugars such as glucose and sucrose, TOR phosphorylates E2FA, which further regulates the transcription of cell–cycle regulating genes such as ORIGIN RECOGNITION COMPLEX (ORC2/6), MINOCHROMOSOME MAINTENANCE (MCM3/5/7), CELL DIVISION CYCLE (CDC6), E2F TARGET GENE1 (ETG1), and PROLIFERATING CELL NUCLEAR ANTIGEN1 (PCNA1) by directly binding to their promoters for maintaining root meristem activation (Xiong et al., 2013). Glucose/sucrose has also been shown to promote LR development in a concentration-dependent manner (Gupta et al., 2015).
The discovery of the Mediator complex has enhanced our understanding of eukaryotic gene expression. Mediator is a megadalton protein complex consisting of four modules named as head, middle, tail, and kinase modules (Malik and Roeder, 2010). It forms a bridge between DNA-bound TF and RNA Pol II to initiate the process of transcription (Poss et al., 2013). Along with transcription initiation, the role of the Mediator has been established in several other processes, such as transcription elongation, chromatin reorganization, mRNA processing and export, and transcription reinitiation (Huang et al., 2012; Hsieh et al., 2015; Tantale et al., 2016). The subunit composition of the Mediator is different in different organisms, such as 25 in yeast, 30 in humans, and 34 in Arabidopsis (Guglielmi et al., 2004; Bäckström et al., 2007; Malik and Roeder, 2010; Nagulapalli et al., 2016). In Arabidopsis, the role of the Mediator has been established in different developmental processes, such as root and shoot development, embryo development, flowering, and senescence (reviewed in Samanta and Thakur, 2015; Malik et al., 2017; Agrawal et al., 2020). In Arabidopsis, some of the Mediator subunits have been shown to have a role in the primary and lateral root development (Raya-González et al., 2014, 2017, 2018, 2021; Ruiz-Aguilar et al., 2020). MED17 is a subunit in the head module. It has been found to be important for the structural stability of the complex (Guglielmi et al., 2004; Nozawa et al., 2017; Maji et al., 2019). The role of MED17 has been established in microRNA biogenesis and thermomorphogenesis (Kim et al., 2011; Agrawal et al., 2022). A recent report demonstrates the role of MED17 in DNA damage response in Arabidopsis (Giustozzi et al., 2022). In one report, MED17 has been indicated to be involved in LR development (Ito et al., 2016). However, the systematic study showing the precise mode of action of MED17 in the development of LR remains unknown.
This study delineates the role of MED17 in primary and lateral root development by establishing a link between auxin and sucrose signaling. We report that MED17 regulates the expression of auxin-responsive target genes such as LBD16, LBD18, LBD29, and LBD33 during LR development. Besides auxin, sucrose also regulates the expression of these genes, and this regulation is dependent on MED17. Interestingly, MED17 also regulates the transcription of ARF7, the TF that activates LBD genes and is involved in LR growth. In addition to LR development, the expression of genes underlying root meristem activation and, thus, the PR development is also controlled by MED17. It is already known that sugars promote the activation of root meristem by regulating the expression of cell cycle genes. In this study, we report that the sucrose-induced upregulation of root meristem genes also depends on MED17. As a matter of fact, the expression of MED17 itself is induced by sucrose via E2F signaling components. Besides this, MED17 physically interacts with E2FA and E2FB. Thus, E2FA/B potentiates the recruitment of MED17 on the target gene promoters to regulate primary and lateral root development.
Results
MED17 is involved in root development
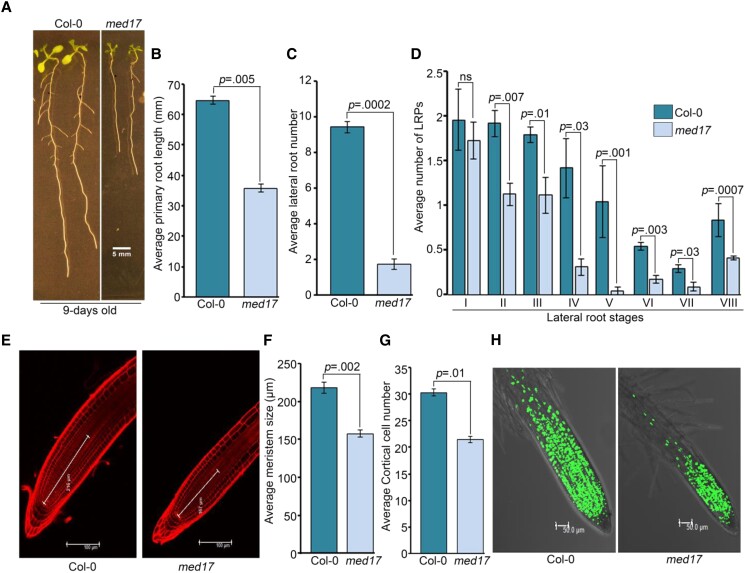
In earlier reports, MED17 has been shown to be required for the overall growth and development of plants (Kim et al., 2011; Giustozzi et al., 2022). So, we measured the root versus shoot biomass ratio in med17 seedlings and compared that with wild type (WT) seedlings. Although statistically insignificant, the root biomass seems to be more affected in med17 seedlings (Supplemental Figure S1A). Next, we looked at the expression of MED17 in Col-0 seedlings and found its higher transcript level in the root as compared to the shoot (Supplemental Figure S1B). Moreover, in one study, LR density in med17 seedlings was shown to be significantly low, indicating a role of MED17 in RSA (Ito et al., 2016). So, we decided to perform an in-depth study to get mechanistic insight into the role of MED17 in Arabidopsis root development. Comparing the roots of 9-day-old Col-0 and med17 seedlings revealed defects in the overall growth of roots in med17 seedlings (Figure 1A). The PRs were shorter (∼40%) in length, and the LRs were less in number in med17 seedlings (Figure 1, B and C). Recently, although studied in different aspects, other researchers have also observed shorter primary roots in med17 mutants (Giustozzi et al., 2022). Overexpression of MED17 under the control of a 35S promoter in med17 mutant has been shown to recover the phenotype to the extent of WT (Agrawal et al., 2022; Giustozzi et al., 2022). Here also, overexpression of MED17 using 35S::YFP-MED17 cassette rescued the defective root phenotype of med17 (Supplemental Figure S1, C–E), confirming the importance of MED17 for root development. The process of LR emergence starts deep inside the PR. This process has been divided into eight stages, which are characterized by regulated cell division (Malamy and Benfey, 1997). Any deregulation in these stages can lead to either fewer LRs or irregular LR development (Vangheluwe and Beeckman, 2021). Since med17 seedlings possess fewer LRs, we wanted to know whether there was any problem in any of these stages of LR development. For this, LR primordia (LRP) at different stages were quantified in 7-day-old Col-0 and med17 seedlings. We found that in med17 seedlings, the number of LRP was significantly less (stage II onwards) as compared to Col-0 (Figure 1D), suggesting the requirement of MED17 during LR development. Root meristem is the primary determinant of PR development and root elongation (Ioio et al., 2008). To investigate the role of MED17 in regulating the root meristem, we looked at its length in Col-0 and med17 seedlings (Figure 1, E and F). We also examined cortical cell numbers (Figure 1, E and G). Indeed, there was a significant reduction in both the length of meristem and the number of cortical cells in med17 seedlings (Figure 1, E–G). In the complementation line of med17/35S::YFP-MED17, this shortened root meristem phenotype of med17 was rescued (Supplemental Figure S1, F–H), suggesting the role of MED17 in the maintenance of root meristem in Arabidopsis. We also examined the entry of root meristematic cells into the S-phase by using 5-ethynyl-29-deoxyuridine (EDU) nucleoside, an analog of thymidine. For this, 4-day-old Col-0 and med17 seedlings were transferred to EDU-containing media for 3 h, and then the fluorescence of the stained nuclei was observed. We found substantially lower number of EDU-stained nuclei in med17 root meristem as compared to Col-0 (Figure 1H). All these results suggest that MED17 is important for root meristem maintenance and LR development in Arabidopsis.
Figure 1.
MED17 promotes root development in Arabidopsis. A, Root phenotype of Col-0 and med17 seedlings. Length of the scale bar in the image is 5 mm. B, Graph showing the length of primary roots in Col-0 and med17 seedlings. C, Graph showing the number of lateral roots in Col-0 and med17 seedlings. D, Graph showing the number of lateral root primordia (LRPs) in different stages of development in Col-0 and med17 seedlings. In (B–D), Col-0 and med17 seedlings were grown on ½ MS for 9 days. Lengths of primary roots were measured using ImageJ. Counting of LRP was done according to Malamy and Benfey (1997). Data shown are average of three independent biological replicates of at least 25 seedlings. Bar plots represent mean values, and error bars denote Se. Statistical difference was assessed by P-value as assessed by one-way ANOVA and Tukey's HSD post hoc test. E, Root tip stained with PI. Lines are showing size of the meristem. Length of the scale bar in the image is 100 µm. F, Graph showing size of root meristems in Col-0 and med17 seedlings. G, Graph showing cortical cell numbers of Col-0 and med17 seedlings. H, Root tip stained with EDU. Length of the scale bar in the image is 50 µm. In (E–H), seedlings were grown on ½ MS for 4 days and processed for confocal imaging. ImageJ was used to measure meristem length and count the cortical cell number. Data shown are average of three independent biological replicates containing at least 10 seedlings. Bar plots represent mean values, and error bars denote Se. Statistical difference has been depicted by P-value as assessed by one-way ANOVA and Tukey's HSD post hoc test. For all the graphs, P-value of 0.05 or lower (P ≤ 0.05) was considered statistically significant, whereas P-value greater than 0.05 (P > 0.05) was considered non-significant (ns).
MED17 influences auxin responses in roots
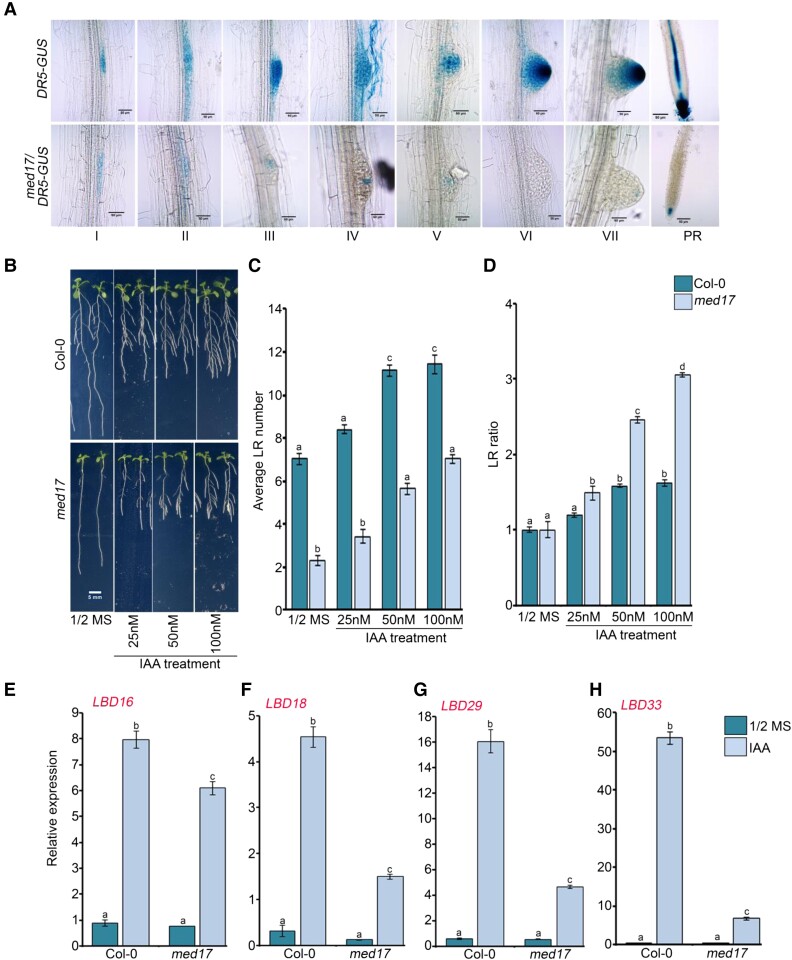
Next, to understand how MED17 regulates RSA, we studied the signaling components involved in root development. Auxin is a critical regulator of RSA as it plays an important role in PR elongation and LR development (Muday and Haworth, 1994). So, fewer LRs in med17 seedlings, as compared to WT, suggest that there is a lower level of auxin at LRPs in med17 (Figure 1, A–D). This was confirmed by the lower expression level of DR5:GUS, an auxin-responsive marker (Ulmasov et al., 1997), in med17 seedlings as compared to WT (Figure 2A). So, in order to know if MED17 is involved in auxin-mediated LR development, we transferred 4-day-old Col-0 and med17 seedlings to different concentrations of IAA and observed the phenotype. The increasing concentration of IAA caused an increase in the number of LRs in Col-0 seedlings (Figure 2, B and C and Supplemental Figure S2, A and B). In med17 also, there was an increase in the emergence of LRs upon auxin treatment, but the total number of LRs remained less in med17 as compared to Col-0 (Figure 2, B and C and Supplemental Figure S2, A and B). Next, we analyzed the ratio of the number of LRs in Col-0 and med17 seedlings in the presence or absence of auxin in the media (Figure 2D and Supplemental Figure S2C). We found that the ratio was higher in med17 as compared to Col-0 plants suggesting that the med17 plants are able to sense and transport auxin provided exogenously. Since the number of LRs is much less in med17 in normal conditions, i.e. in the absence of exogenous auxin, we hypothesized that in normal conditions, the basal auxin level might be impaired in med17 seedlings. During LR development, auxin-responsive TFs activate the transcription of LBD genes in response to auxin (Okushima et al., 2007; Feng et al., 2012). So, we asked whether the expression of auxin-responsive LBD genes (LBD16, LBD18, LBD29, and LBD33) was affected in med17 seedlings. When the seedlings were treated with exogenous auxin, the extent of upregulation of LBD genes was significantly affected in med17 seedlings (Figure 2, E–H and Supplemental Figure S3, A–D). Thus, it seems that although the overall auxin signaling is affected in med17, it is still functional, suggesting that MED17 is important for optimal auxin signaling in Arabidopsis root. Moreover, this also raises a possibility that in addition to auxin, MED17 might be engaging some other signaling as well for LR development.
Figure 2.
MED17 partially regulates auxin responsiveness during root development in Arabidopsis. A, Different stages of lateral root primordia and root tips of 7-day-old seedlings showing DR5::GUS activity. Data shown are representative of three biological replicates (n = 3). I to VII are different stages of lateral root primordium, and in the PR panel, tip of primary root is shown. Length of the scale bar in the image is 50 µm. B, Root phenotype of Col-0 and med17 seedlings after treatment with IAA. Length of the scale bar in the image is 5 mm. C, Graph showing lateral root (LR) number in the Col-0 and med17 after exogenous auxin treatment. D, Graph showing LR ratio in the Col-0 and med17 after exogenous auxin treatment. B–D, Four-day old Col-0 and med17 seedlings were transferred to ½ MS plates containing different concentrations of IAA (25, 50, and 100 nM) for another 4 days, and LR numbers were counted. For plotting LR ratio, the total number of LRs after auxin treatment was divided by the total number of LRs on ½ MS. Data shown are average of three independent biological replicates containing at least 20 seedlings. Bar plots represent mean values, and error bars denote Se. Statistical difference has been depicted by P-value as assessed by one-way ANOVA and Tukey's HSD post hoc test. E–H, RT-qPCR showing expression of LBD genes (LBD16, LBD18, LBD29, and LBD33) after IAA treatment. Seven-day-old Col-0 and med17 seedlings were treated with IAA (1 µM) for 3 h. Gene expression values were calculated as ΔCt. RT-qPCR analysis was performed on three independent biological replicates (n = 3). Bar plots represent mean values, and error bars denote Se. Statistical difference has been depicted by P-value as assessed by one-way ANOVA and Tukey's HSD post hoc test. For all the graphs, P-value of 0.05 or lower (P ≤ 0.05) was considered statistically significant, whereas P-value greater than 0.05 (P > 0.05) was considered non-significant (ns).
MED17 influences sucrose-mediated meristem activation in Arabidopsis root
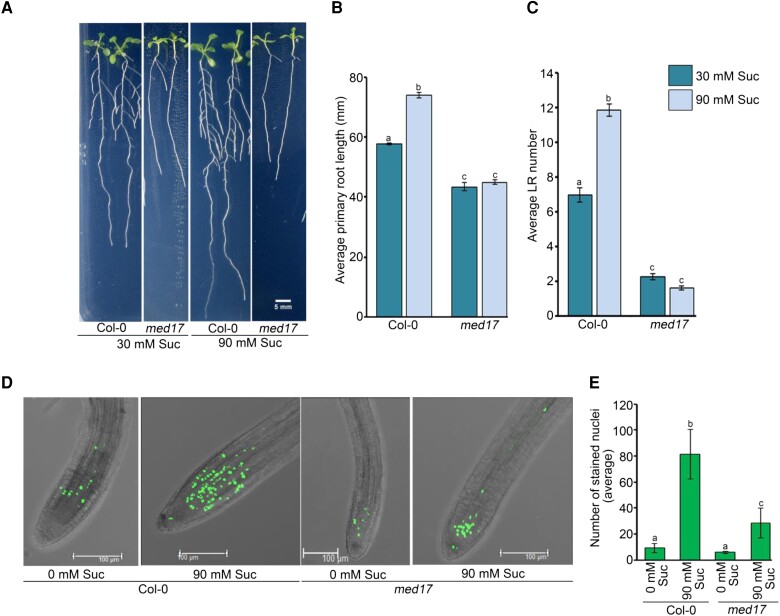
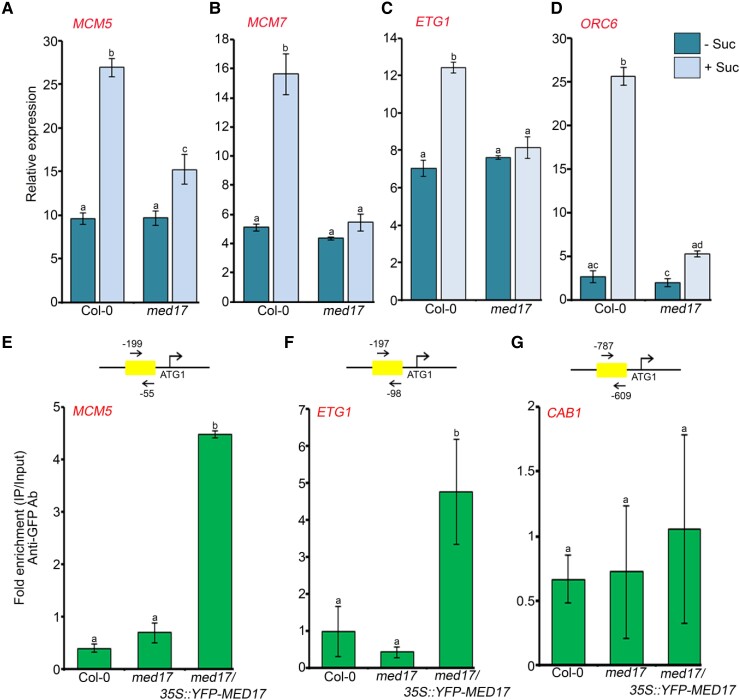
Besides being a structural component, sucrose acts as a signaling molecule that influences cell cycle progression and auxin signaling during plant development (MacGregor et al., 2008; Kircher and Schopfer, 2012; Wang and Ruan, 2013). Moreover, it is already known that sucrose at low concentrations promotes LR development, whereas it is inhibitory at very high concentrations (5%) (Malamy and Ryan, 2001; Gupta et al., 2015). So, we decided to assess the role of MED17 in sucrose-regulated RSA. For this, we grew the Col-0 and med17 seedlings in the low and optimum concentrations of sucrose (30 and 90 mM). We observed that at optimum concentration, Col-0 seedlings had longer PRs and more LRs (Figure 3, A–C). On the contrary, there was no difference in the root architecture of med17 seedlings in the low and optimum concentrations of sucrose (Figure 3A). Thus, neither the length of PRs nor the number of LRs was affected in med17 seedlings at higher sucrose concentration (Figure 3, B and C), suggesting that MED17 is critical for sucrose-triggered changes in RSA in Arabidopsis. Sucrose is known to activate root meristem in Arabidopsis (Xiong et al., 2013). So, we decided to look at the effect of sucrose on meristem activity in med17. For this, Col-0 and med17 seedlings were grown on sucrose-free media for 3 days, followed by sucrose treatment and EDU staining. In accordance with earlier reports, there was no or much less root meristem activity in the absence of sucrose in both Col-0 and med17 seedlings (Figure 3, D and E). However, treatment of seedlings with sucrose caused activation of the root meristem in Col-0 but not so much in the med17 seedlings (Figure 3, D and E). On the other hand, in the complementation line of med17/35S::YFP-MED17, the number of EDU-stained nuclei was similar to Col-0 after the sucrose treatment (Supplemental Figure S4) suggesting the possible involvement of MED17 in the sucrose-regulated activation of the root meristem. Sucrose-mediated root meristem activation involves the upregulation of cell cycle genes MCM3/5/7, ORC6, and ETG1 in a TARGET OF RAPAMYCIN-E2 PROMOTER BINDING FACTOR A (TOR-E2FA)-dependent manner (Xiong et al., 2013). Since sucrose-mediated meristem activation was perturbed in med17, we looked at the expression of these cell cycle genes and found that in response to sucrose treatment, expression of MCM5/7, ORC6, and ETG1 was induced in Col-0 but not in med17 (Figure 4, A–D). So, next, we looked at the occupancy of MED17 on the promoters of these genes, which harbor E2F-binding sites. Indeed, we observed a very good enrichment of MED17 on the promoters of these cell cycle genes (Figure 4, E and F and Supplemental Figure S5, A and B). In addition to the cell cycle genes, we also tested the occupancy of MED17 on the promoter of CHLOROPHYLL A/B BINDING PROTEIN 1 (CAB1), which does not possess any E2F-binding motif and worked as a negative control. There was no enrichment of MED17 on the promoter of the CAB1 promoter (Figure 4G and Supplemental Figure S5D). Thus, all these results suggest that MED17 is involved in the sucrose-triggered activation of the root meristem.
Figure 3.
MED17 is required for sucrose-regulated root development in Arabidopsis. A–C, Root phenotype of Col-0 and med17 seedlings after treatment with sucrose. Four-day-old Col-0 and med17 seedlings were transferred to ½ MS plates containing different concentrations of sucrose (30 or 90 mM) for another 4 days. ImageJ was used to count lateral root (LR) number and measure the PR length. Data shown are average of three independent biological replicates containing at least 15 seedlings. Bar plots represent mean values, and error bars denote Se. Statistical difference has been depicted by P-value as assessed by one-way ANOVA and Tukey's HSD post hoc test. Length of the scale bar in the image is 5 mm. D, Confocal images of root tips showing EDU staining. Length of the scale bar in the image is 100 µm. E, Graphical representation of total number of EDU-stained nuclei. D, E, Four-day-old Col-0 and med17 seedlings were treated with 90 mM sucrose for 3 h followed by EDU staining and confocal imaging. Data shown are average of three independent biological replicates containing at least 10 seedlings. Bar plots represent mean values, and error bars denote Se. Statistical difference has been depicted by P-value as assessed by one-way ANOVA and Tukey's HSD post hoc test. For all the graphs, P-value of 0.05 or lower (P ≤ 0.05) was considered statistically significant, whereas P-value greater than 0.05 (P > 0.05) was considered non-significant (ns).
Figure 4.
MED17 regulates the expression of cell cycle genes during root meristem activation. A–D, RT-qPCR showing expression of cell cycle genes (MCM5/7, ETG1, and ORC6) after sucrose treatment. Four-day-old Col-0 and med17 seedlings were treated with 90 mM sucrose for 3 h. Gene expression values were calculated as ΔCt. RT-qPCR analysis was performed on three independent biological replicates (n = 3). Bar plots represent mean values, and error bars denote Se. Statistical difference has been depicted by P-value as assessed by one-way ANOVA and Tukey's HSD post hoc test. E–G, ChIP-qPCR showing enrichment of YFP-MED17 at the promoters of MCM5, ETG1, and CAB1 in 7-day-old Col-0, med17, and med17/35S:YFP-MED17 seedlings. The promoter regions harboring the E2F-binding elements were amplified. Amplicon positions relative to ATG (translation start site) are shown in the upper panel. Ct values with and without antibody samples were normalized to input control. Untransformed Col-0 and med17 seedlings were taken as negative control. CAB1, which does not possess any E2F-binding motifs in its promoter, was used as a negative control. YFP-MED17 binding on the promoter regions was calculated as fold enrichment. ChIP-qPCR analysis was performed on three technical replicates (n = 3) from a single representative experiment. Experiments were independently repeated twice (biological replicates; n = 2). Bar plots represent mean values, and error bars denote Sd. Statistical difference has been depicted by P-value as assessed by one-way ANOVA and Tukey's HSD post hoc test. For all the graphs, P-value of 0.05 or lower (P ≤ 0.05) was considered statistically significant, whereas P-value greater than 0.05 (P > 0.05) was considered non-significant (ns).
MED17 and E2FA/B regulate sucrose-mediated LR development
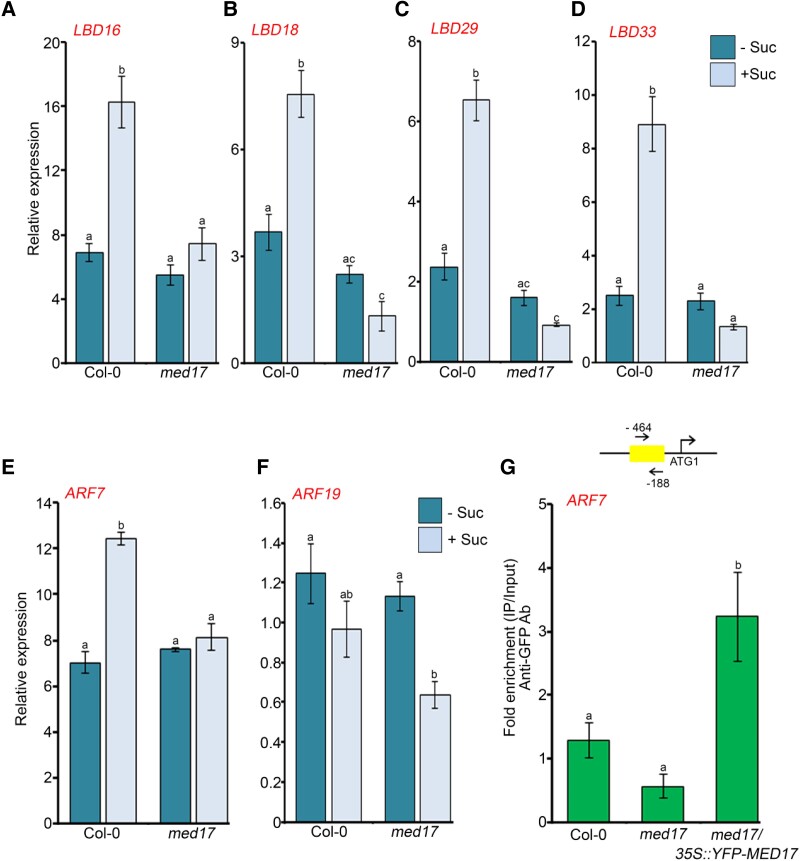
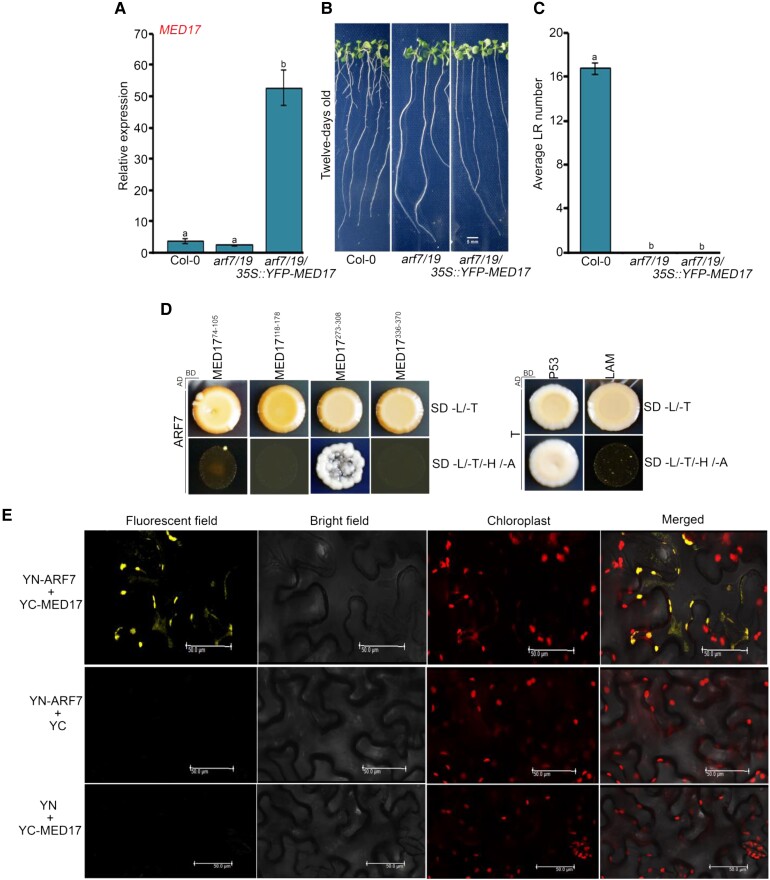
Earlier, it has been shown that the auxin-responsive LBD genes are induced by sugars such as glucose (Gupta et al., 2015). So, in order to find whether sugar-triggered signaling is transduced via MED17, we studied the effect of sucrose on the expression of LBD genes in Col-0 and med17 seedlings. We found that in Col-0 seedlings, the expression of LBD genes was induced significantly after 3 h of sucrose treatment but not in med17 seedlings (Figure 5, A–D). It is known that the auxin-responsive TFs ARF7 and ARF19 interact with the Mediator subunit MED25 to regulate the expression of LBDs in response to auxin during LR development (Ito et al., 2016). However, nothing is known about the regulation of ARF genes by sugars. For this, we asked whether sugars also affect the expression of ARF7 and ARF19 and whether MED17 is involved in this regulation. We observed that the expression of only ARF7 was induced upon sucrose treatment. This induced expression of ARF7 was abolished in med17 seedlings (Figure 5E), suggesting that MED17 regulates the expression of ARF7 in the presence of sugar. Intriguingly, there was no noticeable difference in ARF19 expression in response to sucrose (Figure 5F). Next, we assessed the occupancy of MED17 on the promoter of ARF7. For this, we scanned the 2 kb promoter region of ARF7 and found E2F-binding motifs in it. Since MED17 was found to be enriched at E2F-binding elements present in the promoters of cell cycle genes (Figure 4, E and F), we hypothesized that it might be occupying the same E2F motifs present in the promoter of ARF7. To assess the occupancy of MED17 on the ARF7 promoter, we performed chromatin immunoprecipitation-quantitative real time PCR (ChIP-qPCR) with med17/35S::YFP-MED17 seedlings using an anti-green fluorescent protein (GFP) antibody. Indeed, MED17 was found to occupy the promoter of ARF7 (Figure 5G and Supplemental Figure S5C). Next, to find whether MED17 and ARF7 work in the same genetic pathway in controlling LRs, we overexpressed MED17 in the background of arf7/19 (Figure 6A) and examined the phenotype. We observed that overexpression of MED17 in the arf7/19 background could not rescue the defective LR phenotype of arf7/19 (Figure 6, B and C). This genetic data confirms that MED17 acts upstream of ARFs in regulating the LRs. Moreover, MED17 physically interacts with ARF7 as assessed by yeast two hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) analysis (Figure 6, D and E). All these data suggest that MED17 not only regulates the transcription of ARF7 in response to sucrose but also makes a complex with ARF7 to regulate the transcription of LBD genes.
Figure 5.
MED17 is required for sucrose-triggered expression of auxin signaling genes. A–D, RT-qPCR showing expression of auxin-responsive LBD genes (LBD16, LBD18, LBD29, and LBD33) after sucrose treatment. E, F, RT-qPCR showing expression of auxin-responsive TF genes (ARF7 and ARF19) after sucrose treatment. In (A–F), 7-day-old Col-0 and med17 seedlings were treated with 90 mM sucrose for 3 h. Gene expression values were calculated as ΔCt. RT-qPCR analysis was performed on three independent biological replicates (n = 3). Bar plots represent mean values, and error bars denote Se. Statistical difference has been depicted by P-value as assessed by one-way ANOVA and Tukey's HSD post hoc test. G, ChIP-qPCR showing enrichment of YFP-MED17 at the promoter of ARF7 in 7-day-old Col-0, med17, and med17/35S:YFP-MED17 seedlings. Promoter region of ARF7 harboring E2F-binding element was amplified. Positions of the amplicon relative to ATG are shown in the upper panel. Ct values with and without antibody samples were normalized to input control. Untransformed Col-0 and med17 seedlings were taken as negative control. YFP-MED17 binding on promoters was calculated as fold enrichment. ChIP-qPCR analysis was performed on three technical replicates (n = 3) from a single representative experiment. Experiments were independently repeated twice (biological replicates; n = 2). Bar plots represent mean values, and error bars denote Sd. Statistical difference has been depicted by P-value as assessed by one-way ANOVA and Tukey's HSD post hoc test. For all the graphs, P-value of 0.05 or lower (P ≤ 0.05) was considered statistically significant, whereas P-value greater than 0.05 (P > 0.05) was considered non-significant (ns).
Figure 6.
MED17 regulates LR development in an ARF-dependent manner. A, RT-qPCR showing the expression level of MED17 in Col-0, arf7/19, and arf7/19/35S:YFP-MED17 seedlings. The seedlings were grown on ½ MS for 7 days and then harvested for RNA isolation. Gene expression values were calculated as ΔCt. RT-qPCR analysis was performed on three independent biological replicates (n = 3). Bar plots represent mean values, and error bar denotes Se. Statistical difference has been depicted by P-values as assessed by one-way ANOVA and Tukey's HSD post hoc test. B, Root phenotype of Col-0, arf7/19, and arf7/19/35S::YFP-MED17 seedlings. Length of the scale bar in the image is 5 mm. C, Graphical representation of the average number of lateral roots (LRs) per seedling in Col-0, arf7/19, and arf7/19/35S::YFP-MED17. Data shown are the average of three independent biological replicates containing at least 20 seedlings. Bar plots represent mean values, and error bar denotes Se. Statistical difference has been depicted by P-values as assessed by one-way ANOVA and Tukey's HSD post hoc test. D, Y2H assay to study the interaction of MED17 with ARF7. Interactions were scored by growth of yeast colonies on quadruple drop out (QDO) (-H/-L/-T/-A) selection media. P53-T and LAM-T were used as positive and negative controls, respectively. E, BiFC assay showing interaction of MED17 with ARF7. MED17 and ARF7 were cloned into CD3-1651 and CD3-1648 vectors, respectively. Interaction was confirmed on the basis of YFP signals. No YFP signals were observed between YFP N ARF7–C YFP and YFP N–C YFP MED17. Hence, they served as negative controls. Length of the scale bar in the image is 50 µm. For all the graphs, P-value of 0.05 or lower (P ≤ 0.05) was considered statistically significant, whereas P-value greater than 0.05 (P > 0.05) was considered non-significant (ns).
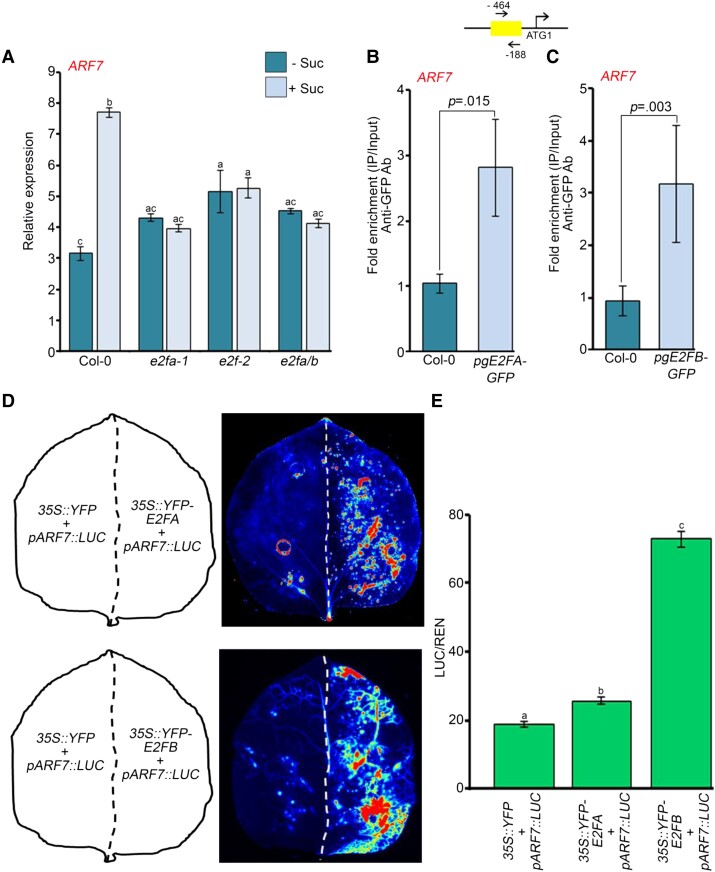
Earlier, transcription factor E2FA was shown to regulate LR development in response to auxin (Berckmans et al., 2011). Since the expression of ARF7 is induced by sucrose (Figure 5E) and it possesses E2F-binding motifs in its promoter region, we wanted to know whether E2Fs regulate the expression of ARF7. For this, we checked the expression of ARF7 in e2fa-1, e2fb-2, and e2fa/b mutants (Magyar et al., 2012; Leviczky et al., 2019; Oszi et al., 2020) in response to sucrose. The expression of ARF7 was induced by sucrose in Col-0 (Figure 7A) but not in any of the e2f mutants (Figure 7A). On the other hand, there was no upregulation of ARF19 after sucrose treatment (Supplemental Figure S6A). The presence of E2F motifs in the promoter of ARF7 led us to test the binding of E2Fs onto the promoter of ARF7. Indeed, we found an enrichment of E2FA and E2FB proteins on the ARF7 promoter, suggesting that E2Fs occupy the promoter of ARF7 and so contribute to the regulation of its expression (Figure 7, B and C and Supplemental Figure S6, D–E). Next, we looked at the expression of a luciferase reporter driven by ARF7 promoter in the presence or absence of E2FA/B. Indeed, both E2FA and B could activate the promoter of ARF7 in Nicotiana benthamiana (N. benthamiana) leaf, the E2FB being more effective than E2FA (Figure 7, D and E). As the promoter of ARF19 does not possess any E2F-binding motif, we could not observe any enrichment of either E2FA or E2FB on its promoter (Supplemental Figure S6, B and C). Thus, ARF19 worked as a negative control in this case. All these data suggest that E2FA and E2FB are important for the sucrose-induced expression of ARF7 to regulate LR development.
Figure 7.
E2F TFs are required for sucrose-induced expression of ARF7. A, RT-qPCR showing expression of ARF7 in Col-0, e2fa-1, e2fb-2, and e2fa/b seedlings after sucrose treatment. Seven-day old Col-0, e2fa-1, e2fb-2, and e2fa/b seedlings were treated with 90 mM sucrose for 3 h. Gene expression values were calculated as ΔCt. RT-qPCR analysis was performed on three independent biological replicates (n = 3). Bar plots represent mean values, and error bars denote Se. Statistical difference has been depicted by P-value as assessed by one-way ANOVA and Tukey's HSD post hoc test. B, ChIP-qPCR showing enrichment of E2FA–GFP at the promoter of ARF7 in 7-day-old Col-0 and pgE2FA-GFP seedlings. C, ChIP-qPCR showing enrichment of E2FB–GFP at the promoter of ARF7 in 7-day-old Col-0 and pgE2FB-GFP seedlings. In (B and C), the promoter region of ARF7 harboring E2F-binding element was amplified. Ct values with and without antibody samples were normalized to input control. Untransformed Col-0 seedlings were taken as negative control. E2FA–GFP and E2FB–GFP binding on promoters were calculated as fold enrichment. ChIP-qPCR analysis was performed on three technical replicates (n = 3) from a single representative experiment. Experiments were independently repeated twice (biological replicates; n = 2). Bar plots represent mean values, and error bars denote Sd. Statistical difference has been depicted by P-value as assessed by paired t test. D, Luciferase activity assay for determining the activation of ARF7 promoter by E2FA and E2FB TFs. Luciferase was cloned under the control of ARF7 promoter. The left halves of the leaves were infiltrated with 35S::YFP and pARF7::LUC plasmids whereas the right halves of the leaves were infiltrated with either 35S::YFP-E2FA or 35S::YFP-E2FB plasmid along with pARF7::LUC plasmid. Activation of ARF7 promoter by E2FA or E2FB was assessed by measuring the luciferase signals. E, Graph showing the quantification of luminescence from pARF7::LUC in the presence or absence of E2FA/B. Relative luminescence was calculated by dividing luminescence of firefly luciferase by luciferase of Renilla. Experiments were independently repeated twice (biological replicates; n = 2, each containing three technical replicates). Bar plots represent mean values, and error bars denote Se. Statistical difference has been depicted by P-value as assessed by one-way ANOVA and Tukey's HSD post hoc test. For all the graphs, P-value of 0.05 or lower (P ≤ 0.05) was considered statistically significant, whereas P-value greater than 0.05 (P > 0.05) was considered non-significant (ns).
MED17 interacts with E2FA and E2FB
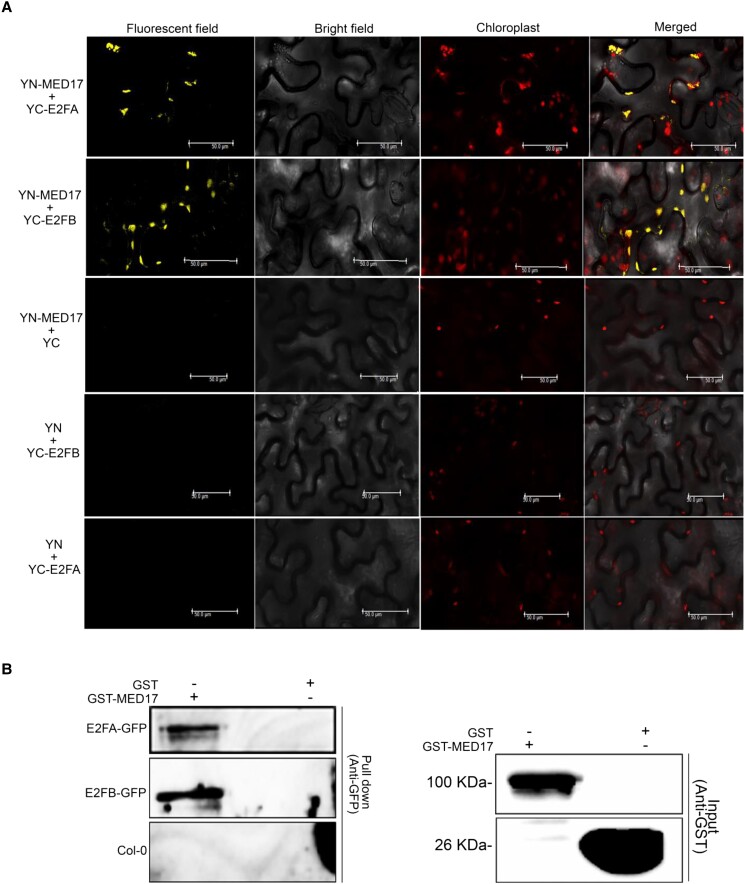
All the above results suggest that MED17 plays an essential role in the E2F-mediated sucrose signaling pathway to regulate the overall RSA of Arabidopsis. More importantly, MED17 occupies the E2F-binding elements present in the promoters of root development-related genes. So, we asked if E2Fs recruit MED17 to these promoters by physically interacting with it. Indeed, MED17 was found to interact with E2FA and E2FB in the BiFC and pull-down experiments (Figure 8, A and B). Thus, it seems that E2FA/B interacts with MED17 to recruit the Mediator complex and transcriptional machinery on the promoter of its target genes to regulate the development of primary and lateral roots in Arabidopsis.
Figure 8.
MED17 interacts with E2FA and E2FB. A, BiFC assay showing interaction of MED17 with E2FA and E2FB. MED17 and E2FA/B were cloned into CD3-1648 and CD3-1651 vectors, respectively. Interactions were confirmed on the basis of YFP signals. No YFP signals were observed between YFP N MED17–C YFP, YFP N–C YFP E2FA, and YFP N–C YFP E2FB. Hence, these were used as negative controls. Length of the scale bar in the image is 50 µm. B, Pull-down assay showing interaction of MED17 with E2FA and E2FB. Whole protein extract of pgE2FA-GFP or pgE2FB-GFP seedling was incubated with purified GST-MED17 or GST alone. Immuno pull-down was performed using anti-GST antibody, and the GFP-tagged proteins remaining on the beads were detected by anti-GFP antibody. Only GST was used as a negative control. Data shown are the representative of two independent biological replicates (n = 2). For all the graphs, P-value of 0.05 or lower (P ≤ 0.05) was considered statistically significant, whereas P-value greater than 0.05 (P > 0.05) was considered non-significant (ns).
Sucrose regulates the expression of MED17 in E2FA/B-dependent manner
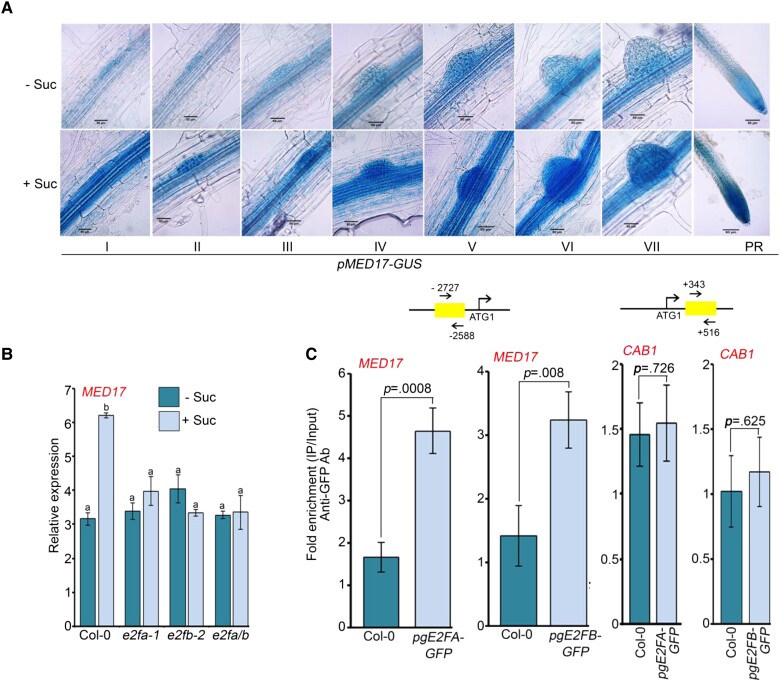
Next, we wanted to know if the expression of MED17 is also regulated by sucrose. For this, we analyzed the activity of GUS in pMED17-GUS transgenic lines. Seven-day-old pMED17-GUS seedlings were kept in the dark in sucrose-free media for 16 h to deplete the internal sucrose. Subsequently, these seedlings were treated with sucrose, and then GUS signals were detected. We found that pMED17-GUS was expressed in all the stages of LR development and also at the PR tip (Figure 9A). Interestingly, the intensity of the GUS signal was higher in sucrose-treated seedlings suggesting that sucrose can induce the expression of MED17 (Figure 9A). Indeed, in Col-0 seedlings, sucrose treatment enhanced the transcript level of MED17 (Figure 9B). In order to get an idea of the transcription factors that regulate the expression of MED17 in response to sucrose, we scanned the promoter region of MED17. Surprisingly, we found E2F-binding motifs in the promoter of the MED17 gene. This finding led us to hypothesize that sucrose might be regulating MED17 expression through E2Fs. So, we analyzed the expression of MED17 in the mutants of e2fa-1, e2fb-2, and e2fa/b. As expected, the sucrose-induced expression of MED17 was lost in the mutants of e2fs, confirming that E2Fs are required for this (Figure 9B). Next, we checked the occupancy of E2FA and E2FB on the promoter of MED17 using pgE2FA::GFP and pgE2FB::GFP transgenic lines, respectively. Indeed, both E2FA and E2FB were found to occupy the promoter of MED17 (Figure 9C and Supplemental Figure S7, A and B). CAB1 served as a negative control (Figure 9C and Supplemental Figure S7, C and D). All these results suggest that in response to sucrose, E2FA and E2FB regulate the expression of MED17 by directly occupying its promoter.
Figure 9.
Sucrose-induced expression of MED17 is regulated by E2FA/B. A, Activity of pMED17-GUS in response to sucrose. Seven-day-old pMED17-GUS seedlings were treated with 90 mM sucrose followed by GUS staining. I to VII are different stages of lateral root primordium and in the PR panel, and tip of primary root is shown. Images shown are representative of three biological replicates (n = 3). Length of the scale bar in the image is 50 µm. B, RT-qPCR showing expression of MED17 in Col-0, e2fa-1, e2fb-2, and e2fa/b seedlings after sucrose treatment. Seven-day-old seedlings were treated with 90 mM sucrose for 3 h. Gene expression values were calculated as ΔCt. RT-qPCR analysis was performed on three independent biological replicates (n = 3). Bar plots represent mean values, and error bars denote Se. Statistical difference has been depicted by P-value as assessed by one-way ANOVA and Tukey's HSD post hoc test. C, ChIP-qPCR to check enrichment of E2FA–GFP and E2FB–GFP at the promoters of MED17 and CAB1 in 7-day-old Col-0, pgE2FA-GFP, and pgE2FB-GFP seedlings. Amplicon positions relative to ATG are shown in the upper panel. Ct values with and without antibody samples were normalized to input control. Untransformed Col-0 seedlings were taken as negative control. CAB1, which does not possess any E2F-binding motifs in its promoter, was used as a negative control. E2FA and E2FB binding on promoters were calculated as fold enrichment. ChIP-qPCR analysis was performed on three technical replicates (n = 3) from a single representative experiment. Experiments were independently repeated twice (biological replicates; n = 2). Bar plots represent mean values, and error bars denote Sd. Statistical difference has been depicted by P-value as assessed by paired t test. For all the graphs, P-value of 0.05 or lower (P ≤ 0.05) was considered statistically significant, whereas P-value greater than 0.05 (P > 0.05) was considered non-significant (ns).
Discussion
Roots not only anchor the plants in the soil but also help in foraging for nutrients and water. Thus, the RSA, which is defined by the spatial distribution of roots, is highly critical for a plant's growth and survival. The RSA is determined by both the downward (primary roots) and sideways (lateral roots) growth of the root. The optimal root development in plants is a complex process that is influenced by various factors, including phytohormones and sugars. In several reports, auxin and sucrose have been established as the key regulators of RSA (Okushima et al., 2007; Xiong et al., 2013; Gupta et al., 2015; Zhang et al., 2016). However, little is known about the factors directly linking these two important signaling pathways during root development. This study highlights the role of MED17, one of the most important subunits of the Mediator complex, as an important integrator of sucrose and auxin signaling for optimal primary and lateral root development.
The findings that RSA in med17 seedlings is related to reduced responsiveness towards auxin but being insensitive towards sucrose reveal a critical role of this Mediator subunit—hitherto undescribed—in the processing of these two signaling pathways during root development. The mutants of med17 possess short PR and fewer LRs (Figure 1, A–C). The shorter primary root in med17 seedlings is due to reduced cell division and elongation (Figure 1, E–H). The lower number of LRs in med17 is due to defects in the formation of LRP (Figure 1D).
The process of LRP development is governed by auxin, as the exogenous application of auxin stimulates lateral root formation in Col-0 and med17 seedlings (Figure 2B), but their number remains significantly less in med17 seedlings (Figure 2, B and C). However, when we plotted the ratio of the number of LRs in Col-0 and med17 seedlings in +/− exogenous auxin treatment, this ratio was higher in med17, indicating that auxin sensitivity and transport are intact in med17 (Figure 2D). This also indicates that there are other factors which might be affecting LR number in med17 seedlings. However, in normal conditions, i.e. in the absence of exogenous auxin, the number of LRs in med17 is lower, suggesting that the basal auxin level is low in the mutant roots. Low auxin level in med17 LR primordia is also supported by low DR5::GUS signals (Figure 2A). This corroborates the fact that even the auxin-induced expression level of auxin-responsive LBD genes remains significantly lower in med17 seedlings (Figure 2, E–H). Thus, MED17 seems to be required for optimal auxin signaling during root development. On the other hand, the non-responsive phenotype of med17 roots in the presence of exogenous sucrose suggests that MED17 is required for sucrose signaling. There is neither elongation of PR nor formation of new LRs in med17 seedlings in response to sucrose (Figure 3, A–C). Activation of root meristem is the key event for root development and is known to be regulated by sucrose (Xiong et al., 2013). Sucrose-mediated activation of root meristem is impaired in the loss of function mutant of MED17, hampering the development of PR and LRs (Figure 3, A–E). Thus, as the roots of med17 mutants show reduced transcript levels of auxin-responsive genes and complete insensitivity towards sucrose, this study positions MED17 as one of the critical integrators of auxin and sugar signals during root development in Arabidopsis.
Sucrose-mediated meristem activation is regulated by TOR-E2F pathways (Xiong et al., 2013; Li et al., 2017), and E2FA and E2FB have been shown to regulate cell cycle progression in Arabidopsis (Berckmans et al., 2011; Magyar et al., 2012; Leviczky et al., 2019). In response to sucrose/glucose, TOR phosphorylates E2FA and E2FB, which further regulate the transcription of cell cycle genes to maintain meristem activation (Xiong et al., 2013). In this study, MED17 has been found to regulate the transcription of the cell cycle genes MCM5/7, ORC6, and ETG1 by occupying their promoters (Figure 4, A–G). Along with the primary root, sucrose also regulates the development of LRs by inducing the transcription of auxin-responsive LBD genes (Figure 5, A–D; Gupta et al., 2015). However, how sucrose regulates the transcription of LBD genes is not yet known. This study reveals that sucrose-induced expression of LBD genes is occluded in med17 seedlings (Figure 5, A–D). Thus, MED17 seems to be very important for sucrose signaling during root development. The LBD genes are known to be regulated by AUXIN RESPONSE FACTORS, ARF7 and ARF19, which are transcriptional activators of auxin response genes in roots (Okushima et al., 2007). Since sucrose-induced transcription of LBDs was impaired in med17, we hypothesized that MED17 might be regulating this through ARF7 and ARF19. Indeed, sucrose-induced transcription of ARF7 is decreased in med17 (Figure 5E). More importantly, MED17 occupies the promoter of ARF7 (Figure 5G), strengthening the possibility of the involvement of MED17 in sucrose-triggered upregulation of ARF7. Additionally, overexpression of MED17 in the arf7/19 background could not rescue the defective LR phenotype of arf7/19, confirming that MED17 functions upstream of ARF7 (Figure 6, A–C). In an earlier study, the number of LRs was found to be less in arf7 seedlings, and the expression of auxin-responsive LBDs was also impaired (Okushima et al., 2005). On the other hand, there was no change in the LR phenotype and the expression of LBDs in the arf19 mutant (Okushima et al., 2005). In this study, we have found that the expression of ARF7 is induced by sucrose, but ARF19 is not responsive to exogenous sucrose. Moreover, the sucrose-induced upregulation of ARF7 is regulated by MED17 (Figure 5, E–G). Thus, in sucrose signaling, MED17 plays a critical role as it is required for sucrose-induced expression of ARF7, which functions as an activator of LBDs. There are about 23 ARF genes in Arabidopsis (Okushima et al., 2005). In this study, we looked at two ARFs, ARF7 and ARF19, and found that sucrose-induced expression of ARF7 requires MED17, whereas ARF19 is unaffected. This suggests that only a subset of ARF genes are induced by sucrose through MED17. Since not all ARFs are under the control of MED17, it indicates the specificity of its function. This could be the reason that we do not see complete abrogation of auxin-induced upregulation of LBD genes and tuning of the RSA in med17 seedlings (Figure 2, D–G). But the question is, why does MED17 regulate only ARF7 and not ARF19? In order to get this answer, we scanned the promoters of ARF7 and ARF19. Interestingly, we found E2F-binding elements in the promoter of only ARF7, not ARF19, and MED17 has been found to be recruited at the E2F-binding sites. Indeed, MED17 was found to be recruited at the promoter of ARF7 (Figure 5G). The presence of E2F-binding elements in the promoter of ARF7 also indicates the possible role of E2Fs in the transcriptional regulation of ARF7. Indeed, E2Fs were found to regulate the expression of ARF7 as its expression in response to sucrose was significantly decreased in the mutants of e2fa-1, e2fb-2, and e2fa/b (Figure 7A). Additionally, both E2FA and E2FB were found to occupy the promoter of ARF7 (Figure 7, B and C) and could activate its transcription (Figure 7, D and E). Since there is no E2F-binding motif in the promoter of ARF19, we could not observe any enrichment of E2FA and E2FB on its promoter, and consequently, there was no change in its transcription in response to sucrose (Supplemental Figure S6, A–C). Overall, both MED17 and E2Fs occupy the promoters of auxin-responsive TF gene ARF7 and work together to regulate its transcription. Mediator complex is required for the recruitment of RNA Pol II, and MED17 is essential for the structural stability of the complex (Maji et al., 2019). So in the absence of MED17, we observed less occupancy of RNA Pol II on the promoters of ARF7 and the cell cycle genes (Supplemental Figure S8, A–D). Reduced enrichment of RNA Pol II on the target promoters in med17 seedlings highlights the importance of MED17 in the functioning of the Mediator complex as the main recruiter of the transcriptional machinery. Thus, this study elucidates the important role of MED17 in sucrose signaling during root morphogenesis. It seems that both MED17 and E2Fs work together on the same set of promoter sequences to regulate sucrose-responsive RSA. Mediator complexes do not directly bind to DNA; they need other proteins to recruit them at the target promoters. Occupancy of MED17 and E2Fs on the same promoter loci triggered us to think that it might be E2FA and E2FB, which are recruiting MED17 (and Mediator complex) at the sucrose-responsive gene promoters. So we tested the physical interaction between E2FA/B and MED17. Indeed, both E2FA and E2FB interact with MED17 (Figure 8, A and B).
The E2F TFs are already known to be regulated by the glucose–TOR pathway, but to date, there has been no report about the regulation of MED17 by sucrose/glucose or other sugars. This study highlights that the expression of MED17 is also induced upon sucrose treatment (Figure 9A). More importantly, this induction is dependent on E2Fs as there was no upregulation of MED17 in loss-of-function mutants of E2Fs after sucrose treatment (Figure 9B). Additionally, both E2FA and E2FB can occupy the promoter of MED17 (Figure 9C), confirming that the transcription of MED17 is also regulated through the sucrose–E2F pathway.
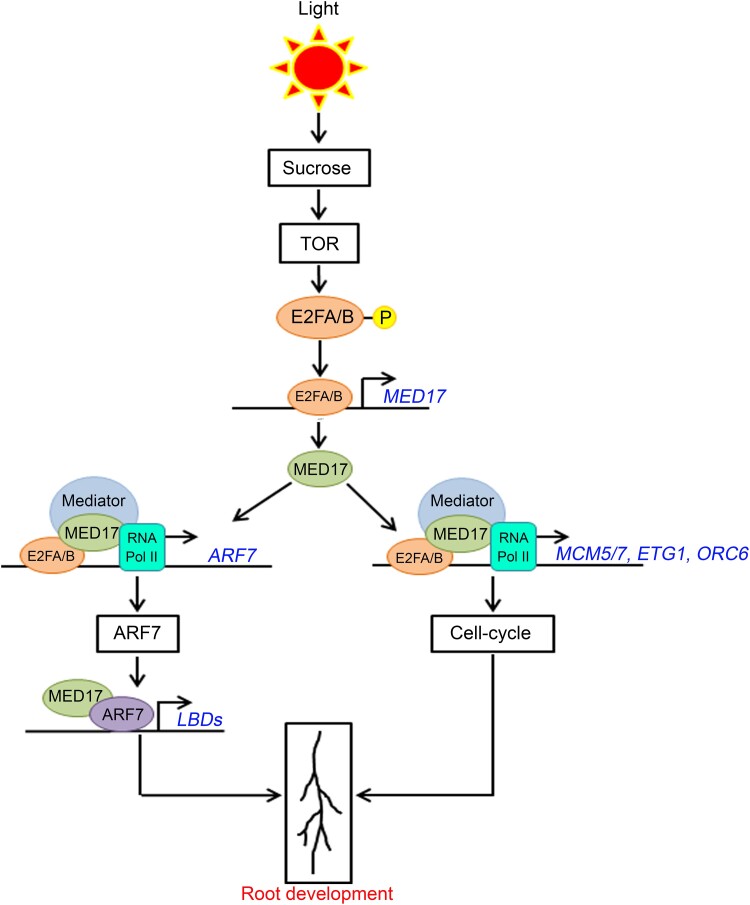
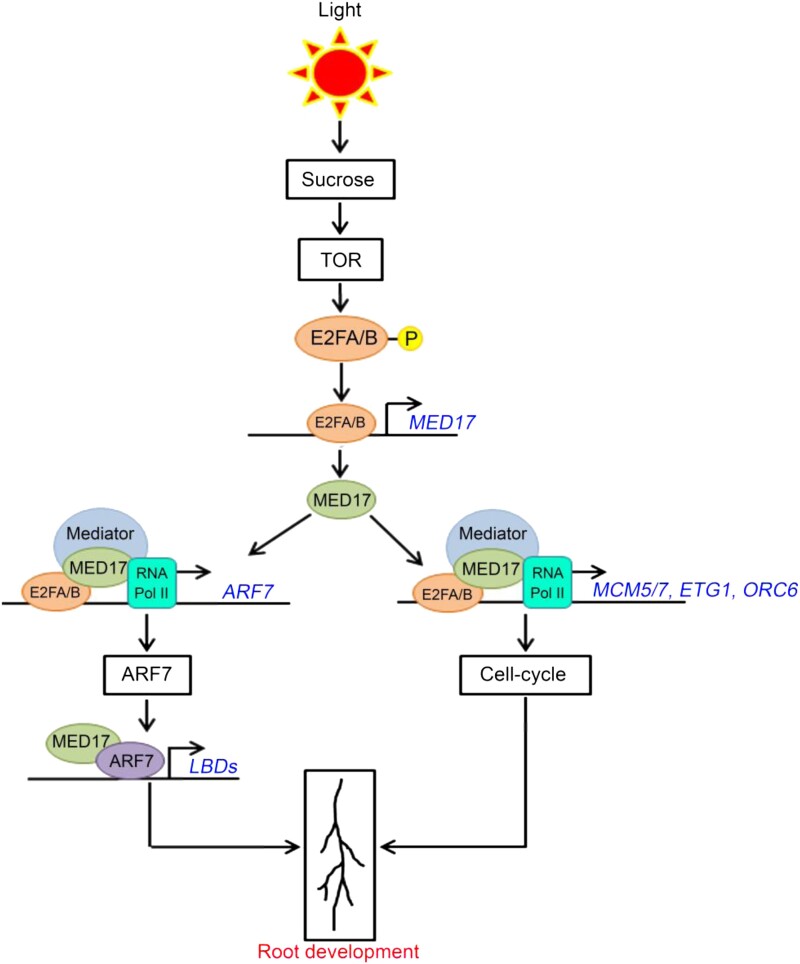
Overall, this study provides an understanding of the tuning of auxin responses by sucrose through the Mediator complex. Overall, the model is as follows (Figure 10). In response to sucrose, TOR activates E2FA and E2FB by phosphorylating them. The active E2FA/B TFs bind to the promoter of MED17 and activate its transcription. The occupancy of both E2FA/B and MED17 on the promoter of ARF7 suggests that E2FA/B can recruit the Mediator complex through their interaction with MED17 protein and induce the transcription of ARF7. After transcriptional induction, MED17 physically interacts with ARF7. The ARF7 TF, in turn, regulates auxin signaling by activating the transcription of LBD genes leading to the development of LRs. On the other hand, the presence of E2FA/B and MED17 on the promoters of cell cycle genes suggests that these genes are also induced by the E2F–Mediator complex leading to the activation of root meristems. Thus, MED17 (and the Mediator complex) coordinates with the sucrose-activated E2FA/B TFs to regulate the effect of auxin signaling on the development of PRs and LRs. As both PRs and LRs constitute the RSA, this study underlines the critical role of the Mediator complex in processing the signals of sugars and hormones for the optimal root system architecture in plants.
Figure 10.
Putative model explaining regulation of root system architecture by Mediator subunit MED17. In the presence of sucrose, TOR phosphorylates and activates TFs E2FA and E2FB (Xiong et al., 2013). The active E2FA/B TFs bind to the promoter of MED17 and induce its transcription. In addition to inducing the transcription of MED17, the E2FA/B TFs physically interact with MED17 protein. Thus, it seems that the E2FA/B TFs recruit the Mediator complex (through their interaction with MED17 subunit) on the promoters of the cell cycle genes MCM5, ETG1, and ORC6 and the auxin-responsive ARF7 TF gene and activate their transcription. In the subsequent step, ARF7 interacts with MED17 and regulates the expression of auxin-responsive LBD genes LBD16, LBD18, LBD29, and LBD33. As these genes (MCM5, ETG1, ORC6, and LBDs) collectively regulate the overall root architecture, MED17 seems to play a critical role in integrating the sucrose and auxin signaling to regulate the root system architecture in Arabidopsis.
Materials and methods
Plant materials and growth condition
All the mutant and overexpression seeds used in the study were in the Col-0 background. The Arabidopsis (Arabidopsis thaliana) mutant seeds of med17 (AT5G20170; salk_102813, previously described in Agrawal et al., 2022), e2fa-1 (AT2G36010; SALK_103138), e2fb-2 (AT5G22220; SALK_120959), and arf7/19 (CS24629) were obtained from The Arabidopsis Biological Resource Centre at Ohio State University (http://arabidopsis.info/). Double mutant line e2fa/b and transgenic lines pgE2FA-GFP and pgE2GB-GFP were obtained from originally published resources. The complementation line of med17 (med17/35S::YFP-MED17) used in this study has been described previously (Agrawal et al., 2022). Despite several attempts, unfortunately, we could not raise transgenics to express MED17 under the control of an endogenous MED17 promoter. So, we had to stick with 35S-driven overexpression of MED17. We also confirmed transgenic line arf7/19/35S::YFP-MED17 by real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR)) showing induced expression of MED17 (Figure 6A). Seeds of wild-type, mutants, and transgenic lines were surface-sterilized and incubated at 4°C for 48 h. Seeds were sown on square petri plates (120 × 120 mm) containing ½ MS medium with 1% (w/v) sucrose (30 mM) and 0.8% (w/v) agar (24 mM) under sterile conditions. Seed germination was carried out in a climate-controlled growth room.
Transgenic plants
To generate the complementation transgenic plants, the full-length coding sequence (CDS) of the Arabidopsis MED17 subunit was amplified from complementary DNA (cDNA) samples prepared from 7-day-old Arabidopsis Col-0 plants using primer pairs described in the Supplemental Table S1. Amplified fragments were cloned into a gateway-derived pENTR/D-TOPO vector. Full-length CDS was confirmed through sequencing and transferred into gateway-derived pSITE-3CA (stock #CD3-1638) binary vector containing CaMV35S promoter fused with enhanced yellow fluorescent protein (eYFP) reporter. The CaMV35S::YFP-MED17 construct was then transformed into Agrobacterium tumefaciens strain GV3101. Agrobacterium harboring CaMV35S::YFP-MED17 construct was transformed into Arabidopsis arf7/19 background plants using the floral dip method (Clough and Bent, 1998). For the selection of transgenic plants, next-generation transgenic seeds were plated onto MS media containing kanamycin. The MED17 overexpression in the arf7/19 background was confirmed by examining the expression of MED17 using RT-qPCR. Transgenic lines proMED7::GUS and med17/35S::YFP-MED17 have already been described (Agrawal et al., 2022).
Measurement of root length and counting of LRP
For quantification of root length, Arabidopsis Col-0, mutant, and transgenic seedlings were directly grown on ½ MS or different concentrations of sucrose (30 and 90 mM) for 9 to 12 days in square petri plates under long-day (16-h light/8-h dark; 60 µmol m−2 s−1 white light intensity) photoperiod. Seedlings were photographed with a digital camera (Nikon). Root lengths were measured using ImageJ. Three biological replicates were taken for each experiment. Each experiment was considered an independent biological replicate containing at least 20 seedlings. For LRP staging, Col-0 and med17 seedlings were grown on ½ MS for 7 days, and counting was done according to Malamy and Benfey (1997).
Propidium iodide
For fluorescent staining with propidium iodide (PI), plants were transferred from the growth medium to 2 mg mL−1 of PI solution on microscope slides. The samples were recorded separately at wavelengths specific to PI fluorescence, 550 nm (excitation) and 650 nm (emission), using a confocal microscope.
EDU staining
EDU staining was performed as described (Kotogány et al., 2010) using an EDU detection cocktail (Invitrogen). Briefly, Col-0, med17, and med17/35S::YFP-MED17 seedlings were grown on ½ MS containing 1% sucrose for 4 days. For the root meristem activation experiment, 4-day-old seedlings were kept in the dark for 16 h for the depletion of internal sucrose leading to the sucrose treatment (90 mM) for 3 h. Seedlings were treated with 10 µM EDU for 3 h. Seedlings were then fixed in 4% (w/v) formaldehyde solution in phosphate buffer saline (PBS), followed by 0.1% Triton X-100 treatment for 30 min. Fixer was washed with PBS (3 × 10 min), and then seedlings were incubated in EDU detection cocktail for 1 h in the dark, followed by PBS wash (3 × 10 min) before observation by confocal microscope. Fluorescence was recorded at specific wavelengths: 491 nm for excitation and 520 nm for emission.
IAA and sucrose treatment
For IAA treatment, 4-day-old seedlings were transferred to different concentrations of IAA (25 nM, 50 nM, 100 nM, 0.5 µM, and 1 µM) for another 4 days. For sucrose treatment, 4-day-old seedlings were transferred to ½ MS containing either 30 or 90 mM sucrose for another 4 days. Treated seedlings were then photographed for PR length and LR number measurement.
Gene expression analysis
The RT-qPCR was performed for gene expression analysis. The seedlings of Col-0, mutants, and transgenics were grown on square petri plates containing 0.5X MS media supplemented with different concentrations of sucrose and 0.5 g L−1 MES hydrate. Medium was solidified with 0.8% (w/v) agar. The plates were kept vertically in the Arabidopsis growth room at 22°C under LD conditions (16-h light/8-h dark; 60 µmol m−2 s−1 white light intensity). For cell cycle gene expression analysis, 4-day-old seedlings were treated with (90 mM) or without (0 mM) sucrose for 3 h. For the expression of ARFs and LBDs, 7-day-old seedlings were treated with (90 mM) or without (0 mM) sucrose for 3 h, or seedlings were kept in either liquid ½ MS or ½ MS supplemented with auxin (10 µM) for 3 h. For the expression of MED17, 7-day-old seedlings were treated with (90 mM) or without (0 mM) sucrose for 3 h. Total RNA was isolated using RNeasy Plant Mini Kits (Qiagen), followed by reverse transcription using Verso cDNA Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer's protocol. One microgram of total RNA was taken for cDNA preparation. Primers for unique sequences were designed using Primer Express (version 3.0; Applied Biosystems). RT-qPCR was performed using QuantStudio™ 6 Flex Real-Time PCR Systems. Gene expression values were calculated as ΔCt. RT-qPCR analysis was performed on three independent biological replicates (n = 3). 18S rRNA and GAPC2 were used as a reference gene to calculate relative expression values (ΔΔCt values). All the primers used in this study are mentioned in Supplemental Table S1.
Chromatin immunoprecipitation assay
ChIP assays were performed as previously described (Saleh et al., 2008) with minor modifications. For the binding of MED17, E2FA, and E2FB on the promoters of genes regulating RSA, 7-day-old 0.5X MS grown pgE2FA-GFP, pgE2FB-GFP, and med17/CaMV35S::YFP-MED17 seedlings were grown under LD conditions. Untransformed Col-0 seedlings were taken as a negative control. Briefly, 2-g tissue of each sample was cross-linked with 1% (v/v) formaldehyde to fix protein–DNA complexes. The samples were crushed in liquid N2 and homogenized in nuclei isolation and nuclei lysis buffers, followed by sonication. Sonication of chromatin was done in 4°C water sonicator (Diagenode Bioruptor Plus). Sonicated samples were first precleared with Protein A Agarose beads (Millipore #16-157) and then mixed with antibodies. Antibodies against GFP were obtained from Abcam (GFP, ab290). The immunocaptured protein–DNA complexes were washed, and precipitated DNA samples were recovered and analyzed by ChIP-qPCR using the primer pairs designed for specific sites (E2F-binding elements). All the primers used for ChIP-qPCR are listed in Supplemental Table S1.
GUS staining
For the GUS assay, 7-day-old homozygous proMED17:GUS transgenic seedlings were treated with (90 mM) or without (0 mM) sucrose for 3 h. For GUS detection in DR5-GUS and med17/DR5-GUS lines, seedlings were grown on ½ MS for 7 days. Seedlings were subjected to GUS solution and kept at 37°C for 4 h under the dark. Following GUS detection, seedlings were bleached with 70% ethanol for the removal of chlorophyll. Images were taken using a Stereo zoom microscope.
In vitro pull-down assay
IP was performed as described in Magyar et al. (2012). For the pull-down experiment, AtMED17 was expressed in bacteria. Total plant protein was extracted from transgenic seedlings pgE2FA-GFP and pgE2FB-GFP. Bacterial expressed GST-MED17 or only GST was incubated with plant protein extracts overnight in the cold room. Immunoprecipitation was done using Glutathione Sepharose® 4 Fast Flow beads (GE17-5132-01). Immunoprecipitated protein samples were run on sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and detected using the anti-GFP antibody (Abcam-ab290). GST-MED17 and only GST were detected using an anti-GST antibody (sc-138).
BiFC
E2FA, E2FB, and MED17 were cloned in CD3-1651 and CD3-1648 vectors, respectively. For the interaction of MED17 with ARF7, MED17 and ARF7 were cloned in CD3-1651 and CD3-1648 vectors, respectively. The plasmids were transformed into Agrobacterium tumefaciens strain GV3101 and infiltrated in N. benthamiana leaf as described previously (Sparkes et al., 2006). Infected N. benthamiana leaves were imaged after 48 h using a Leica SP8 confocal microscope. Along with the YFP (excitation 490 nm and emission 520 nm) channel, a red fluorescent channel (excitation 550 nm and emission 580 nm) was also used to detect the autofluorescence of chloroplasts.
Dual-luciferase reporter assay
To test the transactivation of ARF7, 1 kb promoter region of ARF7 harboring E2F-binding elements was cloned into p635nRRF vector, which contains 35S:REN. TFs E2FA and E2FB were cloned into the pSITE-3CA vector. The reporters were coexpressed with different effector constructs into N. benthamiana leaves, and empty vector served as control. After infiltration, plants were incubated at 22°C for 12 h in the dark and then transferred to light condition for 48 h. ChemiDoc imaging system was used for capturing luciferase (LUC) images. For the measurement of luciferase activities, infiltered leaf disks were ground to a fine powder in liquid nitrogen and mixed with 1× passive lysis buffer provided in the Dual-Luciferase® Reporter Assay System (Promega, USA). Firefly luciferase and Renillia (REN) activities were measured following the manufacturer's instructions (Promega, USA). The luciferase activity was calculated by normalizing the REN expression level.
Quantification and statistical analysis
For the gene expression analysis, values were calculated as ΔΔCt. RT-qPCR analysis was performed on three independent biological replicates (n = 3). For the ChIP-qPCR experiments, analysis was performed on three technical replicates (n = 3) from a single representative experiment. Experiments were independently repeated twice or thrice (or otherwise mentioned in the figure legends). For each ChIP experiment, a second biological replicate is shown in the supplemental figures. All the experiments, including physiological, confocal, and ChIP experiments yielding similar results/trends, were repeated as described in the figure legends. For physiological analysis, each experiment was considered as an independent biological replicate containing at least 20 seedlings. Statistical significance has been denoted by the P-value above the bar graphs as assessed by one-way analysis of variance (ANOVA) and Tukey's honestly significant difference (HSD) post hoc test. For all the graphs, P-value of 0.05 or lower (P ≤ 0.05) is considered statistically significant, whereas P-value greater than 0.05 (P > 0.05) is considered non-significant. All analysis was done using Microsoft excel. All the graphs were made using Microsoft Excel. All the statistical analysis was performed using SigmaPlot GraphPad Prism 5.
Accession numbers
The sequence data for the genes used in this article can be found in the GenBank/EMBL data libraries under accession numbers MED17 (AT5G20170), E2FA (AT2G36010), E2FB (AT5G22220), ARF7 (AT5G20730), ARF19 (AT1G19220), LBD16 (AT2G42430), LBD18 (AT2G45420), LBD29 (AT3G58190), LBD33 (AT5G06080), MCM5 (AT2G07690), MCM7 (AT4G02060), ETG1 (AT2G40550), ORC6 (AT1G26840), and CAB1 (AT1G29930).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Root phenotype of med17 is rescued by its complementation with 35S::YFP-MED17.
Supplemental Figure S2 . Effect of exogenous auxin on lateral roots.
Supplemental Figure S3 . MED17 regulates expression of auxin-responsive LBD genes.
Supplemental Figure S4 . Defective root meristem activation of med17 is rescued by 35S::YFP-MED17.
Supplemental Figure S5 . Second biological replicate showing enrichment of MED17 at the promoters of ARF7 and cell-cycle genes.
Supplemental Figure S6 . Second biological replicate showing enrichment of E2FA and E2FB at the promoters of auxin responsive TF genes.
Supplemental Figure S7 . Second biological replicate showing enrichment of E2FA and E2FB at the promoter of MED17.
Supplemental Figure S8 . MED17 is required for enrichment of RNA Pol II on the promoters of ARF7 and cell cycles genes.
Supplemental Table S1 . Primers used in this study.
Supplementary Material
Acknowledgments
We thank the DBTeLibrary consortium (DeLCON) for all the literatures. Support from all the facilities of NIPGR and ICGEB are acknowledged. Authors are thankful to Dr. Mohan Sharma, Albert-Ludwigs-Universität Freiburg, Germany, for editing the manuscript.
Contributor Information
Rekha Agrawal, Plant Mediator Lab, National Institute of Plant Genome Research, Aruna Asaf Ali Marg, New Delhi 110067, India.
Amrita Singh, Plant Transcription Regulation, International Centre for Genetic Engineering and Biotechnology, Aruna Asaf Ali Marg, New Delhi 110067, India.
Jitender Giri, Plant Nutritional Lab, National Institute of Plant Genome Research, Aruna Asaf Ali Marg, New Delhi 110067, India.
Zoltan Magyar, Molecular Regulation of Plant Development and Adaptation, Institute of Plant Biology, Biological Research Centre, Szeged 6728, Hungary.
Jitendra Kumar Thakur, Plant Mediator Lab, National Institute of Plant Genome Research, Aruna Asaf Ali Marg, New Delhi 110067, India; Plant Transcription Regulation, International Centre for Genetic Engineering and Biotechnology, Aruna Asaf Ali Marg, New Delhi 110067, India.
Data availability
The data that support the findings of this study are available on request from the corresponding author.
Funding
This work was supported by the grant BT/PR13009/BPA/118/71/2015 from the Department of Biotechnology, Government of India. R.A. and A.S. acknowledge fellowships from the Council of Scientific and Industrial Research, Government of India and Department of Biotechnology, Government of India.
References
- Agrawal R, Jiří F, Thakur JK. The kinase module of the Mediator complex: an important signalling processor for the development and survival of plants. J Exp Bot. 2020:72(2):224–240. 10.1093/jxb/eraa439 [DOI] [PubMed] [Google Scholar]
- Agrawal R, Sharma M, Dwivedi N, Maji S, Thakur P, Laxmi A, Thakur JK, Asaf A, Marg A, Asaf A, et al. MEDIATOR SUBUNIT17 integrates jasmonate and auxin signaling pathways to regulate thermomorphogenesis. Plant Physiol. 2022:189(4):2259–2228. 10.1093/plphys/kiac220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckström S, Elfving N, Nilsson R, Wingsle G, Björklund S. Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell. 2007:26(5): 717–729. 10.1016/j.molcel.2007.05.007 [DOI] [PubMed] [Google Scholar]
- Berckmans B, Vassileva V, Schmid SPC, Maes S, Parizot B, Naramoto S, Magyar Z, Lessa Alvim Kamei C, et al. (2011) Auxin-dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell. 23(10): 3671–3683. 10.1105/tpc.111.088377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Heidstra R, Aida M, Palme K, Scheres B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005:433(7021):39–44. 10.1038/nature03184 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998:16(6):735–743. 10.1046/j.1365-313X.1998.00343.x [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Inzé D. The ins and outs of the plant cell cycle. Nat Rev Mol Cell Biol. 2007:8(8): 655–665. 10.1038/nrm2227 [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benková E. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci USA. 2008:105(25): 8790–8794. 10.1073/pnas.0712307105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Zhu J, Du X, Cui X. Effects of three auxin-inducible LBD members on lateral root formation in Arabidopsis thaliana. Planta. 2012:236(4): 1227–1237. 10.1007/s00425-012-1673-3 [DOI] [PubMed] [Google Scholar]
- Gallavotti A. The role of auxin in shaping shoot architecture. J Exp Bot. 2013:64(9): 2593–2608. 10.1093/jxb/ert141 [DOI] [PubMed] [Google Scholar]
- Giustozzi M, Freytes SN, Jaskolowski A, Lichy M, Mateos J, Falcone Ferreyra ML, Rosano GL, Cerdán P, Casati P. Arabidopsis mediator subunit 17 connects transcription with DNA repair after UV-B exposure. Plant J. 2022:110(4):1047–1067. 10.1111/tpj.15722 [DOI] [PubMed] [Google Scholar]
- Guglielmi B, van Berkum NL, Klapholz B, Bijma T, Boube M, Boschiero C, Bourbon HM, Holstege FCP, Werner M. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 2004:32(18): 5379–5391. 10.1093/nar/gkh878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Singh M, Laxmi A. Interaction between glucose and brassinosteroid during the regulation of lateral root development in Arabidopsis. Plant Physiol. 2015:168(1): 307–320. 10.1104/pp.114.256313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh THS, Weiner A, Lajoie B, Dekker J, Friedman N, Rando OJ. Mapping nucleosome resolution chromosome folding in yeast by micro-C. Cell. 2015:162(1): 108–119. 10.1016/j.cell.2015.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li W, Yao X, Lin QJ, Yin JW, Liang Y, Heiner M, Tian B, Hui J, Wang G. Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol Cell. 2012:45(4): 459–469. 10.1016/j.molcel.2011.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram PA, Malamy JE. Root system architecture. Science Direct. 2010:55: 75–117. 10.1016/B978-0-12-380868-4.00002-8 [DOI] [Google Scholar]
- Ioio RD, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. A genetic framework for the control of cell division and differentiation in the root meristem. Science. 2008:322(5906): 1380–1384. 10.1126/science.1164147 [DOI] [PubMed] [Google Scholar]
- Ito J, Fukaki H, Onoda M, Li L, Li C, Tasaka M, Furutani M. Auxin-dependent compositional change in Mediator in ARF7- and ARF19-mediated transcription. Proc Natl Acad Sci USA. 2016:113(23): 6562–6567. 10.1073/pnas.1600739113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov VB, Dubrovsky JG. Longitudinal zonation pattern in plant roots: conflicts and solutions. Trends Plant Sci. 2013:18(5): 237–243. 10.1016/j.tplants.2012.10.002 [DOI] [PubMed] [Google Scholar]
- Iwakawa H, Ueno Y, Semiarti E, Onouchi H, Kojima S, Tsukaya H, Hasebe M, Soma T, Ikezaki M, Machida C, et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002:43(5): 467–478. 10.1093/pcp/pcf077 [DOI] [PubMed] [Google Scholar]
- Ketelaar T, Faivre-Moskalenko C, Esseling JJ, De Ruijter NCA, Grierson CS, Dogterom M, Emons AMC. Positioning of nuclei in Arabidopsis root hairs: an actin-regulated process of tip growth. Plant Cell. 2002:14(11): 2941–2955. 10.1105/tpc.005892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Zheng B, Yu Y, Won SY, Mo B, Chen X. The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 2011:30(5): 814–822. 10.1038/emboj.2011.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Schopfer P. Photosynthetic sucrose acts as cotyledon-derived long-distance signal to control root growth during early seedling development in Arabidopsis. Proc Natl Acad Sci. 2012:109(28): 11217–11221. 10.1073/pnas.1203746109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick DA, Westfall CS, Lee SG, Nanao MH, Dumas R, Hagen G, Guilfoyle TJ, Jez JM, Strader LC. Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci. 2014:111(14): 5427–5432. 10.1073/pnas.1400074111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotogány E, Dudits D, Horváth G V, Ayaydin F. A rapid and robust assay for detection of S-phase cell cycle progression in plant cells and tissues by using ethynyl deoxyuridine. Plant Methods. 2010:6(1): 1–15. 10.1186/1746-4811-6-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leviczky T, Molnár E, Papdi C, Szi E O, Horváth G V, Vizler C, Nagy V, Pauk J, Bögre L, Magyar Z. E2FA and E2FB transcription factors coordinate cell proliferation with seed maturation. Development. 2019:146(22):dev179333. 10.1242/dev.179333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cai W, Liu Y, Li H, Fu L, Liu Z, Xu L, Liu H, Xu T, Xiong Y. Differential TOR activation and cell proliferation in Arabidopsis root and shoot apexes. Proc Natl Acad Sci USA. 2017:114(10): 2765–2770. 10.1073/pnas.1618782114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor DR, Deak KI, Ingram PA, Malamy JE. Root system architecture in Arabidopsis grown in culture is regulated by sucrose uptake in the aerial tissues. Plant Cell. 2008:20(10): 2643–2660. 10.1105/tpc.107.055475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar Z, Bögre L, Ito M. DREAMs make plant cells to cycle or to become quiescent. Curr Opin Plant Biol. 2016:34: 100–106. 10.1016/j.pbi.2016.10.002 [DOI] [PubMed] [Google Scholar]
- Magyar Z, Horváth B, Khan S, Mohammed B, Henriques R, De Veylder L, Bakó L, Scheres B, Bögre L. Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes. EMBO J. 2012:31(6): 1480–1493. 10.1038/emboj.2012.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maji S, Dahiya P, Waseem M, Dwivedi N, Bhat DS, Dar TH, Thakur JK. Interaction map of Arabidopsis Mediator complex expounding its topology. Nucleic Acids Res. 2019:47(8): 3904–3920. 10.1093/nar/gkz122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997:124(1):33–44. 10.1242/dev.124.1.33 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Ryan KS. Environmental regulation of lateral root initiation in Arabidopsis. Plant Physiol. 2001:127(3): 899–909. 10.1104/pp.010406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik N, Agarwal P, Tyagi A. Emerging functions of multi-protein complex Mediator with special emphasis on plants. Crit Rev Biochem Mol Biol. 2017:52(5):475–502. 10.1080/10409238.2017.1325830 [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010:11(11): 761–772. 10.1038/nrg2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muday GK, Haworth P. Tomato root growth, gravitropism, and lateral development: correlation with auxin transport. Plant Physiol Biochem PPB. 1994:32(2): 193–203 [PubMed] [Google Scholar]
- Nagulapalli M, Maji S, Dwivedi N, Dahiya P, Thakur JK. Evolution of disorder in Mediator complex and its functional relevance. Nucleic Acids Res. 2016:44(4): 1591–1612. 10.1093/nar/gkv1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanao MH, Vinos-Poyo T, Brunoud G, Thévenon E, Mazzoleni M, Mast D, Lainé S, Wang S, Hagen G, Li H, et al. Structural basis for oligomerization of auxin transcriptional regulators. Nat Commun. 2014:5(1): 3617. 10.1038/ncomms4617 [DOI] [PubMed] [Google Scholar]
- Nozawa K, Schneider TR, Cramer P. Core Mediator structure at 3.4 Å extends model of transcription initiation complex. Nature. 2017:545(7653): 248–251. 10.1038/nature22328 [DOI] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell Online. 2007:19(1): 118–130. 10.1105/tpc.106.047761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005:17(2): 444–463. 10.1105/tpc.104.028316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oszi E, Papdi C, Mohammed B, Petkó-Szandtner A, Leviczky T, Molnár E, Galvan-Ampudia C, Khan S, Juez EL, Horváth B, et al. E2FB interacts with RETINOBLASTOMA RELATED and regulates cell proliferation during leaf development. Plant Physiol. 2020:182(1): 518–533. 10.1104/pp.19.00212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN. Control of arabidopsis root development. Annu Rev Plant Biol. 2012:63(1): 563–590. 10.1146/annurev-arplant-042811-105501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posner J, Bradley S. Peterson JAR . 基因的改变NIH public access. Bone. 2008:23: 1–7 [Google Scholar]
- Poss ZC, Ebmeier CC, Taatjes DJ. The Mediator complex and transcription regulation. Crit Rev Biochem Mol Biol. 2013:48(6): 575–608. 10.3109/10409238.2013.840259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya-González J, López-Bucio JS, Prado-Rodríguez JC, Ruiz-Herrera LF, Guevara-García ÁA, López-Bucio J. The MEDIATOR genes MED12 and MED13 control Arabidopsis root system configuration influencing sugar and auxin responses. Plant Mol Biol. 2017:95(1–2):141–156. 10.1007/s11103-017-0647-z [DOI] [PubMed] [Google Scholar]
- Raya-González J, Ojeda-Rivera JO, Mora-Macias J, Oropeza-Aburto A, Ruiz-Herrera LF, López-Bucio J, Herrera-Estrella L. MEDIATOR16 Orchestrates local and systemic responses to phosphate scarcity in Arabidopsis roots. New Phytol. 2021:229(3): 1278–1288. 10.1111/nph.16989 [DOI] [PubMed] [Google Scholar]
- Raya-González J, Oropeza-Aburto A, López-Bucio JS, Guevara-García ÁA, de Veylder L, López-Bucio J, Herrera-Estrella L. MEDIATOR18 influences Arabidopsis root architecture, represses auxin signaling and is a critical factor for cell viability in root meristems. Plant J. 2018:96(5): 895–909. 10.1111/tpj.14114 [DOI] [PubMed] [Google Scholar]
- Raya-González J, Ortiz-Castro R, Ruíz-Herrera LF, Kazan K, López-Bucio J. Phytochrome and flowering time1/mediator25: regulates lateral root formation via auxin signaling in Arabidopsis. Plant Physiol. 2014:165(2): 880–894. 10.1104/pp.114.239806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Aguilar B, Raya-González J, López-Bucio JS, de la Cruz H R, Herrera-Estrella L, Ruiz-Herrera LF, Martínez-Trujillo M, López-Bucio J. Mutation of MEDIATOR 18 and chromate trigger twinning of the primary root meristem in Arabidopsis. Plant Cell Environ. 2020:43(8): 1989–1999. 10.1111/pce.13786 [DOI] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc. 2008:3(6):1018–1025. 10.1038/nprot.2008.66 [DOI] [PubMed] [Google Scholar]
- Samanta S, Thakur JK. Importance of Mediator complex in the regulation and integration of diverse signaling pathways in plants. Front Plant Sci. 2015:6: 757. 10.3389/fpls.2015.00757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai B, Reynaga-Peña CG, Springer PS. The Lateral Organ Boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002:129(2): 747–761. 10.1104/pp.010926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc. 2006:1(4): 2019–2025. 10.1038/nprot.2006.286 [DOI] [PubMed] [Google Scholar]
- Tantale K, Mueller F, Kozulic-Pirher A, Lesne A, Victor JM, Robert MC, Capozi S, Chouaib R, Bäcker V, Mateos-Langerak J, et al. A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat Commun. 2016:7(1): 12248. 10.1038/ncomms12248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Creation of a highly active synthetic AuxRE. Society. 1997:9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangheluwe N, Beeckman T. Lateral root initiation and the analysis of gene function using genome editing with CRISPR in Arabidopsis. Genes (Basel). 2021:12(6):884. 10.3390/genes12060884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Ruan YL. Regulation of cell division and expansion by sugar and auxin signaling. Front Plant Sci. 2013:4: 1–9. 10.3389/fpls.2013.00163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell. 2005:123(7): 1337–1349. 10.1016/j.cell.2005.09.042 [DOI] [PubMed] [Google Scholar]
- Wyrzykowska J, Schorderet M, Pien S, Gruissem W, Fleming AJ. Induction of differentiation in the shoot apical meristem by transient overexpression of a retinoblastoma-related protein. Plant Physiol. 2006:141(4): 1338–1348. 10.1104/pp.106.083022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature. 2013:496(7444): 181–186. 10.1038/nature12030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhu JY, Roh J, Marchive C, Kim SK, Meyer C, Sun Y, Wang W, Wang ZY. TOR Signaling promotes accumulation of BZR1 to balance growth with carbon availability in Arabidopsis. Curr Biol. 2016:26(14): 1854–1860. 10.1016/j.cub.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.