Abstract
The genomics and pathways governing metastatic dormancy are critically important drivers of long-term patient survival given the considerable portion of cancers that recur aggressively months to years after initial treatments. Our understanding of dormancy has expanded greatly in the last two decades, with studies elucidating that the dormant state is regulated by multiple genes, microenvironmental (ME) interactions, and immune components. These forces are exerted through mechanisms that are intrinsic to the tumor cell, manifested through cross-talk between tumor and ME cells including those from the immune system, and regulated by angiogenic processes in the nascent micrometastatic niche. The development of new in vivo and 3D ME models, as well as enhancements to decades-old tumor cell pedigree models that span the development of metastatic dormancy to aggressive growth, has helped fuel what arguably is one of the least understood areas of cancer biology that nonetheless contributes immensely to patient mortality. The current review focuses on the genes and molecular pathways that regulate dormancy via tumor-intrinsic and ME cells, and how groups have envisioned harnessing these therapeutically to benefit patient survival.
Keywords: Metastasis, Genomics, Dormancy, Dormancy reawakening
1. Introduction
The vast majority of cancer deaths are associated with a disease that recurs months or years after multiple local and systemic treatment modalities have been used to remove primary tumor lesions and, as well, to mitigate the existence of actively proliferating tumor cell disseminations [1, 2]. As an example, primary breast cancers (BrCa) have been subtyped by the World Health Organization for treatment purposes: molecular (luminal A, luminal B, ERB-B2/HER2-positive and triple-negative (TNBC)) and immunohistological subtypes (estrogen receptor (ER)-positive, progesterone receptor (PR)-positive, ERB-B2 (HER2)-positive). Yet, even though these subtypes have different patterns of aggressiveness and are treated differently, disease recurrence that results from the reactivation of dormant cells can occur in any of these subtypes, and moreover, these relapses are the major contributor to BrCa-associated morbidity [3]. Some differences exist regarding the onset of disease recurrence relative to BrCa subtypes. For example, even though systemic adjuvant endocrine therapy for ER+ cases can improve metastasis-free survival, the risk of disease relapse rises steadily after initial treatments, peaking at about 5 years post-diagnosis, whereas the risk of recurrence in TNBC peaks earlier at about 2 years [4].
It is important to first clarify the sometimes nuanced biological and functional differences in this field that have contributed to cancers being labeled as either “dormant” or “quiescent” or that dormancy can be governed by pathways intrinsic to tumor cells (“cell dormancy”) versus those controlled by ME cells in a metastatic niche (“tumor dormancy”). As elegantly described in the review by Phan and Croucher [5], a dormant tumor is one that arises typically at secondary sites months to years after the identification, treatment, and clinical “cure” of a primary-site tumor. Three possibilities contribute to this: (i) single early-disseminating tumor cells can survive at peripheral sites in a non-proliferative dormant state (“cell dormancy”), or (ii) micrometastases made up of slowly proliferating cells or cells with equal levels of proliferation and death (“tumor dormancy”) disseminate to and are sustained in peripheral tissues. In a strict sense, dormant cells are non-cycling tumor cells isolated from in vivo sites or 3D tumor microenvironment (TME) cultures by virtue of their ability to, for example, retain vital dyes that would otherwise be diluted in proliferating cells. Quiescent cells are dormant cells with reversible cell cycle arrest. Examples of human and mouse breast cancer cell lines that have been used to isolate dormant cells in the lung, bone, and/or brain are listed in Table 1.
Table 1.
In vivo models of dormancy and dormancy reawakening
| Breast cancer models | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lung | Bone | Brain | ||||||||||
| Cell line | Species | Hosta | Subtypeb | Injection routec | Met. | Dormancy (weeks) |
Met. | Dormancy (weeks) |
Met. | Dormancy (weeks) |
Findings | Ref. |
| T47D | Human | NSG, nude, NOD/SCID | ER+, PR+ | IC, SQ, ortho | Yes | Yes (2) | Yes | Yes (8) | N.R. | N.R. | No growing metastases were detected 4–5 weeks after orthotopic injection, whereas ressection of the primary tumor induced growing bone metastases over the next 8 weeks (similar finding with MCF-7 cells). Genes most associated with actively growing bone metastases (vs. primary tumor) include MYC, CLDN1, IL1B, CRSK, and TNFRSF11A | [6-10] |
| T47D-DBM | Human | Nude | ER+, PR+ | IC | N.R. | N.R. | Yes | Yes (6–8) | N.R. | N.R. | Dormancy in DBM cells is associated with the downregulation of mitosis genes whereas reawakening was associated with an increase ECM-encoding genes | [6] |
| MDA-MB-231-SCP6 | Human | Nude | TN | IC | N.R. | N.R. | Yes | Yes (14) | N.R. | N.R. | SCP6, which lacks a signature of active bone metastatic growth (including CXCR4, IL11, CTGF, MMP1, OPN), remained dormant in bones 100 days after IC injection, with 10% of cases then showing progressing bone metastases (resulting in PD lines). Knockdown of VCAM-1, but not IL1B, TFPI2, MAGEB2 or KLRC1, decreased metastatic ability of PD1 cells | [11] |
| MCF-7 | Human | Nude | ER+, PR+ | IV, IC, ortho | Yes | Yes (9) | Yes | Yes (8) | N.R. | N.R. | See data for T47D and D2.0R cells | [8, 12-18] |
| HCC1954-LCC1 | Human | Nude | TN | IC | N.R. | N.R. | N.R. | N.R. | Yes | Yes (12) | Latent LCC1 cells isolated from brains showed SOX9 upregulation and downregulation of MYC and WNT pathways | [19] |
| HMT-3522-T4-2 | Human | NOD/SCID | TN | IC | Yes | Yes (8) | Yes | Yes (8) | Yes | Yes (8) | Notch1 factors secreted by endothelial cells increased the dormancy of HMT-3522-T4-2, whereas POSTN, TNC, VCAN, and FN1 added to co-cultures with endothelial and bone fibroblasts induced reawakening | [20] |
| ZR-75-1 | Human | Nude | ER+, PR+ | IC, ortho | N.R. | N.R. | Yes | Yes (8) | N.R. | N.R. | See data for T47D-DBM cells | [6] |
| 4T07 | Mouse | BALB/cfC3H | N.R. | IV | Yes | Yes (>12) | N.R. | N.R. | N.R. | N.R. | Transduction of retrovirus cDNA library from actively metastasizing 4T1 cells in 4T07 cells induced actively growing lung metastases showed that upregulation of Coco (encoded by Dand5) was sufficient to induce reawakening | [21] |
| D2-0R | Mouse | BALB/c | ER+ | Ortho | Yes | Yes (2, 10) | Yes | N.R. | N.R. | N.R. | D2.0R dormancy in the lungs associated with local proliferation of AQP5+, PDPN+ alveolar type I cells (similar findings with 4T07 and MCF-7 cells). Dormant D2.0R cells isolated from lungs showed increases in the expression of known dormancy (BHLHE41, NR2F1), ECM (COL3A1, POSTN, TNC, LUM), and EMT genes (SNAI1, SNAI2, ZEB2, TWIST1) compared to isolated actively growing D2A1 cells | [14, 16, 22] |
| D2.A1-d | Mouse | BALB/c | TN | IV, IC | Yes | Yes (4) | Yes | Yes (1) | N.R. | N.R. | Compared to the parental D2A1 cells, which produced actively growing lung metastases, D2A1-d showed upregulation of EMT genes encoding Slug, Snail, Twist, and Zeb1. Injection of lipopolysaccharide induced active metastasis formation of D2A1-d cells in the lung and bone | [23] |
| EMT6 | Mouse | BALB/c | ER+ | Ortho | Yes | Yes (2) | N.R. | N.R. | N.R. | N.R. | EMT6 cells disseminate to lymph nodes and lung but fail to form macrometastases. Injection of EMT6-luc into EMT6-tumored BALB/c mice elicits a CD8-mediated anti-tumor immunity | [24] |
NSG, NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ; Nude; Foxn1nu; NOD/SCID, NOD.Cg-Prkdcscid/J
ER, estrogen receptor; PR, progesterone receptor; TN, triple negative
IC, intracardiac; SQ, subcutaneous; ortho, orthotopic mammary fat pad
DBM, dormant bone metastatic; ECM, extracellular matrix; IC, intracardiac; LCC, latency competent cancer; Met., metastasis; N.R., not reported; PD, post dormancy; SCP, single cell population
It should be noted that many papers may use the term “dormancy” too enthusiastically by including genes that generically induce growth arrest, even in tumor cells that fail to show bona fide dormancy in vivo or in 3D-ME cultures. Examples of these include the growth arrest induced by MED12 knockout [25] or PAX5 haploinsufficiency in Raji cells [26]. Thus, the gold standard for dormancy should be cases in which gene expression changes have the ability to selectively cause dormancy in in vivo metastatic or ME culture settings while not necessarily affecting primary tumor growth or even proliferation of cancer cells in 2D culture.
Even though dormant cancer cells share features with stem cells, there are significant differences. These include that only some dormant cancer cells express markers found on slow-cycling cancer stem cells, such as NANOG, SOX2, and NR2F1, and that whereas cancer stem cells can self-renew at early stages of differentiation [27], reactivated dormant cells often mimic the same differentiation stage of their primary tumor [28, 29]. Dormant tumor cells can arise within non-treated primary tumors, where the spontaneous expression of stem-like transcriptional programs leads to in situ quiescence [30]. Additionally, quiescent cells that adopt stem-like phenotypes or that upregulate potentially cytoprotective autophagy programs [31] can be selected for by treatments that ablate actively proliferating tumor cells [32, 33]. In order for tumor cells to establish dormancy at secondary, metastatic sites, they first need to (i) lose cell–cell [34] and cell-extracellular matrix (ECM) adhesiveness [35] at the primary tumor site; (ii) gain increased cell motility, especially in the context of chemotaxis towards gradients of soluble factors shed by distal pre-metastatic niches [36]; (iii) increase focal-adhesion kinase (FAK) mediated intravasation into the bloodstream [37, 38]; (iv) increase resistance of circulating tumor cells (CTC) to anoikis through both tumor-intrinsic Rho-family GTPase mechanisms [39-41] and through the association with or fusion to platelets or macrophages [42-44]; and finally (v) invade and survive at distal sites as so-called disseminated tumor cells (DTC) [45-47].
2. The contribution of early dissemination to metastasis and survival
Certain observations about metastasis progression have been taken for years to mean that metastatic dissemination is a late event. In many types of cancer, larger primary tumors correlate with higher incidences of mortality associated with metastases [48-52]. Moreover, the early surgical removal of primary breast or prostate cancers, for example, translates to fewer metastatic disease relapses and reduced disease-specific mortality [53]. Animal xenograft models showed that macrometastases were formed by rare variant cells [54] which could be enriched by multiple rounds of in vivo selection [55].
However, there is a growing body of work suggesting that dissemination can occur early and that these early DTCs are relevant inducers of recurrent disease at metastatic sites. This includes the identification of gene expression signatures from primary cancers [56] or CTCs [57] that can predict the chance of future metastasis formation, as well as patients with small or non-detectable primary tumors who exhibit progressing metastases [58]. Seminal work from the Klein lab clearly demonstrated the presence of early DTCs with normal karyotypes in the bone marrow of most BrCa patients [59], even those with ductal carcinoma in situ [60], that exhibit HER2 amplification [61]. Indeed, the transfer of bone marrow carrying as few as 80 early-disseminated HER2-driven mouse transgenic BrCa cells was shown to be sufficient to induce lethal metastatic disease [60]. A subpopulation of Her2+p-p38lop-Atf2loTwist1hiE-cadlo cells generated in the MMTV-Her2/neu transgenic mouse model was capable of inducing early dissemination into the blood, bone marrow, and lungs at stages where mammary ducts showed hyperplasia or intraepithelial neoplasia, and though these DTCs were initially dormant, they gave rise to progressing metastatic lesions [62]. It is possible that HER2 promotes dissemination by suppressing the expression of the antioxidant transcription factor NRF2, which normally promotes BrCa progression by upregulating de novo nucleotide synthesis [63]. Whether subsequent dormancy requires HER2 downregulation in DTCs has not been addressed. A follow-up study in 10,307 BrCa cases showed that 27.3% of cases had bone marrow DTCs, correlating with HER2-positivity [64]. Most provocative, though, were analyses that tracked early vs. late dissemination based on comparing the mutational changes in primary tumors, DTCs, and metastatic lesions from the same patient. Even though most so-called founder mutations are shared between primary BrCa and metastases in the same patient [65], many of the predicted driver mutations in DTCs or metastases were not shared with primary tumors, leading Stoecklein and Klein [66] to question the “use of primary tumors as surrogate for the genetics of systemic cancer.” Similar findings were reported in prostate cancer [67, 68] and melanoma [69], with the note that although genetic analysis of the primary tumor could not predict the driver mutations found in metastases in the same patient, they could readily identify incidences of metastasis-to-metastasis seeding. More recent data indicate that the ability to form progressing metastases in melanoma xenografts requires the ability of DTCs to proliferate into local colonies in lymph nodes [69] in response to ME-derived IL-6 [70].
Lastly, there is mainly anecdotal evidence that in rare cases, the surgical removal of a primary tumor results in the bloom of metastatic disease. As reviewed by Tohme et al. [71], this observation is based on evidence from several clinical trials as well as multiple preclinical xenograft models. While this might reflect the outgrowth of early-disseminated dormant cells after the removal of suppressive factors secreted by primary tumors, it cannot be ruled out that these metastatic growths are the consequence of new tumor cells being liberated by specific surgical methods.
3. Molecular pathways controlling dormancy
The initial observations by Aguirre-Ghiso et al. [72] that dormant cells exhibit high relative activation levels of p38-MAPK (MAPK14) concomitant with low ERK1/2 activation levels and that p38MAPK negatively regulates ERK1/2 have held true for many tumor types [73, 74]. Most, if not all, changes to pro-dormancy signaling or expression changes for dormancy-regulating genes have been characterized as downstream manifestations of this p38MAPKhigh/ERKlow axis. For example, among the sixteen transcription factor genes predicted by Adam et al. as being p38MAPK-regulated, the upregulation of TP53 or BHLHE41 (a.k.a., BHLHB3, DEC2 or SHARP1) or the downregulation of JUN and FOXM1 were sufficient to induce quiescence in human HEp3 squamous carcinoma cells [75]. Yang et al. [76] confirmed that BHLHE41 overexpression induced dormancy in salivary adenoid cystic carcinoma cells and that growing metastatic tumors in the lung showed BHLHE41 downregulation. McGrath et al. used a genomics shRNA screen to identify HBP1 and BHLHE41 as p38MAPK-upregulated genes that maintain dormancy of MDA-MB-231 human BrCa cells in 3D cultures recapitulating the bone endosteal niche [77]. Other p38MAPK-induced dormancy-regulating genes include Glypican-3 (GPC3) [78] and ATF6α (ATF6), which promote HEp3 dormancy by upregulating RHEB and AKT-independent mTOR survival signaling [79]. Taken one step further in this signaling cascade, the ability of regucalcin (RGN), which normally regulates intracellular calcium levels [80], to promote in vivo dormancy of prostate cancer cells requires the p38MAPK-mediated downregulation of FOXM1 [81]. Moreover, FOXM1 could directly downregulate eleven genes (MYL9, CNN1, DES, TPM2, COL4A6, KCNMB1, ACTG2, WFDC2, CSRP1, PCP4, TGFB1I1) whose upregulation in prostate cancers with high RGN expression levels can predict longer recurrence-free survival. Thus, this study identifies multiple layers of p38MAPK-regulated feedback networks that control genes that either promote dormancy or reawakening.
A panel of genes previously identified encoding proteins that suppress metastasis, such as KISS1, KAI1, MKK4/7, NME1 (a.k.a., Nm23-H1), or AKAP12 (a.k.a., SSeCKS/Gravin), can also increase metastatic dormancy using various mechanisms and by acting at various stages leading to DTC colonization at metastatic niche sites [82, 83]. For example, MKK4, MKK7, NME1, and AKAP12 block either progression of micrometastases already formed at distal sites either through tumor cell growth arrest or by suppressing neovascularization. In contrast, KAI1 induces dormancy of tumor cells preparing to extravasate at vascular junctions, whereas KISS1 is capable of inducing quiescence of single tumor cells anywhere during the metastatic cascade. Thus, while KAI1 and KISS1 likely induce tumor cells to exit the proliferative cycle (e.g., cellular dormancy), MKK4, MKK7, NME1, and AKAP12 may facilitate dormancy by inducing a balance between proliferation and apoptosis rates (e.g., tumor dormancy). Another such factor, although not previously identified as a metastasis-suppressor, is the mitogen- and stress-activated protein kinase-1 (MSK1). MSK1 seems to specifically promote luminal differentiation and quiescence of ER-positive BrCa at distal sites, likely by upregulating the transcription factors, GATA3 and FOXA1 [6].
In recent years, other tumor-intrinsic genes have been identified that regulate the induction and/or maintenance of dormancy or dormancy reawakening. A complicating factor in the field is that multiple models are used that vary in their use of tumor cells (e.g., human vs. mouse cancer cell lines, patient-derived xenografts [PDX] or patient CTCs), their tissue of origin (e.g., breast, prostate, pancreas), and whether they measured the induction of dormancy vs. the dormancy reawakening (with the latter marked by the active growth of macrometastases). Moreover, tumor cell variants have been developed with increased tropism for specific peripheral organs (e.g., lung, bone, brain) or that had been selected for increased dormancy at specific sites, either spontaneously (after primary xenografting at orthotopic sites) or experimentally (e.g., after intracardiac, intratibial or intravenous injection). Table 1 shows examples of these models and how the regulatory genes or associated gene signatures differ depending on the endpoint biology (e.g., dormancy vs. reawakening) or the organ type studied. Table 2 shows examples of human or mouse breast cancer cell lines which were selected for increased dormancy in vivo and how these models were used to address signaling pathways and genes that either contribute to dormancy or to dormancy reawakening.
Table 2.
Studies identifying dormancy-associated/regulating genomic signatures
| Tumor type | Samples | Host | Primary tumor site | In vivo dormancy model | Genomic analysis | Signatures | References |
|---|---|---|---|---|---|---|---|
| Breast | Human ER+ (MCF7, T47D, Pt. samples); TN (MDA-MB-231, Pt. samples) | SCID | Orthotopic | FACS isolation of Cell Trace Violet-retaining cells from orthotopic tumor | Affimetrix microarray (HG-U133A/B) | Upregulated: ACVR1, ADAM10, AMOT, BHLHE41, COL1A1, COL4A5, CTSD, DDR1, EPHA5, GATA6, HIST1H2BK, IGFBP5, MMP2, NR2F1, P2HA1, SOX9, SREBF1, STAT3, TGFB2, THBS1, TP53, TPM1 Downregulated: APEX1, ASNS, ATF3, ATF4, BUB1, BUB1B, CDKN3, CENPB, CKS2, DNMT1, DTYMK, EGFR, EGR1, ESM1, FOSL1, FOXD1, FOXM1, IGFR1, IL8, JUN, MMP1, NT5E, ODC1, PIK3CB, PLAT, TIMP3, TK1 |
[84] |
| Breast | Human PDXs: HCI-001, HCI-002, HCI-010 | NOD/SCID | Orthotopic | Early-stage (low burden) vs. late-stage (high burden) metastases in lung, LN, BM, liver, brain | Fluidigm 96.96 qPCR DynamicArray chips | Upregulated: TGFR2, BCL2L1, EPHA4, AR, LGR5, IGFBP6, TGFB2, SOX2, BMI1, CXCL12, TWIST1, BCL2, NOTCH4, KRT5, POU4F1, TGFB1, THY1, CDKN1B, WNT2, SKP2, DAND5, PGR, CHEK1, CDJ3, MTOR, TP73, TGFBR3, ESR2, ESR1, MAX, NTRK2, NOTCH3, FIGF, MME, TP63, TP53, MYCN, SNAI2, ITGA6, JAG1, ACTA2 Downregulated: PTEN, TGFBR1, ERBB3, CDH1, CDK2, MUC1, VEGFA, CAV2, MYC, ITGB1, PARP2, EMP1, CD24, VIM |
[85] |
| Breast | Mouse 4TO7 cells | BALB/c | IV | Infection of dormant 4TO7 cells with a retroviral cDNA library from macrometastasis-forming 4T1 tumors or with mouse-specific shRNA library; isolation of lung metastases. | PCR identification of transduced cDNAs or shRNAs in metastatic cells | cDNAs that drive metastasis: Tm4sf1, Dand5, Mrpl3, Malat1 shRNAs that drive metastasis: Cart1, Pcna, Nek4, Cd3g, Itga3, Numb, Pdcd5, Smyd5, Smurf2, Ebrbb4, Gfg2, Gpr34, Il2, Lyn, March5, Sfrs6, Shbg, Src, Yes1 |
[86] |
| Prostate | Human Pt. DTCs | Humans | N.A. | Isolation of EpCAM+/CD45- DTCs from Pts with PSA rise after radical prostatectomy, but no evidence of disease | Agilent microarray array | Upregulated: SMAD7, KRT18, CDKN2C, CDKN2B, CDKN1A, TGIF1, RXRA, NR2F1, ACVR1, COL4A5, DDR1, HIST1H2BK, IGFBP5, P4HA1, SREBF1, STAT3, TGFB2, THBS1, TP53, TPM1, BMP7, BHLHE41, HSPA5, HSP90B1, PDIA3, PPIB Downregulated: APEX1, ATF3, ATF4, BUB1, BUB1B, CDKN3, CEBPG, DNMT1, DTYMK, EGR1, FOXD1, FOXM1, IGF1R, IL8, JUN, NT5E, ODC1, PIK2CB, PLAT, ESM1, MMP1 |
[29] |
| Pancreas | Mouse Ink4a.1 cells | FVB | Orthotopic | Isolation of luciferase-positive liver DTCs or macrometastases after primary tumor resection | 10X Genomics scRNA-seq | Upregulated: Fam167b, Clec1b, Apoa2, Clec4f, Alb, Cldn5, Gm21320, Xcl1, Dnase1l3, Clec14a, Gpr182, Jchain, Ccl5, Ly6d, Gzma, Cd300lg, Apoc1, Adgr14, Robo4, Fabp1, Esam, Kdr, Bmp2, Gm21762, Fam174b, Cd5l, Apoa1, Gpihbp1, Slc40a1, Cyp4b1, Rapgel5, Vsig4, Egfl7, Stab2, btnl9, nkg7, tff2, tspan7, Calcrl, Gimap7, Selenop, Cd55, Apoe, Hspa1a, Gimap6, Mzb1, Ppp1r16b Downregulated: Tagln, Tpm2, Gpx8, Crct1, Prrx2, Timp1, Lox, Grem1, Fn1, Prl2c2, Ak1, Col8a1, Actg2, Ankrd1, Ltbp1, Prkg2, Cthrc1, Tnn2, Ociad2, Col5a2, Phlda3, Prrx1, Cemip |
[87] |
| Multiple sites | Human and mouse | N.A. | N.A. | Analysis of 10 datasets yielding 21 comparisons between cancer vs. dormancy conditions to produce a dormancy-interaction network | multiple platforms | AKAP12, CD44, CDKN1A, DDR1, EPAS1 GSN, HBP1, IL1B, NDRG1, NOTCH3, PLAUR, SMAD3, THBS1, TIMP2, TSC22D3 | [88] |
| Breast | MCF-7, SK-BR-3, MDA, MB-468, MDA-MB-231 | N.A. | N.A. | Selection for quiescent BrCa cells: retention of lipophilic dye Vybrant DiD after 6 passages in vitro | Illumina RNA-seq | CCL5, CCN3, CDH11, COL17A1, FPR3, HMGCS2, IGFBP5, KLK8, MGAM, MUC5AC, OASL, PLAT, PTGFR, PTGS2, PTPRH, PTPRN2, RSAD2, SERPINA1, SERPINA3, SERPINE1, TNFRSF10C, VWA5A | [89] |
| Breast | Mouse PyMT-Bo1; D2A1 & D2.0R | C57BL6/J; BALB/cJ | IC, IT | Isolation of tetracycline-induced H2B-mCherry+ BrCa cells in collagenase-digested bone; single cells isolated by FACS | Illumina scRNA-seq | CFH, GAS6, MME, OGN, POSTN, PDGFRB, DHRS3, MGP, ALDH1A1, ALDH3A1, PRELP, AAK1, THBD, BCL2L11, GLIS1 | [90] |
| Breast | Mouse MMTV-Wnt1/iFG-FRR1 | FVB.rGH-luc-GFP | Orthotopic | Minimal residual disease/quiescence: treatment of tumored mice with the FGFRi, NVP-BGJ398 for 14d. 2 weeks off drug, recurrent tumors isolated 1–4 months later | 10X Chromium scRNA-seq | Dormancy Upregulated: Notch3, Aph1b, Dll1, Mfng, Hes7, Sox9, Snai2, Hes1 Downregulated: Psenen, Got1 Recurrence Upregulated: Rita1, Psen1, Adam17, Kat2b, Tgfb2, Gata2, Snai1 Downregulated: Trp63, Hes1, Fbwx7, Stat1 |
[91] |
N.A., not applicable
Notwithstanding these variables, several upregulated genes (BHLHE41, NR2F1, TGFB2, DAND5, HIST1H2BK, DDR1, IGFBP5, SOX9, HBP1, SNAI2, CDKN1A) are shared by two or more dormancy studies (Table 1), suggesting that they induce or maintain dormancy or prevent reawakening. Interestingly, studies that assessed quiescent cells selected in vitro [89] or after castration-induced prostate cancer dormancy [91] did not share any of these upregulated genes, further suggesting that experimental quiescence may not fully reflect in vivo dormancy.
Another issue is whether these “dormancy genes” either are differentially expressed or function only in the context of the metastatic niche. Indeed, such a restriction has been shown for multiple autophagy-regulating proteins, such as ATG5/7/9B or DIRAS3, which likely induce dormancy by increasing the survival of cancer stem cells in hypoxic metastatic environments (such as in the bone endosteal niche) by providing energy via the recycling of lysosomes through cellular compartments (see Jahangiri and Ishola [31] for a comprehensive review on the role of autophagy in dormancy). Related to this, parathyroid hormone-related protein (PTHrP) was originally thought to induce bone dormancy reawakening of BrCa cell lines through cAMP activation by the PTHrP receptor (PTHR1) [92], yet a more recent study indicated that this dormancy awakening function is PTHR1-independent and is mediated by control of calcium signaling possibly linked to autophagy suppression [93]. La Belle Flynn et al. [94] showed that dormancy reawakening after inhibition of autophagy in BrCa cell lines required the induction and stabilization of 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase-3 (PFKFB3). Lastly, Blessing et al. [95] used the selective dependence of autophagic ovarian cancer cells on the ALK gene to show that dormant cells could be ablated using the ALK inhibitor, crizotinib.
Many other dormancy-regulating genes have been identified by comparing cross-biological dormancy-related systems. For example, the upregulation of F-box/WD repeat-containing protein 7 (FBXW7) likely induces dormancy by engaging the Skp1-Cul1-F box-type (SCF-type) E3 ubiquitin ligase to degrade Cyclin-E and c-Myc, thereby inhibiting proliferative cycling [96]. Interestingly, FBXW7 induces the quiescence and, therefore, survival of hematopoietic stem cells ex vivo [97]. Dormancy induction by the paired-related homeobox transcription factor (PRRX1) correlates with its ability to induce EMT in models of head and neck squamous cell carcinoma (HNSCC) [98]. NR2F1, a retinoid-responsive gene originally identified in in silico screens for genes upregulated in dormant BrCa tumors [75], was shown to suppress the growth of ER-positive luminal MCF-7 tumors in vivo [84] and then shown to induce dormancy in HEp3 squamous carcinoma cells through SOX9- and RARβ-driven quiescence programs [99]. Moreover, hypoxia, a hallmark of many dormancy niches [100], was sufficient to induce NR2F1 expression [101] through the upregulation of the hypoxia regulator CSN8 [102]. The notion that NR2F1 promotes dormancy through tumor cell-intrinsic mechanisms has been questioned by a recent study showing that it is predominantly expressed from cancer-associated fibroblasts [103]. Another controversy stems from data showing that the expression of the long non-coding RNA, NR2F1-AS1, is higher in relapsed ER-positive BrCa and that high expression correlates with lower NR2F1 mRNA expression and worse overall survival [104]. In contrast, Liu et al. showed that NR2F1-AS1 induces a stem-like state in BrCa cells by upregulating NR2F1 translation, which then suppresses transcription of the epithelial-to-mesenchyme (EMT) suppressor ΔNp63 [105]. FBXO8 is an example of a protein that promotes dormancy by upregulating EMT and stem-like markers [106] (a possible role for EMT in metastatic dormancy is addressed in a review by Gooding and Schiemann [107]).
4. Microenvironmental (ME) soluble and adhesion factors regulating dormancy
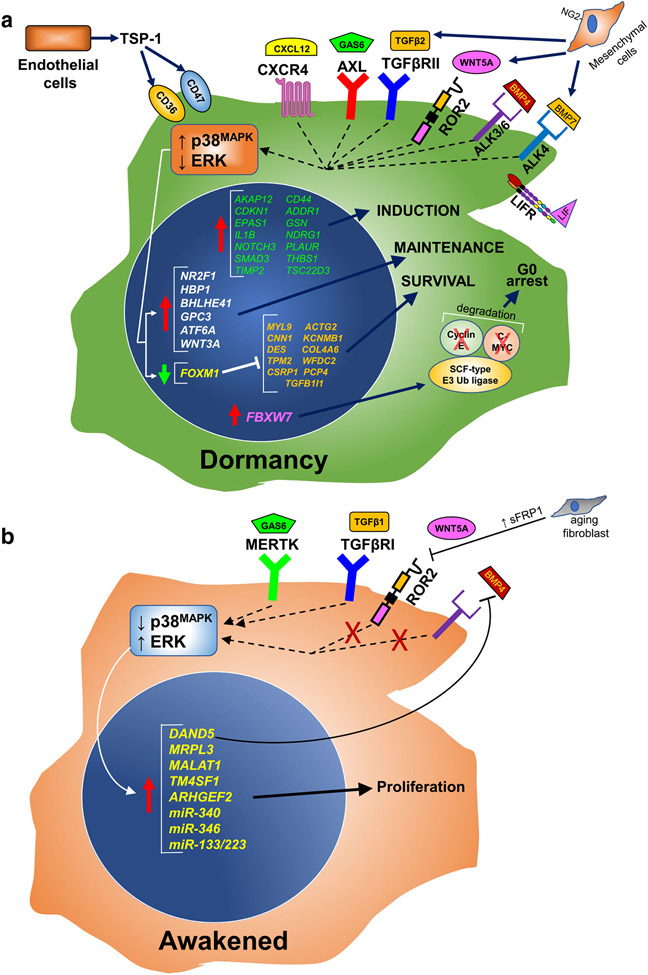
In addition to the tumor-intrinsic dormancy-regulating factors, a host of extrinsic factors produced by ME cells in metastatic niches have been identified that might promote dormancy or reawakening (Fig. 1A and B). For example, the expression of the IL-6 family member, leukemia inhibitory factor (LIF), in the bone ME promotes BrCa dormancy through the tumor expression of the LIF receptor (LIFR). High LIFR expression levels correlate with better overall survival in invasive BrCa cases, and moreover, the forced knockdown of LIFR increased the relative number of progressing bone metastases in mice following intra-cardiac injection of MCF-7 cells [108]. Interestingly, HDAC inhibitors induce LIFR expression and increase BrCa dormancy [109], which may represent an underappreciated clinical benefit of these drugs in advanced BrCa cases. Another example is TGF-β2, whose selective expression by ME cells in the bone marrow, but not in the lung, correlated with increased long-term dormancy of HEp3 cells in the bone through the activation of p38MAPK [110]. This was thought to reflect that for HEp3 cells, the bone (but not the lung) represents a permissive “soil” for dormant colonization, based on Paget’s “seed-and-soil” hypothesis [111].
Fig. 1.
Dormancy and reawakening
A permissive ME that supports tumor cell dormancy involves tumor-ME crosstalk, mediated through secreted factors and/or cell–cell or cell/ECM adhesions. For example, Ruppender et al. [112] demonstrated that cell–cell contact with bone stromal cells facilitated the expression of TGF-β2 by prostate cancer cells. Yumoto et al. [113] showed that this contact-induced dormancy signaling was mediated by AXL, a member of the TAM tyrosine kinase receptor family that responds to GAS6 [114]. Indeed, AXL expression was sufficient to induce in vivo dormancy in human prostate cancer cell lines [115], whereas others report that knockdown of the TAM family member, MERTK, induced bone dormancy after intra-cardiac injection of prostate cancer cell lines [116], suggesting the possibility of opposing regulatory effects by TAM members.
As examples of secreted ME factors that regulate dormancy, other members of the TGF-β family in addition to TGF-β2, such as BMP7 or BMP4, have been shown to regulate ME-mediated dormancy in the bone marrow and lungs, respectively [117, 118], whereas the secretion of TGF-β2 and BMP-7 by NG2+/Nestin+ mesenchymal stem cells in the bone induces dormancy in disseminated BrCa cells [119] (the SMAD proteins engaged by various dormancy-associated TGF-β-family members are reviewed in [120]). Moreover, dormant PC3MM prostate cancer cells in the bone set up a feed-forward loop to induce more dormancy by secreting high levels of SPARC, which induces BMP7 production and secretion by bone marrow stromal cells [121]. A potential mechanism by which BMP7 induces dormancy might be through the upregulation of hTERT and its suppression of telomerase activity [122]. Additionally, the expression of FOXC1 in ME stromal cells correlates with increased dormancy of T and NK lymphomas, although it is not known whether this effect was mediated by soluble or adhesion factors [123]. The co-culture of breast and prostate DTCs with alveolar type-I lung epithelial cells was shown to induce dormancy in in vivo models by inducing the lysosomal accumulation of the pro-survival receptor, EphB6 [124].
Not addressed in this review are the large body of studies showing that extracellular vesicles (e.g., exosomes) from tumor cells can promote metastatic progression (reviewed in [125]). There is also an equally important body of literature showing that vesicles from ME cells can educate colonizing DTC cells towards dormancy (reviewed in [31, 126]) by, for example, transferring miR-23b, which decreases DTC expression of the pro-proliferation and pro-motility gene, MARCKS [127].
Dormant BrCa cells are known to attach to perivascular niche ME cells in the lung and bone marrow that express high thrombospondin-1 (TSP-1) but low levels of TGF-β1 and periostin (POSTN); stimuli, inflammation or injury that activate this microvasculature and subsequently invert the TSP-1 to TGF-β1/POSTN expression ratio, cause dormant tumor cells to reawaken [20]. An additional way is for tumor cells to overexpress a natural competitive BMP4 receptor antagonist such as DAND5 (a.k.a. COCO) [21].
Although WNT signaling through the canonical β-catenin pathway was originally viewed as only promoting metastatic progression, the story has become more nuanced with the elucidation that individual WNT family members can have opposing roles, especially when including analysis of non-canonical hedgehog (Hh) pathways [128]. Indeed, WNT5A expressed from the osteoblastic bone niche can induce prostate cancer cell dormancy by inducing the expression of the β-catenin signaling suppressor, Siah E3 Ubiquitin Protein Ligase 2 (SIAH2) [129]. WNT5A expressed from lung fibroblasts also facilitates the dormancy of melanoma DTCs in the lung, and interestingly, this dormancy circuit wanes as aging lung fibroblasts increasingly express and secrete sFRP1, a natural WNT5A antagonist [130]. Moreover, aged-related dormancy reawakening is associated with an inversion to low AXL and high MERTK expression. Consistent with the finding that WNT3A is required to maintain dormancy of MDA-MB-231 cells in a 3D endosteal niche culture [77], Lee et al. showed that it might also induce dormancy in these cells by upregulating stem-like expression signatures [131] that typically include SOX2 and NANOG [132].
There is a growing appreciation for the role played by nerve cell networks, even specialized neuronal cells such as Schwann cells, in primary tumor progression and treatment response [133-135]. Indeed, the activation of β2-adrenergic receptors on sympathetic nerves increases the bone colonization of MDA-MB-231 BrCa cells [136]. In a similar manner, Dai et al. [137] showed that BrCa DTCs trafficking to the brain concentrated at astrocyte-vascular junctions and that astrocyte-associated laminin-2 (LAMA2) mediates DTC quiescence by preventing the nuclear localization of its cognate receptor, dystroglycan-1 (DAG1), in tumor cells. In another nerve-tumor interaction, the binding of glial cell line-derived neurotrophic factor (GDNF) to its cognate receptor, GFRA1, was shown to activate nerve cells infiltrating gastrointestinal stromal tumors (GIST), resulting in tumor dormancy due to the induction of autophagy pathways [138].
5. Mediators of dormancy through control of stemness
As cited above, specific gene products can affect dormancy induction or reawakening by regulating the stem-like phenotype of cancer cells in distal sites. For example, dormant melanoma cells that escaped tumor-specific immunotherapy are marked by the absence of the glucocorticoid-induced leucine zipper protein, GILZ, and by the expression of stem-like markers; reawakening could be induced by the forced expression of GILZ, which suppressed FOXO3A and its downstream target gene p21CIP1 (CDKN1A), a known cell cycle suppressor [139]. Members of the LOX (lysyl oxidase) and LOXL (lysyl oxidase-like) family, which crosslink elastin and collagen in the ECM, have been implicated in metastatic progression [140, 141], and a recent study by Weidenfeld [142] suggested that low levels of LOXL2 promote EMT and a stem-like phenotype in MCF-7 cells in 3D culture systems whereas high levels promote dormancy reawakening.
6. Transcriptional networks controlling dormancy: single-cell and system analyses
A number of studies have compared the genomics of various cancer types cultured under proliferative versus dormant conditions in order to identify possible mRNA and miRNA signatures of dormancy (see Table 1). Several technical and analytic hurdles need to be overcome in these systems: first, the need to remove panels of proliferation genes, and second, addressing how single dormant cells are isolated from in vivo sites. In regard to the latter, isolation of DTCs from soft tissue sites is facilitated by digestion with ECM-degrading enzymes such as collagenase and/or dispase. However, because many dormant DTCs in the bone are trapped in resistant sites such as the endosteal niche [100], multiple rounds of bone grinding and tissue digestion are needed, followed by gentler isolation techniques such as magnetic-activated cell sorting (MACS) in order to get sufficient DTC yields for genomic analysis [90]. Another issue typically not addressed but which might affect the identification of dormancy signatures is how DTC manipulation during isolation affects cell viability and gene expression.
An early attempt at using functional genomic screens to identify dormancy-regulating gene signatures was performed by Gao et al. [86], using the non-metastatic relative of the 4T1 mouse mammary carcinoma line, 4T07, which spontaneously homes to the lung after orthotopic injection but does not produce macrometastases [143]. By transducing 4T07 cells with lentivirus libraries encoding cDNAs and miRNAs, they identified Coco (Dand5), Mrpl3, the long non-coding RNA Malat1, Tm4sf1, Arhgef2, miR-340, miR-346, or the combination of miR-138 and miR-223 as being capable of inducing lung macrometastases, suggesting that these genes or RNAs were reawakening suppressors. Similarly, McGrath et al. [77] transduced MDA-MB-231 cells with a genomic shRNA library in order to identify genes required for dormancy in a 3D endosteal niche culture. In addition to identifying and confirming roles for BHLHE41, HBP1, and WNT3A, they identified a host of translation/ribogenesis genes whose knockdown induced BrCa proliferation.
By comparing two dormancy-related datasets, one which identified p38MAPK-regulated, dormancy-associated genes [75] and the other consisting of genes downregulated when dormant patient cancers were pushed into fast-growing angiogenic phenotypes [144], Kim et al. [84] identified 49 differentially expressed genes that were shared with 51 BrCa cell lines cultured in quiescent conditions. This analysis identified five genes commonly upregulated in dormant conditions (CTSD, DDR1, IGFBP5, SREBF1, STAT3) as well as nineteen downregulated proliferation or invasiveness genes. Tiram et al. [145] used a similar approach to identify dormancy-associated miRNAs in osteosarcomas. Uzuner et al. [88] gathered ten such analyses, produced as either RNA-seq or microarray studies, across seven cancer types, in order to develop a dormancy-related interaction network (the ten datasets were listed in Supplementary Table S1 in their paper). They identified 139 genes and 1974 inferred protein–protein interactions, eventually identifying fifteen upregulated genes (AKAP12, CD44, CDKN1A, DDR1, EPAS1, GSN, HBP1, IL1B, NDRG1, NOTCH3, PLAUR, SMAD3, THBS1, TIMP2, and TSC22D3) and ten dormancy-associated protein–protein interaction genes (CLU, APP, HIST1H1C, HIST2H2BE, OPTN, FBXO32, CSTB, THBS1, and CDKN2B) that likely induce dormancy through cell stemness or cell cycle inhibitory functions. Quayle et al. [89] identified a 22-gene quiescence signature that included CCL5, CDH11, and IGFBP5 from quiescent BrCa cells that retained the vital lipophilic dye DiD after 6 in vitro passages. Importantly, up to 50% of the dormancy-associated genes are shared between these various studies, which include analyses of liquid and solid cancers and cancer from different organ sites, strongly suggesting the existence of shared dormancy-inducing transcriptional signatures. However, it is unclear whether the non-shared genes reflect differences between cancer types or the techniques used to identify relevant differentially expressed genes. It is also important to note that the gene expression profiles from the studies cited above were derived from cell lines or tumors, but not typically from DTCs isolated from metastatic sites. Thus, at best, these signatures might be predictive of gene expression changes in primary tumor cells that eventually lead to the dormant state.
Several newer studies have applied single-cell RNA-seq analyses on isolated DTCs from metastatic sites in order to more directly analyze how tumor cell heterogeneity influences dormancy-maintenance gene expression signatures. For example, Chéry et al. [29] performed RNA-seq on individual EpCAM-positive/CD45-negative DTCs isolated from the bones of prostate cancer patients who either had no evidence of metastatic disease (following primary treatment for Gleason 6 or 7 lesions) or who showed no evidence of biochemical (PSA) recurrence after initial prostatectomy. After binning the top differentially expressed genes for known p38MAPK regulation, they identified a signature of fifteen upregulated dormancy genes (ABI1, CDC25B, CDK7, CELF1, COX7B2, CUL9, FTSJ1, LBR, MALT1, MLKL, N4BP1, NIPAL3, PSMB5, SETMAR, and TFRC). Ren et al. [90] used a bone-homing pro-dormancy sub-variant line of the MMTV-PyMT mammary carcinoma line, PyMT-Bo1, along with the lipophilic dye DiD to isolate dormant bone DTCs after intra-cardiac injection. Starting with a 15-gene dormancy signature, they showed that the high expression of four genes, Cfh, Gas6, Ogn, and Mme correlated with statistically significant increased overall survival probability in BrCa cases. However, the ectopic overexpression of any one of these genes did not induce quiescence in in vitro 2D cultures or in lungs after intravenous tumor cell injection, suggesting that dormancy might require a combination of genes. Lastly, Owen et al. [146] isolated bone DTCs (based on retention of the vital dye PKH26) after intra-cardiac injection of a bone-homing variant of murine RM1 prostate cancer cells into immunocompetent syngeneic mice [147]. When compared to proliferating RM1 cells in the bone (CFP/Luc2-positive, PHK26-negative), they identified dormancy-related gene clusters by single-cell RNA-seq that included known dormancy genes, Bhlhe41 and Gas6, and that mapped to the re-expression of intrinsic type I interferon pathways. This led to the conclusion that this model may focus more on immune cell mechanisms of dormancy induction. Indeed, Wang et al. [148] reevaluated the dataset from Owen et al. (GSE147150) and confirmed the induction of immune response genes, but also identified downregulated MYC-target cell cycle genes such as Madl21, Ccna2, and Plk1.
7. Therapeutic interventions
Two major modalities have been considered for therapeutic interventions of metastatic dormancy: sustained suppression of reawakening vs. induction of reawakening followed by anti-proliferative therapy. In the first modality, the concept would be to identify drugs that, for example, sustain p38MAPK signaling while simultaneously preventing the reactivation of ERK1/2. In theory, this might first be focused on groups whose primary tumor genomics predict the likelihood of metastatic relapse, as in the case of Oncotype DX in ER-positive BrCa [56]. One of the problems confounding the use of p38-MAPK agonists is that whereas the p38α is involved in the induction of stemness and dormancy [149], other isoforms, such as p38γ or p38δ, are known to induce pro-inflammatory and oncogenic pathways [150]. Even though p38-MAPK agonists exist (e.g., U-46619 is a thromboxane A2 agonist that activates p38α [151]), there would be concern that their long-term use (needed in the adjuvant setting to prevent disease relapse) might have pro-oncogenic effects by activating other p38 isoforms. In the second treatment modality, drugs that would antagonize p38-MAPK or agonize ERK1/2, either of which is sufficient to induce dormancy reawakening, could be given to induce the active proliferation of dormant DTCs as well as reversing their stem-like phenotypes, rendering them susceptible to more conventional treatments. The worry here is that anti-proliferative therapies that may have shown efficacy against the primary tumor in the same patient may be less effective or ineffective based on the acquisition of new genetic drivers.
As theorized by Janowska et al. [152], clinically relevant drugs that target the eventual activation of ERK1/2, such as inhibitors of RAF, MEK or ERK itself, or downstream CDK inhibitors, might show efficacy in sustaining dormancy. However, many monotherapy CDK inhibitor trials have demonstrated disappointing results, especially regarding the prevention of disease relapse [153]. Among the theoretical targets cited in the review of Singh et al. [154], several preclinical studies were mentioned that attempted to prevent the initiation of dormancy using a combination of a thioredoxin reductase inhibitor plus a pan-AKT inhibitor, based on the reliance of these two pathways for DTC establishment. Also cited was the study by Sosa et al. [99] showing that the clinically approved treatments 5-azacytidine (AZA) or all-trans retinoic acid (ATRA) could induce the expression of pro-dormancy genes such as SOX9, NR2F1, RARB, and CDKN1A in prostate cancer and HNSCC cells. A clinical study, NCT03572387, combined AZA and ATRA in prostate cancer cases with biochemical (PSA) recurrence after local treatment and was completed in the summer of 2022. An ever-growing list of clinical trials that either directly target metastatic cancers or that assess effects on both primary-site cancers and their associated metastases is addressed in a recent review by Ramamoorthi et al. [155]. Although dormancy is not a direct criterion per se in these studies, the ability of many of the combination therapies to prevent the formation of macrometastases is cited as a potential avenue targeting DTC colonization and/or reawakening. This raises the issue that clinical trials using dormancy reawakening as endpoints will be costly and difficult because of the need for long-term patient follow-up. Similarly, pre-clinical trials in mice would need to use models that reliably recapitulate clinically relevant dormancy biologies, something not fully achieved to date.
8. Conclusions and perspectives
The advent of more sophisticated analysis techniques such as single-cell DNA and RNA sequencing has increased the number of candidate genes and gene expression signatures that might control the induction, maintenance, and reawakening of dormant cells at distal sites. However, the field remains hampered by a limited number of clinically relevant in vivo models of dormancy. Many of these rely on decades-old murine or human cancer pedigree cell lines selected for gain or loss of specific parameters in the metastatic cascade or on transgenic tumor models where dissemination and dormancy can be studied in short windows before the onset of malignant disease at the primary tumor site. A gold standard for dormancy gene validation will continue to be the demonstration that a single gene expression change can affect dormancy in either 3D ME cultures or in in vivo models that maximize focus on dormancy induced by spontaneous dissemination mechanisms. Indeed, xenograft or 3D culture studies that even partially validate their dormancy genes against data from actual DTCs isolated from patient distal sites, such as the bone marrow or lymph nodes, clearly have the greatest bona fides in the field. However, the ability to harvest these cells from patients is limited in the USA if a therapeutic benefit cannot be identified. More effort to develop cooperative studies that harvest such cells under the shared IRB protocols, the standardization of handling and genomic analysis techniques, and the sharing of the resulting genomic datasets (e.g., through the TCGA) will aid in validating the clinical relevance of identified dormancy genes. This will increase patient survival by identifying actionable targets and/or pathways that can lead to sustained tumor dormancy.
Funding
This work was supported by grants R21-CA235092 (NCI) and W81XWH-20-BCRP-BTA12-2 (DOD) to I.H.G. and by the P30-CA016056 (NCI) Comprehensive Cancer Center grant.
Footnotes
Conflict of interest The author declares no competing interests.
References
- 1.Lambert AW, Pattabiraman DR, & Weinberg RA (2017). Emerging biological principles of metastasis. Cell, 168(4), 670–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguirre-Ghiso JA (2018). How dormant cancer persists and reawakens. Science, 361(6409), 1314–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banys-Paluchowski M, Reinhardt F, & Fehm T (2020). Disseminated tumor cells and dormancy in breast cancer progression. Advances in Experimental Medicine and Biology, 122035, 35–43. [DOI] [PubMed] [Google Scholar]
- 4.Bushnell GG, Deshmukh AP, den Hollander P, Luo M, Soundararajan R, Jia D, Levine H, Mani SA, & Wicha MS (2021). Breast cancer dormancy: Need for clinically relevant models to address current gaps in knowledge. NPJ Breast Cancer, 7(1), 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phan TG, & Croucher PI (2020). The dormant cancer cell life cycle. Nature Reviews Cancer, 20(7), 398–411. [DOI] [PubMed] [Google Scholar]
- 6.Gawrzak S, Rinaldi L, Gregorio S, Arenas EJ, Salvador F , Urosevic J, et al. (2018). MSK1 regulates luminal cell differentiation and metastatic dormancy in ER(+) breast cancer. Nature Cell Biology, 20(2), 211–221. [DOI] [PubMed] [Google Scholar]
- 7.Keydar I, Chen L, Karby S, Weiss FR, Delarea J, Radu M, Chaitcik S, & Brenner HJ (1979). Establishment and characterization of a cell line of human breast carcinoma origin. European Journal of Cancer, 15(5), 659–670. [DOI] [PubMed] [Google Scholar]
- 8.Harrell JC, Dye WW, Allred DC, Jedlicka P, Spoelstra NS, Sartorius CA, & Horwitz KB (2006). Estrogen receptor positive breast cancer metastasis: Altered hormonal sensitivity and tumor aggressiveness in lymphatic vessels and lymph nodes. Cancer Research, 66(18), 9308–9315. [DOI] [PubMed] [Google Scholar]
- 9.Puchalapalli M, Zeng X, Mu L, Anderson A, Hix GL, Zhang M, Sayyad MR, Mosticone WS, Clevenger CV, & Koblinski JE (2016). NSG mice provide a better spontaneous model of breast cancer metastasis than athymic (Nude) mice. PLoS ONE, 11(9), e0163521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lefley D, Howard F, Arshad F, Bradbury S, Brown H, Tulotta C, Eyre R, Alférez D, Wilkinson JM, Holen I, Clarke RB, & Ottewell P (2019). Development of clinically relevant in vivo metastasis models using human bone discs and breast cancer patient-derived xenografts. Breast Cancer Research, 21(1), 130–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu X, Mu E, Wei Y, Riethdorf S, Yang Q, Yuan M, Yan J, Hua Y, Tiede BJ, Lu X, Haffty BG, Pantel K, Massagué J, & Kang Y (2011). VCAM-1 promotes osteolytic expansion of indolent bone micrometastasis of breast cancer by engaging alpha4beta1-positive osteoclast progenitors. Cancer Cell, 20(6), 701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albrengues J, Shields MA, Ng D, Park CG, Ambrico A, Poindexter ME, et al. (2018). Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science, 361(6409), eaao4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wada M, Canals D, Adada M, Coant N, Salama MF, Helke KL, Arthur JS, Shroyer KR, Kitatani K, Obeid LM, & Hannun YA (2017). P38 delta MAPK promotes breast cancer progression and lung metastasis by enhancing cell proliferation and cell detachment. Oncogene., 36(47), 6649–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barkan D, Kleinman H, Simmons JL, Asmussen H, Kamaraju AK, Hoenorhoff MJ, Liu ZY, Costes SV, Cho EH, Lockett S, Khanna C, Chambers AF, & Green JE (2008). Inhibition of metastatic outgrowth from single dormant tumor cells by targeting the cytoskeleton. Cancer Research, 63(15), 6241–6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rucci N, Ricevuto E, Ficorella C, Longo M, Perez M, Di FC, Funari A, Teti A, & Migliaccio S (2004). In vivo bone metastases, osteoclastogenic ability, and phenotypic characterization of human breast cancer cells. Bone, 34(4), 697–709. [DOI] [PubMed] [Google Scholar]
- 16.Sowder ME, & Johnson RW (2018). Enrichment and detection of bone disseminated tumor cells in models of low tumor burden. Science Reports, 3(1), 14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlson P, Dasgupta A, Grzelak CA, Kim J, Barrett A, Coleman IM, Shor RE, Goddard ET, Dai J, Schweitzer EM, Lim AR, Crist SB, Cheresh DA, Nelson PS, Hansen KC, & Ghajar CM (2019). Targeting the perivascular niche sensitizes disseminated tumour cells to chemotherapy. Nature Cell Biology, 21(2), 238–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holen I, Walker M, Nutter F, Fowles A, Evans CA, Eaton CL, & Ottewell PD (2016). Oestrogen receptor positive breast cancer metastasis to bone: Inhibition by targeting the bone microenvironment in vivo. Clinical and Experimental Metastasis, 33(3), 211–224. [DOI] [PubMed] [Google Scholar]
- 19.Malladi S, Macalinao DG, Jin X, He L, Basnet H, Zou Y, De SE, & Massagué J (2016). Metastatic Latency and Immune Evasion through Autocrine Inhibition of WNT. Cell, 165(1), 45–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghajar CM, Peinado H, Mori H, Matei IR, Evason KJ, Brazier H, et al. (2013). The perivascular niche regulates breast tumour dormancy. Nature Cell Biology, 15(7), 807–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao H, Chakraborty G, Lee-Lim AP, Mo Q, Decker M, Vonica A, Shen R, Brogi E, Brivanlou AH, & Giancotti EG (2012). The BMP inhibitor Coco reactivates breast cancer cells at lung metastatic sites. Cell, 150(4), 764–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montagner M, Bhome R, Hooper S, Chakravarty P, Qin X, Sufi J, Bhargava A, Ratcliffe CDH, Naito Y, Pocaterra A, Tape CJ, & Sahai E (2020). Crosstalk with lung epithelial cells regulates Sfrp2-mediated latency in breast cancer dissemination. Nature Cell Biology, 22(3), 289–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Cock JM, Shibue T, Dongre A, Keckesova Z, Reinhardt F, & Weinberg RA (2016). Inflammation triggers Zeb1-dependent escape from tumor latency. Cancer Research, 76(23), 6778–6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piranlioglu R, Lee E, Ouzounova M, Bollag RJ, Vinyard AH, Arbab AS, Marasco D, Guzel M, Cowell JK, Thangaraju M, Chadli A, Hassan KA, Wicha MS, Celis E, & Korkaya H (2019). Primary tumor-induced immunity eradicates disseminated tumor cells in syngeneic mouse model. Nature Communications, 10(1), 1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo XL, Deng CC, Su XD, Wang F, Chen Z, Wu XP, Liang SB, Liu JH, & Fu LW (2018). Loss of MED12 induces tumor dormancy in human epithelial ovarian cancer via downregulation of EGFR. Cancer Research, 78(13), 3532–3543. [DOI] [PubMed] [Google Scholar]
- 26.Liang X, Gu J, Li T, Zhao L, Fu X, Zhang W, Wang J, Shang Z, Huang W, & Zhou J (2018). PAX5 haploinsufficiency induce cancer cell dormancy in Raji cells. Experimental Cell Research, 367(1), 30–36. [DOI] [PubMed] [Google Scholar]
- 27.Kleinsmith LJ, & Pierce GB Jr. (1964). Multipotentiality of single embryonal carcinoma cells. Cancer Research, 24, 1544–1551. [PubMed] [Google Scholar]
- 28.Lawson MA, McDonald MM, Kovacic N, Hua KW, Terry RL, Down J, Kaplan W, Paton-Hough J, Fellows C, Pettitt JA, Neil DT, Van VE, Baldock PA, Rogers MJ, Eaton CL, Vanderkerken K, Pettit AR, Quinn JM, Zannettino AC, … Croucher PI (2015). Osteoclasts control reactivation of dormant myeloma cells by remodelling the endosteal niche. Nature Communications, 6, 8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chery L, Lam HM, Coleman I, Lakely B, Coleman R, Larson S, Aguirre-Ghiso JA, Xia J, Gulati R, Nelson PS, Montgomery B, Lange P, Snyder LA, Vessella RL, & Morrissey C (2014). Characterization of single disseminated prostate cancer cells reveals tumor cell heterogeneity and identifies dormancy associated pathways. Oncotarget, 5(20), 9939–9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sistigu A, Musella M, Galassi C, Vitale I, & De MR (2020). Tuning cancer fate: Tumor microenvironment’s role in cancer stem cell quiescence and reawakening. Frontiers Immunology, 11, 2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jahangiri L, & Ishola T (2022). Dormancy in breast cancer, the role of autophagy, lncRNAs, miRNAs and exosomes. International Journal of Molecular Science, 23(9), 5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korentzelos D, Clark AM, & Wells A (2020). A perspective on therapeutic pan-resistance in metastatic cancer. International Journal of Molecular Science, 21(19), E7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baram T, Rubinstein-Achiasaf L, Ben-Yaakov H, & Ben-Baruch A (2021). Inflammation-driven breast tumor cell plasticity: Stemness/EMT, therapy resistance and dormancy. Frontiers Oncology, 10, 614468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smart JA, Oleksak JE, & Hartsough EJ (2021). Cell Adhesion Molecules in Plasticity and Metastasis. Molecular Cancer Research, 19(1), 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dhaliwal D, & Shepherd TG (2022). Molecular and cellular mechanisms controlling integrin-mediated cell adhesion and tumor progression in ovarian cancer metastasis: A review. Clinical Experimental Metastasis, 39(2), 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stuelten CH, Parent CA, & Montell DJ (2018). Cell motility in cancer invasion and metastasis: Insights from simple model organisms. Nature Reviews Cancer, 18(5), 296–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Genna A, & Gil-Henn H (2018). FAK family kinases: The Yin and Yang of cancer cell invasiveness. Molecular and Cellular Oncology, 5(4), e1449584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zavyalova MV, Denisov EV, Tashireva LA, Savelieva OE, Kaigorodova EV, Krakhmal NV, & Perelmuter VM (2019). Intravasation as a key step in cancer metastasis. Biochemistry (Mosc), 84(7), 762–772. [DOI] [PubMed] [Google Scholar]
- 39.Tajbakhsh A, Rivandi M, Abedini S, Pasdar A, & Sahebkar A (2019). Regulators and mechanisms of anoikis in triple-negative breast cancer (TNBC): A review. Critical Reviews in Oncology and Hematology, 140, 17–27. [DOI] [PubMed] [Google Scholar]
- 40.Adeshakin FO, Adeshakin AO, Afolabi LO, Yan D, Zhang G, & Wan X (2021). Mechanisms for modulating anoikis resistance in cancer and the relevance of metabolic reprogramming. Frontiers Oncology, 11, 626577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khan SU, Fatima K, & Malik F (2022). Understanding the cell survival mechanism of anoikis-resistant cancer cells during different steps of metastasis. Clinical and Experimental Metastasis, 39(5), 715–726. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Zhang Y, Ding Y, & Zhuang R (2021). Platelet-mediated tumor metastasis mechanism and the role of cell adhesion molecules. Critical Reviews in Oncology and Hematology, 167, 103502. [DOI] [PubMed] [Google Scholar]
- 43.Reduzzi C, Vismara M, Gerratana L, Silvestri M, De BF, Raspagliesi F, Verzoni E, Di CS, Locati LD, Cristofanilli M, Daidone MG, & Cappelletti V (2020). The curious phenomenon of dual-positive circulating cells: Longtime overlooked tumor cells. Seminars in Cancer Biology, 60, 344–350. [DOI] [PubMed] [Google Scholar]
- 44.Hamilton G, & Rath B (2017). Circulating tumor cell interactions with macrophages: Implications for biology and treatment. Translational Lung Cancer Research, 6(4), 418–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banys M, Krawczyk N, & Fehm T (2014). The role and clinical relevance of disseminated tumor cells in breast cancer. Cancers (Basel), 6(1), 143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linde N, Fluegen G, & Aguirre-Ghiso JA (2016). The relationship between dormant cancer cells and their microenvironment. Advances in Cancer Reseach, 13, 245–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ring A, Spataro M, Wicki A, & Aceto N (2022). Clinical and biological aspects of disseminated tumor cells and dormancy in breast cancer. Frontiers in Cell and Developmental Biology, 10, 929893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Illyes I, Tokes AM, Kovacs A, Szasz AM, Molnar BA, Molnar IA, Kaszas I, Baranyak Z, Laszlo Z, Kenessey I, & Kulka J (2014). In breast cancer patients sentinel lymph node metastasis characteristics predict further axillary involvement. Virchows Archives, 465(1), 15–24. [DOI] [PubMed] [Google Scholar]
- 49.Walter SD, Chao DL, Feuer W, Schiffman J, Char DF, & Harbour JW (2016). Prognostic implications of tumor diameter in association with gene expression profile for uveal melanoma. JAMA Ophthalmology, 134(7), 734–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wangchinda P, & Ithimakin S (2016). Factors that predict recurrence later than 5 years after initial treatment in operable breast cancer. World Journal of Surgical Oncology, 14(1), 223–0988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sopik V, & Narod SA (2018). The relationship between tumour size, nodal status and distant metastases: On the origins of breast cancer. Breast Cancer Research and Treatments, 170(3), 647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asare EA, Silva-Figueroa A, Hess KR, Busaidy N, Graham PH, Grubbs EG, Lee JE, Williams MD, & Perrier ND (2019). Risk of distant metastasis in parathyroid carcinoma and its effect on survival: A retrospective review from a high-volume center. Annals of Surgical Oncology, 26(11), 3593–3599. [DOI] [PubMed] [Google Scholar]
- 53.Holmberg L, Bill-Axelson A, Helgesen F, Salo JO, Folmerz P, Häggman M, Andersson SO, Spångberg A, Busch C, Nordling S, Palmgren J, Adami HO, Johansson JE, & Norlén BJ (2002). A randomized trial comparing radical prostatectomy with watchful waiting in early prostate cancer. New England Journal of Medicine, 347(11), 781–789. [DOI] [PubMed] [Google Scholar]
- 54.Fidler IJ, & Kripke ML (1977). Metastasis results from preexisting variant cells within a malignant tumor. Science, 197(4306), 893–895. [DOI] [PubMed] [Google Scholar]
- 55.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, & Massagué J (2003). A multigenic program mediating breast cancer metastasis to bone. Cancer Cell, 3(6), 537–549. [DOI] [PubMed] [Google Scholar]
- 56.Va’nt Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, & Friend SH (2002). Gene expression profiling predicts clinical outcome of breast cancer. Nature, 415(6871), 530–536. [DOI] [PubMed] [Google Scholar]
- 57.Patsialou A, Wang Y, Lin J, Whitney K, Goswami S, Kenny PA, & Condeelis JS (2012). Selective gene-expression profiling of migratory tumor cells in vivo predicts clinical outcome in breast cancer patients. Breast Cancer Research, 14(5), R139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kato S, Alsafar A, Walavalkar V, Hainsworth J, & Kurzrock R (2021). Cancer of unknown primary in the molecular era. Trends in Cancer, 7(5), 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmidt-Kittler O, Ragg T, Daskalakis A, Granzow M, Ahr A, Blankenstein TJ, Kaufmann M, Diebold J, Arnholdt H, Muller P, Bischoff J, Harich D, Schlimok G, Riethmuller G, Eils R, & Klein CA (2003). From latent disseminated cells to overt metastasis: Genetic analysis of systemic breast cancer progression. Proceedings of the National Academy of Sciences, USA, 100(13), 7737–7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hüsemann Y, Geigl JB, Schubert F, Musiani P, Meyer M, Burghart E, Forni G, Eils R, Fehm T, Riethmüller G, & Klein CA (2008). Systemic spread is an early step in breast cancer. Cancer Cell, 13(1), 58–68. [DOI] [PubMed] [Google Scholar]
- 61.Schardt JA, Meyer M, Hartmann CH, Schubert F, Schmidt-Kittler O, Fuhrmann C, Polzer B, Petronio M, Eils R, & Klein CA (2005). Genomic analysis of single cytokeratin-positive cells from bone marrow reveals early mutational events in breast cancer. Cancer Cell, 8(3), 227–239. [DOI] [PubMed] [Google Scholar]
- 62.Harper KL, Sosa MS, Entenberg D, Hosseini H, Cheung JF, Nobre R, Avivar-Valderas A, Nagi C, Girnius N, Davis RJ, Farias EF, Condeelis J, Klein CA, & Aguirre-Ghiso JA (2016). Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature, 540588–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fox DB, Garcia NMG, McKinney BJ, Lupo R, Noteware LC, Newcomb R, Liu J, Locasale JW, Hirschey MD, & Alvarez JV (2020). NRF2 activation promotes the recurrence of dormant tumour cells through regulation of redox and nucleotide metabolism. Nature Metabolism, 2(4), 318–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hartkopf AD, Brucker SY, Taran FA, Harbeck N, von Au A, Naume B, et al. (2021). Disseminated tumour cells from the bone marrow of early breast cancer patients: Results from an international pooled analysis. European Journal of Cancer, 154, 128–137. [DOI] [PubMed] [Google Scholar]
- 65.Popawski AB, Jankowski M, Erickson SW, de Díaz ST, Partridge EC, Crasto C, et al. (2010). Frequent genetic differences between matched primary and metastatic breast cancer provide an approach to identification of biomarkers for disease progression. European Journal of Human Genetics, 18(5), 560–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stoecklein NH, & Klein CA (2010). Genetic disparity between primary tumours, disseminated tumour cells, and manifest metastasis. International Journal of Cancer, 126(3), 589–598. [DOI] [PubMed] [Google Scholar]
- 67.Gundem G, Van LP, Kremeyer B, Alexandrov LB, Tubio JM, Papaemmanuil E, et al. (2015). The evolutionary history of lethal metastatic prostate cancer. Nature, 520(7547), 353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cackowski FC, Wang Y, Decker JT, Sifuentes C, Weindorf S, Jung Y, et al. (2019). Detection and isolation of disseminated tumor cells in bone marrow of patients with clinically localized prostate cancer. Prostate, 79(14), 1715–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Werner-Klein M, Scheitler S, Hoffmann M, Hodak I, Dietz K, Lehnert P, et al. (2018). Genetic alterations driving metastatic colony formation are acquired outside of the primary tumour in melanoma. Nature Communications, 9(1), 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Werner-Klein M, Grujovic A, Irlbeck C, Obradovic M, Hoffmann M, Koerkel-Qu H, et al. (2020). Interleukin-6 trans-signaling is a candidate mechanism to drive progression of human DCCs during clinical latency. Nature Communications, 11(1), 4977–18701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tohme S, Simmons RL, & Tsung A (2017). Surgery for cancer: A trigger for metastases. Cancer Research, 77(7), 1548–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aguirre-Ghiso JA, Estrada Y, Liu D, & Ossowski L (2003). ERK(MAPK) activity as a determinant of tumor growth and dormancy; regulation by p38(SAPK). Cancer Research, 63(7), 1684–1695. [PubMed] [Google Scholar]
- 73.Yanagisawa M, Huveldt D, Kreinest P, Lohse CM, Cheville JC, Parker AS, Copland JA, & Anastasiadis PZ (2008). A p120 catenin isoform switch affects Rho activity, induces tumor cell invasion, and predicts metastatic disease. Journal of Biological Chemistry, 283(26), 18344–18354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yumoto K, Eber MR, Berry JE, Taichman RS, & Shiozawa Y (2014). Molecular pathways: Niches in metastatic dormancy. Clinical Cancer Research, 20(13), 3384–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adam AP, George A, Schewe D, Bragado P, Iglesias BV, Ranganathan AC, Kourtidis A, Conklin DS, & Aguirre-Ghiso JA (2009). Computational identification of a p38SAPK-regulated transcription factor network required for tumor cell quiescence. Cancer Research, 69(14), 5664–5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang X, Wu JS, Li M, Zhang WL, Gao XL, Wang EF, et al. (2021). Inhibition of DEC2 is necessary for exiting cell dormancy in salivary adenoid cystic carcinoma. Journal of Experimental Clinical Cancer Research, 40(1), 169–01956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McGrath JE, Panzica L, Ransom R, Withers HG, & Gelman IH (2019). Identification of genes regulating breast cancer dormancy in 3D bone endosteal niche cultures. Molecular Cancer Research, 17(4), 1541–7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guereño M, Delgado PM, Lugones AC, Cercato M, Todaro L, Urtreger A, & Peters MG (2020). Glypican-3 (GPC3) inhibits metastasis development promoting dormancy in breast cancer cells by p38 MAPK pathway activation. European Journal of Cell Biology, 99(6), 151096. [DOI] [PubMed] [Google Scholar]
- 79.Schewe DM, & Aguirre-Ghiso JA (2008). ATF6alpha-Rheb-mTOR signaling promotes survival of dormant tumor cells in vivo. Proceedings of the National Academy of Sciences, USA, 105(30), 10519–10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yamaguchi M (2005). Role of regucalcin in maintaining cell homeostasis and function (review). International Journal of Molecular Medicine, 15(3), 371–389. [PubMed] [Google Scholar]
- 81.Sharma S, Pei X, Xing F, Wu SY, Wu K, Tyagi A, Zhao D, Deshpande R, Ruiz MG, Singh R, Lyu F, & Watabe K (2020). Regucalcin promotes dormancy of prostate cancer. Oncogene, 40(5), 1012–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Horak CE, Lee JH, Marshall JC, Shreeve SM, & Steeg PS (2008). The role of metastasis suppressor genes in metastatic dormancy. APMIS., 116(7–8), 586–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gelman IH (2012). Suppression of tumor and metastasis progression through the scaffolding functions of SSeCKS/Gravin/AKAP12. Cancer Metastasis Review, 31(3–4), 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim RS, Avivar-Valderas A, Estrada Y, Bragado P, Sosa MS, Aguirre-Ghiso JA, & Segall JE (2012). Dormancy signatures and metastasis in estrogen receptor positive and negative breast cancer. PLoS ONE, 7(4), e35569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, Zhou A, Eyob H, Balakrishnan S, Wang CY, Yaswen P, Goga A, & Werb Z (2015). Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature, 526(7571), 131–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gao H, Chakraborty G, Lee-Lim AP, Mavrakis KJ, Wendel HG, & Giancotti FG (2014). Forward genetic screens in mice uncover mediators and suppressors of metastatic reactivation. Proceedings of the National Academy of Sciences, U.S.A, 111(46), 16532–16537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dudgeon C, Harris CR, Chen Y, Ghaddar B, Sharma A, Shah MM, Roberts AI, Casabianca A, Collisson EA, Balachandran VP, Vertino PM, De S, & Carpizo DR (2020). A novel model of pancreatic cancer dormancy reveals mechanistic insights and a dormancy gene signature with human relevance. BioRxiv. 10.1101/2020.04.13.037374 [DOI] [Google Scholar]
- 88.Uzuner D, Akkoyç Y, Peker N, Pir P, Gözüaçik D, & Çakir T (2021). Transcriptional landscape of cellular networks reveal interactions driving the dormancy mechanisms in cancer. Science Reports, 11(1), 15806–94005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Quayle LA, Spicer A, Ottewell PD, & Holen I (2021). Transcriptomic profiling reveals novel candidate genes and signalling programs in breast cancer quiescence and dormancy. Cancers (Basel), 13(16), 3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ren Q, Khoo WH, Corr AP, Phan TG, Croucher PI, & Stewart SA (2022). Gene expression predicts dormant metastatic breast cancer cell phenotype. Breast Cancer Research, 24(1), 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Janghorban M, Yang Y, Zhao N, Hamor C, Nguyen TM, Zhang XH, & Rosen JM (2022). Single-Cell analysis unveils the role of the tumor immune microenvironment and notch signaling in dormant minimal residual disease. Cancer Research, 82(5), 885–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Son J, Lee JH, Kim HN, Ha H, & Lee ZH (2010). cAMP-response-element-binding protein positively regulates breast cancer metastasis and subsequent bone destruction. Biochemical and Biophysical Research Communications, 398(2), 309–314. [DOI] [PubMed] [Google Scholar]
- 93.Johnson RW, Sun Y, Ho PWM, Chan ASM, Johnson FA, Pavlos NJ, Sims NA, & Martin TJ (2018). Parathyroid hormone-related protein negatively regulates tumor cell dormancy genes in a PTHR1/cyclic AMP-independent manner. Frontiers in Endocrinology (Lausanne), 9, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.La Belle FA, Calhoun BC, Sharma A, Chang JC, Almasan A, & Schiemann WP (2019). Autophagy inhibition elicits emergence from metastatic dormancy by inducing and stabilizing Pfkfb3 expression. Nature Communications, 10(1), 3668–11640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Blessing AM, Santiago-O’Farrill JM, Mao W, Pang L, Ning J, Pak D, et al. (2020). Elimination of dormant, autophagic ovarian cancer cells and xenografts through enhanced sensitivity to anaplastic lymphoma kinase inhibition. Cancer, 126(15), 3579–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Onoyama I, & Nakayama KI (2008). Fbxw7 in cell cycle exit and stem cell maintenance: Insight from gene-targeted mice. Cell Cycle, 7(21), 3307–3313. [DOI] [PubMed] [Google Scholar]
- 97.Iriuchishima H, Takubo K, Matsuoka S, Onoyama I, Nakayama KI, Nojima Y, & Suda T (2011). Ex vivo maintenance of hematopoietic stem cells by quiescence induction through Fbxw7a; overexpression. Blood, 117(8), 2373–2377. [DOI] [PubMed] [Google Scholar]
- 98.Jiang J, Zheng M, Zhang M, Yang X, Li L, Wang SS, Wu JS, Yu XH, Wu JB, Pang X, Tang YJ, Tang YL, & Liang XH (2019). PRRX1 regulates cellular phenotype plasticity and dormancy of head and neck squamous cell carcinoma through miR-642b-3p. Neoplasia., 21(2), 216–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sosa MS, Parikh F, Maia AG, Estrada Y, Bosch A, Bragado P, et al. (2015). NR2F1 controls tumour cell dormancy via SOX9- and RARbeta-driven quiescence programmes. Nature Communications, 6, 6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Satcher RL, & Zhang XH (2021). Evolving cancer-niche interactions and therapeutic targets during bone metastasis. Nat Rev Cancer, 22(2), 85–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fluegen G, Avivar-Valderas A, Wang Y, Padgen MR, Williams JK, Nobre AR, et al. (2017). Phenotypic heterogeneity of disseminated tumour cells is preset by primary tumour hypoxic microenvironments. Nature Cell Biology, 19(2), 120–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ju S, Wang F, Wang Y, & Ju S (2020). CSN8 is a key regulator in hypoxia-induced epithelial-mesenchymal transition and dormancy of colorectal cancer cells. Molecular Cancer, 19(1), 168–01285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wu R, Roy AM, Tokumaru Y, Gandhi S, Asaoka M, Oshi M, Yan L, Ishikawa T, & Takabe K (2022). NR2F1, a tumor dormancy marker, is expressed predominantly in cancer-associated fibroblasts and is associated with suppressed breast cancer cell proliferation. Cancers (Basel), 14(12), 2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sanchez CA, Yamamoto T, Kawamura Y, Hironaka-Mitsuhashi A, Ono M, Tsuda H, Shimomura A, Tamura K, Takeshita F, Ochiya T, & Yamamoto Y (2020). Long non-coding NR2F1-AS1 is associated with tumor recurrence in estrogen receptor-positive breast cancers. Molecular Oncology, 14(9), 2271–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu Y, Zhang P, Wu Q, Fang H, Wang Y, Xiao Y, Cong M, Wang T, He Y, Ma C, Tian P, Liang Y, Qin LX, Yang Q, Yang Q, Liao L, & Hu G (2021). Long non-coding RNA NR2F1-AS1 induces breast cancer lung metastatic dormancy by regulating NR2F1 and DNp63. Nature Communications, 12(1), 5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhu X, Wang F, Wu X, Li Z, Wang Z, Ren X, Zhou Y, Song F, Liang Y, Zeng Z, Liao W, Ding Y, Liao W, & Liang L (2020). FBX8 promotes metastatic dormancy of colorectal cancer in liver. Cell Death and Disease, 11(8), 622–02870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gooding AJ, & Schiemann WP (2020). Epithelial-mesenchymal transition programs and cancer stem cell phenotypes: Mediators of breast cancer therapy resistance. Molecular Cancer Research, 18(9), 1257–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Johnson RW, Finger EC, Olcina MM, Vilalta M, Aguilera T, Miao Y, et al. (2016). Induction of LIFR confers a dormancy phenotype in breast cancer cells disseminated to the bone marrow. Nature Cell Biology, 18(10), 1078–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Clements ME, Holtslander L, Edwards C, Todd V, Dooyema SDR, Bullock K, Bergdorf K, Zahnow CA, Connolly RM, & Johnson RW (2021). HDAC inhibitors induce LIFR expression and promote a dormancy phenotype in breast cancer. Oncogene, 40(34), 5314–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bragado P, Estrada Y, Parikh F, Krause S, Capobianco C, Farina HG, Schewe DM, & Aguirre-Ghiso JA (2013). TGF-beta2 dictates disseminated tumour cell fate in target organs through TGF-beta-RIII and p38alpha/beta signalling. Nature Cell Biology, 15(11), 1351–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ribatti D, Mangialardi G, & Vacca A (2006). Stephen Paget and the ‘seed and soil’ theory of metastatic dissemination. Clinical Experimental Medicine, 6(4), 145–149. [DOI] [PubMed] [Google Scholar]
- 112.Ruppender N, Larson S, Lakely B, Kollath L, Brown L, Coleman I, et al. (2015). Cellular adhesion promotes prostate cancer cells escape from dormancy. PLoS ONE, 10(6), e0130565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yumoto K, Eber MR, Wang J, Cackowski FC, Decker AM, Lee E, Nobre AR, Aguirre-Ghiso JA, Jung Y, & Taichman RS (2016). Axl is required for TGF-beta2-induced dormancy of prostate cancer cells in the bone marrow. Sci Reports, 6, 36520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Burstyn-Cohen T, & Maimon A (2019). TAM receptors, phosphatidylserine, inflammation, and cancer. Cell Communication and Signaling, 17(1), 156–0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Axelrod HD, Valkenburg KC, Amend SR, Hicks JL, Parsana P, Torga G, DeMarzo AM, & Pienta KJ (2018). AXL is a putative tumor suppressor and dormancy regulator in prostate cancer. Molecular Cancer Research, 17(2), 356–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cackowski FC, Eber MR, Rhee J, Decker A, Yumoto E , Berry JE, Lee E, Shiozawa Y, Jung Y, Aguirre-Ghiso JA, & Taichman RS (2016). Mer tyrosine kinase regulates disseminated prostate cancer cellular dormancy. Journal of Cell Biochemistry, 118(4), 891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kobayashi A, Okuda H, Xing F, Pandey PR, Watabe M, Hirota S, Pai SK, Liu W, Fukuda K, Chambers C, Wilber A, & Watabe K (2011). Bone morphogenetic protein 7 in dormancy and metastasis of prostate cancer stem-like cells in bone. Journal of Experimental Medicine, 208(13), 2641–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Eckhardt BL, Cao Y, Redfern AD, Chi LH, Burrows AD, Roslan S, Sloan EK, Parker BS, Loi S, Ueno NT, Lau PKH, Latham B, & Anderson RL (2020). Activation of canonical BMP4-SMAD7 signaling suppresses breast cancer metastasis. Cancer Research, 80(6), 1304–1315. [DOI] [PubMed] [Google Scholar]
- 119.Nobre AR, Risson E, Singh DK, Di Martino JS, Cheung JF, Wang J, Johnson J, Russnes HG, Bravo-Cordero JJ, Birbrair A, Naume B, Azhar M, Frenette PS, & Aguirre-Ghiso JA (2021). Bone marrow NG2(+)/Nestin(+) mesenchymal stem cells drive DTC dormancy via TGFb2. Nature Cancer, 2(3), 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Prunier C, Baker D, Ten DP, & Ritsma L (2019). TGF-b family signaling pathways in cellular dormancy. Trends in Cancer, 5(1), 66–78. [DOI] [PubMed] [Google Scholar]
- 121.Sharma S, Xing F, Liu Y, Wu K, Said N, Pochampally R, Shiozawa Y, Lin HK, Balaji KC, & Watabe K (2016). Secreted protein acidic and rich in cysteine (SPARC) mediates metastatic dormancy of prostate cancer in bone. Journal of Biological Chemistry, 291(37), 19351–19363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cassar L, Li H, Pinto AR, Nicholls C, Bayne S, & Liu JP (2008). Bone morphogenetic protein-7 inhibits telomerase activity, telomere maintenance, and cervical tumor growth. Cancer Research, 68(22), 9157–9166. [DOI] [PubMed] [Google Scholar]
- 123.Nahm JH, Yang WI, & Yoon SO (2020). Forkhead Box C1 (FOXC1) Expression in stromal cells within the microenvironment of T and NK Cell Lymphomas: Association with tumor dormancy and activation. Cancer Research and Treatments, 52(4), 1273–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zangrossi M, Romani P, Chakravarty P, Ratcliffe CDH, Hooper S, Dori M, Forcato M, Bicciato S, Dupont S, Sahai E, & Montagner M (2021). EphB6 regulates TFEB-lysosomal pathway and survival of disseminated indolent breast cancer cells. Cancers (Basel), 13(5), 1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Visan KS, Lobb RJ, & Moller A (2020). The role of exosomes in the promotion of epithelial-to-mesenchymal transition and metastasis. Frontiers in Bioscience (Landmark Ed.), 251022–1057. [DOI] [PubMed] [Google Scholar]
- 126.Attaran S, & Bissell MJ (2021). The role of tumor microenvironment and exosomes in dormancy and relapse. Seminars in Cancer Biology, 78, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ono M, Kosaka N, Tominaga N, Yoshioka Y, Takeshita F, Takahashi RU, Yoshida M, Tsuda H, Tamura K, & Ochiya T (2014). Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Science Signaling, 7(332), ra63. [DOI] [PubMed] [Google Scholar]
- 128.Li X, Yang J, Bao M, Zeng K, Fu S, Wang C, & Ye E (2018). Wnt signaling in bone metastasis: Mechanisms and therapeutic opportunities. Life Sciences, 208, 33–45. [DOI] [PubMed] [Google Scholar]
- 129.Ren D, Dai Y, Yang Q, Zhang X, Guo W, Ye L, Huang S, Chen X, Lai Y, Du H, Lin C, Peng X, & Song L (2019). Wnt5a induces and maintains prostate cancer cells dormancy in bone. Journal of Experimental Medicine, 216(2), 428–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fane ME, Chhabra Y, Alicea GM, Maranto DA, Douglass SM, Webster MR, et al. (2022). Stromal changes in the aged lung induce an emergence from melanoma dormancy. Nature, 606(7913), 396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lee E, Yang J, Ku M, Kim NH, Park Y, Park CB, Suh JS, Park ES, Yook JI, Mills GB, Huh YM, & Cheong JH (2015). Metabolic stress induces a Wnt-dependent cancer stem cell-like state transition. Cell Death and Disease, 6, e1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bragado P, Estrada Y, Parikh F, Krause S, Capobianco C, Farina HG, Schewe DM, & Aguirre-Ghiso JA (2013). TGF-b dictates disseminated tumour cell fate in target organs through TGF-b-RIII and p38a signalling. Nature Cell Biology, 15(11), 1351–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hernandez S, Serrano AG, & Solis Soto LM (2022). The role of nerve fibers in the tumor immune microenvironment of solid tumors. Advanced Biology (Weinh), 6(9), e2200046. [DOI] [PubMed] [Google Scholar]
- 134.Erin N, Shurin GV, Baraldi JH, & Shurin MR (2022). Regulation of carcinogenesis by sensory neurons and neuromediators. Cancers (Basel), 14(9), 2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Roda N, Blandano G, & Pelicci PG (2021). Blood vessels and peripheral nerves as key players in cancer progression and therapy resistance. Cancers (Basel), 13(17), 4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mulcrone PL, Campbell JP, Clément-Demange L, Anbinder AL, Merkel AR, Brekken RA, Sterling JA, & Elefteriou F (2017). Skeletal colonization by breast cancer cells is stimulated by an osteoblast and b2AR-dependent neoangiogenic switch. Journal of Bone Mineral Research, 32(7), 1442–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dai J, Cimino PJ, Gouin KH III., Grzelak CA, Barrett A, Lim AR, et al. (2022). Astrocytic laminin-211 drives disseminated breast tumor cell dormancy in brain. Nature Cancer, 3(1), 25–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ni B, Li Q, Zhuang C, Huang P, Xia X, Yang L, Ma X, Huang C, Zhao W, Tu L, Shen Y, Zhu C, Zhang Z, Zhao E, Wang M, & Cao H (2022). The nerve-tumour regulatory axis GDNF-GFRA1 promotes tumour dormancy, imatinib resistance and local recurrence of gastrointestinal stromal tumours by achieving autophagic flux. Cancer Letters, 535, 215639. [DOI] [PubMed] [Google Scholar]
- 139.Touil Y, Segard P, Ostyn P, Begard S, Aspord C, El Machhour MR, et al. (2016). Melanoma dormancy in a mouse model is linked to GILZ/FOXO3A-dependent quiescence of disseminated stem-like cells. Science Reports, 6, 30405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liburkin-Dan T, Toledano S, & Neufeld G (2022). Lysyl oxidase family enzymes and their role in tumor progression. Internatinal Journal of Molecular Science, 23(11), 6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ferreira S, Saraiva N, Rijo P, & Fernandes AS (2021). LOXL2 inhibitors and breast cancer progression. Antioxidants (Basel), 10(2), 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Weidenfeld K, Schif-Zuck S, Abu-Tayeh H, Kang K, Kessler O, Weissmann M, Neufeld G, & Barkan D (2016). Dormant tumor cells expressing LOXL2 acquire a stem-like phenotype mediating their transition to proliferative growth. Oncotarget, 7(44), 71362–71377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Aslakson CJ, & Miller FR (1992). Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Research, 52(6), 1399–1405. [PubMed] [Google Scholar]
- 144.Almog N, Ma L, Raychowdhury R, Schwager C, Erber R, Short S, Hlatky L, Vajkoczy P, Huber PE, Folkman J, & Abdollahi A (2009). Transcriptional switch of dormant tumors to fast-growing angiogenic phenotype. Cancer Research, 69(3), 836–844. [DOI] [PubMed] [Google Scholar]
- 145.Tiram G, Segal E, Krivitsky A, Shreberk-Hassidim R, Ferber S, Ofek P, et al. (2016). Identification of dormancy-associated MicroRNAs for the design of osteosarcoma-targeted dendritic polyglycerol nanopolyplexes. ACS Nanotechnology, 10(2), 2028–2045. [DOI] [PubMed] [Google Scholar]
- 146.Owen KL, Gearing LJ, Zanker DJ, Brockwell NK, Khoo WH, Roden DL, et al. (2020). Prostate cancer cell-intrinsic interferon signaling regulates dormancy and metastatic outgrowth in bone. EMBO Reports, 21(6), e50162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Power CA, Pwint H, Chan J, Cho J, Yu Y, Walsh W, & Russell PJ (2009). A novel model of bone-metastatic prostate cancer in immunocompetent mice. Prostate, 69(15), 1613–1623. [DOI] [PubMed] [Google Scholar]
- 148.Wang X, Yu J, Yan J, Peng K, & Zhou H (2022). Single-cell sequencing reveals MYC targeting gene MAD2L1 is associated with prostate cancer bone metastasis tumor dormancy. BMC Urology, 22(1), 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kudaravalli S, den Hollander P, & Mani SA (2022). Role of p38 MAP kinase in cancer stem cells and metastasis. Oncogene, 41(23), 3177–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Qin JZ, Xin H, Qi XM, & Chen G (2022). Isoform-specific and cell/tissue-dependent effects of p38 MAPKs in regulating inflammation and inflammation-associated oncogenesis. Frontiers in Bioscience (Landmark Ed.), 27(1), 31. [DOI] [PubMed] [Google Scholar]
- 151.Ohkubo S, Nakahata N, & Ohizumi Y (1996). Thromboxane A2-mediated shape change: Independent of Gq-phospholipase C-Ca2+ pathway in rabbit platelets. British Journal of Pharmacology, 117(6), 1095–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Janowska A, Iannone M, Fidanzi C, Romanelli M, Filippi L, Del RM, Martins M, & Dini V (2022). The genetic basis of dormancy and awakening in cutaneous metastatic melanoma. Cancers (Basel), 14(9), 2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Asghar U, Witkiewicz AK, Turner NC, & Knudsen ES (2015). The history and future of targeting cyclin-dependent kinases in cancer therapy. Nature Reviews Drug Discovery, 14(2), 130–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Singh DK, Patel VG, Oh WK, & Aguirre-Ghiso JA (2021). Prostate cancer dormancy and reactivation in bone marrow. Journal of Clinical Medicine, 10(12), 2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ramamoorthi G, Kodumudi K, Gallen C, Zachariah NN, Basu A, Albert G, Beyer A, Snyder C, Wiener D, Costa RLB, & Czerniecki BJ (2021). Disseminated cancer cells in breast cancer: mechanism of dissemination and dormancy and emerging insights on therapeutic opportunities. Seminars in Cancer Biology, 78, 78–89. [DOI] [PubMed] [Google Scholar]