Abstract
Acidic tea (Camellia sinensis) plantation soil usually suffers from magnesium (Mg) deficiency, and as such, application of fertilizer containing Mg can substantially increase tea quality by enhancing the accumulation of nitrogen (N)-containing chemicals such as amino acids in young tea shoots. However, the molecular mechanisms underlying the promoting effects of Mg on N assimilation in tea plants remain unclear. Here, both hydroponic and field experiments were conducted to analyze N, Mg, metabolite contents, and gene expression patterns in tea plants. We found that N and amino acids accumulated in tea plant roots under Mg deficiency, while metabolism of N was enhanced by Mg supplementation, especially under a low N fertilizer regime. 15N tracing experiments demonstrated that assimilation of N was induced in tea roots following Mg application. Furthermore, weighted gene correlation network analysis (WGCNA) analysis of RNA-seq data suggested that genes encoding glutamine synthetase isozymes (CsGSs), key enzymes regulating N assimilation, were markedly regulated by Mg treatment. Overexpression of CsGS1.1 in Arabidopsis (Arabidopsis thaliana) resulted in a more tolerant phenotype under Mg deficiency and increased N assimilation. These results validate our suggestion that Mg transcriptionally regulates CsGS1.1 during the enhanced assimilation of N in tea plant. Moreover, results of a field experiment demonstrated that high Mg and low N had positive effects on tea quality. This study deepens our understanding of the molecular mechanisms underlying the interactive effects of Mg and N in tea plants while also providing both genetic and agronomic tools for future improvement of tea production.
Magnesium transcriptionally regulates the glutamine synthetase gene CsGS1.1 during the enhanced assimilation of nitrogen in tea plants.
Introduction
Tea (Camellia sinensis) is an important economic crop in Asia, and tea plantations have become a pivotal industry that increases the income of farmers. The positive effect of magnesium (Mg) on the growth and quality of tea plants has been widely demonstrated (Ruan et al. 2012; Gerendás and Führs 2013); however, tea plantations are mainly distributed in areas with acidic red soil, where Mg deficiency is common, and the use of Mg fertilizer in tea cultivation is generally rare (Ni et al. 2019). Therefore, the mechanisms involved in Mg nutrition and ways to optimize Mg nutrition management in tea plantations are of considerable interest, to both improve the quality and enhance the efficiency of tea production.
Mg is not only the central atom in chlorophyll but also essential for the activity of many enzymes (Marschner 2012). Numerous researchers found that Mg has an important role in improving crop yield and quality (Marschner 2012; Gerendás and Führs 2013). In the tea plant, Mg content is generally 500 to 4000 mg/kg, and the application of Mg fertilizer can improve both tea yield and quality (Ruan et al. 1998, 1999, 2012; Gerendás and Führs 2013). Application of Mg substantially increases the content of caffeine and aroma components in tea (Ruan et al. 1999) while low supply of Mg caused changes in the levels of other nutrients in mature leaves (Ruan and Gerendá 2015). Jayaganesh and Venkatesan (2010) and Jayaganesh et al. (2011) demonstrated that the input–output ratio of Mg sulfate is higher than magnesite and Mg nitrate. Due to the high mobility of Mg in the phloem, the application of Mg fertilizer on leaves has been demonstrated to substantially increase the yield and quality of a number of crops (Vrataric et al. 2006; Dordas 2009; Rao and Rajput 2011; Wszelaczynska and Poberezny 2011).
N is essential for plant growth and development and a limiting factor for crop yield and quality (Tegeder and Masclaux-Daubresse 2018; Liu et al. 2021). In agricultural production, the application of N fertilizer generally increases plant yield. However, generally, <50% of N fertilizer can be used by cereal crops; most of the remainder is lost to the environment, causing pollution (Zhang et al. 2013; Fernie 2021). However, plant nitrogen use efficiency (NUE) is highly complex, being influenced both by absorption and assimilation of N via roots, its long-distance transport from roots to leaves, and the remobilization of stored N reserves in plants. Ammonium (NH4+) and nitrate (NO3−) are the main inorganic forms of N used by tea plants with the utilization efficiency of NH4+ substantially higher than that of NO3− in tea plant (Tang et al. 2020). NH4+ is assimilated via the glutamine synthetase–glutamate synthase (GS-GOGAT) pathway, which assimilates ammonium either immediately following its uptake or after nitrate has been reduced to nitrite (Cruz et al. 2006; Thomsen et al. 2014). Studies suggest that glutamine synthetase (GS) (EC 6.3.1.2) is a key enzyme regulating tea plant absorption and utilization of N (Chardon et al. 2012; Liu et al. 2019). GS activity is regulated by 2 isoenzymes, cytosolic GS (GS1) and plastidic GS (GS2) (Cren and Hirel 1999). GS1 is encoded by 3 to 5 genes, whereas GS2 is encoded by a single gene (Swarbreck et al. 2011; Thomsen et al. 2014). Moreover, GS1 is important for primary N assimilation in roots and for the re-assimilation of ammonium generated during protein turnover in leaves, whereas the GS2 is predominantly involved in the re-assimilation of photo-respiratory ammonium in chloroplasts as well as the assimilation of ammonium derived from the reduction of nitrate in plastids (Bernard and Habash 2009; Thomsen et al. 2014). Intriguingly, the overexpression of GSs in transgenic tobacco (Nicotiana tabacum) increases the activity of GS and enhances N assimilation under low-N conditions (Wang et al. 2013).
An alternative route is improving the remobilization of stored N since this allows plants to effectively utilize limited N (Staswick 1994; Masclaux-Daubresse et al. 2010). During the sprouting period of spring tea, approximately 70% of N is derived from remobilization of the plants’ own reserves (Okano et al. 1994). Ruan and Gerendá (2015) painted 15N-labelled urea on to mature tea leaves and found that it was mainly transported to the young new shoots. It has been established that amino acids are transported across membranes via amino acid transporters, to regulate N distribution between the source and sink, and that proton-coupled amino acid permeases (AAPs) are involved in this process (Tegeder and Masclaux-Daubresse 2018). In general, the non-proteinogenic amino acid theanine, the free amino acid present at the highest levels in tea, is considered the primary form of stored and transported N in tea plants, being synthesized in the roots and transported to the leaves (Dong et al. 2020).
The cooperative mechanisms involved in crop N and Mg nutrition have been extensively studied (Mulder 1956; Ruan et al. 2000, 2004). The coordination of N and Mg nutrition is an important factor that affects amino acid accumulation (Ruan et al. 2012); however, excessive application of Mg fertilizer can also lead to potassium–Mg antagonism, which may inhibit tea plant biomass and amino acid accumulation (Ruan et al. 1998). Similarly, Ruan et al. (2006) found that soil pH, as well as soil and plant Mg content, decreased substantially with the application of excessive N fertilizer. This is important because Mg fertilizer can considerably increase the activity of amino acid transporter proteins in tea shoots (Jayaganesh and Venkatesan 2010). As such, the effect of Mg nutrition on amino acid accumulation in tea leaves is likely related to the long-distance transport of metabolites in tea plants (Ruan et al. 2006, 2012). However, the mechanism by which Mg nutrition regulates N assimilation and metabolism requires further study. Thus, the purpose of this study was to investigate the molecular mechanisms underlying the interactive effects of Mg and N in tea plants. We present evidence that Mg supplementation increased the amino acid content and N assimilation in tea roots and shoots. Heterologous expression of the CsGS1.1 gene in Arabidopsis (Arabidopsis thaliana) further validated this conclusion. Our study thus deepened our understanding of the molecular mechanisms underlying the interactive effects of Mg and N in tea plants, which also provided a potential fertilization strategy on improving N utilization efficiency by Mg supplementation under low N condition. The recent explosion in genomic and transcriptomic resources (Jiang et al. 2022) means that characterizing tea natural variants following the identification of CsGS1.1 as a candidate gene paves the road for the breeding of enhanced NUE in tea.
Results
Mg deficiency decreases the transport of N from roots to mature leaves, while Mg supplementation enhances N metabolism
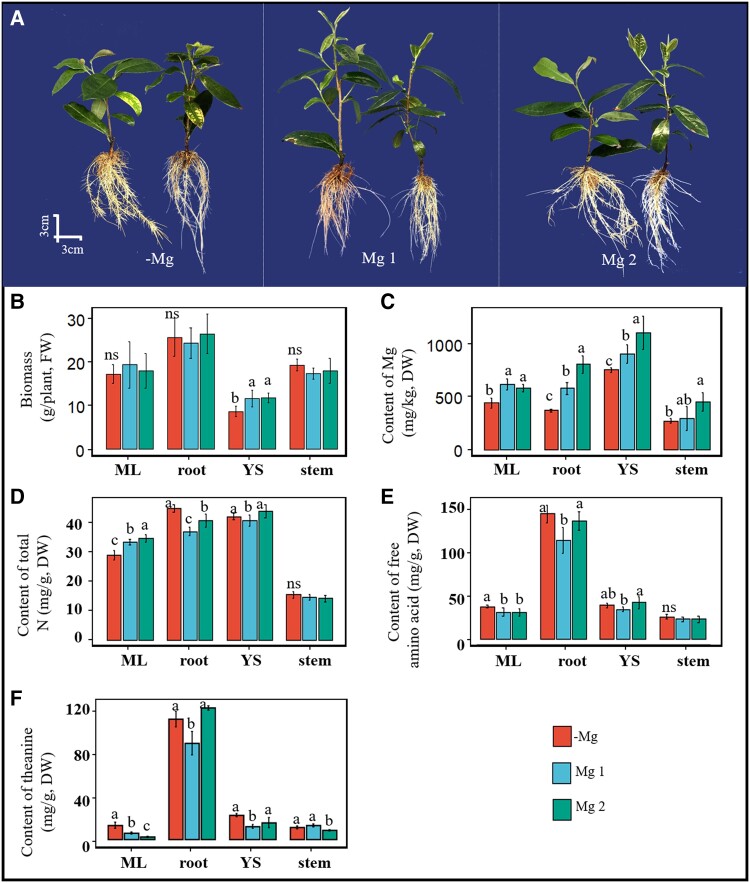
Mature leaves of tea plants exhibited observable chlorosis after 3 weeks of Mg deficiency (−Mg). Although root, mature leaf (ML), and stem biomass did not differ significantly among the 3 Mg treatment groups, young shoot (YS) biomass was significantly decreased by Mg deficiency (in comparison to plants grown under the Mg1 and Mg2 regimes (Fig. 1B), suggesting that Mg deficiency inhibits YS growth. The Mg levels in roots and YS significantly increased with increasing Mg fertilizer, and Mg levels were highest in plants grown under the Mg2 regime (Fig. 1C). Notably, Mg levels in YS were substantially higher than those in the root, ML, and stem (Fig. 1C), suggesting that Mg is a relatively mobile element.
Figure 1.
Changes of biomass, Mg content, total N, free amino acid, and theanine content in tea plants under different Mg application levels. A) Phenotype of tea plants treated with Mg deficiency (−Mg) and Mg application (Mg1 and Mg2, full-strength nutrient solution). The images were digitally extracted for comparison. B) Biomass, C) Mg content, D) total N content, E) amino acid content, and F) theanine content in mature leaf (ML), root, young/new shoot (YS), and stem of tea plants treated with Mg deficiency (−Mg), Mg concentration in the full-strength nutrient solution (Mg1, 0.4 mmol/L), and 2 times the Mg concentration in the full-strength nutrient solution (Mg2). Individual images of plants were digitally extracted for comparison. Error bars show the mean values ± SD (n = 3). Statistical analysis was carried out with Duncan's multiple range test. Different letters indicate significant differences (P < 0.05). DW, dry weight; FW, fresh weight; ns, not significant.
Under the stress of Mg starvation following the −Mg treatment, the content of total N and free amino acid contents in the tea roots highly increased, when compared to those of plants grown under the Mg1 regime (Fig. 1, D and E), suggesting that starvation of Mg may lead to inhibition of N mobilization from the root. That said, in the mature leaves, amino acids accumulated under −Mg conditions, even though the total N content significantly decreased in the −Mg group compared to both other groups. In comparison with Mg sufficiency, Mg deficiency resulted in an increased level of total N in roots but a decrease in the total N in ML (Fig. 1D), whereas an opposite trend was observed for mature-leaf amino acid contents (Fig. 1E). Analysis of gene expression (Supplemental Fig. S1) showed that the amino acid transport gene, amino acid permease gene CsAAP5, was downregulated in the roots under −Mg conditions compared to those in Mg1 conditions, while it was also downregulated in new shoots under −Mg conditions compared to those in Mg2 conditions. Notably, asparagine synthetase gene CsASN and glutamate synthase homologue gene CsGLT, which are associated with N reactivation and reuse/relocation in tea leaves (Fan et al. 2020), were significantly upregulated under −Mg conditions compared with the more Mg replete conditions.
In summary, Mg deficiency has a negative effect on tea plants, while enhanced supplementation of Mg (Mg2) above the normal level of application of Mg (Mg1) resulted in tea plants displaying better performance. Intriguingly, the higher level (Mg2 vs. Mg1) of Mg supplementation led to a higher N content in most tea plant tissues with the exception of the stem (Fig. 1D). Meanwhile, contents of free amino acid and its dominant composition—theanine—were higher in roots and YS following enhanced Mg application (Mg2 vs. Mg1; Fig. 1, E and F). These results thus indicate that enhanced Mg application likely induces N-related metabolism in tea.
15NH4+ assimilation in tea plants is enhanced by increasing Mg supplement
To further assess our hypothesis that increased Mg supplementation enhances N metabolism, we next followed the absorption of 15N in tea plants under a normal Mg level (Mg1) and high Mg supply (Mg2) was conducted (Table 1). To mimic the real condition as the practice of tea production, hydroponic N was provided with a mixture of NH4+ and NO3−. As shown in Table 1, the absorption efficiency was substantially higher for NH4+ than NO3− in tea plants (Table 1), consistent with previous observations (Ishigaki 1974). Additionally, the effect of Mg supplementation on NH4+ absorption was substantially higher than the effect on NO3− absorption. The 15N enrichment (atom percent excess) under Mg1 conditions was much lower than that under Mg2 conditions in the 15NH4+ groups, indicating that NH4+ absorption was enhanced under Mg2 conditions. By contrast, the 15N enrichment under Mg1 conditions was higher than that under Mg2 conditions in the 15NO3− groups, indicating relatively decreased NO3− absorption and utilization under Mg2 conditions. The absorption and utilization of NH4+ differed under different Mg treatment conditions and as culture time increased. The ratio of N levels in samples below and above the ground (root/(mature leaf + shoot + stem)) was also higher under low Mg condition (0.33 and 0.51, respectively) than that under the Mg-supplied condition (0.26 and 0.41, respectively), which suggests that N source transformation from the root to the leaf has been enhanced under higher Mg condition.
Table 1.
15N enrichment (atom percent excess) in mature leaves, roots, shoots, and stems of tea plants treated with low (Mg1, full-strength nutrient solution) and high (Mg2, 2 times of full-strength nutrient solution) Mg application after 15N labeling. Data were presented as mean values ± SD (n = 3)
| Treatments | 15N enrichment in different tissues | |||||
|---|---|---|---|---|---|---|
| Mg | N | Mature leaf | Root | Shoot | Stem | Root/(mature leaf + shoot + stem) |
| Mg1 | K15NO3 | 0.66 ± 0.01 | 0.85 ± 0.02 | 1.12 ± 0.03 | 0.82 ± 0.02 | 0.33 ± 0.01 |
| 15NH4SO4 | 1.63 ± 0.06 | 5.70 ± 0.15 | 6.64 ± 0.17 | 2.87 ± 0.13 | 0.51 ± 0.02 | |
| Mg2 | K15NO3 | 0.53 ± 0.03 | 0.53 ± 0.02 | 0.85 ± 0.04 | 0.59 ± 0.02 | 0.26 ± 0.01 |
| 15NH4SO4 | 2.88 ± 0.11 | 8.39 ± 0.22 | 11.30 ± 0.29 | 5.86 ± 0.16 | 0.41 ± 0.01 | |
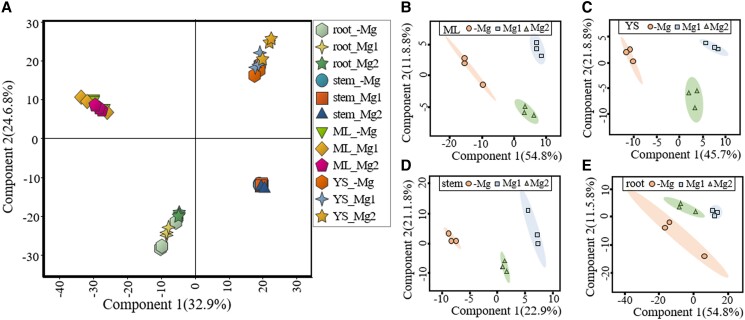
Mg supplementation enhanced accumulation of metabolites related to N metabolism in roots, stems, and leaves of tea plants
To reveal the portion of the global metabolome related to N metabolism that was regulated by Mg application, GC mass spectrometry-based analysis coupled to principal component analysis (PCA) was used for phytochemical profiling (Fig. 2). The PCA score plot (Fig. 2A) clearly distinguished the four tea tissue samples in PC1 (explaining 32.9% of the variance) and PC2 (explaining 24.6% of the variance). The metabolites that significantly differed (P < 0.01) in their abundance among the 3 treatments have been identified based on further PCA analysis performed on individual data of each plant tissue (Fig. 2, B to E). A total of 21 differential metabolites were identified (Supplemental Table S1). The contents of asparagine, glutamine, phenylalanine, and the dipeptide γ-Glu-Leu were significantly higher in the roots grown in the −Mg than those grown the Mg1 regime, consistent with the total amino acid contents. Comparing Mg2 with the Mg1 treatment, increases in phenylalanine and theanine in the roots; asparagine, glutamine, threonine, phenylalanine, theanine, and γ-Glu-Leu in the new shoots; and asparagine and citrulline in the mature leaves could be observed. Further, glucose decreased in the roots in the −Mg group compared to the Mg1 group. In the mature leaves, certain glycoside group-containing metabolites significantly increased both under −Mg and Mg2 conditions, respectively, compared to those under Mg1 conditions, for example, rutin (quercetin-3-rutinoside) and kaempferol-glucoside.
Figure 2.
Global metabolites profiling using principal component analysis (PCA). The PCA score plots of metabolites in A) all four tissues, B) mature leaves (ML), C) young shoots (YS), D) stems, and E) roots from tea plants treated with 3 Mg levels. Each dot represents an individual plant.
Mg application improves the efficiency of N utilization in tea plants under low but not high N fertilization
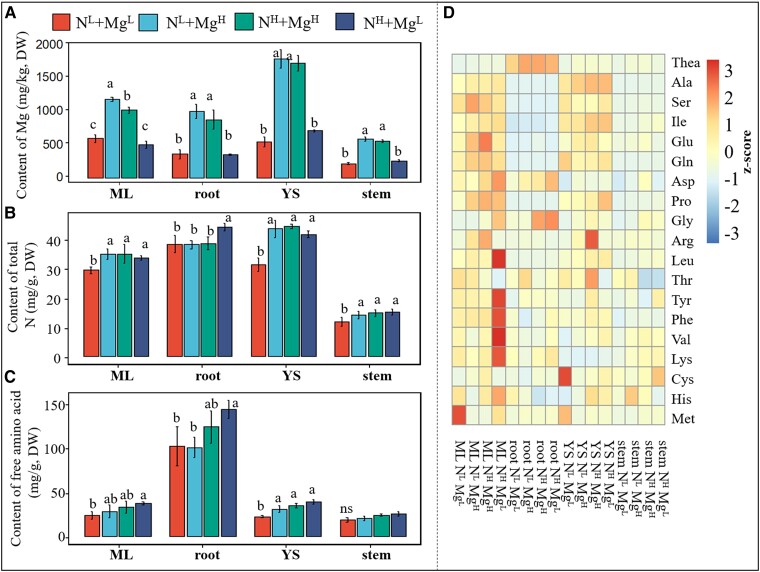
In order to evaluate the interaction effect of Mg and N on tea plants, we carried out hydroponic experiments. In this experiment, endogenous Mg levels were significantly higher in all four tea plant tissues from plants grown under high availability of Mg (MgH) compared to their levels in plants grown under low Mg levels (MgL) (Fig. 3A), with the Mg level being highest in YS compared to roots, ML, and stem (Fig. 3A). In comparison with the MgL condition, MgH exhibited significantly elevated total N contents in YS, ML, and stem under low N conditions (NL), to levels comparable to those observed under high N conditions (NH) (Fig. 3B). However, under MgH condition, there was no significant difference in amino acid content in YS and ML between NL and NH groups (Fig. 3C), but these were lower in the roots in the NL group than the NH group. By contrast, under NH condition, MgL increased N accumulation in the roots (Fig. 3B), which is consistent with the results that Mg deficiency led to N accumulation in the soil-grown root (Fig. 1D). Thus, Mg application was more effective at improving the efficiency of N utilization in tea plants at the NL level than under NH condition.
Figure 3.
The interaction effect of Mg and N on hydroponic tea plants. Changes of A) Mg, B) total N, and C) total free amino acid content. D) Heatmap of free amino acids in the mature leaf (ML), root, young shoot (YS), and stem of tea plants treated with different levels of N and Mg. DW, dry weight. Error bars show the mean values ± SD (n = 3). Statistical analysis was carried out with Duncan's multiple range test. Different letters indicate significant differences (P < 0.05); ns, not significant. Tea seedlings were treated with 2 levels of Mg [1 quarter (MgL = 0.1 mmol/L) and 2 times (MgH = 0.8 mmol/L) concentration in the full-strength nutrient solution] and 2 levels of N [1 quarter (NL) and 2 times (NH) concentration in the full-strength nutrient solution].The 4 treatment groups were as follows: low N and low Mg (NL + MgL), low N and high Mg (NL + MgH), high N and high Mg (NH + MgH), and high N and low Mg (NH + MgL).
Theanine is the most abundant free amino acids in tea plant (Dong et al. 2020) and stable isotope-labeled precursor tracing reveals that L-alanine is converted to L-theanine via L-glutamate not ethylamine in tea plants in vivo (Fu et al. 2021). Its content in the roots was higher than that in YS, ML, and stem, whereas the levels of the other amino acids such as Ala, Ser, Ile, Glu, and Gln were lower in roots than they were in ML and YS (Fig. 3D). Specifically, changes of theanine under the four treatments reflect those of total N, i.e. in ML and YS, it was present at a lower level under MgL supply when plants were cultivated under the NL condition, while its levels were invariant between NL and NH when tea plants were grown under MgH conditions. Given that theanine represents a major N store in tea, this result suggests that Mg supply highly increases the utilization of N under NL supply.
Transcriptomic profiling of roots, stems, and leaves of tea plants under Mg treatment
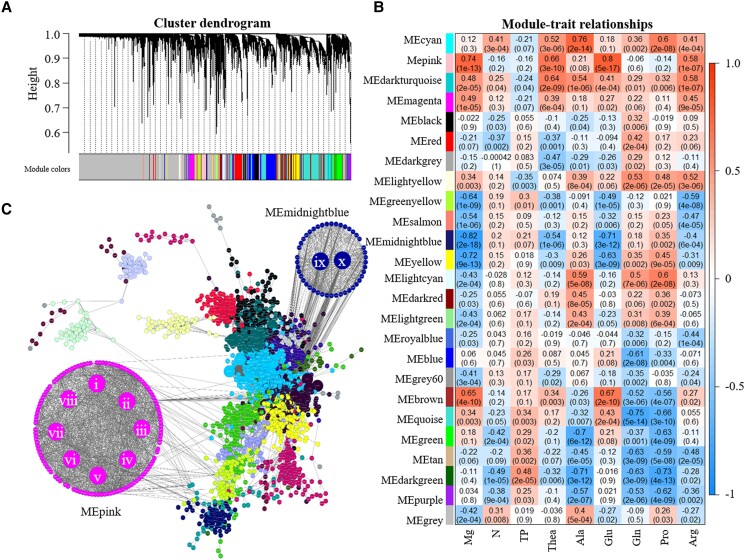
To evaluate the effect of Mg on gene expression in tea, samples from the hydroponic experiment examining the Mg and N interaction were subjected to RNA-seq analysis. A total of 8,490, 8,556, 7,318, and 4,702 unique DEGs were identified in the ML, root, YS, and stem, respectively. GO enrichment analysis revealed that DEGs were most substantially enriched in the extracellular region and transporter activity, signal transduction and transport, cellular amino acid, and derivative metabolic process. Co-expression modules by weighted gene correlation network analysis (WGCNA) were constructed to investigate genes related to the Mg level. Among the 9,576 DEGs, 25 WGCNA modules were identified. Based on the correlation analysis of module–trait, the “MEbrown” and “MEpink” modules positively correlated with Mg content in tea plant (r = 0.65, P = 4.0 × 10−10, and r = 0.74, P = 1.0 × 10−13, respectively) (Fig. 4). In contrary, the “MEmidnightblue” modules negatively correlated with Mg content (r = −0.82, P = 2.0 × 10−18). Genes in the “MEpink” and “MEmidnightblue” modules contained GO terms related to amino acid transport and metabolism and carbohydrate transport and metabolism.
Figure 4.
Identification of genes related to Mg application level. A) Gene modules identified by weighted gene co-expression network analysis (WGCNA). B) Relationships of consensus module genes and content of Mg, free amino acids. C) Interaction network of genes and hub genes in the MEpink and MEmidnightblue modules. The Roman numerals shown in the MEpink and MEmidnightblue modules represent the ten hub genes. The module name is shown on the left side of each cell. Numbers in the table report the correlations of the corresponding module eigengenes and contents, with the P-values printed below the correlations in parentheses. The table is color coded by correlation according to the color legend. Intensity and direction of correlations are indicated on the right side of the heatmap (red, positively correlated; blue, negatively correlated).
To identify candidate genes associated with N and amino acid metabolism that were affected by Mg nutrition, the edge number for each node of the co-expression modules was then analyzed. Eight hub genes were identified from the “MEpink” module (Supplemental Table S2). Among them, four genes were related to amino acid metabolism (CSS0050330, CSS0033052, CSS0007310 and CSS0007224), 2 were glycosyl transferase genes (CSS0001107 and CSS0006318), and 2 were TF genes (Dof domain and Myb-like DNA-binding domain) (Supplemental Table S2). In the “MEmidnightblue” module, 2 genes, UDP-glycosyltransferase (CSS0010308 and CSS0019985), were highly connected with other genes.
Identification of potential CsGSs members regulating Mg-mediated N assimilation
Based on the above WGCNA analysis, among the four genes related to amino acid metabolism, CSS0007310 and CSS0007224 were glutamine synthetase (GS) coding genes directly associated with N assimilation. Considering that NH4+ assimilation in tea plants is enhanced by increasing Mg supply in this study, the potential gene members of the GS family in tea plants that might be involved in the regulation of Mg-mediated N assimilation were identified (Supplemental Fig. S2). There are six CsGS homologues in the tea plant genome (Xia et al. 2019; Supplemental Fig. S2A). Combined with data from our previous study (Tang et al. 2020), there were four gene members that belong to CsGSs, including CsGS2 (CSS0007310), CsGS1.1 (CSS0007224), CsGS1.2 (CSS0015313 or CSS002630), and CsGS1.3 (CSS0034978 or CSS0037306). Tissue specificity analysis revealed that CsGS1.1 is highly expressed in the root and also abundant in the YS and ML, CsGS1.2 is highly abundant in the fruit, and CsGS1.3 is most abundant in the flower, while CsGS2 is most abundant in ML (Supplemental Fig. S2C). This result suggests that CsGS1.1 might be involved in the regulation of Mg-mediated N assimilation.
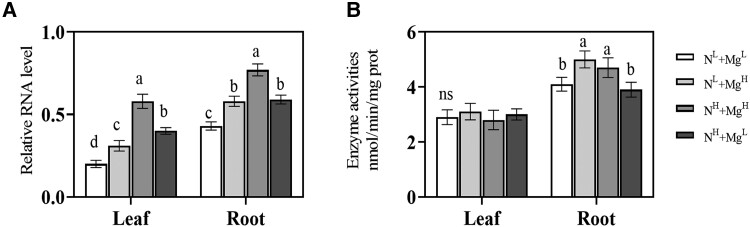
Moreover, under both low and high N conditions, the expression of CsGS1.1 was significantly upregulated in the leaves (1.6- and 1.5-fold, under low and high N treatment, respectively) in response to elevated Mg supply (Fig. 5A), as well as in roots (1.5- and 1.2-fold, under low and high N treatment, respectively). In addition, high N supply also increased the expression of CsGS1.1 under both Mg conditions compared to the low N condition. Notably, low Mg significantly downregulated the activity of GS in the roots but did not affect the GS activity in the leaves (Fig. 5B).
Figure 5.
Changes of CsGS1.1 expression levels and GS activities in hydroponic tea plants cultured with different levels of N and Mg. A)CsGS1.1 relative expression levels and B) GS enzyme activities in the leaf and root of tea plant treated with different levels of N and Mg. Error bars show the mean values ± SD (n = 3). Statistical analysis was carried out with Duncan's multiple range test. Different letters indicate significant differences (P < 0.05). GS, glutamine synthetase; ns, not significant. Tea seedlings were treated with 2 levels of Mg [1 quarter (MgL = 0.1 mmol/L) and 2 times (MgH = 0.8 mmol/L) concentration in the full-strength nutrient solution] and 2 levels of N [1 quarter (NL) and 2 times (NH) concentration in the full-strength nutrient solution].The four treatment groups were as follows: low N and low Mg (NL + MgL), low N and high Mg (NL + MgH), high N and high Mg (NH + MgH), and high N and low Mg (NH + MgL).
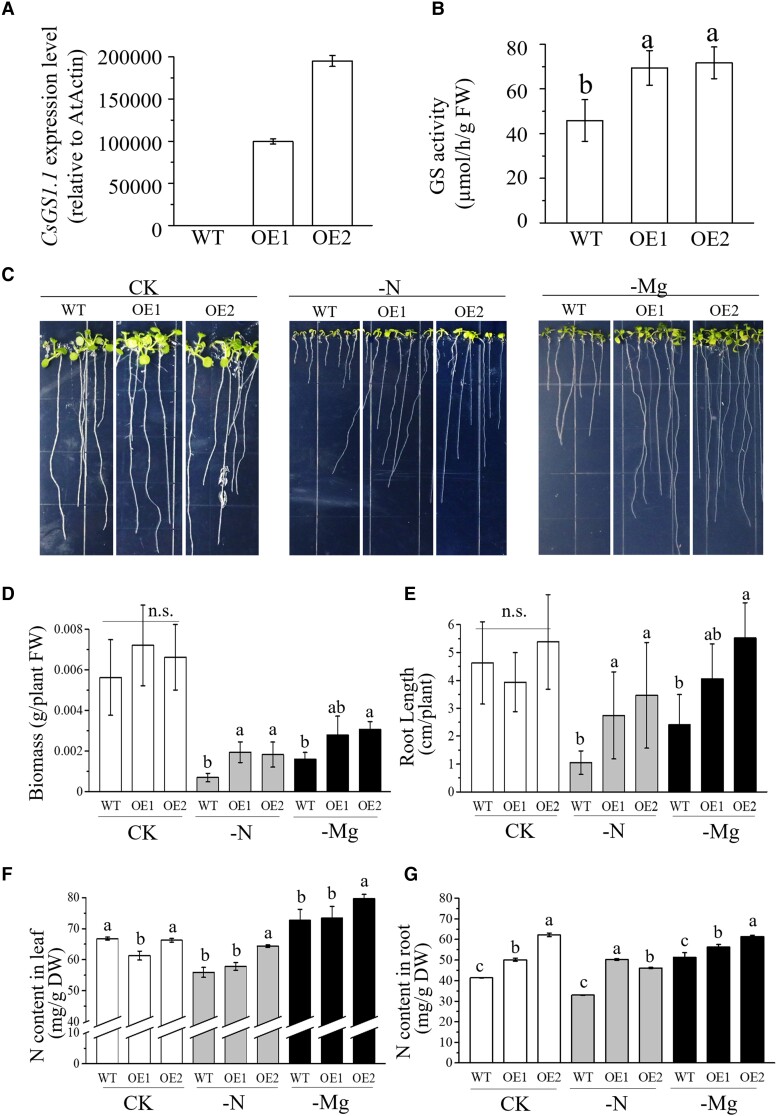
Overexpression of CsGS1.1 in transgenic Arabidopsis promoted the growth of plants and increased N assimilation under Mg deficiency
Given that tea transformation remains highly recalcitrant and only possible in a few labs, therefore, to investigate the function of CsGS1.1 in regulating Mg-mediated N assimilation, we overexpressed CsGS1.1 in wild-type (WT, Col-0) Arabidopsis. High CsGS1.1 expression and GS activity in the transgenic lines were confirmed via RT-qPCR and enzymatic analysis (Fig. 6, A and B). Overexpression of CsGS1.1 in transgenic plants increased their tolerance to N deficiency with higher plant fresh weight and longer root length in transgenic plants (Fig. 6, C to E), indicating that CsGS1.1 had functional roles in N assimilation. The growth of WT plants was significantly inhibited under Mg deficiency compared to that of plants grown under normal 1/2 MS medium (Fig. 6, C to E). The root length of CsGS1.1 overexpressed plants was significantly longer under the Mg deficiency condition than the WT (Fig. 6, C to E), and the N contents in CsGS1.1 overexpressed lines were significantly higher than those of WT (Fig. 6, F and G). That said, CsGS1.1 overexpression effectively promoted the growth of plants and increased N assimilation under Mg deficiency. This provided strong support that CsGS1.1 regulates Mg-mediated N metabolism in tea. Given that a wide range of genomic, transcriptomic, and metabolomic studies have been carried out in tea (Li et al. 2017; Huang et al. 2018; Xia et al. 2019; Liu et al. 2020) we next analyzed published data to investigate the level of allelic and expression variance in CsGS1.1 and if the levels of N-containing metabolites also varied. These analyses revealed that the level of free amino acid, especially theanine and glutamine, is highly associated with the expression of CsGS1.1, suggesting that CsGS1.1 regulates N metabolism in tea plant.
Figure 6.
Phenotypic analysis of transgenic Arabidopsis plants overexpressing CsGS1.1 under Mg deficiency (−Mg) and N deficiency (−N). WT was the Col-0 Arabidopsis and OE1 and OE2 were for CsGS1.1 expression in the WT background. The treatments of Mg deficiency (−Mg) and N deficiency (−N) were plants grown under the standard 1/2MS medium without Mg or N, and the other components were the same as 1/2MS medium (CK). Overexpression of CsGS1.1 (lines OE1 and OE2) resulted in increased A)CsGS1.1 expression and B) GS activity. C) Phenotype, D) plant biomass (FW, fresh weight), E) root length, and F and G) N content of CsGS1.1 overexpression plants grown under −Mg and −N conditions compared to CK. GS, glutamine synthetase; DW, dry weight. Error bars show the mean values ± SD (n = 3). Statistical analysis was carried out with Duncan's multiple range test. Different letters indicate significant differences (P < 0.05); ns, not significant.
Confirmation of the interaction effect of Mg and N on tea plants in the practice of tea production
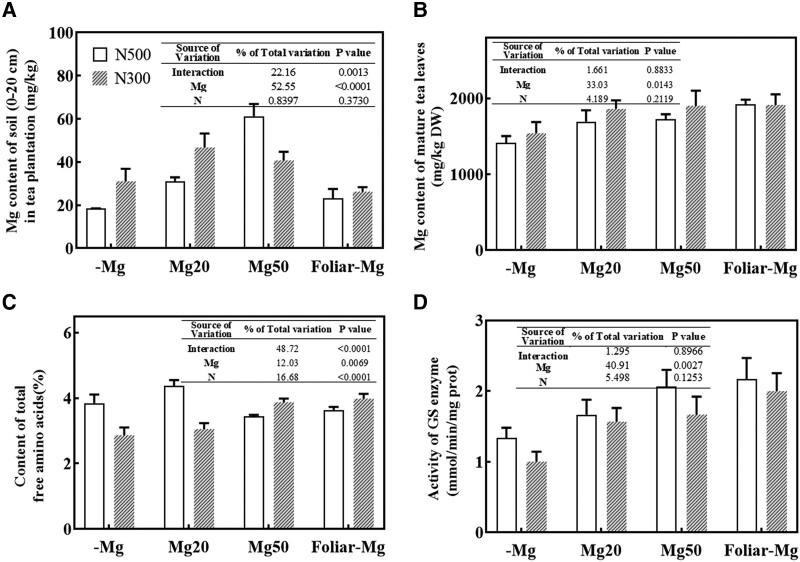
To investigate the interaction effect of Mg and N on tea production, a field experiment was conducted with 2 levels of N supply and 3 levels of Mg application. The results show that Mg levels in the 0 to 20 cm soil of tea plantation increased with the amount of Mg applied under both N conditions (Fig. 7A). The concentration of Mg in mature tea leaves was considerably lower in the −Mg condition than in the plants to which Mg was applied (Mg20 and Mg50) (Fig. 7B). Notably, under low N conditions, both soil and foliar Mg supplementation enhanced the amino acid content in new shoots, compared with control treatment (low Mg conditions, −Mg) (Fig. 7C). Furthermore, the inhibited activity of GS enzyme was observed in leaves under −Mg condition when compared to Mg supply (Fig. 7D). Under high N conditions, a moderate Mg level (Mg20) increased the content of free amino acids but high Mg supplementation (Mg50 or foliar Mg) decreased the content of amino acids (Fig. 7C). Thus, Mg application was more effective at improving the efficiency of N utilization in tea plants at low N levels than under high N conditions. These results have important implications as agriculture attempts to become more sustainable.
Figure 7.
Effects of Mg and N supply on the content of Mg, amino acids, and GS activity in tea plants grown under the field experiment. A) Mg content in the soil of tea plantation. B) Mg content in mature tea leaves. C) Content of total free amino acids in tea leaves. D) GS activity in tea leaves. N300 and N500 represent that the levels of nitrogen fertilizers are provided as 300 kg/ha and 500 kg/ha urea, respectively. −Mg, Mg20, and Mg50 represent that soil Mg fertilizer levels are 0, 20 kg/ha, and 50 kg/ha, respectively. Error bars show the mean values ± SD (n = 4). Statistical analysis was carried out with Duncan's multiple range test.
Discussion
N translocation from roots to leaves is inhibited and consequently reuse of N in mature tea leaves is enhanced by Mg deficiency in tea plants
By measuring phloem sap in tea plants, Ruan et al. (2012) found that Mg supplementation could promote the long-distance transport of amino acids. In this study, high levels of amino acid accumulation were detected in tea roots subjected to Mg deficiency, compared with roots under normal Mg levels (Mg1), while the total N in mature tea leaves significantly decreased under the Mg deficiency group than that in the Mg1 group (Fig. 1, D and E). These data indicate that the partitioning of N between roots and leaves in tea plants is regulated by Mg nutrition, with the long-distance transport of N from roots to leaves seemingly being inhibited by Mg deficiency. Cakmak and Kirkby (2008) found that the long translocation of N from plant roots inhibited by Mg deficiency was associated with the partitioning of carbohydrates between shoots and roots, as there was not enough carbon skeleton for amino acid synthesis and limited N assimilation in roots. In this study, we also found that Mg deficiency resulted in the reduction of carbohydrates in tea roots (Supplemental Table S1). Furthermore, we found that asparagine and glutamine (which possess relatively high N : C ratios, Supplemental Table S1) mainly accumulated in the roots during Mg deficiency, and these amino acids represent the main form of N transport (Herrera-Rodriguez et al. 2006; Avila et al. 2015; Gaufichon et al. 2017), indicating that Mg deficiency inhibits transport of amino acid from roots to leaves in tea plants. The expression of CsAAP5, the homolog of amino acid permeases which have been shown to be highly important in amino acid transport (Tegeder and Masclaux-Daubresse 2018), was downregulated under Mg deficiency compared with normal Mg levels (Supplemental Fig. S1). This downregulation corresponded to the inhibition of N distribution from the roots to mature leaves during Mg deficiency. The function of AAP family genes in tea plants has been reported previously. For example, CsAAP1 was suggested to be highly important in theanine transport (Dong et al. 2020). Using 15N isotope tracer experiments, Fan et al. (2020) identified a significant positive correlation between CsAAP6 expression and 15N accumulation in young shoots, as well as 15N consumption in mature leaves. Considering the general lack of Mg in tea plantations, these findings indicate that Mg deficiency may restrict the NUE of tea plantations. Here, Mg deficiency downregulated CsGS1.1 expression in tea roots (Fig. 5). Considering the amino acid accumulation in the roots, our results indicate that downregulation of genes related to N assimilation, such as CsGS1.1, may be feedback regulated as a consequence of product accumulation. N assimilation requires not only the inorganic N but also the carbon skeleton α-ketoglutarate (Yanagisawa et al. 2004). As such, the level of carbon skeleton production determines the assimilation of N (Foyer and Ferrario 1994; Foyer et al. 2011). It has been reported that the photosynthate transport could be inhibited under Mg deficiency condition (Hermans et al. 2005; Farhat et al. 2016). Here, photosynthates such as glucose decreased in the roots in the −Mg group compared to the Mg1 group, while in the mature leaves, glycoside group-containing metabolites significantly increased under −Mg conditions (Supplemental Table S1), indicating that Mg deficiency might have an influence in the transport of photosynthate in tea plant.
Previous studies have shown that N from the degradation and remobilization of reserves of old leaves is the main N source for reproductive growth (Diaz et al. 2008; Masclaux-Daubresse et al. 2010) and shoot germination (Okano et al. 1994). For example, in walnut (Juglans nigra) trees, approximately 54% of the N in spring shoots is derived from the reuse of stored N (Frak et al. 2002), mainly in the form of arginine, glutamate, and citrulline. In tea plants, approximately 70% of N in spring tea originates from N remobilization from intracellular N reserves (Okano et al. 1994). Here, we observed that, the total N content decreased, while the amino acid content increased, in mature tea leaves under Mg deficiency. Based on the combined expression of genes related to N activation (e.g. CsGLT) and the amino acid content in the young shoots, we propose that N reuse in young shoots, and especially in mature tea leaves, is enhanced under Mg deficiency. Liu et al. (2016) demonstrated that the reuse of stored N was the main reason for differences in amino acid content in young tea shoots. Moreover, 15N tracing has confirmed that mature leaves transport substantial amounts of N to corresponding young shoots (Ruan and Gerendá 2015). The enhanced N reuse observed in this study confirmed that the inhibition of the long-distance transport of amino acids resulted in a depletion of N sources in the aboveground part of the tea plants.
Regulation of Mg supplementation enhanced N assimilation by CsGS1.1
Previous studies have reported that the biological effects of Mg application are closely related to the status of nutrients such as N or potassium (K) (Ruan and Gerendas 2015; Lasa et al. 2000). In tea, Ruan et al. (1998) also demonstrated that Mg application improved N metabolism, leading to an increased synthesis of amino acids. In this study, hydroponic and field experiments demonstrated that Mg supplementation substantially increased the N content and amino acids in the tea plant (Fig. 8), indicating that N assimilation in tea plants was higher under enhanced Mg supply than normal Mg supply. Notably, under low N condition, Mg can substantially improve the N utilization efficiency (improve the N content of mature leaves and shoots) and even partly replace the function of N when regarding to the content of amino acids (the most important quality index) in young shoots (the mainly harvesting tissues of tea). Previous studies have proved that Mg regulates N uptake in plant roots (Peng et al. 2020; Tian et al. 2021). Thus, the effect of Mg on N in plants is not merely at the N uptake level but also at the N metabolism level.
Figure 8.
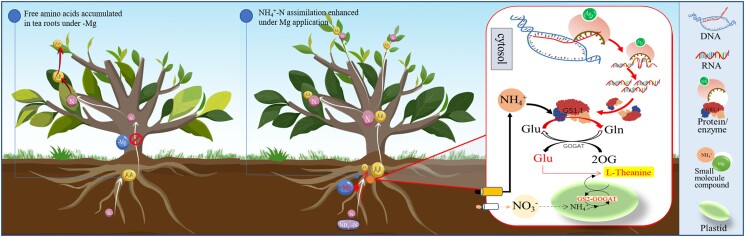
Schematic representation of nitrogen metabolism response to Mg status in tea plant. AA, amino acid; Gln, glutamine; GS1.1, cytosolic glutamine synthetase; Glu, glutamate; GS2-GOGAT, plastidic glutamine synthetase–glutamate synthase; Mg, magnesium; N, nitrogen; NH4+-N, ammonium nitrogen; NO3−, nitrate nitrogen; OG, 2-oxoglutarate; +Mg, magnesium application; −Mg, magnesium deficiency.
Based on co-expression analysis (WGCNA) of genes, we identified CsGS1.1 as a potential key gene that is both regulated by Mg and involved in N assimilation in tea plants. GS potentially represents a major bottleneck during the ligation of free NH4+ into organic carbon skeletons and as such plays a crucial role in plant N assimilation/metabolism in land plants (Gallais and Hirel 2004; Martin et al. 2006; Bernard and Habash 2009; Thomsen et al. 2014). GS1 isogenes are expressed in different tissues and have functional roles in the assimilation of ammonia into glutamine. GS2 encoded by a single gene (Taira et al. 2004) is expressed abundantly in leaf mesophyll cells (Tobin and Yamaya 2001) and participates in ammonium assimilation during the processes of nitrate reduction and photorespiration (Wallsgrove et al. 1987; Lam et al. 1995). Here, we found CsGS1.1 to be highly expressed in tea roots (Supplemental Fig. S2) and that its expression level followed the Mg status (Fig. 5), demonstrating that assimilation of N through CsGS1.1 has been highly affected by Mg provision in tea roots. Correspondingly, the 15N isotope tracer experiments demonstrated that the efficiency of NH4+ assimilation in roots was substantially higher during higher Mg conditions than under normal Mg fertilization. Using a single N source supply (i.e. NO3− or NH4+, not both), Ruan et al. (1998) demonstrated that NO3− absorption and metabolism in tea plants were improved by Mg application, while additional Mg fertilizer also increased nitrate reductase activity. In this study, by simultaneously supplying NO3− and NH4+, we examined the priority of N sources and the role of Mg in N, especially in NH4+-derived N utilization (Table 1). To improve N assimilation, heterologous GS genes have been overexpressed in plants such as tobacco (N. tabacum) (Oliveira et al. 2002), poplar (Populus tremula) trees (Pascual et al. 2008) and rice (Oryza sativa) (Cai et al. 2009). Some studies showed that N assimilation efficiency was increased in transgenic lines overexpressing heterologous GS genes (Man et al. 2005). However, some studies found that both N metabolism and NUE were not affected under the condition of GS overexpression, possibly due to the post-translational regulation of GS enzymes (Lima et al. 2006; Cai et al. 2009). In our study, we demonstrate that overexpression of CsGS1.1 in Arabidopsis enhanced N assimilation in plants grown under −Mg conditions (Fig. 6), suggesting that this overexpression at least partially circumvents the need for Mg supplementation.
Adequate plant Mg status seems to be indispensable for optimal GS activity because Mg is an integral component of GS activity, having both a structural and a catalytic role (Eisenberg et al. 2000). In addition, the GS activity appears to be regulated by end-product feedback inhibition, especially under the high levels of free glutamine (Rhodes et al. 1975) and glutamine-dependent amino acids (Eisenberg et al. 2000). Previous studies have also reported that Mg deficiency negatively impacted the activity of GS in rice and spinach (Spinacia oleracea) (Ding et al. 2006; Yin et al. 2009). Other studies have suggested that Mg deficiency is not necessarily accompanied by decreased GS activity with neither the activity nor the abundance of this enzyme being directly associated to the concentration of Mg in the plant (Jezek et al. 2015). In this study, the GS enzyme activity decreased in the case of Mg deficiency condition when compared to normal condition (Figs. 5B and 7D, Supplemental Fig. S1), suggesting that the supply of Mg is an influencer of GS enzyme activity. However, the GS activity did not correspond linearly with the content of Mg both in the hydroponic (Fig. 1 and Supplemental Fig. S1) and field (Fig. 7, B and D) experiment, especially under the non-deficient conditions of Mg supplementation. Considering that the transcription of the gene CsGS1.1 is highly responsive to the Mg application (Figs. 4, 5A and 6), we suggest that N assimilation regulation by different Mg application levels is mainly due to the Mg transcriptionally regulating CsGS1.1. This suggestion could be validated by the results from transgenic Arabidopsis plants, in which the association of N content and overexpression of CsGS1.1 is sensitive (Fig. 6, A, F, and G), while changes of GS activity are also not always consistent with the variation of N content in OE1 and OE2 Arabidopsis (Fig. 6, B, F, and G). It is well-known that Mg deficiency in plants commonly increase the levels of 2 metabolites (also known as signal molecules), sugar, and ROS (Cakmak and Kirkby 2008). These metabolites transcriptionally fine-tune many intracellular metabolic processes, including circadian clock (Chen et al. 2022). Given that many N uptake and assimilation genes are circadian-regulated, Mg regulation in GS1.1 is possibly through this pathway.
As we know, additional N fertilizer may be needed to maintain tea yield stability and quality; however, the excessive N input involved in current tea production in China has caused great environmental and ecological burdens (Zhang et al. 2013). Nowadays, a major focus of research on tea plants is to develop a strategy that reduces the N fertilization level and increases the quality and yield of tea. Thus, it is meaningful to use a small amount of Mg fertilizer to achieve the same effect of a large amount of N fertilizer. In this study, Mg supplementation improved N utilization efficiency in tea plants, under both low and high N conditions. Considering the effects of Mg both on AAP expression and N transport and the effects of Mg on N distribution, assimilation, and reuse, we propose that increasing the Mg supply could effectively improve N utilization efficiency, and evaluation of combinations of Mg and N levels could provide ways to move towards a more sustainable production of tea. Notably, our hydroponic experiments on N levels and field experiments found that under high Mg conditions, the amino acid content in the new shoots and mature leaves did not differ substantially between the low and high N conditions, which demonstrated the effectiveness of an improved fertilization strategy including lower N but enhanced Mg application. Given the importance of nitrogenous compounds such as, but not limited to, theanine for tea quality, these findings will likely be of great importance to the tea industry.
Materials and methods
Experimental design
Hydroponic experiments
To evaluate the effect of Mg on tea (C. sinensis) plants, hydroponic experiments were conducted with 3 different Mg supply levels. Young tea plants (1-year-old rooted cuttings of “Longjing 43”) were treated with 3 levels of Mg: without Mg (Mg0), 0.4 mmol/L (Mg1), and 0.8 mmol/L (Mg2). All experiments were conducted in an environmentally controlled growth chamber, as described previously (Liu et al. 2017). The pH of all nutrient solutions was titrated to 5.0 with H2SO4 and NaOH.
15N isotope tracer hydroponic experiments
To evaluate the effect of Mg on N assimilation, 15N-labelled ammonium (NH4+) and nitrate (NO3−) were supplied in an independent hydroponic experiment. Tea cuttings were cultured in the same manner as those used for the Mg level experiments. In 1 treatment group, Mg levels were the same as the full-strength nutrient solution (Mg1, 0.4 mmol/L), and in the other, the Mg level was 0.8 mmol/L (Mg2) (Supplemental Fig. S3). Six weeks (42 d) later, the tea cuttings in each group were divided into 2 subgroups, with K15NO3 and 15NH4SO415N-labelled compounds being provided as the N source for each group.
Mg and N interaction experiment
To further evaluate the interaction effect of Mg and N on tea plants, tea seedlings were treated with 2 levels of Mg [1 quarter (MgL = 0.1 mmol/L) and 2 times (MgH = 0.8 mmol/L) concentration in the full-strength nutrient solution] and 2 levels of N [1 quarter (NL) and 2 times (NH) concentration in the full-strength nutrient solution].
Field experiment
For evaluating the application prospect of the interaction effect of Mg and N in tea production, Mg-deficient tea plantation (25 mg/kg of effective soil Mg content, Supplemental Fig. S4) in Shengzhou, Zhejiang (120°82′53″ E, 29°74′83″ N), was chosen for the experiments. In tea production, the N form is usually provided as urea by tea farmers. Here, 2 N levels (N300 and N500), 300 and 500 kg/ha, were used. For each N level, four Mg treatments were used, using a split–plot design, including various soil Mg levels (−Mg, Mg20, Mg50; 0, 20, and 50 kg/ha, respectively), and Mg was also applied directly to the foliage (50 kg/ha). The foliar Mg fertilizers tested in this experiment included 1% (w/v) MgSO4•7H2O and was applied in October twice with a 1-week interval in between applications. In October, the shoots of tea plants stop growing, and samples were collected in spring of the following year when the new shoots grow out. The field experiments began in 2017 and each treatment was replicated four times.
Measurement of Mg and free amino acid levels in tea plants
Tea plant samples (0.1 g) were digested using 5 mL of mixed concentrated acids HNO3 and HClO4 (Ruan et al. 2012) at 140°C in an electric oven. The reaction was diluted to 25 mL with distilled water. Thereafter, the nutrients were analyzed by inductively coupled plasma atomic emission spectroscopy. Free amino acids in leaf samples were measured by high-performance liquid chromatography using a diode array detector (HPLC-DAD; 2695–2998; Waters, Milford, MA, USA), as previously reported by Zhang et al. (2017).
Determination of the total N and 15N content in tea plants
To determine the total N and carbon (C) content in tea leaves and new shoots, 100 mg of each dried sample was used. Measurements were completed automatically using an elemental analyzer (Vario Max CN Analyzer, Elementar Analysensysteme GmbH, Langenselbold, Germany).
Gas chromatography-based metabolomics analysis
Polar primary metabolites were extracted using an adapted form of the method reported by Mokochinski et al. (2018). The extracts were analyzed using the gas chromatography mass spectrometry system, comprising an Agilent 6890 gas chromatograph (Agilent Technologies, Santa Clara, CA, USA) coupled to a Pegasus III TOF-MS instrument (Leco Instruments, Saint Joseph, MI, USA), with hard ionization at 70 eV. Annotations were based on comparing both the spectra and the retention index (RI) to standard compounds previously analyzed using the same system, by referring to the mass spectral databases of NIST (Gaithersburg, MD, USA) and the Max Planck Institute (Golm, Germany) (Kopka et al. 2005; Schauer et al. 2005). The obtained metabolite intensity data were introduced to SIMCA-P V13.0 (Umetrics, Sweden) for principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA).
RNA-seq, data processing, gene annotation, and GO analysis
Total RNA of tea samples from the hydroponic experiment examining the Mg and N interaction was extracted using TRIzol reagent (Invitrogen, USA) following the manufacturer's protocol. Sequencing, data processing, gene annotation, and GO analysis were all performed according to the method described by Liu et al. (2017). The transcriptome data of all the samples were deposited into the NCBI Sequence Read Archive (SRA) database under the accession number SRP423919.
Gene co-expression network, module identification, and gene edge number
The construction of the gene co-expression network, followed by module detection of highly correlated genes, was inferred from the 9,576 differentially expressed genes (DEGs) using weighted gene co-expression network analysis (WGCNA), an R software package (Langfelder and Horvath 2008). WGCNA network construction and module detection were conducted using default settings. A dynamic cut-tree algorithm was used for automatically and precisely identifying modules in a hierarchical clustering dendogram (Li and Horvath 2007). The total number of edges for gene within each module was estimated using as a cut-off a WGCNA edge weight ≥ 0.5. Networks were visualized using Cytoscape 3.4.0 (Saito et al. 2012).
Identification of CsGSs in tea plant
The tea plant genome data and Generic Feature Format version 3 (GFF3) annotation were downloaded from TPIA (Xia et al. 2019). All coding sequence (CDS) of CSS genome was extracted based on GFF3 using Tbtools (Chen et al. 2020). GS genes in Arabidopsis (A. thaliana) were downloaded from PlantTFDB.
Phylogenetic analysis
MEGA-X software (https://www.megasoftware.net/) was used to construct the phylogenetic tree. The protein sequences were obtained in different ways. The protein sequences of CsGSs were extracted from the TPIA website (http://tpia.teaplant.org/download.html) according to the genome ID. GS sequences of Arabidopsis (A. thaliana), rice (O. sativa), and grape (Vitis vinifera) were obtained through a BLAST search on the UniProt website (https://www.uniprot.org/) using the CsGSs protein sequences. GS sequences of poplar (Populus trichocarpa) were extracted from the populous genome (https://genome.jgi.doe.gov/portal/). All these sequences were imported into MEGA-X and aligned using MUSCLE with all default settings. By using the “Find Best DNA Model for ML” function, we choose the “LG + G” model for constructing a maximum likelihood tree. The bootstrap replication was 2,000, and the site coverage cutoff was 85%. The phylogenetic tree was visualized using the ITOL site (https://itol.embl.de).
Construction of transgenic plants
To generate the transgenic plants overexpressing CsGS1.1, the coding region of CsGS1.1 without stop codon was amplified using primers (forward: 5′-GGGGTACCATGTCTTTGCTATCAGA-3′; reverse: 5′-GCTCTAGATGGCTTCCACAGCAGAGTGG-3′) and cloned into the pMD18-T vector (TaKaRa) for sequence verification. The verified plasmid was cleaved and ligated into a modified pCAMBIA1300 vector between Kpn I and Xba I restriction sites under the control of CaMV35S promoter to generate the 35S::CsGS1.1 construct. The resulting construct was transformed into wild-type (WT) plants (Col-0) via Agrobacterium tumefaciens (EHA 105)-mediated floral dip transformation (Clough and Bent 1998). T1 seeds were surface sterilized and plated on 1/2 Murashige Skoog (MS) medium (pH 5.8) supplemented with hygromycin. The resistant plants were transferred to soil and allowed to set seeds (T2). All experiments were performed using plants corresponding to the T4 generation.
Growth assay of transgenic plants under Mg and N treatment
The transgenic lines and WT seeds were sowed in Petri dishes with 1/2 MS medium in the growth chamber under 16 h 24°C : 8 h 22°C, light : night, a light intensity of 200 µmol photons m−2 s−1% and 70% relative humidity. One week later, seedlings with approximately 1 cm primary roots were transferred to Petri dishes with different medium containing no N (−N), no Mg (−Mg), and the same component as the 1/2 MS medium (CK).
Reverse transcription quantitative PCR (Rt-qPCR) analysis
Total RNA was isolated using an RNAplant Plus kit (TIANGEN BIOTECH, Beijing, China). cDNA was synthesized using a PrimeScript RT reagent kit (TaKaRa) and RT-qPCR was performed on a 7300 Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). The primer pairs used for RT-qPCR are the same as in Fan et al (2020) and Tang et al. (2020). Relative transcript levels were then calculated using the 2−ΔΔCt method, with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as the internal control.
Determination of GS activity
To determine the total GS activity (synthetase reaction), 100 mg samples were measured according to Magalhaes and Huber (1989). The extracts were centrifuged at 13,000 × g (4°C) for 25 min, and the supernatants were analyzed for soluble protein (Bradford 1976) and GS activity. To determine GS activity, the supernatants were incubated in a reaction buffer at 37°C for 30 min (Magalhaes and Huber 1989; Husted et al. 2002). Absorbance at 540 nm was recorded with a spectrophotometer.
Statistical analyses
The statistical analyses were performed in R (http://www.r-project.org/). Differences among mean values were tested for statistical significance using one-way analysis of variance (ANOVA), with correction for multiple testing using the false discovery rate (FDR) technique, followed by Tukey's post hoc test for pairwise comparisons. Differences were considered significant when P < 0.01.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers SRP423919 and KY649469.
Conclusion
In our study, hydroponic experiments with Mg and N levels suggested that Mg supplementation increased the amino acid content of tea roots and shoots, especially under low N condition, most likely as a consequence of enhanced NH4+ assimilation. By analyzing the N content, gene expression, and enzyme activity in tea plant by a range of strategies, including whole-plant 15N label tracing, CsGS1.1 has been identified to play a key role in the regulation of NH4+ assimilation and N metabolism, in response to Mg status. This finding was genetically validated by the heterologous expression of CsGS1.1 in Arabidopsis. Field experiments further confirmed that Mg supplementation highly enhanced N assimilation, especially under a low N application. These results on the one hand provide an improved fertilization strategy with reduced N but increased Mg application in tea plantations. On the other hand, they suggest that broader screens of the allelic variance may result in the isolation of genetic tools to improve NUE in tea.
Supplementary Material
Contributor Information
Qunfeng Zhang, Tea Research Institute, Chinese Academy of Agricultural Sciences, Hangzhou 310008, China; Key Laboratory of Biology, Genetics and Breeding of Special Economic Animals and Plants (Ministry of Agriculture and Rural Affairs), Hangzhou 310008, China.
Yutao Shi, Tea Research Institute, Chinese Academy of Agricultural Sciences, Hangzhou 310008, China; College of Tea and Food Science, Wuyi University, Wuyishan 354300, China.
Hao Hu, Department of Botany and Plant Sciences, Institute of Integrative Genome Biology, University of California, Riverside, CA 92521, USA; Key Laboratory for Biology of Horticultural Plants, Ministry of Education, College of Horticulture & Forestry Sciences, Huazhong Agricultural University, Wuhan 430070, China.
Yuanzhi Shi, Tea Research Institute, Chinese Academy of Agricultural Sciences, Hangzhou 310008, China; Key Laboratory of Biology, Genetics and Breeding of Special Economic Animals and Plants (Ministry of Agriculture and Rural Affairs), Hangzhou 310008, China.
Dandan Tang, Tea Research Institute, Chinese Academy of Agricultural Sciences, Hangzhou 310008, China; College of Horticulture, Sichuan Agricultural University, Chengdu 611130, China.
Jianyun Ruan, Tea Research Institute, Chinese Academy of Agricultural Sciences, Hangzhou 310008, China; Key Laboratory of Biology, Genetics and Breeding of Special Economic Animals and Plants (Ministry of Agriculture and Rural Affairs), Hangzhou 310008, China.
Alisdair R Fernie, Max-Planck-Institute of Molecular Plant Physiology, Am Muehlenberg 1, 14476 Potsdam-Golm, Germany.
Mei-Ya Liu, Tea Research Institute, Chinese Academy of Agricultural Sciences, Hangzhou 310008, China; Key Laboratory of Biology, Genetics and Breeding of Special Economic Animals and Plants (Ministry of Agriculture and Rural Affairs), Hangzhou 310008, China.
Author contributions
Q.Z. and M.-Y.L. participated in the study design; Q.Z., Y.S., and M.-Y.L. performed the research; Q.Z. and M.-Y.L. performed data analysis; D.T. and Y.S. provided experimental assistance to Q.Z. and M.-Y.L.; Q.Z., H.H., A.R.F., and M.-Y.L. interpreted the results and drafted the manuscript; M.-Y.L. and J.R. provided funding; M.-Y.L. and A.R.F. finalized the manuscript. All authors read and approved the final manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Relative expression levels of genes associated with N metabolism and enzyme activities of glutamine synthetase (GS) in mature leaf (ML), root, and young/new shoot (YS) of tea plants under different treatments.
Supplemental Figure S2. Identification of potential CsGS members.
Supplemental Figure S3. Phenotype of hydroponic tea plants under different Mg supply conditions.
Supplemental Figure S4. Field experiment investigating the interaction effect of Mg and N in tea production.
Supplemental Table S1. Putatively identified primary metabolites that showed significant changes in relative abundance in tea plants treated with Mg deficiency (−Mg), Mg concentration in the full-strength nutrient solution (Mg1), and 2 times the Mg concentration in the full-strength nutrient solution (Mg2).
Supplemental Table S2. Identified candidate genes associated with N and amino acid metabolism that is affected by Mg nutrition with weighted gene co-expression network analysis.
Funding
The work was financially supported by the Zhejiang Provincial Natural Science Foundation of China (LZ23C160006, LY21C150005), National Natural Science Foundation of China (no. 32072627), and the Chinese Academy of Agricultural Sciences through Agricultural Sciences Innovation Project (CAAS-ASTIP-2020-TRICAAS).
Data availability
All data generated or used during the study appear in the submitted article.
References
- Avila O, Marmagne A, Talbotec J, Krupinska K, Masclaux D. The identification of new cytosolic glutamine synthetase and asparagine synthetase genes in barley (Hordeum vulgare L.), and their expression during leaf senescence. J Exp Bot. 2015:66(7): 2013–2026 10.1093/jxb/erv003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard SM, Habash DZ. The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol. 2009:182(3): 608–620 10.1111/j.1469-8137.2009.02823.x [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein binding. Anal Biochem. 1976:72(1-2): 248–254 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Cai H, Zhou Y, Xiao J, Li X, Zhang Q, Lian X. Overexpressed glutamine synthetase gene modifies nitrogen metaboliasm and abiotic stress responses in rice. Plant Cell Rep. 2009:28(3):527–537 10.1007/s00299-008-0665-z [DOI] [PubMed] [Google Scholar]
- Cakmak I, Kirkby EA. Role of magnesium in carbon partitioning and alleviating photooxidative damage. Physiol Plantarum. 2008:133(4): 692–704 10.1111/j.1399-3054.2007.01042.x [DOI] [PubMed] [Google Scholar]
- Chardon F, Noel V, Masclaux D. Exploring NUE in crops and in Arabidopsis ideotypes to improve yield and seed quality. J Exp Bot. 2012:63(9): 3401–3412 10.1093/jxb/err353 [DOI] [PubMed] [Google Scholar]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020:13(8): 1194–1202 10.1016/j.molp.2020.06.009 [DOI] [PubMed] [Google Scholar]
- Chen CQ, Tian XY, Li J, Bai S, Zhang ZY, Li Y, Cao HR, Chen ZC. Two central circadian oscillators OsPRR59 and OsPRR95 modulate magnesium homeostasis and carbon fixation in rice. Mol Plant. 2022:15(10):1602–1614 10.1016/j.molp.2022.09.008 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998:16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cren M, Hirel B. Glutamine synthetase in higher plants: regulation of gene and protein expression from the organ to the cell. Plant Cell Physiol. 1999:40(12): 1187–1193 10.1093/oxfordjournals.pcp.a029506 [DOI] [Google Scholar]
- Cruz C, Bio AFM, Dominguez-Valdivia MD, Aparicio-Tejo PM, Lamsfus C, Martins-Loucao MA. How does glutamine synthetase activity determine plant tolerance to ammonium. Planta. 2006:223(5): 1068–1080 10.1007/s00425-005-0155-2 [DOI] [PubMed] [Google Scholar]
- Diaz C, Lemaitre T, Christ A, Azzopardi M, Kato Y, Sato F, Morot-Gaudry JF, Le Dily F, Masclaux D. Nitrogen recycling and remobilization are differentially controlled by leaf senescence and development stage in Arabidopsis under low nitrogen nutrition. Plant Physiol. 2008:147(3): 1437–1449 10.1104/pp.108.119040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Luo W, Xu G. Characterisation of magnesium nutrition and interaction of magnesium and potassium in rice. Ann Appl Biol. 2006:149(2): 111–123 10.1111/j.1744-7348.2006.00080.x [DOI] [Google Scholar]
- Dong CX, Li F, Yang TY, Feng L, Zhang SP, Li FD, Li WH, Xu GH, Bao SL, Wan XC, et al. Theanine transporters identified in tea plants (Camellia sinensis L.). Plant J. 2020:101(1): 57–70 10.1111/tpj.14517 [DOI] [PubMed] [Google Scholar]
- Dordas C. Foliar application of calcium and magnesium improves growth, yield, and essential oil yield of oregano (Origanum vulgare ssp. hirtum). Ind Crops Prod. 2009:29(2-3): 599–608 10.1016/j.indcrop.2008.11.004 [DOI] [Google Scholar]
- Eisenberg D, Gill HS, Pfluegl GMU, Rotstein SH. Structure-function relationships of glutamine synthetases. Biochim Biophys Acta. 2000:1477(1-2): 122–145 10.1016/S0167-4838(99)00270-8 [DOI] [PubMed] [Google Scholar]
- Fan K, Zhang QF, Tang DD, Shi YZ, Ma LF, Liu MY, Ruan JY. Dynamics of nitrogen translocation from mature leaves to new shoots and related gene expression during spring shoots development in tea plants (Camellia sinensis L. J Plant Nutr Soil Sci. 2020:183(2): 180–191 10.1002/jpln.201900268 [DOI] [Google Scholar]
- Farhat N, Elkhouni A, Zorrig W. Effects of magnesium deficiency on photosynthesis and carbohydrate partitioning. Acta Physiol Plant. 2016:38(6): 145. 10.1007/s11738-016-2165-z [DOI] [Google Scholar]
- Fernie A. Using landrace transcription factor alleles to increase yield in modern rice under low input agriculture. J Plant Physiol. 2021:258: 153362–153362 10.1016/j.jplph.2021.153362 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Ferrario S. Modulation of carbon and nitrogen metabolism in transgenic plants with a view to improved biomass production. Biochem Soc Trans. 1994:22(4): 909–915 10.1042/bst0220909 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G, Hodges M. Respiration and nitrogen assimilation: targeting mitochondria-associated metabolism as a means to enhance nitrogen use efficiency. J Exp Bot. 2011:62(4): 1467–1482 10.1093/jxb/erq453 [DOI] [PubMed] [Google Scholar]
- Frak E, Millard P, Le Roux X, Guillaumie S, Wendler R. Coupling sap flow velocity and amino acid concentrations as an alternative method to 15N labeling for quantifying nitrogen remobilization by walnut trees. Plant Physiol. 2002:130(2): 1043–1053 10.1104/pp.002139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Liao Y, Cheng S, Deng R, Yang Z. Stable isotope-labeled precursor tracing reveals that L-alanine is converted to L-theanine viaL-glutamate not ethylamine in tea plants in vivo. J Agric Food Chem. 2021:69(50): 15354–15361 10.1021/acs.jafc.1c06660 [DOI] [PubMed] [Google Scholar]
- Gallais A, Hirel B. An approach to the genetics of nitrogen use efficiency in maize. J Exp Bot. 2004:55(396): 295–306 10.1093/jxb/erh006 [DOI] [PubMed] [Google Scholar]
- Gaufichon L, Marmagne A, Belcram K. ASN 1-encoded Asparagine synthetase in floral organs contributes to nitrogen filling in Arabidopsis seeds. The Plant J. 2017:91(3): 371–393 10.1111/tpj.13567 [DOI] [PubMed] [Google Scholar]
- Gerendás J, Führs H. The significance of magnesium for crop quality. Plant Soil. 2013:368(1-2): 101–128 10.1007/s11104-012-1555-2 [DOI] [Google Scholar]
- Hermans C, Bourgis F, Faucher M. Magnesium deficiency in sugar beets alters sugar partitioning and phloem loading in young mature leaves. Planta. 2005:220(4): 541–549 10.1007/s00425-004-1376-5 [DOI] [PubMed] [Google Scholar]
- Herrera-Rodriguez MB, Maldonado JM, Perez-Vicente R. Role of asparagine and asparagine synthetase genes in sunflower (Helianthus annuus) germination and natural senescence. J Plant Physiol. 2006:163(10): 1061–1070 10.1016/j.jplph.2005.10.012 [DOI] [PubMed] [Google Scholar]
- Huang H, Yao Q, Xia E, Gao L. Metabolomics and transcriptomics analyses reveal nitrogen influences on the accumulation of flavonoids and amino acids in young shoots of tea plant (Camellia sinensis L. associated with tea flavor. J Agri Food Chem. 2018:66(37):9828–9838 10.1021/acs.jafc.8b01995 [DOI] [PubMed] [Google Scholar]
- Husted S, Mattsson M, Mollers C, Wallbraun M, Schjoerring JK. Photorespiratory NH4 + production in leaves of wild-type and glutamine synthetase 2 antisense oilseed rape. Plant Physiol. 2002:130(2): 989–998 10.1104/pp.006759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki K. Comparison between ammonium-nitrogen and nitrate-nitrogen on the effect of tea plant growth. Jpn Agr Res Q. 1974:8: 101–105 [Google Scholar]
- Jayaganesh S, Venkatesan S. Impact of magnesium sulphate on biochemical and quality constituents of black tea. Am J Food Technol. 2010:5: 1557–4571 [Google Scholar]
- Jayaganesh S, Venkatesan S, Senthurpandian V. Impact of different sources and doses of magnesium fertilizer on biochemical constituents and quality parameters of black tea. Asian J Biochem. 2011:6(3): 273–281 10.3923/ajb.2011.273.281 [DOI] [Google Scholar]
- Jezek M, Geilfus CM, Mühling KH. Glutamine synthetase activity in leaves of Zea mays L. As influenced by magnesium status. Planta. 2015:242(6): 1309–1319 10.1007/s00425-015-2371-8 [DOI] [PubMed] [Google Scholar]
- Jiang X, Zhang W, Fernie AR, Wen W. Combining novel technologies with interdisciplinary basic research to enhance horticultural crops. The Plant J. 2022:109(1):35–46 10.1111/tpj.15553 [DOI] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, et al. GMD@CSB.DB: the golm metabolome database. Bioinformatics. 2005:21(8):1635–1638 10.1093/bioinformatics/bti236 [DOI] [PubMed] [Google Scholar]
- Lam HM, Coschigano K, Schultz C, Melo-Oliveira R, Tjaden G, Oliveira I, Ngai N, Hsieh MH, Coruzzi G. Use of Arabidopsis mutants and genes to study amide amino acid biosynthesis. Plant Cell. 1995:7(7):887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008:9(1): 1–13 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasa B, Frechilla S, Aleu M. Effects of low and high levels of magnesium on the response of sunflower plants grown with ammonium and nitrate. Plant Soil. 2000:225(1/2): 167–174 10.1023/A:1026568329860 [DOI] [Google Scholar]
- Li A, Horvath S. Network neighborhood analysis with the multi-node topological overlap measure. Bioinformatics. 2007:23(2): 222–231 10.1093/bioinformatics/btl581 [DOI] [PubMed] [Google Scholar]
- Li W, Xiang F, Zhong M, Zhou L, Liu H, Li S, Wang X. Transcriptome and metabolite analysis identifies nitrogen utilization genes in tea plant (Camellia sinensis). Sci Rep. 2017:7(1):1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima L, Seabra A, Melo P, Cullimore J, Carvalho H. Posttranslational regulation of cytosolic glutamine synthetase of Medicago truncatula. J Exp Bot. 2006:57(11): 2751–2761 10.1093/jxb/erl036 [DOI] [PubMed] [Google Scholar]
- Liu MY, Burgos A, Zhang Q, Tang D, Shi Y, Ma L, Yi X, Ruan J. Analyses of transcriptome profiles and selected metabolites unravel the metabolic response to NH4+ and NO3− as signaling molecules in tea plant (Camellia sinensis L.). Sci Hortic. 2017:218:293–303 10.1016/j.scienta.2017.02.036 [DOI] [Google Scholar]
- Liu ZW, Li H, Liu JX, Wang Y, Zhuang J. Integrative transcriptome, proteome, and microRNA analysis reveals the effects of nitrogen sufficiency and deficiency conditions on theanine metabolism in the tea plant (Camellia sinensis). Hortic Res. 2020:7(1):65. 10.1038/s41438-020-0290-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MY, Tang DD, Shi YZ. Short-term inhibition of glutamine synthetase leads to reprogramming of amino acid and lipid metabolism in roots and leaves of tea plant (Camellia sinensis L.). BMC Plant Biol. 2019:19(1): 425. 10.1186/s12870-019-2027-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang H, Jiang Z, Wang W, Xu R, Wang Q, Zhang Z, Li A, Liang Y, Ou S, et al. Genomic basis of geographical adaptation to soil nitrogen in rice. Nature. 2021:590(7847): 600–605 10.1038/s41586-020-03091-w [DOI] [PubMed] [Google Scholar]
- Liu J, Zhang Q, Liu M, Ma L, Shi Y, Ruan J. Metabolomic analyses reveal distinct change of metabolites and quality of green tea during the short duration within single spring season. J Agric Food Chem. 2016:64(16): 3302–3309 10.1021/acs.jafc.6b00404 [DOI] [PubMed] [Google Scholar]
- Magalhaes JR, Huber DM. Ammonium assimilation in different plant species as affected by nitrogen form and pH control in solution culture. Fertil Res. 1989:21(1): 1–6 10.1007/BF01054728 [DOI] [Google Scholar]
- Man H-m, Boriel R, El-Khatib R, Kirby EG. Characterization of transgenic poplar with ectopic expression of pine cytosolic glutamine synthetase under conditions of varying nitrogen availability. New Phytol. 2005:167(1):31–39 10.1111/j.1469-8137.2005.01461.x [DOI] [PubMed] [Google Scholar]
- Marschner P. Marschner's Mineral Nutrition of Higher Plants. 3rd edn. UK: Academic Press, London; 2012 [Google Scholar]
- Martin A, Lee J, Kichey T, Gerentes D, Zivy M, Tatout C, Dubois F, Balliau T, Valot B, Davanture M, et al. Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell. 2006:18(11): 3252–3274 10.1105/tpc.106.042689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A. Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann Bot. 2010:105(7):1141–1157 10.1093/aob/mcq028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokochinski JB, Mazzafera P, Sawaya ACHF, Mumm R, De Vos RCH, Hall RD. Metabolic responses of Eucalyptus species to different temperature regimes. J Integr Plant Biol. 2018:60: 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder E. Nitrogen-magnesium relationships in crop plants. Plant Soil. 1956:7(4): 341–376 10.1007/BF01394322 [DOI] [Google Scholar]
- Ni K, Liao WY, Yi XY, Niu SY, Ma LF, Shi YZ, Zhang QF, Liu MY, Ruan JY. Fertilization status and reduction potential in tea gardens of China. J Plant Nutr. 2019:25: 421–432. (in Chinese) [Google Scholar]
- Okano K, Komaki S, Matsuo K. Remobilization of nitrogen from vegetative parts to sprouting shoots of young tea (Camellia sinensis L.) plants. Jpn J Crop Sci. 1994:63(1): 125–130 10.1626/jcs.63.125 [DOI] [Google Scholar]
- Oliveira IC, Brears T, Knight TJ, Clark A, Coruzzi GM. Overexpression of cytosolic glutamine synthetase. Relation to nitrogen, light, and photorespiration. Plant Physiol. 2002:129(3):1170–1180 10.1104/pp.020013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual MB, Jing ZP, Kirby EG, Cánovas FM, Gallardo F. Response of transgenic poplar overexpressing cytosolic glutamine synthetase to phosphinothricin. Phytochemistry. 2008:69(2):382–389 10.1016/j.phytochem.2007.07.031 [DOI] [PubMed] [Google Scholar]
- Peng WT, Qi WL, Nie MM, Xiao YB, Liao H, Chen ZC. Magnesium supports nitrogen uptake through regulating NRT2.1/2.2 in soybean. Plant Soil. 2020:457(1-2):97–111 10.1007/s11104-019-04157-z [DOI] [Google Scholar]
- Rao BR, Rajput D. Response of palmarosa {Cymbopogon martinii (roxb.) wats. Var. motia burk.} to foliar application of magnesium and micronutrients. Ind Crop Prod. 2011:33(2): 277–281 10.1016/j.indcrop.2010.12.020 [DOI] [Google Scholar]
- Rhodes D, Rendon GA, Stewart GR. Control of glutamine synthetase level in Lemna minor L. Planta. 1975:125(3): 201–211 10.1007/BF00385596 [DOI] [PubMed] [Google Scholar]
- Ruan JY, Gerendas J. Absorption of foliar-applied urea-15N- and the impact of low nitrogen, potassium, magnesium and sulfur nutritional status in tea (Camellia sinensis L.) plants. Soil Sci Plant Nutr. 2015:61(4): 653–663 10.1080/00380768.2015.1027134 [DOI] [Google Scholar]
- Ruan J, Ma L, Shi Y. Aluminium in tea plantations: mobility in soils and plants, and the influence of nitrogen fertilization. Environ Geochem Hlth. 2006:28(6): 519–528 10.1007/s10653-006-9047-z [DOI] [PubMed] [Google Scholar]
- Ruan J, Ma L, Shi Y, Zhang F. Effects of litter incorporation and nitrogen fertilization on the contents of extractable aluminium in the rhizosphere soil of tea plant (Camellia sinensis (L.) O. Kuntze). Plant Soil. 2004:26(1): 283–296 10.1023/B:PLSO.0000047744.44940.96 [DOI] [Google Scholar]
- Ruan J, Ma L, Yang Y. Magnesium nutrition on accumulation and transport of amino acids in tea plants. J Sci Food Agric. 2012:92(7): 1375–1383 10.1002/jsfa.4709 [DOI] [PubMed] [Google Scholar]
- Ruan JY, Wu X, Hardter R. Effects of potassium and magnesium nutrition on the quality components of different types of tea. J Sci Food Agric. 1999:79(1): 47–52 [DOI] [Google Scholar]
- Ruan JY, Wu X, Ye Y, Hardter R. Effect of potassium, magnesium and sulphur applied in different forms of fertilisers on free amino acid content in leaves of tea (Camellia sinensis L). J Sci Food Agric. 1998:76(3): 389–396 [DOI] [Google Scholar]
- Ruan J, Zhang F, Wong MH. Effect of nitrogen form and phosphorus source on the growth, nutrient uptake and rhizosphere soil property of Camellia sinensis L. Plant Soil. 2000:223(1/2): 65–73 10.1023/A:1004882001803 [DOI] [Google Scholar]
- Saito R, Smoot M E, Ono K. A travel guide to Cytoscape plugins. Nat Methods. 2012:9(11):1069. 10.1038/nmeth.2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, Lundgren K, Roessner-Tunali U, Forbes MG, Willmitzer L, et al. GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 2005:579(6):1332–1337 10.1016/j.febslet.2005.01.029 [DOI] [PubMed] [Google Scholar]
- Staswick EP. Storage proteins of vegetative plant tissues. Ann Rev Plant Physiol Plant Mol Biol. 1994:45(1):303–322 10.1146/annurev.pp.45.060194.001511 [DOI] [Google Scholar]
- Swarbreck SM, Defoin-Platel M, Hindle M, Saqi M, Habash DZ. New perspectives on glutamine synthetase in grasses. J Exp Bot. 2011:62(4):1511–1522 10.1093/jxb/erq356 [DOI] [PubMed] [Google Scholar]
- Taira M, Valtersson U, Burkhardt B, Ludwig RA. Arabidopsis thaliana GLN2-encoded glutamine synthetase is dual targeted to leaf mitochondria and chloroplasts. Plant Cell. 2004:16(8):2048–2058 10.1105/tpc.104.022046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang DD, Liu MY, Zhang QF, Ma LF, Shi YZ, Ruan JY. Preferential assimilation of NH4+ over NO3− in tea plant associated with genes involved in nitrogen transportation, utilization and catechins biosynthesis. Plant Sci. 2020:291: 110369. 10.1016/j.plantsci.2019.110369 [DOI] [PubMed] [Google Scholar]
- Tegeder M, Masclaux-Daubresse C. Source and sink mechanisms of nitrogen transport and use. New Phytol. 2018:217(1):35–53 10.1111/nph.14876 [DOI] [PubMed] [Google Scholar]
- Thomsen HC, Eriksson D, Møller IS, Schjoerring JK. Cytosolic glutamine synthetase, a target for improvement of crop nitrogen use efficiency. Trends Plant Sci. 2014:19(10):656–663 10.1016/j.tplants.2014.06.002 [DOI] [PubMed] [Google Scholar]
- Tian XY, He DD, Bai S, Zeng WZ, Wang Z, Wang M, Wu LQ, Chen ZC. Physiological and molecular advances in magnesium nutrition of plants. Plant Soil. 2021:468(1-2):1–17 10.1007/s11104-021-05139-w [DOI] [Google Scholar]
- Tobin AK, Yamaya T. Cellular compartmentation of ammonium assimilation in rice and barley. J Exp Bot. 2001:52(356):591–604 10.1093/jexbot/52.356.591 [DOI] [PubMed] [Google Scholar]
- Vrataric M, Sudaric A, Kovacevic V. Response of soybean to foliar fertilization with magnesium sulfate (epsom salt). Cereal Res Commun. 2006:34(1): 709–712 10.1556/CRC.34.2006.1.177 [DOI] [Google Scholar]
- Wallsgrove RM, Turner JC, Hall NP, Kendall AC, Bright SWJ. Barley mutants lacking chloroplast glutamine synthetasebiochemical and genetic analysis. Plant Physiol. 1987:83(1):155–158 10.1104/pp.83.1.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Fu B, Pan L. Overexpression of Arabidopsis dof1, GS1 and GS2 enhanced nitrogen assimilation in transgenic tobacco grown under low-nitrogen conditions. Plant Mol Biol Rep. 2013:31(4): 886–900 10.1007/s11105-013-0561-8 [DOI] [Google Scholar]
- Wszelaczynska E, Poberezny J. Effect of foliar magnesium fertilisation and storage on some parameters of the nutritive value of carrot storage roots. J Elementol. 2011:16: 635–649 [Google Scholar]
- Xia EH, Li FD, Tong W, Li PH, Wu Q, Zhao HJ, Ge RH, Li RP, Li YY, Zhang ZZ, et al. Tea plant information archive: a comprehensive genomics and bioinformatics platform for tea plant. Plant Biotechnol J. 2019:17(10): 1938–1953 10.1111/pbi.13111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Akiyama A, Kisaka H, Uchimiya H, Miwa T. Metabolic engineering with dof1 transcription factor in plants: improved nitrogen assimilation and growth under low-nitrogen conditions. Proc Natl Acad Sci USA. 2004:101(20):7833–7838 10.1073/pnas.0402267101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin ST, Ze YG, Liu C, Li N, Zhou M, Duan YM, Hong FS. Cerium relieves the inhibition of nitrogen metabolism of spinach caused by magnesium deficiency. Biol Trace Elem Res. 2009:132(1-3): 247–258 10.1007/s12011-009-8392-z [DOI] [PubMed] [Google Scholar]
- Zhang F, Chen X, Vitousek P. Chinese Agriculture: an experiment for the world. Nature. 2013:497(7447): 33–35 10.1038/497033a [DOI] [PubMed] [Google Scholar]
- Zhang Q, Liu M, Ruan J. Metabolomics analysis reveals the metabolic and functional roles of flavonoids in light sensitive tea leaves. BMC Plant Biol. 2017:17(1): 1–10 10.1186/s12870-017-1012-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or used during the study appear in the submitted article.