Abstract
The uses of vegetable oils are determined by functional properties arising from their chemical composition. Soybean oil was previously used in margarines and baked foods after partial hydrogenation to achieve heat and oxidative stability. This process, however, generates trans fats that are now excluded from food use because of cardiovascular health risks. Also present in soybean oil are the anti-oxidant tocopherols, with α-tocopherol (vitamin E) typically present as a minor component compared to γ-tocopherol. Genetic improvement of the fatty acid profile and tocopherol profile is an attractive solution to increase the functional and health qualities of soybean oil. The objective of this research was to develop resources to directly select with molecular markers for the elevated vitamin E trait in soybean oil and to use a molecular breeding approach to combine elevated vitamin E with the high oleic/low linolenic acid seed oil trait that improves oil functionality and nutrition. New soybean germplasm was developed from the molecular breeding strategy that selected for alleles of six targeted genes. Seed oil from the novel soybean germplasm was confirmed to contain increased vitamin E α-tocopherol along with a high oleic acid/low linolenic acid profile.
Keywords: Soybean, Oilseed, Seed composition, Nutrition
Introduction
Oil extracted from soybean seeds accounts for 56% of the vegetable oil consumption in the USA. Soybean oil is a reliable source of many beneficial health compounds for both livestock and humans and naturally contains vitamin E, which is made up of four forms of structurally related tocopherols and tocotrienols, including α-, β-, δ-, and γ-forms (Hunter and Cahoon 2007; SoyStats 2020). These four forms are differentiated by the number and position of methyl groups on the aromatic ring (Fryer 1992; Hunter and Cahoon 2007). Of these forms, α-tocopherol has the highest vitamin E activity in human and livestock nutrition due to its higher bioavailability (Schneider 2005). Tocopherols are lipid-soluble antioxidants produced in chloroplasts, where they protect membrane integrity and prevent oxidative stress through scavenging fatty acid peroxy radicals, improving oil stability (Munné-Bosch and Alegre 2002; Schneider 2005). Plant tocopherols also are crucial for seed germination, early growth, and longevity (Sattler et al. 2004). Photosynthetic tissues contain higher amounts of α-tocopherol, while tocopherols of conventional soybean seeds are comprised of ~ 65% γ-tocopherols but < 10% of α-tocopherols. This composition provides a breeding opportunity for developing soybean seeds with enhanced α-tocopherol content (elevated vitamin E) for improved nutritional quality (Konda et al. 2020; Ujiie et al. 2005).
Plant tocopherols, particularly α-tocopherol, have been identified as beneficial health compounds to enhance immune system function, providing a role in prevention of cancer, cardiovascular diseases, and neurodegenerative diseases (Bramley et al. 2000; Buring and Hennekens 1997). According to one study, only 8% of men and ~ 2% of women in the USA are consuming the estimated average requirements for vitamin E in their diets (Maras et al. 2004). The majority of α-tocopherol consumed in the American diet comes from seed oils, due to the large role that vegetable oils play in culinary culture (Grusak and DellaPenna 1999). In addition to its health benefits for humans, α-tocopherol is commonly added to animal feeds; dietary α-tocopherol has been shown to improve the meat quality of chicken and pigs and improve the sensory perception of pork (Dirinck et al. 1996; Morrissey et al. 1994).
The vast majority of α-tocopherol in the worldwide vitamin E market, 85–88%, is synthetically derived (Subramaniam et al. 2008). However, it only has 50–74% of the biological activity of naturally derived α-tocopherol (Clemente and Cahoon 2009). The remaining 12–15% of worldwide α-tocopherol comes almost entirely from soybean oil processing and typically involves chemical methylation of γ-tocopherol, the predominant tocopherol form in soybean oil, to produce “natural” α-tocopherol (Valentin and Qi 2005). As such, enhancement of α-tocopherol content is a target for improvement of the health-promoting properties of soybean oil and may mitigate the need for chemical methylation (Van Eenennaam et al. 2003). A forward genetics screen in Arabidopsis thaliana identified a tocopherol biosynthetic enzyme VTE3, encoding a 2-methyl-6-phytylbenzoquinol methyltransferase, and when the gene was transformed into soybean lines, there was enhanced conversion of δ- and β-tocopherols to γ- and α-tocopherols (Van Eenennaam et al. 2003). While these efforts resulted in an eight-fold increase in α-tocopherol concentration and a five-fold increase in vitamin E activity, these transgenic soybean lines are subject to regulatory approval prior to commercial release.
In the last 30 years, a number of the genes responsible for the tocopherol biosynthetic steps have been cloned in carrot, Arabidopsis, sunflower, and soybean (Dwiyanti et al. 2011; Garcia et al. 1997; Hass et al. 2006; Keller et al. 1998; Norris et al. 1995; Norris et al. 1998; Van Eenennaam et al. 2003). A key step in tocopherol biosynthesis is the final methylation of δ- and γ-tocopherol by γ-tocopherol methyltransferase, encoded by VTE4 in Arabidopsis and γ-ΤΜΤ3 in soybean, to produce β- and α-tocopherols (Dwiyanti et al. 2011; Kumari et al. 2019). Plant breeders can now characterize variant alleles of these target genes and create new soybean lines through traditional plant breeding and molecular selection; soybean lines with increased ⍺-tocopherol content have already been identified in soybean germplasm collections (Dwiyanti et al. 2011; Kumari et al. 2019).
Conventional soybean breeding has also led to development of soybean varieties with superior oil fatty acid profile (Pham et al. 2012). Edible soybean oil is typically chemically hydrogenated in order to increase the more healthful and stable oleic acid component and to reduce the amount of the oxidatively unstable polyunsaturated fatty acids (PUFAs) linoleic and linolenic acids. While hydrogenation improves the shelf life of soybean oil by reducing the levels of the less oxidatively stable PUFAs, it creates trans fats which have been regulated in foods because of negative effects on health (FDA 2003; FDA 2015). A typical commodity soybean seed contains about 25% oleic acid (18:1) as well as 52% linoleic acid (18:2) and 8% linolenic acid (18:3) in the seed oil (Fehr 2007). We have been able to mine the USDA’s GRIN collection for lines containing alleles encoding increased oleic acid and decreased linoleic and linolenic acids, and combine these alleles for the high oleic/low linolenic acid (HOLL) trait (Pham et al. 2011; Pham et al. 2010; Pham et al. 2012). HOLL soybean germplasm has seeds with over 80% oleic acid, 3–7% linoleic acid, and less than 3% linolenic acid (Pham et al. 2012).
Combining HOLL lines with the increased α-tocopherol trait through molecular-assisted breeding would enable the creation of nutritionally enhanced, improved stability soybean oil with a reduced requirement for processing. Little is known about potential interactions between improved fatty acid profile and increased α-tocopherol composition in seed oil. In this work, we report the development of a novel molecular marker assay to screen for the presence of the elevated vitamin E trait/increased α-tocopherol allele, OE γ-ΤΜΤ3, and a molecular marker-driven breeding strategy that successfully combined the five alleles responsible for a HOLL/elevated vitamin E soybean line along with the seed composition analyses for the novel soybean germplasm.
Results
Analysis of γ-ΤΜΤ3 gene region genomic variation and molecular marker assay development for increased seed α-tocopherol/elevated vitamin E
In order to screen soybean accessions for the presence of the over-expression allele of γ-ΤΜΤ3 (VTE4) originally identified by Dwiyanti et al., the DNA sequence upstream from the γ-ΤΜΤ3 gene (Glyma.09g222800, Wm82.a2.v1; Phytozome start codon position 09:44,342,003) was surveyed in a number of whole genome resequenced soybean accessions using SNPViz (Dwiyanti et al. 2011; Langewisch et al. 2014; Valliyodan et al. 2020; Valliyodan et al. 2016; Zhou et al. 2015). The γ-ΤΜΤ3 promoter region was putatively indicated to be responsible for the gain-of-function mutation resulting in over-expression of γ-ΤΜΤ3 found in soybean accessions with increased α-tocopherol (Dwiyanti et al. 2011; Kumari et al. 2019). There were 38 Plant Introduction (PI) accessions that contained the “G” SNP that created a MYB binding site (“CTGTTA”) present in the promoter region ~ 638 bases upstream from the start codon of γ-ΤΜΤ3 (09:41,625,605A Wm82.a1.v1.1 and 09:44,341,365A Wm82.a2.v1) (Table 1). Herein, we designate the over-expression alleles (OE γ-ΤΜΤ3) as those containing the “G” SNP variant in the promoter at position 09:44,341,365 Wm82.a2.v1. The ratio of α-tocopherol to total tocopherol was determined from field produced seed for a small subset of these accessions along with the reference cultivar Williams 82 (Table 1). The four accessions with the OE γ-ΤΜΤ3 promoter SNP had ratios higher than Williams 82 with wild-type (WT) γ-ΤΜΤ3. The OE γ-ΤΜΤ3 soybean variety Hutcheson was previously demonstrated to contain a high α-tocopherol to total tocopherol ratio (0.12), similar to our results (Britz et al. 2008). The cultivar Williams 82 without the reference γ-ΤΜΤ3 allele was the only accession to have an α-tocopherol to total tocopherol ratio below 0.11.
Table 1.
Soybean reference genome Williams 82 and resequenced plant introduction (PI) accessions with variation for OE γ-TMT3 promoter variant
| Accession | Name | Taxonomy | Origin | Maturity group | Gm09: 44341365 | Ratioa |
|---|---|---|---|---|---|---|
| PI 518671 | “Williams 82” | G. max | Illinois, USA | III | A | 0.06 |
| PI 58955 | Common Yellow Variety | G. max | Shandong Sheng, China | IV | G | |
| PI 68521 -1 | 205 | G. max | China | II | G | |
| PI 70466 -3 | 7336 | G. max | Jilin Sheng, China | IV | G | |
| PI 84637 | S-62 | G. max | Kyonggi, Korea, South | II | G | |
| PI 84973 | Takiya | G. max | Saitama, Japan | III | G | |
| PI 153231 | B-63 | G. max | Unknown | III | G | |
| PI 153262 | Roumanie | G. max | Belgium | 0 | G | 0.22 |
| PI 189873 | Miko Saumon | G. max | France | 0 | G | |
| PI 209334 | No. 9 | G. max | Hokkaidô, Japan | III | G | |
| PI 253661 B | No. 12 | G. max | China | III | G | |
| PI 378680 E | (VNIIMK 9186) | G. max | Russian Federation | I | G | |
| PI 398296 | KAS 173-3 | G. max | Kyonggi, Korea, South | II | G | |
| PI 398614 | KAS 390-9 | G. max | Chungcheongbuk-do, Korea, South | V | G | |
| PI 407131 | RB 1072 | G. soja | Kumamoto, Japan | VI | G | |
| PI 407801 | G. max | Kyonggi, Korea, South | VI | G | 0.11 | |
| PI 408105 A | KAS 633-19 | G. max | Gyeongsangbuk-do, Korea, South | IV | G | |
| PI 424391 | KAS 521-15 | G. max | Jeollabuk-do, Korea, South | VI | G | 0.12 |
| PI 437112 A | VIR 249 | G. max | Russian Federation | II | G | |
| PI 437240 | CSchi 1069 | G. max | Moldova | 0 | G | |
| PI 437662 | Gun’-tszu-lin’ 658 | G. max | China | II | G | |
| PI 437793 | VIR 3024 | G. max | China | II | G | |
| PI 438230 A | VIR 4521 | G. max | China | I | G | |
| PI 438323 | Grignon 53-F-3 | G. max | France | I | G | |
| PI 438336 | Sao 208 | G. max | Algeria | 0 | G | |
| PI 445824 A | Wolfsthaler | G. max | Germany | 0 | G | |
| PI 458538 | G. soja | Heilongjiang Sheng, China | 0 | G | ||
| PI 464927 A | LS-005 | G. soja | Liaoning Sheng, China | 0 | G | |
| PI 468916 | G. soja | Liaoning Sheng, China | III | G | ||
| PI 479752 | GD 50388-2 | G. soja | Jilin Sheng, China | I | G | |
| PI 507467 | Tousan kei F 764 | G. max | Japan | IV | G | |
| PI 507471 | Tousan kei na 16 | G. max | Japan | III | G | |
| PI 518664 | “Hutcheson” | G. max | Virginia, USA | V | G | 0.11 |
| PI 549021 A | Na hei dou | G. max | Liaoning Sheng, China | III | G | |
| PI 603154 | GL 2622 /96 | G. max | Korea, North | V | G | |
| PI 603397 | Hei qi huang da dou | G. max | China | IV | G | |
| PI 603559 | ZDD08590 | G. max | China | IV | G | |
| PI 612611 | “Browngilgun” | G. max | Korea, North | III | G | |
| PI 639550 E | (KSHI 713) | G. max | Moldova | II | G |
aRatio represents seed α-tocopherol/total tocopherol
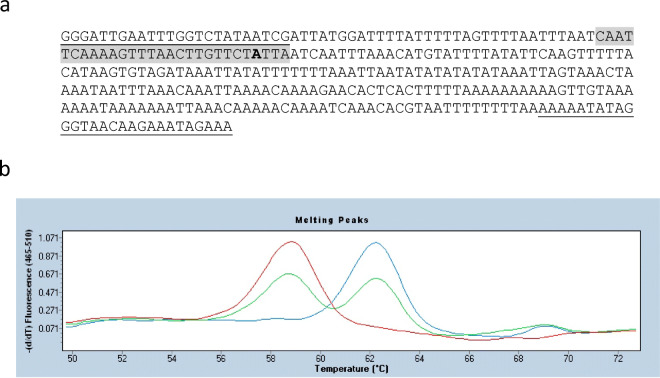
A molecular marker assay based on a SimpleProbe was designed to the OE γ-ΤΜΤ3 promoter SNP (Fig. 1). The molecular marker assay uses real-time PCR and a fluorescent probe to differentiate between DNA samples containing homozygous WT γ-ΤΜΤ3 alleles, heterozygous alleles, and homozygous OE γ-ΤΜΤ3 alleles. Melt curve analysis of the reaction products revealed distinct peaks for each of the two different homozygous alleles and heterozygous individuals: samples containing WT γ-ΤΜΤ3 alleles produced peaks at 62.5 °C, and samples with OE γ-ΤΜΤ3 alleles produced peaks at 58.5 °C, while heterozygous samples produced both peaks (Fig. 1). Soybean accessions Keszthelyi Aproszemu Sarga (PI 209129B) and Dobrudza No. 14 (PI 248397) were previously found to have high α-tocopherol content (Ujiie et al. 2005), and DNA from those accessions produced peaks at 58.5 °C, demonstrating the presence of OE γ-ΤΜΤ3 alleles (data not shown).
Fig. 1.
A SimpleProbe molecular marker assay for the OE γ-ΤΜΤ3 alleles. a The DNA sequence shown represents the WT γ-ΤΜΤ3 promoter 318 bp amplicon on Williams 82 (Wm82.a2.v1) chromosome 09 from position 44,341,285 to 44,341,602. The underlined sequences represent the marker assay forward and reverse primers, the gray-highlighted region represents the SimpleProbe sequence, and the bold “A” base is the polymorphic site at position 44,341,365 that is a “G” base in OE γ-ΤΜΤ3 alleles. b Melting curve results from the OE γ-ΤΜΤ3 molecular marker assay. The negative first derivatives of the disappearance in the florescence with increasing temperature of three samples is shown, with the homozygous WT γ-ΤΜΤ3 allele sample in blue (peak at 58.5 °C), the homozygous OE γ-ΤΜΤ3 sample in red peak at 62.5 °C, and a heterozygous sample in green (both peaks)
Molecular breeding strategy to combine the HOLL seed oil trait with the elevated vitamin E trait
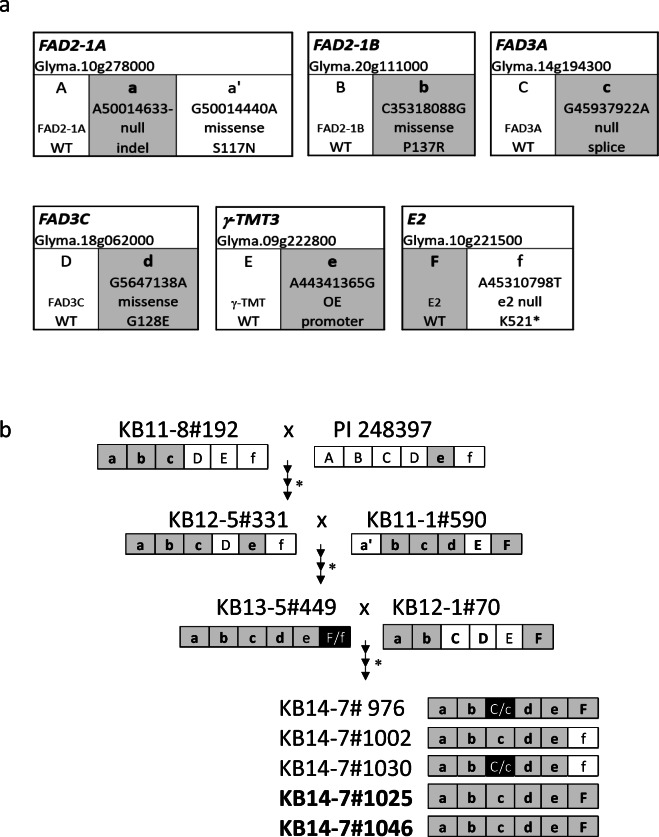
Soybean germplasm lines containing variant alleles for the HOLL seed oil phenotype were selected from our germplasm collection and were combined with the OE γ-ΤΜΤ3 alleles from PI 248397 (Fig. 2). We have previously reported that four genes are responsible for the most dramatic increase in oleic acid and decrease in linolenic acids, and the combination of these four alleles results in ~ 80% oleic acid in the seed oil and reductions in linolenic acid to less than 3%. We targeted variant alleles with the most dramatic improvements in oil profile from our germplasm collection: null alleles of FAD2-1A and FAD3C as well as missense alleles of FAD2-1B and FAD3C (Fig. 2) (Bilyeu et al. 2018). Genotypic selection for these alleles paired with our novel molecular marker assay for the OE γ-ΤΜΤ3 alleles enabled the creation of quintuple mutant lines developed after 3 years of crossing and genotype selection (Fig. 2). The final desired genotype was for the five alleles (FAD2-1A, FAD2-1B, FAD3A, FAD3C, and OE γ-ΤΜΤ3) controlling the seed oil HOLL trait plus the elevated vitamin E trait, in a functional E2 maturity gene background (Fig. 2). A confounding factor was the presence of null alleles for the major maturity gene e2 in the parents (PI 248397 and experimental line KB11-8#192) of the germplasm development scheme (Langewisch et al. 2017; Watanabe et al. 2011). The e2 alleles were originally physically linked to the indel alleles of FAD2-1A (from the PI 603452 donor), but that linkage was broken for the high oleic germplasm line KB12-1#70 (Fig. 2) (Pham et al. 2011; Watanabe et al. 2011). Two of our developed soybean germplasm lines contained the targeted alleles of those six genes: KB14-7#1025 and KB14-7#1046. The other developed germplasm lines either were still segregating for the FAD3C alleles or contained the undesirable nonfunctional e2 alleles (Fig. 2).
Fig. 2.
Molecular marker based breeding scheme to combine the HOLL seed oil trait with the elevated vitamin E trait. a Gene names, identification in Wm82.a2.v1, and allele information for six targeted genes. For simplicity, each targeted gene is coded by an uppercase (functional allele) or lowercase letter (alternate allele). The details of the alleles are provided, and bold plus gray shading represents the targeted allele for this study. b Breeding scheme with experimental line names and allele code for the six targeted genes. The genotypes are listed for the parents of each cross and the selection of lines derived from those crosses indicated by the triple arrows (for three generations of advancement and selection in the F2 or F2 and F3 generation). Arrows represent the advancing generations from each cross, with the asterisks representing the F2 generation molecular selection steps. Shaded and bolded codes represent the desired alleles of each gene. Black boxes with F/f or C/c represent lines that were still segregating for those genes. The two developed soybean lines in bold font were homozygous for the desired alleles of all six of the targeted genes
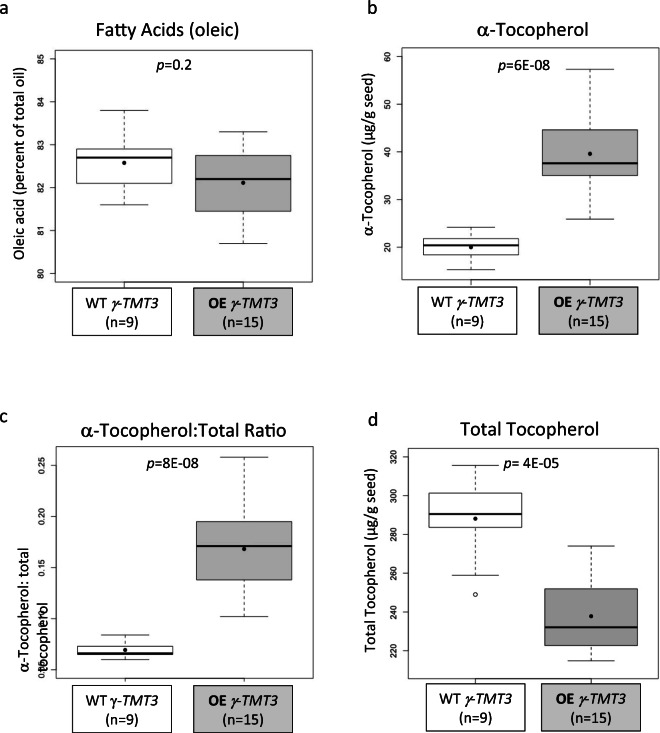
Increased α-tocopherol content does not impact the high oleic trait in soybean seeds
Soybean lines containing the high oleic trait from FAD2-1A indel and FAD2-1B P137R and either WT γ-ΤΜΤ3 or OE γ-ΤΜΤ3 alleles (disregarding alleles for low linolenic acid) were field tested and analyzed for seed oil oleic acid content, elevated α-tocopherol content, the ratio of α-tocopherol to total tocopherol, and total tocopherol (Fig. 3). All of the lines were high in oleic acid (overall mean of 82.3%), and there was no significant difference in seed oil oleic acid between the WT γ-ΤΜΤ3 or the OE γ-ΤΜΤ3 group. The OE γ-ΤΜΤ3 group contained significantly higher seed α-tocopherol content and a significantly higher ratio of α-tocopherol to total tocopherol compared to the WT γ-ΤΜΤ3 group (Fig. 3). The OE γ-ΤΜΤ3 group contained significantly lower seed total tocopherol (Fig. 3).
Fig. 3.
Seed oil phenotypes for control and experimental soybean lines contrasting for either WT or OE γ-ΤΜΤ3 alleles but fixed for the alternate alleles of FAD2-1A and FAD2-1B conditioning the high oleic acid oil trait. For the two categories, the box plots present the 25 to 75% quartiles as the box, whiskers representing the range, a bold horizontal line as the median, and a dot as the mean. The p values are listed from the t tests. There were nine WT γ-ΤΜΤ3 samples and 15 OE γ-ΤΜΤ3 samples (highlighted in gray). a Fatty acid results for oleic acid as a percent of the seed oil for high oleic acid soybean lines with WT or OE γ-ΤΜΤ3 alleles. b Seed α- tocopherol results for high oleic acid soybean lines with WT or OE γ-ΤΜΤ3 alleles. c Results for ratio of α-tocopherol to total tocopherols for high oleic acid soybean lines with WT or OE γ-ΤΜΤ3 alleles
Seed fatty acid profile and tocopherol profile of control and experimental soybean germplasm
Seed oil fatty acid profiles were determined for field-produced seed of the control and experimental soybean germplasm utilized or developed in this study. Palmitic (16:0), stearic (18:0), oleic (18:1), linoleic (18:2), and linolenic (18:3) acid contents are reported as percentages of the seed oil (Table 2). Seeds from lines containing the alleles (ab) for the high oleic acid oil trait showed somewhat variable but significantly lower mean values for palmitic acid, linoleic acid, and linolenic acid compared to lines without those alleles. There were significantly higher mean values for oleic acid for lines containing the alleles (ab) for the high oleic acid oil trait compared to lines without those alleles (Table 2). The control line KB13-15_14-224 (abcdE_) with the HOLL trait alone was not significantly different for mean linolenic acid compared to the three lines with the HOLL trait in combination with the OE γ-ΤΜΤ3 alleles (abcde_) (Table 2). The OE γ-ΤΜΤ3 alleles did not interfere with the HOLL lines’ seed composition achieving the target of > 75% oleic acid and < 3.0% linolenic acid in the seed oil.
Table 2.
Fatty acid components of the seed oil in experimental soybean lines
| Soybean line | γ-TMT3 allele | Genotype | Samples | Fatty acids | ||||
|---|---|---|---|---|---|---|---|---|
| Palmitic (16:0) | Stearic (18:0) | Oleic (18:1) | Linoleic (18:2) | Linolenic (18:3) | ||||
| no. | Percent of total oil | |||||||
| Williams 82 | WT | ABCDEF | 2 | 11.6ba | 3.8bc | 25.1e | 52.5a | 7.1b |
| PI 248397 | OE | ABCDef | 3 | 13.6a | 4.2a | 24.1e | 50.0b | 8.1a |
| KB11-8#192 | WT | abcDEf | 3 | 8.6cd | 3.1e | 82.6ab | 2.7e | 2.9e |
| KB11-1#590A | WT | a'bcdEF | 3 | 8.3de | 3.6cd | 79.5d | 6.3c | 2.3f |
| KB12-1#70 | WT | abCDEF | 3 | 8.0ef | 3.3e | 82.0abc | 2.2ef | 4.5c |
| KB13-15_14-224 | WT | abcdEF | 3 | 7.3g | 3.3de | 83.1a | 3.8d | 2.5f |
| KB14-7#976 | OE | ab(C/c)deF | 3 | 8.5cd | 3.3e | 82.9ab | 1.7f | 3.6d |
| KB14-7#1002 | OE | abcdef | 3 | 8.2de | 3.1e | 82.4abc | 4.1d | 2.3f |
| KB14-7#1030 | OE | ab(C/c)def | 3 | 8.4cd | 3.1e | 81.2c | 4.0d | 3.4d |
| KB14-7#1025 | OE | abcdeF | 3 | 8.8c | 3.1e | 81.7bc | 4.0d | 2.4f |
| KB14-7#1046 | OE | abcdeF | 3 | 7.8f | 3.9b | 82.3abc | 3.8d | 2.2f |
aValues followed by a common lowercase letter are not significantly different according to LSD (p = 0.05)
Similarly, tochopherol profiles were determined for field-produced seed of the control and experimental soybean germplasm utilized or developed in this study. Here we report δ-tocohpherol, γ- and β-tocopherols combined (γ/β), and α-tocopherol as content contributing to the total vitamin E, in μg/g seed dry weight, as well as the ratio of α-tocopherol to total tocopherol (ratio) (Table 3). There were variable mean values for seed δ-, γ/β-, α-tocopherol, and total tocopherol for the lines studied, but the parental line PI 248397 with OE γ-ΤΜΤ3 alleles was significantly higher for mean α-tocopherol and significantly lower for mean δ-tocopherol than all of the other lines investigated. Seeds from soybean lines containing the OE γ-ΤΜΤ3 alleles had generally lower mean γ/β-tocopherol than lines with WT γ-ΤΜΤ3 alleles. The ratio values showed the WT γ-ΤΜΤ3 lines forming a group with no significant difference in ratio values in the 0.06 to 0.08 range. The OE γ-ΤΜΤ3 lines were more variable for ratio values, but all were significantly higher than the WT γ-ΤΜΤ3 lines. The parental line PI 248397 with OE γ-ΤΜΤ3 alleles had the highest ratio value (0.33), and it was significantly higher than all of the experimental lines with OE γ-ΤΜΤ3 alleles. The experimental lines with OE γ-ΤΜΤ3 alleles had ratios in the range of 0.12 to 0.22.
Table 3.
Tocopherol components of the seed in experimental soybean lines
| Soybean line | γ-TMT3 allele | Genotype | Samples | Tocopherols | ||||
|---|---|---|---|---|---|---|---|---|
| δ | γ/βa | α | Total | Ratiob | ||||
| no. | μg/g seed | |||||||
| Williams 82 | WT | ABCDEF | 2 | 89.6ac | 221.6a | 19.8ef | 331.0a | 0.06f |
| PI 248397 | OE | ABCDef | 3 | 38.8g | 135.6de | 86.1a | 260.5bcde | 0.33a |
| KB11-8#192 | WT | abcDEf | 3 | 81.3abc | 171.6bc | 16.9f | 269.7bcd | 0.06f |
| KB11-1#590A | WT | a’bcdEF | 3 | 84.4ab | 172.6bc | 17.5f | 274.4bc | 0.06f |
| KB12-1#70 | WT | abCDEF | 3 | 74.2bcd | 200.7ab | 23.1ef | 298.0ab | 0.08f |
| KB13-15_14-224 | WT | abcdEF | 3 | 88.1a | 188.5b | 20.0ef | 296.6ab | 0.07f |
| KB14-7#976 | OE | ab(C/c)deF | 3 | 70.6cd | 152.5cd | 29.4de | 252.5cdef | 0.12e |
| KB14-7#1002 | OE | abcdef | 3 | 50.8fg | 128.5de | 41.2bc | 220.5f | 0.19bc |
| KB14-7#1030 | OE | ab(C/c)def | 3 | 63.4de | 133.4de | 35.4cd | 232.3def | 0.15d |
| KB14-7#1025 | OE | abcdeF | 3 | 55.7ef | 121.8e | 49.4b | 226.8ef | 0.22b |
| KB14-7#1046 | OE | abcdeF | 3 | 65.4de | 149.0cde | 42.5bc | 256.9cdef | 0.17cd |
aγ/β indicates both γ-tocopherol and β-tocopherol unresolved by HPLC
bRatio represents α-tocopherol/total tocopherol
cValues followed by a common lowercase letter are not significantly different according to LSD (p = 0.05)
Molecular breeding for targeted alleles of six genes resulted in the development of two soybean germplasm lines (KB14-7#1025 and KB14-7#1046) that produced seeds with improved seed composition with the HOLL oil trait in combination with the elevated vitamin E trait.
Discussion
Our research demonstrated that the soybean elevated vitamin E trait resulting from an increase in seed ⍺-tocopherol and conditioned by the OE ɣ-TMT3 alleles can be successfully combined with the four fatty acid desaturase alleles responsible for the HOLL seed oil trait in soybean. Substantial interactions between fatty acid profile of the oil and α-tocopherol content were not detected. These seed composition traits were stacked together to achieve a healthier, more oxidatively stable and nutritionally enhanced soybean. Oil produced from these non-GMO soybean lines would enhance the value of soybean with elevated vitamin E in a more oxidatively stable oil.
While seeds from soybean lines containing combinations of the HOLL alleles with the OE ɣ-TMT3 alleles did show a significant reduction in ⍺-tocopherol when compared to the OE ɣ-TMT3 allele donor PI 248397 with an unmodified fatty acid profile background, the novel soybean germplasm lines (abcdeF) still showed significantly higher seed ⍺-tocopherol levels than the reference WT ɣ-TMT3 line Williams 82 (Table 3). The decrease in ɣ/β tocopherol observed in OE ɣ-TMT3 allele-containing lines was expected as a result of the enhanced activity of the ɣ-TMT3 gene drawing from the pool of ɣ-tocopherol in the biochemical pathway.
It is not clear if the PI 248397 has additional genetic components contributing to the significantly higher seed vitamin E content than all other soybean lines evaluated in this research. Seeds from other soybean lines containing the OE ɣ-TMT3 alleles generally produced about two- to three-fold higher ratios of α-tocopherol to total tocopherol in this study. It was notable that the cultivar Hutcheson was identified as a potential OE ɣ-TMT3 allele donor, since it was shown here to have the OE ɣ-TMT3 alleles and previously ranked at the top of soybean accessions evaluated for ratio (Britz et al. 2008; Buss et al. 1988).
The work of McCord et al. investigated the tocopherol content of seeds from soybean lines with either normal seed oil fatty acid profiles or the ultra-low linolenic acid oil trait (~ 1% of the seed oil). In that study, there was lower total tocopherol in the ultra-low linolenic acid lines that was due to a proportionate decrease in all of the tocopherols except the β-tocopherol component (McCord et al. 2004). We did not design our study to tease apart the differences in total tocopherols for the various seed composition combinations; however, despite high variation, we did observe significantly reduced seed total tocopherols for high oleic acid lines with the OE ɣ-TMT3 alleles compared to high oleic acid lines with WT ɣ-TMT3 alleles (Fig. 3d). When comparing individual soybean lines, it was not possible to discern if this effect was related to the fatty acid profile (Table 3).
Although generating soybean germplasm with the appropriate plant maturity for mid-Missouri environments (maturity group III/IV) was a component of this work, evaluating the effect of temperature during podfill was beyond the scope of this research. Warmer environments have been shown to positively impact seed composition with regard to fatty acid profile and tocopherol content: Carrera et al. (2011) reported that warmer environments favored increased seed ⍺-tocopherol accumulation and improved oleic to linolenic acid ratio in the seed oil of unmodified soybeans. These data suggest that future research into the environmental stability of the HOLL oil plus elevated vitamin E combination should include production across a range of maturity zones in order to establish the stability of the seed composition phenotypes as well as their yield potential so that the value of the grain can be priced appropriately for the production of these novel value-enhanced soybeans.
Materials and methods
SNPViz analysis
The analysis to investigate the haplotypes in the γ-ΤΜΤ3 gene region was initially done with the original SNPViz (Langewisch et al. 2014), but the final analysis utilized an updated SNPViz2.0 and 775 unique resequenced soybean accessions (http://soykb.org/newSNPViz/). The region 1000 bp upstream from the start codon and including the coding sequence of Glyma09g35680/Glyma.09g222800 or the 1000 bp surrounding the promoter SNP were analyzed. Soybean accessions with the promoter SNP (09:41,625,605A Wm82.a1.v1.1and 09:44,341,365A Wm82.a2.v1) were present as a subset of a distinct haplotype from the reference accessions and one other distinct haplotype. The promoter SNP was present in the “non-reference” haplotypes. Within 100 bp upstream of the promoter SNP, there were six additional potential variant sequence positions present in both Wm82.a1.v1.1 and Wm82.a2.v1, but none of those positions correlated perfectly with the defined promoter SNP; the additional potential variant sequence positions present in both data sets that were present between the promoter SNP and the γ-ΤΜΤ3 start codon also did not correlate with the promoter SNP.
Molecular marker assays
The OE γ-ΤΜΤ3 SimpleProbe assay was based on the disassociation kinetics of a SimpleProbe oligonucleotide (XCR Diagnostics, Salt Lake City, UT) to be exactly complimentary to the Williams 82 WT reference sequence. The SimpleProbe consisted of 5′-Fluorescein-SPC-CAATTCAAAAGTTTAACTTGTTCTATTAATCAATT-phosphate-3′. The A > G mutation is indicated by bold font. Genotyping reactions were performed with a 5:2 (reverse to forward) asymmetric mix of primers (5′-GGGATTGAATTTGGTCTATAATCG-3′ at 2 μM final concentration, and 5′-AATTTATCTACACTTATGTAAAAACTTGA-3′ at 5 μM final concentration). Reactions were carried out in 20 μl, containing DNA template, primers, 0.2 μM final concentration of SimpleProbe, buffer (40 mM Tricine-KOH [pH 8.0] 16 mM KCl, 3.5 mM MgCl2, 3.75 μg ml−1 BSA,), 5% DMSO, 200 μM dNTPs, and 0.2X Titanium Taq polymerase (BD Biosciences, Palo Alto, CA). Genotyping reactions were performed using a Lightcycler 480 II real-time PCR instrument (Roche Life Sciences, Indianapolis, IN), using the following PCR parameters: 95 °C for 5 min followed by 40 cycles of 95 °C for 20 s, 60 °C for 20 s, 72 °C for 20 s, and then a melting curve from 50 °C to 70 °C. Reference alleles produced a peak at 62.5 °C, mutant alleles produced a peak at 58.5 °C, and heterozygous samples produced both peaks.
The molecular marker assay for FAD3A was similar to above, with the anti-sense SimpleProbe 5′-Fluorescein-SPC-GTTACCTTGCCGCGATACCA-phosphate-3′ along with 5:2 forward 5′-TTGCATCACCATGGTCATCAT-3′ to reverse 5′-AGCTATTATCTAGCATTAACCTCA-3′ primers. The melting curve was 50 to 75 °C; reference alleles produced a peak at 64 °C, mutant alleles produced a peak at 54 °C, and heterozygous samples produced both peaks.
Likewise, the molecular marker assay for FAD3C had an anti-sense SimpleProbe 5′-Fluorescein-SPC-AGGAACCGACCATCCATGGTATGGTACAAGAAT-phosphate-3′ along with 5:2 forward 5′-GTCCTTTGTTGAACAGCATT-3′ to reverse 5′-CTCCTGCAAAAAATCCATGAGTTGT-3′ primers. The melting curve was 60 to 75 °C; reference alleles produced a peak at 68 °C, mutant alleles produced a peak at 64 °C, and heterozygous samples produced both peaks.
The molecular marker assays for FAD2-1A, FAD2-1B, and E2 were performed as previously described (Langewisch et al. 2017; Pham et al. 2011; Pham et al. 2010; Pham et al. 2012).
Seed composition analyses
Tocopherols were extracted from powdered seeds and content and composition were measured by HPLC with fluorescence detection as previously described (Konda et al. 2020). Fatty acid analyses were conducted on mature seeds as described (Beuselinck et al. 2006).
Breeding scheme for germplasm development and field experiment
Novel soybean germplasm was developed through a molecular breeding approach that consisted of soybean crossing at the South Farm Research Center near Columbia, Missouri, during the annual field season (May–October) and two generations of advancement and genotype selection in a winter nursery. The F1 seeds were sent to a winter nursery in Costa Rica and advanced one cycle to produce F2 seeds. In the second off-season generation, the F2 plants were sampled with Whatman® FTA® cards (Whatman, Clifton, NJ, USA) for genotyping with molecular marker assays for the desired alleles, and selected F2:3 seeds from single plant threshes were returned to Missouri to be planted and used as parents in the subsequent Missouri field season. After true leaves emerged from plants in the Missouri plots, genotyping assays were used to confirm the status of targeted alleles or identify selections that were still segregating from some of the genes. This scheme was followed for crossing in 2012, 2013, and 2014 (as in Fig. 3). The PI 248397 was the OE γ-ΤΜΤ3 promoter variant donor, and several different experimental soybean germplasm lines contributed the targeted alleles for FAD2-1A, FAD2-1B, FAD3A, FAD3C, and E2. The undesirable e2 alleles were physically linked on chromosome 10 to the targeted indel alleles of FAD2-1A in the original donor PI 603452 (Langewisch et al. 2017; Pham et al. 2011).
In the 2015 Missouri field season, control soybean lines and a subset of the PI accessions that were found to contain the OE γ-ΤΜΤ3 promoter variant were grown in 50 seed plots in a random design. The developed experimental soybean lines (F2:3 plants) were grown in approximately 30 seed plots in a random design, and five plants from each plot were single plant threshed for seed composition analyses (F3:4 seeds). Single plant threshed seeds were used for analyses of the control soybean lines, but the PI accessions (Table 1) were bulk harvested by plot. Separate three seed samples from three different single plants per plot were analyzed for fatty acids and tocopherols; a single three seed sample from each plot bulk was used for tocopherol analysis for the PI accessions in Table 1.
Statistics
Box plots were created in R using the boxplot() function. In box plots, points for the means were calculated by colMeans() and added to the plots with points(). The Student t test (two-sample assuming unequal variances) was used in the Excel Data Analysis package for discriminating differences in means for oleic acid, α-tocopherol, and ratio in Fig. 3. For statistical comparative analyses of the seed fatty acid and tocopherol data in Tables 2 and 3, a one-way ANOVA was conducted in R version 3.5.1 using the aov() function. There were three replications per line, except for Williams 82 which had only two replications. To generate LSD values, the R package “agricolae” was used for multiple comparisons with the function LSD.test().
Acknowledgments
The authors acknowledge Christine Cole and Paul Little for essential technical assistance for most aspects of field management and gas chromatography phenotyping. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer.
Authors’ contributions
KB conceived and conducted or directed the experiments except for the tocopherol analyses. JHK conducted the statistical analyses and prepared figures. ARK and EBC conducted the tocopherol analyses and edited the manuscript. KH and KB wrote the manuscript.
Funding
Funding was provided by the USDA/ARS. Support was also provided in part by grants from the Nebraska Soybean Board and the United States Department of Agriculture-National Institute of Food and Agriculture (grant no. 2015-67013-2283) to EBC.
Compliance with ethical standards
The authors declare no conflicts of interest. Code availability is not applicable. Ethics approval is not applicable. Consent to participate is not applicable. All authors consented for publication. Soybean germplasm is available for research with a Material Transfer Agreement. Support was also provided by grants from the Nebraska Soybean Board and the United States Department of Agriculture-National Institute of Food and Agriculture (grant no. 2015-67013-2283) to EBC.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Beuselinck PR, Sleper DA, Bilyeu KD. An assessment of phenotype selection for linolenic acid using genetic markers. Crop Sci. 2006;46:747–750. doi: 10.2135/cropsci2005-04-0041. [DOI] [Google Scholar]
- Bilyeu K, Škrabišová M, Allen D, Rajcan I, Palmquist DE, Gillen A, Mian R, Jo H. The interaction of the soybean seed high oleic acid oil trait with other fatty acid modifications. J Am Oil Chem Soc. 2018;95:39–49. doi: 10.1002/aocs.12025. [DOI] [Google Scholar]
- Bramley PM, Elmadfa I, Kafatos A, Kelly FJ, Manios Y, Roxborough HE, Schuch W, Sheehy PJA, Wagner K-H. Vitamin E. J Sci Food Agric. 2000;80:913–938. doi: 10.1002/(sici)1097-0010(20000515)80:7<913::Aid-jsfa600>3.0.Co;2-3. [DOI] [Google Scholar]
- Britz SJ, Kremer DF, Kenworthy WJ. Tocopherols in soybean seeds: genetic variation and environmental effects in field-grown crops. J Am Oil Chem Soc. 2008;85:931–936. doi: 10.1007/s11746-008-1286-y. [DOI] [Google Scholar]
- Buring JE, Hennekens CH. Antioxidant vitamins and cardiovascular disease. Nutr Rev. 1997;55:S53–S58. doi: 10.1111/j.1753-4887.1997.tb06103.x. [DOI] [PubMed] [Google Scholar]
- Buss G, Camper JHM, Roane C. Registration of ‘Hutcheson’ soybean. Crop Sci. 1988;28:1024–1025. doi: 10.2135/cropsci1988.0011183X002800060042x. [DOI] [Google Scholar]
- Carrera C, Martínez MJ, Dardanelli J, Balzarini M. Environmental variation and correlation of seed components in nontransgenic soybeans: protein, oil, unsaturated fatty acids, tocopherols, and isoflavones. Crop Sci. 2011;51:800–809. doi: 10.2135/cropsci2010.06.0314. [DOI] [Google Scholar]
- Clemente TE, Cahoon EB. Soybean oil: genetic approaches for modification of functionality and total content. Plant Physiol. 2009;151:1030–1040. doi: 10.1104/pp.109.146282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirinck P, De Winne A, Casteels M, Frigg M. Studies on vitamin E and meat quality. 1. Effect of feeding high vitamin E levels on time-related pork quality. J Agric Food Chem. 1996;44:65–68. doi: 10.1021/jf940607x. [DOI] [Google Scholar]
- Dwiyanti MS, Yamada T, Sato M, Abe J, Kitamura K. Genetic variation of γ-tocopherol methyltransferase gene contributes to elevated α-tocopherol content in soybean seeds. BMC Plant Biol. 2011;11:152. doi: 10.1186/1471-2229-11-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA (2003) Food labeling: trans fatty acids in nutrition labeling, nutrient content claims, and health claims. Federal Register Rule Number 68 FR 41433 Session Number 21 CFR Part 101, 68:41433–41506 [PubMed]
- FDA (2015) Final determination regarding partially hydrogenated oils. Federal Register Rule Number 80 FR 34650, 80:34650–34670
- Fehr WR. Breeding for modified fatty acid composition in soybean. Crop Sci. 2007;47:S-72–S-87. doi: 10.2135/cropsci2007.04.0004IPBS. [DOI] [Google Scholar]
- Fryer MJ. The antioxidant effects of thylakoid vitamin E (α-tocopherol) Plant Cell Environ. 1992;15:381–392. doi: 10.1111/j.1365-3040.1992.tb00988.x. [DOI] [Google Scholar]
- Garcia I, Rodgers M, Lenne C, Rolland A, Sailland A, Matringe M. Subcellular localization and purification of a p-hydroxyphenylpyruvate dioxygenase from cultured carrot cells and characterization of the corresponding cDNA. Biochem J. 1997;325:761–769. doi: 10.1042/bj3250761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusak MA, DellaPenna D. Improving the nutrient composition of plants to enhance human nutrition and health. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:133–161. doi: 10.1146/annurev.arplant.50.1.133. [DOI] [PubMed] [Google Scholar]
- Hass CG, Tang S, Leonard S, Traber MG, Miller JF, Knapp SJ. Three non-allelic epistatically interacting methyltransferase mutations produce novel tocopherol (vitamin E) profiles in sunflower. Theor Appl Genet. 2006;113:767–782. doi: 10.1007/s00122-006-0320-4. [DOI] [PubMed] [Google Scholar]
- Hunter SC, Cahoon EB. Enhancing vitamin E in oilseeds: unraveling tocopherol and tocotrienol biosynthesis. Lipids. 2007;42:97–108. doi: 10.1007/s11745-007-3028-6. [DOI] [PubMed] [Google Scholar]
- Keller Y, Bouvier F, d'Harlingue A, Camara B. Metabolic compartmentation of plastid prenyllipid biosynthesis--evidence for the involvement of a multifunctional geranylgeranyl reductase. Eur J Biochem. 1998;251:413–417. doi: 10.1046/j.1432-1327.1998.2510413.x. [DOI] [PubMed] [Google Scholar]
- Konda AR, Nazarenus TJ, Nguyen H, Yang J, Gelli M, Swenson S, Shipp JM, Schmidt MA, Cahoon RE, Ciftci ON, Zhang C, Clemente TE, Cahoon EB. Metabolic engineering of soybean seeds for enhanced vitamin E tocochromanol content and effects on oil antioxidant properties in polyunsaturated fatty acid-rich germplasm. Metab Eng. 2020;57:63–73. doi: 10.1016/j.ymben.2019.10.005. [DOI] [PubMed] [Google Scholar]
- Kumari K, Rai MP, Bansal N, Rama Prashat G, Kumari S, Krishnan V, Srivathsa R, Dahuja A, Sachdev A, Praveen S, Vinutha T. Analysis of γ-Tocopherol methyl transferase3 promoter activity and study of methylation patterns of the promoter and its gene body. Plant Physiol Biochem. 2019;144:375–385. doi: 10.1016/j.plaphy.2019.09.044. [DOI] [PubMed] [Google Scholar]
- Langewisch T, Zhang H, Vincent R, Joshi T, Xu D, Bilyeu K. Major soybean maturity gene haplotypes revealed by SNPViz analysis of 72 sequenced soybean genomes. PLoS One. 2014;9:e94150. doi: 10.1371/journal.pone.0094150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langewisch T, Lenis J, Jiang G-L, Wang D, Pantalone V, Bilyeu K. The development and use of a molecular model for soybean maturity groups. BMC Plant Biol. 2017;17:91. doi: 10.1186/s12870-017-1040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maras JE, Bermudez OI, Qiao N, Bakun PJ, Boody-Alter EL, Tucker KL. Intake of alpha-tocopherol is limited among US adults. J Am Diet Assoc. 2004;104:567–575. doi: 10.1016/j.jada.2004.01.004. [DOI] [PubMed] [Google Scholar]
- McCord KL, Fehr WR, Wang T, Welke GA, Cianzio SR, Schnebly SR. Tocopherol content of soybean lines with reduced linolenate in the seed oil. Crop Sci. 2004;44:772–776. doi: 10.2135/cropsci2004.7720. [DOI] [Google Scholar]
- Morrissey PA, Buckley DJ, Sheehy PJ, Monahan FJ. Vitamin E and meat quality. Proc Nutr Soc. 1994;53:289–295. doi: 10.1079/pns19940034. [DOI] [PubMed] [Google Scholar]
- Munné-Bosch S, Alegre L. The function of tocopherols and tocotrienols in plants. Crit Rev Plant Sci. 2002;21:31–57. doi: 10.1080/0735-260291044179. [DOI] [Google Scholar]
- Norris SR, Barrette TR, DellaPenna D. Genetic dissection of carotenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell. 1995;7:2139–2149. doi: 10.1105/tpc.7.12.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris SR, Shen X, DellaPenna D. Complementation of the Arabidopsis pds1 mutation with the gene encoding p-hydroxyphenylpyruvate dioxygenase. Plant Physiol. 1998;117:1317–1323. doi: 10.1104/pp.117.4.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham A-T, Lee J-D, Shannon JG, Bilyeu K. Mutant alleles of FAD2-1A and FAD2-1B combine to produce soybeans with the high oleic acid seed oil trait. BMC Plant Biol. 2010;10:195. doi: 10.1186/1471-2229-10-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham A-T, Lee J-D, Shannon J, Bilyeu K. A novel FAD2-1A allele in a soybean plant introduction offers an alternate means to produce soybean seed oil with 85% oleic acid content. TAG Theor Appl Genet. 2011;123:793–802. doi: 10.1007/s00122-011-1627-3. [DOI] [PubMed] [Google Scholar]
- Pham AT, Shannon JG, Bilyeu KD. Combinations of mutant FAD2 and FAD3 genes to produce high oleic acid and low linolenic acid soybean oil. Theor Appl Genet. 2012;125:503–515. doi: 10.1007/s00122-012-1849-z. [DOI] [PubMed] [Google Scholar]
- Sattler SE, Gilliland LU, Magallanes-Lundback M, Pollard M, DellaPenna D. Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell. 2004;16:1419–1432. doi: 10.1105/tpc.021360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. Chemistry and biology of vitamin E. Mol Nutr Food Res. 2005;49:7–30. doi: 10.1002/mnfr.200400049. [DOI] [PubMed] [Google Scholar]
- SoyStats (2020) SoyStats: A reference guide to important soybean facts and figures. The American Soybean Association. http://soystats.com/. Accessed (1/14/2020)
- Subramaniam SS, Slater SC, Karberg K, Chen R, Valentin HE, Wong Y-HH (2008) Nucleic acid sequences to proteins involved in tocopherol synthesis. USA Patent 7,420,101 Patent publication date September 2, 2008
- Ujiie A, Yamada T, Fujimoto K, Endo Y, Kitamura K. Identification of soybean varieties with high alpha-Tocopherol content. Breed Sci. 2005;55:123–125. doi: 10.1270/jsbbs.55.123. [DOI] [Google Scholar]
- Valentin HE, Qi Q. Biotechnological production and application of vitamin E: current state and prospects. Appl Microbiol Biotechnol. 2005;68:436–444. doi: 10.1007/s00253-005-0017-7. [DOI] [PubMed] [Google Scholar]
- Valliyodan B, Dan Q, Patil G, et al. Landscape of genomic diversity and trait discovery in soybean. Sci Rep. 2016;6:23598. doi: 10.1038/srep23598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valliyodan B, Brown AV, Cannon SB, Nguye, N H (2020) Data from: Genetic variation among 481 diverse soybean accessions. Ag Data Commons. doi:10.15482/USDA.ADC/1518301 [DOI] [PMC free article] [PubMed]
- Van Eenennaam AL, Lincoln K, Durrett TP, Valentin HE, Shewmaker CK, Thorne GM, Jiang J, Baszis SR, Levering CK, Aasen ED, Hao M, Stein JC, Norris SR, Last RL. Engineering vitamin E content: from Arabidopsis mutant to soy oil. Plant Cell. 2003;15:3007–3019. doi: 10.1105/tpc.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Xia Z, Hideshima R, Tsubokura Y, Sato S, Yamanaka N, Takahashi R, Anai T, Tabata S, Kitamura K, Harada K. A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics. 2011;188:395–407. doi: 10.1534/genetics.110.125062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Jiang Y, Wang Z, Gou Z, Lyu J, Li W, Yu Y, Shu L, Zhao Y, Ma Y, Fang C, Shen Y, Liu T, Li C, Li Q, Wu M, Wang M, Wu Y, Dong Y, Wan W, Wang X, Ding Z, Gao Y, Xiang H, Zhu B, Lee SH, Wang W, Tian Z. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat Biotechnol. 2015;33:408–414. doi: 10.1038/nbt.3096. [DOI] [PubMed] [Google Scholar]