Abstract
Cardiomyocyte differentiation continues throughout murine gestation and into the postnatal period, driven by temporally regulated expression changes in the transcriptome. The mechanisms that regulate these developmental changes remain incompletely defined. Here we used cardiomyocyte specific ChIP-seq of the activate enhancer marker P300 to identify 54,920 cardiomyocyte enhancers at seven stages of murine heart development. These data were matched to cardiomyocyte gene expression profiles at the same stages and Hi-C and H3K27ac HiChIP chromatin conformation data at fetal, neonatal, and adult stages. Regions with dynamic P300 occupancy exhibited developmentally regulated enhancer activity, as measured by massively parallel reporter assays in cardiomyocytes in vivo, and identified key transcription factor binding motifs. These dynamic enhancers interacted with temporal changes of 3D genome architecture to specify developmentally regulated cardiomyocyte gene expression. Our work provides a 3D genome-mediated enhancer activity landscape of murine cardiomyocyte development.
Keywords: Enhancer, Massively parallel reporter assay, Nuclear receptor, Hi-C, HiChIP, Cardiomyocyte maturation
Graphical Abstract

Introduction
Coordinated changes in gene expression, governed by epigenetic mechanisms including the transcriptional regulation by enhancers and three dimensional (3D) chromatin1,2, drive cell differentiation. The developmental roles of enhancer activation and 3D genome organization have been studied extensively in early embryogenesis and lineage specification from pluripotent stem cells3,4. However, less is known about later stages of embryonic and postnatal development, when cell lineages continue to differentiate to their mature state. Indeed, few time courses of 3D genome organization during fetal-to-adult maturation have been reported.
Cardiomyocytes (CMs) are specified from mesoderm between embryonic day (E)7.5-E10.5 of murine development and continue to differentiate throughout gestation. A dramatic change in CM phenotype and transcriptome, CM maturation, occurs during the first postnatal week. CMs largely withdraw from the cell-cycle, become reliant on oxidative phosphorylation, and switch expression of several sarcomere gene isoforms5. Among identified transcriptional regulators of CM maturation are SRF-MRTF6,7, thyroid hormone receptors 8 and peroxisome proliferator-activated receptor gamma coactivator 1 (Ppargc1a).9 The enhancer network that governs CM differentiation and maturation remains incompletely defined. Moreover, studies of developmental changes in CM 3D genome organization have been limited to differentiating pluripotent stem cells (PSC)10,11, which fail to mature5,12. As a result, we lack an understanding of later developmental changes in the CM 3D genome and their contributions to maturation.
Dissection of gene regulatory mechanisms in developing tissues faces several challenges. One is the diversity of cell types in mammalian tissues and their changing distribution during development. For instance, the fraction of CMs in the heart decreases throughout development, dropping from ~80% at E11.513 to ~30–35% in adult murine hearts14. Prior studies using chromatin immunoprecipitation followed by next-generation sequencing (ChIP-seq) of transcription factors (TFs)15 or active enhancer markers H3K27ac16 and P30017 successfully identified cardiac enhancers, but did not account for cellular heterogeneity and the changing frequency of CMs in developing hearts. High throughput measurement of the enhancer activity of enhancers identified by their chromatin features is another challenge. Transient transgenesis is the gold standard approach, but is slow and resource intensive. As a result, less than 200 active murine cardiac enhancers have been identified by this method16,18, and their activity changes over time have not been systematically measured.
Here we established a CM 4D nucleome atlas of CM maturation that encompasses CM-targeted measurement of gene expression, P300 chromatin occupancy, in vivo enhancer activity, and 3D genome architecture between E12.5 through adulthood. Our integrative analysis identified mechanisms that regulate CM gene expression through interactions between dynamic enhancers and 3D chromatin contacts.
Results
The cardiomyocyte enhancer landscape during mouse development
We used a lineage-specific P300 bioChIP-seq system (Fig. 1A) that enables highly sensitive and lineage-targeted identification of active enhancers from tissues and avoids effects of cell dissociation on chromatin state. We tagged endogenous P300, a transcriptional coactivator and a marker of active enhancers17, with C-terminal FLAG and BIO tags (P300flbio)19. The BIO sequence was selectively biotinylated by BirA20, a biotin ligase whose expression from a conditional allele (Rosa26fsBirA; fs=flox-stop) was triggered by CM-specific Myh6-Cre19. Heart expression of BirA and P300 biotinylation required Myh6-Cre (Fig. 1B). We then compared heart bioChIP-seq data obtained with widespread expression of BirA (Pan_P300: germline Cre activation 19), CM selective expression (CM_P300: Myh6-Cre), and endothelium selective expression (EC_P300: CHD5-CreERT219) (Fig. S1A–B). Replicate experiments confirmed the high reproducibility of P300 bioChIP-seq and the differences in P300 chromatin occupancy between CM_P300, EC_P300, and Pan_P300 (Fig. S1C–E). CM_P300 but not EC_P300 demonstrated strong P300 signal flanking CM-specific genes Myl2, Ryr2, and Scn5a (Fig. S1D), whereas EC_P300 but not CM_P300 showed strong signal flanking endothelial cell specific genes Egfl7, Notch1, and S1pr1 (Fig. S1E). Pan_P300 contained both CM and EC P300 signals (Fig. S1D–E).
Figure. 1. Identification of CM P300 enhancers. See related Figure S1.
A. Cre-dependent expression of BirA enabled CM-selective biotinylation of P300. The BIO tag was knocked into the C-terminus of P300.
B. P300 biotinylation was triggered by CM Cre expression. Input: heart extracts. SA, streptavidin.
C. P300 data acquisition.
D. Spearman correlation of P300 signal across the union of P300 regions.
E. Representative images of hearts used in this study, shown at the same scale. Ventricular tissue (below the dashed lines) was used.
F. P300 bioChIP-seq signal at Bmp10, Ppargc1a, and Myh6/Myh7.
G. CM p300 bioChIP-seq test characteristics based on the Vista Enhancer Database. CM, E12.5 and E16.5 ventricular CMs. Heart, E12.5 ventricular tissue. FB, E12.5 forebrain.
H. Heart TF bioChIP-seq signals15 at CM p300 regions. Average of biological duplicates.
We targeted bioChIP-seq to CMs using either Tnnt2-Cre21 (embryonic stages) or Myh6-Cre (postnatal stages) and measured P300 chromatin occupancy at seven developmental stages (E12.5, E16.5, P0, P7, P14, P28, P42; P=postnatal day; Fig. 1C–E and Fig. S1F–G; Data S1). Heart ventricles were sampled in biological duplicate (Fig. S1H), which were highly correlated with one another (Fig. 1D). 54,920 P300 enriched regions were reproducible between replicates (see Methods; Data S1). The majority of P300 regions were distal (>1 kb) to transcriptional start sites (TSSs; Fig. S1H), consistent with prior studies17,19. Occupancy adjacent to developmentally regulated genes dynamically changed during development. For example, P300 occupancy of regions flanking Bmp10 (downregulated), Ppargc1a (upregulated), and Myh6-Myh7 (down- and up-regulated, respectively) genes coincided with their temporal expression patterns (Fig. 1F).
To assess cardiac enhancer detection by CM selective P300 bioChIPseq, we used the Vista enhancer database18, which houses the results of thousands of regions tested for enhancer activity using the transient transgenic assay. Comparison groups were E12.5 heart ventricle Pan_P30019, heart ventricle H3K27ac19, and forebrain P300 bioChIPseq19. Of 1372 murine regions in Vistas, 175 had heart activity. CM p300 bioChIP-seq identified 60% of these (Fig. S1I) with high (91%) specificity (fraction of regions without heart activity that lacked P300 occupancy; Fig. 1G). Pan_P300 and H3K27ac had greater sensitivity (fraction of regions with heart activity that had P300 occupancy; 91% and 90%) and reduced specificity (67% and 63%). As expected, forebrain P300 regions had lower sensitivity (18%) and specificity (60%) for heart enhancer activity. Vista heart enhancers bound by P300 in CMs were highly enriched for GO terms related to heart and cardiac muscle development (Fig. S1J), whereas without P300 in CMs were related to blood vessel development (Fig. S1K). Activity in non-CMs scored as heart enhancer activity in the Vista database likely depressed the calculated sensitivity of CM-selective p300 bioChIP-seq.
P300 occupies genomic DNA indirectly through interactions with DNA-binding TFs, including cardiac transcriptional regulators GATA4, MEF2A, MEF2C, TEAD1, TBX5, SRF, and NKX2–522–26. Fetal and adult genome-wide occupancy data for these TFs15 showed that each TF is highly enriched at E12.5 and adult CM_P300 regions (Fig. 1H). TBX5 was among the most highly enriched at E12.5 but was the least enriched in adult samples, consistent with TBX5 downregulation in adult heart ventricles.
Dynamic landscape of active enhancers reveals key transcription factors that regulate CM differentiation
We focused on P300 regions with dynamic occupancy across the developmental time course. Filtering out low-signal peaks and peaks not found in at least two time points left 43,063 regions, which we ranked by the ratio of each region’s strongest to weakest signal across the time course. The quartiles with the greatest and smallest temporal variation in signal were defined as “Dynamic” and “Static” P300 Regions, respectively (Fig. 2A; Data S1). After exclusion of regions within 1 kb of TSSs, K-means clustering of the dynamic and static regions yielded Early (3960 dynamic regions), Late (6444 dynamic regions), and Static (7050 static regions; Fig. 2B) regions. Most Gene Ontology (GO) terms linked to Early and Late regions did not overlap (Fig. 2C). Among these were “regulation of growth factor stimulus” and “regulation of Wnt receptor signaling”, associated with early regions; “regulation of myeloid cell differentiation” and “response to insulin stimulus”, enriched in late regions; and “RNA processing” and “mitochondrion organization”, enriched in static regions.
Figure 2. Characterization of P300 dynamic enhancers during heart development. See related Figure S1.
A. Flowchart for identification of dynamic distal P300 regions.
B. P300 signal in Early, Static, and Late regions.
C. Top 5 GO terms enriched among Early, Static, and Late P300 regions.
D. Enriched TF motifs in P300-bound regions. OR, odds ratio. NR, nuclear receptor.
E. Cardiac transcription factor (TF) bioChIP-seq signal15 at P300 peaks. Average of biological duplicates.
F. THRA ChIP-seq signal in P15 hearts (Hirose et al., 2019) centered at P300 peaks.
G. Phastcons scores at Early, Static, and Late P300 regions.
To identify TFs that recruit P300 in the three region classes, we scanned for TF motifs enrichment. Motifs of cardiac TFs TEAD1, GATA4, and MEF2 were enriched in all three classes of P300 regions (Fig. 2D). The TBX5 motif was enriched in Early but not Late or Static regions, consistent with TBX5 downregulation and its reduced binding signal in adult P300 regions (Fig. 1H). Among the motifs enriched in Late P300 regions were several nuclear receptors (NRs), such as thyroid hormone receptor A (THRA) and the glucocorticoid receptor, which are known to regulate CM maturation in vivo and in iPSC-derived CMs8,27,28.
Cardiac TF occupancy signals were concordant with P300 occupancy at dynamic and static P300 regions. Early and Late P300 regions had strong TF occupancy at the congruent developmental stage. Static regions showed strong TF occupancy at both stages (Fig. 2E). Again TBX5 showed the most dynamic changes between fetal and adult heart and between Early and Late P300 regions. We also examined THRA occupancy of these regions using a P15 cardiac THRA occupancy data8. THRA occupancy was strongest in Late P300 regions as well as Static P300 regions and weak at Early P300 regions (Fig. 2F).
Evolutionary conservation of enhancers is indicative of biological function that is essential for reproductive fitness. Early, Late, and Static P300 regions were all strongly conserved (Fig. 2G). The aggregate conservation scores were substantially higher than those of the set of heart enhancers marked by H3K27ac and slightly higher than those of heart enhancers bound by multiple cardiac TFs15.
High throughput validation of enhancer activity during in vivo murine cardiomyocyte development
To test if dynamic P300 regions show developmentally regulated enhancer activity, we used adeno-associated virus serotype 9 (AAV)-based MPRA to measure their in vivo activity in CMs. In heart, AAV9 selectively transduces CMs. We designed an MPRA vector in which a minimal promoter is placed upstream of an mCherry reporter (Fig. 3A and S2A). In pilot studies, enhancer-reporter AAV containing one of three Early P300 regions located −5, +14, and +20 kb from the Bmp10 TSS (Fig. 1F) was delivered at E15.5 or P0 (Fig. 3A–B). Poor AAV9 transduction prior to E15.5 made earlier developmental stages experimentally inaccessible. Ventricular mCherry expression driven by each enhancer was present at P0 and weaker at P7 and P28 (Fig. 3C). Quantitative PCR comparing reporter RNA to DNA confirmed this temporal pattern (Fig. 3D–E).
Figure. 3. Enhancer activity of dynamic P300 regions during in vivo CM development. See related Figure S2.
A. AAV vector for testing cardiac enhancer activity.
B. Timeline for MPRA experiments.
C. Representative images of enhancer activity of three Early P300 regions flanking Bmp10 (see Fig. 1F). Bar = 1 mm.
D-E. Quantification of individual enhancer activity. Activity of −5k, +14k and +20k Bmp10 enhancers was normalized to vector lacking enhancer. t-test: **, P<0.01.
F. Design of MPRA to measure Early P300 and Late P300 region activity. Synthesized 400 bp regionswere cloned into the 3’ UTR of a basal promoter-mCherry AAV reporter.
G. MPRA results. Lower line plot indicates the annotation assigned to each region.
H. MPRA activity of Early P300 and Late P300 regions at P0, P7, and P28. Early P300 group, 685 regions; Late P300 group, 1231 regions. n=12 biological replicates at P0; n=28 at P7 and P28. Steel-Dwass. ns, not significant.
I. Activity of three selected Late P300 regions with increasing MPRA activity. Left, P300 bioChIP-signal. Middle, representative images. Right, quantification. t-test: **, P<0.01. Bar = 1 mm.
J-K. Motifs enriched in regions with selective neonatal or adult MPRA activity. Non-redundant motifs with significant enrichment (Neg. log10 Pval > 10) are shown.
Having validated that AAV enhancer-reporters capture dynamic enhancer activity in CMs in vivo, we performed an AAV-MPRA to determine if dynamic P300 binding predicts dynamic enhancer activity. Using pooled oligonucleotide synthesis, we generated a library of 400 bp regions centered on 685 Early and 1231 Late P300 regions (Fig. 3F). This library also contained static regions, regions bound by multiple transcription factors15, P300 regions adjacent to strongly expressed cardiac genes, positive control regions previously validated by AAV-MPRA15 or transient transgenesis18, and negative control regions comprised of brain or embryonic stem cell P300 regions, random intergenic regions, and regions without activity in transgenic assays18. CM P300 signal was strongest in positive controls, weakest in negative controls, and intermediate in the dynamically regulated P300 regions (Fig. S2B). The enhancer library was cloned into the mCherry reporter’s 3’ untranslated region, so that the enhancer drives its own transcription (Fig. 3F) to enable sequencing-based enhancer activity measurement29. The pooled AAV library was delivered to E15.5 embryos or newborn mice, and ventricles were collected at P0, P7, and P28, with 12 to 28 replicates per time point. Although the MPRA assay was performed with variable time between library delivery and enhancer activity measurement, for intervals of 4 days or longer these changes did not significantly alter activity measurements (Fig. S2C–F). Amplicons containing the test regions were quantified by sequencing heart RNA and AAV DNA. Regions with low coverage in the AAV DNA library were excluded (Fig. S2G). Correlation between remaining regions was high across the three time points (Fig. S2H).
Regions were ranked by enhancer activity, the ratio of reads from RNA to AAV DNA (Fig. 3G and Fig. S2I; Data S2). Positive and negative control regions had high and low enhancer activity, respectively (Fig. 3G and S2I). The 95th percentile of the negative control regions defined the 5% false discovery threshold. Among the 685 Early P300 regions tested, 16% (110) were active at P0, 14% (94) at P7, and 8% (57) at P28. Among the 1231 Late P300 regions, 9% (116), 15% (181), and 18% (220) were active at P0, P7, and P28, respectively. The relatively low frequency of P300 regions with activity in this assay suggests that it is relatively less sensitive than transient transgenesis, potentially due to the smaller size of tested elements, and the use of dynamic regions, which tended to have lower P300 signal. We then analyzed the dynamic activity of the subset of regions with activity in at least one time point. Early P300 regions demonstrated significantly reduced enhancer activity at P28 compared to P0 and P7, and Late P300 regions showed significantly elevated enhancer activity at P7 and P28 compared to P0 (Fig. 3H; Fig. S2J). Three Late P300 regions with dynamic enhancer activity in the MPRA assay were individually validated in vivo using AAV enhancer-reporters. Fluorescent imaging and quantitative PCR confirmed that these regions gain enhancer activity between P0 and P28 (Fig. 3I).
We performed motif analysis to define TFs that contribute to temporal regulation of these experimentally validated dynamic regions. Neonatal and Adult regions with the strongest change in enhancer activity (“Neonatal”=Log2 fold-change < −1 (n=554); “Adult”=Log2 fold-change > 1 (n=347); Fig. 3J) were strongly enriched for MEF2, GATA, and TEAD motifs when compared to random background (Fig. 3K). To highlight motifs with strong differential enrichment between Neonatal and Adult MPRA regions, we compared Neonatal foreground to Adult background and vice versa. This demonstrated the strong enrichment of TBX (T-box) and HAND (bHLH) motifs in Neonatal regions, and NR motifs of THRA and ESRRA in Adult regions (Fig. 3K). These data are consistent with the important roles of T-box and HAND factors in heart development 30, and THRA27 and estrogen-related receptor31 for CM maturation.
Nuclear receptor motifs are required for postnatal cardiomyocyte enhancer activation
To begin to assess the contribution of nuclear receptor (NR) motifs to the activity of Late P300 regions, we mutated a NR binding motif in one of the individually validated Late P300 regions (Fig. 3I). The NR motif mutation prevented developmental enhancer activation, resulting in an activity decrease of 50% at P7 and 70% at P28 (Fig. 4A–B).
Fig. 4. The nuclear receptor motif is essential for activation of Late P300 regions. See related Figure S3.
A. Cardiac enhancer activity of NR motif-containing Late P300 regions was measured with or without NR motif mutation.
B. Representative mCherry reporter images and enhancer activity quantification. Bar = 1 mm. t-test: **, P<0.01.
C. MPRA to probe the requirement for nuclear receptor (NR) motifs. NR motif-containing P300 regions were synthesized as wild-type and mutant pairs and inserted into the AAV-MPRA vector.
D. MPRA result measured at P7. RNA/DNA ratio was normalized to the mean value of the negative control group. Activity threshold was set at the negative control 95th percentile. Line plot indicates the annotation assigned to each region.
E. Effect of NR mutation on 337 WT:Mut pairs with detectable MPRA activity at P7. Paired t-test. Marker size indicates thyroid hormone receptor (THRA) ChIP-seq signal in P15 heart (GSE125414). n=14 biological replicates.
F-H. Activity of Late P300 region near Mhrt promoter requires thyroid response element (TRE). F, THRA- and P300-bound TRE within the Myh6/Myh7 locus. G, P300 enhancer activity, from Fig. 3 MPRA. n=12. G'; Enhancer inhibition by TRE mutation, from panel D MPRA. n=14. H, Myh7 silencing, measured by YFP FACS in P28 Myh7YFP CMs with or without somatic mutagenesis of endogenous TRE.
To more systematically validate the importance of the NR motif in Late P300 region activation, we generated an AAV-MPRA library containing WT-Mut pairs of 120 Early regions, 2705 Late P300 regions, and a subset of positive and negative control regions (Fig. 4C). Pooled AAV library was administered at P0 and analyzed at P7 by amplicon sequencing. We excluded regions with low coverage in AAV library DNA (Fig. S3A) and then calculated enhancer activity by the ratio of read counts in RNA versus DNA amplicons. Replicates correlated well (Fig. S3B). Of the 2825 tested dynamic regions, 11.9% (337) had detectable activity in a member of the WT-Mut pair (activity greater than 95% of the negative controls; Fig. 4D; Data S3). This low activity frequency likely reflects the small size (190 bp) of tested regions and their centering on the NR motif. Of the pairs with detectable activity, enhancer activity significantly differed between WT and Mut in 86 pairs (Paired t-test adj. P<0.05 and absolute fold-change>2; Fig. 4E, Data S3). Notably, 97.7% of these were down-regulated by NR motif mutation, indicating that in the large majority of cases these NR motifs promote enhancer activation.
THRA is a NR that is a well established regulator of CM maturation, including the perinatal switch of myosin from Myh7 to Myh68. Overlaying P15 heart THRA occupancy8 and NR mutagenesis MPRA datasets demonstrated that many of the essential NR motifs are bound by THRA in vivo (Fig. 4E). At the Myh6/Myh7 locus, a Late P300 enhancer containing a THRA-bound thyroid hormone response element (TRE) falls in the intergenic region, close the promoter for Mhrt, a long non-coding RNA antisense to Myh7 that contributes to Myh7 repression in adult CMs32. The MPRA data demonstrated that postnatal activation of this enhancer required the TRE (Fig. 4G). Using Cas9 and AAV-mediated somatic mutagenesis33, we ablated the TRE in the native locus at birth. TRE mutagenesis, largely confined to the TRE motif (Fig. S3C), resulted in persistent Myh7 expression, as reported by YFP fluorescence from an Myh7YFP knockin allele (Fig. 4H; Fig. S3D).
Dynamic P300 enhancers are linked to developmental changes in 3D genome architecture
The contribution of 3D chromatin structure to gene regulation during later stages of development has been little studied. We performed Hi-C34 on E12.5, P0, and P42 murine ventricular CMs (Fig. 5A). For each of three replicates for each stage, we obtained more than 500M reads, allowing intra-chromosomal (cis) interactions to be assessed with 5 kb resolution (Fig. S4A). The biological replicates were highly reproducible and intragroup variation was much smaller than variation between time points (Fig. S4B). The reads from each time point were pooled for subsequent analyses. A representative contact map illustrates two developmentally regulated chromatin loops that connect the Synpo2 TSS to dynamic P300 regions within an adjacent gene (Fig. 5B).
Figure 5. Correlation of dynamic P300 binding to chromatin conformation. see also Related Figures S4 and S5.
A. CM samples used to generate HiC data. Scale bar: 1 mm.
B. Representative Hi-C contact map and the corresponding P300 bioChIP-seq tracks.
C. Interaction score of loops between Early (n=104) or Late (n=141) P300 Regions and TSS. Wilcoxon ranked sum test.
D. Percentage of genome in compartments that were stable (90.2%), A→B (4.3%), or B→A (5.5%), between E12.5 and P42.
E. Distribution of P300 regions in stable and switching compartments.
F. Representative region (dashed box, enlarged in tracks below) that underwent B→A switch and the corresponding CM P300 bioChIP-seq tracks.
G. Total number of TAD splits or mergers between E12.5, P0, and P42.
H. Representative example of a TAD split and P300 bioChIP signal in daughter TADs.
I. P300 signal between daughters of split TADs at E12.5 (pre-split) and P42 (post-split). n=314.
We detected 7,991, 8,332, and 9,747 significant loops at E12.5, P0, and P42, respectively (Fig. S4C; Data S4), a comparable number to an independent study of adult mouse CMs35. 40% of loops were shared between all time points, whereas 20% and 40% were found in two or only one time point, respectively (Fig. S4D). Loops were overrepresented for P300 regions (hypergeometric test: P < 1.4E-196). Late P300 regions had stronger loop to promoters in adults compared to fetal CMs (Fig. 5C). However, contact scores of Early P300 region loops to promoters did not significantly differ between stages, despite their difference in P300 occupancy (Fig. 5C).
Based on Hi-C data, the genome can be categorized into active (A) and repressive (B) compartments34. 90.2% of the genome remained in the same compartment at both stages. 4.3% switched from A to B (“A→B chromatin”) and 5.5% from B to A (“B→A chromatin”; Fig. 5D and Fig. S5A; Data S5). Consistent with P300 marking active enhancers, A→B chromatin had 3.1-fold more Early than Late P300 regions and the greatest Early P300 region density (Fig. 5E and Fig. S5B). In contrast, B→A chromatin had 2.4-fold more Late than Early P300 regions and the highest Late P300 region density (Fig. 5E and Fig. S5B). B→A chromatin switching occurred both by spreading at the edge of a pre-existing A domain (Fig. S5C) and by generating A domains de novo inside of B domains (Fig. 5F). Late P300 regions were enriched in both types of B→A switching. P300 deposits H3K27ac, which modulates chromatin folding36. Thus, the correlation between dynamic P300 occupancy and chromatin compartment transitions suggest that P300 binding is a driving force behind chromatin domain switching during CM maturation.
Hi-C data can also be used to identify self-interacting chromatin regions named topologically associating domains (TADs). TADs can “split” or “merge” through strengthening or weakening of boundaries. We utilized our TADsplimer algorithm37 to detect TAD splits and mergers between time points. We detected about 7000 TADs at each time point (Fig. S5D). Consistent with prior studes37, only ~4% of TADs split or merged between developmental stages. The vast majority were TAD splits, and most occurred during postnatal CM maturation (Fig. 5G and Fig. S5E). A representative TAD split demonstrated a predominance of Late P300 regions on one side of the split TAD (Fig 5H). To evaluate whether this is a common feature of split TADs, we compared the ratio of P300 signal on each side of the split TAD at E12.5 (pre-split) and P42 (post-split). There was a greater difference between P300 signal on each side of the TAD post-split than pre-split (Fig. 5I). These data suggest that developmental enforcement of TAD boundaries restricts the spatial domain of P300 activation.
3D genome-mediated regulation of cardiomyocyte developmental gene expression
We profiled developmental changes in CM gene expression by sequencing bulk RNA isolated from purified CMs in biological triplicate at the same seven stages from E12.5 to P42 (Fig. S6A; Data S5). Principal component analysis (PCA) showed clustering of replicates from each time point and separation of time points along the first principal component (Fig. S6B). We identified differentially expressed genes (DEGs) between fetal and adult stages (average of P28 and P42 compared to average of E12.5 and E16.5: Padj<0.01 and absolute Log2FC >=3; Fig. 6A). Genes with known developmental regulation showed the expected temporal patterns (Fig. S6C), supporting the RNA-seq data quality. This was further confirmed by GO analysis: the 493 Adult DEGs were associated with fatty acid metabolism, ion transport, and cellular response to hormone stimulus (Fig. 6B), and the 783 Fetal DEGs were related to cell cycle, heart development, and DNA packaging (Fig. 6B), consistent with well established changes during CM maturation5.
Figure. 6. Interaction between developmental changes in P300 occupancy and 3D chromatin structure modulate CM gene expression. Related to Figure S6.
A. Differentially expressed genes (DEGs; Padj<0.01 and absolute Log2FC ≥ 3) in CMs from E12.5 to P42. Hierarchical clustering.
B. Gene ontology terms enriched among Fetal and Adult DEGs.
C. Expression of genes in chromatin that underwent A and B switches between E12.5 and P42.
D. Expression of genes in left and right sides of split TADs at E12.5, P0, and P42. Wilcoxon ranked sum test. n=314.
E. Representative example of TAD splitting and gene expression changes.
F. ABC scores of Fetal (n=235) and Adult (n=265) DEGs with EP loops at E12.5 and P42. Wilcoxon ranked sum test.
G. Correlation between developmental change in gene expression and change in ABC scores. Gene expression values from E12.5 and P42. Contact scores were from (i) Hi-C data, (ii) genome-wide distance-contact score relationships at E12.5 and P42 (see panel H), (iii) stage swapped distance-contact score relationships, and (iv) stage-swapped Hi-C data.
H. Cumulative distribution function of the genome-wide average frequency of contacts between a region and the TSS as a function of their genomic distance. Kolmogorov–Smirnov (KS) test.
We analyzed the effect of 3D genome changes on developmentally regulated CM gene expression. Genes within A→B chromatin and B→A chromatin were downregulated and upregulated, respectively, between E12.5 and P42 (Fig. 6C). Moreover, developmental splitting of TADs, which increased the difference in mean P300 signal between left and right daughter TADs (Fig. 5I), was mirrored by increased difference in mean gene expression between daughter TADs (Fig. 6D). A representative example demonstrates gain in P300 signal on the right side of a TAD split (blue dotted line; Fig. 6E). Several genes selectively expressed in mature CMs, including Myoz2 and Synpo2, are upregulated in this right-sided daughter TAD. Adult DEGs contained within split TADs were enriched for GO terms including metabolic process, heart development, and regulation of heart rate, while Fetal DEGs within split TADs were enriched for terms including cell cycle and cell division (Fig. S6D), suggesting that TAD splitting promotes CM maturation.
Enhancer regulation of a target gene is a function of enhancer strength and contact frequency between the enhancer and the gene’s promoter. The “activity-by-contact” (ABC) model implements this concept as a score based on the product of enhancer strength (A) and enhancer-promoter contact strength (C), summed across the enhancers linked to each gene38. We calculated ABC scores using stage-specific P300 signals and Hi-C contact scores. As expected, ABC scores of Fetal DEGs were higher in fetal CMs, and scores of Adult DEGs were higher in adult CMs (Fig. 6F). We used the ABC score to investigate the contribution of P300 signals and chromatin looping to developmental gene expression genome-wide. Developmental changes in ABC scores and gene expression significantly correlated (r=0.59, p=1.1E-258, Fig. 6G, panel i). In contrast, ABC scores calculated using stage-matched P300 signals and stage-mismatched Hi-C contact scores only correlated weakly with gene expression changes (r=0.20, Fig 6G, panel iv). This analysis highlights the importance of developmental changes in 3D genome organization, in combination with developmental regulation of enhancers, to maturational changes in gene expression.
We used the Hi-C contact matrixes to analyze the relationship between promoter-distal region contact frequency and genomic distance genome wide. This relationship changed between E12.5 and P42, so that adult promoters tended to contact regions that were closer along the primary DNA sequence (Fig. 6H). Consistent with this result, Adult DEGs were closer to the nearest P300 region than Fetal DEGs (Fig. S6E). We evaluated whether this aggregate contact vs distance relationship could substitute for the actual Hi-C contact frequencies in the ABC model. The aggregate contact frequency vs. distance relationship performed nearly as well (r=0.52, Fig. 6G-ii) as the actual contact data (r=0.59, Fig. 6G-i). However, calculating the aggregate contact frequency using the stage mis-matched contact frequency-distance relationship resulted in much lower correlation (r=0.34, Fig 6J-iii), suggesting that the developmental change in the contact frequency-distance relationship is functionally important.
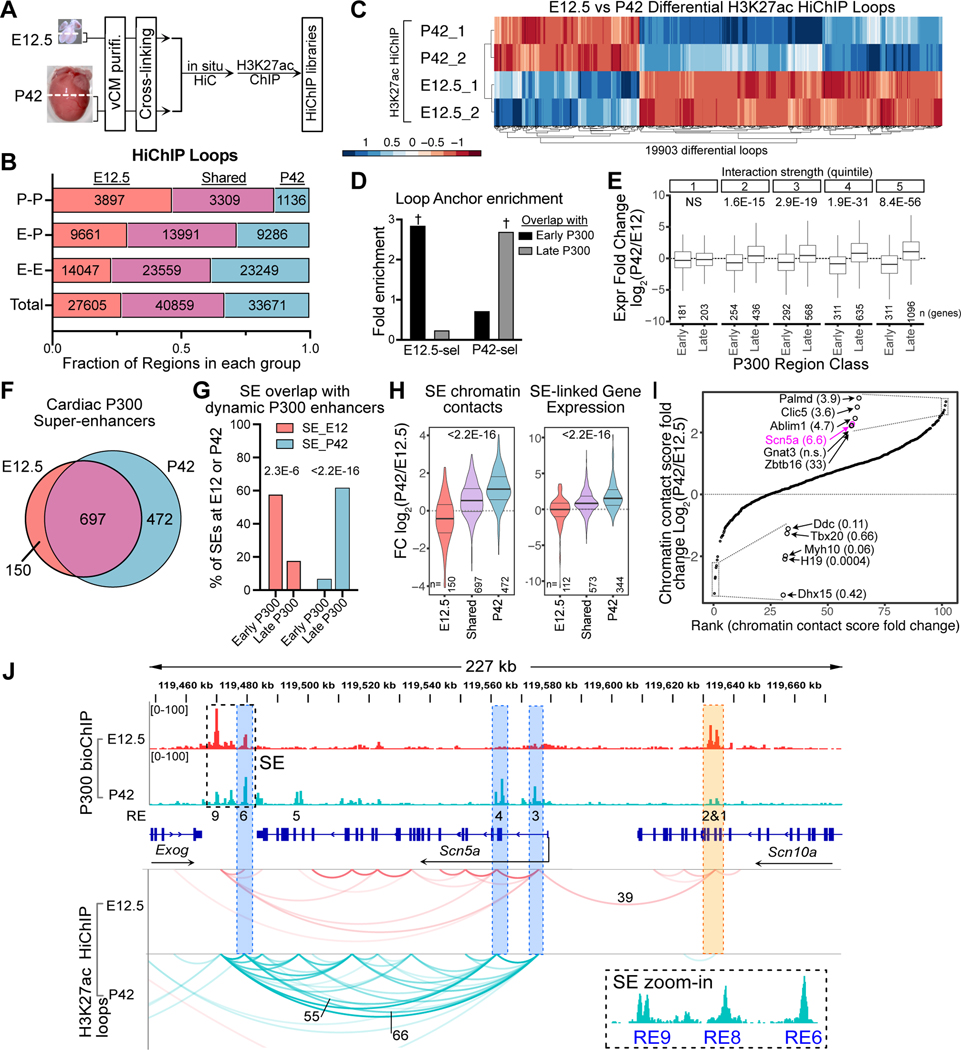
Hi-C provides a global view of the 3D genome but has relatively low sensitivity – the number of called loops (~8700 per time point) greatly underestimates actual chromatin contacts. Therefore we performed H3K27ac HiChIP39 on purified E12.5 and P42 ventricular CMs (Fig. 7A and S7A). Replicates correlated well (Fig. S7B). We identified 68,464 fetal and 74,530 adult reproducible loops (>5 reads in each sample; Table S1). These were categorized as enhancer-enhancer (E-E, 59%), enhancer-promoter (E-P, 33%), and promoter-promoter (P-P, 8%) (Fig. 7B). 27,605 (40.3%) and 33,671 (45.2%) loops were called only at E12.5 or P42, respectively, while 40,859 were shared by both stages (Fig. 7B). Quantitative analysis likewise showed that many loops (19.5%) had significantly different contact scores at E12.5 and P42 (Padj<0.05 and | Log2FC | > 2; Fig. 7C). Genome browser views of Bmp10 and Myh6/Myh7 loci, shown in Fig. S7C–D, illustrate developmentally regulated loops and P300 occupancy. E12.5-selective and P42-selective loops were enriched for Early and Late P300 regions, respectively (Fig. 7D and Fig. S7E). Genes linked to Early and Late P300 regions by strong HiChIP loops were more highly expressed at E12.5 and P42, respectively (Fig. 7E). Conversely, P300 regions with HiChIP loops to fetal or adult DEGs had greater P300 signal at E12.5 and P42, respectively (Fig. S7F).
Fig. 7. Interaction between developmental changes in P300 occupancy, 3D chromatin structure, and gene expression. Related to Figure S7.
A. H3K27ac Hi-ChIP workflow. vCMs, purified ventricular CMs
B. H3K27ac Hi-ChIP loops called at E12.5 and P42. E-E, enhancer-enhancer; E-P, enhancer-promoter; P-P, promoter-promoter.
C. Differential loops (Padj<0.05 and absolute Log2FC>2) between E12.5 and P42.
D. Enrichment of P300 Early or Late enhancers at E12.5- or P42-selective loop anchors, compared to all other loops. †, Fisher P<2.2E-16.
E. Developmental change in expression of genes linked to Early or Late P300 enhancers by E-P loops, categorized by quintile of HiChIP loop strength at E12.5 (Early P300) or P42 (Late P300). Wilcoxon ranked sum test with Holm's multiple testing correction.
F. CM super-enhancers (SEs) identified at E12.5 and P42 by P300 bioChIP-seq signal.
G. Enrichment of Early or Late P300 regions in E12.5 or P42 SEs. Proportions test.
H. Ratio of SE chromatin contacts (left) or expression of genes (right) linked to SEs by HiChIP loops at E12.5 and P42. Kruskal-Wallis test.
I. Variation in chromatin contact scores at E12.5 vs. P42 for SEs shared between E12.5 and P42. Boxed regions are magnified and labeled with SE-associated genes and their expression fold-change (P42/E12.5). Scn5a (magenta) is shown in panel J.
J. SE at the Scn5a locus. Average loop scores for selected loops are labeled. Loops with scores <15 are omitted for clarity.
Typical enhancers cluster into regulatory centers termed super-enhancers (SEs)40. Using P300 occupancy data, we identified 1319 SEs at E12.5 and P42. 150 and 472 were unique to E12.5 or P42, respectively (Fig. 7F; Data S7) and were enriched for Early or Late P300 regions, respectively (Fig. 7G). SE chromatin contact density, calculated from HiChIP, was also developmentally regulated, with E12.5-selective and P42-selective SEs having greater interactions at the congruent stage (Fig. 7H). Indeed, developmental changes in SE P300 signal, SE chromatin contact density, and expression of genes linked to these SEs by chromatin contacts were correlated with one another (Fig. 7H and S7G).
To examine the contribution of SE interaction changes to developmentally regulated gene expression, we focused on SEs shared between E12.5 and P42, which by definition have less developmental change in P300 signal. A subset of Shared SEs exhibited large changes in contact density between stages (Fig. 7I). Those with the greatest changes in contact density were associated with corresponding developmental changes in gene expression, suggesting that changes in SE interactions as well as strength contribute to target gene regulation. A notable example is a previously characterized SE linked to Scn5a41,42, encoding the cardiac sodium channel. The Scn5a locus contains several previously characterized regulatory elements (REs). RE6–9 comprise the SE, which is present at E12.5 and P42 (Fig. 7J). RE1–2 and RE3, 4, and 6 have developmentally down- and up-regulated P300 occupancy, respectively. Correspondingly, RE1–2 lost contact with the Scn5a TSS, whereas contacts between RE3–9, including RE6–9 within the SE, increased with each other and the Scn5a TSS, in mature CMs (Fig. 7J). Thus the 6.6-fold upregulation of Scn5a between E12.5 and P42 is associated with marked increases in SE-promoter contacts.
These results suggest that the interaction of developmental changes in P300 occupancy and 3D chromatin structure contribute to developmentally regulated CM gene expression.
Discussion
We generated a rich data resource for integrative investigation of CM-specific P300 binding, enhancer activity, cis-regulatory motifs, 3D genome, and transcriptome at multiple time points during in vivo CM differentiation and maturation.
A fundamental question in development is how stage-specific enhancers are established and maintained. Our dataset of P300 enhancers across seven developmental stages from fetal to adult CMs suggests that differential binding of TFs mediates the differential P300 binding to different genomic loci during CM development and maturation. TF ChIP-seq and motif enrichment in Fetal P300 regions and P300 regions with neonatal MPRA activity most strongly implicated TBX5, TEAD, and HAND in the early recruitment of P300. At later developmental stages, these analyses implicated NR and MEF2 motifs. There has been growing appreciation of the importance of NRs, including THRA, estrogen-related receptors, and PPARGC1A, in CM maturation 8,9,31,43,44. These NRs bind P300, sometimes in a ternary complex with MEF2 45–47. We used AAV-MPRA to validate the key function of NRs and NR motifs in promoting P300 binding and activation of Late P300 regions, and to experimentally identify 84 dynamic enhancers that required the NR motif for activity. Many of these validated enhancers were bound by THRA, consistent with its important role in CM maturation8. However, other MPRA-validated NR motifs, including many with the strongest effect and nearly all that inhibited enhancer activity, were not bound by THRA at P15, suggesting that these effects are mediated by other NRs.
Current evidence points to causal links between enhancer activation and 3D genome structure. These operate in both directions and are context dependent. For example, genomic rearrangements that disrupt TADs result in ectopic enhancer-promoter interactions and gene dysregulation48. At the same time, TFs mediated the reorganization of chromatin loops and chromatin compartments during cell differentiation49,50. P300 and histone acetylation may mediate some of these effects of TF binding, since histone acetylation modulated chromatin loops and compartmentalization, and ectopic H3K27ac facilitated formation of de novo loops in leukemia cells36. Our data indicate that developmentally regulated binding of TFs and P300 at Early and Late P300 regions modulate chromatin loops, TADs, compartments, and SEs. Reciprocally, changes in chromatin conformation mediated by other factors may influence TF and P300 binding and enhancer activity. Determining the mechanisms by which these different levels of regulation affect each other is an important area for future study.
The contribution of 3D genome organization to the differentiation and maturation of cell lineages has been little studied. We considered three types of 3D genome organization: loops, TADs, and compartments, which differ in scale and underlying structural mechanisms. CM differentiation between E12.5 and adulthood affected ~20% of HiChIP loops, ~4% of TADs, and ~10% of compartments. Developmentally regulated P300 occupancy interacted with all three types of 3D genome organization to influence developmental gene expression in CMs in vivo. At the level of loops, both P300 occupancy signal and contact frequency influenced developmental gene expression. Modeling this with the ABC score demonstrated the importance of stage-specific contact frequencies for optimal correlation with developmental gene expression. This was also seen at SEs, where developmentally regulated SE contacts of a subset of SEs appeared to play a predominant role in target gene regulation.
TADs have been considered to be conserved between different cell types51. We observed that the large majority of TADs were constant between fetal and adult CMs. However, we found that a subset of TADs changed during CM development by altered enforcement of TAD boundaries. Intriguingly, TAD splits greatly outnumbered mergers during CM differentiation. This progressive enforcement of within-TAD boundaries during CM maturation confined domains of chromatin activation and may be a mechanism to constrain gene expression within highly specialized cell types. It will be interesting to determine if this mechanism is deployed in other cell types.
We discovered a surprising global developmental shift in the relationship between genomic distance and promoter-distal region contact frequency, so that promoter-distal region distance tends to be shorter in adult CMs. This correlates with the closer average location of TF binding sites to promoters in adult compared to fetal CMs15. This shift is likely functionally important, because the contact values calculated from the stage matched relationship yielded ABC scores that correlated nearly as well with developmental changes in gene expression as the actual contact data, whereas values calculated from the stage-mismatched relationship yielded poorly correlating ABC scores. Whether similar shifts in this relationship will be observed in other maturing cell types and the underlying mechanisms, will be interesting questions for further study.
Limitations of the Study
Our study had limitations. Mice from both genders were pooled. The MPRA enhancer activity measurements were made in episomes and may differ from regulation at the endogenous locus. Inefficient AAV9 transduction prior to E15.5 precluded enhancer activity early in development by MPRA. MPRA assays measure enhancer activity in limited time points and contexts, so that negative activity measurements do not exclude enhancer activity at other time points or contexts. While Hi-C and H3K27ac HiChIP are state of the art methods to measure 3D genome contacts, they have limited sensitivity and resolution.
STAR★Methods
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact William Pu (william.pu@cardio.chboston.org).
Material availability
Key Plasmids have been deposited to Addgene. Mice are available from public repositories. Other unique materials and reagents generated in this study are available upon request from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
The following public data sources were used for this study: E12.5_heart_P300_bioChIP-seq, E12.5_forebrain_P300_bioChIP-seq, adult_heart_P300_bioChIP-seq and adult_heart-EC_P300_bioChIP-seq data are from GSE88789 19; GATA4 bioChIP-seq data of E12.5 or adult heart apex are from GSE52123 52; Mef2a, Mef2c, Nkx2–5, SRF, Tbx5, and Tead1 bioChIP-seq data of E12.5 or adult heart apex are from GSE124008 15. Original data in this manuscript including P300 bioChIP-seq, bulk RNA-seq, Hi-C and H3K27ac HiChIP data of mouse CMs have been deposited to gene expression omnibus SuperSeries GSE195905.
The custom scripts used in this study are publicly available on github (https://github.com/yanchunzhang/Cardiomyocyte_maturation_HiC). DOI 10.5281/zenodo.7716979.
Additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Mice
All mouse experiments were performed under protocols approved by the Boston Children’s Hospital Institutional Animal Care and Use Committee. P300flbio (Jax #025980), Tnnt2-Cre (Jax #024240), Myh6-Cre (Jax #011038), Rosa26mTmG (Jax #007576) and Cdh5 (Pac)-CreERT2 (Taconic #13073) have been previously described. Rosa26fsBirA was derived from the Rosa26fsTRAP (Jax #022367) mouse by removal of the frt-TRAP-frt cassette using germline Flp recombination 19. Mice were on a C57BL6/129 mixed background. Mixed genetic background mice or Swiss Webster outbred mice were used for AAV injection. Male and female mice were pooled for all assays. Mice were housed in a specific pathogen free facility with 12 hour light and dark cycles.
Intracardiac fetal injections were performed on anesthetized pregnant dams. Portions of the uterus were externalized through a midline abdominal incision. A pulled glass needle was advanced through the uterine wall and the back of the embryo into the heart. Virus was then injected and the needle withdrawn. After injection of multiple embryos, the uterus was returned to the embryo and the abdominal wall was sutured in layers.
Method details
Cardiomyocyte isolation and purification
For E12.5, E16.5, P0, and P7 CMs, ventricular apexes were dissociated using the Neonatal mouse CM isolation kit (Cellutron Life Technologies) and purified using the mouse neonatal CM isolation kit (Miltenyi). P14, P28, and P42 CMs were dissociated and purified from ventricular apexes by Langendorf collagenase perfusion followed by differential sedimentation as described 6. CM preparations were over 90% pure as determined by examining CMs purified from Rosa26mTmG/+;Tnnt2-Cre+ mice with fluorescence microscopy. Purified CMs were used with three biological replicates per group for Hi-C and RNA-seq.
P300 pulldown for Western Blot
For streptavidin pulldown of P300flbio from P7 hearts, p300flbio/+; Rosa26fsBirA/+;Myh6-Cre+ heart ventricles were homogenized in cold PBS and protein lysate was prepared with 0.5% SDS lysis buffer (1xPBS supplemented with 1 mM DTT, protease inhibitor cocktail, and 0.5% SDS). Biotinylated proteins were then pulled down by incubating with Dynabeads M-280 streptavidin (ThermoFisher Scientific, Cat#11206D) at 4°C for one hour. After washing at room temperature 5 times for 5 minutes each with PBS supplemented with 0.2% SDS, bound proteins were eluted from the beads with SDS elution buffer (20 mM Tris-HCl pH 8.0, 1% SDS, and 2 mM ETDA) at 98°C for 10 minutes. Western blotting was performed using standard protocols and antibodies against P300 (Santa Cruz, sc-585) and BirA (Abcam, ab14002).
Immunostaining
Cryosections of E12.5 embryos and P0 hearts were stained with cardiac troponin I (TNNI3, Abcam, ab56357) and imaged by confocal microscopy (Olympus FV3000).
P300 bioChIP-seq of cardiomyocytes
P300 bioChIP-seq was performed as described previously 19. E12.5 to P42 heart ventricles were harvested, homogenized, and crosslinked in 1% formaldehyde in PBS for 10 minutes at room temperature and subsequently quenched with 125 mM glycine for 5 min. Chromatin was fragmented using a microtip sonicator (Qsonica, S-3000) at 30% amplitude for 8 minutes. Biotinylated P300 and bound chromatin was pulled down by incubation with streptavidin beads (ThermoFisher Scientific, 11206D). Bead-bound chromatin was resuspended in SDS elution buffer (50 mM Tris–HCl pH 8,10 mM EDTA, 1% SDS) and incubated overnight at 65 °C to reverse crosslinking. After RNAse A and Proteinase K treatment, ChIP DNA was purified using Qiagen MinElute columns. Libraries were constructed using the KAPA HyperPrep ChIP-seq library preparation kit (Roche, cat# 07962347001). Sequencing (75 nt single end) was performed on an Illumina NextSeq 500.
AAV reporter assays
Wild type or mutant genomic regions were PCR amplified or synthesized (sequences listed in Table S1) and cloned into a self-complementary adeno-associated virus (scAAV) mCherry reporter vector. Adeno-associated virus serotype 9 (AAV9) was generated as described 76. Briefly, HEK293T cells were transfected with ITR plasmid, rep-cap plasmid, and helper plasmid. AAV was purified using OptiPrep density gradient purification (Sigma). Purified virus was delivered to E15.5 embryos by transuterine intracardiac injection (5E10 viral genomes/embryo), or subcutaneous injection in P0 pups (2E11 viral genomes/g body weight). Heart ventricles were collected for RNA and DNA at P0, P7, or P28. RNA was extracted using TRIzol (Life Technologies) and purified with Zymo RNA Clean kit. Viral DNA was recovered with DirectPCR lysis reagent (Viagen, Cat#101-T). Reporter activity was determined by the ratio of mCherry RNA/DNA, normalized to Rplp0 and control genomic regions, respectively.
Massively parallel reporter assay (MPRA)
Enhancers were synthesized by Agilent as an oligonucleotide pool. Each enhancer consisted of two 230 bp single stranded DNA oligos with 20 bp 3′ overlap. Each oligonucleotide’s 5′ end had a 20 bp primer-binding site. Oligonucleotides within the pool were annealed and 3′ ends were extended by PCR to create a library of 400 bp enhancers flanked by 20 bp priming sites. NotI/AscI-restriction sites were added to enhancers in a second round of PCR. The enhancer library was then digested, size selected, and ligated into the multiple cloning site of a self-complementary AAV plasmid containing minimal promoter-mCherry-NotI/AscI-polyA sequences (Addgene #182649). The ligation product was electroporated into Agilent SURE Electrocompetent cells following manufacturer recommendations, spread onto agar plates, and cultured overnight. Approximately 900,000 colonies were harvested and pooled for plasmid maxi-prep. The MPRA library was packaged into AAV9 as described above.
For enhancer activity measurement at P0, AAV9-MPRA library was delivered to wild type CFW E15.5 embryos (5E10 viral genomes/embryo, n=12) by intracardiac injection. Heart ventricles were harvested at P0. For enhancer activity at P7 and P28, AAV9-MPRA library was injected subcutaneously to P0 pups (2E11 viral genomes/g). Hearts were harvested at P7 (n=28) or P28 (n=28). For nuclear receptor WT-Mut pairs, 190 bp regions centered on a candidate nuclear receptor motif and flanked by 20 bp priming sites, were synthesized as a pool (Agilent) and cloned into the AAV9-MPRA vector. NR motifs were mutated by shuffling their DNA sequence and rescanning to confirm loss of the motif. AAV9-MPRA library was injected subcutaneously to P0 pups (2E11 viral genomes/g) and harvested at P7 (n=14).
RNA was isolated from homogenized ventricular apexes via TRIzol extraction and reverse transcribed using a primer recognizing the start of the polyA sequence. NGS adapters and unique indexes were added to each sample in subsequent rounds of PCR amplification. Untransduced AAV DNA from the library pool was also prepared in the same fashion for sequencing in triplicate. Indexed samples were pooled and paired-end (2 × 150 bp) sequenced on a NextSeq500. After removal of adapters53, reads were aligned to the mouse genome54,55, keeping only mates that produced concordant alignments between 395 and 405 bp. On average, each sample contained ~5M alignments passing these criteria. The number of reads for each enhancer in each sample was determined using BedTools 56. The average number of reads for each enhancer within the untransduced AAV DNA was then calculated, and enhancers present at low frequencies (<5 RPM) were excluded. The majority (>90%) of enhancers were successfully created and were detected above the 5 RPM threshold. Read numbers for RNA samples was determined using the same method. RNA:DNA ratio for each region was calculated and used as a readout of enhancer activity.
Regions were defined as “active” when their RNA;DNA ratio exceeded the 95th percentile for the negative control regions. This is a relatively conservative definition of activity, since some negative control regions had relatively high RNA:DNA ratio.
Amplicon-sequencing of somatic mutagenesis target site
Newborn Rosa26fsCas9; Myh7YFP mice were transduced with a mosaic dose of AAV vector carrying Tnnt2-Cre and a gRNA (ACATTGGGTCAGCTTGGGTC) targeting a nuclear receptor motif within the Mhrt promoter. CMs were collected at P28 and genomic DNA was isolated. A 16 bp UMI and partial NGS adapter fused to primers flanking the cut site were added in a single PCR cycle, followed by 25 cycles of amplification which added full length NGS adapters. Three amplicon replicates were paired-end sequenced (2×150 bp) to an average depth of ~1.7M reads per replicate. Read pairs were merged using Paired-End reAd mergeR (PEAR), deduplicated by UMI using AmpUMI, and the distribution of mutations was analyzed via CRISPResso277.
Hi-C with mouse cardiomyocytes
Hi-C was performed as described 10. For each biological replicate, 2M purified CMs were fixed in PBS supplemented with 1% formaldehyde for 10 min at RT and then quenched with 125 mM glycine for 5 min at RT. Nuclei were isolated by treating with a hypotonic buffer (10 mM Tris-HCl, pH 8.0, 10 mM KCl, 1 mM MgCl2, 0.2% Igepal CA630) on ice for 10 min. Pelleted nuclei were permeabilized using 0.5% SDS and incubated for 10 min at 62 °C. Chromatin was digested using the 4-base cutter restriction enzyme MboI (200 units, NEB) at 37 °C overnight. After inactivation of MboI for 20 min at 62 °C, single stranded overhangs were filled with biotinylated-14-dATP (Life Technologies) and dCTP, dGTP, and dTTP using Klenow DNA polymerase (40 units, NEB) for 90 min at 37 °C. DNA was then ligated for 4 h at 16 °C using T4 DNA ligase (2,000 units, NEB). Reverse crosslinking was performed by incubating at 55 °C with proteinase K for 30 min and then 68 °C overnight. Purified DNA was sonicated using an ultra sonicator to 300–700 bp small fragments. After size selection and purification using SPRI beads (Beckman Coulter), biotinylated DNA was isolated using Dynabeads MyOne T1 Streptavidin beads (Life Technologies). Sequencing libraries were prepared on magnetic beads and final PCR amplification was carried out for 6 cycles. Size distribution of bead-purified libraries was checked using a High Sensitivity D1000 ScreenTape on a Tapestation (Agilent) and quantified using Qubit (Life Technologies). Libraries were sequenced on Hiseq 4000 (100 bp PE, Illumina). More than 70% of the cis reads were long-range (>20 kb) intra-chromosomal interactions.
H3K27ac HiChIP of mouse cardiomyocytes
H3K27ac HiChIP libraries were prepared using the Arima HiC+ kit (Arima Genomics). Briefly, ~2 M purified CMs were crosslinked with 2% formaldehyde for 10 min at room temperature. Crosslinked chromatin was digested using the Arima proprietary restriction enzyme cocktail. The fragmented chromatin ends were then filled in with a biotinylated nucleotide. Spatially proximal fragmented ends were ligated. Subsequently, chromatin was fragmented using a QSonica Q800R3 sonicator and incubated with H3K27ac antibody (Catalog No: 39133, Active Motif) at 4 °C overnight. The antibody-bound chromatin was immunoprecipitated using Protein A magnetic beads (Invitrogen) and reverse crosslinked using Arima-HiC+ kit reagents. Sequencing adapters and indexing primers were added using the Swift Biosciences Accel-NGS 2S Plus DNA Library Kit. The libraries were quantified to determine the appropriate number of PCR cycles needed for library amplification using the KAPA Library Quantification Kit (Roche) and then amplified using the KAPA Library Amplification Kit (Roche).
Cardiomyocyte RNA-Seq
RNA was purified using the Purelink RNA mini kit (Life Technologies). Ribosomal RNA was depleted using Ribo-Zero rRNA removal kits (Epicentre). RNA-seq libraries were prepared using ScriptSeq v2 library kit (Epicentre). Libraries were sequenced using an Illumina NextSeq500 with single end 75 bp reads. At least 50 M reads were obtained for each biological replicate.
ChIP-seq data analysis
The raw reads from bioChIP-Seq datasets were mapped to the Mus musculus genome (mm10 build) using Bowtie 78 v1.1.1. Alignments with more than one match were removed. MACS2 57 was used to determine peaks in each replicate with p-value 1e-5 as cutoff. The peak list was further filtered to remove all blacklisted regions (as defined by ENCODE for mm10 ChIP-Seq: http://mitra.stanford.edu/kundaje/akundaje/release/blacklists/mm10-mouse/). The intersection of peaks from the replicates at each time point yielded the reproducible peaks. The intersection and union sets were determined by the Bedtools 56 intersect function. Profile and heatmap plot were generated by ngs.plot 58. To define dynamic P300 regions, Diffbind 79 was used to normalize across samples. Peaks with significant signal in at least two time points were ranked by the ratio of their maximum to minimum values, and classified as described in Fig. 2A. Motifs were determined by scanning a 200 bp window centered on the summits of peaks for all possible matches to a reference PWM file using Homer 60. The reference PWM file was generated from the union of the default motifs provided with Homer and experimentally determined TF heterodimer motifs defined as reported 80. The motifs were then clustered based on their similarity to each other using STAMP 81 and highly redundant motifs were removed.
Characterization of P300 enhancer regions
Conservation of regions was analyzed using precalculated phastCons 60-way vertebrate scores 82. The profiles of dynamic/static P300 enhancer regions were generated by computeMatrix and plotProfile commands from DeepTools 62. P300 signals were quantile-normalized using Danpos283 for comparison between stages.
We evaluated the specificity, sensitivity, and accuracy of P300-occupied regions compared to cardiac enhancer activity experimentally measured by transient transgenesis and compiled in the Vista enhancer database 18. Mouse genomic regions tested in Vista were classified using the database entries into those with or without cardiac activity (cardiac+, cardiac-), where cardiac activity indicates transgene expression in any portion of the heart. These regions were also classified based on P300 occupancy (P300+, P300-). Sensitivity is cardiac+;P300+ / [cardiac+;P300+ + cardiac+;P300-]. Specificity is cardiac-;P300- / [cardiac-;P300+ + cardiac-;P300-]. Accuracy is [cardiac+;P300+ + cardiac-;P300-] / [cardiac+ + cardiac-].
RNA-seq data analysis
Raw reads were mapped to the mm10 reference genome using STAR 63. Raw read counts of all genes were generated by HTseq-count 84. Differential expressed genes (DEGs) were identified by DEseq2 65. For DEGs between E12.5 and P42, cutoffs were set as absolute log2FC >=1 and Padj < 0.05. For DEG heatmap between embryonic stages and adult stages, all replicates of E12.5 and E16.5 were pooled as embryonic and all replicates of P28 and P42 were pooled as adult, with cutoffs as absolute log2FC >=3 and Padj < 0.01. Heatmap of DEGs was generated by R package pheatmap. Gene ontology enrichment analysis was done by R package ClusterProfiler 66.
Hi-C data analysis
Raw reads were mapped with Juicer v2.0 67 to mm10 reference genome with default parameters. Then the paired mapped reads with mapping quality no less than 30 were binned and converted into .cool files by Cooler v0.8.11 68. KR balancing was applied to the .cool file by cooler balance commands. Expected contact matrix at different distances was calculated by cooltools v0.5.0 85 expected-cis command.
Compartment scores were calculated by cooltools eigs-cis command at 100 kb resolution. Then compartment scores were calibrated by GC content and defined as compartment A or B, according to higher or lower GC content. Principal component analysis was implemented by R prcomp function on the whole-genome compartment scores of all replicates to check the reproducibility of replicates. Then replicates were merged into one combined .cool file for further analysis.
Loops were called by SIP v1.6.1 70 at 5 kb and 10 kb resolutions. Then loops from different stages were pooled together for further analysis. Loops were defined as enhancer-promoter (E-P) loops when at least one P300 enhancer located at one anchor of the loop and at least one TSS located at another anchor of the loop. For all E-P loops, the observed/expected value at loop center was deemed as the intensity of the loop.
TAD and TAD split events were defined by TADsplimer v1.1 37 at 10 kb resolution with default parameters. TAD insulation scores were calculated by cooltools diamond with 500 kb window at resolution of 5kb. For all TAD split events, the fold change of the intensity of P300 signal or gene expression level was calculated as the intensity of the stronger side of split sites divided by the intensity of the weaker side. Visualization of contact matrixes was performed using python modules cooltools+matplotlib or Juicebox v1.9.8 67.
To plot the relationship between a region’s distance to TSS and its TSS contact frequency, aggregated intra-chromosomal contact score at each distance (in 5 kb bins) was summed then divided by the total intra-chromosomal contact score. The curve was plotted as a cumulative distribution function.
H3K27ac HiChIP data analysis
Raw sequencing reads were mapped to mm10 reference genome using the HiChIP pipeline (https://hichip.readthedocs.io/en/latest/before_you_begin.html). Briefly, sequencing reads were first mapped to mm10 by bwa86. Then aligned results were processed by pairtools87 to find ligation junctions in the HiChIP libraries, remove duplicates, and generate bam files for further analysis.
To prepare input files for loop calling, H3K27ac peaks were called by MACS2 for each replicate separately and interacting read pairs were extracted from the bam files. Loops were called by hichipper73 for each replicate separately. Then filtered intra-chromosomal loops of all replicates by hichipper were pooled together into a union loop set. Read counts for the union loop set were extracted from ‘one.interaction.bepe’ file from hichipper. Significance of loops in the union set were calculated by mango74. Result files by mango were processed by diffloop75 following the instruction of (https://rpubs.com/caleblareau/diffloop_vignette) to get the final filtered loop sets and normalized read counts. In the diffloop processing step, loops with mango_FDR larger than 0.01 were filtered out.The remaining loops with normalized read counts either in two E12.5 replicates or in two P42 replicates were kept as the final filtered loop set. Differential loops were defined as diffloop_FDR<0.05 and | log2 fold-change| >= 2.
ABC score
For each gene, the total ABC score 38 contributed by P300 enhancer was calculated using the formula below:
For gene G, n is the number of P300 enhancers within 5 Mb of the TSS of gene G, Aek is the Activity of P300 enhancer k (normalized enhancer signal in 400 bp window around enhancer peak), Cegk is the Hi-C contact between P300 enhancer k and the TSS of gene.
Fold change of total ABC score and gene expression of five adjacent genes ranked by fold change of gene expression calculated by DESeq2 were binned together. The fold-change from E12.5 to P42 was calculated as the ratio of the scores at P42 and E12.5. The correlation between fold change of total ABC score and fold change of gene expression was calculated using the Spearman method in R, using binned log2FC values.
For stage mis-matched ABC scores (Fig. 6G, panel iv), P42 and E12.5 contact data were swapped for calculation of ABC scores. For calculated, stage matched contact scores (Fig. 6G, panel ii), we used the genome-wide aggregate contact frequency as a function of distance (Fig. 6H) in place of actual contact data. For calculated, stage mis-matched contact scores (Fig. 6G, panel iii), we swapped the contact frequency-distance relationship between E12.5 and P42.
HiChIP loops were used to analyze expression of genes linked to Early or Late P300 regions. Enhancer-promoter loops that overlapped Early or Late P300 regions linked these regions to genes. Linked genes were stratified by loop score quintile at E12.5 for Early P300, or at P42 for Late P300.
Super-enhancers
SE calling was performed using ROSE 88,89 on E12.5 and P42 P300 bioChIP-seq using default parameters. Stage specific and shared SEs were identified using bedtools 62 intersect. SE-associated genes were linked by H3K27ac HiChIP loops.
To calculate the aggregated interaction score of super-enhancers, typical P300 enhancers within super-enhancers were extracted by bedtools intersect. The interaction scores of all HiChIP loops with anchors overlapping these P300 enhancers were then summed to yield the aggregated super-enhancer interaction score.
Visualization of data with genome browser
ChIP-seq peaks were visualized with normalized RPKM value using IGV Genome Browser 90. H3K27ac HiChIP data were visualized using WashU Epigenome Browser 91. Average loop scores from two replicates for selected loops are labeled for comparison. For Myh6-Myh7 and Scn5a loci, loops with scores under 15 were omitted for clarity.
Quantification and statistical analysis
Statistics
Statistical analysis was performed in Graphpad Prism or R. Bar graphs show mean ± SD, box plots show median, interquartile range, and the maximum and minimum values that are within 1.5 times the interquartile range of the 3rd and 1st quartile. Statistical tests are indicated in figure legends.Supplemental Data Tables
Supplementary Material
Data S2 – MPRA of dynamic cardiomyocyte P300 regions. Related to Fig. 3.
Data S3 – Effect of NR motif ablation on enhancer activity. Related to Fig. 4.
Data S4 – Hi-C loops, compartments, and TAD splits during cardiomyocyte maturation. Related to Fig. 5.
Data S5 – Cardiomyocyte gene expression during maturation. Related to Fig. 6.
Data S6 – HiChIP loops in cardiomyocytes at E12.5 and P42. Related to Fig. 7.
Data S7 – Cardiomyocyte super-enhancers at P42 or E12.5. Related to Fig. 7.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-P300 | Santa Cruz | sc-585 |
| Chicken polyclonal anti-BirA | Abcam | ab14002 |
| Goat polyclonal to Cardiac Troponin I | Abcam | ab56357 |
| Bacterial and virus strains | ||
| SURE Electrocompetent cells | Agilent | 200227 |
| scAAV-Enh-Mlc2v-mCherry (MPRA and individual enhancer testing) | Addgene | 182649 |
| scAAV-Hsp68-mCherry (MPRA vector) | Addgene | 195412 |
| Biological samples | ||
| Maturation MPRA: AAV DNA | This paper | GEO GSM5876202-GSM5876204 |
| Maturation MPRA: E15.5 injection, P0 collection of RNA from ventricles | This paper | GEO GSM5876205-GSM5876216 |
| Maturation MPRA: P0 injection, P7 collection of RNA from ventricles | This paper | GEO GSM5876217-GSM5876244 |
| Maturation MPRA: P0 injection, P28 collection of RNA from ventricles | This paper | GEO GSM5876245-GSM5876272 |
| Nuclear Receptor MPRA: AAV DNA | This paper | GEO GSM5876185-GSM5876187 |
| Nuclear Receptor MPRA: P0 injection, P7 collection of RNA from ventricles | This paper | GEO GSM5876188-GSM5876201 |
| E12.5, E16.5, P0, P7, P14, P28, and P42 mouse ventricles for P300 bioChIP-seq | This paper | N/A |
| E12.5, P0, and P42 mouse ventricular CMs for Hi-C | This paper | N/A |
| E12.5, E16.5, P0, P7, P14, P28, and P42 mouse ventricular CMs for for bulk RNA-seq | This paper | N/A |
| E12.5 and P42 mouse ventricular CMs for H3K27ac Hi-ChIP | This paper | N/A |
| Critical commercial assays | ||
| Dynabeads M-280 Streptavidin | ThermoFisher Scientific | 11205D |
| KAPA Hyper Prep Kit | Kapa Biosystems | KK8502 |
| Mouse neonatal CM isolation kit | Miltenyi Biotec | 130-100-825 |
| Ribo-Zero Plus rRNA Depletion Kit | Illumina | 20040526 |
| TruSeq RNA Library Prep Kit v2 | Illumina | RS-122-2001 |
| Arima-HiC+ kit for HiChIP | Arima Genomics | 201911-792 |
| Experimental models: Organisms/strains | ||
| p300-flbio;Rosa26BirA | The Jackson Laboratory | 025980 |
| Rosa26fsTRAP | The Jackson Laboratory | 022367 |
| Rosa26fsBirA | Derived from Rosa26fsTRAP | N/A |
| B6J.129(B6N)-Gt(ROSA)26Sortm1(CAG-cas9*,-EGFP)Fezh/J mice | The Jackson Laboratory | 026175 |
| Myh7YFP mice | UNC-CH/Oliver Smithies | N/A |
| Swiss Webster (CFW) mice | Charles River | 024 |
| Oligonucleotides | ||
| Oligo pool libraries for MPRA | Agilent SurePrint | N/A |
| A full list of oligos and primers in Table S1 | This paper | N/A |
| Deposited data | ||
| P300 bioChIP-seq of E12.5-P42 murine CMs | This paper | GSE195901 |
| Bulk RNA-seq of E12.5-P42 murine CMs | This paper | GSE195902 |
| Hi-C of E12.5, P0, and P42 murine CMs | This paper | GSE194087 |
| H3K27ac HiChIP of E12.5 and P42 murine CMs | This paper | GSE222160 |
| Massively parallel reporter assay of P300 enhancers | This paper | GSE196346 |
| Massively parallel reporter assay of nuclear receptor-containing enhancers | This paper | GSE196346 |
| ChIP-seq of Mef2A, Mef2C, Nkx2-5, Tbx5, Tead1 in fetal and adult murine heart ventricles | Akerberg et al. 15 | GSE124008 |
| ChIP-seq of GATA4 in fetal and adult murine heart ventricles. | He et al. 52 | GSE52123 |
| THRA ChIP-seq of P15 murine hearts | Hirose et al. 8 | GSE125414 |
| P300 ChIP-seq of E12.5 murine forebrains and hearts, H3K27ac ChIP-seq of E12.5 murine hearts. | Zhou et al. 19 | GSE88789 |
| Uncropped western blots | This paper | https://figshare.com/articles/figure/Uncropped_WB_Fig_1B_pdf/22250587 |
| Software | ||
| Trimmomatic/0.36 | Bolget et al. 53 | http://www.usadellab.org/cms/index.php?page=trimmomatic |
| Bowtie2/2.3.4.3 | Langmead and Salzberg54 | bowtie-bio.sourceforge.net/bowtie2 |
| Samtools/1.9 | Li et al. 55 | http://www.htslib.org/ |
| Bedtools/2.27.1 | Quinlan and Hall56 | bedtools.readthedocs.io |
| MACS2 | Zhang et al.57 | https://pypi.org/project/MACS2/ |
| ngs.plot | Shen et al.58 | https://github.com/shenlab-sinai/ngsplot |
| diffbind | Ross-Innes et al.59 | https://bioconductor.org/packages/release/bioc/html/DiffBind.html |
| Homer | Heinz et al.60 | http://homer.ucsd.edu/homer/ |
| STAMP | Parks et al.61 | https://beikolab.cs.dal.ca/software/STAMP |
| deeptools | Ramierez et al.62 | https://deeptools.readthedocs.io/en/develop/ |
| STAR | Dobin et al.63 | https://github.com/alexdobin/STAR |
| HTSeq-count | Putri et al.64 | https://htseq.readthedocs.io/en/release_0.11.1/index.html |
| DESeq2 | Love et al.65 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| ClusterProfiler | Yu et al.66 | https://guangchuangyu.github.io/software/clusterProfiler/ |
| Juicer v2.0 | Durand67 | https://github.com/aidenlab/juicer |
| Cooler v0.8.11 | Abdennur and Mirny68 | https://github.com/open2c/cooler |
| cooltools v0.5.0 | Open2c69 | https://github.com/open2c/cooltools |
| SIP v1.6.1 | Rowley et al.70 | https://github.com/PouletAxel/SIP |
| TADsplimer v1.1 | Wang et al.37 | https://github.com/GuangyWang/TADsplimer |
| Juicebox v1.9.8 | Durand et al.71 | https://github.com/aidenlab/Juicebox |
| HiC-Pro v3.1.0 | Servant et al.72 | https://github.com/nservant/HiC-Pro |
| hichipper v0.7.7 | Lareau et al.73 | https://github.com/aryeelab/hichipper |
| mango | Phanstiel et al.74 | https://github.com/dphansti/mango |
| diffloop | Lareau et al.75 | https://github.com/aryeelab/diffloop |
| ABC score calculation | This paper | https://github.com/yanchunzhang/Cardiomyocyte_maturation_HiC |
Funding Sources
This study was funded by grants from the National Institutes of Health (PZ: U01HL131003; NJV: T32HL007572, F32HL13423501, K99HL14319; WTP: UM1 HL098166, R01 HL146634; KC: R01GM125632, R01HL148338).
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- 1.Schoenfelder S, and Fraser P. (2019). Long-range enhancer–promoter contacts in gene expression control. Nat. Rev. Genet. 20, 437–455. [DOI] [PubMed] [Google Scholar]
- 2.Stadhouders R, Filion GJ, and Graf T. (2019). Transcription factors and 3D genome conformation in cell-fate decisions. Nature 569, 345–354. [DOI] [PubMed] [Google Scholar]
- 3.Glaser J, and Mundlos S. (2021). 3D or Not 3D: Shaping the Genome during Development. Cold Spring Harb. Perspect. Biol. 10.1101/cshperspect.a040188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng H, and Xie W. (2019). The role of 3D genome organization in development and cell differentiation. Nat. Rev. Mol. Cell Biol. 20, 535–550. [DOI] [PubMed] [Google Scholar]
- 5.Guo Y, and Pu WT (2020). Cardiomyocyte Maturation: New Phase in Development. Circ. Res. 126, 1086–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo Y, Jardin BD, Zhou P, Sethi I, Akerberg BN, Toepfer CN, Ai Y, Li Y, Ma Q, Guatimosim S, et al. (2018). Hierarchical and stage-specific regulation of murine cardiomyocyte maturation by serum response factor. Nat. Commun. 9, 3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo Y, Cao Y, Jardin BD, Sethi I, Ma Q, Moghadaszadeh B, Troiano EC, Mazumdar N, Trembley MA, Small EM, et al. (2021). Sarcomeres regulate murine cardiomyocyte maturation through MRTF-SRF signaling. Proc. Natl. Acad. Sci. U. S. A. 118. 10.1073/pnas.2008861118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirose K, Payumo AY, Cutie S, Hoang A, Zhang H, Guyot R, Lunn D, Bigley RB, Yu H, Wang J, et al. (2019). Evidence for hormonal control of heart regenerative capacity during endothermy acquisition. Science 364, 184–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy SA, Miyamoto M, Kervadec A, Kannan S, Tampakakis E, Kambhampati S, Lin BL, Paek S, Andersen P, Lee D-I, et al. (2021). PGC1/PPAR drive cardiomyocyte maturation at single cell level via YAP1 and SF3B2. Nat. Commun. 12, 1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Li T, Preissl S, Amaral ML, Grinstein JD, Farah EN, Destici E, Qiu Y, Hu R, Lee AY, et al. (2019). Transcriptionally active HERV-H retrotransposons demarcate topologically associating domains in human pluripotent stem cells. Nat. Genet. 51, 1380–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertero A, Fields PA, Ramani V, Bonora G, Yardimci GG, Reinecke H, Pabon L, Noble WS, Shendure J, and Murry CE (2019). Dynamics of genome reorganization during human cardiogenesis reveal an RBM20-dependent splicing factory. Nat. Commun. 10, 1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kannan S, Farid M, Lin BL, Miyamoto M, and Kwon C. (2021). Transcriptomic entropy benchmarks stem cell-derived cardiomyocyte maturation against endogenous tissue at single cell level. PLoS Comput. Biol. 17, e1009305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLaughter DM, Bick AG, Wakimoto H, McKean D, Gorham JM, Kathiriya IS, Hinson JT, Homsy J, Gray J, Pu W, et al. (2016). Single-Cell Resolution of Temporal Gene Expression during Heart Development. Dev. Cell 39, 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinto AR, Ilinykh A, Ivey MJ, Kuwabara JT, D’Antoni ML, Debuque R, Chandran A, Wang L, Arora K, Rosenthal NA, et al. (2016). Revisiting Cardiac Cellular Composition. Circ. Res. 118, 400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akerberg BN, Gu F, VanDusen NJ, Zhang X, Dong R, Li K, Zhang B, Zhou B, Sethi I, Ma Q, et al. (2019). A reference map of murine cardiac transcription factor chromatin occupancy identifies dynamic and conserved enhancers. Nat. Commun. 10, 4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nord AS, Blow MJ, Attanasio C, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer-Frick I, Shoukry M, Afzal V, et al. (2013). Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell 155, 1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. (2009). ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Visel A, Minovitsky S, Dubchak I, and Pennacchio LA (2007). VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 35, D88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou P, Gu F, Zhang L, Akerberg BN, Ma Q, Li K, He A, Lin Z, Stevens SM, Zhou B, et al. (2017). Mapping cell type-specific transcriptional enhancers using high affinity, lineage-specific Ep300 bioChIP-seq. Elife 6. 10.7554/eLife.22039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beckett D, Kovaleva E, and Schatz PJ (1999). A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 8, 921–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiao K, Kulessa H, Tompkins K, Zhou Y, Batts L, Baldwin HS, and Hogan BLM (2003). An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 17, 2362–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai YS, and Markham BE (2001). p300 Functions as a coactivator of transcription factor GATA-4. J. Biol. Chem. 276, 37178–37185. [DOI] [PubMed] [Google Scholar]
- 23.He J, Ye J, Cai Y, Riquelme C, Liu JO, Liu X, Han A, and Chen L. (2011). Structure of p300 bound to MEF2 on DNA reveals a mechanism of enhanceosome assembly. Nucleic Acids Res. 39, 4464–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sartorelli V, Huang J, Hamamori Y, and Kedes L. (1997). Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol. Cell. Biol. 17, 1010–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li T, Li Y-M, Jia Z-Q, Chen P, Ma K-T, and Zhou C-Y (2007). Carboxyl terminus of Nkx2.5 impairs its interaction with p300. J. Mol. Biol. 370, 976–992. [DOI] [PubMed] [Google Scholar]
- 26.He A, Kong SW, Ma Q, and Pu WT (2011). Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc. Natl. Acad. Sci. U. S. A. 108, 5632–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.VanDusen NJ, Lee JY, Gu W, Butler CE, Sethi I, Zheng Y, King JS, Zhou P, Suo S, Guo Y, et al. (2021). Author Correction: Massively parallel in vivo CRISPR screening identifies RNF20/40 as epigenetic regulators of cardiomyocyte maturation. Nat. Commun. 12, 5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parikh SS, Blackwell DJ, Gomez-Hurtado N, Frisk M, Wang L, Kim K, Dahl CP, Fiane A, Tønnessen T, Kryshtal DO, et al. (2017). Thyroid and Glucocorticoid Hormones Promote Functional T-Tubule Development in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Circ. Res. 121, 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnold CD, Gerlach D, Stelzer C, Boryń ŁM, Rath M, and Stark A. (2013). Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339, 1074–1077. [DOI] [PubMed] [Google Scholar]
- 30.Srivastava D, and Olson EN (2000). A genetic blueprint for cardiac development. Nature 407, 221–226. [DOI] [PubMed] [Google Scholar]
- 31.Sakamoto T, Matsuura TR, Wan S, Ryba DM, Kim JU, Won KJ, Lai L, Petucci C, Petrenko N, Musunuru K, et al. (2020). A Critical Role for Estrogen-Related Receptor Signaling in Cardiac Maturation. Circ. Res. 126, 1685–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han P, Li W, Lin C-HH, Yang J, Shang C, Nurnberg ST, Jin KK, Xu W, Lin C-YY, Lin C-JJ, et al. (2014). A long noncoding RNA protects the heart from pathological hypertrophy. Nature 514, 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo Y, VanDusen NJ, Zhang L, Gu W, Sethi I, Guatimosim S, Ma Q, Jardin BD, Ai Y, Zhang D, et al. (2017). Analysis of Cardiac Myocyte Maturation Using CASAAV, a Platform for Rapid Dissection of Cardiac Myocyte Gene Function In Vivo. Circ. Res. 120, 1874–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosa-Garrido M, Chapski DJ, Schmitt AD, Kimball TH, Karbassi E, Monte E, Balderas E, Pellegrini M, Shih T-T, Soehalim E, et al. (2017). High-Resolution Mapping of Chromatin Conformation in Cardiac Myocytes Reveals Structural Remodeling of the Epigenome in Heart Failure. Circulation 136, 1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sungalee S, Liu Y, Lambuta RA, Katanayeva N, Donaldson Collier M, Tavernari D, Roulland S, Ciriello G, and Oricchio E. (2021). Histone acetylation dynamics modulates chromatin conformation and allele-specific interactions at oncogenic loci. Nat. Genet. 53, 650–662. [DOI] [PubMed] [Google Scholar]
- 37.Wang G, Meng Q, Xia B, Zhang S, Lv J, Zhao D, Li Y, Wang X, Zhang L, Cooke JP, et al. (2020). TADsplimer reveals splits and mergers of topologically associating domains for epigenetic regulation of transcription. Genome Biol. 21, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fulco CP, Nasser J, Jones TR, Munson G, Bergman DT, Subramanian V, Grossman SR, Anyoha R, Doughty BR, Patwardhan TA, et al. (2019). Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 51, 1664–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, and Chang HY (2016). HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, and Young RA (2013). Super-enhancers in the control of cell identity and disease. Cell 155, 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Man JCK, Mohan RA, van den Boogaard M, Hilvering CRE, Jenkins C, Wakker V, Bianchi V, Laat W. de, Barnett P, Boukens BJ, et al. (2019). An enhancer cluster controls gene activity and topology of the SCN5A-SCN10A locus in vivo. Nat. Commun. 10, 4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van den Boogaard M, Wong LYE, Tessadori F, Bakker ML, Dreizehnter LK, Wakker V, Bezzina CR, ‘t Hoen PAC, Bakkers J, Barnett P, et al. (2012). Genetic variation in T-box binding element functionally affects SCN5A/SCN10A enhancer. J. Clin. Invest. 122, 2519–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rog-Zielinska EA, Thomson A, Kenyon CJ, Brownstein DG, Moran CM, Szumska D, Michailidou Z, Richardson J, Owen E, Watt A, et al. (2013). Glucocorticoid receptor is required for foetal heart maturation. Hum. Mol. Genet. 22, 3269–3282. [DOI] [PubMed] [Google Scholar]
- 44.Cao Y, Zhang X, Akerberg BN, Yuan H, Sakamoto T, Xiao F, VanDusen NJ, Zhou P, Sweat ME, Wang Y, et al. (2023). In Vivo Dissection of Chamber-Selective Enhancers Reveals Estrogen-Related Receptor as a Regulator of Ventricular Cardiomyocyte Identity. Circulation. 10.1161/CIRCULATIONAHA.122.061955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dowell P, Ishmael JE, Avram D, Peterson VJ, Nevrivy DJ, and Leid M. (1997). p300 functions as a coactivator for the peroxisome proliferator-activated receptor alpha. J. Biol. Chem. 272, 33435–33443. [DOI] [PubMed] [Google Scholar]
- 46.Kuznetsova T, Wang S-Y, Rao NA, Mandoli A, Martens JHA, Rother N, Aartse A, Groh L, Janssen-Megens EM, Li G, et al. (2015). Glucocorticoid receptor and nuclear factor kappa-b affect three-dimensional chromatin organization. Genome Biol. 16, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Luca A, Severino A, De Paolis P, Cottone G, De Luca L, De Falco M, Porcellini A, Volpe M, and Condorelli G. (2003). p300/cAMP-response-element-binding-protein (‘CREB’)-binding protein (CBP) modulates co-operation between myocyte enhancer factor 2A (MEF2A) and thyroid hormone receptor-retinoid X receptor. Biochem. J 369, 477–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. (2015). Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stadhouders R, Vidal E, Serra F, Di Stefano B, Le Dily F, Quilez J, Gomez A, Collombet S, Berenguer C, Cuartero Y, et al. (2018). Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat. Genet. 50, 238–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang R, Chen F, Chen Q, Wan X, Shi M, Chen AK, Ma Z, Li G, Wang M, Ying Y, et al. (2022). MyoD is a 3D genome structure organizer for muscle cell identity. Nat. Commun. 13, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, and Ren B. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He A, Gu F, Hu Y, Ma Q, Ye LY, Akiyama JA, Visel A, Pennacchio LA, and Pu WT (2014). Dynamic GATA4 enhancers shape the chromatin landscape central to heart development and disease. Nat. Commun. 5, 4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bolger AM, Lohse M, and Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen L, Shao N, Liu X, and Nestler E. (2014). ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. (2012). Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parks DH, Tyson GW, Hugenholtz P, and Beiko RG (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke T. (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Putri GH, Anders S, Pyl PT, Pimanda JE, and Zanini F. (2022). Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics 38, 2943–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Love MI, Huber W, and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu G, Wang L-G, Han Y, and He Q-Y (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES, and Aiden EL (2016). Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst 3, 95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abdennur N, and Mirny LA (2020). Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics 36, 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Open2C, Abdennur N, Abraham S, Fudenberg G, Flyamer IM, Galitsyna AA, Goloborodko A, Imakaev M, Oksuz BA, and Venev SV (2022). Cooltools: enabling high-resolution Hi-C analysis in Python. bioRxiv, 2022.10.31.514564. 10.1101/2022.10.31.514564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rowley MJ, Poulet A, Nichols MH, Bixler BJ, Sanborn AL, Brouhard EA, Hermetz K, Linsenbaum H, Csankovszki G, Lieberman Aiden E, et al. (2020). Analysis of Hi-C data using SIP effectively identifies loops in organisms from C. elegans to mammals. Genome Res. 30, 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, and Aiden EL (2016). Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cell Syst 3, 99–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Servant N, Varoquaux N, Lajoie BR, Viara E, Chen C-J, Vert J-P, Heard E, Dekker J, and Barillot E. (2015). HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lareau CA, and Aryee MJ (2018). hichipper: a preprocessing pipeline for calling DNA loops from HiChIP data. Nat. Methods 15, 155–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Phanstiel DH, Boyle AP, Heidari N, and Snyder MP (2015). Mango: a bias-correcting ChIA-PET analysis pipeline. Bioinformatics 31, 3092–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lareau CA, and Aryee MJ (2018). diffloop: a computational framework for identifying and analyzing differential DNA loops from sequencing data. Bioinformatics 34, 672–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang S, Guo Y, and Pu WT (2021). AAV Gene Transfer to the Heart. Methods Mol. Biol. 2158, 269–280. [DOI] [PubMed] [Google Scholar]
- 77.Clement K, Rees H, Canver MC, Gehrke JM, Farouni R, Hsu JY, Cole MA, Liu DR, Joung JK, Bauer DE, et al. (2019). CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37, 224–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. (2012). Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, and Taipale J. (2015). DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527, 384–388. [DOI] [PubMed] [Google Scholar]
- 81.Mahony S, Auron PE, and Benos PV (2007). DNA familial binding profiles made easy: comparison of various motif alignment and clustering strategies. PLoS Comput. Biol. 3, e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. (2005). Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen K, Xi Y, Pan X, Li Z, Kaestner K, Tyler J, Dent S, He X, and Li W. (2013). DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing. Genome Res. 23, 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Anders S, Pyl PT, and Huber W. (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Open2C, Abdennur N, Abraham S, Fudenberg G, Flyamer, Galitsyna AA, Goloborodko A, Imakaev M, Oksuz BA, and Venev SV (2022). Cooltools: enabling high-resolution Hi-C analysis in Python. bioRxiv, 2022.10.31.514564. 10.1101/2022.10.31.514564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li H, and Durbin R. (2010). Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Song F, Xu J, Dixon J, and Yue F. (2022). Analysis of Hi-C Data for Discovery of Structural Variations in Cancer. Methods Mol. Biol. 2301, 143–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, and Young RA (2013). Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, and Young RA (2013). Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol. 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li D, Harrison JK, Purushotham D, and Wang T. (2022). Exploring genomic data coupled with 3D chromatin structures using the WashU Epigenome Browser. Nat. Methods 19, 909–910. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S2 – MPRA of dynamic cardiomyocyte P300 regions. Related to Fig. 3.
Data S3 – Effect of NR motif ablation on enhancer activity. Related to Fig. 4.
Data S4 – Hi-C loops, compartments, and TAD splits during cardiomyocyte maturation. Related to Fig. 5.
Data S5 – Cardiomyocyte gene expression during maturation. Related to Fig. 6.
Data S6 – HiChIP loops in cardiomyocytes at E12.5 and P42. Related to Fig. 7.
Data S7 – Cardiomyocyte super-enhancers at P42 or E12.5. Related to Fig. 7.
Data Availability Statement
The following public data sources were used for this study: E12.5_heart_P300_bioChIP-seq, E12.5_forebrain_P300_bioChIP-seq, adult_heart_P300_bioChIP-seq and adult_heart-EC_P300_bioChIP-seq data are from GSE88789 19; GATA4 bioChIP-seq data of E12.5 or adult heart apex are from GSE52123 52; Mef2a, Mef2c, Nkx2–5, SRF, Tbx5, and Tead1 bioChIP-seq data of E12.5 or adult heart apex are from GSE124008 15. Original data in this manuscript including P300 bioChIP-seq, bulk RNA-seq, Hi-C and H3K27ac HiChIP data of mouse CMs have been deposited to gene expression omnibus SuperSeries GSE195905.
The custom scripts used in this study are publicly available on github (https://github.com/yanchunzhang/Cardiomyocyte_maturation_HiC). DOI 10.5281/zenodo.7716979.
Additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.