Abstract
Fetal bovine serum (FBS) is used commonly in cell culture. Charcoal-stripped FBS (CS-FBS) is used to study androgen responsiveness and androgen metabolism in cultured prostate cancer (CaP) cells. Switching CaP cells from FBS to CS-FBS may reduce activity of androgen receptor (AR), inhibit cell proliferation, or modulate intracellular androgen metabolism. Removal of proteins by charcoal stripping may cause changes in biological functions. Proteins in FBS and CS-FBS were profiled using an ion current-based quantitative platform consisting of reproducible surfactant-aided precipitation/on-pellet digestion, long-column nano-liquid chromatography (LC) separation, and ion current-based analysis (ICan). A total of 143 proteins were identified in FBS, among which 14 proteins including insulin-like growth factor 2 (IGF-2) and IGF binding protein (IGFBP)−2 and −6 were reduced in CS-FBS. IGF1 receptor (IGF1R) and insulin receptor (IR) were sensitized to IGFs in CS-FBS. IGF1 and IGF2 stimulation fully compensated for the loss of AR activity to maintain cell growth in CS-FBS. Endogenous production of IGF and IGFBPs was verified in CaP cells and clinical CaP specimens. This study provided the most comprehensive protein profiles of FBS and CS-FBS, and offered an opportunity to identify new protein regulators and signaling pathways that regulate AR activity, androgen metabolism and proliferation of CaP cells.
Keywords: Prostate cancer, serum, fetal bovine serum, charcoal stripping, growth factors, cell culture, medium, proteomics, IGF1, phosphorylation
INTRODUCTION
Androgen receptor (AR) plays an important role in prostate cancer. AR is activated by testosterone (T) and dihydrotestosterone (DHT), its preferred ligands, to transcribe genes that are critical for proliferation of CaPcells 1–4. CaPcell lines are used as models for studying functions of AR and androgen metabolism that produce T or DHT from various androgen substrates. Two types of fetal bovine serum (FBS) are used in cell culture medium in prostate cancer research. FBS is used for routine propagation of CaPcell lines. Charcoal-stripped FBS (CS-FBS) is used to mimic androgen-free conditions in studies on androgen metabolism and androgen signaling in prostate cancer cells 5–6 because charcoal stripping removes trace amount of androgens in FBS, which are believed to interfere with experiments. CaP cells show reduced AR activity and growth rate and change intracellular androgen metabolism upon transfer from medium supplemented with FBS to medium supplemented with CS-FBS 7. The changes in AR activity and growth rate are attributed often to the removal of androgens from FBS. T is the only androgen detected in FBS using liquid chromatography-tandem mass spectrometry (LC-MS/MS) 7. The amount of T in medium supplemented with 10% FBS is ~0.03 nM, which may minimally activate AR activity. Charcoal stripping may remove other factors that maintain AR activity, sustain cell growth, or modulate androgen metabolism.
FBS provides a broad spectrum of biological molecules, which include growth factors, hormones, carrier proteins, attachment and spreading proteins, protease inhibitors, fatty acids and lipids, and low molecular weight nutrients 8. Charcoal stripping removes hormones and growth factors, certain vitamins and metabolites from FBS 9–10. The effects of removal or reduction of other hormones or growth factors may confound results of cellular response to factors of interest 11. Protein composition of FBS and CS-FBS have not been defined clearly even though both have been used widely in cell culture. Proteomic techniques have been applied to analyze different lots of FBS and approximately 80 proteins were identified 12. No data have been reported with regard to proteins that may be removed using charcoal stripping. Characterization of CS-FBS at the proteomic level will help researchers utilize CS-FBS in a better informed manner, and interpret results with appropriate caution. The acquired profiles of proteins that are reduced in CS-FBS may present protein candidates as modulators of cell signaling pathways that provide new targets to regulate AR activity, cell growth, and/or androgen metabolism.
LC-MS/MS based proteomic techniques enable identification and quantification of proteins on a global scale. Two quantitative strategies are applied commonly in proteomics: i) labeling techniques, such as isobaric tags, for relative and absolute quantification (iTRAQ) 13, tandem mass tags (TMT) 14, and stable isotope labeling using amino acids in cell culture (SILAC) 15, and ii) label-free approaches, such as spectral counting 16–17 and peptide ion current-based analyses 18–20. Increasing use of high-resolution mass spectrometry (MS) instrumentations makes peptide ion current-based methods attractive for their simplicity, cost-effectiveness, and feasibility of analysis of multiple biological samples (N≥10) 21–22. The ion current-based approach developed previously by our group assigns significance to proteins with small changes (i.e. 1.5-fold) and shows better performance than other label-free methods 23.
The present study was designed to obtain protein profiles of FBS and CS-FBS in order to provide a comprehensive view of protein composition of FBS, determine the proteins that are removed using charcoal stripping, and identify proteins that contribute to AR activation or stimulate cell growth. Ion current-based analysis was employed to perform relative quantification among FBS, commercially available CS-FBS (CCS-FBS) and 2 home-made CS-FBS (HCS1/2-FBS). IGF1R signaling-associated proteins were identified and tested for their effect on cell growth and IGF response.
EXPERIMENTAL PROCEDURES
FBS samples
A commercial FBS sample, a commercial CS-FBS (CCS-FBS) sample, and 2 home-made CS-FBS (HCS1/2-FBS) samples were analyzed. The FBS (catalog number S11550) and CCS-FBS (catalog number SH30068.03) were purchased from Atlantic Biologicals (Atlanta, GA) and Hyclone Laboratory (Logan, UT), respectively. HCS-FBS was made from filtration of FBS through activated charcoal as described 7. HCS1-FBS was prepared from the same batch of FBS used in this study, whereas HCS2-FBS was prepared from a different batch of FBS. Sera were stored at −20°C until used.
Reagents and cell culture
Recombinant human insulin-like growth factor (IGF)−1 and −2 (Thermo Fisher Scientific, Waltham, MA) were reconstituted in 0.1% bovine serum albumin cold ethanol fraction (BSA) in phosphate-buffered saline (PBS) (w/v). BSA and human insulin solution were purchased from Sigma-Aldrich (St. Louis, MO). The concentration of insulin (μU/ml) was calculated based on the assumption that 1 international unit (IU) equals 0.0347 mg insulin (World Health Organization document WHO/BS/10.2143). Antibodies for IGF-1 receptor (IGF1R), insulin receptor (IR), phospho-IGF1R, and phospho-IR were used from the Phospho-Insulin/IGF Receptor Antibody Sampler Kit (Cell Signaling Technology, Danvers, MA). Antibodies for Akt and phospho-Akt (S473) were purchased from Cell Signaling Technology. Antibody for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was purchased from EMD Millipore (Billerica, MA). VCaP, LAPC-4, and LNCaP were used as cell models in these experiments because the cell lines are commonly used CaP cell lines that express AR 24. VCaP and LNCaP were purchased from the ATCC (Manassas, VA). LAPC-4 cells were established in Dr. Charles Sawyers laboratory 25.
Cell line authentication was performed in the Roswell Park Comprehensive Cancer Center (Roswell Park) Genetic Shared Resources. DNA profiles were acquired using 15 short tandem repeat (STR) loci and an Amelogenin gender-specific marker. Test and control samples were amplified using the AmpFLSTR® Identifiler® Plus PCR Amplification Kit (Thermo Fisher Scientific) using the Verti 96-well Thermal Cycler (Applied Biosystems, Foster City, CA) in 9600 Emulation Mode (initial denature: 95°C 11 min, 28 cycles of denature: 94°C 20 sec and anneal/extend: 59°C 3 min, final extension: 60°C 10 min, and hold: 12°C). PCR products were evaluated using the 3130xl Genetic Analyzer (Applied Biosystems) and analyzed using GeneMapper v4.0 (Applied Biosystems). Eight of the 15 STRs and Amelogenin from the DNA profile for the cell line(s) were compared to the ATCC STR database (https://www.atcc.org/STR%20Database.aspx?slp=1) and the DSMZ combined Online STR Matching Analysis (http://www.dsmz.de/fp/cgi-bin/str.html). All matches above 80% were considered the same lineage. LNCaP and VCaP cell lines were authenticated and identity verified. Authentication was performed for LAPC-4 but the results were not conclusive due to the lack of commonly accepted standard databases for this cell line.
VCaP cells were maintained in medium A that was DMEM (catalog number 11995–065) (Thermo Fisher Scientific), supplemented with 10% (v/v) FBS, 2 mM L-glutamine (catalog number MT25-005-CI) (Corning Life Sciences, Tewksbury, MA), and 100 U/ml penicillin and 100 μg/ml streptomycin (catalog number MT30-002-CI) (Corning Life Sciences). For cell culture in CS-FBS, VCaP cells were cultured in medium B that was phenol red-free DMEM (catalog number 31053–028) (Thermo Fisher Scientific), supplemented with 10% (v/v) CS-FBS, 6 mM L-glutamine, 1 mM sodium pyruvate (catalog number MT25-000-CI, Corning Life Sciences), and 100 U/ml penicillin and 100 μg/ml streptomycin. L-glutamine and sodium pyruvate were added to the phenol red-free DMEM medium supplemented with 10% CS-FBS in different amounts compared to regular DMEM medium supplemented with 10% FBS in order to ensure the same final concentrations of the ingredients in the culture medium. All media for VCaP culture were prepared freshly and equilibrated for 10–15 minutes in a cell culture incubator at 37°C in an atmosphere with 95% air and 5% CO2 before application to cells. LAPC-4 and LNCaP cells were maintained in RPMI1640 medium (catalog number 11875–093, Thermo Fisher Scientific), supplemented with 10% (v/v) CS-FBS, 2 mM L-glutamine, and 100 U/ml penicillin and 100 μg/ml streptomycin. For cell culture in CS-FBS, LAPC-4 or LNCaP cells were cultured in medium C that was phenol red-free RPMI1640 (catalog number 11835–030, Thermo Fisher Scientific), supplemented with 10% (v/v) CS-FBS, 2 mM L-glutamine, and 100 U/ml penicillin and 100 μg/ml streptomycin. In all experiments that used LAPC-4 cells, cell culture vessels were coated with 1.7 μg/ml poly-D-lysine (Sigma-Aldrich) in H2O at 0.076 ml/cm2 at room temperature for 15 min, followed by aspiration of the coating reagent and overnight air dry.
NanoLC-MS/MS Sample preparation
The whole protein concentrations of sera were determined using the bicinchoninic acid (BCA) Protein Assay (Pierce, Rockford, IL). All serum samples (each containing ~100 μg of total protein) were denatured in an ice-cold lysis buffer (50 mM Tris-formic acid, 150 mM NaCl, 0.5% sodium deoxycholate, 2% SDS, 2% NP-40, pH 8.0) as described 12, 26. Lysis buffer containing strong surfactants improved enzymatic digestion performance in terms of peptide recovery, reproducibility and sensitivity for plasma sample preparation and analysis 27. The mixtures were reduced with 3 mM tris(2-carboxyethyl)phosphine (TCEP) for 10 min and alkylated with 20 mM 2-iodoacetamide (IAM) for 30 min in darkness. An acetone precipitation/on-pellet-digestion procedure was applied to perform precipitation and tryptic digestion for reproducible peptide recovery as reported 28–29.
NanoLC-MS/MS analysis
The peptide mixture was separated using a nano-LC system (Eksigent, Dublin, CA) and analyzed using an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific). The mobile phase A consisted of 0.1% formic acid in 2% acetonitrile. Mobile phase B contained 0.1% formic acid in 88% acetonitrile. The nano-LC column was heated at 52°C to improve chromatographic resolution and reproducibility. Peptide mixture was loaded onto a reversed-phase trap (300 μm ID x0.5 cm) with 1% mobile phase B at flow rate 10 μL/min and washed for 3 min. A series of nanoflow gradients (flow rate 250 nL/min) was used to back-flush the trapped peptides onto the nano-LC column (75 μm ID × 75 cm, packed with 3-μm particles) for separation. A 160-min gradient was applied and gradient slope was adjusted to account for the change of mobile phase B: 0 to 3% over 3 min; 3 to 6% over 5 min; 6 to 28% over 118 min; 28 to 50% over 10 min; 50 to 97% over 1 min; and isocratic at 97% B for 23 min.
LC-MS/MS was acquired using the data-dependent product ion mode. Survey scans (m/z range 400–1500) were performed at a resolution of 120,000 with an automatic gain control (AGC) target of 5 × 105. Tandem mass spectrometry (MS2) was performed using isolation at 1.2 Th with the quadrupole for high energy collision dissociation (HCD) fragmentation. The normalized collision energy was 35% with activation q 0.25 and tandem mass spectra were analyzed using Orbitrap with resolution 15,000 in centroid mode. Monoisotopic precursor selection was used. The MS2 AGC target was set to 5 × 104 and the maximum injection time was 50 ms. Peptide precursors with charge state 2–7 were sampled for MS2. Dynamic exclusion was enabled with the settings: repeat count 1; repeat duration 50 s; exclusion duration 60 s; and mass tolerance ± 10 ppm. The instrument was run in top speed mode with cycle time 3 s. Capillary temperature was 250°C. Each sample was analyzed 5 times.
Database search and validation
Raw files were used to perform database searching against the Swiss-Prot protein database (released January, 2015) using MaxQuant (version 1.5.2.8) 30 that included the Andromeda search engine 31. The bovine database contains 5,993 protein entries. The search parameters used were: Two missed cleavages were permitted for fully tryptic peptides. The minimum required peptide length was 7 amino acids. Mass tolerances for first search peptide tolerance and main search peptide tolerance were 20 ppm and 4.5 ppm, respectively. Mass tolerance for fragment ion masses was 20 ppm. Carbamidomethylation of cysteines was set as a fixed modification. Variable modifications of methionine oxidation and protein N-terminal acetylation were allowed. The false discovery rate (FDR) was determined using a target-decoy search strategy to search against the databases using reversed sequences 32. The maximum FDRs of peptide-spectrum matches (PSM) and protein were set to 1% for identification. The “matching between runs” option in MaxQuant for label-free quantification was selected with match time window 1 min and an alignment time window 20 min.
Protein quantification
Quantitative analysis was performed using the ion current-based approach (Fig. 1). The peak detection and chromatographic alignment based on retention time, m/z and charge states were analyzed using SIEVE® v2.2 (Thermo Fisher Scientific). Quantitative frames/features were defined based on m/z (width 10 ppm) and retention time (width 2.5 min) of peptide precursors in the aligned runs. Peak area under the curve (AUC) was calculated for individual replicates in each frame. The MS2 fragmentation scans associated with each frame were assigned to the peptide/protein identifications from MaxQuant. Frames assigned to multiple peptides and shared peptides assigned to multiple proteins were excluded in the quantitative analysis. LOESS normalization 33 was performed to reduce systematic bias. A value of 1000 was assigned as the baseline quantitative value in case of missing data 21. Intensities of frames with the same sequence were combined to form unique peptide intensity. Intensities of unique peptides of the same protein were combined to form one protein intensity for Grubbs’ test analysis using the ListPOR program (Version 2.2.2104, panomics.pnnl.gov). Minimum dataset presence was 2 and p-value cutoff was 0.01. The relative protein ratio was calculated by comparing the summed abundance values of the protein in each group. Student’s t-test statistics were applied to analyze log-transformed values of protein intensities. Abundance change ≥ 1.5-fold and p-value ≤ 0.05 were used as thresholds to define altered proteins. The label-free quantification (LFQ) intensities from MaxQuant were used for relative quantification and protein ratios were calculated by comparing the LFQ intensities in each group. Results were compared for the ion current-based approach and LFQ quantitation using MaxQuant.
Fig. 1.
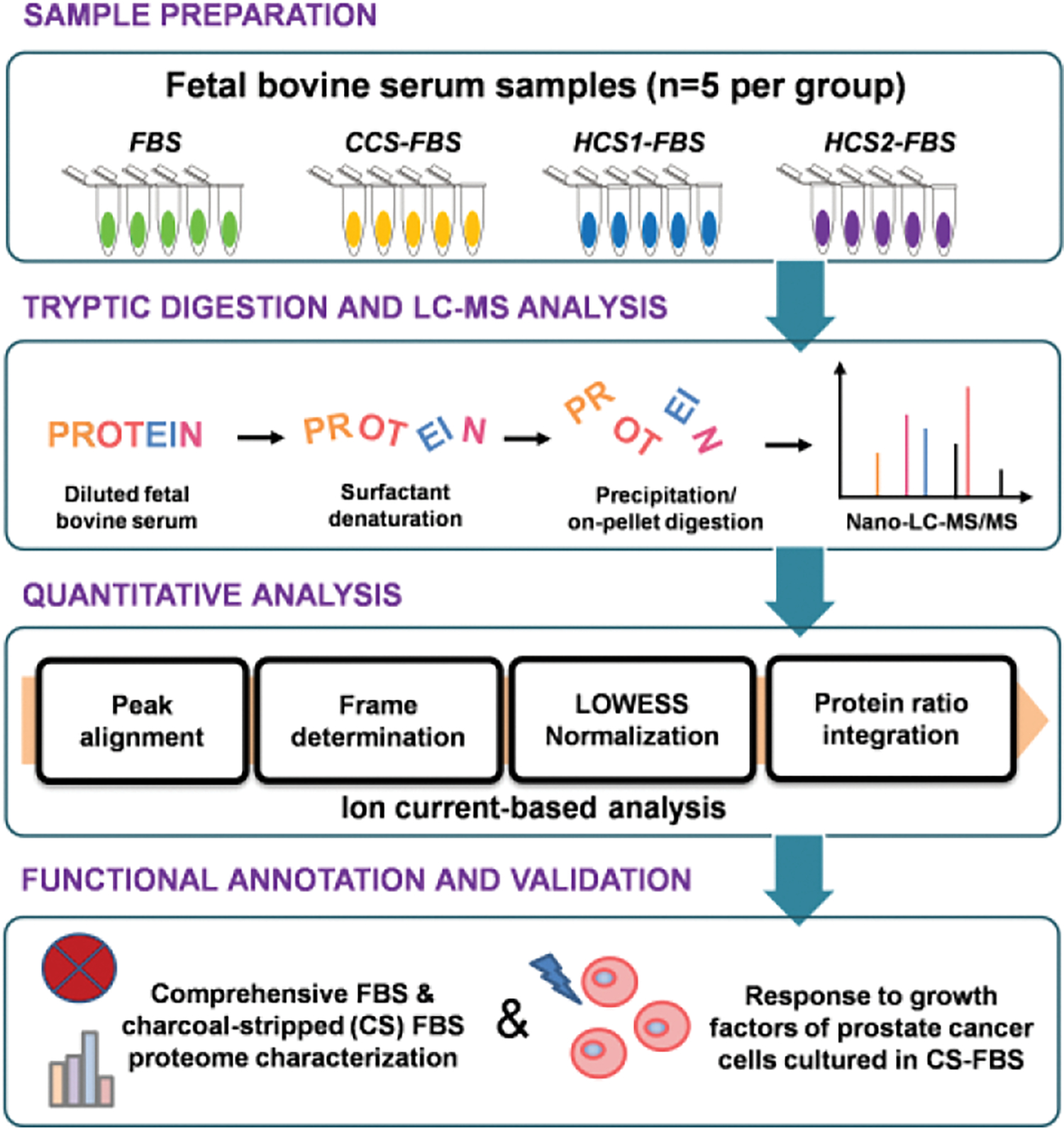
The experimental flowchart of comparative proteomic profiling of charcoal stripped fetal bovine serum (CS-FBS). Fetal bovine serum (FBS), commercial CS-FBS (CCS-FBS), and two homemade CS-FBS (HCS1-FBS and HCS2-FBS) were analyzed using the ion current-based quantitative analysis.
Bioinformatics analysis
Ingenuity Pathway Analysis (IPA, Ingenuity Systems, http://www.lingenuity.com/) was used for protein function and pathway analyses. Ingenuity’s knowledge base was created by manual curation of scientific literature supported by experimental results that were structured into an ontological relational database. Gene Ontology (GO) analysis was performed using High-Throughput GoMiner 34. Hierarchical cluster analysis was performed using Cluster 3.0 35 and displayed using TreeView, which supports tree-based and image-based browsing of hierarchical trees (http://www.eisenlab.org). In the heat map generated using TreeView, the elevated protein intensities were indicated in red, and the decreased protein intensities were indicated in green. The prediction of secreted proteins was performed using SignalP 4.1 Server 36, which discriminates between signal peptides and transmembrane regions.
Cell viability assay
Cell viability was assessed using 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) 37. A stock solution of 5 mg/ml MTT in H2O was prepared and used within 2 weeks of preparation. Cells were cultured and treated in phenol red-free medium. At the end of each treatment, fresh medium corresponding to each respective treatment condition was prepared. MTT stock solution was added to the fresh treatment medium at 20% (v/v). Culture medium was removed from cell culture, and fresh MTT-containing treatment medium was added to the cell culture. Cells were cultured at 37°C in a cell incubator for 4 hr. A solubilization solution (20% SDS in 0.02 M HCl) was added to cell culture at equal volume of the culture medium. Cells were cultured at 37°C overnight. Cell lysate was mixed thoroughly and absorption at 570 nm was measured on a Spectramax 384 Plus plate reader (Molecular Devices, Sunnyvale, CA).
Construction of an AR activity reporter plasmid using a secreted luciferase
Activity of AR was assessed using the androgen-responsive pGL3-ARE3-luciferase promoter reporter vector 38. The pGL3-ARE3-luciferase vector contains 3 tandemly arranged ARE (ARR3) from the rat probasin promoter, an AR-transcribed, androgen-responsive gene 39. A new ARE-luciferase promoter activity reporter vector was constructed for this study. The ARR3 was cloned into a plasmid pEZX-PG02 (GeneCopoeia, Rockville, MD) using the In-Fusion HD Cloning Plus CE Kit (Clonetch Laboratories, Mountain View, CA). Primers for PCR amplification were designed using the respective DNA sequence and following the manual of the In-Fusion kit. Primers for PCR amplification of the ARR3 fragment were: 5’-CAGATCTTGGAATTCAAGCTTGGAGCTTATGATAG-3’ and 5’-CTCGGTACCAAGCTTCAGATCTGCGGCACGCTG-3’. Primers for PCR amplification of the pEZX-PG02 fragment were: 5’-AAGCTTGGTACCGAGCTC-3’ and 5’-GAATTCCAAGATCTGGTTCTATC-3’. PCR was performed using a Platinum Taq DNA Polymerase High Fidelity kit (Thermo Fisher Scientific) on a MyCycler thermal cycler (Bio-Rad, Hercules, CA). The pEZX-PG02 fragment was amplified using the conditions: 94°C for 1 min, 30 cycles of 94°C for 15 sec, 55°C for 30 sec, and 68°C for 6 min, followed by holding at 4°C. PCR conditions for amplification of ARR3 fragment were: 94°C, 1 min, 30 cycles of 94°C for 15 sec, 60°C for 30 sec, and 68°C for 1 min, followed by holding at 4°C. The newly constructed ARE-luciferase vector was named pEZX-PG02-ARE, which contained the gene encoding the secretory Gaussia luciferase. The luciferase is expressed and secreted into the medium upon AR activation; activity of the luciferase in the medium indicates activity of AR.
AR activity assay
Cells were transfected with the pEZX-PG02-ARE plasmid using Lipofectamine 2000 (Thermo Fisher Scientific). Transfected cells were replated at 50,000 cells per well on 24-well plates in culture medium supplemented with 10% CCS-FBS for 24 hr, then treated with fresh treatment medium for 24 hr. At the end of treatment, 10 μl of culture medium was collected. Luciferase activity was assessed using the Secrete-Pair Gaussia Luciferase Assay Kit (GeneCopoeia). Luciferase assay was performed on a Clarity Luminescence Microplate Reader (BioTek Instruments, Inc., Winooski, VT). The viability of cells from the same culture was measured using the MTT assay. Luciferase activity was normalized for each replicate using the reading at OD570.
Western blotting
Sample preparation, protein determination, SDS-PAGE and Western blotting were described 40. Cells in culture were rinsed 3 times with PBS, and lysed in cell lysis buffer (Cell Signaling Technology) and supplemented with 1 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich), 50 mM NaF, and 1 tablet/7 mL of Mini Complete Protease Inhibitor (Roche Applied Science, Indianapolis, IN). Protein concentration of the lysate was determined using the Bicinchoninic Acid Protein Assay Kit (Pierce). Cell lysate was mixed with one-third volume of 4X SDS sample buffer [200 mmol/L Tris-HCl (pH 6.8), 8% SDS, 0.4% bromophenol blue, 40% glycerol, 60 μL/mL β-mercaptoethanol] and heated at 100°C for 10 minutes. Polyvinylidene difluoride (PVDF) membranes containing the transferred proteins were blocked with 5% nonfat dry milk in TBST buffer [10 mmol/L Tris-HCl (pH 7.5), 150 mmol/L NaCl, 0.1% Tween 20] at room temperature for 1 hr before probing with the primary antibodies and the horseradish peroxidase (HRP)-conjugated secondary antibodie (BioRad). Protein bands were visualized using the SuperSignal West Pico Chemiluminescent Substrate kit (Pierce) or the Luminata Crescendo Western HRP Substrate (EMD Millipore).
Growth factor antibody array analysis
VCaP cells were seeded at 3×106 per 10-cm tissue culture dish and cultured for 6 days in 10 ml culture medium supplemented with 10% FBS. Cell culture was rinsed 3 times with 10 ml fresh culture medium, and cultured for 3 days in 10 ml fresh culture medium with 10% FBS or 10% HCS-FBS. The remainder of each fresh medium was stored at −80°C. Culture medium was collected and centrifuged to remove floating cells at 400x for 5 min. Supernatant was stored at −80°C before antibody array analysis. Cells were lysed in cell lysis buffer following manufacturer’s instruction (RayBiotech, Norcross, GA). Growth factors in the stored fresh medium and the cell culture medium were analyzed using RayBio C-Series Human Growth Factor Antibody Array C1 (RayBiotech).
CaP tissue specimens and tissue microarray (TMA)
Sections with thickness 4 μm of an Formalin-fixed, paraffin-embedded (FFPE) CaP tissue block was obtained from Roswell Park Comprehensive Cancer Center (Roswell Park) Pathology Network Shared Resource (PNSR). A TMA block was constructed by Roswell Park PNRS. The TMA block contained matched CaP tissue and adjacent benign prostate tissue from 10 patients. Each tissue specimen had 3 cores. Serial sections with thickness 4 μm from the TMA block were prepared by Roswell Park PNSR. The use of the TMA and CaP FFPE sections was covered under an institutional comprehensive tissue procurement protocol “Roswell Park Remnant Clinical Biospecimen Storage, Collection and Distribution for Research Purposes”.
Immunohistochemistry (IHC) staining
FFPE sections were de-paraffinized, rehydrated under an alcohol gradient, and antigen retrieved using Reveal Decloaker (Biocare Medical, Concord, CA) for 30 min at 110°C and 5.5 – 6.0 psi. Sections were blocked for endogenous peroxidase activity using 3% H2O2 in dd-H2O for 15 min at room temperature, washed in 10 mM Tris-HCl (pH 7.8), and blocked with Background Punisher (Biocare Medical) for 10 minutes at room temperature to reduce nonspecific staining. IGF1R antibody (LSBio, Seattle, WA), InsR antibody (LSBio), and non-immuned control mouse IgG (Sigma-Aldrich) were diluted in Renoir Red Diluent (Biocare Medical) and used at 0.001667 mg IgG/ml, 0.005 mg IgG/ml, or 0.005 mg IgG/ml, respectively. Sections were incubated overnight with the diluted antibodies at 4°C. The sections were incubated with the Biocare MACH4 mouse probe and MACH 4 HRP polymer (Biocare Medical) at room temperature for 15 minutes. Immunostaining was developed using diaminobenzidine (Sigma-Aldrich) and the sections were counterstained with hematoxylin (Vector Laboratories, Burlingame, CA). Sections were dehydrated and mounted using Cytoseal 60 permanent mounting medium (Richard-Allen Scientific, Kalamazoo, MI). Sections were scanned using Aperio ScanScope XT (Leica Biosystems, Buffalo Grove, IL), and images were organized using Aperio eSlide Manager.
In situ RNA hybridization
In situ RNA hybridization was performed to assess expression of genes of interest using the RNAScope 2.5 HD Brown Assay (Advanced Cell Diagnostics Inc., Hayward, CA). Catalog numbers of the probes used for detection of mRNA of human IGF-1, IGF-2, IGFBP-2, IGFBP-3, IGFBP6, and IGFBP-7 were 313031, 594361, 313061, 313051, 496061, and 316681, respectively. RNAScope was performed following the manufacturer’s instruction. Sections were scanned on an Aperio ScanScope XT. Images were acquired and managed using Aperio eSlide Manager.
RESULTS
Removal of T from FBS with charcoal-stripping had limited impact on AR activity and growth of CaP cells.
AR activity assay was performed using multiple human CaP cell lines to test if T removal from FBS using charcoal-stripping impacted AR activity. AR activity varied in the 3 cell lines that were cultured in FBS (Fig. 2A). AR activity was diminished in cells cultured in CS-FBS without T. Addition of 0.03 nM T in medium supplemented with CS-FBS did not restore AR activity to the levels found in medium supplemented with FBS. Addition of 1 nM T stimulated AR activity. In VCaP and LAPC-4 cells, 1 nM T-stimulated AR activity surpassed AR activity in FBS. Charcoal-stripping of FBS impaired AR activity. The data suggested that factors other than T played a more significant role in stimulating or maintaining AR activity in FBS.
Fig. 2.
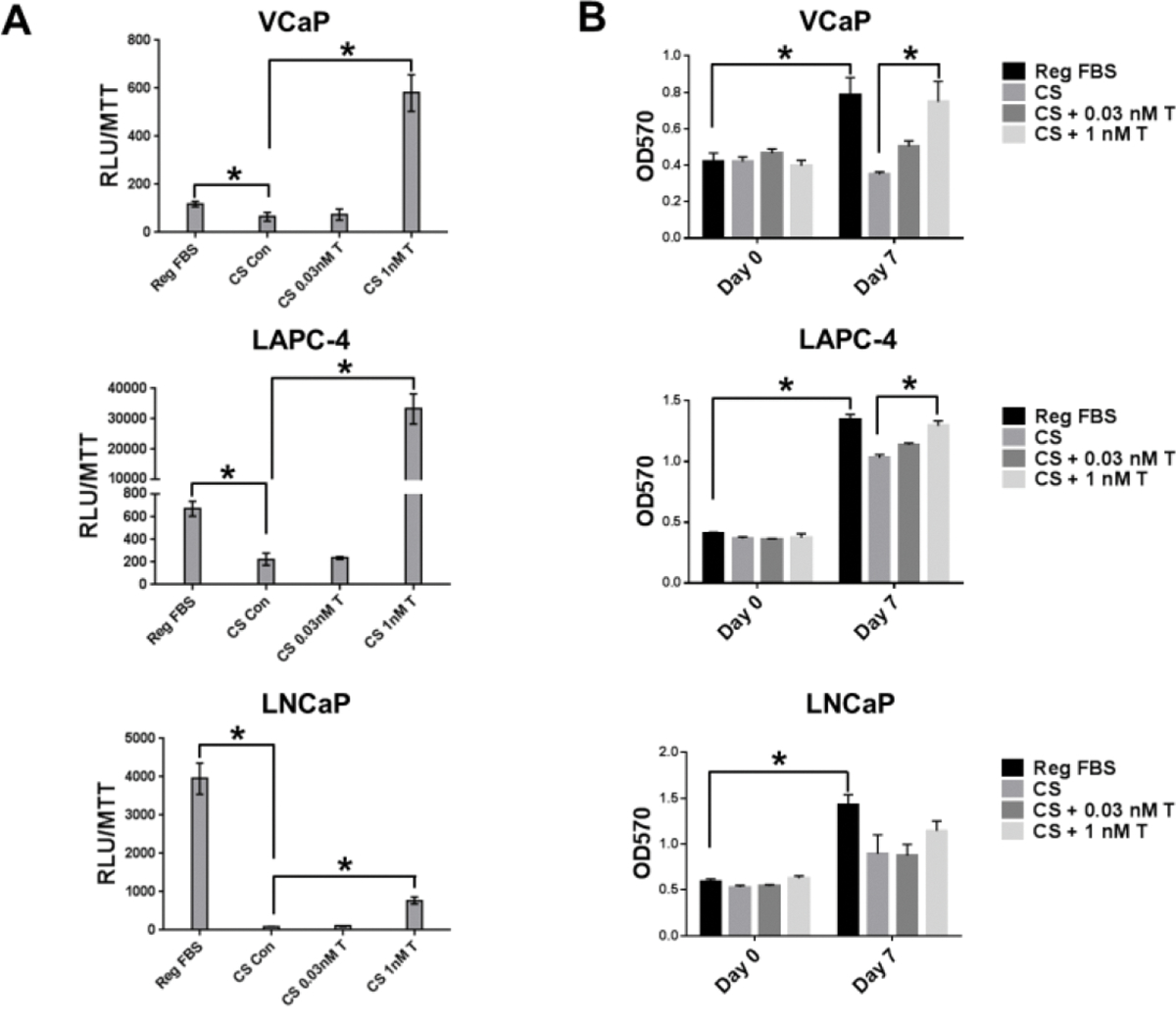
Limited impact of removing testosterone (T) from FBS on (A) AR activity; and (B) CaP cell growth. AR activity assay using Secrete Paired luciferase assay. Experiments were set up in 3 (n=3) or 4 (n=4) replicates for (A) or (B), respectively. RLU/MTT – relative light units per MTT reading at OD570; Reg FBS-medium supplemented with 10% FBS; CS Con-10% HCS1-FBS without T; CS 0.03 nM T- 10% HCS1-FBS and 0.03 nM T; CS 1 nMT −10% HCS1-FBS and 1 nM T. *: p < 0.05.
MTT assay revealed that charcoal-stripping of FBS affected cell proliferation (Fig. 2B). Cell lines multiplied 2–3 fold in medium supplemented with FBS; VCaP cells stopped growing in medium supplemented with CS-FBS, and viability was reduced on Day 7 compared to Day 0. T at 0.03 nM maintained viability on day 7 at levels similar to Day 0. T at 1 nM restored growth to levels comparable to FBS. LAPC-4 and LNCaP cells maintained growth in medium supplemented with CS-FBS, although the growth rate was lower than that in medium supplemented with CS-FBS, Overall growth response to T was weaker in LNCaP and LAPC-4 cells than VCaP cells. Growth stimulatory effect of T at 1 nM in all cell lines was consistent with the stimulation of AR activity. The reason for viability of VCaP cells maintained by 0.03 nM T remains unclear. Growth rate of the cell lines was reduced in medium supplemented with CS-FBS, and adding 0.03 nM T to the CS-FBS did not restore growth and AR activity. Charcoal-stripping must have removed factors that acted via mechanisms that were independent of the AR signaling pathway to maintain cell growth. VCaP cell line was selected as the cell model for in vitro experiments for proteomic study of FBS and CS-FBS
Protein profiles of FBS
A total of 143 protein groups with at least 2 distinct peptides were identified (Supplemental Table S1) using MaxQuant with FDR 1% at both PSM and protein level. The physicochemical characteristics of the identified proteins were further analyzed. Most proteins (>70%) in the FBS proteome had molecular weight (MW) < 60 kDa (Fig. 3A). Vast majority of the identified proteins in FBS are hydrophilic; 95.8% of proteins had negative grand average of hydropathy (GRAVY) values (Fig. 3B). Most proteins (102, 71.3%) were secreted proteins as predicted by Singal P 4.1. Gene Ontology (GO)–cellular component analysis showed that 113 and 16 proteins were assigned to the extracellular region and cell surface, respectively (Fig. 3C). Protein absolute quantitative values (PAQV), calculated as protein abundance values divided by the number of quantified unique peptides, were used to determine the ranking order of proteins in FBS. Fetuin-A (alpha-2-HS-glycoportein) was the most abundant, followed by serum albumin and alpha-1-antitrypsin (Fig. 3D). The top 20 proteins in FBS accounted for 89.9% of protein abundance. Fetuin-A and albumin PAQV were 32.0% and 28.2% of the total PAQV values, respectively. Alpha-fetoprotein (AFP), a glycoprotein produced early in gestation, was the 10th most abundant protein in FBS.
Fig. 3.
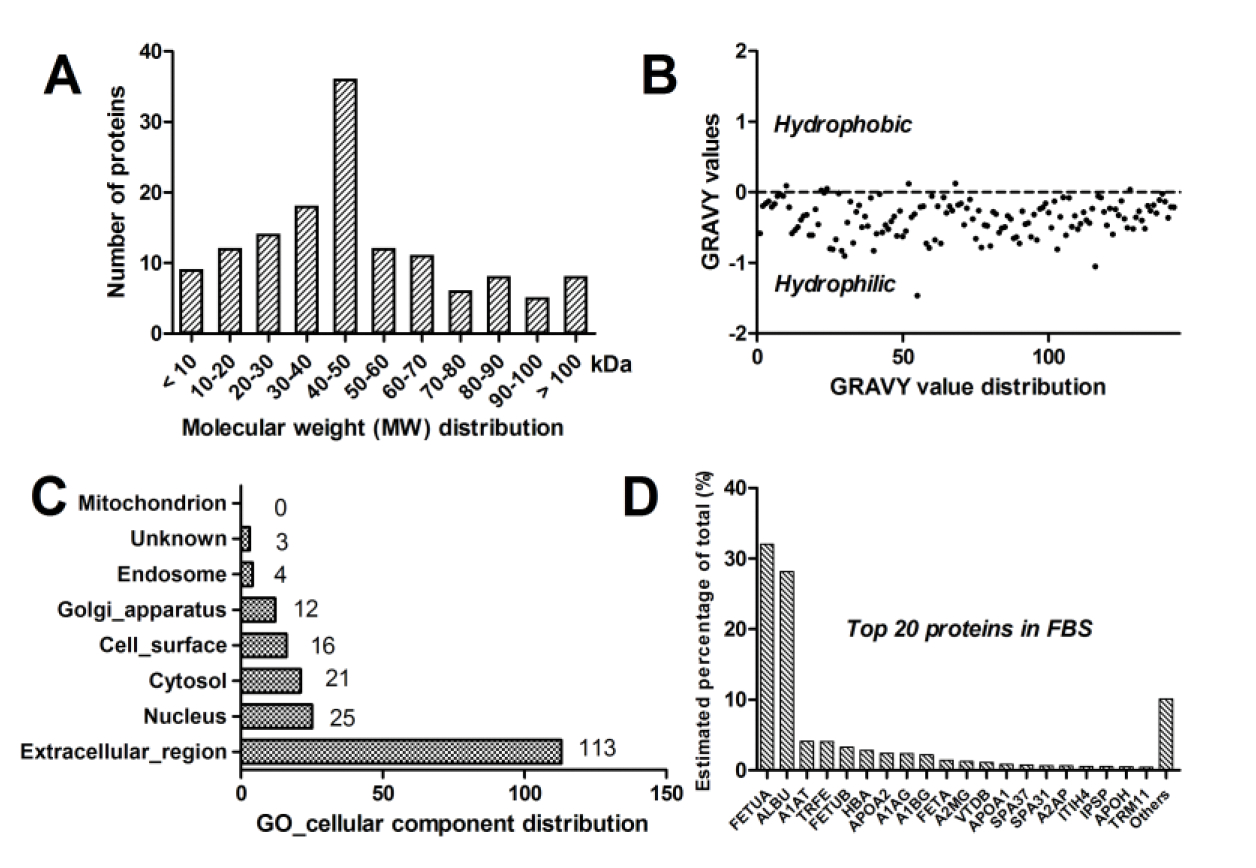
Characteristics of the fetal bovine serum proteins. (A) Molecular weight (MW) distribution; (B) Hydrophobicity (as expressed by the GRAVY values) distribution; (C) GO cellular component distribution; and (D) Top 20 proteins in fetal bovine serum (FBS).
Proteomic comparison of FBS and CS-FBS
Peak alignment scores ranged from 0.83 to 0.96, which indicated excellent reproducibility of the 20 LC-MS/MS analyses (Fig. 4A). R-squared values of linear regression analysis between any 2 randomly selected replicates from the same group were all larger than 0.999. The linear correlation was almost perfect through a dynamic range of more than 6 orders of magnitude (Fig. 4B). The coefficients of variations (CVs, calculated as the ratios of standard deviation to mean) of quantified proteins among 5 replicates in each group were calculated. The median CV values for all quantified proteins were 6.7%, 7.2%, 8.3% and 7.3%, respectively for FBS, CCS-FBS, HCS1-FBS and HCS2-FBS. The excellent peak alignment and reproducibility improved the quantitative accuracy and sensitivity. The percentage of missing values (%) at protein levels across all the 20 LC-MS/MS analyses was calculated as: Percentage of missing values (%) = Total number of missing values in all analyses / (the number of identified proteins × 20) × 100%. Only ~0.1% of missing values across 20 LC-MS/MS analyses were presented in analyses, while ~31.3% of missing values were observed using the MaxQuant label-free quantification method (Fig. 4C). Since FBS and HCS1-FBS are paired samples that prepared from the same bottle of sera, theoretically no proteins with increased expression levels should be observed in HCS1-FBS. A volcano plot used log10-transferred p-values in the x-axis and log2-transferred ratios in the y-axis (Fig. 4D). The expression of twenty proteins were decreased and only 1 was increased in HCS1-FBS compared to FBS among the 21 proteins that were changed.
Fig. 4.
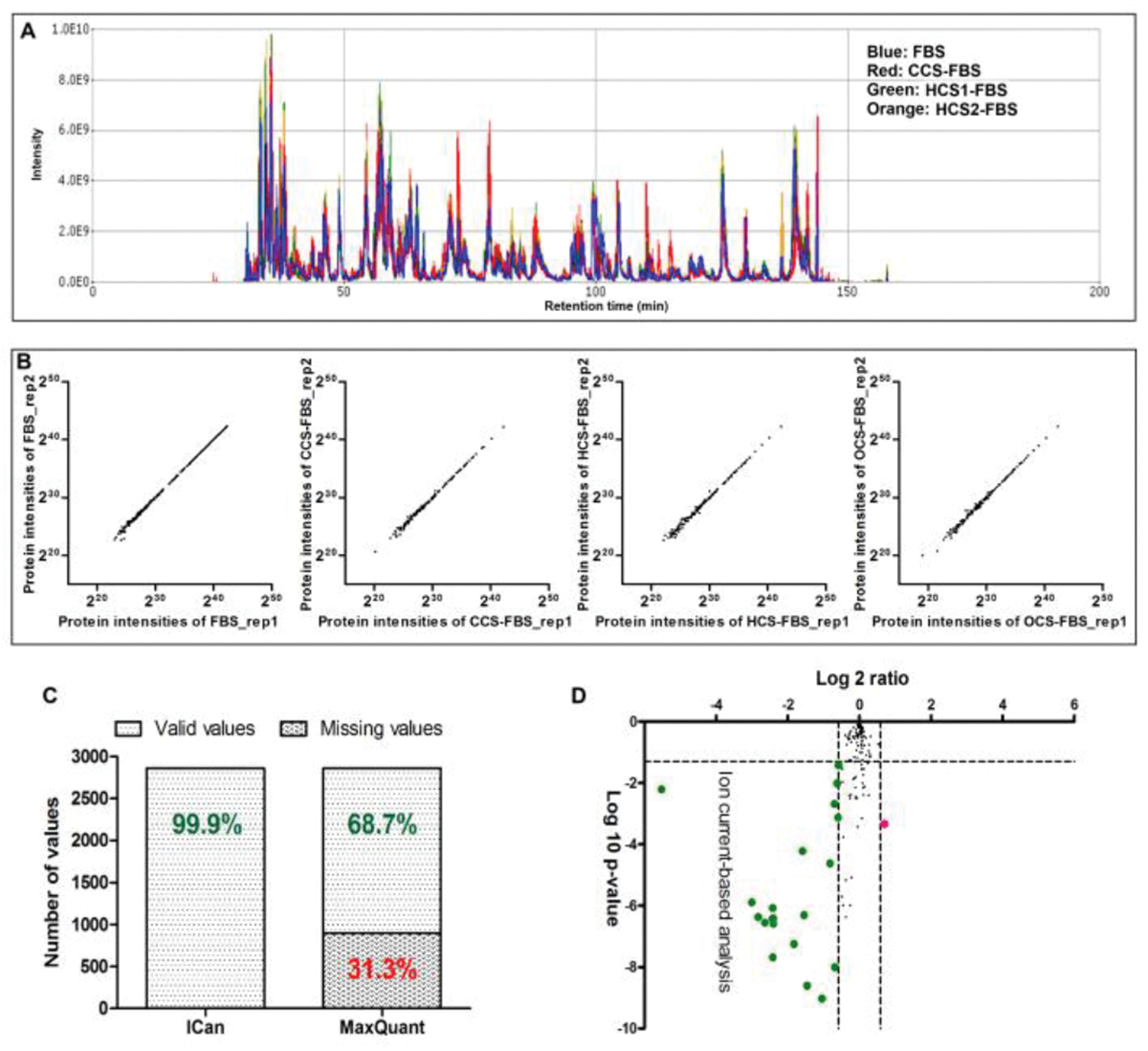
Evaluation of ion current-based quantitative analysis. (A) Peak alignment using SIEVE package (Thermo Fisher Scientific) for the twenty LC-MS analyses; (B) Protein quantitative values between 2 replicates within the same group; (C) Missing data by ion current-based analysis relative to MaxQuant; and (D) Volcano plot analysis (log10-transferred p-value versus log2-transferred ratio) of the comparison between FBS and paired HCS1-FBS. Thresholds were p value 0.05 and fold of change 1.5.
A total of 59 altered proteins were identified using thresholds ≥ 1.5-fold change and p-values ≤ 0.05 in comparisons between CCS-FBS versus FBS, HCS1-FBS versus FBS, and HCS2-FBS versus FBS. FBS, and CSS-, HCS1- and HCS2-FBS classified into distinctive clusters using hierarchical cluster analysis, which indicated sera from different sources had different protein profiles (Fig. 5A). The similar heat maps of the 5 LC-MS/MS analyses for the same serum sample confirmed the reproducibility of the quantitative platform used. Fourteen among the 59 altered proteins were removed consistently from all 3 CS-FBS samples (Fig. 5B). The 14 proteins were characterized using detailed physiochemical and functional annotations including GO annotation, isoelectric point (pI), GRAVY, protein domain analysis (data not shown) and MW distribution (Table 1). The only common feature was that all 14 proteins were low-molecular weight proteins (average weight 16 kDa) (Fig. 5C). This was consistent with previous studies that found that activated charcoal removed smaller proteins from larger proteins efficiently41. Activated charcoal treatment might remove selectively some low-molecular weight proteins in addition to small molecules including T and other steroids. Of the 14 proteins, 8 proteins are positive regulators and 4 proteins are negative regulators of cell proliferation. Insulin-like growth factor 2 (IGF-2), IGF binding protein 2 (IGFBP-2), and IGF binding protein 6 (IGFBP-6) were associated with the IGF signaling pathway. IGF-associated proteins were selected for additional studies, since IGF signaling pathways have been reported to be important in CaP.
Fig. 5.
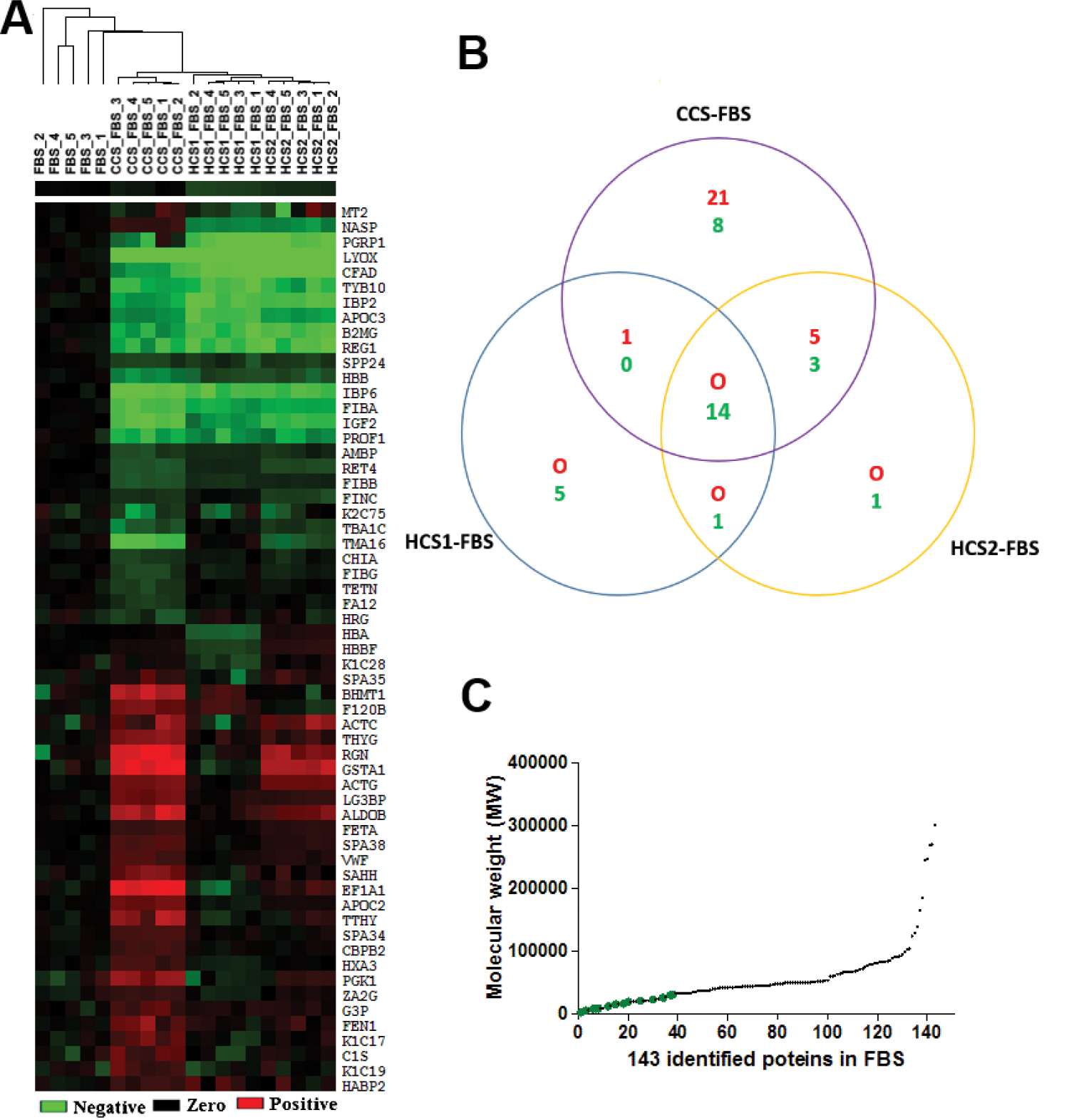
Comparative proteomic profiles of FBS and CS-FBS. (A) Clustering and heatmap analysis; (B) Shared changes among 3 analyses of CCS-FBS, HCS1-FBS and HCS2-FBS respectively compared to FBS; and (C) The molecular weight of the 14 consistently changed proteins. Red indicated elevated expression level and green indicated decreased expression level in CS-FBS relative to FBS.
Table I.
Fourteen proteins in fetal bovine serum commonly depleted charcoal-stripping.
| Protein name | Gene name | Relative ratios | MW (kDa) /pI | GO-Cellular Component | Cell Proliferation Related Reports | References | ||
|---|---|---|---|---|---|---|---|---|
| FBS/ HCS1 | FBS/ HCS2 | FBS/ CCS | ||||||
| Protein-lysine 6-oxidase | LOX | 46.1 | 55.2 | 60.7 | 29/5.9 | Extracellular region | Stimulator | Shih YH et al., Head Neck 2013, 35: 250–256. |
| Complement factor D | CFD | 8.0 | 13.6 | 3.4 | 25/6.9 | Extracellular region | Stimulator | Corrales L et al., J Immunol 2012, 189: 4674–4683. |
| Peptidoglycan recognition protein 1 | PGLYRP1 | 7.1 | 9.2 | 1.5 | 19/9.4 | Extracellular region | Stimulator | De Marzi MC et al., Immunology 2015, 145: 429–442. |
| Insulin-like growth factor-binding protein 2 | IGFBP2 | 6.2 | 5.5 | 3.3 | 31/6.9 | Extracellular region | Stimulator | Han S et al., Br J Cancer 2014, 111: 1400–1409. |
| Thymosin beta-10 | TMSB10 | 5.4 | 4.1 | 4.0 | 5/6.5 | Cytoplasm | Inhibitor | Lee SH et al., Cancer Res 2005, 65: 137–148. |
| Beta-2- microglobulin | B2M | 5.4 | 5.6 | 3.4 | 12/8.0 | Extracellular region | Stimulator/ Inhibitor | Min R et al., Br J Haematol 2002, 118: 495–505; Li Let al., Chin Med J (Engl) 2016, 129:448–455. |
| Apolipoprotein C-III | APOC3 | 5.3 | 3.3 | 2.8 | 8/4.7 | Extracellular region | Stimulator | Li H et al., Cardiovasc Res. 2015, 107: 579–589. |
| Insulin-like growth factor-binding protein 6 | IGFBP6 | 5.3 | 5.1 | 9.0 | 22/8.5 | Extracellular region | Inhibitor | Kim EJ et al., J Cell Physiol 2002, 190: 92–100. |
| Regakine-1 | N/A | 5.3 | 6.1 | 3.9 | 8/8.8 | Extracellular region | N/A | N/A |
| Fibrinogen alpha chain | FGA | 3.6 | 3.7 | 5.6 | 2/7.7 | Extracellular region | Stimulator | Palumbo JS et al., Blood 2000, 96: 3302–3309. |
| Profilin-1 | PFN1 | 3.0 | 3.0 | 3.9 | 15/8.5 | Extracellular region | Inhibitor | Zou L et al., J Cell Physiol 2010, 223: 623–9. |
| Insulin-like growth factor II | IGF2 | 2.9 | 4.6 | 6.3 | 8/6.4 | Extracellular region | Stimulator | Pollak M et al., Nat Rev Cancer 2008, 8: 915–928. |
| Hemoglobin subunit beta | HBB | 1.8 | 1.8 | 2.7 | 16/7.0 | Extracellular region | Inhibitor | Maman S et al., Cancer Res 2017, 77: 14–26. |
| Secreted phosphoprotein 24 | SPP2 | 1.5 | 1.5 | 1.6 | 20/7.9 | Extracellular region | Inhibitor | Lao L et al., Anticancer Res 2016, 36: 5773–5780. |
IGF-1 stimulated phosphorylation of IGF1R more efficiently in medium supplemented with CS-FBS than in medium supplemented with FBS.
Phosphorylation of IGF1R and IR was low in non-treated control cells cultured in medium supplemented with FBS and CS-FBS across all time points (Fig. 6A). Phospho-IGF1R and phospho-IR levels were similar in control cells cultured in medium supplemented with CS-FBS versus FBS. The reduction of bovine IGF-2 and IGFBPs by charcoal-stripping appeared to have little if any effect on IGF-1R and IR. Response of phosphorylation of IGF1R and IR to IGF-1, insulin and IGF-2 was dose-dependent. Phosphorylation of IGF-1 and IR at all tested sites in cells treated with 1 nM IGF-1 was higher in medium supplemented with CS-FBS than FBS at the 6 hr time point. The same was true for phosphorylation at IGF1R (Tyr1135/1136)/IR (Tyr1150/1151) and IGF1R (Tyr1135) of the 4 tested phosphorylation sites in cells treated with lower doses of insulin and IGF-2. Lower dose growth factors more effectively stimulated phosphorylation of IGF1R and IR at the phosphorylation sites in medium supplemented with CS-FBS compared to FBS. The increased sensitivity to lower-dose growth factors was sustained for 24 hrs at IGF1R (Tyr1135/1136)/IR (Tyr1150/1151) for all 3 growth factors, and at IGF1R (Tyr1131)/IR (Tyr1146) for IGF-1 and insulin. The higher efficiency of the lower-dose growth factors in medium supplemented with CS-FBS was diminished at 48 hr, whereas phosphorylation stimulated by higher-dose growth factors was sustained. Intensity of Western blotting signal of the bands in the presented Western blots were quantitated using densitometry and presented in Supplementary Data Fig S1.
Fig. 6.
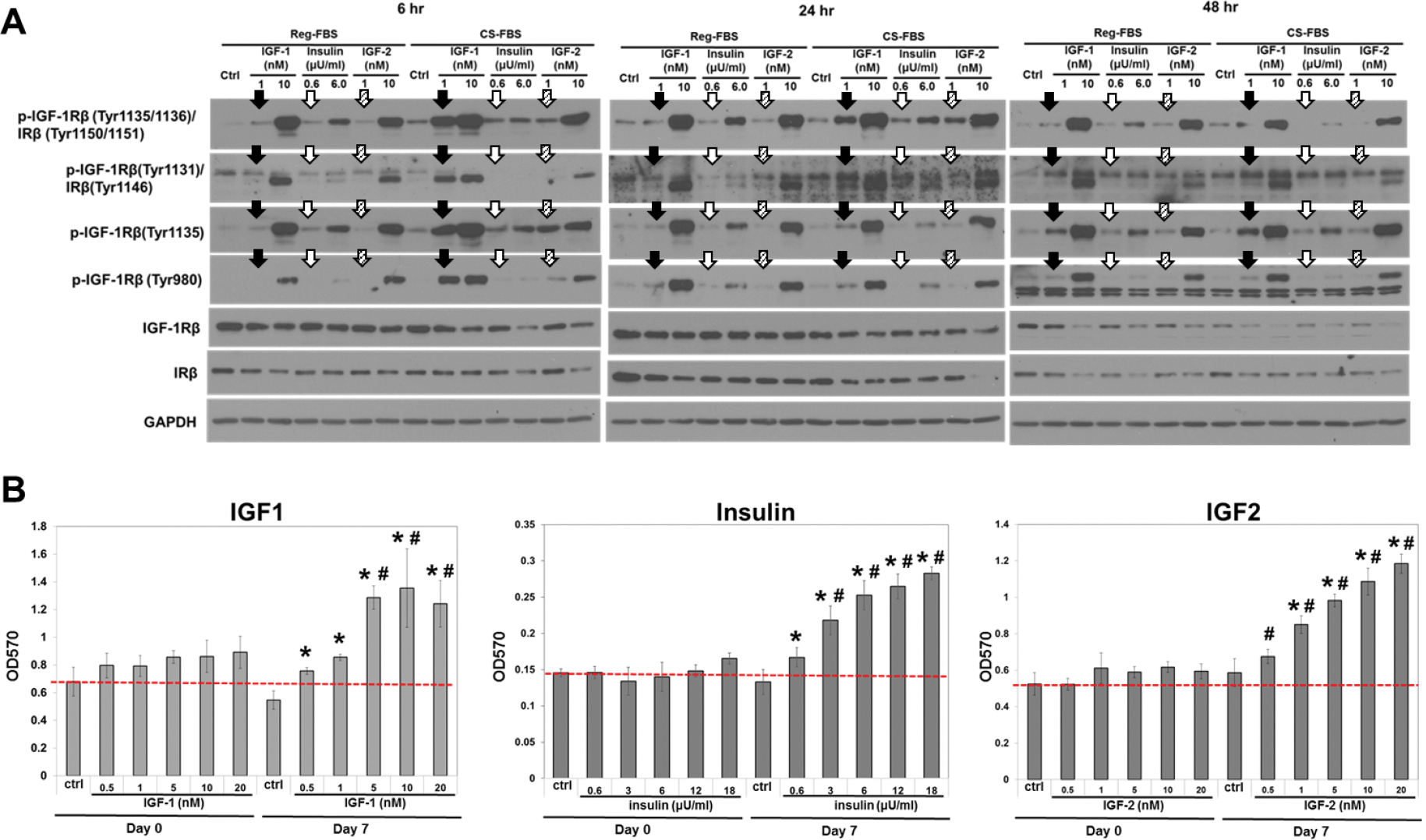
Effects of growth factors on IGF1R/IR signaling and cell growth of VCaP cell line. (A) Phosphorylation of IGF1R and IR, same type of arrows indicated the bands in comparison in the same treatment conditions in each time point; and (B) MTT assay for cell proliferation. Dotted line indicated the value of control on day 0. Experiments for growth (Fig 6B) were set up in triplicates (n=3). *: p < 0.05 compared to Day 7 control; #: p < 0.05 compared to Day 0 respective treatment.
IGF1, insulin, and IGF2 stimulated cell growth in a dose-dependent manner in medium supplemented with CS-FBS (Fig. 6B). Adding IGF2 back to CS-FBS sustained growth of VCaP cells.
VCaP cells secreted IGFBP.
VCaP cells were cultured in medium supplemented with 10% FBS or 10% CS-FBS for 3 days. Growth factor antibody arrays were used to examine growth factors in the freshly prepared medium (pre-culture medium) and the medium collected after 3 days of culture of VCaP cells (post-culture medium). Growth factors were not detected in either pre-culture medium supplemented with FBS or CS-FBS, except for a trace amount of IGF2 (Fig. 7A and B). Negative reaction of the blank and the negative controls ruled out non-specific binding. Profiles of growth factors secreted into the culture medium were similar between medium supplemented with FBS or CS-FBS (Fig. 7C and D). The most abundant growth factors or growth factor-associated proteins that were secreted by VCaP cells included hepatocyte growth factor (HGF), IGFBP-2, and platelet-derived growth factor (PDGF)-AA, -AB, and –BB. Production of IGFPB-2 by VCaP cells was of particular interest for 2 reasons: 1) Proteomic analysis revealed reduction of the bovine version of IGFBP-2 in CS-FBS compared to FBS; and 2) the change in cell response to low dose growth factors over time coincided with the production of IGFBP-2. IGFBP-2 may regulate IGF1R signaling in the presence of the growth factors at lower doses.
Fig. 7.
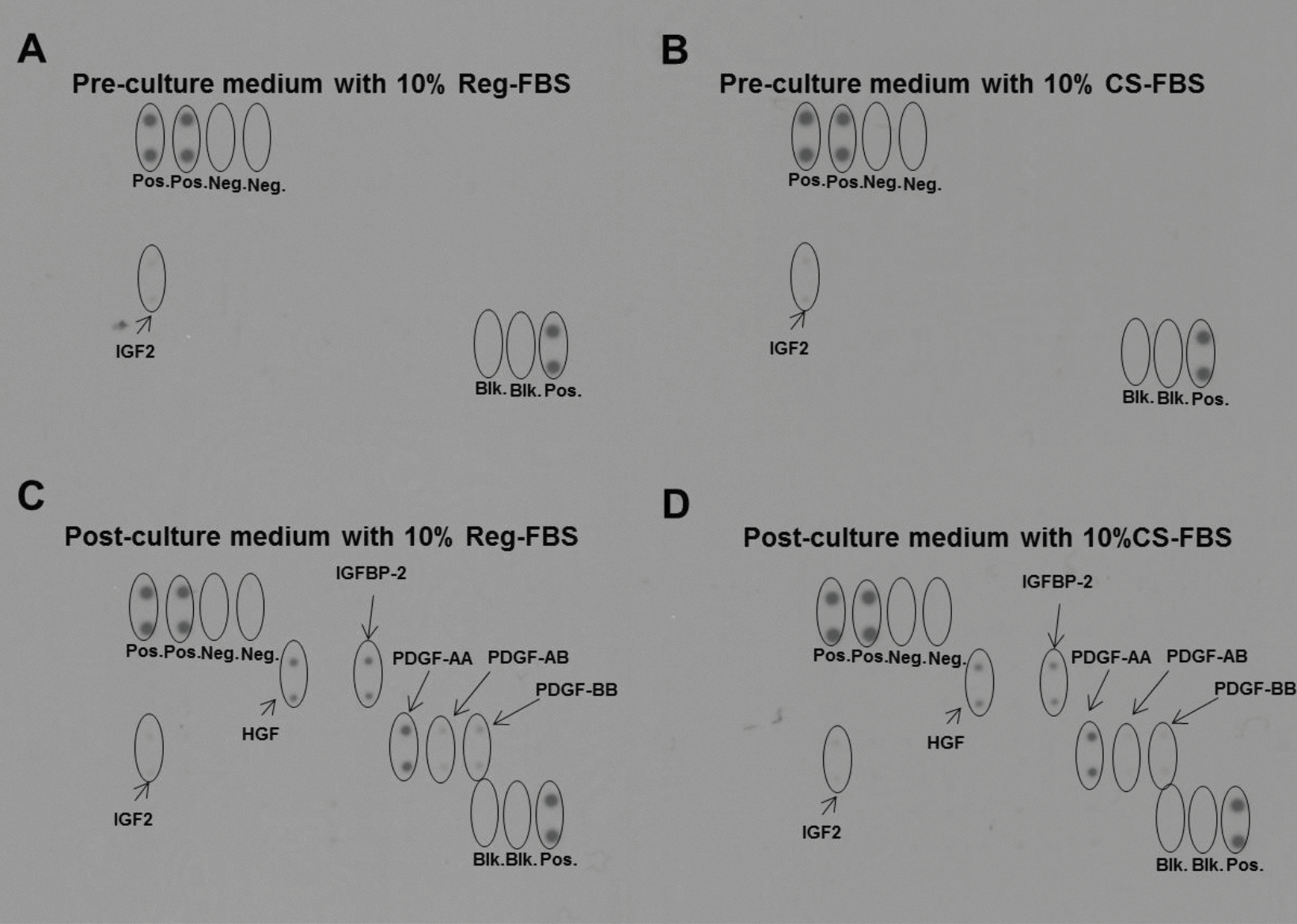
Analysis of growth factor production by VCaP cell line using a growth factor antibody array. Pre-culture medium (A and B) and post-culture medium (C and D). Pos., positive control spots with controlled amount of biotinylated antibody printed on the array. Neg., negative control spots with buffer (no antibodies) printed on the array. Blk., blank spots with nothing printed on the array.
Local production of IGF1 and IGFPB in CaP tissue
Expression of IGF1, IGF2, IGFBP-2, IGFBP-3, IGFBP6, and IGFBP7 at the mRNA levels was examined in serial sections of an FFPE human CaP tissue (Fig. 8). IGF1, IGF2, IGFBP-2, IGFBP-3 and IGFBP-6 were expressed predominantly in benign epithelial cells and malignant cells, whereas IGFBP-7 was expressed predominantly in stroma. The expression of IGF2 and IGFBP-6 seemed restricted in some niches but not immediate neighboring locations. The data demonstrated potential local production of IGF1R/IR signaling-associated proteins by benign epithelial cells, malignant cells, and stromal cells. Expression of IGF1R and IR was examined using IHC in matched tissue specimens of benign prostate and CaP in the TMA block (Fig. 9). No IHC staining was detected on the FFPE sections of the matched tissue specimens using non-immuned IgG as negative control. Expression of IGF1R and IR was evident in benign prostate tissue and CaP.
Fig. 8.
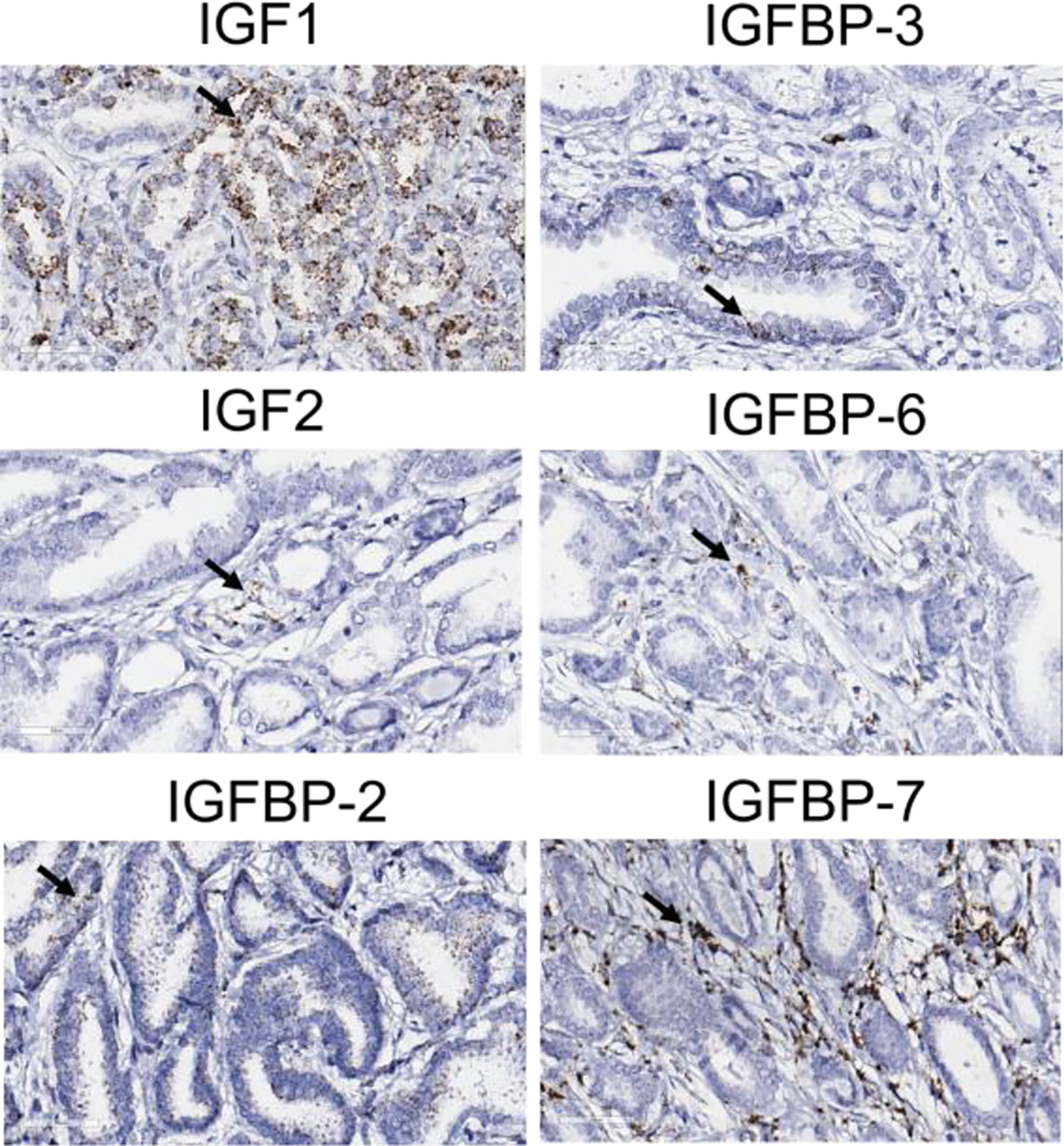
Expression of IGF and IGFBP genes at mRNA levels using RNAScope. Arrows indicated positive cells.
Fig. 9.
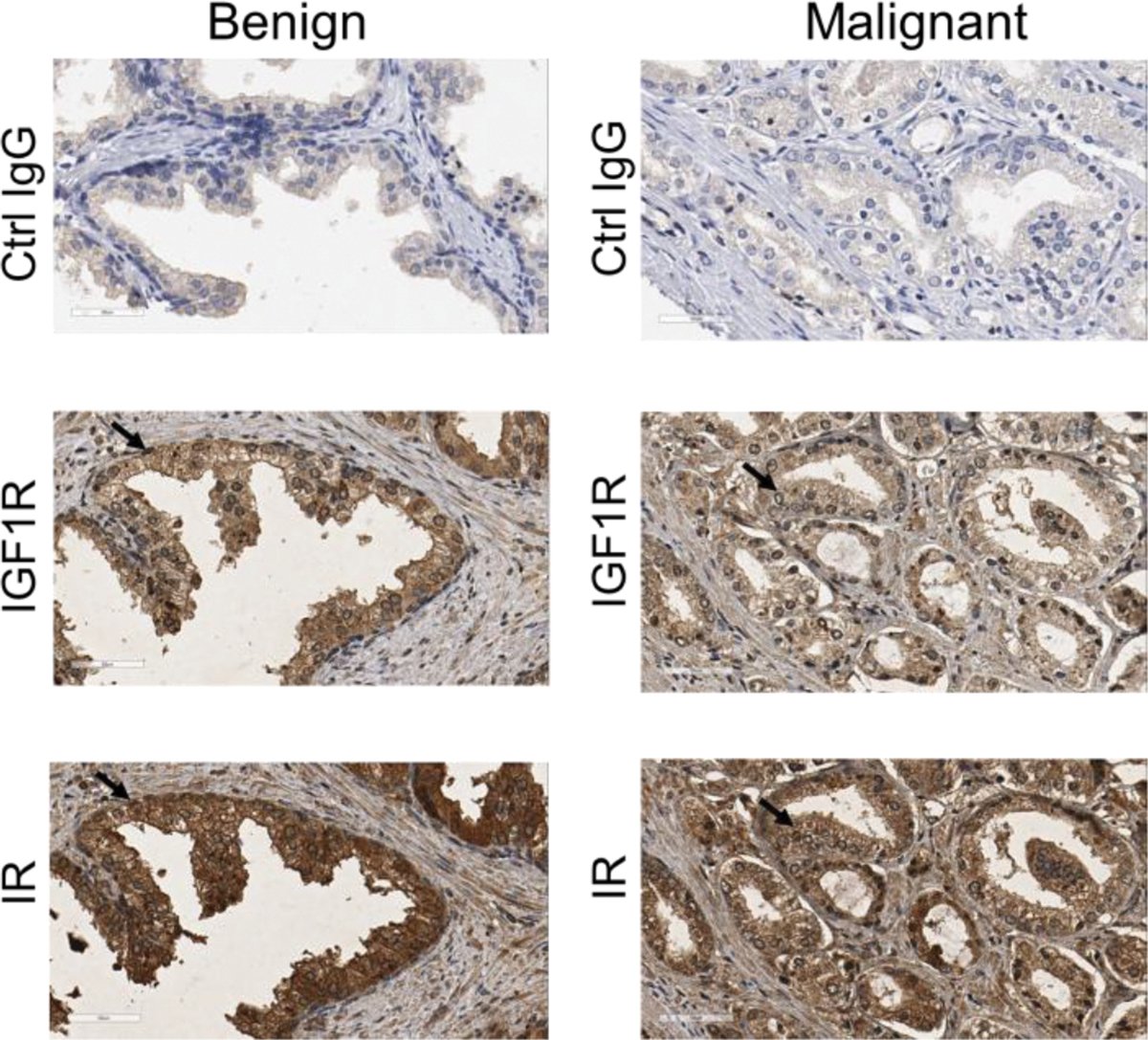
Expression of IGF1R and IR in prostate benign and malignant tissue using immunohistochemistry (IHC). Control IgG, a non-immuned mouse IgG was diluted to match the IgG concentration of IGF1R and IR antibodies for negative control. Arrows indicated positive cells.
DISCUSSION
There are 3 key findings in the present study. First, factors in addition to T maintained AR activity and supported CaP cell growth in FBS. Second, 143 proteins were identified in FBS, of which 14 were reduced consistently in all CS-FBS samples. Third, IGF2 was reduced in CS-FBS, addition of IGF2 in CS-FBS sustained growth of CaP cells independent of AR activity, and IGF1R/IR were more sensitive to stimulation by IGF in medium supplemented with CS-FBS.
CS-FBS is the most commonly used FBS derivative to supplement cell culture media for steroid hormone related experiments. A widely accepted argument for using CS-FBS is that charcoal-stripping removes the trace amount of steroids to provide a cleaner background, but preserves other constituents that may be important for cell growth or viability. FBS T levels measured using LC-MS/MS have been reported to range from 0.3 to 0.5 nM, and other androgens including dihydrotestosterone (DHT), androstenedione (ASD), and dehydroepiandrosterone (DHEA) were undetectable 7, 42–43. FBS that was used in the current study was analyzed using an LC-MS/MS method described in previous studies 7, 42–43. FBS contained 0.0695 ng/ml, or 0.214 nM T. DHT, ASD, and DHEA were undetectable. Charcoal-stripping removed all androgens from FBS; none of the above androgens were detected in CS-FBS (Supplementary Data Table S2). Final concentration of T in tissue culture medium supplemented with 10% FBS was 0.0214 nM. However, the stimulatory effect of T on AR activity and CaP cell growth was marginal when 0.03 nM T was added to CS-FBS to restore the T levels found in medium supplemented with FBS. These results questioned the impact of the trace amount of T in FBS on AR signaling and CaP cell growth. On the other hand, the data showed that AR activity and cell growth were impaired by removal of factors in addition to the trace amount of steroid when cells were switched from FBS- to CS-FBS-containing medium. The growth of CaP cells in FBS was inhibited when cells were treated with bicalutamide, an AR antagonist 43, which suggested that AR activity is important for cell growth in FBS, and unidentified factors may sensitize AR to the trace amount of T in FBS. On the other hand, IGF1, IGF2 and insulin did not increase AR activity when CaP cells were cultured in medium supplemented with 10% CS-FBS and 0.03 nM T (Supplementary Data, Fig S2), indicating the growth factors restored cancer cell growth independent of AR activity.
Androgen metabolism may be affected when cells are switched from FBS to CS-FBS. For example, endogenous T production in LAPC-4 cells was higher in FBS compared to CS-FBS, and the difference was more pronounced after 6 days than after 1 day in culture 7. Some of the proteins that were decreased in CS-FBS and the signaling cascades they initiated may have been responsible for maintaining AR activity using the trace amount of T as found in FBS and/or modulation of androgen metabolic pathway for intracellular T production. The protein profiles that were revealed in the proteomic study provided candidates for follow-up studies.
Proteomic study identified IGF2, IGFBP2, and IGFBP6 in FBS, and the proteins were reduced >3-fold in CS-FBS. IGF2 has been reported in FBS 12, 44. Bovine IGF2 was found to be functionally equipotent as human IGF2 44. IGF1 or IGF2 stimulated growth of VCaP cells in CS-FBS without androgens. Androgen-dependent CaP cells used the IGF1R signaling pathway to replace the AR signaling pathway for growth in the absence of androgen. IGF and IGFBP are among the most abundant growth factors and growth factor-related proteins in the circulation. They also are involved in a wide range of normal functions in normal cells and diseases. The importance of IGF and IGFBP in cancer including CaP has been studied extensively 45–48. The relevance of the findings was supported by the IHC and RNAScope staining of key components of the IGF1R signaling pathways in CaP specimens. IHC data also revealed that not only the prostate and CaP tissues could locally generate IFG-1 and IGFBPs, benign prostate epithelial cells and cancer cells could also be subjects to the regulation of the locally produced IGF-1and IGFBPs. IGF2R is unrelated to IGF1R and insulin receptor, and is a non-signaling receptor that regulates the bioavailability of circulating IGF2 49–55. IGF2R is also the mannose-6-phosphate (M6P) receptor, which has high affinity for binding with IGF2 and M6P-containing molecules 56–59. On the other hand, IGF1R and insulin receptors are the main receptors that relay the signals from IGF1, IGF2, and insulin. The focus of the present study therefore was not on IGF2R due to the unique characteristics of IGF2R in relation to the growth factors.
Most proteins in FBS should be hydrophilic since serum is an aqueous fluid. As expected the majority of proteins (95.8%) in FBS are hydrophilic indicated by negative GRAVY values, consistent with previous studies in plasma proteomes28, 60. However, MW distribution in FBS is different from that in adult plasma proteomes. More than 60% of plasma/serum proteins have MW > 60 kDa in human and swine 28, 60–61, whereas < 40% of cellular proteins have MW > 60 kDa 62–63. The difference in MW distribution could be caused by glomerular filtration-mediated removal of proteins < 60 kDa from the circulation of adult mammals 61. Less than 30% of FBS proteins had MW > 60 kDa; the MW distribution was different in plasma/serum proteins from adult mammals, and similar to cellular proteins. Contamination of FBS with cellular proteins was unlikely since most of the FBS proteins were secreted proteins or cell surface proteins, which was observed in proteomes of human plasma/serum 60, 64. MW distribution of FBS proteins may indicate incomplete filtration by the kidney in the developing bovine fetus as reported in human fetus compared to older infants and children 65–66.
Concentrations of FBS proteins distributed in a wide range. The top 20 proteins were 89.9% of total protein abundance. This concentration distribution was similar to other serum/plasma samples 60, 64. Fetuin-A was more abundant than albumin in FBS, which agreed with a previous report 67. The high abundance of fetuin-A in FBS is unique because serum albumin is found usually as the most abundant protein in sera of human and other mammals 28, 61. Moreover, alpha-fetoprotein (AFP) and fetuin-B also were high abundant proteins in FBS, although the 2 proteins usually disappear after birth 67–68.
A total of 56 proteins were reduced in the 3 CS-FBS samples compared to the FBS sample, and 14 proteins were reduced consistently (Table I). Protein-lysine 6-oxidase (LOX), complement factor D (CFD), peptidoglycan recognition protein 1 (PGLYRP1), IGFBP2, apolipoprotein C-III (APOC3), fibrinogen alpha chain (FGA), and IGF2 have been reported to promote cell proliferation. Thymosin beta-10 (TMSB10), IGFBP6, profilin-1, hemoglobin subunit beta (HBB) and secreted phosphoprotein 24 (SPP2) have been reported to inhibit cell proliferation. β−2-microglobulin, a component of MHC class I molecules, is a positive or negative growth regulator in different cancer cells 69–70. Regakine-1, a novel CC chemokine recently identified in bovine serum, could synergize with other proinflammatory chemokines and enhance the inflammatory response to infection 71. LOX was the most depleted protein (~50-fold change) in CS-FBS. LOX is a copper-dependent enzyme that cross-links collagen and elastin in the extracellular region matrix. LOX can induce cell proliferation and angiogenesis in oral squamous cell carcinoma 72 and suppression of LOX results in lower cell motility in collagen gel and reduces metastasis in mice 73. Another highly depleted protein was CFD, which plays a key role in the first line of defense against microbes through the initial alternative pathway C3 convertase 74. Recent studies showed that complement activation promoted tumor growth and angiogenesis, while C3 or C5aR-deficient mice showed decreased tumor growth relative to wild-type mice 75–76. Overexpression of TMSB10 inhibited vascular endothelial growth factor-induced endothelial cell proliferation, migration and invasion through its interaction with Ras 77. SPP2, a binding protein of bone morphogenetic proteins (BMPs), inhibited the growth of human cancer cells such as prostate and pancreatic cancer cells, induced by BMP-2 78–79. IGF2 is a major fetal growth factor, which stimulates cell proliferation in many different tissues during gestation 80. The binding of IGF2 and IGF1R induced the phosphorylation of insulin receptor substrates (IRSs), which promoted protein translation and cell proliferation through the PI3K/Akt pathway 81. The ligand bioavailability of IGF2 is regulated by IGFBP proteins, which are the dominant regulators of the IGF signaling pathway 82. Exogenous IGFBP-2 promotes cell proliferation, invasion, and chemoresistance in glioma cells via integrin β1/ERK signaling 83. IGFBP-6 is an inhibitor of IGF2 actions and has been recognized as a tumor suppressor 84.
FBS has been the most commonly used supplement for tissue culture medium for a long time. Serum proteins including protein hormones and growth factors have been considered the most important components in FBS, but comprehensive profiling of the proteins and examination of their effects on cell function become possible only recently with the emergence of high quality proteomic approaches. The present study demonstrated the feasibility of proteomic characterization of FBS and CS-FBS using ion current-based LC-MS/MS analysis. The identification and functional validation of the proteins in the IGF1R signaling pathway provides an example of utility of proteomic profiling to identify biologically important proteins in FBS and CS-FBS. The identified proteins, particularly the 14 proteins that were reduced in CS-FBS, present candidates that may lead to discovery of new molecular mechanisms that help CaP cells manage androgen metabolism to maintain AR activity.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported, in part, by NIH grant 1R21CA191895-01 (YW), Department of Defense Post-doctoral Training Award W81XWH-15-1-0409 (MVF), NIH grants HD071594 (JQ), GM121174 (JQ), AI125746 (JQ), Dongguan University of Technology innovation team startup KCYXPT2016004 (DM) and National Cancer Institute (NCI) grant P30CA016056 involving the use of Roswell Park Comprehensive Cancer Center’s Pathology Network Shared Resource.
ABBREVIATIONS
- AFP
alpha-fetoprotein
- AGC
automatic gain control
- APOC3
apolipoprotein C-III
- AR
androgen receptor
- ASD
androstenedione
- AUC
area under the curve
- BCA
bicinchoninic acid
- BMP
bone morphogenetic proteins
- CaP
prostate cancer
- CCS-FBS
commercially available CS-FBS
- CFD
complement factor D
- CS-FBS
charcoal-stripped FBS
- DHEA
dehydroepiandrosterone
- DHT
dihydrotestosterone
- FBS
fetal bovine serum
- FDR
false discovery rate
- FFPE
Formalin-fixed, paraffin-embedded
- FGA
fibrinogen alpha chain
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GO
Gene Ontology
- HBB
hemoglobin subunit beta
- HCD
high energy collision dissociation
- HCS1/2-FBS
home-made CS-FBS
- HGF
hepatocyte growth factor
- HRP
horseradish peroxidase
- ICan
ion current-based analysis
- IGF1R
IGF1 receptor
- IGF-2
insulin-like growth factor 2
- IGFBP-2
IGF binding protein 2
- IGFBP-6
IGF binding protein 6
- IHC
immunohistochemistry
- IR
insulin receptor
- IRSs
insulin receptor substrates
- IU
international unit
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- LFQ
label-free quantification
- LOX
protein-lysine 6-oxidase
- PAQV
protein absolute quantitative values
- PDGF
platelet-derived growth factor
- PGLYRP1
peptidoglycan recognition protein 1
- PNSR
Pathology Network Shared Resource
- PSM
peptide-spectrum matches
- Roswell Park
Roswell Park Comprehensive Cancer Center
- SILAC
stable isotope labeling using amino acids
- SPP2
secreted phosphoprotein 24
- STR
short tandem repeat
- T
testosterone
- TCEP
tris(2-carboxyethyl)phosphine
- TMA
tissue microarray
- TMSB10
thymosin beta-10
- TMT
tandem mass tags
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, Department of Defense, National Cancer Institute, Roswell Park Comprehensive Cancer Center and Dongguan University of Technology.
SUPPORTING INFORMATION
The following supporting information is available free of charge at ACS website http://pubs.acs.org.
Supplementary Table S1: Quantitation information of 143 proteins identified from fetal bovine serum (FBS) and charcoal-stripped FBS using ion current-based analysis.
Supplementary Table S2: T concentrations in regular FBS and home-made CS-FBS measured using LC-MS/MS.
Supplementary Figure S1: Densitometry of the bands of phosphorylated IGF1R or IR on the Western blot presented in Fig 6A.
Supplementary Figure S2: AR activity in LAPC-4 and VCaP cells treated with growth factors in medium supplemented with 10% CS-FBS in the presence or absence of 0.03 nM T.
DATA AVAILABILITY
Raw mass spectrometry data have been deposited in Chorus (http://chorusproject.org) with accession Project ID 1401: Charcoal stripped FBS.
References
- 1.Mohler JL, Concept and viability of androgen annihilation for advanced prostate cancer. Cancer 2014, 120 (17), 2628–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Askew EB; Gampe RT Jr.; Stanley TB; Faggart JL; Wilson EM, Modulation of androgen receptor activation function 2 by testosterone and dihydrotestosterone. J Biol Chem 2007, 282 (35), 25801–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou ZX; Lane MV; Kemppainen JA; French FS; Wilson EM, Specificity of ligand-dependent androgen receptor stabilization: receptor domain interactions influence ligand dissociation and receptor stability. Mol Endocrinol 1995, 9 (2), 208–18. [DOI] [PubMed] [Google Scholar]
- 4.Heinlein CA; Chang C, Androgen receptor in prostate cancer. Endocr Rev 2004, 25 (2), 276–308. [DOI] [PubMed] [Google Scholar]
- 5.Rothermund CA; Gopalakrishnan VK; Vishwanatha JK, Androgen signaling and post-transcriptional downregulation of Bcl-2 in androgen-unresponsive prostate cancer. Prostate Cancer Prostatic Dis 2004, 7 (2), 158–64. [DOI] [PubMed] [Google Scholar]
- 6.Soifer HS; Souleimanian N; Wu S; Voskresenskiy AM; Collak FK; Cinar B; Stein CA, Direct regulation of androgen receptor activity by potent CYP17 inhibitors in prostate cancer cells. J Biol Chem 2012, 287 (6), 3777–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiandalo MV; Wilton JH; Mantione KM; Wrzosek C; Attwood KM; Wu Y; Mohler JL, Serum-free complete medium, an alternative medium to mimic androgen deprivation in human prostate cancer cell line models. Prostate 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gstraunthaler G, Alternatives to the use of fetal bovine serum: serum-free cell culture. Altex 2003, 20 (4), 275–81. [PubMed] [Google Scholar]
- 9.Cao Z; West C; Norton-Wenzel CS; Rej R; Davis FB; Davis PJ, Effects of resin or charcoal treatment on fetal bovine serum and bovine calf serum. Endocr Res 2009, 34 (4), 101–8. [DOI] [PubMed] [Google Scholar]
- 10.Krycer JR; Brown AJ, Does changing androgen receptor status during prostate cancer development impact upon cholesterol homeostasis? PloS One 2013, 8 (1), e54007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang ZC; Lowik CW, Removal of serum factors by charcoal treatment promotes adipogenesis via a MAPK-dependent pathway. Mol Cell Biochem 2005, 268 (1–2), 159–67. [DOI] [PubMed] [Google Scholar]
- 12.Zheng X; Baker H; Hancock WS; Fawaz F; McCaman M; Pungor E Jr., Proteomic analysis for the assessment of different lots of fetal bovine serum as a raw material for cell culture. Part IV. Application of proteomics to the manufacture of biological drugs. Biotechnol Prog 2006, 22 (5), 1294–300. [DOI] [PubMed] [Google Scholar]
- 13.Ross PL; Huang YN; Marchese JN; Williamson B; Parker K; Hattan S; Khainovski N; Pillai S; Dey S; Daniels S; Purkayastha S; Juhasz P; Martin S; Bartlet-Jones M; He F; Jacobson A; Pappin DJ, Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 2004, 3 (12), 1154–69. [DOI] [PubMed] [Google Scholar]
- 14.Thompson A; Schafer J; Kuhn K; Kienle S; Schwarz J; Schmidt G; Neumann T; Johnstone R; Mohammed AK; Hamon C, Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem 2003, 75 (8), 1895–904. [DOI] [PubMed] [Google Scholar]
- 15.Ong SE; Blagoev B; Kratchmarova I; Kristensen DB; Steen H; Pandey A; Mann M, Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 2002, 1 (5), 376–86. [DOI] [PubMed] [Google Scholar]
- 16.Gao J; Friedrichs MS; Dongre AR; Opiteck GJ, Guidelines for the routine application of the peptide hits technique. J Am Soc Mass Spectrom 2005, 16 (8), 1231–8. [DOI] [PubMed] [Google Scholar]
- 17.Liu H; Sadygov RG; Yates JR 3rd, A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 2004, 76 (14), 4193–201. [DOI] [PubMed] [Google Scholar]
- 18.Wiener MC; Sachs JR; Deyanova EG; Yates NA, Differential mass spectrometry: a label-free LC-MS method for finding significant differences in complex peptide and protein mixtures. Anal Chem 2004, 76 (20), 6085–96. [DOI] [PubMed] [Google Scholar]
- 19.Bondarenko PV; Chelius D; Shaler TA, Identification and relative quantitation of protein mixtures by enzymatic digestion followed by capillary reversed-phase liquid chromatography-tandem mass spectrometry. Anal Chem 2002, 74 (18), 4741–9. [DOI] [PubMed] [Google Scholar]
- 20.Tu C; Li J; Jiang X; Sheflin LG; Pfeffer BA; Behringer M; Fliesler SJ; Qu J, Ion-current-based proteomic profiling of the retina in a rat model of Smith-Lemli-Opitz syndrome. Mol Cell Proteomics 2013, 12 (12), 3583–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tu C; Li J; Sheng Q; Zhang M; Qu J, Systematic assessment of survey scan and MS2-based abundance strategies for label-free quantitative proteomics using high-resolution MS data. J Proteome Res 2014, 13 (4), 2069–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neilson KA; Ali NA; Muralidharan S; Mirzaei M; Mariani M; Assadourian G; Lee A; van Sluyter SC; Haynes PA, Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics 2011, 11 (4), 535–53. [DOI] [PubMed] [Google Scholar]
- 23.Tu C; Sheng Q; Li J; Shen X; Zhang M; Shyr Y; Qu J, ICan: an optimized ion-current-based quantification procedure with enhanced quantitative accuracy and sensitivity in biomarker discovery. J Proteome Res 2014, 13 (12), 5888–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sobel RE; Sadar MD, Cell lines used in prostate cancer research: a compendium of old and new lines--part 2. J Urol 2005, 173 (2), 360–72. [DOI] [PubMed] [Google Scholar]
- 25.Klein KA; Reiter RE; Redula J; Moradi H; Zhu XL; Brothman AR; Lamb DJ; Marcelli M; Belldegrun A; Witte ON; Sawyers CL, Progression of metastatic human prostate cancer to androgen independence in immunodeficient SCID mice. Nat Med 1997, 3 (4), 402–8. [DOI] [PubMed] [Google Scholar]
- 26.Tu C; Mammen MJ; Li J; Shen X; Jiang X; Hu Q; Wang J; Sethi S; Qu J, Large-scale, ion-current-based proteomics investigation of bronchoalveolar lavage fluid in chronic obstructive pulmonary disease patients. J Proteome Res 2014, 13 (2), 627–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.An B; Zhang M; Johnson RW; Qu J, Surfactant-aided precipitation/on-pellet-digestion (SOD) procedure provides robust and rapid sample preparation for reproducible, accurate and sensitive LC/MS quantification of therapeutic protein in plasma and tissues. Anal Chem 2015, 87 (7), 4023–9. [DOI] [PubMed] [Google Scholar]
- 28.Tu C; Li J; Young R; Page BJ; Engler F; Halfon MS; Canty JM Jr.; Qu J, Combinatorial peptide ligand library treatment followed by a dual-enzyme, dual-activation approach on a nanoflow liquid chromatography/orbitrap/electron transfer dissociation system for comprehensive analysis of swine plasma proteome. Anal Chem 2011, 83 (12), 4802–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tu C; Bu Y; Vujcic M; Shen S; Li J; Qu M; Hangauer D; Clements JL; Qu J, Ion Current-Based Proteomic Profiling for Understanding the Inhibitory Effect of Tumor Necrosis Factor Alpha on Myogenic Differentiation. J Proteome Res 2016, 15 (9), 3147–57. [DOI] [PubMed] [Google Scholar]
- 30.Cox J; Mann M, MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008, 26 (12), 1367–72. [DOI] [PubMed] [Google Scholar]
- 31.Cox J; Neuhauser N; Michalski A; Scheltema RA; Olsen JV; Mann M, Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 2011, 10 (4), 1794–805. [DOI] [PubMed] [Google Scholar]
- 32.Elias JE; Haas W; Faherty BK; Gygi SP, Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods 2005, 2 (9), 667–75. [DOI] [PubMed] [Google Scholar]
- 33.Smyth GK; Speed T, Normalization of cDNA microarray data. Methods 2003, 31 (4), 265–73. [DOI] [PubMed] [Google Scholar]
- 34.Zeeberg BR; Qin H; Narasimhan S; Sunshine M; Cao H; Kane DW; Reimers M; Stephens RM; Bryant D; Burt SK; Elnekave E; Hari DM; Wynn TA; Cunningham-Rundles C; Stewart DM; Nelson D; Weinstein JN, High-Throughput GoMiner, an ‘industrial-strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID). BMC Bioinformatics 2005, 6, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Hoon MJ; Imoto S; Nolan J; Miyano S, Open source clustering software. Bioinformatics 2004, 20 (9), 1453–4. [DOI] [PubMed] [Google Scholar]
- 36.Petersen TN; Brunak S; von Heijne G; Nielsen H, SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 2011, 8 (10), 785–6. [DOI] [PubMed] [Google Scholar]
- 37.Li S; Zhou Y; Wang R; Zhang H; Dong Y; Ip C, Selenium sensitizes MCF-7 breast cancer cells to doxorubicin-induced apoptosis through modulation of phospho-Akt and its downstream substrates. Mol Cancer Ther 2007, 6 (3), 1031–8. [DOI] [PubMed] [Google Scholar]
- 38.Wu Y; Chhipa RR; Zhang H; Ip C, The antiandrogenic effect of finasteride against a mutant androgen receptor. Cancer Biol Ther 2011, 11 (10), 902–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rennie PS; Bruchovsky N; Leco KJ; Sheppard PC; McQueen SA; Cheng H; Snoek R; Hamel A; Bock ME; MacDonald BS; et al. , Characterization of two cis-acting DNA elements involved in the androgen regulation of the probasin gene. Mol Endocrinol 1993, 7 (1), 23–36. [DOI] [PubMed] [Google Scholar]
- 40.Wu Y; Zhang H; Dong Y; Park YM; Ip C, Endoplasmic reticulum stress signal mediators are targets of selenium action. Cancer Res 2005, 65 (19), 9073–9. [DOI] [PubMed] [Google Scholar]
- 41.Stone MT; Kozlov M, Separating proteins with activated carbon. Langmuir 2014, 30 (27), 8046–55. [DOI] [PubMed] [Google Scholar]
- 42.Wu Y; Godoy A; Azzouni F; Wilton JH; Ip C; Mohler JL, Prostate cancer cells differ in testosterone accumulation, dihydrotestosterone conversion, and androgen receptor signaling response to steroid 5alpha-reductase inhibitors. Prostate 2013, 73 (13), 1470–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu Y; Chhipa RR; Cheng J; Zhang H; Mohler JL; Ip C, Androgen receptor-mTOR crosstalk is regulated by testosterone availability: implication for prostate cancer cell survival. Anticancer Res 2010, 30 (10), 3895–901. [PMC free article] [PubMed] [Google Scholar]
- 44.Honegger A; Humbel RE, Insulin-like growth factors I and II in fetal and adult bovine serum. Purification, primary structures, and immunological cross-reactivities. J Biol Chem 1986, 261 (2), 569–75. [PubMed] [Google Scholar]
- 45.Baxter RC, IGF binding proteins in cancer: mechanistic and clinical insights. Nat Rev Cancer 2014, 14 (5), 329–41. [DOI] [PubMed] [Google Scholar]
- 46.Rowlands MA; Gunnell D; Harris R; Vatten LJ; Holly JM; Martin RM, Circulating insulin-like growth factor peptides and prostate cancer risk: a systematic review and meta-analysis. Int J Cancer 2009, 124 (10), 2416–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Travis RC; Appleby PN; Martin RM; Holly JMP; Albanes D; Black A; Bueno-de-Mesquita HBA; Chan JM; Chen C; Chirlaque MD; Cook MB; Deschasaux M; Donovan JL; Ferrucci L; Galan P; Giles GG; Giovannucci EL; Gunter MJ; Habel LA; Hamdy FC; Helzlsouer KJ; Hercberg S; Hoover RN; Janssen J; Kaaks R; Kubo T; Le Marchand L; Metter EJ; Mikami K; Morris JK; Neal DE; Neuhouser ML; Ozasa K; Palli D; Platz EA; Pollak M; Price AJ; Roobol MJ; Schaefer C; Schenk JM; Severi G; Stampfer MJ; Stattin P; Tamakoshi A; Tangen CM; Touvier M; Wald NJ; Weiss NS; Ziegler RG; Key TJ; Allen NE, A Meta-analysis of Individual Participant Data Reveals an Association between Circulating Levels of IGF-I and Prostate Cancer Risk. Cancer Res 2016, 76 (8), 2288–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arcaro A, Targeting the insulin-like growth factor-1 receptor in human cancer. Front Pharmacol 2013, 4, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brahmkhatri VP; Prasanna C; Atreya HS, Insulin-like growth factor system in cancer: novel targeted therapies. BioMed Res International 2015, 2015, 538019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiess W; Greenstein LA; White RM; Lee L; Rechler MM; Nissley SP, Type II insulin-like growth factor receptor is present in rat serum. Proc Natl Acad Sci U S A 1987, 84 (21), 7720–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valenzano KJ; Remmler J; Lobel P, Soluble insulin-like growth factor II/mannose 6-phosphate receptor carries multiple high molecular weight forms of insulin-like growth factor II in fetal bovine serum. J Biol Chem 1995, 270 (27), 16441–8. [DOI] [PubMed] [Google Scholar]
- 52.Zaina S; Squire S, The soluble type 2 insulin-like growth factor (IGF-II) receptor reduces organ size by IGF-II-mediated and IGF-II-independent mechanisms. J Biol Chem 1998, 273 (44), 28610–6. [DOI] [PubMed] [Google Scholar]
- 53.Costello M; Baxter RC; Scott CD, Regulation of soluble insulin-like growth factor II/mannose 6-phosphate receptor in human serum: measurement by enzyme-linked immunosorbent assay. J Clin Endocrinol Metab 1999, 84 (2), 611–7. [DOI] [PubMed] [Google Scholar]
- 54.MacDonald RG; Tepper MA; Clairmont KB; Perregaux SB; Czech MP, Serum form of the rat insulin-like growth factor II/mannose 6-phosphate receptor is truncated in the carboxyl-terminal domain. J Biol Chem 1989, 264 (6), 3256–61. [PubMed] [Google Scholar]
- 55.Xu Y; Papageorgiou A; Polychronakos C, Developmental regulation of the soluble form of insulin-like growth factor-II/mannose 6-phosphate receptor in human serum and amniotic fluid. J Clin Endocrinol Metab 1998, 83 (2), 437–42. [DOI] [PubMed] [Google Scholar]
- 56.Oates AJ; Schumaker LM; Jenkins SB; Pearce AA; DaCosta SA; Arun B; Ellis MJ, The mannose 6-phosphate/insulin-like growth factor 2 receptor (M6P/IGF2R), a putative breast tumor suppressor gene. Breast Cancer Res Treat 1998, 47 (3), 269–81. [DOI] [PubMed] [Google Scholar]
- 57.Morgan DO; Edman JC; Standring DN; Fried VA; Smith MC; Roth RA; Rutter WJ, Insulin-like growth factor II receptor as a multifunctional binding protein. Nature 1987, 329 (6137), 301–7. [DOI] [PubMed] [Google Scholar]
- 58.Oshima A; Nolan CM; Kyle JW; Grubb JH; Sly WS, The human cation-independent mannose 6-phosphate receptor. Cloning and sequence of the full-length cDNA and expression of functional receptor in COS cells. J Biol Chem 1988, 263 (5), 2553–62. [PubMed] [Google Scholar]
- 59.Braulke T, Type-2 IGF receptor: a multi-ligand binding protein. Horm Metab Res 1999, 31 (2–3), 242–6. [DOI] [PubMed] [Google Scholar]
- 60.Tu CJ; Dai J; Li SJ; Sheng QH; Deng WJ; Xia QC; Zeng R, High-sensitivity analysis of human plasma proteome by immobilized isoelectric focusing fractionation coupled to mass spectrometry identification. J Proteome Res 2005, 4 (4), 1265–73. [DOI] [PubMed] [Google Scholar]
- 61.Schenk S; Schoenhals GJ; de Souza G; Mann M, A high confidence, manually validated human blood plasma protein reference set. BMC Medical Genomics 2008, 1, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie X; Yi Z; Bowen B; Wolf C; Flynn CR; Sinha S; Mandarino LJ; Meyer C, Characterization of the Human Adipocyte Proteome and Reproducibility of Protein Abundance by One-Dimensional Gel Electrophoresis and HPLC-ESI-MS/MS. J Proteome Res 2010, 9 (9), 4521–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taylor SW; Fahy E; Zhang B; Glenn GM; Warnock DE; Wiley S; Murphy AN; Gaucher SP; Capaldi RA; Gibson BW; Ghosh SS, Characterization of the human heart mitochondrial proteome. Nature Biotechnol 2003, 21 (3), 281–6. [DOI] [PubMed] [Google Scholar]
- 64.Anderson NL; Anderson NG, The human plasma proteome: history, character, and diagnostic prospects. Molecular & cellular proteomics : MCP 2002, 1 (11), 845–67. [DOI] [PubMed] [Google Scholar]
- 65.Haycock GB, Development of glomerular filtration and tubular sodium reabsorption in the human fetus and newborn. British J Urol 1998, 81 Suppl 2, 33–8. [DOI] [PubMed] [Google Scholar]
- 66.Moniz CF; Nicolaides KH; Bamforth FJ; Rodeck CH, Normal reference ranges for biochemical substances relating to renal, hepatic, and bone function in fetal and maternal plasma throughout pregnancy. J Clin Pathol 1985, 38 (4), 468–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jahnen-Dechent W; Heiss A; Schafer C; Ketteler M, Fetuin-A regulation of calcified matrix metabolism. Circ Res 2011, 108 (12), 1494–509. [DOI] [PubMed] [Google Scholar]
- 68.Gabant P; Forrester L; Nichols J; Van Reeth T; De Mees C; Pajack B; Watt A; Smitz J; Alexandre H; Szpirer C; Szpirer J, Alpha-fetoprotein, the major fetal serum protein, is not essential for embryonic development but is required for female fertility. Proc Natl Acad Sci U S A 2002, 99 (20), 12865–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Min R; Li Z; Epstein J; Barlogie B; Yi Q, Beta(2)-microglobulin as a negative growth regulator of myeloma cells. Br J Haematol 2002, 118 (2), 495–505. [DOI] [PubMed] [Google Scholar]
- 70.Li L; Dong M; Wang XG, The Implication and Significance of Beta 2 Microglobulin: A Conservative Multifunctional Regulator. Chinese Med J 2016, 129 (4), 448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Struyf S; Stoops G; Van Coillie E; Gouwy M; Schutyser E; Lenaerts JP; Fiten P; Van Aelst I; Proost P; Opdenakker G; Van Damme J, Gene cloning of a new plasma CC chemokine, activating and attracting myeloid cells in synergy with other chemoattractants. Biochemistry 2001, 40 (39), 11715–22. [DOI] [PubMed] [Google Scholar]
- 72.Shih YH; Chang KW; Chen MY; Yu CC; Lin DJ; Hsia SM; Huang HL; Shieh TM, Lysyl oxidase and enhancement of cell proliferation and angiogenesis in oral squamous cell carcinoma. Head Neck 2013, 35 (2), 250–6. [DOI] [PubMed] [Google Scholar]
- 73.Nishioka T; Eustace A; West C, Lysyl oxidase: from basic science to future cancer treatment. Cell Struct Func 2012, 37 (1), 75–80. [DOI] [PubMed] [Google Scholar]
- 74.Nilsson B; Nilsson Ekdahl K, The tick-over theory revisited: is C3 a contact-activated protein? Immunobiology 2012, 217 (11), 1106–10. [DOI] [PubMed] [Google Scholar]
- 75.Nunez-Cruz S; Gimotty PA; Guerra MW; Connolly DC; Wu YQ; DeAngelis RA; Lambris JD; Coukos G; Scholler N, Genetic and pharmacologic inhibition of complement impairs endothelial cell function and ablates ovarian cancer neovascularization. Neoplasia 2012, 14 (11), 994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Corrales L; Ajona D; Rafail S; Lasarte JJ; Riezu-Boj JI; Lambris JD; Rouzaut A; Pajares MJ; Montuenga LM; Pio R, Anaphylatoxin C5a creates a favorable microenvironment for lung cancer progression. J Immunol 2012, 189 (9), 4674–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee SH; Son MJ; Oh SH; Rho SB; Park K; Kim YJ; Park MS; Lee JH, Thymosin {beta}(10) inhibits angiogenesis and tumor growth by interfering with Ras function. Cancer Res 2005, 65 (1), 137–48. [PubMed] [Google Scholar]
- 78.Lao L; Shen J; Tian H; Yao Q; Li Y; Qian L; Murray SS; Wang JC, Secreted Phosphoprotein 24 kD Inhibits Growth of Human Prostate Cancer Cells Stimulated by BMP-2. Anticancer Res 2016, 36 (11), 5773–5780. [DOI] [PubMed] [Google Scholar]
- 79.Li CS; Tian H; Zou M; Zhao KW; Li Y; Lao L; Brochmann EJ; Duarte ME; Daubs MD; Zhou YH; Murray SS; Wang JC, Secreted phosphoprotein 24 kD (Spp24) inhibits growth of human pancreatic cancer cells caused by BMP-2. Biochem Biophys Res Commun 2015, 466 (2), 167–72. [DOI] [PubMed] [Google Scholar]
- 80.Burns JL; Hassan AB, Cell survival and proliferation are modified by insulin-like growth factor 2 between days 9 and 10 of mouse gestation. Development 2001, 128 (19), 3819–30. [DOI] [PubMed] [Google Scholar]
- 81.Pollak M, Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer 2008, 8 (12), 915–28. [DOI] [PubMed] [Google Scholar]
- 82.Tian D; Mitchell I; Kreeger PK, Quantitative analysis of insulin-like growth factor 2 receptor and insulin-like growth factor binding proteins to identify control mechanisms for insulin-like growth factor 1 receptor phosphorylation. BMC Syst Biol 2016, 10, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Han S; Li Z; Master LM; Master ZW; Wu A, Exogenous IGFBP-2 promotes proliferation, invasion, and chemoresistance to temozolomide in glioma cells via the integrin beta1-ERK pathway. British J Cancer 2014, 111 (7), 1400–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim EJ; Kang YH; Schaffer BS; Bach LA; MacDonald RG; Park JH, Inhibition of Caco-2 cell proliferation by all-trans retinoic acid: role of insulin-like growth factor binding protein-6. J Cell Physiol 2002, 190 (1), 92–100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw mass spectrometry data have been deposited in Chorus (http://chorusproject.org) with accession Project ID 1401: Charcoal stripped FBS.


