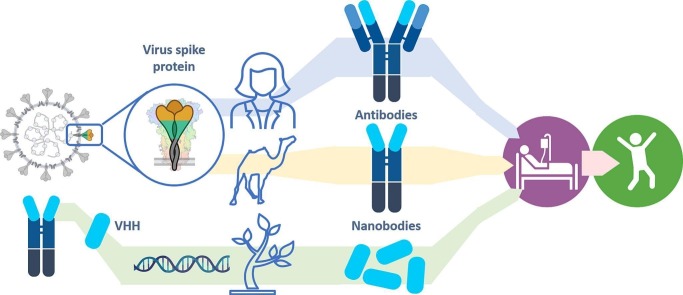
Abstract
Data show a decrease in the risk of hospitalization and death from COVID-19. To date, global vaccinations for SARS-CoV-2 protections are underway, but additional treatments are urgently needed to prevent and cure infection among naïve and even vaccinated people. Neutralizing monoclonal antibodies are very promising for prophylaxis and therapy of SARS-CoV-2 infections. However, traditional large-scale methods of producing such antibodies are slow, extremely expensive and possess a high risk of contamination with viruses, prions, oncogenic DNA and other pollutants. The present study is aimed at developing an approach of producing monoclonal antibodies (mAbs) against SARS-CoV-2 spike (S) protein in plant systems which offers unique advantages, such as the lack of human and animal pathogens or bacterial toxins, relatively low-cost manufacturing, and ease of production scale-up. We selected a single N-terminal domain functional camelid-derived heavy (H)-chain antibody fragments (VHH, AKA nanobodies) targeted to receptor binding domain of SARS-CoV-2 spike protein and developed methods of their rapid production using transgenic plants and plant cell suspensions. Isolated and purified plant-derived VHH antibodies were compared with mAbs produced in traditional mammalian and bacterial expression systems. It was found that plant generated VHH using the proposed methods of transformation and purification possess the ability to bind to SARS-CoV-2 spike protein comparable to that of monoclonal antibodies derived from bacterial and mammalian cell cultures. The results of the present studies confirm the visibility of producing monoclonal single-chain antibodies with a high ability to bind the targeted COVID-19 spike protein in plant systems within a relatively shorter time span and at a lower cost when compared with traditional methods. Moreover, similar plant biotechnology approaches can be used for producing monoclonal neutralizing antibodies against other types of viruses.
Keywords: SARS-CoV-2, Spike protein, Plant antibodies, Nanobodies, Heavy chain antibodies, Single domain antibody
Graphical abstract
1. Introduction
The coronavirus disease 2019 (COVID-19) pandemic has created the greatest global public health crisis. The pandemic was caused by severe acute respiratory distress syndrome coronavirus 2 (SARS-CoV-2) which was first identified in Wuhan, Hubei province China in 2019. This coronavirus was isolated and sequenced in 2020 [1,2]. Although a large number of approaches to the treatment of coronavirus have now been developed and tested, vaccination (mainly by RNA-based vaccines) is the preferred approach to prevent the disease and limit its spread in the population [3]. The main task of vaccination against COVID-19 is the initiation of an immune response and develop the acquired long term immunity (without provoking an actual disease) to form an immune memory in order to mobilize a strong immune response after following an actual infection [4]. However, this vaccination may not be very effective for patients that cannot mount a required immune response (e.g., immunocompromised individuals) or even cannot be applied for individuals that have comorbidities or medical conditions with a high risk of developing complications after immunization. For such patients, the treatment with external long-acting antibodies option may provide an additional approach for protecting against COVID-19 [5]. While vaccines are now available, they do not work for everyone and new effective therapeutics are urgently needed to slow down the spread of SARS-CoV-2 and save lives. Neutralizing monoclonal antibodies (mAbs) have been developed as therapies for treating many viral infections, including RSV, Influenza, SARS, Ebola, HIV, HCMV, and Rabies. They play an essential role as a passive immunotherapy method to fight against various types of infection [5]. Consequently, there is a high demand for the production of effective, safe and inexpensive antibodies for the prevention and treatment of COVID-19.
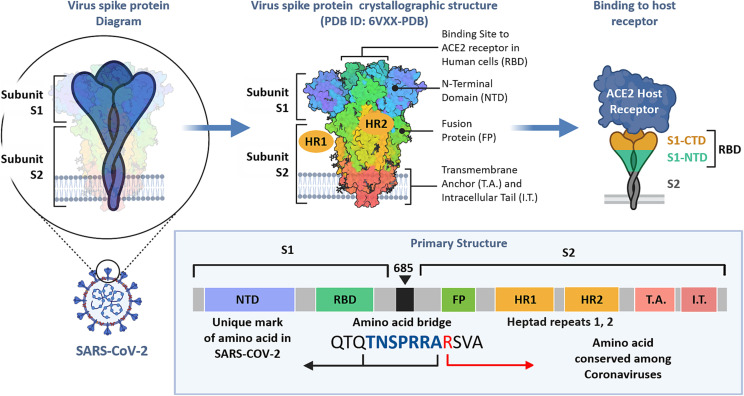
Based on the structure of COVID-19 (as well as other coronaviruses), its mechanisms of action and lifespan, the spike protein represents the most attractive target for developing therapeutic monoclonal antibodies [[6], [7], [8]]. The spike protein of the virus corona consists of two subunits (S1 and S2 in Fig. 1 ). It is the subunit S1 that is responsible for the recognition and binding the angiotensin-converting enzyme 2 (ACE2) receptors, which initiates the uptake of the virus inside the host cells. In turn, the S1 subunit has a receptor binding domain (RBD) that specifically binds to the host receptors. The majority of the primary structure (predominately in the S2 subunit) is conserved among various coronaviruses. At the same time, the S1 subunit (in the N-terminal domain, NTD) contains a unique amino acid sequence that is specific to the COVID-19 viruses.
Fig. 1.
Structure of the SARS-CoV2 spike glycoprotein. Composed using data from [[6], [7], [8]].
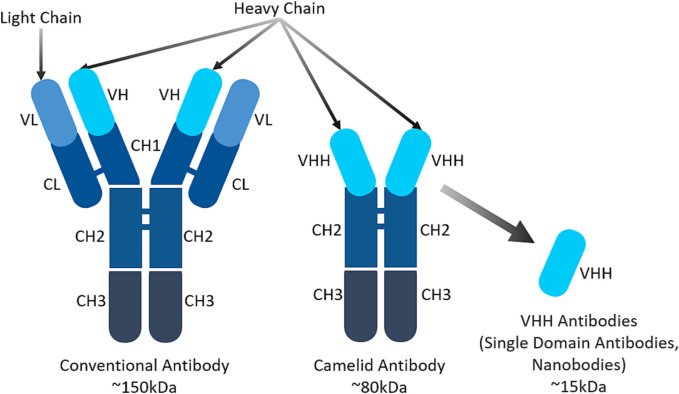
The conventional antibody has a complex structure that consists of heavy and light chains (Fig. 2 ). This structure can retain the antibody's antigen-binding specificity, but it will often reduce its affinity and solubility. Due to the emergence of recombinant DNA technology many antibody domains and fragments have been developed as attractive alternatives to conventional antibodies [[9], [10], [11], [12], [13], [14]]. Due to their small size, superior properties and ease of manufacturing they became excellent candidates for the development of antibodies against COVID-19.
Fig. 2.
A schematic representation of conventional human, camelid and single domain VHH antibodies (nanobodies).
Recently, camelid antibodies have been extensively studied as alternatives to conventional human antibodies [[15], [16], [17], [18], [19]]. In addition to conventional heterotetrameric antibodies, serum in Camelids (dromedaries, Bactrian camels, and llamas) as well as cartilaginous fish contains unique single N-terminal domain functional heavy (H)-chain antibodies (Fig. 2). The recombinant single variable domain camelid-derived heavy chain antibody fragments (VHH, also known as nanobody and often abbreviated as VHH, VNAR, sdAb) possess exclusive properties. The VHH can bind to antigens without requiring domain pairing and can also recognize antigenic sites that are normally not recognized by conventional antibodies [20]. Single-domain antibodies consist of a single monomeric variable antibody domain and do not contain the light chain and CH domain of the conventional Fab region's heavy chain. Unique characteristics of VHH, such as small size, high affinity/specificity to antigens, stability and relatively low production cost, make them an attractive alternative to traditional antibodies as well as antibody fragments in therapeutic applications [15,[21], [22], [23], [24]]. In addition, these small antibodies benefit from improved penetration to sites of infection. They might be very efficient for the treatment of respiratory diseases and particularly for COVID-19 due to the possibility of alternative routes of administration, especially nasal and inhalation routes. For example, these antibodies can be nebulized and administered via an inhaler directly to infected lungs [25]. Consequently, VHH specific to COVID-19 spike protein can form a basis for an effective and innovative approach to the treatment of pandemic caused by the SARS-CoV-2 virus.
Due to strong market demand, the production of anti-COVID-19 antibodies requires industrial-scale production. Traditional large-scale manufacturing processes are based on mammalian cell systems, which require high initial investments and substantial production costs. Additionally, the high risk of contamination with viruses, prions, oncogenic DNA and other pollutants requires additional efforts leading to a further increase in cost. Other complications caused by bioreactors such as development time, scalability, yield, and authenticity make the development of alternative production platforms more compelling [26]. Unlike most full-length mAbs which can only be efficiently produced in mammalian cells, nanobodies can be produced in bacterial system, because they do not required glycosylation. E. coli's rapid growth in inexpensive media and well-establish genetics make it popular system for nanobody expression. However bacterial system has several disadvantages such as formation of inclusion bodies, protease and endotoxin contamination. Therefore, there is an urgent need for alternative production platforms. In recent years, plants have been increasingly explored as bioreactors to produce recombinant vaccines, antibodies, enzymes, hormones, and other proteins [[27], [28], [29], [30]]. Plants offer unique advantages as antibody production system, such as the lack of human/animal pathogens or bacterial toxins, relatively low-cost manufacturing, and ease of production scale-up [[31], [32], [33], [34]]. Unlike other expression systems, plant systems can be implemented in different forms, including the cultivation of whole plants producing recombinant therapeutics in fields, transient expression based on viral vectors, and plant cell suspension culture growing under aseptic conditions. Plant cell suspension cultures are commonly cultivated in simple inexpensive liquid medium compared to mammalian cells and can offer significant advantages, including high safety, low production cost and easy scale-up. The potential of a plant cell suspension system for rapid production at a commercial scale has been demonstrated by the marketing of taliglucerase alfa (Elelyso®) for Gaucher disease that is produced in transgenic carrot cell suspension cultures [[35], [36], [37]]. Plant rapid production systems can quickly respond to pandemic crisis like COVID-19, as demonstrated during the Ebola outbreak by the manufacture of ZMapp, a cocktail of tobacco-produced monoclonal antibodies that confer protection against the Ebola virus [38].
Previously, we successfully developed, characterized, and tested a different recombinant vaccine against SARS-CoV-1 by expressing the N-terminal fragment of SARS-CoV-1 S protein (S1) in tomato and low-nicotine tobacco plants, which demonstrated its immunogenicity after oral and parenteral administration [34]. Based on the close similarity of the S protein structure between SARS-CoV-1 and SARS-CoV-2 viruses, we hypothesized that similar plant biotechnology approaches can be used for producing monoclonal neutralizing antibodies against the COVID-19 virus. The present investigation is aimed at developing of single-domain monoclonal antibodies against SARS-CoV-2 using mammalian, bacterial, and plant-based expression systems while comparing the resulting products based on their characteristics, affinity, and stability to propose a suitable method for the production of VHH nanobodies for the treatment of SARS-CoV-2 infection. The specific aim of this investigation is to produce transgenic plants and plant cell suspensions that express SARS-CoV-2 antibodies targeted the receptor-binding domain on the spike protein. For production of such antibodies, we focused on human heavy chain variable domain VHH, that shows potent neutralization activity and specificity against SARS-CoV-2 in vitro and in animal models [39].
2. Materials and methods
2.1. Preparation of chemically competent E. Coli cells for VHH cloning
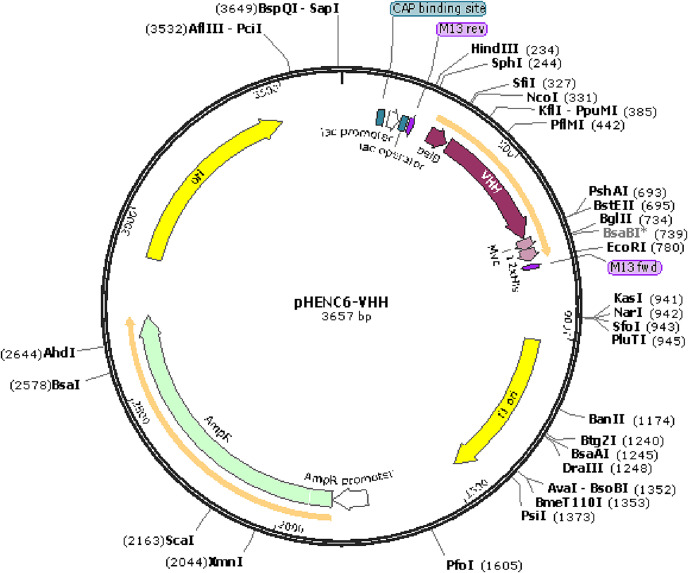
Using standard cloning techniques, the DNA sequences encoding VHH for SARS-CoV-2 (GenBank, MT943599) [40] were designed, codon-optimized for bacterial expression, and cloned into the pHEN6c bacterial expression vector with a C-terminal 12xHis-tag using NcoI and EcoRI restriction sites (this plasmid was obtained from Dr. Muyldermans, Cellular and Molecular Immunology Laboratory, Vrije Universiteit Brussel, Brussels, Belgium). The amino acid sequence of used SARS-CoV-2 VHH and the structure of the plasmid with an inserted sequence is shown in Table 1 and Fig. 3 , respectively. Cellular Myelocytomatosis (c-Myc) and protein affinity 12xHis-tag sequences were incorporated in the vectors allowing detection and purification of the resulting VHH protein. The cloned vectors were transformed into WK6 Chemically Competent E. coli Cells (Thermo Fisher Scientific, Boston, MA) using the recommended transformation protocol [41]. Briefly, 5 μL of WK6 cells were spread on LB agar (without antibiotic) plates for 14 h or overnight. Then, colonies of the overnight culture were inoculated into 100 mL LB media, and the culture was grown to the optical density at 600 nm of 0.6 units (after 4 h). After that, the cells were incubated on ice for 10 min. Following this step, the bacteria were pelleted at 4000 g for 20 min at 4 °C. The pellet was resuspended in 50 mM CaCl2 and placed on ice for 30 min. Next, the cell suspensions were centrifuged as explained above, and pellets were dissolved in 15 mL CaCl2 (50 mM). At this stage, the plate was centrifuged at 4 °C at 4000g for 10 min, and the supernatant was discarded. Again, the pellet was resuspended in 1 mL ice-cooled 50 mM CaCl2, and 50 μL aliquots were prepared and stored at −80 °C for further experiments.
Table 1.
The amino acid sequences of the SARS-CoV-2 VHH inserted into a plasmid. The highlighted letters show the positions of the added c-Myc (red), 12× His-tag (blue).
Fig. 3.
The SARS-CoV-2 VHH-cMYC-12xHis-tag sequence was inserted into the pHEN6c (This plasmid was obtained from Dr. Serge Muyldermans, Cellular and Molecular Immunology Laboratory, Vrije Universiteit Brussel, Brussels, Belgium) using NcoI and EcoRI restriction sites.
2.2. Competent E. Coli cells transformation
WK6 Chemically Competent E. coli Cells transformations were performed as follows; 50 μL of thawed competent bacteria and 5 μL (1 pg-100 ng) of pDNA or pHEN6c-VHH sample (2 μL stock pDNA +18 μL CaCl2) were added to the pre-chilled tube and pipetted gently. Then, the suspensions were kept on ice for 30 min, and after that, the mixture was heated at 42 °C for 45 s. Following the heat-shock step, the tubes were kept on ice for another 2 min. Then 950 μL of Recovery Medium (SOC, Sigma Aldrich, St. Louis, MO) was added, and the suspension was incubated in a shaker at 37 °C for 1 h Next, 10 μL of the transformation was spread on prewarmed selection plates (containing 100 μg/mL Carbenicilin), and the plates were incubated overnight at 37 °C. The next day, 11 colonies were selected and expanded in 5 mL of LB Broth (containing 100 μg/mL Carbenicilin), and the peptide expression was induced by IPTG (0.4 mM) at the OD 600 of three units. Then the colonies were checked for VH ab8 protein production using SDS-PAGE (NuPAGE™ 4 to 12%, Bis-Tris, 1.0 mm, Mini Protein Gel) and PageBlue™ Protein staining (Thermo Fisher Scientific, Boston, MA) based on manufacturer protocol. The suitable colony was expanded in 500 mL LB Broth (containing 100 μg/mL Carbenicilin), and the peptide expression was induced by IPTG (0.4 mM) at the optic density at 600 nm of three units. Bacterial cells were grown for 4 h post-induction. Then the bacterial solution was centrifuged (6000 xg, 10 min). The resulting bacterial pellet was stored at −80 °C for further experiments.
2.3. Mammalian cell culture conditions
EXPI293F™ cells were grown in Expi293 medium (Thermo Fisher Scientific, Boston, MA) at 37 °C, 8% CO2 atmosphere in plastic Corning™ Cell Culture Treated Flasks (Thermo Fisher Scientific, Boston, MA). A CO2 Resistant orbital shaker (Thermo Fisher Scientific, Boston, MA) was used for the incubation. The cells were split to 0.3 × 106 viable cells/mL when they reached a 3 × 106 /mL density. Cell density was determined with a Scepter cell counter (Millipore, Burlington, MA) during the expansion and maintenance phase. One day before transfection, the medium was replaced by spinning down the cells at 300 x g for 5 min at room temperature and then carefully decanting or pipetting away all cell media before resuspending the cells in the prewarmed fresh Expi293 medium.
2.4. Mammalian cell transfection
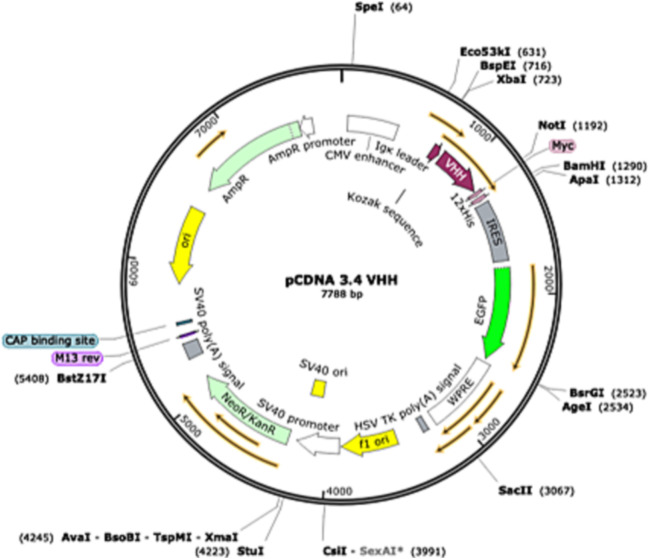
On transfection day, the cells were spun at 300 xg for 5 min at room temperature. The medium was removed by aspiration. Then, the cells were resuspended in the culture medium at a final concentration of 2 × 106/mL. The cells were transfected with a pcDNA3.4-VHH mammalian expression vector with a C-terminal 12xHis-tag by using Expi293™ Expression System Kit (Thermo Fisher Scientific, Boston, MA) following the manufacturer's protocol. The amino acid sequence of used SARS-CoV-2 VH ab8 VHH and the structure of the plasmid with an inserted sequence are shown in Table 2 and Fig. 4 , respectively. After 24 h incubation at 37 °C, 8% CO2 atmosphere, the cells were transferred to a larger flask and diluted with prewarmed Expi293™ medium to a concentration of 0.3 × 106/mL. The transfected cells were incubated for 7–10 days at the condition mentioned above, and the cell viability was regularly checked every second day.
Table 2.
The amino acid sequences of the SARS-CoV-2 VHH recombinant vectors. The highlighted letters show the positions of the added c-Myc (red), 12× His-tag (blue). Sequences of added linkers are underlined.
Fig. 4.
The SARS-CoV-2 VHH-cMYC-12xHis-tag sequence was inserted into the pCDNA 3.4 (Thermo Fisher Scientific, Boston, MA) using XbaI and AgeI restriction sites.
2.5. Purification of the SARS-CoV-2 VHH from bacterial and mammalian cells
For purification of the SARS-CoV-2 VHH from WK6 Chemically Competent E. coli Cells, the bacterial pellet was lysed using the lysis buffers A and B (Table 3 ). For every gram of bacterial pellet, 10 mL of lysis buffer A and 15 mL of lysis buffer B were used. The slurry was vigorously stirred at 4 °C for 1 h, centrifuged at 20,000 g (4 °C) for 1 h, followed by passing through a 0.2 μL PES membrane filter column (VWR, Radnor, PA) using a vacuum-driven filtration. In the next step, Ni-NTA resin (1 mL resin per 30 g of the pellet) was added to the filtered suspension, and the sample was incubated with the Ni-NTA resin for 1 h at room temperature while rocking gently. The mix was then passed through the Econo-Pac® Chromatography Columns (Bio-Rad, Hercules, CA) followed by washing the column with 50 mL of Imidazole 10 mM in the first step and then 50 mL of Imidazole 30 mM. For purification of the SARS-CoV-2 VHH from EXPI293F™ cells, the cells were spun at 300 xg for 5 min. Then the supernatant was vacuum filtered using a 0.2 μL PES membrane filter column (VWR, Radnor, PA). In the next step, 500 μL Ni-NTA resin was added to the filtered suspension, and the sample was incubated with the Ni-NTA resin for 1 h at room temperature while rocking gently. The mix was then passed through the Econo-Pac® Chromatography Columns (Bio-Rad, Hercules, CA). The VHH antibody was eluted with 500 μL of the elution buffer and then mixed with 500 μL of glycerol and stored at −20 °C for further experiments.
Table 3.
The compositions of lysis and elution buffers (pH = 7) for protein purification.
| Reagents | Lysis Buffer A | Lysis Buffer B | Elution Buffer |
|---|---|---|---|
| NaH2PO4 | – | – | 20 mM |
| Tris | 24.22 g/L | 8.1 g/L | – |
| NaCl | – | – | 500 mM |
| Imidazole | – | – | 500 mM |
| EDTA | 0.2 g/L | 0.05 g/L | – |
| Sucrose | 170 g/L | 43 g/L | – |
2.6. Sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Western Blot
Before SDS-PAGE, VHH was quantitated using a BCA Protein Assay Kit (Thermo Fisher Scientific, Boston, MA) based on manufacturer protocol [42]. To analyze the VHH antibody's purity, SDS-PAGE (4–20% Mini-PROTEAN® TGX™ Precast Protein Gels, Bio-Rad, Hercules, CA) and PageBlue™ Protein staining (Thermo Fisher Scientific, Boston, MA) was used based on manufacturer protocol. To confirm the expression of the VHH antibody, the Western blot was used. Briefly, 100 μg of samples were denatured at 95 °C for 10 min and transferred to a 4–20% Mini-PROTEAN® TGX™ Precast Protein Gels (Bio-Rad, Hercules, CA), and electrophoresis was performed at 120 V for 60 min. Then, the band was transferred to a 0.2 μm cellulose nitrate membrane (Thermo Fisher Scientific, Boston, MA). The filters were incubated in 5% BSA TBST at room temperature for 2 h with a shaker to avoid unspecific binding. Subsequently, filters were incubated with DyLight™ 680 Conjugated Affinity Purified anti-His-tag antibody (Rockland Inc., Pottstown, PA) at 1:10000 dilution, at 4 °C overnight. Afterward, the immunoreactive bands were detected by ChemiDoc MP Imaging System (Bio-Rad, Hercules, CA). Quantitative western blot was performed by a regression analysis of a calibration curve created using purified bacteria VHH protein in a 2× serial dilution. Concentration of plant-derived VHH protein was determined by analyzing band intensity.
2.7. The Enzyme-Linked Immunosorbent Assay (ELISA)
ELISA was performed to evaluate the ability of the VHH antibody to bind to the SARS-COV2 spike protein. For this purpose, 100 μL of 1 μg/mL recombinant human coronavirus SARS-CoV-2 Spike Glycoprotein RBD (Abcam, Boston, MA) was coated onto an Immuno-Maxi Absorbance 96-well plate (Thermo Fisher Scientific, Boston, MA) and incubated overnight at 4 °C. Each well was washed three times with phosphate-buffered saline (PBS) plus 0.1% Tween (PBS-T) and subsequently blocked by 2% bovine serum albumin (BSA, VWR, Radnor, PA) in PBS-T for 2 h at room temperature. Then, different concentrations of VHH ranging from 0 to 100 μg/mL were added to each SARS-CoV-2 Spike Glycoprotein RBD-coated well and incubated for 30 min at room temperature. Each well was washed three times by PBS-T and then incubated with 100 μL of the HRP-conjugated Anti-c-Myc antibody (Abcam, Boston, MA) for 60 min at room temperature. Samples were washed six times by PBS-T, followed by the addition of 50 μL 3,3′,5,5′-tetramethylbenzidine (TMB), and incubation for 15 min at room temperature. Finally, the reaction was stopped by 50 μL of 2 M sulfuric acid (Thermo Fisher Scientific, Boston, MA), and the absorbance was measured using a Tecan Infinite M200 Pro Nanoquant microplate reader (Männedorf, Switzerland) at 450 nm.
2.8. Plant material and propagation in vitro
Seeds of tobacco (Nicotiana tabacum) cultivar Wisconsin (obtained from the Victory Seed Company, Molalla, OR) were surface sterilized by immersion in 70% ethanol for 1 min, followed by soaking in 1.5% sodium hypochlorite for 10 min. After rinsing three times in sterile distilled water, and blotting dry with sterile filter paper, seeds were placed on germination medium MSG containing MS macro- and microelements [43] with 1% sucrose and 0.7% agar (see Table 4 for details on media compositions). In vitro cultures were maintained by transferring 1- cm-long stem segments with axillary buds at 5–6 weeks intervals onto fresh MSP propagation medium.
Table 4.
Media for tissue culture and transformation experiments.
| Media | Composition |
|---|---|
| MSG | Basic MS basal medium with 1% sucrose, 0.7% agar |
| MSP | MS with 3% sucrose, 0.7% agar |
| MSA | MS with 3% sucrose, 0.7% agar, 100 μM acetosyringone |
| MSRT | MS with 3% sucrose, 0.7% agar, 2 mg/L BAP, 0.1 mg/L NAA, 300 mg/L timentin, 100 mg/L kanamycin |
| MST | MS with 3% sucrose, 0.7% agar, 300 mg/L timentin, 100 mg/L kanamycin |
| MSCIT | MS with 3% sucrose, 0.7% agar, 1 mg/L 2,4 D, 0.5 mg/L NAA, 0.5 mg/L BAP, 300 mg/L timentin, 100 mg/L kanamycin |
| MSCT | MS with 3% sucrose, 0.7% agar, 2 mg/L 2.4-D, 200 mg/L timentin, 100 mg/L kanamycin |
| MSST | MS with 3% sucrose 2 mg/L 2.4-D 100 mg/L timentin, 100 mg/L kanamycin |
2.9. Generation of transgenic plants
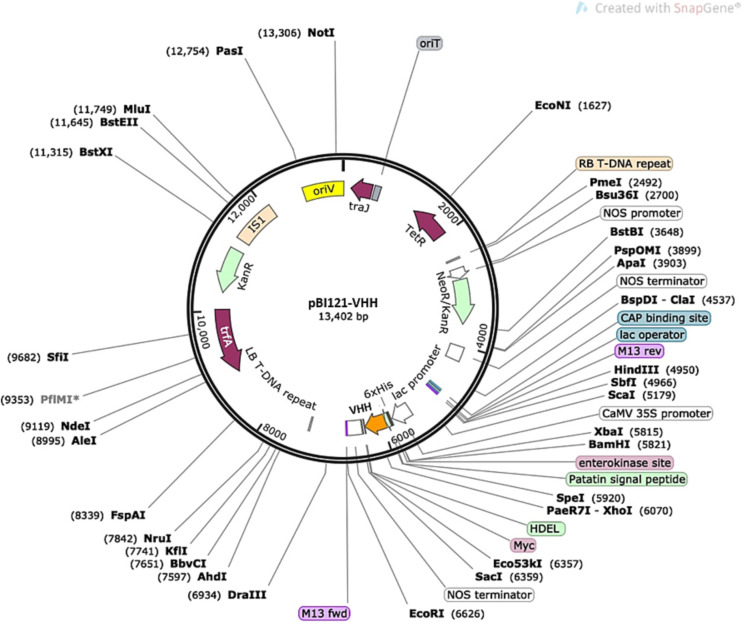
For plant transformation, DNA sequences encoding VHH specific for SARS-CoV-2 (GenBank, MT943599, VH ab8) were codon-optimized for plant expression. A synthetic cDNA encoding the VHH protein was cloned into the modified pBI121 binary vector [44] (Table 5 and Fig. 5 ) under a strong CaMV- 35S promoter. The transformation vector included the nptII gene for kanamycin selection of the transgenic plants and HDEL. For purification of recombinant protein 6xHistidine and C-Myc tags were added. Vector pBI121-VHH was introduced into Agrobacterium tumefaciens strain LBA4404 for plant transformation experiments. Tobacco cv Wisconsin was transformed by Agrobacterium-mediated method according to Horsch [45] with some modifications. Agrobacterium tumefaciens strain LBA4404 was grown on solid LB media supplemented with 50 mg/L kanamycin and 20 mg/L rifampicin. For transformation experiments, the agrobacterial cell suspension was prepared by inoculating 20 mL of the liquid LB medium with a single agrobacterial colony and grown for 2 days at 150 rpm on a rotary shaker at 25 °C. The suspension of Agrobacterium was diluted with a liquid MS medium to obtain optical density of the solution at 600 nm (OD600) close to 0.5 units. Leaf segments excised from 1 to 1.5-month-old tobacco plants were inoculated with Agrobacterium suspension with OD600 = 0.5 for 10 min. After blotting dry with sterile filter paper, leaf segments were transferred to MSTC co-cultivation medium and incubated in the dark for 2 days at 23 °C. Then tobacco explants were transferred to regeneration selection medium (MSTS, Table 4) containing 100 mg/L kanamycin. After 4–5 weeks of cultivation on MSTS medium, kanamycin-resistant green shoots (putative transformants) were formed. Healthy green shoots (1–2 cm) were excised and transferred to Phytatrays containing MST selection medium with 100 mg/L kanamycin.
Table 5.
The amino acid sequences of SARS-CoV-2 VHH inserted into pCDNA used for plant transfection. The highlighted letters show the positions of the added c-Myc (red), 12× His-tag (blue) and linkers (underlined).
Fig. 5.
Vector pBI121-VHH that was used for plant transformation.
2.10. Callus induction and cell suspensions
For callus induction, leaf segments were placed in 100 × 15 mm Petri dishes containing MSCI callus induction medium. Plates were incubated in the dark at 23 °C. Well-developed tobacco callus tissues were selected and transferred to the callus propagation MSCP medium (Table 4) and incubated in the dark. For initiation of cell suspension, approximately 1 g of friable callus tissue was transferred into the sterile 250 mL conical flasks containing 50 mL of liquid MSCS medium. Cell cultures were grown in the dark at 23 °C, on a rotary shaker at 110 rpm. Maintenance of the cell suspension was carried out on the MSCS medium with 10-day intervals for subcultures.
2.11. Protein extraction from plants and plant cell suspensions
Proteins were extracted from transgenic tobacco leaf and plant cell suspensions that were engineered to express VHH nanobodies of interest. Respective samples (frozen tobacco leaves and cells) were ground to a fine dry powder in the presence of liquid nitrogen. Samples were resuspended in phosphate buffered saline (PBS) from Sigma Aldrich (Saint Louis, MO) and centrifuged at 20,000 xg for 30 min. The supernatant was then vacuum filtered through a poly(ethersulfone) 0.2 μm pore size filtration cup (VWR, Radnor, PA). Samples were then frozen at −20 °C until further analysis.
2.12. DNA isolation and real-time polymerase chain reaction (RT-PCR)
For extraction of the Tobacco cultivar Wisconsin plant DNA, DNeasy® Plant Mini Kit was used (Qiagen, Germantown, MD). Briefly, around 100 mg wet weight of plant tissues were disrupted using a mortar and pestle then the resulted materials were transferred to a microtube containing 400 μL of lysis buffer. Then, 4 μL RNase A was added to the solution, vortexed, and incubated for 10 min at 65 °C. After the incubation, 130 μL of binding buffer was added to the mixture and incubated for 5 min on ice. In the next step, the lysate was centrifuged at 20,000 xg for 5 min. Then, the lysate was pipet into a QIA shredder spin column placed in a 2 mL collection tube and centrifuged at 20,000 xg for 2 min. In the next step, the flow-through solution was transferred into a new tube followed by the addition of 1.5 volumes of washing buffer. Then, 650 μL of the mixture was transferred into a DNeasy® Mini spin column placed in a 2 mL collection tube. The mixture was centrifuged at 6000 xg for 1 min, the flow-through liquid was discarded and the procedure was repeated for the remaining sample. The spin column was then placed into a new 2 mL collection tube and washed two times with 500 μL of fresh washing buffer by centrifugation at 6000 xg for 1 min and 20,000 x g for 2 min. Then, the spin columns were transferred to a new 1.5 mL microcentrifuge tubes, 100 μL of elution buffer were added and the mixture was incubated at room temperature for 5 min. Finally, DNA was eluted at 6000 x g for 1 min and the resulting DNA concentration was measured by Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Boston, MA).
To perform PCR the following primers from IDT, Coralville, IA were used: 5’-GAAGTACAACTTGTTGAATCCGGTG-3′ (forward) and 3’-TGAAGATACTGTCACGGTAGTACCT-5′ (reverse). The primers were designed to amplify the 372-base pair (bp) sequence of the target gene excluding tags and stop codon. The PCR amplification of the V H ab8 gene was done using DreamTaq Green PCR Master Mix according to the manufacturer protocol (Thermo Fisher Scientific, Boston, MA). Cycling conditions were 95 °C for 1 min, followed by 35 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min and final extension at 72 °C for 7 min. Then, 10 μL of amplified DNA sample were loaded on 1% Agarose gel containing 0.4 μg/mL ethidium bromide (EtBr) and after electrophoresis bands were detected by ChemiDoc MP Imaging System (Bio-Rad, Hercules, CA).
2.13. Statistical analysis
Data were analyzed using descriptive statistics, single-factor analysis of variance (ANOVA), and presented as mean values ± standard deviation (SD). The comparison among groups was performed by the independent sample student's t-test. The difference between variants was considered significant if P < 0.05.
3. Results
A VHH antibody that can bind to SARS-CoV-2 spike protein was produced in the WK6 bacterial system. For this purpose, the VHH genome sequence (DOI: https://doi.org/10.1016/j.cell.2020.09.007) with added cMYC and 12xHis-tag elements was inserted into the pHEN6c vector using NcoI and EcoRI restriction sites. The resulting plasmid was transformed into the WK6 bacterial systems using a heat-shock method [46]. The transfected bacteria were cultured in a culture plate with a selection marker (Carbenicillin 100 μg/mL), and selected colonies were checked for the expression of VHH-cMYC-12xHis-tag using SDS-PAGE (Fig. 6 ). Based on obtained results suitable colonies were selected for expansion. After expanding the selected colonies in LB Broth (containing 100 μg/mL Carbenicilin), the expression of the VHH was initiated by the addition of IPTG (0.4 mM). Then the protein was extracted and purified.
Fig. 6.
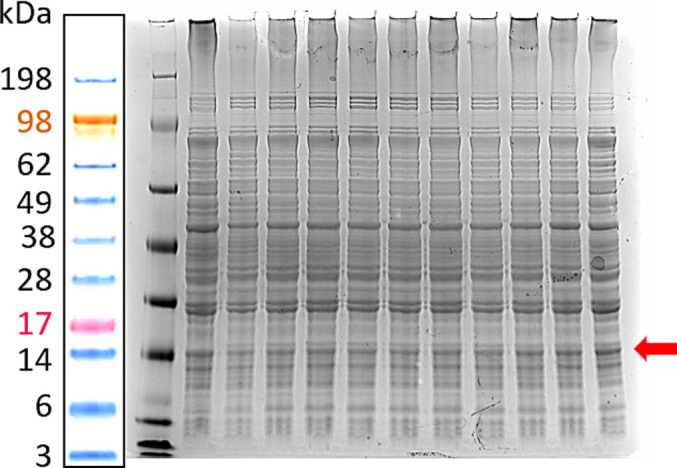
Confirmation of the SARS-CoV-2 VHH expression in WK6 bacterial systems. The total protein expression of various bacterial colonies was tested using SDS-PAGE after the expression was induced by IPTG (0.4 mM). The representative image of the gel before the purification of VHH protein is shown. The red arrow points to VHH lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
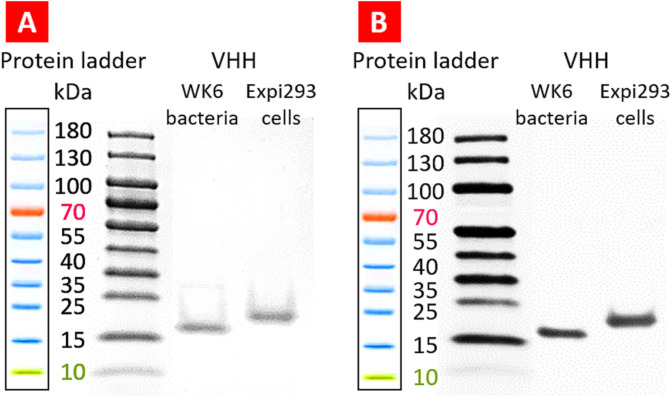
In the second step of this study, a single-chain VHH antibody that can bind to SARS-COV2 spike protein was produced in the EXPI293F™ cells. For this purpose, the VHH genome sequence (DOI: https://doi.org/10.1016/j.cell.2020.09.007, VH ab8) with added cMYC and 12xHis-tag elements were inserted into the pCDNA3.4™ vector using XbaI and AgeI restriction sites (Table 2 and Fig. 4). The produced proteins were extracted based on the affinity of 12 Histag to the Ni-NTA resin column. To evaluate the purity of extracted recombinant proteins, SDS-PAGE and PageBlue™ protein staining methods were used. The result showed high purity of the product (Fig. 7 , A). To confirm the SARS-CoV-2 VHH-cMYC-12xHis-tag antibody expression, Western blot analysis was performed as previously described [47], and the results confirmed the expression of the desired product (Fig. 7, B).
Fig. 7.
Evaluation of the purity and expression of VHH antibody. Representative images of SDS-PAGE (A) Western blot (B) are shown.
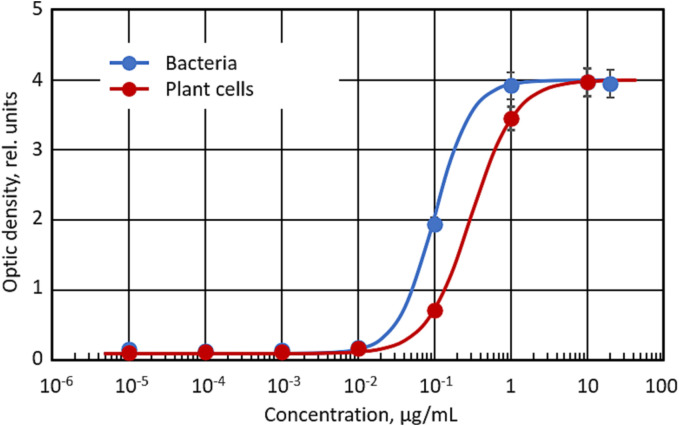
The ability of the VHH antibodies to bind the SARS-CoV-2 spike antigen was evaluated by ELISA. The results confirmed that the bacteria-derived VHH antibody was able to effectively bind the COVID-19 spike protein (Fig. 8 ). Similar results were obtained for VHH antibodies obtained in mammalian cells (data are not shown).
Fig. 8.
The ability of bacteria and plant cells-derived SARS-CoV-2 VHH antibody to bind SARS-COV2 spike protein. Means ± SD are shown.
For plant transformation experiments seeds of tobacco (Nicotiana tabacum) cultivar Wisconsin were surface sterilized and placed on germination plant medium MSG (Table 4). Tobacco plants developed from the seeds were grown in Phytatrays™ (MilliporeSigma, Burlington, MA) under aseptic conditions. Leaves from one month old plants were used as explants for transformation experiments. For tobacco transformation, DNA sequences encoding VH ab8 specific for SARS-CoV-2 (GenBank, MT943599) were codon-optimized for plant expression. A synthetic cDNA encoding the VH ab8 protein was cloned into the modified pBI121 binary vector (Fig. 5) under CaMV-35 promoter, which widely used as a strong constitutive promoter for transgene expression. The transformation vector also included the nptII gene for kanamycin selection of the transgenic plants and plant-specific endoplasmic reticulum retention signal (HDEL). For purification of recombinant protein 6xHistidine and C-Myc tags were added.
For stable transformation of tobacco, we used the Agrobacterium-mediated method, the most efficient for many plant species that use the agrobacteria as the biological vector to transfer exogenous T-DNA into the plant cell. The vector pBI121-VHH was introduced into Agrobacterium tumefaciens strain LBA4404 and used for tobacco transformation. Tobacco leaf segments were co-cultivated with Agrobacterium suspension on MSR medium and then transferred to regeneration selection MSRT medium (Table 4) containing 100 mg/L kanamycin and plant growth regulators for stimulation of shoot regeneration (2 mg/L 6-benzylaminopurine and 0.2 mg/L naphthaleneacetic acid). After 4–5 weeks of cultivation kanamycin-resistant shoots were developed (Fig. 9A) and transferred for elongation and root development in a hormone-free MST medium (Table 4). Kanamycin-resistant tobacco plants demonstrated good growth and development on the selection medium (Fig. 9B) whereas non-transgenics were not able to grow, bleached, and died.
Fig. 9.
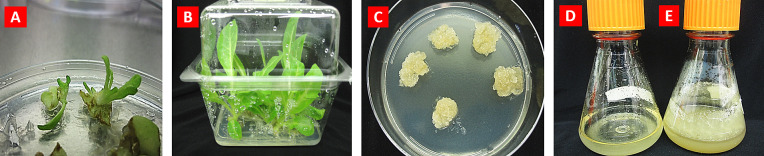
Transgenic tobacco plants expressing COVID-19 VHH antibody. (A) Transgenic shoot formation; (B) Propagation of transgenic plants in the presence of selective agent kanamycin; (C) Callus tissues growing on MSCT medium; (D-E) Transgenic tobacco cell suspension growing in MSC liquid medium: D - 1 day, E - 10 days.
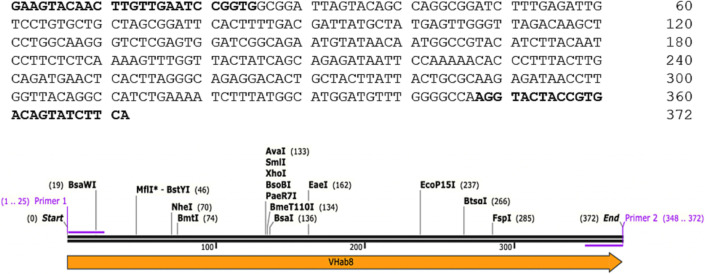
Eight independent tobacco kanamycin-resistant plants have been tested by PCR for the presence of the V H ab8 gene in genomic DNA. The incorporation of the VHH-antibody gene into plant genomes was confirmed by PCR analysis. The sequence of the PCR product is shown in Fig. 10 . All eight transgenic plants showed the presence of the antibody's gene in plant DNA, whereas the control (non-transgenic plant) did not demonstrate the occurrence of this gene (Fig. 11 ).
Fig. 10.
Structure of VHH PCR primers and product (the sequence of primers is highlighted in bold).
Fig. 11.

Confirmation of the SARS-CoV-2 VHH gene expression in tobacco plants. DNA was extracted from eight independent transgenic and one non-transgenic (negative control, NTC) tobacco plants. The gene was amplified using VHH-specific primers and visualized by agarose gel electrophoresis. A representative image of gel electrophoresis is shown. Transgenic tobaco plant DNA demonstrated a resulting PCR product with an expected 372 bp length. In contrast, no-transfected plant DNA does not show such an amplified band.
It should be stressed that transgenic tobacco plants expressing VHH have been produced in the very short period of the time, approximately in 2 months. Transgenic plants producing antibodies, vaccines and other recombinant proteins via molecular farming are considered as bioreactors or factories; cultivation of them can be expanded to the agricultural scale and has the potential to fulfill emergency demands, such as in the present situation of the COVID-19 pandemic.
Another plant bioreactor is the cell suspension, which produces VHH antibody in sterile conditions. For the development of a cell suspension culture bioreactor, transgenic tobacco leaf explants were placed on callus induction media, in darkness. Various concentrations of plant growth regulators were tested for callus induction: 2,4 -dichlorophenoxyacetic acid (2,4-D; 1–2 mg/L), naphthaleneacetic acid (NAA; 0.4–0.5 mg/L) and benzylaminopurine (BAP; 0.3–0.5 mg/L) Tobacco callus tissues (mass of undifferentiated cells) have been initiated after 4–5 weeks of incubation. The most efficient medium for callus initiation was MSCI, contained 2 mg/L 2,4-D, 0.5 mg/L NAA and 0.5 mg/L BAP (Table 4). Then, primary callus tissues were transferred to callus propagation medium MSCT (Table 4). Fig. 9C illustrates callus grown on the propagation medium. For initiation of cell suspension, fresh friable callus tissues were transferred into the sterile flasks containing liquid MSCS medium (Table 4). Cell suspensions were grown in the dark at 23 °C, on a rotary shaker at 110 rpm. Maintenance of transgenic cell suspensions producing recombinant VHH antibodies was carried out on the MSCS medium with 10-day intervals for subcultures. Very fast-growing transgenic tobacco cell suspension was produced: about a 10-fold increase in cell volume was achieved in 9–10 days (Fig. 9D-E). Such efficient growth of cell suspension can provide a very efficient system for the production of monoclonal antibodies, vaccines and other recombinant proteins.
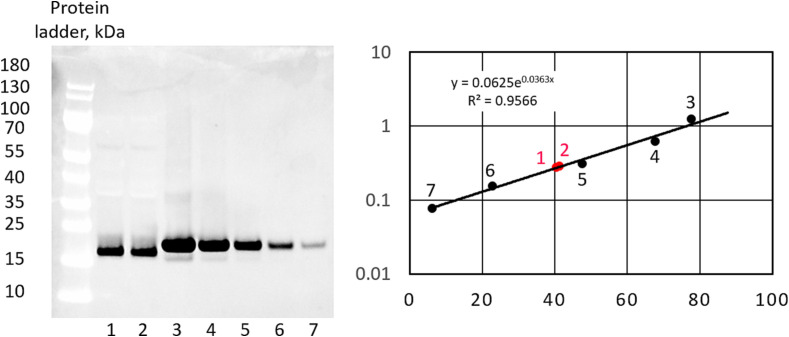
Western blot analysis of several transgenic lines (plant and cell suspension) confirmed the presence of recombinant VHH antibodies with the expected molecular size at variable levels. The 15 kDa protein was detectable in several independent transgenic plant lines (Fig. 12 ). The quantitative analysis of Western blots estimated the final concentration of VHH in the stock solution of 16.6 ± 2.3 mg/mL.
Fig. 12.
Expression of VHH proteun in tobacco plants. (A) Representative image of Western blot of SARS-CoV-2 spike protein VHH antibodies purified from two independent transgenic plant lines. (B) Quantitative Western blot image and regression analysis. 1, 2 – VHH samples isolated from the transgenic plants. 3–7 – Serial dilutions of VHH protein standard.
A high level of expression of recombinant VHH protein was observed in cell suspensions. Several transgenic plants as well as cell suspensions were tested by ELISA. The binding of tobacco-derived VHH antibody to SARS-COV2 spike protein was confirmed by ELISA. Plan-derived antibodies demonstrated a similar binding activity to the COVID-19 spike protein when compared with antibodies derived from bacterial and mammalian cells (Fig. 8).
4. Discussion
Single domain antibody, also identified as VHH, VNAR, sdAb, or VHH, is a kind of antibody fragment that is consists of a single monomeric variable antibody domain. It also does not have the light chain and CH domain of the conventional Fab region's heavy chain. In recent years, the VHH application has expanded in medicine because of its beneficial biochemical and economic properties such as size, affinity, specificity, stability, and production cost [24]. One prospective application for VHH is for the treatment of the COVID-19 pandemic caused by the SARS-CoV-2 virus.
The overall aim of the present work is to develop of single-domain monoclonal antibodies against SARS-CoV-2 using a plant system and compare plan-derived antibodies with those produced in bacterial and mammalian cell systems. Consequently, we developed three VHH containing vectors capable of transfecting bacteria, mammalian, and plant cells based on pHEN6c, pcDNA™ and pBI121, respectively. In this work, we used WK6 bacteria and EXPI293F™ cells got the production of the SARS-CoV-2 VHH antibody. The produced proteins were extracted based on the affinity of 12×-Histag to the Ni-NTA resin column, and their production and purity were confirmed using SDS-Page and Western blot analysis. We also showed the ability of produced antibodies to bind to SARS-COV2 spike protein.
The most popular method for producing monoclonal antibodies is based on Chinese hamster ovary (CHO) cells, which can be grown in large bioreactors. However, this process is slow and can take many months to determine productive CHO cell culture and scale it up in large bioreactors. In addition, the process of growing these cell cultures, as well as the research to identify and develop the most effective antibodies can make such therapies extremely expensive. For example, Hernandez et al. examined the prices of all monoclonal antibodies approved by the US Food and Drug Administration (FDA) in the recent 20 years to find that the average annual price of monoclonal antibody therapies was $96,731 [48].
Plants have become a prospective replacement bioreactor for currently available production systems to manufacture biopharmaceuticals [49,50]. Moreover, plants offer several advantages as a mAb production system, such as the lack of human pathogens, relatively low-cost manufacturing, and ease of production scale-up and are inherently safe because no human pathogens can replicate in plants.
Transgenic tobacco plants were produced by the proposed method very quickly, within 2 months, and stable transgenic plants constantly expressed VHH antibodies. In our experiments stable transgenic tobacco plants constantly expressing VHH antibodies were produced by the proposed method very quickly compared to a longer span of months required to other production systems. For large-scale production, tobacco plants can be mass propagated in a greenhouse under controlled environmental conditions. Furthermore, transgenic plant cultivation can be expanded into the fields where tobacco plant needs only sun, water and fertilizer. Tobacco is the most popular plant species due to high amount of leaf biomass and easy genetic manipulations. Tobacco can generate up to 170 tons/ha of green inexpensive biomass more efficiently, than almost any agricultural crop. Due to rapid production on an industrial scale, tobaco-based nanobodies have the potential to fulfill emergency demand during the COVID-19 pandemic.
Another plant bioreactor is the cell suspension which produces VHH antibodies in sterile conditions with a lack of human/animal pathogens, relatively low-cost manufacturing, and ease of production scale-up. Plant cell cultures are based on plant cells growing in a fully controlled environment, and offer the advantage of producing proteins in bioreactors under the conditions of the good manufacturing practice. Suspension cell cultures have the same advantages of sterility and containment similar to other cell culture expression systems. The ability to use low-cost plant growth medium represents an additional advantage over expensive mammalian medium. Similar plant biotechnology approaches can be used for producing monoclonal neutralizing antibodies against the other types of coronaviruses.
As the portal of entry for SARS-CoV-2 is the respiratory tract, direct delivery of small VHH antibodies to the nasal mucosa and lungs can improve antibody efficacy. Based on our previous extensive experience in the delivery of various drugs, nucleic acids and peptides directly to the lungs via inhalation [[51], [52], [53], [54], [55], [56], [57]], currently, we are investigating the inhalation delivery of plant-derived VHH anti-COVID-19 antibodies bound to several different nanoscale-based delivery systems.
Since large capital cost is associated with traditional mammalian produced recombinant proteins, plant-based technology could represent the breakthrough need to make low-cost antibodies for markets.
5. Conclusions
In the face of a pandemic infectious disease outbreak, new approaches should be explored to enable the most rapid and cost-effective methodologies for the production and evaluation of antibodies for passive immunization and treatment. The fastest timeline from lead mAb identification to phase I studies is an important goal for all companies and patients. Technological advancements in plant-based therapeutics have strengthened the approach and it is now a promising production system for fighting the COVID-19 pandemic where plant-based antibodies can be produced quickly and at low cost compare to mammalian-based antibodies. In the present investigation, we carried out proof-of-concept experiments for producing single chain VHH antibodies against the spike protein of the COVID-19 virus and showed the viable production in plant systems of antibodies with a high ability to bind the targeted virus protein. As the portal of entry for SARS-CoV-2 is the respiratory tract, direct delivery of small VHH antibodies to the nasal mucosa and lungs can improve antibody efficacy.
CRediT authorship contribution statement
Andrew M. Shen: Investigation. Obeid M. Malekshah: Investigation, Methodology, Writing – original draft, Writing – review & editing. Natalia Pogrebnyak: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review & editing. Tamara Minko: Conceptualization, Data curation, Formal analysis, Funding acquisition, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing.
Acknowledgments
This work was supported in part by the R01 CA238871 grant from the National Institute of Health.
Data availability
Data will be made available on request.
References
- 1.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Majumder J., Minko T. Recent developments on therapeutic and diagnostic approaches for COVID-19. AAPS J. 2021;23:14. doi: 10.1208/s12248-020-00532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pulendran B., Ahmed R. Immunological mechanisms of vaccination. Nat. Immunol. 2011;12:509–517. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shanmugaraj B., Siriwattananon K., Wangkanont K., Phoolcharoen W. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for coronavirus disease-19 (COVID-19) Asian Pac. J. Allergy Immunol. 2020;38:10–18. doi: 10.12932/AP-200220-0773. [DOI] [PubMed] [Google Scholar]
- 6.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jain S., Venkataraman A., Wechsler M.E., Peppas N.A. Messenger RNA-based vaccines: past, present, and future directions in the context of the COVID-19 pandemic. Adv. Drug Deliv. Rev. 2021;179 doi: 10.1016/j.addr.2021.114000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;183:1735. doi: 10.1016/j.cell.2020.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng Y., Meshaw R., McDougald D., Zhou Z., Zhao X.G., Jannetti S.A., Reiman R.E., Pippen E., Marjoram R., Schaal J.L., Vaidyanathan G., Zalutsky M.R. Evaluation of an (131)I-labeled HER2-specific single domain antibody fragment for the radiopharmaceutical therapy of HER2-expressing cancers. Sci. Rep. 2022;12:3020. doi: 10.1038/s41598-022-07006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghani S., Deravi N., Pirzadeh M., Rafiee B., Gatabi Z.R., Bandehpour M., Yarian F. Antibody fragment and targeted colorectal cancer therapy: a global systematic review. Curr. Pharm. Biotechnol. 2022;23:1061–1071. doi: 10.2174/1389201022666210810104226. [DOI] [PubMed] [Google Scholar]
- 11.Haist C., Poschinski Z., Bister A., Hoffmann M.J., Grunewald C.M., Hamacher A., Kassack M., Wiek C., Scheckenbach K., Hanenberg H. Engineering a single-chain variable fragment of cetuximab for CAR T-cell therapy against head and neck squamous cell carcinomas. Oral Oncol. 2022;129 doi: 10.1016/j.oraloncology.2022.105867. [DOI] [PubMed] [Google Scholar]
- 12.Minenkova O., Santapaola D., Milazzo F.M., Anastasi A.M., Battistuzzi G., Chiapparino C., Rosi A., Gritti G., Borleri G., Rambaldi A., Dental C., Viollet C., Pagano B., Salvini L., Marra E., Luberto L., Rossi A., Riccio A., Merlo Pich E., Santoro M.G., De Santis R. Human inhalable antibody fragments neutralizing SARS-CoV-2 variants for COVID-19 therapy. Mol. Ther. 2022;30:1979–1993. doi: 10.1016/j.ymthe.2022.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munoz-Lopez P., Ribas-Aparicio R.M., Becerra-Baez E.I., Fraga-Perez K., Flores-Martinez L.F., Mateos-Chavez A.A., Luria-Perez R. Single-chain fragment variable: recent Progress in cancer diagnosis and therapy. Cancers (Basel) 2022;14 doi: 10.3390/cancers14174206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Velazquez E.J., Cress J.D., Humpherys T.B., Mortimer T.O., Bellini D.M., Skidmore J.R., Smith K.R., Robison R.A., Weber S.K., O’Neill K.L. Selection of human single domain antibodies (sdAb) against thymidine kinase 1 and their incorporation into sdAb-fc antibody constructs for potential use in cancer therapy. PLoS One. 2022;17 doi: 10.1371/journal.pone.0264822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D. Könning, S. Zielonka, J. Grzeschik, M. Empting, B. Valldorf, S. Krah, C. Schröter, C. Sellmann, B. Hock, H. Kolmar. Camelid and shark single domain antibodies: structural features and therapeutic potential. Curr. Opin. Struct. Biol. 2017, 45, 10–16, doi:https://doi.org/ 10.1016/j.sbi.2016.10.019. [DOI] [PubMed]
- 16.Danis C., Dupre E., Zejneli O., Caillierez R., Arrial A., Begard S., Mortelecque J., Eddarkaoui S., Loyens A., Cantrelle F.X., Hanoulle X., Rain J.C., Colin M., Buee L., Landrieu I. Inhibition of tau seeding by targeting tau nucleation core within neurons with a single domain antibody fragment. Mol. Ther. 2022;30:1484–1499. doi: 10.1016/j.ymthe.2022.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ezzikouri S., Nourlil J., Tsukiyama-Kohara K., Kohara M., El Ossmani H., Windisch M.P., Benjelloun S. Nanobodies: an unexplored opportunity to combat COVID-19. J. Biomol. Struct. Dyn. 2022;40:3129–3131. doi: 10.1080/07391102.2020.1845801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wani N.A., Praveen Kumar K., Hong S., Umer M.A. Telomere length in dromedary camels (Camelus dromedarius) produced by somatic cell nuclear transfer (SCNT) and their age-matched naturally produced counterparts. Theriogenology. 2022;177:151–156. doi: 10.1016/j.theriogenology.2021.10.012. [DOI] [PubMed] [Google Scholar]
- 19.Zhang T., Wang T., You F., Li Z., Chen D., Zhang K., Tian S., Sheng B., Wu H., Jiang L., Ma R., An G., Meng H., Yang L. Nanobody-based anti-CD22-chimeric antigen receptor T cell immunotherapy exhibits improved remission against B-cell acute lymphoblastic leukemia. Transpl. Immunol. 2022;71 doi: 10.1016/j.trim.2022.101538. [DOI] [PubMed] [Google Scholar]
- 20.Harmsen M.M., De Haard H.J. Properties, production, and applications of camelid single-domain antibody fragments. Appl. Microbiol. Biotechnol. 2007;77:13–22. doi: 10.1007/s00253-007-1142-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jovčevska I., Muyldermans S. The therapeutic potential of nanobodies. BioDrugs. 2020;34:11–26. doi: 10.1007/s40259-019-00392-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freise A.C., Wu A.M. In vivo imaging with antibodies and engineered fragments. Mol. Immunol. 2015;67:142–152. doi: 10.1016/j.molimm.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steeland S., Vandenbroucke R.E., Libert C. Nanobodies as therapeutics: big opportunities for small antibodies. Drug Discov. Today. 2016;21:1076–1113. doi: 10.1016/j.drudis.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 24.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 25.Haga K., Takai-Todaka R., Matsumura Y., Song C., Takano T., Tojo T., Nagami A., Ishida Y., Masaki H., Tsuchiya M., Ebisudani T., Sugimoto S., Sato T., Yasuda H., Fukunaga K., Sawada A., Nemoto N., Murata K., Morimoto T., Katayama K. Nasal delivery of single-domain antibody improves symptoms of SARS-CoV-2 infection in an animal model. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spadiut O., Capone S., Krainer F., Glieder A., Herwig C. Microbials for the production of monoclonal antibodies and antibody fragments. Trends Biotechnol. 2014;32:54–60. doi: 10.1016/j.tibtech.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma J.K., Chikwamba R., Sparrow P., Fischer R., Mahoney R., Twyman R.M. Plant-derived pharmaceuticals--the road forward. Trends Plant Sci. 2005;10:580–585. doi: 10.1016/j.tplants.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 28.Margolin E., Chapman R., Williamson A.L., Rybicki E.P., Meyers A.E. Production of complex viral glycoproteins in plants as vaccine immunogens. Plant Biotechnol. J. 2018;16:1531–1545. doi: 10.1111/pbi.12963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marsian J., Lomonossoff G.P. Molecular pharming - VLPs made in plants. Curr. Opin. Biotechnol. 2016;37:201–206. doi: 10.1016/j.copbio.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 30.Moustafa K., Makhzoum A., Trémouillaux-Guiller J. Molecular farming on rescue of pharma industry for next generations. Crit. Rev. Biotechnol. 2016;36:840–850. doi: 10.3109/07388551.2015.1049934. [DOI] [PubMed] [Google Scholar]
- 31.Pogrebnyak N., Markley K., Smirnov Y., Brodzik R., Bandurska K., Koprowski H., Golovkin M. Collard and cauliflower as a base for production of recombinant antigens. Plant Sci. 2006;171:677–685. doi: 10.1016/j.plantsci.2006.06.017. [DOI] [Google Scholar]
- 32.Brodzik R., Glogowska M., Bandurska K., Okulicz M., Deka D., Ko K., van der Linden J., Leusen J.H., Pogrebnyak N., Golovkin M., Steplewski Z., Koprowski H. Plant-derived anti-Lewis Y mAb exhibits biological activities for efficient immunotherapy against human cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8804–8809. doi: 10.1073/pnas.0603043103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Golovkin M., Spitsin S., Andrianov V., Smirnov Y., Xiao Y., Pogrebnyak N., Markley K., Brodzik R., Gleba Y., Isaacs S.N., Koprowski H. Smallpox subunit vaccine produced in planta confers protection in mice. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6864–6869. doi: 10.1073/pnas.0701451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pogrebnyak N., Golovkin M., Andrianov V., Spitsin S., Smirnov Y., Egolf R., Koprowski H. Severe acute respiratory syndrome (SARS) S protein production in plants: development of recombinant vaccine. Proc. Natl. Acad. Sci. 2005;102:9062–9067. doi: 10.1073/pnas.0503760102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leonart L.P., Fachi M.M., Boger B., Silva M.R.D., Szpak R., Lombardi N.F., Pedroso M.L.A., Pontarolo R. A systematic review and meta-analyses of longitudinal studies on drug treatments for Gaucher disease. Ann. Pharmacother. 2022;10600280221108443 doi: 10.1177/10600280221108443. [DOI] [PubMed] [Google Scholar]
- 36.Maharjan P.M., Choe S. Plant-based COVID-19 vaccines: current status, design, and development strategies of candidate vaccines. Vaccines (Basel) 2021;9 doi: 10.3390/vaccines9090992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Titievsky L., Schuster T., Wang R., Younus M., Palladino A., Quazi K., Wajnrajch M.P., Hernandez B., Becker P.S., Weinreb N.J., Chambers C., Mansfield R., Taylor L., Tseng L.J., Kaplan P. Safety and effectiveness of taliglucerase alfa in patients with Gaucher disease: an interim analysis of real-world data from a multinational drug registry (TALIAS) Orphanet J Rare Dis. 2022;17:145. doi: 10.1186/s13023-022-02289-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hiatt A., Pauly M., Whaley K., Qiu X., Kobinger G., Zeitlin L. The emergence of antibody therapies for Ebola. Hum Antibod. 2015;23:49–56. doi: 10.3233/HAB-150284. [DOI] [PubMed] [Google Scholar]
- 39.Fischer R., Scillberg S., editors. Molecular Farming: Plant-made Pharmaceutical and Technical Proteins. Wiley-VCH Verlag GmbH&Co; Weinheim: 2004. [Google Scholar]
- 40.Li W., Schäfer A., Kulkarni S.S., Liu X., Martinez D.R., Chen C., Sun Z., Leist S.R., Drelich A., Zhang L., Ura M.L., Berezuk A., Chittori S., Leopold K., Mannar D., Srivastava S.S., Zhu X., Peterson E.C., Tseng C.T., Mellors J.W., Falzarano D., Subramaniam S., Baric R.S., Dimitrov D.S. High potency of a bivalent human V(H) domain in SARS-CoV-2 animal models. Cell. 2020;183(429–441) doi: 10.1016/j.cell.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Froger A. Vol. 7. 2007. Transformation of Plasmid DNA into E. coli using the Heat Shock Method; p. 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeng J., Meng X., Zhou P., Yin Z., Xie Q., Zou H., Shen N., Ye Z., Tang Y. Interferon-α exacerbates neuropsychiatric phenotypes in lupus-prone mice. Arthritis Res Ther. 2019;21:019–1985. doi: 10.1186/s13075-019-1985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.T. Murashige, F. Skoog. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497, doi:https://doi.org/ 10.1111/j.1399-3054.1962.tb08052.x. [DOI]
- 44.Jefferson R.A., Kavanagh T.A., Bevan M.W. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horsch R.B., Klee H.J., Stachel S., Winans S.C., Nester E.W., Rogers S.G., Fraley R.T. Analysis of Agrobacterium tumefaciens virulence mutants in leaf discs. Proc. Natl. Acad. Sci. U. S. A. 1986;83:2571–2575. doi: 10.1073/pnas.83.8.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rahimzadeh M., Sadeghizadeh M., Najafi F., Arab S., Mobasheri H. Impact of heat shock step on bacterial transformation efficiency. Mol Biol Res Commun. 2016;5:257–261. [PMC free article] [PubMed] [Google Scholar]
- 47.Majumder J., Minko T. Multifunctional lipid-based nanoparticles for codelivery of anticancer drugs and siRNA for treatment of non-small cell lung cancer with different level of resistance and EGFR mutations. Pharmaceutics. 2021;13 doi: 10.3390/pharmaceutics13071063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hernandez I., Bott S.W., Patel A.S., Wolf C.G., Hospodar A.R., Sampathkumar S., Shrank W.H. Pricing of monoclonal antibody therapies: higher if used for cancer? Am. J. Manag. Care. 2018;24:109–112. [PubMed] [Google Scholar]
- 49.Lico C., Santi L., Twyman R.M., Pezzotti M., Avesani L. The use of plants for the production of therapeutic human peptides. Plant Cell Rep. 2012;31:439–451. doi: 10.1007/s00299-011-1215-7. [DOI] [PubMed] [Google Scholar]
- 50.Dhama K., Natesan S., Iqbal Yatoo M., Patel S.K., Tiwari R., Saxena S.K., Harapan H. Plant-based vaccines and antibodies to combat COVID-19: current status and prospects. Hum Vaccin Immunother. 2020;16:2913–2920. doi: 10.1080/21645515.2020.1842034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taratula O., Kuzmov A., Shah M., Garbuzenko O.B., Minko T. Nanostructured lipid carriers as multifunctional nanomedicine platform for pulmonary co-delivery of anticancer drugs and siRNA. J. Control. Release. 2013;171:349–357. doi: 10.1016/j.jconrel.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen A.M., Minko T. Pharmacokinetics of inhaled nanotherapeutics for pulmonary delivery. J. Control. Release. 2020;326:222–244. doi: 10.1016/j.jconrel.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuzmov A., Minko T. Nanotechnology approaches for inhalation treatment of lung diseases. J. Control. Release. 2015;219:500–518. doi: 10.1016/j.jconrel.2015.07.024. [DOI] [PubMed] [Google Scholar]
- 54.Garbuzenko O.B., Saad M., Pozharov V.P., Reuhl K.R., Mainelis G., Minko T. Inhibition of lung tumor growth by complex pulmonary delivery of drugs with oligonucleotides as suppressors of cellular resistance. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10737–10742. doi: 10.1073/pnas.1004604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garbuzenko O.B., Mainelis G., Taratula O., Minko T. Inhalation treatment of lung cancer: the influence of composition, size and shape of nanocarriers on their lung accumulation and retention. Cancer Biol Med. 2014;11:44–55. doi: 10.7497/j.issn.2095-3941.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garbuzenko O.B., Kbah N., Kuzmov A., Pogrebnyak N., Pozharov V., Minko T. Inhalation treatment of cystic fibrosis with lumacaftor and ivacaftor co-delivered by nanostructured lipid carriers. J. Control. Release. 2019;296:225–231. doi: 10.1016/j.jconrel.2019.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garbuzenko O.B., Ivanova V., Kholodovych V., Reimer D.C., Reuhl K.R., Yurkow E., Adler D., Minko T. Combinatorial treatment of idiopathic pulmonary fibrosis using nanoparticles with prostaglandin E and siRNA(s) Nanomedicine. 2017;13:1983–1992. doi: 10.1016/j.nano.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.