Abstract
There is a large global unmet need for the development of countermeasures to combat hundreds of viruses known to cause human disease and for the establishment of a therapeutic portfolio for future pandemic preparedness. Most approved antiviral therapeutics target proteins encoded by a single virus, providing a narrow spectrum of coverage. This, combined with the slow pace and high cost of drug development, limits the scalability of this direct-acting antiviral (DAA) approach. Here, we summarize progress and challenges in the development of broad-spectrum antivirals that target either viral elements (proteins, genome structures, and lipid envelopes) or cellular proviral factors co-opted by multiple viruses via newly discovered compounds or repurposing of approved drugs. These strategies offer new means for developing therapeutics against both existing and emerging viral threats that complement DAAs.
Introduction
Over 200 viruses are known to cause disease in humans, yet currently approved antiviral drugs are available to treat only about 10 of these viral infections. The past decade has underscored the global threat posed by emerging viruses. Spillovers from animals to humans have resulted in several Ebola virus disease (EVD) outbreaks, the Middle East respiratory syndrome (MERS) outbreak, and possibly the current coronavirus disease 2019 (COVID-19) pandemic. Global warming, increased urbanization, and air travel have contributed to the spread of vector-borne viruses endemic to various parts of the world, including dengue virus (DENV), estimated to infect 400 million people in over 128 countries, and Zika virus (ZIKV), the causative agent of a 2015 outbreak. Moreover, political instability in various parts of the world continues to pose risks to our military forces and civilians from potential spread of biothreat agents, such as poxviruses — whose natural spread caused the ongoing monkeypox virus (MPXV) outbreak — and Venezuelan equine encephalitis virus (VEEV) (1, 2). There is thus a huge unmet need for the development of effective therapeutics for the treatment of existing and newly emerging viral infections.
Most approved antivirals target viral enzymes, particularly proteases and polymerases (Figure 1). Such direct-acting antivirals (DAAs) have shown tremendous utility for the treatment of hepatitis C virus (HCV) and human immunodeficiency virus type 1 (HIV-1) infections, and more recently COVID-19. However, this approach to drug development has several major limitations. First, the spectrum of coverage provided is typically narrow, ranging from a single viral genotype to a few related viruses at best. Moreover, this approach is not scalable to address the large unmet need. It takes, on average, an 8- to 12-year timeline (3) and an average cost of over $2 billion to develop a new drug. Thus, targeting viruses individually is expensive and slow. While timely, effective efforts were noted during the COVID-19 outbreak, the rapid rollout of nirmatrelvir, for example, was enabled by accelerated derivatization of an existing series of SARS-CoV-1 main protease (Mpro) inhibitors. No such DAAs are, however, currently available for the majority of viral families. The inability to predict the next emerging viral infection is another limitation, hampering adequate global health protection and national security preparedness. Lastly, when used individually, treatment with conventional DAAs often results in rapid emergence of drug resistance, complicating monotherapy regimens for HIV, HCV, and influenza A virus (IAV). In the case of SARS-CoV-2, escape mutations conferring high-level resistance to remdesivir and nirmatrelvir have already been selected in vitro and identified in circulating strains (4, 5). While combining drugs that target distinct viral functions can overcome viral resistance, as exemplified by HIV and HCV treatment, developing such “cocktail” regimens for multiple acute infections is not feasible.
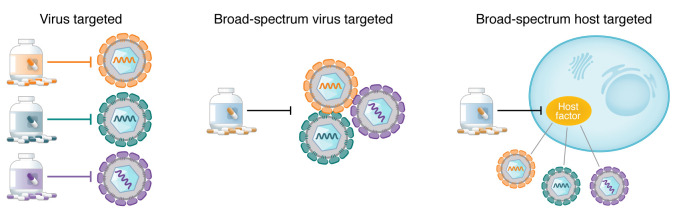
Figure 1. Toward broad-spectrum antivirals.
Antiviral drugs that selectively inhibit unique viral proteins typically provide a narrow-spectrum solution (left), whereas broad-spectrum drugs can restrict multiple viruses by inhibiting either common viral functions or structures (middle) or host factors commonly required by several viruses (right). Adapted with permission from Science (204).
An alternative solution is the development of broad-spectrum antiviral drugs. One advantage of this approach is reduced time and cost associated with the early stages of drug development per approved indication. It can also diminish the clinical risks in more advanced stages of development. The off-label use of approved antivirals against new viral indications can provide further economic incentives, as these drugs were already rigorously tested for toxicity, pharmacokinetics, pharmacodynamics, dosing, etc. These advantages have been recently demonstrated by the repurposing of remdesivir and molnupiravir — originally developed to treat EVD and VEEV, respectively — for the treatment of COVID-19 (6, 7). Importantly, this approach can facilitate readiness for future outbreaks of newly emerging pathogens. Broad-spectrum antivirals could also be used to treat rare viral infections for which no drug is available. Lastly, a broad-spectrum antiviral could be administered before a viral threat has been accurately diagnosed, increasing the likelihood of viral control, with implications for front-line health care providers and military personnel.
Broad-spectrum antiviral activity can be achieved by targeting of viral components or cellular factors required for the replication of multiple viruses (Figure 1). The latter approach could complement DAAs, such as by conferring synergistic antiviral effects, as recently demonstrated by a combination of molnupiravir (DAA) with camostat mesylate (host-targeted) (8). Here, we summarize recent efforts to characterize the therapeutic potential and biological rationale of representative approaches under these categories. Notably, we define broad-spectrum coverage as activity against viruses from at least two unrelated viral families.
Broad-spectrum DAAs
Most virally encoded proteins show extensive sequence and structural diversity. Thus, the spectrum of coverage typically provided by DAAs is narrow, ranging from several serotypes or variants of the same virus to a few related viruses at most, as exemplified by paritaprevir and Paxlovid — HCV and SARS-CoV-2 inhibitors, respectively. Accordingly, the number of DAA classes showing promise in preclinical and clinical studies has been limited to date (Figure 2 and Table 1).
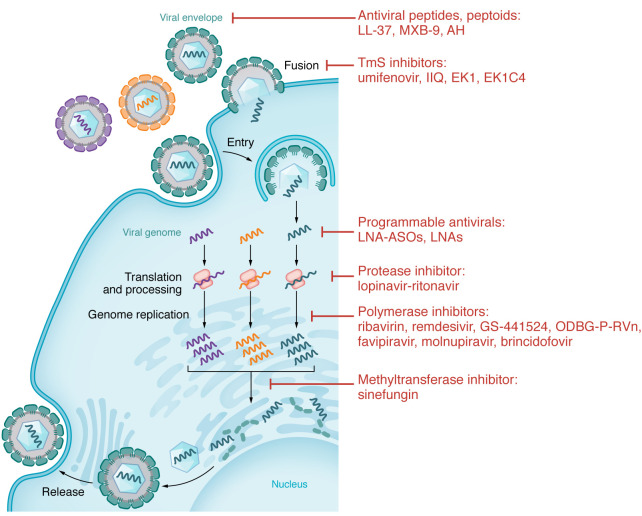
Figure 2. Approved and experimental direct-acting compounds with broad-spectrum antiviral activity.
Depicted here is a generic viral life cycle. Examples of classes of inhibitors with broad-spectrum antiviral activity are connected to the specific stages of the viral life cycle or cellular process they target.
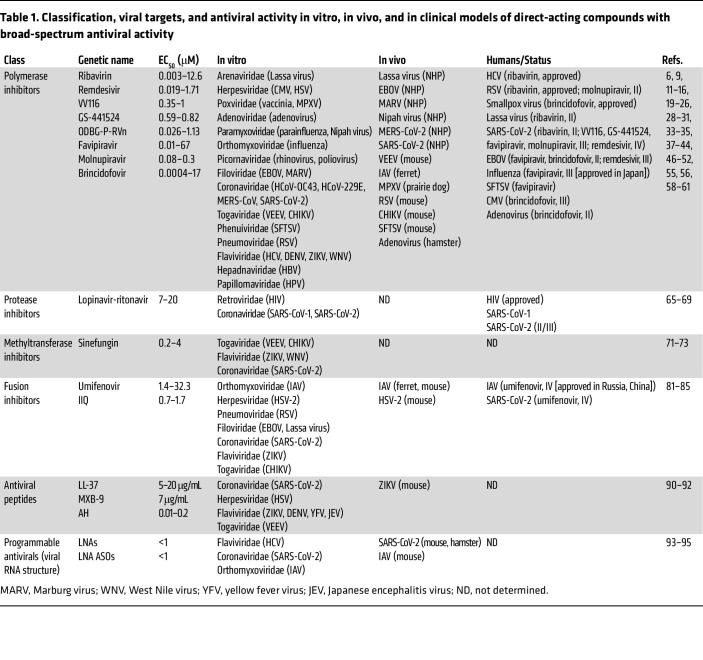
Table 1. Classification, viral targets, and antiviral activity in vitro, in vivo, and in clinical models of direct-acting compounds with broad-spectrum antiviral activity.
Targeting viral polymerases.
The structure of the catalytic units of most RNA-dependent RNA polymerases is highly conserved across viral families, making them attractive targets for broad-spectrum antivirals (9). Discovered in the 1970s, the nucleoside analog ribavirin introduced the concept of broad-spectrum antivirals. Several mechanisms of ribavirin’s antiviral action have been demonstrated, including inhibition of viral RNA or DNA synthesis (10). Ribavirin was shown to suppress the replication of multiple viruses in vitro and to confer protection from multiple emerging viral pathogens, including filo- and arenaviruses, in nonhuman primates (NHPs) (11, 12). Ribavirin is approved for the treatment of HCV infection in combination drug regimens (13) and of respiratory syncytial virus (RSV) infection in immunocompromised patients (14). Moreover, ribavirin reduced mortality when tested in over 1,800 patients infected with Lassa virus, albeit the comparative arm was historic controls (15). Ribavirin treatment, however, did not impact COVID-19 outcomes (16), and its clinical utility for other viral infections remains to be determined.
In the past decade, several chemically distinct, next-generation nucleotide and nucleoside analogs have demonstrated broad-spectrum antiviral activity (reviewed in ref. 17). One example is remdesivir, an intravenously administered nucleotide analog prodrug that suppresses viral RNA replication via delayed chain termination (18). Remdesivir was initially developed for treatment of EVD after demonstrating effective suppression of viral replication in human primary cells and 100% protection from lethality in NHPs (6). Contrastingly, however, in a randomized multi-intervention trial (the PALM study) in 681 EVD patients, remdesivir treatment did not reduce viremia and in fact increased mortality rate relative to monoclonal antibodies (19). Remdesivir has shown activity against other hemorrhagic viruses, including Nipah virus, albeit thus far in preclinical models only (20). Remdesivir has also shown utility for the treatment of respiratory viruses, suppressing replication and/or tissue injury in NHP models of RSV, and coronaviruses (21–23). Remdesivir was therefore one of the first repurposed agents to be tested clinically for COVID-19 treatment. Following inconclusive studies (24, 25), in a phase III trial (Adaptive Covid-19 Treatment Trial [ACTT-1]) involving 1,062 hospitalized patients with SARS-CoV-2 pneumonia, remdesivir shortened the median recovery time and reduced mortality rate relative to placebo without causing severe side effects (26). Based on these findings and its prior de-risking in human trials, remdesivir was the first drug to receive FDA approval for COVID-19 treatment. Nevertheless, the need to deliver remdesivir intravenously has somewhat limited its global application, prompting the design of analogs for oral delivery (27). VV116, one such analog, potently suppresses SARS-CoV-2 replication and improves oral bioavailability (28). In a phase III trial, VV116 demonstrated comparable time to clinical recovery to Paxlovid and a favorable safety profile (29). Other oral analogs of remdesivir, such as GS-441524 (30), are undergoing development.
Favipiravir (T-705) is a nucleoside analog whose active form gets incorporated into the nascent viral RNA strand, inducing lethal mutagenesis (31, 32). In cell culture models, favipiravir has demonstrated moderate antiviral activity against IAV and VEEV, and weak activity against SARS-CoV-2 and Ebola virus (EBOV) (EC50 values over 60 μM) (33–35). While high concentrations are required to achieve therapeutic levels in humans, by inhibiting its own metabolism, favipiravir increases its cellular uptake (reviewed in ref. 36). Favipiravir was approved for flu treatment in Japan in 2014 and for the treatment of COVID-19 in China and India after demonstrating some benefits in early studies (37–39). However, in prospective randomized COVID-19 studies, favipiravir showed no clinical benefit over placebo (40, 41). Beyond respiratory viral infections, favipiravir protected EBOV-infected mice from lethality (33). Nevertheless, while it reduced viral load and prolonged survival in a retrospective EBOV study, it showed no benefit in a phase II trial (42, 43). Conversely, favipiravir increased viral clearance and reduced mortality rate in a trial involving 145 patients infected with a different hemorrhagic virus: the phlebovirus severe fever with thrombocytopenia syndrome virus (SFTSV) (44). The mutagenesis pattern of SFTSV in serum samples was comparable to that observed in preclinical models, confirming favipiravir’s mechanism of action (44).
Molnupiravir is another orally bioavailable nucleoside analog whose incorporation into the viral genome causes lethal mutagenesis (45). Designed to inhibit VEEV (7), molnupiravir is rapidly distributed to brain tissue and protects mice from a lethal VEEV challenge (46). Molnupiravir demonstrated activity in animal models of EBOV and respiratory viral infections, including IAV and pandemic coronaviruses (47–50). Yet, whereas in earlier phase II and III trials in mild-to-moderate COVID-19 patients, molnupiravir accelerated SARS-CoV-2 clearance and reduced mortality (51, 52), prompting its Emergency Use Authorization as a second-line COVID-19 treatment, in a more recent phase II trial, molnupiravir’s antiviral effect was inconclusive (53).
DNA-dependent DNA polymerases have also been shown to be amenable to broad-spectrum inhibition. Brincidofovir is an oral nucleoside analog, prodrug of cidofovir, whose incorporation into the elongating viral DNA by the viral polymerase interrupts DNA replication via chain termination and/or direct inhibition (54). Brincidofovir has demonstrated in vitro and in vivo activity against multiple DNA viruses (55). Based on efficacy data in animal models, brincidofovir was approved for the treatment of smallpox in 2021 (56). Nevertheless, brincidofovir showed no virologic benefit in patients infected with MPXV in a retrospective observational study, and treatment was complicated by liver toxicity (57). In phase II and III trials in allogeneic hematopoietic cell transplant recipients, brincidofovir reduced adenovirus viremia and prevented cytomegalovirus (CMV) viremia (58–60). Yet a trend toward reduced mortality was observed in adenovirus viremic patients only, and treatment was complicated by acute graft-versus-host disease (58, 60). Independent of polymerase inhibition, suppression of EBOV replication in vitro by brincidofovir is thought to be mediated by its lipid side chain (61), yet its clinical utility for this indication remains to be determined, as it has been studied only anecdotally to date (62, 63).
These and other examples highlight the broad-spectrum potential of polymerase inhibitors.
Targeting other viral enzymes.
While the unique substrate preference of viral (versus cellular) proteases can facilitate relatively selective inhibition, their large diversity across viral families has limited their potential as targets for broad-spectrum antivirals. Approved for the treatment of HIV-1 infection, lopinavir-ritonavir combination (Kaletra) was shown to bind the substrate-binding pocket of SARS-CoV-1’s main protease (Mpro) (64) and suppress SARS-CoV-2 replication in vitro (65) — somewhat surprising findings since coronaviruses encode cysteine proteases whereas HIV-1 encodes an aspartic protease. However, while potential benefit in reducing lung injury was demonstrated in a retrospective study in SARS-CoV-1–infected patients treated with a combination of lopinavir-ritonavir and ribavirin (66), no such benefit was observed in SARS-CoV-2–infected ferrets and humans (67–69). Thus, the overall broad-spectrum utility of viral protease inhibitors to date has been limited.
Targeting of viral methyltransferases (MTases) — enzymes essential for capping the mRNA 5′ ends of some viruses for efficient translation and evasion of immune responses — has also been explored (70). Competition with S-adenosyl-l-methionine (SAM) on MTase binding, such as by sinefungin, was shown to suppress MTases of alphaviruses, flaviviruses, and SARS-CoV-2 in vitro (71–73), yet severe toxicity in preclinical models, attributed to lack of selectivity, hampered the clinical development of this approach (74). Greater selectivity achieved by targeting of conserved pockets near the SAM-binding site, combined with conservation of MTase structure within viral families, has enabled the discovery of investigational pan-flaviviral inhibitors with reduced toxicity, yet the feasibility of developing MTase inhibitors with activity across viral families is low (75–78). Similarly, the broad-spectrum potential of inhibitors targeting other viral enzymes including exonucleases and helicases remains to be defined.
Targeting viral fusion proteins, lipid envelope, and genome.
Targeting class I fusion glycoproteins of enveloped viruses is another strategy explored for its broad-spectrum potential. The transmembrane subunit (TmS) of these proteins is highly conserved and thus an attractive target for broad viral inhibition (reviewed in ref. 79). Umifenovir (Arbidol), one example of such a strategy, binds to a hydrophobic pocket in the stem region of the TmS of IAV hemagglutinin, thereby blocking viral fusion with endosomal membranes (80). Umifenovir has shown efficacy in cell culture and animal models of IAV infection (81), and in a phase IV trial in 359 flu patients (82), leading to its approval for flu treatment in Russia and China. Umifenovir suppresses replication of other RNA viruses in vitro, albeit with moderate EC50 values (5.7–32.3 μM) (81). Whereas an open-label study suggested potential benefit of umifenovir treatment in 100 COVID-19 patients (83), a retrospective study showed increased mortality in severe COVID-19 patients (84), and the results of a phase IV randomized study are unavailable (ClinicalTrials.gov NCT04260594), making it difficult to draw conclusions. Beyond small molecules, suppression of viral fusion by α-helical lipopeptides that disrupt α-helix–mediated interactions of the TmS is another strategy that shows broad-spectrum potential. IIQ, one such candidate, suppresses the replication of multiple RNA viruses in vitro and achieves good exposure levels in rats (85). EK1 and EK1C4, peptides that target the heptad repeat-1 (HR1) domain of TmS of human coronaviruses, have shown prophylactic and therapeutic effects when administered intranasally to mice infected with coronaviruses (86). However, the broad-spectrum potential of these and other fusion-suppressing peptides demonstrating activity against specific viruses (87, 88) remains to be defined.
The viral envelope is another emerging target for broad-spectrum antiviral interventions. The utilization of antimicrobial peptides has been challenged by cytotoxicity resulting from a lack of selectivity to the viral lipid envelope and by rapid degradation by cellular proteases. Nevertheless, recent efforts indicate that harnessing differences between the membrane curvature of viral particles and that of cells can achieve selectivity, and that modifying peptides — such as by stapling or designing synthetic peptidomimetics that resist proteolytic degradation (peptoids) — can improve biostability. Indeed, various amphipathic, α-helical (AH) peptides and self-assembling peptoids have demonstrated effective viral membrane lysis and abrogation of infectivity without impacting cellular viability (89, 90). In a mouse model, an AH peptide suppressed ZIKV infection and reduced inflammation and blood-brain barrier injury (91). LL-37 and MXB-9, with activity against multiple viruses in cultured cells and/or mice infected with SARS-CoV-2 pseudovirus (90, 92), provide additional proof of concept for the potential utility of this approach.
Targeting of the viral genome as a broad-spectrum antiviral approach has also shown promise recently. “Programmable antivirals,” such as locked nucleic acids (LNAs) and LNA antisense oligonucleotides (LNA ASOs) targeting highly conserved viral RNA structures involved in viral packaging or replication, are one example of this approach. Such LNAs and LNA ASOs suppressed replication of HCV, IAV, and SARS-CoV-2 in vitro (93–95), and reduced mortality, viral load, and/or transmission (94, 95) in mice infected with SARS-CoV-2 and IAV (94).
Taken together, while the design of DAAs with activity across viral families is overall challenged by the extensive sequence and structural diversity of virally encoded proteins, targeting of viral polymerases and non-enzymatic viral functions holds promise.
Host-targeted broad-spectrum antiviral approaches
The cellular machineries co-opted to support the life cycle of viruses are often conserved across viral families, representing attractive targets for broad-spectrum antiviral strategies. With approximately 20,000 proteins, the human proteome offers a much larger repertoire of candidate targets than a viral proteome. Indeed, the discovery of such proviral factors required by multiple viruses has been the subject of fruitful research. Aided by breakthroughs in multi-omics approaches, these efforts have led to the discovery of numerous druggable proviral factors. Some examples are discussed below.
Beyond a larger target repertoire, an important advantage of the host-targeted approach is its higher barrier to viral resistance. Since cellular targets are not under genetic control of a virus, the likelihood that escape mutations will emerge is lower than with DAAs. This advantage was demonstrated in cell culture models, such as with inhibitors targeting various cellular kinases (96–98), and in animal models, such as DENV-infected mice treated with α-glucosidase inhibitors (99). In patients, cyclophilin inhibitors and other host-targeted approaches have demonstrated longer time to resistance and lower levels of resistance than DAAs (100).
Targeting cellular functions can also provide opportunities not only to suppress viral replication but also to moderate deleterious host responses, which play key roles in the pathogenesis of multiple viral infections, including dengue, EVD, and COVID-19. Targeting p38 MAPK or ErbBs, for example, as we and others have demonstrated in preclinical models, can reduce inflammation and protect from tissue injury beyond suppression of viral replication (98, 101). Another example is enhancement of type I interferon responses contributing to the protective effect of tamoxifen treatment in vesicular stomatitis virus–infected (VSV-infected) mice (102). Lastly, since most approved drugs target cellular functions, there is an opportunity to repurpose existing drugs for antiviral indications, as was extensively explored during the COVID-19 outbreak (reviewed in ref. 103).
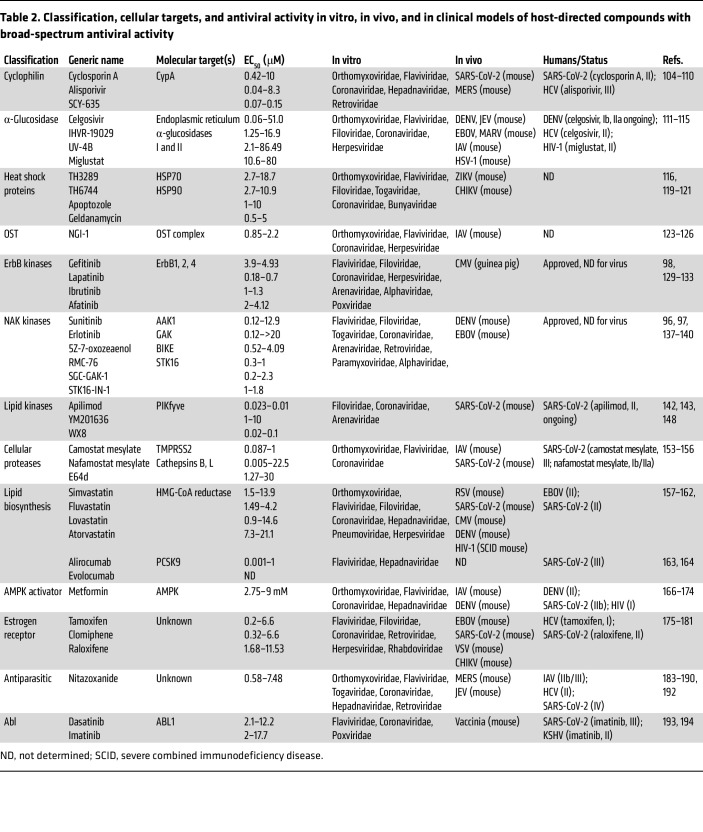
Below are examples of classes of host-targeted approaches that show some promise (Figure 3 and Table 2).
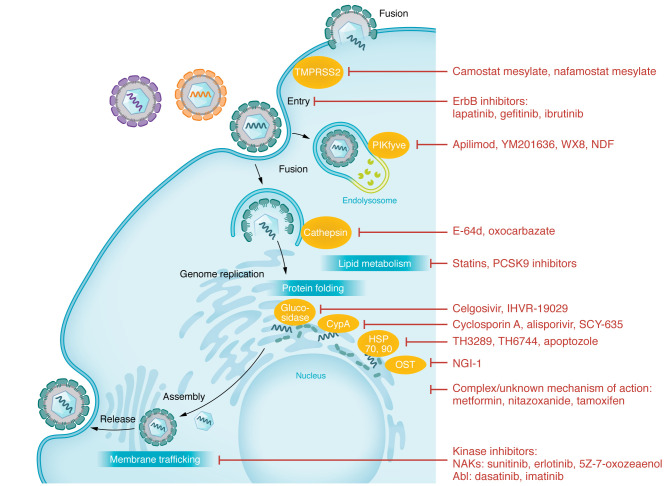
Figure 3. Approved and experimental host-targeted compounds with broad-spectrum antiviral activity.
Depicted here is a generic viral life cycle. Examples of classes of inhibitors with broad-spectrum antiviral activity are connected to the specific stage(s) of the viral life cycle or cellular process they target.
Table 2. Classification, cellular targets, and antiviral activity in vitro, in vivo, and in clinical models of host-directed compounds with broad-spectrum antiviral activity.
Targeting protein folding and transport.
Cyclosporin A (CsA) and experimental non-immunosuppressive inhibitors of cyclophilin A (CypA) — a cellular factor involved in protein folding — such as alisporivir (Debio-025) and SCY-635, suppress the replication of multiple viruses in vitro (104). Blockage of interactions between CypA and the HIV-1 nucleocapsid and HCV NS5A proteins is thought to mediate the antiviral effect (105, 106). Other mechanisms of antiviral action were reported, including suppression of HBV binding to its entry receptor (107), of coronaviral RNA synthesis (104), and of nuclear import of IAV genome (108). The effect of these compounds in mouse models has been variable (100), yet prevention of disease progression was demonstrated in mice infected with coronaviruses (109). Accordingly, transplant recipients receiving CsA treatment for their underlying condition experienced reduced morbidity and mortality upon SARS-CoV-2 infection (110). Whereas alisporivir significantly reduced viremia in chronically infected HCV patients, a phase III trial was terminated due to toxicity.
α-Glucosidase is another protein required for proper folding of proteins — including viral glycoproteins — that serves as a broad-spectrum antiviral target. Celgosivir and other iminosugars are competitive substrates for α-glucosidases with activity against multiple viruses in cultured cells (111). These inhibitors have demonstrated efficacy in murine models of RNA and DNA viruses (111, 112). The utility of celgosivir for the treatment of dengue infection is currently being explored, although safety but little or no efficacy have been documented to date in a dengue pilot study and in patients infected with HCV or HIV-1 (113–115).
The molecular chaperones heat shock protein 70 (HSP70) and HSP90, involved in protein folding and transport, are also broadly required factors shown to function at temporally distinct stages of viral life cycles (116, 117). Stabilization and transport of viral proteins were among the proposed underlying mechanisms (117, 118). Pharmacological inhibition of HSP70 by TH3289 blocked replication of flaviviruses, coronaviruses, and Crimean-Congo hemorrhagic fever virus in vitro (116, 119). In murine models of ZIKV and Chikungunya virus (CHIKV) infections, small-molecule inhibitors of these chaperones reduced viral titers, inflammation, and/or mortality (120, 121). While thus far demonstrated with tool compounds only, these examples provide evidence for the potential utility of targeting HSPs.
Oligosaccharyltransferase (OST), an endoplasmic reticulum protein complex that catalyzes N-glycosylation, was discovered as a candidate antiviral target via CRISPR screens for flaviviral proviral factors (122). OST subunits interact with DENV nonstructural proteins and are required for viral RNA replication (122). NGI-1, a small-molecule inhibitor of OST, has shown antiviral activity against flaviviruses and more recently HSV-1 and SARS-CoV-2 (123–125). Interestingly, whereas the anti-DENV activity is independent of the canonical role of OST in N-linked glycosylation, the anti-IAV effect is associated with reduced hemagglutinin (HA) and neuraminidase (NA) glycosylation (123). A concern was recently raised that glycome-modified viruses generated upon NGI-1 treatment can reduce antibody responses in IAV-infected mice and requires further investigation (126).
Targeting cellular kinases.
Multiple cellular kinases are hijacked by viruses, representing candidate targets for broad-spectrum antivirals (127). The epidermal growth factor receptor family of tyrosine kinases (ErbB1, 2, 4) is one example. A requirement for ErbBs was documented in the entry and/or post-entry stages of multiple viruses (128). Several anticancer ErbB inhibitors, including gefitinib, demonstrate activity against HCV, human cytomegalovirus (HCMV), poxvirus, and Lassa virus in cultured cells (129–133), and CMV in guinea pigs (132). In human lung and brain organoid models of SARS-CoV-2 and VEEV infections, respectively, we have recently shown that, beyond suppressing viral replication, lapatinib, an anticancer pan-ErbB inhibitor, protects from virus-induced activation of pathways implicated in non-infectious tissue injury downstream of ErbBs, proinflammatory cytokine production, and epithelial or blood-brain barrier injury (98). Moreover, we have validated ErbB inhibition as the mechanism of antiviral action (98). Remarkably, ibrutinib, a BTK inhibitor with potent pan-ErbB activity (134), has demonstrated protection from progression to severe COVID-19, albeit in a small number of patients (135), highlighting that clinical evaluation of these ErbB inhibitors is warranted.
The numb-associated (serine/threonine) kinases (NAKs) — AAK1, BIKE, GAK, and STK16 — have also been studied as targets for broad-spectrum antivirals. We have demonstrated a requirement for NAKs in the regulation of intracellular cotrafficking of specific cellular cargo adaptor proteins with viral particles during entry, assembly, and/or release of HCV, DENV, EBOV, and SARS-CoV-2 (96, 97, 136, 137). Approved anticancer drugs with potent anti-NAK activity, including sunitinib-erlotinib combinations, 5Z-7-oxozeaenol, and chemically distinct more selective inhibitors, demonstrate broad-spectrum antiviral activity against eight viral families in vitro (96, 97, 137–139). A combination treatment with sunitinib-erlotinib was shown to protect mice from DENV and EBOV challenges (96, 138). Inhibition of intracellular membrane trafficking regulated by NAKs was validated as an important mechanism of antiviral action (96, 97, 140). The safety and efficacy of NAK inhibition for the treatment of viral infections in humans remain to be determined.
Lipid kinases have also been shown to be required for effective replication of multiple viruses. For example, the endosomal phosphatidylinositol-3-phosphate 5-kinase (PIKfyve) (141) has been implicated in the entry of filoviruses, Lassa virus, and coronaviruses (142). The PIKfyve inhibitors apilimod and YM201636 suppress trafficking and maturation of endolysosomes, preventing viral fusion and/or egress (142, 143). Apilimod is currently being studied as a COVID-19 therapeutic (NCT04446377). Whereas a suboptimal pharmacokinetic profile (144, 145) limits its development, the excellent safety profile demonstrated with apilimod in clinical trials for inflammatory diseases has de-risked PIKfyve as a target (146, 147). While two chemically distinct small molecules with anti-PIKfyve activity were recently shown to increase SARS-CoV-2–induced pathology in a mouse model, since their selectivity has not been reported, it is possible that other targets have mediated this effect (148). Further evaluation of the potential of PIKfyve inhibition in other animal models and ideally human organoid models is therefore warranted. Pharmacological inhibition of other lipid and protein kinases by approved and investigational compounds has also shown promise in vitro with variable results in animal models (reviewed in ref. 149).
Targeting cellular proteases.
Proteases are another group of cellular enzymes co-opted by viruses. Influenza viruses and coronaviruses, for example, rely on proteases, such as TMPRSS2 and cathepsins, for cleavage and activation of their surface glycoproteins (150, 151). Among cellular protease inhibitors showing antiviral activity, camostat mesylate and nafamostat mesylate, oral serine protease inhibitors approved for the treatment of chronic pancreatitis and other conditions (152), have shown TMPRSS2-dependent suppression of viral fusion in vitro (151) and protection in mouse models of IAV and coronaviral infections (153, 154). However, when studied for the treatment of COVID-19 patients, these compounds had no significant impact on clinical outcomes (155, 156). Thus, the evaluation of other strategies targeting cellular proteases for the treatment of viral infections is warranted.
Targeting lipid metabolism.
Cholesterol-lowering drugs, like statins, have demonstrated in vitro activity against HCV, attributed to their effect on lipid biosynthesis. Indeed, antiviral activity in cells was reversed upon addition of mevalonate or geranylgeraniol, and resistance to these drugs coincided with an increase in HMG-CoA reductase level — statins’ target (157). Nevertheless, a variable, modest, and short-lived effect was demonstrated in HCV patients when statins were combined with peginterferon-ribavirin (158). Beyond HCV, statins have demonstrated efficacy in animal models of multiple viral infections, including respiratory viruses, CMV, HIV-1, and DENV (159, 160). Owing to their ability to restore endothelial stability, statins were used, albeit in a non-formal study, in combination with an angiotensin receptor blocker for treating EVD, an infection whose pathogenesis is associated with endothelial dysfunction — showing reduced mortality in 100 patients (161). Recently, reduced morbidity and mortality were documented also in COVID-19 patients with statin prescriptions, albeit in observational studies only (162). Inhibitors of proprotein convertase subtilisin kexin type 9 (PCSK9), such as the monoclonal antibodies alirocumab and evolocumab, represent another class of lipid-lowering agents shown to suppress DENV replication in vitro and reduce mortality and inflammation in severe COVID-19 patients (163, 164). Whereas statins showed no antiviral activity in dengue patients (165), PCSK9 inhibitors may offer greater protection given the recent discovery that PCSK9 expression is induced by DENV infection in cells residing in physiologically hypoxic conditions and is increased in severe dengue patients, reducing cholesterol uptake and dampening susceptibility to statins (163).
Host-targeted approaches with complex mechanisms of action.
Metformin, an approved oral drug for the treatment of diabetes, has demonstrated potent antiviral activity against multiple viruses in vitro. Activation of AMP-activated protein kinase–dependent (AMPK-dependent) type I interferon signaling was proposed as an underlying mechanism in DENV and HCV infections (166, 167). Metformin reduced morbidity and mortality in mice infected with DENV and IAV, but not ZIKV (168, 169). Diabetic patients on metformin treatment were found to have lower morbidity and mortality upon influenza virus infection (170) and a trend toward reduced mortality when infected with SARS-CoV-2 (171). Contrastingly, metformin showed no clinical benefit in nondiabetic COVID-19 patients (172). The therapeutic potential of metformin in reducing HIV-1 reservoirs and combating DENV infection is currently being studied clinically (173, 174).
Tamoxifen and other inhibitors of the estrogen receptor (ER) approved for the treatment of breast cancer inhibit the replication of multiple RNA and DNA viruses in vitro (175). The proposed mechanisms of antiviral action include blockage of a chloride channel required for HSV-1 entry; endosomal/lysosomal proteins required for EBOV entry; SARS-CoV-2 spike-mediated membrane fusion (176, 177); and binding of ER to HCV and CHIKV polymerases (178, 179). In rodent models of VSV, EBOV, CHIKV, and SARS-CoV-2 infections, treatment with ER antagonists reduced viral titers, inflammation, and/or mortality (175, 179, 180). Treatment with ER antagonists in humans shortened the time of SARS-CoV-2 shedding (181), reduced HCV viremia but not the resulting liver inflammation (NCT00749138), and did not impact HIV-1 viremia (182). Thus, further studies are required to define the clinical utility of ER antagonists as antivirals.
Nitazoxanide, approved for the treatment of parasitic infections, is another candidate drug for repurposing with a complex mechanism of antiviral action. Nitazoxanide suppresses replication of multiple RNA viruses in vitro and in vivo (183, 184). While the precise target remains unknown, several mechanisms of action have been proposed, such as blocking of the maturation of the influenza hemagglutinin (185) and the coronaviral spike proteins (186, 187) and, in the case of HCV and HBV infections, blocking of protein kinase R–mediated phosphorylation of eIF2α (183, 188). Nitazoxanide modestly reduced the time to resolution of flu symptoms in a phase II trial and is currently being evaluated in a phase III trial for this indication (189). Whereas the addition of nitazoxanide to peginterferon-ribavirin improved sustained virologic responses in HCV patients in a phase II trial (190), no such improvement was observed in a phase III trial in genotype 4–infected patients (191). In a recent randomized, double-blind pilot study in 50 COVID-19 patients, nitazoxanide shortened hospitalization, accelerated viral clearance, and reduced inflammatory cytokine production (192), warranting a larger-scale study.
Ongoing challenges and future perspectives
Collectively, these examples highlight the potential held in expanding the repertoire of candidate targets from viral proteins to other viral elements and to cellular functions, and provide proof of concept for the potential utility of broad-spectrum antiviral strategies. Nevertheless, major challenges remain to be overcome to expand the clinical applications of these strategies.
Toxicity is a major concern, particularly in targeting cellular factors, requiring careful safety investigations. For example, dasatinib, an inhibitor of the Src and c-Abl kinases, has demonstrated broad-spectrum antiviral activity in cultured cells, yet in a murine model of vaccinia virus, it induced immunosuppression rather than protection (193, 194). Nevertheless, since all non-infectious human diseases are treated with drugs targeting cellular functions, the increased risk posed by host-targeted antivirals is theoretical and can be potentially mitigated by the identification of a therapeutic window within which a drug level is sufficient to suppress viral replication without causing cellular toxicity. Directing the use of host-targeted approaches toward acute viral infections requiring shorter duration of treatment should further help limit toxicity. Indeed, chronically infected HCV patients receiving alisporivir unexpectedly developed fatal cases of pancreatitis during a phase III trial, albeit after several months of treatment (195). Broad-spectrum DAAs are also not devoid of toxicity: brincidofovir administration to patients infected with MPXV was complicated by liver toxicity (57), and caution is needed with favipiravir and molnupiravir treatment due to teratogenicity (196). Significant toxicity caused by lack of selectivity to the viral targets has hampered the clinical development of some DAAs, such as sinefungin targeting cellular MTases and nucleoside analogs targeting mitochondrial RNA polymerase (74, 197).
Another challenge of host-targeted approaches is that the mechanism of antiviral action is often elusive and the molecular targets underlying the antiviral effect are unvalidated. This challenge is driven in part by the complex network of interactions in which cellular proteins function and the limited selectivity of some of their inhibitors. For example, whereas the effect of erlotinib on HCV infection was first attributed solely to its effect on its cancer target, EGFR, inhibition of GAK, another target of erlotinib, was then shown to play a role (96, 129). The mechanism of antiviral action of some drugs, such as nitazoxanide and tamoxifen, is even less clear and is often pathogen specific (176–179, 185–188).
But the greatest challenge of all antiviral approaches is the limited translatability of protective effects observed in preclinical models into clinical benefit in humans. While this limitation would be predicted to impact primarily host-targeted approaches owing to potential differences in the sequence and/or structure of proviral factors across species, this does not appear to be the case. The translation of broad-spectrum DAAs seems to be comparably impacted. For example, remdesivir showed excellent protection from EVD in NHPs, yet no benefit in EBOV-infected patients (19). The narrow window of opportunity for therapeutic interventions in the case of acute viral infections undoubtedly contributes to these low clinical translation rates.
These challenges underscore the need to consider revising the procedures currently in place to assess antivirals. Preclinically, careful consideration of differences in pharmacological properties including pharmacokinetics and tissue distribution between species may improve the success rate of clinical translation. The use of more biologically relevant human organoids and organ-on-chip models to mimic human tissue architecture may also help address this challenge. Indeed, the use of such models is now being encouraged by the FDA (198). On the clinical front, the design of clinical studies, particularly those conducted in the setting of outbreaks, could be considerably improved. The adaptive platform design — adapted from clinical studies in cancer (199) and approved by the FDA (NCT02380625) (200) — is one solution showing promise during the COVID-19 pandemic (NCT04280705) (reviewed in ref. 103). Improving patient selection in clinical trials by targeting treatment to patients more likely to develop severe outcomes may further enhance the resolution of clinical studies. Recent breakthroughs in omics approaches and machine learning algorithms enabling the discovery of clinically usable biomarkers — such as those we and others have identified to predict progression to severe dengue infection and other severe viral infections (201–203) — may aid with this effort.
Taken together, while much progress has been achieved in the field of broad-spectrum antivirals, the need to establish a therapeutic portfolio for future pandemic preparedness is far from being met. Developing and stocking host-targeted broad-spectrum antivirals as the first line of defense, and in parallel developing DAAs for representative viruses from each major viral family — efforts currently supported by US government funding — should bring us closer to achieving this goal.
Acknowledgments
SE is a Chan Zuckerberg Biohub San Francisco Investigator who is also supported by National Institute of Allergy and Infectious Diseases grants RO1AI158569 and 1 U19 AI171421-01, Defense Threat Reduction Fundamental Research to Counter Weapons of Mass Destruction grant HDTRA11810039, Investigator Initiated Awards W81XWH1910235 and W81XWH2210283 and Expansion Award W81XWH2110456 from the Department of Defense office of the Congressionally Directed Medical Research Programs/Peer Reviewed Medical Research Program, and a Transformational Award from the Dr. Ralph and Marian Falk Medical Research Trust. MK is supported by a Postdoctoral Fellowship in Translational Medicine from the PhRMA Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors acknowledge all the contributions in the field that could not be included in this Review.
Version 1. 06/01/2023
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2023, Karim et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: J Clin Invest. 2023;133(11):e170236. https://doi.org/10.1172/JCI170236.
Contributor Information
Marwah Karim, Email: mkarim@stanford.edu.
Chieh-Wen Lo, Email: cwlo@stanford.edu.
Shirit Einav, Email: seinav@stanford.edu.
References
- 1.Leffel EK, Reed DS. Marburg and Ebola viruses as aerosol threats. Biosecur Bioterror. 2004;2(3):186–191. doi: 10.1089/bsp.2004.2.186. [DOI] [PubMed] [Google Scholar]
- 2.Weaver SC, et al. Venezuelan equine encephalitis. Annu Rev Entomol. 2004;49:141–174. doi: 10.1146/annurev.ento.49.061802.123422. [DOI] [PubMed] [Google Scholar]
- 3. Tufts Center for the Study of Drug Development, Tufts University. Cost to Develop and Win Marketing Approval for a New Drug Is $2.6 Billion. https://f.hubspotusercontent10.net/hubfs/9468915/TuftsCSDD_June2021/pdf/pr-coststudy.pdf Updated November 18, 2014. Accessed April 20, 2023.
- 4.Szemiel AM, et al. In vitro selection of remdesivir resistance suggests evolutionary predictability of SARS-CoV-2. PLoS Pathog. 2021;17(9):e1009929:e1009929. doi: 10.1371/journal.ppat.1009929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heilmann E, et al. SARS-CoV-2 3CLpro mutations selected in a VSV-based system confer resistance to nirmatrelvir, ensitrelvir, and GC376. Sci Transl Med. 2022;15(678):eabq7360. doi: 10.1126/scitranslmed.abq7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warren TK, et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531(7594):381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willyard C. How antiviral pill molnupiravir shot ahead in the COVID drug hunt. Nature. doi: 10.1038/d41586-021-02783-1. [published online October 8, 2021]. [DOI] [PubMed] [Google Scholar]
- 8.Wagoner J, et al. Combinations of host- and virus-targeting antiviral drugs confer synergistic suppression of SARS-CoV-2. Microbiol Spectr. 2022;10(5):e03331–22. doi: 10.1128/spectrum.03331-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lang DM, et al. Highly similar structural frames link the template tunnel and NTP entry tunnel to the exterior surface in RNA-dependent RNA polymerases. Nucleic Acids Res. 2013;41(3):1464–1482. doi: 10.1093/nar/gks1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graci JD, Cameron CE. Mechanisms of action of ribavirin against distinct viruses. Rev Med Virol. 2006;16(1):37–48. doi: 10.1002/rmv.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lingas G, et al. Lassa viral dynamics in non-human primates treated with favipiravir or ribavirin. PLoS Comput Biol. 2021;17(1):e1008535. doi: 10.1371/journal.pcbi.1008535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alfson KJ, et al. Determination and therapeutic exploitation of Ebola virus spontaneous mutation frequency. J Virol. 2016;90(5):2345–2355. doi: 10.1128/JVI.02701-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feld JJ, et al. Treatment of HCV with ABT-450/r–ombitasvir and dasabuvir with ribavirin. N Engl J Med. 2014;370(17):1594–1603. doi: 10.1056/NEJMoa1315722. [DOI] [PubMed] [Google Scholar]
- 14.Shah DP, et al. Impact of aerosolized ribavirin on mortality in 280 allogeneic haematopoietic stem cell transplant recipients with respiratory syncytial virus infections. J Antimicrob Chemother. 2013;68(8):1872–1880. doi: 10.1093/jac/dkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCormick JB, et al. Lassa fever. Effective therapy with ribavirin. N Engl J Med. 1986;314(1):20–26. doi: 10.1056/NEJM198601023140104. [DOI] [PubMed] [Google Scholar]
- 16.Li H, et al. Efficacy of ribavirin and interferon-α therapy for hospitalized patients with COVID-19: a multicenter, retrospective cohort study. Int J Infect Dis. 2021;104:641–648. doi: 10.1016/j.ijid.2021.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adalja A, Inglesby T. Broad-spectrum antiviral agents: a crucial pandemic tool. Expert Rev Anti Infect Ther. 2019;17(7):467–470. doi: 10.1080/14787210.2019.1635009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tchesnokov EP, et al. Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11(4):326. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulangu S, et al. A randomized, controlled trial of Ebola virus disease therapeutics. N Engl J Med. 2019;381(24):2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo MK, et al. Remdesivir (GS-5734) protects African green monkeys from Nipah virus challenge. Sci Transl Med. 2019;11(494):eaau9242. doi: 10.1126/scitranslmed.aau9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown AJ, et al. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antiviral Res. 2019;169:104541. doi: 10.1016/j.antiviral.2019.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williamson BN, et al. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature. 2020;585(7824):273–276. doi: 10.1038/s41586-020-2423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackman RL, et al. Prodrugs of a 1′-CN-4-aza-7,9-dideazaadenosine C-nucleoside leading to the discovery of remdesivir (GS-5734) as a potent inhibitor of respiratory syncytial virus with efficacy in the African green monkey model of RSV. J Med Chem. 2021;64(8):5001–5017. doi: 10.1021/acs.jmedchem.1c00071. [DOI] [PubMed] [Google Scholar]
- 24.Grein J, et al. Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med. 2020;382(24):2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395(10236):1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beigel JH, et al. Remdesivir for the treatment of Covid-19 — final report. N Engl J Med. 2020;383(19):1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cully M. A tale of two antiviral targets — and the COVID-19 drugs that bind them. Nat Rev Drug Discov. 2022;21(1):1813–1826. doi: 10.1038/d41573-021-00202-8. [DOI] [PubMed] [Google Scholar]
- 28.Xie Y, et al. Design and development of an oral remdesivir derivative VV116 against SARS-CoV-2. Cell Res. 2021;31(11):1212–1214. doi: 10.1038/s41422-021-00570-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao Z, et al. VV116 versus nirmatrelvir-ritonavir for oral treatment of Covid-19. N Engl J Med. 2022;388(5):406–417. doi: 10.1056/NEJMoa2208822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo MK, et al. Broad-spectrum in vitro antiviral activity of ODBG-P-RVn: an orally-available, lipid-modified monophosphate prodrug of remdesivir parent nucleoside (GS-441524) Microbiol Spectr. 2021;9(3):e0153721. doi: 10.1128/Spectrum.01537-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baranovich T, et al. T-705 (favipiravir) induces lethal mutagenesis in influenza A H1N1 viruses in vitro. J Virol. 2013;87(7):3741–3751. doi: 10.1128/JVI.02346-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sangawa H, et al. Mechanism of action of T-705 ribosyl triphosphate against influenza virus RNA polymerase. Antimicrob Agents Chemother. 2013;57(11):5202–5208. doi: 10.1128/AAC.00649-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oestereich L, et al. Successful treatment of advanced Ebola virus infection with T-705 (favipiravir) in a small animal model. Antiviral Res. 2014;105:17–21. doi: 10.1016/j.antiviral.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 34.Furuta Y, et al. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc Jpn Acad Ser B Phys Biol Sci. 2017;93(7):449–463. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang M, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joshi S, et al. Role of favipiravir in the treatment of COVID-19. Int J Infect Dis. 2021;102:501–508. doi: 10.1016/j.ijid.2020.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai Q, et al. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering (Beijing) 2020;6(10):1192–1198. doi: 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen C, et al. Favipiravir versus arbidol for clinical recovery rate in moderate and severe adult COVID-19 patients: a prospective, multicenter, open-label, randomized controlled clinical trial. Front Pharmacol. 2021;12:683296:683296. doi: 10.3389/fphar.2021.683296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shinkai M, et al. Efficacy and safety of favipiravir in moderate COVID-19 pneumonia patients without oxygen therapy: a randomized, phase III clinical trial. Infect Dis Ther. 2021;10(4):2489–2509. doi: 10.1007/s40121-021-00517-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szabo BG, et al. Favipiravir treatment does not influence disease progression among adult patients hospitalized with moderate-to-severe COVID-19: a prospective, sequential cohort study from Hungary. Geroscience. 2021;43(5):2205–2213. doi: 10.1007/s11357-021-00452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bosaeed M, et al. Efficacy of favipiravir in adults with mild COVID-19: a randomized, double-blind, multicentre, placebo-controlled clinical trial. Clin Microbiol Infect. 2022;28(4):602–608. doi: 10.1016/j.cmi.2021.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bai C-Q, et al. Clinical and virological characteristics of Ebola virus disease patients treated with favipiravir (T-705)-Sierra Leone, 2014. Clin Infect Dis. 2016;63(10):1288–1294. doi: 10.1093/cid/ciw571. [DOI] [PubMed] [Google Scholar]
- 43.Sissoko D, et al. Experimental treatment with favipiravir for Ebola virus disease (the JIKI Trial): a historically controlled, single-arm proof-of-concept trial in Guinea. PLoS Medicine. 2016;13(3):e1001967. doi: 10.1371/journal.pmed.1001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, et al. Clinical effect and antiviral mechanism of T-705 in treating severe fever with thrombocytopenia syndrome. Signal Transduct Targeted Ther. 2021;6(1):145. doi: 10.1038/s41392-021-00541-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kabinger F, et al. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat Struct Mol Biol. 2021;28(9):740–746. doi: 10.1038/s41594-021-00651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Painter GR, et al. The prophylactic and therapeutic activity of a broadly active ribonucleoside analog in a murine model of intranasal Venezuelan equine encephalitis virus infection. Antiviral Res. 2019;171:104597. doi: 10.1016/j.antiviral.2019.104597. [DOI] [PubMed] [Google Scholar]
- 47.Toots M, et al. Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci Transl Med. 2019;11(515):eaax5866. doi: 10.1126/scitranslmed.aax5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bluemling GR, et al. The prophylactic and therapeutic efficacy of the broadly active antiviral ribonucleoside N4-hydroxycytidine (EIDD-1931) in a mouse model of lethal Ebola virus infection. Antiviral Res. 2023;209:105453. doi: 10.1016/j.antiviral.2022.105453. [DOI] [PubMed] [Google Scholar]
- 49.Sheahan TP, et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med. 2020;12(541):eabb5883. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wahl A, et al. SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature. 2021;591(7850):451–457. doi: 10.1038/s41586-021-03312-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fischer WA, et al. A phase 2a clinical trial of molnupiravir in patients with COVID-19 shows accelerated SARS-CoV-2 RNA clearance and elimination of infectious virus. Sci Transl Med. 2021;14(628):eabl7430. doi: 10.1126/scitranslmed.abl7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jayk Bernal A, et al. Molnupiravir for oral treatment of Covid-19 in nonhospitalized patients. N Engl J Med. 2021;386(6):509–520. doi: 10.1056/NEJMoa2116044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khoo SH, et al. Molnupiravir versus placebo in unvaccinated and vaccinated patients with early SARS-CoV-2 infection in the UK (AGILE CST-2): a randomised, placebo-controlled, double-blind, phase 2 trial. Lancet Infect Dis. 2023;23(2):183–195. doi: 10.1016/S1473-3099(22)00644-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chamberlain JM, et al. Cidofovir diphosphate inhibits adenovirus 5 DNA polymerase via both nonobligate chain termination and direct inhibition, and polymerase mutations confer cidofovir resistance on intact virus. Antimicrob Agents Chemother. 2018;63(1):e01925–18. doi: 10.1128/AAC.01925-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lanier R, et al. Development of CMX001 for the treatment of poxvirus infections. Viruses. 2010;2(12):2740–2762. doi: 10.3390/v2122740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hutson CL, et al. Pharmacokinetics and efficacy of a potential smallpox therapeutic, brincidofovir, in a lethal monkeypox virus animal model. mSphere. 2021;6(1):e00927. doi: 10.1128/mSphere.00927-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adler H, et al. Clinical features and management of human monkeypox: a retrospective observational study in the UK. Lancet Infect Dis. 2022;22(8):1153–1162. doi: 10.1016/S1473-3099(22)00228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grimley MS, et al. Brincidofovir for asymptomatic adenovirus viremia in pediatric and adult allogeneic hematopoietic cell transplant recipients: a randomized placebo-controlled phase II trial. Biol Blood Marrow Transplant. 2017;23(3):512–521. doi: 10.1016/j.bbmt.2016.12.621. [DOI] [PubMed] [Google Scholar]
- 59.Hiwarkar P, et al. Brincidofovir is highly efficacious in controlling adenoviremia in pediatric recipients of hematopoietic cell transplant. Blood. 2017;129(14):2033–2037. doi: 10.1182/blood-2016-11-749721. [DOI] [PubMed] [Google Scholar]
- 60.Marty FM, et al. A randomized, double-blind, placebo-controlled phase 3 trial of oral brincidofovir for cytomegalovirus prophylaxis in allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2019;25(2):369–381. doi: 10.1016/j.bbmt.2018.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McMullan LK, et al. The lipid moiety of brincidofovir is required for in vitro antiviral activity against Ebola virus. Antiviral Res. 2016;125:71–78. doi: 10.1016/j.antiviral.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 62.Florescu DF, et al. Administration of brincidofovir and convalescent plasma in a patient with Ebola virus disease. Clin Infect Dis. 2015;61(6):969–973. doi: 10.1093/cid/civ395. [DOI] [PubMed] [Google Scholar]
- 63.Dunning J, et al. Experimental treatment of Ebola virus disease with brincidofovir. PLoS One. 2016;11(9):e0162199. doi: 10.1371/journal.pone.0162199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nukoolkarn V, et al. Molecular dynamic simulations analysis of ritonavir and lopinavir as SARS-CoV 3CL(pro) inhibitors. J Theor Biol. 2008;254(4):861–867. doi: 10.1016/j.jtbi.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Choy KT, et al. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 2020;178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chu CM, et al. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59(3):252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cao B, et al. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N Engl J Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.RECOVERY Collaborative Group. Lopinavir-ritonavir in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet. 2020;396(10259):1345–1352. doi: 10.1016/S0140-6736(20)32013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park SJ, et al. Antiviral efficacies of FDA-approved drugs against SARS-CoV-2 infection in ferrets. mBio. 2020;11(3):e01114–20. doi: 10.1128/mBio.01114-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramanathan A, et al. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016;44(16):7511–7526. doi: 10.1093/nar/gkw551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brecher M, et al. Novel broad spectrum inhibitors targeting the flavivirus methyltransferase. PLoS One. 2015;10(6):e0130062. doi: 10.1371/journal.pone.0130062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mudgal R, et al. Inhibition of Chikungunya virus by an adenosine analog targeting the SAM-dependent nsP1 methyltransferase. FEBS Lett. 2020;594(4):678–694. doi: 10.1002/1873-3468.13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pearson L-A, et al. Development of a high-throughput screening assay to identify inhibitors of the SARS-CoV-2 guanine-N7-methyltransferase using rapidfire mass spectrometry. SLAS Discov. 2021;26(6):749–756. doi: 10.1177/24725552211000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zweygarth E, et al. Evaluation of sinefungin for the treatment of Trypanosoma (Nannomonas) congolense infections in goats. Trop Med Parasitol. 1986;37(3):255–257. [PubMed] [Google Scholar]
- 75.Dong H, et al. Structural and functional analyses of a conserved hydrophobic pocket of flavivirus methyltransferase. J Biol Chem. 2010;285(42):32586–32595. doi: 10.1074/jbc.M110.129197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lim SP, et al. Small molecule inhibitors that selectively block dengue virus methyltransferase. J Biol Chem. 2011;286(8):6233–6240. doi: 10.1074/jbc.M110.179184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Krafcikova P, et al. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat Commun. 2020;11(1):3717. doi: 10.1038/s41467-020-17495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kottur J, et al. High-resolution structures of the SARS-CoV-2 N7-methyltransferase inform therapeutic development. Nat Struct Mol Biol. 2022;29(9):850–853. doi: 10.1038/s41594-022-00828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu L, et al. Antivirals with common targets against highly pathogenic viruses. Cell. 2021;184(6):1604–1620. doi: 10.1016/j.cell.2021.02.013. [DOI] [PubMed] [Google Scholar]
- 80.Kadam RU, Wilson IA. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc Natl Acad Sci U S A. 2017;114(2):206–214. doi: 10.1073/pnas.1617020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shi L, et al. Antiviral activity of arbidol against influenza A virus, respiratory syncytial virus, rhinovirus, coxsackie virus and adenovirus in vitro and in vivo. Arch Virol. 2007;152(8):1447–1455. doi: 10.1007/s00705-007-0974-5. [DOI] [PubMed] [Google Scholar]
- 82.Pshenichnaya NY, et al. Clinical efficacy of umifenovir in influenza and ARVI (study ARBITR) Ter Arkh. 2019;91(3):56–63. doi: 10.26442/00403660.2019.03.000127. [DOI] [PubMed] [Google Scholar]
- 83.Nojomi M, et al. Effect of Arbidol (umifenovir) on COVID-19: a randomized controlled trial. BMC Infect Dis. 2020;20(1):954. doi: 10.1186/s12879-020-05698-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou X, et al. Arbidol is associated with increased in-hospital mortality among 109 patients with severe COVID-19: a multicenter, retrospective study. J Glob Health. 2021;11:05017. doi: 10.7189/jogh.11.05017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang C, et al. De novo design of α-helical lipopeptides targeting viral fusion proteins: a promising strategy for relatively broad-spectrum antiviral drug discovery. J Med Chem. 2018;61(19):8734–8745. doi: 10.1021/acs.jmedchem.8b00890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xia S, et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv. 2019;5(4):eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pancera M, et al. Crystal structures of trimeric HIV envelope with entry inhibitors BMS-378806 and BMS-626529. Nat Chem Biol. 2017;13(10):1115–1122. doi: 10.1038/nchembio.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.de Wispelaere M, et al. Inhibition of flaviviruses by targeting a conserved pocket on the viral envelope protein. Cell Chem Biol. 2018;25(8):1006–1016. doi: 10.1016/j.chembiol.2018.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tabaei SR, et al. Single vesicle analysis reveals nanoscale membrane curvature selective pore formation in lipid membranes by an antiviral α-helical peptide. Nano Lett. 2012;12(11):5719–5725. doi: 10.1021/nl3029637. [DOI] [PubMed] [Google Scholar]
- 90.Diamond G, et al. Potent antiviral activity against HSV-1 and SARS-CoV-2 by antimicrobial peptoids. Pharmaceuticals (Basel) 2021;14(4):304. doi: 10.3390/ph14040304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jackman JA, et al. Therapeutic treatment of Zika virus infection using a brain-penetrating antiviral peptide. Nat Mater. 2018;17(11):971–977. doi: 10.1038/s41563-018-0194-2. [DOI] [PubMed] [Google Scholar]
- 92.Wang C, et al. Human cathelicidin inhibits SARS-CoV-2 infection: killing two birds with one stone. ACS Infect Dis. 2021;7(6):1545–1554. doi: 10.1021/acsinfecdis.1c00096. [DOI] [PubMed] [Google Scholar]
- 93.Mutso M, et al. RNA interference-guided targeting of hepatitis C virus replication with antisense locked nucleic acid-based oligonucleotides containing 8-oxo-dG modifications. PLoS One. 2015;10(6):e0128686. doi: 10.1371/journal.pone.0128686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hagey RJ, et al. Programmable antivirals targeting critical conserved viral RNA secondary structures from influenza A virus and SARS-CoV-2. Nat Med. 2022;28(9):1944–1955. doi: 10.1038/s41591-022-01908-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhu C, et al. An intranasal ASO therapeutic targeting SARS-CoV-2. Nat Commun. 2022;13(1):4503:4503. doi: 10.1038/s41467-022-32216-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bekerman E, et al. Anticancer kinase inhibitors impair intracellular viral trafficking and exert broad-spectrum antiviral effects. J Clin Invest. 2017;127(4):1338–1352. doi: 10.1172/JCI89857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pu S, et al. BIKE regulates dengue virus infection and is a cellular target for broad-spectrum antivirals. Antiviral Res. 2020;184:104966. doi: 10.1016/j.antiviral.2020.104966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Saul S, et al. Discovery of pan-ErbB inhibitors protecting from SARS-CoV-2 replication, inflammation, and lung injury by a drug repurposing screen [preprint]. Posted on bioRxiv June 15, 2021. [DOI]
- 99.Warfield KL, et al. Inhibition of endoplasmic reticulum glucosidases is required for in vitro and in vivo dengue antiviral activity by the iminosugar UV-4. Antiviral Res. 2016;129:93–98. doi: 10.1016/j.antiviral.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lin K, Gallay P. Curing a viral infection by targeting the host: the example of cyclophilin inhibitors. Antiviral Res. 2013;99(1):68–77. doi: 10.1016/j.antiviral.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wei D, et al. Roles of p38 MAPK in the regulation of the inflammatory response to swine influenza virus-induced acute lung injury in mice. Acta Virol. 2014;58(4):374–379. doi: 10.4149/av_2014_04_374. [DOI] [PubMed] [Google Scholar]
- 102.Cham LB, et al. Tamoxifen protects from vesicular stomatitis virus infection. Pharmaceuticals (Basel) 2019;12(4):142. doi: 10.3390/ph12040142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Saul S, Einav S. Old drugs for a new virus: repurposed approaches for combating COVID-19. ACS Infect Dis. 2020;6(9):2304–2318. doi: 10.1021/acsinfecdis.0c00343. [DOI] [PubMed] [Google Scholar]
- 104.De Wilde AH, et al. Coronaviruses and arteriviruses display striking differences in their cyclophilin A-dependence during replication in cell culture. Virology. 2018;517:148–156. doi: 10.1016/j.virol.2017.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Billich A, et al. Mode of action of SDZ NIM 811, a nonimmunosuppressive cyclosporin A analog with activity against human immunodeficiency virus (HIV) type 1: interference with HIV protein-cyclophilin A interactions. J Virol. 1995;69(4):2451–2461. doi: 10.1128/jvi.69.4.2451-2461.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Colpitts CC, et al. Hepatitis C virus exploits cyclophilin A to evade PKR. Elife. 2020;9:e52237. doi: 10.7554/eLife.52237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Watashi K, et al. Cyclosporin A and its analogs inhibit hepatitis B virus entry into cultured hepatocytes through targeting a membrane transporter, sodium taurocholate cotransporting polypeptide (NTCP) Hepatology. 2014;59(5):1726–1737. doi: 10.1002/hep.26982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ma C, et al. Discovery of cyclosporine A and its analogs as broad-spectrum anti-influenza drugs with a high in vitro genetic barrier of drug resistance. Antiviral Res. 2016;133:62–72. doi: 10.1016/j.antiviral.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sauerhering L, et al. Cyclosporin A reveals potent antiviral effects in preclinical models of SARS-CoV-2 infection. Am J Respir Crit Care Med. 2022;205(8):964–968. doi: 10.1164/rccm.202108-1830LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Demir E, et al. COVID-19 in kidney transplant recipients: a multicenter experience in Istanbul. Transpl Infect Dis. 2020;22(5):e13371. doi: 10.1111/tid.13371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ma J, et al. Enhancing the antiviral potency of ER α-glucosidase inhibitor IHVR-19029 against hemorrhagic fever viruses in vitro and in vivo. Antiviral Res. 2018;150:112–122. doi: 10.1016/j.antiviral.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Warfield KL, et al. The iminosugar UV-4 is a broad inhibitor of influenza A and B viruses ex vivo and in mice. Viruses. 2016;8(3):71. doi: 10.3390/v8030071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fischl MA, et al. The safety and efficacy of combination N-butyl-deoxynojirimycin (SC-48334) and zidovudine in patients with HIV-1 infection and 200-500 CD4 cells/mm3. J Acquir Immune Defic Syndr (1988) 1994;7(2):139–147. [PubMed] [Google Scholar]
- 114.Durantel D. Celgosivir, an α-glucosidase I inhibitor for the potential treatment of hepatitis C virus infection. Curr Opin Investig Drugs. 2009;10(8):860–870. [PubMed] [Google Scholar]
- 115.Low JG, et al. Efficacy and safety of celgosivir in patients with dengue fever (CELADEN): a phase 1b, randomised, double-blind, placebo-controlled, proof-of-concept trial. Lancet Infect Dis. 2014;14(8):706–715. doi: 10.1016/S1473-3099(14)70730-3. [DOI] [PubMed] [Google Scholar]
- 116.Taguwa S, et al. Defining Hsp70 subnetworks in dengue virus replication reveals key vulnerability in flavivirus infection. Cell. 2015;163(5):1108–1123. doi: 10.1016/j.cell.2015.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Geller R, et al. Broad action of Hsp90 as a host chaperone required for viral replication. Biochim Biophys Acta. 2012;1823(3):698–706. doi: 10.1016/j.bbamcr.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Taguwa S, et al. Zika virus dependence on host Hsp70 provides a protective strategy against infection and disease. Cell Rep. 2019;26(4):906–920. doi: 10.1016/j.celrep.2018.12.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tampere M, et al. Novel broad-spectrum antiviral inhibitors targeting host factors essential for replication of pathogenic RNA viruses. Viruses. 2020;12(12):1423. doi: 10.3390/v12121423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang J, et al. Small molecule inhibitor of ATPase activity of HSP70 as a broad-spectrum inhibitor against flavivirus infections. ACS Infect Dis. 2020;6(5):832–843. doi: 10.1021/acsinfecdis.9b00376. [DOI] [PubMed] [Google Scholar]
- 121.Rathore AP, et al. Chikungunya virus nsP3 & nsP4 interacts with HSP-90 to promote virus replication: HSP-90 inhibitors reduce CHIKV infection and inflammation in vivo. Antiviral Res. 2014;103:7–16. doi: 10.1016/j.antiviral.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 122.Marceau CD, et al. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature. 2016;535(7610):159–163. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Puschnik AS, et al. A small-molecule oligosaccharyltransferase inhibitor with pan-flaviviral activity. Cell Rep. 2017;21(11):3032–3039. doi: 10.1016/j.celrep.2017.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lu H, et al. Targeting STT3A-oligosaccharyltransferase with NGI-1 causes herpes simplex virus 1 dysfunction. FASEB J. 2019;33(6):6801–6812. doi: 10.1096/fj.201802044RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Huang HC, et al. Targeting conserved N-glycosylation blocks SARS-CoV-2 variant infection in vitro. EBioMedicine. 2021;74:103712. doi: 10.1016/j.ebiom.2021.103712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Alymova IV, et al. Aberrant cellular glycosylation may increase the ability of influenza viruses to escape host immune responses through modification of the viral glycome. mBio. 2022;13(2):e0298321. doi: 10.1128/mbio.02983-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Keating JA, Striker R. Phosphorylation events during viral infections provide potential therapeutic targets. Rev Med Virol. 2012;22(3):166–181. doi: 10.1002/rmv.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kuroda M, et al. HER2-mediated enhancement of Ebola virus entry. PLoS Pathog. 2020;16(10):e1008900. doi: 10.1371/journal.ppat.1008900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lupberger J, et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med. 2011;17(5):589–595. doi: 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Diao J, et al. Hepatitis C virus induces epidermal growth factor receptor activation via CD81 binding for viral internalization and entry. J Virol. 2012;86(20):10935–10949. doi: 10.1128/JVI.00750-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Langhammer S, et al. Inhibition of poxvirus spreading by the anti-tumor drug gefitinib (Iressa) Antiviral Res. 2011;89(1):64–70. doi: 10.1016/j.antiviral.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 132.Schleiss M, et al. Protein kinase inhibitors of the quinazoline class exert anti-cytomegaloviral activity in vitro and in vivo. Antiviral Res. 2008;79(1):49–61. doi: 10.1016/j.antiviral.2008.01.154. [DOI] [PubMed] [Google Scholar]
- 133.Mizuma K, et al. The Pan-ErbB tyrosine kinase inhibitor afatinib inhibits multiple steps of the mammarenavirus life cycle. Virology. 2022;576:83–95. doi: 10.1016/j.virol.2022.09.005. [DOI] [PubMed] [Google Scholar]
- 134.Chen J, et al. Ibrutinib inhibits ERBB receptor tyrosine kinases and HER2-amplified breast cancer cell growth. Mol Cancer Ther. 2016;15(12):2835–2844. doi: 10.1158/1535-7163.MCT-15-0923. [DOI] [PubMed] [Google Scholar]
- 135.Treon SP, et al. The BTK inhibitor ibrutinib may protect against pulmonary injury in COVID-19-infected patients. Blood. 2020;135(21):1912–1915. doi: 10.1182/blood.2020006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schor S, et al. The cargo adapter protein CLINT1 is phosphorylated by the Numb-associated kinase BIKE and mediates dengue virus infection. J Biol Chem. 2022;298(6):101956. doi: 10.1016/j.jbc.2022.101956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Karim M, et al. Numb-associated kinases are required for SARS-CoV-2 infection and are cellular targets for antiviral strategies. Antiviral Res. 2022;204:105367. doi: 10.1016/j.antiviral.2022.105367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pu S-Y, et al. Optimization of isothiazolo[4,3-b]pyridine-based inhibitors of cyclin G associated kinase (GAK) with broad-spectrum antiviral activity. J Med Chem. 2018;61(14):6178–6192. doi: 10.1021/acs.jmedchem.8b00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Verdonck S, et al. Synthesis and structure-activity relationships of 3,5-disubstituted-pyrrolo[2,3-b]pyridines as inhibitors of adaptor-associated kinase 1 with antiviral activity. J Med Chem. 2019;62(12):5810–5831. doi: 10.1021/acs.jmedchem.9b00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Neveu G, et al. AP-2-associated protein kinase 1 and cyclin G-associated kinase regulate hepatitis C virus entry and are potential drug targets. J Virol. 2015;89(8):4387–4404. doi: 10.1128/JVI.02705-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cai X, et al. PIKfyve, a class III PI kinase, is the target of the small molecular IL-12/IL-23 inhibitor apilimod and a player in Toll-like receptor signaling. Chem Biol. 2013;20(7):912–921. doi: 10.1016/j.chembiol.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kang YL, et al. Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117(34):20803–20813. doi: 10.1073/pnas.2007837117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jefferies HB, et al. A selective PIKfyve inhibitor blocks PtdIns(3,5)P2 production and disrupts endomembrane transport and retroviral budding. EMBO Rep. 2008;9(2):164–170. doi: 10.1038/sj.embor.7401155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Harb WA, et al. Phase 1 clinical safety, pharmacokinetics (PK), and activity of apilimod dimesylate (LAM-002A), a first-in-class inhibitor of phosphatidylinositol-3-phosphate 5-kinase (PIKfyve), in patients with relapsed or refractory B-cell malignancies. Blood. 2017;130:4119 [Google Scholar]
- 145.Ikonomov OC, et al. Small molecule PIKfyve inhibitors as cancer therapeutics: translational promises and limitations. Toxicol Appl Pharmacol. 2019;383:114771. doi: 10.1016/j.taap.2019.114771. [DOI] [PubMed] [Google Scholar]
- 146.Sands BE, et al. Randomized, double-blind, placebo-controlled trial of the oral interleukin-12/23 inhibitor apilimod mesylate for treatment of active Crohn’s disease. Inflamm Bowel Dis. 2010;16(7):1209–1218. doi: 10.1002/ibd.21159. [DOI] [PubMed] [Google Scholar]
- 147.Krausz S, et al. A phase IIA, randomized, double-blind, placebo-controlled trial of apilimod mesylate, an IL-12/IL-23 inhibitor, in patients with rheumatoid arthritis. Arthritis Rheum. 2012;64(6):1750–1755. doi: 10.1002/art.34339. [DOI] [PubMed] [Google Scholar]
- 148.Logue J, et al. PIKfyve-specific inhibitors restrict replication of multiple coronaviruses in vitro but not in a murine model of COVID-19. Commun Biol. 2022;5(1):808. doi: 10.1038/s42003-022-03766-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Schor S, Einav S. Combating intracellular pathogens with repurposed host-targeted drugs. ACS Infect Dis. 2018;4(2):88–92. doi: 10.1021/acsinfecdis.7b00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Simmons G, et al. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci U S A. 2005;102(33):11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Aoyama T, et al. Pharmacological studies of FUT-175, nafamstat mesilate. I. Inhibition of protease activity in in vitro and in vivo experiments. Jpn J Pharmacol. 1984;35(3):203–227. doi: 10.1254/jjp.35.203. [DOI] [PubMed] [Google Scholar]
- 153.Lee M, et al. Evaluation of anti-influenza effects of camostat in mice infected with non-adapted human influenza viruses. Arch Virol. 1996;141(10):1979–1989. doi: 10.1007/BF01718208. [DOI] [PubMed] [Google Scholar]
- 154.Li K, et al. The TMPRSS2 inhibitor nafamostat reduces SARS-CoV-2 pulmonary infection in mouse models of COVID-19. mBio. 2021;12(4):e0097021. doi: 10.1128/mBio.00970-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kinoshita T, et al. A multicenter, double-blind, randomized, parallel-group, placebo-controlled study to evaluate the efficacy and safety of camostat mesilate in patients with COVID-19 (CANDLE study) BMC Med. 2022;20(1):342. doi: 10.1186/s12916-022-02518-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Quinn TM, et al. Randomised controlled trial of intravenous nafamostat mesylate in COVID pneumonitis: Phase 1b/2a experimental study to investigate safety, pharmacokinetics and pharmacodynamics. EBioMedicine. 2022;76:103856. doi: 10.1016/j.ebiom.2022.103856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Simon TG, Butt AA. Lipid dysregulation in hepatitis C virus, and impact of statin therapy upon clinical outcomes. World J Gastroenterol. 2015;21(27):8293–8303. doi: 10.3748/wjg.v21.i27.8293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ikeda M, et al. Different anti-HCV profiles of statins and their potential for combination therapy with interferon. Hepatology. 2006;44(1):117–125. doi: 10.1002/hep.21232. [DOI] [PubMed] [Google Scholar]
- 159.Del Real G, et al. Statins inhibit HIV-1 infection by down-regulating Rho activity. J Exp Med. 2004;200(4):541–547. doi: 10.1084/jem.20040061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Teixeira L, et al. Simvastatin downregulates the SARS-CoV-2-induced inflammatory response and impairs viral infection through disruption of lipid rafts. Front Immunol. 2022;13:820131. doi: 10.3389/fimmu.2022.820131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fedson DS, et al. Treating the host response to Ebola virus disease with generic statins and angiotensin receptor blockers. mBio. 2015;6(3):e00716. doi: 10.1128/mBio.00716-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Santosa A, et al. Protective effects of statins on COVID-19 risk, severity and fatal outcome: a nationwide Swedish cohort study. Sci Rep. 2022;12(1):12047. doi: 10.1038/s41598-022-16357-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gan ES, et al. Dengue virus induces PCSK9 expression to alter antiviral responses and disease outcomes. J Clin Invest. 2020;130(10):5223–5234. doi: 10.1172/JCI137536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Navarese EP, et al. PCSK9 inhibition during the inflammatory stage of SARS-CoV-2 infection. J Am Coll Cardiol. 2023;81(3):224–234. doi: 10.1016/j.jacc.2022.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Whitehorn J, et al. Lovastatin for the treatment of adult patients with dengue: a randomized, double-blind, placebo-controlled trial. Clin Infect Dis. 2016;62(4):468–476. doi: 10.1093/cid/civ949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tsai WL, et al. Metformin activates type I interferon signaling against HCV via activation of adenosine monophosphate-activated protein kinase. Oncotarget. 2017;8(54):91928–91937. doi: 10.18632/oncotarget.20248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Soto-Acosta R, et al. DENV up-regulates the HMG-CoA reductase activity through the impairment of AMPK phosphorylation: a potential antiviral target. PLoS Pathog. 2017;13(4):e1006257. doi: 10.1371/journal.ppat.1006257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Alwarawrah Y, et al. Targeting T-cell oxidative metabolism to improve influenza survival in a mouse model of obesity. Int J Obes (Lond) 2020;44(12):2419–2429. doi: 10.1038/s41366-020-00692-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Farfan-Morales CN, et al. The antiviral effect of metformin on Zika and dengue virus infection. Sci Rep. 2021;11(1):8743. doi: 10.1038/s41598-021-87707-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yen FS, et al. Metformin use before influenza vaccination may lower the risks of influenza and related complications. Vaccines (Basel) 2022;10(10):1752. doi: 10.3390/vaccines10101752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Luo P, et al. Metformin treatment was associated with decreased mortality in COVID-19 patients with diabetes in a retrospective analysis. Am J Trop Med Hyg. 2020;103(1):69–72. doi: 10.4269/ajtmh.20-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Reis G, et al. Effect of early treatment with metformin on risk of emergency care and hospitalization among patients with COVID-19: the TOGETHER randomized platform clinical trial. Lancet Reg Health Am. 2022;6:100142. doi: 10.1016/j.lana.2021.100142. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 173.Nguyen NM, et al. Metformin as adjunctive therapy for dengue in overweight and obese patients: a protocol for an open-label clinical trial (MeDO) Wellcome Open Res. 2021;5:160. doi: 10.12688/wellcomeopenres.16053.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Planas D, et al. LILAC pilot study: effects of metformin on mTOR activation and HIV reservoir persistence during antiretroviral therapy. EBioMedicine. 2021;65:103270. doi: 10.1016/j.ebiom.2021.103270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Madrid PB, et al. Evaluation of Ebola virus inhibitors for drug repurposing. ACS Infect Dis. 2015;1(7):317–326. doi: 10.1021/acsinfecdis.5b00030. [DOI] [PubMed] [Google Scholar]
- 176.Fan H, et al. Selective inhibition of Ebola entry with selective estrogen receptor modulators by disrupting the endolysosomal calcium. Sci Rep. 2017;7(1):41226. doi: 10.1038/srep41226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zu S, et al. Tamoxifen and clomiphene inhibit SARS-CoV-2 infection by suppressing viral entry. Signal Transduct Target Ther. 2021;6(1):435. doi: 10.1038/s41392-021-00853-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Watashi K, et al. Anti-hepatitis C virus activity of tamoxifen reveals the functional association of estrogen receptor with viral RNA polymerase NS5B. J Biol Chem. 2007;282(45):32765–32772. doi: 10.1074/jbc.M704418200. [DOI] [PubMed] [Google Scholar]
- 179.Mudgal R, et al. Selective estrogen receptor modulators limit alphavirus infection by targeting the viral capping enzyme nsP1. Antimicrob Agents Chemother. 2022;66(3):e0194321. doi: 10.1128/aac.01943-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Miao G, et al. Antiviral efficacy of selective estrogen receptor modulators against SARS-CoV-2 infection in vitro and in vivo reveals bazedoxifene acetate as an entry inhibitor. J Med Virol. 2022;94(10):4809–4819. doi: 10.1002/jmv.27951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nicastri E, et al. A phase 2 randomized, double-blinded, placebo-controlled, multicenter trial evaluating the efficacy and safety of raloxifene for patients with mild to moderate COVID-19. EClinicalMedicine. 2022;48:101450. doi: 10.1016/j.eclinm.2022.101450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Scully EP, et al. Impact of Tamoxifen on vorinostat-induced human immunodeficiency virus expression in women on antiretroviral therapy: AIDS Clinical Trials Group A5366, the MOXIE Trial. Clin Infect Dis. 2022;75(8):1389–1396. doi: 10.1093/cid/ciac136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rossignol JF. Nitazoxanide: a first-in-class broad-spectrum antiviral agent. Antiviral Res. 2014;110:94–103. doi: 10.1016/j.antiviral.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Shi Z, et al. Nitazoxanide inhibits the replication of Japanese encephalitis virus in cultured cells and in a mouse model. Virol J. 2014;11(1):10. doi: 10.1186/1743-422X-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Rossignol JF, et al. Thiazolides, a new class of anti-influenza molecules targeting viral hemagglutinin at the post-translational level. J Biol Chem. 2009;284(43):29798–29808. doi: 10.1074/jbc.M109.029470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Cao J, et al. A screen of the NIH Clinical Collection small molecule library identifies potential anti-coronavirus drugs. Antiviral Res. 2015;114:1–10. doi: 10.1016/j.antiviral.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Riccio A, et al. Impairment of SARS-CoV-2 spike glycoprotein maturation and fusion activity by nitazoxanide: an effect independent of spike variants emergence. Cell Mol Life Sci. 2022;79(5):227. doi: 10.1007/s00018-022-04246-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Korba BE, et al. Nitazoxanide, tizoxanide and other thiazolides are potent inhibitors of hepatitis B virus and hepatitis C virus replication. Antiviral Res. 2008;77(1):56–63. doi: 10.1016/j.antiviral.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 189.Haffizulla J, et al. Effect of nitazoxanide in adults and adolescents with acute uncomplicated influenza: a double-blind, randomised, placebo-controlled, phase 2b/3 trial. Lancet Infect Dis. 2014;14(7):609–618. doi: 10.1016/S1473-3099(14)70717-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Rossignol JF, et al. Improved virologic response in chronic hepatitis C genotype 4 treated with nitazoxanide, peginterferon, and ribavirin. Gastroenterology. 2009;136(3):856–862. doi: 10.1053/j.gastro.2008.11.037. [DOI] [PubMed] [Google Scholar]
- 191.Kohla MA, et al. Impact of nitazoxanide on sustained virologic response in Egyptian patients with chronic hepatitis C genotype 4: a double-blind placebo-controlled trial. Eur J Gastroenterol Hepatol. 2016;28(1):42–47. doi: 10.1097/MEG.0000000000000492. [DOI] [PubMed] [Google Scholar]
- 192.Blum VF, et al. Nitazoxanide superiority to placebo to treat moderate COVID-19 — a pilot prove of concept randomized double-blind clinical trial. EClinicalMedicine. 2021;37:100981. doi: 10.1016/j.eclinm.2021.100981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Reeves PM, et al. Variola and monkeypox viruses utilize conserved mechanisms of virion motility and release that depend on abl and SRC family tyrosine kinases. J Virol. 2011;85(1):21–31. doi: 10.1128/JVI.01814-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.de Wispelaere M, et al. The small molecules AZD0530 and dasatinib inhibit dengue virus RNA replication via Fyn kinase. J Virol. 2013;87(13):7367–7381. doi: 10.1128/JVI.00632-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Gallay PA, Lin K. Profile of alisporivir and its potential in the treatment of hepatitis C. Drug Des Devel Ther. 2013;7:105–115. doi: 10.2147/DDDT.S30946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wattana K, et al. Potential drug interaction between favipiravir and warfarin in patients with COVID-19: a real-world observational study. J Clin Pharmacol. 2022;63(3):338–344. doi: 10.1002/jcph.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Arnold JJ, et al. Sensitivity of mitochondrial transcription and resistance of RNA polymerase II dependent nuclear transcription to antiviral ribonucleosides. PLoS Pathog. 2012;8(11):e1003030. doi: 10.1371/journal.ppat.1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wadman M. FDA no longer needs to require animal tests before human drug trials. Science News. 2023;379:6628. doi: 10.1126/science.adg6276. [DOI] [PubMed] [Google Scholar]
- 199.Barker A, et al. I-SPY 2: an adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. Clin Pharmacol Ther. 2009;86(1):97–100. doi: 10.1038/clpt.2009.68. [DOI] [PubMed] [Google Scholar]
- 200.Berry SM, et al. A response adaptive randomization platform trial for efficient evaluation of Ebola virus treatments: a model for pandemic response. Clin Trials. 2016;13(1):22–30. doi: 10.1177/1740774515621721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Robinson M, et al. A 20-gene set predictive of progression to severe dengue. Cell Rep. 2019;26(5):1104–1111. doi: 10.1016/j.celrep.2019.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Liu YE, et al. An 8-gene machine learning model improves clinical prediction of severe dengue progression. Genome Med. 2022;14(1):33. doi: 10.1186/s13073-022-01034-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Shojaei M, et al. Multi-site validation of a host response signature for predicting likelihood of bacterial and viral infections in patients with suspected influenza. Eur J Clin Invest. 2023;53(5):e13957. doi: 10.1111/eci.13957. [DOI] [PubMed] [Google Scholar]
- 204.Bekerman E, Einav S. Infectious disease. Combating emerging viral threats. Science. 2015;348(6232):282–283. doi: 10.1126/science.aaa3778. [DOI] [PMC free article] [PubMed] [Google Scholar]