Abstract
Objectives
To characterize a novel acquired MBL, BIM-1, in a Pseudomonas #2 (subgroup P. guariconensis) strain isolated from the Aurá river located in the Brazilian Amazon hydrographic basin.
Methods
WGS using an Illumina® MiSeq System was used to characterize the genome of Pseudomonas sp. IEC33019 strain. Southern blotting/hybridization assays were performed to confirm the location of the MBL-encoding gene, blaBIM-1 (Belém Imipenemase). Antimicrobial susceptibility testing, cloning, and biochemical and phenotypic characterization were performed to determine BIM-1 kinetics.
Results
The IEC33019 strain showed high resistance rates to β-lactams, ciprofloxacin and aminoglycosides, being susceptible only to polymyxins and susceptible, increased exposure to aztreonam. WGS analysis revealed a novel acquired MBL-encoding gene, blaBIM-1, found as a gene cassette inserted into a class 1 integron (In1326) that also carried qnrVC1 and aadA11e. In1326 was located in a complex transposon, Tn7122, carried by a 52.7 kb conjugative plasmid (pIEC33019) with a toxin/antitoxin system (vapB/vapC). BIM-1 belongs to the molecular subgroup B1 and shares 70.2% and 64.9% similarity with SIM-1 and IMP-1, respectively. Kinetics analysis of BIM-1 showed hydrolytic activity against all β-lactams tested.
Conclusions
BIM-1 is a novel acquired MBL encoded by a gene carried by mobile genetic elements, which can be transferred to other Gram-negative bacilli (GNB). Because the IEC33019 strain was recovered from a river impacted by a populous metropolitan region with poor basic sanitation and served by limited potable freshwater, it would be important to establish the role of the BIM-1-producing GNB as nosocomial pathogens and/or as colonizers of the riverside population in this geographical region.
Introduction
Carbapenemases probably originated some million years ago1. However, they emerged in clinical settings in the mid-1980s,2 spreading among Gram-negative bacilli (GNB) over the following decades,3,4 as a predictable consequence of increased use of carbapenems against cephalosporin-resistant infections.1 Carbapenemases constitute a heterogenic and versatile group of hydrolytic enzymes classified into Ambler molecular classes A (functional subgroup 2f); B (also called MBLs), which are subdivided into subgroups B1 to B3 (functional subgroups 3a and 3b); and D (functional subgroup 2df).2,4 The acquired MBLs represent a public health threat,3 because these Zn2+-dependent carbapenemases confer broad-spectrum β-lactam resistance, except for monobactams,5 and are not inhibited by any β-lactamase inhibitors currently used in clinical practice, including the novel β-lactamase inhibitors like avibactam, relebactam and vaborbactam.5,6 Hence, the therapeutic options for treatment of infections caused by MBL-producing GNB isolates are very limited. Furthermore, acquired MBL-encoding genes are located in mobile genetic elements (MGEs),4,5 and can be transferred to other bacterial species, partially explaining their successful spread in the environment.3,5 To date, 14 acquired MBL groups have been reported worldwide (Beta-Lactamase DataBase), with most belonging to the B1 molecular subclass and very few to subclass B3.7 Although most of these groups are restricted to certain geographical regions,3,8 at least three of them (IMP, VIM and NDM) have been reported globally, representing an important medical problem.8
To date, Pseudomonas aeruginosa has evolved as the main MBL producer in nosocomial settings.3,8 Many factors are involved in this phenomenon, such as its genetic plasticity and ubiquitous behaviour,9 which favour the acquisition of antimicrobial resistance genes (ARGs) from other environmental Pseudomonas species, mainly those included in the P. putida group,10 which inhabit the same ecological niche and serve as reservoirs of conjugative plasmid-mediated resistance markers.11 Although most species belonging to the P. putida group are rarely involved in human infections, P. putida has been considered as an emergent aetiological agent of healthcare-associated infections.12,13 Similarly, MBL-producing P. putida strains have also been recovered from temperate soil and aquatic environmental matrices, reinforcing its broad metabolic versatility and role as the natural host for these β-lactam resistance determinants.10,11
Here we report the biochemical and genetic characterization of a newly acquired MBL, named BIM-1 (Belém Imipenemase), in a Pseudomonas #2 (subgroup P. guariconensis) strain isolated from a river located in the Amazon hydrographic basin. This river has been impacted by anthropogenic activities of a nearby populous metropolitan area in Brazil, which is served by limited potable freshwater.14 This scenario is worrisome due to the poor local basic sanitation15,16 associated with the hydrography of the region, which can serve to maintain GNB carrying ARGs in the environment that further contribute to human colonization.
Materials and methods
Water sample analysis and bacterial identification
In April 2015, a carbapenem-resistant Pseudomonas sp. IEC33019 strain was recovered from a surface water sample of the Aurá river, which is located near the Belém Metropolitan Region (BMR) in Pará state, Brazilian Amazon region (Figure S1, available as Supplementary data at JAC Online). The BMR comprises six municipalities (Figure S1), including the city of Belém (state capital), with an estimated resident population of 2 505 242 inhabitants and a territorial area of 6890 km².14 The Aurá river is an important tributary of the Guamá River and Guajará Bay, which serve as a water supply to the BMR. However, due to the proximity to uncontrolled landfill waste from the BMR (Figure S1), the Aurá river water is not suitable for conventional treatment for public water supply.14–16 For microbiological analysis, surface water samples (1 L) were collected from the Aurá river at low tide and filtered through 0.22 μm cellulose acetate membrane filters (Merck KGaA, Darmstadt, Germany). Subsequently, the filters were shaken in 20 mL PBS (Sigma-Aldrich, St Louis, MO, USA) for 1 h and centrifuged at 500 rpm for 10 min. Aliquots of 200 µL supernatant were inoculated into brain heart infusion (BHI) broth (Merck KGaA, Darmstadt, Germany) containing 10 µg meropenem discs (Thermo Fisher Scientific, Basingstoke, UK) and incubated at 35 ± 2°C for 24 h. Later, BHI broth was seeded onto MacConkey agar (Merck KGaA, Darmstadt, Germany) plates. The carbapenem-resistant IEC33019 strain was initially identified at the species level as P. putida by the VITEK 2 automated system (bioMérieux, Marcy l’Etoile, France) and confirmed as Pseudomonas sp. #2, a novel species that belongs to the subgroup P. guariconensis (Average nucleotide identity by BLAST 91.47%), as proposed by Girard and colleagues using a phylogenetic tree of the P. putida group based on the rpoD gene.17
Susceptibility testing and MBL screening test
MICs of piperacillin/tazobactam, ceftazidime, cefepime, aztreonam, imipenem, meropenem, ciprofloxacin, amikacin, gentamicin, tobramycin, colistin and polymyxin B (Sigma-Aldrich, St Louis, MO, USA) were determined for the IEC33019 strain by CAMHB microdilution according to the Brazilian Committee on Antimicrobial Susceptibility Testing (BrCAST)/EUCAST guidelines (http://brcast.org.br/). Additionally, the MICs of ceftolozane/tazobactam, levofloxacin and moxifloxacin were determined by Etest® gradient strips (bioMérieux, Marcy l’Etoile, France), following the manufacturer’s recommendations. The MICs obtained were interpreted according to BrCAST/EUCAST guidelines among those drugs for which breakpoints are available. Moreover, MBL phenotype detection was performed for the IEC33019 strain using the double-disc synergy test (DDST) using discs of meropenem 10 µg (Thermo Fisher Scientific, Basingstoke, UK) and meropenem 10 µg + 100 mM EDTA (Sigma-Aldrich, St Louis, MO, USA). Acquired MBL-encoding genes were screened by PCR followed by sequencing using specific primers, as previously published.18
Pseudomonas #2 IEC33019 DNA extraction, WGS and bioinformatics analysis
Total genomic DNA was extracted from Pseudomonas #2 IEC33019 strain using the Wizard® Genomic DNA purification kit (Promega, Madison, WI, USA), according to the manufacturer’s protocol. Subsequently, libraries were constructed using the Nextera® Mate Pair library preparation kit (Illumina Inc., San Diego, CA, USA). The sequencing was carried out on a MiSeqTM system 2 × 250 bp in paired-end mode (Illumina Inc., San Diego, CA, USA), with V2 kit, 500 cycles. This achieved ∼120× coverage with the short reads determined using raspberry-v0.3.19 The contigs were ordered and scaffolded using SSPACE,20 and the gaps were closed using Gapfiller.21 The assembly and the gaps were curated manually using Geneious,22 and prediction and automatic annotation was performed using Prokka.23 Plasmids, ARG determinants and virulence markers were identified using the ABRicate-v0.8.10 (https://github.com/tseemann/abricate) tool utilizing the PlasmidFinder (https://cge.food.dtu.dk/services/PlasmidFinder/), ARG-ANNOT (https://ifr48.timone.univ-mrs.fr/blast/arg-annot_nt.html), Comprehensive Antibiotic Resistance Database—CARD (https://card.mcmaster.ca/) and Virulence Factor DataBase—VFDB,24 respectively. In silico MLST was done using mlst-v2.15.1 (https://github.com/tseemann/mlst), and the new alleles and respective STs were designated using PubMLST P. putida database.25 Additionally, the presence of ISs and class 1 integrons was identified using the ISfinder26 platform and INTEGRALL—The Integron Database,27 respectively. For novel transposon recording, we used the LSTM Transposon Registry.28
Cloning of blaBIM-1
For cloning experiments, blaBIM-1 was amplified by PCR using the specific primers BIM-1_NcoI_FW1 (5′-GGTGGTCCATGGGCAGAATATCGTTAGCTTTATGCTTG-3′) and BIM-1_XhoI_rev (5′-CCACCACTCGAGTTATTTGCTGAGCTGTGATGTTTTTTT-3′), which contained the NcoI and XhoI restriction sites (Thermo Fisher Scientific, Waltham, MA, USA), respectively. PCR was performed using the Phusion® High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA, USA). Both amplicon fragment and pET-28b(+) vector (Merck KGaA, Darmstadt, Germany) were digested with NcoI and XhoI and then incubated with T4 DNA ligase (Thermo Fisher Scientific, Waltham, MA, USA). The recombinant plasmids (Figure S2) were transformed by heat-shock into the chemically competent Novagen®E. coli DH5α strain (Merck KGaA, Darmstadt, Germany). The recombinant pET-28b(+)-blaBIM-1 plasmid was purified using QIAprep Spin Miniprep Kit (Qiagen, Hilden, Germany).
BIM-1 expression and purification
The purified pET-28b(+)-blaBIM-1 plasmid (Figure 2S) was inserted (50 ηg) into 200 µL of chemically competent Novagen®E. coli BL21(Dε3) strain (Merck KGaA, Darmstadt, Germany) by heat-shock transformation, and incubated in 800 µL of LB broth at 37°C for 1 h. Transformant E. coli BL21(Dε3) cells were selected onto LB agar plates supplemented with 50 mg/L kanamycin. The production of BIM-1 enzyme was induced in 1.5 L of LB broth supplemented with 50 mg/L kanamycin and 0.20 mM IPTG, followed by incubation at 20°C overnight. The cells were harvested by centrifugation at 4.388 g for 30 min at 4°C, and the pellet obtained was resuspended in a hypertonic solution (40:1 v/v) containing 40% sucrose and 50 mM Tris-HCl buffer (pH 8.0), and incubated for 15 min in an ice bath. Subsequently, a 1/4,5 v/v dilution was performed using a hypotonic solution of 5 mM MgCl2, and the mixture was incubated at room temperature for 30 min, followed by centrifugation at 4.388 g for 30 min. The supernatant was filtered in a 0.22 µm Millipore membrane (Merck KGaA, Darmstadt, Germany) and subjected to a purification step by size exclusion chromatography, using a Superdex® 75 (10/300) column (Sigma-Aldrich, St Louis, MO, USA) coupled to an Äkta purifier system (GE Healthcare, Orsay, France). The column was previously equilibrated with 50 mM Tris-HCl buffer (pH 8.0) and 150 mM NaCl (Merck KGaA, Darmstadt, Germany), and the obtained fractions were submitted to a hydrolysis test by spectrophotometry, with a wavelength of 486 nm using 20 mM nitrocefin (Sigma-Aldrich, St Louis, MO, USA) as substrate. The tubes that showed hydrolysis activity were submitted to 12% SDS-PAGE gel (Figure S2) and those samples that presented >90% purity were concentrated using an Amicon Ultra-15 centrifugal filter device (Merck KGaA, Darmstadt, Germany) of 10 000 nominal molecular weight limit. The BIM-1 recombinant (rBIM-1) quantification was performed by spectrophotometry at 280 nm on a Nanodrop spectrometer (Thermo Fisher Scientific, Waltham, MA, USA), assuming an ɛ value of 46 410 M−1 cm−1 as calculated by ProtParam of the ExPASy proteomics server (https://web.expasy.org). In addition, the β-lactam MICs for E. coli BL21(Dε3) strain, its transformant containing only the vector pET-28b(+) or pET-28b(+)-blaBIM-1, were determined by CAMHB microdilution supplemented with 0.20 mM IPTG.
Figure 2.
Amino acid sequence alignment of BIM-1 with other MBLs representative of subclasses B1 (IMP-1; ABK27309.1), B2 (CphA; CAA40386.1) and B3 (L1; CAA52968.1). Conserved residues in the first and second Zn2+ coordination sites are shown in red boxes, and dashes correspond to the sequence gaps. Numbering is according to the updated BBL scheme.32 The figure was obtained using MegAlign (Lasergene software package; DNAStar, Madison, WI, USA).
Kinetics measurements
Purified BIM-1 was used for the determination of kinetics parameters (kcat and KM) for the β-lactam hydrolysis reactions using a cuvette 10 mm SpectraMax® M2 Microplate Reader (Molecular Devices LLC, San José, CA, USA) at 30°C in 50 mM HEPES buffer (pH 7.5). The wavelengths and changes in extinction coefficients of β-lactams were determined through the slope of the difference in absorbance of intact and degraded molecules of each β-lactam tested: ampicillin, cefalotin, ceftazidime, cefepime, imipenem and meropenem (Sigma-Aldrich, St Louis, MO, USA). The results underwent linear regression analysis by GraFit Data Analysis software version 5.0 (Erithacus Software; http://www.erithacus.com/grafit/) using Eadie–Hofstee plots and the Lineweaver–Burk linearization of the Henri–Michaelis–Menten equation. The final values were obtained by calculating the mean of three independent measurements.
Nucleotide sequence accession number
The nucleotide sequences described in this study have been assigned under accession numbers CP016634 (Pseudomonas #2 IEC33019 genome) and CP016446 (plasmid pIEC33019) in the GenBank® genetic sequence database.
Results and discussion
Antimicrobial susceptibility profile and genomic analysis of IEC33019 strain
The Pseudomonas #2 IEC33019 strain was resistant to ceftazidime, cefepime, imipenem, meropenem, ciprofloxacin, amikacin and tobramycin (Table 1). In addition, the IEC3309 strain showed a MIC of 4 mg/L for gentamicin. The polymyxins and aztreonam (susceptible, increased exposure) were the only antimicrobials tested that showed activity against Pseudomonas #2 IEC33019 strain. The DDST results using meropenem/meropenem + EDTA discs (6 mm versus 18 mm, respectively) suggested that Pseudomonas #2 IEC33019 was phenotypically an MBL producer. However, PCR screening results indicated that the IEC33019 strain was negative for any known acquired MBL-encoding gene. The chromosome of Pseudomonas #2 IEC33019 strain was estimated to be 6 156 701 bp with an average G + C content of 62.33% (39.58%–71.84%). A total of 5547 coding sequences were found in the genome of the IEC33019 strain including 1027 hypothetical proteins, 57 repeat regions, 74 tRNAs and 22 rRNAs. The IEC33019 strain belonged to the singleton ST137 that showed a unique allele profile (71-90-122-84-105-104-46-106).25 Besides, genes offering resistance to β-lactams (ampC), aminoglycosides (strA-strB), macrolides (macA-macB), bicyclomycin (bcr) and tetracyclines (tetA) were found in the IEC33019 genome. Furthermore, resistance determinants to tellurite (terB) and to quaternary ammonium compounds (sugE) were also found.
Table 1.
MICs for Pseudomonas #2 IEC33019, E. coli BL21(Dε3) and for its transformant carrying pET-28b(+)-blaBIM-1
| Antimicrobial | MIC (mg/L) | ||
|---|---|---|---|
| Pseudomonas #2 IEC33019 | E. coli a | ||
| BL21(Dε3) [pET-28b(+)-blaBIM-1] | BL21(Dε3) | ||
| Piperacillin/tazobactam | 16 | — | — |
| Ceftazidime | >128 | 1 | 0.015 |
| Ceftolozane/tazobactamb | >256 | — | — |
| Cefepime | >128 | 0.5 | 0.0075 |
| Aztreonam | 8 | 0.03 | 0.00375 |
| Imipenem | 64 | 0.125 | 0.06 |
| Meropenem | 64 | 0.03 | ≤0.000937 |
| Ciprofloxacin | 64 | — | — |
| Levofloxacinb | 16 | — | — |
| Moxifloxacinb | >32 | — | — |
| Amikacin | 16 | — | — |
| Gentamicin | 4 | — | — |
| Tobramycin | 4 | — | — |
| Colistin | 2 | — | — |
| Polymyxin B | 2 | — | — |
The results of MICs for transformants ≥2 dilutions compared with the strain BL21 alone were considered significant and are marked in bold. The concentration ranges of each β-lactam tested varied from 0.000937 to 128 mg/L. Dash (—) indicates antimicrobial not tested.
MICs determined by Etest® gradient strips (bioMérieux).
Genetic context of blaBIM-1
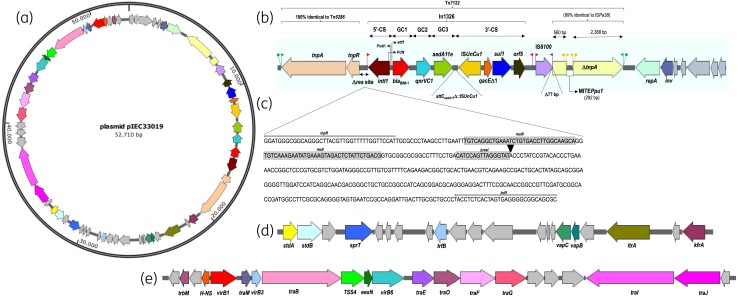
WGS analysis also demonstrated that Pseudomonas #2 IEC33019 strain harboured a plasmid (pIEC33019) with an estimated size of 52 710 bp comprising an average G + C content of 59.17% and 62 coding sequences distributed among three domains (Figure 1a). In the first domain (Figure 1b), a unique class 1 integron, named In1326,27 was found. In1326 contained a cassette array (Figure 1b) comprising a new MBL-encoding gene named blaBIM-1 (attCIMP-1/132 bp) at first position (738 bp), followed by the quinolone resistance determinant gene qnrVC1 (attCqnrVC1/132 bp) from Vibrio cholerae, and the aminoglycoside-modifying enzyme-encoding gene aadA11e. The qnrVC1 gene was first described in a V. cholerae O1 clinical strain recovered from the Amazon region during the Brazilian cholera epidemic that occurred from 1991 to 2000.29 In addition, the attC of aadA11e (ΔattCaadA11/56 bp) in In1326 was truncated with ISUnCu1 (IS110 family), which was inserted in the opposite transcriptional direction in the 3′-CS region (Figure 1b). In1326 was flanked upstream by the transposition module tnpAR-res similar to Tn6286 (accession number KU130294.1). Interestingly, Tn6286 was previously described carrying another integron-borne MBL-encoding gene, blaDIM-2, reported in a P. putida strain isolated in China.30 In Tn6286, the module tnpAR-res comprised tnpA and tnpR that encode for a transposase and resolvase, respectively, and a truncated res site (ΔresI) of 119 bp rather than the 107 bp found in In1326 (Figure 1c). Although a 25 bp Invert Repeat transposase (IRt) was found downstream of orf5, the respective Invert repeat integrase (IRi) was detected upstream of In1326 (Figure 1b—red flags), but in the reverse complementary sequence to that usually found.31 It seems that IRi and IRt facilitate the mobilization of class 1 integrons, when the tnpAR-res transposition module is present, defining its structural borders during the transposition process.32 Downstream of In1326 was found an IS6100 copy (IS6-like family)26 flanked by IRs truncated with a tnpA from ISPa38 (Tn3-like family)26 from P. aeruginosa, which led to the loss of the first 77 bp of the transposase gene. Interestingly, the ΔtnpA from ISPa38 was also disrupted by a novel miniature inverted-repeat transposon element (MITE) 292 bp in length, designated MITEPpu1 (IS481 family),26 flanked by 26 bp imperfect IRs and 5 bp DRs (Figure 1b). In addition, imperfect 26 bp IRs flanked by 5 bp DRs were found upstream of the Tn6286 transposition module (tnpA-tnpR) and downstream of the IS6100/ΔtnpA::MITEPpu1 genetic structure (Figure 1b), indicating that this 15 318 bp MGE (tnpAR-Δres-In1326-IS6100-ΔtnpA::MITEPpu1) moved to its present location by transposition. This complex transposon was designated Tn7122.28 A second domain of pIEC33019 (Figure 1d) was shown to contain genes encoding a type II toxin-antitoxin system (vapB/vapC), zinc metalloprotease (sprT), fimbrial adhesins (operon stdAB), plasmid transcriptional repressor (trfB), multifunctional group II intron reverse transcriptase/maturase (ltrA) and a transcriptional autoregulator KfrA-type (kfrA). The mobilization/conjugation module was found in the third domain of pIEC33019 (Figure 1e), where the traMBEOFGIJ, type IV secretion system (TSS4), and plasmid conjugal transfer family protein-encoding genes virB1, virB3 and virB6, were located. Interestingly, traG, virB3 and the partial conjugal transfer protein apparatus composed of trbBCEJLFGI gene clusters were identified in the chromosome of the Pseudomonas #2 IEC33019 strain, which might indicate a remnant fragment of what was once a suicide integrative plasmid. The WGS results were corroborated by Southern blotting/hybridization experiments performed twice, confirming that blaBIM-1 was located on a ∼55 kb plasmid (data not shown). Despite many attempts, pIEC33019 harbouring blaBIM-1 was not successfully transferred by conjugation or electroporation using as recipients E. coli J53 AzdR, P. aeruginosa PAO1, P. aeruginosa ATCC® 27853™ and electrocompetent E. coli DH5α strains, respectively (data not shown).
Figure 1.
Genetic structure of pIEC33019 conjugative plasmid carrying blaBIM-1 in Pseudomonas #2 IEC33019 strain. (a) Circular map of pIEC33019. (b) Antimicrobial resistance domain containing In1326 (dashed box). (c)Truncated res site (ΔresI) with In1326 of module tnpAR-res. (d) Virulence-mediated domain. (e) Mobilization/conjugation domain. Grey arrows indicate genes encoding for hypothetical proteins.
Identification of BIM-1 as an MBL
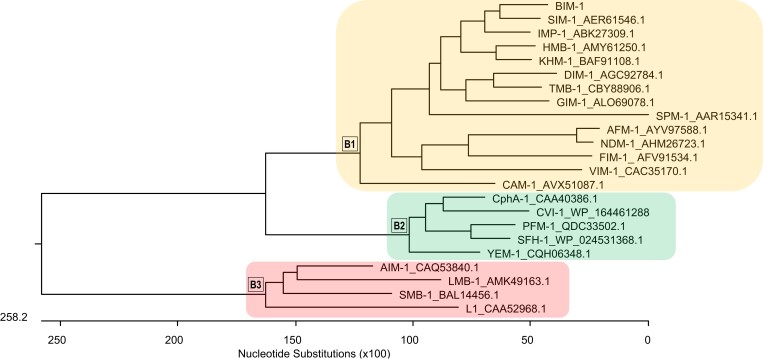
The blaBIM-1 gene encoded an enzyme with 245 amino acids (Figure 2), a molecular weight of 27 524.46 (Figure 3S) and an estimated isoelectric point of 7.42. The novel MBL was named BIM-1 (Belém Imipenemase) and possessed the conserved motifs characteristic of class B enzymes (Figure 2), including the consensus Zn+2-binding conserved motif His116-X-His118-X-Asp120, His196, Cys221 and His263 (Figure 2), according to the Class B Beta-Lactamase (BBL) nomenclature.32 BIM-1 shares 70.2% similarity with SIM-1 (AER61546.1), 64.9% with IMP-1 (ABK27309.1) and 29.8% with SPM-1 (AAR15341.1), as shown in Figure 3. Kinetics analysis revealed that BIM-1 efficiently hydrolysed all β-lactams tested (kcat/KM varying from 22.4 to 266.1 mM s−1). A relative affinity for cephalosporins and carbapenems was observed (KM varying from 11.4 to 17.1 µM), except for cefepime (KM = 42.1 µM). The best substrate for BIM-1 was cefalotin, followed by imipenem and meropenem (Table 2). Additionally, the MICs of ceftazidime and cefepime increased 64-fold in the transconjugant strain compared with strain BL21(Dε3) with no cloning vector, whereas for meropenem, cefuroxime and ceftriaxone they increased four-, eight- and four-fold, respectively (Table 1). In contrast, a slight two-fold increase was observed for imipenem, cefalotin and ampicillin MICs (Table 1). For the detection of blaBIM-1 in clinical and environmental bacterial strains, we recommend using the primers BIM_F (5′-TCAGGGAAATATTTATGAAAAACATGG-3′) and BIM_R (5′-TTACTAGGTATCCACACGACCTC-3′) that amplify a 375 bp inner fragment, according to the following thermocycling conditions: initial denaturation 95°C at 10 min; then 40 cycles of denaturation at 95°C for 30 s, annealing at 53°C for 30 s, and extension at 72°C for 1 min; and a final extension at 72°C for 10 min.
Figure 3.
Phylogenetic tree constructed based on the ClustalV alignment of BIM-1 and 16 acquired MBL groups described to date using MegAlign (Lasergene software package; DNAStar, Madison, WI, USA).
Table 2.
Steady-state kinetic parameters of purified MBL BIM-1a
| Compound | k cat (s−1) | K M (µM) | k cat/KM (mM−1 s−1) |
|---|---|---|---|
| Ampicillin | 6 | 267 | 22.4 |
| Cefalotin | 3.3 | 12.4 | 266.1 |
| Ceftazidime | 1.3 | 11.4 | 114 |
| Cefepime | 4.4 | 42.1 | 104.8 |
| Imipenem | 2.8 | 11.7 | 239.3 |
| Meropenem | 7.5 | 17.1 | 228.1 |
Values represent the mean of three independent experiments. SDs were within 10% of the means.
Conclusions
BIM-1 is a novel acquired MBL belonging to subclass B1 identified in an MDR Pseudomonas #2 strain recovered from a river highly impacted by anthropogenic action in the Brazilian Amazon region. blaBIM-1 is mediated by MGEs that also carry other ARG- and virulence-encoding genes. The risk of blaBIM-1 spread to different bacterial hosts and human gut microbiota is very worrisome. Poor basic sanitation conditions associated with a lack of health infrastructure in this Brazilian geographical region can serve to maintain GNB carrying ARGs in the environment and further contribute to human colonization.
Supplementary Material
Acknowledgements
We are grateful to Fernanda F. Santos for technical support during the genomic analysis. We also would like to thank Thomás Jové from INTEGRALL,27 Patricia Siguier from ISfinder,26 Kohei Ogura from PubMLST Pseudomonas putida databases,25 and Adam Roberts from The Transposon Registry.28 This work was presented in part as a 1 h ePoster Review (3743) and was included in the top-rated abstracts at the 31st European Congress of Clinical Microbiology and Infectious Diseases (ECCMID), 2021.
Contributor Information
Cintya O Souza, Seção de Bacteriologia e Micologia, Instituto Evandro Chagas (IEC), Ananindeua, PA, Brazil.
Rodrigo Cayô, Universidade Federal de São Paulo (UNIFESP), Laboratório ALERTA, Disciplina de Infectologia, Departamento de Medicina, Escola Paulista de Medicina (EPM), São Paulo, SP, Brazil; Universidade Federal de São Paulo (UNIFESP), Laboratório de Bacteriologia e Imunologia (LIB), Setor de Biologia Molecular, Microbiologia e Imunologia, Departamento de Ciências Biológicas (DCB), Instituto de Ciências Ambientais, Químicas e Farmacêuticas (ICAQF), Diadema, SP, Brazil.
Karla Valéria B Lima, Seção de Bacteriologia e Micologia, Instituto Evandro Chagas (IEC), Ananindeua, PA, Brazil.
Danielle M Brasiliense, Seção de Bacteriologia e Micologia, Instituto Evandro Chagas (IEC), Ananindeua, PA, Brazil.
Ana Paula Streling, Universidade Federal de São Paulo (UNIFESP), Laboratório ALERTA, Disciplina de Infectologia, Departamento de Medicina, Escola Paulista de Medicina (EPM), São Paulo, SP, Brazil.
André V Siqueira, Universidade Federal de São Paulo (UNIFESP), Laboratório ALERTA, Disciplina de Infectologia, Departamento de Medicina, Escola Paulista de Medicina (EPM), São Paulo, SP, Brazil.
Felipe Alberto-Lei, Universidade Federal de São Paulo (UNIFESP), Laboratório ALERTA, Disciplina de Infectologia, Departamento de Medicina, Escola Paulista de Medicina (EPM), São Paulo, SP, Brazil.
Josiane T Leal, Universidade Federal de São Paulo (UNIFESP), Laboratório ALERTA, Disciplina de Infectologia, Departamento de Medicina, Escola Paulista de Medicina (EPM), São Paulo, SP, Brazil.
Carolina S Nodari, Universidade Federal de São Paulo (UNIFESP), Laboratório ALERTA, Disciplina de Infectologia, Departamento de Medicina, Escola Paulista de Medicina (EPM), São Paulo, SP, Brazil.
Paula Juliana Pérez-Chaparro, Department of Clinical Analysis, School of Pharmacy (FCF), Universidade de São Paulo (USP), São Paulo, SP, Brazil.
Luana N G C Lima, Seção de Bacteriologia e Micologia, Instituto Evandro Chagas (IEC), Ananindeua, PA, Brazil.
Marcelo O Lima, Seção de Meio Ambiente, Instituto Evandro Chagas (IEC), Ananindeua, PA, Brazil; Programa de Pós-Graduação em Epidemiologia e Vigilância em Saúde (PPGEVS), Instituto Evandro Chagas (IEC), Ananindeua, PA, Brazil; Universidade Federal do Pará (UFPA), Programa de Pós-Graduação em Ecologia Aquática e Pesca (PPGEAP), Belém, PA, Brazil.
Brenda Natasha S Costa, Seção de Meio Ambiente, Instituto Evandro Chagas (IEC), Ananindeua, PA, Brazil; Universidade Federal do Pará (UFPA), Programa de Pós-Graduação em Ecologia Aquática e Pesca (PPGEAP), Belém, PA, Brazil.
Thais Karolina L De Queiroz, Seção de Meio Ambiente, Instituto Evandro Chagas (IEC), Ananindeua, PA, Brazil; Universidade Federal do Pará (UFPA), Programa de Pós-Graduação em Ecologia Aquática e Pesca (PPGEAP), Belém, PA, Brazil.
Paola J S N Silva, Universidade Federal de São Paulo (UNIFESP), Laboratório de Enzimologia, Department of Biophysics, Escola Paulista de Medicina (EPM), São Paulo, SP, Brazil.
Elza M Mamizuka, Department of Clinical Analysis, School of Pharmacy (FCF), Universidade de São Paulo (USP), São Paulo, SP, Brazil.
Marcelo F Marcondes, Universidade Federal de São Paulo (UNIFESP), Laboratório de Enzimologia, Department of Biophysics, Escola Paulista de Medicina (EPM), São Paulo, SP, Brazil.
John Anthony Mcculloch, Department of Clinical Analysis, School of Pharmacy (FCF), Universidade de São Paulo (USP), São Paulo, SP, Brazil.
Ana Cristina Gales, Universidade Federal de São Paulo (UNIFESP), Laboratório ALERTA, Disciplina de Infectologia, Departamento de Medicina, Escola Paulista de Medicina (EPM), São Paulo, SP, Brazil.
Funding
We are grateful to Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for providing grants to A.P.S. and C.S.N., and to the National Council for Science and Technological Development (CNPq) for providing grants to A.V.S. and A.C.G. (process number: 312066/2019).
Transparency declarations
A.C.G. has recently received research funding and/or consultation fees from bioMérieux, Eurofarma, MSD, Sandoz, Pfizer and United Medical. Other authors have nothing to declare. This study was not financially supported by any diagnostic/pharmaceutical company.
Supplementary data
Figures S1 and S3 are available as Supplementary data at JAC Online.
References
- 1. Perry J, Waglechner N, Wright G. The prehistory of antibiotic resistance. Cold Spring Harb Perspect Med 2016; 6: a025197. 10.1101/cshperspect.a025197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bush K. Past and present perspectives on β-lactamases. Antimicrob Agents Chemother 2018; 62: e01076-18. 10.1128/AAC.01076-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bonomo RA, Burd EM, Conly Jet al. Carbapenemase-producing organisms: a global scourge. Clin Infect Dis 2018; 66: 1290–7. 10.1093/cid/cix893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bush K, Bradford PA. Epidemiology of β-lactamase-producing pathogens. Clin Microbiol Rev 2020; 33: e00047-19. 10.1128/CMR.00047-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Palzkill T. Metallo-β-lactamase structure and function. Ann N Y Acad Sci 2013; 1277: 91–104. 10.1111/j.1749-6632.2012.06796.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bush K, Bradford PA. Interplay between β-lactamases and new β-lactamase inhibitors. Nat Rev Microbiol 2019; 17: 295–306. Erratum in: Nat Rev Microbiol 2019; 17: 459-60. 10.1038/s41579-019-0159-8 [DOI] [PubMed] [Google Scholar]
- 7. Naas T, Oueslati S, Bonnin RAet al. Beta-lactamase database (BLDB) – structure and function. J Enzyme Inhib Med Chem 2017; 32: 917–19. 10.1080/14756366.2017.1344235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. López C, Ayala JA, Bonomo RAet al. Protein determinants of dissemination and host specificity of metallo-β-lactamases. Nat Commun 2019; 10: 3617. 10.1038/s41467-019-11615-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Botelho J, Grosso F, Peixe L. Antibiotic resistance in Pseudomonas aeruginosa – mechanisms, epidemiology and evolution. Drug Resist Updat 2019; 44:100640. 10.1016/j.drup.2019.07.002 [DOI] [PubMed] [Google Scholar]
- 10. Kivisaar M. Narrative of a versatile and adept species Pseudomonas putida. J Med Microbiol 2020; 69: 324–38. 10.1099/jmm.0.001137 [DOI] [PubMed] [Google Scholar]
- 11. Meireles C, Costa G, Guinote Iet al. Pseudomonas putida are environmental reservoirs of antimicrobial resistance to β-lactamic antibiotics. World J Microbiol Biotechnol 2013; 29: 1317–25. 10.1007/s11274-013-1295-3 [DOI] [PubMed] [Google Scholar]
- 12. Aumeran C, Paillard C, Robin Fet al. Pseudomonas aeruginosa and Pseudomonas putida outbreak associated with contaminated water outlets in an oncohaematology paediatric unit. J Hosp Infect 2007; 65: 47–53. 10.1016/j.jhin.2006.08.009 [DOI] [PubMed] [Google Scholar]
- 13. Gilarranz R, Juan C, Castillo-Vera Jet al. First detection in Europe of the metallo-β-lactamase IMP-15 in clinical strains of Pseudomonas putida and Pseudomonas aeruginosa. Clin Microbiol Infect 2013; 19: E424–7. 10.1111/1469-0691.12248 [DOI] [PubMed] [Google Scholar]
- 14. Siqueira GW, Aprile F, Darwich Aet al. A. Environmental diagnostic of the Aurá river basin (Pará, Brazil): water pollution by uncontrolled landfill waste. Arch Curr Res Int 2016; 5: 1–13. 10.9734/ACRI/2016/28249 [DOI] [Google Scholar]
- 15. Oliveira RS, Kiyatake DM, Harada MLet al. Sanitary quality of the public groundwater supply for the municipality of Belém in northern Brazil. Cad Saúde Colet 2013; 21: 377–83. 10.1590/S1414-462X2013000400004 [DOI] [Google Scholar]
- 16. Mourão FV, Pereira JAR. Urban expansion impacts the surface water source of the water supply system in Belém and Ananindeua, Brazil. Res Soc Dev 2020; 9: 1–16. 10.33448/rsd-v9i7.4098 [DOI] [Google Scholar]
- 17. Girard L, Lood C, Höfte Met al. The ever-expanding Pseudomonas genus: description of 43 new species and partition of the Pseudomonas putida group. Microorganisms 2021; 9: 1766. 10.3390/microorganisms9081766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brasiliense D, Cayô R, Streling APet al. Diversity of metallo-β-lactamase-encoding genes found in distinct species of Acinetobacter isolated from the Brazilian Amazon region. Mem Inst Oswaldo Cruz 2019; 114: e190020. 10.1590/0074-02760190020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Katta MAVSK, Khan AW, Doddamani Det al. NGS-QCbox and raspberry for parallel, automated, and rapid quality control analysis of large-scale next generation sequencing (Illumina) data. PLoS One 2015; 10: e0139868. 10.1371/journal.pone.0139868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boetzer M, Henkel CV, Jansen HJet al. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011; 27: 578–9. 10.1093/bioinformatics/btq683 [DOI] [PubMed] [Google Scholar]
- 21. Nadalin F, Vezzi F, Policriti A. Gapfiller: a de novo assembly approach to fill the gap within paired reads. BMC Bioinformatics 2012; 13: S8. 10.1186/1471-2105-13-S14-S8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kearse M, Moir R, Wilson Aet al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012; 28: 1647–9. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014; 30: 2068–9. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 24. Liu B, Zheng D, Jin Qet al. Vfdb 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 2019; 47: D687–92. 10.1093/nar/gky1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ogura K, Shimada K, Miyoshi-Akiyama T. A multilocus sequence typing scheme of Pseudomonas putida for clinical and environmental isolates. Sci Rep 2019; 9: 13980. 10.1038/s41598-019-50299-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Siguier P, Perochon J, Lestrade Let al. Isfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 2006; 34: D32–6. 10.1093/nar/gkj014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moura A, Soares M, Pereira Cet al. Integrall: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 2009; 25: 1096–8. 10.1093/bioinformatics/btp105 [DOI] [PubMed] [Google Scholar]
- 28. Tansirichaiya S, Rahman MA, Roberts AP. The transposon registry. Mob DNA 2019; 10: 40. 10.1186/s13100-019-0182-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fonseca EL, Dos Santos Freitas F, Vieira VVet al. New qnr gene cassettes associated with superintegron repeats in Vibrio cholerae O1. Emerg Infect Dis 2008; 14: 1129–31. 10.3201/eid1407.080132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sun F, Zhou D, Wang Qet al. Genetic characterization of a novel blaDIM-2-carrying megaplasmid p12969-DIM from clinical Pseudomonas putida. J Antimicrob Chemother 2016; 71: 909–12. 10.1093/jac/dkv426 [DOI] [PubMed] [Google Scholar]
- 31. Stokes HW, Nesbø CL, Holley Met al. Class 1 integrons potentially predating the association with tn402-like transposition genes are present in a sediment microbial community. J Bacteriol 2006; 188: 5722–30. 10.1128/JB.01950-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garau G, García-Sáez I, Bebrone Cet al. Update of the standard numbering scheme for class B β-lactamases. Antimicrob Agents Chemother 2004; 48: 2347–9. 10.1128/AAC.48.7.2347-2349.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.