Abstract
Polyglutamine diseases are a group of neurodegenerative disorders caused by an abnormal expansion of CAG repeat tracts in the codifying regions of nine, otherwise unrelated, genes. While the protein products of these genes are suggested to play diverse cellular roles, the pathogenic mutant proteins bearing an expanded polyglutamine sequence share a tendency to self-assemble, aggregate and engage in abnormal molecular interactions. Understanding the shared paths that link polyglutamine protein expansion to the nervous system dysfunction and the degeneration that takes place in these disorders is instrumental to the identification of targets for therapeutic intervention. Among polyglutamine diseases, spinocerebellar ataxias (SCAs) share many common aspects, including the fact that they involve dysfunction of the cerebellum, resulting in ataxia. Our work aimed at exploring a putative new therapeutic target for the two forms of SCA with higher worldwide prevalence, SCA type 2 (SCA2) and type 3 (SCA3), which are caused by expanded forms of ataxin-2 (ATXN2) and ataxin-3 (ATXN3), respectively. The pathophysiology of polyglutamine diseases has been described to involve an inability to properly respond to cell stress. We evaluated the ability of GTPase-activating protein-binding protein 1 (G3BP1), an RNA-binding protein involved in RNA metabolism regulation and stress responses, to counteract SCA2 and SCA3 pathology, using both in vitro and in vivo disease models. Our results indicate that G3BP1 overexpression in cell models leads to a reduction of ATXN2 and ATXN3 aggregation, associated with a decrease in protein expression. This protective effect of G3BP1 against polyglutamine protein aggregation was reinforced by the fact that silencing G3bp1 in the mouse brain increases human expanded ATXN2 and ATXN3 aggregation. Moreover, a decrease of G3BP1 levels was detected in cells derived from patients with SCA2 and SCA3, suggesting that G3BP1 function is compromised in the context of these diseases. In lentiviral mouse models of SCA2 and SCA3, G3BP1 overexpression not only decreased protein aggregation but also contributed to the preservation of neuronal cells. Finally, in an SCA3 transgenic mouse model with a severe ataxic phenotype, G3BP1 lentiviral delivery to the cerebellum led to amelioration of several motor behavioural deficits. Overall, our results indicate that a decrease in G3BP1 levels may be a contributing factor to SCA2 and SCA3 pathophysiology, and that administration of this protein through viral vector-mediated delivery may constitute a putative approach to therapy for these diseases, and possibly other polyglutamine disorders.
Keywords: G3BP1, stress granules, spinocerebellar ataxia, neurodegeneration
Koppenol et al. show that overexpression of G3BP1 in cell models of SCA2 and SCA3 leads to a reduction in ataxin-2 and ataxin-3 aggregation. G3BP1 lentiviral delivery reduces motor deficits and neuropathology in preclinical models, suggesting that G3BP1 may be a potential therapeutic target for polyQ disorders.
Introduction
Polyglutamine (polyQ) diseases are a group of hereditary neurodegenerative disorders that includes Huntington’s disease, spinal bulbar muscular atrophy, dentatorubral-pallidoluysian atrophy and several spinocerebellar ataxias (SCA1, 2, 3, 6, 7 and 17). These diseases are characterized by abnormal expansions of the trinucleotide CAG in coding regions of each disease-associated gene, which encode for an expanded polyQ tract in the respective proteins. A central feature of these diseases is the aggregation of the mutant protein, which promotes aberrant interactions with other proteins and mRNAs, leading to the impairment of several cellular pathways and organelles.1 Nevertheless, the complete picture of the molecular events leading to selective neurodegeneration of specific brain regions is yet to be fully understood. Moreover, until now there have been no therapies that could stop or delay the disease progression that culminates in the premature death of patients with polyQ diseases.
SCA2 and SCA3 (or Machado–Joseph disease, MJD) are two of the most prevalent SCAs and are both characterized by a neurodegenerative profile that mainly affects the cerebellum and the brainstem. SCA2 is caused by the abnormal expansion of the ATXN2 gene above 31–33 CAG repeats, resulting in an overexpanded ataxin-2 (ATXN2) protein.2 SCA3 is caused by mutant forms of the ATXN3 gene bearing above 44–45 CAG repeats, causing the ataxin-3 (ATXN3) protein to be abnormally expanded.3,4 Both mutant ATXN2 and ATXN3 are prone to aggregate and form large inclusions capable of sequestering other proteins. Although large inclusions are often reported as hallmarks of these diseases, whether they are directly leading to toxicity is still a matter of debate.5–8
The pathological aggregation of polyQ proteins and the abnormal interactions in which they engage can result in significant changes in the cellular stress response pathways.9,10 To cope with stress, cells display several mechanisms that promote survival, including the assembly of stress granules (SGs). These are transiently formed foci that act in the triage and regulation of RNA during stress periods.11 Recently, SG dysregulation has been suggested to underlie the pathogenesis of several diseases, including neurodegenerative disorders.12 SGs are dynamic, phase-separated structures that can have different compositions depending on the type of stress and type of cell, although RNA-binding proteins (RBPs) are their main components. One of these components, which is also a core nucleator and marker of SGs, is the GTPase-activating protein-binding protein 1 (G3BP1).13,14 G3BP1 has an important role in mRNA stabilization, degradation and in splicing modulation.15–19 Structurally, G3BP1 has at least two important domains, an RNA-recognition motif (RRM) domain and a nuclear transport factor 2-like domain (NTF2-like). The former is crucial for G3BP1 ability to bind mRNAs, whereas the latter is involved in the import of proteins through the nuclear pore complex.19 Additionally, phosphorylation of G3BP1 at its serine 149 residue has been referred to be important for SG assembly,14 although recent studies do not support this hypothesis.20 Importantly, the role of these domains and this phosphorylation site in specific steps of RNA metabolism is yet to be elucidated.13,14
Therefore, in this work, we aimed to investigate the involvement of G3BP1 in the pathogenesis of SCA2 and SCA3, and its suitability as a target for therapy in the context of these diseases. We determined that G3BP1 overexpression led to a significant reduction in the number of cells with aggregates and in the levels of ATXN2 and ATXN3 proteins. The NTF2-like domain and S149 residue seem important in this mechanism of action of G3BP1. Moreover, we found that G3BP1 levels are reduced in SCA2 and SCA3 patients’ samples. Importantly, in lentiviral mouse models of SCA2 and SCA3, the knockdown of G3BP1 increased the number of aggregates, highlighting the functional role of this protein in the context of SCA2 and SCA3 pathogenic alterations. Conversely, re-establishment of G3BP1 levels in the same models reduced neuropathological anomalies associated with the expression of mutant polyQ proteins. In the same line, G3BP1 expression significantly reduced behaviour and neuropathological deficits in a transgenic SCA3 mouse model. Altogether, the results of this work identify G3BP1 as a relevant target in the context of SCA2 and SCA3 pathogenesis, whose modulation could be explored as a putative therapeutic strategy for both diseases.
Materials and methods
Plasmid vectors
Plasmids encoding for human ATXN3 contain 28 glutamines (pEGFP-C1-Ataxin3Q28; #22122; Addgene) or 84 glutamines (pEGFP-C1-Ataxin3Q84; #22123; Addgene) were a gift from Henry Paulson and both are fused with a GFP protein at the N terminus.21 Plasmids encoding for human ATXN2 containing 22 glutamines (pEGFP-Ataxin2Q22) or 104 glutamines (pEGFP-Ataxin2Q104) were kindly provided by Professor Stefan Pulst.22 The LacZ gene was cloned in our laboratory under the control of a phosphoglycerate kinase promoter,23 and the GFP construct was cloned as previously described.24 The plasmid encoding for human G3BP1 (GeneBank accession DQ893058.2) purchased from Source Bioscience, was cloned into a lentiviral vector backbone using the GatewayTM LR ClonaseTM II Enzyme Mix, Invitrogen, according to the manufacturer’s instructions. The G3BP1-ΔNTF2 (G3BP1 deleted at the site 11–133) and G3BP1-ΔRRM (G3BP1 deleted at the site 340–415) constructs were synthesized by GeneScript and cloned into the vector pcDNA3.1 + N-MYC. A validated short-hairpin RNA (shRNA) targeting mouse G3bp1 (#MSH031039-LVRU6MP-b) and an shRNA scramble, as control (with no known target, #CSHCTR001-LVRU6MP) were acquired from GeneCopoeia.
G3BP1 mutagenesis of the serine149 residue
Site-directed mutagenesis was performed using the NZY Mutagenesis kit (NZYTech) according to the manufacturer’s instructions. In the human variant of G3BP1 (GeneBank accession DQ893058.2), a serine was changed by alanine or aspartate at the site 149 to generate a G3BP1 phospho-dead mutant (G3BP1_S149A) or a G3BP1-phosphomimetic mutant (G3BP1_S149D), respectively. The pair of primers used to induce the substitution S149A were: 5′–CT GAG CCT CAG GAG GAG GCT GAA GAA GAA GTA GAG–3′ and 5′–CT CTA CTT CTT CTT CAG CCT CCT CCT GAG GCT CAG–3′. The pair of primers used to induce the substitution S149D were: 5′–CT GAG CCT CAG GAG GAG GAT GAA GAA GAA GTA GAG–3′ and 5′–CTC TAC TTC TTC TTC ATC CTC CTC CTG AGG CTC AG–3′. The mutations S149A and S149D were confirmed by DNA sequencing (Eurofins Genomics).
Neuroblastoma culture and transfection
A mouse neuroblastoma cell line (Neuro2a cells) acquired from the American Type Culture Collection cell biology bank (CCL-131), was cultured in Dulbecco's modified Eagle medium, supplemented with 10% (v/v) foetal bovine serum, 100 U/ml penicillin and 100 µg/ml streptomycin. Cells were seeded onto 12- or 6-multiwell plates. After 24 h of growth, cells were transfected with 0.5–1 µg of DNA per well, using polyethyleneimine reagent (PEI; PEI MAX Polysciences, Inc.), following the manufacturer’s instructions. For SG induction experiments, cells were treated with sodium arsenite (SA) (Sigma-Aldrich 10 µg/ml) to a final concentration of 0.05 M 1 h before harvest.
Human fibroblast culture
Patients’ fibroblasts from SCA2, SCA3 and healthy individuals were obtained from Coriell Institute or kindly provided by collaborators,25,26 and are fully characterized for CAG expansions: SCA2 (Patient 1, 22/41; Patient 2, 20/44); SCA3 (Patient 1, 18/79; Patient 2, 22/77; Patient 3, 23/80; Patient 4, 23/71; Patient 5, 24/74); healthy controls (1, 14/19; 2, 14/23; 3, 22/23; 4, 22/23). Fibroblasts were kept in culture in Dulbecco's modified Eagle medium, supplemented with 15% (v/v) foetal bovine serum, 100 U/ml penicillin and 100 µg/ml streptomycin. All cell cultures were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Translation rate assay:SUnSET protocol
We used a method that allows the monitoring and quantification of global protein synthesis based on the incorporation of puromycin during translation.27 N2a cells were plated into multiwell plates and transfected with lacZ or G3BP1. Twenty-four hours post-transfection, cells were incubated with 10 μg/ml of puromycin (Sigma) for 15 min, and then collected for western blot processing. As a positive control for the translation inhibition, some cells were incubated with 10 mM of cycloheximide (Sigma) for 15 min, and then incubated with 10 mg/ml of puromycin (Sigma) for an additional 15 min. For the SG condition, cells were treated for 1 h with 0.05 M SA and then incubated with 10 mg/ml of puromycin (Sigma) for an additional 15 min. Additional controls of non-treated cells were also used.
Human brain tissue
Post-mortem striatal and cerebellar tissues from clinically and genetically confirmed SCA2 patients were obtained from the NIH NeuroBioBank. Control striatal and cerebellar tissues from healthy individuals, without neurological conditions diagnosed, were obtained from NIH NeuroBioBank. Tissues were preserved in a 4% (w/v) paraformaldehyde solution, dehydrated in 30% (w/v) sucrose/phosphate buffer solution (PBS) for 48 h and cryoprotected at −80°C. Tissues were sectioned in 40-µm thick slices using a cryostat (Cryostar NX50, Thermo Fisher Scientific), and stored at 4°C, free-floating in 0.02% (w/v) sodium azide/PBS.
Animals
Adult C57BL/6J wild-type animals and transgenic SCA3 mice,23 bred in the animal facility of the Universidade do Algarve, were used. Animals were maintained in a temperature-controlled room on a 12 h light/12 h dark cycle. Food and water were dispensed ad libitum. All experiments were carried out according to the European Community Council directive (86/609/EEC) for the care and use of laboratory animals. The researchers received certified training (FELASA course) and approval to perform the experiments from the Portuguese authorities (Direcção Geral de Alimentação e Veterinária) in the project Neuropath (421/2019).
Lentiviral vectors
The cDNA encoding for human G3BP1, GFP, ATXN2MUT and for ATXN3MUT were cloned in a self-inactivating lentiviral vector under the control of the phosphoglycerate kinase promoter, as described previously.28 Lentiviral vectors were produced in human embryonic kidney 293T cells using a four-plasmid system described previously.29 Viral productions were quantified using a RetroTek HIV-1 p24 Antigen Enzyme-Linked Immunoabsorbent Assay (ZeptoMetrix), according to the manufacturer’s instructions.
In vivo injection of lentiviral vectors
For the stereotaxic injection of lentiviral vectors, concentrated viral stocks were thawed on ice and homogenized. Animals were anaesthetized by intraperitoneal injection of a mixture of ketamine (75 mg/kg, Nimatek, Dechra) with medetomidine (0.75 mg/kg, DOMITOR®, Esteve). For the SCA2 lentiviral mouse model, mice (10–12 weeks old) were injected with lentiviral particles encoding for human ATXN2MUT containing 82 glutamines or encoding for ATXN2MUT and G3BP1 at the left and right hemispheres of the striatum, respectively. Brain coordinates were used relative to bregma as follows: antero-posterior (+0.6 mm), medial-lateral (±1.8 mm) and dorsal-ventral (−3.3 mm).30 Lentiviruses were injected at a concentration of 400 ng p24/µl, at a rate of 0.20 µl/min. For the SCA3 lentiviral mice, viral particles encoding for human ATXN3MUT containing 72 glutamines or encoding for ATXN3MUT and G3BP1, were injected into the mouse striatum (left and right hemispheres, respectively) at a concentration of 400 ng of p24/µl, using the same coordinates described before. To perform safety assays, wild-type C57/BL6 mice (10–12 weeks old) were injected into the striatum with lentiviral particles encoding for G3BP1 at a concentration of 400 ng p24/µl of lentivirus, using the same coordinates described before. For the transgenic animals, lentiviral particles encoding for G3BP1 or GFP, as respective controls, were injected into mice cerebella (4 weeks old), at a concentration of 800 ng p24/µl of lentiviruses at the coordinates: −1.6 mm rostral to lambda, 0.0 mm midline and −1.0 mm ventral to the skull surface, with the mouth bar set at −3.3.24 For the G3bp1 silencing studies in SCA2, wild-type C57/BL6 mice (10–12 weeks old) were injected in the striatum with lentiviral particles encoding for human ATXN2MUT containing 82 glutamines and with lentiviral particles encoding for an shRNA scramble, whereas in the contralateral hemisphere the animals were injected with lentiviral particles encoding for human ATXN2MUT containing 82 glutamines and with lentiviral particles encoding for an shRNA targeting mouse G3bp1. For SCA3, the procedure was similar, but we used lentiviral particles encoding for human ATXN3MUT containing 72 glutamines. The lentiviral particles were injected at a concentration of 400 ng p24/µl of lentiviruses, using the same coordinates described before.
All stereotaxic injections were performed with an automatic injector (Stoelting Co.) using a 34-gauge blunt-tip needle linked to a Hamilton syringe. Mice were sacrificed for posterior analysis, a few weeks after surgery, according to the model: SCA2 lentiviral mice, 4 weeks and 12 weeks; SCA3 lentiviral mice, 4 weeks; G3BP1-injected mice, 4 weeks and SCA3 transgenic mice, 9 weeks.
Behavioural testing
Transgenic mice were subjected to several motor behaviour tests starting before the stereotaxic injection (4 weeks of age), every 3 weeks until 9 weeks post-injection. Motor and gait coordination were blindly assessed by the rotarod and footprint tests, following the same procedure described before.24 In the footprint test analysis, steps taken by mice at the beginning and at the end of the walking test were not considered for the measures. Swimming performance was assessed by placing mice at one end of a rectangular tank (100 × 10.5 × 20 cm) filled with water at room temperature. Mice freely swam for 1 m until they reached a platform and the time taken to transverse the tank was recorded. Mice performed the trial three times, with an interval of 15–20 min per trial. The mean of the time taken to cross the tank in the three trials was used for statistical analysis.
Tissue processing
For immunohistochemical assays, animals were sacrificed by sodium pentobarbital overdose and transcardially perfused with 0.1 M PBS and a 4% (w/v) paraformaldehyde fixative solution (Sigma-Aldrich). For quantitative PCR (qPCR) and western blot analysis, mice were euthanized by cervical dislocation and striatal punches were performed using a Harris Core pen with 2.5 mm diameter for tissue harvesting. The brains collected were post-fixed in 4% paraformaldehyde for 24 h, dehydrated in a 20% sucrose/0.1 M PBS for 48 h and cryoprotected at −80°C. Sagittal or coronal brain sections of 30 and 25 µm thicknesses, respectively, were obtained using the cryostat-microtome model CryoStar NX50 (Thermo Fisher). For preservation, brain sections were stored at 4°C, free-floating in 0.05% (w/v) sodium azide PBS.
Cresyl violet staining
To stain brain sections with cresyl violet, they were mounted in gelatin-coated microscope slides. Brain sections were sequentially submerged in water, ethanol 96% (v/v), ethanol 100% (v/v), xylene, ethanol 75% (v/v) and the 0.1% (w/v) cresyl violet solution. To wash slices, brain sections were sequentially submerged in water, ethanol 75% (v/v), ethanol 96% (v/v), ethanol 100% (v/v) and xylene. Finally, brain sections were mounted with Eukitt (Sigma-Aldrich). Images were acquired with a 10× objective in a Zeiss Axio Imager Z2.
Immunocytochemistry
For the immunocytochemical procedure, cells were fixed using a 4% (w/v) paraformaldehyde fixative solution for 20 min and washed with 0.1 M PBS. Samples were then incubated in PBS containing 0.1% TritonTM X-100 for 10 min. Blocking in PBS with 3% of bovine serum albumin (BSA) (Sigma) was performed for 1 h. Samples were incubated with the primary antibody overnight in the proper dilution at 4°C and with the secondary antibody (1:200) for 2 h at room temperature. The secondary antibody was coupled to a fluorophore (Alexa Fluor®, Invitrogen). Finally, the coverslips were mounted on microscope slides using Fluoromount-G mounting media with DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen).
Immunohistochemistry
The immunohistochemical procedure, for light imaging, began with the incubation of brain sections in phenylhydrazine diluted in PBS (1:1000; 15 min, 37°C). For the human brain sections, an additional step with a Tris-buffered saline (TBS) pH 9 antigen retrieval method (30 min, 95°C) was performed. Brain sections were blocked in a 10% (v/v) normal goat serum in 0.1% (v/v) TritonTM X-100/PBS solution (1 h, room temperature) and incubated with the respective primary (overnight at 4°C) and secondary biotinylated antibodies (2 h at room temperature) diluted in blocking solution, followed by a reaction with the Vectastain elite avidin-biotin-peroxidase kit and the 3,3′-diaminobenzidine substrate (both from Vector Laboratories). Then, the sections were assembled on microscope slides, dehydrated in ethanol solutions with increasing concentrations (75%, 96% and 100%) and xylene, and finally cover slipped using the mounting medium Eukitt (O. Kindler GmbH & Co). For fluorescence immunohistochemistry procedures, brain sections were incubated in the previously described blocking solution, followed by incubation of primary and secondary antibodies. Brain sections were mounted on microscope slides with Fluoromount-G mounting medium with DAPI (Invitrogen).
Immunochemical antibodies
For immunochemical procedures, the following primary antibodies were used: mouse anti-ataxin-2 (1:1000, ref. 611378, BD Biosciences); mouse anti-ubiquitin (1:1000, ref. 3936S, Cell Signaling) rabbit anti-DARPP-32 (1:1000, ref. AB10518, Merck Millipore); rabbit anti-G3BP1 (1:1000, ref. 07-1801, Millipore); mouse anti-human G3BP1 (1:1000, ref. 611126, BD Biosciences); anti-G3BP1 (1:1000, ref. 05-1938; Sigma-Aldrich); mouse anti-GFAP (1:1000, ref. 644702, BioLegend); rabbit anti-HA (1:1000, ref. Ab9110, Abcam); mouse anti-calbindin D-28K (1:1000, ref. C9848, Sigma-Aldrich); mouse anti-PABP-1 (1:1000, ref. 04-1467, Millipore) and mouse β-Gal (14B7) (1:500, ref. 2372, Cell Signaling Technology). For fluorescence immune procedures, the following antibodies were used: anti-mouse AlexaFluor 647 (1:200, ref. A21235, Invitrogen), anti-mouse AlexaFluor 488 (1:200, ref. A11001, Invitrogen), anti-rabbit AlexaFluor 488 (1:200, ref. A11008, Invitrogen), anti-mouse AlexaFluor 594 (1:200, ref. A11005, Invitrogen) and anti-rabbit AlexaFluor 594 (1:200, ref. A11012, Invitrogen). For light imaging, the following antibodies were used: anti-mouse biotinylated (1:200, ref. BA-9200, Vector Laboratories) and anti-rabbit biotinylated (1:200, ref. BA-1000, Vector Laboratories).
Image quantitative analysis and data processing
Immunocytochemistry images were acquired in a Zeiss Axio Imager Z2 for quantification and in a Zeiss LSM710 confocal microscope for representative images. Quantitative analysis was blindly performed by counting the number of cells with aggregates within 100 transfected cells, using the 40× or 63× objective for each condition in each independent experiment. Immunohistochemistry images from the lentiviral mouse models were acquired with 20× objective in a Zeiss Axio Imager Z2 and Axio Scan.Z1 Slide Scanner microscopes. For quantification of ATXN2 aggregates and DARPP-32 staining loss, 18 coronal sections per animal were analysed in ZEN lite software (Zeiss), so that a complete rostrocaudal picture of the striatum was obtained. ATXN2 inclusions were manually counted in all animals. Neuronal lesion was manually assessed for all animals by measuring the area of DARPP-32 staining loss, according to the formula: volume = d*(a1 + a2 + a3), where d is the distance between serial sections (200 μm) and a1 + a2 + a3 are depleted areas for each individual section. For the transgenic mice, eight sagittal sections covering the entire cerebellum were used, spanning 280 μm between them, for immunohistochemistry procedures. These sections were stained with anti-HA, anti-Calbindin and DAPI, and images were acquired using a Zeiss Axio Imager Z2 microscope using a 20× objective. For each section, the number of cells with HA-positive aggregates and Purkinje cells number was blindly counted in all cerebellar lobules using image analysis software (ZEN 2.1 lite, Zeiss).
Western blot
Samples were either lysed in 1× RIPA solution (Merck Millipore) when using cell extracts, or homogenized in a urea/DTT solution when using mouse striatal punches: both containing a cocktail of protease inhibitors (Roche). Following that, samples were then subjected to five cycles of ultrasound sonication of 30 s on and 30 s off (Bioruptor Pico). Protein concentration was determined using the Pierce™ BCA Protein Assay Kit (Thermo Scientific) for cell lysates and the NZYBradford reagent (Nzytech) for mouse samples. Equal amounts of protein extracts were resolved in sodium dodecyl sulphate–polyacrylamide gels (7.5 and 12%), followed by protein transfer to PVDF membranes (Merck Millipore), membrane blocking in 0.1% (v/v) Tween-20/TBS containing 3% of BSA or 5% of milk, and antibody probing overnight at 4°C for primary and 2 h at room temperature for secondary. The following antibodies were used: mouse anti-ataxin-2 (1:1000, ref. 611378, BD Biosciences); mouse anti-ataxin-3(1H9) (1:1000, ref. MAB5360, BD Biosciences); rabbit anti-G3BP1 (1:1000, ref. 07-1801, Millipore); mouse anti-G3BP1 (1:1000, ref. 05-1938; Sigma-Aldrich); mouse anti-β-actin (1:5000, ref. A5316, Sigma-Aldrich) mouse anti-β-tubulin (1:5000, ref. T7816, Sigma); mouse anti-puromycin (1:250, ref. MABE343, Millipore); mouse anti-GFP (1:1000, ref. 668205, BioLegend); mouse anti-β-Gal (14B7) (1:500, ref. 2372, Cell Signaling Technology); rabbit anti-Phospho-eIF-2α (1:1000, ref. 07-760-I, Millipore) and rabbit anti-eIF-2α (1:1000, ref. 324, Cell Signalling). The following secondary antibodies were used: anti-mouse ECL Horseradish peroxidase (1:10 000, ref. NA931V, GE Healthcare), and anti-rabbit ECL Horseradish peroxidase (1:10 000, ref. NA934, GE Healthcare). Membranes were imaged using Enhanced Chemiluminescence (GE Healthcare) and scanned with a ChemiDoc™ XRS + (Bio-Rad). Optical densitometric analysis was carried out using ImageJ software.
Reverse transcription quantitative PCR
Total RNA from mouse striatal punches started with TRIzol (Invitrogen) tissue dissociation and RNA/DNA/protein separation using chloroform. Then, both mouse and cell samples were extracted with NZY Total RNA Isolation kit (Nzytech). RNA concentration and purity were determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). cDNA molecules were obtained from 1 µg of RNA using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer’s recommendations. Reverse transcription quantitative PCR (RT-qPCR) was performed with the SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad), using primers made in-house for the gene of interest and for the human GAPDH housekeeping gene as control, and performed in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). We determined mRNA expression levels using amplification values and levels were normalized to the mRNA expression levels of the housekeeping gene. The following primers were used: human ATXN2 (QT01852480) and human ATXN3 (QT00094927) from QuantiTect Primer Assays, Qiagen. Human G3BP1 (Forward 5′–GAA ATC CAA GAG GAA AAG CC–3′; Reverse 5′–CCC AAG AAA ATG TCC TCA AG), human GAPDH (Forward 5′–ACA GTT GCC ATG TAG ACC–3′; Reverse 5′–TTG AGC ACA GGG TAC TTT A–3′) and mouse Hprt (Forward 5′–AGG GAT TTG AAT CAC GTT TG–3′; Reverse 5′–TTT ACT GGC AAC ATC AAC AG–3′) from KiCqStart Pre-designed Primers, Sigma-Aldrich.
Statistical analysis
Statistical analysis was performed using either Student’s t-test or one-way ANOVA complemented with Bonferroni multiple comparisons test, resorting to GraphPad software (La Jolla).
Data availability
The data that support the findings of this study are available from the corresponding author, on reasonable request.
Results
Stress granule assembly does not alter the levels of ATXN2 and ATXN3 proteins
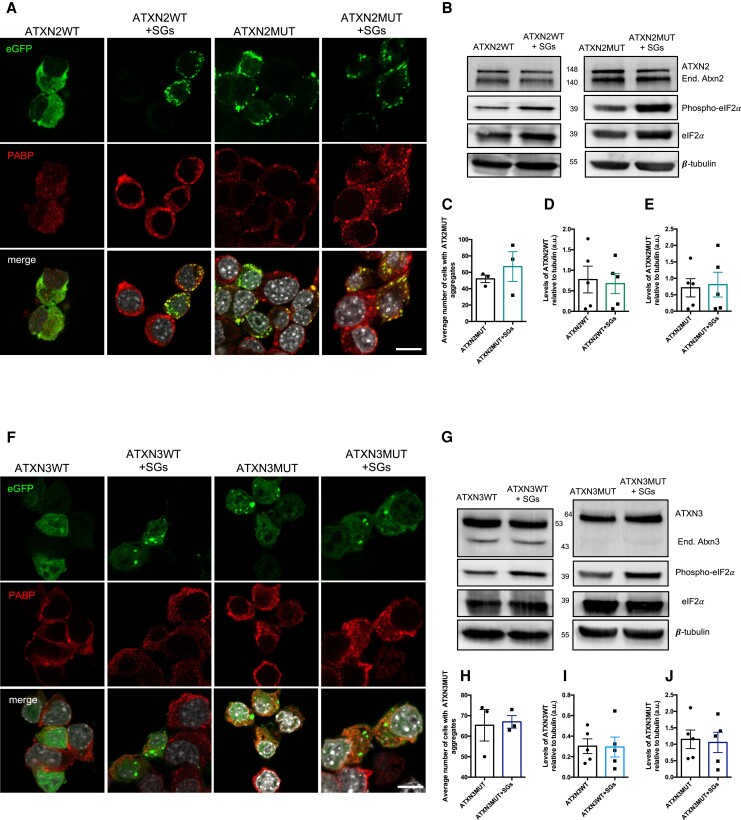
G3BP1 is a core nucleator of SGs, a type of cellular foci formed in response to stress in which mRNAs, translation factors and RBPs coalesce together to prevent cellular damage.31,32 Therefore, we aimed to investigate the impact of SG assembly on ATXN2 and ATXN3 protein dynamics, both for their pathological (ATXN2MUT and ATXN3MUT) and non-pathological forms (ATXN2WT and ATXN3WT). For that, in Neuro2a cells expressing ATXN2 (ATXN2WT: pEGFP-ATXN2-Q22 or ATXN2MUT: pEGFP-ATXN2-Q104) or ATXN3 (ATXN3WT: pEGFP-ATXN3-Q24 or ATXN3MUT: pEGFP-ATXN3-Q844), SG assembly was pharmacologically induced using SA (Supplementary Fig. 1A). As previously reported, ATXN2 was recruited to SGs,33 detected as polyadenylate-binding protein 1 (PABP)-positive foci; however, ATXN3 was not. As expected, the mutant forms of both proteins accumulated in the form of aggregates (Fig. 1A and F). However, stimulation of SG assembly did not alter the number of cells with ATXN2MUT or ATXN3MUT aggregates, compared to the control conditions (ATXN2MUT and ATXN3MUT, respectively), in which the stress stimulus was not induced (Fig. 1Cand H). While the non-pathological forms of ATXN2 or ATXN3 did not form aggregates, the induction of SG assembly led to the formation of aggregate-like structures in both the ATXN2WT- and ATXN3WT-expressing conditions (Fig. 1A and F).
Figure 1.
SG assembly mediated by SA does not alter the number of aggregates or protein levels of ATXN2 and ATXN3. (A) Confocal microscopy representative images from Neuro2a cells expressing pathological and non-pathological forms of ATXN2, fused with an eGFP tag (top row), treated with SA to induce SG assembly and stained with antibodies against PABP, an SG marker (middle row). (B) Representative western blot of Neuro2a lysates expressing pathological and non-pathological forms of ATXN2 and treated SA to induce SG assembly. Western blots were labelled using ATXN2, phospho-eIF2α, and β-tubulin antibodies. (C) The number of cells with ATXN2 aggregates was not significantly altered on SG assembly (n = 3 independent experiments). (D) The levels of non-pathological ATXN2 (ATXN2WT) protein were not significantly altered on SG assembly. (E) The levels of pathological ATXN2 (ATXN2MUT) protein were not significantly altered on SG assembly (n = 5 independent experiments). (F) Confocal microscopy representative images from Neuro2a cells expressing pathological and non-pathological forms of ATXN3, fused with an eGFP tag (top row), treated with SA to induce SG assembly and stained with antibodies against PABP1. (G) Representative western blot of Neuro2a lysates expressing pathological and non-pathological forms of ATXN3 and treated SA to induce SG assembly. Western blots were labelled using ATXN3, eIF2α, phospho-eIF2α and β-tubulin antibodies. (H) The number of cells with ATXN3 aggregates was not significantly altered on SG assembly (n = 3 independent experiments). (I) The levels of non-pathological ATXN3 (ATXN3WT) protein were not significantly altered on SG assembly. (J) The levels of pathological ATXN3 (ATXN3MUT) protein were not significantly altered on SG assembly (n = 5 independent experiments). Values are expressed as mean ± SEM. Scale bar = 10 µm.
SG assembly is accompanied by the phosphorylation of eIF2α, and translation inhibition,34 leading to a reduction in overall protein synthesis (Supplementary Fig. 1Band C). Thus, we next investigated whether the levels of ATXN2 and ATXN3 proteins were altered on SG assembly, by analysing their levels through western blot (Fig. 1B and G). In line with the results obtained for the aggregates, we did not observe alterations in the levels of ATXN2 or ATXN3 when SG formation was induced, compared to controls, neither in the non-pathological (Fig. 1D and I) nor in the pathological (Fig. 1E and J) protein forms.
Altogether, these results show that, although SG assembly reduces overall protein translation, it does not interfere with the expression of ATXN2 and ATXN3 proteins nor with their aggregation.
G3BP1 overexpression reduces the number of cells with aggregates and the levels of ATXN2 and ATXN3 proteins
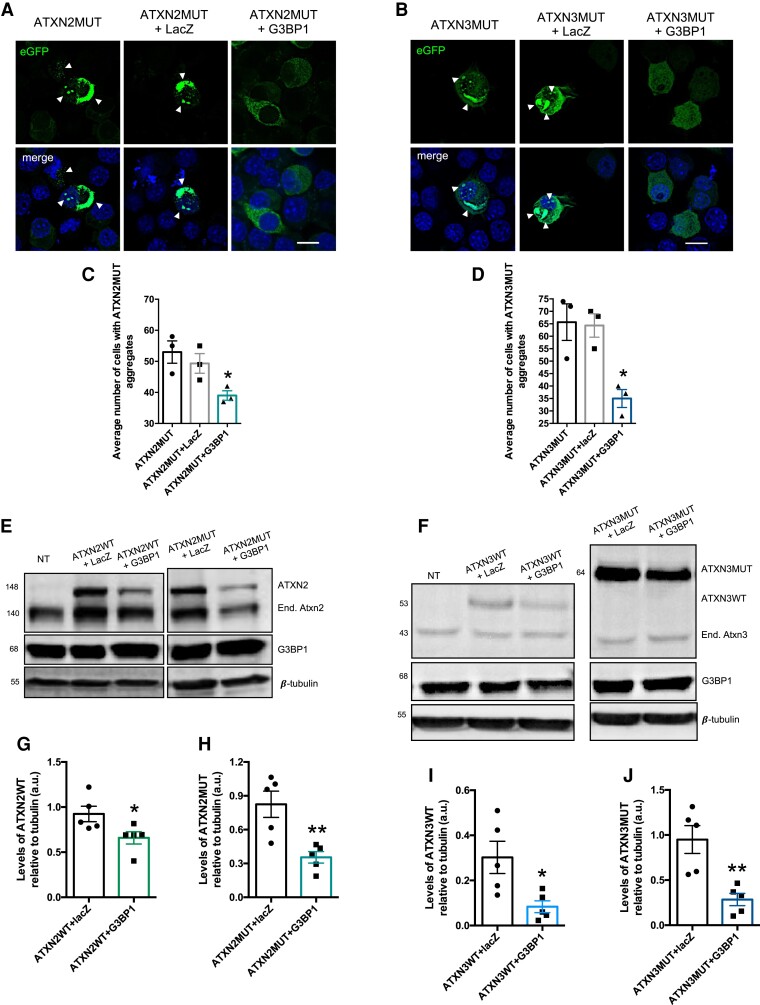
As observed for SGs, G3BP1 overexpression also leads to an inhibition of protein synthesis, although at a lower degree (Supplementary Fig. 1B and D). The increase in G3BP1 protein levels is around 60%, compared to non-transfected controls (Supplementary Fig. 2A and B). Taking this into consideration, we next aimed to investigate the impact of G3BP1 overexpression on ATXN2MUT and ATXN3MUT protein aggregation. To achieve this goal, we co-transfected Neuro2a cells with ATXN2MUT or ATXN3MUT and G3BP1, and as controls cells were co-transfected with ATXN2MUT or ATXN3MUT and lacZ or transfected only with ATXN2MUT or ATXN3MUT (Supplementary Fig. 4). As described before, expression of the mutant forms of both proteins led to the formation of aggregates, which are a hallmark of polyQ diseases (Fig. 2A and B). We found that G3BP1 overexpression was able to significantly reduce the number of cells with aggregates of both ATXN2MUT (ATXN2MUT + G3BP1: 0.39 ± 0.0153 versus ATXN2MUT: 0.53 ± 0.036, n = 3, P = 0.0233) and ATXN3MUT (ATXN3MUT + G3BP1: 0.35 ± 0.036 versus ATXN3MUT: 0.66 ± 0.073, n = 3, P = 0.0201), compared to control conditions (Fig. 2C and D).
Figure 2.
The expression of G3BP1 reduces the number of cells with aggregates of ATXN2MUT and ATXN3MUT. (A) Confocal microscopy representative images depicting Neuroa2 cells expressing ATX2MUT in three different experimental conditions. (B) Confocal microscopy representative images depicting Neuroa2 cells expressing ATX3MUT in three different experimental conditions. (C) The number of cells with aggregates of ATX2MUT per 100 transfected cells was significantly reduced on G3BP1 expression, compared to both control conditions. (D) The number of cells with aggregates of ATX3MUT per 100 transfected cells was significantly reduced on G3BP1 expression, compared to both control conditions (n = 3 independent experiments; *P < 0.05; one-way ANOVA followed by post hoc Bonferroni multiple comparisons test). Values are expressed as mean ± SEM. Scale bar = 10 µm. (E) Representative western blot of Neuro2a lysates expressing pathological and non-pathological forms of ATXN2, co-transfected with lacZ or with G3BP1. Western blots were labelled using ATXN2, G3BP1 and β-tubulin
antibodies. (F) Representative western blot of Neuro2a lysates expressing pathological and non-pathological forms of ATXN3, co-transfected with lacZ or with G3BP1. Western blots were labelled using ATXN3, G3BP1 and β-tubulin
antibodies. (G) The levels of ATXN2WT protein are significantly reduced on G3BP1 expression, compared with control cells co-expressing lacZ. (H) The levels of ATXN2MUT protein are significantly reduced on G3BP1 expression, compared with control cells co-expressing lacZ. (I) The levels of ATXN3WT protein are significantly reduced on G3BP1 expression, compared with control cells co-expressing lacZ. (J) The levels of ATXN3MUT protein are significantly reduced on G3BP1 expression, compared with control cells co-expressing lacZ (n = 5 independent experiments; *P < 0.05; **P < 0.01; Student’s t-test).
Next, we investigated whether the observed reduction in aggregation on G3BP1 overexpression (Supplementary Fig. 2) could be associated with a reduction in the protein levels of ATXN2MUT and ATXN3MUT (Fig. 2E and F). Furthermore, we also analysed the impact of G3BP1 overexpression on the levels of the non-pathological forms of the proteins ATXN2WT and ATXN3WT, respectively (Fig. 2E and F). We found that G3BP1 overexpression was able to significantly reduce the expression levels of both ATXN2WT (ATXN2WT + G3BP1: 0.65 ± 0.06 versus ATXN2WT + lacZ: 0.692 ± 0.08, n = 5, P = 0.04) and ATXN2MUT (ATXN2MUT + G3BP1: 0.35 ± 0.1343 versus ATXN2MUT + lacZ: 0.82 ± 0.116 n = 5, P = 0.0076) (Fig. 2G and H). In the same line, we also observed a significant reduction in ATXN3WT and ATXN3MUT levels on G3BP1 overexpression, compared to control conditions (ATXN3WT + lacZ: 0.608 ± 0.026 versus ATXN3WT + G3BP1: 0.3 ± 0.071, n = 5, P = 0.02 and ATXN3MUT + G3BP1: 0.28 ± 0.067 versus ATXN3MUT + lacZ: 0.95 ± 0.154 n = 5, P = 0.004, respectively) (Fig. 2I and J). However, no alteration was observed in mouse endogenous levels (Neuro2a cells) of ATXN2 and ATXN3 on G3BP1 overexpression (Supplementary Fig. 3). Moreover, in an additional control experiment, GFP levels were not altered when G3BP1 was overexpressed (Supplementary Fig. 4). Altogether, these results show that G3BP1 overexpression reduces the levels of ATXN2 and ATXN3 proteins and decreases aggregation of their mutant forms.
Overexpression of core components of SGs, including G3BP1, can induce the formation of these foci.14,35 However, we observed that, in Neuro2a cells, G3BP1 overexpression alone is less effective at inducing SG formation than when combined with a SA stimulus (Supplementary Fig. 5). In the same line, in fibroblasts from SCA2 and SCA3 patients, G3BP1 has a diffuse expression, which is also observed in healthy fibroblasts. On the contrary, G3BP1 condensates in PABP-positive foci on SA treatment (Supplementary Fig. 6).
The NTF2-like domain is important for G3BP1 action on ATXN2 and ATXN3 mutant proteins
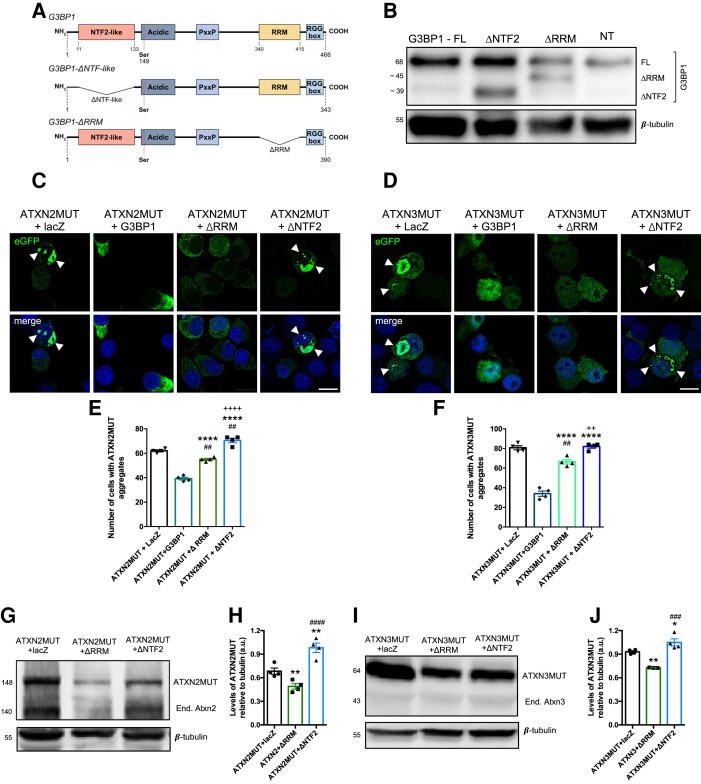
G3BP1 is an RBP with several molecular and biological functions, including mRNA binding, DNA binding36 and helicase activity, and has important roles in immune response.37 Overall, RBPs, including G3BP1, interact with mRNAs through specific RNA-binding domains.16,38 The RRM of G3BP1 is known for interacting with target RNA sequences.39 G3BP1 also harbours a NTF2-like domain that is involved in the nuclear shuttling of proteins through the nuclear pore complex,40 facilitates protein–protein interactions,41 mediates G3BP1 dimerization and is important in SGs formation.14 Therefore, to better understand G3BP1 action on mutant ATXN2 and ATXN3 aggregation and protein levels, we developed two different forms of the protein, one in which the NTF2-like domain was deleted (G3BP1-ΔNTF2) and the other with a deletion of the RRM domain (G3BP1-ΔRRM) (Fig. 3A and B). Next, we co-transfected Neuro2a cells with the ATXN2MUT or ATXN3MUT and either G3BP1-ΔNTF2 or G3BP1-ΔRRM, or, alternatively, with full-length G3BP1 or lacZ, as controls (Fig. 3C and D). Co-expression of G3BP1-ΔRRM led to a significant decrease in the number of cells with aggregates of ATXN2MUT and ATXN3MUT, compared to the lacZ control condition (ATXN2MUT + G3BP1-ΔRRM: 55 ± 0.815 versus ATXN2MUT + lacZ: 62 ± 0.814, n = 4, P < 0.001 and ATXN3MUT + G3BP1-ΔRRM: 66.5 ± 2.305 versus ATXN3MUT + lacZ: 80.7 ± 2.37, n = 4, P < 0.001, respectively) (Fig. 3E and F). However, when compared to the expression of the full-length G3BP1, G3BP1-ΔRRM led to a significant increase in the number of cells with aggregates of ATXN2MUT and ATXN3MUT. On the contrary, the expression of G3BP1-ΔNTF2 led to an increase in the number of cells with aggregates of ATXN2MUT and ATXN3MUT, compared to both lacZ and full-length G3BP1 conditions (Fig. 3E and F).
Figure 3.
The NFT2-like domain of G3BP1 is important in the modulation of aggregation and protein levels of ATXN2MUT and ATXN3MUT. (A) Schematic representation of G3BP1 structural domains and the respective constructs with deleted NFT2 domain and deleted RRM domain. The delta symbol indicates deletion; PxxP = proline-rich region; RGG box = arginine and glycine rich box. (B) Neuro2a cells were transfected with full-length G3BP1, G3BP1-ΔRRM or G3BP1-ΔNTF2, and non-transfected (NT). Protein lysates were analysed through western blot depicting the expression of G3BP1 truncated forms with different molecular weight. Western blots were labelled using G3BP1, and β-tubulin antibodies. (C) Confocal microscopy representative images depicting Neuroa2 cells expressing ATXN2MUT and lacZ or G3BP1 or G3BP1-ΔNTF2 or G3BP1-ΔRRM. The expression of ATXN2MUT leads to the formation of aggregates (arrowheads). (D) Confocal microscopy representative images depicting Neuro2a cells expressing ATXN3MUT and lacZ or G3BP1 or G3BP1-ΔNTF2 or G3BP1-ΔRRM. The expression of ATXN3MUT leads to the formation of aggregates (arrowheads). Scale bar = 10 µm. (E) The number of cells with aggregates of ATX2MUT per 100 transfected cells with G3BP1-ΔRRM was significantly reduced compared with the lacZ control condition and increased compared to full-length G3BP1 condition. The expression of G3BP1-ΔNTF2 leads to a significant increase in the number of cells with aggregates compared to all the other experimental conditions (n = 4 independent experiments; ##P < 0.01 to ATXN2MUT + lacZ; ****P < 0.0001 to ATXN2MUT + G3BP1; ++++P < 0.0001 to ATXN2MUT + G3BP1-ΔRRM; one-way ANOVA, followed by post hoc Bonferroni multiple comparisons test). (F) The number of cells with aggregates of ATX3MUT per 100 transfected cells with G3BP1-ΔRRM was significantly reduced compared with the lacZ control condition and increased compared to full-length G3BP1 condition. The expression of G3BP1-ΔNTF2 leads to a significant increase in the number of cells with aggregates compared to ATXN3MUT + G3BP1 and ATXN3MUT + G3BP1-ΔRRM (n = 4 independent experiments; ##P < 0.01 to ATXN3MUT + lacZ; ****P < 0.0001 to ATXN3MUT + G3BP1; ++P < 0.01 to ATXN3MUT + G3BP1-ΔRRM; one-way ANOVA, followed by post hoc Bonferroni multiple comparisons test). (G) Representative western blot of Neuro2a lysates expressing ATXN2MUT, co-transfected with lacZ or G3BP1-ΔRRM or G3BP1-ΔNTF2. (H) The levels of ATXN2MUT protein are significantly reduced on G3BP1-ΔRRM expression compared to the other experimental conditions, whereas the expression of G3BP1-ΔNTF2 leads to a significant increase in ATXN2MUT protein levels, compared to the other conditions (n = 4 independent experiments; **P < 0.01 to ATXN2MUT + lacZ; ####P < 0.0001 to ATXN2MUT + G3BP1-ΔRRM; one-way ANOVA, followed by post hoc Bonferroni multiple comparisons test). (I) Representative western blot of Neuro2a lysates expressing ATXN3MUT, co-transfected with lacZ or G3BP1-ΔRRM or G3BP1-ΔNTF2. (J) The levels of ATXN3MUT protein are significantly reduced on G3BP1-ΔRRM expression compared to the other experimental conditions, whereas the expression of G3BP1-ΔNTF2 leads to a significant increase in ATXN3MUT protein levels, compared to ATXN3MUT + G3BP1-ΔRRM (n = 4 independent experiments; *P < 0.05 to ATXN3MUT + lacZ; **P < 0.01 to ATXN3MUT + lacZ; ###P < 0.001 to ATXN3MUT + G3BP1-ΔRRM; one-way ANOVA, followed by post hoc Bonferroni multiple comparisons test). Values are expressed as mean ± SEM.
Next, we analysed the levels of ATXN2MUT and ATXN3MUT on expression of both truncated forms of G3BP1 (Fig. 3G and I). We found that G3BP1-ΔRRM led to a significant reduction of the levels of ATXN2MUT and ATXN3MUT, compared to control (ATXN2MUT + G3BP1-ΔRRM: 0.48 ± 0.035 versus ATXN2MUT + lacZ: 0.64 ± 0.013, n = 4, P < 0.001, and ATXN3MUT + G3BP1-ΔRRM: 0.725 ± 0.001 versus ATXN3MUT + lacZ: 0.93 ± 0.012, n = 4, P < 0.001, respectively) (Fig. 3H and J). Conversely, the expression of G3BP1-ΔNTF2 led to a significant increase in the levels of ATXN2MUT and ATXN3MUT proteins (Fig. 3H and J).
Altogether, these results point to a relevant role of the NTF2-like domain in the molecular mechanism that mediates G3BP1 action on mutant ATXN2 and mutant ATXN3 protein levels and propensity to aggregate.
The Ser149 phosphorylation site is important for G3BP1 action on ATXN2 and ATXN3 mutant proteins
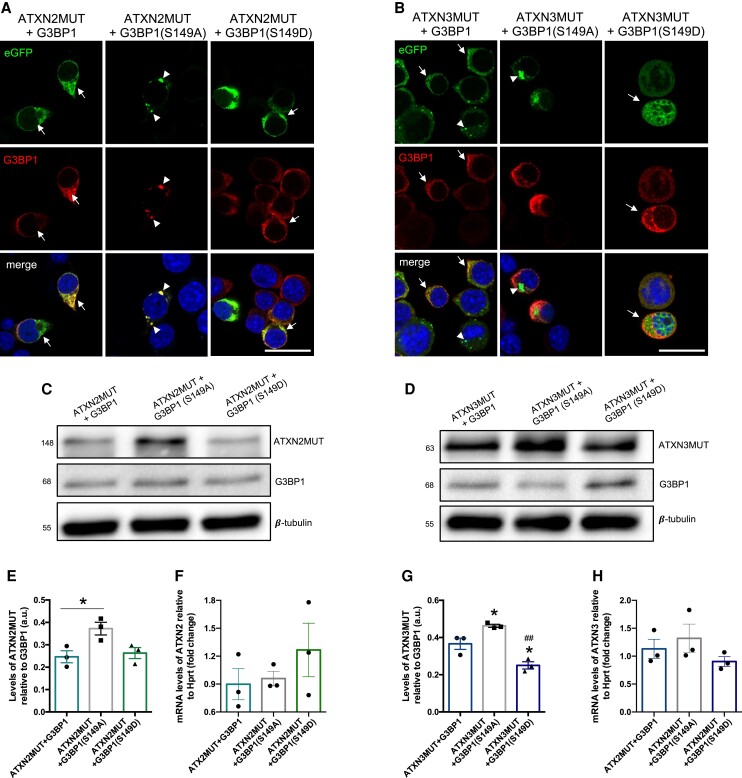
The NTF2-like domain of G3BP1 is in close proximity to the S149 phosphorylation site, which has been described to have an important functional role.15–17 As we observed that G3BP1-ΔRRM was able to reduce the levels and aggregation of ATXN2MUT and ATXN3MUT, although to a lesser extent than the full-length G3BP1, we next aimed to investigate whether S149 modification was also important in this context. For that, we developed two phosphomutants of G3BP1: a phosphomimetic S149D mutant and a non-phosphorylatable S149A mutant (Supplementary Fig. 7). Neuro2a cells were co-transfected with ATXN2MUT or ATXN3MUT and with G3BP1 (S149D) or G3BP1 (S140A). Using confocal imaging, we observed that cells expressing wild-type G3BP1 showed no aggregates of ATXN2MUT or ATXN3MUT (Fig. 4A and B, arrows). The same pattern was observed on expression on the phosphomimetic G3BP1 (S149D) mutant. On the contrary, we observed aggregates of ATXN2MUT and ATXN3MUT in cells expressing the phospho-dead G3BP1(S149A) mutant (Fig. 4A and B, arrowheads).
Figure 4.
The Ser149 phosphorylation site is important in G3BP1 action on ATXN2 and ATXN3 mutant proteins. (A) Confocal microscopy representative images depicting Neuroa2 cells expressing ATXN2MUT and wild-type G3BP1 or G3BP1(S149A) or G3BP1(S149D). In the cells expressing wild-type G3BP1 or the phosphomimetic G3BP1(S149D), there are no aggregates of ATXN2MUT (arrows), contrasting with the cells expressing the phospho-dead G3BP1(Ser149A), where ATXN2MUT aggregates are observed (arrowheads). (B) Confocal microscopy representative images depicting Neuro2a cells expressing ATXN3MUT and wild-type G3BP1 or G3BP1(S149A) or G3BP1(S149D). In the cells expressing wild-type G3BP1 or the phosphomimetic G3BP1 (S149D) there are no aggregates of ATXN3MUT (arrows), contrasting with the cells expressing the phospho-dead G3BP1(Ser149A), where ATXN3MUT aggregates are observed (arrowheads). Scale bar = 20 µm. (C) Representative western blot of Neuro2a lysates expressing ATXN2MUT, co-transfected with wild-type G3BP1 or G3BP1(S149A) or G3BP1(S149D). Western blots were labelled using ATXN2, G3BP1 and β-tubulin antibodies. (D) Representative western blot of Neuro2a lysates expressing ATXN3MUT, co-transfected with wild-type G3BP1 or G3BP1(S149A) or G3BP1(S149D). Western blots were labelled using ATXN3, G3BP1 and β-tubulin antibodies. (E) The levels of ATXN2MUT protein are significantly increased in cells expressing the phospho-dead G3BP1(Ser149A), compared to ATX2MUT + G3BP1 (n = 3 independent experiments; *P < 0.05; one-way ANOVA, followed by post hoc Bonferroni multiple comparisons test). (F) No significant alterations were found in the ATX2MUT mRNA levels between all the experimental conditions. (G) The levels of ATXN3MUT protein are significantly increased in cells expressing the phospho-dead G3BP1(Ser149A), compared to the other two conditions, whereas the cells expressing the phosphomimetic G3BP1(Ser149D) are significantly reduced, compared to the other two conditions. (H) No significant alterations were found in the ATX3MUT mRNA levels between all the experimental conditions (n = 3 independent experiments; *P < 0.05 to ATXN3MUT + G3BP1; ##P < 0.01 to ATXN3MUT + G3BP1(Ser149A); one-way ANOVA, followed by post hoc Bonferroni multiple comparisons test). Values are expressed as mean ± SEM.
Next, we investigated the impact of the expression of the two phosphomutants on the protein levels of ATXN2MUT and ATXN3MUT (Fig. 4C and D). We found that the protein levels of ATXN2MUT were significantly increased with G3BP1 (S149A) expression (ATXN2MUT + G3BP1: 0.24 ± 0.026 versus ATXN2MUT + G3BP1(S149A): 0.37 ± 0.028, n = 3, P < 0.05) (Fig. 4E). On the other hand, the protein levels of ATXN2MUT were similar between cells co-expressing wild-type G3BP1 or the phosphomimetic G3BP1 (S149D) mutant (Fig. 4E). Consistently, the protein levels of ATXN3MUT are increased on G3BP1(S149A) expression, compared to the wild-type G3BP1 and G3BP1(S149D) expressing conditions (Fig. 4G). The expression of G3BP1(S149D) also led to a significant reduction in the levels of ATXN3MUT, as compared to the wild-type G3BP1 condition (ATXN3MUT + G3BP1: 0.36 ± 0.03 versus ATXN3MUT + G3BP1(S149D): 0.25 ± 0.01, n = 3, P < 0.05).
On wild-type G3BP1 expression, we observed that there was a significant reduction in the mRNA levels of ATXN2MUT and ATXN3MUT, compared to control conditions (Supplementary Fig. 8). However, no differences were observed in the mRNA levels of ATXN2MUT and ATXN3MUT on the expression of the two phosphomutants, compared to the wild-type G3BP1 condition (Fig. 4F and H). Altogether, these results indicate that the S149 phosphorylation site is important for G3BP1 modulatory activity on the aggregation and expression of mutant ATXN2 and mutant ATXN3.
G3BP1 levels are reduced in SCA2 and SCA3, and its silencing increases protein aggregation
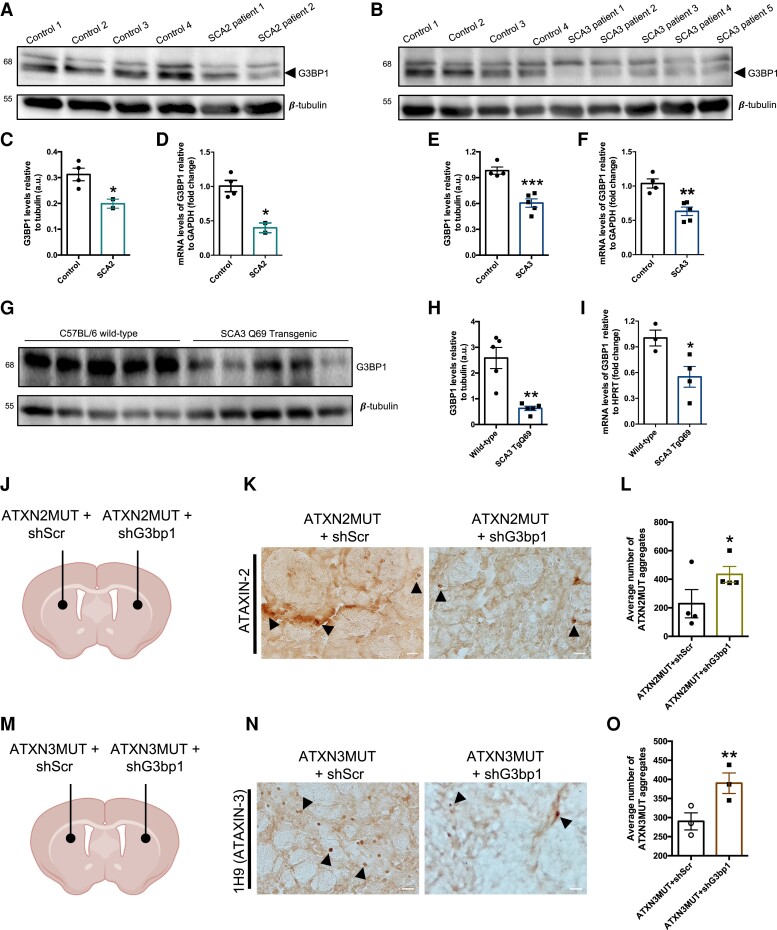
Previous studies have reported that mutant polyQ proteins can dysregulate the expression of several genes.1,42 In fact, we have shown that the expression of mutant ATXN3 drives an abnormal reduction of wild-type ATXN2 levels.43 Taking this into consideration, we next assessed whether the levels of G3BP1 were altered in samples from SCA2 and SCA3 patients and disease models. In post-mortem brain samples of SCA2 patients, we detected a reduction in the immunodetection of G3BP1 comparing with healthy individuals, both in the striatum and the cerebellum (Supplementary Fig. 9). Furthermore, in fibroblasts from SCA2 patients, we detected a significant reduction in the protein (Fig. 5A and C) and mRNA levels (Fig. 5D) of G3BP1, compared to fibroblasts from healthy controls. Similarly, in fibroblasts from SCA3 patients we observed a decrease in G3BP1 protein (Fig. 5B and E) and mRNA levels (Fig. 5F), compared to fibroblasts form healthy controls. The same reduction was also observed in cerebellar samples from a transgenic mouse model of SCA3, which we used in this study (Fig. 5G–I). G3BP1 protein and mRNA levels were significantly reduced in the transgenic SCA3 animals, compared to wild-type C57BL/6 mice. This transgenic mouse expresses a truncated form of ATXN3 with 69 glutamines in the Purkinje cells of the cerebellum. In fact, microscopy analysis of cerebellar sections demonstrated that the reduction of G3BP1 in the transgenic animals was particularly evident in these cells (Supplementary Fig. 10).
Figure 5.
G3BP1 mRNA and protein levels are reduced in the SCA2 and SCA3, and its silencing in the mouse brain increases aggregation. (A) Representative western blot for protein lysates of fibroblasts from SCA2 patients and healthy controls. (B) Representative western blot for protein lysates from fibroblast of SCA3 patients and healthy controls. (C) The levels of G3BP1 protein and (D) mRNA are significantly reduced in SCA2, compared to controls. (E) The levels of G3BP1 protein and (F) mRNA are significantly reduced in SCA3, compared to controls (healthy controls n = 3; SCA2 n = 2; SCA3 n = 5). (G) Representative western blot for protein lysates of cerebella from a transgenic SCA3 mouse model. (H) The levels of G3BP1 protein and (I) mRNA are significantly reduced in the SCA3 transgenic mice, compared to C57BL/6 wild-type animals (n = 3–5). Western blots were labelled using G3BP1, and β-tubulin antibodies. (J) Schematic representation of the injection site for the SCA2 model. Briefly, lentiviral vectors encoding ATXN2MUT and an shRNA scramble were co-injected in one hemisphere of the striatum, and in the contralateral hemisphere was co-injected ATXN2MUT and an shRNA targeting G3bp1. (K) At 4 weeks post-injection the animals were euthanized, and brain sections labelled with ATXN2 to highlight the presence of pathological aggregates. (L) The average number of ATXN2MUT aggregates is significantly increased on shG3bp1 expression, as compared to the control hemisphere. (M) Schematic representation of the injection site for the SCA2 model. Briefly, lentiviral vectors encoding ATXN3MUT and a shRNA scramble were co-injected in one hemisphere of the striatum and in the contralateral hemisphere it was co-injected ATXN2MUT and a shRNA targeting G3bp1. (N) At 4 weeks post-injection, the animals were euthanized and brain sections labelled with ATXN3 to highlight the presence of pathological aggregates. (O) The average number of ATXN3MUT aggregates is significantly increased on shG3bp1 expression, as compared to the control hemisphere. (*P < 0.05; **P < 0.01; ***P < 0.001; Student’s t-test). Values are expressed as mean ± SEM.
To investigate the functional impact of G3BP1 levels reduction on mutant ATXN2- or mutant ATXN3-induced pathology, we next injected lentiviral vectors encoding a validated shRNA targeting murine G3bp1 (shG3bp1) (Supplementary Fig. 11) into the lentiviral models of SCA2 and SCA344,45 (Fig. 5I and L). Briefly, one hemisphere of the striatum was co-injected with lentiviral vectors encoding for ATXN2MUT (or ATXN3MUT) and shG3bp1, while in the contralateral hemisphere, as control, we injected ATXN2MUT (or ATXN3MUT) and a scramble shRNA (shScr). At 4 weeks post-injection we sacrificed the animals, and the striatum was histologically analysed for the presence of aggregates of ATXN2MUT and ATXN3MUT (Fig. 5J and M). We found that the silencing of G3bp1 led to a significant increase in the average number of aggregates of ATXN2MUT (ATXN2MUT + shG3bp1: 434 ± 55.62 versus ATXN2MUT + shScr: 228 ± 98.85, n = 4, P < 0.01) and ATXN3MUT (ATXN3MUT + shG3bp1: 390 ± 26.89 versus ATXN3MUT + shScr: 290 ± 22.37, n = 3, P < 0.05) (Fig. 5K and N).
Altogether, these results highlight that G3BP1 mRNA and protein levels are reduced in the context of SCA2 and SCA3, and that its decrease may play a role on disease pathogenesis.
Restoring G3BP1 levels alleviates neuropathology in SCA2 and SCA3 lentiviral mouse models
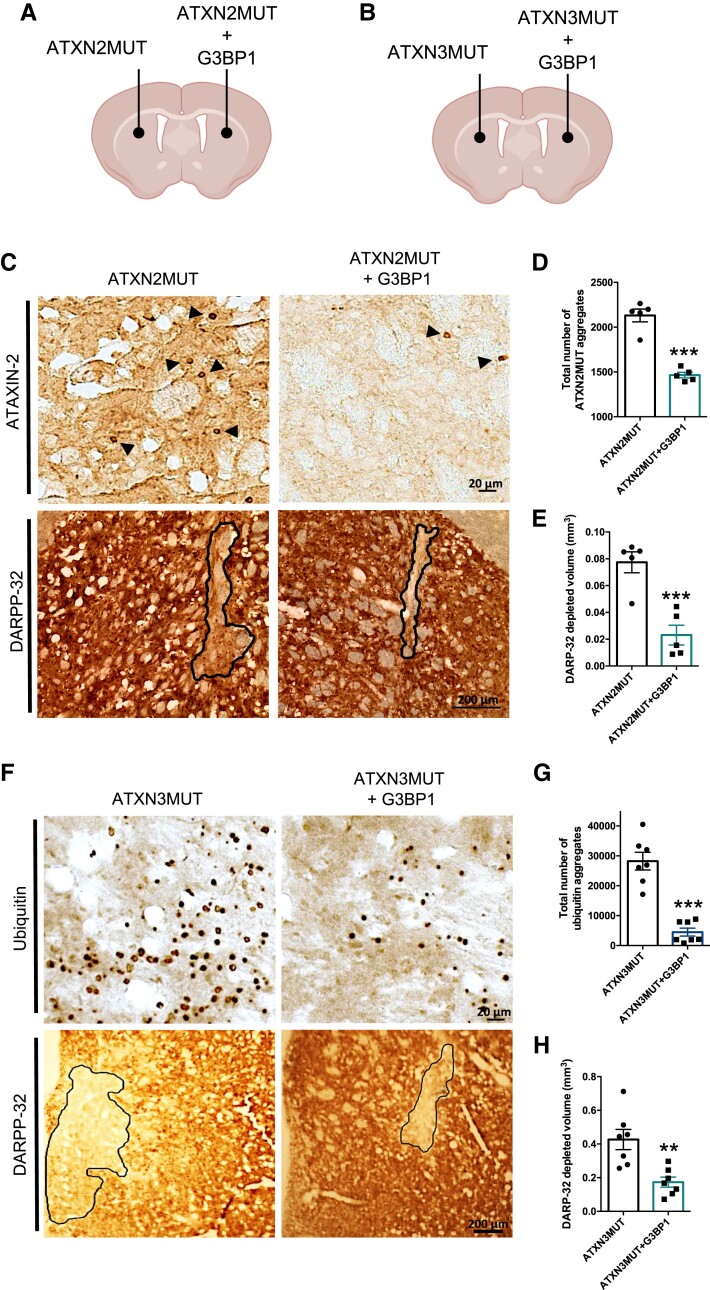
The expression of ATXN2MUT and ATXN3MUT mediated by lentiviral vectors leads to the formation of intraneuronal aggregates and to the loss of neuronal markers,44,45 which are neuropathological signs also found in post-mortem SCA2 and SCA3 brain tissue.46–48 Thus, we next aimed to investigate whether restoring G3BP1 levels could improve abnormalities induced by ATXN2MUT and ATXN3MUT expression in vivo. For that, we co-expressed lentiviral vectors encoding ATXN2MUT (or ATXN3MUT) and human G3BP1 in one hemisphere of the striatum and, as a control, in the contralateral hemisphere, we injected lentiviral vectors encoding ATXN2MUT (or ATXN3MUT) (Fig. 6A and B). At 12 weeks post-injection for the SCA2 lentiviral mouse model, and at 4 weeks post-injection for the SCA3 lentiviral mouse model, animals were sacrificed and the striatum was histologically analysed. In both models, the expression of G3BP1 was able to significantly reduce the number of mutant protein aggregates (ATXN2MUT + G3BP1: 1466 ± 31.13, n = 5 versus ATXN2MUT: 2131 ± 71.04, n = 5, P = 0.0002; ATXN3MUT + G3BP1: 6066 ± 1958 versus ATXN3MUT: 30 076 ± 2717, n = 7, P < 0.0001) (Fig. 6C–D and F–G). The mRNA and soluble protein levels of ATXN2 and ATXN3 were also analysed in a group of animals at 4 weeks post-injection (Supplementary Fig. 12). In the SCA2 lentiviral model, no significant differences were observed in mRNA and protein levels of ATXN2MUT on G3BP1 expression (Supplementary Fig. 12A and B). On the other hand, in the SCA3 lentiviral model, there is a robust reduction in ATXN3MUT protein levels in the hemisphere expressing G3BP1 (ATXN3MUT + G3BP1: 0.285 ± 0.04 versus ATXN3MUT: 0.413 ± 0.08, n = 4, P = 0.054) (Supplementary Fig. 12D and E). No alterations in ATXN3MUT mRNA levels were observed between hemispheres (Supplementary Fig. 12C and F).
Figure 6.
G3BP1 expression reduces the number of aggregates and the loss of neuronal markers in lentiviral mouse models of SCA2 and SCA3. Mice were stereotaxically injected into the striatum either with lentiviral particles encoding for mutant forms of ATXN2 or ATXN3, or co-injected with lentiviral particles encoding for the mutant form and G3BP1. (A) Schematic representation of the injection site and lentiviral vectors injected in the mouse model of SCA2. Animals were bilaterally injected and euthanized 12 weeks after the injection for tissue collection. (B) Schematic representation of the injection site and lentiviral vectors injected in the mouse model of SCA3. Animals were bilaterally injected and euthanized 4 weeks after the injection for tissue collection. (C) Brain sections from the lentiviral mouse model of SCA2 were analysed through immunohistochemistry using ATXN2 and DARPP-32 antibodies. Images show aggregates of ATXN2MUT (arrowheads; scale bar = 20 µm) and the loss of staining (line) of the neuronal marker DARPP-32 (scale bar = 200 µm). (D) The hemisphere expressing G3BP1 presented a reduced number of ATXN2MUT aggregates, compared to the control hemisphere (n = 5; ***P < 0.0001; Student’s t-test). (E) G3BP1 expression rescues neuronal marker loss, compared to the contralateral hemisphere only expressing ATXN2MUT (n = 5; ***P < 0.0001; Student’s t-test). (F) Representative images of immunohistochemistry brain sections, from the lentiviral mouse model of SCA3. The figures show ubiquitinated ATXN3MUT aggregates (dots; scale bar = 20 µm) and the neuronal marker DARPP-32 loss of staining (scale bar = 200 µm). (G) The expression of G3BP1 led to a significant reduction in the number of ubiquitinated ATXN3MUT aggregates, compared to the control condition (n = 7; ***P < 0.0001; Student’s t-test). (H) G3BP1 expression rescues neuronal marker loss, as compared to controls (n = 7; ***P < 0.0001; Student’s t-test). Values are expressed as mean ± SEM.
Consistently, G3BP1 expression led to a preservation of the neuronal marker dopamine- and cAMP-regulated neuronal phosphoprotein 32 (DARPP-32) in both models, as compared to the control hemispheres (ATXN2MUT + G3BP1: 0.02 ± 0.0078 versus ATXN2MUT: 0.08 ± 0.0078, n = 5, P = 0.001; ATXN3MUT + G3BP1: 0.19 ± 0.0291 versus ATXN3MUT: 0.45 ± 0.0647, n = 7, P = 0.0072) (Fig. 6C, E, F and H). Altogether, these results show that G3BP1 expression in the striatum promotes neuroprotection in the context of ATXN2- and ATXN3-induced neuropathology pathology, reducing protein aggregation and loss of neuronal markers.
Overexpression of G3BP1 in the brain of wild-type mice did not produce neuronal loss or astrogliosis
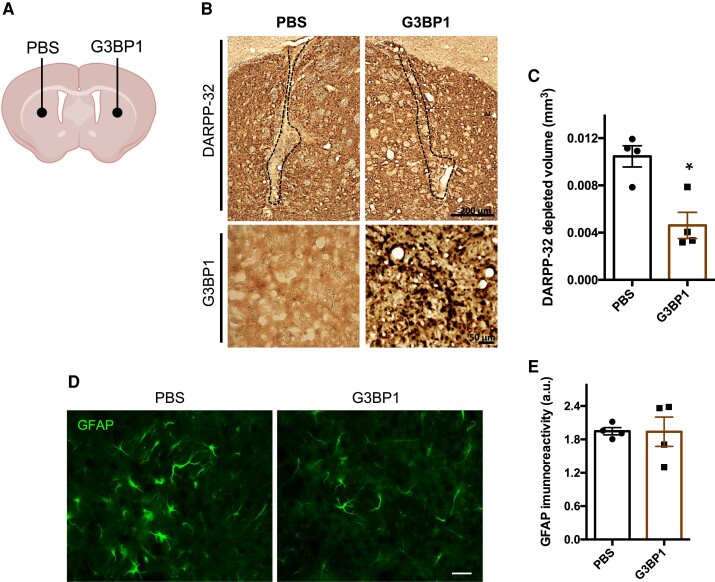
We next aimed to evaluate whether G3BP1 expression in the brain could have a detectable negative impact, translated in the form of neuronal loss or astrogliosis. For that, lentiviral particles encoding G3BP1 were injected in one hemisphere of the striatum of wild-type C57BL/6 mice, while the contralateral hemisphere was injected with PBS, as control (Fig. 7A). At 4 weeks post-injection, the loss of the neuronal marker DARPP-32 (Fig. 7B) in the hemisphere injected with G3BP1 was significantly lower compared to the control hemisphere injected with PBS (G3BP1: 0.003 ± 0.0014 versus PBS: 0.01 ± 0.0011, n = 4 Student’s t-test, P = 0.035) (Fig. 7B and C). In fact, in the G3BP1-injected animals the lesion area was restricted to the injection site. Astrocytes activation was analysed by detection of the GFAP marker and compared between the G3BP1-injected hemisphere and the PBS-injected control hemisphere (Fig. 7D). No differences were found in the immunoreactivity of GFAP between hemispheres (Fig. 7E). Altogether, these results suggest that G3BP1 overexpression in the normal brain does not produce gross histological changes.
Figure 7.
Overexpression of lentiviral vectors encoding G3BP1 in the brain of wild-type mice did not produce neuronal marker loss or inflammation. Mice at 8–12 weeks of age were stereotaxically injected into the striatum (bilaterally) either with PBS or with lentiviral particles encoding for human G3BP1 and euthanized for tissue collection 4 weeks after injection. (A) Schematic representation of the injection site in the striatum. (B) Immunohistochemistry images analysis of DARPP-32 depletion volume (dashed line; top; scale bar = 200 µm) and G3BP1 (bottom; scale bar = 50 µm) labelling in brain sections from mice injected with PBS and in the contralateral hemisphere injected with lentiviral particles encoding for G3BP1. (C) The total area of DARPP-32 depletion, in mice brain sections, was reduced on injection of lentiviral particles encoding for G3BP1 when compared to the injection with PBS (n = 4; *P < 0.05; Student's t-test). (D) We also labelled GFAP, a marker of astrogliosis, in brain sections from mice injected with PBS and in the contralateral hemisphere injected with lentiviral vectors encoding G3BP1. (E) The quantification of GFAP immunoreactivity did not detect significant differences between both hemispheres. Scale bar = 200 µm. Values are expressed as mean ± SEM.
Restoring G3BP1 levels mitigates behaviour deficits and neuropathological abnormalities
PolyQ SCAs are characterized by progressive neuronal loss and motor dysfunctionality. Thus, to model this phenotype, we used the previously mentioned transgenic mouse line expressing a truncated form of mutant ATXN3 with 69 glutamines in cerebellar Purkinje cells, which displays severe motor dysfunctions and neurodegeneration with an early onset.49 This line can also be envisioned as a relevant general model of polyQ diseases, considering that it only contains a small region of the ATXN3 protein and a significant tract of glutamines, sufficient for causing pathology as observed in other polyQ diseases.1,49 We aimed to investigate the impact of G3BP1 expression on this transgenic mouse model, which we determined to present reduced levels of G3BP1 (Fig. 5G–I). For that, at 4 weeks of age, animals were stereotaxically injected into the cerebellum50 with lentiviral vectors encoding for G3BP1, while control animals were injected with lentiviral vectors encoding for GFP. A third group of non-injected animals was also included in the experiments.
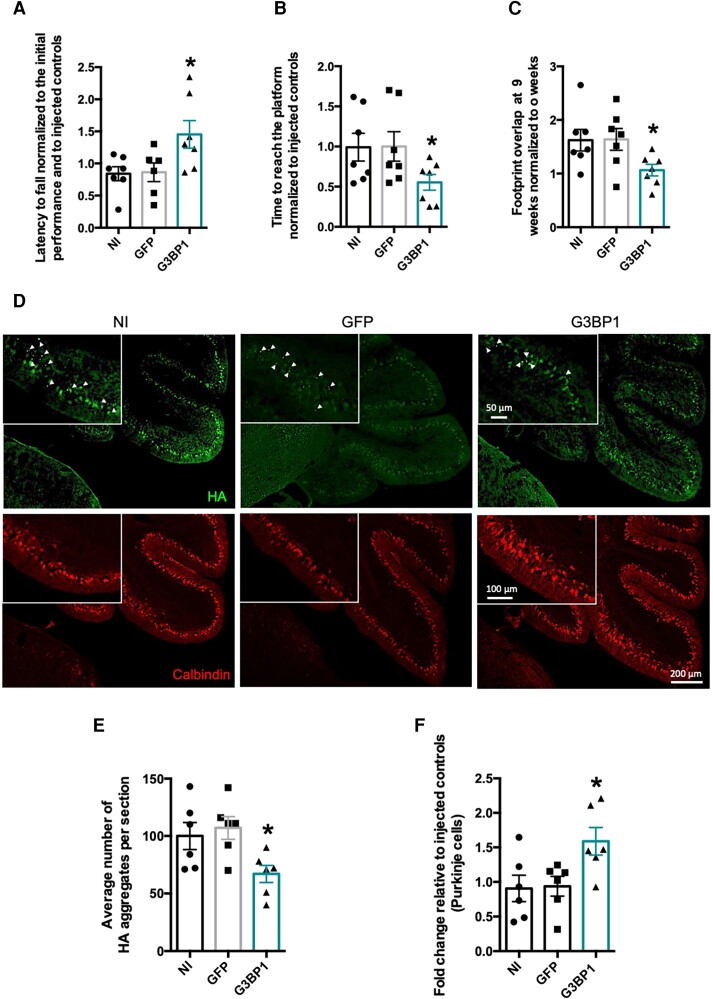
Animals were subjected to a battery of behaviour tests every 3 weeks, until 9 weeks post-injection. We observed that, at this final time point, G3BP1-injected animals stayed for longer times in the rotating rotarod compared to the control animals, thus showing an amelioration of motor deficits (G3BP1: 1.45 ± 0.0124, n = 7 versus non-injected: 0.84 ± 0.1082, n = 7, P = 0.0254) (Fig. 8A). Consistently, at 9 weeks post-injection, compared to control animals, G3BP1-injected animals took less time to complete a swimming test in which the mice had to cross a pool to reach a safe platform (G3BP1: 0.55 ± 0.0974, n = 7 versus non-injected: 0.99 ± 0.173, n = 7, P = 0.0476) (Fig. 8B). Finally, in a footprint patterns test that analyses diverse gait parameters extracted from the prints that mice leave when crossing a white sheet tunnel with their paws painted, the animals injected with G3BP1 had a better footprint overlap, as compared to control animals, which suggests a mitigation of motor deficits (G3BP1: 1.06 ± 0.1081, n = 7 versus non-injected: 1.62 ± 0.1997, n = 7, P = 0.0297) (Fig. 8C). Altogether, these results show that G3BP1 expression in the cerebellum can ameliorate motor deficits.
Figure 8.
G3BP1 expression mitigates motor deficits and neuropathological abnormalities in an SCA3 transgenic mouse model. Transgenic mice animals expressing a truncated form of the ATXN3 protein containing 69 glutamines were stereotaxically injected in the cerebellum with lentiviral particles encoding for GFP (control group) or with G3BP1 (treated group). Mice, at 4 weeks of age, were first tested 1–2 days before injection and then repeatedly tested every 3 weeks until 9 weeks post-injection, to be euthanized at 10 weeks post-surgery. (A–C) Representative plots of mice motor performance at 9 weeks post-injection. (A) Mice injected with G3BP1 significantly improved motor performance (assessed by the rotarod test), as they remain more time at the rotating rod compared to control mice treated with GFP or non-injected (NI) mice. (B) Mice injected with G3BP1 significantly reduced the time needed to cross the water-filled tank and reach the platform, compared to control mice treated with GFP or non-injected animals. (C) Footprint analysis showed that mice injected with G3BP1 improved overlap measures, compared to controls treated with GFP or non-injected animals. (D) Representative images of immunohistochemistry brain sections from mice cerebellum either injected with lentiviral particle encoding for GFP (control) or with G3BP1. Upper panel: ATXN3 aggregates assessed by HA-tag immunoreactivity (arrowhead; scale bars = 50, 100 and 200 µm). Lower panel: Purkinje cells were assessed by calbindin immunoreactivity (scale bars = 100 and 200 µm). (E) G3BP1 expression significantly reduced the number of HA-ATXN3 aggregates (arrowhead), compared to control mice injected with GFP or non-injected. (F) G3BP1 expression significantly preserved the number of Purkinje cells within lobe IX, compared to non-injected and GFP injected controls (n = 6–7; *P < 0.05; one-way ANOVA followed by post hoc Bonferroni multiple comparisons test). Values are expressed as mean ± SEM.
Neuropathologically, this mouse model is characterized by the formation of aggregates in cerebellar Purkinje cells, a severe reduction in the number of these cells and a marked disarrangement of cerebellar layers architecture.49,51 Thus, we next aimed to evaluate the impact of G3BP1 expression on these neuropathological abnormalities (Fig. 8D). In agreement with the improvements observed in motor deficits, we found that animals injected with G3BP1 exhibited a significantly reduced number of pathological aggregates (HA-tagged), compared to controls (G3BP1: 63.12 ± 10.17, n = 6 versus non-injected: 101.2 ± 15.29, n = 6, P = 0.0397) (Fig. 8E). We also found that, compared to controls, G3BP1-injected animals showed a preservation in the number of Purkinje cells, imaged using calbindin as a marker (G3BP1: 1.62 ± 0.2405, n = 6 versus non-injected: 0.91 ± 0.1904, n = 6, P = 0.0437) (Fig. 8F). Importantly, in non-transduced lobes, no differences were found between the experimental groups regarding the number of pathological aggregates (G3BP1: 44.45 ± 7.169, n = 6 versus non-injected: 49.7 ± 9.385, n = 6) or the number of cerebellar Purkinje cells (G3BP1: 1 ± 0.1457, n = 6 versus non-injected: 0.97 ± 0.1988 n = 6) (Supplementary Fig. 13). As these transgenic animals present a strong atrophy of the cerebellum, we next analysed the thickness of cerebellar layers. We found that the molecular layer thickness of transduced lobules (II/III) was significantly wider in G3BP1-injected animals than non-injected controls (G3BP1: 64.99 ± 3.189, n = 6 versus non-injected: 56.03 ± 1.824, n = 6, P < 0.0118), while no difference was found in non-transduced lobules (Supplementary Fig. 14).
Altogether, these results show that promoting G3BP1 expression in the cerebellum of a polyQ SCA disease model significantly reduced motor behaviour impairments and neuropathological abnormalities.
Discussion
The development of therapies for neurodegenerative diseases faces many challenges, some of which derive from the fact that the understanding of pathogenesis is often incomplete. This is also true for disorders that are described as monogenic, as is the case of the nine polyQ diseases. Although in these cases the disease is clearly associated with a CAG expansion of designated genes, the cascade of molecular events that culminates in nervous system dysfunction and cell loss remains to be clearly elucidated. In the case of SCAs associated with polyQ protein expansion, it is puzzling that a shared mutation in otherwise unrelated genes chiefly affects the same region of the nervous system—the cerebellum—and produces a set of common symptoms that include the hallmark ataxia. These commonalities afford the exploration of disease mechanisms that are common among these diseases, and which may be explored with therapy development in mind. The current work identified G3BP1 as a putative therapeutic target for the most common SCAs caused by polyQ expansion mutations, SCA2 and SCA3.
G3BP1 is an RBP involved in stress responses, and an inability to keep up with cellular stress (in particular, proteotoxic and oxidative stress) has been repeatedly suggested to play a part in the molecular pathophysiology of polyQ diseases.12,52 We have determined that G3BP1 expression is diminished in fibroblasts from SCA2 and SCA3 patients (Fig. 5A–F), in brain samples from an SCA3 transgenic mouse model (Fig. 5G–I) and, importantly, the same was suggested by observations from post-mortem brain samples of two SCA2 patients (Supplementary Fig. 9). This suggests that the reduction of G3BP1 levels may constitute a contributing factor to SCA2 and SCA3 disease courses, possibly causing, or adding to, the impairment of cell stress responses. How expanded ATXN2 and ATXN3 may lead to a decrease in G3BP1 levels remains to be explored, however. ATXN2 is an RBP that has been described to regulate protein translation,43,53–55 and ATXN3 has been functionally implicated in transcription regulation and protein degradation.1,4 The alteration of these specific roles in a disease context or more general aberrations deriving from protein aggregation and abnormal interactions may help explain the decrease.
In line with the hypothesis that a decrease in G3BP1 levels contributes to SCA2 and SCA3, we observed that silencing endogenous G3bp1 exacerbated aggregation of human mutant ATXN2 and mutant ATXN3 expressed in the mouse brain (Fig. 5J–O). Aggregation is a crucial step in disease pathogenesis; although large macromolecular inclusions containing polyQ proteins are currently regarded as not being the primary drivers of cell toxicity, the tendency of polyQ-containing proteins to self-assemble and engage in abnormal interactions is regarded as a common mechanism responsible for the intrinsic toxicity of these proteins.56,57 Our results indicate that G3BP1 is able to counter mutant protein aggregation to an extent, suggesting that G3BP1 has a protective role against the toxicity of these disease-associated proteins.
G3BP1 overexpression was demonstrated to ameliorate SCA2 and SCA3 phenotypes, both in vitro (Fig. 2) and in vivo (Fig. 6). In cell cultures and in animal models, G3BP1 overexpression was shown to decrease protein aggregation, in line with what was suggested by the endogenous G3bp1 silencing experiment (Fig. 5J–O). While this effect can be hypothesized to result from a G3BP1-mediated alteration of expanded protein turnover or aggregation dynamics, our results suggest that the decrease in aggregation may be explained by a decrease in ATXN2 and ATXN3 levels caused by G3BP1 overexpression. The fact that the reduction of ATXN2 and ATXN3 expression was detected at the mRNA level (Supplementary Fig. 8) as well as at the protein level (Fig. 2) suggests that it may stem from a decrease in protein translation, which is in accordance with G3BP1 role as an RBP involved in the regulation of RNA metabolism. Our results also suggest that the effect of G3BP1 may be independent from SG formation, considering that chemical induction of SG formation did not produce the same effects on mutant protein expression and aggregation (Fig. 1), and that G3BP1 overexpression in Neuro2a cells by itself was insufficient to induce SG formation (Supplementary Fig. 5).
Our observations further suggest that the effect of G3BP1 on mutant protein level regulation and aggregation is mediated by the NTF2 domain (Fig. 3) and the S149 phosphorylation site of G3BP1 (Fig. 4). Future studies could investigate whether a truncated G3BP1 form comprising just these two functional regions could produce the same ameliorating effects. The experiments using G3BP1 S149 phosphomutants suggest that S149 phosphorylation is required for G3BP1-mediated decrease in mutant protein levels and aggregation, since blocking phosphorylation abrogated this effect, while mimicking the modification preserved the alteration observed with the wild-type G3BP1 (Fig. 4C–E and G). At the mRNA level, ATXN2 and ATXN3 decrease was observed on wild-type G3BP1 expression, and no differences were observed between this condition and the ones expressing the S149 phosphomutants (Fig. 4F and H). This variation may suggest that the effect of G3BP1 S149 phosphorylation on polyQ mutant protein expression and aggregation results from an effect at the protein level rather than an alteration of ATXN2 and ATXN3 translation. Alternatively, G3BP1 S149 phosphorylation could affect the translation of other proteins that in turn contribute to ATXN2 and ATXN3 degradation, also explaining the reduction in aggregation.
In animal models, we observed that the protective effect of G3BP1 overexpression against mutant ATXN2 and ATXN3 toxicity extends to the preservation of neuronal cells, in lentiviral mouse models of both SCA2 and SCA3 (Fig. 6C, E, Fand H), and in a transgenic SCA3 mouse model (Fig. 8Dand F). Furthermore, in the latter, lentiviral delivery of G3BP1 to the cerebellum improved the motor behavioural phenotype (Fig. 8A–C), which is considerably severe from an early time point. This indicates that G3BP1 overexpression can elicit wide-ranging amelioration of the polyQ SCA phenotype, counteracting neuropathological changes associated with mutant protein expression but also the behavioural signs that correlate with human symptomatology. The fact that G3BP1 expression in wild-type animals did not originate gross alterations indicative of toxicity (Fig. 7) suggests that viral delivery of G3BP1 may constitute a reasonable strategy to treat SCA2 and SCA3, worthy of further exploration.
Overall, our work provides a contribution to the understanding of SCA2 and SCA3 pathogenesis and to the development of a potential therapy for these diseases. On one hand, we determined that a decrease in the levels of the G3BP1 protein may constitute an important element of the pathogenic cascade of events that are responsible for SCA2 and SCA3. On the other, we demonstrated that overexpression of G3BP1 has a promising ameliorating effect in diverse models of these disorders. Considering that polyQ diseases share many aspects beyond the causative mutation responsible for each of them, it will be important to explore whether these observations have parallels in other polyQ diseases, and whether G3BP1 may constitute a therapeutic target for the nine disorders of this group.
Supplementary Material
Acknowledgements
We acknowledge the Light Microscopy Unit of the Algarve Biomedical Center Research Institute, which is partially supported by the project PPBI-POCI-01-0145-FEDER-022122. We also acknowledge ViraVector, namely Rui Nobre and Carina Henriques for the viral vectors production. We extended our thank to the Município de Loulé for the support in the publication.
Contributor Information
Rebekah Koppenol, ABC-RI, Algarve Biomedical Center Research Institute, 8005-139 Faro, Portugal; PhD Program in Biomedial Sciences, Faculdade de Medicina e Ciências Biomédicas, Universidade do Algarve, 8005-139 Faro, Portugal; Faculdade de Medicina e Ciências Biomédicas, Universidade do Algarve, 8005-139 Faro, Portugal; Center for Neuroscience and Cell Biology (CNC), University of Coimbra, 3004-504 Coimbra, Portugal.
André Conceição, ABC-RI, Algarve Biomedical Center Research Institute, 8005-139 Faro, Portugal; PhD Program in Biomedial Sciences, Faculdade de Medicina e Ciências Biomédicas, Universidade do Algarve, 8005-139 Faro, Portugal; Faculdade de Medicina e Ciências Biomédicas, Universidade do Algarve, 8005-139 Faro, Portugal.
Inês T Afonso, ABC-RI, Algarve Biomedical Center Research Institute, 8005-139 Faro, Portugal; Faculdade de Medicina e Ciências Biomédicas, Universidade do Algarve, 8005-139 Faro, Portugal.
Ricardo Afonso-Reis, ABC-RI, Algarve Biomedical Center Research Institute, 8005-139 Faro, Portugal; Faculdade de Medicina e Ciências Biomédicas, Universidade do Algarve, 8005-139 Faro, Portugal.
Rafael G Costa, ABC-RI, Algarve Biomedical Center Research Institute, 8005-139 Faro, Portugal; Faculdade de Medicina e Ciências Biomédicas, Universidade do Algarve, 8005-139 Faro, Portugal.
Sandra Tomé, Center for Neuroscience and Cell Biology (CNC), University of Coimbra, 3004-504 Coimbra, Portugal.
Diogo Teixeira, ABC-RI, Algarve Biomedical Center Research Institute, 8005-139 Faro, Portugal.
Joana Pinto da Silva, ABC-RI, Algarve Biomedical Center Research Institute, 8005-139 Faro, Portugal.
José Miguel Côdesso, ABC-RI, Algarve Biomedical Center Research Institute, 8005-139 Faro, Portugal; PhD Program in Biomedial Sciences, Faculdade de Medicina e Ciências Biomédicas, Universidade do Algarve, 8005-139 Faro, Portugal; Faculdade de Medicina e Ciências Biomédicas, Universidade do Algarve, 8005-139 Faro, Portugal; Center for Neuroscience and Cell Biology (CNC), University of Coimbra, 3004-504 Coimbra, Portugal.
David V C Brito, ABC-RI, Algarve Biomedical Center Research Institute, 8005-139 Faro, Portugal.
Liliana Mendonça, Center for Neuroscience and Cell Biology (CNC), University of Coimbra, 3004-504 Coimbra, Portugal.
Adriana Marcelo, ABC-RI, Algarve Biomedical Center Research Institute, 8005-139 Faro, Portugal; Faculdade de Medicina e Ciências Biomédicas, Universidade do Algarve, 8005-139 Faro, Portugal; Center for Neuroscience and Cell Biology (CNC), University of Coimbra, 3004-504 Coimbra, Portugal.
Luís Pereira de Almeida, Center for Neuroscience and Cell Biology (CNC), University of Coimbra, 3004-504 Coimbra, Portugal; Faculty of Pharmacy, University of Coimbra, 3000-548 Coimbra, Portugal.
Carlos A Matos, ABC-RI, Algarve Biomedical Center Research Institute, 8005-139 Faro, Portugal; Faculdade de Medicina e Ciências Biomédicas, Universidade do Algarve, 8005-139 Faro, Portugal.
Clévio Nóbrega, ABC-RI, Algarve Biomedical Center Research Institute, 8005-139 Faro, Portugal; Faculdade de Medicina e Ciências Biomédicas, Universidade do Algarve, 8005-139 Faro, Portugal; Champalimaud Research Program, Champalimaud Center for the Unknown, 1400-038 Lisbon, Portugal.
Funding
This work was funded by the Portuguese Science and Technology Foundation (FCT) project (ALG-01- 0145-FEDER-29480) ‘SeGrPolyQ’, with CRESC ALGARVE 2020 cofunding and the French Muscular Dystrophy Association (AFM-Téléthon) project #22424. SFRH/BD/148533/2019 Rebekah Koppenol’s PhD Fellowship, DFA/BD/7892/2020 André Conceição’s PhD Fellowship and SFRH/BD/133192/2017 Adriana Marcelo’s PhD Fellowship were funded by FCT.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Matos CA, Almeida LP, Nóbrega C. Machado–Joseph disease/spinocerebellar ataxia type 3: Lessons from disease pathogenesis and clues into therapy. J Neurochem. 2019;148:8–28. [DOI] [PubMed] [Google Scholar]
- 2. Pulst SM, Nechiporuk A, Nechiporuk T. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat Genet. 1996;14:269–276. [DOI] [PubMed] [Google Scholar]
- 3. Kawaguchi Y, Okamoto T, Taniwaki M, et al. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat Genet. 1994;8:221–228. [DOI] [PubMed] [Google Scholar]
- 4. Matos C, Macedo-Ribeiro S, Carvalho A. Polyglutamine diseases: The special case of ataxin-3 and Machado–Joseph disease. Prog Neurobiol. 2011;95:26–48. [DOI] [PubMed] [Google Scholar]
- 5. Matos C, Pereira de Almeida L, Nobrega C. Proteolytic cleavage of polyglutamine disease-causing proteins: Revisiting the toxic fragment hypothesis. Curr Pharm Des. 2017;23:753–775. [DOI] [PubMed] [Google Scholar]
- 6. Huynh DP, Figueroa K, Hoang N, Pulst SM. Nuclear localization or inclusion body formation of ataxin-2 are not necessary for SCA2 pathogenesis in mouse or human. Nat Genet. 2000;26:44–50. [DOI] [PubMed] [Google Scholar]
- 7. Onodera O, Idezuka J, Igarashi S, et al. Progressive atrophy of cerebellum and brainstem as a function of age and the size of the expanded CAG repeats in the MJD1 gene in Machado-Joseph disease. Ann Neurol. 1998;43:288–296. [DOI] [PubMed] [Google Scholar]
- 8. Todd TW, Lim J. Aggregation formation in the polyglutamine diseases: Protection at a cost? Mol Cells. 2013;36:185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cowan KJ, Diamond MI, Welch WJ. Polyglutamine protein aggregation and toxicity are linked to the cellular stress response. Hum Mol Genet. 2003;12:1377–1391. [DOI] [PubMed] [Google Scholar]
- 10. Takahashi A, Ohnishi T. Molecular mechanisms involved in adaptive responses to radiation, UV light, and heat. J Radiat Res. 2009;50:385–393. [DOI] [PubMed] [Google Scholar]
- 11. Kedersha N, Anderson P. Stress granules: Sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30(Pt 6):963–969. [DOI] [PubMed] [Google Scholar]
- 12. Marcelo A, Koppenol R, de Almeida L, Matos C, Nóbrega C. Stress granules, RNA-binding proteins and polyglutamine diseases: Too much aggregation? Cell Death Dis. 2021;12:592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sahoo PK, Lee SJ, Jaiswal PB, et al. Axonal G3BP1 stress granule protein limits axonal mRNA translation and nerve regeneration. Nat Commun. 2018;9:3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tourrière H, Chebli K, Zekri L, et al. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160:823–831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15. Gallouzi IE, Parker F, Chebli K, et al. A novel phosphorylation-dependent RNase activity of GAP-SH3 binding protein: A potential link between signal transduction and RNA stability. Mol Cell Biol. 1998;18:3956–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alam U, Kennedy D. Rasputin a decade on and more promiscuous than ever? A review of G3BPs. Biochim Biophys Acta Mol Cell Res. 2019;1866:360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tourrière H, Gallouzi IE, Chebli K, et al. RasGAP-associated endoribonuclease G3BP: Selective RNA degradation and phosphorylation-dependent localization. Mol Cell Biol. 2001;21:7747–7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Martin S, Bellora N, González-Vallinas J, et al. Preferential binding of a stable G3BP ribonucleoprotein complex to intron-retaining transcripts in mouse brain and modulation of their expression in the cerebellum. J Neurochem. 2016;139:349–368. [DOI] [PubMed] [Google Scholar]
- 19. Winslow S, Leandersson K, Larsson C. Regulation of PMP22 mRNA by G3BP1 affects cell proliferation in breast cancer cells. Mol Cancer. 2013;12:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Panas MD, Kedersha N, Schulte T, Branca RM, Ivanov P, Anderson P. Phosphorylation of G3BP1-S149 does not influence stress granule assembly. J Cell Biol. 2019;218:2425–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chai Y, Shao J, Miller VM, Williams A, Paulson HL. Live-cell imaging reveals divergent intracellular dynamics of polyglutamine disease proteins and supports a sequestration model of pathogenesis. Proc Natl Acad Sci U S A. 2002;99:9310–9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huynh DP. Expansion of the polyQ repeat in ataxin-2 alters its Golgi localization, disrupts the Golgi complex and causes cell death. Hum Mol Genet. 2003;12:1485–1496. [DOI] [PubMed] [Google Scholar]
- 23. Nóbrega C, Conceição A, Costa RG, et al. The cholesterol 24-hydroxylase activates autophagy and decreases mutant huntingtin build-up in a neuroblastoma culture model of Huntington’s disease. BMC Res Notes. 2020;13:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nóbrega C, Nascimento-Ferreira I, Onofre I, et al. Overexpression of mutant ataxin-3 in mouse cerebellum induces ataxia and cerebellar neuropathology. Cerebellum. 2013;12:441–455. [DOI] [PubMed] [Google Scholar]
- 25. Xia G, Santostefano K, Hamazaki T, et al. Generation of human-induced pluripotent stem cells to model spinocerebellar ataxia type 2 in vitro. J Mol Neurosci. 2013;51:237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Onofre I, Mendonça N, Lopes S, et al. Fibroblasts of Machado Joseph disease patients reveal autophagy impairment. Sci Rep. 2016;6:28220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6:275–277. [DOI] [PubMed] [Google Scholar]
- 28. Déglon N, Tseng JL, Bensadoun JC, et al. Self-inactivating lentiviral vectors with enhanced transgene expression as potential gene transfer system in Parkinson’s disease. Hum Gene Ther. 2000;11:179–190. [DOI] [PubMed] [Google Scholar]
- 29. De AL, Ross CA, Zala D, Aebischer P, Déglon N. Lentiviral-mediated delivery of mutant huntingtin in the striatum of rats induces a selective neuropathology modulated by polyglutamine repeat size, huntingtin expression levels, and protein length. J Neurosci. 2002;22:3473–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Simões AT, Gonçalves N, Koeppen A, et al. Calpastatin-mediated inhibition of calpains in the mouse brain prevents mutant ataxin 3 proteolysis, nuclear localization and aggregation, relieving Machado-Joseph disease. Brain. 2012;135(Pt 8):2428–2439. [DOI] [PubMed] [Google Scholar]
- 31. Anderson P, Kedersha N. Stressful initiations. J Cell Sci. 2002;115:3227–3234. [DOI] [PubMed] [Google Scholar]
- 32. Mahboubi H, Stochaj U. Cytoplasmic stress granules : Dynamic modulators of cell signaling and disease. BBA Mol Basis Dis. 2017;1863:884–895. [DOI] [PubMed] [Google Scholar]
- 33. Nonhoff U, Ralser M, Welzel F, Piccini I. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell. 2007;18:1385–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kedersha N, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Protter DSW, Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;xx:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu ZS, Cai H, Xue W, et al. G3BP1 promotes DNA binding and activation of cGAS. Nat Immunol. 2019;20:18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim SSY, Sze L, Lam KP. The stress granule protein G3BP1 binds viral dsRNA and RIG-I to enhance interferon-β response. J Biol Chem. 2019;294:6430–6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lunde BM, Moore C, Varani G. RNA-binding proteins: Modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nagai K, Oubridge C, Ito N, Avis J, Evans P. The RNP domain: A sequence-specific RNA-binding domain involved in processing and transport of RNA. Trends Biochem Sci. 1995;20:235–240. [DOI] [PubMed] [Google Scholar]
- 40. Vognsen T, Møller IR, Kristensen O. Crystal structures of the human G3BP1 NTF2-like domain visualize FxFG nup repeat specificity. PLoS ONE. 2013;8:e80947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kennedy D, French J, Guitard E, Ru K, Tocque B, Mattick J. Characterization of G3BPs: Tissue specific expression, chromosomal localisation and rasGAP120 binding studies. J Cell Biochem. 2002;84:173–187. [DOI] [PubMed] [Google Scholar]
- 42. Pflieger LT, Dansithong W, Paul S, et al. Gene co-expression network analysis for identifying modules and functionally enriched pathways in SCA2. Hum Mol Genet. 2017;26:3069–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nóbrega C, Carmo-Silva S, Albuquerque D, et al. Re-establishing ataxin-2 downregulates translation of mutant ataxin-3 and alleviates Machado-Joseph disease. Brain. 2015;138:3537–3554. [DOI] [PubMed] [Google Scholar]
- 44. Alves S, Régulier E, Nascimento-ferreira I, et al. Striatal and nigral pathology in a lentiviral rat model of Machado-Joseph disease. Hum Mol Genet. 2008;17:2071–2083. [DOI] [PubMed] [Google Scholar]
- 45. Marcelo A, Afonso IT, Afonso-Reis R, et al. Autophagy in spinocerebellar ataxia type 2, a dysregulated pathway, and a target for therapy. Cell Death Dis. 2021;12:1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Estrada R, Galarraga J, Orozco G, Nodarse A, Auburger G. Spinocerebellar ataxia 2 (SCA2): Morphometric analyses in 11 autopsies. Acta Neuropathol. 1999;97:306–310. [DOI] [PubMed] [Google Scholar]
- 47. Paulson HL, Perez MK, Trottier Y, et al. Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron. 1997;19:333–344. [DOI] [PubMed] [Google Scholar]
- 48. Figiel M, Szlachcic WJ, Switonski PM, Gabka A, Krzyzosiak WJ. Mouse models of polyglutamine diseases: Review and data table. Part I. Mol Neurobiol. 2012;46:393–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Torashima T, Koyama C, Iizuka A, et al. Lentivector-mediated rescue from cerebellar ataxia in a mouse model of spinocerebellar ataxia. EMBO Rep. 2008;9:393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nóbrega C, Nascimento-ferreira I, Onofre I, et al. Silencing mutant ataxin-3 rescues motor deficits and neuropathology in Machado-Joseph disease transgenic mice. PLoS ONE. 2013;8:e52396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Oue M, Mitsumura K, Torashima T, et al. Characterization of mutant mice that express polyglutamine in cerebellar Purkinje cells. Brain Res. 2009;1255:9–17. [DOI] [PubMed] [Google Scholar]
- 52. Duennwald ML. Cellular stress responses in protein misfolding diseases. Future Sci OA. 2015;1:FSO42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Inagaki H, Hosoda N, Tsuiji H, Hoshino SI. Direct evidence that ataxin-2 is a translational activator mediating cytoplasmic polyadenylation. J Biol Chem. 2020;295:15810–15825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lastres-Becker I, Nonis D, Eich F, et al. Mammalian ataxin-2 modulates translation control at the pre-initiation complex via PI3K/mTOR and is induced by starvation. Biochim Biophys Acta. 2016;1862:1558–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Carmo-silva S, Nobrega C, de Almeida LP, Cavadasi C. Unraveling the role of ataxin-2 in metabolism. Trends in Endocrinol Metab. 2017;28:309–318. [DOI] [PubMed] [Google Scholar]
- 56. Mier P, Andrade-Navarro MA. Between interactions and aggregates: The PolyQ balance. Genome Biol Evol. 2021;13:evab246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lee D, Lee YI, Lee YS, Lee SB. The mechanisms of nuclear proteotoxicity in polyglutamine spinocerebellar ataxias. Front Neurosci. 2020;14:489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, on reasonable request.