Abstract
Over the past 10 years, the drive to improve outcomes from epilepsy surgery has stimulated widespread interest in methods to quantitatively guide epilepsy surgery from intracranial EEG (iEEG). Many patients fail to achieve seizure freedom, in part due to the challenges in subjective iEEG interpretation. To address this clinical need, quantitative iEEG analytics have been developed using a variety of approaches, spanning studies of seizures, interictal periods, and their transitions, and encompass a range of techniques including electrographic signal analysis, dynamical systems modeling, machine learning and graph theory. Unfortunately, many methods fail to generalize to new data and are sensitive to differences in pathology and electrode placement.
Here, we critically review selected literature on computational methods of identifying the epileptogenic zone from iEEG. We highlight shared methodological challenges common to many studies in this field and propose ways that they can be addressed. One fundamental common pitfall is a lack of open-source, high-quality data, which we specifically address by sharing a centralized high-quality, well-annotated, multicentre dataset consisting of >100 patients to support larger and more rigorous studies. Ultimately, we provide a road map to help these tools reach clinical trials and hope to improve the lives of future patients.
Keywords: epilepsy, neurosurgery, intracranial-EEG, data-sharing
Bernabei et al. provide an update on quantitative methods for guiding epilepsy surgery using intracranial EEG. They identify challenges which have prevented successful clinical translation of these methods, and offer potential solutions, including the release of a new dataset with more than 100 patients to support larger and more rigorous studies.
Epilepsy surgery and traditional localization of the epileptogenic zone
Clinicians plan epilepsy surgery by localizing the regions which causally generate seizures known as the epileptogenic zone (EZ), a process which often involves using intracranial EEG (iEEG)1 (Fig. 1A) to record seizures directly over a 1–2 week period. Teams of epileptologists analyse the temporal, spatial, and spectral characteristics of these events to select the areas from which they most likely originate. The insights from iEEG are then combined with findings from the patient’s history, seizure semiology, scalp EEG, neuropsychological testing, as well as structural and functional neuroimaging to determine the ultimate surgical strategy.
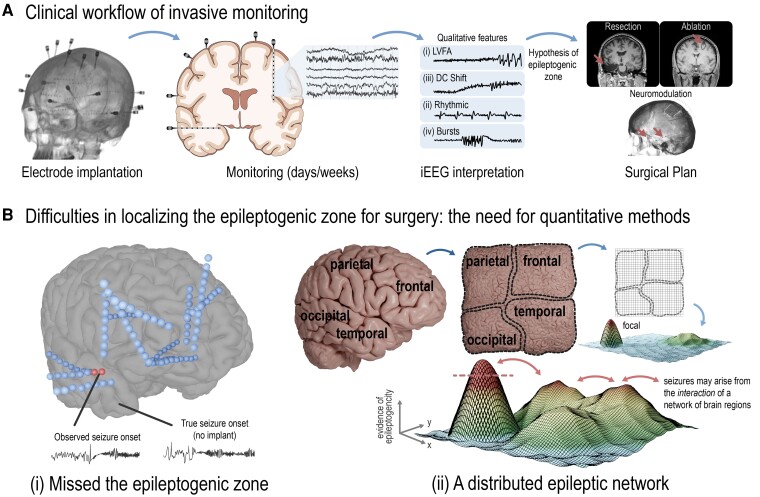
Figure 1.
Clinical workflow and the need for quantitative methods to localize the EZ. (A) Clinicians often localize the EZ for surgery using iEEG. Qualitative recognition of specific seizure onset patterns are primary used, including the identification of (i) low voltage fast activity; (ii) ‘DC shift’; (iii) preictal rhythmic spiking; and (iv) bursts of polyspikes or spike-and-wave activity (B) Difficulties in localizing the EZ may arise due to many factors. These can include (i) the implant somehow missed the region driving seizures; and (ii) the EZ is not a singular focus per se, but rather, seizures arise from a distributed interaction of brain regions in some patients. This is called the distributed epileptic network hypothesis.2,3 Given relevant iEEG and other clinical data, a singular focus may present itself as the EZ (red dashed line, adapted from Khambati et al.2 and Revell et al.3 with permission); however, other regions across the brain are involved in seizure generation, too. Removal of the primary seizure focus may not result in complete seizure freedom. Thus, there is the need for quantitative methods to (i) better localize seizure onset given imperfect implantation schemes; and (ii) quantify if a patient may be better treated with broader neuromodulation over focal intervention.
The primary method of EZ identification from iEEG is the qualitative recognition of specific seizure onset patterns that indicate a well-localized onset.4,5 Common patterns include (i) low voltage fast activity; (ii) ‘DC shift’ or ‘diffuse electrodecremental event’; (iii) preictal rhythmic spiking of low frequency and high amplitude; and (iv) bursts of polyspikes or spike-and-wave activity (Fig. 1A). Such onset patterns are known to vary by anatomical location and aetiology of epilepsy,4 which aids clinicians in their approach to localization. Occasionally, typical onset patterns are not observed and instead poorly localized, lower frequency or ‘propagated’ patterns are seen, leaving clinicians to wonder if the implant has somehow missed the region driving seizures, or if the EZ is ‘distributed’,2,3 and better treated with broader neuromodulation than focal intervention (Fig. 1B).
Traditional methods of EZ localization lead the majority of patients to become seizure free. However, even when clinicians feel that epileptogenic networks have been well defined, many patients relapse after surgery.6,7 These poor outcomes may reflect inherent challenges in qualitative iEEG interpretation, limited spatial sampling, as well as our incomplete understanding of how seizures arise from brain networks in epilepsy. Furthermore, concerns about financial cost, potential neurologic morbidity, and the dependence on referral to a limited number of highly experienced clinicians restrict the access of epilepsy surgery to a small fraction of potential candidates.8-10 Overall, there is a substantial need to leverage quantitative tools to improve surgical decision making while reducing cost and morbidity and increasing access.
In this article, we briefly review prior efforts to build quantitative tools intended to guide iEEG evaluation, identify barriers and potential solutions to clinical translation, and publicly release a large, multicentre dataset to accelerate research and clinical translation. Intracranial EEG, including microelectrode arrays have also provided substantial progress towards seizure prediction11-14 uncovering the mechanisms underlying seizure generation15-17; however, an in-depth review of these topics is beyond our current scope. Here, we focus on the potential of quantitative iEEG as a tool for precision medicine in epilepsy.
Quantitative localization of the epileptogenic zone
Interictal methods
Methods of identifying epileptogenic surgical targets using interictal data could significantly reduce the need for long hospital stays waiting for unpredictable seizures to occur and increase the diagnostic yield for the patients who do not have seizures while implanted with iEEG. Many promising methods have been developed, including those studying high-frequency oscillations (HFOs; Fig. 2B), interictal spikes,18-26 resting-state signal analysis, and functional connectivity.
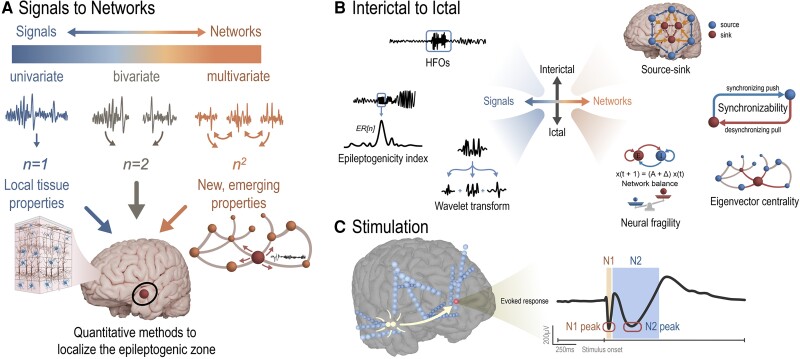
Figure 2.
Quantitative methods to localize the EZ. (A) Methods of iEEG analysis range from univariate signal processing analyses of individual channels to bivariate analyses on pairs of channels, and to networks in which connectivity between all pairs of channels are assessed and emergent network properties are studied. (B) Various methods of quantitative seizure onset zone localization have been studied using both interictal and ictal data (C) Stimulation-based mapping is an effective method of probing the dynamics of epileptogenic networks.
HFOs are transient oscillatory events in the frequency range of 80 to 500 Hz, often separated into ripples (80–250 Hz) and fast ripples (250–500 Hz), that stand out from background activity.18,20,27 It has been reported that high rates of HFOs are present in epileptic tissue and that removal of regions with high HFO rates results in more favorable outcomes.28-31 However, as HFO analysis has moved to automatic detection due to the poor inter-rater reliability of visual review,32-35 recent prospective studies36 and meta-analyses37,38 have reported little advantage in using HFOs for surgical planning. The performance of HFOs in predicting surgical outcome may be biased by several factors. First, HFOs are present physiologically, and this confounds the distinction of pathological HFOs.39-41 Efforts to create an HFO atlas42 and to assess HFO rates during cognitive tasks43 may help distinguish pathological tissues. Second, though fast ripples were shown to be more specific to epileptogenic tissues,44 clinical viability may rely on fast ripple detection combined with epileptic spikes or post-resection recordings.45,46 However, the mechanistic theory that HFOs are network-driven phenomena may explain this discrepancy in fast ripple specificity.45,47,48
Interictal spikes are brief, abnormal electrical discharges seen in epileptic patients during seizure-free intervals. The mechanistic relationship between spikes and seizures is still unknown, although they are temporally related in many patients49 and both are observed to manifest similar multi-day cycles.50 Given that spikes occur more frequently than seizures, they have been extensively studied for their ability to guide epilepsy surgery. Examining their location in frequency in iEEG has revealed that their spatial distribution fluctuates over time,26 but that good surgical outcome is associated with the resection of regions which generate the highest frequency of spikes.51,52 Furthermore, gamma activity preceding spikes provides additional sensitivity for discharges which mark the EZ.53 However, spikes often arise outside regions of seizure onset,54 complicating the presumed relationship between spike and seizure generation. These areas remain under active investigation, spanning research protocols that vary widely across institutions.
With epilepsy increasingly conceptualized as a network disorder, various network-based measures have been proposed to characterize the connectivity patterns in the epileptic brain network.55-61 Recent studies suggest that the epileptic network not only demonstrates abnormalities during seizures but also at rest55,62,63 and information about epileptogenic regions can be gleaned from the resting-state network.57,58,64 Several studies have demonstrated increased synchronization in seizure-onset regions.56,65 and a high influence of epileptogenic regions on the brain network during interictal periods.55,57,58,62,64,66 To construct iEEG networks, graph-theoretic measures computed from an adjacency matrix that represents pairwise dependencies (correlation or coherence) between iEEG channels56,64,67 are often used. These metrics are not always easily interpretable as different networks can result in identical metrics.68 To overcome this, others have derived dynamical models of the iEEG signals,57,58,69 which are designed to capture the underlying dynamical properties of the epileptic network responsible for seizure generation. A recent study implemented a time-varying autoregressive model to conceptualize source and sink nodes (regions) in the epileptic network where the sinks, regions being inhibited by sources, are correlated to epileptogenic regions during interictal periods.58,70
Ictal methods
Quantitative evaluation of ictal recordings can provide additional localizing value beyond clinician recognition of canonical seizure onset patterns. Many such approaches analyse the spectral characteristics of seizure onset patterns to determine which channels are most critically involved in seizure generation. One method leverages a wavelet transform (Fig. 2B) of the iEEG signals recorded on each individual channel in patients that became seizure free, to identify a ‘fingerprint of the epileptogenic zone’.71,72 Approaches from computer vision were used to extract features from the time-frequency plot, which were then classified as epileptogenic using a support vector machine. Another widely studied, non-linear univariate metrics is the epileptogenicity index (EI; Fig. 2B),73 which quantifies clinically-observable patterns based on both the spectral and temporal delay patterns of iEEG. To compute EI, two specific metrics are calculated over a sliding window: (i) the ‘ER’, or the signal energy ratio between the beta and gamma bands compared to the theta and alpha bands; and (ii) a cumulative sum algorithm used over the ‘ER’ signal to determine when it significantly changes, marking a shift from low frequency to high frequency activity. Additionally, cross-frequency coupling has proved to be an important characteristic of epileptogenic tissues. For example, channels within the ictal core contain a high degree of phase locking between the high-gamma band and lower frequencies.74 Studies have shown that using this method to localize epileptogenic networks accurately predicts surgical outcomes better than experts marking the seizure onset zone alone.74
Connectivity-based, or ‘network’ approaches for analysing ictal activity generally estimate connectivity values between channels, via either a bivariate, or multivariate statistical approach75 (Fig. 2A). Analysing eigenvector centrality (EC), a measure of a node’s influence within a network, (Fig. 2B) of the iEEG correlation graph in frequency space shows a network transition from the interictal to ictal state.76 A similar analysis correlates the EC with the clinically hypothesized epileptogenic regions of a retrospective pool of patients.59 Analysing the network during ictal periods may also help predict surgical outcomes.77 Assuming the network is estimated sufficiently, virtual resection in the context of the model may help predict surgical outcomes.65,78 In a recent study, analysis of time-varying dynamical connectivity enabled computation of ‘neural fragility’ metric (Fig. 2B) that predicted surgical outcome in a large retrospective multicentre cohort of 91 patients.79-81 The study showed this by conditioning on the clinically hypothesized epileptogenic regions, indirectly suggesting that neural fragility could be useful for localization. Interestingly, neural fragility was also shown to increase across the course of epileptogenesis in a small animal study,82 implying that it provides not only localizing but perhaps mechanistic implications towards seizure generation.
Stimulation-based mapping
Investigations of the EZ using electrical stimulation (Fig. 2C) have undergone a resurgence in the past two decades since the first pioneering experiments by Penfield, Jasper, Ojemann and others.83-85 Specifically, single pulse electrical stimulation (SPES) has gained popularity as a measure of effective connectivity, where brief (150–300 µs per phase) pulses evoke responses in local and remote regions to indicate a structural or functional connection to the stimulation site.86-88 These cortico-cortical evoked potentials (CCEPs) have been used to map interregional connectivity in vivo in the language network,89 motor cortex,89 limbic network,90 parietal-frontal connections,91 and deep brain structures.87,92 SPES is safe,93 reveals local and distant functional networks,89,94,95 and provides complementary information when combined with other neuroimaging modalities.96
To delineate seizure networks, differences in features of the CCEP waveform have been associated with increased cortical excitability.90,97,98 The amplitude of the N1 response (typically 10–50 ms post-stimulus) is often greater in seizure onset regions and early spread regions when compared to healthy tissue,90,97-99 and stimulating the seizure onset region produces larger remote responses with increased connectivity.99-101 More recently, greater cortico-cortical spectral responses and induced high-frequency activity have been shown to localize epileptogenic tissue.102-106 Additionally, ‘delayed responses’, neuronal activities that resemble spikes or slow waves that occur 100 ms to 1 s after stimulation onset, are more frequently observed in seizure onset zone regions,107,108 and removal of these areas result in improved outcomes.107,109
In contrast to these signal properties, systems-level analysis of CCEP data to localize the epileptogenic network has been recently proposed.110-113 In a recent study, authors tested the hypothesis that dynamical network models derived from CCEPs can reveal epileptic network connections and the underlying dynamics of seizure generation. Specifically, they posit that brain regions where small periodic inputs produce large amplitude oscillations in the intracranial EEG correspond to the seizure onset zone. Such responses occur when there is a resonant frequency of the brain network, and this frequency can be detected by a sharp peak in the frequency response curve.113
Several factors that affect the CCEP waveform remain open questions, impeding the widespread use of SPES in surgical planning for epilepsy. SPES and CCEPs were first defined with electrocorticography (ECoG) grids, but the increased use of stereo-EEG (SEEG) raises questions about optimal stimulation parameters, volume of activated tissue, and artifact considerations.85,114-116 The CCEP waveform depends heavily on location of the stimulating and response electrodes,66 whether in grey or white matter117,118 or in highly functional regions.119 Larger validation studies answering these questions will ease implementation of stimulation-based investigations of seizure networks in the clinical workflow.
In a broader context, electrical stimulation has been used to evoke seizures to aid in seizure network inference.113,120,121 There are relatively few studies that directly examine the improvement in surgical outcome when using clinical information derived from stimulation-induced seizures,122 but a general trend towards adoption of stimulation-derived investigations of seizure networks provides a unique and timely opportunity for further investigation.113
Challenges and opportunities
In this section, we review a series of challenges that have prevented most quantitative methods of localizing the EZ from becoming clinical tools which routinely impact clinical practice for patients undergoing epilepsy surgery (Fig. 3). For each of these challenges, we propose ways of adapting future studies to mitigate their effects on results and clinical translatability.
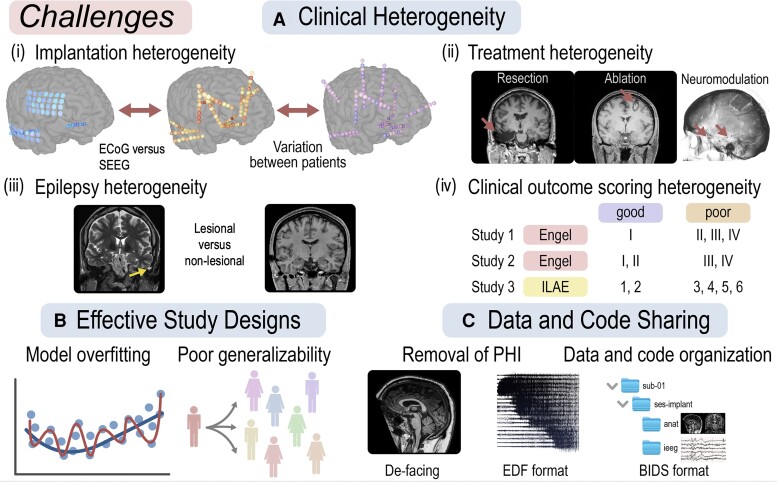
Figure 3.
Challenges and opportunities. (A) Clinical heterogeneity is a primary challenge in deploying quantitative iEEG methods in epilepsy surgery, and encompasses variability in implant type, therapy, underlying pathology, and outcome metrics. (B) Effective study design should include development, validation, and test cohorts to minimize overfitting and maximize generalizability. (C) Data and code sharing should be a standard practice in iEEG studies in epilepsy and should incorporate de-identification and standardized formatting of raw data, imaging, and metadata such as using the BIDS framework.
Challenge 1: clinical heterogeneity
One of the primary barriers to deploying quantitative methods to guide epilepsy surgery is the vast clinical heterogeneity that exists across patients with epilepsy (Fig. 3A). We identify the main sources of heterogeneity which may influence network models as (i) variability in electrode implantation types and placement, such as ECoG and SEEG; (ii) variability in treatment approaches, such as resection, ablation, and neurostimulation; (iii) variability in types of epilepsy, aetiology and underlying genetic and pathological substrates; and (iv) variation in the methods of assessing clinical outcome.
The type of data recorded from ECoG and SEEG are inherently different. ECoG records from the surface of the brain capturing grey matter often with a uniform spacing between contacts on a grid electrode array. SEEG depth electrodes capture white matter in addition to grey matter, which differ in both qualitative and quantitative properties during interictal and seizure periods. These differences in implants are known to impact the utility of network models in each type of implant123 and the effects are different across different network statistics.124,125 We propose that future studies validate their methods using SEEG subjects when possible.
Treatment type is also an important factor because resections typically cover a larger (and possibly even different) portion of the brain than the clinically hypothesized epileptogenic region. Laser ablations target less tissue but are typically only used if clinicians have a strong hypothesis for a focal epileptogenic network. Finally, implanted neurostimulators do not remove tissue at all, but rather stimulate to suppress activity in regions hypothesized as epileptogenic or exert functional influence over it (e.g. thalamic stimulation).126 Understanding possible treatment and selection bias is vital to conducting large, controlled multicentre studies involving multiple treatment modalities.127 We propose that future efforts control for the type of surgical intervention, and exercise caution when cohorts contain both subjects that underwent surgery with curative intent (resection, ablation) and those with palliative intent (e.g. neurostimulation).
Epilepsy is a clinically heterogeneous disorder which encompasses a variety of underlying aetiologies, pathologies and anatomic distributions. Aetiologies of focal epilepsy include both clearly delineated lesions, such as mesial temporal sclerosis, focal cortical dysplasias, and cavernomas. Poorly localized or multi-focal processes can also cause epilepsy, including traumatic brain injury, stroke, or infection. As seizure onset patterns differ between different lesions and aetiologies, even for the same brain region,5 it follows that the mechanisms underlying seizure generation and therefore quantitative variability could also differ and affect the accuracy of quantitative models. Furthermore, distinct anatomic distributions of hypothesized epileptogenic lesions lead to different network sampling and therefore distinct quantitative properties evident on iEEG. We suggest that future models document both performance on mesial temporal lobe epilepsy and neocortical epilepsy where possible.
Finally, there is substantial bias in quantitative iEEG studies as a result of different clinical assessments of surgical outcome. Criteria for defining which patients achieve ‘good’ versus ‘poor’ outcome are not universally accepted, and even the term ‘seizure free’ may or may not encompass non-disabling auras. Additionally, these metrics change over time as patients relapse, so the results of a single study may only hold valid at a single point in time. Finally, appropriate outcome scales do not exist for neuromodulatory devices or surgical interventions intended to be palliative. We propose that studies document both early and late outcomes when feasible, and that patients with true seizure freedom are treated separately from those with a favorable clinical outcome but remaining auras.
Overall, few single centres have a sufficient clinical volume to study a clinically homogeneous cohort. Rigorous documentation of subject-level clinical metadata will increase the interpretability of future studies, and renewed efforts towards cross-centre collaboration and data-sharing will permit the aggregation of subjects with similar implants, surgery, and pathology to more finely probe the factors which drive seizure freedom and relapse.
Challenge 2: effectively designing retrospective studies
Another significant barrier to clinical translation is variability in retrospective study design (Fig. 3B). We propose that methods should be developed on a training cohort, optimized on a validation cohort, and performance quantified on a held-out test cohort to prevent overfitting to small datasets.128 Ideally, the set of clinically hypothesized regions are well within the surgically resected/ablated areas in training data to provide the most accurate representation of epileptogenicity possible.
Additionally, most retrospective studies aim to localize tissue that should be removed, but do not explicitly identify regions that should be preserved. In clinical practice, the selection of surgical approach and extent weighs the benefit of seizure freedom with the risk of neurologic morbidity and patient preference. Besides the avoidance of eloquent cortex, regions which have a desynchronizing effect on the epileptogenic network also may be important to preserve for an optimal seizure outcome. Practically, future studies should indicate which subjects had resection strategies limited by their EZ localized to eloquent cortex and models should take neuro-cognitive testing and cortical mapping into account for symptom avoidance.
A recent trend in designing studies to validate localization algorithms is to predict surgical outcomes instead of identifying the EZ, since these regions are not observed. However, one should be aware of non-causal covariates that may bias results to be overly optimistic. For example, if the number of channels is higher in patients with surgical outcomes Engel II, or greater, and a prediction algorithm adds the number of channels to its set of features then the prediction algorithm leverages a spurious correlation of the number of channels to predict outcome, which has no real clinical utility.129 Designing effective studies that fairly evaluate a proposed biomarker of epileptogenicity requires a deep understanding of the data and statistical challenges.
Finally, some models have a high predictive value but low explanatory value—addressing this trade-off is another challenge. For example, quantitative models that have high physiological plausibility may be easier for physicians to understand and use but they may not generalize well. In contrast, some deep-learning models might generalize well on new data but could be challenging to interpret. Indeed, many of the best tools for seizure prediction apply machine learning to quantitative iEEG features rather than model underlying electrophysiologic phenomena. On the other hand, mechanistic studies of epilepsy may reveal novel approaches to treating seizures including with drugs and neuromodulation. As our understanding of epilepsy grows, it is critical to advance predictive and explanatory studies together.
Challenge 3: data and code sharing
Data and code sharing is fundamental to moving computational epilepsy studies towards clinical translation. We advocate for proper de-identification of protected health information, unified formatting of datasets, long-term public storage of the data, and the release of open-source code (Fig. 3C). Many groups choose to not share raw data due to institutional regulations, privacy laws, or burdensome data wrangling. However, misaligned incentives against data-sharing exist such as competition for publications, grants and potential for monetization.130,131 We propose that sufficiently de-identifying iEEG datasets and including complementary imaging and clinical metadata is feasible, such that quality data sharing should be required by funding agencies and academic journals. Many software packages exist that facilitate removing identifying information in common recording formats, such as EDF.132-134 For neuroimaging, defacing software135 can automatically remove the face part of the images and storage of Nifti (.nii) files facilitates anonymization. With these advances, it is possible to share EEG-recording and imaging data along with non-identifiable metadata to facilitate community-driven solutions for epileptogenic network localization.
However, it is not enough simply to ‘share’ the data as many formats for both raw EEG data and metadata are difficult for outside users to effectively parse. The Brain Imaging Data Structure (BIDS) is a community-driven format for storing and sharing de-identified data,135-137 supports all common EEG recording formats and has an open-source specification for metadata storage. Open-source web portals, such as OpenNeuro, can automatically check data to determine BIDS-compliance. Other platforms such as Pennsieve, which is developing an Epilepsy Data Ecosystem to integrate with other platforms, does similar data aggregation and data sharing for the NIH SPARC program.138 Finally, sharing high-quality code and documentation (such as on GitHub) can allow researchers to easily benchmark their proposed algorithms against prior work.
Introduction of a diverse, multicentre dataset
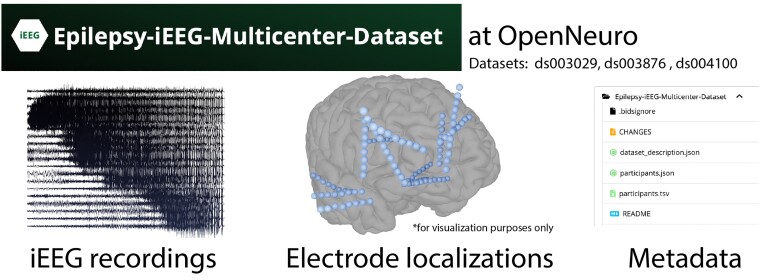
One of the primary barriers to appropriately developing and testing robust methods of EZ localization which can handle the vast clinical heterogeneity that exists among patients with epilepsy is a lack of enough high-quality data. To address this need, we document a publicly available dataset including recordings from 122 patients who underwent iEEG implantation and subsequent surgery for drug resistant epilepsy (Fig. 4). The data are organized in the standardized BIDS format on OpenNeuro, in projects ds003029, ds003876, and ds004100. Each dataset includes clinical metadata, such as implant type, surgery type, surgical outcome, electrode labels and standardized coordinates, as well as which electrode contacts were targeted by surgery. The electrophysiologic data is recorded and stored in referential montages and is unprocessed. Seizures were identified by board-certified epileptologists, and interictal clips were selected to minimize the presence of artefacts. Project ds003029 contains seizure recordings (mean per patient 3 ± 1.3), ds003876 contains interictal recordings (one to four clips per patient), while ds004100 contains both (mean seizures per patient 5.6 ± 1.5 with one interictal clip each). Full descriptions are present in the ReadMe files contained on OpenNeuro. Our datasets do not include clips where stimulation-based mapping was performed. Portions of each dataset have been used and published as part of previous studies58,63,65,79; however, we release additional subjects, recordings, electrode localizations, and metadata that were previously unpublished.
Figure 4.
Multicentre open-source iEEG datasets. We have released our data on OpenNeuro to aid in development, validation, and testing of new methods of seizure onset zone localization.
Using our dataset as well as other high-quality iEEG cohorts,139,140 future studies may effectively develop, validate, and test a wide variety of methods to quantitatively localize the EZ. For example, subsets of our dataset can be used to study more clinically homogeneous populations, such as those which were only implanted with sEEG, or only underwent MTL laser ablations. Alternatively, our dataset could serve as a ‘test set’ to quantify performance of algorithms developed on private cohorts. Finally, we encourage others to release de-identified data from their centre in the iEEG BIDS format so that further studies may achieve even better statistical rigor, and our group currently has NIH funding to help aggregate more data across epilepsy centres to accelerate research.
Prospective studies
We posit that multiple methods, each which offer complementary information, may be integrated into a single software package that provides spatially-localized probabilities of epileptogenicity.70 Ideally, these tools may reach a high level of accuracy using only interictal recording or stimulation data alone, reducing the lengthy and costly stays in epilepsy monitoring units for recording seizures. While we foresee that some proprietary algorithms may arise, we believe that it is critical to keep scientific discovery in this field open-source so that all centres may have equitable access to tools which can improve care. We further expect that clinical quantitative iEEG models will complement insights from neuroimaging-based EZ localization tools leveraging structural, diffusion-weighted, and functional MRI. Ultimately, we foresee that the quantitative insight that these tools provide will augment, rather than replace, the expertise of clinicians in surgical planning.
Incorporating retrospectively validated computational models in prospective clinical trials will require careful study design. Decision making at the patient level could include predictions from quantitative models in an unblinded, randomized fashion to help clinicians decide between two equally probable clinical hypotheses (e.g. medial temporal versus medial + temporal neocortical onsets), decide between therapeutic options in very specific epilepsy surgery decisions (e.g. standard temporal lobectomy versus medial temporal ablation), or to advise surgical teams regarding the extent of intervention in cases where several options are present. While there are precedents for all of these types of trials, a careful, multicentre approach will likely be necessary to establish protocols, standards for data collection, annotation, analysis and interpretation, with sufficient power to assess the value of these methods. Certainly non-destructive therapies, such as neuromodulation, which can be tested in different portions of the epileptic network in the same patient, provide an interesting alternative to larger surgical trials, but current hardware limitations in the number of contacts and leads make this somewhat invasive, as switching between different implanted leads currently requires repeat surgery and changing connections to implanted devices.
With our description and consolidation of a high-quality multicentre dataset, we hope that researchers will benchmark their proposed algorithms using this cohort in a sound statistical fashion, similar to our prior efforts to benchmark seizure detection and prediction.11,141 We encourage researchers to de-identify their data using the tools described and release their data in an open access and easily shareable format. These collective efforts will continue to move the field towards a robust epileptogenic network localization algorithm.
Supplementary Material
Contributor Information
John M Bernabei, Department of Bioengineering, School of Engineering & Applied Science, University of Pennsylvania, Philadelphia, PA 19104, USA; Center for Neuroengineering & Therapeutics, University of Pennsylvania, Philadelphia, PA 19104, USA.
Adam Li, Department of Computer Science, Columbia University, New York, NY 10027, USA.
Andrew Y Revell, Department of Bioengineering, School of Engineering & Applied Science, University of Pennsylvania, Philadelphia, PA 19104, USA.
Rachel J Smith, Department of Electrical and Computer Engineering, University of Alabama at Birmingham, Birmingham, AL 35294, USA; Neuroengineering Program, University of Alabama at Birmingham, Birmingham, AL 35294, USA.
Kristin M Gunnarsdottir, Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD 21218, USA; Institute for Computational Medicine, Johns Hopkins University, Baltimore, MD 21218, USA.
Ian Z Ong, Department of Bioengineering, School of Engineering & Applied Science, University of Pennsylvania, Philadelphia, PA 19104, USA.
Kathryn A Davis, Center for Neuroengineering & Therapeutics, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Neurology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Nishant Sinha, Center for Neuroengineering & Therapeutics, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Neurology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Sridevi Sarma, Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD 21218, USA; Institute for Computational Medicine, Johns Hopkins University, Baltimore, MD 21218, USA.
Brian Litt, Department of Bioengineering, School of Engineering & Applied Science, University of Pennsylvania, Philadelphia, PA 19104, USA; Center for Neuroengineering & Therapeutics, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Neurology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Neurosurgery, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Funding
J.M.B. and A.Y.R. received support from NINDS T32: NS091006. A.L. received support by NIH T32 EB003383, the NSF GRFP (DGE-1746891), the ARCS Scholarship, Whitaker Fellowship, the Chateaubriand Fellowship and National Science Foundation under CIFellows 2021 grant #: 2127309 to the Computing Research Association for the CIFellows Project. N.S. acknowledges funding from NINDS R01NS116504 and American Epilepsy Society (953257). R.J.S. received support from an NIH IRACDA program, ASPIRE at JHU. K.M.G. was supported by a grant from the American Epilepsy Society. K.D. acknowledges funding from NINDS R01NS116504 and 2R56NS099348–05A1. S.S. received support from NIH R01NS125897 and NIH R01NS122927. B.L. received support from DP1NS122038, 2R56NS099348-05A1, The Mirowski Family Foundation, and Jonathan and Bonnie Rothberg.
Competing interests
The authors have no conflicts of interest to disclose.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Rosenow F, Lüders H. Presurgical evaluation of epilepsy. Brain. 2001;124:1683–1700. [DOI] [PubMed] [Google Scholar]
- 2. Khambhati AN, Davis KA, Lucas TH, Litt B, Bassett DS. Virtual cortical resection reveals push-pull network control preceding seizure evolution. Neuron. 2016;91:1170–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Revell AY, Silva AB, Mahesh D, et al. White matter signals reflect information transmission between brain regions during seizures. bioRxiv. [Preprint]https://doi.org/2021.09.15.460549 [Google Scholar]
- 4. Lagarde S, Buzori S, Trebuchon A, et al. The repertoire of seizure onset patterns in human focal epilepsies: Determinants and prognostic values. Epilepsia. 2019;60:85–95. [DOI] [PubMed] [Google Scholar]
- 5. Perucca P, Dubeau F, Gotman J. Intracranial electroencephalographic seizure-onset patterns: Effect of underlying pathology. Brain. 2014;137:183–196. [DOI] [PubMed] [Google Scholar]
- 6. Tellez-Zenteno JF, Dhar R, Hernandez-Ronquillo L, Wiebe S. Long-term outcomes in epilepsy surgery: Antiepileptic drugs, mortality, cognitive and psychosocial aspects. Brain. 2007;130:334–345. [DOI] [PubMed] [Google Scholar]
- 7. Jobst BC, Cascino GD. Resective epilepsy surgery for drug-resistant focal epilepsy: A review. JAMA. 2015;313:285–293. [DOI] [PubMed] [Google Scholar]
- 8. Khoo A, Tisi J, Mannan S, et al. Reasons for not having epilepsy surgery. Epilepsia. 2021;62:2909–2919. [DOI] [PubMed] [Google Scholar]
- 9. Engel J Jr. What can we do for people with drug-resistant epilepsy? The 2016 Wartenberg lecture. Neurology. 2016;87:2483–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Steinbrenner M, Kowski AB, Holtkamp M. Referral to evaluation for epilepsy surgery: Reluctance by epileptologists and patients. Epilepsia. 2019;60:211–219. [DOI] [PubMed] [Google Scholar]
- 11. Kuhlmann L, Karoly P, Freestone DR, et al. Epilepsyecosystem.org: Crowd-sourcing reproducible seizure prediction with long-term human intracranial EEG. Brain. 2018;141:2619–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Freestone DR, Karoly PJ, Cook MJ. A forward-looking review of seizure prediction. Curr Opin Neurol. 2017;30:167–173. [DOI] [PubMed] [Google Scholar]
- 13. Gadhoumi K, Lina JM, Mormann F, Gotman J. Seizure prediction for therapeutic devices: A review. J Neurosci Methods. 2016;260:270–282. [DOI] [PubMed] [Google Scholar]
- 14. Mormann F, Andrzejak RG, Elger CE, Lehnertz K. Seizure prediction: The long and winding road. Brain. 2007;130:314–333. [DOI] [PubMed] [Google Scholar]
- 15. Parvizi J, Kastner S. Promises and limitations of human intracranial electroencephalography. Nat Neurosci. 2018;21:474–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jobst BC, Bartolomei F, Diehl B, et al. Intracranial EEG in the 21st century. Epilepsy Curr. 2020;20:180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chari A, Thornton RC, Tisdall MM, Scott RC. Microelectrode recordings in human epilepsy: A case for clinical translation. Brain Commun. 2020;2:fcaa082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Frauscher B, Bartolomei F, Kobayashi K, et al. High-frequency oscillations: The state of clinical research. Epilepsia. 2017;58:1316–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thomschewski A, Hincapié A-S, Frauscher B. Localization of the epileptogenic zone using high frequency oscillations. Front Neurol. 2019;10:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Remakanthakurup Sindhu K, Staba R, Lopour BA. Trends in the use of automated algorithms for the detection of high-frequency oscillations associated with human epilepsy. Epilepsia. 2020;61:1553–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Khadjevand F, Cimbalnik J, Worrell GA. Progress and remaining challenges in the application of high frequency oscillations as biomarkers of epileptic brain. Curr Opin Biomed Eng. 2017;4:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee S, Issa NP, Rose S, et al. DC Shifts, high frequency oscillations, ripples and fast ripples in relation to the seizure onset zone. Seizure. 2020;77:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park CJ, Hong SB. High frequency oscillations in epilepsy: Detection methods and considerations in clinical application. J. Epilepsy Res. 2019;9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roehri N, Bartolomei F. Are high-frequency oscillations better biomarkers of the epileptogenic zone than spikes? Curr Opin Neurol. 2019;32:213–219. [DOI] [PubMed] [Google Scholar]
- 25. Dworetzky BA, Reinsberger C. The role of the interictal EEG in selecting candidates for resective epilepsy surgery. Epilepsy Behav. 2011;20:167–171. [DOI] [PubMed] [Google Scholar]
- 26. Conrad EC, Tomlinson SB, Wong JN, et al. Spatial distribution of interictal spikes fluctuates over time and localizes seizure onset. Brain. 2020;143:554–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jacobs J, Staba R, Asano E, et al. High-frequency oscillations (HFOs) in clinical epilepsy. Prog Neurobiol. 2012;98:302–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Klink NEC, van’t Klooster MA, Zelmann R, et al. High frequency oscillations in intra-operative electrocorticography before and after epilepsy surgery. Clin Neurophysiol. 2014;125:2212–2219. [DOI] [PubMed] [Google Scholar]
- 29. Kerber K, Dümpelmann M, Schelter B, et al. Differentiation of specific ripple patterns helps to identify epileptogenic areas for surgical procedures. Clin Neurophysiol. 2014;125:1339–1345. [DOI] [PubMed] [Google Scholar]
- 30. Cho JR, Koo DL, Joo EY, et al. Resection of individually identified high-rate high-frequency oscillations region is associated with favorable outcome in neocortical epilepsy. Epilepsia. 2014;55:1872–1883. [DOI] [PubMed] [Google Scholar]
- 31. Haegelen C, Perucca P, Châtillon CE, et al. High-frequency oscillations, extent of surgical resection, and surgical outcome in drug-resistant focal epilepsy. Epilepsia. 2013;54:848–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nariai H, Wu JY, Bernardo D, et al. Interrater reliability in visual identification of interictal high-frequency oscillations on electrocorticography and scalp EEG. Epilepsia Open. 2018;3:127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Spring AM, Pittman DJ, Aghakhani Y, et al. Interrater reliability of visually evaluated high frequency oscillations. Clin Neurophysiol. 2017;128:433–441. [DOI] [PubMed] [Google Scholar]
- 34. Spring AM, Pittman DJ, Aghakhani Y, et al. Generalizability of high frequency oscillation evaluations in the ripple band. Front Neurol. 2018;9:510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gardner AB, Worrell GA, Marsh E, Dlugos D, Litt B. Human and automated detection of high-frequency oscillations in clinical intracranial EEG recordings. Clin Neurophysiol. 2007;118:1134–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jacobs J, Wu JY, Perucca P, et al. Removing high-frequency oscillations: A prospective multicenter study on seizure outcome. Neurology. 2018;91:e1040–e1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gloss D, Nolan SJ, Staba R. The role of high-frequency oscillations in epilepsy surgery planning. Cochrane Database Syst Rev. 2017;10:CD010235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Höller Y, Kutil R, Klaffenböck L, et al. High-frequency oscillations in epilepsy and surgical outcome. A meta-analysis. Front Hum. Neurosci. 2015;9:574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cimbalnik J, Brinkmann B, Kremen V, et al. Physiological and pathological high frequency oscillations in focal epilepsy. Ann Clin Transl Neurol. 2018;5:1062–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Matsumoto A, Brinkmann BH, Matthew Stead S, et al. Pathological and physiological high-frequency oscillations in focal human epilepsy. J Neurophysiol. 2013;110:1958–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alkawadri R, Gaspard N, Goncharova II, et al. The spatial and signal characteristics of physiologic high frequency oscillations. Epilepsia. 2014;55:1986–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Frauscher B, von Ellenrieder N, Zelmann R, et al. High-frequency oscillations in the normal human brain. Ann Neurol. 2018;84:374–385. [DOI] [PubMed] [Google Scholar]
- 43. Kucewicz MT, Cimbalnik Jan, Matsumoto JY, et al. High frequency oscillations are associated with cognitive processing in human recognition memory. Brain. 2014;137:2231–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bragin A, Engel JJR, Wilson CL, Fried I, Buzsáki G. High-frequency oscillations in human brain. Hippocampus. 1999;9:137–142. [DOI] [PubMed] [Google Scholar]
- 45. van ‘t Klooster MA, van Klink NEC, Zweiphenning WJEM, et al. Tailoring epilepsy surgery with fast ripples in the intraoperative electrocorticogram. Ann Neurol. 2017;81:664–676. [DOI] [PubMed] [Google Scholar]
- 46. Roehri N, Pizzo F, Lagarde S, et al. High-frequency oscillations are not better biomarkers of epileptogenic tissues than spikes. Ann Neurol. 2018;83:84–97. [DOI] [PubMed] [Google Scholar]
- 47. Li X, Zhang H, Lai H, Wang J, Wang W, Yang X. High-Frequency oscillations and epileptogenic network. Curr Neuropharmacol. 2022;20:1687–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zweiphenning WJEM, van ‘t Klooster MA, van Diessen E, et al. High frequency oscillations and high frequency functional network characteristics in the intraoperative electrocorticogram in epilepsy. Neuroimage Clin. 2016;12:928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Karoly PJ, Freestone DR, Boston R, et al. Interictal spikes and epileptic seizures: Their relationship and underlying rhythmicity. Brain. 2016;139:1066–1078. [DOI] [PubMed] [Google Scholar]
- 50. Baud MO, Kleen JK, Mirro EA, et al. Multi-day rhythms modulate seizure risk in epilepsy. Nat Commun. 2018;9:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim DW, Kim HK, Lee SK, Chu K, Chung CK. Extent of neocortical resection and surgical outcome of epilepsy: Intracranial EEG analysis. Epilepsia. 2010;51:1010–1017. [DOI] [PubMed] [Google Scholar]
- 52. Azeem A, Ellenrieder N, Hall J, et al. Interictal spike networks predict surgical outcome in patients with drug-resistant focal epilepsy. Ann Clin Transl Neurol. 2021;8:1212–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Thomas J, Kahane P, Abdallah C, et al. A subpopulation of spikes predicts successful epilepsy surgery outcome. Ann Neurol. Published online 13 November 2022. doi: 10.1002/ana.26548 [DOI] [PubMed] [Google Scholar]
- 54. Marsh ED, Peltzer B, Brown MW III, et al. Interictal EEG spikes identify the region of electrographic seizure onset in some, but not all, pediatric epilepsy patients. Epilepsia. 2010;51:592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Korzeniewska A, Cervenka MC, Jouny CC, et al. Ictal propagation of high frequency activity is recapitulated in interictal recordings: Effective connectivity of epileptogenic networks recorded with intracranial EEG. Neuroimage. 2014;101:96–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shah P, Bernabei JM, Kini LG, et al. High interictal connectivity within the resection zone is associated with favorable post-surgical outcomes in focal epilepsy patients. Neuroimage Clin. 2019;23:101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sinha N, Dauwels J, Kaiser M, et al. Predicting neurosurgical outcomes in focal epilepsy patients using computational modelling. Brain. 2017;140:319–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gunnarsdottir KM, Li A, Smith RJ, et al. Source-sink connectivity: A novel interictal EEG marker for seizure localization. Brain. 2022;145:3901–3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li A, Chennuri B, Subramanian S, et al. Using network analysis to localize the epileptogenic zone from invasive EEG recordings in intractable focal epilepsy. Netw Neurosci. 2018;2:218–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sinha N, Johnson GW, Davis KA, Englot DJ. Integrating network neuroscience into epilepsy care: Progress, barriers, and next steps. Epilepsy Curr. 2022;22:272–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stacey W, Kramer M, Gunnarsdottir Ket al. Emerging roles of network analysis for epilepsy. Epilepsy Res. 2020;159:106255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Taylor PN, Papasavvas CA, Owen TW, et al. Normative brain mapping of interictal intracranial EEG to localise epileptogenic tissue. Brain. 2022;145:939–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bernabei JM, Sinha N, Arnold TC, et al. Normative intracranial EEG maps epileptogenic tissues in focal epilepsy. Brain. 2022;145:1949–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Narasimhan S, Kundassery KB, Gupta K, et al. Seizure-onset regions demonstrate high inward directed connectivity during resting-state: An SEEG study in focal epilepsy. Epilepsia. 2020;61:2534–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kini LG, Bernabei JM, Mikhail F, et al. Virtual resection predicts surgical outcome for drug-resistant epilepsy. Brain. 2019;142:3892–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hays MA, Coogan C, Crone NE, Kang JY. Graph theoretical analysis of evoked potentials shows network influence of epileptogenic mesial temporal region. Hum Brain Mapp. 2021;42:4173–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cimbalnik J, Klimes P, Sladky V, et al. Multi-feature localization of epileptic foci from interictal, intracranial EEG. Clin Neurophysiol. 2019;130:1945–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chung, J, Bridgeford EW, Arroyo J, et al. Statistical Connectomics. OSF Preprints. [Preprint] doi: 10.31219/osf.io/ek4n3 [DOI]
- 69. Li A, Sarma SV, Fitzgerald Z, et al. Virtual cortical stimulation mapping of epilepsy networks to localize the epileptogenic zone. Annu Int Conf IEEE Eng Med Biol Soc 2019:2328–2331. [DOI] [PubMed] [Google Scholar]
- 70. Sinha N. Localizing epileptogenic tissues in epilepsy: Are we losing (the) focus? Brain. 2022;145:3735–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Grinenko O, Li J, Mosher JC, et al. A fingerprint of the epileptogenic zone in human epilepsies. Brain. 2018;141:117–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Li J, Grinenko O, Mosher JC, Gonzalez-Martinez J, Leahy RM, Chauvel P. Learning to define an electrical biomarker of the epileptogenic zone. Hum Brain Mapp. 2020;41:429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bartolomei F, Chauvel P, Wendling F. Epileptogenicity of brain structures in human temporal lobe epilepsy: A quantified study from intracerebral EEG. Brain. 2008;131:1818–1830. [DOI] [PubMed] [Google Scholar]
- 74. Weiss SA, Lemesiou A, Connors R, et al. Seizure localization using ictal phase-locked high gamma: A retrospective surgical outcome study. Neurology. 2015;84:2320–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bassett DS, Sporns O. Network neuroscience. Nat Neurosci. 2017;20:353–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Burns SP, Santaniello S, Yaffe RB, et al. Network dynamics of the brain and influence of the epileptic seizure onset zone. Proc Natl Acad Sci U S A. 2014;111:E5321–E5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Goodfellow M, Rummel C, Abela E, Richardson MP, Schindler K, Terry JR. Estimation of brain network ictogenicity predicts outcome from epilepsy surgery. Sci Rep. 2016;6:29215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Scheid BH, Bernabei JM, Khambhati AN, et al. Intracranial electroencephalographic biomarker predicts effective responsive neurostimulation for epilepsy prior to treatment. Epilepsia. 2022;63:652–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Li A, Huynh C, Fitzgerald Z, et al. Neural fragility as an EEG marker of the seizure onset zone. Nat Neurosci. 2021;24:1465–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li A, Inati S, Zaghloul K, Sarma S. Fragility in epileptic networks: The epileptogenic zone. In: 2017 American control conference (ACC). IEEE; 2017. [Google Scholar]
- 81. Li A, Gunnarsdottir KM, Inati S, et al. Linear time-varying model characterizes invasive EEG signals generated from complex epileptic networks. Annu Int Conf IEEE Eng Med Biol Soc. 2017:2802–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ehrens D, Li A, Aeed F, Schiller Y, Sarma SV. Network fragility for seizure genesis in an acute in vivo model of epilepsy. In: 2020 42nd annual international conference of the IEEE engineering in medicine & biology society (EMBC); 2020. [DOI] [PubMed] [Google Scholar]
- 83. Penfield W, Jasper H. Epilepsy and the functional anatomy of the human brain. JAMA. 1954;155:86. [Google Scholar]
- 84. Ojemann GA. Individual variability in cortical localization of language. J Neurosurg. 1979;50:164–169. [DOI] [PubMed] [Google Scholar]
- 85. Grande KM, Ihnen SKZ, Arya R. Electrical stimulation mapping of brain function: A comparison of subdural electrodes and stereo-EEG. Front Hum Neurosci. 2020;14:611291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hays MA, Smith RJ, Haridas B, Coogan C, Crone NE, Kang JY. Effects of stimulation intensity on intracranial cortico-cortical evoked potentials: A titration study. Clin Neurophysiol. 2021;132:2766–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Matsumoto R, Kunieda T, Nair D. Single pulse electrical stimulation to probe functional and pathological connectivity in epilepsy. Seizure. 2017;44:27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Miller KJ, Müller KR, Hermes D. Basis profile curve identification to understand electrical stimulation effects in human brain networks. Biorxiv. [Preprint] doi: 10.1101/2021.01.24.428020 [DOI] [PMC free article] [PubMed]
- 89. Matsumoto R, Nair DR, LaPresto E, et al. Functional connectivity in the human language system: A cortico-cortical evoked potential study. Brain. 2004;127:2316–2330. [DOI] [PubMed] [Google Scholar]
- 90. Enatsu R, Piao Z, O’Connor T, et al. Cortical excitability varies upon ictal onset patterns in neocortical epilepsy: A cortico-cortical evoked potential study. Clin Neurophysiol. 2012;123:252–260. [DOI] [PubMed] [Google Scholar]
- 91. Matsumoto R, Nair DR, Ikeda A, et al. Parieto-frontal network in humans studied by cortico-cortical evoked potential. Hum Brain Mapp. 2012;33:2856–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Mégevand P, Groppe DM, Bickel S, et al. The hippocampus and amygdala are integrators of neocortical influence: A CorticoCortical evoked potential study. Brain Connect. 2017;7:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kobayashi K, Matsumoto R, Usami K, et al. Cortico-cortical evoked potential by single-pulse electrical stimulation is a generally safe procedure. Clin Neurophysiol. 2021;132:1033–1040. [DOI] [PubMed] [Google Scholar]
- 94. Keller CJ, Honey CJ, Mégevand P, et al. Mapping human brain networks with cortico-cortical evoked potentials. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Matsumoto R, Nair DR, LaPresto E, et al. Functional connectivity in human cortical motor system: A cortico-cortical evoked potential study. Brain. 2007;130:181–197. [DOI] [PubMed] [Google Scholar]
- 96. Crocker B, Ostrowski L, Williams ZM, et al. Local and distant responses to single pulse electrical stimulation reflect different forms of connectivity. Neuroimage. 2021;237:118094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lega B, Dionisio S, Flanigan P, et al. Cortico-cortical evoked potentials for sites of early versus late seizure spread in stereoelectroencephalography. Epilepsy Res. 2015;115:17–29. [DOI] [PubMed] [Google Scholar]
- 98. Iwasaki M, Enatsu R, Matsumoto R, et al. Accentuated cortico-cortical evoked potentials in neocortical epilepsy in areas of ictal onset. Epileptic Disord. 2010;12:292–302. [DOI] [PubMed] [Google Scholar]
- 99. Zhao C, Liang Y, Li C, et al. Localization of epileptogenic zone based on cortico-cortical evoked potential (CCEP): A feature extraction and graph theory approach. Front Neuroinform. 2019;13:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Guo ZH, Zhao BT, Toprani S, et al. Epileptogenic network of focal epilepsies mapped with cortico-cortical evoked potentials. Clin Neurophysiol. 2020;131:2657–2666. [DOI] [PubMed] [Google Scholar]
- 101. Parker CS, Clayden JD, Cardoso MJ, et al. Structural and effective connectivity in focal epilepsy. Neuroimage Clin. 2018;17:943–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Mitsuhashi T, Sonoda M, Iwaki H, Luat AF, Sood S, Asano E. Effects of depth electrode montage and single-pulse electrical stimulation sites on neuronal responses and effective connectivity. Clin Neurophysiol. 2020;131:2781–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Mouthaan BE, van ‘t Klooster MA, Keizer D, et al. Single pulse electrical stimulation to identify epileptogenic cortex: Clinical information obtained from early evoked responses. Clin Neurophysiol. 2016;127:1088–1098. [DOI] [PubMed] [Google Scholar]
- 104. van ‘t Klooster MA, Zijlmans M, Leijten FS, et al. Time-frequency analysis of single pulse electrical stimulation to assist delineation of epileptogenic cortex. Brain. 2011;134:2855–2866. [DOI] [PubMed] [Google Scholar]
- 105. Mălîia MD, Donos C, Barborica A, et al. High frequency spectral changes induced by single-pulse electric stimulation: Comparison between physiologic and pathologic networks. Clin Neurophysiol. 2017;128:1053–1060. [DOI] [PubMed] [Google Scholar]
- 106. Kobayashi K, Matsumoto R, Matsuhashi M, et al. High frequency activity overriding cortico-cortical evoked potentials reflects altered excitability in the human epileptic focus. Clin Neurophysiol. 2017;128:1673–1681. [DOI] [PubMed] [Google Scholar]
- 107. Valentín A, Alarcon G, Garcia-Seoane JJ, et al. Single-pulse electrical stimulation identifies epileptogenic frontal cortex in the human brain. Neurology. 2005;65:426–435. [DOI] [PubMed] [Google Scholar]
- 108. Valentín A, Anderson M, Alarcón G, et al. Responses to single pulse electrical stimulation identify epileptogenesis in the human brain in vivo. Brain. 2002;125:1709–1718. [DOI] [PubMed] [Google Scholar]
- 109. Flanagan D, Valentín A, García Seoane JJ, Alarcón G, Boyd SG. Single-pulse electrical stimulation helps to identify epileptogenic cortex in children. Epilepsia. 2009;50:1793–1803. [DOI] [PubMed] [Google Scholar]
- 110. Kamali G, Smith RJ, Hays M, et al. Localizing the seizure onset zone from single pulse electrical stimulation responses using transfer function models. Annu Int Conf IEEE Eng Med Biol Soc. 2020:2524–2527. [DOI] [PubMed] [Google Scholar]
- 111. Kamali G, Smith RJ, Hays M, et al. Transfer function models for the localization of seizure onset zone from cortico-cortical evoked potentials. Front Neurol. 2020;11:579961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Smith RJ, Kamali G, Hays M, et al. State-space models of evoked potentials to localize the seizure onset zone. Annu Int Conf IEEE Eng Med Biol Soc. 2020:2528–2531. [DOI] [PubMed] [Google Scholar]
- 113. Smith RJ, Hays MA, Kamali G, et al. Stimulating native seizures with neural resonance: A new approach to localize the seizure onset zone. Brain. 2022;145:3886–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Prime D, Rowlands D, O’Keefe S, Dionisio S. Considerations in performing and analyzing the responses of cortico-cortical evoked potentials in stereo-EEG. Epilepsia. 2018;59:16–26. [DOI] [PubMed] [Google Scholar]
- 115. Trebaul L, Rudrauf D, Job AS, et al. Stimulation artifact correction method for estimation of early cortico-cortical evoked potentials. J Neurosci Methods. 2016;264:94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Prime D, Woolfe M, O’Keefe S, Rowlands D, Dionisio S. Quantifying volume conducted potential using stimulation artefact in cortico-cortical evoked potentials. J Neurosci Methods 2020;337:108639. [DOI] [PubMed] [Google Scholar]
- 117. Mohan UR, Watrous AJ, Miller JF, et al. The effects of direct brain stimulation in humans depend on frequency, amplitude, and white-matter proximity. Brain Stimul. 2020;13:1183–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Greene P, Li A, González-Martínez J, Sarma SV. Classification of stereo-EEG contacts in white matter vs. Gray matter using recorded activity. Front Neurol. 2020;11:605696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Basu I, Robertson MM, Crocker B, et al. Consistent linear and non-linear responses to invasive electrical brain stimulation across individuals and primate species with implanted electrodes. Brain Stimul. 2019;12:877–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kovac S, Kahane P, Diehl B. Seizures induced by direct electrical cortical stimulation–mechanisms and clinical considerations. Clin Neurophysiol. 2016;127:31–39. [DOI] [PubMed] [Google Scholar]
- 121. Kämpfer C, Racz A, Quesada CM, Elger CE, Surges R. Predictive value of electrically induced seizures for postsurgical seizure outcome. Clin Neurophysiol. 2020;131:2289–2297. [DOI] [PubMed] [Google Scholar]
- 122. Oderiz CC, von Ellenrieder N, Dubeau F, et al. Association of cortical stimulation–induced seizure with surgical outcome in patients with focal drug-resistant epilepsy. JAMA Neurol. 2019;76:1070–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Bernabei JM, Arnold TC, Shah P, et al. Electrocorticography and stereo EEG provide distinct measures of brain connectivity: Implications for network models. Brain Commun. 2021;3:fcab156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Conrad EC, Bernabei JM, Kini LG, et al. The sensitivity of network statistics to incomplete electrode sampling on intracranial EEG. Netw Neurosci. 2020;4:484–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Conrad EC, Bernabei JM, Sinha N, et al. Addressing spatial bias in intracranial EEG functional connectivity analyses for epilepsy surgical planning. J. Neural Eng. 2022;19:056019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Thomas GP, Jobst BC. Critical review of the responsive neurostimulator system for epilepsy. Med Devices. 2015;8:405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Ye Y, Kaskutas LA. Using propensity scores to adjust for selection bias when assessing the effectiveness of alcoholics anonymous in observational studies. Drug Alcohol Depend 2009;104:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Hosseini M, Powell M, Collins J, et al. I tried a bunch of things: The dangers of unexpected overfitting in classification of brain data. Neurosci Biobehav Rev. 2020;119:456–467. [DOI] [PubMed] [Google Scholar]
- 129. Simon HA. Spurious correlation: A causal interpretation*. Causal Mod Soc Sci. 2017:7–22. [Google Scholar]
- 130. Hulsen T. Sharing is caring—Data sharing initiatives in healthcare. Int J Environ Res Public Health. 2020;17:3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Chiruvella V, Guddati AK. Ethical issues in patient data ownership. Interact J Med Res. 2021;10:e22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Goldberger AL, Amaral LA, Glass L, et al. Physiobank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation. 2000;101:E215–E220. [DOI] [PubMed] [Google Scholar]
- 133. Appelhoff S, Sanderson M, Brooks T, et al. MNE-BIDS: Organizing electrophysiological data into the BIDS format and facilitating their analysis. J Open Source Softw. 2019;4:1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Delorme A, Makeig S. EEGLAB: An open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J Neurosci Methods. 2004;134:9–21. [DOI] [PubMed] [Google Scholar]
- 135. Gulban OF, Nielson D, Poldrack R, et al. poldracklab/pydeface: v2.0.0. Accessed 1 September 2022. https://zenodo.org/record/3524401#.Y-o2GXbP1D8 [Google Scholar]
- 136. Gorgolewski KJ, Auer T, Calhoun VD, et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci Data. 2016;3:160044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Holdgraf C, Appelhoff S, Bickel S, et al. iEEG-BIDS, extending the brain imaging data structure specification to human intracranial electrophysiology. Sci Data. 2019;6:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Markiewicz CJ, Gorgolewski KJ, Feingold F, et al. The OpenNeuro resource for sharing of neuroscience data. Elife. 2021;10:e71774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Fedele T, Burnos S, Boran E, et al. Resection of high frequency oscillations predicts seizure outcome in the individual patient. Sci Rep. 2017;7:13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Demuru M, van Blooijs D, Zweiphenning W, et al. A practical workflow for organizing clinical intraoperative and long-term iEEG data in BIDS. Neuroinformatics. 2022;20:727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Baldassano SN, Brinkmann BH, Ung H, et al. Crowdsourcing seizure detection: Algorithm development and validation on human implanted device recordings. Brain. 2017;140:1680–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.