Abstract
Aims
Although highly heritable, the genetic etiology of calcific aortic stenosis (AS) remains incompletely understood. The aim of this study was to discover novel genetic contributors to AS and to integrate functional, expression, and cross-phenotype data to identify mechanisms of AS.
Methods and results
A genome-wide meta-analysis of 11.6 million variants in 10 cohorts involving 653 867 European ancestry participants (13 765 cases) was performed. Seventeen loci were associated with AS at P ≤ 5 × 10−8, of which 15 replicated in an independent cohort of 90 828 participants (7111 cases), including CELSR2–SORT1, NLRP6, and SMC2. A genetic risk score comprised of the index variants was associated with AS [odds ratio (OR) per standard deviation, 1.31; 95% confidence interval (CI), 1.26–1.35; P = 2.7 × 10−51] and aortic valve calcium (OR per standard deviation, 1.22; 95% CI, 1.08–1.37; P = 1.4 × 10−3), after adjustment for known risk factors. A phenome-wide association study indicated multiple associations with coronary artery disease, apolipoprotein B, and triglycerides. Mendelian randomization supported a causal role for apolipoprotein B-containing lipoprotein particles in AS (OR per g/L of apolipoprotein B, 3.85; 95% CI, 2.90–5.12; P = 2.1 × 10−20) and replicated previous findings of causality for lipoprotein(a) (OR per natural logarithm, 1.20; 95% CI, 1.17–1.23; P = 4.8 × 10−73) and body mass index (OR per kg/m2, 1.07; 95% CI, 1.05–1.9; P = 1.9 × 10−12). Colocalization analyses using the GTEx database identified a role for differential expression of the genes LPA, SORT1, ACTR2, NOTCH4, IL6R, and FADS.
Conclusion
Dyslipidemia, inflammation, calcification, and adiposity play important roles in the etiology of AS, implicating novel treatments and prevention strategies.
Keywords: Aortic stenosis, Genome-wide association study, Mendelian randomization, Phenome-wide association study, Gene expression, Genetic risk score
Structured Graphical Abstract
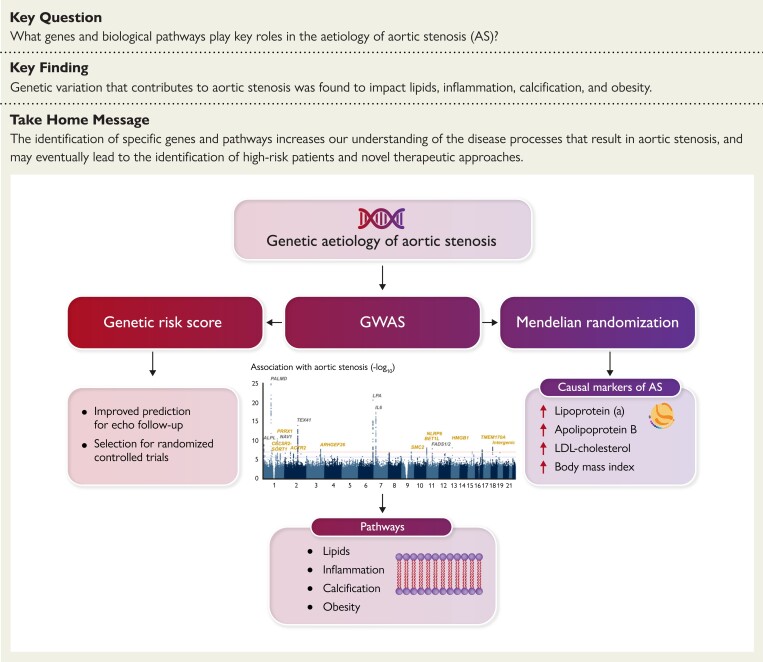
Structured Graphical Abstract.
Genetic etiology of aortic stenosis. This study meta-analyzed 13 765 AS cases vs. 640 102 controls and confirmed 15 genetic loci associated with AS. Downstream analyses implicated additional candidate genes involved in dyslipidemia, inflammation, calcification, and adiposity. The Manhattan plot shows variants with P ≥ 1 × 10−25, for improved visualization. Genetic loci in grey were previously identified, and those in gold are new discoveries. Abbreviations: AS, aortic stenosis; GWAS, genome-wide association study; LDL, low-density lipoprotein.
See the editorial comment for this article ‘The journey towards identification of actionable molecular pathways in calcific aortic valve stenosis’, by S. Thériault et al., https://doi.org/10.1093/eurheartj/ehad134.
Translational perspective.
This large-scale genetic study of calcific aortic stenosis (AS) in 653 867 European ancestry participants (13 765 cases) identified 15 robustly replicated genetic loci, including SORT1–CELSR2, involved in lipid metabolism, and NLRP6, involved in the inflammatory response. We provided evidence in favor of a causal association for apolipoprotein B, lipoprotein(a), body mass index, and low-density lipoprotein cholesterol. A genetic risk score including all identified loci was associated with both AS and aortic valve calcium and improved the classification of AS when added to risk factors. The differential expression of several genes in relevant tissues highlights their role in the etiology of AS. Together, these findings provide candidates for therapeutic targeting and highlight the use of a genetic risk score to improve clinical risk assessment.
Introduction
Calcific aortic stenosis (AS) is the leading form of incident valvular heart disease in high-income populations.1 In individuals over 75 years, the prevalence of AS is 10%–15% but is expected to more than double by 2040.2 Although a replacement of the aortic valve is effective for severe AS cases,3,4 there are no treatments to prevent progression to valve replacement. Furthermore, it remains unclear which patients are at high risk of a severe prognosis.
A genetic component contributes to the etiology of AS, as siblings of AS patients have more than four-fold the risk of AS.5 Several families have also been identified with many affected members, including 1 extended family with 48 cases of severe AS.6 Previous genome- and transcriptome-wide association efforts have identified seven genetic loci associated with AS,7–11 including LPA7 [which codes for the apolipoprotein(a) moiety of lipoprotein(a)], IL610 (which codes for interleukin-6), and the FADS1/2 gene cluster11 (which codes for desaturases involved in fatty acid biosynthesis). Additionally, Mendelian randomization studies supported a causal contribution of low-density lipoprotein cholesterol (LDL-C),12 non-high-density lipoprotein cholesterol,9 lipoprotein(a),13 arachidonic acid11, and body mass index (BMI)14 to AS, suggesting susceptibility to AS is partially mediated by lipid metabolism and inflammation.
Identifying additional genetic loci for AS could provide novel targets for therapeutic intervention and improve risk stratification. Accordingly, we combined genome-wide association study (GWAS) results from 10 cohorts to identify novel loci. We conducted functional analyses for significant variants, examined their association with biomarkers and other diseases, assessed cross-ancestry transferability of variants, and developed an AS genetic risk score to assess its association with diagnosed AS. Finally, we investigated whether individual genes or gene sets were predicted to be differentially expressed in AS cases.
Methods
Cohorts and case definition
The cohorts included in the meta-analysis are listed in Table 1. A full description of the cohorts and their case definition for identifying AS is in the Supplementary material online.
Table 1.
Cohorts in the genome-wide meta-analysis
| Cohort | Country | Aortic stenosis definition | No. of cases | No. controls | Median age (Quartile 1, Quartile 3) | Genotyping array | Imputation reference panel | No. of variants |
|---|---|---|---|---|---|---|---|---|
| Vanderbilt University Biobank | USA | her | 759 | 7555 | 68 (62, 75) | Illumina Multi-Ethnic Genotyping Array | HRC version r1.1 | 10 689 407 |
| CAVS-France1 | France | Echocardiography | 1261 | 1305 | 75 (7079)a | Affymetrix Axiom Genome-Wide CEU-1 Array | HRC version r1.1 | 10 395 306 |
| CAVS-France2 | France | Echocardiography | 1495 | 2707 | 77 (70, 83)a | Affymetrix Axiom Genome-Wide Precision Medicine Research Array | HRC version r1.1 | 10 031 533 |
| CAVS-France3 | France | Echocardiography | 367 | 2519 | 74 (67,79)a | Affymetrix Axiom Genome-Wide Precision Medicine Research Array | HRC version r1.1 | 9 884 426 |
| deCODE | Iceland | her | 2464 | 351 068 | 77 (68, 83)a | Illumina Chips | Icelandic reference panel by deCODE Genetics | 9 812 907 |
| GERA | USA | EHR | 3469 | 51 723 | 66 (61, 74) | Affymetrix Axiom Genome-Wide EUR | HRC version r1.1 | 10 578 354 |
| Malmö Diet and Cancer Study | Sweden | her | 464 | 4878 | 58 (53, 63) | Illumina Human Omni Express Exome BeadChip | HRC version r1.1 | 6 130 056 |
| Penn Medicine Biobank | USA | EHR | 1593 | 4550 | 71 (62, 79) | Illumina Quad Omni Genotyping Chip | HRC version r.1.1 | 11 016 108 |
| UK Biobankb | UK | her | 1675 | 213 361 | 63 (59, 66) | Affymetrix UK Biobank Axiom Array | HRC version r1.1 and UK10K + 1000 Genomes Project Phase 3 | 9 927 329 |
| Umeå University | Sweden | Surgery | 218 | 436 | 60 (60, 66) | Affymetrix UK Biobank Axiom Array r3 | HRC version r1.1 | 8 810 694 |
In cases only.
Provided are the number of cases and controls in the genome-wide association study for aortic stenosis. For analyses performed using UK Biobank data for the polygenic risk scores, phenome-wide association study, and Mendelian randomization, an updated dataset consisting of 2213 cases and 255 018 controls was used. See the UK Biobank cohort description for more details.
Abbreviations: CAVS, calcific aortic valve stenosis; EHR, electronic health records; EUR, European; GERA, Genetic Epidemiology Research on Adult Health and Aging; HRC, Haplotype Reference Consortium.
Genome-wide meta-analysis for aortic stenosis
We performed centralized, cohort-specific quality control of genome-wide summary statistics for prevalent AS from 10 cohorts totaling 653 867 European ancestry participants (13 765 cases) (Table 1). With the exception of the Malmö Diet and Cancer Study, which excluded variants with >5% genotype missingness, minor allele frequency ≤ 1%, and Hardy–Weinberg equilibrium test P ≥ 1 × 10−4, and deCODE, which excluded variants not found in the Haplotype Reference Consortium version r1.1 panel, quality control used unified criteria. For all cohorts, we included bi-allelic variants with non-ambiguous strands (i.e. no C/G or A/T allele pairs), imputation quality score ≥0.3, and minor allele frequency ≥0.001 and whose associations with AS had standard errors less than or equal to the median standard error plus five times the interquartile range, calculated from all summary statistics from each cohort. Using PLINK version 1.9, we performed inverse variance-weighted, fixed-effects meta-analysis for 11 591 806 variants with summary statistics that passed quality control and the allele frequency threshold in at least two cohorts. For independent (r2 ≤ 0.01) and genome-wide significant (P ≤ 5 × 10−8) variants, i.e. index variants and variants in high linkage disequilibrium (LD) (r2 ≥ 0.95) in European ancestry individuals of the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort or the 1000 Genomes Project Phase 3,15 we used the University of California Santa Cruz16 GRCh37 assembly for genomic location.
Among 90 828 participants (7111 AS cases) from the Copenhagen Hospital Biobank or the Danish Blood Donor Study, we evaluated replication of the association with AS for all index variants. Associations were modeled using logistic regression adjusted for age, sex, and 10 principal components, with a P ≤ 0.05 considered as replication. For all index variants, we also performed inverse variance-weighted, fixed-effects meta-analysis of summary statistics from the discovery and replication cohorts. For additional cohort details, see the Supplementary material online.
Region, gene-based, and functional analysis of variants
We used Annotate Variation (ANNOVAR)17 to extract predicted function and pathogenicity of variants, including scores generated by Combined Annotation Dependent Depletion (CADD),18 Deleterious Annotation of genetic variants using Neural Networks (DANN),19 Linear INSIGHT (LINSIGHT),20 Eigen-Principal Component (EIGEN-PC),21 and Functional Analysis Through Hidden Markov Models-Multiple Kernel Learning (FATHMM-MKL) non-coding.22 From Genotype-Tissue Expression Project (GTEx) version 8,23 we identified significant expression quantitative trait loci in the aorta, left ventricle, liver, and whole blood. To identify gene regions associated with AS, we employed the single-nucleotide polymorphism (SNP)-wise mean approach of Multi-marker Analysis of GenoMic Annotation (MAGMA)24 to test for association. We employed MetaXcan25 and coloc26 to identify variants whose effects on AS may be mediated by gene expression. We also applied the Genotype Imputed Gene Set Enrichment Analysis (GIGSEA)27 approach, to assess whether sets of genes with shared function or regulation demonstrate differential predicted expression. We examined gene sets defined by the Kyoto Encyclopedia of Genes and Genomes (KEGG),28 genes in the same pathway; Gene Ontology (GO),29 genes with related functions; Functional ANnoTation Of the Mammalian genome version 5 (FANTOM5),30 genes with shared transcription factor binding sites; and miRBase,31 genes with the same microRNA seed sequence in their 3’ untranslated region.
Genetic risk scores
We constructed three separate genetic risk scores. The first two used PLINK version 2.032 with weights estimated from a meta-analysis excluding the UK Biobank: a GRS18, using all 18 genome-wide significant index variants from the discovery analysis, as well as a GRS559 including the 559 variants at P < 1 × 10−4. We assessed the association of GRS18 and GRS559 with AS in 220 159 unrelated White British participants in the UK Biobank aged 55 years or older (3091 cases) (see Supplementary material online) using a logistic regression model adjusted for age2 and sex and then further adjusted for diabetes, LDL-C, systolic blood pressure, smoking (ever/never), BMI, and coronary artery disease (CAD). The UK Biobank participants in these and subsequent analyses differed slightly from the discovery analysis as an updated version of the data became available. In addition, we performed a polygenic risk score (PRS) analysis with LDpred233 in 244 641 UK Biobank participants (3410 cases). We also assessed the association of all the risk scores with the presence of aortic valve calcium (AVC) in the Multi-Ethnic Study of Atherosclerosis (MESA), using logistic regression adjusted for age and sex and then fully adjusted for fasting glucose, LDL-C, systolic blood pressure, smoking (ever/never), BMI, and coronary artery calcium. The PLINK-derived risk scores used 17 and 550 variants as neither variants nor proxies were available for some SNPs in this dataset. For the GRS17 and GRS550 risk scores, 2440 unrelated European participants (381 cases of AVC >0) were analyzed. The PRS analysis with LDpred2 included 2205 unrelated European participants (355 cases with AVC >0) of MESA. The area under the receiver operating characteristic curve (AUC) for the null hypothesis of no risk score was compared to other models using DeLong’s test for two correlated ROC curves from the pROC R package.
Cross-ancestry and cross-phenotype associations
In the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium, we extracted summary statistics7 for the association of the index variants with prevalent AVC in an inverse variance-weighted fixed-effects meta-analysis of three independent cohorts totaling 6942 European participants (2245 cases), adjusting for age and sex. Variants that were not available were replaced with one in high LD (r2 ≥ 0.8) when available.
To examine the association of index variants with AS in other ancestries, we analyzed 1917 African American participants (86 cases) and 3482 Latin American participants (159 cases) of the GERA cohort, adjusted for age2 and sex.
We performed a phenome-wide association study of the index variants with 58 diseases, serum biomarkers, and physiological measurements, among 257 231 unrelated White British UK Biobank participants aged 55 years or older. Disease cases were identified using hospital diagnosis codes, procedure codes, and causes of death. Levels of alkaline phosphatase, C-reactive protein, gamma-glutamyl transferase, lipoprotein(a), and triglycerides were natural logarithm transformed. Logistic and linear regression models were adjusted for age2 and sex, except for breast cancer (analyzed only in women). A false discovery rate correction of 5% was applied across phenotypes for each variant tested.
For six traits associated with multiple variants in the phenome-wide association study, we performed two-sample Mendelian randomization to assess the causal contribution to AS. We performed a GWAS for each trait in unrelated UK Biobank White British participants (up to 383 533 participants). We constructed a genetic instrument for each biomarker using genome-wide significant variants (331 ≤ n ≤ 702) which were independent (r2 ≤ 0.01) with imputation quality ≥0.3 and minor allele frequency ≥0.001. We used the R package MendelianRandomization version 0.4.234 to estimate the inverse variance-weighted association with AS, using summary statistics from our discovery meta-analysis. The summary statistics for AS were generated with meta-analysis results that did not include UK Biobank so that instruments and outcomes were from non-overlapping cohorts. In secondary analyses, we also applied the contamination mixture, penalized weighted median, and Egger approaches.
Correlation and conditional analyses
We estimated the genetic correlation of AS with 157 cardiovascular biomarkers, risk factors, and diseases using the LD score regression method35 as implemented on LD Hub.36 We selected GWAS or meta-analyses which had been performed in European populations and applied a 5% false discovery rate correction.
To identify additional variants associated with AS, we used the conditional and joint analysis method37 from the Genome-wide Complex Trait Analysis (GCTA) software38 to re-estimate the summary statistics from our genome-wide meta-analysis conditioned upon the index variants. Variants not genome-wide significant in the original meta-analysis but which (i) became genome-wide significant in the conditional analysis and (ii) were independent (r2 ≤ 0.01) would be deemed to be novel associations.
Results
Genome-wide meta-analysis identifies 10 novel loci for aortic stenosis
We performed a genome-wide meta-analysis for AS using summary statistics from 10 European ancestry cohorts totaling 653 867 participants (13 765 cases) (Table 1). Estimates for each of 11 591 806 variants were combined in an inverse variance-weighted, fixed-effects model (see Figure 1 for study design overview). We observed no evidence of strongly inflated test statistics in the meta-analysis (genomic inflation factor [λ] = 1.04 and LD score regression intercept = 1.020; Supplementary material online, Figure S1).
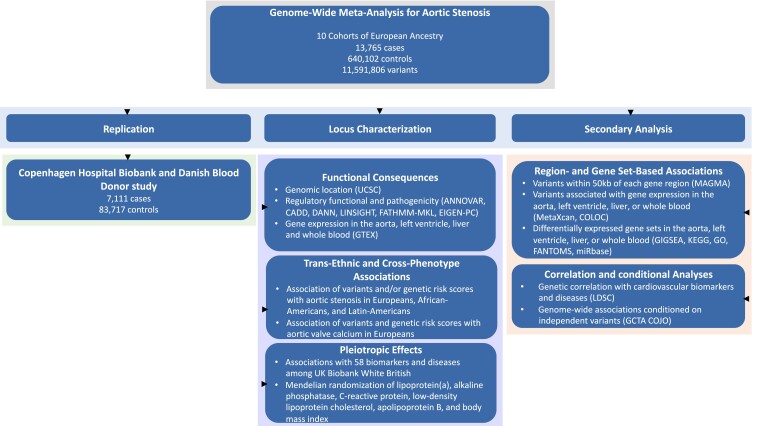
Figure 1.
Design of the genome-wide meta-analysis and follow-up analyses. Abbreviations: ANNOVAR, Annotate Variation; CADD, Combined Annotation Dependent Depletion; DANN, Deleterious Annotation of genetic variants using Neural Networks; EIGEN-PC, Eigen-Principal Component; FANTOM5, Functional Annotation of the Mammalian Genome version 5; FATHMM-MKL, Functional Analysis Through Hidden Markov Models-Multiple Kernel Learning; GCTA COJO, Genome-wide Complex Trait Analysis Conditional and Joint Association Analysis; GIGSEA, Genotype Imputed Gene Set Enrichment Analysis; GO, Gene Ontology; GTEx, Genotype-Tissue Expression project; KEGG, Kyoto Encyclopedia of Genes and Genomics; LDSC, Linkage Disequilibrium Score Regression; LINSIGHT, Linear INSIGHT; MAGMA, Multi-marker Analysis of Genomic Annotation; UCSC, University of California Santa Cruz Genome Browser.
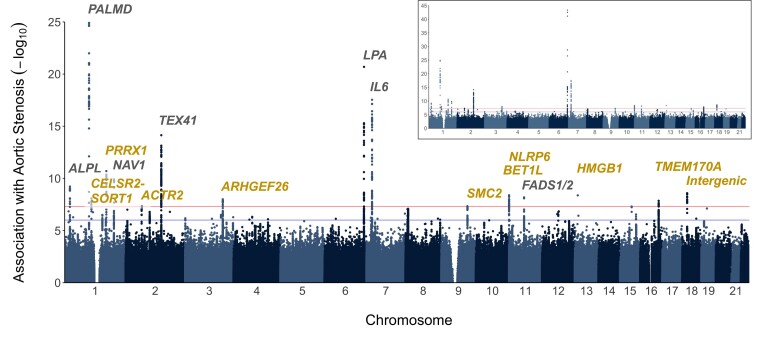
We identified 17 genetic loci containing one or more independent variants (r2 ≤ 0.01) which exceeded a genome-wide significance threshold (P ≤ 5 × 10−8) for association with AS (Figure 2). We confirmed all 7 known loci and identified 10 loci not previously reported to be genome-wide significant. After pruning for variants in LD, we identified 18 independent variants (r2 ≤ 0.01) (see meta-analysis results in Table 2 and forest plots in Supplementary material online, Figure S2). The association with AS of variants surrounding the index variants is provided in Supplementary material online, Figure S3).
Figure 2.
P for the association of 11 591 806 variants with aortic stenosis in the meta-analysis. The inset shows all associations, while the main plot shows variants with P ≥ 1 × 10−25, for improved visualization. Genetic loci in grey were previously identified and those in gold are new discoveries.
Table 2.
Association with aortic stenosis of new and previously identified genetic loci for aortic stenosis in the discovery and replication cohorts
| Chr. | Locus | Variant | Minor allele | Discovery | Replication | Discovery and replication | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF | Odds ratio per minor allele (95% CI) | P | I2% (95% CI) | Imputation quality score range | MAF | Odds ratio per minor allele (95% CI) | P | odds ratio per minor allele (95% CI) | P | ||||
| New loci | |||||||||||||
| 1 | CELSR2–SORT1 | rs12740374 | T | 0.21 | 0.90 (0.87, 0.94) | 8.4 × 10−09 | 0 (0, 0) | 0.77 to 1.00 | 0.23 | 0.95 (0.91, 0.99) | 0.016 | 0.92 (0.90, 0.95) | 2.4 × 10−09 |
| 1 | PRRX1 | rs61817383 | T | 0.26 | 1.11 (1.08, 1.14) | 2.0 × 10−11 | 10 (0, 66) | 0.94 to 1.00 | 0.26 | 1.06 (1.02, 1.10) | 8.1 × 10−03 | 1.09 (1.06, 1.12) | 3.7 × 10−12 |
| 2 | ACTR2 | rs62139062 | T | 0.28 | 1.09 (1.06, 1.12) | 4.2 × 10−08 | 32 (0, 68) | 0.92 to 1.00 | 0.26 | 1.08 (1.04, 1.12) | 2.3 × 10−04 | 1.08 (1.06, 1.11) | 4.4 × 10−11 |
| 3 | ARHGEF26 | rs6794263 | C | 0.11 | 0.88 (0.84, 0.92) | 1.0 × 10−08 | 39 (0, 71) | 0.98 to 1.00 | 0.11 | 0.93 (0.88, 0.99) | 0.013 | 0.90 (0.87, 0.93) | 1.5 × 10−09 |
| 9 | SMC2 | rs55909255 | C | 0.39 | 1.08 (1.05, 1.11) | 4.7 × 10−08 | 53 (3, 77) | 0.99 to 1.00 | 0.36 | 1.05 (1.01, 1.09) | 0.014 | 1.07 (1.04, 1.09) | 5.1 × 10−09 |
| 11 | BET1L | rs73386631 | T | 0.043 | 1.22 (1.14, 1.31) | 2.8 × 10−08 | 20 (0, 62) | 0.37 to 1.00 | 0.038 | 1.06 (0.97, 1.17) | 0.19 | 1.16 (1.10, 1.23) | 1.9 × 10−07 |
| 11 | NLRP6 | rs17156153 | T | 0.085 | 1.16 (1.10, 1.22) | 4.2 × 10−09 | 0 (0, 56) | 0.73 to 1.00 | 0.075 | 1.16 (1.09, 1.24) | 1.3 × 10−05 | 1.16 (1.11, 1.21) | 2.6 × 10−13 |
| 13 | HMGB1 | rs181753401 | A | 2.4 × 10−03 | 2.29 (1.74, 3.02) | 4.2 × 10−09 | 36 (0, 73) | 0.40 to 1.00 | 3.9 × 10−03 | 1.23 (0.92, 1.63) | 0.16 | 1.69 (1.38, 2.06) | 2.1 × 10−07 |
| 16 | TMEM170A | rs11643207 | C | 0.38 | 0.92 (0.89, 0.95) | 1.4 × 10−08 | 50 (0, 76) | 0.98 to 1.00 | 0.39 | 0.92 (0.89, 0.96) | 8.6 × 10−06 | 0.92 (0.90, 0.94) | 5.6 × 10−13 |
| 18 | Intergenic (18q11.2) | rs551520 | T | 0.23 | 0.91 (0.88, 0.94) | 2.6 × 10−09 | 27 (0, 65) | 0.99 to 1.01 | 0.24 | 0.94 (0.90, 0.98) | 2.8 × 10−03 | 0.92 (0.90, 0.94) | 6.8 × 10−11 |
| Previously identified loci | |||||||||||||
| 1 | ALPL | rs6696066 | A | 0.48 | 0.92 (0.89, 0.94) | 6.1 × 10−10 | 0 (0, 6.3) | 0.99 to 1.00 | 0.47 | 0.93 (0.89, 0.96) | 2.3 × 10−05 | 0.92 (0.90, 0.94) | 7.1 × 10−14 |
| 1 | PALMD | rs6702619 | G | 0.49 | 1.16 (1.13, 1.19) | 1.2 × 10−25 | 59 (18, 80) | 0.97 to 1.00 | 0.51 | 1.17 (1.13, 1.21) | 7.1 × 10−19 | 1.16 (1.14, 1.19) | 8.7 × 10−43 |
| 1 | NAV1 | rs631556 | A | 0.41 | 1.10 (1.07, 1.13) | 1.4 × 10−10 | 0 (0, 44) | 0.98 to 1.00 | 0.38 | 1.08 (1.04, 1.12) | 5.3 × 10−05 | 1.09 (1.07, 1.12) | 4.1 × 10−14 |
| 2 | TEX41 | rs7593336 | G | 0.38 | 1.12 (1.09, 1.15) | 7.3 × 10−15 | 59 (17, 79) | 0.95 to 1.00 | 0.41 | 1.14 (1.10, 1.18) | 2.7 × 10−12 | 1.12 (1.10, 1.15) | 1.7 × 10−25 |
| 6 | LPA | rs10455872 | G | 0.069 | 1.42 (1.35, 1.49) | 4.6 × 10−44 | 28 (0, 65) | 0.76 to 1.00 | 0.081 | 1.50 (1.41, 1.60) | 1.1 × 10−33 | 1.45 (1.39, 1.51) | 1.4 × 10−75 |
| rs140570886 | C | 0.014 | 1.55 (1.40, 1.73) | 5.1 × 10−16 | 24 (0, 64) | 0.83 to 1.00 | 0.010 | 1.31 (1.10, 1.56) | 3.0 × 10−03 | 1.48 (1.35, 1.62) | 2.3 × 10−17 | ||
| 7 | IL6 | rs1800797 | A | 0.45 | 1.13 (1.10, 1.16) | 2.9 × 10−18 | 45 (0, 73) | 0.88 to 1.00 | 0.46 | 1.12 (1.08, 1.16) | 9.3 × 10−11 | 1.13 (1.10, 1.15) | 1.9 × 10−27 |
| 11 | FADS1/2 | rs174533 | A | 0.34 | 0.91 (0.88, 0.94) | 6.7 × 10−09 | 0 (0, 54) | 0.93 to 1.00 | 0.34 | 0.93 (0.89, 0.96) | 4.8 × 10−05 | 0.92 (0.89, 0.94) | 2.0 × 10−12 |
Abbreviation: MAF, minor allele frequency.
Subsequently, we performed a replication study of the association for each of the 10 previously unreported loci with AS in 90 828 individuals (7111 AS cases) from the Danish Blood Donor Study and Copenhagen Hospital Biobank, and variants at 8 of these 10 loci were nominally associated with AS with concordant direction of effect (P ≤ 0.05) (Table 2). The two least common variants, BET1L rs73386631 and HMGB1 rs181753401, were not associated with AS in this cohort, and meta-analysis of the discovery and replication cohorts did not achieve genome-wide significance (Table 2).
When we re-estimated the association of variants with AS in the genome-wide meta-analysis, conditioned upon the 18 index variants, the PLG variant rs191108153 became genome-wide significant [odds ratio (OR) per T allele, 1.57; 95% CI, 1.34–1.83; P = 9.6 × 10−9]. Given the proximity of this variant to LPA, we tested the association of this variant with AS conditioned on genetically predicted levels of lipoprotein(a) and observed substantial attenuation (OR per T allele, 1.20; 95% CI, 1.03–1.40; P = 0.019).
We examined publicly available databases for functional effects of the index variants and their proxies in high LD (r2 ≥ 0.95) (see Supplementary material online, Tables S1 and S2). ARGHEF26 rs6794263 was in high LD (r2 = 0.99)39 with the missense variant rs13096373 (p.Phe203Ser). This substitution was predicted by the CADD software40 to be in the 5% most deleterious substitutions (scaled C-score = 14.5). Variants in the CELSR2–SORT1, PRRX1, NLRP6, PALMD, and IL6 loci and the intergenic region at 18q11.2 had high CADD scores, and variants at the ACTR2 and NLRP6 loci were in the 5% most pathogenic variants according to DANN19 (ranked score ≥ 0.95). Variants at the CELSR2–SORT1, PRRX1 PALMD, IL6, FADS1/2, and 18q11.2 regions were classified as deleterious using the FATHMM-MKL non-coding approach (P ≥ 0.5).22
Identified variants are also associated with aortic valve calcium
From a previous meta-analysis for AVC involving 6942 European participants from three cohorts in the CHARGE consortium (2245 participants with AVC >0),7 we identified fixed-effects associations with prevalent AVC for the index variants. For unavailable variants, a proxy in high LD (r2 ≥ 0.8) was used, but no proxies were found for LPA rs140570886 and HMGB1 rs181753401. Five variants were nominally associated with the presence of AVC in the same direction of effect as for AS (P ≤ 0.05): PRRX1 rs61817383 (OR per minor allele, 1.10; 95% CI, 1.01–1.21; P = 0.035), ACTR2 rs62139062 (OR per minor allele, 1.11; 95% CI, 1.01–1.21; P = 0.029), LPA rs10455872 (OR per minor allele, 2.05; 95% CI, 1.63 to 2.57; P = 9.0 × 10−10), NLRP6 rs17156153 (OR per minor allele, 1.17; 95% CI, 1.02–1.35; P = 0.028), and FADS1/2 rs174533 (OR per minor allele, 0.91; 95% CI, 0.83–0.99; P = 0.024) (see Supplementary material online, Table S3).
Genetic risk scores are associated with aortic stenosis and aortic valve calcium
The prevalence of AS increased across GRS18 risk score tertiles, ranging from 0.97% in the lowest tertile to 1.92% in the highest tertile (see Supplementary material online, Figure S4). Each 1 standard deviation (SD) higher GRS18 was associated with 37% higher odds of AS (OR per SD, 1.37; 95% CI, 1.32–1.41; P = 3.0 × 10−72) with an AUC of 0.697, when adjusted for age2 and sex, with a similar OR after additional adjustment for diabetes, LDL-C, systolic blood pressure, smoking, BMI, and CAD (OR per SD, 1.31; 95% CI, 1.26–1.35; P = 2.6 × 10−51) (see Supplementary material online, Table S4). The AUC for this full model was 0.824, the addition of the genetic risk score modestly improving the AUC compared to a model restricted to non-genetic cardiovascular risk factors only (AUC = 0.818; PAUC difference = 5.9 × 10−6). The GRS559 demonstrated stronger effect sizes and higher discrimination in the fully adjusted model (OR 1.53 per 1-SD GRS559, 95% CI 1.48–1.58; P = 1.54 × 10−141; AUC = 0.829; PAUC difference = 3.2 × 10−9). In an additional sensitivity analysis, we dropped the six LPA region SNPs from the GRS559, and this resulted in an OR of 1.50 per 1-SD for AS (CI 1.45–1.55; P = 7.21 × 10−130). Using the LDpred2 approach, we observed similar results with an OR of 1.45 per 1-SD (95% CI 1.41, 1.50; P = 7.44 × 10−122) and an AUC of 0.706 when adjusting only for age2 and sex. In the model also adjusting for cardiovascular risk factors, we observed an OR of 1.40 (95% CI 1.36, 1.45; P = 1.19 × 10−84) and an AUC of 0.827 (see Supplementary material online, Table S4).
When we applied the GRS17 to MESA, the prevalence of AVC was 15.2%, 13.9%, and 17.7% in the first, second, and third tertiles, respectively (see Supplementary material online, Figure S4). After adjustment for age and sex, each 1 SD higher genetic risk score was associated with 22% higher odds of AVC (OR per SD, 1.22; 95% CI, 1.08–1.37; P = 1.44 × 10−3; AUC = 0.796), and this association persisted after additional adjustment for fasting glucose, LDL-C, systolic blood pressure, smoking, BMI, and coronary artery calcium (OR per SD, 1.23; 95% CI, 1.09–1.39; P = 1.10 × 10−3; AUC = 0.815). The GRS550 demonstrated weaker effects and worse discrimination with AVC when adjusted for age and sex (OR 1.15, 95% CI 1.02–1.29; P = 0.022; AUC = 0.794) as well as in the fully adjusted model (OR 1.15, 95% CI 1.02–1.30; P = 0.023; AUC = 0.814). Without the six LPA region SNPs, the OR was 1.11 per 1-SD for AVC (CI 0.98–1.25; P = 0.093). However, we observed stronger effects and better discrimination with LDpred2, an OR of 1.27 (1.12, 1.44; P = 1.34 × 10−9) and an AUC of 0.799 when adjusted for age and sex. In the fully adjusted model, we observed an OR of 1.32 (1.16, 1.50; P = 2.24 × 10−5) and an AUC of 0.823 (see Supplementary material online, Table S4).
A subset of the identified variants replicates in African and Latin Americans
When we examined the index variants among 1917 African American participants (86 cases) and 3482 Latin American participants (159 cases) from the GERA cohort, we observed replication (P ≤ 0.05 for AS with concordant direction of effects) for CELSR2–SORT1 rs12740374 in both ancestries, for ALPL rs6696066 and NLRP6 rs17156153 in Latin Americans, and for LPA rs10455872 in African Americans (see Supplementary material online, Table S5).
Region-based analysis identifies additional associations with aortic stenosis
For each of 18 539 protein-coding genes, we used MAGMA24 to examine the joint contribution of all variants in a gene region. We tested the mean association with AS of variants within 50 kb of each gene, correcting for LD between variants. We identified 95 regions associated with AS after Bonferroni correction (see Supplementary material online, Table S6), including regions spanning 11 of the 17 loci identified with single variant analysis (ALPL, CELSR2, PRRX1, NAV1, ARHGEF26, LPA, IL6, BET1L, NLRP6, FADS1/2, and TMEM170A). The TMEM170A gene region had a level of significance similar to an overlapping region defined by CFDP1 (P = 2.9 × 10−10 for CFDP1 vs. P = 4.5 × 10−10 for TMEM170A). The three most significant regions not identified in the single variant-based approaches were LDLR (P = 2.3 × 10−10), AGO2 (P = 5.9 × 10−10), and XKR6 (P = 9.8 × 10−10). Although this approach did not account for LD between regions, these three regions were >100 Mb away from another association.
Expression-based analyses
We used MetaXcan25 to analyze gene expression and expression quantitative trait loci (eQTL) extracted from GTEx project data.23 We examined four tissues which may be involved in the AS disease process: aorta, left ventricle, liver, and whole blood. With a false discovery rate of 5%, we identified 42 genes with predicted expression that differed between cases and controls (see Supplementary material online, Figure S5). Increased LPA expression in the liver was predicted for AS cases (Z score = 5.25; P = 1.5 × 10−7). Expression of IL6R was inferred to be increased in the blood of cases (Z score = 4.66; P = 3.1 × 10−6). NOTCH4 mRNA in the aorta was predicted to be decreased in cases (Z score = −4.07; P = 4.7 × 10−5). In contrast to these single tissue effects, inferred RPS23 expression was lower in the blood, left ventricle, and aorta in AS cases (P ≤ 3.3 × 10−5). The most significant, predicted differential expression was decreased ZEB2 expression in the aorta of individuals with AS (Z score = −7.14; P = 9.2 × 10−13). Additional GIGSEA27 were performed to infer the differential expression of genes (see Supplementary material online, Table S7).
As identified in GTEx,23 the index variants at the ACTR2 and BET1L loci were in high LD (r2≥ 0.98) with the most significant eQTL for their respective genes in the aorta. The index variant at the ARHGEF26 locus was in high LD (r2 = 0.96) with the most significant eQTL for ARHGEF26 in the left ventricle. In addition, rs55909255 was associated with the expression of SMC2 in the left ventricle, and rs140570886 was associated with the expression of LPA in the liver. Index variants and proxies at the CELSR2–SORT1, IL6, and FADS1/2 loci were all eQTL for multiple genes in three to four tissues (see Supplementary material online, Table S2). Notably, the index variant at the CELSR2–SORT1 locus was the most significant hepatic eQTL for CELSR2 and SORT1, and the index variant at the FADS1/2 locus was in high LD (r2 ≥ 0.96) with the most significant eQTL for FADS1 in the aorta and liver. Neither the index variant at the IL6 locus nor any variants in high LD (r2 ≥ 0.95) were associated with IL6 expression in the tissues examined. However, the index variant was in high LD with the most significant eQTL for IL6 antisense RNA 1 (IL6-AS1) in the blood (r2 = 0.96) (see Supplementary material online, Table S2). Several of these results were confirmed by additional colocalization analyses using MetaXcan and coloc (see Supplementary material online, Figure S5, and Supplementary material online, Table S8). Notably, we identified high posterior probabilities for colocalization for SORT1 expression in the liver for rs6702619, as well as ACTR2 expression in the aorta for rs7556894 and FADS1 in whole blood and left ventricle for rs174535.
AS is genetically correlated with adiposity
Using LD score regression35 as implemented on LD Hub,36 we computed the genetic correlation of AS with 157 traits related to cardiovascular risk factors, metabolites, and immunological diseases using GWAS summary statistics. With a false discovery rate of 5%, we observed 32 (18.5%) traits that were genetically correlated with AS, most of which involved adiposity, glycemia, or lipids (see Supplementary material online, Table S9). Measures of adiposity were positively correlated with AS and represented seven of the 10 most significant correlations. The only inverse correlations were with high-density lipoprotein particles. The total lipid content of chylomicrons and large very low-density lipoprotein particles shared the highest absolute genetic correlation with AS (rg, 0.31; 95% CI, 0.13 to 0.48; P = 6.0 × 10−4).
Pleiotropic effects of variants associated with aortic stenosis
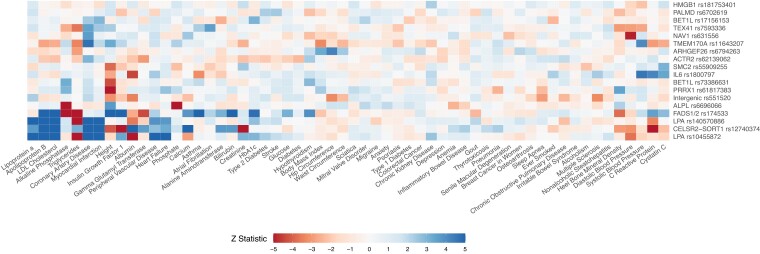
We assessed the association of the index variants with 58 biomarkers, physiological measurements, and diseases among 257 231 UK Biobank White British participants. Traits were selected to reflect cardiovascular, cerebrovascular, immune, hepatic, and adiposity phenotypes. We observed 98 associations with false discovery rate-adjusted P ≤ 0.05 and the five most significant associations involved apolipoprotein B, lipoprotein(a), LDL-C, or triglycerides (see Supplementary material online, Table S10). The traits with the greatest number of associations were height (eight variants); CAD, albumin, and triglycerides (six variants each); C-reactive protein and apolipoprotein B (five variants each); and heel bone mineral density, diastolic blood pressure, and LDL-C cholesterol (four variants each) (Figure 3; Supplementary material online, Table S10).
Figure 3.
Association of aortic stenosis variants with biomarkers, physiological measurements, and diseases. Variants were ordered to reflect similarity in their associations with traits and vice versa. The strength and direction of associations are represented by cells of different colors, with blue cells indicating positive effects on the odds of aortic stenosis and red cells indicating inverse associations. For ease of visualization, Z statistics >5 or less than −5 were rounded to 5 and −5, respectively.
Blood-based biomarkers are causal contributors to aortic stenosis
We performed Mendelian randomization for six traits associated with several variants in the phenome-wide association study. We constructed genetic instruments using summary statistics from GWAS performed on UK Biobank (White British) and assessed the association of these instruments with AS. Using the IVW method, genetically elevated levels of apolipoprotein B, lipoprotein(a), LDL-C, and BMI were associated with increased odds of AS [OR per g/L of apolipoprotein B, 3.85; 95% CI, 2.90–5.12; P = 2.1 × 10−20; OR per natural logarithm of lipoprotein(a), 1.20; 95% CI, 1.17–1.23; P = 3.0 × 10−53; OR per mmol/L of LDL-C, 1.57; 95% CI, 1.44–1.71; P = 1.5 × 10−25; OR per kg/m2 of BMI, 1.07; 95% CI, 1.05, 1.09; P = 1.9 × 10−12, respectively], with no evidence of pleiotropy (all Egger intercepts P > 0.05) (see Supplementary material online, Table S11). The associations between AS and higher genetically predicted levels of both C-reactive protein and alkaline phosphatase were pleiotropic (Egger intercept P-values of 0.024 and 0.003), and MR results were not consistent across methods limiting causal inference (see Supplementary material online, Table S11).
Discussion
In this genome-wide meta-analysis combining summary statistics from 10 cohorts of European ancestry, we confirmed association for seven previously identified loci and identified 10 loci not previously reported to be genome-wide significant for AS. For newly discovered variants, we observed modest changes in AS odds (9%–55% increased odds per risk allele). In independent replication, 8 of the 10 previously unreported variants were associated with AS, with the exception of BET1L rs73386631 and HMGB1 rs181753401 which had low allele frequencies. Our work brings the total number of loci robustly associated with AS to 15. Several of these variants were associated with AS in other ancestries, and a risk score composed of all identified variants was a predictor of AS independent of other risk factors. Further analysis using region- and gene-based methods identified additional gene regions associated with AS, including LDLR and NOTCH4, as well as differential expression of co-regulated groups of genes. Finally, Mendelian randomization supported a causal contribution of lipoprotein(a), apolipoprotein B, LDL-C, and BMI to AS.
Our findings support four key etiological mechanisms for AS: calcification, lipid metabolism, adiposity, and inflammation (Structured Graphical Abstract). The variants at the PRRX1, ACTR2, LPA, NLRP6, and FADS1/2 loci were also associated with aortic valve calcification assessed by computed tomography. We observed associations of the index variants at the CELSR2–SORT1, PRRX1, TEX41, and FADS1/2 loci with heel bone mineral density, suggesting systemic effects on calcification. The paired-related homeobox protein 1, the product of the locus PRRX1, is a transcription factor required for osteoblast differentiation by TNF-α41 and is an inducer of the epithelial–mesenchymal transition,42 a key process in early calcification. Interestingly, zinc finger e-box binding homeobox 2, coded by ZEB2, is a repressor of this process.43 Predicted ZEB2 expression in the aorta was the most significant differentially expressed gene. Consistent with a potential role in calcification, earlier work demonstrated that TEX41 variants may be associated with AS through long-range chromatin interactions with the ZEB2 promoter region, including in the aorta9 implicating TEX41, ZEB2, and PRRX1 in inducing early calcification and subsequent valve stenosis.
Located in the 3’ untranslated region of CELSR2, rs12740374 affects expression of SORT1,44 which has been reported to decrease hepatic excretion of apolipoprotein B and increase catabolism of LDL-C.45 Studies have reported the association of lower LDL-C46,47 and lower odds of CAD,48,49 and in a study involving two cohorts participating in the current study, a variant in perfect LD with rs12740374 was associated with AS after Bonferroni correction (P = 3.4 × 10−4).9 The present analysis identified genome-wide significance in the association of the CELSR2–SORT1 variant with AS. Consistent with its effects on LDL-C and apolipoprotein B, the minor allele conferred a reduction in the odds of AS. Additional evidence that lipid metabolism is a causal mechanism for AS was provided by Mendelian randomization, which confirmed the role of LDL-C12 and identified a contribution of apolipoprotein B, which extends this association to all apolipoprotein B-containing lipoprotein particles. The findings were consistent with work demonstrating an association of a non-high-density lipoprotein cholesterol genetic risk score with AS.9 In addition to these lipoprotein-mediated effects, sortilin, coded by SORT1, is a regulator of vascular calcification and is associated with increased aortic calcification in mice.50 Furthermore, sortilin expression is associated with increased expression of ALPL,50 a known AS locus replicated in this study.
Both overall and abdominal obesity have been previously associated with AS,51 and a Mendelian randomization study provided support for the causality of BMI.14 Our Mendelian randomization analyses replicate and extend these findings by observing genetic correlations between AS and multiple measures of adiposity, including BMI, waist and hip circumferences, and obesity. However, only one of our genome-wide significant variants, ARHGEF26 rs6794263, which was in high LD with the missense variant rs13096373 (p.Phe203Ser), was associated with BMI and hip circumference in our phenome-wide analysis. Another missense variant rs12493885 (p.Val29Leu) in ARHGEF26 was previously associated with CAD,52 mediated by a gain of function for ARHGEF26 that may lead to increased transendothelial migration, greater adhesion of leukocytes, and proliferation of vascular smooth muscle cells.52 However, this variant was independent of ARHGEF26 rs6794263 (r2 = 0.018)39 and was not associated with AS in our meta-analysis (OR per T allele, 1.02; 95% CI, 0.98–1.06; P = 0.44 for rs1713812, which is in perfect LD with rs1249388539). Conversely, the allele of rs6794263 that confers a lower odds of AS has also been associated with lower odds of CAD, but not at genome-wide significance (OR per C allele, 0.96; 95% CI, 0.92–1.00; P = 0.037).53 The presence of two independent, missense mutations in ARHGEF26 with discordant effects on AS and CAD suggests pleiotropic effects of this locus.
The accumulation of inflammatory cells in the aortic valve is associated with remodeling and fibrosis,54 highlighting the role for inflammation in disease progression. We confirmed that the FADS1/2 locus was associated with AS.11 Both FADS1 and FADS2 encode key enzymes in the conversion of dietary n-6 fatty acids to arachidonic acid, a precursor of pro-inflammatory leukotrienes and prostaglandins.55 In addition, we also identified a novel variant at the locus for NLRP6, which assembles an inflammasome that plays a role in immunity to bacterial infection as well as proinflammatory responses to other stimuli, including fatty acids.56,57 We replicated that the association previously reported between an IL6 variant and AS10 and the IL6 variant rs2069832 (r2 = 0.95 with our index variant) colocalizes with IL6-AS1 expression.10 Notably, the risk-increasing alleles are associated with increased expression of both IL6 and IL6-AS1 in fibroblasts in GTEx.23 Our analyses therefore indicate a pro-inflammatory association with AS in the IL6 region but also higher predicted IL6R expression in the blood cells of AS cases. Thus, several orthogonal signals support the association of pro-inflammatory pathways with AS.
The present study represents the largest genome-wide meta-analysis of AS, a common condition with no available medical treatment, and included 653 867 participants (13 765 cases) in which we applied variant-, gene-, and gene set-based analyses to identify additional risk loci and disease mechanisms. Our work highlights novel mechanisms and pathways which may have important clinical and future research implications. First, our analyses point to several possible therapeutic interventions using both lifestyle and novel drugs. Our findings of lipoproteins and adiposity as key drivers of aortic stenosis suggest that maintenance of healthy lifestyle and adherence to lipid-lowering recommendations may reduce the incidence of aortic stenosis. Furthermore, lipoprotein(a)-lowering therapies currently in development58,59 may provide a new avenue for prevention of disease progression. Whether novel hypoglycemics, which also lead to marked weight loss and metabolic improvements,60 and anti-inflammatory agents (e.g. that inhibit IL-6 signaling61) could reduce AS requires randomized trials. Finally, our observation of a robust association between a genetic risk score and AS may allow identification of patients at risk for rapid disease progression in clinical practice, who may require echocardiographic follow-up, as well as permit targeted enrolment of such at-risk patients in future randomized trials.
Despite the strengths of the study, there are several limitations. Since our discovery cohorts were of European ancestry, the transferability of our results to other ancestries may be limited.62–64 Although we attempted cross-ancestry replication of genome-wide significant variants, the observed limited reproducibility may be due to the low numbers of African and Latin American participants. Future studies should focus on non-Europeans. Our analyses also made exclusive use of bioinformatic methods to identify genetic loci. These results require confirmation using complementary approaches. Lastly, cases were selected using various criteria (international classification of diseases (ICD) codes, surgical AVR, etc.) and not solely by echocardiography. Therefore we could not entirely exclude bicuspid aortic valves; however, these likely represent a small proportion of the cases.
In conclusion, our results identify novel genetic contributors to AS and identify specific contributions to disease etiology that are characterized by the effects of calcification, altered lipid metabolism, adiposity, and inflammation. An AS genetic risk score was an independent predictor of both clinical and subclinical disease, providing additional discrimination when added to clinical risk factors. Established and novel genetic loci warrant investigation as potential therapeutic targets to prevent the initiation of aortic calcification and progression to stenosis.
Supplementary Material
Acknowledgements
We would like to acknowledge the valuable contributions of the investigators, staff, and participants from all cohorts. Analyses conducted using the UK Biobank Resource were performed under Application Numbers 11537, 41025, and 24281.
Contributor Information
Hao Yu Chen, Division of Experimental Medicine, McGill University, 1001 Decarie Blvd., Room EM1.2218, Montreal, Quebec H4A 3J1, Canada; Preventive and Genomic Cardiology, McGill University Health Centre and Research Institute, 1001 Decarie Blvd., Room D05.5120, Montreal, Quebec H4A 3J1, Canada.
Christian Dina, Nantes Université, CHU Nantes, CNRS, INSERM, l’institut du thorax, 8 Quai Moncousu, Nantes F-44000, France.
Aeron M Small, Department of Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, USA.
Christian M Shaffer, Vanderbilt Translational and Clinical Cardiovascular Research Center, Vanderbilt University Medical Center, Nashville, USA.
Rebecca T Levinson, Vanderbilt Translational and Clinical Cardiovascular Research Center, Vanderbilt University Medical Center, Nashville, USA.
Anna Helgadóttir, deCODE genetics/Amgen Inc., Reykjavik, Iceland.
Romain Capoulade, Nantes Université, CHU Nantes, CNRS, INSERM, l’institut du thorax, 8 Quai Moncousu, Nantes F-44000, France.
Hans Markus Munter, McGill University and Genome Quebec Innovation Centre, Montreal, Canada.
Andreas Martinsson, Department of Cardiology, Clinical Sciences, Lund University, Sweden and Skåne University Hospital, Lund, Sweden; The Wallenberg Laboratory/Department of Molecular and Clinical Medicine, Institute of Medicine, Gothenburg University and the Department of Cardiology, Sahlgrenska University Hospital, Gothenburg, Sweden.
Benjamin J Cairns, MRC Population Health Research Unit, Clinical Trial Service Unit and Epidemiological Studies Unit, Nuffield Department of Population Health, University of Oxford, Oxford, UK.
Linea C Trudsø, Laboratory for Molecular Cardiology, Department of Cardiology, Copenhagen University Hospital, Rigshospitalet, Copenhagen, Denmark.
Mary Hoekstra, Division of Experimental Medicine, McGill University, 1001 Decarie Blvd., Room EM1.2218, Montreal, Quebec H4A 3J1, Canada; Preventive and Genomic Cardiology, McGill University Health Centre and Research Institute, 1001 Decarie Blvd., Room D05.5120, Montreal, Quebec H4A 3J1, Canada.
Hannah A Burr, Division of Experimental Medicine, McGill University, 1001 Decarie Blvd., Room EM1.2218, Montreal, Quebec H4A 3J1, Canada; Preventive and Genomic Cardiology, McGill University Health Centre and Research Institute, 1001 Decarie Blvd., Room D05.5120, Montreal, Quebec H4A 3J1, Canada.
Thomas W Marsh, Preventive and Genomic Cardiology, McGill University Health Centre and Research Institute, 1001 Decarie Blvd., Room D05.5120, Montreal, Quebec H4A 3J1, Canada; Department of Human Genetics, McGill University, Montreal, Canada.
Scott M Damrauer, Department of Surgery, Perelman School of Medicine, University of Pennsylvania, Philadelphia, USA.
Line Dufresne, Preventive and Genomic Cardiology, McGill University Health Centre and Research Institute, 1001 Decarie Blvd., Room D05.5120, Montreal, Quebec H4A 3J1, Canada.
Solena Le Scouarnec, Nantes Université, CHU Nantes, CNRS, INSERM, l’institut du thorax, 8 Quai Moncousu, Nantes F-44000, France.
David Messika-Zeitoun, Department of Cardiology, Assistance Publique - Hôpitaux de Paris, Bichat Hospital, Paris, France; Division of Cardiology, University of Ottawa Heart Institute, Ottawa, Ontario, Canada.
Dilrini K Ranatunga, Division of Research, Kaiser Permanente of Northern California, Oakland, USA.
Rachel A Whitmer, Department of Public Health Sciences, University of California Davis, Davis, USA.
Amélie Bonnefond, University Lille, Inserm, CNRS, CHU Lille, Institut Pasteur de Lille, UMR1283-8199 EGID, Lille, France; Department of Metabolism, Imperial College London, London, UK.
Garðar Sveinbjornsson, deCODE genetics/Amgen Inc., Reykjavik, Iceland.
Ragnar Daníelsen, Internal Medicine and Emergency Services, Landspitali—The National University Hospital of Iceland, Reykjavik, Iceland.
David O Arnar, deCODE genetics/Amgen Inc., Reykjavik, Iceland; Internal Medicine and Emergency Services, Landspitali—The National University Hospital of Iceland, Reykjavik, Iceland; School of Health Sciences, Faculty of Medicine, University of Iceland, Reykjavik, Iceland.
Gudmundur Thorgeirsson, deCODE genetics/Amgen Inc., Reykjavik, Iceland; School of Health Sciences, Faculty of Medicine, University of Iceland, Reykjavik, Iceland.
Unnur Thorsteinsdottir, deCODE genetics/Amgen Inc., Reykjavik, Iceland; School of Health Sciences, Faculty of Medicine, University of Iceland, Reykjavik, Iceland.
Daníel F Gudbjartsson, deCODE genetics/Amgen Inc., Reykjavik, Iceland; School of Engineering and Natural Sciences, University of Iceland, Reykjavik, Iceland.
Hilma Hólm, deCODE genetics/Amgen Inc., Reykjavik, Iceland.
Jonas Ghouse, Laboratory for Molecular Cardiology, Department of Cardiology, Copenhagen University Hospital, Rigshospitalet, Copenhagen, Denmark.
Morten Salling Olesen, Laboratory for Molecular Cardiology, Department of Cardiology, Copenhagen University Hospital, Rigshospitalet, Copenhagen, Denmark.
Alex H Christensen, Laboratory for Molecular Cardiology, Department of Cardiology, Copenhagen University Hospital, Rigshospitalet, Copenhagen, Denmark; Department of Cardiology, Herlev-Gentofte Hospital, Copenhagen, Denmark.
Susan Mikkelsen, Department of Clinical Immunology, Aarhus University Hospital, Aarhus, Denmark.
Rikke Louise Jacobsen, Department of Clinical Immunology, Copenhagen University Hospital, Rigshospitalet, Copenhagen, Denmark.
Joseph Dowsett, Department of Clinical Immunology, Copenhagen University Hospital, Rigshospitalet, Copenhagen, Denmark.
Ole Birger Vesterager Pedersen, Department of Clinical Immunology, Zealand University Hospital, Naestved, Denmark.
Christian Erikstrup, Department of Clinical Immunology, Aarhus University Hospital, Aarhus, Denmark; Department of Clinical Medicine, Aarhus University, Aarhus, Denmark.
Sisse R Ostrowski, Department of Clinical Immunology, Copenhagen University Hospital, Rigshospitalet, Copenhagen, Denmark; Department of Clinical Medicine, University of Copenhagen, Copenhagen, Denmark.
Regeneron Genetics Center, Regeneron Genetics Center, New York, USA.
Christopher J O’Donnell, National Heart, Lung, and Blood Institute's and Boston University's Framingham Heart Study, Boston, USA.
Matthew J Budoff, The Lundquist Institute for Biomedical Innovation at Harbor-UCLA Medical Center, Torrance, USA.
Vilmundur Gudnason, Faculty of Medicine, University of Iceland, Reykjavík, Iceland.
Wendy S Post, Division of Cardiology, Department of Medicine, The Johns Hopkins University School of Medicine, Baltimore, USA.
Jerome I Rotter, The Institute for Translational Genomics and Population Sciences, Department of Pediatrics, The Lundquist Institute for Biomedical Innovation at Harbor-UCLA Medical Center, Torrance, USA.
Mark Lathrop, McGill University and Genome Quebec Innovation Centre, Montreal, Canada; Department of Human Genetics, McGill University, Montreal, Canada.
Henning Bundgaard, Department of Cardiology, Copenhagen University Hospital, Rigshospitalet, Copenhagen, Denmark.
Bengt Johansson, Department of Public Health and Clinical Medicine, Umeå University, Umeå, Sweden.
Johan Ljungberg, Department of Public Health and Clinical Medicine, Umeå University, Umeå, Sweden.
Ulf Näslund, Department of Public Health and Clinical Medicine, Umeå University, Umeå, Sweden.
Thierry Le Tourneau, Nantes Université, CHU Nantes, CNRS, INSERM, l’institut du thorax, 8 Quai Moncousu, Nantes F-44000, France.
J Gustav Smith, Department of Cardiology, Clinical Sciences, Lund University, Sweden and Skåne University Hospital, Lund, Sweden; Wallenberg Center for Molecular Medicine and Lund University Diabetes Center, Lund, Sweden; The Wallenberg Laboratory/Department of Molecular and Clinical Medicine, Institute of Medicine, Gothenburg University and the Department of Cardiology, Sahlgrenska University Hospital, Gothenburg, Sweden.
Quinn S Wells, Vanderbilt Translational and Clinical Cardiovascular Research Center, Vanderbilt University Medical Center, Nashville, USA.
Stefan Söderberg, Department of Public Health and Clinical Medicine, Umeå University, Umeå, Sweden.
Kári Stefánsson, deCODE genetics/Amgen Inc., Reykjavik, Iceland; School of Health Sciences, Faculty of Medicine, University of Iceland, Reykjavik, Iceland.
Jean-Jacques Schott, Nantes Université, CHU Nantes, CNRS, INSERM, l’institut du thorax, 8 Quai Moncousu, Nantes F-44000, France.
Daniel J Rader, Departments of Genetics and Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, USA.
Robert Clarke, MRC Population Health Research Unit, Clinical Trial Service Unit and Epidemiological Studies Unit, Nuffield Department of Population Health, University of Oxford, Oxford, UK.
James C Engert, Division of Experimental Medicine, McGill University, 1001 Decarie Blvd., Room EM1.2218, Montreal, Quebec H4A 3J1, Canada; Preventive and Genomic Cardiology, McGill University Health Centre and Research Institute, 1001 Decarie Blvd., Room D05.5120, Montreal, Quebec H4A 3J1, Canada; Department of Human Genetics, McGill University, Montreal, Canada.
George Thanassoulis, Division of Experimental Medicine, McGill University, 1001 Decarie Blvd., Room EM1.2218, Montreal, Quebec H4A 3J1, Canada; Preventive and Genomic Cardiology, McGill University Health Centre and Research Institute, 1001 Decarie Blvd., Room D05.5120, Montreal, Quebec H4A 3J1, Canada.
Author contributions
Concept and design: Chen, Engert, and Thanassoulis. Acquisition, analysis, or interpretation of data: Chen, Dina, Small, Shaffer, Levinson, Helgadóttir, Capoulade, Munter, Martinsson, Cairns, Trudsø, Hoekstra, Burr, Marsh, Dufresne, Messika-Zeitoun, Le Scouarnec, Ghouse, Olesen, Christensen, Mikkelsen, Jacobsen, Dowsett, Pedersen, Erikstrup, Ostrowski, Budoff, Post, Rotter, Bundgaard, Le Tourneau, Smith, Hólm, Söderberg, Schott, Engert, and Thanassoulis. Drafted the manuscript: Chen. Critical revision of the manuscript for important intellectual content: Helgadóttir, Capoulade, Martinsson, Cairns, Trudsø, Hoekstra, Marsh, Dufresne, Hólm, Christensen, Mikkelsen, Erikstrup, Ostrowski, Post, Rotter, Bundgaard, Johansson, Ljungberg, Näslund, Smith, Söderberg, Schott, Clarke, Engert, and Thanassoulis. Obtained funding: Gudnason, Näslund, Post, Rotter, Lathrop, Le Tourneau, Messika-Zeitoun, Smith, Söderberg, Schott, Engert, and Thanassoulis. Administrative, technical, or material support: Ranatunga, Whitmer, Bonnefond, Lathrop, Ljungberg, Näslund, Le Tourneau, Smith, Söderberg, Schott, Engert, and Thanassoulis. Supervision: Damrauer, Budoff, Gudnason, Rotter, Johansson, Smith, Rader, Clarke, Engert, and Thanassoulis.
Supplementary data
Supplementary data is available at European Heart Journal online.
Data availability
The summary level statistics from the meta-GWAS that support the findings of this study are available online @ Zenodo.org (https://doi.org/10.5281/zenodo.7505361).
Funding
Thomas W. Marsh was supported by a research award from the Fonds de Recherche du Québec—Santé. Scott M. Damrauer is supported by the US Department of Veteran Affairs (IK2-CX00180). This work does not represent the views of the US Government or the US Department of Veterans Affairs. Stefan Söderberg was supported by Hjärt-Lungfonden (grant numbers 20140799, 20120631, and 20100635), the County Council of Västerbotten (ALF, VLL-548791), Umeå University, and the Heart Foundation of Northern Sweden. J. Gustav Smith was supported by grants from the Swedish Heart-Lung Foundation (2016-0134, 2016-0315, and 2019-0526), the Swedish Research Council (2017-02554), the European Research Council (ERC-STG-2015-679242), the Crafoord Foundation, Skåne University Hospital, the Scania County, governmental funding of clinical research within the Swedish National Health Service, a generous donation from the Knut and Alice Wallenberg foundation to the Wallenberg Center for Molecular Medicine in Lund, and funding from the Swedish Research Council (Linnaeus grant Dnr 349-2006-237, Strategic Research Area Exodiab Dnr 2009-1039) and the Swedish Foundation for Strategic Research (Dnr IRC15-0067) to the Lund University Diabetes Center. George Thanassoulis is a Chercheur Boursier Clinicien—Senior from the Fonds de Recherche du Québec—Santé. George Thanassoulis and James C. Engert hold an operating grant from the Canadian Institutes for Health Research, and George Thanassoulis holds operating grants from the Heart and Stroke Foundation of Canada and the National Institutes of Health (R01 HL128550). This study was funded by grants from the Fonds de Recherche du Québec—Santé, Canadian Institutes for Health Research, Heart and Stroke Foundation of Canada, and National Institutes of Health (R01 HL128550). The Kaiser Permanente Research Program on Genes, Environment, and Health was funded by the Ellison Medical Foundation, Robert Wood Johnson Foundation, Wayne and Gladys Valley Foundation, Kaiser Permanente Northern California, and the Kaiser Permanente National and Regional Community Benefit Programs. The GERA cohort was funded by a grant from the National Institutes of Health. The University of Oxford MRC Population Health Research Unit is funded through a strategic partnership between the Medical Research Council and the University of Oxford. Staff were also supported by the British Heart Foundation and British Heart Foundation Oxford Centre for Research Excellence (RE/18/3/34214). A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org. Multi-Ethnic Study of Atherosclerosis and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts 75N92020D00001, HHSN268201500003I, N01-HC-95159, 75N92020D00005, N01-HC-95160, 75N92020D00002, N01-HC-95161, 75N92020D00003, N01-HC-95162, 75N92020D00006, N01-HC-95163, 75N92020D00004, N01-HC-95164, 75N92020D00007, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, UL1-TR-001420. Funding for SHARe genotyping was provided by NHLBI Contract N02-HL-64278. Genotyping was performed at Affymetrix (Santa Clara, California, USA) and the Broad Institute of Harvard and MIT (Boston, Massachusetts, USA) using the Affymetrix Genome-Wide Human SNP Array 6.0. Also supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR001881, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. This research was supported by R01 HL071739. Infrastructure for the CHARGE consortium is supported in part by the NHLBI grant R01HL105756 and also supported in part by National Institute of Health contract R01HL146860. Vanderbilt University Medical Center’s BioVU projects are supported by numerous sources: institutional funding, private agencies, and federal grants. These include the National Institutes of Health Grant S10RR025141; Clinical and Translational Science Award grants UL1TR002243, UL1TR000445, and UL1RR024975. Genomic data are also supported by investigator-led projects that include U01HG004798, R01NS032830, RC2GM092618, P50GM115305, U01HG006378, U19HL065962, and R01HD07471. This work was supported by an ANR & FRM grant [13-BSV6-0011 and DCV20070409278] to Jean-Jacques Schott, by a Fédération Française de Cardiologie, a Fondation Coeur et Recherche and an Inserm Translational Research grant to Thierry Le Tourneau. Romain Capoulade is supported by a ‘Connect Talent’ research chair from Région Pays de la Loire. Cohort PREGI set up was supported by the French Regional Council of Pays-de-la-Loire (VaCaRMe program). We thank Marie Marrec and Guénola Coste for their contribution to clinical data collection and the Genomics and Bioinformatics Core Facility of Nantes (GenoBiRD, Biogenouest) for its technical support. The D.E.S.I.R. study (providing a set of controls) has been supported by INSERM contracts with CNAMTS, Lilly, Novartis Pharma, and Sanofi-Aventis; by INSERM (Réseaux en Santé Publique, Interactions entre les déterminants de la santé), Cohortes Santé TGIR, the Association Diabète Risque Vasculaire, the Fédération Française de Cardiologie, La Fondation de France, ALFEDIAM, Société francophone du diabète, ONIVINS, Abbott, Ardix Medical, Bayer Diagnostics, Becton Dickinson, Cardionics, Merck Santé, Novo Nordisk, Pierre Fabre, Roche, and Topcon. The D.E.S.I.R. Study Group. INSERM U1018: B. Balkau, P. Ducimetière, E. Eschwège; INSERM U367: F. Alhenc-Gelas; CHU D’Angers: A. Girault; Bichat Hospital: F. Fumeron, M. Marre, R Roussel; CHU de Rennes: F. Bonnet; CNRS UMR8090, Lille: A. Bonnefond, P. Froguel; Univ Paris Descartes, UMR1153, Paris: F.Rancière Centres d’Examens de Santé: Alençon, Angers, Blois, Caen, Chateauroux, Chartres, Cholet, Le Mans, Orléans, Tours; Institute de Recherche Médecine Générale: J. Cogneau; General practitioners of the Region; Institute inter-Regional pour la Santé: C. Born, E. Caces, M. Cailleau, O Lantieri, J.G. Moreau, F. Rakotozafy, J. Tichet, S. Vol. We would like to specially thank the team of the Centre d'Investigation Clinique (Xavier Duval), the Centre de Ressources Biologiques (Sarah Tubiana), Christophe Aucan from the Assistance Publique—Hôpitaux de Paris, Département de la Recherche Clinique et du Développement (DRCD) and Estelle Marcault from the Unité de Recherche Clinique Paris Nord for their help and support during all these years. The COFRASA (clinicalTrial.gov number NCT 00338676) and GENERAC (clinicalTrial.gov number NCT00647088) studies are supported by grants from the Assistance Publique—Hôpitaux de Paris (PHRC National 2005 and 2010, and PHRC regional 2007). The Copenhagen Hospital Biobank and The Danish Blood Donor Study were supported by the Novo Nordisk Foundation (grant numbers NNF14CC0001 and NNF17OC0027594). This work was further supported by The Innovation Fund Denmark (PM Heart) Nord-Forsk (project no. 90580) and the Capital Regions Research Council (No. A5920). None of the funding sources were involved in the collection, analysis, and interpretation of data, nor the writing of this paper or the decision to submit the paper for publication.
References
- 1. Andell P, Li X, Martinsson A, Andersson C, Stagmo M, Zoller B, et al. Epidemiology of valvular heart disease in a Swedish nationwide hospital-based register study. Heart 2017;103:1696–1703. 10.1136/heartjnl-2016-310894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Danielsen R, Aspelund T, Harris TB, Gudnason V. The prevalence of aortic stenosis in the elderly in Iceland and predictions for the coming decades: the AGES-Reykjavik study. Int J Cardiol 2014;176:916–922. 10.1016/j.ijcard.2014.08.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vahanian A, Beyersdorf F, Praz F, Milojevic M, Baldus S, Bauersachs J, et al. 2021 ESC/EACTS guidelines for the management of valvular heart disease. Eur Heart J 2022;43:561–632. 10.1093/eurheartj/ehab395 [DOI] [PubMed] [Google Scholar]
- 4. Members WC, Otto CM, Nishimura RA, Bonow RO, Carabello BA, Erwin JP III, et al. 2020 ACC/AHA guideline for the management of patients with valvular heart disease: a report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. J Am Coll Cardiol 2021;77:e25–e197. 10.1016/j.jacc.2020.11.018 [DOI] [PubMed] [Google Scholar]
- 5. Martinsson A, Li X, Zoller B, Andell P, Andersson C, Sundquist K, et al. Familial aggregation of aortic valvular stenosis: a nationwide study of sibling risk. Circ Cardiovasc Genet 2017;10:e001742. 10.1161/CIRCGENETICS.117.001742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Probst V, Le Scouarnec S, Legendre A, Jousseaume V, Jaafar P, Nguyen JM, et al. Familial aggregation of calcific aortic valve stenosis in the western part of France. Circulation 2006;113:856–860. 10.1161/CIRCULATIONAHA.105.569467 [DOI] [PubMed] [Google Scholar]
- 7. Thanassoulis G, Campbell CY, Owens DS, Smith JG, Smith AV, Peloso GM, et al. Genetic associations with valvular calcification and aortic stenosis. N Engl J Med 2013;368:503–512. 10.1056/NEJMoa1109034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Theriault S, Gaudreault N, Lamontagne M, Rosa M, Boulanger MC, Messika-Zeitoun D, et al. A transcriptome-wide association study identifies PALMD as a susceptibility gene for calcific aortic valve stenosis. Nat Commun 2018;9:988. 10.1038/s41467-018-03260-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Helgadottir A, Thorleifsson G, Gretarsdottir S, Stefansson OA, Tragante V, Thorolfsdottir RB, et al. Genome-wide analysis yields new loci associating with aortic valve stenosis. Nat Commun 2018;9:987. 10.1038/s41467-018-03252-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Theriault S, Dina C, Messika-Zeitoun D, Le Scouarnec S, Capoulade R, Gaudreault N, et al. Genetic association analyses highlight IL6, ALPL, and NAV1 as 3 new susceptibility genes underlying calcific aortic valve stenosis. Circ Genom Precis Med 2019;12:e002617. 10.1161/CIRCGEN.119.002617 [DOI] [PubMed] [Google Scholar]
- 11. Chen HY, Cairns BJ, Small AM, Burr HA, Ambikkumar A, Martinsson A, et al. Association of FADS1/2 locus variants and polyunsaturated fatty acids with aortic stenosis. JAMA Cardiol 2020;5:694–702. 10.1001/jamacardio.2020.0246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Smith JG, Luk K, Schulz CA, Engert JC, Do R, Hindy G, et al. Association of low-density lipoprotein cholesterol-related genetic variants with aortic valve calcium and incident aortic stenosis. JAMA 2014;312:1764–1771. 10.1001/jama.2014.13959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arsenault BJ, Boekholdt SM, Dube MP, Rheaume E, Wareham NJ, Khaw KT, et al. Lipoprotein(a) levels, genotype, and incident aortic valve stenosis: a prospective Mendelian randomization study and replication in a case-control cohort. Circ Cardiovasc Genet 2014;7:304–310. 10.1161/CIRCGENETICS.113.000400 [DOI] [PubMed] [Google Scholar]
- 14. Kaltoft M, Langsted A, Nordestgaard BG. Obesity as a causal risk factor for aortic valve stenosis. J Am Coll Cardiol 2020;75:163–176. 10.1016/j.jacc.2019.10.050 [DOI] [PubMed] [Google Scholar]
- 15. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM; 1000 Genomes Project Consortium , et al. A global reference for human genetic variation. Nature 2015;526:68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gonzalez JN, Zweig AS, Speir ML, Schmelter D, Rosenbloom KR, Raney BJ, et al. The UCSC genome browser database: 2021 update. Nucleic Acids Res 2021;49:D1046–D1057. 10.1093/nar/gkaa1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res 2019;47:D886–D894. 10.1093/nar/gky1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Quang D, Chen Y, Xie X. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics 2015;31:761–763. 10.1093/bioinformatics/btu703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang YF, Gulko B, Siepel A. Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nat Genet 2017;49:618–624. 10.1038/ng.3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ionita-Laza I, McCallum K, Xu B, Buxbaum JD. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat Genet 2016;48:214–220. 10.1038/ng.3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shihab HA, Gough J, Mort M, Cooper DN, Day IN, Gaunt TR. Ranking non-synonymous single nucleotide polymorphisms based on disease concepts. Hum Genomics 2014;8:11. 10.1186/1479-7364-8-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. GTEx Consortium . The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348:648–660. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 2015;11:e1004219. 10.1371/journal.pcbi.1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barbeira AN, Dickinson SP, Bonazzola R, Zheng J, Wheeler HE, Torres JM, et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun 2018;9:1825. 10.1038/s41467-018-03621-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, et al. Bayesian Test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 2014;10:e1004383. 10.1371/journal.pgen.1004383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu S, Qian T, Hoshida Y, Shen Y, Yu J, Hao K. GIGSEA: genotype imputed gene set enrichment analysis using GWAS summary level data. Bioinformatics 2019;35:160–163. 10.1093/bioinformatics/bty529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. The Gene Ontology Consortium . The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res 2019;47:D330–D338. 10.1093/nar/gky1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lizio M, Harshbarger J, Shimoji H, Severin J, Kasukawa T, Sahin S, et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol 2015;16:22. 10.1186/s13059-014-0560-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res 2019;47:D155–D162. 10.1093/nar/gky1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015;4:7. 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Prive F, Arbel J, Vilhjalmsson BJ. LDpred2: better, faster, stronger. Bioinformatics 2020;36:5424–5431. 10.1093/bioinformatics/btaa1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yavorska OO, Burgess S. Mendelianrandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol 2017;46:1734–1739. 10.1093/ije/dyx034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J; Schizophrenia Working Group of the Psychiatric Genomics Consortium , et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015;47:291–295. 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC, et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 2017;33:272–279. 10.1093/bioinformatics/btw613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang J, Ferreira T, Morris AP, Medland SE; Genetic Investigation of ANthropometric Traits (GIANT) Consortium; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium , et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet 2012;44:369–375, S361–363. 10.1038/ng.2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011;88:76–82. 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 2015;31:3555–3557. 10.1093/bioinformatics/btv402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014;46:310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lu X, Beck GR Jr, Gilbert LC, Camalier CE, Bateman NW, Hood BL, et al. Identification of the homeobox protein Prx1 (MHox, prrx-1) as a regulator of osterix expression and mediator of tumor necrosis factor alpha action in osteoblast differentiation. J Bone Miner Res 2011;26:209–219. 10.1002/jbmr.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ocana OH, Corcoles R, Fabra A, Moreno-Bueno G, Acloque H, Vega S, et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell 2012;22:709–724. 10.1016/j.ccr.2012.10.012 [DOI] [PubMed] [Google Scholar]
- 43. Vandewalle C, Comijn J, De Craene B, Vermassen P, Bruyneel E, Andersen H, et al. SIP1/ZEB2 Induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res 2005;33:6566–6578. 10.1093/nar/gki965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature 2010;466:714–719. 10.1038/nature09266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Strong A, Ding Q, Edmondson AC, Millar JS, Sachs KV, Li X, et al. Hepatic sortilin regulates both apolipoprotein B secretion and LDL catabolism. J Clin Invest 2012;122:2807–2816. 10.1172/JCI63563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hoffmann TJ, Theusch E, Haldar T, Ranatunga DK, Jorgenson E, Medina MW, et al. A large electronic-health-record-based genome-wide study of serum lipids. Nat Genet 2018;50:401–413. 10.1038/s41588-018-0064-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet 2009;41:56–65. 10.1038/ng.291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. van der Harst P, Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res 2018;122:433–443. 10.1161/CIRCRESAHA.117.312086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, et al. A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 2015;47:1121–1130. 10.1038/ng.3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Goettsch C, Hutcheson JD, Aikawa M, Iwata H, Pham T, Nykjaer A, et al. Sortilin mediates vascular calcification via its recruitment into extracellular vesicles. J Clin Invest 2016;126:1323–1336. 10.1172/JCI80851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Larsson SC, Wolk A, Hakansson N, Back M. Overall and abdominal obesity and incident aortic valve stenosis: two prospective cohort studies. Eur Heart J 2017;38:2192–2197. 10.1093/eurheartj/ehx140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Klarin D, Zhu QM, Emdin CA, Chaffin M, Horner S, McMillan BJ, et al. Genetic analysis in UK Biobank links insulin resistance and transendothelial migration pathways to coronary artery disease. Nat Genet 2017;49:1392–1397. 10.1038/ng.3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 2011;43:333–338. 10.1038/ng.784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cote N, Mahmut A, Bosse Y, Couture C, Page S, Trahan S, et al. Inflammation is associated with the remodeling of calcific aortic valve disease. Inflammation 2013;36:573–581. 10.1007/s10753-012-9579-6 [DOI] [PubMed] [Google Scholar]
- 55. Schmitz G, Ecker J. The opposing effects of n-3 and n-6 fatty acids. Prog Lipid Res 2008;47:147–155. 10.1016/j.plipres.2007.12.004 [DOI] [PubMed] [Google Scholar]
- 56. Zheng D, Kern L, Elinav E. The NLRP6 inflammasome. Immunology 2021;162:281–289. 10.1111/imm.13293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lee HJ, Yeon JE, Ko EJ, Yoon EL, Suh SJ, Kang K, et al. Peroxisome proliferator-activated receptor-delta agonist ameliorated inflammasome activation in nonalcoholic fatty liver disease. World J Gastroenterol 2015;21:12787–12799. 10.3748/wjg.v21.i45.12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Koren MJ, Moriarty PM, Baum SJ, Neutel J, Hernandez-Illas M, Weintraub HS, et al. Preclinical development and phase 1 trial of a novel siRNA targeting lipoprotein(a). Nat Med 2022;28:96–103. 10.1038/s41591-021-01634-w [DOI] [PubMed] [Google Scholar]
- 59. Tsimikas S, Karwatowska-Prokopczuk E, Gouni-Berthold I, Tardif JC, Baum SJ, Steinhagen-Thiessen E, et al. Lipoprotein(a) reduction in persons with cardiovascular disease. N Engl J Med 2020;382:244–255. 10.1056/NEJMoa1905239 [DOI] [PubMed] [Google Scholar]
- 60. Malik IO, Petersen MC, Klein S. Glucagon-like peptide-1, glucose-dependent insulinotropic polypeptide, and glucagon receptor poly-agonists: a new era in obesity pharmacotherapy. Obesity (Silver Spring) 2022;30:1718–1721. 10.1002/oby.23521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hafiane A, Daskalopoulou SS. Targeting the residual cardiovascular risk by specific anti-inflammatory interventions as a therapeutic strategy in atherosclerosis. Pharmacol Res 2022;178:106157. 10.1016/j.phrs.2022.106157 [DOI] [PubMed] [Google Scholar]
- 62. Han Y, Dorajoo R, Chang X, Wang L, Khor CC, Sim X, et al. Genome-wide association study identifies a missense variant at APOA5 for coronary artery disease in multi-ethnic cohorts from Southeast Asia. Sci Rep 2017;7:17921. 10.1038/s41598-017-18214-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Adeyemo A, Bentley AR, Meilleur KG, Doumatey AP, Chen G, Zhou J, et al. Transferability and fine mapping of genome-wide associated loci for lipids in African Americans. BMC Med Genet 2012;13:88. 10.1186/1471-2350-13-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liu CT, Raghavan S, Maruthur N, Kabagambe EK, Hong J, Ng MC, et al. Trans-ethnic meta-analysis and functional annotation illuminates the genetic architecture of fasting glucose and insulin. Am J Hum Genet 2016;99:56–75. 10.1016/j.ajhg.2016.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The summary level statistics from the meta-GWAS that support the findings of this study are available online @ Zenodo.org (https://doi.org/10.5281/zenodo.7505361).