Abstract
Challenging enantio- and diastereoselective cobalt-catalyzed C–H alkylation has been realized by an innovative data-driven knowledge transfer strategy. Harnessing the statistics of a related transformation as the knowledge source, the designed machine learning (ML) model took advantage of delta learning and enabled accurate and extrapolative enantioselectivity predictions. Powered by the knowledge transfer model, the virtual screening of a broad scope of 360 chiral carboxylic acids led to the discovery of a new catalyst featuring an intriguing furyl moiety. Further experiments verified that the predicted chiral carboxylic acid can achieve excellent stereochemical control for the target C–H alkylation, which supported the expedient synthesis for a large library of substituted indoles with C-central and C–N axial chirality. The reported machine learning approach provides a powerful data engine to accelerate the discovery of molecular catalysis by harnessing the hidden value of the available structure-performance statistics.
Subject terms: Asymmetric catalysis, Synthetic chemistry methodology
The design of efficient and selective catalysts is a formidable challenge in chemical science. Here the authors design a data-driven workflow to achieve the digitalized knowledge transfer between the synthetically relevant transformations, which was demonstrated in the prediction of chiral carboxylic acid co-catalyst for the cobalt-catalyzed asymmetric C–H alkylation of indoles.
Introduction
The design of efficient and selective catalysts is a formidable challenge in chemical science. Because of the magnificent molecular universe and the transformation-dependent catalysis property, the complexity of the structure-performance relationship (SPR) in molecular catalysis is beyond imagination. As a revolutionary change to the classic experience-driven strategy of catalyst development, machine learning (ML) has recently emerged as a powerful approach for exploring the high-dimensional SPR1,2. A series of breakthroughs have realized the accurate and efficient ML predictions of new catalysts and transformations3–7, Fig. 1a highlights the general workflow of the current data-driven exploration of chemical space. Relying on the statistics of the target catalysis, ML is able to create an SPR model, which drives the subsequent data acquisition. This data acquisition is essentially an optimization problem, and greedy search8 (Top-k method) or Bayesian optimization9 are the representative engines for providing the candidate reaction designs. Experimental evaluations of these ML designs offer new data sources to improve the ML model, which completes a feedback loop until the target synthetic performance is achieved. This process, in principle, does not involve human intervention and can be accelerated by automatic synthesis. Landmark studies by Cronin8, Cooper10, Doyle9, Jensen11, Denmark3 and others12,13 have highlighted that this data-driven workflow can discover powerful catalysis conditions starting from zero knowledge of the target transformation.
Fig. 1. Data-driven discovery of molecular catalysis.

a General workflow of current machine learning-assisted reaction optimization. b Cp*Co(III)/CCA-catalyzed asymmetric C−H functionalization of indoles. c Designed knowledge transfer model for predicting new CCAs of asymmetric C−H functionalization of indoles.
Despite the remarkable success of ML-assisted reaction optimizations, it should be noted that this logic of optimization starting from zero knowledge or data is fundamentally different as compared to the way that human chemists are typically practicing. It is extremely rare to design a catalyst that has absolutely no related knowledge available. In the typical scenario, the chemist’s catalyst design is based on the careful evaluation of related SPR data and the judicious chemical innovation of a given compound14–16. This is essentially a knowledge transfer process where the explored chemical space facilitated the rational expansion of the known SPR to new catalyst. In recent years, the concept of knowledge transfer has also been applied to the data-driven modeling in synthetic chemistry, which has shown great potential in addressing the problem of limited sample size. By leveraging innovative modeling strategies, knowledge transfer modeling can connect chemically related data and reduce the data demand for target domain. For example, through the unsupervised ML that increases the model’s differentiation ability of phosphine ligands, Schoenebeck and co-workers17 were able to achieve the successful prediction of dinuclear palladium catalyst with only five labeled data. We recently developed a hierarchical learning approach which can select appropriate datasets for layered modeling based on the proximity in chemical space, thereby improving the predictive performance of the ML model18,19. These knowledge transfer models not only improve the efficiency of catalyst design but also help expand the known chemical space in a data-driven fashion. Therefore, the integration of knowledge transfer modeling into data-driven synthetic discovery is of great significance for advancing the field of catalysis and beyond.
Over the last years, cobalt-catalyzed asymmetric C–H functionalization has garnered significant attention20,21. The groups of Yoshikai22–24, Dong25, Lautens26, Yang27, Wencel-Delord28, and Shi29 have successively combined low-valent cobalt catalysts with chiral ligands to achieve stereoselective C–H functionalization, while Cramer and co-workers developed chiral CpxCo(III) complexes for this purpose30–32. In addition, achiral Cp*Co(III)/chiral carboxylic acid (CCA) systems33–41 have also been widely deployed to catalyze asymmetric C–H alkylation of indoles39–41 (Fig. 1b). However, the use of synthetically demanding chiral acids requires laborious multi-step synthesis, limiting the potential of these transformations33–40. In 2018, Ackermann and coworkers achieved the enantioselective cobalt-catalyzed C–H alkylation by a designed C2-symmetric CCA that can be easily synthesized41. This CCA-based chiral catalysis serves as a powerful platform41,42, and the engineering of the CCA structure is of great potential for the enrichment of asymmetric derivatization of indoles.
Axial chirality is of major importance for modern pharmaceutical industry43,44, and synthetically challenging C–N axially chiral indoles are privileged motifs in drug design, crop protection, and material science45,46. Thus, the efficient synthesis of these compounds has become a rapidly expanding field47,48. However previous studies mainly relied on the use of noble 4d and 5d transition metal catalysts49–52, while sustainable 3d-metal-catalyzed transformation53,54 remains underdeveloped. Therefore, the development of an efficient CCA co-catalyst for cobalt-catalyzed C–H activation to enable the assembly of atropisomeric compounds bearing C–N axial chirality, and simultaneously construct C-centered chirality with high stereoselectivity is a tremendously important and unrealized challenge.
In light of the critical knowledge transfer for catalyst development, we envisioned that the digitalization of the knowledge transfer process can serve as an innovative data-driven strategy for catalyst design. This requires the ML model to capture the key differences between the given transformation and the target reaction, so that the available statistics of the given transformation can serve as a knowledge source and guide the design of the target reaction.
Herein we report the development of a data-driven transfer learning workflow to achieve the ML prediction of catalytic performance using related synthetic data (Fig. 1c). Demonstrated in the discovery of new CCA catalyst, our ML model provided a powerful CCA prediction that realized the challenging enantio- and diastereoselective C–H alkylation of indoles utilizing earth-abundant cobalt catalyst. The ML-predicted CCA catalyst enabled the target transformation that can simultaneously control both the C-centered and the C–N axial chirality, providing the atropisomeric indoles with excellent diastereo- and enantioselectivities (Fig. 1c). This work offered a paradigm-shifting tool for the discovery of molecular catalyst, which is expected to serve as a powerful data engine to support the innovation of catalysis science.
Results and discussions
Design of knowledge transfer model
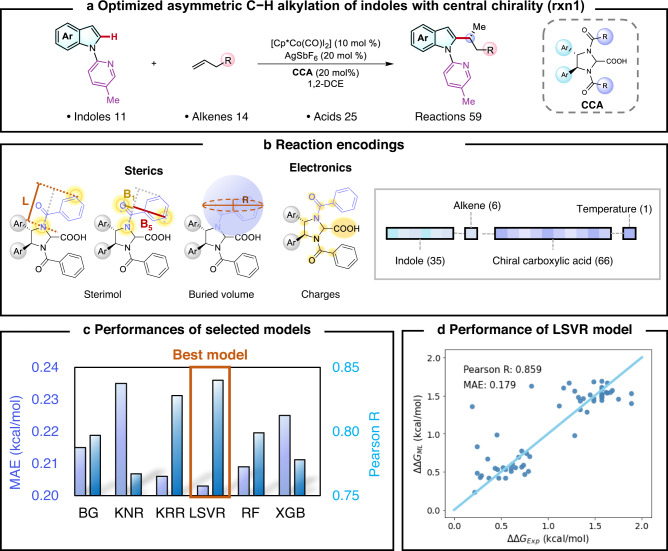
To achieve the desired knowledge transfer, the first step is to create a reliable SPR model using the available statistics of the optimized transformation (Fig. 2). The already optimized Cp*Co(III)/CCA-catalyzed asymmetric C−H alkylation of indoles (rxn1) does not involve the control of the axial chirality, which was previously discovered by the Ackermann group; 59 SPR data of rxn1 were accumulated during the catalysis screening, involving the variations of 11 indoles, 14 alkenes and 25 CCAs41 (Fig. 2a). The detailed data distribution is provided in the Supplementary Information (Supplementary Fig. 2). Inspired by recent data-driven selectivity prediction studies using physical organic descriptors55,56, we applied a series of steric (i.e. Sterimol parameters) and electronic (i.e. charge) features to describe the influence of the N-substituent of CCA; the entire catalysis encoding is a 108-dimensional physical organic space containing 35 descriptors for indoles, 6 descriptors for alkenes, 66 descriptors for CCAs and 1 descriptor for temperature (Fig. 2b). Based on the regression performances in the 10-fold cross-validation, linear support vector regression57 emerged as the most suitable algorithm with a Pearson R of 0.859 and MAE of 0.179 kcal/mol; the detailed regression results are shown in Fig. 2c, in which a nice correlation between the ML-predicted and the experimental enantioselectivities was identified. The detailed results of all tested ML models are provided in the Supplementary Information (Supplementary Table 4).
Fig. 2. Machine learning enantioselectivity prediction for the Cp*Co(III)/CCA-catalyzed C–H alkylation of indoles with central chirality.
a Overview of accumulated data of Cp*Co(III)/CCA-catalyzed asymmetric C−H alkylation of indoles with central chirality. b Reaction encodings and highlighted descriptors of steric and electronic effect using CCA as an example. c Regression performances of various algorithms in 10-fold cross validation. d Performance of enantioselectivity predictions using LSVR model. Source data are presented in the Source_Data.
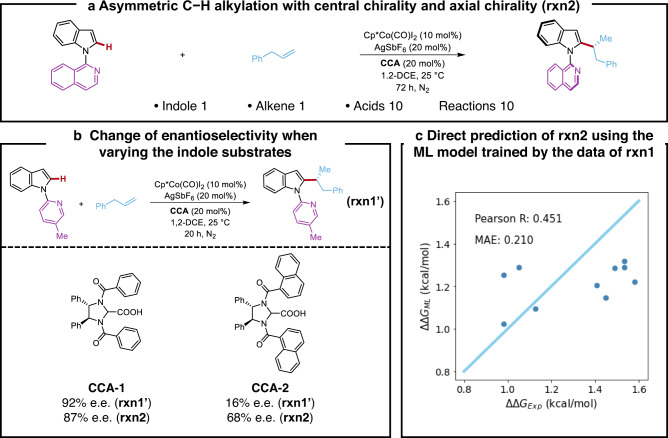
With the ML model of rxn1 in hand, we tested its direct application in the target C−H alkylation with axial chirality (rxn2). Among the tested CCAs for rxn1, ten representative ones were experimentally evaluated for rxn2 with the axial chirality challenge (Fig. 3a). The selection of representative CCAs were based on the diversity of their chemical structures and enantioselectivities. Due to the introduction of isoquinoline moiety in the indole substrate, the two transformations do not follow the exact same SPR. Figure 3b showed two highlighted examples: the optimized CCA-1 for rxn1' achieved a 92% e.e. for this transformation, while its application in the atroposelective rxn2 delivered a 87% e.e., which was one of the major motivations for the data-driven design of new CCAs; in addition, the naphthyl CCA-2 only achieved a 16% e.e. in rxn1', but the corresponding rxn2 has the enantioselectivity of 68% e.e. This non-intuitive perturbation of SPR widely exists in molecular catalysis, which results in the unsatisfying prediction performance of the trained ML model in rxn2; the Pearson R is only 0.451, which is in sharp contrast to its performance in rxn1 (Fig. 3c vs. Fig. 2d).
Fig. 3. Enantioselectivity prediction of the Cp*Co(III)/CCA-catalyzed C–H alkylation of indoles with central and axial chirality using machine learning modelling without knowledge transfer.
a Overview of the target C–H alkylation of indoles with central and axial chirality. b Enantioselectivity change of the Cp*Co(III)/CCA-catalyzed C–H alkylation when varying the indole substrates. c Enantioselectivity predictions of the target C–H alkylation of indoles with central and axial chirality using the machine learning model trained by the statistics of the C–H alkylation without axial chirality. Source data are presented in the Source_Data.
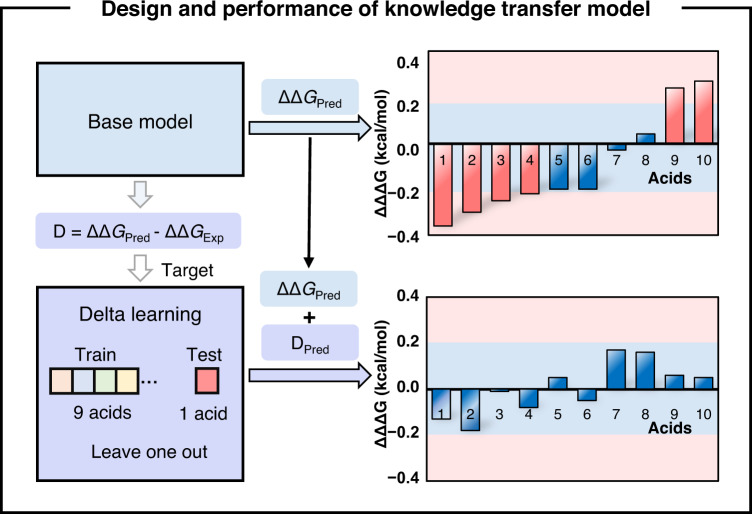
We next trained a delta ML model to capture the SPR perturbation, in order to correct the enantioselectivity predictions of the rxn1 model. For the ten evaluated CCAs in rxn2, each CCA has the experimentally measured enantioselectivity (ΔΔGexp) as well as the predicted value (ΔΔGpred) from the rxn1 model (Fig. 4). The differences between the two values (D = ΔΔGpred − ΔΔGexp) provided a limited but valuable data source for the delta learning. Using the same physical organic encodings, the leave-one-out (LOO) training provided the delta learning model, which significantly improved the predictions of the rxn1 model (Fig. 4); the MAE decreases from 0.210 kcal/mol to 0.095 kcal/mol, and the outliner predictions (highlighted in red) were all eliminated. Therefore, the final prediction of rxn2 is the sum of the rxn1 model’s prediction and the delta model’s prediction. This ML approach represents the digitalized knowledge transfer. The training of rxn1 model harnessed the SPR from the available data of related catalysis screening, and subsequent delta learning corrected the understanding of rxn1 using the limited data from the experimental reoptimization of rxn2, which mimics the logic of a human chemist.
Fig. 4. Design and performance of knowledge transfer model for making accurate predictions of the target C–H alkylation of indoles with central and axial chirality.
Source data are presented in the Source_Data.
Using the established knowledge transfer model, we performed the virtual screening of CCAs to identify the highly selective catalyst for the atroposelective rxn2. Considering the synthetic access of the derivatized CCAs, 4 representative aryl substituents with different steric hindrance and electronic effects were evaluated for the CCA backbone, and a selection of 90 variations was explored for the N-substitution (including aromatic and heteroaromatic rings with different electronic effects and sterically hindered substituents, as well as alkyl substituents), which allowed the thorough evaluation of the CCA candidates (Fig. 5a). A few highlighted examples of the 90 substituents are provided in Fig. 5. The combination of considered substitutions together created 360 candidate C2-symmetric CCAs including the 10 CCAs that have been used in the knowledge transfer modeling, and their predicted enantioselectivities for rxn2 are summarized in Fig. 5b. 13 out of the 360 have a predicted selectivity below 40%; 280 were predicted to have an enantioselectivity between 40 and 80%; 67 have the predicted enantioselectivity >80%. Figure 5c shows the chemical structures of the predicted Top-3 CCAs. It is interesting that the furan moiety was identified as a privileged choice of the N-substitution. Both the 2-furyl and the 3-furyl substituted CCA-3 and CCA-4 were predicted to have an 89% enantioselectivity, which ranked the first and the second of the 360 predictions. The third CCA has the para-OMe-phenyl substituent, whose predicted enantioselectivity was 88%. It is worth noticing that these three substitutions are all electron-rich aryl moieties with limited steric repulsions, which indicated that the chirality control may involve non-covalent interactions with the N-substitution. Subsequently, the predicted Top-3 CCAs were synthesized and evaluated for rxn2. Excellent enantioselectivities were found for all three cases, with the 3-furyl substituted CCA-4 as the optimal catalyst. This CCA achieved a 94% enantioselectivity for rxn2, which highlighted the predictive power of the data-driven knowledge transfer approach. We want to emphasize that the naïve training with all the enantioselective C–H alkylation data (59 data of rxn1 and 10 data of rxn2) without the usage of delta learning led to a significantly reparametrized model. The virtual screening using this reparametrized naïve model provided a reshuffled ranking, and the 3-furyl substituted CCA-4 was predicted to have an 81% enantioselectivity with a ranking of 98, which is in sharp contrast to the outcome of the knowledge transfer model.
Fig. 5. Virtual screening of highly selective CCAs using the knowledge transfer model and experimental verifications.
a Designed structures of the 360 candidate CCAs. b Distribution of the predicted enantioselectivities. c Predicted and experimental enantioselectivities of the Top-3 CCAs. d Predicted and experimental enantioselectivities of CCAs with medium performances. Source data are presented in the Source_Data.
In order to further validate the accuracy of the model’s prediction for the entire value range and its discriminative ability for CCA’s catalytic performance, we selected a series of CCAs with medium to low predicted performances and conducted experimental synthesis and verification. Figure 5d shows the prediction and experimental results of the four tested CCAs (CCA-6 to CCA-9), with a maximum error of only 14% e.e. These results further demonstrated the predictive ability of the developed knowledge transfer model, indicating that it can effectively discriminate the enantioselectivities of the candidate CCAs and uncover the useful catalysts with superior performance. To ensure the reliability of the training and prediction of the knowledge transfer model, we also evaluated the model predictions with five additional delta data. Using a total of 15 delta data to retrain the knowledge transfer model, we compared the prediction results of the seven experimentally verified CCAs (CCA-3 to CCA-9) with those obtained by training with the 10 delta data. The two sets of prediction values were highly correlated (Pearson R = 0.961, Supplementary Fig. 8), which indicated that the additional five data had a relatively small impact on the modeling. To confirm that the success of knowledge transfer model is not accidental in substrate 1a, we also performed the same knowledge transfer learning process on substrate 1k; the delta learning achieved similarly effectiveness in correcting the base model’s predictions (Supplementary Fig. 10). These comparisons further validated the knowledge transfer approach, highlighting the effectiveness of the hierarchical usage of synthetic data based on chemical heuristics.
Substrate scope for cobalt-catalyzed asymmetric C–H alkylation
After locating the optimal CCA by the data-driven knowledge transfer, the substrate scope was explored under the optimized reaction conditions to delineate the potential of this transformation (Fig. 6). A variety of indole substrates were investigated (Fig. 6a). Both electron-withdrawing and electron-donating groups at the 4-, 5- or 6-position of the indole ring were tolerated to afford the desired products 3a–3j in good yields with excellent diastereo- and enantioselectivities (94:6– > 95:5 d.r., 92–95% e.e.). The atropostability of the products is conserved even for the less hindered methyl-substituted product 3k, although with a slight decrease in stereoselectivity. A broad range of alkenes bearing different substituents on para-, meta- or ortho-position of the arene were well tolerated and gave the desired products 3l–3t in high yields and high levels of stereocontrol (all >95:5 d.r., 87–93% e.e.) (Fig. 6b). Additionally, 2-allylnaphthalene, 1-allylnaphthalene and allylpentafluorobenzene efficiently underwent the cobalt-catalysis providing the target products 3u–3w with good stereoselectivities (all >95:5 d.r., up to 91% e.e.). The absolute configuration of the alkylation products was unambiguously confirmed by single-crystal X-ray diffraction analysis of 3c and 3w.
Fig. 6. Substrate scope for asymmetric C-H alkylation.
a Scope of indoles. b Scope of alkenes. Reaction conditions: 1 (0.1 mmol), 2 (0.3 mmol), Cp*Co(CO)I2 (10 mol%), AgSbF6 (20 mol%), and CCA-4 (20 mol%) in 1,2-DCE (0.5 mL) at 25 °C for 72 h under N2. Unless noted, the ratio of b:l is >95:5. Yields are those of the isolated products. The diastereomeric ratio (d.r.) were determined by 1H NMR spectroscopy. The enantiomeric excess (e.e.) was determined by HPLC.
In conclusion, we have designed a data-driven workflow to achieve the digitalized knowledge transfer between the synthetically relevant transformations, which was demonstrated in the prediction of chiral carboxylic acid co-catalyst for the asymmetric C–H alkylation of indoles with atropselectivity challenge utilizing non-precious cobalt catalyst. Using the available catalysis screening data of a related asymmetric cobalt-catalyzed C–H alkylation, the physical organic descriptors and linear support vector regression algorithm provided a predictive machine learning model. This model serves as the knowledge base, whose predictions were further corrected using the delta learning method. The delta learning method only requires a handful of selectivity data of the target atroposelective transformation, which captures the perturbation of the structure-performance relationship between the two synthetically relevant transformations and enabled the desired data-driven knowledge transfer.
The designed data-driven knowledge transfer model enabled a powerful virtual screening of 360 candidate chiral carboxylic acids for the target atroposelective C–H alkylation of indoles. The top-3 predicted acids were synthesized and experimentally evaluated. The three predicted chiral carboxylic acids featured good to excellent experimental enantioselectivities, with the 3-furyl substituted one presenting the highest selectivity. These successful predictions and the identification of the suitable N-substituent provided strong support for the effectiveness of the designed knowledge transfer approach. The robustness of the enantio- and diastereoselective cobalt-catalyzed C–H alkylation promoted by the predicted chiral carboxylic acid was further explored, leading to the assembly of a large family of substituted indoles in good yields and with excellent stereoselectivities. This work provides a new data-driven strategy for knowledge transfer of synthetic chemistry. The established machine learning model was able to capture the non-intuitive perturbation of structure-performance relationship and make useful predictions in the few-shot learning scenario of synthetic optimization, which provides a powerful smart engine to accelerate the discovery of molecular catalysis.
Methods
General procedure for cobalt-catalyzed asymmetric C–H alkylation
To a flame-dried and N2-purged Schlenk tube were added indole substrate 1 (0.1 mmol), Cp*Co(CO)I2 (0.01 mmol, 10 mol%, 4.8 mg), AgSbF6 (0.02 mmol, 20 mol%, 6.9 mg), and chiral carboxylic acid CCA-4 (0.02 mmol, 20 mol%, 9.1 mg). The vial was then sealed, purged and backfilled with N2 three times before adding alkene substrate 2 (0.3 mmol) and 1,2-dichloroethane (0.5 mL) at room temperature. The resulting solution was then stirred at 25 °C for 72 h. The resulting solution was diluted with dichloromethane (2.0 mL), filtered through a pad of Celite (eluted with dichloromethane), then the solvent was removed in vacuo. The diastereomeric ratio was determined by 1H NMR analysis of the crude reaction mixture. The residue was purified by column chromatography on silica gel (n-hexane: ethyl acetate = 15:1) to afford the desired product 3.
Supplementary information
Acknowledgements
The authors gratefully acknowledge the support from the ERC Advanced Grant (no. 101021358) and the DFG (SPP2363), the Alexander-von-Humboldt Foundation (fellowship to Z.-J.Z.), the National Key R&D Program of China (2022YFA1504301, X.H.), the National Natural Science Foundation of China (22122109 and 22271253, X.H.; 22103070, S.-Q.Z.), Zhejiang Provincial Natural Science Foundation of China under Grant No. LDQ23B020002 (X.H.), the Starry Night Science Fund of Zhejiang University Shanghai Institute for Advanced Study (SN-ZJU-SIAS-006, X.H.), Beijing National Laboratory for Molecular Sciences (BNLMS202102, X.H.), the Center of Chemistry for Frontier Technologies and Key Laboratory of Precise Synthesis of Functional Molecules of Zhejiang Province (PSFM 2021-01, X.H.), the State Key Laboratory of Clean Energy Utilization (ZJUCEU2020007, X.H.), Fundamental Research Funds for the Central Universities (226-2022-00140, 226-2022-00224 and 226-2023-00115, X.H.) and CAS Youth Interdisciplinary Team (JCTD-2021-11, X.H.). Calculations were performed on the high-performance computing system at Department of Chemistry, Zhejiang University. The authors thank Dr. Christopher Golz (University of Göttingen) for assistance with the X-ray diffraction analysis.
Source data
Author contributions
L.A. and X.H. conceived the project. Z.-J.Z. and Y.L. performed and analyzed the experimental studies. T.R. assisted in the synthesis of chiral carboxylic acids. S.-W.L. performed the machine learning modelings and analyzed the results. J.C.A.O., X.C., S.-Q.Z. and L.-C.X. assisted in data processing and machine learning modeling. All authors were involved in the discussions and manuscript writing.
Peer review
Peer review information
Nature Communications thanks Jason Stevens, Naohiko Yoshikai, and the other, anonymous, reviewer for their contribution to the peer review of this work.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The data that support the findings of this study are available within the main text, the Supplementary Information and https://github.com/Shuwen-Li/FindBestChiralAcid58. Source data are presented in the Source_Data. Details about materials and methods, experimental procedures, characterization data, NMR and HPLC spectra are available in the Supplementary Information, and all other data are available from the corresponding author upon request. Crystallographic data are available free of charge under Cambridge Crystallographic Data Centre (CCDC) reference numbers 2176897 (3c), 2176898 (3w) [www.ccdc.cam.ac.uk/data_request/cif]. Source data are provided with this paper.
Code availability
All codes needed to run this model are available at https://github.com/Shuwen-Li/FindBestChiralAcid58.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zi-Jing Zhang and Shu-Wen Li.
Contributor Information
Xin Hong, Email: hxchem@zju.edu.cn.
Lutz Ackermann, Email: Lutz.Ackermann@chemie.unigoettingen.de.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-023-38872-0.
References
- 1.Rinehart NI, Zahrt AF, Henle JJ, Denmark SE. Dreams, false starts, dead ends, and redemption: A chronicle of the evolution of a chemoinformatic workflow for the optimization of enantioselective catalysts. Acc. Chem. Res. 2021;54:2041–2054. doi: 10.1021/acs.accounts.0c00826. [DOI] [PubMed] [Google Scholar]
- 2.Gromski PS, Henson AB, Granda JM, Cronin L. How to explore chemical space using algorithms and automation. Nat. Rev. Chem. 2019;3:119–128. doi: 10.1038/s41570-018-0066-y. [DOI] [Google Scholar]
- 3.Zahrt AF, et al. Prediction of higher-selectivity catalysts by computer-driven workflow and machine learning. Science. 2019;363:eaau5631. doi: 10.1126/science.aau5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu K, Doyle AG. Parameterization of phosphine ligands demonstrates enhancement of nickel catalysis via remote steric effects. Nat. Chem. 2017;9:779–784. doi: 10.1038/nchem.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen MK, Ahneman DT, Riera O, Doyle AG. Deoxyfluorination with sulfonyl fluorides: Navigating reaction space with machine learning. J. Am. Chem. Soc. 2018;140:5004–5008. doi: 10.1021/jacs.8b01523. [DOI] [PubMed] [Google Scholar]
- 6.Henle JJ, et al. Development of a computer-guided workflow for catalyst optimization. Descriptor validation, subset selection, and training set analysis. J. Am. Chem. Soc. 2020;142:11578–11592. doi: 10.1021/jacs.0c04715. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, et al. Electro-descriptors for the performance prediction of electro-organic synthesis. Angew. Chem. Int. Ed. 2021;60:4199–4207. doi: 10.1002/anie.202014072. [DOI] [PubMed] [Google Scholar]
- 8.Granda JM, Donina L, Dragone V, Long DL, Cronin L. Controlling an organic synthesis robot with machine learning to search for new reactivity. Nature. 2018;559:377–381. doi: 10.1038/s41586-018-0307-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shields BJ, et al. Bayesian reaction optimization as a tool for chemical synthesis. Nature. 2021;590:89–96. doi: 10.1038/s41586-021-03213-y. [DOI] [PubMed] [Google Scholar]
- 10.Burger B, et al. A mobile robotic chemist. Nature. 2020;583:237–241. doi: 10.1038/s41586-020-2442-2. [DOI] [PubMed] [Google Scholar]
- 11.Reizman BJ, Jensen KF. Feedback in flow for accelerated reaction development. Acc. Chem. Res. 2016;49:1786–1796. doi: 10.1021/acs.accounts.6b00261. [DOI] [PubMed] [Google Scholar]
- 12.Meuwly M. Machine learning for chemical reactions. Chem. Rev. 2021;121:10218–10239. doi: 10.1021/acs.chemrev.1c00033. [DOI] [PubMed] [Google Scholar]
- 13.Zhu, Q. et al. An all-round AI-Chemist with scientific mind. Natl. Sci. Rev. 10.1093/nsr/nwac190 (2022). [DOI] [PMC free article] [PubMed]
- 14.Poree C, Schoenebeck F. A holy grail in chemistry: computational catalyst design: feasible or fiction? Acc. Chem. Res. 2017;50:605–608. doi: 10.1021/acs.accounts.6b00606. [DOI] [PubMed] [Google Scholar]
- 15.Houk KN, Cheong PH. Computational prediction of small-molecule catalysts. Nature. 2008;455:309–313. doi: 10.1038/nature07368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahn S, Hong M, Sundararajan M, Ess DH, Baik MH. Design and optimization of catalysts based on mechanistic insights derived from quantum chemical reaction modeling. Chem. Rev. 2019;119:6509–6560. doi: 10.1021/acs.chemrev.9b00073. [DOI] [PubMed] [Google Scholar]
- 17.Hueffel JA, et al. Accelerated dinuclear palladium catalyst identification through unsupervised machine learning. Science. 2021;374:1134–1140. doi: 10.1126/science.abj0999. [DOI] [PubMed] [Google Scholar]
- 18.Xu LC, et al. Towards data-driven design of asymmetric hydrogenation of olefins: database and hierarchical learning. Angew. Chem. Int. Ed. 2021;60:22804–22811. doi: 10.1002/anie.202106880. [DOI] [PubMed] [Google Scholar]
- 19.Xu, L.-C., et al. Enantioselectivity Prediction of Pallada-Electrocatalysed C–H Activation Using Transition State Knowledge in Machine Learning. 10.1038/s44160-022-00233-y (2023).
- 20.Pellissier H, Clavier H. Enantioselective cobalt-catalyzed transformations. Chem. Rev. 2014;114:2775–2823. doi: 10.1021/cr4004055. [DOI] [PubMed] [Google Scholar]
- 21.Gao K, Yoshikai N. Low-valent cobalt catalysis: new opportunities for C–H functionalization. Acc. Chem. Res. 2014;47:1208–1219. doi: 10.1021/ar400270x. [DOI] [PubMed] [Google Scholar]
- 22.Yang J, Yoshikai N. Cobalt-catalyzed enantioselective intramolecular hydroacylation of ketones and olefins. J. Am. Chem. Soc. 2014;136:16748–16751. doi: 10.1021/ja509919x. [DOI] [PubMed] [Google Scholar]
- 23.Lee P-S, Yoshikai N. Cobalt-catalyzed enantioselective directed C−H alkylation of indole with styrenes. Org. Lett. 2015;17:22–25. doi: 10.1021/ol503119z. [DOI] [PubMed] [Google Scholar]
- 24.Yang J, Rérat A, Lim YJ, Gosmini C, Yoshikai N. Cobalt-catalyzed enantio- and diastereoselective intramolecular hydroacylation of trisubstituted alkenes. Angew. Chem. Int. Ed. 2017;56:2449–2453. doi: 10.1002/anie.201611518. [DOI] [PubMed] [Google Scholar]
- 25.Kim DK, Riedel J, Kim RS, Dong VM. Cobalt catalysis for enantioselective cyclobutanone construction. J. Am. Chem. Soc. 2017;139:10208–10211. doi: 10.1021/jacs.7b05327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whyte A, et al. Cobalt-catalyzed enantioselective hydroarylation of 1,6-enynes. J. Am. Chem. Soc. 2020;142:9510–9517. doi: 10.1021/jacs.0c03246. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Wang J, Yang S-D. Enantioselective cobalt-catalyzed reductive cross-coupling for the synthesis of axially chiral phosphine-olefin ligands. ACS Catal. 2021;11:14008–14015. doi: 10.1021/acscatal.1c04128. [DOI] [Google Scholar]
- 28.Jacob N, Zaid Y, Oliveira JCA, Ackermann L, Wencel-Delord J. Cobalt-catalyzed enantioselective C–H arylation of indoles. J. Am. Chem. Soc. 2022;144:798–806. doi: 10.1021/jacs.1c09889. [DOI] [PubMed] [Google Scholar]
- 29.Yao Q-J, Chen J-H, Song H, Huang F-R, Shi B-F. Cobalt/salox-catalyzed enantioselective C–H functionalization of arylphosphinamides. Angew. Chem. Int. Ed. 2022;61:e202202892. doi: 10.1002/anie.202202892. [DOI] [PubMed] [Google Scholar]
- 30.Ozols K, Jang Y-S, Cramer N. Chiral cyclopentadienyl cobalt(III) complexes enable highly enantioselective 3d-metal-catalyzed C−H functionalizations. J. Am. Chem. Soc. 2019;141:5675–5680. doi: 10.1021/jacs.9b02569. [DOI] [PubMed] [Google Scholar]
- 31.Ozols, K., Onodera, S., Woźniak, Ł. & Cramer, N. Cobalt(III)-catalyzed enantioselective intermolecular carboamination by C−H functionalization. Angew. Chem. Int. Ed. 60, 655-659 (2021). [DOI] [PubMed]
- 32.Herraiz AG, Cramer N. Cobalt(III)-catalyzed diastereo- and enantioselective three-component C−H functionalization. ACS Catal. 2021;11:11938–11944. doi: 10.1021/acscatal.1c03153. [DOI] [Google Scholar]
- 33.Zell D, Bursch M, Mgller V, Grimme S, Ackermann L. Full selectivity control in cobalt(III)-catalyzed C−H alkylations by switching of the C−H activation mechanism. Angew. Chem. Int. Ed. 2017;56:10378–10382. doi: 10.1002/anie.201704196. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y-H, et al. Cp*Co(III)/MPAA-catalyzed enantioselective amidation of ferrocenes directed by thioamides under mild conditions. Org. Lett. 2019;21:1895–1899. doi: 10.1021/acs.orglett.9b00511. [DOI] [PubMed] [Google Scholar]
- 35.Fukagawa S, et al. Enantioselective C(sp3)−H amidation of thioamides catalyzed by a cobaltIII/chiral carboxylic acid hybrid system. Angew. Chem. Int. Ed. 2019;58:1153–1157. doi: 10.1002/anie.201812215. [DOI] [PubMed] [Google Scholar]
- 36.Sekine D, et al. Chiral 2-aryl ferrocene carboxylic acids for the catalytic asymmetric C(sp3)−H activation of thioamides. Organometallics. 2019;38:3921–3926. doi: 10.1021/acs.organomet.9b00407. [DOI] [Google Scholar]
- 37.Yuan W-K, Shi B-F. Synthesis of chiral spirolactams via sequential C−H olefination/asymmetric [4+1] spirocyclization under a simple CoII/chiral spiro phosphoric acid binary system. Angew. Chem. Int. Ed. 2021;60:23187–23192. doi: 10.1002/anie.202108853. [DOI] [PubMed] [Google Scholar]
- 38.Hirata Y, et al. Cobalt(III)/chiral carboxylic acid-catalyzed enantioselective synthesis of benzothiadiazine-1-oxides via C–H activation. Angew. Chem. Int. Ed. 2022;61:e202205341. doi: 10.1002/anie.202205341. [DOI] [PubMed] [Google Scholar]
- 39.Kurihara T, Kojima M, Yoshino T, Matsunaga S. Cp*CoIII/chiral carboxylic acid-catalyzed enantioselective 1,4-addition reactions of indoles to maleimides. Asian J. Org. Chem. 2020;9:368–371. doi: 10.1002/ajoc.201900565. [DOI] [Google Scholar]
- 40.Liu Y-H, et al. Cp*Co(III)-catalyzed enantioselective hydroarylation of unactivated terminal alkenes via C−H activation. J. Am. Chem. Soc. 2021;143:19112–19120. doi: 10.1021/jacs.1c08562. [DOI] [PubMed] [Google Scholar]
- 41.Pesciaioli F, et al. Enantioselective cobalt(III)-catalyzed C−H activation enabled by chiral carboxylic acid cooperation. Angew. Chem. Int. Ed. 2018;57:15425–15429. doi: 10.1002/anie.201808595. [DOI] [PubMed] [Google Scholar]
- 42.Dhawa U, Connon R, Oliveira JCA, Steinbock R, Ackermann L. Enantioselective ruthenium-catalyzed C−H alkylations by a chiral carboxylic acid with attractive dispersive interactions. Org. Lett. 2021;23:2760–2765. doi: 10.1021/acs.orglett.1c00615. [DOI] [PubMed] [Google Scholar]
- 43.LaPlante SR, et al. Assessing atropisomer axial chirality in drug discovery and development. J. Med. Chem. 2011;54:7005–7022. doi: 10.1021/jm200584g. [DOI] [PubMed] [Google Scholar]
- 44.Toenjes ST, Gustafson JL. Atropisomerism in medicinal chemistry: challenges and opportunities. Future Med. Chem. 2018;10:409–422. doi: 10.4155/fmc-2017-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang M-Z, Chen Q, Yang G-F. A review on recent developments of indole-containing antiviral agents. Eur. J. Med. Chem. 2015;89:421–441. doi: 10.1016/j.ejmech.2014.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sravanthi TV, Manju SL. Indoles-a promising scaffold for drug development. Eur. J. Pharm. Sci. 2016;91:1–10. doi: 10.1016/j.ejps.2016.05.025. [DOI] [PubMed] [Google Scholar]
- 47.Rodríguez-Salamanca P, Fernández R, Hornillos V, Lassaletta JM. Asymmetric synthesis of axially chiral C–N atropisomers. Chem. Eur. J. 2022;28:e202104442. doi: 10.1002/chem.202104442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu Y-J, Liao G, Shi B-F. Stereoselective construction of atropisomers featuring a C–N chiral axis. Green. Synth. Catal. 2022;3:117–136. doi: 10.1016/j.gresc.2021.12.005. [DOI] [Google Scholar]
- 49.He C, Hou M, Zhu Z, Gu Z. Enantioselective synthesis of indole-based biaryl atropoisomers via palladium-catalyzed dynamic kinetic intramolecular C–H cyclization. ACS Catal. 2017;7:5316–5320. doi: 10.1021/acscatal.7b01855. [DOI] [Google Scholar]
- 50.Li T-Z, Liu S-J, Tan W, Shi F. Catalytic asymmetric construction of axially chiral indole-based frameworks: an emerging area. Chem. Eur. J. 2020;26:15779–15792. doi: 10.1002/chem.202001397. [DOI] [PubMed] [Google Scholar]
- 51.Li Y, Liou Y-C, Oliveira JCA, Ackermann L. Ruthenium(II)/imidazolidine carboxylic acid-catalyzed C−H alkylation for central and axial double enantio-induction. Angew. Chem. Int. Ed. 2022;61:e202212595. doi: 10.1002/anie.202212595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Newton CG, Wang S-G, Oliveira CC, Cramer N. Catalytic enantioselective transformations involving C−H bond cleavage by transition-metal complexes. Chem. Rev. 2017;117:8908–8976. doi: 10.1021/acs.chemrev.6b00692. [DOI] [PubMed] [Google Scholar]
- 53.Loup J, Dhawa U, Pesciaioli F, Wencel-Delord J, Ackermann L. Enantioselective C–H activation with earth-abundant 3d transition metals. Angew. Chem. Int. Ed. 2019;58:12803–12818. doi: 10.1002/anie.201904214. [DOI] [PubMed] [Google Scholar]
- 54.Woźniak Ł, Cramer N. Enantioselective C–H bond functionalizations by 3d transition-metal catalysts. Trends Chem. 2019;1:471–484. doi: 10.1016/j.trechm.2019.03.013. [DOI] [Google Scholar]
- 55.Gallegos LC, Luchini G, John PCS, Kim S, Paton RS. Importance of engineered and learned molecular representations in predicting organic reactivity, selectivity, and chemical properties. Acc. Chem. Res. 2021;54:827–836. doi: 10.1021/acs.accounts.0c00745. [DOI] [PubMed] [Google Scholar]
- 56.Liu Y, Yang Q, Li Y, Zhang L, Luo S. Application of machine learning in organic chemistry. Chin. J. Org. Chem. 2020;40:3812–3827. doi: 10.6023/cjoc202006051. [DOI] [Google Scholar]
- 57.Cortes C, Vapnik V. Support-vector networks. Mach. Learn. 1995;20:273–297. doi: 10.1007/BF00994018. [DOI] [Google Scholar]
- 58.Zhang, Z.-J. et al. Data-driven Design of New Chiral Carboxylic Acid for Construction of Indoles with C-central and C–N Axial Chirality via Cobalt Catalysis.10.5281/zenodo.7855048 (2023). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available within the main text, the Supplementary Information and https://github.com/Shuwen-Li/FindBestChiralAcid58. Source data are presented in the Source_Data. Details about materials and methods, experimental procedures, characterization data, NMR and HPLC spectra are available in the Supplementary Information, and all other data are available from the corresponding author upon request. Crystallographic data are available free of charge under Cambridge Crystallographic Data Centre (CCDC) reference numbers 2176897 (3c), 2176898 (3w) [www.ccdc.cam.ac.uk/data_request/cif]. Source data are provided with this paper.
All codes needed to run this model are available at https://github.com/Shuwen-Li/FindBestChiralAcid58.