Summary
Background
To evaluate whether circulating proteins are associated with survival after lung cancer diagnosis, and whether they can improve prediction of prognosis.
Methods
We measured up to 1159 proteins in blood samples from 708 participants in 6 cohorts. Samples were collected within 3 years prior to lung cancer diagnosis. We used Cox proportional hazards models to identify proteins associated with overall mortality after lung cancer diagnosis. To evaluate model performance, we used a round-robin approach in which models were fit in 5 cohorts and evaluated in the 6th cohort. Specifically, we fit a model including 5 proteins and clinical parameters and compared its performance with clinical parameters only.
Findings
There were 86 proteins nominally associated with mortality (p < 0.05), but only CDCP1 remained statistically significant after accounting for multiple testing (hazard ratio per standard deviation: 1.19, 95% CI: 1.10–1.30, unadjusted p = 0.00004). The external C-index for the protein-based model was 0.63 (95% CI: 0.61–0.66), compared with 0.62 (95% CI: 0.59–0.64) for the model with clinical parameters only. Inclusion of proteins did not provide a statistically significant improvement in discrimination (C-index difference: 0.015, 95% CI: −0.003 to 0.035).
Interpretation
Blood proteins measured within 3 years prior to lung cancer diagnosis were not strongly associated with lung cancer survival, nor did they importantly improve prediction of prognosis beyond clinical information.
Funding
No explicit funding for this study. Authors and data collection supported by the US National Cancer Institute (U19CA203654), INCA (France, 2019-1-TABAC-01), Cancer Research Foundation of Northern Sweden (AMP19-962), and Swedish Department of Health Ministry.
Keywords: Lung cancer, Lung cancer survival, Protein biomarkers, Lung cancer prognosis
Research in context.
Evidence before this study
The use of blood-based biomarkers is often proposed to improve prediction of survival among lung cancer patients. However, because of inadequate study designs, measurement techniques, and interpretation of the data, most of the proposed biomarkers for predicting lung cancer prognosis have proven unsuccessful in independent validation studies.
Added value of this study
We used data from the INTEGRAL program to perform a broad scan of protein biomarkers measured up to three years before diagnosis for lung cancer survival. Surprisingly, we did not identify strong and consistent associations between circulating proteins and lung cancer survival, nor any improvement in the prediction of prognosis beyond standard clinical information (difference in C-index: 0.015, 95% CI: −0.003 to 0.035). Only CDCP1 showed a stable association with survival, but with a weak effect size.
Implications of all the available evidence
Protein biomarkers did not improve prediction of lung cancer prognosis beyond standard clinical information. Future biomarker studies for lung cancer survival should prioritize the use of blood samples collected from well-characterized newly diagnosed lung cancer cases and apply rigorous study designs with separate development and validation.
Introduction
Lung cancer is the leading cause of cancer-related death worldwide1 with 5-year survival less than 20% in most countries.2 Predicted prognosis is an important factor in clinical decision-making and communication with the patient.3 The primary information used to predict prognosis is clinical stage, along with age and performance status.4 Five-year survival ranges from 60 to 90% in stage I lung cancer to around 10% for stage IV patients.5,6 However, there is considerable heterogeneity in outcomes within each stage group, and better prognostic tools are needed.7, 8, 9
A potential approach to improve prediction of lung cancer survival is to use blood-based biomarkers such as proteins, mRNAs, epigenetic alterations, and circulating molecules.10, 11, 12, 13 However, most proposed biomarkers for predicting lung cancer prognosis have proven unsuccessful in independent validation studies due to inadequate study designs, measurement techniques, and interpretation of the data.14 For example, in the late 1990s, several studies reported that high levels of circulating vascular endothelial growth factor (VEGF) were associated with poorer survival for patients with lung cancer; however, the sample matrix (plasma or serum) and assay sensitivities differed across the studies, which affected the estimation of the magnitude of the association.15,16 Later studies suggested that VEGF may not be an independent prognostic factor for lung cancer when controlling for clinical factors.17,18
We previously launched large-scale proteomics analyses within the integrative Analysis of Lung Cancer Etiology and Risk (INTEGRAL) program, with the primary goal of identifying protein markers for early lung cancer detection. We measured over 1000 proteins among more than 700 pairs of lung cancer cases and controls selected from people who currently or formerly smoked in 6 prospective cohorts worldwide.19 Blood samples were pre-diagnostic, collected up to 3 years before lung cancer diagnosis. The study identified 36 circulating proteins as robustly associated with imminent lung cancer diagnosis, including CEACAM5, MUC-16, MMP12, WFDC2, and CDCP1.20
In the current study, we re-analysed the INTEGRAL data with the aim to identify and characterize potential protein biomarkers for survival after lung cancer diagnosis. We carried out a broad scan of proteins, then combined a small number of markers into a prediction algorithm and evaluated whether it improved prediction of lung cancer prognosis beyond available clinical information. Importantly, we leveraged the large size of the INTEGRAL study to build independent validation directly into our analytic design.
Methods
Study design and sample
Ethics
This study was conducted in the context of the INTEGRAL project, which was previously described by Robbins et al.19 and approved by the Ethics Committee of the International Agency for Research on Cancer. The ethics approval title was “Biomarkers of lung cancer risk (LC3)” (No. 11–13). Informed consent from all participants was obtained in each cohort.
In brief, INTEGRAL is an ongoing research effort to develop and validate a protein panel for early detection of lung cancer among current and former smokers. In an initial full discovery phase, 1161 proteins were measured on nested case-control pairs from 2 cohorts. Subsequently, a targeted discovery phase re-measured 392–484 proteins on an additional 4 cohorts. A later validation phase will evaluate the custom panel in additional cohorts.
In this study, to investigate potential biomarkers for lung cancer survival, we analysed data from participants diagnosed with lung cancer from the full and targeted discovery phases of INTEGRAL. Included participants were those who developed incident lung cancer (ICD code: C34) during the follow-up and had their blood sample drawn within 3 years prior to diagnosis. Participants were excluded if lung cancer was identified at death, or no protein measurements were available. We therefore included 708 participants with lung cancer from the Cancer Prevention Study II (CPS-II, USA, n = 115), the Trøndelag Health Study (HUNT, Norway, n = 154), the Melbourne Collaborative Cohort Study (MCCS, Australia, n = 105), the Singapore Chinese Health Study (SCHS, Singapore, n = 88), the European Investigation into Cancer and Nutrition (EPIC, Europe, n = 183), and the Northern Sweden Health and Disease Study (NSHDS, Sweden, n = 63). EPIC and NSHDS comprised the full discovery phase, while CPS-II, HUNT, MCCS, and SCHS comprised the targeted discovery phase. Details regarding each of these cohorts, including inclusion and exclusion criteria, recruitment strategies, and time period of enrolment, are provided in Robbins et al.19
We assessed all-cause mortality (rather than lung cancer specific mortality) as the primary outcome due to the likelihood of differences in cause-of-death ascertainment across the 6 included cohorts. This is a reasonable approach because the vast majority of deaths among individuals with lung cancer are caused by lung cancer. Nevertheless, in a sensitivity analysis conducted in 4 cohorts with cause-of-death information (EPIC, CPS-II, MCCS, and SCHS), we re-analysed protein associations while using lung cancer specific mortality as the outcome.
Proteomics assays
We used the Olink proteomics platform (Olink, Uppsala, Sweden) to measure circulating proteins. The Olink platform provides high-throughput semi-quantitative concentration measurements of annotated proteins. Proteins were measured in 14 panels categorized by biological function including inflammation, immuno-oncology, cell regulation, immune response, metabolism, etc. Additional details are provided in Robbins et al.19 Protein measurements are expressed as normalized protein expression (NPX) values which are log-base-2 transformed.19 We replaced protein values below the limit of detection (LOD) with the LOD divided by the square root of 2 and rescaled each protein to have a mean of 0 and a standard deviation (SD) of 1 within each cohort. We excluded 2 of the 1161 proteins which were not measured in EPIC and NSHDS (ADGRB3 and LTBP3) and performed an initial overall scan of 1159 proteins, including 678 measured only in EPIC and NSHDS and 481 measured in at least 5 cohorts (including EPIC and NSHDS).
To develop and validate a survival prediction model, we considered the 299 proteins measured in all 6 cohorts and missing in less than 10% of participants as the candidate proteins. We imputed missing protein values as the mean value within each cohort. IL-24 was missing for 20 participants, while each of the other proteins was missing in 2 or fewer participants.
Statistical analysis
For survival analysis, the time origin was the date of lung cancer diagnosis, and the time metric was time since diagnosis. Participants entered on the date of diagnosis and exited at the first of death, end of registry follow-up, or 5 years. We truncated follow-up at 5 years, because survival after 5 years was highly heterogeneous across the cohorts and could be subject to differences in ascertainment. Throughout analyses, we visually confirmed linear associations with continuous variables, and confirmed that the proportional hazards assumption for Cox models was fulfilled for the key variables of interest. We used the Kaplan–Meier method to estimate the overall probability of survival at 1, 3, and 5 years and its 95% confidence interval (CI) by treating death as the event of interest. We also used the Kaplan–Meier method to estimate the median follow-up time and its IQR (25%–75%) range by treating alive as the event, and death as censoring.
Overall scan of 1159 protein markers for lung cancer survival
We first evaluated the association between each protein and overall mortality after lung cancer diagnosis using Cox proportional hazards models with adjustment for age at diagnosis, sex, year of blood draw, cohort, and smoking status. To account for differences across participants in lead time between blood draw and diagnosis, we additionally adjusted for lead time. We accounted for multiple testing using effective-number-of-tests (ENT) statistical significance. The ENT method accounts for multiple testing by applying a Bonferroni correction, but determines the number of independent tests as the number of principal components needed to explain 95% of the variance in protein abundance.21
Prediction model development and validation
To examine the potential predictive utility of proteins for lung cancer survival, we used a round-robin, leave-one-cohort-out method. Six times, each time omitting 1 cohort, we used 5 cohorts to develop a protein-based model, a clinical model, and an integrated (protein + clinical) model to predict 5-year survival after lung cancer diagnosis. Then, each time, we validated the models in the omitted cohort.
Specifically, in the development sets (5 cohorts), we first selected proteins for the protein-based model. Among the 299 proteins measured in all 6 cohorts, we applied LASSO Cox proportional hazards models (“glmnet” package in R version 4.0.4) adjusted for age at diagnosis, sex, year of blood draw, cohort, smoking status, and lead time between blood draw and diagnosis. In each set we set the shrinkage parameter so that 5 proteins were selected. Using the selected proteins, we fit the protein-based model as a Cox proportional hazards model with adjustment for year of blood draw, cohort, and lead time. We separately fit the clinical model using age, sex, smoking status (former and current), TNM stage (I–II, III, IV, and missing), histology (adenocarcinoma, small cell carcinoma, squamous cell carcinoma, other/missing), year of blood draw, and cohort. We fit the integrated (protein + clinical) model by combining all parameters included in either model.
For each leave-one-cohort-out set, we internally validated the 3 models in the 5 development cohorts using 500 bootstrap samples to correct for optimism. Then, we externally validated the 3 models in the omitted cohort. Finally, we pooled the predicted risks from external validation across all 6 cohorts to obtain one summarized C-index for each model. We used Harrell’s C-index in the “survival” package to evaluate model discrimination and created calibration plots using the “rms” package, including observed and bias-corrected estimates of predicted vs. observed survival. We used a bootstrap with 1000 iterations to estimate a confidence interval for the difference in C-indices between different models.
Sample size
The sample size was 708 participants, and 587 (83%) events accumulated during 5 years of follow-up. Assuming an ENT p-value threshold of 0.0005, this sample size provides at least 80% power to identify protein markers with a hazard ratio (per 1-SD increment) above 1.20.22
Sensitivity analyses
For comparison with the primary analysis, which included a simple adjustment for lead time between blood draw and diagnosis, we also fit models with an interaction term between lead time and the protein measures. This makes the protein effect interpretable as the effect expected if measured at diagnosis. As a second approach, we restricted the analysis to participants whose blood sample was collected less than 1 year prior to lung cancer diagnosis. Finally, as mentioned above, we estimated protein associations using lung cancer specific mortality as the outcome after restricting to the 4 cohorts with cause-of-death information.
Role of funders
The funders had no role in study design, data analysis, data interpretation, or writing of this report.
Results
Among the 708 current and former smoking participants in our study, 54% smoked at the time of blood draw, 33% self-reported as female, and the mean age at lung cancer diagnosis was 66 years (SD 9.1 years) (Table 1). Information on TNM stage was unavailable for 54% of participants, due largely to the MCCS cohort (75% missing) and SCHS cohort (100% missing). Among participants with known stage, 76% were diagnosed at stage III–IV and 24% at stage I–II. Adenocarcinoma was the most common histological subtype (35%), followed by squamous cell carcinoma (22%). The lead time between blood draw and diagnosis was less than 1 year for 31% of cases.
Table 1.
Characteristics of 708 participants with lung cancer from cohort studies in Europe, North America, Asia, and Australia.
| Overall | EPIC | NSHDS | MCCS | CPS-II | HUNT | SCHS | |
|---|---|---|---|---|---|---|---|
| Lung cancer cases (N) | 708 | 183 | 63 | 105 | 115 | 154 | 88 |
| Location | Europe | Sweden | Australia | USA | Norway | Singapore | |
| Years of blood draw | 1991–2002 | 1988–2016 | 1990–1994 2003–2007 |
1998–2001 | 1995–1997 2006–2008 |
1994–2005 | |
| Age at diagnosis, years (mean ± SD) | 66 (9.1) | 61 (8.7) | 59 (5.8) | 68 (7.5) | 72 (5.0) | 68 (9.6) | 70 (6.5) |
| Current (vs. former) smokers | 381 (54%) | 124 (68%) | 38 (60%) | 41 (39%) | 21 (18%) | 96 (62%) | 61 (69%) |
| Females (vs. males) | 234 (33%) | 59 (32%) | 31 (49%) | 33 (31%) | 42 (37%) | 58 (38%) | 10 (11%) |
| Time between blood draw and diagnosis | |||||||
| <1 year | 217 (31%) | 53 (29%) | 15 (24%) | 40 (38%) | 30 (26%) | 53 (34%) | 26 (30%) |
| 1–1.9 years | 229 (32%) | 54 (29%) | 20 (32%) | 35 (33%) | 45 (39%) | 47 (31%) | 28 (32%) |
| 2–3 years | 262 (37%) | 76 (42%) | 28 (44%) | 30 (29%) | 40 (35%) | 54 (35%) | 34 (38%) |
| Lung cancer stagea | |||||||
| I–II | 78 (24%) | 18 (28%) | 12 (24%) | 11 (41%) | 19 (18%) | 18 (23%) | – |
| III–IV | 250 (76%) | 47 (72%) | 37 (76%) | 16 (59%) | 89 (82%) | 61 (77%) | – |
| Unknown/missing | 380 | 118 | 14 | 78 | 7 | 75 | 88 |
| Histology | |||||||
| Adenocarcinoma | 238 (35%) | 56 (31%) | 23 (47%) | 47 (45%) | 38 (33%) | 51 (34%) | 23 (29%) |
| Small cell carcinoma | 114 (17%) | 35 (19%) | 6 (12%) | 20 (19%) | 16 (14%) | 23 (15%) | 14 (18%) |
| Squamous cell carcinoma | 149 (22%) | 33 (18%) | 13 (27%) | 19 (18%) | 27 (23%) | 34 (23%) | 23 (29%) |
| Other/NOS | 179 (26%) | 58 (32%) | 7 (14%) | 19 (18%) | 34 (30%) | 41 (28%) | 20 (25%) |
| Missing | 28 | 1 | 14 | 0 | 0 | 5 | 8 |
EPIC: European Investigation into Cancer and Nutrition; NSHDS: Northern Sweden Health and Disease Study; MCCS: Melbourne Collaborative Cohort Study; CPS-II: Cancer Prevention Study II; HUNT: Trøndelag Health Study; SCHS: Singapore Chinese Health Study; SD: standard deviation; NOS: not otherwise specified.
For CPS-II, stage used SEER categories. We classified localized as stage I–II, regional as stage III, and distant/systemic as stage IV.
There were 587 deaths over 5 years and 652 deaths over the full follow-up (Table 2). The median follow-up time, when disregarding deaths and not truncating follow-up at 5 years, was 14.3 years (interquartile range (IQR) = 14.0–14.6 years). Overall survival of participants with lung cancer was 44% (95% CI: 41–48%) at 1 year, 22% (95% CI: 19–25%) at 3 years, and 17% (95% CI: 14–20%) at 5 years. Across the 6 cohorts, survival was highest in CPS-II (USA) (28%, 95% CI: 21–37%) and lowest in SCHS (Singapore) (2%, 95% CI: 0.6–9%). Including the truncation of follow-up at 5 years, 17% (121/708) of participants were censored, including 98% (119/121) who were censored because they reached 5 years and 2% (2/121) who were censored earlier because they reached the end of mortality registry follow-up.
Table 2.
Follow-up time and survival among 708 participants with lung cancer from cohort studies in Europe, North America, Asia, and Australia.
| Overall | EPIC | NSHDS | MCCS | CPS-II | HUNT | SCHS | |
|---|---|---|---|---|---|---|---|
| Number of cases | 708 | 183 | 63 | 105 | 115 | 154 | 88 |
| Median follow-up years (IQR)a | 14.3 (14.0–14.6) | 13.8 (12.6–14.0) | 14.0 (13.3–17.0) | 8.3 (7.7–NA) | 15.5 (15.2–16.0) | 11.2 (11.0–12.1) | NA |
| Number of deaths throughout follow-up (%) | 652 (92) | 162 (89) | 53 (84) | 98 (93) | 107 (93) | 144 (94) | 88 (100) |
| Number of deaths within 5 years (%) | 587 (83) | 155 (85) | 48 (76) | 87 (83) | 82 (71) | 130 (84) | 85 (97) |
| Probability of survival at 1 year (%, 95% CI) | 44 (41–48) | 48 (41–55) | 44 (34–59) | 44 (35–54) | 60 (52–70) | 36 (29–44) | 27 (19–38) |
| Probability of survival at 3 years (%, 95% CI) | 22 (19–25) | 20 (15–27) | 28 (19–42) | 20 (14–29) | 34 (26–44) | 19 (14–27) | 7 (3–15) |
| Probability of survival at 5 years (%, 95% CI) | 17 (14–20) | 15 (11–22) | 22 (13–35) | 17 (11–26) | 28 (21–37) | 16 (11–23) | 2 (0.6–9) |
IQR: interquartile range. NA: unable to estimate because of too many events.
The median survival time was estimated using the Kaplan–Meier method, defining alive or loss to follow-up as the event and treating death as censoring. In the SCHS cohort, the median follow-up time could not be calculated because all participants died by the end of follow-up.
Overall scan of protein markers for lung cancer survival
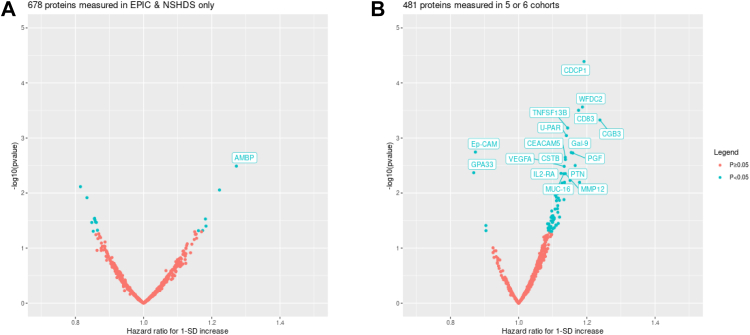
When analysing all participants by pooling data across the cohorts, among the 1159 proteins analysed, 86 proteins were nominally significantly associated with overall mortality in participants with lung cancer (p < 0.05) (Fig. 1). However, after accounting for multiple testing, only CDCP1 remained statistically significant (hazard ratio [HR] per standard deviation increase = 1.19, 95% CI: 1.10–1.30, unadjusted p = 0.00004). Overall, the hazard ratios per standard deviation increase in protein measurements were modest, ranging from 0.8 to 1.3 across all proteins.
Fig. 1.
Associations between pre-diagnostic protein concentrations and overall mortality after lung cancer diagnosis among 708 participants with lung cancer from 6 population cohorts in Europe, North America, Asia, and Australia. Proteins with p < 0.005 are labelled. This arbitrary threshold was chosen for illustration only. Panel A shows results for 678 proteins which were only measured in 2 cohorts (EPIC: European Investigation into Cancer and Nutrition; NSHDS: Northern Sweden Health and Disease Study), while panel B shows results for 481 proteins which were additionally measured in one or more of MCCS: Melbourne Collaborative Cohort Study; CPS-II: Cancer Prevention Study II; HUNT: Trøndelag Health Study; SCHS: Singapore Chinese Health Study.
Table 3 shows HRs for the 18 proteins with p-values less than 0.005, an arbitrary threshold set for descriptive purposes only. It also compares the results obtained in the primary analysis, with simple adjustment for lead time (continuous, ranging from 0 to 3 years), vs. the sensitivity analysis which includes an interaction between the protein measurement and lead time. Addition of the interaction term, which makes the HR for each protein interpretable as the predicted HR if the protein had been measured at time of diagnosis, tended to move HRs farther from the null compared with the main analysis. For CDCP1, for example, the HR increased from 1.19 in the main analysis to 1.34 with the interaction term.
Table 3.
Associations between pre-diagnostic protein measurements and overall mortality after lung cancer diagnosis, comparing two methods to account for pre-diagnostic lead time.
| Protein | Examined sample size | Cox model with main effects only |
Cox model with interaction |
|||
|---|---|---|---|---|---|---|
| Hazard ratio (95% CI) | p-value for protein | Hazard ratio (95% CI) | p-value for protein | p-interactiona | ||
| CDCP1 | 707 | 1.19 (1.10–1.30) | 0.00004 | 1.34 (1.12–1.59) | 0.001 | 0.15 |
| WFDC2 | 708 | 1.19 (1.08–1.30) | 0.0003 | 1.22 (1.01–1.48) | 0.04 | 0.76 |
| CD83 | 707 | 1.18 (1.08–1.28) | 0.0003 | 1.06 (0.89–1.27) | 0.51 | 0.19 |
| CGB3 | 551 | 1.24 (1.10–1.40) | 0.0005 | 1.38 (1.15–1.64) | 0.0004 | 0.13 |
| TNFSF13B | 708 | 1.14 (1.06–1.24) | 0.0007 | 1.04 (0.89–1.22) | 0.63 | 0.18 |
| U-PAR | 708 | 1.14 (1.06–1.23) | 0.0009 | 1.17 (1.01–1.37) | 0.04 | 0.66 |
| Ep-CAM | 708 | 0.87 (0.80–0.95) | 0.002 | 0.82 (0.69–0.97) | 0.02 | 0.38 |
| Gal-9 | 707 | 1.15 (1.05–1.26) | 0.002 | 1.25 (1.04–1.49) | 0.02 | 0.33 |
| PGF | 706 | 1.16 (1.06–1.27) | 0.002 | 1.20 (1.00–1.43) | 0.05 | 0.67 |
| CEACAM5 | 708 | 1.14 (1.05–1.23) | 0.002 | 1.24 (1.09–1.42) | 0.001 | 0.10 |
| CSTB | 708 | 1.14 (1.05–1.24) | 0.002 | 1.17 (1.00–1.38) | 0.05 | 0.65 |
| PTN | 706 | 1.17 (1.05–1.29) | 0.003 | 1.12 (0.94–1.34) | 0.19 | 0.61 |
| AMBP | 246 | 1.27 (1.08–1.49) | 0.003 | 1.35 (0.97–1.89) | 0.08 | 0.68 |
| VEGFA | 708 | 1.13 (1.04–1.23) | 0.003 | 1.25 (1.07–1.47) | 0.005 | 0.14 |
| GPA33 | 551 | 0.87 (0.79–0.96) | 0.004 | 0.79 (0.65–0.95) | 0.02 | 0.25 |
| IL2-RA | 708 | 1.12 (1.04–1.22) | 0.004 | 1.08 (0.91–1.28) | 0.36 | 0.60 |
| MMP12 | 707 | 1.14 (1.04–1.24) | 0.004 | 1.13 (0.99–1.29) | 0.08 | 0.87 |
| MUC-16 | 707 | 1.13 (1.04–1.23) | 0.004 | 1.26 (1.11–1.44) | 0.0004 | 0.04 |
The table includes proteins with p < 0.005 (arbitrary threshold) in the model with main effects only, listed in order of p-value. Both models are adjusted for lead time between blood draw and diagnosis, age at diagnosis, sex, year of blood draw, cohort, and smoking status. The model with interaction additionally includes an interaction term between lead time and the protein measurement, which changes the interpretation of the protein hazard ratio to be the predicted effect if the protein were measured at the time of diagnosis.
CI: confidence interval.
p-value for the interaction term in the model.
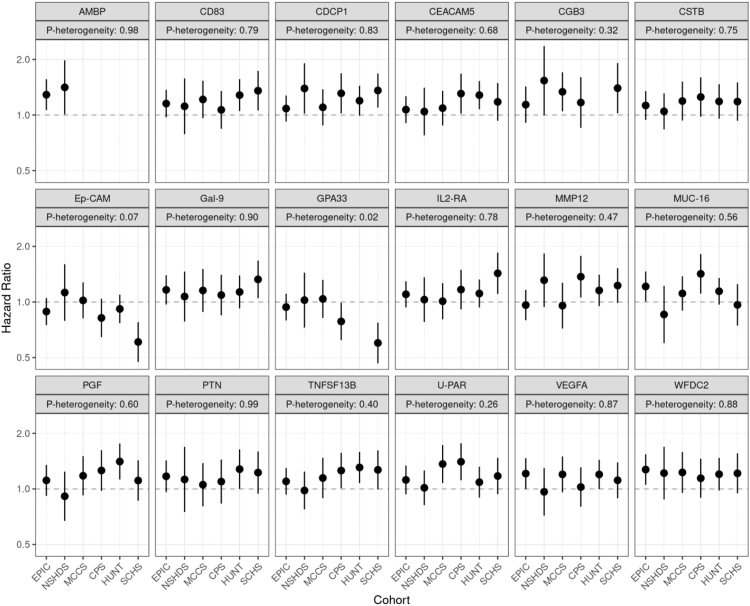
Among the 18 proteins with p < 0.005, there was no evidence for heterogeneity in associations across participants from the 6 cohorts, with the possible exception of GPA33 (p-heterogeneity = 0.02) (Fig. 2). Similarly, there was no evidence for heterogeneity in associations by TNM stage or histological type (Supplementary Table S1).
Fig. 2.
Stratified results by cohort for the association between highly ranked proteins and overall mortality after lung cancer diagnosis. EPIC: European Investigation into Cancer and Nutrition; NSHDS: Northern Sweden Health and Disease Study; MCCS: Melbourne Collaborative Cohort Study; CPS-II: Cancer Prevention Study II; HUNT: Trøndelag Health Study; SCHS: Singapore Chinese Health Study.
Development and validation of prediction models for lung cancer survival
We applied LASSO Cox regression in each of the 6 leave-one-cohort-out sets, each time to select 5 proteins for prediction of mortality after lung cancer diagnosis (Table 4). Across the 6 sets, CDCP1 was always selected, Ep-CAM and TNFSF13B were selected 5 times, U-PAR was selected 4 times, CD83 and WFDC2 were selected twice, and TNFRSF6B, IFN-gamma, MUC-16, CEACAM5, ERBB4, CCL25, and IL-12B were each selected once. Although the selected proteins were different across the 6 sets, they showed similar internal C-indices, ranging from 0.60 to 0.63 (Supplementary Table S2). In the clinical models, consistent predictors of survival included age, sex, and TNM stage, and internal C-indices ranged from 0.64 to 0.66 across the 6 sets. The external validation gave a summarized C-index of 0.58 (95% CI: 0.56–0.61) for the protein-based model and 0.62 (95% CI: 0.59–0.64) for the clinical model (Table 4). Combining the protein markers with the clinical factors gave a C-index of 0.63 (95% CI: 0.61–0.66) which did not represent a statistically significant improvement in discrimination over the clinical model (difference in C-index between integrated vs. clinical model = 0.015, 95% CI: −0.003 to 0.035) (Table 4). Calibration plots for 5-year survival are shown in Supplementary Fig. S2.
Table 4.
Integrated models for overall mortality after lung cancer diagnosis, and assessment of the utility of protein measurements beyond clinical factors, using a round-robin, leave-one-cohort-out method for independent training and testing among 708 participants from 6 population cohorts.
| Omit CPS | Omit EPIC | Omit HUNT | Omit MCCS | Omit NSHDS | Omit SCHS | |
|---|---|---|---|---|---|---|
| CDCP1 | 1.11 (1.01–1.23) | 1.24 (1.11–1.38) | 1.15 (1.03–1.28) | 1.19 (1.08–1.31) | 1.11 (1.00–1.22) | 1.14 (1.03–1.26) |
| Ep-CAM | 0.88 (0.80–0.97) | 0.89 (0.81–0.99) | 0.86 (0.78–0.96) | 0.88 (0.80–0.96) | 0.85 (0.78–0.93) | |
| TNFSF13B | 1.05 (0.96–1.15) | 1.06 (0.94–1.19) | 1.11 (1.02–1.21) | 1.08 (0.97–1.20) | 1.04 (0.93–1.16) | |
| U-PAR | 1.06 (0.95–1.20) | 1.06 (0.95–1.17) | 1.07 (0.96–1.19) | 0.97 (0.86–1.09) | ||
| CD83 | 1.07 (0.96–1.20) | 1.06 (0.95–1.18) | ||||
| WFDC2 | 1.13 (1.00–1.28) | 1.13 (1.00–1.28) | ||||
| TNFRSF6B | 1.07 (0.97–1.18) | |||||
| IFN-gamma | 1.04 (0.95–1.14) | |||||
| MUC-16 | 1.13 (1.03–1.24) | |||||
| CEACAM5 | 1.17 (1.06–1.28) | |||||
| ERBB4 | 0.89 (0.81–0.98) | |||||
| CCL25 | 0.87 (0.79–0.97) | |||||
| IL-12B | 0.84 (0.76–0.94) | |||||
| Age, 5-year increment | 1.04 (0.98–1.11) | 1.03 (0.96–1.11) | 1.07 (0.99–1.16) | 1.04 (0.97–1.10) | 1.04 (0.97–1.11) | 1.05 (0.99–1.13) |
| Sex | ||||||
| Male | ref | ref | ref | ref | ref | ref |
| Female | 0.86 (0.70–1.07) | 0.87 (0.69–1.10) | 0.90 (0.71–1.13) | 0.84 (0.68–1.04) | 0.84 (0.68–1.03) | 0.82 (0.67–1.00) |
| Smoking status | ||||||
| Former | ref | ref | ref | ref | ref | ref |
| Current | 1.00 (0.82–1.21) | 1.11 (0.88–1.39) | 0.91 (0.70–1.17) | 1.06 (0.86–1.31) | 1.05 (0.85–1.29) | 1.06 (0.85–1.33) |
| TNM stage | ||||||
| I–II | ref | ref | ref | ref | ref | ref |
| III | 2.64 (1.70–4.10) | 1.67 (1.07–2.59) | 1.84 (1.16–2.92) | 1.99 (1.32–3.00) | 1.81 (1.20–2.72) | 2.07 (1.41–3.04) |
| IV | 4.48 (2.91–6.92) | 3.61 (2.39–5.46) | 3.99 (2.59–6.15) | 4.26 (2.84–6.38) | 3.43 (2.31–5.08) | 4.07 (2.81–5.89) |
| Missing | 3.10 (2.09–4.60) | 2.51 (1.66–3.80) | 2.53 (1.65–3.88) | 2.95 (2.00–4.34) | 2.40 (1.66–3.46) | 2.76 (1.94–3.94) |
| Histology | ||||||
| Adenocarcinoma | ref | ref | ref | ref | ref | ref |
| Small Cell Carcinoma | 1.12 (0.86–1.46) | 1.11 (0.83–1.5) | 1.37 (1.04–1.82) | 1.34 (1.02–1.76) | 1.36 (1.05–1.75) | 1.37 (1.05–1.79) |
| Squamous Cell Carcinoma | 0.76 (0.58–0.98) | 0.72 (0.55–0.95) | 0.81 (0.62–1.07) | 0.87 (0.67–1.13) | 0.78 (0.61–1.00) | 0.78 (0.60–1.01) |
| Other/missing | 0.88 (0.70–1.12) | 1.07 (0.83–1.39) | 1.12 (0.87–1.44) | 1.04 (0.82–1.31) | 1.07 (0.85–1.34) | 0.94 (0.74–1.18) |
| Internal C-index | 0.66 (0.64–0.69) | 0.68 (0.65–0.71) | 0.68 (0.65–0.70) | 0.68 (0.66–0.70) | 0.66 (0.64–0.69) | 0.67 (0.64–0.70) |
| External C-indices | Overall |
| Protein-based model | 0.58 (0.56–0.61) |
| Clinical model | 0.62 (0.59–0.64) |
| Integrated model | 0.63 (0.61–0.66) |
| Difference in C-index, integrated vs. clinical model | 0.015 (−0.003 to 0.035) |
To implement the leave-one-cohort-out method, in each of 6 iterations, we used 5 cohorts to train the model and the remaining (omitted) cohort for independent testing. The table shows the integrated models, their internal C-indices, and the external C-indices for the protein-based, clinical, and integrated models (see Methods). The detailed parameters and internal C-indices for the protein-based models and clinical models are shown in Supplementary Table S2. Internal indices were estimated using 500 bootstrap iterations in the training set. Models were adjusted by cohort and year of blood draw.
Sensitivity analyses
When using Cox models with an interaction term between protein measurements and lead time, the list of top-ranked proteins (based on lowest p-value) differed from the main analysis with a simple adjustment for lead time (Supplementary Table S3). In this sensitivity analysis, the high-ranking proteins tended to show strong interactions between lead time and protein measurements. For example, MCP-4 was not related to mortality in the primary analysis (HR = 1.03, 95% CI: 0.94–1.12), but was associated after addition of the interaction term (HR = 1.38, 95% CI: 1.15–1.64, p-interaction = 0.0002).
When restricting the analysis to 217 participants whose blood sample was collected less than 1 year prior to lung cancer diagnosis, the observed associations with mortality were stronger than when including longer lead times (HRs ranging from 0.6 to 2.0, compared with 0.8 to 1.3 in the primary analysis) (Supplementary Fig. S1). There were 6 proteins with statistically significant associations after adjusting for multiple testing, including APBB1IP, FLI1, MAX, NUB1, and PIgR in EPIC and NSHDS only, and CHI3L1 in the larger sample. There were 36 proteins with p < 0.005 compared to 18 in the main analysis. CDCP1, WFDC2, CGB3, U-PAR, Gal-9, CEACAM5, and PGF were highly ranked both in the <1 year lead time sample and in the primary analysis (Supplementary Fig. S1).
Finally, when using lung cancer specific mortality as the outcome and analysing 4 cohorts with cause-of-death information (EPIC, CPS-II, MCCS, and SCHS), results for most proteins resembled those for overall mortality (Supplementary Table S4).
Discussion
We evaluated the association between pre-diagnostic blood measurements of 1159 circulating proteins and lung cancer survival in 6 cohorts and assessed whether proteins could improve prediction of lung cancer prognosis. When analysing blood samples drawn up to 3 years prior to lung cancer diagnosis, we found little evidence for strong associations between circulating proteins and survival after lung cancer diagnosis. Only one marker, CDCP1, seemed to show a stable association, but with a relatively weak effect size. When we included proteins in a model to predict mortality after lung cancer diagnosis, there was no evidence to support an important improvement in prediction over standard clinical information.
Previous research on circulating proteins and lung cancer survival has examined much smaller sets of proteins than our study, and mostly focused on immune and inflammatory markers.23, 24, 25, 26 In an Italian study including 84 short-term and 157 long-term surviving patients diagnosed with non-small cell lung cancer (NSCLC) at stage I–II, Bodelon et al. found CCL15 [chemokine (C-C motif) ligand 15] to be most strongly associated with survival among 77 immune and inflammatory markers. The highest quartile of CCL15 showed a 5-fold increase in the odds of short survival compared with the lowest quartile, with other associated markers including IL-8, C-reactive protein (CRP), IL-2Ra, TNF-α, IL-6, TRAIL, and IL-6R.23 Another study examined 33 inflammatory proteins among 129 US patients with stage I adenocarcinoma and found shorter survival among those with elevated levels of IL-6 and IL-17A.25 A blood-based proteomic signature test called VeriStrat® has been proposed to divide patients with lung cancer into predicted high and low survival groups by examining a protein signature (based on mass spectrometry features) associated with a chronic inflammatory disease state and aggressive cancer.27, 28, 29 A meta-analysis reported worse survival with higher circulating CRP and IL-6 levels, but no significant association for IL-8 or TNF-α.24 Among the most frequently studied proteins, specifically CCL15, CRP, IL-8, IL-6, and TNF-α, our study did not measure CRP or TNF-α. Among the others, we found that IL-8 was associated with lung cancer survival but was not statistically significant after accounting for multiple comparisons, while no association was found for CCL15 and IL-6 (Supplementary Table S4).
CDCP1 was the only protein clearly associated with overall survival among patients with lung cancer after accounting for multiple testing in our study. Notably, CDCP1 is also associated with increased risk of incident lung cancer.30 CDCP1 is a transmembrane noncatalytic receptor involved in the loss of anchorage in epithelial cells during mitosis31 and has been shown to be involved in the pathway of tumor invasion and metastasis in lung cancer cells, which could provide a potential mechanism for the association we observed.32,33 Of note, well-known markers such as CEACAM5/CEA and CA-125/MUC-16 which are associated with incident lung cancer also showed nominal associations with mortality after lung cancer diagnosis in our study.34 We note that the proteins highlighted in our study are unlikely to be specific to lung cancer, and might be related to several diseases. CDCP1, for example, was associated with lung cancer survival in our study but is also upregulated in malignancies of the breast, colorectum, ovary, kidney, liver, pancreas, and hematopoietic system.35 Our prior work focused on early detection identified 36 proteins robustly associated with lung cancer onset, among which only 1 protein was predominantly expressed by lung tumor tissue.20
Most studies have aimed to identify cancer prognostic biomarkers by using blood samples collected at the time of diagnosis. However, this approach can be influenced by cancer-related lifestyle changes, treatment, or other interventions around the time of diagnosis.36 Therefore, there are potential advantages of our approach which used pre-diagnostic protein measurements. We tried multiple approaches to account for the pre-diagnostic lead time, including direct adjustment, additional inclusion of an interaction term between protein measurements and lead time, and restriction to participants whose blood was collected less than 1 year before diagnosis. In each approach, we identified proteins with apparent associations with survival, but the ranking of proteins was unstable across the approaches. We did observe somewhat stronger associations after restricting to blood collected within 1 year of diagnosis. Therefore, it is still possible that robust protein markers for lung cancer survival may exist, but our results suggest that they are unlikely to be very strong or highly predictive. Future research should prioritize use of samples collected from well-characterized, newly diagnosed lung cancer cases, with robust independent discovery and validation phases.
One limitation of our study, deriving from its design as a consortium of population cohorts, is that we lacked complete and detailed clinical information on the participants with lung cancer. Information on clinical stage and histological type had high missingness, and we had no information on other factors that might importantly influence survival such as lung cancer treatment, other comorbidities, access to care, and social support. However, we consider it unlikely that accounting for additional predictors would change our conclusion, because this would further improve the performance of the clinical model, rendering any added contribution of the protein markers more difficult to demonstrate. The second limitation is that HUNT and NSHDS were lack of cause of death information, we were unable to analyse lung cancer mortality in the full dataset. We also acknowledge that our sensitivity analysis using an interaction term assumes that associations between protein measurements and mortality change linearly with lead time prior to diagnosis, which may be incorrect. Only a subset of 299 among the 1159 total proteins could be considered for inclusion in the prediction models, but these included proteins with strong associations (CDCP1, WFDC2, and CD83). The key strength of our study is its large size, both in terms of the number of proteins examined and the number of participants. It also benefitted from a diverse set of cohorts from Europe, North America, Asia, and Australia.
Conclusion
Our study aimed to evaluate whether circulating proteins are associated with survival after lung cancer diagnosis, and whether they can improve prediction of prognosis. However, we did not identify strong associations with lung cancer survival among 1159 protein markers studied. Similarly, proteins did not offer improvement beyond clinical factors such as age, sex, and clinical stage for prediction of mortality. Nonetheless, our results do not exclude the possibility that prognostic protein markers for lung cancer may exist, and future studies should prioritize the use of blood samples collected from well-characterized newly diagnosed lung cancer cases.
Contributors
HAR, MaJ, DM, and XF conceived the study idea and designed the study.
HAR, MaJ, DM, and XF contributed to the methodology.
JMY, WPK, RW, RLM, JKB, AL, KH, VLS, YW, MiJ, AT, and RT contributed samples and data.
XF, HZ, KA, FG, and KSB contributed to the data extraction.
XF and HZ performed the statistical analysis.
XF, HZ, and KA have verified the underlying data.
XF, DM, MS, HAR, and MaJ interpreted the analysis.
XF and HAR drafted the manuscript.
MaJ and HAR supervised the project.
All authors read and approved the final version of the manuscript.
XF takes responsibility for the content of the manuscript, including the data and analysis.
Data sharing statement
Access to data from the Lung Cancer Cohort Consortium (LC3) is governed by the LC3 Access Policy, which is available at the following link: https://www.iarc.who.int/wp-content/uploads/2021/12/LC3_Access_Policy.pdf. Interested investigators are encouraged to contact Dr Johansson or Dr Robbins.
Disclaimer
Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer/World Health Organization.
Declaration of interests
Jian-Min Yuan has a declaration on NIH grant funding, and the other authors have no conflicts of interest.
Acknowledgements
The Trøndelag Health Study (HUNT) is a collaboration between HUNT Research Centre (Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology NTNU), Trøndelag County Council, Central Norway Regional Health Authority, and the Norwegian Institute of Public Health.
The Singapore Chinese Health Study was supported by the US National Institutes of Health Grant No. R01CA080205, R01CA144034 and UM182876.
Melbourne Collaborative Cohort Study (MCCS) cohort recruitment was funded by by VicHealth and Cancer Council Victoria. The MCCS was further augmented by Australian National Health and Medical Research Council grants 209057, 396414 and 1074383 and by infrastructure provided by Cancer Council Victoria.
The authors express sincere appreciation to all Cancer Prevention Study-II participants, and to each member of the study and biospecimen management group. The authors would like to acknowledge the contribution to this study from central cancer registries supported through the Centers for Disease Control and Prevention's National Program of Cancer Registries and cancer registries supported by the National Cancer Institute's Surveillance Epidemiology and End Results Program.
We thank the Biobank Research Unit at Umeå University, Västerbotten Intervention Programme, the Northern Sweden MONICA study, the Mammography Study and Region Västerbotten for providing data and samples and acknowledge the contribution from Biobank Sweden, supported by the Swedish Research Council (VR 2017-00650).
The coordination of EPIC was financially supported by Direction Générale de la Santé (French Ministry of Health) (Grant GR-IARC-2003-09-12-01), the European Commission (Directorate General for Health and Consumer Affairs), International Agency for Research on Cancer (IARC) and by the Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London with additional infrastructure support provided by the NIHR Imperial Biomedical Research Centre (BRC). The national cohorts are supported by: Danish Cancer Society (Denmark); Ligue Contre le Cancer, Institut Gustave-Roussy, Mutuelle Générale de l'Education Nationale, Institut National de la Santé et de la Recherche Médicale (INSERM) (France); German Cancer Aid, German Cancer Research Center (DKFZ), German Institute of Human Nutrition Potsdam-Rehbruecke (DIfE), Federal Ministry of Education and Research (BMBF) (Germany); Associazione Italiana per la Ricerca sul Cancro-AIRC-Italy, Compagnia di San Paolo and National Research Council (Italy); Dutch Ministry of Public Health, Welfare and Sports (VWS), Netherlands Cancer Registry (NKR), LK Research Funds, Dutch Prevention Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF), Statistics Netherlands (The Netherlands); Health Research Foundation (FIS) - Instituto de Salud Carlos III (ISCIII), Regional Governments of Andalucía, Asturias, Basque Country, Murcia and Navarra, and the Catalan Institute of Oncology - ICO (Spain); Swedish Cancer Society, Swedish Research Council and County Councils of Skåne and Västerbotten (Sweden); Cancer Research UK (14136 to EPIC-Norfolk; C8221/A29017 to EPIC-Oxford), Medical Research Council (1000143 to EPIC-Norfolk; MR/M012190/1 to EPIC-Oxford) (United Kingdom). We thank the National Institute for Public Health and the Environment (RIVM), Bilthoven, the Netherlands, for their contribution and ongoing support to the EPIC Study.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104623.
Contributor Information
Xiaoshuang Feng, Email: fengx@iarc.who.int.
Hilary A. Robbins, Email: robbinsh@iarc.who.int.
Appendix A. Supplementary data
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Allemani C., Matsuda T., Di Carlo V., et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391(10125):1023–1075. doi: 10.1016/S0140-6736(17)33326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Detterbeck F.C., Lewis S.Z., Diekemper R., Addrizzo-Harris D., Alberts W.M. Executive summary: diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest. 2013;143(5 Suppl):7s–37s. doi: 10.1378/chest.12-2377. [DOI] [PubMed] [Google Scholar]
- 4.Oken M.M., Creech R.H., Tormey D.C., et al. Toxicity and response criteria of the Eastern Cooperative Oncology Group. Am J Clin Oncol. 1982;5(6):649–655. [PubMed] [Google Scholar]
- 5.Chansky K., Detterbeck F.C., Nicholson A.G., et al. The IASLC Lung Cancer Staging Project: external validation of the revision of the TNM stage groupings in the eighth edition of the TNM classification of lung cancer. J Thorac Oncol. 2017;12(7):1109–1121. doi: 10.1016/j.jtho.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 6.Sui X., Jiang W., Chen H., Yang F., Wang J., Wang Q. Validation of the stage groupings in the eighth edition of the TNM classification for lung cancer. J Thorac Oncol. 2017;12(11):1679–1686. doi: 10.1016/j.jtho.2017.07.028. [DOI] [PubMed] [Google Scholar]
- 7.Aramini B., Casali C., Stefani A., et al. Prediction of distant recurrence in resected stage I and II lung adenocarcinoma. Lung Cancer. 2016;101:82–87. doi: 10.1016/j.lungcan.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Torok J.A., Gu L., Tandberg D.J., et al. Patterns of distant metastases after surgical management of non-small-cell lung cancer. Clin Lung Cancer. 2017;18(1):e57–e70. doi: 10.1016/j.cllc.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 9.Grosu H.B., Manzanera A., Shivakumar S., Sun S., Noguras Gonzalez G., Ost D.E. Survival disparities following surgery among patients with different histological types of non-small cell lung cancer. Lung Cancer. 2020;140:55–58. doi: 10.1016/j.lungcan.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Tang H., Wang S., Xiao G., et al. Comprehensive evaluation of published gene expression prognostic signatures for biomarker-based lung cancer clinical studies. Ann Oncol. 2017;28(4):733–740. doi: 10.1093/annonc/mdw683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puderecki M., Szumiło J., Marzec-Kotarska B. Novel prognostic molecular markers in lung cancer. Oncol Lett. 2020;20(1):9–18. doi: 10.3892/ol.2020.11541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapeleris J., Kulasinghe A., Warkiani M.E., et al. The prognostic role of circulating tumor cells (CTCs) in lung cancer. Front Oncol. 2018;8:311. doi: 10.3389/fonc.2018.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robles A.I., Arai E., Mathé E.A., et al. An integrated prognostic classifier for stage I lung adenocarcinoma based on mRNA, microRNA, and DNA methylation biomarkers. J Thorac Oncol. 2015;10(7):1037–1048. doi: 10.1097/JTO.0000000000000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Šutić M., Vukić A., Baranašić J., et al. Diagnostic, predictive, and prognostic biomarkers in non-small cell lung cancer (NSCLC) management. J Pers Med. 2021;11(11) doi: 10.3390/jpm11111102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu P., Liu W., Wang L., Yang M., Du J. High circulating VEGF level predicts poor overall survival in lung cancer. J Cancer Res Clin Oncol. 2013;139(7):1157–1167. doi: 10.1007/s00432-013-1425-1. [DOI] [PubMed] [Google Scholar]
- 16.Guo S., Martin M.G., Tian C., et al. Evaluation of detection methods and values of circulating vascular endothelial growth factor in lung cancer. J Cancer. 2018;9(7):1287–1300. doi: 10.7150/jca.22020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakra M., Pujol J.L., Lamy P.J., et al. Circulating serum vascular endothelial growth factor is not a prognostic factor of non-small cell lung cancer. J Thorac Oncol. 2008;3(10):1119–1126. doi: 10.1097/JTO.0b013e318187464e. [DOI] [PubMed] [Google Scholar]
- 18.Hegde P.S., Jubb A.M., Chen D., et al. Predictive impact of circulating vascular endothelial growth factor in four phase III trials evaluating bevacizumab. Clin Cancer Res. 2013;19(4):929–937. doi: 10.1158/1078-0432.CCR-12-2535. [DOI] [PubMed] [Google Scholar]
- 19.Robbins H.A., Alcala K., Moez E.K., et al. Design and methodological considerations for biomarker discovery and validation in the integrative analysis of lung cancer etiology and risk (INTEGRAL) Program. Ann Epidemiol. 2023;77:1–12. doi: 10.1016/j.annepidem.2022.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The Lung Cancer Cohort Consortium (LC3) The blood proteome of imminent lung cancer diagnosis. Nat Commun. 2023 doi: 10.1038/s41467-023-37979-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galwey N.W. A new measure of the effective number of tests, a practical tool for comparing families of non-independent significance tests. Genet Epidemiol. 2009;33(7):559–568. doi: 10.1002/gepi.20408. [DOI] [PubMed] [Google Scholar]
- 22.Hsieh F.Y., Lavori P.W. Sample-size calculations for the Cox proportional hazards regression model with nonbinary covariates. Control Clin Trials. 2000;21(6):552–560. doi: 10.1016/s0197-2456(00)00104-5. [DOI] [PubMed] [Google Scholar]
- 23.Bodelon C., Polley M.Y., Kemp T.J., et al. Circulating levels of immune and inflammatory markers and long versus short survival in early-stage lung cancer. Ann Oncol. 2013;24(8):2073–2079. doi: 10.1093/annonc/mdt175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao C., Yu Z., Guo W., et al. Prognostic value of circulating inflammatory factors in non-small cell lung cancer: a systematic review and meta-analysis. Cancer Biomark. 2014;14(6):469–481. doi: 10.3233/CBM-140423. [DOI] [PubMed] [Google Scholar]
- 25.Meaney C.L., Zingone A., Brown D., Yu Y., Cao L., Ryan B.M. Identification of serum inflammatory markers as classifiers of lung cancer mortality for stage I adenocarcinoma. Oncotarget. 2017;8(25):40946–40957. doi: 10.18632/oncotarget.16784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaes R.D.W., Reynders K., Sprooten J., et al. Identification of potential prognostic and predictive immunological biomarkers in patients with stage I and stage III non-small cell lung cancer (NSCLC): a prospective exploratory study. Cancers. 2021;13(24):6259. doi: 10.3390/cancers13246259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leal T.A., Argento A.C., Bhadra K., et al. Prognostic performance of proteomic testing in advanced non-small cell lung cancer: a systematic literature review and meta-analysis. Curr Med Res Opin. 2020;36(9):1497–1505. doi: 10.1080/03007995.2020.1790346. [DOI] [PubMed] [Google Scholar]
- 28.Gregorc V., Novello S., Lazzari C., et al. Predictive value of a proteomic signature in patients with non-small-cell lung cancer treated with second-line erlotinib or chemotherapy (PROSE): a biomarker-stratified, randomised phase 3 trial. Lancet Oncol. 2014;15(7):713–721. doi: 10.1016/S1470-2045(14)70162-7. [DOI] [PubMed] [Google Scholar]
- 29.Taguchi F., Solomon B., Gregorc V., et al. Mass spectrometry to classify non-small-cell lung cancer patients for clinical outcome after treatment with epidermal growth factor receptor tyrosine kinase inhibitors: a multicohort cross-institutional study. J Natl Cancer Inst. 2007;99(11):838–846. doi: 10.1093/jnci/djk195. [DOI] [PubMed] [Google Scholar]
- 30.Dagnino S., Bodinier B., Guida F., et al. Prospective identification of elevated circulating CDCP1 in patients years before onset of lung cancer. Cancer Res. 2021;81(13):3738–3748. doi: 10.1158/0008-5472.CAN-20-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hooper J.D., Zijlstra A., Aimes R.T., et al. Subtractive immunization using highly metastatic human tumor cells identifies SIMA135/CDCP1, a 135 kDa cell surface phosphorylated glycoprotein antigen. Oncogene. 2003;22(12):1783–1794. doi: 10.1038/sj.onc.1206220. [DOI] [PubMed] [Google Scholar]
- 32.Uekita T., Fujii S., Miyazawa Y., et al. Oncogenic Ras/ERK signaling activates CDCP1 to promote tumor invasion and metastasis. Mol Cancer Res. 2014;12(10):1449–1459. doi: 10.1158/1541-7786.MCR-13-0587. [DOI] [PubMed] [Google Scholar]
- 33.Zeng X.J., Wu Y.H., Luo M., Cong P.G., Yu H. Inhibition of pulmonary carcinoma proliferation or metastasis of miR-218 via down-regulating CDCP1 expression. Eur Rev Med Pharmacol Sci. 2017;21(7):1502–1508. [PubMed] [Google Scholar]
- 34.Fahrmann J.F., Marsh T., Irajizad E., et al. Blood-based biomarker panel for personalized lung cancer risk assessment. J Clin Oncol. 2022;40(8):876–883. doi: 10.1200/JCO.21.01460. Jco2101460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khan T., Kryza T., Lyons N.J., He Y., Hooper J.D. The CDCP1 signaling hub: a target for cancer detection and therapeutic intervention. Cancer Res. 2021;81(9):2259–2269. doi: 10.1158/0008-5472.CAN-20-2978. [DOI] [PubMed] [Google Scholar]
- 36.Tollosa D.N., Holliday E., Hure A., Tavener M., James E.L. Multiple health behaviors before and after a cancer diagnosis among women: a repeated cross-sectional analysis over 15 years. Cancer Med. 2020;9(9):3224–3233. doi: 10.1002/cam4.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.