Abstract
The SARS-CoV-2 spike protein (S) represents an important viral component that is required for successful viral infection in humans owing to its essential role in recognition of and entry to host cells. The spike is also an appealing target for drug designers who develop vaccines and antivirals. This article is important as it summarizes how molecular simulations successfully shaped our understanding of spike conformational behavior and its role in viral infection. MD simulations found that the higher affinity of SARS-CoV-2-S to ACE2 is linked to its unique residues that add extra electrostatic and van der Waal interactions in comparison to the SARS-CoV S. This illustrates the spread potential of the pandemic SARS-CoV-2 relative to the epidemic SARS-CoV. Different mutations at the S-ACE2 interface, which is believed to increase the transmission of the new variants, affected the behavior and binding interactions in different simulations. The contributions of glycans to the opening of S were revealed via simulations. The immune evasion of S was linked to the spatial distribution of glycans. This help the virus to escape the immune system recognition. This article is important as it summarizes how molecular simulations successfully shaped our understanding of spike conformational behavior and its role in viral infection. This will pave the way to us preparing for the next pandemic as the computational tools are tailored to help fight new challenges.
Keywords: SARS-CoV-2, Spike, Glycosylation, ACE2, Structural bioinformatics, MD simulation
1. Introduction
Molecular simulations became a robust method for studying molecular systems [1]. Furthermore, biomolecular simulations solved different human-related health problems as the algorithms and hardware developed rapidly during the last two decades [1,2]. For example, the human immunodeficiency virus (HIV) and hepatitis C virus (HCV) were successful case studies that heavily relied on computer simulations to find potential inhibitors [[3], [4], [5], [6], [7]]. The pandemic SARS-CoV-2 represents the best recent example of utilizing molecular simulations to investigate human-health-related problems saving time, effort, money, and life [8]. Many studies were conducted on SARS-CoV-2 that use computational tools to find possible drug targets, repurpose old medicines or natural compounds previously suggested/tested against different viral proteins, and develop a vaccine against the virus [[9], [10], [11], [12], [13]].
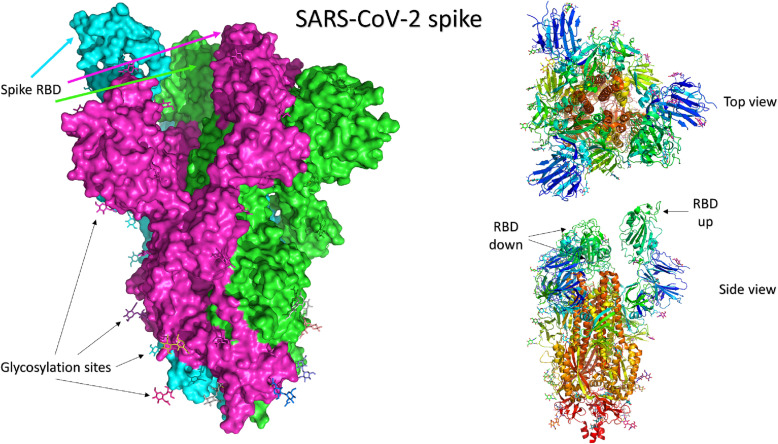
Severe acute respiratory syndrome coronavirus (SARS-CoV) (2003) and SARS-CoV-2 (2019) are two pathogenic viruses of the β-coronaviruses (β-CoVs) lineage. CoVs are spherical-shaped, enveloped viruses with their genetic material as a positive-sense single-stranded RNA [14]. The S protein is the main component responsible for viral-host binding and fusion to the cell membrane (Fig. 1 ). Structurally, S is a homo-trimeric protein that decorates the SARS-CoV-2 surface and binds to the host cell receptor, angiotensin-converting enzyme 2 (ACE2), to facilitate viral entry [15]. Each monomer of the spike protein consists of a signal peptide (residues: 1–13) at the N-terminal domain (NTD), S1 subunit (14–685), and S2 subunit (686–1273). The subunits S1 and S2 are responsible for receptor binding and cell fusion, respectively [15,16]. S1 is composed of two domains, a receptor binding domain (RBD) and NTD.
Fig. 1.
The structure of SARS-CoV-2 spike protein in its trimeric form (PDB ID: 6VYB) [36]. The three monomers in the left panel are shown in the surface representation of different colors (green, magenta, and cyan). The carbohydrate moieties are shown in colored sticks. The right panel shows the top view (upper) and side view (bottom) of the cartoon representation of the same structure. The receptor binding domains (RBD) are shown with only one in the up configuration (chain B) and two in the down state. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The binding of the virus to the human cell receptor, ACE2, is initiated by interactions between RBD and ACE2 (Fig. 2 ) [17]. The ACE2 peptidase domain (ACE2-PD) contains seven N-linked and one O-linked glycans at N53, N90, N103, N322, N432, N546, N690, and S155 positions [18]. On the other hand, the RBD of S contains the receptor-binding motif (RBM), a crucial functional segment as it contributes directly to the attachment to ACE2 and has physiochemical properties that affect the binding affinity [19]. The RBM consists of amino acids in the range T425:Q492 in SARS-CoV and S438:Q506 in SARS-CoV-2, with 16 and 17 residues for SARS-CoV and SARS-CoV-2 [20]. These motifs were reported to be within 4 Å of ACE2 [18]. It is well known that S of other coronaviruses undergoes a series of conformational changes during binding to ACE2, such as the slipping of S1 and refolding of S2 [21,22].
Fig. 2.
The structure of the complex between SARS-CoV-2 spike RBD (green) and the human receptor angiotensin-converting enzyme 2 (ACE2) (cyan). The left panel shows the cartoon representation, while the right panel shows the surface representation of the complex. The carbohydrate moieties are shown in magenta on both panels. The dashed-red circle indicates the cyclic region C480-C488. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The SARS-CoV-2 spike 3D structure was modeled, and its binding was predicted against different host-cell receptors [[23], [24], [25]]. Later on, in 2020, several solved structures of spike were deposited in the Protein Data Bank (PDB IDs: 6VXX, 7CAB, 6UL7, 6VYB, etc.), which shows that at least one monomer of SARS-CoV-2 spike RBD is in its “up” state [[26], [27], [28]]. Due to its pivotal role in the infection process, several studies targeted S with small molecules in an attempt to prevent the infection by blocking S-ACE2 binding [29,30].
Adem et al. studied the binding affinity of caffeic acid derivatives against SARS-CoV-2 proteins, including the spike. They used combined docking and dynamics simulations in their analysis [31]. Abosheasha et al. studied the potential use of antiplatelets against the viral spike and main protease using computational methods [32]. Basal et al. studied the Chaga medicinal mushroom Inonotus obliquus (Agaricomycetes) terpenoids against the spike using molecular docking [33]. Sarfraz et al. reviewed the studied polyphenol compounds that show potential therapeutic efficacy against SAR-CoV and SARS-CoV-2 proteins, including the spike protein. They reported Naringenin, Epigallocatechin gallate (EGCG), and Curcumin as potential interest for the SARS-CoV-2 spike [30]. Singh et al. studied plant-based bioactive compounds as inhibitors of the spike using computational methods. They found that Dicaffeoylquinic acid interacts strongly with the spike RBD at the interface with ACE2 [34]. Computational methods are also used in studying spike mutations and their interaction with ACE2 [35]. These methods include molecular docking, molecular dynamics simulation, and MM/GBSA approach.
In this review article, we will mine several studies that mainly utilized molecular simulation (at least 100 ns) to study the dynamics of the SARS-CoV-2 spike during the last two years to discuss how molecular simulations successfully elucidate the protein's behavior and function. Mutation effects, glycosylation types, and immune evasion have all been investigated by MD simulations as well as by experimental protocols. The contribution of mutations at S of different variants is also discussed based on the MD simulation findings. The current study sheds light on the effectiveness of using computational tools to fight against the COVID-19 pandemic, which can be improved to prepare for the next pandemic.
2. SARS-CoV versus SARS-CoV-2
2.1. SARS-CoV-2 versus SARS-CoV in binding ACE2
The trimeric S glycoprotein (∼150 kDa) comprises two subunits, S1 and S2, responsible for receptor binding and viral membrane fusion with host-cell membrane, respectively [37]. The work of Yan et al. revealed a similarity percentage of almost 80 % between the receptor binding domains (RBD) of SARS-CoV and SARS-CoV-2 upon binding to ACE2, with some differences in their structures at the interface [38]. Afterward, Zhang et al. presented a structure-based sequence alignment between SARS-CoV and SARS-CoV-2 RBDs and their differences in residues, with a shared sequence identity between RBDs and RBMs of both of them, is about 73.2 % and 50 % respectively [39,40]. The RBD structure of SARS-CoV-2 is more open and flexible and has a larger solvent-accessible surface area than SARS-CoV RBD. Several molecular dynamics simulations accompanied by experimental studies have been performed to understand the binding affinity differences between the two complexes. Viral infectivity and spread rates of viruses and how evolving mutations of SARS-CoV-2 variants affect its binding affinity to ACE2 have been studied [24,[41], [42], [43], [44]].
Yan et al. conducted one of these studies, which reported Cryo-electron microscopy (CryoEM) structures of the full-length human ACE2 bound with the RBD of the S protein of SARS-CoV and SARS-CoV-2 individually (PDB ID: 2AJF & 6 M17, respectively) [38]. Superimposition of the RBD in the two complexes showed a similarity with a root mean square deviation (RMSD) of 0.68 Å. There were many sequence variations at the interface with ACE2 (summarized in Table 1 ). They concluded that some of these variations might strengthen the binding affinity between SARS-CoV-2-RBD and ACE2, but others may decrease its binding affinity to ACE2 concerning SARS-CoV-RBD/ACE2 interaction. The alteration from Val404 to Lys317 may result in stronger interaction due to the formation of a salt bridge between Lys317 and Asp30 of ACE2. Moreover, due to the change of Leu472 in SARS-CoV-RBD to Phe486 in SARS-CoV-2-RBD, more hydrophobic interactions with Met82 of ACE2 have been generated which enhances the overall interaction by van der Waals forces [45]. On the other hand, the change from Arg426 to Asn439 has reduced the overall interaction by removing the salt bridge with Asp329 of ACE2 [38].
Table 1.
The structural and conformational interaction changes between SARS-CoV-RBD and SARS-CoV2-RBD upon the association interface with ACE2, according to Yan et al. 2020.
| Changing position | SARS-CoV-RBD | SARS-CoV-2-RBD |
|---|---|---|
| N terminus of α1 | Arg426 | Asn439 |
| Tyr484 | Gln498 | |
| Thr487 | Asn501 | |
| In the middle | Val404 | Lys417 |
| Tyr442 | Leu455 | |
| Leu443 | Phe456 | |
| Phe460 | Tyr473 | |
| Asn479 | Gln493 | |
| C terminus of α1 | Leu472 | Phe486 |
Similar to most viral entry scenarios, S interacts with ACE2 via non-covalent interactions where residues vary widely in terms of their contributions to the attraction or repulsion of the partner protein [46]. Rodriguez used a fragment-based quantum-chemical method to evaluate different residues' attraction and repulsion contributions at ACE2 and S surfaces. This method depends on dividing the interacting surfaces of RBD into four-residue fragments called quartets. Two fragments on ACE2 (E37, N330, K353, Q42) and (E329, N330, K353, G354) are essential in binding to viral proteins as they promote intermolecular attraction for SARS-CoV-2 and SARS-CoV, respectively. The former quartet was also found to attract S of SARS-CoV, while (D30, K31, N33, and H34) residues show strong, attractive interactions toward SARS-CoV-2 and weak repulsive ones against SARS-CoV RBD [46].
2.2. Mutations impact on spike binding to ACE2
Mutations' effects on S binding to ACE2 was a hot topic of research that adopted different computational approaches, such as computational alanine mutagenesis, thermodynamic integration, and deep learning-based statistical models [[40], [41], [42], [43], [44]]. Monte Carlo and MD simulations have investigated the binding interactions between mutant SARS-CoV-2 (or SARS-CoV) and ACE2 [47]. Based on the CryoEM structure of S-ACE2 (PDB ID: 6M17), several mutations were constructed (V404K, R426N, Y442L, L443F, F460Y, L472F, N479Q, D480S, Y484Q, and T487N) and the electrostatic potential of each mutant structure was calculated using Adaptive Poisson−Boltzmann Solver (APBS). Enhanced electrostatic interactions were a major factor for the increased binding affinities of SARS-CoV-2 S compared to the spike of SARS-CoV [47]. Much of these electrostatic interactions come from the salt bridges between S and ACE2; R426:E329 in SARS-CoV and K417:D30 in SARS-CoV-2 by a factor of 1.54. The results also showed that these mutations of SARS-CoV-2 had caused sophisticated structural changes that further enhanced the electrostatic and van der Waals energies. Much of these electrostatic interactions come from the salt bridges between S and ACE2; R426:E329 in SARS-CoV and K417:D30 in SARS-CoV-2 [47].
The observations of stronger SARS-CoV-2 S binding to ACE2 are supported by the findings of Nguyen and associates who combined coarse-grained and all-atom steered molecular dynamics (SMD) simulations to evaluate the interaction pattern and KD values [48]. Coarse-grained simulations showed the KD value of the SARS-CoV-2 RBD - ACE2 complex three times less than that of the SARS-CoV RBD-ACE2. The SMD simulations revealed that a higher rupture force and nonequilibrium work (Wneq) are required to unbind SARS-CoV-2 RBD from ACE2 compared to that SARS-CoV. The calculated binding energies between the two complexes revealed that electrostatic interaction dominates the van der Waals interaction. These results are consistent with previous studies that inferred a significant contribution of electrostatic interactions to SARS-CoV-2/ACE2 binding [49].
Indeed, computational alanine scanning and the molecular mechanics-generalized Born surface area (MM/GBSA) method of ACE2 complex with either SARS-CoV RBD and SARS-CoV-2 RBD were employed to assess binding affinities and hotspot residues. The calculations also revealed that Q24, K31, H34, Y41, Y83, and K353 as the most important hotspot residues [42]. Hotspot residues are defined as amino acids that contribute with an increase in the difference between ∆G upon alanine mutatgenesis and ∆G of the wild type (∆∆Gtotal), with a minimum cutoff of −1.5 kcal/mol [50]. The calculated binding free energies showed that SARS-CoV-2 had higher affinity than SARS-CoV (−36.2 ± 1.1 versus −33.6 ± 1.6 kcal/mol, respectively) [49]. This increase in binding energy is likely due to more polar residues at the interface region of SARS-CoV-2 RBD (∼ 45 % more than in SARS-CoV) as reported by Delgado and colleagues [40]. Interestingly, a single mutation could account for a substantial difference in binding energy. For instance, an increase in the binding free energy by −4.3 kcal/mol was found upon the introduction of the N501Y mutation at SARS-CoV-2 (Fig. 3 ) [49]. This was explained by the enhanced van der Waals interaction energy between the Y501 of the mutant SARS-CoV-2 and ACE2. In addition, the thermodynamic integration method revealed that the total binding energy was increased by −3.1 kcal/mol due to this mutation. Experimental validation of these computational approaches was done by Wrapp et al., who showed that SARS-CoV-2 has an association affinity toward ACE2 10–20 times higher than SARS-CoV [26]. Furthermore, Walls et al. reported that SARS-CoV-2 has a dissociation constant (KD) against ACE2 about four times less than SARS-CoV [42]. It should be mentioned that the weaker binding affinity of SARS-CoV RBD may be attributed to its higher flexibility in comparison to SARS-CoV-2 RBD as evidenced by higher root-mean-square of fluctuations (RMSF) experienced by SARS-CoV RBD [48].
Fig. 3.
Important mutaations on the S of SARS-CoV-2. Based on the full-length model made publicly available by Casalino et al. [60] under Creative Commons licenses.
Furthermore, Chen et al. have conducted a new deep learning method called “TopNetTree” to analyze the existing RBD mutations' impacts on the spike protein's binding free energies toward the ACE2. They systematically screened 3686 possible future mutations on all 194 residues of the RBD and then classified them according to the possibility of existence. In addition, the infectivity of SARS-CoV-2 mutations was analyzed and compared to SARS-CoV. They found that most of the mutations had small positive changes in the binding free energy, while just a small number had significant changes. In addition, by combining sequence alignment, binding free energy analysis, and probability estimation, they have found that 452, 489, 500, 501, and 505 residues of SARS-CoV-2 RBM has a high probability of mutating into more infectious strains [51].
2.3. Binding of new variants to ACE2
As in other RNA viruses, SARS-CoV-2 underwent several mutations, and variants are being reported from different regions globally. The new variants (alpha, beta, gamma, delta, and omicron) exhibit higher infectivity, disease severity, and mortality rates [[52], [53], [54], [55], [56], [57], [58]]. Several mutations in these variants affect the RBD binding to the human receptor ACE2. The mutations can be in the RBD or the NTD of the spike protein. Using computational methods, this section inspects why some variants bind more strongly to ACE2 than others.
Three mutations, N439K, S477N, and T478K, were reported to have higher binding affinity and infectivity rates (Fig. 3). Using MD simulation, protein-protein docking, and binding free energy calculations, the change in the binding interactions between SARS-CoV-2 RDB and ACE2 receptor was investigated for these mutations. First, mutants and wild-type S were docked with ACE2 receptor using HADDOCK and HDOCK algorithms. The binding affinities were calculated using MM/GBSA approach. The results showed that N439K, S477N, and T478K variants have a higher binding affinity toward the ACE2 with higher numbers of salt bridges, hydrogen bonds, and non-bonded contacts. The increased binding affinities of the mutants were attributed to better van der Waals, electrostatic, and non-polar solvation energies. The MM/GBSA calculation revealed a two-fold increase in the binding energies of the variants compared to that of the wild-type. The total binding energies of the complexes were −31.86 kcal/mol for the wildtype-ACE2 complex, −69.82 kcal/mol for S477N mutation, −69.64 kcal/mol for T478K mutation, and −67.85 kcal/mol for N439K mutation [59].
N501Y mutation had gained special interest due to its potential role in the transmission of new variants [[61], [62], [63]]. Socher et al. studied the wild-type's conformational stability, linear interaction energies, and mutated B.1.1.7 (N501Y) RBD-ACE2 complexes. MD simulation of the trimeric spike protein and RBD-ACE2 complex showed reduced flexibility for R78, L249, and T250 after the deletion of amino acids 69, 70, and 144 of the NTD of the B.1.1.7 variant. The C-terminal of the S2 cleavage site and the fusion peptide showed an increase in flexibility with an overall increase of the whole structure size of up to 3 Å compared to the wild-type. Furthermore, an increase in flexibility of the fusion peptide was related to salt bridge rearrangements made by the D614G mutation in B.1.1.7. The results also showed that insertion of N501Y mutation excluded E498 from the RBD–ACE2 interface and reduced its electrostatic interaction toward ACE2. This could explain the significant decrease in the linear interaction energy and binding affinity for B.1.1.7 variant toward ACE2 [61]. However, this mutation increases the van der Waals interaction of K356.
In contrast to the previous finding, Luan et al. investigated how the N501Y mutation within the RBD of the spike protein of SARS-CoV-2 could enhance the binding affinity toward the ACE2 receptor via MD simulations and free energy perturbation method. The simulated ACE2- RBD interface revealed how this mutation could cause conformational changes affecting the interface's binding energy. Simulations were carried out for both the RBD and ACE2-RBD complex. It is worth mentioning that the production run in this work was carried out with only residues far away from the RBD constrained to prevent rotation of the complex from the water box. They concluded that the naturally occurring N501Y mutation could enhance RBD binding affinity toward ACE2 and lead to avoidance of antibody neutralization. It was assumed that Y501 could favor the closed form of the spike protein to evade antibodies before interacting with ACE2. Moreover, RBD Y501 could form hydrophobic π-π stacking interactions with Y41 and hydrophobic interactions with the side chain of K353 of ACE2 [62]. These findings are supported by cell surface-binding assay, single-molecule force microscopy, kinetics study, and molecular dynamics simulations that found N501Y mutation to trigger a stronger interaction of SARS-CoV-2 RBD to ACE2. In addition, the MD simulations of the complex indicated that the N501Y mutation provides additional π-π and π-cation interactions that raise the binding affinity [63].
Additionally, Ou et al. studied the most dominant mutations of SARS-CoV-2 and focused on how the V367F mutation enhances the binding affinity toward the ACE2. The combined sequence alignment, MD simulations, and binding free energy calculation using MM/PBSA proved that V367F mutation decreased the ΔG energy to about 25 % compared to the wild-type spike. Also, the calculated KD of the V367F mutant was estimated to be 0.11 nM, two times lower than that of the wild-type spike. This indicates an increase in the affinity of V367F mutated RBD to the ACE2. This can be attributed to the enhanced structural stability of the RBD beta-sheet scaffold. In addition, this finding was further verified using pseudotyped virus assays, surface plasmon resonance, and receptor-ligand binding enzyme-linked immunosorbent assay [64].
Laffeber et al. have provided experimental evidence about how some SARS-CoV-2 variants bind more strongly to ACE2 than the wild type [65]. They concluded that B.1.1.7 variant (which has N501Y mutation) has an RBD with a binding affinity toward ACE2 seven times stronger than the wild type. On the other hand, the B.1.351 variant (which contains N501Y, E484K, and K417N mutations in the RBD) binds three times stronger to ACE2 than the wild type but two times weaker than B.1.1.7 variant. This is because the E484K mutation enhances the binding affinity toward ACE2 only slightly, while K417N reduces it. They also found that E484K/N501Y variant binds even stronger than B.1.1.7 variant.
3. Conformational dynamics, accessibility, and energetics of S
During viral attachment to cellular receptors, confirmational changes are experienced by at least one partner to form a stable and efficient binding. To investigate the dynamics events in S-ACE2 binding, Ping and associates performed MD simulations for 100 ns on two structures (PDB IDs): 2AJF [66] and 6M0J [67,68]. At the time of the study, the ACE2-bound SARS-CoV-2 trimer was not available yet, so they used four available structures for the ACE2-bound SARS-CoV trimers (PDB ID: 6ACG, 6ACJ, 6ACK, and 6CS2), in which all of them have one RBD up with different angles ranging from 54.8° to 84.6°, as templates to build SARS-CoV-2 trimer. This RBD angle was defined by residues D405–V622–V991. The results revealed that the SARS-CoV-2 sustained a higher binding affinity to ACE2 than the SARS-CoV, regardless of the RBD angle within the specified range.
By decomposing the ΔG to the contributing residues, they found that SARS-CoV-2 had three more residues interacting with ACE2 in comparison to SARS-CoV with a ΔG ≤ −1.0 kcal/mol, namely Y449, Q493, and G496. The contribution of Q493 was found to be −2.64 kcal/mol of the overall ΔG, while the corresponding residue in SARS-CoV N479 has no energy contribution. This shed light on the vital role of the residue Q493 variations on the binding affinity to ACE2. They mutated the interface of SARS-CoV with ACE2 (18 residues) to be like the corresponding residues of SARS-CoV-2 to examine the importance of residue variations between SARS-CoV-2 and SARS-CoV. The mutated SARS-CoV had a much stronger binding affinity than the wild-type SARS-CoV and nearly the same binding affinity as SARS-CoV-2. This result means that we can quantitatively attribute the stronger binding affinity of SARS-CoV-2 against ACE2 to these residue variations.
3.1. Spike conformational dynamics
Cryo-EM studies of S overexpressed in Z cells have identified two states; pre-fusion and post-fusion. The majority of S (97 %) were found in the pre-fusion state that has three isoforms (closed RBD (31 %), one RBD open (55 %), and two RBD open (14 %)) [69]. The pre-fusion and post-fusion isoforms both are flexible over the viral membrane due to their sparsity, giving a tilt angle between 0o and 90o. Hence, antibodies can access the stalk region that lacks high glycosylation [70].
As time passes, most of the published work focuses on the contribution of the RBD to the infection process. However, other studies showed that targeting the NTD in both MERS-CoV and SARS-CoV-2 with monoclonal antibodies 7D10 and 4A8, respectively, gave a neutralizing effect without blocking the RBD-ACE2 binding. This gave more insights into the possible role of NTD in the RBD functional conformational changes, thus might help control viral infection.
Li et al. proposed other geometrical metrics (angle and distances) to describe the movement of NTD and RBD during the simulation. These metrics were; RBD angle (θ r), the tilting poses between RBD and any other conformation; RBD distance (d r), the distance between the RBD and the center of the spike protein; and NTD distance (d n), the distance between the NTD and the center of the spike protein [16]. Four different states of S protein have been revealed with different orientations of RBDs and varying binding affinities to the ACE2 receptor. First, a closed state in which all the three RBDs in the down direction, so they cannot access the ACE2 (inactive), a partially open state with one RBD flipped upward, a semi-open state with two RBD flipped upward, and finally, the open state in which all the three RBDs flipped upward, and hence are accessible to ACE2. To reveal the functional role of the NTD, each state was simulated for 1 μs using accelerated MD with five repeats to sample the relative motion between the NTD and the RBDs. Their findings suggested that the upward RBD is not favored and tends to incline to reach the more stable favored downward orientation. The NTD tends to either interact with the RBD as a wedge to prevent this motion or detach from the RBD, allowing its transition between the upward and downward positions.
Based on the work of Peng et al., the role of the entire spike protein in determining the binding affinity was highlighted [71]. Depending on MD simulation and binding energy calculations, they studied the conformational distribution of the two spike proteins of SARS-CoV-2 and SARS-CoV, the energy barriers between the two conformations of ACE2-accessible and inaccessible, and the ACE2 accessibility and binding strength toward the S-proteins at different RBD angles. They have confirmed tha t SARS-CoV-2 RBD binds more strongly to ACE2 than SARS-CoV RBD because of the higher electrostatic interactions. With four different angles of RBD of SARS-CoV-2, which were built according to ACE2-bound SARS-CoV-S trimmers retrieved from the solved structure, they found that they all have binding affinity for ACE2 higher than that of SARS-CoV regardless of the RBD angle. In addition, they have computed the contribution of each residue of RBD to the ΔG energy. They have concluded that a SARS-CoV structure with 18 specific residue mutations is approaching the binding affinity of SARS-CoV-2 to ACE2. According to the conformational change of the pathway of SARS-CoV-2-S trimer between the down and up states, 52.2° is considered the smallest accessible RBD angle of SARS-CoV-2-S to ACE2, and with increasing the angle, the binding interaction becomes stronger. However, they found that SARS-CoV-2-S has less accessible conformations to ACE2 than SARS-CoV-S, and the inaccessible conformations cannot easily shift to the accessible state as SARS-CoV-S does. Therefore, the total binding affinity of the entire SARS-CoV-2-S protein is similar to or even lower than SARS-CoV-S toward ACE2. This is referred to as the less accessible conformations of SARS-CoV-2-S and the more difficult shift from inaccessible to accessible conformations in the solution [71].
Using insect cells as expression systems, Toelzer et al. experimentally proved that the closed state is predominant and is stabilized by the binding to linoleic acid (LA), an essential fatty acid [72]. The interaction affinity of LA against S trimer and S RBD alone was computationally assessed through repeated MD simulations. The results showed a stable LA-S trimer association. Additionally, the affinity of LA to the S trimer was higher than its binding to the RBD alone. Tian and Tao used 10 μs MD simulations to study the transition between the two S1 subunits, closed and partially open states. The RMSD values in the two states are 5.9 Å and 10.6 Å, respectively. They clarified that the closed state is relatively stable compared to the partially open state, which is dynamic, and undergoes conformational changes after one μs but stabilizes on reaching the sixth microsecond. Their analyses provided more insights into the closed-open transition probability, and a complete pathway was identified between the closed and open states [73].
3.2. The role of C480-C488 in spike dynamics
Grishin et al. discussed the role of disulfide bonds (S—S) in the stability and function of Spike protein [74]. SARS-CoV-2 S protein has 16 disulfide bridges at specific sites, which play an essential role in the stability of the protein. RBD has four S—S bonds; C480-C488 is in the loop that binds to the ACE2 receptor, while the remaining three bonds, C379-C432, C336-C361, and C391-C525, are found on the opposite side of the RBD domain with no contribution in ACE2 interaction. The MD simulations showed that the flexibility of the RBD increases when one S—S (C480-C488) or all of the disulfide bonds are reduced. This was done by simulating three systems (i) RBD that has no reduced S—S bond, (ii) RBD(−1SS) with only C480-C488 reduced, and (iii) RBD(−4SS) with all the S—S bonds reduced.
Firstly, RMSD values of RBD (−1SS) and RBD(−4SS) changed by three folds (1.0–2.5 vs 3.5–6.0 Å) in the RBD (Fig. 4 ). Secondly, RMSF values showed that the regions adjacent to the S—S bonds have higher flexibility. One of the crucial findings was that the ACE2 binding loop remains relatively stable (closed) as long as the C480-C488 is present. In the absence of C480-C488, a series of structural transitions between open states were identified and none of them was capable of ACE2 binding. By increasing the simulation temperature, efficient conformational sampling can be achieved, as they elevated the temperature to 77 °C and allowed the three systems to be simulated for two μs each. They reported an opening of the ACE2-binding loop in all three trajectories. This happened at the beginning of the simulation in both RBD(−1 SS) and RBD(−4 SS), but it happened after one μs in the non-reduced RBD as the loop kept its conformation along half of the trajectory. This confirmed the effect of S—S bonds on the stability of the RBD and its affinity toward ACE2. Experimental techniques confirmed these results by using reducing agents that break the S—S bonds. As a result, the secondary structures are affected, and the melting temperature is reduced from 52 °C to as low as 36–39 °C.
Fig. 4.
Snapshots of the RBD ACE2 binding loop at the surface conformation, residues 454–492, and their RMSD along six 37 °C molecular dynamics runs. (A) RMSD of Cα Coordinates (nm) against time (ps). RBD with 4 S-S bonds, RBD lacking C480-C488 bond – RBD (−1 SS), and RBD with all four bonds reduced – RBD (−4 SS) are colored blue, green, and red, respectively. For each of them, two curves correspond to two repeats of the simulations. (B) to the left is a ribbon representation of the Spike RBD – ACE2 complex crystal structure focusing on the loop. We showed here only a part of the structure, in olive, the Spike RBD domain is colored with wheat-colored ACE2. Cyan carbon and yellow sulfur atoms in a wire representation of The C480-C488 S—S bond. To the right are the MD simulations snapshots, which present the different ACE2 – binding loop conformations and are labeled accordingly. Copyrights were acquired from the publisher with license number 5503040177103. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Glucose-regulated protein 78 (GRP78) was reported before to be another host-cell receptor that can bind to the SARS-CoV-2 spike [25,75]. This was predicted by protein-protein docking and later confirmed experimentally [76,77]. The recognition site on the spike that was associated with GRP78 substrate binding domain β (SBDβ) was the C480-C488. The recognition of GRP78 to SARS-CoV-2 was predicted to be not specific for that strain of human coronaviruses (HCoVs) but also for other members of HCoVs [78]. Additionally, this region of the host-cell chaperone (SBDβ) was targeted with natural compounds to inhibit viral recognition by the cell-surface protein GRP78 [[79], [80], [81], [82]]. Additionally, the C480-C488 region of the spike of SARS-CoV-2 was also targeted by small molecule inhibitors [29,[31], [32], [33]].
The use of MD simulations to understand SARS-CoV-2 continued to unveil more insights about spike conformational dynamics. Williams et al. studied the role of the flexible loop (namely loop3) at the interface of spike RBD against ACE2 using MD simulations [83]. The RBD binding interface is usually comprised of four loops that tend to be flexible in both unbound and bound states; Loops 1 (438–450), 2 (455–470), 3 (471–491), and 4 (495–508). It is known that loops 3 and 4 are the most flexible regions in the RBD, while the three residues in loop 3 (F486, N487, and Y489) are known to have a stabilizing effect on the binding to ACE2. RMSD analysis following four μs-long molecular dynamics simulations showed that the RBD remains in a stable equilibrium conformation along the trajectories, with an average RMSD value of 1.39 Å. In addition, increased flexibility in residues 369–373 and the Loop 3 region from ∼471 to 491 was observed, which are part of the large ACE2 binding interface. Loop 3 is centralized around the conserved disulfide bond C480-C488. Four structures containing naturally occurring mutants, namely T478I, S477N, V483A, and G476S, were subjected to two μs-long MD simulations for each mutated structure. The backbone RMSD analysis revealed that they all remain stable relative to their starting structure on average with RMSD values: G476S: 1.65 Å; S477N: 1.48 Å; T478I: 1.46 Å; and V483A: 1.44 Å. Also, the per-residue RMSF confirmed the same, leaving us with one remark that loop3 flexibility is resilient to single mutations.
The S2 subunit of the spike is composed of a hydrophobic fusion peptide, two heptad repeats, a transmembrane region, and a cytoplasmic C-terminal tail (1242–1273). Despite the extensive work on solving spike protein 3d structures, these structures mainly showed the S1 subunit, with S2 not included or missing electron density at the C-terminal tail. However, being the least studied region of S protein, the C-terminal tail contains a conserved ER retrieval signal (KKXX), making it essential to study. In addition to its sub-cellular localization role, deletions in the cytoplasmic C-terminal tail affected the infectivity of the virus [84,85]. Despite the microseconds-long MD simulations at different temperatures, the C-terminal tail of S remained disordered [86]. This unstructured nature was also reported by circular dichroism spectroscopy.
4. Role of glycans on S and ACE2
4.1. Nature of SARS-CoV-2 S glycosylation
Glycosylation is a post-translational modification that confers additional capabilities on the modified protein. Viruses evolved to exploit the cellular machinery to add glycans to their proteins, facilitating many roles during the viral replication cycle [87,88]. Indeed, events of viral entry are initiated by molecular recognition and specific interactions between viral and host proteins (glycoproteins) [88]. Moreover, the glycosylation of viral envelope proteins plays a pivotal role in successful infection, and virion integrity since the folding and trafficking of the viral protein are affected by glycosylation. For instance, tunicamycin-mediated inhibition of spike glycosylation in SARS-CoV-2 resulted in spike-deficient virions due to the disruption of proper protein folding [42]. In various enveloped viruses, glycosylated S proteins evade the host's innate and adaptive immune responses by shielding the amino acids at the immunogenic epitopes from molecular recognition by the host's immune system [69,88].
The glycosylation of proteins is classified into two groups, depending on the linked sugar moiety to the amino acids: N-glycans and O-glycans. The former is established by covalent bonding to the amide nitrogen atom of an asparagine residue, while the O-glycosylation takes place on an oxygen atom at the side chain of serine or threonine residues. N-glycosylation sites are usually found in a sequence of NXT/S, where X is any amino acid except proline [87,88].
SARS-CoV-2 is not an exception. Its trimeric S protein has been found to harbor numerous glycosylation sequons, and 40 % of the protein is shielded by glycans [89,90]. On each protomer of the SARS-CoV-2 S protein, 22 N-glycosylation sites have been predicted, and 17 have already been confirmed experimentally (Fig. 5 ) [91,92]. Additional O-glycosylation sites were also found on the Receptor Binding Domain (RBD) [91,92]. It is worth mentioning that the nature of glycosylation of the S1 subunit varies by the host cell type and growth conditions adopted during the experimentation [91,93,94]. For instance, tandem mass spectrometry studies revealed that S1 domains of spike expressed in human embryonic kidney (HEK) 293 cells were predominantly glycosylated by the complex type N-glycans [93,94]. Another study that used the same expression system reported different findings where high-mannose glycans were the main N-glycans type [91]. However, spikes expressed in HEK293 cells have high-mannose and complex types as the dominant types, while the hybrid glycans were only found in a small proportion [90]. In the baculovirus-infected insect system, however, most glycans were high-mannose type, while hybrid glycans constituted only a small proportion [93].
Fig. 5.
Surface rendering of the glycosylated S protein (PDB id 6VYB). The spike is in open conformation where the RBD of chain B (colored in light brown) is up, and the RBD of chain C (white) is in the down conformation. Glycans are shown in blue. The upper panel is a cutaway view of the top region showing the N-glycans at positions N234 (green), N165 (cyan), and N343 (purple). Reproduced from [36] under Creative Commons licenses. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4.2. Structural roles of glycans
The spike protein needs to undergo moderate structural changes in its RBD to bind to its cellular receptor ACE2. Several MD simulation studies have suggested pivotal roles for spike glycans to mediate such structural changes. Microsecond-long MD simulations of the pre-fusion full-length glycosylated S protein elucidated N-glycans' contributions in modulating the spike's conformational dynamics [60]. During the conformational change of the RBD from the closed state “down” to the open state “up”, one or more of the RBDs emerge up to the exterior of the spike, leaving a vacant pocket that leads to the interior of the trimeric spike [95]. If water molecules fill the exposed pocket after RBD opening, the structural stability of S is believed to be markedly disrupted [96]. The N-glycans attached to the neighboring ‘down’ protomer at N165, N234 (on the NTD), and N343 (on the closed RBD) play the most critical roles regarding the structural stability of S protein in its “up” conformation [36,60,97]. Upon RBD opening, the large oligomannose at N234 has been found to fill up the exposed pocket and maintain the integrity of S via multiple hydrogen bonding with the N-terminal domain (NTD), the central helix, and the open conformation of RBD [36,60]. In the closed state, the N165 glycan is located above the RBD, while the glycan of N234 is located underneath.
Replacement of large glycans (Man7 & Man9) on N234 with a paucimannose (a Man3 short glycan) resulted in an unstable RBD opening and ∼ 60 % reduction in binding to ACE2 as a response to inefficient interactions, specifically with the disordered loop of the receptor-binding motif (RBM) [36,60]. N-glycan at N165 was also found either to fill the same pocket (filled by N234) or to make extensive interactions by acting as a bridge between NTD and the opened RBD [60]. These observations were supported by additional MD simulations of non-glycosylated mutants (N165A and N234A) in which the opened RBD explored a large conformational space compared to the glycosylated wild-type structure [60]. Principal component and angle analyses of the RBD movement in the mentioned mutants suggested a regulatory role of N-glycans in opening and closing the structure. The absence of glycans at N165 and N234 destabilized the open conformation by permitting its disordered motions. Furthermore, another MD simulation study that adopted a weighted ensemble path-sampling strategy identified N343 glycan as well as three residues (D405, R408, and D427) as major players in the RBD opening process [98]. N343 glycan was also found to initiate spike transition from closed to open conformation by acting as a gate that pushes the RBD from the ‘down’ to the ‘up’ state via intercalation (within 3.5 Å) between F456, R457, Y489, and F490 on the RBM [98]. Alanine mutagenesis of N165 and N234 or N343 prior to biolayer interferometry analysis showed a significant decrease (50–90 %) in the binding response of the mutated spike to ACE2 [60]. Additional evidence for glycans' roles in the spike structural stability was obtained through micro-second MD simulations of de-glycosylated S1 constructs (heads only) that showed an early reversion of open conformation to its closed form during the first 100 ns [97].
4.3. Glycans affect receptor binding
ACE2 is also heavily glycosylated, mostly by complex-type sugars, especially near the region interacting with the RBD of SARS-CoV-2 S glycoprotein [99]. The spike-ACE2 binding interaction has been studied computationally and experimentally, focusing on the glycans' contribution [93,97,99,100]. As mentioned previously, the stability of the RBD by glycans at N165 and N234 allows efficient binding to ACE2. Experimental evidence of such suggestions was obtained via biolayer interferometry that detected a significant reduction in binding response to ACE2 when glycans at N234, N156, or N343 were abolished by alanine mutations [60,98]. This reduction was attributed to the RBD conformational shift toward the closed state based on the increased population of spikes with the closed RBD state. These observations from MD simulation studies explain, at the atomic level, the roles of glycans in the RBD opening and the inefficient entry of virions lacking N-glycans to human cell models [101].
The nature and extent of glycans' contributions to the binding energy are not entirely clear. It is currently believed that glycans play partial roles in binding the RBD to human ACE2. The MD simulation-based comparisons of glycosylated wild-type and omicron variants in terms of binding energies have shown comparable results [102,103]. However, real-time surface plasmon resonance assays showed different dissociation constant KD values (range: 2.05–23.9 nM) for S1-ACE2 obtained from different expression systems (different glycosylation patterns), including Baculovirus-infected insect, Chinese hamster ovarian, and HEK 293 cells [93].
Steered MD simulations and biolayer interferometry experiments revealed that S glycans flexibility allows H-bonds to be formed and relaxed in a catch-slip behavior in which a Hydrogen bond is broken, and another takes its place at a larger distance [104]. These H-bonds are formed between the spike's glycans and ACE2 amino acids (glycan-protein) and/or S glycans and ACE2 glycans (glycan-glycan) [104]. On the other side, the glycan at N322 on ACE2 strengthens the binding of RBD to the receptor by −19.12 to −47.80 kcal/mol [105]. This strong binding is attributed to the architecture of the N322 conserved pocket on the RBD, which is composed of a hydrophobic core surrounded by polar and charged residues [105]. These findings explain the genetic data regarding the failure of coronaviruses to bind to receptors lacking N322 glycosylation. Additionally, N90 glycan interacts tightly with the glycans of the RBD (−9.56 to −35.85 kcal/mol), especially with the high-mannose type on N322 [105].
Strikingly, glycans near the receptor-binding regions may negatively affect the binding process. The de-glycosylated complexes showed stronger binding affinities in plasmon resonance assays as well as in MM/PBSA free energy calculations after 100–200 ns MD simulations [93]. These variations in binding affinity were linked to the observed steric hindrance between the glycosylated N-terminal regions of S1 domains and the ACE2 [93]. Moreover, high molecular size N-glycans caused more steric clashes with the cellular receptor. Furthermore, the Coulomb repulsions were also found between negatively charged residues on ACE2 and the sialic acid moieties on complex glycans [93]. Nonetheless, the glycans on coronaviruses are fewer than seen in other viruses, which is an advantage to the virus for efficient receptor binding [88].
Acharya et al. used MD simulations and binding affinity calculations to reveal the role of ACE2 glycans in the binding of SARS-CoV and SARS-CoV-2. They found that glycans at N357 and N330 positions of SARS-CoV RBD block the interaction between ACE2 glycan at N322 and SARS-CoV RBD. In contrast, the absence of a glycan at N357 of SARS-CoV RBD (N370 in SARS-CoV-2) enhances its binding to N322 glycan on ACE2 via exposing additional sites for interaction and increased stability of the RBD open conformation. Therefore, the absence of the glycan at N370 in the SARS-CoV-2 RBD may be an evolutionary step toward stronger binding to ACE2. Additionally, the N53 glycan on ACE2 plays a significant role in stabilizing the homodimer interface of ACE2. On the contrary, mutations of SARS-CoV-2 may reduce ACE2 binding affinity according to the position of the mutation. For example, the spike mutations N439 & G504 but not G404 & N437 are found in the S RBD–ACE2 interface, and both decrease ACE2 binding [106].
4.4. Immune evasion and antigenicity
The glycans on the spike are believed to shield protein epitopes from the effector immune cells and molecules; however, it is unclear how effective this shielding is on immune evasion [87,88]. Antibodies against different spike epitopes are readily elicited, and the currently effective vaccines are based on S [107]. Computational probing of different epitopes on a spike has been utilized to evaluate the effect of glycosylation on antibody binding possibilities (Fig. 6 ) [89,97]. Different structures of glycosylated spikes with small glycans (paucimannose, M3) were subjected to calculations of accessible surface area (ASA). The results showed that 69 % of the protein surface is accessible for antibodies. However, others have found complex glycans to shield approximately 44 % of the spike (56 % accessible) [89]. The glycans at the stalk region of the spike are more prevalent than in the S1 subunit and provide a more shielding effect. It is worth mentioning that most glycans on the SARS-CoV-2 spike are highly flexible, with a wide conformational space covering most of the spike's surface [97]. Indeed, the superimposition of RBD-antibody complexes on glycosylated spike proteins has shown only a few cases of potential clashes and steric hindrances [97].
Fig. 6.
The protein surface is colored according to antibody accessibility. The RBD region in the open conformation is circled in blue. Spaces occupied by glycans are shown as moss surface from MD simulations. The glycans are shown in ball-and-stick representation: M9 (green), M5 (dark yellow), hybrid (orange), complex (pink). Reproduced from [42] under Creative Commons licenses. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Four types of RBD-antibodies (B38, CR3022, H11-D4, and S309) were used to interrogate the efficiency of glycans on fully-glycosylated models to occlude epitopes on RBD [97]. The epitopes of B38 and CR3022 are irrelevant to the glycans. B38 can only bind if the open state of the RBD is attained; otherwise, it is precluded by the neighboring RBDs. However, CR3022 requires all RBDs on the spike to be in the “up” conformation. In contrast, the H11-D4 binds easily to the open RBD; however, in the closed state, the residue, N165, has weak steric clashes with the antibody but can be overcome, as seen in the experimentally solved structure (PBD ID: 6Z43). Four glycans are located around the epitope of S309; two (N331 and N343) are on the same RBD, while the other (N122 and N165) are attached to the nearby NTD. Only N343 and N122 were found to experience minor and severe steric clashes with the S309 antibody, respectively [97]. Furthermore, the N343 glycan shielding decreases during the RBD transition to the ‘up’ state, whereas the shielding role of glycans at N165 and N234 is sustained during the opening process [98]. Such observations support the belief that the glycans are not only for shielding purposes but also participate in structural and functional processes.
For different reasons, these results of antigenicity evaluation should be received with caution. First, sphere-based probing of antibody accessible surface area is expected to overestimate the contributions of glycans shielding of S protein from immune recognition as antigen-binding sites on the antibody are not necessarily spherical. Protruded regions (i.e., antigen-binding sites on the antibody) might be able to access deep epitopes. Second, the effects of glycosylation heterogeneity on lectins' abilities to recognize the spike are still unknown (see Fig. 7 ).
Fig. 7.
Accessibility of neutralizing antibody epitopes. (A) Distribution of glycans when S head structures in multiple snapshots are aligned as mosses (left). Four different epitopes targeted by neutralizing antibodies are shown in different colors (right). (B) H11-D4 epitope (green) in the RBD (left: open, right: closed), N165 glycan (red) on the neighboring NTD, and N343 glycan (violet) on the neighboring RBD. Reproduced from [97] under creative common license. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Lastly, the glycans could be recognized as “self” since human enzymes synthesized and modified them in human host cells. Nonetheless, glycan-dependent neutralizing antibodies have been identified in SARS-CoV-2 as parts of epitopes [107] as well as in human immunodeficiency virus [108].
Based on these findings, computational methods, including MD simulation, MM-GBSA, and protein-protein docking, can intensely study viral protein dynamics. This can help us understand molecular recognition and its changes due to mutations to the viral proteins. Fabricating a vaccine or designing inhibitors to that recognition can help the host cell from being a shelter for new infectious virions. Furthermore, it could be helpful in future pandemics, which could be more ferocious to humans than COVID-19.
5. Conclusion
This review article summarizes the effort spent on simulating the SARS-CoV-2 spike by many groups worldwide and how it shapes our understanding of the spike's role in viral infectivity. The receptor binding domain of the spike is the region of interest that recognizes host-cell receptors and facilitates viral entry. Loop dynamics affect the recognition by the host-cell receptors, where mutation may have a great impact. The carbohydrate moieties that decorate the spike is crucial for host-cell recognition and conformational transitions. Additionally, mutations in the RBD affect ACE2 and GRP78 binding and hence, viral recognition. The new strains have different patterns of increasing or decreasing viral recognition by affecting the spike RBD binding affinities against the host-cell receptors. Experimental data support the simulation findings throughout the lifespan of the pandemic. This paves the way for finding tangible ways to fight against future pandemics.
CRediT authorship contribution statement
J.A., A.Elg., and A.S. wrote the first draft. A.Ez. and A.Elf. revised the manuscript. All authors approve the final form.
Ethics approval and consent to participate.
Not applicable.
Consent for publication
All authors approve the final form and approve the submission.
Funding
Not applicable.
Declaration of competing interest
All the authors affirm no conflict of interest in this work.
Acknowledgments
Not applicable.
Data availability
Not applicable.
References
- 1.Leach A. 2nd edition. Prentice Hall; 2001. Molecular Modelling: Principles and Applications. [Google Scholar]
- 2.Schlick T. Springer; 2010. Molecular Modeling and Simulation: An Interdisciplinary Guide. [Google Scholar]
- 3.Dey D., Biswas P., Paul P., Mahmud S., Ema T.I., Khan A.A., et al. Natural flavonoids effectively block the CD81 receptor of hepatocytes and inhibit HCV infection: a computational drug development approach. Mol. Divers. 2022 doi: 10.1007/s11030-022-10491-9. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 4.Beerenwinkel N., Sing T., Lengauer T., Rahnenfuhrer J., Roomp K., Savenkov I., et al. Computational methods for the design of effective therapies against drug resistant HIV strains. Bioinformatics. 2005;21(21) doi: 10.1093/bioinformatics/bti654. 3943-50. [DOI] [PubMed] [Google Scholar]
- 5.Elfiky A.A. Novel guanosine derivatives as anti-hcv ns5b polymerase: a QSAR and molecular docking study. Medicinal Chemistry. 2019;15(2):130–137. doi: 10.2174/1573406414666181015152511. (Shariqah (United Arab Emirates)) [DOI] [PubMed] [Google Scholar]
- 6.Elfiky A.A., Elshemey W.M., Gawad W.A. 2′-Methylguanosine prodrug (IDX-184), Phosphoramidate prodrug (Sofosbuvir), Diisobutyryl prodrug (R7128) are better than their parent nucleotides and ribavirin in hepatitis C virus inhibition: A molecular modeling study. J. Comput. Theor. Nanosci. 2015;12(3):376–386. [Google Scholar]
- 7.Ezat A.A., Elshemey W.M. A comparative study of the efficiency of HCV NS3/4A protease drugs against different HCV genotypes using in silico approaches. Life Sci. 2019;217:176–184. doi: 10.1016/j.lfs.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Hufsky F., Lamkiewicz K., Almeida A., Aouacheria A., Arighi C., Bateman A., et al. Computational strategies to combat COVID-19: useful tools to accelerate SARS-CoV-2 and coronavirus research. Brief. Bioinform. 2021;22(2):642–663. doi: 10.1093/bib/bbaa232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shehroz M., Zaheer T., Hussain T. Computer-aided drug design against spike glycoprotein of SARS-CoV-2 to aid COVID-19 treatment. Heliyon. 2020;6(10) doi: 10.1016/j.heliyon.2020.e05278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248 doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogunyemi O.M., Gyebi G.A., Elfiky A.A., Afolabi S.O., Ogunro O.B., Adegunloye A.P., et al. Alkaloids and flavonoids from African phytochemicals as potential inhibitors of SARS-Cov-2 RNA-dependent RNA polymerase: an in silico perspective. Antivir Chem Chemother. 2020;28 doi: 10.1177/2040206620984076. 2040206620984076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abraham Peele K., Srihansa T., Krupanidhi S., Ayyagari V.S., Venkateswarulu T.C. Design of multi-epitope vaccine candidate against SARS-CoV-2: a in-silico study. J. Biomol. Struct. Dyn. 2021;39(10):3793–3801. doi: 10.1080/07391102.2020.1770127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elshemey W.M., Elfiky A.A., Ibrahim I.M., Elgohary A.M. Interference of Chaga mushroom terpenoids with the attachment of SARS-CoV-2; in silico perspective. Comput. Biol. Med. 2022;145 doi: 10.1016/j.compbiomed.2022.105478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakraborty H., Bhattacharjya S. Mechanistic insights of host cell fusion of SARS-CoV-1 and SARS-CoV-2 from atomic resolution structure and membrane dynamics. Biophys. Chem. 2020;265 doi: 10.1016/j.bpc.2020.106438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y., Wang T., Zhang J., Shao B., Gong H., Wang Y., et al. Exploring the regulatory function of the N-terminal domain of SARS-CoV-2 spike protein through molecular dynamics simulation. Advanced Theory and Simulations. 2021;4(10):2100152. doi: 10.1002/adts.202100152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malik Y.A. Properties of coronavirus and SARS-CoV-2. The Malaysian journal of pathology. 2020;42(1):3–11. [PubMed] [Google Scholar]
- 18.Acharya A., Lynch D.L., Pavlova A., Pang Y.T., Gumbart J.C. ACE2 glycans preferentially interact with SARS-CoV-2 over SARS-CoV. Chem. Commun. (Camb.) 2021;57(48):5949–5952. doi: 10.1039/d1cc02305e. [DOI] [PubMed] [Google Scholar]
- 19.Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S., et al. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24(8):1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J., Xiao T., Cai Y., Chen B. Structure of SARS-CoV-2 spike protein. Curr Opin Virol. 2021;50:173–182. doi: 10.1016/j.coviro.2021.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nassar A., Ibrahim I.M., Amin F.G., Magdy M., Elgharib A.M., Azzam E.B., et al. A review of human Coronaviruses’ receptors: the host-cell targets for the crown bearing viruses. Molecules. 2021;26(21):6455. doi: 10.3390/molecules26216455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., Elfiky A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Inf. Secur. 2020;80(5):554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Jr., et al. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369(6511):1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., Elfiky A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 2020;80(5):554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henderson R., Edwards R.J., Mansouri K., Janowska K., Stalls V., Gobeil S.M.C., et al. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol. 2020;27(10):925–933. doi: 10.1038/s41594-020-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., et al. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science. 2020;369(6504):650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamdi A.H., S, A Elfiky A. Chitosan, from crayfish wastes, as a possible therapeutic option against COVID-19; an in-silico perspective. Egyptian Journal of Aquatic Biology and Fisheries. 2022;26(2):429–441. [Google Scholar]
- 30.Sarfraz I., Rasul A., Adem Ş., Ucak I., Sarfraz A., Bukhari S.A., et al. In: Coronavirus Drug Discovery. Egbuna C., editor. Elsevier; 2022. Chapter 8 - dietary polyphenols as therapeutic agents to combat COVID-19; pp. 203–215. [Google Scholar]
- 31.Adem S., Eyupoglu V., Sarfraz I., Rasul A., Zahoor A.F., Ali M., et al. Caffeic acid derivatives (CAFDs) as inhibitors of SARS-CoV-2: CAFDs-based functional foods as a potential alternative approach to combat COVID-19. Phytomedicine. 2021;85 doi: 10.1016/j.phymed.2020.153310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abosheasha M.A., El-Gowily A.H., Elfiky A.A. Potential antiviral properties of antiplatelet agents against SARS-CoV-2 infection: an in silico perspective. J. Thromb. Thrombolysis. 2022;53(2):273–281. doi: 10.1007/s11239-021-02558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basal W.T., Elfiky A., Eid J. Chaga medicinal mushroom Inonotus obliquus (Agaricomycetes) Terpenoids may interfere with SARS-CoV-2 spike protein recognition of the host cell: A molecular docking study. Int J Med Mushrooms. 2021;23(3):1–14. doi: 10.1615/IntJMedMushrooms.2021037942. [DOI] [PubMed] [Google Scholar]
- 34.Singh R., Bhardwaj V.K., Sharma J., Kumar D., Purohit R. Identification of potential plant bioactive as SARS-CoV-2 spike protein and human ACE2 fusion inhibitors. Comput. Biol. Med. 2021;136 doi: 10.1016/j.compbiomed.2021.104631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aljindan R.Y., Al-Subaie A.M., Al-Ohali A.I., Kumar D.T., Doss C.G., Kamaraj B. Investigation of nonsynonymous mutations in the spike protein of SARS-CoV-2 and its interaction with the ACE2 receptor by molecular docking and MM/GBSA approach. Comput. Biol. Med. 2021;135 doi: 10.1016/j.compbiomed.2021.104654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harbison A.M., Fogarty C.A., Phung T.K., Satheesan A., Schulz B.L., Fadda E. Fine-tuning the spike: role of the nature and topology of the glycan shield in the structure and dynamics of the SARS-CoV-2 S. Chem. Sci. 2022;13(2):386–395. doi: 10.1039/d1sc04832e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen H.L., Lan P.D., Thai N.Q., Nissley D.A., O’Brien E.P., Li M.S. Does SARS-CoV-2 bind to human ACE2 more strongly than does SARS-CoV? J. Phys. Chem. B. 2020;124(34):7336–7347. doi: 10.1021/acs.jpcb.0c04511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Z.B., Xia Y.L., Shen J.X., Du W.W., Fu Y.X., Liu S.Q. Mechanistic origin of different binding affinities of SARS-CoV and SARS-CoV-2 spike RBDs to human ACE2. Cells. 2022;11(8) doi: 10.3390/cells11081274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delgado J.M., Duro N., Rogers D.M., Tkatchenko A., Pandit S.A., Varma S. Molecular basis for higher affinity of SARS-CoV-2 spike RBD for human ACE2 receptor. Proteins. 2021;89(9):1134–1144. doi: 10.1002/prot.26086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J., Xiao T., Cai Y., Chen B. Structure of SARS-CoV-2 spike protein. Current Opinion in Virology. 2021;50:173–182. doi: 10.1016/j.coviro.2021.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2) doi: 10.1016/j.cell.2020.02.058. 281-92 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henderson R., Edwards R.J., Mansouri K., Janowska K., Stalls V., Gobeil S.M.C., et al. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol. 2020;27(10):925–933. doi: 10.1038/s41594-020-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Juraszek J., Rutten L., Blokland S., Bouchier P., Voorzaat R., Ritschel T., et al. Stabilizing the closed SARS-CoV-2 spike trimer. Nat. Commun. 2021;12(1):244. doi: 10.1038/s41467-020-20321-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopez-Cortes G.I., Palacios-Perez M., Zamudio G.S., Velediaz H.F., Ortega E., Jose M.V. Neutral evolution test of the spike protein of SARS-CoV-2 and its implications in the binding to ACE2. Sci. Rep. 2021;11(1):18847. doi: 10.1038/s41598-021-96950-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez J.H. Attractive and repulsive residue fragments at the interface of SARS-CoV-2 and hACE2. Sci. Rep. 2021;11(1):12567. doi: 10.1038/s41598-021-91877-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amin M., Sorour M.K., Kasry A. Comparing the binding interactions in the receptor binding domains of SARS-CoV-2 and SARS-CoV. The journal of physical chemistry letters. 2020;11(12):4897–4900. doi: 10.1021/acs.jpclett.0c01064. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen H.L., Lan P.D., Thai N.Q., Nissley D.A., O’Brien E.P., Li M.S. Does SARS-CoV-2 bind to human ACE2 more strongly than does SARS-CoV? J. Phys. Chem. B. 2020;124(34):7336–7347. doi: 10.1021/acs.jpcb.0c04511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Z., Zhang J.Z.H. Quantitative analysis of ACE2 binding to coronavirus spike proteins: SARS-CoV-2 vs. SARS-CoV and RaTG13. Physical Chemistry Chemical Physics : PCCP. 2021;23(25):13926–13933. doi: 10.1039/d1cp01075a. [DOI] [PubMed] [Google Scholar]
- 50.Simoes I.C., Costa I.P., Coimbra J.T., Ramos M.J., Fernandes P.A. New parameters for higher accuracy in the computation of binding free energy differences upon alanine scanning mutagenesis on protein-protein interfaces. J. Chem. Inf. Model. 2017;57(1):60–72. doi: 10.1021/acs.jcim.6b00378. [DOI] [PubMed] [Google Scholar]
- 51.Chen J., Wang R., Wang M., Wei G.W. Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol. 2020;432(19):5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krause P.R., Fleming T.R., Longini I.M., Peto R., Briand S., Heymann D.L., et al. SARS-CoV-2 variants and vaccines. N. Engl. J. Med. 2021;385(2):179–186. doi: 10.1056/NEJMsr2105280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elfiky A.A., Ibrahim I.M. Host-cell recognition through Cs-GRP78 is enhanced in the new omicron variant of SARS-CoV-2, in silico structural point of view. J. Inf. Secur. 2022;84(5):722–746. doi: 10.1016/j.jinf.2022.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winger A., Caspari T. The spike of concern—the novel variants of SARS-CoV-2. Viruses. 2021;13(6):1002. doi: 10.3390/v13061002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie X., Liu Y., Liu J., Zhang X., Zou J., Fontes-Garfias C.R., et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021;27(4):620–621. doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- 57.Meng B., Kemp S.A., Papa G., Datir R., Ferreira I., Marelli S., et al. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the alpha variant B.1.1.7. Cell Rep. 2021;35(13) doi: 10.1016/j.celrep.2021.109292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang R., Chen J., Gao K., Wei G.W. Vaccine-escape and fast-growing mutations in the United Kingdom, the United States, Singapore, Spain, India, and other COVID-19-devastated countries. Genomics. 2021;113(4):2158–2170. doi: 10.1016/j.ygeno.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suleman M., Yousafi Q., Ali J., Ali S.S., Hussain Z., Ali S., et al. Bioinformatics analysis of the differences in the binding profile of the wild-type and mutants of the SARS-CoV-2 spike protein variants with the ACE2 receptor. Comput. Biol. Med. 2021;138 doi: 10.1016/j.compbiomed.2021.104936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Casalino L., Gaieb Z., Goldsmith J.A., Hjorth C.K., Dommer A.C., Harbison A.M., et al. Beyond shielding: the roles of Glycans in the SARS-CoV-2 spike protein. ACS Cent Sci. 2020;6(10):1722–1734. doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Socher E., Conrad M., Heger L., Paulsen F., Sticht H., Zunke F., et al. Mutations in the B.1.1.7 SARS-CoV-2 spike protein reduce receptor-binding affinity and induce a flexible link to the fusion peptide. Biomedicines. 2021;9(5) doi: 10.3390/biomedicines9050525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luan B., Wang H., Huynh T. Enhanced binding of the N501Y-mutated SARS-CoV-2 spike protein to the human ACE2 receptor: insights from molecular dynamics simulations. FEBS Lett. 2021;595(10):1454–1461. doi: 10.1002/1873-3468.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tian F., Tong B., Sun L., Shi S., Zheng B., Wang Z., et al. N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. Elife. 2021;10 doi: 10.7554/eLife.69091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ou J., Zhou Z., Dai R., Zhang J., Zhao S., Wu X., et al. V367F mutation in SARS-CoV-2 spike RBD emerging during the early transmission phase enhances viral infectivity through increased human ACE2 receptor binding affinity. J. Virol. 2021;95(16) doi: 10.1128/JVI.00617-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laffeber C., de Koning K., Kanaar R., Lebbink J.H.G. Experimental evidence for enhanced receptor binding by rapidly spreading SARS-CoV-2 variants. J. Mol. Biol. 2021;433(15) doi: 10.1016/j.jmb.2021.167058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 67.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 68.Peng C., Zhu Z., Shi Y., Wang X., Mu K., Yang Y., et al. Computational insights into the conformational accessibility and binding strength of SARS-CoV-2 spike protein to human angiotensin-converting enzyme 2. The journal of physical chemistry letters. 2020;11(24):10482–10488. doi: 10.1021/acs.jpclett.0c02958. [DOI] [PubMed] [Google Scholar]
- 69.Ismail A.M., Elfiky A.A. SARS-CoV-2 spike behavior in situ: a Cryo-EM images for a better understanding of the COVID-19 pandemic. Signal Transduct Target Ther. 2020;5(1):252. doi: 10.1038/s41392-020-00365-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588(7838):498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peng C., Zhu Z., Shi Y., Wang X., Mu K., Yang Y., et al. Computational insights into the conformational accessibility and binding strength of SARS-CoV-2 spike protein to human angiotensin-converting enzyme 2. The journal of physical chemistry letters. 2020;11(24):10482–10488. doi: 10.1021/acs.jpclett.0c02958. [DOI] [PubMed] [Google Scholar]
- 72.Toelzer C., Gupta K., Yadav S.K.N., Borucu U., Davidson A.D., Kavanagh Williamson M., et al. Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein. Science. 2020;370(6517):725–730. doi: 10.1126/science.abd3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tian H., Tao P. Deciphering the protein motion of S1 subunit in SARS-CoV-2 spike glycoprotein through integrated computational methods. J. Biomol. Struct. Dyn. 2021;39(17):6705–6712. doi: 10.1080/07391102.2020.1802338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grishin A.M., Dolgova N.V., Landreth S., Fisette O., Pickering I.J., George G.N., et al. Disulfide bonds play a critical role in the structure and function of the receptor-binding domain of the SARS-CoV-2 spike antigen. J. Mol. Biol. 2022;434(2) doi: 10.1016/j.jmb.2021.167357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Elfiky A.A., Ibrahim I.M., Amin F.G., Ismail A.M., Elshemey W.M. In: Coronavirus Disease - COVID-19. Rezaei N., editor. Springer International Publishing; Cham: 2021. COVID-19 and cell stress; pp. 169–178. [Google Scholar]
- 76.Carlos A.J., Ha D.P., Yeh D.W., Van Krieken R., Tseng C.C., Zhang P., et al. The chaperone GRP78 is a host auxiliary factor for SARS-CoV-2 and GRP78 depleting antibody blocks viral entry and infection. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shin W.J., Ha D.P., Machida K., Lee A.S. The stress-inducible ER chaperone GRP78/BiP is upregulated during SARS-CoV-2 infection and acts as a pro-viral protein. Nat. Commun. 2022;13(1):6551. doi: 10.1038/s41467-022-34065-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Elfiky A.A. SARS-CoV-2 spike-heat shock protein A5 (GRP78) recognition may be related to the immersed human coronaviruses. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.577467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Elfiky A.A. Natural products may interfere with SARS-CoV-2 attachment to the host cell. J. Biomol. Struct. Dyn. 2021;39(9):3194–3203. doi: 10.1080/07391102.2020.1761881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Elgohary A.M., Elfiky A.A., Barakat K. GRP78: A possible relationship of COVID-19 and the mucormycosis; in silico perspective. Comput. Biol. Med. 2021;139 doi: 10.1016/j.compbiomed.2021.104956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Palmeira A., Sousa E., Köseler A., Sabirli R., Gören T., Türkçüer İ., et al. Preliminary virtual screening studies to identify GRP78 inhibitors which may interfere with SARS-CoV-2 infection. Pharmaceuticals. 2020;13(6):132. doi: 10.3390/ph13060132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abu-Mahfouz A., Ali M., Elfiky A. Anti-breast cancer drugs targeting cell-surface glucose-regulated protein 78: a drug repositioning in silico study. J. Biomol. Struct. Dyn. 2022;1-15 doi: 10.1080/07391102.2022.2125076. [DOI] [PubMed] [Google Scholar]
- 83.Williams J.K., Wang B., Sam A., Hoop C.L., Case D.A., Baum J. Molecular dynamics analysis of a flexible loop at the binding interface of the SARS-CoV-2 spike protein receptor-binding domain. Proteins. 2022;90(5):1044–1053. doi: 10.1002/prot.26208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Giri R., Bhardwaj T., Shegane M., Gehi B.R., Kumar P., Gadhave K., et al. Understanding COVID-19 via comparative analysis of dark proteomes of SARS-CoV-2, human SARS and bat SARS-like coronaviruses. Cellular and Molecular Life Sciences : CMLS. 2021;78(4):1655–1688. doi: 10.1007/s00018-020-03603-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dieterle M.E., Haslwanter D., Bortz R.H., 3rd, Wirchnianski A.S., Lasso G., Vergnolle O., et al. A replication-competent vesicular stomatitis virus for studies of SARS-CoV-2 spike-mediated cell entry and its inhibition. Cell Host Microbe. 2020;28(3):486–96 e6. doi: 10.1016/j.chom.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kumar P., Bhardwaj T., Garg N., Giri R. Microsecond simulations and CD spectroscopy reveals the intrinsically disordered nature of SARS-CoV-2 spike-C-terminal cytoplasmic tail (residues 1242-1273) in isolation. Virology. 2022;566:42–55. doi: 10.1016/j.virol.2021.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vigerust D.J., Shepherd V.L. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 2007;15(5):211–218. doi: 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Watanabe Y., Bowden T.A., Wilson I.A., Crispin M. Exploitation of glycosylation in enveloped virus pathobiology. Biochimica et Biophysica Acta General Subjects. 2019;1863(10):1480–1497. doi: 10.1016/j.bbagen.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Grant O.C., Montgomery D., Ito K., Woods R.J. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Sci. Rep. 2020;10(1):14991. doi: 10.1038/s41598-020-71748-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369(6501):330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shajahan A., Supekar N.T., Gleinich A.S., Azadi P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 2020;30(12):981–988. doi: 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sanda M., Ahn J., Kozlik P., Goldman R. Analysis of site and structure specific core fucosylation in liver cirrhosis using exoglycosidase-assisted data-independent LC-MS/MS. Sci. Rep. 2021;11(1):23273. doi: 10.1038/s41598-021-02838-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang N., Perez P., Kato T., Mikami Y., Okuda K., Gilmore R.C., et al. SARS-CoV-2 infection of the oral cavity and saliva. Nat. Med. 2021;27(5):892–903. doi: 10.1038/s41591-021-01296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang L., Richards A., Barrasa M.I., Hughes S.H., Young R.A., Jaenisch R. Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proc. Natl. Acad. Sci. U. S. A. 2021;118(21) doi: 10.1073/pnas.2105968118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2) doi: 10.1016/j.cell.2020.02.058. 281-92.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wrobel A.G., Benton D.J., Xu P., Roustan C., Martin S.R., Rosenthal P.B., et al. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 2020;27(8):763–767. doi: 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Choi A., Koch M., Wu K., Chu L., Ma L., Hill A., et al. Safety and immunogenicity of SARS-CoV-2 variant mRNA vaccine boosters in healthy adults: an interim analysis. Nat. Med. 2021;27(11):2025–2031. doi: 10.1038/s41591-021-01527-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sztain T., Ahn S.H., Bogetti A.T., Casalino L., Goldsmith J.A., Seitz E., et al. A glycan gate controls opening of the SARS-CoV-2 spike protein. Nat. Chem. 2021;13(10):963–968. doi: 10.1038/s41557-021-00758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gong S.Y., Chatterjee D., Richard J., Prevost J., Tauzin A., Gasser R., et al. Contribution of single mutations to selected SARS-CoV-2 emerging variants spike antigenicity. Virology. 2021;563:134–145. doi: 10.1016/j.virol.2021.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mehdipour A.R., Hummer G. Dual nature of human ACE2 glycosylation in binding to SARS-CoV-2 spike. Proc. Natl. Acad. Sci. U. S. A. 2021;118(19) doi: 10.1073/pnas.2100425118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang L., Liu S., Liu J., Zhang Z., Wan X., Huang B., et al. COVID-19: immunopathogenesis and Immunotherapeutics. Signal Transduct Target Ther. 2020;5(1):128. doi: 10.1038/s41392-020-00243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nguyen L.H., Joshi A.D., Drew D.A., Merino J., Ma W., Lo C.H., et al. Self-reported COVID-19 vaccine hesitancy and uptake among participants from different racial and ethnic groups in the United States and United Kingdom. Nat. Commun. 2022;13(1):636. doi: 10.1038/s41467-022-28200-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Coderc de Lacam E.G., Blazhynska M., Chen H., Gumbart J.C., Chipot C. When the dust has settled: calculation of binding affinities from first principles for SARS-CoV-2 variants with quantitative accuracy. J. Chem. Theory Comput. 2022;18(10):5890–5900. doi: 10.1021/acs.jctc.2c00604. [DOI] [PubMed] [Google Scholar]
- 104.Huang J., Liu J., Tian R., Liu K., Zhuang P., Sherman H.T., et al. Correction: Huang et al. a next generation sequencing-based protocol for screening of variants of concern in autism spectrum disorder. Cells. 2022;11(20) doi: 10.3390/cells11010010. 3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mehdipour A.R., Hummer G. Dual nature of human ACE2 glycosylation in binding to SARS-CoV-2 spike. Proc. Natl. Acad. Sci. 2021;118(19) doi: 10.1073/pnas.2100425118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Acharya A., Lynch D.L., Pavlova A., Pang Y.T., Gumbart J.C. ACE2 glycans preferentially interact with SARS-CoV-2 over SARS-CoV. Chem. Commun. 2021;57(48):5949–5952. doi: 10.1039/d1cc02305e. [DOI] [PubMed] [Google Scholar]
- 107.Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583(7815):290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 108.Blazkova J., Refsland E.W., Clarridge K.E., Shi V., Justement J.S., Huiting E.D., et al. Glycan-dependent HIV-specific neutralizing antibodies bind to cells of uninfected individuals. J. Clin. Invest. 2019;129(11):4832–4837. doi: 10.1172/JCI125955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.