Abstract
The first oral drug for the treatment of COVID-19, Paxlovid, has been authorized; however, nirmatrelvir, a major component of the drug, is reported to be associated with some side effects. Moreover, the appearance of many novel variants raises concerns about drug resistance, and designing new potent inhibitors to prevent viral replication is thus urgent. In this context, using a hybrid approach combining machine learning (ML) and free energy simulations, 6 compounds obtained by modifying nirmatrelvir were proposed to bind strongly to SARS-CoV-2 Mpro. The structural modification of nirmatrelvir significantly enhances the electrostatic interaction free energy between the protein and ligand and slightly decreases the vdW term. However, the vdW term is the most important factor in controlling the ligand-binding affinity. In addition, the modified nirmatrelvir might be less toxic to the human body than the original inhibitor.
Graphical abstract
1. Introduction
The outbreak of SARS-CoV-2 caused the COVID-19 pandemic, which has created a huge issue for community health and the global economy since December 2019 [1,2]. The virus rapidly transmits among humans through aerosols [3,4]. Some vaccines were authorized for preventing infection [5]; however, the vaccine effectiveness was significantly reduced with the appearance of SARS-CoV-2 novel variants [[6], [7], [8], [9], [10], [11]]. Discovering novel compounds to treat SARS-CoV-2 is thus of great interest. SARS-CoV-2 main protease (Mpro or 3CL pro) is a high-profile drug target for preventing viral replication and proliferation [[12], [13], [14], [15], [16], [17], [18], [19], [20]]. Nirmatrelvir, which was approved for use as an oral drug for COVID-19 treatment by the U.S. Food and Drug Administration (FDA) [21], is a SARS-CoV-2 Mpro inhibitor. Although the drug is highly effective, there are concerns about its effectiveness against novel SARS-CoV-2 variants.
Currently, searching for potential inhibitors for an enzymatic target can be rapidly and accurately completed via computational work [[22], [23], [24], [25], [26], [27], [28]]. The binding free energy between protein‒ligand, , is an important quantity to obtain because this metric relates to the experimental inhibition constant via formula , where is the absolute temperature and is the gas constant. Several approaches have been designed to calculate the value. Among them, the double-annihilation binding free method [29] is known as one of the most accurate methods. The approach is based on free energy perturbation (FEP) calculations [30] in which the ligand is annihilated twice in two systems, including the solvated complex and ligand in solution. However, FEP calculation is very expensive in terms of computing resources. In addition, the fast pulling of ligand (FPL) approach [31], an affordable method, is employed to rapidly rank the binding affinity of SARS-CoV-2 Mpro inhibitors with a high correlation coefficient [32,33]. A combination of FPL and FEP can rapidly and accurately evaluate a potential inhibitor for SARS-CoV-2 Mpro. Moreover, traditionally, modifying the chemical structure of a current inhibitor can lead to a stronger binding compound. In computational work, the chemical structure alteration of an inhibitor in binding mode with a protein target can be predicted via a deep learning model [34] with the expectation that the ligand-binding affinity will be improved.
It should be noted that DeepFrag, FPL, and FEP are often used independently to construct and evaluate compounds, but combining them to design and evaluate inhibitors for the SARS-CoV-2 Mpro was not previously done. Therefore, in this context, we proposed to use a combination of a deep learning model and atomistic simulations to modify the chemical structure of nirmatrelvir in the binding mode with SARS-CoV-2 Mpro with the hope of finding a modified compound with a stronger ligand-binding affinity. DeepFrag [34,35], a deep learning model based on the convolutional neural network, was thus employed to modify nirmatrelvir. The ligand-binding affinity of the altered compounds was then refined via the FPL approach [31]. Ligands with a stronger ligand-binding affinity than nirmatrelvir were continuously modified via DeepFrag. The ligand-binding affinity to SARS-CoV-2 Mpro was refined via the FPL approach [31]. The process was repeated until no improved ligand was found. The stronger ligand-binding affinity was finally confirmed by using FEP calculations [30,36]. Overall, 6/83 modified compounds were found to be able to bind to SARS-CoV-2 Mpro with a stronger binding affinity than nirmatrelvir.
2. Materials and methods
2.1. Modifying the ligand structure
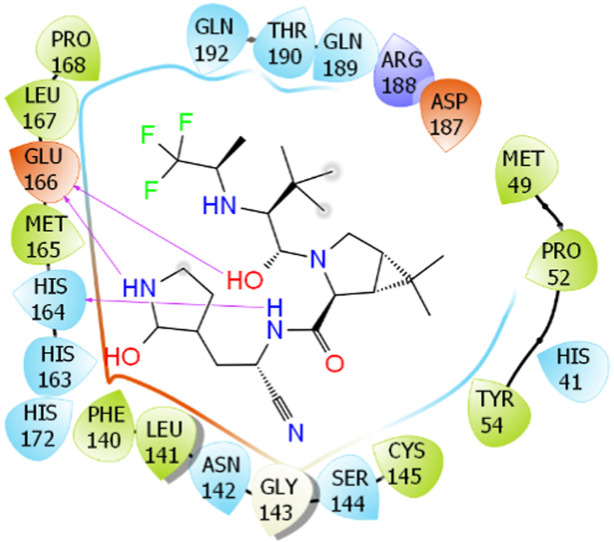
DeepFrag [34,35], a deep learning model based on a convolutional neural network, was utilized to change the ligand structure such that the ligand-binding affinity was improved. In particular, the complex structure of SARS-CoV-2 Mpro with nirmatrelvir was obtained from the Protein Data Bank (PDB) with ID 7VH8 [37] and MD simulations, respectively. The complexed structure was employed as an initial conformation of the DeepFrag model. The PDB files of SARS-CoV-2 Mpro and nirmatrelvir/modified nirmatrelvir were uploaded to the DeepFrag webserver (https://durrantlab.pitt.edu/deepfrag/), as shown in Fig. 1 . Among these, the inhibitor atoms were designated to estimate that the ligand-binding affinity can be enhanced if they are replaced by another chemical group. A probable modification was kept if the score of the DeepFrag model was larger than 0.90 [38].
Fig. 1.
Binding pose of nirmatrelvir to SARS-CoV-2 Mpro (PDB ID: 7VH8) shown by the Deepfrag web application. The yellow ball indicates the selected carbon that will be estimated for the potential of chemical replacement. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.2. Molecular dynamics simulation
The conventional MD simulations were performed using GROMACS version 2019.6 with NVIDIA GPUs supported via CUDA [39]. The Amber99SB-iLDN force field [40], TIP3P [41], and GAFF [42] were employed to parameterize for protein + ion, water molecule, and ligand, respectively. The ligand parameter was obtained using AmberTools18 [43] and ACPYPE [43] approaches. In particular, ligand information was provided by quantum chemical calculations using the hybrid functional B3LYP with 6–31G (d,p) at the level of theory. The quantum calculation was performed using the implicit solvent, . The atomic charges of the ligand were obtained from the outcome of quantum calculation via the restrained electrostatic potential method [42].
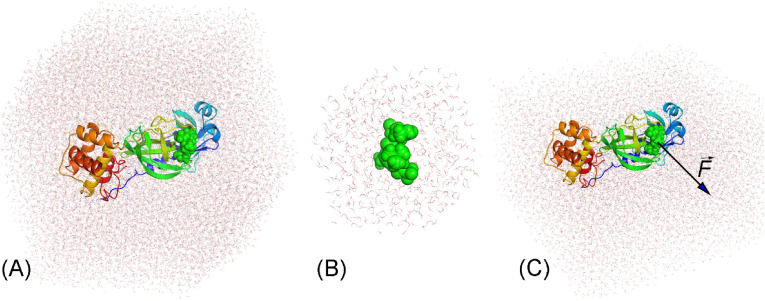
The SARS-CoV-2 Mpro + inhibitor complex was inserted into a dodecahedron and rectangular periodic boundary condition (d/rPBC) box with volumes of ca. 673 and 503 nm3, respectively. The corresponding systems consist of ca. 66 700 and 50 200 atoms (cf. Fig. 2 A and C), respectively. Moreover, the inhibitor was also inserted into a dPBC box with a volume of ca. 25 nm3 (cf. Fig. 2B). The solvated ligand system comprises ca. 2400 atoms. The MD simulations were carried out with the parameters referred to in previous work [32].
Fig. 2.
Initial conformation of simulated systems. (A) The SARS-CoV-2 Mpro + inhibitor complex was inserted into the dPBC box. (B) The inhibitor was inserted into the dPBC box. The equilibrium structures of the SARS-CoV-2 Mpro + inhibitor and individual inhibitor in dPBC box systems were then used as the initial conformation of the FEP calculations. (C) The SARS-CoV-2 Mpro + inhibitor complex was inserted into the rPBC box. The equilibrium shapes of the SARS-CoV-2 Mpro + inhibitor in the rPBC box were then used as the initial conformation of the FPL calculations. In particular, the ligand was then forced to mobilize out of the binding cavity via external force during SMD simulations.
In the first step, the solvated system was minimized via the steepest descent method. In the second step, the minimized systems in the d/rPBC box were then relaxed via 100 ps of NVT and 100 ps/20 ns of NPT simulations. The atoms were positionally restrained during relaxation simulations. Finally, MD simulations with lengths of 20 ns and 5 ns were then performed to obtain equilibrium snapshots of the SARS-CoV-2 Mpro + inhibitor in the dPBC box and isolated inhibitor, respectively. The simulations were repeated 4 times to guarantee sufficient sampling.
2.3. Fast pulling of ligand (FPL) scheme
The last snapshot of the SARS-CoV-2 Mpro + inhibitor in the rPBC box was utilized as the initial shape of the steered-MD simulations. An inhibitor was dissociated from the binding site of SARS-CoV-2 Mpro via an external harmonic force. Among these, the force cantilever spring constant and pulling velocity were set to 600 kJ mol-1 nm-2 and 0.005 nm ps-1, respectively, according to previous work [33]. The pulling work was calculated for predicting the ligand-binding affinity because it is related to the binding free energy through the Jarzynski equality [44]. is calculated as follows:
| (1) |
The predicted binding free energy can be estimated via the following formula:
| (2) |
where the slope and the intersection were obtained using a linear regression for 11 complexes in the previous work [33].
2.4. FEP calculation
The last snapshot of the solvated complex in the dPBC box and ligand in solution systems were used as initial conformations for FEP calculations according to previous work [32]. In particular, the ligand was removed from two systems including the solvated complex and ligand in solution via -alteration simulations [29,30]. The coupling parameter changes from 0, corresponding to the bound state, to 1, corresponding to the unbound state. In particular, 8 values of , including 0.00, 0.10, 0.20, 0.35, 0.50, 0.65, 0.80, and 1.00, were used to modify the electrostatic interactions. Nine values of , involving 0.00, 0.10, 0.25, 0.35, 0.50, 0.65, 0.75, 0.90, and 1.00, were used to alter the van der Waals (vdW) interactions. The free energy alteration, , matches the work of the ligand-demolishing process. The value can be calculated using the Bennett acceptance ratio (BAR) method [45]. The between SARS-CoV-2 Mpro and the inhibitor is thus obtained as follows:
| (3) |
2.5. Analysis tools
The ChemAxon webserver, www.chemicalize.com, was used to predict the ligand protonation states [46]. RMSD value and clustering calculation were computed through the GROMACS tool “gmx rms” and “gmx cluster”, respectively [39]. The 2D ligand interaction diagram was generated via the free version of Maestro [47]. The toxicity of ligands was predicted via the PreADMET webserver [48].
3. Results and discussion
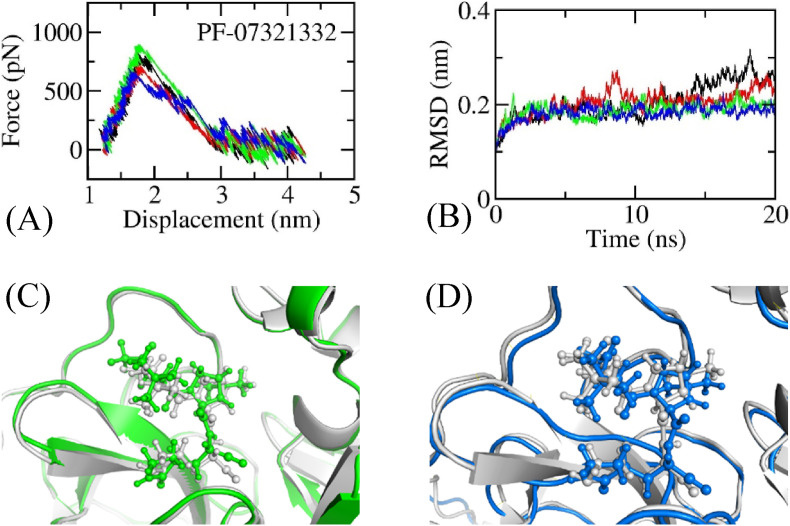
As mentioned above, altering the chemical structure of the current inhibitor can lead to a stronger binding compound. We thus used computational approaches, including physics- and knowledge-based methods, to change the chemical structure of nirmatrelvir with the expectation that the ligand-binding affinity would be enhanced. Atomistic simulations were employed to study the binding process of nirmatrelvir to SARS-CoV-2 Mpro (Fig. 3 ), and its calculation results were used as a positive control. Initially, nirmatrelvir was dissociated from the binding cavity of the protease via the FPL scheme to assess the ligand-binding affinity (Fig. 3A). The FEP calculations were then performed to calculate the absolute binding free energy, in which the MD simulations were carried out to reach the equilibrium states of the solvated complex (Fig. 3B). The complex structure changed only slightly during atomistic simulations (Fig. 3C + D), implying the stability of the simulation results. The calculated ligand-binding affinities were reported in Table 1 . In particular, the binding free energy of nirmatrelvir to the protease predicted by FPL simulations was −11.32 kcal mol-1 via Eq. (2) [33], whereas the rupture force and pulling work over 4 independent trajectories were 788.3 ± 38.1 pN and 103.7 ± 3.6 kcal mol-1, respectively. Moreover, the absolute binding free energy was also calculated as −12.62 ± 1.96 kcal mol-1 via FEP calculation. The obtained results are in good agreement with the experimental binding free energy of nirmatrelvir to SARS-CoV-2 Mpro, which is −13.53 kcal mol-1 ( nM) [49]. Furthermore, is underestimated because the linear fit of Eq. (2) was estimated by using an approximation that is equal to .
Fig. 3.
The equilibrium structure of the SARS-CoV-2 Mpro + inhibitor in the rPBC box was used as the initial conformation of the FPL calculation. In particular, pulling force versus displacement of nirmatrelvir over unbinding pathways (A), in which four colors imply the various trajectories; The SARS-CoV-2 Mpro + inhibitor in dPBC box was relaxed over MD simulations, in which all-atom RMSD of the complex SARS-CoV-2 Mpro + nirmatrelvir over 4 MD trajectories (B); (C) Superposition between experimental (gray) and SMD-initial (green) structures of SARS-CoV-2 Mpro + nirmatrelvir; (D) Superposition between experimental (gray) and MD-refined (blue) structures of SARS-CoV-2 Mpro + nirmatrelvir. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Table 1.
Top-lead compounds showed the largest binding affinity to SARS-CoV-2 Mpro by FPL, FEP, and toxicity calculations.a
| N0 | Name | b | Toxicity | |||||
|---|---|---|---|---|---|---|---|---|
| 1 | PF_3_4_62 | 1032.2 ± 33.5 | 146.5 ± 5.2 | −13.71 | −2.33 | −11.65 | −13.98 ± 1.07 | Low |
| 2 | PF_3_4_32 | 978.9 ± 57.6 | 144.5 ± 4.1 | −13.60 | −5.48 | −10.60 | −16.08 ± 1.33 | Low |
| 3 | PF_3_4_33_32 | 927.2 ± 39.3 | 142.2 ± 6.2 | −13.47 | −4.44 | −10.46 | −14.91 ± 1.08 | Low |
| 4 | PF_3_4_33_31a | 908.0 ± 63.5 | 134.7 ± 8.1 | −13.05 | −2.73 | −9.78 | −12.51 ± 0.34 | Low |
| 5 | PF_12_9_31 | 940.2 ± 15.4 | 133.5 ± 2.2 | −12.99 | −1.53 | −10.34 | −11.86 ± 0.85 | Low |
| 6 | PF_3_4_7 | 957.6 ± 44.0 | 131.2 ± 3.0 | −12.86 | −4.30 | −8.87 | −13.17 ± 0.38 | Low |
| 7 | PF_9a_33 | 882.6 ± 56.9 | 130.1 ± 7.3 | −12.80 | −4.15 | −10.67 | −14.82 ± 1.13 | Low |
| 8 | PF_3_4_33 | 922.9 ± 28.1 | 128.5 ± 3.4 | −12.70 | −2.65 | −10.39 | −13.04 ± 1.44 | Low |
| 9 | PF_12_9_31_32 | 836.6 ± 28.1 | 126.8 ± 3.9 | −12.61 | −3.80 | −10.52 | −14.32 ± 0.95 | Low |
| 10 | PF_3_4_32_32 | 911.3 ± 24.8 | 126.3 ± 3.8 | −12.58 | −4.34 | −11.07 | −15.41 ± 0.21 | Low |
| 11 | PF_12_9_3 | 906.7 ± 34.0 | 125.8 ± 4.6 | −12.55 | −5.39 | −12.69 | −18.08 ± 1.30 | Low |
| 12 | PF_12_33_3 | 882.7 ± 12.2 | 125.4 ± 5.5 | −12.53 | −7.07 | −10.19 | −17.26 ± 0.64 | Low |
| 13 | PF_3_4_31 | 908.5 ± 30.7 | 124.5 ± 4.6 | −12.48 | −2.04 | −9.70 | −11.74 ± 1.21 | Low |
| 14 | PF_3_4_31_32 | 863.1 ± 45.1 | 123.3 ± 6.1 | −12.42 | 0.78 | −9.96 | −9.18 ± 1.55 | Low |
| 15 | PF-07321332 | 788.3 ± 38.1 | 103.7 ± 3.6 | −11.32 | −1.41 | −11.21 | −12.62 ± 1.96 | Medium |
The units of force and energy are pN and kcal mol-1, respectively.
Predicted binding free energy can be calculated via Eq. (2).
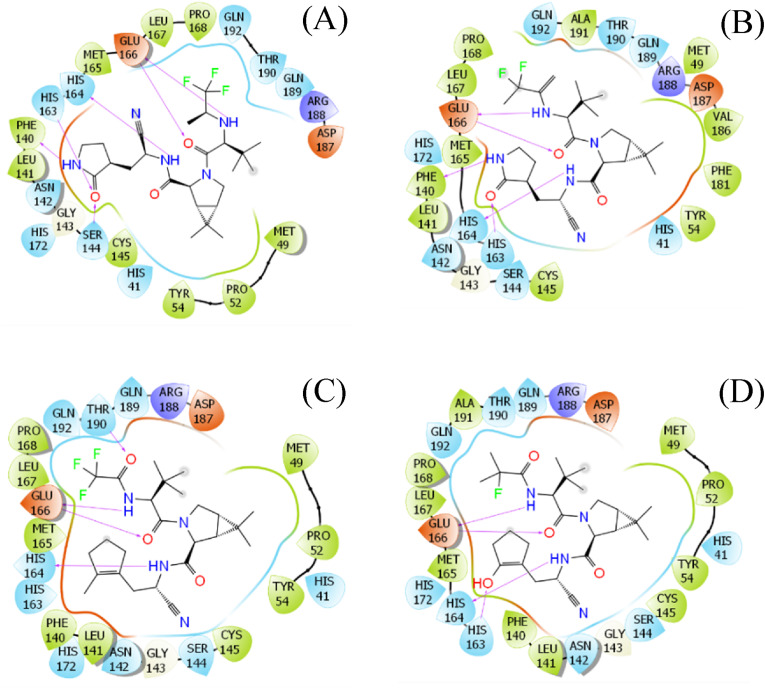
DeepFrag calculations were designed to predict the structural change of the inhibitor that may enhance the ligand-binding affinity [34,35]. In particular, the ML model was successfully applied to the SARS-CoV-2 Mpro + inhibitor [38]. The ligand-binding affinity was then confirmed via FPL simulations [38], which revealed a correlation coefficient with the respective experiment of [32]. The hybrid approaches were thus utilized to modify the structure of nirmatrelvir, which may lead to a novel compound forming stronger binding affinity to the protease. In the first round, the experimental structure of SARS-CoV-2 Mpro + nirmatrelvir was used as an initial shape for DeepFrag calculation. Nine chemical groups were suggested to be modified (Tables S1 and S7 of the Supporting Information - SI file). The ligand-binding affinity of nine compounds was then computed over 4 independent FPL trajectories. The obtained results are reported in Table S1 of the SI file, in which the recorded pulling force over the ligand displacement is described in Table S6 of the SI file. Five compounds were found to be able to form a larger binding affinity, which ranges from −11.52 to −12.35 kcal mol-1, compared to the original inhibitor. Moreover, the clustering method was employed to characterize the representative conformation of the complex over 4 final snapshots of NPT simulations with a cutoff of 0.2 nm. The 2D interaction diagram between the protease and ligand was then prepared and is shown in Fig. 4 and Table S7 of the SI file. The diagrams reveal that the vdW interaction may dominate over electrostatic interactions in the ligand binding process because a larger number of residues formed vdW interactions than hydrogen bonds (HBs).
Fig. 4.
Ligand interaction diagrams of MD-refined structures of SARS-CoV-2 Mpro + PF_9b (A), PF_9a_33 (B), PF_3_4_62 (C), and PF_3_4_33_32 (D). The diagram was prepared via Maestro free version.
Based on five modified compounds, DeepFrag/FPL calculations were continuously performed, and 18 modified compounds were thus generated (cf. Tables S2 and S6). The interaction diagram of these ligands with SARS-CoV-2 Mpro was also analyzed and is described in Table S7. The interaction picture is consistent with the 9 compounds above. Interestingly, 5/18 compounds showed a strong binding affinity, ranging from −12.03 to −12.80 kcal mol-1 (Table S2). These ligands were selected as templates for the next round of DeepFrag/FPL calculations. Therefore, in the next step, 23 altered ligands were estimated to be able to adopt a strong binding affinity to SARS-CoV-2 Mpro. The results of FPL simulations were obtained and mentioned in Tables S3 and S6 of the SI file. Among these, 8/23 compounds adopted strong binding affinity with , which ranges from −12.48 to −13.71 kcal mol-1. Furthermore, the representative conformation of the complex was also characterized, and then the ligand-interaction diagram was produced (cf. Table S7) to clarify the nature of binding. The interaction picture was not changed in comparison with the ligands considered above. The ligand-interaction maps of the representative ligands are shown in Fig. 4, in which the electrostatic interaction is only a weak driving force in the ligand-binding process.
In the next step, 29 modified compounds (Tables S4 and S6) were generated based on 8 compounds obtained in the previous step (Table S3). The ligand-binding affinities of these ligands were also calculated via FPL simulations. Stable conformations of the complexes during simulations were characterized via the clustering method. The 2D ligand-interaction diagrams of these compounds were then produced and are described in Table S7. There are only 2/29 modified compounds that can form a strong binding affinity to SARS-CoV-2 Mpro compared to nirmatrelvir (PF-07321332) (Table S4). Four altered compounds were suggested by the DeepFrag model in the next steps; however, FPL simulations imply that these compounds adopted a weaker binding affinity (Tables S5 and S6 of the SI file). The binding pose of 33 compounds to SARS-CoV-2 Mpro was also analyzed and is shown in Table S7. Therefore, the improvement process was stopped. In total, 14 out of 83 modified compounds were generated, which can inhibit SARS-CoV-2 Mpro with a value of ranging from −12.42 to −13.71 kcal mol-1.
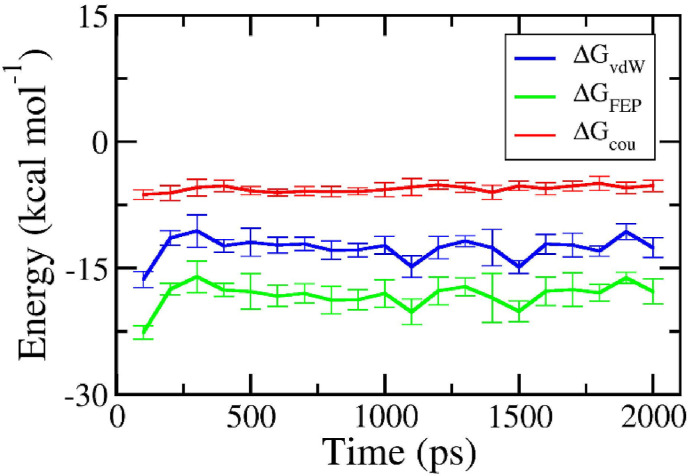
Although the FPL simulations formed an appropriate outcome compared with the respective experiment since [32], perturbation simulations were also performed to confirm the obtained outcome. It should be noted that the binding free energy obtained by FEP calculation correlates strongly with the respective experiment with a value of [32]. As mentioned above, the solvated complex and ligand in solution systems were mimicked over 20 and 5 ns of MD simulations. Both systems reach equilibrium after half of an MD trajectory, and the final snapshots of the simulations were thus used as the starting shape for the -alteration simulations. The ligand was changed from the bound state, fully interacting with surrounding molecules, to the unbound state, having no interaction with neighboring atoms. The free energies of two annihilation processes were thus obtained and described in Fig. 5 . Because the system changed from the full-interaction state to the modified-interaction state, the annihilation free energy fluctuates greatly over the beginning of the -alteration simulations. However, the free energy reaches a stable point within half of the MD simulations. The binding free energy is thus calculated over the equilibrium interval of the -alteration simulations.
Fig. 5.
The interaction free energy between PF_12_9_3 and SARS-CoV-2 Mpro via FEP calculations. Each circle corresponds to the average over 4 independent trajectories of the free energy computed via the BAR method over 100 ps of -alteration simulations. The absolute binding free energy is the sum of and . The error is the standard error of the average.
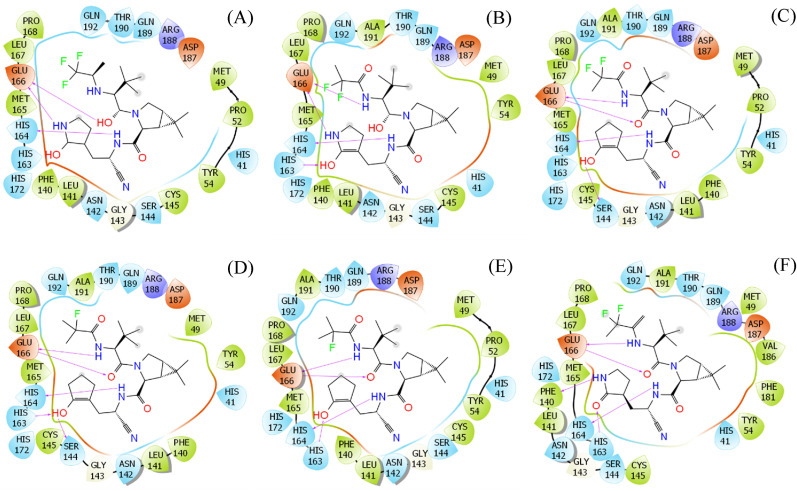
The binding free energy of the top-lead compounds ranges from −9.18 to −18.08 kcal mol-1 (cf. Table 1). In particular, the average electrostatic and vdW interaction free energies are −3.39 and −10.54 kcal mol-1, respectively. The obtained results are consistent with the data of nirmatrelvir, with corresponding values of −1.41 and −11.21 kcal mol-1. Moreover, the absolute binding free energy of nirmatrelvir to the protease is −12.62 ± 1.96 kcal mol-1, which is in good agreement with previous work ( kcal mol-1) [49]. Furthermore, among these, 6 compounds adopted a stronger binding affinity to SARS-CoV-2 Mpro than the original nirmatrelvir. Over these compounds, the average electrostatic and vdW interaction energies are −5.15 and −10.95 kcal mol-1, respectively. The structural change in the proposed ligands significantly increases the electrostatic interaction between the protein and ligand. The vdW interaction among molecules was slightly decreased with an amount of 0.26 kcal mol-1. Although the vdW interaction still dominates over coulomb interactions, the structural change of ligands reduces the energy gap. Furthermore, the increase in electrostatic interaction energy can be explained by the increase in the number of HBs. HB is normally adopted between a partially negatively charged atom and a partially positively charged atom. Therefore, an increase in the number of HBs implies that the electrostatic interaction becomes stronger [50]. Indeed, the 6 modified compounds form a 4.17 ± 0.28 HBs to the protease, which is significantly larger than that of the original nirmatrelvir. This observation is in good agreement with the ligand-interaction diagram (cf. Fig. 6 ).
Fig. 6.
The ligand-interaction diagram between PF_12_9_3 (A), PF_12_33_3 (B), PF_3_4_32 (C), PF_3_4_32_32 (D), PF_3_4_33_32 (E), and PF_9a_33 (F). The diagram was prepared via Maestro free version.
Although nirmatrelvir is very effective in decreasing death or hospitalization compared to placebo, it can cause severe side effects. Consistently, nirmatrelvir was shown to be medium in terms of predicted toxicity (Table 1). Fortunately, all of the top-lead compounds might be less toxic to the human body than the original one (Table 1). More details on the toxicity prediction of the modified and original nirmatrelvir can be accessed in Table S8 of the SI file.
4. Conclusions
In this work, a combined DeepFrag/atomistic simulation approach was proposed to predict the structural change of the compound PF-07321332 to increase the ligand-binding affinity to SARS-CoV-2 Mpro. In particular, the DeepFrag model was used to predict the structural change with the expectation that the ligand-binding affinity would be enhanced. The FPL simulations were then applied to refine the ML predictions. The FEP calculations were finally utilized to confirm the ML/FPL results.
Our ML/FPL calculations suggested that 14/83 modified compounds could inhibit SARS-CoV-2 Mpro with a stronger ligand-binding affinity than nirmatrelvir. The obtained results for were the range of −12.42 to −13.71 kcal mol-1. FEP calculations then confirmed that 6 modified compounds exhibit a smaller binding free energy, including PF_12_9_3, PF_12_33_3, PF_3_4_32, PF_3_4_32_32, PF_3_4_33_32, and PF_9a_33. The ranges from −14.82 to −18.08 kcal mol-1, with an average value of −16.09 kcal mol-1.
The structural modification of nirmatrelvir significantly enhances the electrostatic interaction free energy between the protein and ligand from −1.41 to −5.15 kcal mol-1 and slightly decreases the vdW term by 0.26 kcal mol-1. However, the vdW term is the most important factor in controlling the ligand-binding affinity.
In addition, the proposed compounds might be less toxic to the human body than the original one, according to PreADMET prediction.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was funded by Vietnam National Foundation for Science & Technology Development (NAFOSTED) grant #02/2020/ĐX.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmgm.2023.108535.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.WHO . Nat Med. World Health Organization; 2021. Coronavirus disease (COVID-19) outbreak. 2021. [Google Scholar]
- 2.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan J.F.W., Yuan S.F., Kok K.H., To K.K.W., Chu H., Yang J., et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.COVID-19 Vaccines. FDA, FDA, 2021, Vol. 2021.
- 6.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 7.Hoffmann M., Hofmann-Winkler H., Krüger N., Kempf A., Nehlmeier I., Graichen L., et al. SARS-CoV-2 variant B.1.617 is resistant to Bamlanivimab and evades antibodies induced by infection and vaccination. bioRxiv. 2021:2021. doi: 10.1016/j.celrep.2021.109415. 05.04.442663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cele S., Gazy I., Jackson L., Hwa S.-H., Tegally H., Lustig G., et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021 doi: 10.1038/s41586-021-03471-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 11.Gobeil S.M.-C., Janowska K., McDowell S., Mansouri K., Parks R., Stalls V., et al. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Science. 2021:eabi6226. doi: 10.1126/science.abi6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin Z., Zhao Y., Sun Y., Zhang B., Wang H., Wu Y., et al. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat. Struct. Mol. Biol. 2020;27:529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan H.T.H., Moesser M.A., Walters R.K., Malla T.R., Twidale R.M., John T., et al. Discovery of SARS-CoV-2 Mpro peptide inhibitors from modelling substrate and ligand binding. Chem. Sci. 2021;12:13686–13703. doi: 10.1039/d1sc03628a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rathnayake A.D., Zheng J., Kim Y., Perera K.D., Mackin S., Meyerholz D.K., et al. 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV–infected mice. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc5332. eabc5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vuong W., Khan M.B., Fischer C., Arutyunova E., Lamer T., Shields J., et al. Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus peplication. Nat. Commun. 2020;11:4282. doi: 10.1038/s41467-020-18096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hegyi A., Ziebuhr J. Conservation of substrate specificities among coronavirus main proteases. J. Gen. Virol. 2002;83:595–599. doi: 10.1099/0022-1317-83-3-595. [DOI] [PubMed] [Google Scholar]
- 18.Anand K., Ziebuhr J., Wadhwani P., Mesters Jeroen R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 19.Cannalire R., Cerchia C., Beccari A.R., Di Leva F.S., Summa V. Targeting SARS-CoV-2 proteases and polymerase for COVID-19 treatment: state of the art and future opportunities. J. Med. Chem. 2022;65:2716–2746. doi: 10.1021/acs.jmedchem.0c01140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chauhan M., Bhardwaj V.K., Kumar A., Kumar V., Kumar P., Enayathullah M.G., et al. Theaflavin 3-gallate inhibits the main protease (Mpro) of SARS-CoV-2 and reduces its count in vitro. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-17558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coronavirus (COVID-19) Update . U.S. Food and Drug Administration; 2021. FDA Authorizes First Oral Antiviral for Treatment of COVID-19. [Google Scholar]
- 22.Muhseen Z.T., Hameed A.R., Al-Hasani H.M.H., Ahmad S., Li G. Computational determination of potential multiprotein targeting natural compounds for rational drug design against SARS-COV-2. Molecules. 2021;26:674. doi: 10.3390/molecules26030674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang C.-H., Stone E.A., Deshmukh M., Ippolito J.A., Ghahremanpour M.M., Tirado-Rives J., et al. Potent noncovalent inhibitors of the main protease of SARS-CoV-2 from molecular sculpting of the drug perampanel guided by free energy perturbation calculations. ACS Cent. Sci. 2021;7:467–475. doi: 10.1021/acscentsci.1c00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramos-Guzmán C.A., Ruiz-Pernía J.J., Tuñón I. Computational simulations on the binding and reactivity of a nitrile inhibitor of the SARS-CoV-2 main protease. Chem. Commun. 2021;57:9096–9099. doi: 10.1039/d1cc03953a. [DOI] [PubMed] [Google Scholar]
- 25.Li Z., Li X., Huang Y.-Y., Wu Y., Liu R., Zhou L., et al. Identify potent SARS-CoV-2 main protease inhibitors via accelerated free energy perturbation-based virtual screening of existing drugs. Proc. Natl. Acad. Sci. U.S.A. 2020;117:27381–27387. doi: 10.1073/pnas.2010470117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen T.H., Zhou H.-X., Minh D.D.L. Using the fast fourier transform in binding free energy calculations. J. Comput. Chem. 2018;39:621–636. doi: 10.1002/jcc.25139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tam N.M., Pham M.Q., Ha N.X., Nam P.C., Phung H.T.T. Computational estimation of potential inhibitors from known drugs against the main protease of SARS-CoV-2. RSC Adv. 2021;11:17478–17486. doi: 10.1039/d1ra02529e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ngo S.T., Nguyen T.H., Tung N.T., Vu V.V., Pham M.Q., Mai B.K. Characterizing the ligand-binding affinity toward SARS-CoV-2 Mpro via physics- and knowledge-based approaches. Phys. Chem. Chem. Phys. 2022;24:29266–29278. doi: 10.1039/d2cp04476e. [DOI] [PubMed] [Google Scholar]
- 29.Fujitani H., Tanida Y., Ito M., Jayachandran G., Snow C.D. Direct calculation of the binding free energies of FKBP ligands. J. Chem. Phys. 2005;123 doi: 10.1063/1.1999637. [DOI] [PubMed] [Google Scholar]
- 30.Zwanzig R.W. High‐temperature equation of state by a perturbation method. I. Nonpolar gases. J. Chem. Phys. 1954;22:1420–1426. [Google Scholar]
- 31.Ngo S.T., Hung H.M., Nguyen M.T. Fast and accurate determination of the relative binding affinities of small compounds to HIV-1 protease using non-equilibrium work. J. Comput. Chem. 2016;37:2734–2742. doi: 10.1002/jcc.24502. [DOI] [PubMed] [Google Scholar]
- 32.Ngo S.T., Tam N.M., Pham M.Q., Nguyen T.H. Benchmark of popular free energy approaches revealing the inhibitors binding to SARS-CoV-2 Mpro. J. Chem. Inf. Model. 2021;61:2302–2312. doi: 10.1021/acs.jcim.1c00159. [DOI] [PubMed] [Google Scholar]
- 33.Pham M.Q., Vu K.B., Han Pham T.N., Thuy Huong L.T., Tran L.H., Tung N.T., et al. Rapid prediction of possible inhibitors for SARS-CoV-2 main protease using docking and FPL simulations. RSC Adv. 2020;10:31991–31996. doi: 10.1039/d0ra06212j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Green H., Koes D.R., Durrant J.D. DeepFrag: a deep convolutional neural network for fragment-based lead optimization. Chem. Sci. 2021;12:8036–8047. doi: 10.1039/d1sc00163a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Green H., Durrant J.D. DeepFrag: an open-source browser app for deep-learning lead optimization. J. Chem. Inf. Model. 2021;61:2523–2529. doi: 10.1021/acs.jcim.1c00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ngo S.T., Nguyen T.H., Tung N.T., Nam P.C., Vu K.B., Vu V.V. Oversampling free energy perturbation simulation in determination of the ligand-binding free energy. J. Comput. Chem. 2020;41:611–618. doi: 10.1002/jcc.26130. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Y., Fang C., Zhang Q., Zhang R., Zhao X., Duan Y., et al. Protein Cell; 2021. Crystal Structure of SARS-CoV-2 Main Protease in Complex with Protease Inhibitor PF-07321332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tam N.M., Pham D.-H., Hiep D.M., Tran P.-T., Quang D.T., Ngo S.T. Searching and designing potential inhibitors for SARS-CoV-2 Mpro from natural sources using atomistic and deep-learning calculations. RSC Adv. 2021;11:38495–38504. doi: 10.1039/d1ra06534c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 40.Aliev A.E., Kulke M., Khaneja H.S., Chudasama V., Sheppard T.D., Lanigan R.M. Motional timescale predictions by molecular dynamics simulations: case study using proline and hydroxyproline sidechain dynamics. Proteins: Struct., Funct., Bioinf. 2014;82:195–215. doi: 10.1002/prot.24350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 42.Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 43.Case D.A., Ben-Shalom I.Y., Brozell S.R., Cerutti D.S., Cheatham T.E.C., III, Darden V.W.D., III, et al. University of California; San Francisco: 2018. AMBER 18. [Google Scholar]
- 44.Park S., Schulten K. Calculating potentials of mean force from steered molecular dynamics simulations. J. Chem. Phys. 2004;120:5946–5961. doi: 10.1063/1.1651473. [DOI] [PubMed] [Google Scholar]
- 45.Bennett C.H. Efficient estimation of free energy differences from Monte Carlo data. J. Comput. Phys. 1976;22:245–268. [Google Scholar]
- 46.Chemicalize Was Used for Prediction of Chemical Properties.
- 47.Schrödinger Llc P. Maestro; 2020. Schrödinger Release 2020-4. [Google Scholar]
- 48.Lee S.K., Lee I.H., Kim H.J., Chang G.S., Chung J.E., No K.T. Blackwell Publishing; Maldenh, MA: 2003. The PreADME Approach: Web-Based Program for Rapid Prediction of Physico-Chemical, Drug Absorption and Drug-like Properties, EuroQSAR 2002 Designing Drugs and Crop Protectants: Processes, Problems and Solutions; pp. 418–420. [Google Scholar]
- 49.Owen Dafydd R., Allerton Charlotte M.N., Anderson Annaliesa S., Aschenbrenner L., Avery M., Berritt S., et al. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374:1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 50.van der Lubbe S.C.C., Fonseca Guerra C. The nature of hydrogen bonds: a delineation of the role of different energy components on hydrogen bond strengths and lengths. Chem. Asian J. 2019;14:2760–2769. doi: 10.1002/asia.201900717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.