Abstract
Laminin-α4 (LAMA4) is an extracellular matrix protein implicated in the regulation of adipocyte differentiation and function. Prior research describes a role for LAMA4 in modulating adipocyte thermogenesis and uncoupling protein-1 (UCP1) expression in white adipose; however, the mechanisms involved are poorly understood. Here, we describe that Lama4 knockout mice (Lama4−/−) exhibit heightened mitochondrial biogenesis and peroxisome proliferator-activated receptor γ coactivator-1 (PGC-1) expression in subcutaneous white adipose tissue (sWAT). Furthermore, the acute silencing of LAMA4 with small interfering RNA in primary murine adipocytes was sufficient to upregulate the expression of thermogenic markers UCP1 and PR domain containing 16 (PRDM16). Silencing also resulted in an upregulation of PGC1-α and adenosine 5′-monophosphate–activated protein kinase (AMPK)-α expression. Subsequently, we show that integrin-linked kinase (ILK) is downregulated in the sWAT of Lama4−/− mice, and its silencing in adipocytes similarly resulted in elevated expression of UCP1 and AMPKα. Last, we demonstrate that treatment of human induced pluripotent stem cell–derived thermogenic adipocytes with LAMA4 (LN411) inhibited the expression of thermogenic markers and AMPKα. Overall, our results indicate that LAMA4 negatively regulates a thermogenic phenotype and pathways involving mitochondrial biogenesis in adipocytes through the suppression of AMPKα.
Keywords: laminins, extracellular matrix, adipose tissue, adipocyte, thermogenesis, beige
In recent years, there has been increasing evidence to support the beneficial metabolic effects of beige adipocytes, including improved insulin sensitivity, increased fatty acid oxidation, and reduced adiposity (1). This has driven an extraordinary interest in the study of thermogenic regulators that could be targeted for therapeutic application in metabolic diseases like obesity and type 2 diabetes mellitus. While a number of pathways involved in adipocyte thermogenesis have been identified to date and more are discovered every year, there is still much remaining to be understood about extracellular factors within adipose tissue that regulate the thermogenic program and adipocyte differentiation.
Adipose tissue exists in 3 forms: white, brown, and beige. White adipose tissue (WAT) is known for being energy storing, while brown adipose is rich in mitochondria and expends energy through the process of thermogenesis to generate heat. The third type, beige or brite, is found in white adipose depots but exhibits a higher mitochondrial density and a capacity for thermogenesis. Mitochondria are essential organelles that produce adenosine triphosphate (ATP) through oxidative phosphorylation (2). In beige or brown adipose, compared to other cell types, there is a greater amount of uncoupling protein-1 (UCP1). UCP1 resides in the inner membrane of mitochondria and facilitates proton leak, which diffuses the proton gradient required for ATP generation, leading to heat production instead (3). This process is known as thermogenesis and results in increased energy expenditure. For this reason, promoting thermogenesis in white adipose through beige adipocyte induction and enhanced mitochondrial content could be applied to treat obesity. Although many drugs have been proposed to stimulate beiging, there is increasing interest in how adipocyte fate and function may be modulated through extracellular matrix (ECM) dynamics.
The cellular microenvironment plays a pivotal role in directing differentiation in all tissue types. This modulation can occur through environmental stiffness and mechanosensation, through direct contact of specific extracellular proteins with cell surface receptors, and through the sequestration or release of signaling molecules and factors within the extracellular space (4). The differing environmental requirements of specific cell types is reflected in the diverse composition of extracellular niches. Recent publications have even illustrated the important effects of the ECM composition and density on mitochondrial dynamics and bioenergetics in human mesenchymal stem cells and cancer cells, particularly through integrin-mediated signaling (5, 6). While ECM proteins, such as collagens, have been studied in great depth in their relation to adipose tissue and metabolic disease, there are many types of proteins beyond collagens that contribute to adipocyte differentiation and behavior (4). As research in this field expands, other ECM protein families, such as laminins, have come into focus as important regulators of adipocyte thermogenesis.
Laminins are a family of ECM proteins located in the cellular basement membrane that contribute to cell migration, support, and communication of extracellular cues (7). The many isoforms of laminin each contain an α-, β-, and γ-chain assembled into a cross-like structure. The α-chain is especially important as it contains domains in the C-terminus that interact with numerous cell surface receptors (8). During white adipocyte differentiation, cells express and secrete increasing levels of the laminin-α4 (LAMA4) chain, suggesting a role for this protein unique to white adipogenesis (9, 10). Our group has shown that mice deficient in LAMA4 display elevated UCP1 expression in subcutaneous white adipose tissue (sWAT) compared to wild-type (WT) mice, and are protected from diet-induced obesity (11, 12). More recently, we have published findings showing that differentiation of murine adipose-derived stem cells (ADSCs) to beige adipocytes on LAMA4-coated surfaces impairs Ucp1 expression (13). LAMA4 expression is also significantly elevated in the sWAT of humans with obesity and mice with diet-induced obesity (45% high-fat diet) (14). Our results thus far indicate a correlation between LAMA4 and adiposity by proposing that LAMA4 can suppress thermogenic signaling pathways or differentiation toward a beige adipocyte phenotype.
For this study, we sought to identify the effects of LAMA4 on adipocyte thermogenesis and mitochondrial biogenesis and to understand how this regulation may occur. First, we studied the influence of LAMA4 on mitochondrial biogenesis and found that the deficiency of LAMA4 in vivo and acute silencing in vitro lead to an upregulation of peroxisome proliferator-activated receptor γ coactivator-1 (PGC-1)α expression in adipocytes. We next show that the acute silencing of LAMA4 in vitro is sufficient to enhance thermogenic gene expression in ADSCs during differentiation to beige adipocytes. Our investigation of associated pathways uncovered a major signaling protein that appears to be involved in this LAMA4 signal transduction: adenosine 5′-monophosphate–activated protein kinase (AMPK). We also show that LAMA4 negatively regulates thermogenesis in a human adipocyte model using human induced pluripotent stem cells (hiPSCs) differentiated to thermogenic adipocytes and treated with human recombinant laminin-411 during differentiation. These results denote a critical function for LAMA4 in negatively regulating adipocyte thermogenesis through AMPK, which could be targeted in the future for therapeutic purposes.
Materials and Methods
Animal Model, Care, and Housing
Animal procedures and numbers were approved by the University of Chicago Institutional Animal Care and Use Committee. All experiments were conducted in accordance with accepted standards of humane animal care. The generation of Lama4−/− mice has been previously described (15). WT and Lama4−/− male C57BL/6J mice were given regular chow diet (Teklad 2918; Harlan Laboratories) ad libitum and housed at room temperature. Female mice were not used in this particular study because female Lama4−/− mice do not exhibit the same degree of enhanced sWAT thermogenesis as do male Lama4−/− mice. Mice were humanely euthanized between ages 14 and 17 weeks for tissue collection and cell isolation.
Murine Adipose-Derived Stem Cell Culture and Differentiation
Primary adipose derived stem cells were isolated from 14- to 17-week-old male mice as previously described (11). Cells were expanded and plated into 6-well plates for experiments, and subsequently differentiated to beige adipocytes following a previously published protocol (16). Induction medium containing complete medium (10% fetal bovine serum [FBS], 1% Pen/Strep, Dulbecco’s modified Eagle’s medium [DMEM]/F-12) supplemented with 5 μM dexamethasone, 125 nM indomethacin, 0.5 mM isobutylmethylxanthine, 850 nM insulin, 1 nM 3,5,3′-triiodothyronine (T3), 0.5 μM rosiglitazone was added to cells to initiate differentiation. On day 2 cells were given maintenance medium containing complete medium with 850 nM insulin, 1 nM T3, and 0.5 uM rosiglitazone. Refeeding then occurred every other day with maintenance medium until day 16. We chose to use a 16-day differentiation timeline to match the timeline of the human iPSC model and to allow for silencing only after adipocytes had fully matured (∼ day 8-10).
Human Induced Pluripotent Stem Cell Culture and Differentiation
An iPSC cell line (Coriell catalog No. GM25256, RRID:CVCL_Y803, Origin: Human, Male) was obtained from The Coriell Institute for Medical Research and cultured in feeder-free conditions on Matrigel (Corning; 354277) in mTeSR1 (Stem Cell Technologies [SCT]; 85850). Cells were routinely tested for pluripotency using STEMdiff Trilineage Differentiation Kit (SCT; 05230) and for mycoplasma (PromoKine; PK-CA20-700-20).
We generated beige adipocytes by modifying a previously published protocol by Guénantin et al (17). hiPSCs were cultured on Matrigel and dissociated and passaged as aggregates when cells were 80% confluent using ReLeSR (SCT; 05872). Differentiation was performed on 6-well tissue culture plates. Mesoderm induction was started on day 0 when cells were 90% confluent by changing media to STEMPro34 (Gibco; 10639011), 1:100 Glutamax (Gibco; 35050061), 50 µg/mL L-ascorbic acid (Sigma; A4544), 10 ng/Ml BMP4 (R&D; 314-BP-010/CF), and 25 ng/mL Activin A (R&D; 338-AC-050/CF). Mesoderm induction media was replaced every other day. On day 4, adipocyte differentiation was induced by replacing mesoderm induction media with DMEM/F-12, HEPES (Gibco; 11330032) media containing 10% FBS-D (Hyclone; SH30070.03), 10 µg/mL insulin (Sigma; I9278), 500 µM IBMX (Sigma; I5879), 1 µM dexamethasone (Sigma; A4902), and 50 µM indomethacin (Sigma; I7378). Media was replaced every 2 days. On day 10 adipocyte maturation was started by switching cells to DMEM/F12, 10% FBS-D, 850 nM insulin, 1 nM T3 (Sigma; T6397), and 0.5 µM rosiglitazone (Cayman; 71740). Recombinant human laminin-411 (BioLamina; LN411) was included in the media for treated cells from day 4 through day 16 of differentiation. Cells were collected on day 16 of differentiation.
Small Interfering RNA Transfection and Treatments
Fully mature beige adipocytes derived from murine adipose-derived stem cells were transfected on days 10 and 14 of differentiation. Preparation of lipofectamine RNAiMAX with small interfering (siRNA) was carried out following the manufacturer's instructions for a 6-well volume (Thermo Fisher Scientific; 13778075). Per well: 9 μL of lipofectamine RNAiMAX was diluted in 150 μL of Opti-MEM (Thermo Fisher; 31985062) and 3 μL (integrin-linked kinase [ILK] siRNA, Santa Cruz Biotechnology [SCBT]; sc-35667) or 10 μL (Lama4 siRNA, SCBT; sc-43148) of 10 μM siRNA was diluted in 150 μL of Opti-MEM. Control siRNA A (SCBT; sc-37007) was also used at the respective concentrations per condition. The diluted lipofectamine RNAiMAX and diluted siRNA were then mixed in equal parts and incubated at room temperature for 15 minutes. A total of 250 μL of the lipofectamine-siRNA mix was pipetted onto the cells and then they were placed for 20 minutes at 37 °C in a cell culture incubator, following which 2.5 mL of maintenance media was added per well. Cells were collected on day 16 of differentiation. For AMPK inhibitor experiments, adipocytes were treated with 10 μM of compound C (Dorsomorphin dihydrochloride) (Tocris; 3093) for 24 hours before collection.
Immunoblotting
Samples were lysed using cold 1× radioimmunoprecipitation assay buffer (Sigma; 20-188) containing 1× phosphatase and protease inhibitor cocktail tablets (Sigma; 4906845001 and 05892970001) and a dispersion-based homogenizer (VWR VDI 12). After incubating on ice for 30 minutes, samples were briefly sonicated (Sonics Vibra-cell) and spun at 1000g for 10 minutes at 4 °C to separate the lipid layer. The supernatant, excluding lipid layer, was collected and spun again at 10 000g for 10 minutes at 4 °C and supernatant from this spin was collected and stored at −80 °C. Protein concentration was determined using the Pierce BCA Protein Assay Kit (Thermo Fisher; 23227).
Samples were diluted in water and 4× Laemmli Sample Buffer (Bio-Rad; 1610747) and run on 4-15% or 10% SDS-PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis) Criterion TGX gels (Bio-Rad), then transferred to Immobilon-P PVDF membranes (Sigma; IPVH00010). Membranes were blocked with 5% PhosphoBLOCKER Blocking Reagent (Cell Biolabs Inc; AKR-103) or 5% nonfat dry milk (LabScientific; M0841) in TBST for 1 hour. Blots were incubated overnight at 4 °C in 1% blocking solution with antibody. Membranes were then incubated with secondary antibody (LI-COR Biosciences catalog No. 926-68171, RRID:AB_10956389) for 1 hour. Immunodetection was performed using near-infrared Odyssey CLx System (LI-COR). Analysis was performed using LI-COR Image Studio Lite (RRID:SCR_013715). All samples were normalized to their paired control or, for in vivo studies, the average of group/littermate controls to account for variation between mice.
Antibodies used in this study are as follows: Cell Signaling Technology: GAPDH (glyceraldehyde-3-phosphate dehydrogenase; Cat# 2118, RRID:AB_561053), βActin (catalog No. 4970, RRID:AB_2223172), Vinculin (catalog No. 13901, RRID:AB_2728768), AMPKα (catalog No. 2532, RRID:AB_330331); voltage-dependent anion channel (VDAC; catalog No. 4661, RRID:AB_10557420), ILK1 (catalog No. 3862, RRID:AB_2127050); SDHA (Complex II component) (catalog No. 11998, RRID:AB_2750900); UQCRFS1/RISP (Complex III component) (catalog No. 95231, RRID:AB_2922784); Thermo Fisher Scientific: Phospho-AMPK alpha-1,2 (Thr183, Thr172) (catalog No. 44-1150G, RRID:AB_2533585); Abcam: ATP5A (Complex V component) (catalog No. ab176569, RRID:AB_2801536), PGC1 alpha + beta (catalog No. ab72230, RRID:AB_1640773); Human UQCRC2 (catalog No. ab14745, RRID:AB_2213640), UCP1 (catalog No. ab209483, RRID:AB_2722676); R&D Systems: Human UCP1 (catalog No. MAB6158, RRID:AB_10572490); Millipore Sigma: LAMA4 (catalog No. SAB4501719, RRID:AB_10744529), and Complex 1-75 kD (catalog No. ABN302, RRID:AB_2915902).
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction
RNA was isolated using the E.Z.N.A Total RNA Kit II (Omega Biotek; R6934) for murine studies or RNeasy Mini Kit (Qiagen; 74104) for hiPSC studies following the manufacturer's instructions. Tissue samples were disrupted using a dispersion-based homogenizer (VWR VDI 12). The RNA samples were reverse transcribed using Quanta QScript Master Mix (VWR; 95048) and 500 ng RNA per 20-μL sample reaction volume. Quantitative real-time polymerase chain reaction (PCR) was performed with SYBR green using a Bio-Rad CFX Connect Real-Time PCR Detection System. Primers were purchased from IDT or Qiagen; IDT primer sequences can be found in Table 1. For all samples, GAPDH was used as the housekeeping gene. Gene expression was evaluated by ddCT methods.
Table 1.
Quantitative real-time polymerase chain reaction primers
| Forward | Reverse | |
|---|---|---|
| Human | ||
| GAPDH | TGCACCACCAACTGCTTAGC | GGCATGGACTGTGGTCATGAG |
| PPARG | QIAGEN No. 330001 PPH00461F-200 | |
| ADIPOQ | QIAGEN No. 330001 PPH00441F-200 | |
| UCP1 | QIAGEN No. 330001 PPH2223A-200 | |
| PRDM16 | QIAGEN No. 330001 PPH19483C-200 | |
| CIDEA | QIAGEN No. 330001 PPH00899C-200 | |
| Mouse | ||
| Gapdh | CAATGTGTCCGTCGTGGATCTGA | GAGTTGCTGTTGAAGTCGCAGGA |
| Lama4 | GGAATACCTGAACGTGCACATGAGA | GTGCCATCTGCCATCACAGAGATTCT |
| Cpt1a | CTATGCGCTACTCGCTGAAGG | GGCTTTCGACCCGAGAAGA |
| Cpt1b | TGGGACTGGTCGATTGCATC | TCAGGGTTTGTCGGAAGAAGAA |
| mt-Nd1 | TGCACCTACCCTATCACTCA | GGCTCATCCTGATCATAGAATGG |
| mt-Nd2 | CGCCCCATTCCACTTCTGATTACC | TTAAGTCCTCCTCATGCCCCTATG |
| Prdm16 | GATGGGAGATGCTGACGGAT | TGATCTGACACATGGCGAGG |
| Tbx1 | AGCGAGGCGGAAGGGA | CCTGGTGACTGTGCTGAAGT |
| Cidea | CGGGAATAGCCAGAGTCACC | TGTGCATCGGATGTCGTAGG |
| Ppargc1a | ATGAATGCAGCGGTCTTAGC | AACAATGGCAGGGTTTGTTC |
| Ilk | GTGGCTGGACAACACAGAGA | ATCCCCACGATTCATCACAT |
| Prkaa1 | TGAGAAGTTCGAGTGTTCGGA | TGGTGTTTGGATTTCTGTGG |
| Ucp1 | QIAGEN No. 330001 PPM05164B | |
| Tfam | CAAGTCAGCTGATGGGTATGG | TTTCCCTGAGCCGAATCATCC |
| Nrf-1 | GCACCTTTGGAGAATGTGGT | CTGAGCCTGGGTCATTTTGT |
Abbreviations: ADIPOQ, adiponectin; Cidea, cell death activator CIDE-A; CPT1, carnitine palmitoyltransferase 1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Ilk, integrin-linked kinase; Lama4, Laminin-α4; Nrf-1, nuclear respiratory factor 1; PPARG, peroxisome proliferator activated receptor γ; PRDM16, PR domain containing 16; Tbx1, T-box 1; Tfam, transcription factor A; Ucp1, uncoupling protein-1.
Statistics
Statistical tests for all in vitro studies where n was equal between groups and samples were paired, a paired t test was performed for all data. In studies where samples were paired, data were normalized to the paired control, which was assigned a value of 1.0, to account for variability in differentiation efficiency between experiments, and in expression of LAMA4 between primary cells derived from different mice. In all studies involving whole tissue from mice, comparisons between WT and Lama4−/− mice were assessed with a Welch unequal variances t test as variance between mice was expected to be high among the WT because of differences in LAMA4 expression and low among the Lama4−/− mice. In all cases, P less than or equal to .05 was considered statistically significant.
Results
Laminin-α4 Deficiency Promotes Mitochondrial Biogenesis
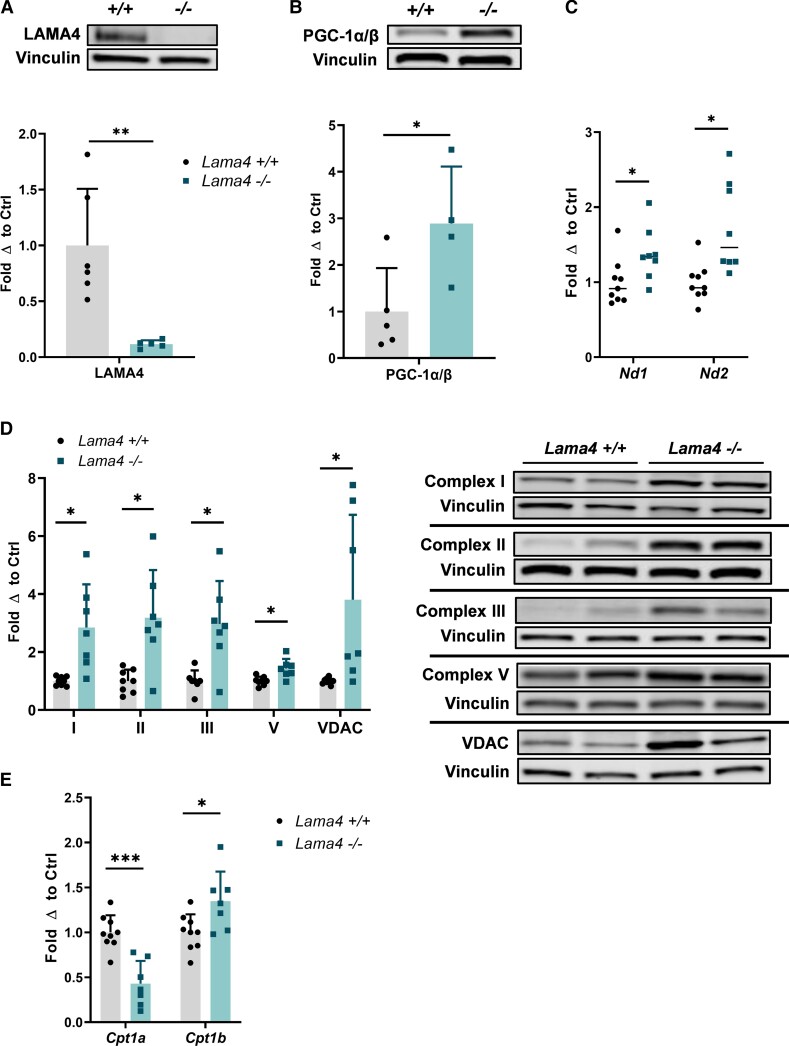
Previously, we had shown that LAMA4 deficiency in vivo leads to elevated thermogenesis through increased expression of UCP1 in the sWAT of male mice (11). Following these results, we wished to better understand how these observed changes in thermogenesis may extend to changes in mitochondrial biogenesis. Initially, we determined that the protein expression of PGC-1, a master regulator of mitochondrial biogenesis, was significantly upregulated in the sWAT of Lama4−/− male mice compared to WT (Fig. 1A and 1B). Next, to assess differences in mitochondrial gene expression, we analyzed the messenger RNA (mRNA) expression of mitochondrial respiratory complex I subunits 1 and 2 (Nd1 and Nd2). We found the mRNA levels of both genes to be significantly elevated in Lama4−/− sWAT (Fig. 1C). Furthermore, the protein expression of mitochondrial markers, including mitochondrial respiratory chain complexes I, II, III, and V, in addition to VDAC, a mitochondrial porin, were significantly elevated in the sWAT of Lama4−/− male mice compared to WT (Fig. 1D). The increased expression of multiple mitochondrial markers indicates a larger mitochondrial load and is suggestive of enhanced mitochondrial biogenesis in sWAT lacking LAMA4, as driven by elevated PGC-1 expression.
Figure 1.
LAMA4 deficiency elevates mitochondrial biogenesis. A, Protein expression of LAMA4 in subcutaneous white adipose tissue (sWAT) of wild-type (WT) (n = 6) and Lama4−/− (n = 5) male mice was assessed by Western blot. Reported as fold change with respect to average of WT mice after normalization to loading control (Vinculin) signal. *, **, and *** indicates P less than or equal to .05, .01, and .001, respectively. Data are means + SD. B, Protein expression of PGC-1α/β in sWAT of WT (n = 5) and Lama4−/− (n = 4) male mice was assessed by Western blot. Vinculin was used as the loading control for normalization. C, Relative messenger RNA (mRNA) expression of mitochondrial genome genes Nd1 and Nd2 (subunits of complex I) in sWAT of WT (n = 9) and Lama4−/− (n = 8) male mice. D, Protein expression of mitochondrial markers: mitochondrial respiratory chain complexes I, II, III, and V, and VDAC in the sWAT of WT (n = 8) and Lama4−/− (n = 7) male mice was assessed by Western blot. Vinculin was used as the loading control for normalization. E, Relative Cpt1a and Cpt1b mRNA expression in sWAT of WT (n = 9) and Lama4−/− (n = 7) male mice.
In addition to mitochondrial content markers, we identified a change in carnitine palmitoyltransferase 1 (CPT1) isoform expression. CPT1 shuttles long-chain fatty acids across the mitochondrial membrane to supply substrate for the rate-limiting step in fatty acid oxidation. The CPT1A isoform is more abundantly expressed in murine and human WAT, although expression of CPT1 overall is quite low in WAT. The CPT1B the isoform is expressed in energetic tissues like muscle and brown adipose tissue (18). We found that the expression Cpt1a was reduced, while the expression of Cpt1b was elevated in Lama4−/− sWAT compared to controls (Fig. 1E). This shift toward the Cpt1b isoform is another indicator of a more brown-like, or thermogenic, adipocyte population in the absence of LAMA4.
Acute Silencing of Laminin-α4 in Adipocytes Enhances Thermogenic Gene Expression
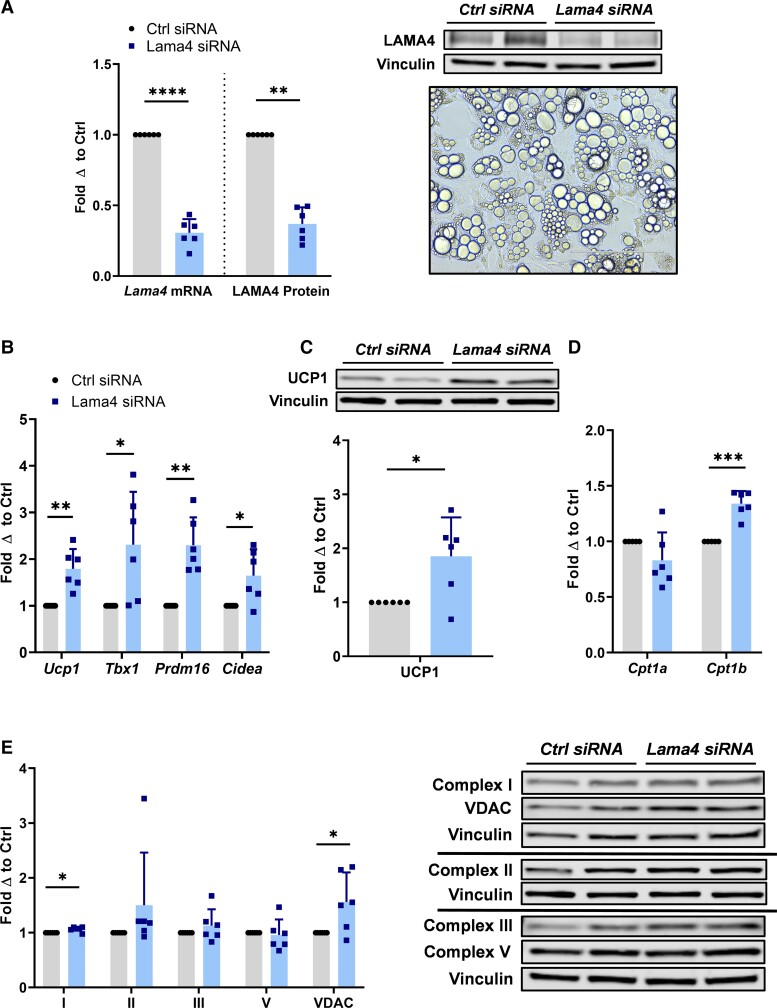
During our in vivo investigation, we noted variability in our results, particularly with WT mice, that seemed to be due to the wide range of LAMA4 expression that can occur in adipose of WT mice, a phenomenon described in our most recent publication (14). To study the effect of LAMA4 signal transduction in a more controlled manner, we therefore next pursued in vitro silencing experiments. This would also aid us in isolating intracellular signaling changes occurring as a result of an acute downregulation of LAMA4 rather than a complete genetic knockout model, which may be more therapeutically insightful. We used WT murine ADSCs differentiated to beige adipocytes for this study as they lipid-load robustly and show an improved capacity for thermogenesis compared to 3T3-L1 lines. We transfected the cells with siRNA targeting Lama4 once the adipocytes were fully matured, on day 10 and 14 of the 16-day differentiation program, and yielded an average of 60% to 70% silencing efficiency (Fig. 2A).
Figure 2.
Acute silencing of LAMA4 in murine adipocytes enhances thermogenic gene expression. A, Relative Lama4 mRNA expression and LAMA4 protein expression in adipocytes treated with control small interfering RNA (siRNA) (n = 6) and Lama4 siRNA (n = 6). Reported as fold change with respect to paired control. *, **, ***, and **** indicates P less than or equal to .05, .01, .001, and .0001, respectively. Data are means + SD. Includes representative image at 20× magnification of differentiated adipocytes on day 16. B, Relative Ucp1, Tbx1, Prdm16, and Cidea messenger RNA (mRNA) expression in adipocytes treated with control siRNA (n = 6) and Lama4 siRNA (n = 6). C, Protein expression of UCP1 in adipocytes treated with control siRNA (n = 6) and Lama4 siRNA (n = 6) was assessed by Western blot. Vinculin was used as the loading control for normalization. D, Relative Cpt1a and Cpt1b mRNA expression in adipocytes treated with control siRNA (n = 6) and Lama4 siRNA (n = 6). E, Protein expression of mitochondrial respiratory chain complexes I, II, III, and V, and VDAC in adipocytes treated with control siRNA (n = 6) and Lama4 siRNA (n = 6) was assessed by Western blot. Vinculin was used as the loading control for normalization.
The beige adipocyte markers Ucp1, T-box 1 (Tbx1), PR domain containing 16 (Prdm16), and cell death activator CIDE-A (Cidea) were all significantly upregulated in the LAMA4 siRNA-transfected adipocytes compared to control siRNA-transfected adipocytes (Fig. 2B). Protein level expression of UCP1 was also significantly elevated by around 85% when LAMA4 was silenced (P = .03) (Fig. 2C). Similar to the in vivo results, we also found a significant increase in the expression of Cpt1b on LAMA4 silencing in vitro, again indicating a more brown-like phenotype (Fig. 2D). Cpt1a expression trended down in the silenced group, but this change was not significant.
The expression of mitochondrial markers remained largely unchanged following silencing, with the exception of mitochondrial respiratory complex I and VDAC, which showed small but statistically significant upward trends in the LAMA4-silenced group (Fig. 2E). The relatively small changes in mitochondrial protein expression may be due to the acute nature of the silencing or the fact that some LAMA4 protein expression remained (∼ 30%). ECM proteins are more stable and less frequently replaced than intracellular proteins, which could feasibly limit the true window of silencing at the protein level to less than 6 days (19). This acute silencing illustrates that, although enhanced beige marker expression can be achieved within a short time frame, changes in mitochondrial content may require a longer period of silencing or the complete absence of LAMA4 to be visualized.
Laminin-α4 Silencing Upregulates Expression of the AMPK-PGC-1α Pathway
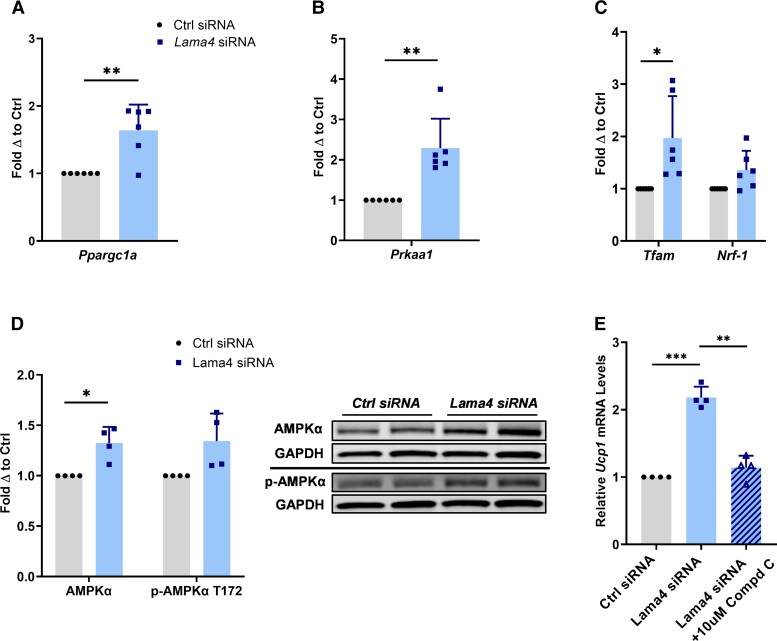
Following these results, we chose to assess intracellular signaling pathways associated with beiging and mitochondrial biogenesis that might be altered. Although we did not observe significant upregulation in mitochondrial content, we did find that LAMA4 silencing resulted in significantly elevated expression of the PGC-1α gene, Ppargc1α (Fig. 3A). In our prior publication, we reported that differentiation of murine ADSCs to beige adipocytes on LAMA4-coated surfaces leads to diminished PGC-1α expression (13). Considering this relationship across multiple studies and models, we elected to investigate AMPK, a serine/threonine kinase that has been implicated in adipocyte beiging and mitochondrial biogenesis (20). AMPK lies upstream of PGC-1α activation and has also been associated with the regulation of the cell surface proteome, specifically β1-integrins (21). Previous research has shown that silencing of AMPKα in the adipose tissue of mice produces diminished thermogenic capacity, while activation of AMPK in adipose enhances thermogenic gene expression (20).
Figure 3.
The AMPK-PGC-1α pathway is upregulated when LAMA4 is silenced. A, Relative Ppargc1a messenger RNA (mRNA) expression (PGC-1α) in adipocytes treated with control siRNA (n = 6) and Lama4 siRNA (n = 6). Reported as fold change with respect to paired control. * and ** indicate P less than or equal to .05 and .01, respectively. Data are means + SD. B, Relative Prkaa1 mRNA expression (AMPKα) in adipocytes treated with control small interfering (siRNA) (n = 6) and Lama4 siRNA (n = 6). C, Relative Tfam and Nrf1 mRNA expression in adipocytes treated with control siRNA (n = 6) and Lama4 siRNA (n = 6). D, Protein expression of AMPKα and p-AMPKα T172 in adipocytes treated with control siRNA (n = 4) and Lama4 siRNA (n = 4) was assessed by Western blot. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as the loading control for normalization. E, Relative Ucp1 mRNA expression in adipocytes treated with control siRNA (n = 4), Lama4 siRNA (n = 5), and Lama4 siRNA + 10 μM of compound C, an AMPK inhibitor (n = 4).
We discovered a significant increase in the mRNA levels of the AMPKα gene, Prkaa1, when LAMA4 was silenced (Fig. 3B). Furthermore, genes downstream of AMPK activation and involved in initiating mitochondrial biogenesis, such as mitochondrial transcription factor A (Tfam) and nuclear respiratory factor 1 (Nrf-1), were either significantly elevated or trended higher in the LAMA4-silenced adipocytes (Fig. 3C) (22). Immunoblotting revealed that total protein levels of AMPKα were also increased, as was the phosphorylation of AMPKα at T172, suggesting that the downregulation of LAMA4 leads to an elevation in AMPK activity, predominantly through an upregulation in AMPKα transcription (Fig. 3D). Last, when adipocytes were treated with an AMPK inhibitor following LAMA4 silencing, the observed elevation in Ucp1 expression was lost (Fig. 3E). This indicates that the thermogenic upregulation we observe due to LAMA4 absence is unable to occur without the activity of AMPK. Overall, these results suggest that the AMPK-PGC-1α pathway is upregulated and drives thermogenic gene expression in the absence of LAMA4 in adipocytes.
Laminin-α4 Suppresses Thermogenic Gene Expression and AMPK in Human Adipocytes
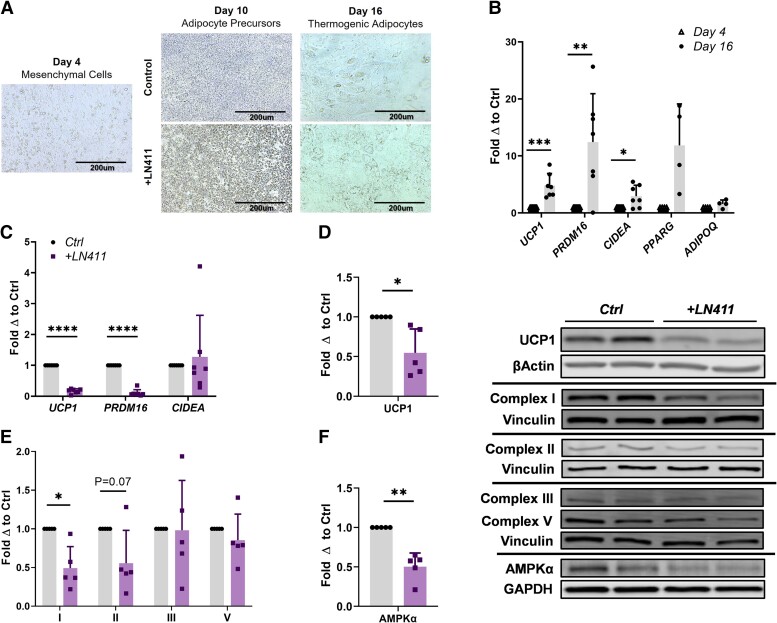
Having shown that the acute silencing of LAMA4 could elevate thermogenic markers and AMPKα expression, we wanted to perform the converse experiment to determine if LAMA4 could directly inhibit thermogenesis and AMPKα expression. In past studies, we have demonstrated that the culture of murine Lama4−/− adipocytes on Lama4+/+ derived ECM could quell the observed increases in Ucp1 expression, suggesting that LAMA4 may be a direct negative regulator of adipocyte beiging (11). Furthermore, we have shown more recently that differentiation of murine ADSCs to beige adipocytes in the presence of LAMA4 diminishes Ucp1 expression (13). However, these experiments did not investigate signaling intermediates such as AMPKα and were limited to murine thermogenic adipocyte models. It was not yet understood if this potential regulatory role would be applicable to human adipocyte models.
To explore this possibility, we took advantage of direct differentiation of thermogenic adipocytes derived from hiPSCs (Fig. 4A). hiPSCs were successfully differentiated to beige adipocytes by day 16 as shown through the elevation of beige markers UCP1, PRDM16, and CIDEA, and adipocyte markers PPARG and adiponectin (ADIPOQ) (Fig. 4B). When human recombinant laminin-411 (LN411), which is composed of the laminin-α4, laminin-β1, and laminin-γ1 chain, was included during differentiation, significant reductions in mRNA levels of thermogenic markers UCP1 and PRDM16 were observed (Fig. 4C). In addition to the reduction in UCP1 mRNA levels, a significant decrease in UCP1 protein expression is detected in the LN411 treated adipocytes (P = .02) (Fig. 4D). We assessed mitochondrial respiratory chain complex protein expression and found that complex I was significantly reduced in expression in the LN411-treated adipocytes (P = .01) and that complex II trended down (P = .07), while expression of complexes III and V appeared unaffected by the treatment (Fig. 4E). Last, we found that the presence of LN411 resulted in a significant decrease in total AMPKα levels (P = .003), further supporting the assertion that LAMA4 is involved in regulating AMPKα (see Fig. 4D). These results suggest that LAMA4 directly inhibits a thermogenic phenotype in human adipocytes through the suppression of AMPKα.
Figure 4.
LAMA4 negatively regulates thermogenic gene expression in human adipocytes. A, Representative images outlining differentiation and treatment trajectory of human induced pluripotent stem cells (hiPSCs) to beige adipocytes. Images were taken at 20× magnification. B, Relative messenger RNA (mRNA) expression of thermogenic genes UCP1, PRDM16, and CIDEA (n = 7), and adipogenic genes PPARG and ADIPOQ (n = 4) in hiPSCs on day 4 and day 16 of beige adipocyte differentiation. Reported as fold change with respect to paired control. *, **, ***, and **** indicate P less than or equal to .05, .01, and .001, 0.0001, respectively. Data are means + SD. C, Relative mRNA expression of thermogenic genes UCP1, PRDM16, and CIDEA (n = 7) in human beige adipocytes treated with or without LN411 during differentiation. D, Protein expression of UCP1 in human beige adipocytes treated with or without LN411 during differentiation (n = 5). Beta actin was used as the loading control for normalization. E, Protein expression of mitochondrial respiratory chain complexes I, II, III, and V in human beige adipocytes treated with or without LN411 during differentiation (n = 5). Vinculin was used as the loading control for normalization. F, Protein expression of AMPKα in human beige adipocytes treated with or without LN411 during differentiation (n = 5). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as the loading control for normalization.
Silencing of Integrin Linked Kinase Mimics the Effects of Laminin-α4 Silencing in Adipocytes
Our data suggested a direct link between LAMA4 and the negative regulation of thermogenesis via the AMPK-PGC-1α pathway; however, the connection between these pathways was unknown. As the last part of this study, we wanted to identify potential mediators of this regulation for future investigation. In previous publications we established that integrin signaling is reduced in LAMA4-deficient adipose, specifically through the downregulation of integrin alpha 7 (ITGA7) and integrin beta 1 (ITGB1) (13). Additionally, the silencing of ITGA7 in vitro was shown to be sufficient to induce thermogenic gene expression in adipocytes, suggesting that a potential LAMA4-integrin interaction might regulate adipocyte thermogenesis (13). Integrins are a major class of transmembrane receptors that operate via heterodimers of an α and β subunit and are heavily implicated in ECM binding and cell signaling. Adaptor proteins reside downstream of integrins and participate in outside-in signaling cascades that alter intracellular function (23). One adaptor protein in particular, ILK, has been tied to laminin signaling in multiple cell types across numerous studies in the literature, and even directly to LAMA4 in human diseases and human endothelial cell differentiation (24, 25). Based on these literature connections and our knowledge that LAMA4 signal transduction may involve integrins, we decided to investigate whether ILK was altered in LAMA4-deficient adipocytes.
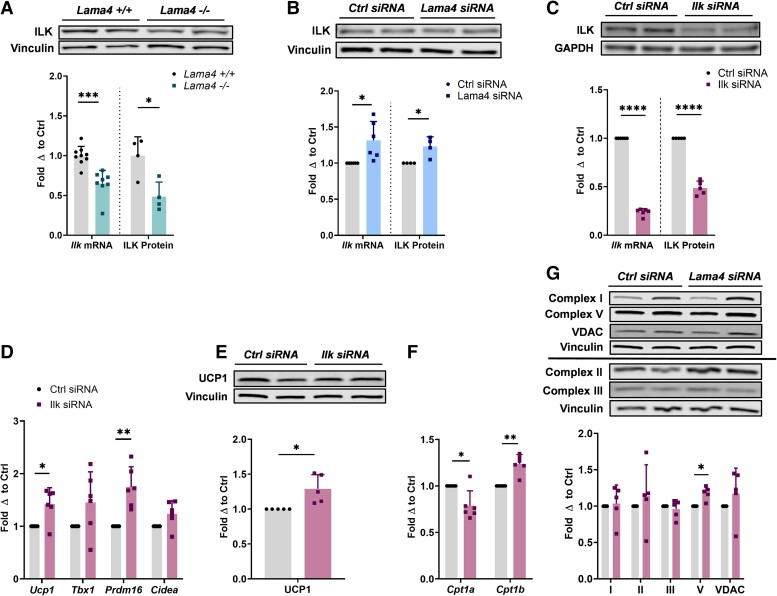
We hypothesized that LAMA4 may engage with integrins on the adipocyte surface, in turn stimulating ILK and leading to suppression of the AMPK-PGC-1α pathway and thermogenic induction. Therefore, the absence of LAMA4, which leads to reductions in ITGA7 and ITGB1, might result in decreased integrin-mediated signaling and decreased activation of ILK, lifting the suppression of the AMPK-PGC-1α pathway. To investigate this hypothesis, we began by assessing ILK expression in our in vivo mouse model to determine if the complete and long-term absence of LAMA4 signaling would alter ILK levels. We found that the expression of ILK was significantly diminished on both the mRNA and protein level in the sWAT of Lama4−/− mice compared to controls (Fig. 5A). Conversely, when we measured ILK expression following the acute silencing of LAMA4 in vitro, we found a very small but statistically significant upregulation in expression (Fig. 5B).
Figure 5.
Silencing of ILK in murine adipocytes elevates thermogenic gene expression. A, Relative Ilk messenger RNA (mRNA) expression (WT n = 9, Lama4−/− n = 8) and ILK protein expression (WT n = 4, Lama4−/− n = 4) in sWAT of male mice. Reported as fold change with respect to average of WT mice after normalization. *, **, ***, and **** indicate P less than or equal to .05, .01, .001, and .0001, respectively. Data are means + SD. B, Relative Ilk mRNA expression (n = 6) and ILK protein expression (n = 4) in adipocytes treated with control small interfering RNA (siRNA) and Lama4 siRNA. Reported as fold change with respect to paired control. C, Relative Ilk mRNA expression (n = 6) and ILK protein expression (n = 5) in adipocytes treated with control siRNA and Ilk siRNA. Reported as fold change with respect to paired control. D, Relative Ucp1, Tbx1, Prdm16, and Cidea mRNA expression in adipocytes treated with control siRNA (n = 6) and Ilk siRNA (n = 6). E, Protein expression of UCP1 in adipocytes treated with control siRNA (n = 5) and Ilk siRNA (n = 5) was assessed by Western blot. Vinculin was used as the loading control for normalization. F, Relative Cpt1a and Cpt1b mRNA expression in adipocytes treated with control siRNA (n = 6) and Ilk siRNA (n = 6). G, Protein expression of mitochondrial respiratory chain complexes I, II, III, and V, and VDAC in adipocytes treated with control siRNA (n = 5) and Ilk siRNA (n = 5) was assessed by Western blot. Vinculin was used as the loading control for normalization.
The difference in outcome may be due to the different models applied in the investigation: long-term knockout vs acute partial silencing. One possibility is that an acute response to LAMA4 loss is the elevation of ILK and mediators of this signaling pathway. Moreover, these results were not necessarily representative of ILK activity. ILK is a serine-threonine kinase that transduces extracellular signals through the formation of signaling complexes. It is not fully understood how ILK is activated under different circumstances, and its activity as a kinase or pseudokinase is highly debated because of unusual aspects of the kinase domain (24). It has been suggested that even if ILK does possess kinase capacity, the kinase and signaling activities may be independent of one another (26). Therefore, based on these data alone we could not determine whether ILK signaling was altered when LAMA4 was absent.
As we were unable to clearly assess ILK signaling activity through these models, we decided to directly silence Ilk gene expression in adipocytes to recapitulate the phenotype discovered in our in vivo model and to determine if reductions in ILK could modulate thermogenesis in adipocytes. We transfected adipocytes with siRNA targeting Ilk in the same manner as previously described and achieved efficient silencing on the mRNA and protein level (Fig. 5C). Markers of beige adipocytes were again upregulated compared to the control siRNA condition, including a nearly 30% elevation in UCP1 protein expression (P = .03) (Fig. 5D and 5E). We also observed the same CPT1 isoform shift as previously seen, a significant downregulation of Cpt1a and upregulation of Cpt1b expression, when ILK was silenced (Fig. 5F). Although we did not observe an elevation in the expression of most mitochondrial proteins when ILK was silenced, we did observe a small but statistically significant increase in the expression of the mitochondrial respiratory chain complex V (Fig. 5G).
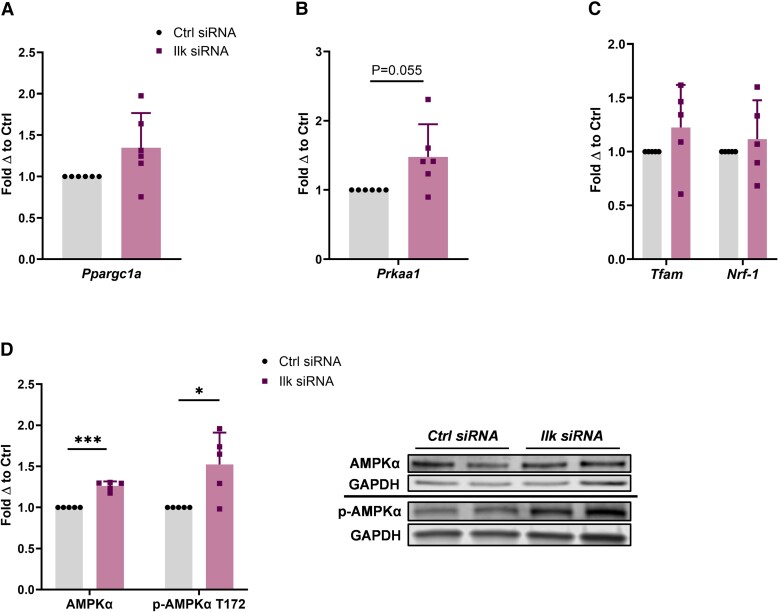
Last, we found that ILK silencing led to a slight upregulation in Ppargc1a expression, although not significant, and a near-significant upregulation in AMPKα expression at the mRNA level (P = .055) (Fig. 6A and 6B). The expression of Tfam and Nrf-1 trended upward, but this change was not significant (Fig. 6C). However, at the protein level, we found that both total and phospho-AMPKα levels and were significantly elevated when ILK was silenced (P = .0005 and .03, respectively) (Fig. 6D).
Figure 6.
AMPKα expression increases when ILK is silenced. A, Relative Ppargc1a messenger RNA (mRNA) expression (PGC-1α) in adipocytes treated with control small interfering RNA (siRNA) (n = 6) and Ilk siRNA (n = 6). Reported as fold change with respect to paired control. *, **, and *** indicate P less than or equal to .05, .01, and .001, respectively. Data are means + SD. B, Relative Prkaa1 mRNA expression (AMPKα) in adipocytes treated with control siRNA (n = 6) and Ilk siRNA (n = 6). C, Relative Tfam and Nrf1 mRNA expression in adipocytes treated with control siRNA (n = 5) and Ilk siRNA (n = 5). D, Protein expression of AMPKα and p-AMPKα T172 in adipocytes treated with control siRNA (n = 5) and Ilk siRNA (n = 5) was assessed by Western blot. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as the loading control for normalization.
Discussion
During the course of this study, we investigated the inhibitory effect of the ECM protein LAMA4 on adipocyte thermogenesis and mitochondrial biogenesis in multiple models. We showed that the acute silencing of LAMA4 during beige adipocyte differentiation allowed for a greater elevation in thermogenic gene expression, and that the treatment of human adipocytes with LN411 during beige differentiation inhibited UCP1 expression. We also found that both the complete genetic knockout and the acute silencing of LAMA4 led to elevations in adipocyte expression of PGC-1α, a master regulator of mitochondrial biogenesis. In an effort to identify potential signaling mediators through which this regulation may occur, we investigated ILK expression in our models. We discovered that ILK was downregulated in the sWAT of Lama4−/− mice and that silencing of ILK in adipocytes resulted in elevated thermogenic gene expression, conflicting outcomes that led us to directly silence ILK in adipocytes to study its relationship with thermogenesis. Interestingly, both LAMA4 and ILK silencing led to the elevation of AMPKα expression, while treatment of human beige adipocytes with LN411 suppressed AMPKα expression. Overall, the work described in this study indicates that LAMA4 negatively regulates the AMPK-PGC-1α pathway to inhibit adipocyte beiging. Although we identify a role for ILK in the regulation of adipocyte thermogenesis and AMPKα expression, the connection to LAMA4 requires further investigation.
One of the main outcomes of this study was the identification of LAMA4 as a negative regulator of AMPKα levels. AMPK is a serine-threonine kinase involved in maintaining cellular energy levels and is known to negatively regulate adipogenesis and promote a brown adipocyte phenotype (27). AMPK becomes active when AMP and adenosine diphosphate levels are high in comparison to ATP, and functions to minimize energy intensive processes while increasing ATP production. When activated, AMPK increases fatty acid uptake and oxidation in addition to promoting the expression and activity of PGC-1α, leading to elevations in mitochondrial biogenesis (28). Previous studies have shown that the direct activation of AMPKα in murine sWAT promotes beiging and improves metabolic parameters, while silencing AMPKα in sWAT compromises adaptive thermogenesis (20). Our results suggest that LAMA4 acts to suppress AMPKα levels, likely for the promotion of white adipogenesis, while the silencing of LAMA4 relieves this inhibition and allows for the development of a more thermogenic phenotype.
Interestingly, AMPK is also known to enhances CPT1 activity by phosphorylating and inhibiting acetyl-CoA carboxylases (20). The majority of work studying shifts in CPT1 isoform expression has taken place in cardiomyocytes and suggests that shifting isoform expression can lead to changes in long-chain fatty acid oxidation rates. Specifically, that overexpression of CPT1a leads to reductions in long-chain fatty acid oxidation rates (29). As we saw an increase in the ratio of Cpt1b to Cpt1a expression with LAMA4 and ILK silencing, it would be interesting to investigate the basis for this shift and to understand how the change may connect to the observed elevated AMPKα expression in our models.
As part of this study, we also identified a role for ILK in mediating adipocyte thermogenesis and identified expression changes in ILK related to LAMA4 deficiency. However, were unable to determine a definitive link between ILK activity and LAMA4 signaling at this time. ILK possesses several means of activation, few of which are well understood. It can execute downstream functions through enzymatic activity, protein-protein interactions, or localization to specific regions of the cell (30). Historically, ILK's function as a kinase or a pseudokinase has been controversial, including how its various behaviors are stimulated. ILK has been proposed to be autophosphorylated at Ser-343, but it is unknown exactly how this contributes to its activity. It can also be phosphorylated at Thr-173 and Ser-246, but again, little is known about how these phosphorylation states or other posttranslational modifications alter activity (24, 30). In addition, multiple isoforms of ILK exist and purportedly display differing characteristics, although ILK1 is the most commonly studied and typically the isoform that most commercial ILK antibodies recognize (24). As interest in ILK grows, and tools to study its activity and various isoforms are more available, we may develop an understanding of whether LAMA4 modulates ILK function and signaling in the future.
Within the scope of this study, we showed that ILK expression is downregulated in sWAT in vivo when LAMA4 is absent. We also found that silencing of ILK leads to many of the same downstream changes observed when LAMA4 was silenced, including elevations in thermogenic gene expression and AMPKα expression, in addition to a shift in CPT1 isoform expression. Unfortunately, we were unable to assess direct changes in ILK activity or signaling in response to LAMA4 silencing in this study. More studies will be needed in the future to determine if the suppression of thermogenesis by LAMA4 is definitively modulated through an integrin-ILK signaling axis.
Last, another main conclusion we drew from this study was the applicability of this LAMA4 regulatory function in human adipocytes. Our results established that LAMA4 acts to negatively regulate the thermogenic program in human adipocytes, which is an important finding for therapeutic and tissue engineering–related applications. As laminins are becoming more frequently incorporated into 3-dimensional scaffolds for tissue growth, it is important to be conscious of which α chains may be functional in supporting specific outcomes, either in stem cell differentiation or therapeutics (31-33). Additionally, functional blocking of the LAMA4 chain may be a viable future therapeutic target to promote the beiging of WAT and combat obesity. Further work is needed to assess how the downregulation of LAMA4 signaling might affect adipose in vivo, and to determine ideal silencing time frames to achieve effective enhancements in energy expenditure.
Acknowledgments
We would like to acknowledge the Diabetes Research Training Center (DRTC) at the University of Chicago for supporting our work on the study of LAMA4 and providing training and guidance on the culture, maintenance, and differentiation of hiPSCs.
Abbreviations
- ADSCs
adipose-derived stem cells
- ADIPOQ
adiponectin
- AMPK
adenosine 5′-monophosphate–activated protein kinase
- ATP
adenosine triphosphate
- Cidea
cell death activator CIDE-A
- CPT1
carnitine palmitoyltransferase 1
- DMEM
Dulbecco’s modified Eagle’s medium
- ECM
extracellular matrix
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- hiPSC
human induced pluripotent stem cell
- Ilk
integrin-linked kinase
- ITGA7
integrin alpha 7
- ITGB1
integrin beta 1
- Lama4
Laminin-α4
- mRNA
messenger RNA
- Nrf-1
nuclear respiratory factor 1
- PCR
polymerase chain reaction
- PGC-1
peroxisome proliferator-activated receptor γ coactivator-1
- PPARG
peroxisome proliferator-activated receptor γ
- PRDM16
PR domain containing 16
- siRNA
small interfering RNA
- sWAT
subcutaneous white adipose tissue
- T3
3,5,3′-triiodothyronine
- Tbx1
T-box 1
- Tfam
transcription factor A
- Ucp1
uncoupling protein-1
- VDAC
voltage-dependent anion channel
- WAT
white adipose tissue
- WT
wild-type
Contributor Information
Anna Goddi, Committee on Molecular Metabolism and Nutrition, The University of Chicago, Chicago, Illinois 60637, USA.
Alanis Carmona, Section of Endocrinology, Diabetes, and Metabolism, The University of Chicago, Chicago, Illinois 60637, USA.
Soo-Young Park, Section of Endocrinology, Diabetes, and Metabolism, The University of Chicago, Chicago, Illinois 60637, USA.
Gokhan Dalgin, Section of Endocrinology, Diabetes, and Metabolism, The University of Chicago, Chicago, Illinois 60637, USA.
Maria A Gonzalez Porras, Department of Biomedical Engineering and Chemical Engineering, The University of Texas at San Antonio, San Antonio, Texas 78249, USA.
Eric M Brey, Department of Biomedical Engineering and Chemical Engineering, The University of Texas at San Antonio, San Antonio, Texas 78249, USA.
Ronald N Cohen, Committee on Molecular Metabolism and Nutrition, The University of Chicago, Chicago, Illinois 60637, USA; Section of Endocrinology, Diabetes, and Metabolism, The University of Chicago, Chicago, Illinois 60637, USA.
Financial Support
This work was supported, in part, by the University of Chicago DRTC (National Institutes of Health grant No. P30 DK020595). Maria A. Gonzalez Porras is supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health, under award No. F32-0DK122754.
Author Contributions
A.G., A.C., G.D., and R.N.C. designed the experiments, which were performed by A.G., A.C., S.Y.P., and G.D.; M.G.P. and E.M.B. provided expertise and consulted on experimental design and results; S.Y.P. and G.D. supported procedures related to hiPSC culture, differentiation, and treatment; M.G.P. supported siRNA transfection protocol optimization; A.G., A.C., and G.D. performed all data analysis; A.G. wrote the manuscript with contributions from A.C.; and R.N.C. and E.M.B. supervised the project.
Disclosures
The authors have nothing to disclose.
Data Availability
All generated data included in this publication can be found in this article or in the data repository listed in “References.”
References
- 1. Cheng L, Wang J, Dai H, et al. Brown and beige adipose tissue: a novel therapeutic strategy for obesity and type 2 diabetes mellitus. Adipocyte. 2021;10(1):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee JH, Park A, Oh KJ, Lee SC, Kim WK, Bae KH. The role of adipose tissue mitochondria: regulation of mitochondrial function for the treatment of metabolic diseases. Int J Mol Sci. 2019;20(19):4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ricquier D. Uncoupling protein 1 of brown adipocytes, the only uncoupler: a historical perspective. Front Endocrinol (Lausanne). 2011;2:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J Cell Sci. 2010;123(Pt 24):4195–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Urra FA, Fuentes-Retamal S, Palominos C, Rodríguez-Lucart YA, López-Torres C, Araya-Maturana R. Extracellular matrix signals as drivers of mitochondrial bioenergetics and metabolic plasticity of cancer cells during metastasis. Front Cell Dev Biol. 2021;9:751301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen K, Wang Y, Deng X, Guo L, Wu C. Extracellular matrix stiffness regulates mitochondrial dynamics through PINCH-1- and kindlin-2-mediated signalling. Curr Res Cell Biol. 2021;2:100008. [Google Scholar]
- 7. Aumailley M. The laminin family. Cell Adhes Migr. 2013;7(1):48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Suzuki N, Yokoyama F, Nomizu M. Functional sites in the laminin alpha chains. Connect Tissue Res. 2005;46(3):142–152. [DOI] [PubMed] [Google Scholar]
- 9. Niimi T, Kumagai C, Okano M, Kitagawa Y. Differentiation-dependent expression of laminin-8 (α4β1γ1) mRNAs in mouse 3T3-L1 adipocytes. Matrix Biol. 1997;16(4):223–230. [DOI] [PubMed] [Google Scholar]
- 10. Noro A, Sillat T, Virtanen I, et al. Laminin production and basement membrane deposition by mesenchymal stem cells upon adipogenic differentiation. J Histochem Cytochem. 2013;61(10):719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vaicik MK, Blagajcevic A, Ye H, et al. The absence of laminin α4 in male mice results in enhanced energy expenditure and increased beige subcutaneous adipose tissue. Endocrinology. 2018;159(1):356–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vaicik MK, Kortesmaa JT, Movérare-Skrtic S, et al. Laminin α4 deficient mice exhibit decreased capacity for adipose tissue expansion and weight gain. PLoS One. 2014;9(10):e109854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gonzalez Porras MA, Stojkova K, Vaicik MK, et al. Integrins and extracellular matrix proteins modulate adipocyte thermogenic capacity. Sci Rep. 2021;11(1):5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goddi A, Carmona A, Schroedl L, et al. Laminin-α4 is upregulated in both human and murine models of obesity. Front Endocrinol (Lausanne). 2021;12:886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thyboll J, Kortesmaa J, Cao R, et al. Deletion of the laminin α4 chain leads to impaired microvessel maturation. Mol Cell Biol. 2002;22(4):1194–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ikeda K, Kang Q, Yoneshiro T, et al. UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nat Med. 2017;23(12):1454–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guénantin AC, Briand N, Capel E, et al. Functional human beige adipocytes from induced pluripotent stem cells. Diabetes. 2017;66(6):1470–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Warfel JD, Vandanmagsar B, Dubuisson OS, et al. Examination of carnitine palmitoyltransferase 1 abundance in white adipose tissue: implications in obesity research. Am J Physiol Integr Comp Physiol. 2017;312(5):R816–R820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cox TR, Erler JT. Remodeling and homeostasis of the extracellular matrix: implications for fibrotic diseases and cancer. Dis Model Mech. 2011;4(2):165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu L, Zhang L, Li B, et al. AMP-activated protein kinase (AMPK) regulates energy metabolism through modulating thermogenesis in adipose tissue. Front Physiol. 2018;9:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ross E, Ata R, Thavarajah T, et al. AMP-activated protein kinase regulates the cell surface proteome and integrin membrane traffic. PLoS One. 2015;10(5):e0128013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garcia-Roves PM, Osler ME, Holmström MH, Zierath JR. Gain-of-function R225Q mutation in AMP-activated protein kinase γ3 subunit increases mitochondrial biogenesis in glycolytic skeletal muscle. J Biol Chem. 2008;283(51):35724–35734. [DOI] [PubMed] [Google Scholar]
- 23. Takada Y, Ye X, Simon S. The integrins. Genome Biol. 2007;8(5):215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Górska A, Mazur AJ. Integrin-linked kinase (ILK): the known vs. the unknown and perspectives. Cell Mol Life Sci. 2022;79(2):100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hall ML, Givens S, Santosh N, Iacovino M, Kyba M, Ogle BM. Laminin 411 mediates endothelial specification via multiple signaling axes that converge on β-catenin. Stem Cell Reports. 2022;17(3):569–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dobner S, Amadi OC, Lee RT. Cardiovascular mechanotransduction. Muscle. 2012;1:173–186. [Google Scholar]
- 27. Ahmad B, Serpell CJ, Fong IL, Wong EH. Molecular mechanisms of adipogenesis: the anti-adipogenic role of AMP-activated protein kinase. Front Mol Biosci. 2020;7:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jeon SM. Regulation and function of AMPK in physiology and diseases. Exp Mol Med. 2016;48(7):e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lewandowski ED, Fischer SK, Fasano M, et al. Acute L-CPT1 overexpression recapitulates reduced palmitate oxidation of cardiac hypertrophy. Circ Res. 2013;112(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hannigan GE, McDonald PC, Walsh MP, Dedhar S. Integrin-linked kinase: not so ‘pseudo’ after all. Oncogene. 2011;30(43):4375–4385. [DOI] [PubMed] [Google Scholar]
- 31. Junka R, Valmikinathan CM, Kalyon DM, Yu X. Laminin functionalized biomimetic nanofibers for nerve tissue engineering. J Biomater Tissue Eng. 2013;3(4):494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baskapan B, Callanan A. Electrospinning fabrication methods to incorporate laminin in polycaprolactone for kidney tissue engineering. Tissue Eng Regen Med. 2022;19(1):73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li G, Chen K, You D, et al. Laminin-coated electrospun regenerated silk fibroin mats promote neural progenitor cell proliferation, differentiation, and survival in vitro. Front Bioeng Biotechnol. 2019;7:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All generated data included in this publication can be found in this article or in the data repository listed in “References.”