Abstract
Extracellular vesicles (EVs) are nanosized vesicles with a lipid bilayer that are released from cells of the cardiovascular system, and are considered important mediators of intercellular and extracellular communications. Two types of EVs of particular interest are exosomes and microvesicles, which have been identified in all tissue and body fluids and carry a variety of molecules including RNAs, proteins, and lipids. EVs have potential for use in the diagnosis and prognosis of cardiovascular diseases and as new therapeutic agents, particularly in the setting of myocardial infarction and heart failure. Despite their promise, technical challenges related to their small size make it challenging to accurately identify and characterize them, and to study EV-mediated processes. Here, we aim to provide the reader with an overview of the techniques and technologies available for the separation and characterization of EVs from different sources. Methods for determining the protein, RNA, and lipid content of EVs are discussed. The aim of this document is to provide guidance on critical methodological issues and highlight key points for consideration for the investigation of EVs in cardiovascular studies.
Keywords: Exosomes, Microvesicles, Cardiovascular diseases, Biodistribution, Therapeutics, Blood, Heart, Extracellular vesicle composition
1. Pathophysiological relevance of EVs in the cardiovascular field
In recent years, extracellular vesicles (EVs) such as exosomes and microvesicles have gained significant interest as mediators of intercellular communication in both the healthy physiological state and during pathophysiological stress.1–4 All cell types in the cardiovascular system release EVs.5 However, most mechanistic studies use cell culture-derived EVs. EVs are also detected in plasma, where they are derived primarily from erythrocytes, platelets, endothelial, and immune cells.6 The plasma EV content responds to environmental changes and can regulate pro-inflammatory and innate immune responses, coagulation pathways, and atherogenic interactions.7 It is therefore of interest to understand the function of EVs in the cardiovascular system.
Several characteristics make EVs promising biomarkers for cardiovascular pathologies.1 For example, EVs are secreted into body fluids such as blood, lymph, and pericardial fluid, and EV molecular cargo reflects the state of the cell of origin. Therefore, by purifying EVs it is possible to enrich for diagnostic markers that may otherwise be obscured by the large quantity of proteins present in the fluid.3 For example, acute coronary syndrome results in the rapid appearance of EVs in plasma that can be purified, aiding the identification of specific microRNAs (miRNAs),8 in comparison to the detection of cardiac miRNAs in total plasma, which is inferior to high-sensitivity assays for traditional markers of damaged myocardium such as troponins.9,10 Cardiac allograft rejection can be predicted with an accuracy of 86% based on the concentration and contents of EVs released by the transplanted heart into the blood, potentially eliminating the need for endomyocardial biopsy.11 miRNA signatures in circulating large EVs, in contrast to freely circulating miRNAs, predicted the occurrence of cardiovascular events in patients with coronary artery disease (CAD),12 highlighting the prognostic potential of EV-miRNA expression pattern.
In certain situations, EVs can contribute to the mechanism of cardiovascular diseases (CVDs). For example, sEVs contribute to the development of pulmonary arterial hypertension13,14 and to vascular calcification.15,16 Adipocyte-derived extracellular vesicles and their ceramide content have an impact on cardiac mortality in advanced atherosclerosis.16,17 Endothelial EVs released during myocardial infarction (MI) can mobilize splenic neutrophils and monocytes following their transcriptional activation and could contribute to attenuated cardiac function.18,19 Therefore, EVs are emerging as key players in different stages of disease development of CVD and metabolic syndrome (reviewed in Boulanger et al.20 Martínez and Andriantsitohaina,21 and Jansen et al.22).
EVs are also promising therapeutic agents for treating CVD. They have been shown to mediate various beneficial effects of conditioned medium from stem cells.23,24 EVs can be separated from tissue-culture medium ‘conditioned' by the growth of cells, and there is growing interest in using such EVs for treating a variety of cardiovascular pathologies.5 For example, EVs purified from medium conditioned by exosomes cardiac progenitor cells, but not from normal dermal fibroblasts, are cardioprotective and proangiogenic in models of MI and chemotherapy-induced cardiotoxicity,25,26 and stimulate cardiovascular cell proliferation following MI.27 Similarly, platelet-derived EVs in endothelial progenitor cell cultures contributed to their proangiogenic activity.28,29 In another example, EV coating of stents accelerated their re-endothelialization and reduced in-stent restenosis compared with drug-eluting and bare-metal stents in mice.30
Currently, there are more than 250 clinical trials registered to use EVs in a range of diseases (ClinicalTrials.gov), as either biomarkers for response to drug treatment or as direct therapeutic mediators. It is, therefore, crucial that appropriate methods are used to separate, validate, and characterize EVs, both to improve their clinical application, and to provide fundamental insights and in-depth analyses of their mechanism of action. The aim of this document is to provide guidance on these critical methodological issues and highlight key points for consideration in the design of experiments using EVs. Some of the methods described can be applied generally to all studies using EVs, but we provide CV-specific methods where relevant.
1.1. Definition of extracellular vesicles and use of terminology
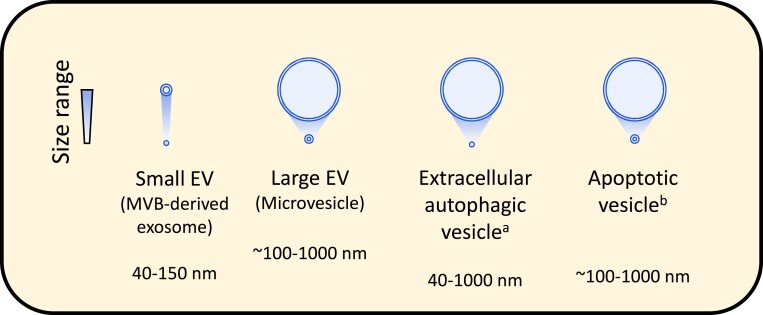
Three main classes of EVs can be distinguished by their mechanism of production: exosomes, microvesicles, and apoptotic bodies (Figure 1). Microvesicles and apoptotic bodies are released directly via outward budding of the plasma membrane in living or dying cells, respectively, and carry proteins, lipids, nucleic acids, and other active components that can affect target cells and modify their behaviour.4,5,32 Exosomes are produced by inward budding of late-stage endosomes, thereby forming intraluminal vesicles in multi-vesicular bodies (MVBs), which are released upon fusion of the limiting membrane of the MVB with the cell membrane.33 The formation of MVBs and subsequent fusion with the plasma membrane is a highly orchestrated mechanism involving the endosomal sorting complexes required for transport (ESCRT) machinery, which includes the proteins hepatocyte growth factor-regulated tyrosine kinase substrate, tumour susceptibility gene 101 protein (TSG101), signal transducing adapter molecule 1, and programmed cell death 6-interacting protein (PDCD6IP or ALIX), although ESCRT-independent mechanisms have also been reported.33 Precisely how cargo is sorted into exosomes is unclear, although some binding motifs have been suggested.33
Figure 1.
The typical size range of the major lipid-bilayer EVs up to 1000 nm diameter. aAs reported by Jeppesen et al.31, bthe size of apoptotic vesicles/bodies can range up to 5 μm in diameter. Please be aware that the diameter of EVs depends on the detection method used.
The umbrella term ‘EVs' encompasses various types of membrane-enclosed vesicles, including exosomes, microvesicles, extracellular autophagic vesicles, and apoptotic bodies, and these can have overlapping size ranges (Figure 1). However, there is no consensus on specific markers that can distinguish EV types. Consequently, and since it is challenging to isolate individual EV types with high purity, it is preferable to refer to the separated vesicles simply as ‘EVs' and report the purification methods used for their separation and characterization. The International Society of Extracellular Vesicles in their position paper, MISEV2018 strongly recommended the use of operational terms based on size [e.g. small (s), medium (m), or large (l) EVs], density range (e.g. low-, medium-, or high-density EVs), biochemical composition (e.g. CD63+ve EVs or Annexin 5+ve EVs), or culture- or cell type of origin (e.g. hypoxic EVs, cardiomyocyte-derived EVs, etc.), unless the biogenesis of the EVs was determined.32 However, it must be recognized that many of these terms are protocol-dependent and relative, so it is important that their use is clearly defined. Here, we use the term ‘sEVs' to refer to purified samples enriched in small EVs and MVB-derived exosomes, and ‘lEVs' to refer to preparations enriched in larger EVs and shed microvesicles.
2. Source of EVs
For investigations of cardiovascular EV function, primary cells, blood, or explanted cardiac tissue may be preferred. When the aim is to develop EVs as therapeutic agents, and large quantities are required, readily expandable cells, or cell lines may be preferable. Mesenchymal stromal cells (MSCs) are a popular source as they are cytoprotective, can improve cardiac contractility and calcium handling and have beneficial immunomodulatory effects including in the setting of atherosclerosis and pulmonary hypertension.14,34–36
EVs from many different sources have been shown to improve cardiac function following MI, including cardiac stem cells,37 cardiovascular progenitor cells (CPCs),38 endothelial progenitor cells,39 cardiosphere-derived cells,40 embryonic stem cells,41 and iPSC-derived cardiomyocytes42 (reviewed in Sluijter et al.5). EVs from the epicardium can promote the proliferation of cardiomyocytes.43 EVs can also be beneficial against other forms of injury such as doxorubicin/trastuzumab-induced cardiac toxicity.26 On the other hand, EVs can be detrimental, for example contributing to vascular smooth muscle cell calcification.15,16 As yet, there is little consensus on the ideal source of EVs; however, one head-to-head comparison suggests CPC may be more efficacious than bone marrow-derived mesenchymal stem cell.25
Certain stimuli can alter EV production and function, in a cell-type dependent manner, including calcium,44 hypoxia/ischaemia,45 shock wave therapy,46 atorvastatin,47 and exercise.48,49 Conversely, CVD can alter EV production and function. For example, MI increase EV release,50 EV-miR-mediated vascular intercellular communication is altered in patients with CAD and chronic kidney disea (CKD), promoting CKD-induced endothelial dysfunction,51 and diabetes mellitus impairs EV function.52,53
Cells can be cultured in standard tissue-culture flasks, or bioreactor flasks or hollow-fibre reactors may be used to maximize production. However, it is important to realize that culture conditions can affect sEV contents and activity significantly.54
3. Methods of separation
The optimal method for separating EVs depends on which biofluid or tissue is used as a source.
3.1. Separation of EVs from the cell culture medium
Several techniques have been developed for the separation of EVs from the cell culture medium, each with its advantages and disadvantages (Table 1). Most procedures are based on separation by size, and/or density, although many other extracellular particles may share these characteristics with EVs. A protocol of differential centrifugation or ultracentrifugation published by Thery et al.55 is commonly used to separate both sEVs and lEVs (Box 1). A subsequent density-gradient separation using sucrose or, preferably, iodixanol, further improves EV purity.56 Size-exclusion chromatography has become popular since it effectively removes part of the contaminating soluble protein, and columns can be readily made or purchased (Figure 2D).57,58 Precipitation of sEVs is possible using polyethylene glycol (PEG)-based reagents, for example in HEK293 or MSC cultures,59 but the purity obtained is generally inferior to other techniques.56,60 Ultrafiltration is more commonly used as an initial clean-up step to remove larger (e.g. >0.8 μM) contaminants because membranes can become blocked when filtering large volumes and because of concerns that high pressures may damage the membranes of lEVs. Affinity isolation, typically using antibodies, provides highly pure isolates although at the expense of yield, and only a subset of EVs might be isolated.31 Furthermore, the procedure to recover EVs from antibodies could affect their functionality and requires testing.61 Diafiltration, asymmetric flow field-flow fractionation (AF4)62 and tangential flow filtration63 purify and concentrate sEV fractions and are scalable, but AF4 requires specialized and expensive equipment.
Table 1.
Potential advantages and disadvantages of the main methods used to purify sEVs
| Method of purification | Disadvantages | Advantages |
|---|---|---|
| Affinity-based methods |
|
|
| Diafiltration |
|
|
| Centrifugation (pelleting) |
|
|
| Density gradient centrifugation |
|
|
| Field-flow fractionation |
|
|
| Precipitation |
|
|
| Size-exclusion chromatography |
|
|
| Tangential flow filtration |
|
|
| Ultrafiltration through a membrane |
|
|
Box 1. The standard differential ultracentrifugation protocol for EV isolation, originally published by Thery et al.55.
Centrifuge sample at 300 g for 10 min, at 4°C (remove cells and cell debris).
Centrifuge supernatant at 2000 g for 10 min, at 4°C (remove larger complexes).
Centrifuge supernatant at 10 000 g for 30 min, at 4°C (microvesicles are in the pellet).
Centrifuge supernatant at 100 000 g for 70 min, at 4°C in ultracentrifuge (EVs are in the pellet).
Re-suspend the pellet containing EVs and contaminating proteins.
Centrifuge 100 000 g 70 min, 4°C in ultracentrifuge to wash (sEVs/exosomes are in the pellet).
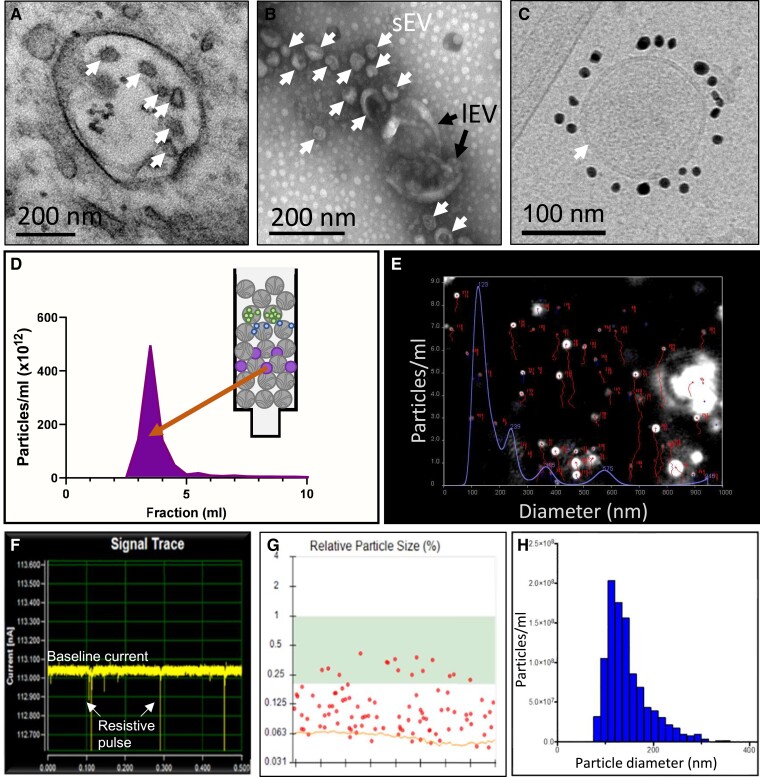
Figure 2.
Representative images of different techniques of EV characterization. (A) Transmission electron micrography (TEM) of multi-vesicular body (MVB) containing exosomes (arrows) in primary HUVECs. (B) Transmission electron micrography (TEM) of negative-stained EVs isolated from HUVECs (sEV = small EVs, lEV = large EVs). (C) Cryo-TEM of a single CD81 + EV from iPS-derived cardiovascular progenitor cells.38 The lipid bilayer is clearly resolved (arrow). (D) Fractionation of sEVs (purple) from proteins (green, blue) by size-exclusion chromatography. (E) Single frame from NTA of an sEV sample under constant flow, showing particle tracks (red) and particle size distribution (blue). (F) Representative trace of EV sample obtained using resistive pulse sensing (RPS). (G) Individual particles detected by RPS, with size determined relative to calibration beads of a known size. (H) Size distribution of EVs obtained by RPS.
Several head-to-head comparisons of EV separation procedures have been published,56,60,64,65 for human plasma, urine, and also specific cardiac-derived progenitor cells, but ultimately, the optimal method and obtained quantity depends on the source of the biofluid, the amount of available biofluid and the intended use. For clinical analyses of thousands of blood samples for EV-associated biomarkers, rapid precipitation might be sufficient but for mechanistic studies, purer EVs are essential. The use of cell culture medium as a source of EVs allows for more rigorously controlled conditions for EV production, but the cell culture environment differs from in vivo physiology. Given the challenge of removing contaminating serum EVs, protein and lipoproteins, when highly pure EVs are required for omics analysis or functional investigation, it is advisable to harvest EVs from cells grown in chemically defined medium rather than EV-depleted serum or serum-replacement supplements. However, control experiments must be in place to assess cell viability and contents of contaminating apoptotic bodies, when removing serum. EV-depleted serum may be used but still contains large quantities of proteins and lipoproteins which can co-isolate with EVs and are common contaminants of EVs, and procedural controls are necessary to check for the potential contaminant.66
3.2. Separation of EVs from blood
A critical consideration when separating EVs from blood is the pre-analytical procedures (Table 2).67,68 For instance, EVs can be separated from either plasma or serum, but serum preparation causes platelet activation, which releases large numbers of platelet-derived EVs, and the thrombus formed traps some of the EVs.69 The yield of EVs separated from plasma can be affected by the type of anticoagulant used and requires great care to prevent platelet activation and haemolysis. It is possible to use any of the methods described above to separate EVs from platelet-free plasma. Plasma contains only ∼108–1010 sEVs/mL and ∼106 lEVs/mL compared with ∼1016 lipoprotein particles/mL and large quantities of albumin, globulins, and other proteins and substances, which greatly complicates the isolation of EVs.70,71 However, by combining several orthogonal methods it is possible to improve both yield and purity of EVs.72 Given the many variables that can substantially influence EV yield and purity, it is essential that all pre-analytical procedures and residual contaminants are comprehensively reported alongside the separation method.73
Table 2.
Major factors to consider when isolating EVs from sources relevant to cardiovascular studies
| Source of EVs | Major factors to consider | Potential solutions |
|---|---|---|
| Cell culture conditioned medium containing serum |
|
|
| Cell culture conditioned medium without serum |
|
|
| Plasma |
|
|
| Serum |
|
|
| Tissue (e.g. myocardium) |
|
|
The importance of these points will vary depending on the intended use of the EVs, and must be evaluated separately for each experiment.
As noted in the main text, these solutions can introduce problems of their own.For example, EV removal from serum also removes other components, and it is probably not possible to remove 100% of the EVs. Serum-free medium may negatively affect cell health and EV quality.
3.3. Separation of EVs from tissue
The isolation of EVs from tissues has considerable scientific interest for understanding their local and remote roles in CVD development. Their presence should first be confirmed in situ, e.g. electron microscopy can identify the presence of vesicle structures in pathological samples such as human atherosclerotic plaques, ischaemic heart and muscles, or the brain.74,75 EV separation from fresh tissues represents a challenging task as the method used should ensure that isolated vesicles come from the extracellular space and do not result from tissue homogenization (cell death, membrane self-assembly; Table 2). Gentle mechanical disruption of tissue, optionally followed by enzymatic treatment, can be used to release EVs.44 EVs have been released by collagenase perfusion of Langendorff-perfused rat hearts followed by differential centrifugation.76,77 Appropriate controls should be considered to estimate the effects of the procedure. Therefore, using tissues from genetically modified models and processing healthy tissues or tissues from sham animal models in parallel to pathological samples might help evaluate the direct effect of tissue homogenization.50,74,78 Furthermore, the effect of the enzymatic cocktail on EV numbers and protein expression also requires investigation.76
4. General principles for EV identification and characterization
A number of recommendations have been published regarding how to characterize and confirm the identity, yield, and purity of EVs,2,5 but the most authoritative are the minimal information for studies of extracellular vesicles (MISEV) guidelines published by the International Society for Extracellular Vesicles (ISEV).32 A key overriding principle of the guidelines is that multiple, complementary techniques should be used to characterize EVs. Other guidelines have made quantifiable metrics to define the identity of MSC-sEV preparations, and facilitate stratification and comparison of different MSC-sEV preparations for therapeutic purposes.79
First, it is important to quantify the number of EVs relative to the total lipid or protein content of EV preparations obtained. The yield of EVs should be measured relative to the amount of starting material (e.g. number of secreting cells, volume of biofluid, or mass of tissue). This calculation should be performed every time EVs are isolated since it can vary significantly. Second, the presence of at least three positive protein markers of EVs (described below) is strongly suggested. Third, it is preferable to evaluate the presence of non-vesicular co-isolated components, e.g. apolipoproteins A1, A2, and B (APOA1, APOA2, and APOB), and albumin from plasma/serum isolates. Fourth, the presence of individual EVs should be demonstrated using, for example, electron microscopy or scanning probe microscopy. If an image with a single vesicle is shown then a wide-field image should also be shown, which helps to illustrate the purity. The most appropriate technique for characterization depends on the type of EV (large or small), as discussed below.
4.1. Techniques for identifying EVs
The most widely used techniques for quantifying EVs include light scattering techniques such as dynamic light scattering, nanoparticle tracking analysis (NTA), and resistive pulse sensing (RPS) (Figures 2E–H). However, the robustness and comparability of measurements is hampered by the lack of standardization, and the quantification of EVs is less straightforward than it seems.80 For example, each technology has different limitations and potential biases towards certain size ranges. An important limitation of most widely used techniques is that they measure all particles, and cannot distinguish between sEVs and lipoprotein particles, protein aggregates, EV aggregates, or other contaminants. Consequently, less pure isolates can paradoxically give the false impression of containing greater numbers of EVs. For this reason, it is preferable to use additional measurements such as total protein and/or lipid content to indicate the yield and purity.81 Alternatively, quantification of EV marker proteins by ELISA (enzyme-linked immunosorbent assay) or western blot (semi-quantitative) can be useful for comparing yields.
Since one of the defining features of exosomes is their size, this is another informative parameter to report when separating sEVs, although this is not specifically recommended in the MISEV2018 guidelines. The size distribution of EVs can be obtained using NTA or RPS, calculated from electron microscope images, or using another technique. A second defining feature of MVB-derived exosomes is that they contain proteins involved in MVB formation and/or exosome release (e.g. CD9, CD63, CD81, ALIX/PDCD6IP, TSG101).31 These can be used as positive protein markers to indicate the enrichment of MVB-derived exosomes within the separated EVs. The presence of at least three markers should be demonstrated.31,32 Notably, acetylcholinesterase is no longer considered a generic marker of exosomes.82
Large EVs have a less well-defined size range but can be analysed using similar techniques as for sEVs, or using flow cytometry, which is described below.2,80
4.2. Electron microscopy
Transmission electron microscopy (TEM) allows imaging at the single EV level, visualizing their size and morphology, as well as detecting the presence of contaminants. Negative staining with uranyl acetate is the most common method. Of note, drying during preparation results in a typical ‘collapsed vesicle' or ‘cup-shaped' appearance (Figure 2B).55 Nowadays, the gold-standard method for imaging biological objects is cryo-TEM, which preserves their native hydrated structure via rapid freezing. Cryo-TEM presents several major advantages, including the better capacity to distinguish bona fide EVs from non-vesicular particles and to determine the actual EV size, and to characterize heterogeneous EV samples, particularly the presence of EV aggregates either contained in the original sample or induced by isolation procedures. Combining EM with immuno-gold labelling aids with phenotyping of EVs in complex media, such as pure plasma or heterogeneous media (Figure 2C).83 Other techniques, including single EV-microarray and atomic force microscopy, can provide images of single EVs, as well as information on their biomechanical properties and size.84
4.3. Flow cytometry
Flow cytometry is an attractive technique for EV analysis, as flow cytometers are robust platforms, widely available and designed for high throughput quantitative analysis of single particles based on light scattering and fluorescence. However, flow cytometers are designed to analyse cells and several requirements need to be met to improve the rigour and reproducibility of EV analysis.85 Flow cytometric analysis of sEVs (<300 nm size) is particularly challenging due to their dim fluorescence and scatter signals.85 In this respect, it is extremely important to calibrate flow cytometers, confirm the detection of single EVs and be aware of the sensitivity of the platform used and potential interference by unbound fluorescent probes.86,87 Nevertheless, the use of single EV flow cytometric analysis has reached a level where reproducible comparisons of EV concentration measurements can be nearly performed, for example of circulating EVs in patients with CVD.88–90 Marker proteins of interest for cardiovascular studies include those such as CD61 and CD144 for platelets and endothelium, respectively, CD147 (SIRPα) for cardiomyocytes, CD235a for erythroid-derived EVs and leucocyte/lymphocyte- and monocyte-derived EVs (CD45/CD3 and CD14).88–91 The MIFlowCyt-EV Framework, drafted by an EV flow cytometry working group of ISEV–ISAC–ISTH (www.evflowcytometry.org), provided a consensus report for EV flow cytometric studies,86 advising the minimal experimental information that should be reported.
4.4. Functional analysis of EVs
Ideally, the functional activity of EVs would be assayed using a simple, in vitro potency assay as a surrogate for their in vivo functionality, but no single, universal method has been identified. In the cardiovascular field, EV function is commonly assessed using an assay of in vitro angiogenesis, cell viability, contractility, or combinations thereof. Commonly used in vitro assays of angiogenesis include the scratch assay,91 Boyden chamber migration assay,92,93 endothelial tube formation,94 and vessel sprouting assays.45,95,96 An accurate measure of sEV quantity and purity is important when conducting dose–response experiments of their functionality. At present, there is no consensus on which measure of quantity (particle number, protein content, quantity of starting cells, etc.) is preferable,32 but whichever normalization technique is used (preferably more than one) it should be reported and justified. Furthermore, appropriate (procedural) controls should be included to proof that effects are EV-mediated. For the use of EVs as therapeutic tools, in vitro potency assays are required to predict the effectiveness of EV preparations for clinical use, but this depends on the ability to convincingly identify the mechanism of action and quantify the biological activity.97
4.5. Reporting methodology
Finally, to aid reproducibility and transparency, isolation and characterization methodology should be reported in public databases and repositories such as EV-TRACK, a crowdsourcing knowledgebase (http://evtrack.org) that centralizes EV biology and methodology with the goal of stimulating authors, reviewers, editors, and funders to put experimental guidelines into practice.98
5. Methods for determining the protein content of EVs
5.1. Total protein content
Total protein content in an EV preparation can be estimated using standard protein assays such as bicinchoninic acid assay or Bradford assay, or variations thereof, optimized for low protein concentrations. Quantification of total protein in an EV sample and comparison with particle counts may give an indication of its purity. It has been suggested that pure sEV isolates contain concentrations of <1 µg protein/1010 EV particles,81 although this is not necessarily universally applicable, because there are not yet methods available that can measure all EVs.
5.2. Antibody-based techniques to identify specific proteins
There may be subpopulations of EVs with different protein content that can be detected using antibodies. Some can be used as marker proteins to identify the cell type of origin within the cardiovascular system (see section Separation of EVs from tissue). In addition to EV marker proteins, hundreds of additional proteins can be identified, which may be either genuine EV components or co-isolated proteins. The most common approaches to detect and quantify the relative levels of EV proteins are antibody-based experimental methods (Table 3).32 All antibody-based techniques require the use of appropriate controls to confirm antibody specificity.99
Table 3.
Advantages and disadvantages of common techniques used for EV detectiona
| Detection method | Advantages | Disadvantages |
|---|---|---|
| Capillary electrophoresis immunoassayb |
|
|
| DELFIAb |
|
|
| Dot blottingb |
|
|
| Flow cytometry |
|
|
| Imaging cytometerb |
|
|
| Immunoelectron microscopy (TEM or Cryo-TEM)b |
|
|
| Mass spectrometry |
|
|
| Sandwich ELISAb |
|
|
| Transmission electron microscopy (TEM) |
|
|
| Cryo-transmission electron microscopy (Cryo-TEM) |
|
|
| Western blottingb |
|
|
An important overarching consideration is whether isolation of EVs is necessary for subsequent analysis steps. For example, some analysis techniques such as flow cytometry can be optimized to work in the presence of (diluted) plasma or serum, negating the need for purification and its attendant limitations and inherent variability.
All techniques using antibodies require validation of antibody specificity and optimization of their concentrations and blocking reagents.
Western blotting can identify proteins that are associated or co-isolated with EVs and provide useful information about the yield and purity of an EV preparation.64 Importantly, it can also confirm the molecular weight of the target protein. Compared with cell lysates, a disadvantage of EV samples is the lack of reference (‘house-keeping') proteins to use for normalization purposes in immunoblotting experiments. Therefore, equal protein amount, volume from which EVs are separated or particle number are commonly used. Inclusion of the original sample, the EV-depleted sample and procedural control samples are required to draw firm conclusions about the enrichment of proteins in the EV isolate (or depletion of contaminants). Western blotting can be challenging since it requires relatively large quantities of EVs for sufficient sensitivity. Alternative versions such as dot blotting or capillary electrophoresis immunoassays can provide considerably higher sensitivity.100
The question of which proteins should be investigated as potential contaminants is debated, but the best guideline is provided by MISEV.32 Depending on the source of EVs, it can be useful to verify the removal of lipoproteins (e.g. APOB, APOA1, APOA2) and serum albumin (Figure 3), and proteins from the endoplasmic reticulum or plasma membrane.
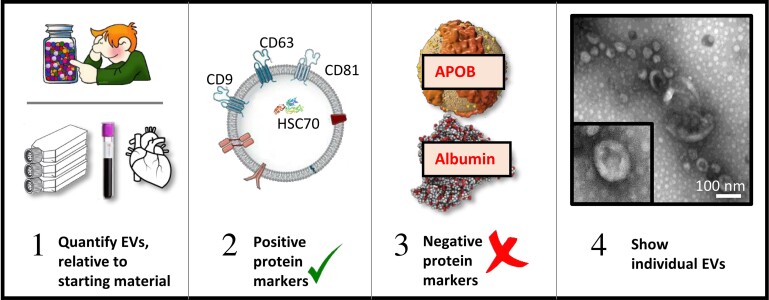
Figure 3.
Steps towards EV characterization, adapted from MISEV2018 guidelines.32 (i) Determine the quantity of EVs obtained, relative to the amount of starting material. (ii) Verify the presence of at least three positive protein markers of small EVs, including one transmembrane or GPI-anchored protein (e.g. CD9, CD63, CD81, NT5E/CD73), and one cytosolic, luminal protein (e.g. ALIX/PDCD6IP, HSC70). For large EVs, a wide range of surface markers such as integrins from the cell of origin may be used. (iii) Preferably, demonstrate the relative abundance of significant contamination by non-vesicular, co-isolated components such as lipoproteins (APOB, APOA1, APOA2) or albumin. (iv) Characterize individual EVs, with images of single EVs (both wide-field and close-up).
ELISA is a well-established technique that can provide sensitive antibody-based detection in multi-well formats. A sandwich ELISA format (combining separate capture and detection antibodies) is likely to be required when using enzyme-linked or fluorescent detection, but a highly sensitive immunoassay variant based on time-resolved fluorescence called DELFIA (dissociation-enhanced lanthanide fluorescence immunoassay) is able to detect EV-associated molecules using a single detection antibody.64,101 Similar to dot blots, immunoassays provide good sensitivity for small sample amounts, but require thoroughly validated antibodies and do provide information to validate the molecular weight.
EV flow cytometry can be used to detect surface protein markers as indicated above. Immuno-gold labelling can be performed for visualization using TEM or cryo-TEM, although it is not quantitative, and it is mostly used to label EV membrane proteins. Detection of immuno-gold label on non-EV particles in the sample may indicate that the target is only a contaminant in the EV isolate.
Novel antibody-based approaches such as surface plasmon resonance102 and interferometric imaging103 have also been used for EV protein characterization, but they usually require expensive specialized equipment and consumables which limit their widespread use.
5.3. Mass spectrometry of the EV proteome
Proteomic analysis of EV samples by mass spectrometry (MS) provides the most comprehensive analysis of the EV protein cargo (Table 3), and does not rely on an a priori selection of proteins based on the availability of antibodies or other affinity reagents for specific proteins.31,104 MS approaches, however, have an inherently lower sensitivity compared with antibody-based techniques. This is mainly due to the excess amounts of highly abundant proteins (e.g. albumin) in the EV preparations which mask the presence of low-abundant EV proteins.105 To address this, MS can be combined with better isolation techniques for EVs that result in less contamination. It is recommended to compare the EV proteome to the tissue or cell source of the EV sample to identify the degree of enrichment/depletion of proteins. For EVs separated from cell cultures in which media are supplemented with xenogenous components (e.g. bovine serum), it is also recommended to searches against databases of other organisms. Bovine serum proteins are a common contaminant in EVs isolated from cell cultures, unless cells are grown in serum-free media. Finally, independent validation with an antibody-based technique is advisable since MS detects peptides, which can originate from both intact and fragmented proteins. Most journals require that EV proteomic data are deposited in online databases.106
5.4. Intraluminal vs. membrane proteins
Determining whether a protein is intraluminal, membrane, or external to the EVs is of great importance for understanding the structure, origin, and function.32 Mixing a broad-range protease (e.g. proteinase K) with an EV-containing sample in the presence or absence of detergent can help to establish whether a protein is intraluminal or present on the surface/outside of the EVs. Notably, EV subtypes have different sensitivities to detergents.107 Detergents will also disrupt other lipid structures such as lipoproteins, another common contaminant in EV preparations. Protease treatment can also determine the topology of membrane proteins or the degree of contamination of an EV sample,108 but proteases will digest the extracellular domains of EV membrane proteins. Alternatively, surface labelling can be performed to enrich for EV membrane proteins and distinguish them from intraluminal cargo.109
6. Methods for determining the RNA content of EVs
EVs carry various species of RNA, including miRNA, circular RNA, vault RNA, small nuclear RNA, small nucleolar RNA, Y RNA, transfer RNA, long non-coding RNA, and messenger RNA, as well as fragments thereof.31 EV subtypes differ in their RNA cargo profile, according to parent cell type and environment, as well as stochastic principles, and the method of isolation used.110 Although most attention has focused on the miRNA content of EVs, miRNAs might only represent a minor constituent of EVs relative to other RNA species.111 The mechanism for sorting RNAs to EVs might include association with RNA-binding proteins, specific RNA motifs and RNA modifications.112,113
6.1. RNA analyses by qRT–PCR and RNA-sequencing
At first, RNA cargo of EVs was based solely on the use of Taqman miR-PCRs focused on individual miRNAs, and it was a challenge finding ways to normalize data. Data normalization was usually implemented by spiking in an exogenous miRNA supposedly not expressed in mammalian species, such as Caenorhabditis elegans miRNA-39 (Cel-39) before RNA extraction. More recently, several quantitative PCR (qRT–PCR) and digital PCR protocols are available to detect the miRNA cargo of EVs.114
Advances in RNA-sequencing technologies have enabled the identification of EV-derived RNAs in nearly all human biofluids,115 and associated with pathophysiological phenotypes.116 The use of RNA-sequencing approaches has provided a better understanding of the diversity of the EV-embedded RNAs.31,47,117
Certain pre-analytic confounders are well known, e.g. heparin can interfere with PCR analyses of RNAs,118 but can be overcome by heparinase treatment. The presence of certain miRNAs is suggestive of haemolysis of blood samples (e.g. miR-486-5p, miR-451, miR-92a, and miR-16), or the presence of contaminating calf serum (e.g. miR-122, miR-451a, and miR-1246).119–121 Lipoprotein contamination can also create difficulties in data analyses and interpretation since they can also carry miRNAs.122 To prevent contamination of EV preparation by RNAs carried by lipoproteins and extra-EV Argonaute proteins, the use of proteinase K and RNase A digestion can be implemented before proceeding to RNA extraction.112 It is useful to include a negative control without enzymatic treatment and positive control samples containing RNA, to confirm complete digestion of non-exosomal RNAs.
In order to compare data, several manually curated databases were developed: Vesiclepedia (http://www.microvesicles.org/) and Exo-carta (http://www.exocarta.org/) include RNAs, lipids, and proteins identified in different classes of EVs. More recently, the extracellular RNA (ExRNA) communication consortium (https://commonfund.nih.gov/exrna) was created by the NIH to establish foundational knowledge and technologies for ExRNA research (https://exrna-atlas.org/).123
6.2. How to evaluate the functional role of EV-RNA
Despite the numerous examples of studies suggesting important roles of EV-mediated RNA transfer on target cell behaviour, e.g. the regenerative potential of epicardium-derived extracellular vesicles mediated by conserved miRNA transfer, assessing the true (patho-)physiological role of such transfer is a formidable challenge, not least because of the relatively low EV-RNA concentrations. For investigations into general mechanisms underlying EV-mediated RNA transfer, sensitive reporter systems have been developed that allow the study of EV-RNA transfer at the single-cell level.124,125 However, to prove a direct effect of endogenous RNA species on EV target cells, additional challenges need to be addressed and important control experiments are required. These include demonstrating that the RNA of interest: (i) full length is present inside EVs; (ii) shows increased levels in recipient cells upon delivery (in the absence of up-regulated expression); and (iii) directly mediates a particular response in target cells, by interfering with its presence or function without affecting the content of EVs or recipient cells in any other way. Recently published reporting guidelines on EV-RNA studies should help to ensure reproducibility and to critically evaluate past and future studies claiming EV-RNA-induced physiological and pathological responses.112
7. Methods for determining EV lipid content
7.1. Lipid content
The phospholipid (PC) bilayer membrane of EVs consists primary of phosphatidylcholine, in addition to phosphatidylethanolamine and phosphatidylserine.62,126 The sEV membrane is relatively rigid due to its enrichment in sphingomyelin and cholesterol, and contains domains with an ordered lipid phase (‘lipid rafts'; reviewed in Record et al.127).
Notably, EVs also carry lipids involved in signalling such as eicosanoids together with functional phospholipases and enzymes of the prostaglandin pathway.128 The lipid composition of lEVs is closer to that of the plasma membrane, which they originate from.126 Translocation of phosphatidylserine to the outer leaflet upon cellular activation has been suggested to be a prerequisite for lEV biogenesis.127 EVs with externalized phosphatidylserine are highly procoagulant, leading to venous thrombosis, particularly in the presence of tissue factor (TF).129
Total lipid content can be easily measured using a sensitive assay.130 The total protein-to-lipid ratio of an EV sample can then be used as an indication of EV concentration and purity.130,131 However, like protein assays, lipid assays are affected by the presence of contaminating lipoproteins.
MS is increasingly used to determine the complete lipidomic profile of EV samples.62,126 Furthermore, targeted lipidomic strategies can be developed based on the results of untargeted MS-based lipidomics. Newer techniques include total reflection Fourier-transform infrared spectroscopy132 and Raman spectroscopy.133 Raman spectroscopy reveals the chemical composition of single sEVs, and can identify different subpopulations of EVs based on their overall biochemical composition, including cholesterol content, phospholipids-to-cholesterol ratio, and surface protein expression.133
Most lipidomic studies of sEVs show an enrichment from cells to sEVs for cholesterol and sphingomyelin (representing ∼40–50% and 10–20% of total sEV lipids, respectively).134 Phosphatidylcholine and phosphatidylserine are in general the most abundant glycerophospholipids while phosphatidic acid, phosphatidylglycerol and phosphatidylinositol tend to be lower. Compared with cells, the content of phosphatidylcholine and phosphatidylinositol is generally lower in sEVs, while sphingolipids are increased. Certain lipids such as triacylglycerols and cholesteryl esters are found in lipoproteins and lipid droplets, and a high content of these lipids in EV preparations might be indicative for co-isolated or contaminating particles. There is evidence that the sphingolipid composition of circulating EVs is altered after myocardial ischaemia.135 Of note, ceramide content in adipocyte-derived EVs regulate vascular redox state in obese patients and is associated with cardiovascular mortality.17 EV lipid composition is also dependent on EV type. MVB-derived sEVs have a higher cholesterol content than EV types released from the plasma membrane.131 In line with this, sEVs show the highest resistance to detergent lysis among EVs.107
A subset of circulating EVs display oxidation-specific epitopes (OSE), which are immunogenic adducts derived from (phospho)lipid peroxidation.136 Thus, OSE+ EVs may be practical markers of pathology-associated oxidative stress and may reflect pathological conditions better than EVs. Several different types of OSE can be identified using specific antibodies, including malondialdehyde, 4-hydroxynonenal, and phosphocholine-containing oxidized PCs.137
8. Measurement of enzymatic activities carried by EVs
EVs harbour active enzymes on their membrane. Most surface enzymes are not easily detectable although the functional activity of EVs can still be measured due to the amplification of the detection signal through the enzymatic process for such enzymes, including the generation of factor Xa (FXa).138 Moreover, in most cases, both activators and inhibitors of a biological process are present at the same EV membrane. The overall functional activity of EVs will reflect the combined effects of these molecules.
8.1. Procoagulant activity
Large EVs possess procoagulant activities. This is mainly determined by the exposure of anionic PCs, especially phosphatidylserine which allows the binding of coagulation factors to the EV surface, as well as the exposure of active TF on some subsets of EVs.139 Assays measuring the functional capacity of EVs to generate FXa, thrombin, or a fibrin clot have been developed.140
Phosphatidylserine contributions can be evaluated by measuring a PC-dependent coagulation time after EV dilution in a PC-depleted plasma and activation with FXa and calcium.141 Other assays combine solid-phase capture of EVs by annexin V and thrombin generation.
The second group of assays focuses on the measurement of TF-dependent procoagulant activity of EVs. Thrombin generation in platelet-free plasma or purified EVs spiked in EV-free plasma is initiated in the presence of PCs without TF. High concentrations of TF-EVs are necessary for detection with this assay. Other studies evaluating the value of EVs as a biomarker of thrombosis have measured procoagulant EVs with FXa generation assays, using either EVs captured on coated plate or EV isolation using ultracentrifugation (UC).142,143 A more global assay also monitors fibrin generation after incubating plasma EVs isolated by UC in the presence of anti-TF or anti-FXII blocking antibodies.144
In clinical practice, all these assays are currently limited either by a lack of specificity, a low sensitivity, or irreproducibility when UC is used to isolate EVs. For example, measurement of TF by flow cytometry remains challenging because of the low levels of TF and some concerns about anti-TF antibody specificity.145 To tackle such issues, a new EV-TF activity assay was recently developed using a new inhibitory anti-TF antibody and a more sensitive protocol.146
Comparisons of assays measuring EV-TF activity suggest that FXa generation assays are more sensitive than the Zymuphen assay,147 and a poor correlation was found between results of the FXa generation assay and the fibrin generation test.148 ISTH initiated a new collaborative project to compare the analytical performance of different assays measuring EV-TF in plasma samples149 to progress towards an optimal method to measure EV procoagulant activity in plasma samples.
8.2. Fibrinolytic activity
EVs have ambivalent functions in haemostasis since they also possess fibrinolytic activity. A subset of EVs may indeed vector plasminogen activators such as urokinase.150 Just as for procoagulant assays, the use of UC can result in poor reproducibility of fibrinolytic assays. To overcome this limitation, a hybrid assay combining specific capture of EVs and measurement of their plasmin generation capacity has been developed.151 High-resolution laser scanning confocal microscopy could be also used to detect EV enzymatic activity using fluorescent reporters.152 However, throughput is limited.
8.3. Enzymatic activities
The presence of acetylcholinesterase is no longer used as a reliable EV marker; neurons and red blood cells produce this activity in abundance, whereas it is almost undetectable in other cell types and often associated with non-vesicular structures.82 Several metalloproteases, e.g. disintegrin metalloproteases and tissue inhibitor of metalloproteases have been reported in different EV preparations; these activities could confer on EVs the capacity to promote cell proliferation and remodelling of the microenvironment, which could contribute to EV therapeutic potential.153 However, it remains crucial to demonstrate that the enzymatic activity is associated with EVs and not with soluble mediators, and does not result from co-isolation during the purification procedure.
9. Methodologies for functional characterization of EVs
Due to the variable quality of the tools and technologies used to study EVs, complete and accurate reporting of methods is essential. These include the above-mentioned isolation and characterization techniques, but to understand the functional interaction and potential of different EV preparations, other points should be taken into consideration.
In addition to EV purification and isolation, ‘EV-depleted' samples and quality and procedural controls (e.g. unconditioned cell culture medium processed in the same way) can help to determine true EV-mediated responses. GW4869, an inhibitor of neutral sphingomyelinase 2 (nSMase2) and sEV release, is sometimes used as a control, but care is required in its use, as it is unlikely to be specific for exosome release.32,154
Co-purified and bound molecules might affect functional assays;155 therefore, it is best to avoid low-specificity methods such as general precipitation (PEG, ‘salting out, ' the basis of many commercial ‘exosome isolation' kits), unless these methods are combined with additional separation steps.
The biological nature of EV preparations makes normalization between conditions essential but there is no clear consensus on the best way forward. Some alternatives include: starting volume or the number of producing cells; the total number of EVs; protein content; lipid content; metabolite content; or specific markers such as levels of tetraspanins or other putative house-keeping proteins or RNA species.156 It is recommended to have two to three different approaches, and to clearly describe each, to allow potential differences in functional outcomes to be explored.
For clinical therapeutic interventions, the identity of the EV preparations can be defined using quantifiable metrics.79
In classical dose–response experiments, the relationship between the concentration of a ligand/drug and a measured outcome parameter is investigated. Such experiments should be considered to understand the dose-dependency of effects, and to understand the biological relevance of the quantity of EVs used. In many published works, the dose relative to physiological concentration is unclear.
Profiling of the EVs proteome and RNAome also will help to characterize their origin and also potential functional activities.157
9.1. Uptake and biodistribution studies
To understand the specific uptake of EV species or how different EV subpopulations are produced, several potent inhibitors are commonly used, including chloroquine, nSMase2 inhibitors, or genetic removal of Rab-protein family members.27,158,159 Inhibitors of micropinocytosis, endocytosis (clathrin, caveolin, or lipid-raft dependent), phagocytosis, or membrane fusion are also suggested to decipher in vitro the different routes and mechanisms of EV uptake by target cells.160 Since these suggested compounds lack specificity, it is important to keep in mind that they only suggest potential mechanisms. No EV-specific interventions have been reported thus far.
It is challenging to document the in vivo biodistribution of EVs. Many studies first isolate and tag EVs before injecting them in vivo, but these exogenous EVs may not reflect the same fate as endogenously released EVs. In addition, the presence of residual contaminants from the isolation procedure, the route of administration, the type of label used, the animal model, and the detection method may all affect in vivo biodistribution. If fluorescent dyes are used for EV labelling they should be carefully selected. Many dyes, particularly lipophilic dyes, can form dye aggregates or micelles that are of similar size to EVs, or may bind to contaminants present in the isolate, such as lipoproteins and certain proteins.161 Furthermore, lipophilic dyes might dissociate from the labelled EV and be incorporated into cellular membranes in vivo, where long dye half-life may lead to incorrect assumptions about EV distribution and longevity and diffuse freely. Genetic approaches crossing ROSAmTmG mice with models expressing Cre recombinase in a cell-specific manner have opened new avenues for quantifying uncommon populations of EV, such as cardiomyocyte-derived EVs in the circulation.162 On the other hand, protein-based labels added using genetic approaches (e.g. GFP) can be susceptible to proteolysis and cannot be used on samples derived from human tissues and fluids. Therefore, careful control experiments are required to ensure the signal is specific and to monitor the influence of any free dye. Cell–cell interaction studies and paracrine activity of secreted exosomes can be studied by co-culture assays of different cell types. Some examples are reported where (direct) EV-cargo loading is used to detect EV-molecule transfer, but indirect effects and reduced EV functionality are examples of possible limitations of these methods.163 Possible controls include comparison with the biodistribution of free label (no EVs) or of EVs that have been physically disrupted.164
Investigation of endogenous EV biodistribution requires genetic labelling strategies, such as degron-tagged reporters or pH-sensitive fluorophores, which provide a stronger EV labelling than that of the parent cell.165,166 However, these approaches might be restricted to one specific subset of endogenous EVs. The EV-mediated transfer of Cre recombinase into floxed reporter cells appears to be an elegant method to study in vivo EV distribution and uptake.166 Another technique is to detect tissue uptake of a miRNA unique to the EVs, such as a foreign miRNA that the EVs have been engineered to express.25
In conclusion, all current approaches to assess EV in vivo biodistribution (see Table 4 for examples) have their strengths and limitations, which must be carefully considered when designing experiments.
Table 4.
Examples of EV labelling for direct transfer and biodistribution studies
| Method of EV labelling | (Animal) models | Observations | Advantages | Disadvantages | References |
|---|---|---|---|---|---|
| Lipophilic dyes (e.g. PKH26, PKH67, DiD) |
|
|
|
|
Maring et al.27 and Takov et al.161 |
|
|
|
|
|
Barile et al.25 and Valadi et al.167 |
| EV siRNA loading |
|
|
|
Alvarez-Erviti et al.168 | |
| Fusion proteins |
|
|
|
|
Verweij et al.166, Luo et al.169 and Neckles et al.170 |
| Degron reporters |
|
|
|
|
Beer et al.16 |
10. Methodologies for clinical use of EVs in CVDs
Potential regenerative/reparative effects of EVs in the cardiovascular system have been observed in both post-infarction and non-ischaemic chemotherapy-induced cardiomyopathy models.1,23,26,38,40,43,171 Although EV biodistribution and direct cellular uptake still needs much attention, preclinical meta-analyses indicate that stem cell-derived EV administration is associated with improvements of left ventricular ejection fraction, fractional shortening, and a reduction of infarct size. These benefits are seen largely irrespective of the type of stem cell, timing of injection, route of delivery, dosage of delivery, or follow-up period.172,173 On the other hand, not unique to EV studies, there is a potential risk of positive publication bias.172,173 While these positive data suggest that clinical studies may be warranted, there are a number of important issues to address including those related to upscaling of EV preparation processes in GMP-quality facilities using non-xenogeneic culture conditions, as well as ethical and regulatory approvals.5 Even with the optimization of EV separation and characterization, several practical hurdles must be overcome to maximize the therapeutic potential of EVs. In addition to regenerative potential, however, EVs can play detrimental roles, for example potentially by causing thrombotic complications or forming microcalcifications that destabilize atherosclerotic plaques.174 The therapies preventing this deteriorating effect are under investigation.
10.1. Production and storage effects on the quality of EV preparations
Prior to in vivo application, it is essential to assess the reproducibility of EV content, purity and functionality in batch preparations. These measures should include evaluation of ingredients and potential co-isolations of culture medium, while also keeping in mind that these might mediate part of the observed functional effects. The production of EV preparations for use in the cardiovascular system is not uniquely different from those for use in other systems. The manufacturing of MSC-sEV preparations for therapeutic applications is currently the most advanced with several preparations in clinical trials, as highlighted elsewhere.175
For the isolation of EVs secreted by cells in culture, several cell culture factories are available, including multi-layered culture flasks,63 hollow-fibre bioreactors,176 and microcarriers.177 Before these systems are used; however, their impact on EV production and bioactivity must be determined. Isolated EVs are believed to be stable and can be frozen, but extensive studies are warranted to confirm that EV functionality is retained following freeze–thaw cycles and long-term storage.178 Multiple additional considerations are essential for handling blood-derived EVs,73 including pre-analytical methods, and quality controls.
10.2. Delivery strategies and biodistribution of EVs
Efficient EV delivery to the target organ/cells may be necessary to achieve full therapeutic potential, but it should also be considered that the primary target may not be the diseased tissue if EVs function indirectly. Both systemic and intra-organ delivery is possible and close monitoring of EV biodistribution is needed since cellular uptake of EVs might not be accurately reflected by the tracking labels used. Due to the small size of EVs, myocardial retention might be severely hampered since even stem cells, which are much larger than EVs, are immediately washed out from the myocardium after injection.179 EVs delivered intravenously are rapidly cleared (within minutes) and mainly distribute to the liver.180 Biodistribution studies, in which EVs are labelled with fluorescently linked lipid or amine dyes,181 radiolabels,182 or iron oxide particles,183 are highly warranted for mechanistic understanding of their effects. To facilitate long-term exposure of EV therapeutics, slow-release systems in which EVs are loaded and slowly exposed to the targeted tissue are key. Both natural184 and synthetic181 delivery systems have been developed and display enhanced beneficial effects for cardiac repair,39 with the caveat that they may require a direct intra-myocardial delivery whose invasiveness may hamper their clinical acceptance. An alternative approach that has been successfully used to promote cardiac repair following MI is thus to inject the EV-producing stem cells into a semi-permeable chamber, which is then inserted subcutaneously to release EVs (and other factors) over time.37
10.3. Loading therapeutics into EVs
For successful intra-myocardial delivery, many limitations and barriers have to be overcome,185 whereas bioengineered EVs with surface and/or cargo modifications might present unique advantages. Engineered therapeutic nanoparticles include: (i) vesicle-mimetics produced from cells by serial extrusion or cell membrane-cloaked nanoparticles, which have substantially greater yield and an easy purification process;186 (ii) EV-liposome hybrids, produced using simple incubation or freeze–thaw cycles, for easier uptake by target cells and for enhanced delivery; and (iii) synthetic EVs, which are based on liposomes with a composition similar to EVs.
EVs have been modified to deliver small molecules, therapeutic RNA, proteins, lipids, and different types of imaging molecules.187,188 Materials can be loaded into EVs via both passive loading (e.g. incubation with EVs or with EV-producing cells) or active loading (e.g. sonication, membrane permeabilization, electroporation, antibody binding of EVs, or transfection of EV-producing cells). EVs can be labelled on the surface or intraluminally.164 However, the labelling and loading procedure may alter the physical, chemical, and therapeutic properties of EVs or EV-mimetics. Moreover, therapeutic loading might be overestimated as observed for electroporation procedures that cause siRNA aggregate formation in the EV preparation.189 Therefore, a thorough in vitro and in vivo evaluation of their uptake, stability, efficacy, and toxicity is necessary to develop suitable methods for future clinical studies. Recent research suggests that EVs of various sizes can naturally carry intact viruses used in therapeutics such as adeno-associated viruses (reviewed in Sahoo et al.157,185 and may thereby be able to circumvent antibody neutralization.
11. Conclusion
In conclusion, researchers are gradually developing a better understanding of the role of endogenously formed EVs in cardiovascular pathophysiology, how they may be sampled as biomarkers of CVD, and how exogenously administered EVs might be used therapeutically. Basic procedures and principles for their purification, characterization, analysis, and modification are in progress, which will facilitate the detailed future mechanistic investigation. However, there are critical caveats at each step, and it is essential to bypass these pitfalls in order to avoid major setbacks and succeed in clinical translation (Tables 1–3). While relatively impure EV preparations may be shown to contain a desired biological activity useful for clinical applications, mechanistic studies may be hampered by the presence of unknown contaminants. This is essential, since the approval of EVs for clinical use is likely to necessitate an effective potency assay (or an array matrix consisting of several potency assays), which would ideally reflect a proven mechanism of action.97 Apart from better separation techniques, characterization of EV preparations is needed using orthogonal and complementary methods to define the purity of the preparations and will reveal potential sources of contamination. With the wide interest in EVs from both academia and the pharmaceutical industry, there is no doubt that methods will continually evolve and improve, which will help to advance EVs studies in cardiovascular science.
Contributor Information
Sean M Davidson, The Hatter Cardiovascular Institute, University College London, WC1E 6HX London, UK.
Chantal M Boulanger, Université Paris Cité, Paris-Cardiovascular Research Center, INSERM, Paris, France.
Elena Aikawa, Department of Medicine, Center for Excellence in Vascular Biology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115, USA.
Lina Badimon, Cardiovascular Science Program-ICCC, IR-Hospital de la Santa Creu i Santa Pau-IIBSantPau, CiberCV, Autonomous University of Barcelona, Barcelona, Spain.
Lucio Barile, Laboratory for Cardiovascular Theranostics, Istituto Cardiocentro Ticino, Ente Ospedaliero Cantonale and Faculty of Biomedical Sciences, Università Svizzera italiana, 6900 Lugano, Switzerland.
Christoph J Binder, Department of Laboratory Medicine, Medical University of Vienna, Vienna, Austria.
Alain Brisson, Molecular Imaging and NanoBioTechnology, UMR-5248-CBMN, CNRS-University of Bordeaux-IPB, Bat. B14, Allée Geoffroy Saint-Hilaire, 33600 Pessac, France.
Edit Buzas, Department of Genetics, Cell- and Immunobiology, Semmelweis University, HCEMM-SU and ELKH-SE Immune Proteogenomics Extracellular Vesicle Research Group, Budapest, Hungary.
Costanza Emanueli, National Heart and Lung Institute, Imperial College London, Hammersmith Campus, London W12 0NN, UK.
Felix Jansen, Department of Internal Medicine II, Heart Center, University Hospital Bonn, Bonn, Germany.
Miroslava Katsur, The Hatter Cardiovascular Institute, University College London, WC1E 6HX London, UK.
Romaric Lacroix, Aix Marseille University, INSERM 1263, Institut National de Recherche pour l’Agriculture, l’Alimentation et l’Environnement (INRAE), Centre de Recherche en CardioVasculaire et Nutrition (C2VN), Marseille, France; Department of Haematology and Vascular Biology, CHU La Conception, APHM, Marseille, France.
Sai Kiang Lim, Institute of Medical Biology and Institute of Molecular and Cell Biology, Agency for Science, Technology and Research, Singapore, Singapore; Department of Surgery, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore.
Nigel Mackman, Department of Medicine, UNC Blood Research Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Manuel Mayr, King's College London British Heart Foundation Centre, School of Cardiovascular Medicine and Sciences, London, UK.
Philippe Menasché, Department of Cardiovascular Surgery, Hôpital Européen Georges Pompidou, Paris, France; Laboratory of Experimental Cardiology, Department of Cardiology, UMC Utrecht Regenerative Medicine Center and Circulatory Health Laboratory, Utrecht University, University Medical Center Utrecht, Utrecht, The Netherlands.
Rienk Nieuwland, Vesicle Observation Center, Amsterdam UMC, University of Amsterdam, Amsterdam, The Netherlands; Laboratory of Experimental Clinical Chemistry, Amsterdam UMC, University of Amsterdam, Amsterdam, The Netherlands.
Susmita Sahoo, Cardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Kaloyan Takov, King's College London British Heart Foundation Centre, School of Cardiovascular Medicine and Sciences, London, UK.
Thomas Thum, Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, Hannover, Germany; Fraunhofer Institute of Toxicology and Experimental Medicine, Hannover, Germany.
Pieter Vader, Université Paris Cité, Paris-Cardiovascular Research Center, INSERM, Paris, France; CDL Research, University Medical Center Utrecht, Heidelberglaan 100, 3584 CX Utrecht, The Netherlands.
Marca H M Wauben, Faculty of Veterinary Medicine, Department of Biomolecular Health Sciences, Utrecht University, Yalelaan 2, Utrecht, The Netherlands.
Kenneth Witwer, Department of Molecular and Comparative Pathobiology, Johns Hopkins University School of Medicine, Baltimore, MD, USA; Department of Neurology, Johns Hopkins University School of Medicine, Baltimore, MD, USA.
Joost P G Sluijter, Laboratory of Experimental Cardiology, Department of Cardiology, UMC Utrecht Regenerative Medicine Center and Circulatory Health Laboratory, Utrecht University, University Medical Center Utrecht, Utrecht, The Netherlands.
Authors’ contributions
All co-authors contributed to the draft of the document; S.D., J.S. and C.M.B. synthesized all contributions and handled the revision of the paper.
Funding
This work was supported by the Hatter Foundation (to S.M.D.), the British Heart Foundation (PG/18/44/33790 to S.M.D.); by the Project EVICARE (no. 725229) of the European Research Council (ERC) and PPS grant (no. 2018B014) to J.P.G.S./P.V., the Dutch Ministry of Economic Affairs, Agriculture and Innovation and the Netherlands CardioVascular Research Initiative (CVON): the Dutch Heart Foundation to J.P.G.S.; by INSERM, the French National Agency for Research (ANR-16-CE92-0032-02) and the Fondation pour la Recherche Médicale (FRM EQU202003010767 to C.M.B.). M.M. is a BHF Chair Holder (CH/16/3/32406) with BHF programme grant support (RG/16/14/32397), and a holder of a BHF Special Project grant to participate in the ERA-CVD Transnational Grant ‘MacroERA: Non-coding RNAs in cardiac macrophages and their role in heart failure'; by the Austrian Science Fund (SFB-54 ‘InThro' to C.J.B.); it is funded by the EU Horizon 2020 project COVIRNA (grant agreement no. 101016072), the Spanish Ministry of Economy and Competitiveness of Science (PID2019-107160RB-I00), the Carlos III Institute of Health (CIBERCV CB16/11/00411 and RICORS 2021—TERAV) cofounded by FEDER; and the Fundación Investigación Cardiovascular-Fundación Jesus Serra (to L.B.); by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—JA 2351/2-1 and Project-ID 397484323—TRR 259 and the Corona Foundation (F.J.), by National Institutes of Health grant numbers R01HL136431, R01HL147095, and R01HL141917 (to E.A.); by the EU Horizon 2020 project Cardioregenix (GA 825670 to T.T.) and Deutsche Forschungsgemeinschaft (Transregio TRR 267 to T.T.); by the US NIH National Cancer Institute (NCI to K.W.) and Office of the Director (UG3CA241694 to K.W.); by Higher Education Institutional Excellence Programme—Therapeutic development (NKFIH OTKA120237, NVKP_16-1-2016-0017 to E.B.; VEKOP-2.3.2-16-2016-00002, VEKOP-2.3.3-15-2016-00016, H2020-MSCA-ITN-2017-722148 TRAIN EV to E.B.); EU’s Horizon 2020 research and innovation programme under grant agreement (739593 to E.B.).
Data availability
No new data were generated or analysed in support of this research.
References
- 1. Davidson SM, Andreadou I, Barile L, Birnbaum Y, Cabrera-Fuentes HA, Cohen MV, Downey JM, Girao H, Pagliaro P, Penna C, Pernow J, Preissner KT, Ferdinandy P. Circulating blood cells and extracellular vesicles in acute cardioprotection. Cardiovasc Res 2019;115:1156–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ridger VC, Boulanger CM, Angelillo-Scherrer A, Badimon L, Blanc-Brude O, Bochaton-Piallat ML, Boilard E, Buzas EI, Caporali A, Dignat-George F, Evans PC, Lacroix R, Lutgens E, Ketelhuth DFJ, Nieuwland R, Toti F, Tunon J, Weber C. Microvesicles in vascular homeostasis and diseases. Position Paper of the European Society of Cardiology (ESC) Working Group on Atherosclerosis and Vascular Biology. Thromb Haemost 2017;117:1296–1316. [DOI] [PubMed] [Google Scholar]
- 3. Properzi F, Logozzi M, Fais S. Exosomes: the future of biomarkers in medicine. Biomark Med 2013;7:769–778. [DOI] [PubMed] [Google Scholar]
- 4. Lv Y, Tan J, Miao Y, Zhang Q. The role of microvesicles and its active molecules in regulating cellular biology. J Cell Mol Med 2019;23:7894–7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sluijter JPG, Davidson SM, Boulanger CM, Buzás EI, de Kleijn DPV, Engel FB, Giricz Z, Hausenloy DJ, Kishore R, Lecour S, Leor J, Madonna R, Perrino C, Prunier F, Sahoo S, Schiffelers RM, Schulz R, Van Laake LW, Ytrehus K, Ferdinandy P. Extracellular vesicles in diagnostics and therapy of the ischaemic heart: position Paper from the Working Group on Cellular Biology of the Heart of the European Society of Cardiology. Cardiovasc Res 2018;114:19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Loyer X, Vion AC, Tedgui A, Boulanger CM. Microvesicles as cell-cell messengers in cardiovascular diseases. Circ Res 2014;114:345–353. [DOI] [PubMed] [Google Scholar]
- 7. Badimon L, Suades R, Fuentes E, Palomo I, Padró T. Role of platelet-derived microvesicles as crosstalk mediators in atherothrombosis and future pharmacology targets: a link between inflammation, atherosclerosis, and thrombosis. Front Pharmacol 2016;7:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vanhaverbeke M, Attard R, Bartekova M, Ben-Aicha S, Brandenburger T, de Gonzalo-Calvo D, Emanueli C, Farrugia R, Grillari J, Hackl M, Kalocayova B, Martelli F, Scholz M, Wettinger SB, Devaux Y, CA EU-CCA . Peripheral blood RNA biomarkers for cardiovascular disease from bench to bedside: a position paper from the EU-CardioRNA COST action CA17129. Cardiovasc Res 2023;118:3183–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deddens JC, Vrijsen KR, Colijn JM, Oerlemans MI, Metz CH, van der Vlist EJ, Nolte-’t Hoen EN, den Ouden K, Jansen Of Lorkeers SJ, van der Spoel TI, Koudstaal S, Arkesteijn GJ, Wauben MH, van Laake LW, Doevendans PA, Chamuleau SA, Sluijter JP. Circulating extracellular vesicles contain miRNAs and are released as early biomarkers for cardiac injury. J Cardiovasc Transl Res 2016;9:291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Emanueli C, Shearn AI, Laftah A, Fiorentino F, Reeves BC, Beltrami C, Mumford A, Clayton A, Gurney M, Shantikumar S, Angelini GD. Coronary artery-bypass-graft surgery increases the plasma concentration of exosomes carrying a cargo of cardiac MicroRNAs: an example of exosome trafficking out of the human heart with potential for cardiac biomarker discovery. PLoS One 2016;11:e0154274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Castellani C, Burrello J, Fedrigo M, Burrello A, Bolis S, Di Silvestre D, Tona F, Bottio T, Biemmi V, Toscano G, Gerosa G, Thiene G, Basso C, Longnus SL, Vassalli G, Angelini A, Barile L. Circulating extracellular vesicles as non-invasive biomarker of rejection in heart transplant. J Heart Lung Transplant 2020;39:1136–1148. [DOI] [PubMed] [Google Scholar]
- 12. Jansen F, Yang X, Proebsting S, Hoelscher M, Przybilla D, Baumann K, Schmitz T, Dolf A, Endl E, Franklin BS, Sinning JM, Vasa-Nicotera M, Nickenig G, Werner N. MicroRNA expression in circulating microvesicles predicts cardiovascular events in patients with coronary artery disease. J Am Heart Assoc 2014;3:e001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aliotta JM, Pereira M, Amaral A, Sorokina A, Igbinoba Z, Hasslinger A, El-Bizri R, Rounds SI, Quesenberry PJ, Klinger JR. Induction of pulmonary hypertensive changes by extracellular vesicles from monocrotaline-treated mice. Cardiovasc Res 2013;100:354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aliotta JM, Pereira M, Wen S, Dooner MS, Del Tatto M, Papa E, Goldberg LR, Baird GL, Ventetuolo CE, Quesenberry PJ, Klinger JR. Exosomes induce and reverse monocrotaline-induced pulmonary hypertension in mice. Cardiovasc Res 2016;110:319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kapustin AN, Chatrou ML, Drozdov I, Zheng Y, Davidson SM, Soong D, Furmanik M, Sanchis P, De Rosales RT, Alvarez-Hernandez D, Shroff R, Yin X, Muller K, Skepper JN, Mayr M, Reutelingsperger CP, Chester A, Bertazzo S, Schurgers LJ, Shanahan CM. Vascular smooth muscle cell calcification is mediated by regulated exosome secretion. Circ Res 2015;116:1312–1323. [DOI] [PubMed] [Google Scholar]
- 16. Durham AL, Speer MY, Scatena M, Giachelli CM, Shanahan CM. Role of smooth muscle cells in vascular calcification: implications in atherosclerosis and arterial stiffness. Cardiovasc Res 2018;114:590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Akawi N, Checa A, Antonopoulos AS, Akoumianakis I, Daskalaki E, Kotanidis CP, Kondo H, Lee K, Yesilyurt D, Badi I, Polkinghorne M, Akbar N, Lundgren J, Chuaiphichai S, Choudhury R, Neubauer S, Channon KM, Torekov SS, Wheelock CE, Antoniades C. Fat-secreted ceramides regulate vascular redox state and influence outcomes in patients with cardiovascular disease. J Am Coll Cardiol 2021;77:2494–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Akbar N, Digby JE, Cahill TJ, Tavare AN, Corbin AL, Saluja S, Dawkins S, Edgar L, Rawlings N, Ziberna K, McNeill E, Oxford Acute Myocardial Infarction (OxAMI) Study, Johnson E, Aljabali AA, Dragovic RA, Rohling M, Belgard TG, Udalova IA, Greaves DR, Channon KM, Riley PR, Anthony DC, Choudhury RP. Endothelium-derived extracellular vesicles promote splenic monocyte mobilization in myocardial infarction. JCI Insight 2017;2:e93344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Akbar N, Braithwaite AT, Corr EM, Koelwyn GJ, van Solingen C, Cochain C, Saliba AE, Corbin A, Pezzolla D, Moller Jorgensen M, Bæk R, Edgar L, De Villiers C, Gunadasa-Rohling M, Banerjee A, Paget D, Lee C, Hogg E, Costin A, Dhaliwal R, Johnson E, Krausgruber T, Riepsaame J, Melling GE, Shanmuganathan M, Oxford Acute Myocardial Infarction Study (OxAMI), Bock C, Carter DRF, Channon KM, Riley PR, Udalova IA, Moore KJ, Anthony DC, Choudhury RP. Rapid neutrophil mobilization by VCAM-1 + endothelial cell-derived extracellular vesicles. Cardiovasc Res 2023;118:246–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boulanger CM, Loyer X, Rautou PE, Amabile N. Extracellular vesicles in coronary artery disease. Nat Rev Cardiol 2017;14:259–272. [DOI] [PubMed] [Google Scholar]
- 21. Martínez MC, Andriantsitohaina R. Extracellular vesicles in metabolic syndrome. Circ Res 2017;120:1674–1686. [DOI] [PubMed] [Google Scholar]
- 22. Jansen F, Li Q, Pfeifer A, Werner N. Endothelial- and immune cell-derived extracellular vesicles in the regulation of cardiovascular health and disease. JACC Basic Transl Sci 2017;2:790–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Timmers L, Lim SK, Arslan F, Armstrong JS, Hoefer IE, Doevendans PA, Piek JJ, El Oakley RM, Choo A, Lee CN, Pasterkamp G, de Kleijn DP. Reduction of myocardial infarct size by human mesenchymal stem cell conditioned medium. Stem Cell Res 2008;1:129–137. [DOI] [PubMed] [Google Scholar]
- 24. Lai RC, Arslan F, Tan SS, Tan B, Choo A, Lee MM, Chen TS, Teh BJ, Eng JK, Sidik H, Tanavde V, Hwang WS, Lee CN, El Oakley RM, Pasterkamp G, de Kleijn DP, Tan KH, Lim SK. Derivation and characterization of human fetal MSCs: an alternative cell source for large-scale production of cardioprotective microparticles. J Mol Cell Cardiol 2010;48:1215–1224. [DOI] [PubMed] [Google Scholar]
- 25. Barile L, Cervio E, Lionetti V, Milano G, Ciullo A, Biemmi V, Bolis S, Altomare C, Matteucci M, Di Silvestre D, Brambilla F, Fertig TE, Torre T, Demertzis S, Mauri P, Moccetti T, Vassalli G. Cardioprotection by cardiac progenitor cell-secreted exosomes: role of pregnancy-associated plasma protein-A. Cardiovasc Res 2018;114:992–1005. [DOI] [PubMed] [Google Scholar]
- 26. Milano G, Biemmi V, Lazzarini E, Balbi C, Ciullo A, Bolis S, Ameri P, Di Silvestre D, Mauri P, Barile L, Vassalli G. Intravenous administration of cardiac progenitor cell-derived exosomes protects against doxorubicin/trastuzumab-induced cardiac toxicity. Cardiovasc Res 2020;116:383–392. [DOI] [PubMed] [Google Scholar]
- 27. Maring JA, Lodder K, Mol E, Verhage V, Wiesmeijer KC, Dingenouts CKE, Moerkamp AT, Deddens JC, Vader P, Smits AM, Sluijter JPG, Goumans MJ. Cardiac progenitor cell-derived extracellular vesicles reduce infarct size and associate with increased cardiovascular cell proliferation. J Cardiovasc Transl Res 2019;12:5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pula G, Mayr U, Evans C, Prokopi M, Vara DS, Yin X, Astroulakis Z, Xiao Q, Hill J, Xu Q, Mayr M. Proteomics identifies thymidine phosphorylase as a key regulator of the angiogenic potential of colony-forming units and endothelial progenitor cell cultures. Circ Res 2009;104:32–40. [DOI] [PubMed] [Google Scholar]
- 29. Prokopi M, Pula G, Mayr U, Devue C, Gallagher J, Xiao Q, Boulanger CM, Westwood N, Urbich C, Willeit J, Steiner M, Breuss J, Xu Q, Kiechl S, Mayr M. Proteomic analysis reveals presence of platelet microparticles in endothelial progenitor cell cultures. Blood 2009;114:723–732. [DOI] [PubMed] [Google Scholar]
- 30. Hu S, Li Z, Shen D, Zhu D, Huang K, Su T, Dinh PU, Cores J, Cheng K. Exosome-eluting stents for vascular healing after ischaemic injury. Nat Biomed Eng 2021;5:1174–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, Fissell WH, Patton JG, Rome LH, Burnette DT, Coffey RJ. Reassessment of exosome composition. Cell 2019;177:428–445.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach JM, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina Zavec A, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, Borras FE, Bosch S, Boulanger CM, Breakefield X, Breglio AM, Brennan MA, Brigstock DR, Brisson A, Broekman ML, Bromberg JF, Bryl-Górecka P, Buch S, Buck AH, Burger D, Busatto S, Buschmann D, Bussolati B, Buzas EI, Byrd JB, Camussi G, Carter DR, Caruso S, Chamley LW, Chang YT, Chen C, Chen S, Cheng L, Chin AR, Clayton A, Clerici SP, Cocks A, Cocucci E, Coffey RJ, Cordeiro-da-Silva A, Couch Y, Coumans FA, Coyle B, Crescitelli R, Criado MF, D’Souza-Schorey C, Das S, Datta Chaudhuri A, de Candia P, De Santana EF, De Wever O, Del Portillo HA, Demaret T, Deville S, Devitt A, Dhondt B, Di Vizio D, Dieterich LC, Dolo V, Dominguez Rubio AP, Dominici M, Dourado MR, Driedonks TA, Duarte FV, Duncan HM, Eichenberger RM, Ekström K, El Andaloussi S, Elie-Caille C, Erdbrügger U, Falcón-Pérez JM, Fatima F, Fish JE, Flores-Bellver M, Försönits A, Frelet-Barrand A, Fricke F, Fuhrmann G, Gabrielsson S, Gámez-Valero A, Gardiner C, Gartner K, Gaudin R, Gho YS, Giebel B, Gilbert C, Gimona M, Giusti I, Goberdhan DC, Görgens A, Gorski SM, Greening DW, Gross JC, Gualerzi A, Gupta GN, Gustafson D, Handberg A, Haraszti RA, Harrison P, Hegyesi H, Hendrix A, Hill AF, Hochberg FH, Hoffmann KF, Holder B, Holthofer H, Hosseinkhani B, Hu G, Huang Y, Huber V, Hunt S, Ibrahim AG, Ikezu T, Inal JM, Isin M, Ivanova A, Jackson HK, Jacobsen S, Jay SM, Jayachandran M, Jenster G, Jiang L, Johnson SM, Jones JC, Jong A, Jovanovic-Talisman T, Jung S, Kalluri R, Kano SI, Kaur S, Kawamura Y, Keller ET, Khamari D, Khomyakova E, Khvorova A, Kierulf P, Kim KP, Kislinger T, Klingeborn M, Klinke DJ II, Kornek M, Kosanoviá MM, Kovács AF, Kramer-Albers EM, Krasemann S, Krause M, Kurochkin IV, Kusuma GD, Kuypers S, Laitinen S, Langevin SM, Languino LR, Lannigan J, Lässer C, Laurent LC, Lavieu G, Lázaro-Ibáñez E, Le Lay S, Lee MS, Lee YXF, Lemos DS, Lenassi M, Leszczynska A, Li IT, Liao K, Libregts SF, Ligeti E, Lim R, Lim SK, Line A, Linnemannstöns K, Llorente A, Lombard CA, Lorenowicz MJ, Lörincz AM, Lotvall J, Lovett J, Lowry MC, Loyer X, Lu Q, Lukomska B, Lunavat TR, Maas SL, Malhi H, Marcilla A, Mariani J, Mariscal J, Martens-Uzunova ES, Martin-Jaular L, Martinez MC, Martins VR, Mathieu M, Mathivanan S, Maugeri M, McGinnis LK, McVey MJ, Meckes DG, Jr., Meehan KL, Mertens I, Minciacchi VR, Möller A, Møller Jørgensen M, Morales-Kastresana A, Morhayim J, Mullier F, Muraca M, Musante L, Mussack V, Muth DC, Myburgh KH, Najrana T, Nawaz M, Nazarenko I, Nejsum P, Neri C, Neri T, Nieuwland R, Nimrichter L, Nolan JP, Nolte-’t Hoen EN, Noren Hooten N, O’Driscoll L, O’Grady T, O’Loghlen A, Ochiya T, Olivier M, Ortiz A, Ortiz LA, Østeikoetxea X, Ostergaard O, Ostrowski M, Park J, Pegtel DM, Peinado H, Perut F, Pfaffl MW, Phinney DG, Pieters BC, Pink RC, Pisetsky DS, Pogge von Strandmann E, Polakovicova I, Poon IK, Powell BH, Prada I, Pulliam L, Quesenberry P, Radeghieri A, Raffai RL, Raimondo S, Rak J, Ramirez MI, Raposo G, Rayyan MS, Regev-Rudzki N, Ricklefs FL, Robbins PD, Roberts DD, Rodrigues SC, Rohde E, Rome S, Rouschop KM, Rughetti A, Russell AE, Saa P, Sahoo S, Salas-Huenuleo E, Sanchez C, Saugstad JA, Saul MJ, Schiffelers RM, Schneider R, Schoyen TH, Scott A, Shahaj E, Sharma S, Shatnyeva O, Shekari F, Shelke GV, Shetty AK, Shiba K, Siljander PR, Silva AM, Skowronek A, Snyder OL II, Soares RP, Sodar BW, Soekmadji C, Sotillo J, Stahl PD, Stoorvogel W, Stott SL, Strasser EF, Swift S, Tahara H, Tewari M, Timms K, Tiwari S, Tixeira R, Tkach M, Toh WS, Tomasini R, Torrecilhas AC, Tosar JP, Toxavidis V, Urbanelli L, Vader P, van Balkom BW, van der Grein SG, Van Deun J, van Herwijnen MJ, Van Keuren-Jensen K, van Niel G, van Royen ME, van Wijnen AJ, Vasconcelos MH, Vechetti IJ, Jr., Veit TD, Vella LJ, Velot E, Verweij FJ, Vestad B, Vinas JL, Visnovitz T, Vukman KV, Wahlgren J, Watson DC, Wauben MH, Weaver A, Webber JP, Weber V, Wehman AM, Weiss DJ, Welsh JA, Wendt S, Wheelock AM, Wiener Z, Witte L, Wolfram J, Xagorari A, Xander P, Xu J, Yan X, Yanez-Mo M, Yin H, Yuana Y, Zappulli V, Zarubova J, Zekas V, Zhang JY, Zhao Z, Zheng L, Zheutlin AR, Zickler AM, Zimmermann P, Zivkovic AM, Zocco D, Zuba-Surma EK. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 2018; 7: 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol 2019;21:9–17. [DOI] [PubMed] [Google Scholar]
- 34. Zhao J, Li X, Hu J, Chen F, Qiao S, Sun X, Gao L, Xie J, Xu B. Mesenchymal stromal cell-derived exosomes attenuate myocardial ischaemia-reperfusion injury through miR-182-regulated macrophage polarization. Cardiovasc Res 2019;115:1205–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Takafuji Y, Hori M, Mizuno T, Harada-Shiba M. Humoral factors secreted from adipose tissue-derived mesenchymal stem cells ameliorate atherosclerosis in Ldlr-/- mice. Cardiovasc Res 2019;115:1041–1051. [DOI] [PubMed] [Google Scholar]
- 36. Mayourian J, Ceholski DK, Gorski PA, Mathiyalagan P, Murphy JF, Salazar SI, Stillitano F, Hare JM, Sahoo S, Hajjar RJ, Costa KD. Exosomal microRNA-21-5p Mediates mesenchymal stem cell paracrine effects on human cardiac tissue contractility. Circ Res 2018;122:933–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kompa AR, Greening DW, Kong AM, McMillan PJ, Fang H, Saxena R, Wong RCB, Lees JG, Sivakumaran P, Newcomb AE, Tannous BA, Kos C, Mariana L, Loudovaris T, Hausenloy DJ, Lim SY. Sustained subcutaneous delivery of secretome of human cardiac stem cells promotes cardiac repair following myocardial infarction. Cardiovasc Res 2021;117:918–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Correa BL, Harane NE, Gomez I, Rachid Hocine H, Vilar J, Desgres M, Bellamy V, Keirththana K, Guillas C, Perotto M, Pidial L, Alayrac P, Tran T, Tan S, Hamada T, Charron D, Brisson A, Renault NK, Al-Daccak R, Menasche P, Silvestre JS. Extracellular vesicles from human cardiovascular progenitors trigger a reparative immune response in infarcted hearts. Cardiovasc Res 2021;117:292–307. [DOI] [PubMed] [Google Scholar]
- 39. Chen CW, Wang LL, Zaman S, Gordon J, Arisi MF, Venkataraman CM, Chung JJ, Hung G, Gaffey AC, Spruce LA, Fazelinia H, Gorman RC, Seeholzer SH, Burdick JA, Atluri P. Sustained release of endothelial progenitor cell-derived extracellular vesicles from shear-thinning hydrogels improves angiogenesis and promotes function after myocardial infarction. Cardiovasc Res 2018;114:1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gallet R, Dawkins J, Valle J, Simsolo E, de Couto G, Middleton R, Tseliou E, Luthringer D, Kreke M, Smith RR, Marban L, Ghaleh B, Marban E. Exosomes secreted by cardiosphere-derived cells reduce scarring, attenuate adverse remodelling, and improve function in acute and chronic porcine myocardial infarction. Eur Heart J 2017;38:201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Khan M, Nickoloff E, Abramova T, Johnson J, Verma SK, Krishnamurthy P, Mackie AR, Vaughan E, Garikipati VN, Benedict C, Ramirez V, Lambers E, Ito A, Gao E, Misener S, Luongo T, Elrod J, Qin G, Houser SR, Koch WJ, Kishore R. Embryonic stem cell-derived exosomes promote endogenous repair mechanisms and enhance cardiac function following myocardial infarction. Circ Res 2015;117:52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gao L, Wang L, Wei Y, Krishnamurthy P, Walcott GP, Menasche P, Zhang J. Exosomes secreted by hiPSC-derived cardiac cells improve recovery from myocardial infarction in swine. Sci Transl Med 2020;12:eaay1318. [DOI] [PubMed] [Google Scholar]
- 43. Del Campo CV, Liaw NY, Gunadasa-Rohling M, Matthaei M, Braga L, Kennedy T, Salinas G, Voigt N, Giacca M, Zimmermann WH, Riley PR. Regenerative potential of epicardium-derived extracellular vesicles mediated by conserved miRNA transfer. Cardiovasc Res 2021;118:597–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Savina A, Furlán M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem 2003;278:20083–20090. [DOI] [PubMed] [Google Scholar]
- 45. Ribeiro-Rodrigues TM, Laundos TL, Pereira-Carvalho R, Batista-Almeida D, Pereira R, Coelho-Santos V, Silva AP, Fernandes R, Zuzarte M, Enguita FJ, Costa MC, Pinto-do-Ó OP, Pinto MT, Gouveia P, Ferreira L, Mason JC, Pereira P, Kwak BR, Nascimento DS, Girão H. Exosomes secreted by cardiomyocytes subjected to ischaemia promote cardiac angiogenesis. Cardiovasc Res 2017;113:1338–1350. [DOI] [PubMed] [Google Scholar]
- 46. Gollmann-Tepeköylü C, Pölzl L, Graber M, Hirsch J, Nagele F, Lobenwein D, Hess MW, Blumer MJ, Kirchmair E, Zipperle J, Hromada C, Mühleder S, Hackl H, Hermann M, Al Khamisi H, Förster M, Lichtenauer M, Mittermayr R, Paulus P, Fritsch H, Bonaros N, Kirchmair R, Sluijter JPG, Davidson S, Grimm M, Holfeld J. miR-19a-3p containing exosomes improve function of ischaemic myocardium upon shock wave therapy. Cardiovasc Res 2020;116:1226–1236. [DOI] [PubMed] [Google Scholar]
- 47. Huang P, Wang L, Li Q, Tian X, Xu J, Xu J, Xiong Y, Chen G, Qian H, Jin C, Yu Y, Cheng K, Qian L, Yang Y. Atorvastatin enhances the therapeutic efficacy of mesenchymal stem cells-derived exosomes in acute myocardial infarction via up-regulating long non-coding RNA H19. Cardiovasc Res 2020;116:353–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hou Z, Qin X, Hu Y, Zhang X, Li G, Wu J, Li J, Sha J, Chen J, Xia J, Wang L, Gao F. Longterm exercise-derived exosomal miR-342-5p: a novel exerkine for cardioprotection. Circ Res 2019;124:1386–1400. [DOI] [PubMed] [Google Scholar]
- 49. Bei Y, Xu T, Lv D, Yu P, Xu J, Che L, Das A, Tigges J, Toxavidis V, Ghiran I, Shah R, Li Y, Zhang Y, Das S, Xiao J. Exercise-induced circulating extracellular vesicles protect against cardiac ischemia-reperfusion injury. Basic Res Cardiol 2017;112:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Loyer X, Zlatanova I, Devue C, Yin M, Howangyin KY, Klaihmon P, Guerin CL, Kheloufi M, Vilar J, Zannis K, Fleischmann BK, Hwang DW, Park J, Lee H, Menasché P, Silvestre JS, Boulanger CM. Intra-cardiac release of extracellular vesicles shapes inflammation following myocardial infarction. Circ Res 2018;123:100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zietzer A, Steffen E, Niepmann S, Dusing P, Hosen MR, Liu W, Jamme P, Al-Kassou B, Goody PR, Zimmer S, Reiners KS, Pfeifer A, Bohm M, Werner N, Nickenig G, Jansen F. MicroRNA-mediated vascular intercellular communication is altered in chronic kidney disease. Cardiovasc Res 2020;118:316–333. [DOI] [PubMed] [Google Scholar]
- 52. Davidson SM, Riquelme JA, Takov K, Vicencio JM, Boi-Doku C, Khoo V, Doreth C, Radenkovic D, Lavandero S, Yellon DM. Cardioprotection mediated by exosomes is impaired in the setting of type II diabetes but can be rescued by the use of non-diabetic exosomes in vitro. J Cell Mol Med 2018;22:141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang X, Huang W, Liu G, Cai W, Millard RW, Wang Y, Chang J, Peng T, Fan GC. Cardiomyocytes mediate anti-angiogenesis in type 2 diabetic rats through the exosomal transfer of miR-320 into endothelial cells. J Mol Cell Cardiol 2014;74:139–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Palviainen M, Saari H, Kärkkäinen O, Pekkinen J, Auriola S, Yliperttula M, Puhka M, Hanhineva K, Siljander PR. Metabolic signature of extracellular vesicles depends on the cell culture conditions. J Extracell Vesicles 2019;8:1596669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol 2006;Chapter 3: Unit 3 22. [DOI] [PubMed] [Google Scholar]
- 56. Paolini L, Zendrini A, Di Noto G, Busatto S, Lottini E, Radeghieri A, Dossi A, Caneschi A, Ricotta D, Bergese P. Residual matrix from different separation techniques impacts exosome biological activity. Sci Rep 2016;6:23550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Böing AN, van der Pol E, Grootemaat AE, Coumans FA, Sturk A, Nieuwland R. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J Extracell Vesicles 2014;3:23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nordin JZ, Lee Y, Vader P, Mäger I, Johansson HJ, Heusermann W, Wiklander OP, Hällbrink M, Seow Y, Bultema JJ, Gilthorpe J, Davies T, Fairchild PJ, Gabrielsson S, Meisner-Kober NC, Lehtiö J, Smith CI, Wood MJ, El Andaloussi S. Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomedicine 2015;11:879–883. [DOI] [PubMed] [Google Scholar]
- 59. Ludwig AK, De Miroschedji K, Doeppner TR, Börger V, Ruesing J, Rebmann V, Durst S, Jansen S, Bremer M, Behrmann E, Singer BB, Jastrow H, Kuhlmann JD, El Magraoui F, Meyer HE, Hermann DM, Opalka B, Raunser S, Epple M, Horn PA, Giebel B. Precipitation with polyethylene glycol followed by washing and pelleting by ultracentrifugation enriches extracellular vesicles from tissue culture supernatants in small and large scales. J Extracell Vesicles 2018;7:1528109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dong L, Zieren RC, Horie K, Kim CJ, Mallick E, Jing Y, Feng M, Kuczler MD, Green J, Amend SR, Witwer KW, de Reijke TM, Cho YK, Pienta KJ, Xue W. Comprehensive evaluation of methods for small extracellular vesicles separation from human plasma, urine and cell culture medium. J Extracell Vesicles 2020;10:e12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kang YT, Kim YJ, Bu J, Cho YH, Han SW, Moon BI. High-purity capture and release of circulating exosomes using an exosome-specific dual-patterned immunofiltration (ExoDIF) device. Nanoscale 2017;9:13495–13505. [DOI] [PubMed] [Google Scholar]
- 62. Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, Mark MT, Molina H, Martin AB, Bojmar L, Fang J, Rampersaud S, Hoshino A, Matei I, Kenific CM, Nakajima M, Mutvei AP, Sansone P, Buehring W, Wang H, Jimenez JP, Cohen-Gould L, Paknejad N, Brendel M, Manova-Todorova K, Magalhaes A, Ferreira JA, Osorio H, Silva AM, Massey A, Cubillos-Ruiz JR, Galletti G, Giannakakou P, Cuervo AM, Blenis J, Schwartz R, Brady MS, Peinado H, Bromberg J, Matsui H, Reis CA, Lyden D. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol 2018;20:332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Andriolo G, Provasi E, Lo Cicero V, Brambilla A, Soncin S, Torre T, Milano G, Biemmi V, Vassalli G, Turchetto L, Barile L, Radrizzani M. Exosomes from human cardiac progenitor cells for therapeutic applications: development of a GMP-grade manufacturing method. Front Physiol 2018;9:1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Takov K, Yellon DM, Davidson SM. Comparison of small extracellular vesicles isolated from plasma by ultracentrifugation or size-exclusion chromatography: yield, purity and functional potential. J Extracell Vesicles 2019;8:1560809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mol EA, Goumans MJ, Doevendans PA, Sluijter JPG, Vader P. Higher functionality of extracellular vesicles isolated using size-exclusion chromatography compared to ultracentrifugation. Nanomedicine 2017;13:2061–2065. [DOI] [PubMed] [Google Scholar]
- 66. Lehrich BM, Liang Y, Fiandaca MS. Foetal bovine serum influence on in vitro extracellular vesicle analyses. J Extracell Vesicles 2021;10:e12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yuana Y, Böing AN, Grootemaat AE, van der Pol E, Hau CM, Cizmar P, Buhr E, Sturk A, Nieuwland R. Handling and storage of human body fluids for analysis of extracellular vesicles. J Extracell Vesicles 2015;4:29260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lacroix R, Judicone C, Mooberry M, Boucekine M, Key NS, Dignat-George F, The ISSCW . Standardization of pre-analytical variables in plasma microparticle determination: results of the International Society on Thrombosis and Haemostasis SSC Collaborative workshop. J Thromb Haemost 2013; 11: 1190–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Palviainen M, Saraswat M, Varga Z, Kitka D, Neuvonen M, Puhka M, Joenvaara S, Renkonen R, Nieuwland R, Takatalo M, Siljander PRM. Extracellular vesicles from human plasma and serum are carriers of extravesicular cargo-implications for biomarker discovery. PLoS One 2020;15:e0236439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Vicencio JM, Yellon DM, Sivaraman V, Das D, Boi-Doku C, Arjun S, Zheng Y, Riquelme JA, Kearney J, Sharma V, Multhoff G, Hall AR, Davidson SM. Plasma exosomes protect the myocardium from ischemia-reperfusion injury. J Am Coll Cardiol 2015;65:1525–1536. [DOI] [PubMed] [Google Scholar]
- 71. Simonsen JB. What are we looking at? Extracellular vesicles, lipoproteins, or both? Circ Res 2017;121:920–922. [DOI] [PubMed] [Google Scholar]
- 72. Zhang X, Borg EGF, Liaci AM, Vos HR, Stoorvogel W. A novel three step protocol to isolate extracellular vesicles from plasma or cell culture medium with both high yield and purity. J Extracell Vesicles 2020;9:1791450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Clayton A, Boilard E, Buzas EI, Cheng L, Falcón-Perez JM, Gardiner C, Gustafson D, Gualerzi A, Hendrix A, Hoffman A, Jones J, Lässer C, Lawson C, Lenassi M, Nazarenko I, O’Driscoll L, Pink R, Siljander PR, Soekmadji C, Wauben M, Welsh JA, Witwer K, Zheng L, Nieuwland R. Considerations towards a roadmap for collection, handling and storage of blood extracellular vesicles. J Extracell Vesicles 2019;8:1647027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Leroyer AS, Ebrahimian TG, Cochain C, Récalde A, Blanc-Brude O, Mees B, Vilar J, Tedgui A, Levy BI, Chimini G, Boulanger CM, Silvestre JS. Microparticles from ischemic muscle promotes postnatal vasculogenesis. Circulation 2009;119:2808–2817. [DOI] [PubMed] [Google Scholar]
- 75. Perrotta I, Aquila S. Exosomes in human atherosclerosis: an ultrastructural analysis study. Ultrastruct Pathol 2016;40:101–106. [DOI] [PubMed] [Google Scholar]
- 76. Crescitelli R, Lässer C, Lötvall J. Isolation and characterization of extracellular vesicle subpopulations from tissues. Nat Protoc 2021;16:1548–1580. [DOI] [PubMed] [Google Scholar]
- 77. Claridge B, Rai A, Fang H, Matsumoto A, Luo J, McMullen JR, Greening DW. Proteome characterisation of extracellular vesicles isolated from heart. Proteomics 2021;21:e2100026. [DOI] [PubMed] [Google Scholar]
- 78. Leroyer AS, Isobe H, Lesèche G, Castier Y, Wassef M, Mallat Z, Binder BR, Tedgui A, Boulanger CM. Cellular origins and thrombogenic activity of microparticles isolated from human atherosclerotic plaques. J Am Coll Cardiol 2007;49:772–777. [DOI] [PubMed] [Google Scholar]
- 79. Witwer KW, Van Balkom BWM, Bruno S, Choo A, Dominici M, Gimona M, Hill AF, De Kleijn D, Koh M, Lai RC, Mitsialis SA, Ortiz LA, Rohde E, Asada T, Toh WS, Weiss DJ, Zheng L, Giebel B, Lim SK. Defining mesenchymal stromal cell (MSC)-derived small extracellular vesicles for therapeutic applications. J Extracell Vesicles 2019;8:1609206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. van der Pol E, Coumans FA, Grootemaat AE, Gardiner C, Sargent IL, Harrison P, Sturk A, van Leeuwen TG, Nieuwland R. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J Thromb Haemost 2014;12:1182–1192. [DOI] [PubMed] [Google Scholar]
- 81. Webber J, Clayton A. How pure are your vesicles? J Extracell Vesicles 2013;2. doi: 10.3402/jev.v2i0.19861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liao Z, Jaular LM, Soueidi E, Jouve M, Muth DC, Schøyen TH, Seale T, Haughey NJ, Ostrowski M, Théry C, Witwer KW. Acetylcholinesterase is not a generic marker of extracellular vesicles. J Extracell Vesicles 2019;8:1628592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Arraud N, Linares R, Tan S, Gounou C, Pasquet JM, Mornet S, Brisson AR. Extracellular vesicles from blood plasma: determination of their morphology, size, phenotype and concentration. J Thromb Haemost 2014;12:614–627. [DOI] [PubMed] [Google Scholar]
- 84. Ridolfi A, Brucale M, Montis C, Caselli L, Paolini L, Borup A, Boysen AT, Loria F, van Herwijnen MJC, Kleinjan M, Nejsum P, Zarovni N, Wauben MHM, Berti D, Bergese P, Valle F. AFM-based high-throughput nanomechanical screening of single extracellular vesicles. Anal Chem 2020;92:10274–10282. [DOI] [PubMed] [Google Scholar]
- 85. Nolan JP. Flow cytometry of extracellular vesicles: potential, pitfalls, and prospects. Curr Protoc Cytom 2015;73, 13.14.11–13.14.16. [DOI] [PubMed] [Google Scholar]
- 86. Welsh JA, Van Der Pol E, Arkesteijn GJA, Bremer M, Brisson A, Coumans F, Dignat-George F, Duggan E, Ghiran I, Giebel B, Gorgens A, Hendrix A, Lacroix R, Lannigan J, Libregts S, Lozano-Andrés E, Morales-Kastresana A, Robert S, De Rond L, Tertel T, Tigges J, De Wever O, Yan X, Nieuwland R, Wauben MHM, Nolan JP, Jones JC. MIFlowCyt-EV: a framework for standardized reporting of extracellular vesicle flow cytometry experiments. J Extracell Vesicles 2020;9:1713526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Libregts S, Arkesteijn GJA, Nemeth A, Nolte-’t Hoen ENM, Wauben MHM. Flow cytometric analysis of extracellular vesicle subsets in plasma: impact of swarm by particles of non-interest. J Thromb Haemost 2018;16:1423–1436. [DOI] [PubMed] [Google Scholar]
- 88. Amabile N, Cheng S, Renard JM, Larson MG, Ghorbani A, McCabe E, Griffin G, Guerin C, Ho JE, Shaw SY, Cohen KS, Vasan RS, Tedgui A, Boulanger CM, Wang TJ. Association of circulating endothelial microparticles with cardiometabolic risk factors in the Framingham Heart Study. Eur Heart J 2014;35:2972–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kränkel N, Strässler E, Uhlemann M, Müller M, Briand-Schumacher S, Klingenberg R, Schulze PC, Adams V, Schuler G, Lüscher TF, Möbius-Winkler S, Landmesser U. Extracellular vesicle species differentially affect endothelial cell functions and differentially respond to exercise training in patients with chronic coronary syndromes. Eur J Prev Cardiol 2021;28:1467–1474. [DOI] [PubMed] [Google Scholar]
- 90. Koganti S, Eleftheriou D, Gurung R, Hong Y, Brogan P, Rakhit RD. Persistent circulating platelet and endothelial derived microparticle signature may explain on-going pro-thrombogenicity after acute coronary syndrome. Thromb Res 2021;206:60–65. [DOI] [PubMed] [Google Scholar]
- 91. Anselmo A, Frank D, Papa L, Viviani Anselmi C, Di Pasquale E, Mazzola M, Panico C, Clemente F, Soldani C, Pagiatakis C, Hinkel R, Thalmann R, Kozlik-Feldmann R, Miragoli M, Carullo P, Vacchiano M, Chaves-Sanjuan A, Santo N, Losi MA, Ferrari MC, Puca AA, Christiansen V, Seoudy H, Freitag-Wolf S, Frey N, Dempfle A, Mercola M, Esposito G, Briguori C, Kupatt C, Condorelli G. Myocardial hypoxic stress mediates functional cardiac extracellular vesicle release. Eur Heart J 2021;42:2780–2792. [DOI] [PubMed] [Google Scholar]
- 92. Boyden S. The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes. J Exp Med 1962;115:453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Takov K, He Z, Johnston HE, Timms JF, Guillot PV, Yellon DM, Davidson SM. Small extracellular vesicles secreted from human amniotic fluid mesenchymal stromal cells possess cardioprotective and promigratory potential. Basic Res Cardiol 2020;115:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc 2007;2:329–333. [DOI] [PubMed] [Google Scholar]
- 95. Todorova D, Simoncini S, Lacroix R, Sabatier F, Dignat-George F. Extracellular vesicles in angiogenesis. Circ Res 2017;120:1658–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Baker M, Robinson SD, Lechertier T, Barber PR, Tavora B, D’Amico G, Jones DT, Vojnovic B, Hodivala-Dilke K. Use of the mouse aortic ring assay to study angiogenesis. Nat Protoc 2012;7:89–104. [DOI] [PubMed] [Google Scholar]
- 97. Gimona M, Brizzi MF, Choo ABH, Massimo D, Grillari J, Davidson SM, Hermann DM, Hill AF, de Kleijn D, Lai RC, Lai C, Lim R, Monguió-Tortajada M, Muraca M, Ochiya T, Ortiz LA, Toh WS, Yi YW, Witwer KW, Giebel B, Lim SK. Critical considerations for the development of potency tests for therapeutic applications of mesenchymal stromal cell (MSC)-derived small extracellular vesicles. Cytotherapy 2021;23:373–380. [DOI] [PubMed] [Google Scholar]
- 98. Consortium E-T, Van Deun J, Mestdagh P, Agostinis P, Akay O, Anand S, Anckaert J, Martinez ZA, Baetens T, Beghein E, Bertier L, Berx G, Boere J, Boukouris S, Bremer M, Buschmann D, Byrd JB, Casert C, Cheng L, Cmoch A, Daveloose D, De Smedt E, Demirsoy S, Depoorter V, Dhondt B, Driedonks TA, Dudek A, Elsharawy A, Floris I, Foers AD, Gartner K, Garg AD, Geeurickx E, Gettemans J, Ghazavi F, Giebel B, Kormelink TG, Hancock G, Helsmoortel H, Hill AF, Hyenne V, Kalra H, Kim D, Kowal J, Kraemer S, Leidinger P, Leonelli C, Liang Y, Lippens L, Liu S, Lo Cicero A, Martin S, Mathivanan S, Mathiyalagan P, Matusek T, Milani G, Monguió-Tortajada M, Mus LM, Muth DC, Németh A, Nolte-’t Hoen EN, O’Driscoll L, Palmulli R, Pfaffl MW, Primdal-Bengtson B, Romano E, Rousseau Q, Sahoo S, Sampaio N, Samuel M, Scicluna B, Soen B, Steels A, Swinnen JV, Takatalo M, Thaminy S, Théry C, Tulkens J, Van Audenhove I, van der Grein S, Van Goethem A, van Herwijnen MJ, Van Niel G, Van Roy N, Van Vliet AR, Vandamme N, Vanhauwaert S, Vergauwen G, Verweij F, Wallaert A, Wauben M, Witwer KW, Zonneveld MI, De Wever O, Vandesompele J, Hendrix A. EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat Methods 2017;14:228–232. [DOI] [PubMed] [Google Scholar]
- 99. Ghosh R, Gilda JE, Gomes AV. The necessity of and strategies for improving confidence in the accuracy of western blots. Expert Rev Proteom 2014;11:549–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Nelson GM, Guynn JM, Chorley BN. Procedure and key optimization strategies for an automated capillary electrophoretic-based immunoassay method. J Vis Exp 2017;10:55911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Welton JL, Webber JP, Botos LA, Jones M, Clayton A. Ready-made chromatography columns for extracellular vesicle isolation from plasma. J Extracell Vesicles 2015;4:27269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R, Lee H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat Biotechnol 2014;32:490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Daaboul GG, Gagni P, Benussi L, Bettotti P, Ciani M, Cretich M, Freedman DS, Ghidoni R, Ozkumur AY, Piotto C, Prosperi D, Santini B, Ünlü MS, Chiari M. Digital detection of exosomes by interferometric imaging. Sci Rep 2016;6:37246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Théry C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci USA 2016;113:E968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Karimi N, Cvjetkovic A, Jang SC, Crescitelli R, Hosseinpour Feizi MA, Nieuwland R, Lötvall J, Lässer C. Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cell Mol Life Sci 2018;75:2873–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, Bond VC, Borràs FE, Breakefield X, Budnik V, Buzas E, Camussi G, Clayton A, Cocucci E, Falcon-Perez JM, Gabrielsson S, Gho YS, Gupta D, Harsha HC, Hendrix A, Hill AF, Inal JM, Jenster G, Krämer-Albers EM, Lim SK, Llorente A, Lötvall J, Marcilla A, Mincheva-Nilsson L, Nazarenko I, Nieuwland R, Nolte-’t Hoen EN, Pandey A, Patel T, Piper MG, Pluchino S, Prasad TS, Rajendran L, Raposo G, Record M, Reid GE, Sánchez-Madrid F, Schiffelers RM, Siljander P, Stensballe A, Stoorvogel W, Taylor D, Thery C, Valadi H, van Balkom BW, Vazquez J, Vidal M, Wauben MH, Yáñez-Mó M, Zoeller M, Mathivanan S. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol 2012;10:e1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Osteikoetxea X, Sódar B, Németh A, Szabó-Taylor K, Pálóczi K, Vukman KV, Tamasi V, Balogh A, Kittel A, Pállinger E, Buzás EI. Differential detergent sensitivity of extracellular vesicle subpopulations. Org Biomol Chem 2015;13:9775–9782. [DOI] [PubMed] [Google Scholar]
- 108. Foers AD, Chatfield S, Dagley LF, Scicluna BJ, Webb AI, Cheng L, Hill AF, Wicks IP, Pang KC. Enrichment of extracellular vesicles from human synovial fluid using size exclusion chromatography. J Extracell Vesicles 2018;7:1490145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Mayr M, Grainger D, Mayr U, Leroyer AS, Leseche G, Sidibe A, Herbin O, Yin X, Gomes A, Madhu B, Griffiths JR, Xu Q, Tedgui A, Boulanger CM. Proteomics, metabolomics, and immunomics on microparticles derived from human atherosclerotic plaques. Circ Cardiovasc Genet 2009;2:379–388. [DOI] [PubMed] [Google Scholar]
- 110. Lässer C, Shelke GV, Yeri A, Kim DK, Crescitelli R, Raimondo S, Sjostrand M, Gho YS, Van Keuren Jensen K, Lötvall J. Two distinct extracellular RNA signatures released by a single cell type identified by microarray and next-generation sequencing. RNA Biol 2017;14:58–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK, Gallichotte EN, Pogosova-Agadjanyan EL, Morrissey C, Stirewalt DL, Hladik F, Yu EY, Higano CS, Tewari M. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci USA 2014;111:14888–14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Mateescu B, Kowal EJ, van Balkom BW, Bartel S, Bhattacharyya SN, Buzás EI, Buck AH, de Candia P, Chow FW, Das S, Driedonks TA, Fernández-Messina L, Haderk F, Hill AF, Jones JC, Van Keuren-Jensen KR, Lai CP, Lässer C, Liegro ID, Lunavat TR, Lorenowicz MJ, Maas SL, Mäger I, Mittelbrunn M, Momma S, Mukherjee K, Nawaz M, Pegtel DM, Pfaffl MW, Schiffelers RM, Tahara H, Théry C, Tosar JP, Wauben MH, Witwer KW, Nolte-’t Hoen EN. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. J Extracell Vesicles 2017;6:1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zietzer A, Hosen MR, Wang H, Goody PR, Sylvester M, Latz E, Nickenig G, Werner N, Jansen F. The RNA-binding protein hnRNPU regulates the sorting of microRNA-30c-5p into large extracellular vesicles. J Extracell Vesicles 2020;9:1786967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Bellingham SA, Shambrook M, Hill AF. Quantitative analysis of exosomal miRNA via qPCR and digital PCR. Methods Mol Biol 2017;1545:55–70. [DOI] [PubMed] [Google Scholar]
- 115. Godoy PM, Bhakta NR, Barczak AJ, Cakmak H, Fisher S, MacKenzie TC, Patel T, Price RW, Smith JF, Woodruff PG, Erle DJ. Large differences in small RNA composition between human biofluids. Cell Rep 2018;25:1346–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Veziroglu EM, Mias GI. Characterizing extracellular vesicles and their diverse RNA contents. Front Genet 2020;11:700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Li S, Li Y, Chen B, Zhao J, Yu S, Tang Y, Zheng Q, Li Y, Wang P, He X, Huang S. exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res 2018;46:D106–D112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Boeckel JN, Thomé CE, Leistner D, Zeiher AM, Fichtlscherer S, Dimmeler S. Heparin selectively affects the quantification of microRNAs in human blood samples. Clin Chem 2013;59:1125–1127. [DOI] [PubMed] [Google Scholar]
- 119. Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, Tait JF, Tewari M. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 2012;5:492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Wei Z, Batagov AO, Carter DR, Krichevsky AM. Fetal bovine serum RNA Interferes with the cell culture derived extracellular RNA. Sci Rep 2016;6:31175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Tosar JP, Cayota A, Eitan E, Halushka MK, Witwer KW. Ribonucleic artefacts: are some extracellular RNA discoveries driven by cell culture medium components? J Extracell Vesicles 2017;6:1272832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Ben-Aicha S, Escate R, Casani L, Padró T, Peña E, Arderiu G, Mendieta G, Badimon L, Vilahur G. High-density lipoprotein remodelled in hypercholesterolaemic blood induce epigenetically driven down-regulation of endothelial HIF-1alpha expression in a preclinical animal model. Cardiovasc Res 2020;116:1288–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Das S, Extracellular RNACC, Ansel KM, Bitzer M, Breakefield XO, Charest A, Galas DJ, Gerstein MB, Gupta M, Milosavljevic A, McManus MT, Patel T, Raffai RL, Rozowsky J, Roth ME, Saugstad JA, Van Keuren-Jensen K, Weaver AM, Laurent LC, Abdel-Mageed AB, Adamidi C, Adelson PD, Akat KM, Alsop E, Ansel KM, Arango J, Aronin N, Avsaroglu SK, Azizian A, Balaj L, Ben-Dov IZ, Bertram K, Bitzer M, Blelloch R, Bogardus KA, Breakefield XO, Calin GA, Carter BS, Charest AL, Chen CC, Chitnis T, Coffey RJ, Courtright-Lim A, Das S, Datta A, DeHoff P, Diacovo TG, Erle DJ, Etheridge A, Ferrer M, Franklin JL, Freedman JE, Galas DJ, Galeev T, Gandhi R, Garcia A, Gerstein MB, Ghai V, Ghiran IC, Giraldez MD, Goga A, Gogakos T, Goilav B, Gould SJ, Guo P, Gupta M, Hochberg F, Huang B, Huentelman M, Hunter C, Hutchins E, Jackson AR, Kalani MYS, Kanlikilicer P, Karaszti RA, Van Keuren-Jensen K, Khvorova A, Kim Y, Kim H, Kim TK, Kitchen R, Kraig RP, Krichevsky AM, Kwong RY, Laurent LC, Lee M, L’Etoile N, Levy SE, Li F, Li J, Li X, Lopez-Berestein G, Lucero R, Mateescu B, Matin AC, Max KEA, McManus MT, Mempel TR, Meyer C, Milosavljevic A, Mondal D, Mukamal KJ, Murillo OD, Muthukumar T, Nickerson DA, O’Donnell CJ, Patel DJ, Patel T, Patton JG, Paul A, Peskind ER, Phelps MA, Putterman C, Quesenberry PJ, Quinn JF, Raffai RL, Ranabothu S, Rao SJ, Rodriguez-Aguayo C, Rosenzweig A, Roth ME, Rozowsky J, Sabatine MS, Sakhanenko NA, Saugstad JA, Schmittgen TD, Shah N, Shah R, Shedden K, Shi J, Sood AK, Sopeyin A, Spengler RM, Spetzler R, Srinivasan S, Subramanian SL, Suthanthiran M, Tanriverdi K, Teng Y, Tewari M, Thistlethwaite W, Tuschl T, Urbanowicz KK, Vickers KC, Voinnet O, Wang K, Weaver AM, Wei Z, Weiner HL, Weiss ZR, Williams Z, Wong DTW, Woodruff PG, Xiao X, Yan IK, Yeri A, Zhang B, Zhang H-G. The extracellular RNA communication consortium: establishing foundational knowledge and technologies for extracellular RNA research. Cell 2019;177:231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Zomer A, Maynard C, Verweij FJ, Kamermans A, Schäfer R, Beerling E, Schiffelers RM, de Wit E, Berenguer J, Ellenbroek SIJ, Wurdinger T, Pegtel DM, van Rheenen J. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 2015;161:1046–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. de Jong OG, Murphy DE, Mäger I, Willms E, Garcia-Guerra A, Gitz-Francois JJ, Lefferts J, Gupta D, Steenbeek SC, van Rheenen J, El Andaloussi S, Schiffelers RM, Wood MJA, Vader P. A CRISPR-Cas9-based reporter system for single-cell detection of extracellular vesicle-mediated functional transfer of RNA. Nat Commun 2020;11:1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Durcin M, Fleury A, Taillebois E, Hilairet G, Krupova Z, Henry C, Truchet S, Trotzmuller M, Köfeler H, Mabilleau G, Hue O, Andriantsitohaina R, Martin P, Le Lay S. Characterisation of adipocyte-derived extracellular vesicle subtypes identifies distinct protein and lipid signatures for large and small extracellular vesicles. J Extracell Vesicles 2017;6:1305677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Record M, Silvente-Poirot S, Poirot M, Wakelam MJO. Extracellular vesicles: lipids as key components of their biogenesis and functions. J Lipid Res 2018;59:1316–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Subra C, Grand D, Laulagnier K, Stella A, Lambeau G, Paillasse M, De Medina P, Monsarrat B, Perret B, Silvente-Poirot S, Poirot M, Record M. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J Lipid Res 2010;51:2105–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Rautou PE, Mackman N. Microvesicles as risk markers for venous thrombosis. Expert Rev Hematol 2013;6:91–101. [DOI] [PubMed] [Google Scholar]
- 130. Visnovitz T, Osteikoetxea X, Sódar BW, Mihaly J, Lőrincz P, Vukman KV, Tóth EA, Koncz A, Székács I, Horváth R, Varga Z, Buzás EI. An improved 96 well plate format lipid quantification assay for standardisation of experiments with extracellular vesicles. J Extracell Vesicles 2019;8:1565263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Osteikoetxea X, Balogh A, Szabó-Taylor K, Németh A, Szabó TG, Pálóczi K, Sódar B, Kittel A, György B, Pállinger E, Matkó J, Buzás EI. Improved characterization of EV preparations based on protein to lipid ratio and lipid properties. PLoS One 2015;10:e0121184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Szentirmai V, Wacha A, Németh C, Kitka D, Rácz A, Héberger K, Mihály J, Varga Z. Reagent-free total protein quantification of intact extracellular vesicles by attenuated total reflection Fourier transform infrared (ATR-FTIR) spectroscopy. Anal Bioanal Chem 2020;412:4619–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Smith ZJ, Lee C, Rojalin T, Carney RP, Hazari S, Knudson A, Lam K, Saari H, Ibanez EL, Viitala T, Laaksonen T, Yliperttula M, Wachsmann-Hogiu S. Single exosome study reveals subpopulations distributed among cell lines with variability related to membrane content. J Extracell Vesicles 2015;4:28533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Skotland T, Sagini K, Sandvig K, Llorente A. An emerging focus on lipids in extracellular vesicles. Adv Drug Deliv Rev 2020;159:308–321. [DOI] [PubMed] [Google Scholar]
- 135. Burrello J, Biemmi V, Dei Cas M, Amongero M, Bolis S, Lazzarini E, Bollini S, Vassalli G, Paroni R, Barile L. Sphingolipid composition of circulating extracellular vesicles after myocardial ischemia. Sci Rep 2020;10:16182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Tsiantoulas D, Perkmann T, Afonyushkin T, Mangold A, Prohaska TA, Papac-Milicevic N, Millischer V, Bartel C, Hörkkö S, Boulanger CM, Tsimikas S, Fischer MB, Witztum JL, Lang IM, Binder CJ. Circulating microparticles carry oxidation-specific epitopes and are recognized by natural IgM antibodies. J Lipid Res 2015;56:440–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Binder CJ, Papac-Milicevic N, Witztum JL. Innate sensing of oxidation-specific epitopes in health and disease. Nat Rev Immunol 2016;16:485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Nieuwland R, Gardiner C, Dignat-George F, Mullier F, Mackman N, Woodhams B, Thaler J. Toward standardization of assays measuring extracellular vesicle-associated tissue factor activity. J Thromb Haemost 2019;17:1261–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Key NS, Mackman N. Tissue factor and its measurement in whole blood, plasma, and microparticles. Semin Thromb Hemost 2010;36:865–875. [DOI] [PubMed] [Google Scholar]
- 140. Lacroix R, Vallier L, Bonifay A, Simoncini S, Mege D, Aubert M, Panicot-Dubois L, Dubois C, Dignat-George F. Microvesicles and cancer associated thrombosis. Semin Thromb Hemost 2019;45:593–603. [DOI] [PubMed] [Google Scholar]
- 141. Exner T, Joseph J, Low J, Connor D, Ma D. A new activated factor X-based clotting method with improved specificity for procoagulant phospholipid. Blood Coagul Fibrinolysis 2003;14:773–779. [DOI] [PubMed] [Google Scholar]
- 142. Hisada Y, Alexander W, Kasthuri R, Voorhees P, Mobarrez F, Taylor A, McNamara C, Wallen H, Witkowski M, Key NS, Rauch U, Mackman N. Measurement of microparticle tissue factor activity in clinical samples: a summary of two tissue factor-dependent FXa generation assays. Thromb Res 2016;139:90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Hisada Y, Mackman N. Measurement of tissue factor activity in extracellular vesicles from human plasma samples. Res Pract Thromb Haemost 2019;3:44–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Berckmans RJ, Sturk A, van Tienen LM, Schaap MC, Nieuwland R. Cell-derived vesicles exposing coagulant tissue factor in saliva. Blood 2011;117:3172–3180. [DOI] [PubMed] [Google Scholar]
- 145. Poncelet P, Robert S, Bailly N, Garnache-Ottou F, Bouriche T, Devalet B, Segatchian JH, Saas P, Mullier F. Tips and tricks for flow cytometry-based analysis and counting of microparticles. Transfus Apher Sci 2015;53:110–126. [DOI] [PubMed] [Google Scholar]
- 146. Vallier L, Bouriche T, Bonifay A, Judicone C, Bez J, Franco C, Guervilly C, Hisada Y, Mackman N, Houston R, Poncelet P, Dignat-George F, Lacroix R. Increasing the sensitivity of the human microvesicle tissue factor activity assay. Thromb Res 2019;182:64–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Tatsumi K, Antoniak S, Monroe DM III, Khorana AA, Mackman N, Subcommittee on Hemostasis and Malignancy of the Scientific and Standardization Committee of the International Society on Thrombosis and Hemostasis . Evaluation of a new commercial assay to measure microparticle tissue factor activity in plasma: communication from the SSC of the ISTH. J Thromb Haemost 2014;12:1932–1934. [DOI] [PubMed] [Google Scholar]
- 148. van Es N, Hisada Y, Di Nisio M, Cesarman G, Kleinjan A, Mahe I, Otten HM, Kamphuisen PW, Berckmans RJ, Buller HR, Mackman N, Nieuwland R. Extracellular vesicles exposing tissue factor for the prediction of venous thromboembolism in patients with cancer: a prospective cohort study. Thromb Res 2018;166:54–59. [DOI] [PubMed] [Google Scholar]
- 149. Lacroix R, Thaler J. ISTH SSC Vascular Biology Project 5: Comparison of the sensitivity and the specificity of assays to measure TF-EVs in plasma samples; 2019. https://cdn.ymaws.com/www.isth.org/resource/resmgr/subcommittees/isth_vascular_biology_ssc_pr.pdf
- 150. Vallier L, Cointe S, Lacroix R, Bonifay A, Judicone C, Dignat-George F, Kwaan HC. Microparticles and fibrinolysis. Semin Thromb Hemost 2017;43:129–134. [DOI] [PubMed] [Google Scholar]
- 151. Cointe S, Harti Souab K, Bouriche T, Vallier L, Bonifay A, Judicone C, Robert S, Armand R, Poncelet P, Albanese J, Dignat-George F, Lacroix R. A new assay to evaluate microvesicle plasmin generation capacity: validation in disease with fibrinolysis imbalance. J Extracell Vesicles 2018;7:1494482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Briens A, Gauberti M, Parcq J, Montaner J, Vivien D, Martinez de Lizarrondo S. Nano-zymography using laser-scanning confocal microscopy unmasks proteolytic activity of cell-derived microparticles. Theranostics 2016;6:610–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Shimoda M. Extracellular vesicle-associated MMPs: a modulator of the tissue microenvironment. Adv Clin Chem 2019;88:35–66. [DOI] [PubMed] [Google Scholar]
- 154. Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem 2010;285:17442–17452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Sódar BW, Kittel A, Pálóczi K, Vukman KV, Osteikoetxea X, Szabó-Taylor K, Németh A, Sperlágh B, Baranyai T, Giricz Z, Wiener Z, Turiák L, Drahos L, Pallinger E, Vékey K, Ferdinandy P, Falus A, Buzás EI. Low-density lipoprotein mimics blood plasma-derived exosomes and microvesicles during isolation and detection. Sci Rep 2016;6:24316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Gouin K, Peck K, Antes T, Johnson JL, Li C, Vaturi SD, Middleton R, de Couto G, Walravens AS, Rodriguez-Borlado L, Smith RR, Marbán L, Marbán E, Ibrahim AG. A comprehensive method for identification of suitable reference genes in extracellular vesicles. J Extracell Vesicles 2017;6:1347019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Sahoo S, Adamiak M, Mathiyalagan P, Kenneweg F, Kafert-Kasting S, Thum T. Therapeutic and diagnostic translation of extracellular vesicles in cardiovascular diseases: roadmap to the clinic. Circulation 2021;143:1426–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Ortega FG, Roefs MT, de Miguel Perez D, Kooijmans SA, de Jong OG, Sluijter JP, Schiffelers RM, Vader P. Interfering with endolysosomal trafficking enhances release of bioactive exosomes. Nanomedicine 2019;20:102014. [DOI] [PubMed] [Google Scholar]
- 159. Ibrahim AG, Cheng K, Marbán E. Exosomes as critical agents of cardiac regeneration triggered by cell therapy. Stem Cell Rep 2014;2:606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles 2014;3. doi: 10.3402/jev.v3.24641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Takov K, Yellon DM, Davidson SM. Confounding factors in vesicle uptake studies using fluorescent lipophilic membrane dyes. J Extracell Vesicles 2017;6:1388731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Hegyesi H, Pallinger E, Mecsei S, Hornyák B, Kovácsházi C, Brenner GB, Giricz Z, Pálóczi K, Kittel A, Tóvári J, Turiak L, Khamari D, Ferdinandy P, Buzás EI. Circulating cardiomyocyte-derived extracellular vesicles reflect cardiac injury during systemic inflammatory response syndrome in mice. Cell Mol Life Sci 2022;79:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Han Y, Jones TW, Dutta S, Zhu Y, Wang X, Narayanan SP, Fagan SC, Zhang D. Overview and update on methods for cargo loading into extracellular vesicles. Processes (Basel) 2021;9:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. de Abreu RC, Fernandes H, da Costa Martins PA, Sahoo S, Emanueli C, Ferreira L. Native and bioengineered extracellular vesicles for cardiovascular therapeutics. Nat Rev Cardiol 2020;17:685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Beer KB, Fazeli G, Judasova K, Irmisch L, Causemann J, Mansfeld J, Wehman AM. Degron-tagged reporters probe membrane topology and enable the specific labelling of membrane-wrapped structures. Nat Commun 2019;10:3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Verweij FJ, Revenu C, Arras G, Dingli F, Loew D, Pegtel DM, Follain G, Allio G, Goetz JG, Zimmermann P, Herbomel P, Del Bene F, Raposo G, van Niel G. Live tracking of inter-organ communication by endogenous exosomes in vivo. Dev Cell 2019;48:573–589.e4. [DOI] [PubMed] [Google Scholar]
- 167. Valadi H, Ekstrom K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654–659. [DOI] [PubMed] [Google Scholar]
- 168. Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol 2011;29:341–345. [DOI] [PubMed] [Google Scholar]
- 169. Luo W, Dai Y, Chen Z, Yue X, Andrade-Powell KC, Chang J. Spatial and temporal tracking of cardiac exosomes in mouse using a nano-luciferase-CD63 fusion protein. Commun Biol 2020;3:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Neckles VN, Morton MC, Holmberg JC, Sokolov AM, Nottoli T, Liu D, Feliciano DM. A transgenic inducible GFP extracellular-vesicle reporter (TIGER) mouse illuminates neonatal cortical astrocytes as a source of immunomodulatory extracellular vesicles. Sci Rep 2019;9:3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Lai RC, Arslan F, Lee MM, Sze NS, Choo A, Chen TS, Salto-Tellez M, Timmers L, Lee CN, El Oakley RM, Pasterkamp G, de Kleijn DP, Lim SK. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res 2010;4:214–222. [DOI] [PubMed] [Google Scholar]
- 172. Yang L, Zhu J, Zhang C, Wang J, Yue F, Jia X, Liu H. Stem cell-derived extracellular vesicles for myocardial infarction: a meta-analysis of controlled animal studies. Aging (Albany NY) 2019;11:1129–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Zwetsloot PP, Végh AM, Jansen of Lorkeers SJ, van Hout GP, Currie GL, Sena ES, Gremmels H, Buikema JW, Goumans MJ, Macleod MR, Doevendans PA, Chamuleau SA, Sluijter JP. Cardiac stem cell treatment in myocardial infarction: a systematic review and meta-analysis of preclinical studies. Circ Res 2016;118:1223–1232. [DOI] [PubMed] [Google Scholar]
- 174. Goettsch C, Hutcheson JD, Aikawa M, Iwata H, Pham T, Nykjaer A, Kjolby M, Rogers M, Michel T, Shibasaki M, Hagita S, Kramann R, Rader DJ, Libby P, Singh SA, Aikawa E. Sortilin mediates vascular calcification via its recruitment into extracellular vesicles. J Clin Invest 2016;126:1323–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Gimona M, Brizzi MF, Choo ABH, Dominici M, Davidson SM, Grillari J, Hermann DM, Hill AF, de Kleijn D, Lai RC, Lai CP, Lim R, Monguió-Tortajada M, Muraca M, Ochiya T, Ortiz LA, Toh WS, Yi YW, Witwer KW, Giebel B, Lim SK. Critical considerations for the development of potency tests for therapeutic applications of mesenchymal stromal cell-derived small extracellular vesicles. Cytotherapy 2021;23:373–380. [DOI] [PubMed] [Google Scholar]
- 176. Gobin J, Muradia G, Mehic J, Westwood C, Couvrette L, Stalker A, Bigelow S, Luebbert CC, Bissonnette FS, Johnston MJW, Sauve S, Tam RY, Wang L, Rosu-Myles M, Lavoie JR. Hollow-fiber bioreactor production of extracellular vesicles from human bone marrow mesenchymal stromal cells yields nanovesicles that mirrors the immuno-modulatory antigenic signature of the producer cell. Stem Cell Res Ther 2021;12:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Fuzeta MdA, Bernardes N, Oliveira FD, Costa AC, Fernandes-Platzgummer A, Farinha JP, Rodrigues CAV, Jung S, Tseng RJ, Milligan W, Lee B, Castanho M, Gaspar D, Cabral JMS, da Silva CL. Scalable production of human mesenchymal stromal cell-derived extracellular vesicles under serum-/xeno-free conditions in a microcarrier-based bioreactor culture system. Front Cell Dev Biol 2020;8:553444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Wu JY, Li YJ, Hu XB, Huang S, Xiang DX. Preservation of small extracellular vesicles for functional analysis and therapeutic applications: a comparative evaluation of storage conditions. Drug Deliv 2021;28:162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. van der Spoel TI, Vrijsen KR, Koudstaal S, Sluijter JP, Nijsen JF, de Jong HW, Hoefer IE, Cramer MJ, Doevendans PA, van Belle E, Chamuleau SA. Transendocardial cell injection is not superior to intracoronary infusion in a porcine model of ischaemic cardiomyopathy: a study on delivery efficiency. J Cell Mol Med 2012;16:2768–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Morishita M, Takahashi Y, Nishikawa M, Takakura Y. Pharmacokinetics of exosomes-an important factor for elucidating the biological roles of exosomes and for the development of exosome-based therapeutics. J Pharm Sci 2017;106:2265–2269. [DOI] [PubMed] [Google Scholar]
- 181. Mol EA, Lei Z, Roefs MT, Bakker MH, Goumans MJ, Doevendans PA, Dankers PYW, Vader P, Sluijter JPG. Injectable supramolecular ureidopyrimidinone hydrogels provide sustained release of extracellular vesicle therapeutics. Adv Healthc Mater 2019;8:e1900847. [DOI] [PubMed] [Google Scholar]
- 182. Smyth T, Kullberg M, Malik N, Smith-Jones P, Graner MW, Anchordoquy TJ. Biodistribution and delivery efficiency of unmodified tumor-derived exosomes. J Control Release 2015;199:145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Hu L, Wickline SA, Hood JL. Magnetic resonance imaging of melanoma exosomes in lymph nodes. Magn Reson Med 2015;74:266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Hernandez MJ, Gaetani R, Pieters VM, Ng NW, Chang AE, Martin TR, van Ingen E, Mol EA, Sluijter JPG, Christman KL. Decellularized extracellular matrix hydrogels as a delivery platform for MicroRNA and extracellular vesicle therapeutics. Adv Ther (Weinh) 2018;1:1800032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Sahoo S, Kariya T, Ishikawa K. Targeted delivery of therapeutic agents to the heart. Nat Rev Cardiol 2021;18:389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Ilahibaks NF, Lei Z, Mol EA, Deshantri AK, Jiang L, Schiffelers RM, Vader P, Sluijter JPG. Biofabrication of cell-derived nanovesicles: a potential alternative to extracellular vesicles for regenerative medicine. Cells 2019;8:1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Vader P, Mol EA, Pasterkamp G, Schiffelers RM. Extracellular vesicles for drug delivery. Adv Drug Deliv Rev 2016;106:148–156. [DOI] [PubMed] [Google Scholar]
- 188. Mackie AR, Klyachko E, Thorne T, Schultz KM, Millay M, Ito A, Kamide CE, Liu T, Gupta R, Sahoo S, Misener S, Kishore R, Losordo DW. Sonic hedgehog-modified human CD34 + cells preserve cardiac function after acute myocardial infarction. Circ Res 2012;111:312–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Kooijmans SAA, Stremersch S, Braeckmans K, de Smedt SC, Hendrix A, Wood MJA, Schiffelers RM, Raemdonck K, Vader P. Electroporation-induced siRNA precipitation obscures the efficiency of siRNA loading into extracellular vesicles. J Control Release 2013;172:229–238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated or analysed in support of this research.